Abstract
Regional differences in large-scale connectivity have been proposed to underlie functional specialization along the anterior-posterior axis of the medial temporal lobe (MTL), including the hippocampus (HC) and the parahippocampal gyrus (PHG). However, it is unknown whether functional connectivity (FC) can be used reliably to parcellate the human MTL. The current study aimed to differentiate subregions of the HC and the PHG based on patterns of whole-brain intrinsic FC. FC maps were calculated for each slice along the longitudinal axis of the PHG and the HC. A hierarchical clustering algorithm was then applied to these data in order to group slices according to the similarity of their connectivity patterns. Surprisingly, three discrete clusters were identified in the PHG. Two clusters corresponded to the parahippocampal cortex (PHC) and the perirhinal cortex (PRC), and these regions showed preferential connectivity with previously described posterior-medial and anterior-temporal networks, respectively. The third cluster corresponded to an anterior PRC region previously described as area 36d, and this region exhibited preferential connectivity with auditory cortical areas and with a network involved in visceral processing. The three PHG clusters showed different profiles of activation during a memory-encoding task, demonstrating that the FC-based parcellation identified functionally dissociable sub-regions of the PHG. In the hippocampus, no sub-regions were identified via the parcellation procedure. These results indicate that connectivity-based methods can be used to parcellate functional regions within the MTL, and they suggest that studies of memory and high-level cognition need to differentiate between PHC, posterior PRC, and anterior PRC.
Keywords: functional connectivity, hierarchical clustering algorithm, parcellation, hippocampus, parahipppocampal gyrus
1. Introduction
The medial temporal lobe (MTL) region is known to be essential for episodic memory formation (Scoville and Milner, 1957; Mishkin, 1978; Zola-Morgan et al., 1989b). Studies in humans and animal models have distinguished between memory processes supported by different MTL sub-regions, including the hippocampus (HC) and the adjacent parahippocampal gyrus (PHG) (Brown and Aggleton, 2001; Davachi, 2006; Diana et al., 2007; Eichenbaum et al., 2007; Aminoff et al., 2013). It has further been suggested that the functional differences among the MTL sub-regions are due to their participation in different large-scale brain networks (Kahn et al., 2008; Libby et al., 2012, Ranganath and Ritchey et al., 2012). The perirhinal cortex (PRC), in the anterior PHG, is extensively interconnected with higher-order visual areas (e.g., area TE and area TEO), the insular cortex, the orbitofrontal cortex, and the amygdala. The parahippocampal cortex (PHC) in the posterior PHG, is extensively interconnected with early visual areas (e.g., V4 and V3) in addition to the higher-order visual areas, auditory association areas (e.g. superior temporal gyrus), the retrosplenial cortex, and the posterior parietal cortex. Researchers have also proposed distinctions between the HC regions, given evidence that dorsal/posterior HC is more extensively interconnected with the mammillary bodies, the PHC, and the medial band of the ERC, whereas ventral/anterior HC is more extensively interconnected with the amygdala, the medial prefrontal cortex, the PRC, and the lateral band of the ERC (Moser and Moser, 1998; Fanselow and Dong, 2010; Poppenk et al., 2013; Strange et al., 2014).
Accurately identifying the MTL sub-regions (i.e., the PRC and PHC, posterior and anterior HC) in living human brains is one of the major obstacles in understanding human MTL function. In animal models, researchers have discriminated between the PRC and the PHC based on cytoarchitectonics, selective lesions, and anatomical connectivity (Zola-Morgan et al., 1989b; Burwell et al., 1995; Burwell and Amaral 1998a; Burwell and Amaral, 1998b; Suzuki and Amaral 1994a; Suzuki and Amaral 1994b; Baxter and Murray, 2001; Lavenex et al., 2002; Lavenex et al., 2004). In humans, magnetic resonance imaging (MRI) has been extensively used to understand MTL function in vivo, and conclusions drawn from structural and functional MRI studies depend critically on the ability to accurately identify homologs of the MTL sub-regions in human subjects. Currently used guidelines for distinguishing MTL sub-regions are based on visible landmarks on MRI images, based on cytoarchitectonic studies from small postmortem samples (Insausti, et al., 1998; Pruessner et al., 2002; Franko et al., 2012).
Although landmark-based segmentation protocols have been helpful for ROI-based analyses, particularly in high-resolution imaging studies of the hippocampal subfields (Zeineh et al., 2000; Duvernoy et al., 2005; Ding and Hoesen, 2015), these approaches do not account for variability in structure-function mapping among different subject groups. Furthermore, these approaches are relatively insensitive to small-scale anatomical boundaries and transitions in the cytoarchitecture between regions in standard MRI images at conventional field strengths. For these and other reasons, visible cortical landmarks identified in postmortem samples can only coarsely localize functionally distinct MTL sub-regions in healthy subjects.
As an alternative to approaches based purely on structural morphology, many researchers have begun to use analyses of intrinsic functional connectivity (FC) to noninvasively parcellate functional subdivisions of the human brain. Many researchers have argued that, within the neocortex, functional specialization is determined largely, if not entirely, by a region's unique pattern of connectivity, or “connectional fingerprint” (Passingham et al., 2002; Cohen et al. 2008; Barnes et al., 2010; Mishra et al., 2014). Therefore, regions that exhibit similar patterns of intrinsic FC could be considered as parts of the same functional unit. Intrinsic FC is computed by correlating low-frequency fluctuations of hemodynamic signals across different voxels in a functional magnetic resonance imaging (fMRI) time-series. The resulting FC patterns reveal brain networks comprised of regions that tend to be co-active over time, and this co-activity is thought to reflect direct and indirect connections between these structures. Many FC-based parcellation methods have been developed to differentiate cortical regions or cortical brain networks in humans (Cohen et al. 2008; Yeo et al. 2011; Wig et al., 2013; Nelson et al, 2013; Gordon et al., 2014). A few studies have utilized intrinsic FC to examine connectivity patterns for the MTL regions (Kahn et al., 2008; Lacy and Stark, 2012; Libby et al., 2012; Poppenk et al., 2013; Maass et al., 2015; Navarro Schroder et al., 2015). These studies revealed evidence to suggest that MTL sub-regions, defined by structural landmarks visible on MRI, exhibit different patterns of whole-brain FC. However, it is still unclear whether intrinsic FC analyses can be used to accurately and reliably parcellate functionally distinct MTL sub-regions.
In the current report, we addressed this question with a data-driven approach, in which hierarchical clustering analyses of whole-brain FC patterns were used to identify functional subdivisions of the HC and PHG. Because studies in animal models indicate that the HC and PHG exhibit functional differentiation along the longitudinal axis, we identified seed regions in successive coronal slices for these regions. The goal of our hierarchical clustering analysis was to identify groups (“clusters”) of slices that exhibit similar whole-brain FC, and to test whether these correspond to functionally distinct MTL sub-regions. Results revealed new and surprising evidence to suggest that the PHG could be subdivided into three sub-regions: one corresponding to the PHC and the other two corresponding to posterior and anterior PRC. Notably, the distinction between the anterior and posterior PRC strongly parallels results from previous anatomical studies of rodents and monkeys (Suzuki and Amaral, 1994b; Burwell and Amaral, 1998b; Burwell 2001; Lavenex et al., 2004), but to the best of our knowledge, it has been overlooked in studies of human MTL function. Finally, we further validated the PHG parcellation by analyzing activity in these regions during a memory-encoding task. In contrast to the PHG, we did not identify any sub-regions in the HC, but as described below, there was a trend for FC differences between the hippocampal head and the hippocampal body and tail.
2. Materials and Methods
2.1 Overview
The parcellation scheme aimed to separate the HC and PHG into functionally specialized sub-regions according to variations in their intrinsic FC patterns. Building on the idea that a region's function is determined by its connectivity, the FC patterns within a functional region should be homogeneous and the FC patterns among different functional regions should be heterogeneous. By detecting similarities among the FC maps for seed regions of the HC and PHG, we should be able to separate the HC and PHG into different functional clusters. In this report, we began by computing the functional connectivity between each coronal slice of the HC and PHG (i.e., segments along the longitudinal axis) and all gray matter voxels in the rest of the brain (Fig.1A). The “connectivity similarity” of two slices was measured by computing the correlation (r) between their whole-brain FC maps. The matrix comprised all the connectivity similarity values for the HC or the PHG was a connectivity homogeneity matrix (Fig.1B). A hierarchical clustering algorithm was applied to cluster coronal slices into a dendrogram according to the dissimilarity of their FC maps, or “connectivity distance” (1-r) (Fig.1C). Slices were successively merged together in branches representing connectivity distances, and permutation tests were used to identify significant clusters. To further investigate the parcellation, we compared the whole-brain FC maps for each cluster identified via our parcellation scheme. Additionally, we conducted a task-related analysis to investigate functional activations of the clusters during a memory test.
Figure 1. Overview of the parcellation procedure.
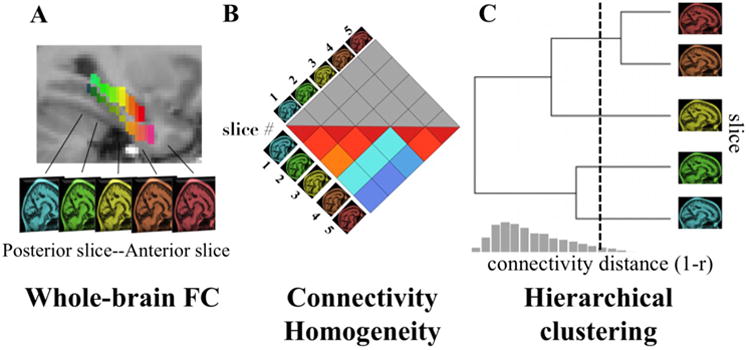
A) The first step in the parcellation procedure was computing whole-brain FC for each coronal slice of the HC and PHG. In the brain image, each coronal slice in the HC and the PHG is labeled in different color. A whole-brain image represents the whole-brain FC map for each coronal slice. B) The second step was to identify similarity among the whole-brain FC maps of coronal slices. We used Pearson's correlation coefficients (r) to measure similarity between FC maps. Within HC and PHG, the FC similarity values were compiled into “connectivity homogeneity matrices”. Each column/row in the matrix contained the pair-wise connectivity similarity values for each coronal slice in the HC or the PHG. Because the matrix is symmetrical, we display only half of the matrix in figure B. C) Finally, significant clusters of coronal slices were determined in the dendrogram by cutting the dendrogram at a connectivity distance threshold (1-r). On the dendrogram, leaves correspond to coronal slices in a brain region and lengths of the branches represented connectivity distance. The distribution under the dendrogram represents the null distribution constructed from permutation tests, which were performed to determine a connectivity distance threshold for identifying significant clusters. The dashed line represents the 5th percentile of the null distribution.
2.2 Image Acquisition & Pre-Processing
The data for this study were drawn from a previously described dataset (Ritchey et al. 2014) that included resting-state and task fMRI data from 19 young adults (11 female; ages 19-30). Participants completed a 7-minute pre-learning resting-state scan, three 10-minute task scans, a 7-minute post-learning resting state scan, and a post-scan behavioral test (see Ritchey et al. 2014 for more details). During the resting state scans, the computer screen was black with a white fixation cross at center, and subjects were instructed to stay awake with their eyes open.
Scanning was performed on a Siemens Skyra 3T scanner system with a 32-channel head coil. High-resolution T1-weighted structural images were acquired using a magnetization prepared rapid acquisition gradient echo (MPRAGE) pulse sequence (Field of view = 25.6 cm, image matrix = 256 × 256, 208 axial slices with 1.0 mm thickness). Functional images were acquired using a gradient echo planar imaging (EPI) sequence (TR = 2000ms; TE = 25 ms; FOV = 20.5 × 21.14 cm; image matrix = 64 × 66; flip angle = 90; 34 interleaved axial slices; voxel size = 3.2 × 3.2 × 3.2 mm; phase-encoding direction: posterior-anterior). Field maps were also collected and used to correct for geometric distortions due to magnetic field inhomogeneities.
SPM8 was used for image pre-processing, including slice-timing correction, realignment, field map correction, normalization (via DARTEL), and smoothing. For parcellation analyses, functional images from both resting-state scans were smoothed with a 3.0-mm FWHM (full-width at half-maximum) Gaussian kernel to account for subject co-registration error. Because the aim of this analysis was to parcellate coronal slices into different clusters according to their FC patterns, the smoothing parameter was relatively narrow to avoid biasing the whole-brain FC patterns of neighboring slices. Seed region voxels (i.e., the HC or PHG slices) were extracted from unsmoothed functional images to have better signal separation among the slices within the same brain region.
2.3 Parcellation procedures
2.3.1 Seed regions: coronal slices of the HC and the PHG
HC and PHG masks were generated by manually tracing on the group-averaged MNI space MPRAGE image following previously- published guidelines (Insausti et al. 1998; Franko et al. 2012). In brief, the anterior limits of the hippocampus were defined by the hippocampal-amygdaloid transition area and the posterior limits were defined as where the gray matter disappears near the lateral ventricle. The PHG mask contained gray matter voxels along the banks of the collateral sulcus (Supplementary Figure 1). Lengthwise, the mask started at the level of the limen insular and stopped at 4 mm posterior to the end of the hippocampus. Widthwise, for slices anterior to the onset of the entorhinal cortex (ERC), the mask contained both banks of the collateral sulcus. For slices containing the ERC, the mask contained the entire lateral bank of the collateral sulcus and extended to the midpoint of the medial bank of the sulcus. For slices posterior to the ERC, the mask included only the medial bank of the collateral sulcus and extended medially to the HC or to the calcarine sulcus.
2.3.2 Intrinsic FC preprocessing and analysis (Fig.1A, B)
Intrinsic FC was calculated using in-house scripts in MATLAB (The MathWorks, Inc., USA). For each subject, functional time-series from both resting-state scans (420 functional images total) were extracted from each coronal slice of the HC and the PHG (unsmoothed) and from a group-averaged mask of gray matter, individual masks of white matter (WM) and cerebrospinal fluid (CSF) (smoothed). The HC had 21 coronal slices with 11 slices in the left HC and 10 slices in the right HC. The PHG had 28 coronal slices with 14 slices in each hemisphere. The first three scans in each resting-state scan were removed to allow for T1 equilibration effects. Time-series were corrected for linear trends. Time-points that were seed outliers or suspected of motion contamination were scrubbed from the time series. Seed outliers were time-points in which the seed (each coronal slice) signal deviated more than three SD from its mean. Other suspect time-points were identified via the artifact detection tools (http://www.nitrc.org/projects/artifact_detect), defined as time-points marked by greater than 1mm in movement or 2% global mean signal change. Data at these time-points were interpolated and then the time-series were band pass filtered for frequencies of .01 to .1 Hz. After band pass filtering, the interpolated time-points were removed from the time series. For each ROI coronal slice, pair-wise correlations (Pearson's r) were computed to correlate mean time-series of each coronal slice with the time-series of all the gray matter voxels over the entire brain controlling for WM mean time-series, CSF mean time-series, six motion parameters, and session means.
To obtain group-level connectivity maps, the FC map for each slice was averaged across subjects. This procedure resulted in a group-level whole-brain gray matter FC map for each coronal slice of the HC and PHG. To define connectivity similarity, pairwise correlation coefficients (Pearson's r) were computed between the group-level FC maps for all coronal slice pairs in the HC and the PHG (Fig.1B). This procedure resulted in a 21 × 21 connectivity homogeneity matrix for the HC and a 28 × 28 matrix for the PHG. Each column on the connectivity homogeneity matrix contained the connectivity similarities (r) between a given slice and all the other coronal slices in the PHG or HC region.
2.3.3 Hierarchical clustering and dendrograms (Fig.1C)
We used the hierarchical clustering algorithm, UPGMA (Unweighted Pair Group Method with Arithmetic Mean), implemented in MATLAB, to successively merge clusters of the coronal slices based on similarities among their FC maps. Connectivity distance was calculated for each coronal slice pair by one minus connectivity similarity (1-r). Connectivity distance was entered into the hierarchical clustering algorithm. This procedure resulted in two respective dendrograms representing the hierarchical relationships of the connectivity distance for the coronal slices. On the dendrograms, leaves correspond to coronal slices of a brain region (i.e. the HC or the PHG) and lengths of the branches represented connectivity distance (1-r). The longer a given branch was, the more dissimilar the FC patterns between two slices/clusters were.
To determine significant clusters in the dendrogram, a connectivity distance threshold (1-r) was calculated via permutation tests. The null hypothesis was that the FC patterns for the coronal slices in the HC or the PHG were not heterogeneous enough to separate the coronal slices into different functional regions, and thus, there were no sub-clusters in the HC or the PHG. Permutation tests were used to determine a connectivity distance threshold at which the dendrogram was partitioned into disjoint clusters. If the connectivity distance between two sets of slices was under this threshold, this would mean that their FC patterns were no more dissimilar than what would be expected by chance, in this case, the two sets of slices would be grouped together into one functional cluster. In contrast, if the connectivity distance between two sets of slices was above the distance threshold, then their FC patterns were more dissimilar than what would be expected by chance, in which case the null hypothesis would be rejected and these two sets of slices would be separated into different functional clusters.
The data were permuted 10,000 times by randomly assigning 1 or -1 labels to each subject's FC data each time. On each permutation, after assigning random 1 or -1 weights to the dataset, we calculated group-level FC maps and connectivity distances (1-r) as described above. The mean of the connectivity distances in a given brain region (i.e. the HC or the PHG) were calculated to represent the overall degree of heterogeneity in the data for each time. The collection of the distance means from all permutations constructed the null distribution of expected connectivity distances. The distance threshold was defined as the 5th percentile of the null distribution, and thus denotes the level of dissimilarity that would be expected to occur by chance only 5% of the time. Leaves attached to the branches that intersected the cut-off line were grouped together as one functional cluster.
2.4 Intrinsic FC profiles for clusters: t-tests
The purpose for this analysis was to better understand the whole-brain FC patterns associated with each PHG cluster that was identified in the hierarchical clustering analysis. In these analyses, whole-brain FC maps associated with each cluster were obtained by averaging FC maps across all slices within each cluster. These analyses followed the same procedures described above, except that: (1) Instead of calculating intrinsic FC for only gray matter voxels, pair-wise correlations (Pearson's r) were computed for all voxels over the entire brain to obtain a smoothly varying group map, and the FC maps were Fisher's r-to-z transformed. (2) Functional images were smoothed with a 6.0 mm Gaussian kernel for this analysis. A larger smoothing kernel was used for these analyses in order to facilitate accurate correction for multiple comparisons (i.e., because we conducted statistical analyses to identify regions that showed supra-threshold FC values for each cluster and significant between-cluster differences) and to better account for inter-subject anatomical variability. Note that, because the regions of interest (ROIs) were clusters that had been identified in the precious parcellation analysis, there was less concern about FC biasing from neighboring slices.
To determine the connectivity profiles that were associated with different clusters, subject-level whole-brain FC maps were entered into one-sample t-tests. For simplicity, only positive functional connectivity values were evaluated and displayed in Figure 3, although the parcellation procedure incorporated all connectivity values across the whole brain. Differences in the intrinsic FC profiles of different clusters were identified via paired t-tests. The t-maps were thresholded at p < .001, one-tailed, with a 38-voxel cluster extent threshold. This combination of thresholds corresponds to cluster-corrected p< .05 according to simulations implemented in 3dClustSim of AFNI (http://afni.nimh.nih.gov/).
Figure 3. Networks connected with the three PHG clusters and the one HC cluster.
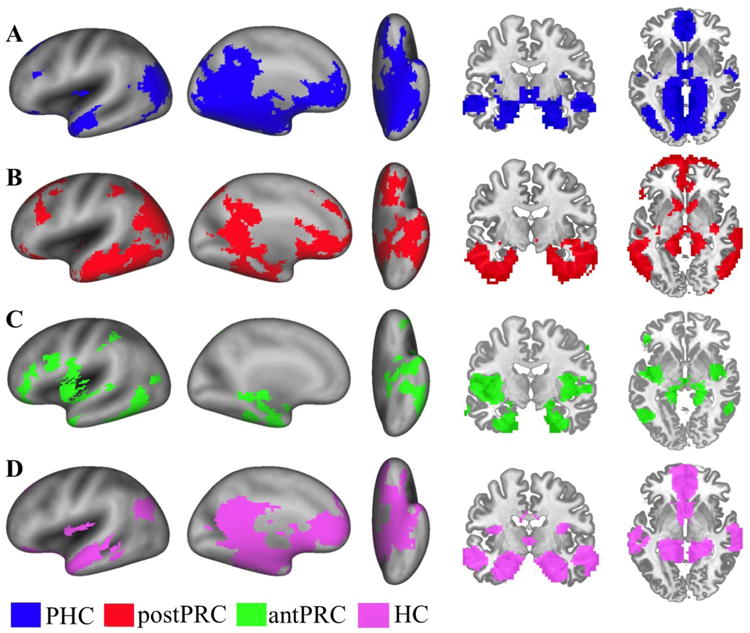
One-sample t-tests were conducted to identify voxels that showed suprathreshold FC values associated with different clusters (voxel-wise p<.001, cluster-corrected p<.05). A) Significant voxels connected with the PHC cluster (blue). B) Significant voxels connected with the postPRC cluster (red). C) Significant voxels connected with the antPRC cluster (green). D) Significant voxels connected with the HC cluster (purple).
2.5 Task activity for clusters: univariate activation analyses
In these analyses, we investigated activations within each clusters during a memory-encoding task. While in the scanner, participants completed an incidental associative encoding task using sentences pairing an object and a fact about either its appearance, its situational context, or its spatial location. For example, one possible sentence might read, “The apple is in the lecture hall.” The post-scan behavioral test consisted of an associative memory test for each sentence, and encoding trials were classified as hits (remembered) or misses (forgotten) based on subsequent memory performance (see Ritchey et al. 2014 for more details).
Task activation was evaluated through a general linear model implemented in SPM8. Functional images from the task scans were normalized and smoothed with a 6.0 -mm Gaussian kernel. Event-related stick-function regressors were used to model trials corresponding to one of six trial types: appearance hits, situational hits, spatial hits, appearance misses, situational misses, and spatial misses. Six motion parameter regressors and a regressor for no-response trials were also included in the model. Nuisance regressors were also included to model time-points identified as ART suspects. For each subject, contrasts were estimated for each of the six trial types, relative to implicit baseline activity. Mean contrast estimates were extracted from three PHG clusters. For each of the three conditions, the misses contrast was subtracted from the hits contrast to estimate activity related to the difference in memory (Dm). A repeated measures analysis of variance (ANOVA) was conducted to test the overall differences among memory effects, conditions, and ROIs. A two (memory effects: hit and miss) × three (conditions: appearance, situational context, and spatial location) × three (ROIs: PHC, postPRC, and antPRC) analysis of variance (ANOVA) was calculated on participants' contrast estimates.
2.6 PHG clusters connectivity along HC long axis
We next analyzed FC between each of the three PHG clusters and each of the HC coronal slice. Functional images were smoothed with a 6.0 mm Gaussian kernel. Functional time-series from both resting-state scans were extracted from each of the three PHG clusters and each of the HC coronal slices, individual masks of white matter (WM) and cerebrospinal fluid (CSF). FC pre-processing procedures and parameters were the same as the aforementioned FC procedures. Mean functional time-series for each of the three PHG clusters were correlated with the mean time-series of each of the HC coronal slice. These procedures result in a 3 by 21 matrix of connectivity between the PHG clusters (the PHC cluster, the postPRC cluster, and the antPRC cluster) and each of the HC coronal slices (left HC, 11 slices; right HC, 10 slices).
3. Results
3.1 PHG parcellation results in three clusters
In the PHG, we expected to identify two clusters, one in anterior PHG and one in posterior PHG, corresponding to the PRC and the PHC, respectively. Figure 2A shows the 28×28 connectivity homogeneity matrix of the PHG, which depicts the similarity of whole-brain FC maps for all slice pairs in left and right PHG. On the matrix, entries in a given column/row represented the connectivity similarity values for a given slice with all other slices in a brain region (i.e. left and right PHG). Surprisingly, from visual inspection of the connectivity homogeneity matrix, slices within each hemisphere appeared to segregate into three clusters, rather than two clusters. Slices in each cluster showed strong inter-correlations across left and right hemispheres and relatively low correlations between slices in different clusters.
Figure 2. Connectivity homogeneity matrices and dendrograms for the HC and PHG.
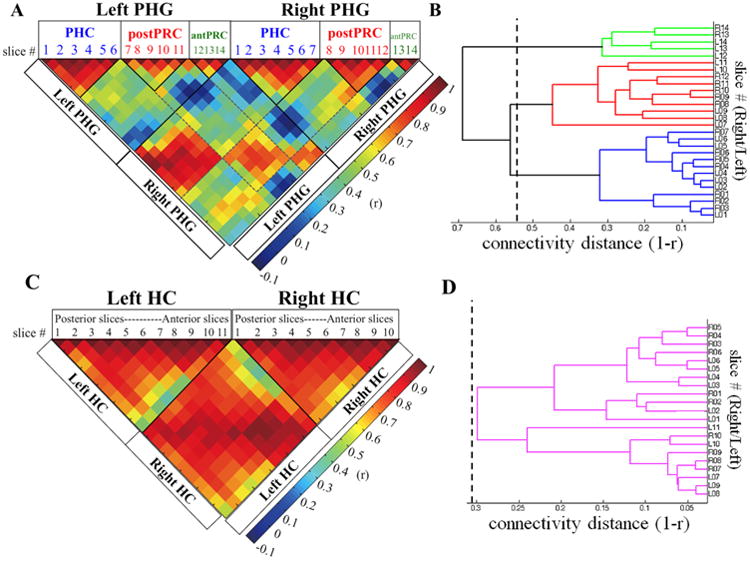
A) and C) are the connectivity homogeneity matrices for the PHG and HC. The entries for each column/row in the matrix are connectivity similarity values (r) between a given slice and all other slices in the PHG or HC. Solid lines indicate significant clusters identified in the dendrogram and dashed lines show the connectivity similarity for homologous clusters across left and right hemisphere. B) and D) are dendrograms for the PHG and HC. The x-axis is “connectivity distance” (1-r). The y-axis is number of the coronal slice, which indicates the physical location of a given slice in a brain region. The numbers correspond to column/row number labeled on the connectivity homogeneity matrix. L stands for left hemisphere and R stands for right hemisphere. The dashed lines on the dendrograms represent the connectivity distance threshold. Clusters to the right of the threshold are the significant clusters, highlighted in different colors (PHC: blue, postPRC: red, antPRC: green, HC: purple). A) The PHG connectivity homogeneity matrix. In the matrix, the first 14 columns/rows contain the connectivity similarity values for each of the left PHG coronal slice. The connectivity similarity values for right PHG coronal slices start from the 15th column/row. Three clusters were identified in the PHG: PHC (blue), postPRC (red), and antPRC (green). Boxes above the matrix contain slice numbers, which are separated into three clusters accordingly. B) The PHG dendrogram, showing the hierarchical relationships among coronal slices based on their connectivity distance. The dashed line on the dendrogram represents the connectivity distance threshold for the PHG (1-r = 0.5572, p< .05). Clusters to the right of the distance threshold are the three significant clusters: PHC (blue), postPRC (red), and antPRC (green). C) HC connectivity homogeneity matrix. In the matrix, the first 11 columns/rows are left HC coronal slices arranging from posterior to anterior along the long axis. The right HC coronal slices start at the 12th column/row and end at the 21st column/row. D) The HC dendrogram. The dashed line on the dendrogram represents the connectivity distance threshold for the HC (1-r = 0.3444, p< .05).
To quantitatively define clusters based on the connectivity homogeneity matrix, a hierarchical clustering algorithm was used. Three significant clusters were identified, such that these sets of slices were less similar to each other than what we would expect by chance, as defined by the connectivity distance threshold (1-r = 0.5572, p< .05) (Fig. 2B). The slice number on the dendrogram indicates to the column number on the connectivity homogeneity matrix. The blue cluster on the dendrogram corresponded well with slices that would be expected to comprise the PHC. This cluster consisted of 6 (y=-42 to y = -26) and 7 (y = -42 to y = -23) coronal slices in the left and right PHG respectively. The same cluster could be visually identified on the connectivity homogeneity matrix. In the matrix, the first 6 columns in the left PHG and the first 7 columns in the right PHG represent the connectivity similarity profiles for the coronal slices in the blue cluster, which corresponds to what would be identified as the PHC. The red and green clusters on the dendrogram, in turn, corresponded anatomically to what would be collectively identified as the PRC in neuroimaging studies. The anatomical transition between the blue and red clusters corresponded well with the anatomical boundary between the PHC and the PRC. This transition was one and two slices posterior to the gyrus intralimbicus (HC head) in the right and left PHG, respectively. The red cluster corresponded to slices in a posterior portion of the PRC (the postPRC cluster). The postPRC cluster consisted of five coronal slices in both left and right PHG (y = -25 to y = -10 and y = -22 to y = - 8) following the slices of the PHC cluster. In the connectivity homogeneity matrix, the five columns following the columns for the PHC cluster represent the connectivity similarity profiles for the coronal slices in the red cluster. Like the PHC cluster, these columns could be visually identified as a group based on homogeneity among their connectivity similarity profiles. Finally, the green cluster on the dendrogram consisted of the remaining anterior slices in each hemisphere, which anatomically corresponded with slices in the remaining anterior portion of the PRC (the antPRC cluster). The left antPRC cluster consisted of the three most anterior coronal slices (y = -9 to y = -2), whereas the right antPRC cluster consistent of the two most anterior slices (y = -7 to y = -2). The transition between the postPRC and antPRC clusters was around the hippocampal- amygdaloid transition area (HATA), at the most anterior end of the HC. In the left PHG, the transition between these two clusters was two slices posterior to the HATA. In the right PHG, the transition was one slice anterior to the HATA.
We performed a control analysis to confirm that the parcellation results were not driven by changes of the PHG masks along the longitudinal axis (Supplementary Figures 1 & 2). We generated a control mask that included only voxels in the lateral half of the medial bank of the collateral sulcus (from the fundus of the collateral sulcus to the midpoint of the medial bank of the collateral sulcus). This region is relatively consistent across the entire PHG longitudinal axis, and it is included in anatomical definitions of both the PRC and PHC. We applied the parcellation scheme described above to parcellate the PHG areas covered by this control mask based on similarity of the FC patterns. The aim was to see whether this control region could be separated into clusters similar to what we found by using the full PHG mask. Two significant clusters were identified in the control region (Supplementary Figure 2B). One cluster corresponded well with the antPRC cluster mentioned above (control-antPRC). The other cluster, control-postPHG, contained two major sub-clusters, each of which corresponded well with the postPRC and PHC clusters. Based on the control analysis, we can affirm that the three PHG clusters identified via the parcellation scheme were unlikely driven by differences in the PHG mask along the longitudinal axis. Finally, additional analyses ruled out that the parcellation was driven by differences in number of voxels or signal-to-noise ratio of each coronal slice (Supplementary Figure 3 & 4).
3.2 Networks connected with the three PHG clusters
Following the parcellation results, we examined the underlying brain networks that were associated with each cluster. Each cluster was used as a ROI in a new FC analysis, and a one-sample t-test was used to identify voxels that showed supra-threshold FC values associated with these ROIs (Fig. 3A-C). The PHC cluster and the postPRC cluster were associated with networks that have been previously identified in studies that examined FC for anatomically-defined PHC and PRC sub-regions (Kahn et al., 2008; Libby et al., 2012). The PHC cluster showed preferential connectivity with the posterior cingulate cortex (BA 23 and BA31), the retrosplenial cortex (BA 30), the precuneus (BA7), the inferior temporal gyrus (BA20), early visual areas (e.g. V1, V2, V3), and the thalamus (Fig. 3A). The PHC cluster also showed significant FC with voxels in the anterior cingulate gyrus (BA32 and BA24), the ventromedial superior frontal gyrus (BA10), the ventromedial prefrontal cortex (BA10), the dorsal lateral prefrontal cortex (BA9), the premotor cortex (BA6), the frontal eye field (BA8), the angular gyrus (BA39), fusiform gyrus, and the superior temporal gyrus (BA22).
The postPRC cluster preferentially connected to voxels in the anterior cingulate cortex (BA24), rostromedial prefrontal cortex (BA10), and the orbitofrontal cortex (BA11) (Fig. 3B). The postPRC cluster also showed significant FC with voxels in the posterior cingulate (BA31 and BA23), the retrosplenial cortex (BA30), the visual cortex V3, the angular gyrus (BA39), the premotor cortex (BA6), and the frontal eye field (BA8). In addition, the postPRC cluster had extensive temporal lobe connections, including the superior temporal gyrus (BA22), the middle temporal gyrus (BA21), the inferior temporal gyrus (BA20), and the fusiform gyrus (BA37).
Finally, the antPRC cluster exhibited high FC with a distinct fronto-insular network (Fig. 3C) consisting of the insula, the planum temporale, the orbitofrontal cortex (BA47), the postcentral gyrus (BA43), and the ventrolateral prefrontal cortex (BA45). The antPRC cluster also connected to ventral part of the premotor cortex (BA6), the amygdala, the anterior HC, and the ventral temporal lobe (BA20).
Paired t-tests (cluster corrected p< .05) were conducted to identify regions that showed preferential FC with each cluster (Fig. 4). As in the previous studies, voxels in the posterior-medial network, including the posterior cingulate, the retrosplenial cortex, and the precuneus, showed stronger connectivity with the PHC cluster than with the postPRC and antPRC clusters (Fig. 4A, B). Voxels in orbitofrontal areas and ventral temporopolar cortex, in turn, exhibited stronger FC with the postPRC than with the PHC and the antPRC clusters (Fig. 4A, C). Voxels in the insula and temporal pole showed stronger connectivity with the antPRC than with the PHC and the postPRC clusters (Fig. 4B, C). The PRC clusters (i.e. the postPRC and the antPRC clusters) had stronger connectivity with the ventral parietal cortex, the premotor cortex, the orbitofrontal areas, and the ventral temporopolar cortex. The antPRC cluster was more connected with the insular areas than the postPRC cluster. In the temporal lobe, both the postPRC and the antPRC clusters connected to the amygdala, the anterior HC, and the ventral temporal lobe. However, their connections to these regions followed their topographical relationships such that the postPRC cluster connected to more posterior portion of these regions and the antPRC cluster connected to relatively more anterior portion of these regions.
Figure 4. Differences among the networks of the three PHG clusters.
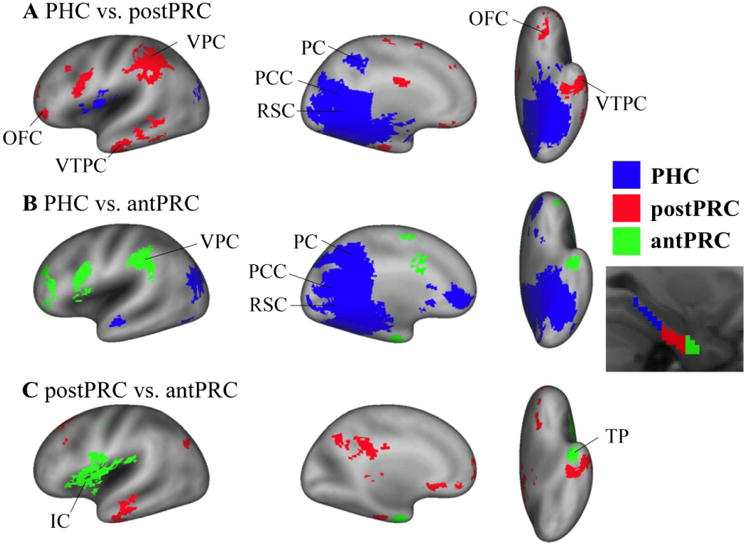
Paired t-tests were conducted to identify significant differences among the three networks for the three PHG clusters (i.e. PHC, postPRC, and antPRC) (cluster corrected p< .05). A) Differences between the PHC network and the postPRC network. Blue voxels represent significant voxels in the PHC network comparing with the postPRC network (PHC > postPRC). Conversely, red voxels represent significant voxels in the postPRC network comparing with the PHC network (postPRC > PHC). B) Differences between the PHC network and the antPRC network. Blue voxels represent significant voxels in the PHC network comparing with the antPRC network (PHC > antPRC). Green voxels represent significant voxels in the antPRC network comparing with the PHC network (antPRC > PHC). C) Differences between the postPRC network and antPRC network. Red voxels are the significant voxels in the postPRC network comparing with the antPRC network (postPRC > antPRC). Green voxels are the significant voxels in the antPRC network comparing with the postPRC network (antPRC > postPRC). IC: insular cortex. OFC: orbitofrontal cortex. PC: precuneus. PCC: posterior cingulated cortex. RSC: retrosplenial cortex. TP: temporal pole. VPC: ventral parietal cortex. VTPC: ventral temporopolar cortex.
3.3 The three PHG clusters had distinct subsequent memory effects
The previous analyses identified three regions that exhibited markedly different patterns of FC during the resting-state scans. Although the result is consistent with the idea that the PHG could be subdivided into three regions, it is unclear whether these FC distinctions are functionally meaningful or informative. To address this question, we interrogated activity in the three PHG clusters during a memory-encoding task (Fig. 5) that was performed by the same participants between the two resting state scans. During the task, participants incidentally encoded sentences describing an object's appearance, situational context, or spatial location (see Ritchey et al. 2014 for further details), and following the scan session, they were tested on memory for each association. Results of the clustering analysis described above were used to identify bilateral ROIs for the antPRC, postPRC, and PHC clusters, and activity in each ROI was separately examined for subsequently remembered associations and forgotten association. Results, summarized in (Fig. 5), revealed that the three PHG clusters showed different patterns of activation during memory encoding. A three-way ANOVA (Memory (hit, miss) × Condition (appearance, situational context, spatial location) × ROI (PHC, postPRC, antPRC), p< .05) analysis revealed that there was a significant two-way interaction between Condition and ROI (F(4,72)= 18.0551, p<.001) and a significant three-way interaction between Memory, Condition, and ROI (F(4,72)= 4.3469, p = 0.003). Putting the results together, we can see three different profiles of activation during memory encoding: the PHC cluster was preferentially involved in encoding of spatial associations, the antPRC cluster was preferentially involved in encoding of item-appearance associations, and the postPRC cluster was involved in encoding both spatial and item-appearance associations.
Figure 5. The three PHG clusters had distinct subsequent memory effects.
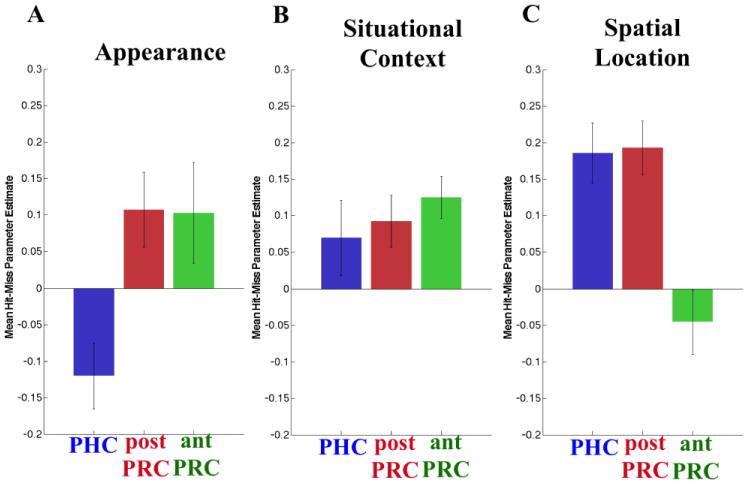
Subsequent memory (Dm) effects were compared for the three clusters (PHC (blue), postPRC (red), and antPRC (green)) across three task conditions: appearance encoding (A), situational context encoding (B), and spatial location encoding (C). Error bars denote the standard error of the mean parameter estimate.
3.4 HC parcellation results in a single cluster
The same parcellation procedure was applied to identify functionally different clusters along the longitudinal axis of the HC. A 21 × 21 connectivity homogeneity matrix was generated representing connectivity similarity among the whole-brain FC maps of all the coronal slices in the HC (Fig. 2C). In contrast to the PHG, the connectivity patterns were relatively homogeneous across the longitudinal axis of the HC. When we applied the hierarchical clustering algorithm to quantitatively determine significant clusters, all left and right HC slices were grouped into a single cluster. The connectivity distance threshold (1-r = 0.3444, p< .05) was larger than, but close to, the largest connectivity distance on the HC dendrogram (Fig. 2D). This result indicated that the FC maps of the HC coronal slices were not different enough to justify separating the slices into distinct clusters. Figure 3D illustrates the whole-brain FC map connected to the HC cluster (one-sample t-test, cluster corrected p< .05). Like the PHC posterior-medial network, the HC showed high FC with voxels in the posterior cingulate cortex (BA 23 and BA31) and the retrosplenial cortex (BA30). In the anterior-medial part of the brain, the HC cluster had connections with the anterior cingulate gyrus (BA12 and BA32), the ventromedial superior frontal gyrus (BA10), and the medial prefrontal cortex (BA10 and BA11). The HC cluster also connected to the superior temporal gyrus (BA22), the middle temporal gyrus, the angular gyrus (BA39), and part of the primary auditory cortex.
Although we did not identify any significant sub-clusters in the HC, there were two major branches dividing the HC into an anterior and a posterior part on the dendrogram (Fig. 2D). Slices in the anterior part of the HC corresponded anatomically to the hippocampal head, and slices in the posterior part of the HC corresponded to the hippocampal body and tail according to previously-published parcellation guidelines (Duvernoy et al., 2005; Franko et al. 2012). We did another set of one-sample t-test analyses to identify the associative brain networks for the anterior and posterior HC (Fig. 6A). Basically, the results revealed that both of the anterior and posterior HC showed high FC with the same brain network described above, with subtle topographic differences. The FC maps for the anterior HC appeared to show a larger extent of supra-threshold voxels in the anterior part of the brain, including the anterior-medial superior frontal gyrus (BA10) and the dorsal lateral prefrontal cortex (BA9), as well as in the insula and the superior and middle temporal gyrus (BA21 and BA22). The posterior HC, in turn, appeared to show a larger extent of supra-threshold voxels in the posterior cingulate cortex (BA23). Altogether, in contrast to the three PHG clusters, the anterior and posterior HC clusters appeared to be more similar to one another than they were different.
Figure 6. The MTL and whole-brain connectivity for the HC.
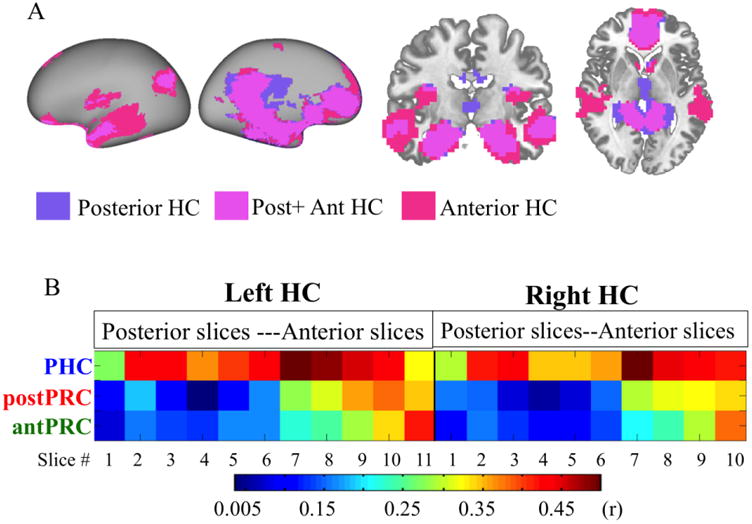
A ) Networks connected with the anterior and posterior HC. A one-sample t-test was conducted to identify voxels that showed suprathreshold FC values associated with different anterior (pink) and posterior (purple) HC (voxel-wise p<.001, cluster-corrected p<.05). Voxels significantly connected with both anterior and posterior HC are in magenta. B) FC between the three PHG clusters and the HC coronal slices. Entries in the matrix are FC values (r). Rows represent each of the three clusters in the PHG (PHC, postPRC, and antPRC). Columns represent each of the coronal slices in the left and right HC ranging from posterior to anterior.
3.5 FC of PHG clusters along long axis of the hippocampus
Although the whole-brain FC maps for the anterior and posterior HC were generally similar, prior work has shown that the intrinsic connectivity between the PHG and the HC might be heterogeneous along the HC longitudinal axis (Libby et al., 2012; Maass et al., 2015). We therefore investigated FC between the three PHG clusters and the HC coronal slices (Fig. 6B). Consistent with previous results, the PHC cluster exhibited high FC with the entire HC, whereas the adjacent postPRC cluster showed preferential connectivity with the anterior part of the HC. Although both of the two PRC clusters preferentially connected to the anterior part of the HC, the antPRC cluster had strong FC limited to the most anterior portion of the HC and the postPRC had a relatively broader anterior HC connectivity. Finally, the homologous brain regions shared similar HC connectivity across left and right hemisphere.
4. Discussion
In this study, we used FC-based parcellation to characterize the functional organization of the human PHG and hippocampus. Surprisingly, our results revealed that the PHG was subdivided into three clusters that could be distinguished on the basis of whole-brain FC and on the basis of activation profiles during memory encoding. Two of these clusters corresponded closely to the PRC and PHC, but the third antPRC cluster has not been previously considered in theoretical accounts of human MTL function. In contrast to the PHG, we did not observe significant evidence that the HC could be functionally subdivided, although we did observe a trend for differences between anterior and posterior HC regions. Below, we consider the implications of these findings for understanding MTL contributions to cognition.
4.1 Three sub-regions of the human PHG
The main finding in this study was that the PHG could be subdivided into three sub-regions along the longitudinal axis based on connectivity differences. The most posterior cluster exhibited high functional connectivity with a posterior-medial network that included the posterior cingulate cortex, the retrosplenial cortex, the precuneus, and occipital areas. This pattern of results corresponds well to previous studies that examined resting-state FC profiles for the posterior PHC, as defined on the basis of structural landmarks (Kahn et al., 2008; Libby et al., 2012). A second, “postPRC” cluster exhibited strong connectivity with an anterior-temporal network that included the orbitofrontal cortex, the superior temporal gyrus, the middle temporal gyrus, and the inferior temporal gyrus. This connectivity pattern corresponds well to what would be expected based on the anatomical connectivity of the PRC (Suzuki and Naya, 2014), and to some extent, with previous analyses of FC profiles for the PRC (Kahn et al., 2008; Libby et al., 2012). The third, “antPRC” cluster exhibited strong connectivity with an fronto-insular network that included the insula, the planum temporale, the orbitofrontal cortex, the postcentral region, and the ventrolateral prefrontal cortex. Critically, we found that FC maps for the left and right homologous clusters were much more similar than for non-homologous clusters, serving as an internal replication of the results.
To validate the FC-based parcellation, we examined activation profiles in the three PHG regions during a memory-encoding task. Results revealed dissociable profiles of encoding-related activity across the three regions. Whereas activity in the antPRC cluster was specifically associated with successful encoding of appearance associations, activity in the PHC cluster was associated with successful encoding of spatial location associations. Finally, activity in the postPRC cluster was associated with both appearance and spatial location encoding. During situational context encoding, we saw progressive changes along the longitudinal axis of the PHG, with the antPRC cluster showing the largest memory effect among the three clusters. The PHC cluster showed the smallest memory effect and the memory effect for the postPRC cluster was in the between of the antPRC and PHC clusters. These results demonstrate that the FC differences between the three clusters are indicative of meaningful functional differences. These differences would not have been apparent in a traditional analysis of activity in structurally-defined PRC and PHC sub-regions, and as such, the results highlight the added value of the FC-based parcellation approach introduced here.
4.2 The antPRC cluster: a homologue of area 36d?
Given the traditional view that the MTL neocortex can be subdivided into two regions, the PHC and the PRC, it might be tempting to dismiss the current results by assuming that the antPRC module corresponds instead to “temporopolar cortex”, a poorly understood region that is sometimes treated as separate from the PRC (Kondo et al., 2003; Olson et al., 2007). A closer look at the anatomy of rodent and monkey PRC, however, indicates that the antPRC cluster exhibits compelling parallels with a region known as area 36d. Anatomical studies of rats and monkeys have consistently distinguished between area 36d, located in the anterior-dorsal part of the PRC, and adjacent PRC regions (i.e., the rest of the area 36 and area 35). In rats, cytoarchitectonic and histochemical analyses revealed unique radially oriented cells and deep layers of myelinated fibers that distinguished area 36d from the rest of the area 36 (Burwell, 2001). Further, a clustering analysis divided retrograde tracer injection sites in the rat area 36 into a dorsal and a ventral clusters based on their neocortical connectivity. The injection sites in the dorsal cluster, approximately corresponding to the rat area 36d, show strong connections with the primary auditory regions and the ventral temporal association areas (Burwell and Amaral 1998b; Burwell, 2001). Other unique intrinsic and extrinsic connection patterns of the rat dorsal area 36 also separated it from the rest of the PRC, including their strong connectivity with the PHC, and the dorsal-ventral connectivity gradient with the ERC and with itself (Burwell & Amaral 1998a; Burwell & Amaral 1998b). In nonhuman primates, area 36d has strong internal connectivity but limited connections with the rest of the PRC (Lavenex et al., 2004). In monkeys, the major cortical inputs to the area 36d originated from the rostral superior temporal gyrus, an auditory processing area (Suzuki & Amaral 1994b). In contrast, the rest of the monkey PRC has strong cortical inputs from the visual areas TE and TEO and has weaker connections with the dorsal bank of the superior temporal sulcus (STS), the insular cortex, and the orbitofrontal areas.
In sum, the human antPRC cluster identified in the current study may be the homologous region of the area 36d in animals. First, the physical location of the antPRC cluster was close to the anterior PRC in monkeys and dorsal PRC in rats, which are where area 36d locates in both animals. Further, features of intrinsic, whole-brain, and PHC connectivity for the antPRC corresponds well with the connectivity patterns of monkey area 36d and the rat dorsal area 36 (Suzuki & Amaral 1994b; Burwell & Amaral, 1998a; Burwell & Amaral 1998b; Kondo et al., 2003; Lavenex et al., 2004).
The present results have implications for understanding the functions of human PRC. Almost every model of PRC function emphasizes its role in memory for objects, with some models placing more emphasis on visual object perception, and others broadening the functions to encompass representations of “items” or “entities” (Meunier et al., 1993; Brown and Aggleton, 2001; Murray et al., 2001; Bussey et al., 2002; Brown et al., 2010; Graham et al., 2010). Based on the present results, one could speculate that these descriptions only apply to the postPRC, whereas the antPRC might instead encode information related to inputs conveyed by auditory association areas, interoceptive information conveyed by the insula, and information about goals and task context conveyed by regions in lateral prefrontal cortex (Murray et al., 2001; Petrides, 2005). Alternatively, it is possible that an “item” is separately and differentially processed by antPRC and postPRC might, with postPRC preferentially emphasizing visual properties and antPRC preferentially emphasizing auditory properties, personal significance, and relevance for action selection (Belin et al. 2000; Belin at al. 2002; Olson et al., 2007; Petkov et al. 2008; Munoz-Lopez et al., 2015). These ideas are of course speculative, and further research is needed to better understand how the antPRC, postPRC, and PHC separately and collectively encode the attributes of an event.
4.3 Differences in the HC MTL and whole-brain connectivity
Despite strong evidence for anatomical and functional differences between the dorsal and ventral HC in rodents (Moser and Moser, 1998; Fanselow and Dong, 2010; Strange et al., 2014), we did not observe strong evidence for a parallel connectivity-based dissociation in humans. Although we only identified a single cluster in the HC, there was a weak trend for differences between anterior and posterior HC (Fig. 2D). The pattern of HC connectivity with neocortical areas within and outside of the MTL reflected this ambiguity (Fig. 6). In the MTL, the two PRC clusters heavily connected to the anterior part of the HC whereas the PHC cluster heavily connected to the entire HC with a preference in the hippocampal head and tail (Fig. 6B). In contrast, FC with neocortical areas outside of the MTL was relatively homogeneous along the longitudinal axis of the HC (Fig. 6A). Both the anterior and posterior HC showed high connectivity with regions in the default network, including the posterior cingulate cortex, the ventral anterior cingulate cortex, and the ventromedial prefrontal cortex. Consistent with findings in rodents (Jones and Witter, 2007), there were minor connectivity differences, such that the anterior HC showed more extensive FC with voxels in the dorsal prefrontal cortex and lateral temporal lobe, whereas the posterior HC showed slightly more extensive HC with voxels in the posterior cingulate cortex and the precuneus (Fig. 6A). These differences were relatively small, however, relative to the visually apparent distinctions in FC profiles associated with the three PHG clusters.
Although our study did not reveal strong evidence for functional heterogeneity in the HC, there are several reasons why one should be careful in interpreting this null result. First, neuroanatomy studies suggest that functional differences along the longitudinal axis of the HC should differ across subfields (Malykhin et al., 2010; Chase et al., 2015), with the strongest gradients to be seen in CA1 and subiculum. This possibility could be assessed in an analysis of high-resolution fMRI data using an approach that would allow parcellation both along the longitudinal and transverse axes of the hippocampus. Alternatively, it is possible that, during the resting state, hippocampal regions function in unison, but during performance of tasks that differentially engage targets of anterior and posterior HC, differences would become apparent. For instance, one might see large FC differences between anterior and posterior HC during navigation in empty spatial contexts or during processing of emotionally arousing objects (Poppenk et al., 2013; Strange et al., 2014), in contrast to the more homogenous pattern of FC seen during rest.
4.4 Limitations
There are some limitations to the current study. First, the parcellation reported here, like most previously reported cortical parcellations (Cohen et al. 2008; Yeo et al. 2011; Wig et al., 2013; Nelson et al, 2013; Gordon et al., 2014), is based on a group-level analysis. Researchers are starting to develop parcellation schemes for identifying cortical systems at single subject level (Wang et al., 2015; Gordon et al., 2015), but single-subject parcellation requires a large amount of resting-state data. For instance, Wang et al. (2015) collected an hour of resting- scan data to evaluate intrinsic functional connectivity in single-subjects. It is also worth noting that these studies used previously identified group-level cortical networks in order to guide the single subject analysis.
As we mentioned above, the hippocampus could not be subdivided into multiple clusters. In the current study, hippocampal subfields were collapsed within each coronal slice, and thus, the proportions of each subfield in each coronal slice varied. The intrinsic FC patterns we obtained for each coronal slice should be a mixed result combining connectivity patterns for different subfields at different longitudinal levels. Thus, high-resolution fMRI data might be necessary to identify functional subdivisions in the hippocampus, and the parcellation would be best performed at the level of voxels, rather than using coronal slices as seed regions.
5. Conclusion
In the current report, three functionally different clusters were identified via the parcellation procedure in the PHG. Our results suggest that the PHC, postPRC, and antPRC each affiliate with different large-scale neocortical association networks, providing a possible substrate for their role in associating different kinds of information during memory formation. The hippocampus, in turn, is positioned to integrate information across the three networks and to modulate the flow of information within each network. Although further research is needed to better understand the how FC is related to the anatomical and functional organization of the MTL, our results are sufficient to establish the feasibility and validity of FC-based parcellation of the MTL. Furthermore, by revealing new information about the distinction between antPRC and postPRC, the present study indicates that the use of FC in combination with anatomy could be a more effective means of identifying MTL subdivisions than traditional approaches based solely on anatomical landmarks. This is an important advance because the ability to accurately and noninvasively identify human MTL sub-regions is a prerequisite for understanding the neural basis of memory and cognition in healthy individuals and clinical populations.
Supplementary Material
Highlights.
Functional regions within the MTL were identified based on connectivity patterns
The PHG divided into three distinct clusters along the longitudinal axis
No significantly different subregions were identified in the hippocampus
Connectivity-based clusters were functionally dissociable in a memory encoding task
Acknowledgments
We would like to thank Szu-Wen Fang for assistance with figure preparation. Funding was provided by the National Institutes of Health Grant R01MH083734 to C.R., K99MH103401 to M.R., and by a National Security Science and Engineering Faculty Fellowship to C.R. (Office of Naval Research Grant N00014-15-1-0033). Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Institutes of Health, the Office of Naval Research, or the U.S. Department of Defense.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aminoff EM, Kveraga K, Bar M. The role of the parahippocampal cortex in cognition. Trends Cogn Sci. 2013;17(8):379–390. doi: 10.1016/j.tics.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes KA, Cohen AL, Power JD, Nelson SM, Dosenbach YB, Miezin FM, Schlaggar BL et al. Identifying Basal Ganglia divisions in individuals using resting-state functional connectivity MRI. Front Syst Neurosci. 2010;418 doi: 10.3389/fnsys.2010.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter MG, Murray EA. Opposite relationship of hippocampal and rhinal cortex damage to delayed nonmatching-to-sample deficits in monkeys. Hippocampus. 2001;11(1):61–71. doi: 10.1002/1098-1063(2001)11:1<61∷AID-HIPO1021>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Belin P, Zatorre RJ, Ahad P. Human temporal-lobe response to vocal sounds. Brain Res Cogn Brain Res. 2002;13(1):17–26. doi: 10.1016/s0926-6410(01)00084-2. [DOI] [PubMed] [Google Scholar]
- Belin P, Zatorre RJ, Lafaille P, Ahad P, Pike B. Voice-selective areas in human auditory cortex. Nature. 2000;403(6767):309–312. doi: 10.1038/35002078. [DOI] [PubMed] [Google Scholar]
- Brown MW, Aggleton JP. Recognition memory: what are the roles of the perirhinal cortex and hippocampus? Nat Rev Neurosci. 2001;2(1):51–61. doi: 10.1038/35049064. [DOI] [PubMed] [Google Scholar]
- Brown MW, Warburton EC, Aggleton JP. Recognition memory: material, processes, and substrates. Hippocampus. 2010;20(11):1228–1244. doi: 10.1002/hipo.20858. [DOI] [PubMed] [Google Scholar]
- Burwell RD. Borders and cytoarchitecture of the perirhinal and postrhinal cortices in the rat. J Comp Neurol. 2001;437(1):17–41. doi: 10.1002/cne.1267. [DOI] [PubMed] [Google Scholar]
- Burwell RD, Amaral DG. Cortical afferents of the perirhinal, postrhinal, and entorhinal cortices of the rat. J Comp Neurol. 1998a;398(2):179–205. doi: 10.1002/(sici)1096-9861(19980824)398:2<179::aid-cne3>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- Burwell RD, Amaral DG. Perirhinal and postrhinal cortices of the rat: interconnectivity and connections with the entorhinal cortex. J Comp Neurol. 1998b;391(3):293–321. doi: 10.1002/(sici)1096-9861(19980216)391:3<293::aid-cne2>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Burwell RD, Witter MP, Amaral DG. Perirhinal and postrhinal cortices of the rat: a review of the neuroanatomical literature and comparison with findings from the monkey brain. Hippocampus. 1995;5(5):390–408. doi: 10.1002/hipo.450050503. [DOI] [PubMed] [Google Scholar]
- Bussey TJ, Saksida LM, Murray EA. Perirhinal cortex resolves feature ambiguity in complex visual discriminations. Eur J Neurosci. 2002;15(2):365–374. doi: 10.1046/j.0953-816x.2001.01851.x. [DOI] [PubMed] [Google Scholar]
- Chase HW, Clos M, Dibble S, Fox P, Grace AA, Phillips ML, Eickhoff SB. Evidence for an anterior-posterior differentiation in the human hippocampal formation revealed by meta-analytic parcellation of fMRI coordinate maps: focus on the subiculum. Neuroimage. 2015;113:44–60. doi: 10.1016/j.neuroimage.2015.02.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen AL, Fair DA, Dosenbach NU, Miezin FM, Dierker D, Van Essen DC, Petersen SE et al. Defining functional areas in individual human brains using resting functional connectivity MRI. Neuroimage. 2008;41(1):45–57. doi: 10.1016/j.neuroimage.2008.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diana RA, Yonelinas AP, Ranganath C. Imaging recollection and familiarity in the medial temporal lobe: a three-component model. Trends Cogn Sci. 2007;11(9):379–386. doi: 10.1016/j.tics.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Ding SL, Van Hoesen GW. Organization and detailed parcellation of human hippocampal head and body regions based on a combined analysis of Cyto- and chemoarchitecture. J Comp Neurol. 2015;523(15):2233–2253. doi: 10.1002/cne.23786. [DOI] [PubMed] [Google Scholar]
- Duvernoy HM. The Human Hippocampus: Functional Anatomy, Vascularization and Serial Sections with MRI. Berlin Heidelberg: Springer Verlag; 2015. [Google Scholar]
- Eichenbaum H, Yonelinas AP, Ranganath C. The medial temporal lobe and recognition memory. Annu Rev Neurosci. 2007;30:123–152. doi: 10.1146/annurev.neuro.30.051606.094328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanselow MS, Dong HW. Are the dorsal and ventral hippocampus functionally distinct structures? Neuron. 2010;65(1):7–19. doi: 10.1016/j.neuron.2009.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franko E, Insausti AM, Artacho-Perula E, Insausti R, Chavoix C. Identification of the human medial temporal lobe regions on magnetic resonance images. Hum Brain Mapp. 2014;35(1):248–256. doi: 10.1002/hbm.22170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon EM, Laumann TO, Adeyemo B, Huckins JF, Kelley WM, Petersen SE. Generation and Evaluation of a Cortical Area Parcellation from Resting-State Correlations. Cereb Cortex. 2014 doi: 10.1093/cercor/bhu239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon EM, Laumann TO, Adeyemo B, Petersen SE. Individual Variability of the System-Level Organization of the Human Brain. Cerebral Cortex. 2015:bhv239–14. doi: 10.1093/cercor/bhv239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham KS, Barense MD, Lee AC. Going beyond LTM in the MTL: a synthesis of neuropsychological and neuroimaging findings on the role of the medial temporal lobe in memory and perception. Neuropsychologia. 2010;48(4):831–853. doi: 10.1016/j.neuropsychologia.2010.01.001. [DOI] [PubMed] [Google Scholar]
- Insausti R, Juottonen K, Soininen H, Insausti AM, Partanen K, Vainio P, Pitkanen A, et al. MR volumetric analysis of the human entorhinal, perirhinal, and temporopolar cortices. AJNR Am J Neuroradiol. 1998;19(4):659–671. [PMC free article] [PubMed] [Google Scholar]
- Jones BF, Witter MP. Cingulate cortex projections to the parahippocampal region and hippocampal formation in the rat. Hippocampus. 2007;17(10):957–976. doi: 10.1002/hipo.20330. [DOI] [PubMed] [Google Scholar]
- Kahn I, Andrews-Hanna JR, Vincent JL, Snyder AZ, Buckner RL. Distinct cortical anatomy linked to subregions of the medial temporal lobe revealed by intrinsic functional connectivity. J Neurophysiol. 2008;100(1):129–139. doi: 10.1152/jn.00077.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo H, Saleem KS, Price JL. Differential connections of the temporal pole with the orbital and medial prefrontal networks in macaque monkeys. J Comp Neurol. 2003;465(4):499–523. doi: 10.1002/cne.10842. [DOI] [PubMed] [Google Scholar]
- Lacy JW, Stark CE. Intrinsic functional connectivity of the human medial temporal lobe suggests a distinction between adjacent MTL cortices and hippocampus. Hippocampus. 2012;22(12):2290–2302. doi: 10.1002/hipo.22047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavenex P, Suzuki WA, Amaral DG. Perirhinal and parahippocampal cortices of the macaque monkey: projections to the neocortex. J Comp Neurol. 2002;447(4):394–420. doi: 10.1002/cne.10243. [DOI] [PubMed] [Google Scholar]
- Lavenex P, Suzuki WA, Amaral DG. Perirhinal and parahippocampal cortices of the macaque monkey: Intrinsic projections and interconnections. J Comp Neurol. 2004;472(3):371–394. doi: 10.1002/cne.20079. [DOI] [PubMed] [Google Scholar]
- Libby LA, Ekstrom AD, Ragland JD, Ranganath C. Differential connectivity of perirhinal and parahippocampal cortices within human hippocampal subregions revealed by high-resolution functional imaging. J Neurosci. 2012;32(19):6550–6560. doi: 10.1523/JNEUROSCI.3711-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maass A, Berron D, Libby LA, Ranganath C, Duzel E. Functional subregions of the human entorhinal cortex. Elife. 2015;4 doi: 10.7554/eLife.06426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malykhin NV, Lebel RM, Coupland NJ, Wilman AH, Carter R. In vivo quantification of hippocampal subfields using 4.7 T fast spin echo imaging. Neuroimage. 2010;49(2):1224–1230. doi: 10.1016/j.neuroimage.2009.09.042. [DOI] [PubMed] [Google Scholar]
- Meunier M, Bachevalier J, Mishkin M, Murray EA. Effects on visual recognition of combined and separate ablations of the entorhinal and perirhinal cortex in rhesus monkeys. J Neurosci. 1993;13(12):5418–5432. doi: 10.1523/JNEUROSCI.13-12-05418.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra A, Rogers BP, Chen LM, Gore JC. Functional connectivity-based parcellation of amygdala using self-organized mapping: a data driven approach. Hum Brain Mapp. 2014;35(4):1247–1260. doi: 10.1002/hbm.22249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser MB, Moser EI. Functional differentiation in the hippocampus. Hippocampus. 1998;8(6):608–619. doi: 10.1002/SICI)1098-1063(1998)8:6&lt;608∷AID-HIPO3&gt;3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Munoz-Lopez M, Insausti R, Mohedano-Moriano A, Mishkin M, Saunders RC. Anatomical pathways for auditory memory II: information from rostral superior temporal gyrus to dorsolateral temporal pole and medial temporal cortex. Front Neurosci. 2015;9158 doi: 10.3389/fnins.2015.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray EA, Richmond BJ. Role of perirhinal cortex in object perception, memory, and associations. Curr Opin Neurobiol. 2001;11(2):188–193. doi: 10.1016/s0959-4388(00)00195-1. [DOI] [PubMed] [Google Scholar]
- Navarro Schroder T, Haak KV, Zaragoza Jimenez NI, Beckmann CF, Doeller CF. Functional topography of the human entorhinal cortex. Elife. 2015;4 doi: 10.7554/eLife.06738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson SM, McDermott KB, Wig GS, Schlaggar BL, Petersen SE. The critical roles of localization and physiology for understanding parietal contributions to memory retrieval. Neuroscientist. 2013;19(6):578–591. doi: 10.1177/1073858413492389. [DOI] [PubMed] [Google Scholar]
- Olson IR, Plotzker A, Ezzyat Y. The Enigmatic temporal pole: a review of findings on social and emotional processing. Brain. 2007;130(Pt 7):1718–1731. doi: 10.1093/brain/awm052. [DOI] [PubMed] [Google Scholar]
- Passingham RE, Stephan KE, Kotter R. The anatomical basis of functional localization in the cortex. Nat Rev Neurosci. 2002;3(8):606–616. doi: 10.1038/nrn893. [DOI] [PubMed] [Google Scholar]
- Petkov CI, Kayser C, Steudel T, Whittingstall K, Augath M, Logothetis NK. A voice region in the monkey brain. Nat Neurosci. 2008;11(3):367–374. doi: 10.1038/nn2043. [DOI] [PubMed] [Google Scholar]
- Petrides M. Lateral prefrontal cortex: architectonic and functional organization. Philos Trans R Soc Lond B Biol Sci. 2005;360(1456):781–795. doi: 10.1098/rstb.2005.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poppenk J, Evensmoen HR, Moscovitch M, Nadel L. Long-axis specialization of the human hippocampus. Trends Cogn Sci. 2013;17(5):230–240. doi: 10.1016/j.tics.2013.03.005. [DOI] [PubMed] [Google Scholar]
- Pruessner JC, Kohler S, Crane J, Pruessner M, Lord C, Byrne A, Evans AC, et al. Volumetry of temporopolar, perirhinal, entorhinal and parahippocampal cortex from high-resolution MR images: considering the variability of the collateral sulcus. Cereb Cortex. 2002;12(12):1342–1353. doi: 10.1093/cercor/12.12.1342. [DOI] [PubMed] [Google Scholar]
- Ranganath C, Ritchey M. Two cortical systems for memory-guided behaviour. Nat Rev Neurosci. 2012;13(10):713–726. doi: 10.1038/nrn3338. [DOI] [PubMed] [Google Scholar]
- Ritchey M, Yonelinas AP, Ranganath C. Functional connectivity relationships predict similarities in task activation and pattern information during associative memory encoding. J Cogn Neurosci. 2014;26(5):1085–1099. doi: 10.1162/jocn_a_00533. [DOI] [PubMed] [Google Scholar]
- Scoville WB, Milner B. Loss of recent memory after bilateral hippocampal lesions. J Neurol Neurosurg Psychiatry. 1957;20(1):11–21. doi: 10.1136/jnnp.20.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strange BA, Witter MP, Lein ES, Moser EI. Functional organization of the hippocampal longitudinal axis. Nat Rev Neurosci. 2014;15(10):655–669. doi: 10.1038/nrn3785. [DOI] [PubMed] [Google Scholar]
- Suzuki WA, Amaral DG. Perirhinal and parahippocampal cortices of the macaque monkey: cortical afferents. J Comp Neurol. 1994a;350(4):497–533. doi: 10.1002/cne.903500402. [DOI] [PubMed] [Google Scholar]
- Suzuki WA, Amaral DG. Topographic organization of the reciprocal connections between the monkey entorhinal cortex and the perirhinal and parahippocampal cortices. J Neurosci. 1994b;14(3 Pt 2):1856–1877. doi: 10.1523/JNEUROSCI.14-03-01856.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki WA, Naya Y. The perirhinal cortex. Annu Rev Neurosci. 2014;37:39–53. doi: 10.1146/annurev-neuro-071013-014207. [DOI] [PubMed] [Google Scholar]
- Wang D, Buckner RL, Fox MD, Holt DJ, Holmes AJ, Stoecklein S, et al. Parcellating cortical functional networks in individuals. 18(12):1853–1860. doi: 10.1038/nn.4164. n.d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wig GS, Laumann TO, Cohen AL, Power JD, Nelson SM, Glasser MF, Petersen SE et al. Parcellating an individual subject's cortical and subcortical brain structures using snowball sampling of resting-state correlations. Cereb Cortex. 2014;24(8):2036–2054. doi: 10.1093/cercor/bht056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo BT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M, Buckner RL, et al. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol. 2011;106(3):1125–1165. doi: 10.1152/jn.00338.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeineh MM, Engel SA, Bookheimer SY. Application of cortical unfolding techniques to functional MRI of the human hippocampal region. Neuroimage. 2000;11(6 Pt 1):668–683. doi: 10.1006/nimg.2000.0561. [DOI] [PubMed] [Google Scholar]
- Zola-Morgan S, Squire LR, Amaral DG, Suzuki WA. Lesions of perirhinal and parahippocampal cortex that spare the amygdala and hippocampal formation produce severe memory impairment. J Neurosci. 1989b;9(12):4355–4370. doi: 10.1523/JNEUROSCI.09-12-04355.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


