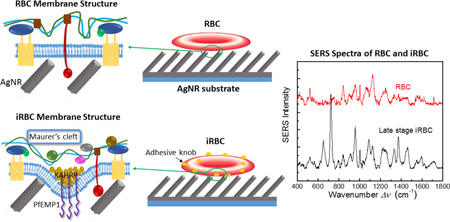
Abstract
Surface enhanced Raman spectra (SERS) of normal red blood cells (RBCs) and Plasmodium falciparum infected RBCs (iRBCs) at different post invasion time were obtained based on silver nanorod array substrates. Distinct spectral differences were observed due to the cell membrane modification of RBCs during malaria infection. The SERS spectra of ring stage iRBCs had a characteristic Raman peak at Δν = 1599 cm−1 as compared to those of normal RBCs, while the trophozoite and schizoid stages had identical SERS spectra with a characteristic peak at Δν = 723 cm−1, which is significantly different from ring stage iRBCs, consistent with ongoing modification of the iRBC membrane. Since ring stage iRBCs of P. falciparum are found circulating in blood, such a difference provides a new strategy for rapid malaria detection. The limit of detection as well as the ability to detect a mixed iRBC and RBC solution were also investigated.
Keywords: SERS, Malaria, diagnostics, cell membrane, Red blood cell, Biosensors
Graphical abstract
Differentiable SERS spectra for malaria infected red blood cells (iRBCs) at different post invasion time were obtained for the first time. This provide a new method for malaria detection.
The peak assignment for iRBCs at different post invasion time is consistent with ongoing modification of the iRBC membrane.
Such a finding demonstrates that SERS can be used as a research/analytic tool to track the host cell membrane evolution for Eukaryotic pathogen infection.
Background
Malaria is a global health problem, an estimated 3.2 billion people in 97 countries and territories are at risk of being infected with malaria and developing disease, and 1.2 billion are at high risk (> 1 in 1000 chance of getting malaria in a year). According to the latest estimation, 198 million cases of malaria occurred globally in 2013 (uncertainty range 124–283 million) and the disease led to 584 000 deaths (uncertainty range 367 000–755 000)1.
Malaria diagnosis is crucial for malaria control and prevention. Current diagnostic methods including blood smear examination2, rapid diagnostic tests (RDTs)2,3, polymerase chain reaction (PCR)4,5, fluorescent microscopy6, antigen detection7, and new method like hemozoin (malaria pigment, by-product of malaria infection in red blood cell) detection8–11. Among those methods, blood smear examination (microscopy method) and RDT are considered as the gold standard, World Health Organization (WHO) recommends that all persons of all ages in all epidemiological settings with suspected malaria should receive a parasitological confirmation of diagnosis by either microscopy or RDT. Blood smear examination requires well trained personnel to achieve high sensitivity and specificity. RDTs can rapid detect malaria, but none of the rapid tests are currently as sensitive as blood smear examination, nor as cheap3. Highly sensitive method like PCR are criticized by its high cost and complicated procedure. Therefore, a low-cost, fast and sensitive strategy is still needed for malaria detection.
Surface enhanced Raman spectroscopy (SERS) is a highly sensitive optical spectroscopic detection technique based on the Raman signal enhancement from metallic (usually silver or gold) nanostructured substrates. Raman spectroscopy and SERS have long been attempted for malaria detection8,12–15, but for Raman spectroscopy, works are focused on plasma detection12,15 while for SERS, the strategy is dominated by hemozoin detection8,13,14. Wood et al. obtained the Raman spectra of hemozoin within the food vacuole of Plasmodium falciparum (P. falciparum) trophozoites, enabling the investigation of hemozoin within its natural environment for the first time16. Yuen et al. used magnetic field enriched Surface-enhanced resonance Raman spectroscopy (SERRS) to detect hemozoin and reached a detection limit of 30 parasites/µl8, which was suggested to be used in early infection stage. However, diagnosis based on hemozoin detection are hindered by the fact that only the earliest stages of infected RBCs (iRBCs), referred to as ring stage iRBC, are circulating in peripheral blood17, and ring stage iRBCs contain very little hemozoin. Ring stage parasites mature further to the trophozoite stage, which is characterized by the appearance of hemozoin17, but it also expresses the cytoadherence protein PfEMP1 on the host cell surface, which will adhere to endothelial cells and results in the sequestration of iRBCs in peripheral blood18–20. This mechanism will make later stage iRBCs less practicable in clinical detection, which can only detect circulating iRBCs in a blood sample. In addition, for most SERS works mentioned above, the SERS substrates are noble metal (i.e. Ag or Au) nanoparticles, which usually lack reproducibility due to the variation in particle size distribution, non-uniform particle aggregation, and other factors.
In fact, the reproducibility and sensitivity of SERS substrates can be solved by the unique silver nanorod (AgNR) substrates fabricated by oblique angle deposition (OAD) method21. The AgNR substrates are highly sensitive and reproducible with a SERS enhancement factor of >108 and a batch to batch variation of <15%21,22. Our previous work and other’s work have demonstrated that SERS can be used to differentiate bacteria species, serotypes, and strains23,24,25. The origin of such a differentiation is due to cell membrane difference for different bacteria. Since in malaria the host RBC membrane undergoes significant structural and biochemical modification regulated by complex molecular activities of the parasites, it is expected that the SERS spectra of normal RBCs and iRBCs will be different. Such a difference can be used to specifically identify malaria infection. Therefore, a new strategy for malaria detection is to directly measure the SERS spectral difference between RBCs and iRBCs. We have previously demonstrated that AgNR substrate has excellent reproducibility for SERS measurements of bacteria and viruses23,22. Therefore, we expect that to use AgNR substrate to perform SERS measurement of RBCs and iRBCs could successfully achieve the detection of malaria infection.
In this work, we have performed detailed SERS measurement of normal RBCs and P. falciparum infected RBCs in blood at different stages. The spectra showed significant difference for RBCs and iRBCs at different stages. Based on a simple chemometric method, the principle component analysis (PCA), we could differentiate three different stages, ring, trophozoite, and schizont stage iRBCs, from uninfected RBCs. The SERS peak changes for iRBC spectra compared with RBC spectra are consistent with the membrane component changes of iRBC reported in literature. Trophozoite and schizont stage iRBCs showed more obvious spectra change compared with ring stage iRBCs due to the rapid parasite growth and heavily modification of host cells. We had also explored the limit of detection for RBCs and late stage iRBCs, as well as the ability to detect a mixture of iRBCs and RBCs.
Methods
AgNR substrates fabrication
The AgNR substrates were fabricated by an oblique angle deposition (OAD) method. The detailed fabrication procedure was introduced in previous reports21,25,26. Briefly, glass microscopic slides were cut into 1 × 1 cm2 pieces, and were cleaned by piranha solution (80% sulfuric acid, 20% hydrogen peroxide v/v) before being loaded into an electron beam deposition system. A 20-nm titanium film and then a 200-nm silver film were first deposited at a rate of 0.2 nm/s and 0.3 nm/s, respectively, monitored in-situ by a quartz crystal microbalance (QCM). The substrates were then tilted to 86° with respect to the incident vapor, and AgNRs were grown at a deposition rate of 0.3 nm/s until a total QCM thickness reading of 2000 nm was reached.
Blood and reagents
Human O+ RBCs were either purchased from Interstate Blood Bank, Inc. or donated by healthy volunteers. This research was approved by the Institutional Review Board (IRB) at the University of Georgia (no. 2013102100); all donors signed consent forms. Unless otherwise noted, all chemicals and reagents for this study were either purchased from Sigma Aldrich or Fisher Scientific.
Parasite culture and synchronization
P. falciparum CS2 parasites were maintained in continuous culture according to routine methods27. Parasites were cultured at 4% hematocrit in O+ RBCs. Cultures were maintained in 25 cm2 or 75 cm2 tissue culture flasks at 37°C under a gas mixture of 90% nitrogen/5% oxygen/5% carbon dioxide and in complete culture medium made up of RPMI containing 25 mM HEPES, 0.05 mg/mL hypoxanthine, 2.2 mg/mL NaHCO3 (J.T. Baker), 0.5% Albumax I (Gibco), 2 g/L glucose, and 0.01 mg/mL gentamicin. Primarily ring-stage cultures were treated routinely with 5% D-Sorbitol to achieve synchronous cultures.
Culture health and stage of parasites were assessed daily and at each time point. Thin film blood smears were fixed with methanol, and stained with Giemsa for analysis by light microscopy. Images were acquired using a Nikon Eclipse E400 microscope fitted with a Nikon Digital Sight DS-5M-L1 camera (Nikon Instruments Inc.).
SERS measurement and data analysis
Infection time dependent SERS measurement of iRBCs for different infection stages was performed. The 48-hour life cycle of iRBC was divided into 6 time periods with 8 h duration: early ring stage (T = 8 h); mid-late ring stage (T = 16 h); trophozoite stage (T = 24 h); mid-late trophozoite (T = 32 h); schizont stage (T = 40 h) and very early ring stage mixed with some very late schizont stage (T = 48 h).
Since later stage iRBCs could be easily enriched using Percoll gradient method, thus, for SERS measurements, 2µL of RBCs or enriched iRBCs solutions (at concentrations from C = 3×108/ml to 3×106/ml) were drop casted on to the substrate surface, and allowed to dry under ambient conditions. All SERS spectra were acquired by an Enwave ProRaman-L-785A2 Raman analyzer (Enwave Optronics, Irvine, CA) equipped with a Leica DM300 microscope (with objectives of 4×, 10×, 40× and 100×), with a 785 nm near-IR diode laser as the excitation source, and two dimensional temperature regulated CCD arrays with 16 bit digitization. All spectra ranged from 400 – 1,800 cm−1 with a spectral resolution of 6 cm−1 were taken for an acquisition time of 30 s, regardless of the objectives used. A 40× objective was used to measure the SERS spectra of concentration dependent RBCs and late stage iRBCs. Such an objective would focus the laser beam into a ~ 20 µm diameter spot and the laser power was set to be 30 mW. SERS spectra were acquired from nine randomly selected spots, the final spectrum presented was averaged from 9 spectra.
However, the parasitemia of ring stage iRBCs from infected human blood is often less than 1%, and it cannot be enriched by Percoll gradient which was used for late stage iRBC parasitemia enrichment. In order to obtain the SERS spectra of ring stage iRBCs, we decided to measure the SERS spectra of single RBC/iRBC cells from the sample. Since the spectra of normal RBCs have well characteristic features, the cells give different SERS spectra would be due to ring stage iRBCs. This will also be confirmed with dye experiments. A 100× objective was used to better focus laser on single cells. The diameter of the laser spot was about 8 µm, which is comparable to the size of a RBC, and the excitation power was set to 8 mW. The SERS spectra were obtained by manually parking the laser spot on top of a single cell under the microscope. For this case, at least 200 cells were measured. In order to make sure the unique spectra did come from ring stage iRBCs, a confirmation experiment was performed based on cell staining. After the cells were stained with Giemsa, the iRBCs can be distinguished by the bluish staining of the parasite nucleus, while the RBCs remained transparent allowing easy differentiation by microscope. The SERS spectra of iRBCs and RBCs after staining were measured separately and showed significant difference, and both were distinct from the spectra of mixed dye.
Data analysis was performed using Origin software 9.0 version (OriginLab Corporation, Northampton, MA). PCA was conducted with Matlab 2000b (The MathWorks, Inc., Natick, MA) using the PLS toolbox (Eigenvector Research, Inc., Wenatchee, WA). Savitzky–Golay derivation, normalization, and mean center process were used to treat the raw spectra data prior to PCA analysis.
Results
Differentiating uninfected and infected RBCs at different stages
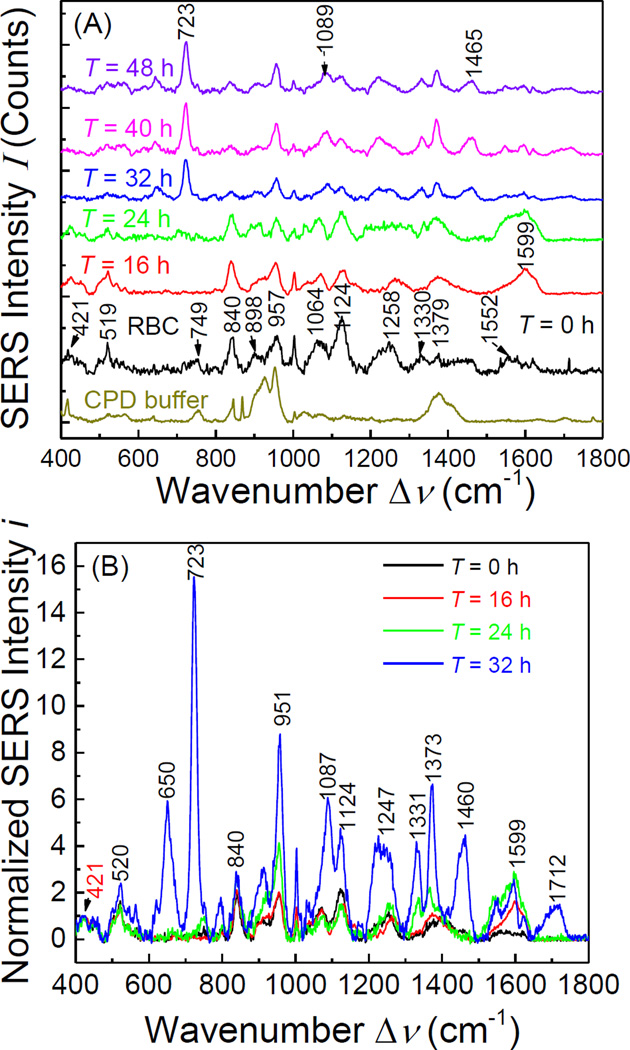
Figure 1 shows representative SERS spectra of RBCs and iRBCs at different infection stages. The overall concentrations of RBCs and late stage iRBCs (Trophozoite and Schizont stages, T ≥ 32 h) are about 3×108/ml, with parasitemia of 9%. However, for the early stage infected samples (T = 16 and 24 h), the spectra shown in Figure. 1A were obtained from single cell measurements. Normal RBC membrane contains 40% lipid, 52% protein and 8% Carbohydrate, thus the SERS characteristic peaks of RBCs shown in Figure. 1 are from these major membrane components, and the detailed peak assignments are listed in Table 1. The characteristic peaks at Δν = 421, 519, 957, 1064, 1124, 1330 and 1379 cm−1 are assigned to the vibrational mode of lipids28–31, peaks at Δν = 749, 1258, 1552 and 1618 cm−1 represent vibrational modes of membrane proteins30,32,33, and peaks at Δν = 840 and 898 cm−1 could be assigned to saccharide bands34.
Figure. 1.
(A) The SERS spectra of CPD buffer alone, RBC, and iRBCs with different infection time (from ring stage to schizont stage); and (B) the normalized spectra for RBC and iRBC with different infection time.
Table 1.
Peak assignment for SERS spectra of RBCs and iRBCs at different infection time.
| Observed Raman Shift Δν (cm−1) |
RBC | ring stage iRBC (16h) |
ring stage iRBC (24h) |
late stage iRBC (32h) |
Vibrational Mode Assignment |
|---|---|---|---|---|---|
| 421 | s | s | m | vw | Cholesterol |
| 520 | s | s | m | w | S-S disulfide stretching in proteins |
| 650 | w | s | C-C twisting mode of tyrosine |
||
| 723 | vw | vw | vw | vs | characteristic for phospholipids, lipid assignment |
| 748 | w | w | m | w | Symmetric breathing of tryptophan (protein assignment) |
| 840 | vs | vs | s | w | Glucose-saccharide band |
| 951 | s | s | vs | vs | ν3(CH3) of proteins |
| 1087 | m | m | m | s |
ν(C- C) skeletal of acyl backbone in lipid |
| 1124 | vs | s | m | m | Proteins, lipids: C–N and C–C stretch |
| 1247 | m | m | m | m | Amide III (collagen assignment) |
| 1373 | m | m | s | s | The most pronounced saccharide band |
| 1554 | m | ν(C=C), tryptophan (protein assignment), ν(C=C), porphyrin |
|||
| 1599 | vs | vs | w | Amide I band of proteins-Due to C=O stretching |
|
| 1712 | m | ν(C=O)OH (amino acids aspartic & glutamic acid) |
Note the abbreviation represents the peak strength: vw – very weak, w – weak, m – medium, s – strong, vs – very strong.
The three major components of RBC membrane were significantly modified during the 48-hour intra-erythrocytic cycle by complex molecular activities of the parasites. The size, deformability, and stability of RBC membrane was also changed. During intra-RBC development of Plasmodium falciparum, the host cell plasma membranes display a significantly decreased content of cholesterol and sphingomyelin, an increased ratio of phospholipid/cholesterol35,36. And the parasite extensively modifies the host cell through the export of a large number of proteins17,35–38. It is expected that the change of SERS spectra as a function of infectious time T shall reflects this overall trend.
At very early ring stage (T = 8 h), the SERS spectra of iRBC (not shown) showed no difference from the spectra of RBCs, which indicated that the RBC membrane components did not experience a significant change during early infection. At the late ring stage (T = 16 h), the SERS peak positions of iRBC spectrum, compared to RBC spectrum, began to show clear changes at some wavenumbers. By analyzing the relative peak intensity ratios and considering the spectra variation from the measurements (11.71%), we found that the mutual SERS peak intensity ratios at Δν = 421, 520, 840, 951, and 1064 cm−1 remained almost unchanged as compared to the corresponding ratios in RBC spectrum, but the peaks at Δν = 1124, 1258, 1379, and 1599 cm−1 had obvious changes. To better compare the relative changes among different spectra, we noticed that the intensity ratio of Δν = 421 and 520 cm−1 peaks did not change for all the SERS spectra from different iRBC stages. Therefore, we replot SERS spectra of different iRBC stages in Figure. 1B by normalizing them with their respective peak intensity at Δν = 421 cm−1. Since the peak at Δν = 421 cm−1 originated from the Cholesterol in the membrane, which has been demonstrated to decrease with T17,35–38, the normalized spectra, iT, in Figure. 1B only represent the relative intensity change for different T. As shown in Figure. 1B, for T = 16 h, since i16 at Δν = 520, 840, 951, and 1064 cm−1 did no changed as compared to i0, which indicated the relative amount of S-S and C-H bonds from the proteins with respect to cholesterol did not change significantly. But at Δν = 1124 cm−1 decreased, while and at Δν = 1370 and 1599 cm−1 increased, and especially there was a significant increase of ≈ 0.4 to ≈ 1.8. This indicated that the relative amount of amino acids on membrane with respect to cholesterol increased. According to the membrane protein evolution during malaria infection, those changes could result from Maurer’s clefts evolution and His-rich protein KAHRP expression39.
When the time post invasion reached T = 24 h, similar to the spectrum at T = 16 h, the majority of the SERS peak positions did not change. However, i24 at Δν = 1064 and 1124 cm−1 decreased slightly compared to i0 while i24 at Δν = 840, 951, 1370, and 1599 cm−1 increased significantly. In particular, increased to ~ 2.8, which indicated the further increase of the relative amount of amino acids on membrane. Note that compared to ring stage at T = 16 h, Maurer’s clefts continue to evolve and KAHRP, as well as PfEMP1, proteins continue to assemble into knobs on the RBC membrane39. Thus dramatically change in membrane proteins is expected and the SERS peaks at Δν = 840, 951, and 1370, especially at Δν = 1599 cm−1, reflect such a change.
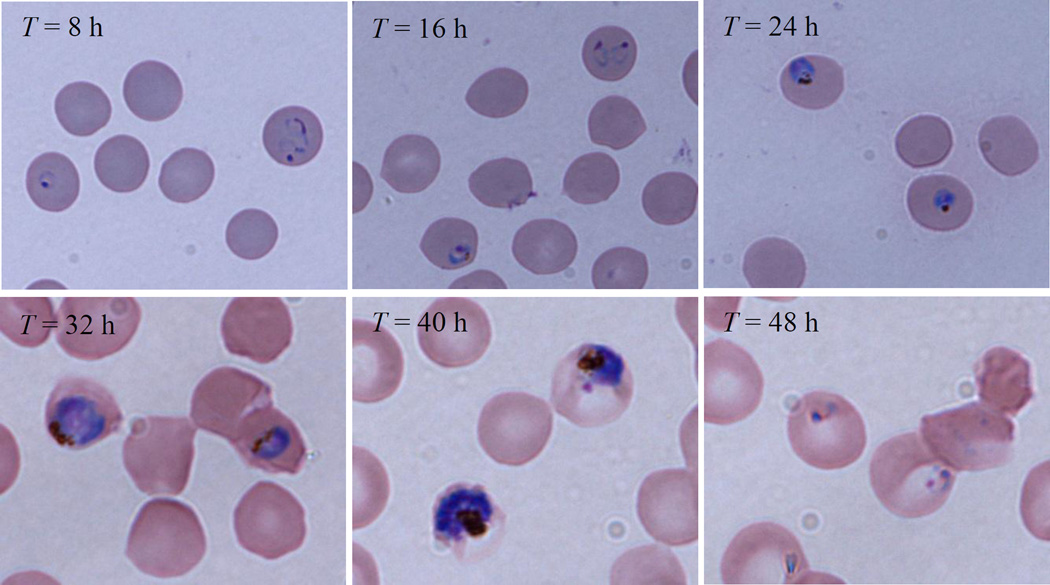
When T ≥ 32 h, all the later stage iRBCs, i.e., trophozoite and schizont stages, have the same SERS spectra. This came as no surprise, because the major membrane components change are completed in the trophozoite stage during malaria infection, and the RBC membrane is dominated by the protein knobs with parasite adhesion protein PfEMP1 (P. falciparum erythrocyte membrane protein 1)39. The most significant changes were the appearance of four new peaks at Δν = 646, 723, 1460, and 1712 cm−1. The Δν = 646 and 723 cm−1 peaks are characteristic peaks for lipid according to Drescher40, while the Δν = 1460 and 1712 cm−1 peaks result from vibrational modes in proteins. In addition, i32 also increased significantly at peaks Δν = 951, 1089, 1124, 1247, 1330 and 1377 cm−1, which represent the structure change of membrane lipids and proteins28,30,33,41. However, (≈ 2.6) stayed almost the same as , which indicated that the peak at Δν = 1599 cm−1 is the most appropriate SERS peak to characterize the ring stage iRBCs. Figure 2 shows the microscope images of each post invasion time.
Figure. 2.
Microscope images of iRBCs for each post invasion time.
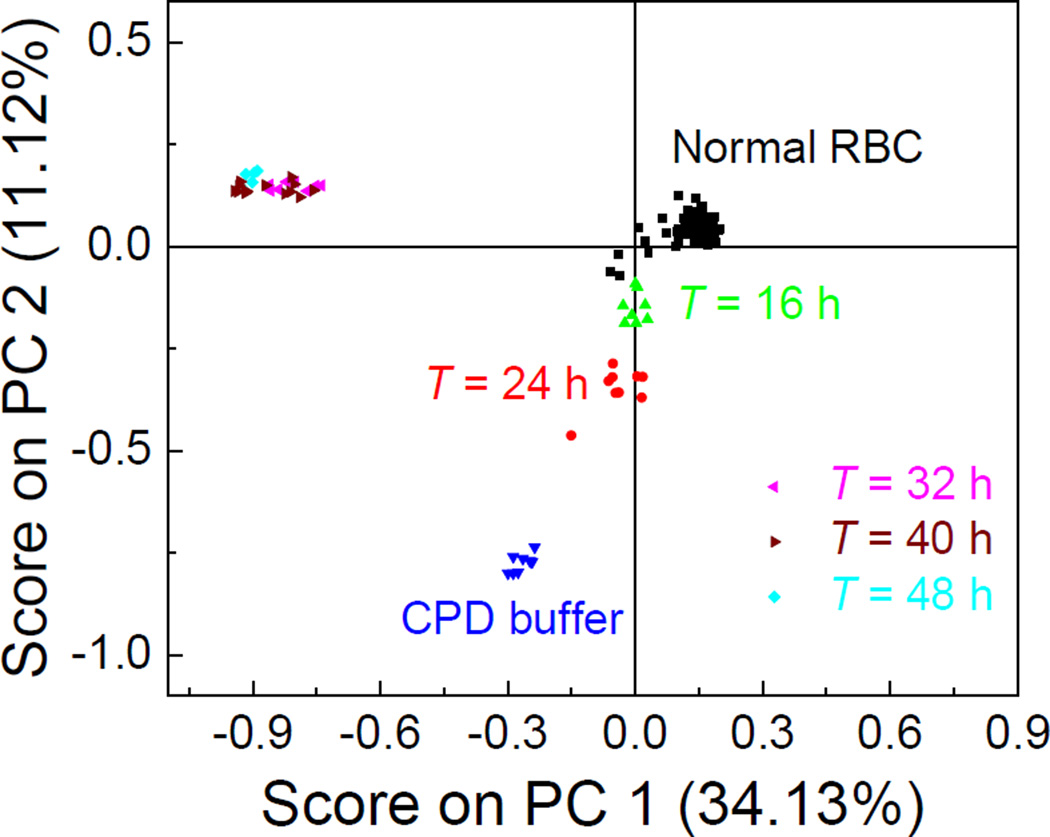
This surface protein evolution for different iRBC stages by SERS spectra can be further illustrated by using a chemometric method, the principle component analysis (PCA), to analyze the SERS spectra. The PCA method is a statistical tool to reduce multidimensional data to lower dimensions while retaining most of the information, and is widely used for spectral analysis42. Figure 3 plots the scores of the principle component (PC), PC 1 and PC 2, for RBC, CPD buffer, and iRBCs at different stages. The spectra of uninfected RBC and iRBCs at different stages were well separated into different clusters, which indicated those spectra are essentially different, PCA gives a more intuitive results than directly analysis spectral difference by raw spectra. As shown in Figure. 3, around 200 RBC spectra were gathered in one close cluster, indicating the consistency and reproducibility of the SERS spectra of RBC. For iRBC samples, though the T = 16 h spectra formed one cluster, it was very close to the cluster of RBCs. However, when T increased to 24 h, the iRBC cluster separated well away from the RBC cluster, but the T = 16 h and T = 24 h clusters were still close, which demonstrated the gradual changes in the spectra, as demonstrated in Figure. 1B. When T ≥ 32 h, as expected, all the spectra were grouped into one cluster, which means the SERS spectra from late stage iRBCs (trophozoite and schizont stages) were dominated by the membrane surfaces.
Figure. 3.
PCA score plot for RBCs and different stages of iRBCs based on their SERS spectra.
Limit of detection for uninfected RBC and infected RBC
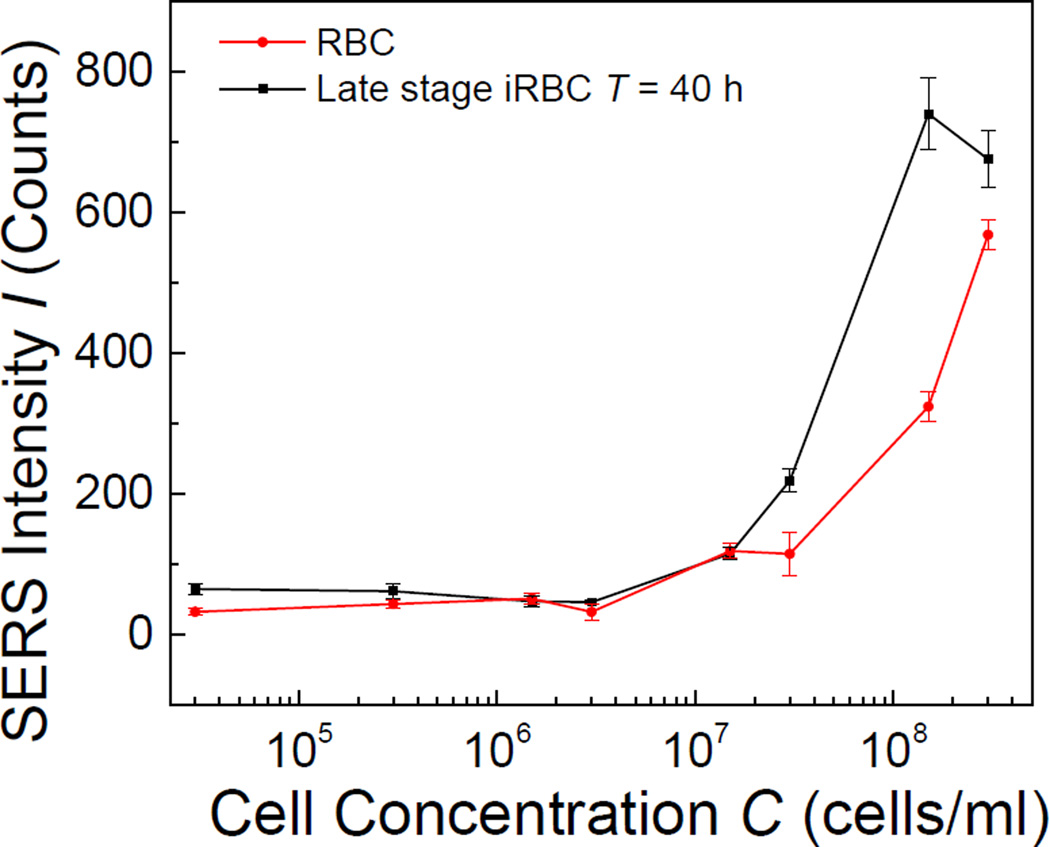
Concentration dependent SERS measurements of both RBCs and late stage iRBCs (T = 40 h) were also performed by the portable Raman system with the 40× objective. Such a measurement could be used to determine the limit of detection (LOD) of bulk RBCs and iRBCs for SERS measurement. The SERS peak intensity I1258 for RBCs at Δν = 1258 cm−1, which was assigned to amide III vibration band of proteins33, and I723 for iRBCs at Δν = 723 cm−1, which represents C-C-N+ symmetric stretching in phosphatidylcholine (lipid assignment)43 and C-S stretching (protein assignment)29, were plotted as a function of cell concentration C in Figure. 4. The corresponding SERS intensities at Δν = 1258 cm−1 and Δν = 723 cm−1 of the CPD buffer were used as reference for C = 0. As shown in Figure. 4, for both RBCs and iRBCs, the corresponding intensity of characterization peak increased monotonically with C. The LOD is defined as the lowest RBC/iRBC concentration at which the peak intensities I1258 or I723 are significantly different from that of CPD buffer (control). Thus, for Δν = 1258 cm−1, the threshold value was 76 counts, and for Δν = 723 cm−1, the threshold was 62 counts. Therefore, we determined that the LOD for both RBCs and iRBCs was the same, CLOD = 1.5×107/ml.
Figure. 4.
The plot of SERS peak intensities I1258 or I723 for RBCs and schizont stage iRBCs versus the cell concentration.
Those values are consistent with theoretical calculation. Ideally the presence of RBCs/iRBCs in a solution can be detected if at least one RBC/iRBC cell is within the laser spot of the Raman instrument. By considering the laser diameter d ≈ 20 µm, a sample volume of V = 2 µL spread uniformly on AgNR surface forming a sample spot with a diameter D ≈ 2.1 mm. Assuming that the cells were uniformly distributed on the sample spot, then the lowest concentration to have the laser spot seeing at least one cell is calculated to be Cmin = 2.2×107/ml, which is close to the value measured from experiments.
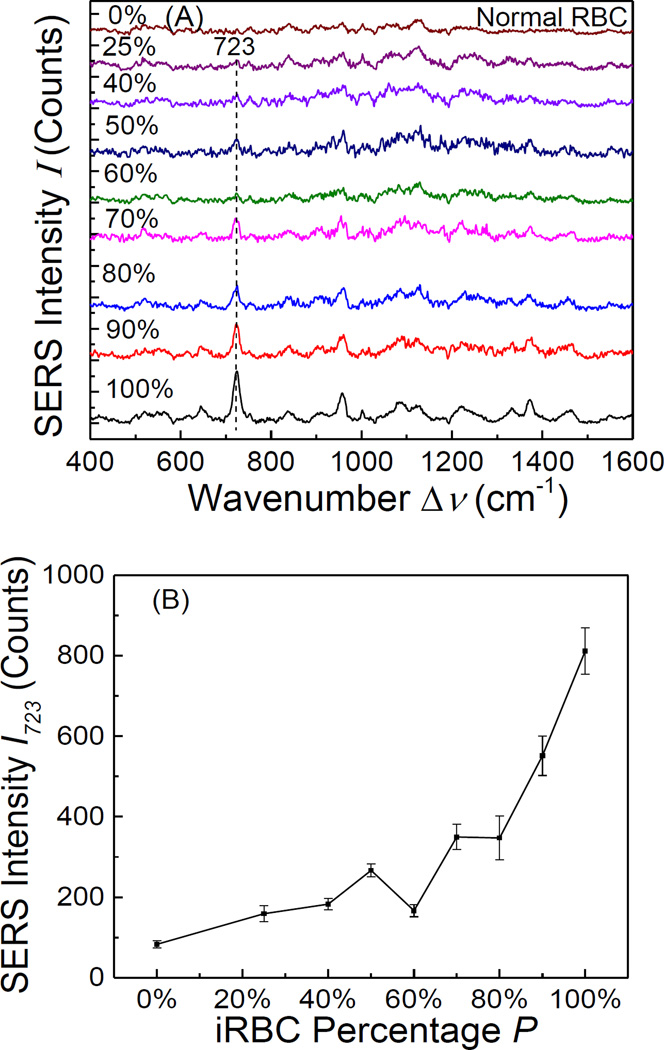
For practical sensing, the iRBCs and RBCs are mixed together in a real blood sample. Though later stage iRBCs are not circulating in blood, and only ring stage iRBCs exist in real blood sample, experimentally one could not design different ring stage iRBC mixtures due to the purification difficulty of ring stage iRBC. Thus, to test the detection capability of SERS, we performed SERS measurements of mixures of RBCs and later stage iRBCs (T = 40 h, since they can be easily separated and purified) with different iRBC percentage P from 90% to 22.5%. Figure 5A shows the average SERS spectrum for different P. The SERS intensity of the dominant peak of iRBCs, Δν = 723 cm−1, increased almost monotonically with P, which is clearly demonstrated in Figure. 5B. Similarly, if the SERS spectra of RBCs were used as the reference (P = 0), one could apply the 3σ rule to determine the minimum P that can be detected. From the data shown in Figure. 5B, such a threshold is P = 60 %. This is a fairly high iRBC percentage. From practical point of view, the percentage of circulating ring stage iRBCs in blood is typically less than 1%, which is significantly lower than 60%. Thus, directly spreading the blood droplet on a SERS substrate to perform the malaria detection is not practical. However, as shown in our experiments, if one can measure the SERS signal one cell by one cell, one can identify late ring stage iRBC unambiguously. Thus, to apply SERS for practical malaria detection, or other related detection associated with cell membrane changes, a fast and portable SERS imaging instrument is needed.
Figure. 5.
(A) The SERS spectra of the mixtures of RBCs and schizont stage iRBCs at a fixed cell concentration of 3×108/ml but with different mixing percentage P; and (B) the plot of SERS peak intensity I723 of the mixtures versus the iRBC percentage P.
Discussion
For the first time, we demonstrated that the SERS spectra of RBCs and iRBCs can be used to differentiate different stages of Plasmodium infected RBCs, in particular the SERS spectra from ring stage iRBCs were different from those of RBCs and late stage iRBCs. This provides a new potential strategy to directly detect ring stage iRBCs. The change of the SERS signals in different iRBC stages is consistent with the evolution of RBC cell membrane after merozoite invasion, especially the membrane proteins expressed during the Plasmodium life cycle in RBC host. Such a finding demonstrates that SERS can be used as a research/analytic tool to track the host cell membrane evolution for Eukaryotic pathogen infection. In addition, the SERS measurements have an ultimate sensitivity of a single cell, and to make the SERS malaria detection a practical method, a reliable SERS imaging technique needs to be developed.
Acknowledgments
This work was supported in part by the National Science Foundation (CBET-1064228), and the National Institutes of Health (2T32AI060546-06).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors whose names are listed above certify that they have NO affiliations with or involvement in any organization or entity with any financial interest, or non-financial interest in the subject matter or materials discussed in this manuscript.
Reference
- 1.2014-World Malaria Report. 2014;2013:2013. [Google Scholar]
- 2.Wongsrichanalai C, Barcus MJ, Muth S, Sutamihardja A, Wernsdorfer WH. A review of malaria diagnostic tools: Microscopy and rapid diagnostic test (RDT) Am. J. Trop. Med. Hyg. 2007;77:119–127. [PubMed] [Google Scholar]
- 3.Moody a. Rapid Diagnostic Tests for Malaria Parasites Rapid Diagnostic Tests for Malaria Parasites. Clin. Microbiol. Rev. 2002;15:66–78. doi: 10.1128/CMR.15.1.66-78.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hermsen CC, et al. Detection of Plasmodium falciparum malaria parasites in vivo by real-time quantitative PCR. Mol. Biochem. Parasitol. 2001;118:247–251. doi: 10.1016/s0166-6851(01)00379-6. [DOI] [PubMed] [Google Scholar]
- 5.Snounou G, et al. High sensitivity of detection of human malaria parasites by the use of nested polymerase chain reaction. Mol. Biochem. Parasitol. 1993;61:315–320. doi: 10.1016/0166-6851(93)90077-b. [DOI] [PubMed] [Google Scholar]
- 6.Hänscheid T. Diagnosis of malaria: A review of alternatives to conventional microscopy. Clin. Lab. Haematol. 1999;21:235–245. doi: 10.1046/j.1365-2257.1999.00220.x. [DOI] [PubMed] [Google Scholar]
- 7.Beadle C, et al. Diagnosis of malaria by detection of Plasmodium falciparum HRP-2 antigen with a rapid dipstick antigen-capture assay. Lancet. 1994;343:564–568. doi: 10.1016/s0140-6736(94)91520-2. [DOI] [PubMed] [Google Scholar]
- 8.Yuen C, Liu Q. Magnetic field enriched surface enhanced resonance Raman spectroscopy for early malaria diagnosis. J. Biomed. Opt. 2012;17:017005. doi: 10.1117/1.JBO.17.1.017005. [DOI] [PubMed] [Google Scholar]
- 9.Wood BR, et al. within a Sectioned Erythrocyte. Nano. 2011;11:1868–1873. doi: 10.1021/nl103004n. [DOI] [PubMed] [Google Scholar]
- 10.Peng WK, et al. Micromagnetic resonance relaxometry for rapid label-free malaria diagnosis. Nat. Med. 2014;20:1069–1073. doi: 10.1038/nm.3622. [DOI] [PubMed] [Google Scholar]
- 11.Delahunt C, Horning MP, Wilson BK, Proctor JL, Hegg MC. Limitations of haemozoin-based diagnosis of Plasmodium falciparum using dark-field microscopy. Malar. J. 2014;13:147. doi: 10.1186/1475-2875-13-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bilal M, et al. Optical diagnosis of malaria infection in human plasma using Raman spectroscopy. J. Biomed. Opt. 2015;20:017002. doi: 10.1117/1.JBO.20.1.017002. [DOI] [PubMed] [Google Scholar]
- 13.Wood BR, et al. Resonance Raman microscopy in combination with partial dark-field microscopy lights up a new path in malaria diagnostics. Analyst. 2009;134:1119–1125. doi: 10.1039/b822603b. [DOI] [PubMed] [Google Scholar]
- 14.Garrett NL, et al. Bio-sensing with butterfly wings: naturally occurring nano-structures for SERS-based malaria parasite detection. Phys. Chem. Chem. Phys. 2015 doi: 10.1039/c4cp04930f. [DOI] [PubMed] [Google Scholar]
- 15.Hobro AJ, Konishi A, Coban C, Smith NI. Raman spectroscopic analysis of malaria disease progression via blood and plasma samples. Analyst. 2013;138:3927–3933. doi: 10.1039/c3an00255a. [DOI] [PubMed] [Google Scholar]
- 16.Wood BR, et al. Raman imaging of hemozoin within the food vacuole of Plasmodium falciparum trophozoites. FEBS Lett. 2003;554:247–252. doi: 10.1016/s0014-5793(03)00975-x. [DOI] [PubMed] [Google Scholar]
- 17.Grüring C, et al. Development and host cell modifications of Plasmodium falciparum blood stages in four dimensions. Nat. Commun. 2011;2:165. doi: 10.1038/ncomms1169. [DOI] [PubMed] [Google Scholar]
- 18.Kriek N, et al. Characterization of the pathway for transport of the cytoadhereince-mediating protein, PfEMP1, to the host cell surface in malaria parasite-infected erythrocytes. Mol. Microbiol. 2003;50:1215–1227. doi: 10.1046/j.1365-2958.2003.03784.x. [DOI] [PubMed] [Google Scholar]
- 19.Bonnefoy S, Ménard R. Deconstructing Export of Malaria Proteins. Cell. 2008;134:20–22. doi: 10.1016/j.cell.2008.06.040. [DOI] [PubMed] [Google Scholar]
- 20.Miller LH, Baruch DI, Marsh K, Doumbo OK. The pathogenic basis of malaria. Nature. 2002;415:673–679. doi: 10.1038/415673a. [DOI] [PubMed] [Google Scholar]
- 21.Chaney SB, Shanmukh S, Dluhy Ra, Zhao YP. Aligned silver nanorod arrays produce high sensitivity surface-enhanced Raman spectroscopy substrates. Appl. Phys. Lett. 2005;87:20–22. [Google Scholar]
- 22.Driskell JD, et al. The use of aligned silver nanorod arrays prepared by oblique angle deposition as surface enhanced raman scattering substrates. J. Phys. Chem. C. 2008;112:895–901. [Google Scholar]
- 23.Wu X, Xu C, Tripp Ra, Huang Y, Zhao Y. Detection and differentiation of foodborne pathogenic bacteria in mung bean sprouts using field deployable label-free SERS devices. Analyst. 2013;138:3005–3012. doi: 10.1039/c3an00186e. [DOI] [PubMed] [Google Scholar]
- 24.Jarvis RM, Goodacre R. Characterisation and identification of bacteria using SERS. Chem. Soc. Rev. 2008;37:931–936. doi: 10.1039/b705973f. [DOI] [PubMed] [Google Scholar]
- 25.Chu H, Huang Y, Zhao Y. Silver nanorod arrays as a surface-enhanced Raman scattering substrate for foodborne pathogenic bacteria detection. Appl. Spectrosc. 2008;62:922–931. doi: 10.1366/000370208785284330. [DOI] [PubMed] [Google Scholar]
- 26.Shanmukh S, et al. Rapid and sensitive detection of respiratory virus molecular signatures using a silver nanorod array SERS substrate. Nano Lett. 2006;6:2630–2636. doi: 10.1021/nl061666f. [DOI] [PubMed] [Google Scholar]
- 27.Trager W, Jensen JB. Human Malaria Parasites in Continuous Culture. Journal of Parasitology. 1976;91:484–486. doi: 10.1645/0022-3395(2005)091[0484:HMPICC]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 28.Malini R. Oral Tissue: A Raman Spectroscopy Study. Biopolymers. 2006;83:297–312. doi: 10.1002/bip.20398. [DOI] [PubMed] [Google Scholar]
- 29.Stone N, Kendall C, Smith J, Crow P, Barr H. Raman spectroscopy for identification of epithelial cancers. Faraday Discuss. 2004;126:141–157. doi: 10.1039/b304992b. discussion 169–183. [DOI] [PubMed] [Google Scholar]
- 30.Cheng WT, Liu MT, Liu HN, Lin SY. Micro-Raman spectroscopy used to identify and grade human skin pilomatrixoma. Microsc. Res. Tech. 2005;68:75–79. doi: 10.1002/jemt.20229. [DOI] [PubMed] [Google Scholar]
- 31.Jyothi Lakshmi R, et al. Tissue Raman spectroscopy for the study of radiation damage: brain irradiation of mice. Radiat. Res. 2002;157:175–182. doi: 10.1667/0033-7587(2002)157[0175:trsfts]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 32.Huang Z, et al. Near-infrared Raman spectroscopy for optical diagnosis of lung cancer. Int. J. Cancer. 2003;107:1047–1052. doi: 10.1002/ijc.11500. [DOI] [PubMed] [Google Scholar]
- 33.Chan JW, et al. Micro-Raman spectroscopy detects individual neoplastic and normal hematopoietic cells. Biophys. J. 2006;90:648–656. doi: 10.1529/biophysj.105.066761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krafft C, Neudert L, Simat T, Salzer R. Near infrared Raman spectra of human brain lipids. Spectrochim. Acta - Part A Mol. Biomol. Spectrosc. 2005;61:1529–1535. doi: 10.1016/j.saa.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 35.Cooke BM, Mohandas N, Coppel RL. Malaria and the red blood cell membrane. Semin. Hematol. 2004;41:173–188. doi: 10.1053/j.seminhematol.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 36.Hsiao LL, Howard RJ, Aikawat M, Taraschi TF. Modification of host cell membrane lipid composition by the intra-erythrocytic human malaria parasite. 1991;274:121–132. doi: 10.1042/bj2740121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maguire Pa, Sherman IW. Phospholipid composition. 1990;38 doi: 10.1016/0166-6851(90)90210-d. [DOI] [PubMed] [Google Scholar]
- 38.Wunderlich F, Fiebig S, Vial H, Kleinig H. Distinct lipid compositions of parasite and host cell plasma membranes from Plasmodium chabaudi-infected erythrocytes. Mol. Biochem. Parasitol. 1991;44:271–278. doi: 10.1016/0166-6851(91)90013-v. [DOI] [PubMed] [Google Scholar]
- 39.Maier AG, Cooke BM, Cowman AF, Tilley L. Malaria parasite proteins that remodel the host erythrocyte. Nat. Rev. Microbiol. 2009;7:341–354. doi: 10.1038/nrmicro2110. [DOI] [PubMed] [Google Scholar]
- 40.Drescher D, Büchner T, McNaughton D, Kneipp J. SERS reveals the specific interaction of silver and gold nanoparticles with hemoglobin and red blood cell components. Phys. Chem. Chem. Phys. 2013;15:5364–5373. doi: 10.1039/c3cp43883j. [DOI] [PubMed] [Google Scholar]
- 41.Sigurdsson S, et al. Detection of skin cancer by classification of Raman spectra. IEEE Trans. Biomed. Eng. 2004;51:1784–1793. doi: 10.1109/TBME.2004.831538. [DOI] [PubMed] [Google Scholar]
- 42.Law J, Jolliffe IT. Principal Component Analysis. Stat. 2002;36:432. [Google Scholar]
- 43.Notingher I, et al. Discrimination between ricin and sulphur mustard toxicity in vitro using Raman spectroscopy. J. R. Soc. Interface. 2004;1:79–90. doi: 10.1098/rsif.2004.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]