Abstract
Cell signaling relies extensively on dynamic pools of redox-inactive metal ions such as sodium, potassium, calcium, and zinc, but their redox-active transition metal counterparts such as copper and iron have been studied primarily as static enzyme cofactors. Here we report that copper is an endogenous regulator of lipolysis, the breakdown of fat, which is an essential process in maintaining the body's weight and energy stores. Utilizing a murine model of genetic copper misregulation, in combination with pharmacological alterations in copper status and imaging studies in a 3T3-L1 white adipocyte model, we demonstrate that copper regulates lipolysis at the level of the second messenger, cyclic AMP (cAMP), by altering the activity of the cAMP-degrading phosphodiesterase PDE3B. Biochemical studies of the copper-PDE3B interaction establish copper-dependent inhibition of enzyme activity and identify a key conserved cysteine residue within a PDE3-specific loop that is essential for the observed copper-dependent lipolytic phenotype.
INTRODUCTION
Metals are essential for every form of life, and all cells actively regulate the spatial and temporal distribution of their metals for physiological function1. Redox-inactive metal ions such as Na+, K+, Ca2+, and Zn2+ in labile forms – defined as dynamic and loosely bound stores that undergo facile ligand exchange – are widely recognized to play ubiquitous roles in cell signaling1. In contrast, redox-active transition metals such as copper and iron have been studied largely as static enzymatic cofactors, in part because of their propensity in labile forms to aberrantly trigger oxidative stress and free radical damage1–6. However, more recent studies reveal regulatory roles for copper in diverse signaling networks such as the mitogen activated protein kinase (MAPK) pathway in tumor growth7,8 and both stimulated and spontaneous neural activity9,10, presaging broader contributions of this redox-active metal beyond its traditional functions as a metabolic cofactor11–13.
Against this backdrop, we became interested in the relationship between copper and lipids, a major form of energy storage in the body whose misregulation contributes to obesity and diseases where obesity is a risk factor, including diabetes14, cancer15, and cardiovascular diseases16. Indeed, lipid stores can be altered by dietary copper17,18, as diet-induced copper deficiency can modulate the levels of plasma cholesterol and lipoproteins19,20. Along similar lines, alterations in lipid metabolism are also observed in both patients and murine models with genetic defects in copper metabolism16,17,21–23. One such genetic disorder is Wilson’s disease, in which mutations in the copper export protein ATP7B result in systemic copper misregulation and copper overload in the liver24. Despite these intriguing observations, the mechanisms underlying the roles of copper in lipid metabolism remain insufficiently understood. Here we report that copper is an endogenous regulator of lipolysis, an essential process for breakdown of fat in the body, in which triglycerides are degraded into fatty acids and glycerol through a canonical 3',5'-cyclic AMP (cAMP) pathway. We demonstrate that the aberrant elevation of liver copper levels exhibited by an ATP7B knockout mouse is accompanied by reciprocal decreases in copper levels and lipolytic activity in white adipose tissue. Functional and imaging assays in a cellular model of white adipocytes, coupled with biochemical studies on purified protein, point to a molecular mechanism by which copper modulates cAMP signaling via reversible copper-dependent inhibition of the cAMP-degrading phosphodiesterase PDE3B through a key conserved cysteine residue. Taken together, the data identify an essential role for copper-dependent lipid homeostasis in the body and contribute to an emerging paradigm of the involvement of transition metals in cell signaling.
RESULTS
Atp7b−/− mouse adipose tissue exhibits altered lipolysis
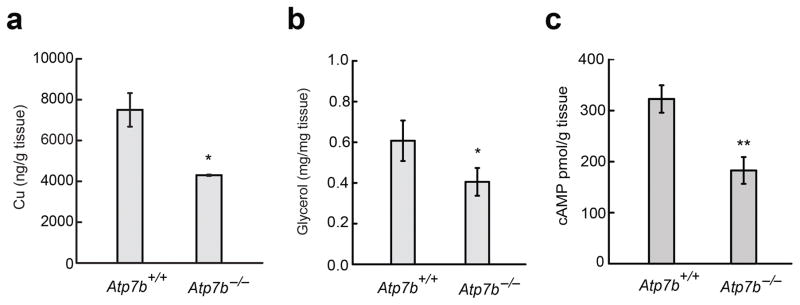
We first investigated the relationship between systemic copper misregulation and lipid metabolism using the ATP7B knockout mouse as a model of genetically induced copper misregulation24,25. Although the hallmark of loss of ATP7B function is copper hyperaccumulation in the liver, alterations in copper status are also observed in other tissues, such as copper increases in brain and kidney26 and impaired copper trafficking and reduction in activity of copper-dependent enzymes27 in the adrenal gland. Given that the liver and adipose tissue are key players in lipid metabolism, we examined the effects of altered copper status on lipid homeostasis in these tissues. Histochemical staining of liver sections with Oil Red O, along with triglyceride measurements, reveal a reduction in lipids in livers of Atp7b−/− mice compared to control mice (Supplementary Results, Supplementary Fig. 1). These data, which correlate aberrantly high copper levels with low lipid levels in the liver, are consistent with observations that dietary copper deficiency can lead to lipid accumulation and cause non-alcoholic fatty liver disease (NAFLD)28. In contrast to the liver, the white adipose tissue (WAT) of Atp7b−/− mice has reduced copper levels compared to wildtype, as measured by atomic absorption spectroscopy (Fig. 1a).
Fig. 1.
Genetically induced copper misregulation affects lipid metabolism in vivo. (a) Copper content of WAT from Atp7b+/+ and Atp7b−/− mice. (n = 3). (b) Glycerol release from Iso-stimulated explant WAT from Atp7b+/− and Atp7b−/− mice (Het n = 5, KO n = 3) (c) cAMP content of Iso-stimulated explant WAT from Atp7b+/− and Atp7b−/− mice (Het n = 5, KO n = 4). Values are shown as mean ± s.e.m. * p < 0.05. ** p < 0.01.
We next sought to evaluate the functional consequences of this copper depletion in Atp7b−/− mouse WAT by treatment with isoproterenol (Iso) – a broad-spectrum β-adrenergic receptor agonist that induces lipolysis through a cAMP-dependent pathway29 – and monitoring release of glycerol, one of the lipolytic end products. Indeed, we observed that explant WAT from the Atp7b−/− mice releases less glycerol (Fig. 1b) compared to control mice. Consistent with this observation, we also found Iso-stimulated levels of cAMP to be lower in the WAT from Atp7b−/− mice relative to control mice (Fig. 1c), suggesting that adipose copper status influences cAMP levels. We are cognizant of the fact that copper levels in tissues are controlled by a systemic homeostatic network that involves communication between multiple organs30; despite this complexity, these data show that alterations in tissue copper levels in vivo indeed influence lipid metabolism. The results prompted us to further investigate this connection in a simplified cellular model system to allow more precise control over copper status.
Copper status affects lipolysis in 3T3-L1 adipocytes
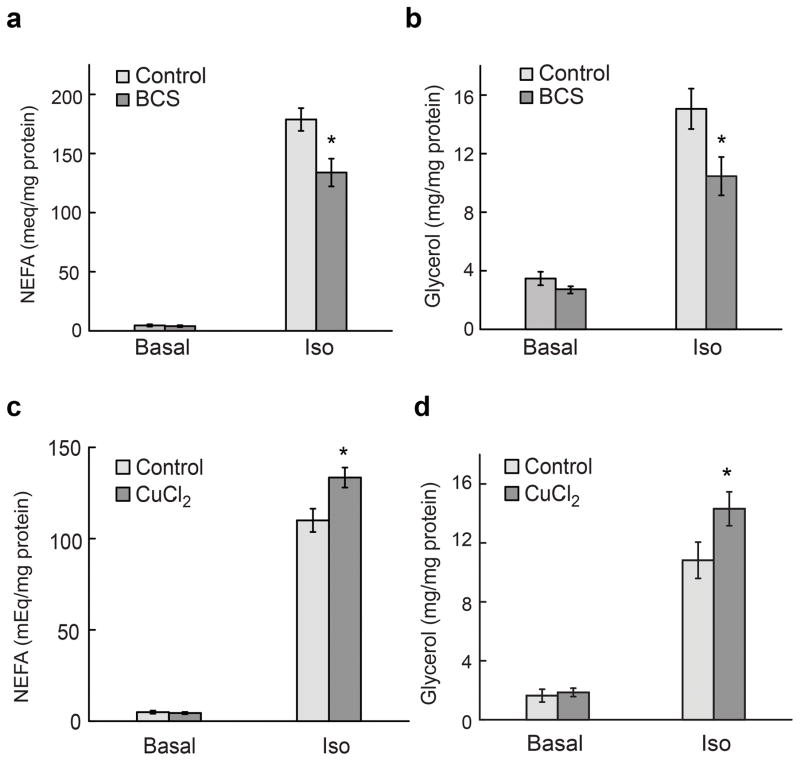
To elucidate a functional relationship between copper and lipolysis, we turned our attention to an established 3T3-L1 adipocyte cellular model for WAT. We altered endogenous labile copper pools through pharmacological depletion with the copper-specific chelator bathocuproine disulfonate (BCS) or elevation with copper chloride, and then monitored the effects of these treatments on lipolytic release of glycerol and non-esterified fatty acids (NEFA). BCS-treated adipocytes showed a dose-dependent attenuation of NEFA and glycerol release upon Iso-stimulated lipolysis, establishing that endogenous copper pools are indeed key to maintaining normal lipolytic responses (Fig. 2a, b and Supplementary Fig. 2). Conversely, copper-supplemented adipocytes displayed a dose-dependent enhancement of NEFA and glycerol release upon Iso stimulation, showing that increases in intracellular copper can promote lipolysis (Fig. 2c, d and Supplementary Fig. 3). We further confirmed that the chelation/supplementation treatments did not affect cellular viability (Supplementary Fig. 4), and we demonstrated that the observed regulation was copper-specific, as iron or zinc supplementation had no effect on lipolytic response (Supplementary Fig. 5). These data show that cellular copper levels can regulate the lipolytic response in 3T3-L1 white adipocytes in both directions, where copper elevation leads to an increase, and copper depletion a decrease, in lipolytic activity.
Fig. 2.
Labile copper pools alter cAMP-dependent lipolysis. For copper chelation experiments (a,b), 3T3-L1 adipocytes were treated with 500 μM BCS for 20 h in DMEM with 2% BSA. For copper supplementation experiments (c,d), cells were incubated for 20 h in DMEM with 2% BSA followed by treatment with 50 μM copper for 1 h in DMEM alone; copper supplementation was performed in the absence of BSA as BSA binds Cu2+ tightly. For both treatments, cells were then stimulated with 100 nM Iso for 60 min in DMEM with 2% BSA. Culture media were harvested for analysis of (a,c) NEFA and (b,d) glycerol release. Values are shown as mean ± s.e.m. (n = 3). * p < 0.05.
CSR1 enables real-time imaging of cellular labile copper
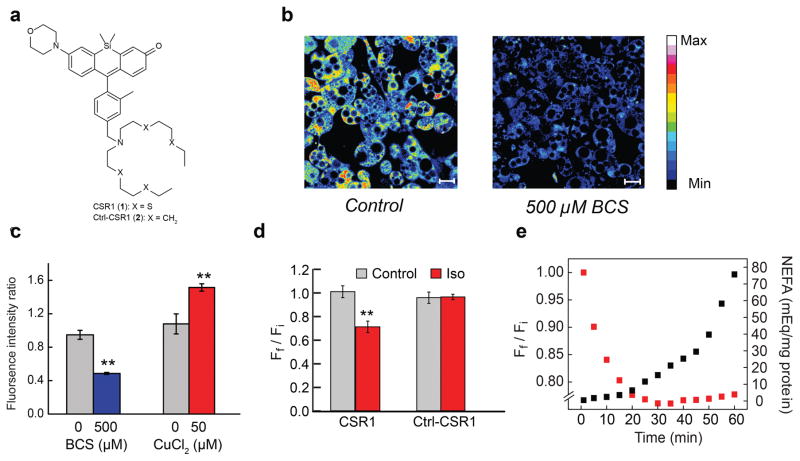
After establishing that genetic and pharmacological alterations in copper status can modulate lipolytic activity, we sought to test whether labile copper pools are mobilized in the lipolytic signaling pathway, analogous to the copper dynamics observed in stimulated neural activity9. To this end, we developed a pair of reagents based on a photostable and hydrophilic silicon rhodol platform, for real-time monitoring of changes in labile copper pools. Copper Silicon Rhodol 1 [CSR1 (1), Fig. 3a and Supplementary Scheme 1] is a Cu+-selective fluorescent sensor that displays high specificity for Cu+, a 1:1 Cu+:probe binding stoichiometry, and an apparent Kd of 2 × 10–13 M (Supplementary Fig. 6). Control Copper Silicon Rhodol 1 [Ctrl-CSR1 (2), Supplementary Scheme 2] is a matched control dye using an identical fluorescent scaffold but lacking a copper-responsive receptor (Supplementary Table 1)10. In vitro spectroscopic measurements confirmed the ability of CSR1 to detect Cu+ in the presence of various cellular components and in cell lysates (Supplementary Fig. 7). We also confirmed that CSR1 fluorescence is not altered by pH changes in both in vitro and in cellulo measurements in physiologically relevant ranges for this adipocyte lipolysis model, but note that like any chemical probe it must be validated for proper use in any new biological applications (Supplementary Fig. 8). Finally, CSR1 exhibits improved photostability in 3T3-L1 cells compared to the previously reported BODIPY-based copper sensor, CS3 (Supplementary Fig. 9)9,11,13. Indeed, the photostability of CSR1 enables expanded capabilities for real-time visualization of labile copper dynamics.
Fig. 3.
Molecular imaging reveals that lipolysis alters labile copper. (a) Structure of CSR1. (b) Images of 3T3-L1 cells supplemented with vehicle or 500 μM BCS in the growth medium for 24 h, stained with 2 μM CSR1 for 10 min at 37 °C in DMEM and then imaged. Scale bar; 20 μm. (c) Quantification of images of 3T3-L1 cells stained with 2 μM CSR1 in DMEM at 37 °C under conditions of copper-supplementation and BCS treatment. The ratio of fluorescent intensity of control cells and treated cells is shown. Values are shown as mean ± s.e.m. (n = 3). (d) 3T3-L1 adipocytes were stained with CSR1 or Ctrl-CSR1 and then stimulated with 100 nM Iso for 60 min. Quantification of images obtained represent Ff /Fi values for control and Iso-treated cells stained with CSR1 or Ctrl-CSR1, respectively.(e) Graph displays data points for concomitant changes in CSR1 fluorescence (red squares) and NEFA release (black squares) obtained every 5 min during lipolysis over 60 min. Values are shown as mean ± s.e.m. (n = 3). ** p < 0.01.
In order to evaluate the ability of CSR1 to detect real-time changes in labile copper, we treated CSR1-stained adipocytes with copper or with the membrane-permeable copper chelator, tris((ethylthio)ethyl)amine (TEMEA). TEMEA has been shown previously by our lab to more rapidly deplete the intracellular labile copper pool accessible to the fluorescent sensor compared to membrane-impermeable chelators such as BCS9,11,13. As expected, on-stage treatment with TEMEA triggered a marked reduction in intracellular fluorescence (Supplementary Fig. 10 and Supplementary Video 1), whereas copper supplementation led to an increase in fluorescence (Supplementary Fig. 11 and Supplementary Video 2). These data support the notion that CSR1 can respond to changes in endogenous, basal levels of labile copper and fluxes in cellular copper pools. Moreover, cells stained with Ctr1-CSR1 exhibited no changes in fluorescence upon copper chelation or supplementation (Supplementary Fig. 9 and Supplementary Video 3; Supplementary Fig. 11 and Supplementary Video 4).
We next utilized these newly developed copper sensors to establish that the copper supplementation and chelation conditions used in the lipolysis experiments can alter labile copper pools. We also evaluated the total cellular copper levels under these conditions using inductively coupled plasma mass spectrometry (ICP-MS) as a direct measure of copper, for comparison with our molecular imaging data. Molecular imaging with CSR1 revealed that copper chelation (Fig. 3b and c and Supplementary Fig. 12) and supplementation treatments (Fig. 3c and Supplementary Fig. 13) showed lower and higher levels of cellular fluorescence relative to control, respectively, as expected for alterations in labile copper pools. Moreover, ICP-MS analysis directly confirmed that BCS treatment reduced and copper supplementation increased total copper content in 3T3-L1 adipocytes, under the conditions used for lipolysis studies (Supplementary Fig. 14). Taken together, these multiple, complementary approaches showcase the utility of molecular copper probes to aid in detecting changes in labile copper pools.
We then proceeded to evaluate the effect of Iso treatment on both labile and total copper content. Stimulation of 3T3-L1 adipocytes with Iso triggered a rapid decrease in CSR1 fluorescence in a dose-dependent manner (Fig. 3d, Supplementary Fig. 15 and Supplementary Video 5). This decrease could be temporally correlated to functional lipolysis assays within the same time frame (Fig. 3e). In contrast, Ctrl-CSR1 was unresponsive to Iso stimulation, supporting that the observed acute change in CSR1 fluorescence is copper-dependent (Fig. 3d and Supplementary Video 6). Because lipolytic stimulation did not alter total copper content as measured by ICP-MS (Supplementary Fig. 16), the observed fluorescence changes could result from altered localization, sequestration, and/or redox balance of cellular labile copper pools. Finally, subsequent addition of copper to CSR1-stained, Iso-stimulated cells triggered a gradual increase in intracellular fluorescence, strongly suggesting that the probe retains its ability to sense labile Cu+ after Iso treatment and that the observed decrease in CSR1 fluorescence upon Iso stimulation was not due to irreversible photobleaching or dye leakage from the cell (Supplementary Fig. 17). The molecular imaging data, combined with the observations of altered lipolysis in adipose tissue and adipocytes, prompted us to further investigate the involvement of copper in regulation of lipolysis.
Copper affects lipolysis at the level of PDEs
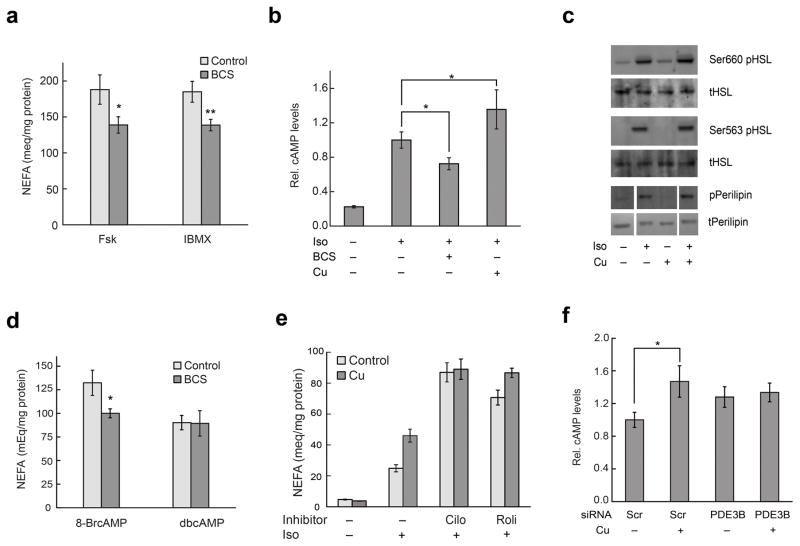
With data showing that labile copper is an endogenous regulator of lipolysis, we next sought to elucidate molecular details of its action. Iso and related catecholamines stimulate lipolysis through binding β-adrenergic receptors to activate adenylyl cyclase (AC), triggering a subsequent increase in the central second messenger cAMP and activation of cAMP-dependent protein kinase A (PKA)31. PKA proceeds to promote lipolysis by phosphorylation of downstream targets such as hormone-sensitive lipase (HSL) and perilipin. We first tested how endogenous copper pools influence the lipolytic response to other agents that act through raising cAMP levels, including forskolin (Fsk), an activator of AC, and 3-isobutyl-1-methylxanthine (IBMX), an inhibitor of cAMP-degrading phosphodiesterase (PDE) enzymes. As observed for the aforementioned Iso stimulation studies, depletion of endogenous labile copper pools with BCS treatment results in lower levels of lipolytic NEFA release induced by Fsk or IBMX compared to control samples (Fig. 4a). In addition, CSR1 imaging shows that both Fsk and IBMX also trigger acute decreases in labile copper (Supplementary Fig. 18). Along these same lines, treatment with the PKA inhibitor H-8932 blocks both NEFA release and changes in labile copper stores as measured by CSR1 fluorescence upon Iso stimulation. In contrast, treatment with the HSL inhibitor CAY10499 has no effect on the change in labile copper induced by Iso but does attenuate downstream NEFA release (Supplementary Fig. 19). These results suggest that changes in labile copper are downstream of PKA but upstream of HSL.
Fig. 4.
Labile copper pools alter cAMP levels by regulating activity of PDE3B. (a) 3T3-L1 adipocytes were treated with 500 μM BCS for 20 h and then stimulated with 1 μM Fsk or 0.5 mM IBMX for 60 min. Culture media were harvested for analysis of NEFA. Values are shown as mean ± s.e.m. (n = 3). (b) 3T3-L1 adipocytes (control, treated with 500 μM BCS for 18–20 h or 50 μM CuCl2 for 1 h) were stimulated with 100 nM Iso for 30 min prior to lysis and assessment of cAMP by enzyme immunoassay. Iso-stimulated cAMP levels are ~10 pmol/mg protein. Values are shown as mean ± s.e.m. (n = 3). (c) Cells were treated with copper, stimulated with 100 nM Iso for 15 mins as described above, lysed and analyzed using Western blot for pHSL and pPerilipin. Representative blots from one of two independent experiments are shown. See Supplementary Figure 20 for full images of blots. (d) Cells were treated with BCS, stimulated with 1 mM 8-bromo-cAMP or dibutyryl cyclic AMP for 60 min and analyzed for NEFA release. Values are shown as mean ± s.e.m. (n = 3). (e) Cells were treated with 10 μM cilostamide or 50 μM rolipram for 5 h and then incubated with 50 μM CuCl2 along with the inhibitors for an additional 1 h. Cells were stimulated with 1 nM Iso for 60 min and analyzed for NEFA release. Values are shown as mean ± s.e.m. (n = 6). Graph shown is a representative data set from three independent biological experiments. (f) 3T3-L1 adipocytes were transfected with scrambled or PDE3B siRNA for 66 h and then incubated in DMEM with BSA for additional 6 h. Cells were stimulated with 100 nM Iso for 25 min prior to lysis and assessment of cAMP. Values are shown as mean ± s.e.m. (n = 3). * p < 0.05. ** p < 0.01.
Given that endogenous copper pools modulate lipolytic responses to cAMP-elevating agents, we then directly compared the levels of cAMP generated in Iso-stimulated 3T3-L1 adipocytes under normal, copper-depleted (e.g., BCS-treated), or copper-supplemented conditions. BCS treatment and copper supplementation resulted in lower and higher relative levels of cAMP, respectively, upon Iso stimulation compared to control adipocytes, establishing that endogenous pools of labile copper contribute to regulating lipolysis at the level of cAMP (Fig. 4b). We further confirmed the role of copper in amplifying the cAMP signal by examining the phosphorylation of HSL and perilipin, which are both phosphorylated by PKA during Iso-stimulated lipolysis; indeed, Western blot analysis showed that HSL and perilipin exhibited increased phosphorylation in the copper-supplemented cells (Fig. 4c and Supplementary Fig. 20). Furthermore, it is known that Iso stimulation leads to increased ERK activity which promotes lipolysis33. Given that copper can affect the activity of ERK in cancer models7,8, we assessed the contributions of ERK in the copper-dependent modulation of lipolysis in adipocytes by Western blot analysis. Unlike the phosphorylation status of HSL, ERK phosphorylation is not enhanced by copper supplementation under conditions that amplify lipolysis in these adipocyte models (Supplementary Fig. 21). These results provide further validation that copper can regulate lipolysis via cAMP and its downstream targets.
A plausible hypothesis for the observed dependence of cAMP levels and phosphorylation levels of PKA substrates on copper is that copper regulates cAMP degradation by phosphodiesterases (PDEs). Therefore, we investigated the involvement of cAMP-degrading PDEs in this copper-dependent signaling cascade by comparing lipolysis stimulated by a hydrolyzable cAMP analog 8-bromo-cAMP to the poorly hydrolyzable cAMP analog dibutyryl-cAMP34–36. Indeed, we observed that BCS treatment attenuates the lipolytic response to 8-Br-cAMP but not dibutyryl-cAMP-induced lipolysis, suggesting that endogenous labile copper regulates lipolysis through cAMP by altering PDE activity (Fig. 4d).
PDE3B is a specific target of copper in adipocytes
3T3-L1 adipocytes express many PDE isoforms, including PDE1, PDE2, PDE3B, and PDE4, but only PDE3B and PDE4 are known to strongly influence lipolysis in these cells37. Therefore, isoform-specific PDE3 (cilostamide) and PDE4 (rolipram) inhibitors were employed to identify the specific PDE target of copper at sub-saturating doses of Iso by measuring NEFA levels of the inhibitor-treated cells in the presence or absence of copper. We observed that treatment with cilostamide, but not rolipram, abrogated the regulation of lipolysis by copper, indicating that PDE3B is likely responsible for the copper-dependent phenotype (Fig. 4e). To further confirm PDE3B as the target, we utilized siRNA to knock down PDE3B specifically in 3T3-L1 cells and then measured cAMP levels in response to Iso stimulation in the presence or absence of copper supplementation. Compared to the cells transfected with scrambled siRNA, the PDE3B siRNA-transfected cells did not exhibit copper-dependent increases in cAMP levels (Fig. 4f and Supplementary Fig. 22). Moreover, we observed that Iso stimulation does not trigger changes in CSR1 fluorescence in 3T3-L1 adipocytes treated with PDE3B siRNA in contrast to cells treated with scrambled siRNA, linking changes in labile copper pools to PDE3B status (Supplementary Fig. 22). Taken together, the results obtained from the murine animal and cell culture models identify a novel role for copper in regulating lipolysis at the level of cAMP through the activity of PDE3B.
Purified PDE3B binds and is inhibited by Cu+ in vitro
In order to probe the interaction of PDE3B with copper at the protein level, we overexpressed and purified a full-length, C-terminally FLAG-tagged PDE3B, as well as the soluble catalytic domain (residues 630–1100, termed ΔN-PDE3B), from insect cells38. cAMP hydrolysis activity of these enzymes was measured anaerobically in the presence of up to 4 equiv. Cu+, showing that both full-length and ΔN-PDE3B are inhibited fully by 2 equiv. Cu+ (Supplementary Fig. 23). We therefore focused on the catalytic domain alone for detailed characterization of Cu+ binding. For these studies, ΔN-PDE3B was expressed in E. coli, refolded from insoluble inclusion bodies, and purified by a custom affinity column using a PDE3-specific inhibitor (Supplementary Fig. 24)39. Compared to the protein expressed in insect cells, the refolded protein had a specific activity several hundred-fold higher (12 μmol/min/mg at 30 °C, comparable to the highest reported for a PDE3B39); consequently, the refolded protein was used for further studies. Activity assays of ΔN-PDE3B, carried out in the presence of catalytic metal Mg2+ (10 mM) and 0–4 equiv. Cu+, revealed copper-dependent inhibition (Fig. 5a and Supplementary Fig. 25). This inhibition could be prevented by co-incubation with BCS but not with weaker copper chelators such as bicinchoninic acid and ferrozine (Supplementary Fig. 26). Importantly, the onset of inhibition occurs only after 1–2 equiv. Cu+, suggesting the presence of one or two tighter or more kinetically accessible Cu+ sites that do not affect activity under our assay conditions. Furthermore, addition of Cu+ above 3 equivalents does not result in complete inactivation (see below).
Fig. 5.
The catalytic domain of PDE3B binds Cu+ at a conserved Cys residue that is implicated in the cellular copper-dependent lipolytic phenotype. (a) Representative cAMP hydrolysis activity assay of wt ΔN-PDE3B, performed anaerobically in the presence of 0–4 equiv. Cu+ (10 mM MgCl2, 0.5 mM cAMP). When included, 20 μM BCS was added to protein prior to addition of Cu+. (b) Apo-subtracted, volume-corrected difference spectra from anaerobic spectrophotometric titration of ΔN-PDE3B (1.5 μM) with Cu+, showing CT bands associated with Cu+-protein interactions. (c) Representative anaerobic spectrophotometric titrations of ΔN-PDE3B in 0 mM (black squares) or 10 mM (red circles) MgCl2. Absorbance change at 265 nm, normalized to protein concentration, is shown as a function of equiv. Cu+ added. Here and in Fig. 6e, data points after the titration endpoint have been corrected for a residual increase due to Cu+-buffer interactions. (d) Crystal structure of the catalytic domain of human PDE3B (PDB code: 1SO2). The catalytic site, consisting of two Mg2+ ions (green spheres) ligated by His, Asp/Glu (sticks), and water molecules, is at right. The 44-amino acid, partially disordered loop is in salmon. The three Cys residues mutated in this study are in sticks (the human PDE3B equivalent to mouse C769 is Ser). (e) Representative anaerobic spectrophotometric titrations of purified wt and mutant ΔN-PDE3Bs in the absence of Mg2+. (f) cAMP levels (ratio of levels in copper-treated vs. untreated cells) in Iso-stimulated 3T3-L1 adipocytes overexpressing full-length wt, C769S, C768S/C769S, and C769S/C777S PDE3B. Copper and Iso treatments were performed as described in Fig. 2. Values are shown as mean ± s.e.m. (n = 5). *p < 0.05. (g) Proposed model for copper as an endogenous regulator of lipolysis via inhibition of PDE3B.
To assess Cu+:PDE3B stoichiometry and ligation, we titrated ΔN-PDE3B with Cu+ under anaerobic conditions and monitored Cu+ binding using UV-vis spectrophotometry. Absorbance in the <300 nm region derives from charge transfer (CT) between Cu+ and its ligands,40–42 chiefly Cys but also other ligands such as His. In particular, Cys ligation manifests as a band at 265 nm. Titration of ΔN-PDE3B with Cu+ reveals a shoulder at 265 nm (Fig. 5b), suggesting the presence of at least one Cu+-Cys interaction. The absorbance change saturates at 4.2 ± 0.5 equiv. Cu+ in the absence of Mg2+ (Supplementary Fig. 27), but at 3.1 ± 0.2 equiv. Cu+ in the presence of 10 mM Mg2+ (Fig. 5c, Table 1). This result is consistent with our activity data showing Cu+-dependent inhibition of ΔN-PDE3B plateauing at and above 3 equivalents. Together with experiments indicating that binding of two Mg2+ ions is required for full ΔN-PDE3B activity (Supplementary Fig. 28)43,44, the titration data suggest that one Cu+ ion (likely the third equivalent) may directly compete with the weaker of two Mg2+ ions at the catalytic site but does not displace the more tightly bound Mg2+ under these experimental conditions.
Table 1.
Cu+ binding stoichiometry and associated absorbance change for anaerobic titrations of wt and mutant ΔN-PDE3Bs with Cu+ (mean ± s.d. for 2–4 independent experiments).
| ΔN-PDE3B | [Mg], mM | Equiv. Cu(I) | ΔA265/μM protein |
|---|---|---|---|
| WT | 0 | 4.2 ± 0.5 | 0.021 ± 0.003 |
| C769S | 0 | 3.8 ± 0.3 | 0.020 ± 0.002 |
| C768S/C769S | 0 | 3.3 ± 0.1 | 0.017 ± 0.001 |
| C769S/C777S | 0 | 2.9 ± 0.5 | 0.013 ± 0.002 |
| C768S/C769S/C777S | 0 | 2.1 ± 0.4 | 0.009 ± 0.002 |
| WT | 10 | 3.1 ± 0.2 | 0.014 ± 0.004 |
The presence of the Cys-Cu+ CT band suggests a possible location of copper binding for the equivalents that are not accounted for by putative active site binding. Mouse ΔN-PDE3B contains 8 Cys residues, including three (C768, C769, and C777, Fig. 5d) on a flexible 44-amino acid insert unique to PDE3s45; C768 and C777 are conserved in all sequenced PDE3Bs (Supplementary Fig. 29). The catalytic function of this loop, which is partially disordered in the crystal structure of human ΔN-PDE3B44, remains insufficiently understood46. We mutated the three candidate Cys residues to Ser, singly and in combination, and carried out activity assays and UV-vis titrations with Cu+. The mutants exhibited activity on the same order as wild type (Supplementary Fig. 30). Titration of the C768S/C769S/C777S mutant in the absence of Mg2+ revealed only ~2 equiv. Cu+ binding (Table 1), indicating that 2 equiv. Cu+ bind to one or more Cys residues in this loop. To identify the residue(s) involved, we first mutated the nonconserved C769 to S and then mutated the conserved C768 and C777 to S in this background. Titrations (Fig. 5e, Table 1) suggest that C768 and C777, but not C769, are ligands to independent Cu+ sites. Furthermore, the C768S mutation (but not C777S) abolishes the delayed onset of Cu+ inhibition observed in wildtype ΔN-PDE3B, suggesting that C768 is the tightest or most accessible Cu+ binding site (Supplementary Fig. 30). The results indicate that, in vitro, C768 and C777 in the PDE3-specific loop are Cu+ ligands.
C768 mediates regulation of lipolysis by copper
After identifying C768 and C777 as critical residues for Cu+ binding in vitro, we sought to validate their physiological relevance on lipolysis in cells. We overexpressed wildtype, C769S, C768S/C769S, and C769S/C777S (full-length and ΔN) PDE3Bs in 3T3-L1 cells and assessed iso-stimulated cAMP levels following treatment with 0 or 50 μM copper (Supplementary Fig. 31 and Fig. 32). In both full-length (Fig. 5f) and ΔN-PDE3B (Supplementary Fig. 32) experiments, the C768S/C769S mutant alone displayed a significant ablation of the copper effect. Unlike the full-length protein, the catalytic domain is constitutively active, supporting the notion that PDE3B itself, and not the kinases (such as PKA) and phosphatases regulating PDE3B activity, is the primary target of copper in this pathway. Whereas both C768 and C777 bind Cu+ in vitro, the observation that C768 alone is important in cells may be explained by our evidence that C768 is the tightest Cu+ binding site in vitro (see Discussion and Supplementary Fig. 33). Taken together with the cellular data, these biochemical data support a model in which copper can reversibly regulate cAMP-dependent lipolysis by inhibiting PDE3B activity via the key residue C768 (Fig. 5g).
DISCUSSION
Numerous studies have linked dietary and genetic alterations in copper to dysregulation of lipid metabolism in animal models17–22 and in patients with Wilson’s disease23. Our findings identify that one of the mechanisms by which copper alters lipid metabolism may be the modulation of lipolysis via inhibition of PDE3B, a key regulator of this pathway. Indeed, copper status alters cAMP levels and lipolysis in white adipose tissue and in an adipocyte cell culture model, supported by molecular imaging of labile copper pools using a newly developed fluorescent sensor. Moreover, pharmacological and genetic studies converge on PDE3B as a major target of copper. Biochemical characterization of the soluble catalytic domain of PDE3B indicates that Cu+, likely the prevalent oxidation state of labile copper in the cell, binds to PDE3B and inhibits activity. Finally, we show that a single, conserved Cys residue (C768) in a PDE3-specific motif is responsible for the observed Cu-dependent effect on lipolysis through expression of wildtype and mutant constructs in cells. Together, the data point to a role of copper as a reversible mediator of signaling in this pathway.
PDE3s are unique among phosphodiesterases as they contain a 44-amino acid insert (residues 734–777) in the catalytic domain. This PDE3-specific insert is critical for activity for reasons that remain insufficiently understood47. Despite the clear importance of the insert residue C768 in Cu+-mediated inhibition of PDE3B within cells, Cu+ binding to C768 and C777 as well as the active site is observed in vitro. One potential explanation is that, whereas binding of Cu+ to the active site decreases kcat of the enzyme, binding of Cu+ to C768 may affect Km. Indeed, studies on PDE3A46 show that mutagenesis of certain residues in the insert leads to modulation of either kcat or Km of the enzyme. Of particular interest is the effect of mutation of the C768 equivalent in PDE3A, Y807. Y807C has kinetic parameters similar to the wildtype enzyme, but the Y807A mutant displays a Km 20 times higher, suggesting the importance of a hydrogen bond donor at that position. Furthermore, Y807 is modified by a substrate analog that irreversibly inactivates the enzyme, suggesting that Y807 is involved in substrate binding. With these data in mind, we hypothesize that Cu+ binding to C768 either directly or indirectly increases the Km of the enzyme such that cAMP degradation is hindered at its physiological levels (Supplementary Fig. 33). Such an effect may have been masked by the super-physiological substrate concentrations used in our in vitro experiments. Current efforts are directed toward characterizing the Cu+-binding sites in more detail to further probe the mechanism of inhibition. The above considerations suggest that PDE3B is unique within the PDE family in its ability to be modulated by copper.
This work contributes to the emerging paradigm supporting Nature’s utilization of copper ions7–11, and transition metal ions more generally12,13, as mediators of cellular signaling processes but leaves many open questions. Perhaps the most important outstanding issue is how cells may exploit the unique properties of transition metals like copper to contribute to signaling compared to their alkali and alkaline earth counterparts. In addition to differences in hard-soft acid-base and coordination geometry preferences, one tempting hypothesis is that the intrinsic redox activity of copper is critical to its function. Indeed, our identification of a cysteine residue as the locus for copper-dependent modulation of lipolysis offers the intriguing possibility that synergies exist between transition metal and redox signaling. For example, C768 has also been identified as a residue that can be S-nitrosated in adipocytes, potentially with the effect of inhibiting PDE3B activity48. We are currently pursuing studies to investigate this possibility.
The centrality of lipolysis in energy homeostasis and the involvement of copper in its regulation presage opportunities to target the homeostasis of this redox-active transition metal to pharmacologically alter lipid metabolism, particularly applied to obesity, diabetes, cancer, and heart disease. More broadly, PDE3B is also expressed in specialized cell types within diverse tissues, such as brown fat, heart, pancreas, and brain, and is a target of therapeutic interest for type 2 diabetes and heart disease49,50. Exploration of the potential roles of copper in modulating PDE3B-dependent pathways in these cell types will likely uncover new metallophysiologies as well as lead to new avenues for therapeutic intervention.
Online Methods
Commercial reagents for in vitro and in cellulo assays
Dulbecco’s modified Eagle’s medium (DMEM), GlutaMAX, sodium pyruvate and non-essential amino acids were obtained from Life Technologies. Fatty acid-free, ultra-pure BSA and protease inhibitor cocktail were obtained from Roche. BCS, copper chloride, 8-bromo-cAMP, dibutyryl cAMP, IBMX, isoproterenol, and free glycerol reagent were obtained from Sigma-Aldrich. BDH Aristar Ultra concentrated nitric acid was obtained from VWR. NEFA kits were obtained from Wako. H-89 dihydrochloride, cilostamide, and rolipram were obtained from EMD Biosciences. CAY10499 and cyclic AMP assay kits were obtained from Cayman Chemical. Restriction endonucleases, Phusion High-Fidelity DNA polymerase, T4 DNA ligase, and Gibson Assembly Master Mix were obtained from New England Biolabs. TaKaRa ExTaq DNA polymerase was purchased from Clontech. Primers were purchased from Integrated DNA Technologies. All other materials used in biochemical assays were purchased from Sigma unless otherwise noted. Baculoviral particles were generated using the BaculoGold™ transfection kit from BD Biosciences. All DNA constructs were verified by sequencing by Quintara Biosciences, and all plasmids used for transfections were prepared using the EndoFree Maxi Kit (Qiagen).
Triglyceride measurements and Oil Red O staining
Wild type, Atp7b+/−, and Atp7b−/− mice were maintained on strain C57BL × 129S6/SvEv. The animals were housed at the Johns Hopkins University School of Medicine Animal Care Facility according to National Institutes of Health guidelines. Liver tissue was dissected from 6- to 8-week-old, male and female, control and Atp7b−/− mice after whole animal perfusion with phosphate-buffered saline (pH 7.4; PBS), frozen in liquid N2, and stored at 80 °C until use. A piece of the liver (~100 mg in weight) was placed in an ice-chilled glass homogenizer and homogenized in HB buffer (50 mM HEPES, 0.1% Igepal, 150 mM NaCl, 1 mM AEBSF, 250 mM sucrose, pH 7.4) containing EDTA-free protease inhibitor cocktail, using the loose (25 times) and tight (25 times) pestles. Then the contents were transferred into an eppendorf tube and triglycerides were measured using the InfinityTM reagent (Thermo Scientific) following the manufacturer’s protocol. Oil Red O staining was performed using standard protocols.
Isolation of adipose tissue and atomic absorption analysis
The entire amount of intra-peritoneal fat was obtained from the age-matched 6- to 8-week-old, male and female, control and Atp7b−/−mice and weighed. For copper measurements, all equipment was cleaned with nitric acid overnight and the rinsed with Milli-Q water; and the process was repeated twice to remove traces of copper. Adipose tissue weighing 182–190 mg was transferred to a 50-mL conical tube and digested with 2 mL concentrated nitric acid at 90 ºC on a heat block for 2 h. The samples are mixed by swirling after every hour. When the sample was completely dissolved, contents of the flask were carefully removed to a 15-mL tube containing 7 mL water using a Pasteur pipette, taking care to avoid lipids. The final volume was made up to 10 mL. The samples were further diluted, when necessary, to fit the linear range of the calibration curve and measured using flame atomic absorption spectrometry using a Shimadzu 6650 graphite furnace atomic absorption spectrophotometer equipped with an ASC-6100 autosampler. Statistical analyses were performed with a two-tailed Student’s t-test in Excel.
Explant WAT lipolysis and cAMP assays
For lipolysis and cAMP assays, visceral adipose tissue was isolated from age-matched 9- to 10-week-old female control and Atp7b−/− mice. Lipolysis assays were performed in explants from freshly removed epididymal fat pads (~25 mg)51. Samples were incubated at 37 °C in 500 μL Krebs-Ringer buffer (12 mM HEPES, 121 NaCl, 4.9 mM KCl, 1.2 mM MgSO4 and 0.33 mM CaCl2) with 3.0% fatty acid-free BSA and 3 mM glucose, with 100 nM Iso. Supernatants were collected after 60 min of stimulation, snap-frozen in liquid N2, and analyzed for glycerol release using free glycerol reagent. Measurements were normalized to the total weight of the explant. One outlier among the Atp7b−/− samples was identified using the Grubbs outlier test and excluded from subsequent analysis. For cAMP analysis, 50 mg of tissue was incubated with 100 nM Iso for 30 min. The tissue was then quickly rinsed in PBS and snap-frozen for further analysis. The explants were then prepared for cAMP enzyme immunoassay (Cayman) according to the manufacturer's protocol. Briefly, 5 mL/g 5% trichloroacetic acid (w/v) in water was added to the pre-weighed samples immediately upon removal from the -80 °C freezer. The samples were homogenized on ice using a battery-powered Argos Pellet Mixer (20 strokes). The samples were centrifuged at 1500 × g, 10 °C for 10 min, and the infranatant was transferred to a new eppendorf tube. TCA was removed by extraction three times using 5 volumes water-saturated ether. Residual ether was removed by heating in a 70 °C sand bath for 5 min. The sample was then analyzed by cAMP enzyme immunoassay, without acetylation, at 8-fold dilution. Statistical analyses were performed with a two-tailed Student’s t-test in Excel.
Cell culture and maintenance
3T3-L1 preadipocytes (CL 173, ATCC, authenticated by short tandem repeat analysis and tested to confirm absence of Mycoplasma by Hoechst staining) were cultured in DMEM containing 10% newborn calf serum and grown to confluence. Cells were then differentiated by the addition of insulin, dexamethasone, and IMBX as described52. Adipocytes were maintained at 37 °C and 5% CO2. For all experiments, cells from P1 to P4 were used at days 10 to 14 post-differentiation. For lipolysis experiments, the cells were serum-starved in high-glucose DMEM without phenol red supplemented with 2 mM GlutaMAX, 1 mM sodium pyruvate, 1x non-essential amino acids (“Supplemented DMEM”) with or without 2% BSA.
Cellular lipolysis assays
3T3-L1 adipocytes were incubated in supplemented DMEM containing 2% BSA for 18–20 h with or without 500 μM BCS. The media was then replaced with fresh media containing 2% BSA with or without BCS and incubated for an additional hour prior to stimulation of lipolysis. For copper supplementation experiments, cells were incubated with BSA-free medium overnight followed by the addition of 10, 20, or 50 μM CuCl2 on the next day. After 1 h of incubation with CuCl2, the media was then replaced with fresh media containing 2% BSA with or without CuCl2 immediately prior to the stimulation of lipolysis. Lipolysis was assessed by measuring the NEFA and glycerol release in the culture medium. The NEFA assay was performed according to the manufacturer’s instructions. Briefly, aliquots of cell culture medium (3–30 μL) were added to 30 μL reagent A and incubated at 37 °C for 10 min. This was followed by the addition of 60 μL reagent B and the absorbance was read at 560 nm. Glycerol release into the media was measured by adding 100 μL free glycerol reagent to the media and incubating for 5 min at room temperature before the absorbance at 540 nm was measured. All assays were performed in 96-well plates and measured using a SpectraMax M2 plate reader (Molecular Devices) or Synergy M2 plate reader (BioTek Instruments). Both NEFA and glycerol release were normalized to total protein content using the BCA assay. For H-89 and CAY10499 inhibition experiments, cells were incubated with 100 μM H-89 for 1 h or 1 μM CAY10499 for 5 min prior to the addition of Iso. For inhibitor treatments, cells were incubated with 10 μM rolipram or 50 μM cilostamide for 5 h in 2% BSA media, followed by 1 h in presence of inhibitor and 0 or 50 μM CuCl2 in media without BSA. Cells were then stimulated for 1 h with 1 nM Iso in 2% BSA media in the presence of the inhibitor. Each experiment was carried out at least 3 times, each condition was in sextuplicate, and each well was assayed in triplicate. Statistical analyses were performed with a two-tailed Student’s t-test in Excel. Statistical analyses for multiple comparisons were performed using one-way ANOVA with the Bonferroni correction in the statistical analysis software, R.
Synthesis of CSR1 and Ctrl-CSR1
See Supplementary Note.
Spectroscopic Characterization of CSR1 and Ctrl-CSR1
The probe was stored and handled as previously described10. Milli-Q water was used to prepare all aqueous solutions. Absorption spectra were recorded using a Varian Cary 50 spectrophotometer, and fluorescence spectra were recorded using a Photon Technology International Quanta Master 4 L-format scan spectrofluorometer equipped with an LPS-220B 75-W xenon lamp and power supply, A-1010B lamp housing with integrated igniter, switchable 814 photocounting/analog photomultiplier detection unit, and MD5020 motor driver. Samples for absorption and emission measurements were contained in 1-cm × 1-cm quartz cuvettes, septum sealed for anaerobic experiments (1.4-mL or 3.5-mL volume; Starna Cells). Fluorescence quantum yields, binding affinities, partition coefficients, and metal selectivity were determined as previously described10. The characterizations of CSR1 and Ctrl-CSR1 in in vitro cellular models were carried out in 25 mM HEPES, pH 7.4. All manipulations involving GSH and cell lysates were carried out in an anaerobic chamber (VAC glovebox, Pd catalyst, atmosphere 10% H2 with N2 balance). For GSH experiments, the buffer (25 mM Hepes, pH 7.0) was deoxygenated on a Schlenk line by multiple rounds of freeze-pump-thaw before being brought into the anaerobic chamber. The responses of CSR1 and Ctrl-CSR1 in model liposomes, proteins, cell lysates were determined as described10,53. For the cell lysate experiments, the fluorescence turn-on response was normalized to the acetonitrile vehicle control.
pH profile of CSR1
a. In vitro measurements
The pH profiles for fluorescence emission of CSR1 and Ctrl-CSR1 were determined by recording the fluorescence intensity before and after the addition of 1.25 equiv. Cu+ to CSR1 or Ctrl-CSR1 (2 μM). The solutions were prepared by diluting the stock solution of the probe (2 mM in DMSO) with the appropriate pH buffer (50 mM acetate for pH 4.0–5.5, 50 mM MES for pH 5.5–6.7, and 50 mM HEPES for pH 6.7–8.4; all buffers were brought to the appropiate pH by aqueous NaOH).
b. Effect of pH on CSR1 fluorescence in 3T3-L1 cells
Cells were grown in Lab-Tek II 4-well chamber slides (Thermo Scientific) and 2 wells were stained with 2 μM of CSR1 in DMEM for 10 min. Cells were then washed twice with 500 μL Buffer A (20 mM MOPS, 140 mM NaCl, 150 mM KCl, 1.8 mM CaCl2, 1 mM MgCl2, pH 7.4 at 37 °C) and then allowed to equilibrate for 30 min at 37 °C in Buffer A containing 10 μM nigericin and 10 μM valinomycin. Confocal microscopy images (4 positions per well) were acquired on a Zeiss LSM 710 laser scanning microscope with a 20X objective lens. CSR1 was excited at 594 nm with a He-Ne laser, and emission was collected using a META detector between 610 and 700 nm. The buffer in one well was changed to Buffer A (pH 7.3) containing nigericin and valinomycin, whereas the buffer in the second well was changed to Buffer A containing nigericin and valinomycin, but at pH 6.7. After equilibration at 37 °C for ~30 min, the same positions were imaged again. Control experiments using the pH indicator 2',7'-bis-(2-carboxyethyl)-5-(and-6)-carboxyfluorescein, acetoxymethyl ester (BCECF-AM, Life Technologies) showed that this was sufficient time for intracellular pH to stabilize. Images were analyzed using ImageJ (National Institutes of Health), with the initial and final fluorescence for each position compared. The experiment was carried out twice.
Confocal microscopy
Cells were grown on Lab-Tek II chamber slides (Thermo Scientific) and stained with 2 μM of CSR1 or Ctrl-CSR1 in DMEM for 10 min. Cells were then washed with fresh DMEM (2 × 450 μL) and allowed to equilibrate at 37 °C for at least 20 min prior to treatment. A solution of 10x strength stimulant (50 μL) was then applied. During the first ~3 min after treatment, an initial decrease in fluorescence intensity (~4%) that we attribute to dye washout was observed; however, the signal stabilizes after ~3 min as indicated by vehicle control experiments. A time-lapse movie was then acquired for 1 h with images taken every 1 min. Cells were treated with Iso, BCS, or CuCl2, stained, and imaged. Confocal microscopy images were acquired on a Zeiss LSM 710 laser scanning microscope with a 40X water-immersion objective lens. CSR1 and Ctrl-CSR1 was excited at 594 nm with a He-Ne laser, and emission was collected using a META detector between 610 and 700 nm. CS3 was excited at 543 nm with a He-Ne laser, and emission was collected using a META detector between 550 and 700 nm. CF3 was excited at 543 nm with a He-Ne laser, and emission was collected using a META detector between 550 and 700 nm. Images were analyzed using ImageJ (National Institutes of Health). Each experiment was carried out at least three times. Statistical analyses were performed with a two-tailed Student’s t-test in Excel. Statistical analyses for multiple comparisons were performed using one-way ANOVA with the Bonferroni correction in the statistical analysis software, R.
Inductively coupled plasma-mass spectrometry (ICP-MS)
Cells cultured in 6-well plates were treated with either BCS or 10–50 μM copper as described above. The plates were then washed 3 times with PBS containing 1 mM EDTA (to remove non-specifically bound copper) followed by the addition of 215 μL concentrated nitric acid (BDH Aristar Ultra). The plates were sealed with parafilm and incubated overnight. After poking a single hole into the caps of 1.5 mL tubes (Sarstedt) using an 18-gauge needle, the digested contents of each well were transferred into the tubes and the samples were boiled at 95 °C for 1 h. Samples (150 μL) were further diluted in 2 mL 2% nitric acid (made freshly from concentrated nitric acid and Milli-Q water) and analyzed on a Thermo Fisher iCAP Qc ICP mass spectrometer in Kinetic Energy Discrimination (KED) mode against a calibration curve of known copper and phosphorus concentrations, with Ga (20 μg/L, Inorganic Ventures) as an internal standard. Each experiment was carried out twice and each condition was repeated in at least triplicate.
cAMP analysis
3T3-L1 cells cultured in 6-well plates were treated with either BCS or CuCl2 as described above. After 30 min of treatment with 100 nM Iso, cells were washed twice with ice-cold PBS and lysed by the addition of 400 μL 0.1 M HCl. After 1–2 min at room temperature, the plates were stored at -80 °C until further analysis. Upon thawing, cells were centrifuged at 5000 × g for 5 min, and the supernatant was assayed for cAMP by enzyme immunoassay, following acetylation, according to the manufacturer’s protocol. cAMP levels were normalized to protein content, by assaying the supernatant using the BCA assay. Each experiment was carried out at least 3 times, each condition was in triplicate, and each well was assayed in duplicate.
Western blotting
Protein samples were denatured in Laemmli buffer, and loaded onto SDS-PAGE gels and transferred to PVDF membranes (Millipore, Billerica, MD). Membranes were then incubated overnight at 4 °C in wash buffer (10 mM Tris, pH 7.5, 100 mM NaCl, 0.1% Tween-20) containing 1% BSA with the primary antibody. Antibodies were obtained from Cell Signaling Technology [pHSL Ser660 (Cat. 4126), pHSL Ser563 (Cat. 4139), HSL (Cat. #4107), pERK1/2 (Cat. 4370), and ERK 1/2 (Cat. 4695)], Santa Cruz Biotechnology [Actin (Cat. #SC69879)], Vala Biosciences [p-perilipin (Cat. 4854), perilipin (Cat. 4856)], and Sigma [M2 anti-FLAG antibody (Cat. F1804)]. The membrane was then rinsed with wash buffer and incubated for 1 h at room temperature in wash buffer containing 1% BSA with 1:2500 HRP-conjugated secondary antibody (Santa Cruz Biotechnology) or 1:2500 fluorophore-conjugated secondary antibody (Life Technologies). After further washing, membranes were visualized using enhanced chemiluminescence (Western Lightning, Perkin-Elmer) or imaged directly for fluorescent secondary antibodies and recorded on a BioRad ChemiDoc MP imaging station.
PDE3B knockdown and RNA extraction
PDE3B knockdown was performed in adipocytes according to the protocol as described38. Briefly, PDE3B specific siRNA (cat. no. L-043781–00) and control non-targeting siRNA (cat. no. D-001810–10)) were purchased from Dharmacon. siRNA (100 nM) was transfected using MBS transfection reagent (Stratagene) and incubated for 12 h. The media was exchanged with fresh media and incubated for an additional 50–56 h. Cells were then incubated for 6 h in DMEM+2% BSA media before the addition of 100 nM Iso for 25 min. The cells were then analyzed for cAMP content. Specific PDE3B knockdown was confirmed using quantitative real-time RT (reverse transcriptase)–PCR as described38. Briefly, cells in 12-well plates were lysed with 166 uL of Trizol reagent and the RNA was extracted using RNeasy mini kit. Total RNA (100 ng) in triplicate was subjected to quantitative RT-PCR, using the Bio-Rad CFX96 and QuantiTect SYBR green RT-PCR kit (Qiagen), according to manufacturer’s protocols. PDE3B was normalized using cyclophilin A as internal control. The primers used were PDE3B_RT1, PDE3B_RT2, CycA_RT1, and CycA_RT2 (Supplementary Table 2).
Cloning of ΔN-PDE3B insect cell expression vector
The catalytic domain of the mPDE3B (630–1100) was amplified from pAcSG2-PDE3B plasmid (Supplementary Table 3, a gift from Dr. V. C. Manganiello)54 using the primers ΔN-PDE3B_for and ΔN-PDE3B_rev (Supplementary Table 2), in which the forward primer carried a new start codon. After digestion by XhoI, the PCR product was ligated using T4 DNA ligase into the pAcSG2 expression vector. The pAcSG2 expression vector was obtained by digestion of pAcSG2-PDE3B plasmid with XhoI followed by gel purification. Plasmids were sequenced in the both forward and the reverse direction using Baculo_for and Baculo_rev primers (Supplementary Table 2) to verify the cloned sequences.
Expression and purification of PDE3B and Δ-PDE3B from insect cells
Overexpression of FLAG-tagged full-length PDE3B and PDE3B residues 630–1100 (ΔN-PDE3B) was achieved via baculoviral infection of Sf21 cells. For purification of ΔN-PDE3B, T75-flasks containing ~1.2 × 106 cells were infected with recombinant virus for 48 h at 27 °C. For a single purification, 6 L of culture was combined by centrifugation and the resulting pellet was resuspended in 20 mL of 50 mM HEPES, 100 mM NaCl, 1 mM EDTA, pH 7.5 buffer that was supplemented with protease inhibitors (Buffer B). Cells were then homogenized using a 50 mL dounce homogenizer (30 strokes), sonicated (2 × 30 s pulses, 40% amplitude) and centrifuged using an Optima XLN-100 ultracentrifuge (100,000 × g, 1 h, 4 °C, 70Ti rotor) to obtain the solubilized fraction. The soluble fraction was passed 5 times through a 2 mL anti-FLAG M2 affinity gel column (Sigma), and the column was then washed with 50 mL of Buffer B. The bound protein was eluted by passing 2 mL of FLAG peptide (100 μg/mL and 500 μg/mL) in 50 mM HEPES, 100 mM NaCl, 10% (v/v) glycerol, pH 7.5, 3 times through the affinity column. The expression and purity of the recombinant protein were verified by Western blot and SDS-PAGE gels, respectively, which showed a single band at ~65 kDa (calculated MW: 55 kDa) (Supplementary Fig. 23). Purification of the full-length protein was conducted according to the previously described protocol38. Expression of full-length PDE3B was verified by Western blotting.
Construction of pET24a-mΔ-PDE3B
pAcSG2-ΔN-PDE3B (2 μg) was digested with 20 U XhoI and, following gel electrophoresis, the 1.6 kb PDE3B fragment was excised and purified using the Zymoclean Gel DNA Recovery Kit (Genesee Scientific). This fragment was used as template for a PCR reaction using primers ΔN-PDE3B_NdeI_for and ΔN-PDE3B_XhoI_rev. After purification, the resulting product was digested with NdeI and XhoI and ligated into pET24a digested with NdeI and XhoI, using T4 DNA ligase (high-concentration) and 3:1 insert:vector, according to the manufacturer’s protocol. Transformants were screened for insert by colony PCR and sequenced. Colonies containing pET24a-mΔN-PDE3B were isolated but contained an F700S (TTT→TCT) mutation present in the template. This mutation was corrected by site-directed mutagenesis using primers PDE-TCT→TTT_for and PDE-TCT→TTT_rev using the Quikchange method with Phusion as polymerase.
Bacterial expression of Δ-PDE3B and inclusion body isolation
BL21-CodonPlus (DE3)-RIPL chemically competent cells (Agilent) were transformed with pET-24a-mΔN-PDE3B and plated on LB-Agar plates containing 25μg/mL kanamycin and 25μg/mL chloramphenicol. Expression cultures were inoculated at 1:20 dilution from saturated overnight cultures and grown with 25μg/mL kanamycin and 25μg/mL chloramphenicol at 37 ºC to OD600~1.0. Protein expression was induced with 1 mM IPTG at 37 ºC for 3 h. The cultures were centrifuged at 4 ºC and 4000 × g for 5 min, yielding ~4 g/L cell paste, which was frozen in liquid N2.
Inclusion bodies containing ΔN-PDE3B were purified similarly to a published protocol55. The cell paste was suspended in 5 mL/g of 50 mM sodium phosphate, 10% glycerol, 1 mM PMSF, pH 7.6, and lysed by passage once through a French pressure cell at 14000 psi. The lysate was incubated at 25 ºC with 5 U/g DNase for 15 min and centrifuged at 15000 × g for 20 min at 4 °C. The pellet was resuspended in 5 mL/g 100 mM Tris, 4% (v/v) Triton X-100, 2 M urea, pH 8.0, and centrifuged at 15,000 × g for 20 min. The resulting pellet was resuspended and centrifuged once more. Finally, the pellet was washed twice via resuspension and centrifugation in 5 mL/g water. (Addition of 10% Buffer B improved pelleting of the inclusion bodies.) The isolation yielded ~0.1 g inclusion bodies/g of cell paste, which were stored at −20ºC.
Purification of mouse Δ-PDE3B
a. Affinity column synthesis
The inhibitor SK&F 93505 – (5R)-6-(4-aminophenyl)-5-methyl-4,5-dihydropyridazin-3(2H)-one – was synthesized as described56. Compound A was prepared from SK&F 93505 as described39. Compound A was linked to Affi-Gel 10 resin (BioRad) following the manufacturer’s protocol. Briefly, Affi-Gel 10 resin (6 mL) was loaded into a 1.5 × 10 cm fritted column and washed with 2 column volumes (CV) isopropanol, followed by 2 CV dimethylsulfoxide (DMSO). The resin was transferred to a 15 mL Falcon tube. Compound A was dissolved in DMSO to 100 mg/mL and 150 μL of this solution were added to the resin. The derivatization reaction proceeded for 2.5 h at room temperature with nutation. Excess sites on the resin were blocked by addition of a slight excess of ethanolamine (7 μL) and incubation at room temperature for 15 min. The resin was returned to the column and washed with several CV of isopropanol. Compound A was stored for extended periods in DMSO at -20 °C and the column was stored in isopropanol at 4 °C.
b. Inclusion body solubilization and refolding
Inclusion bodies were solubilized with agitation at ~5 mg/mL in 40 mL 50 mM Tris, 7 M guanidine-HCl, pH 7.4, containing 0.1 M dithiothreitol (DTT), for 2 h at 4 ºC. After passage of the suspension through a frit, the solubilized inclusion bodies were added dropwise into a stirring 2 L solution of 630 mM Tris, 20 mM MgCl2, 5 mM MnCl2, 40 μM ZnSO4, 0.5 M arginine, 5% glycerol, 10.5 mM NaCl, 0.5 mM KCl, 10 mM DTT, pH 7.4 (MnCl2 and DTT were added ~1 h prior to refolding). The refolding proceeded for 16–20 h at 4 °C. The refolding solution was concentrated to ~300–400 mL using a Millipore Ultracel 30 kDa MWCO Ultrafiltration disc (76 mm).
c. ΔN-PDE3B affinity purification from refolded protein
The Compound A ligand affinity column (4 mL) was equilibrated with 10 CV of Buffer C (50 mM Tris, pH 7.5, 2 mM MgCl2, 0.5 mM EDTA, 50 mM NaCl, 0.5 M L-arginine, 1 mM DTT, 5% glycerol, pH 7.5). Concentrated refolded ΔN-PDE3B was loaded onto the affinity column. The column was washed with 10 CV of Buffer C, and non-specifically bound protein was eluted with 5 CV of Buffer C + 2 M NaCl. The column was once again washed with 5 CV Buffer C, and ΔN-PDE3B was eluted from the column with 30 mL 7 mM IBMX in Buffer C (5% DMSO, diluted from a 140 mM stock in DMSO). The eluate was diluted 2-fold in Buffer C containing 10 mM DTT, concentrated to ~2 mL using an Amicon Ultra YM10 centrifugation device, and frozen in liquid N2.
d. Size exclusion chromatography
The concentrated affinity column eluate was thawed on ice and loaded onto a pre-equilibrated (20 mM HEPES, 10 mM NaCl, 2 mM MgCl2, 0.5 mM EDTA, 5% Glycerol, 5 mM DTT) Sephadex G-50 size exclusion column (2.5 × 32 cm, 160 mL). Eluted protein was collected in 2 mL fractions. Protein-containing fractions were identified by A280nm, pooled, and concentrated to ~350 μL. The procedure yielded ~4 mg from 3 refoldings, each from 200 mg inclusion bodies.
Cloning, expression, and purification of C769S-, C768S/C769S-, C769S/C777S-, and C768S/C769S/C777S-Δ-PDE3B
pET24a-mΔN-PDE3B (1 μg) was digested for 1 h with 20 U XbaI according to the manufacturer’s protocol. Following gel electrophoresis, the larger band was excised and purified using the Zymoclean Gel DNA Recovery Kit (Genesee Scientific). The C→S mutations were inserted as gBlocks via the Gibson assembly57. The gBlocks consisted of the ~0.7 kb XbaI fragment of pET24a-mΔN-PDE3B, with sequences overlapping the vector appended (21 nt at the 5' end and 22 nt at the 3' end of the gBlock); the appropriate Cys codons were mutated to Ser (AGC). After the assembly reaction, performed according to the manufacturer’s protocol, transformants were grown up and sequenced to verify successful mutagenesis. Mutants were expressed in BL21 CodonPlus(DE3)-RIPL cells, refolded, and purified as for the wt protein, except that a 2.5 × 10 cm (50 mL) Sephadex G-50 column was used and 1.5 mL fractions were collected, screening for protein-containing fractions using Bradford reagent. Yields were 2–3 mg protein per g inclusion bodies.
General procedures for biochemical assays
Protein assays were conducted in an anaerobic chamber (Vacuum Atmospheres Company) with 90% nitrogen, 10% hydrogen atmosphere (<1 ppm O2), unless otherwise noted. Buffers and other solutions used for biochemical assays were deoxygenated by three cycles of evacuation (>30 min) and refilling with argon or nitrogen and stored in the anaerobic chamber.
All assays were performed in 50 mM MOPS, 100 mM NaCl, pH 7.5 (Buffer D). We found that treatment of this buffer with Chelex100 (50–100 mesh, sodium form, BioRad) according to the manufacturer’s instructions (5 g/100 mL buffer, stirring for 1 h), followed by readjustment of pH), was essential for reproducible and time-dependent activity measurements. The buffer was stored in acid-washed glassware.
WT or mutant ΔN-PDE3Bs refolded from inclusion bodies (~30 μM, ~100 μL) were desalted using Zeba Spin columns (0.5 mL, 7K MWCO). Columns were buffer exchanged according to the manufacturer’s protocol into Chelex-treated Buffer B. Protein was desalted by passage through two spin columns (2 × 1.5 min, 1500 × g, 4 °C) in order to ensure complete removal of DTT in the protein storage buffer. The protein was transferred to a pear-shaped flask and immediately deoxygenated on a Schlenk line with 4 cycles of evacuation and refilling with N2. The protein was brought into the glovebox and could be stored for up to 2 weeks at 4 °C without significant loss of activity.
Activity assays of PDE3B purified from insect cells
Reduced apo protein was obtained by incubating samples with 1 mM TCEP and 5 mM EDTA for 1 h at 4 °C, followed by buffer exchange into pre-chilled Buffer D containing 5 mM MgCl2 using Amicon 0.5 mL ultra-centrifugal filters (EMD Millipore). Apo protein samples (1 μM for ΔN-PDE3B, 2 μM for full-length PDE3B) were titrated with increasing amounts of Cu+ and incubated for 1 h at 25 °C. A 100x stock solution of Cu+ ([(CH3CN)4Cu+][PF6]) was made in 30% acetonitrile/water (v/v) and further diluted into working concentrations in assay buffer (Buffer D containing 5 mM MgCl2) and added to the protein. Following incubation with metal, 50 μM cAMP was added to the samples and incubated for an additional hour at 37 °C after which the reactions were quenched by addition of acetonitrile to a final concentration of 50% (v/v). Precipitated protein and salt were removed by centrifuging at 13000 rpm for 5 min and the resulting supernatant was dried and resuspended in 30 μL of Milli-Q water. Samples were analyzed on an Agilent 1200 HPLC using an Agilent Eclipse XDB-C18 column (3.5 μm, 3.0 × 150 mm). The method used a 100 mM potassium phosphate, 4 mM tetrabutylammonium chloride, pH 6.0, aqueous mobile phase, a flow rate of 0.5 mL/min, and linear gradient of 0–20% methanol (7 min), followed by 50% methanol (1 min), and 0% methanol (4 min). The nucleotide absorbance was detected at 260 nm. Activity of the protein was normalized to the activity detected in the absence of any copper.
Activity assays of the bacterially expressed Δ-PDE3B
Copper solutions for titrations and activity assays were prepared freshly each day. In an anaerobic chamber, ~5 mg CuCl (stored in the chamber) was dissolved in 400 μL 0.1 M HCl, 1 M NaCl to give a ~70–100 mM Cu+ stock solution42,58. The concentration of CuCl in this stock solution was calculated spectrophotometrically by addition of excess BCS using ε483nm = 13.0 mM−1 cm−1 59.
For a typical activity assay, performed in the anaerobic chamber, a 25 μL reaction contained: 0.1 μM ΔN-PDE3B, 10 mM MgCl2, 0.5–1 mM cAMP, and 0, 0.1, 0.2, 0.3, or 0.4 μM CuCl, in Buffer D. The temperature of the anaerobic chamber was 27–30 °C. Buffer D was mixed with 2.5 μL 100 mM MgCl2, prepared in Buffer D, and WT or mutant ΔN-PDE3B (stock concentration ~30 μM), and the solution was incubation for 10 min at ambient temperature.
Immediately before use, the CuCl stock solution was diluted to 35 μM Cu+ in 0.1 M HCl, 1 M NaCl, and this solution was further diluted to 3.5 μM Cu+ in Buffer D. This solution (0–4 equiv. Cu) was mixed with the protein solution and incubated further for 10 min. In assays in which BCS was used, it was mixed with the protein solution prior to copper addition. Assays were initiated by addition of cAMP to 0.5 or 1 mM from a ~15 mM stock solution. The concentration of the cAMP stock solution, prepared in Buffer D, was determined using ε259nm = 13.8 mM−1 cm−1 60. The assays were quenched at 3–6 min by addition of 25 μL acetonitrile. (Initial experiments indicated that, under these conditions, product formation is linear with time.)
The samples were removed from the glovebox, centrifuged briefly, and evacuated overnight in a vacuum desiccator. The next day, the desiccated samples were resuspended in 40 μL 50 mM HEPES, pH 7.6 and analyzed by HPLC as described above. 5'-AMP eluted at ~6 min and cAMP eluted at ~10 min. Specific activities were determined by comparison of integrated peak area with that of a 50 μM 5'-AMP standard.
Determination of the EC50 values of Δ-PDE3B for Mg2+
Activity assays were carried out anaerobically as described above, using the following conditions: 50 nM ΔN-PDE3B (affinity purified following refolding from inclusion bodies), pre-incubated with the indicated concentration of MgCl2 for 10 min prior to substrate addition, and 1 mM cAMP. The reactions were quenched in acetonitrile after 6 min. Product formation was assayed by HPLC and specific activities were determined by comparison of integrated peak area with that of a 50 μM 5'-AMP standard. In order to extract EC50 values, the data were fitted to the following equation:
UV-vis titrations to probe Cu+:Δ-PDE3B stoichiometry
Copper solutions for titrations and activity assays were prepared freshly each day. Immediately prior to use, the 70–100 mM CuCl stock solution was diluted 100-fold in 0.1 M HCl, 1 M NaCl, and the concentration of CuCl in this stock solution was calculated spectrophotometrically by addition of excess BCS using ε483nm = 13.0 mM−1 cm−1 59. Immediately prior to the titration, this solution was diluted 10-fold into Buffer D and loaded into a 50 μL Hamilton syringe fitted with a repeat dispenser. The syringe was inserted into a septum-sealed anaerobic cuvette (micro volume, Starna Cells) containing, in a total volume of 350 μL, Buffer D, ~1.5 μM protein (2.5 μM for C768S/C769S/C777S), and 35 μL 0.1 M HCl, 1 M NaCl. An initial spectrum was acquired on a Cary 60 UV-visible spectrophotometer between 240 and 700 nm, with 1 nm steps. The protein was titrated with 1 or 2 μL Cu+ solution, up to ~40 μL, with 1–2 min between copper addition and spectrum acquisition. Initial experiments demonstrated that absorbance changes stabilized within this time.
Titrations were analyzed as described in Supplementary Figure 27. Briefly, the average absorbance between 650–700 nm was subtracted from the spectra, and the spectra were corrected for volume change during the course of the titration. Following these corrections, the absorbance at 265 nm was plotted against Cu+ (calculated from the concentration of the ~75–100 μM Cu+ solution used for the titration, determined using BCS). For the wildtype protein, the endpoint of the titration could usually be determined by the change in slope of this A265nm/Cu+ plot, but for the mutants, which displayed smaller total absorbance changes, this was challenging. However, upon subtraction of a control titration in the absence of protein, the endpoint became apparent. The slope of the control titration plot (ΔA265nm/μM Cu+) was typically ~0.003/μM. From this information, the Cu-binding stoichiometry and ΔA265nm/μM protein were calculated.
No features in the 400–700 nm range, which could be ascribed to Cu2+-His interactions, were observed during the course of the titrations, although beyond 1.25–1.5 equiv. Cu+, precipitation and increased baseline was commonly seen.
Construction of plasmids for overexpression of full-length and Δ-PDE3B in mammalian cells
pmaxCloning (2 μg) was digested with 10 U KpnI according to the manufacturer’s protocol and purified. The linearized plasmid was used as the template for a PCR reaction in order to install strong (GCCACCATGG, where the start codon is underlined), adequate (GAGGCCATGA), or weak (GAGCCCATGA) Kozak sequences at the 5' end of the PDE3B gene. The reaction used PDE3B-Gib_1 and either PDE3B-Gib_strong, PDE3B-Gib_ad, or PDE3B-Gib_weak as primers and ExTaq DNA polymerase (TaKaRa). The products were DpnI-digested, gel extracted (Zymoclean DNA Gel Recovery Kit), and ligated to the 3.3 kb, PDE3B-encoding XhoI fragment of pAcSG2-mPDE3B using the Gibson assembly according to the manufacturer’s protocol (50 °C, 1 h), with equimolar concentrations of vector and insert. The reaction mix was used to transform XL10 Gold chemically competent cells, which were plated on LB-agar containing 50 μg/mL kanamycin. Transformants were screened by colony PCR (GoTaq Green, Promega) using the primers pmax-for and pmax-rev, and colonies containing the ~3 kb insert were miniprepped (Qiagen). The correct insertion was confirmed by sequencing the 5' and 3' regions of the PDE3B gene using the primers pmax_for and pmax_rev.
Preliminary experiments using these constructs showed that higher PDE3B expression in the construct containing the strong Kozak sequence led to reduced 3T3-L1 adipocyte viability after transfection, and the construct containing the weak Kozak sequence (pmax-PDE3B, Supplementary Table 3), which did not exhibit significantly lower transfection yield compared to a pmaxCloning control, was used for further experiments.
The plasmid for overexpression of ΔN-PDE3B (pmax-ΔN-PDE3B) was constructed analogously to the above procedure, except 1) the primers used to PCR amplify the KpnI-digested pmaxCloning were PDE3B-Gib_1 and PDE3B-Gib_ad (adequate Kozak sequence: GAGCCCATGG), and 2) the insert was the 1.6 kb XhoI fragment of pAcSG2-ΔN-PDE3B encoding ΔN-PDE3B.
Construction of C769S, C768S/C769S, and C769S/C777S mutants of pmax-PDE3B and pmax-Δ-PDE3B
pmax-PDE3B (3 μg) was digested sequentially with 40 U BstBI (65 °C, 90 min) and 20 U PvuI-HF (37 °C, 1 h). The gel extracted vector/PDE3B fragment was ligated to gBlocks containing the appropriate C→S mutations via the Gibson assembly. The gBlocks consisted of the ~0.45 kb BstBI/PvuI fragment of mPDE3B, flanked by 25 nt at the 5' end and 21 nt at the 3' end of the gBlock, overlapping the vector; the appropriate Cys codons were mutated to Ser (AGC). After the assembly reaction, performed according to the manufacturer’s protocol, transformants were grown up and sequenced to verify mutagenesis. The analogous mutants of pmax-ΔN-PDE3B were constructed by the same procedure.
Transfection of 3T3-L1 adipocytes
3T3-L1 cells were used on day 11–12 after initiation of differentiation for transfection. Cells were transfected with full-length PDE3B constructs (2 μg for WT PDE3B and 4 μg for PDE3B mutants, 4 μg of ΔN-PDE3B constructs) using the Amaxa nucleofector Kit V (Lonza, Walkersville, MD) according to the manufacturer’s protocols, and they were incubated for 18 h. Cells were then incubated for 5 h in DMEM + 2% BSA media, washed once with media without BSA, and then incubated with 0 or 50 μM CuCl2 for 1 h as described above. The media was removed, and media with 2% BSA, 100 nM Iso, and 0 or 50 μM CuCl2 was added. After 25 min, cells were harvested for cAMP assays and analyzed as described above.
Supplementary Material
Acknowledgments
We thank the US National Institutes of Health (GM 79465 to C.J.C., GM067166 and GM101502 to S.L.) for providing funding for this work. C.J.C. is an Investigator and L.K. is a Postdoctoral Research Associate with the Howard Hughes Medical Institute. J.A.C. is supported by a postdoctoral fellowship from the Jane Coffin Childs Memorial Fund for Medical Research. J.C. was supported by a postdoctoral fellowship from the Human Frontiers Science Program. A.T.A. was supported by a National Science Foundation Graduate Research fellowship. C.M.A. was supported by a Hertz Foundation Graduate Fellowship. L.P.S. was supported by the German National Academic Foundation with an international scholarship. S.L.F. was supported by scholarships from Amgen and Merage Foundation for the American Dream Scholarship. E.J.N. was supported by a fellowship from the Royal Commission for the Exhibition of 1851. We thank Ann Fischer, Xiaozhu Zhang, Alison Killilea and Carissa Tosta (UCB Tissue Culture Facility), and Christopher Mangels (Johns Hopkins University) for expert technical assistance; Vincent Manganiello (Laboratory of Biochemical Physiology, NIH) for mPDE3B plasmids; Joern Larsen (Department of Earth Sciences, Lawrence Berkeley National Laboratory) for use of an ICP-MS instrument; and Maeran Uhm (University of Michigan) for advice regarding PDE3B overexpression.
Footnotes
Author contributions:
L.K. J.A.C., S.L., and C.J.C. designed research; L.K and J.A.C. performed most experiments. J.C., S.J., A.T.A., L.P.S., and S.L.F. synthesized and characterized copper probes. J.C. performed cellular imaging experiments. H.K. purified and characterized PDE3B expressed in insect cells. A.M. and V.S.P. performed the animal experiments. C.M.A. performed ICP-MS and other metal analysis experiments. M.N.V. synthesized Compound A for affinity purification of PDE3B. T.G. assisted with bacterial expression and purification of PDE3B. E.J.N. designed and performed preliminary experiments. L.K., J.A.C., and C.J.C. wrote the paper; S.L., J.C., and S.J. provided valuable input on the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Lippard SJ, Berg JM. Principles of Bioinorganic Chemistry. University Science Books; Mill Valley, CA: 1994. [Google Scholar]
- 2.Barnham KJ, Masters CL, Bush AI. Neurodegenerative diseases and oxidative stress. Nat Rev Drug Discov. 2004;3:205–14. doi: 10.1038/nrd1330. [DOI] [PubMed] [Google Scholar]
- 3.Rae TD, Schmidt PJ, Pufahl RA, Culotta VC, O'Halloran TV. Undetectable intracellular free copper: the requirement of a copper chaperone for superoxide dismutase. Science. 1999;284:805–8. doi: 10.1126/science.284.5415.805. [DOI] [PubMed] [Google Scholar]
- 4.Robinson NJ, Winge DR. Copper metallochaperones. Annu Rev Biochem. 2010;79:537–62. doi: 10.1146/annurev-biochem-030409-143539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banci L, et al. Affinity gradients drive copper to cellular destinations. Nature. 2010;465:645–8. doi: 10.1038/nature09018. [DOI] [PubMed] [Google Scholar]
- 6.Boal AK, Rosenzweig AC. Structural biology of copper trafficking. Chem Rev. 2009;109:4760–79. doi: 10.1021/cr900104z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brady DC, et al. Copper is required for oncogenic BRAF signalling and tumorigenesis. Nature. 2014;509:492–6. doi: 10.1038/nature13180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turski ML, et al. A novel role for copper in Ras/mitogen-activated protein kinase signaling. Mol Cell Biol. 2012;32:1284–95. doi: 10.1128/MCB.05722-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dodani SC, et al. Calcium-dependent copper redistributions in neuronal cells revealed by a fluorescent copper sensor and X-ray fluorescence microscopy. Proc Natl Acad Sci U S A. 2011;108:5980–5. doi: 10.1073/pnas.1009932108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dodani SC, et al. Copper is an endogenous modulator of neural circuit spontaneous activity. Proc Natl Acad Sci U S A. 2014;111:16280–5. doi: 10.1073/pnas.1409796111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cotruvo JA, Jr, Aron AT, Ramos-Torres KM, Chang CJ. Synthetic fluorescent probes for studying copper in biological systems. Chem Soc Rev. 2015;44:4400–14. doi: 10.1039/c4cs00346b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang CJ. Searching for harmony in transition-metal signaling. Nat Chem Biol. 2015;11:744–7. doi: 10.1038/nchembio.1913. [DOI] [PubMed] [Google Scholar]
- 13.Aron AT, Ramos-Torres KM, Cotruvo JA, Jr, Chang CJ. Recognition- and reactivity-based fluorescent probes for studying transition metal signaling in living systems. Acc Chem Res. 2015;48:2434–42. doi: 10.1021/acs.accounts.5b00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kahn SE, Hull RL, Utzschneider KM. Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature. 2006;444:840–6. doi: 10.1038/nature05482. [DOI] [PubMed] [Google Scholar]
- 15.Khandekar MJ, Cohen P, Spiegelman BM. Molecular mechanisms of cancer development in obesity. Nat Rev Cancer. 2011;11:886–95. doi: 10.1038/nrc3174. [DOI] [PubMed] [Google Scholar]
- 16.Van Gaal LF, Mertens IL, De Block CE. Mechanisms linking obesity with cardiovascular disease. Nature. 2006;444:875–80. doi: 10.1038/nature05487. [DOI] [PubMed] [Google Scholar]
- 17.Burkhead JL, Lutsenko S. The role of copper as a modifier of lipid metabolism. In: Baez RV, editor. Lipid Metabolism. InTech; 2013. [DOI] [Google Scholar]
- 18.Engle TE. Copper and lipid metabolism in beef cattle: a review. J Anim Sci. 2011;89:591–6. doi: 10.2527/jas.2010-3395. [DOI] [PubMed] [Google Scholar]
- 19.al-Othman AA, Rosenstein F, Lei KY. Copper deficiency alters plasma pool size, percent composition and concentration of lipoprotein components in rats. J Nutr. 1992;122:1199–204. doi: 10.1093/jn/122.6.1199. [DOI] [PubMed] [Google Scholar]
- 20.Carr TP, Lei KY. High-density lipoprotein cholesteryl ester and protein catabolism in hypercholesterolemic rats induced by copper deficiency. Metabolism. 1990;39:518–24. doi: 10.1016/0026-0495(90)90011-z. [DOI] [PubMed] [Google Scholar]
- 21.Huster D, et al. High copper selectively alters lipid metabolism and cell cycle machinery in the mouse model of Wilson disease. J Biol Chem. 2007;282:8343–55. doi: 10.1074/jbc.M607496200. [DOI] [PubMed] [Google Scholar]
- 22.Huster D, Lutsenko S. Wilson disease: not just a copper disorder. Analysis of a Wilson disease model demonstrates the link between copper and lipid metabolism. Mol Biosyst. 2007;3:816–24. doi: 10.1039/b711118p. [DOI] [PubMed] [Google Scholar]
- 23.Seessle J, et al. Alterations of lipid metabolism in Wilson disease. Lipids Health Dis. 2011;10:83. doi: 10.1186/1476-511X-10-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huster D, et al. Consequences of copper accumulation in the livers of the Atp7b−/− (Wilson disease gene) knockout mice. Am J Pathol. 2006;168:423–34. doi: 10.2353/ajpath.2006.050312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lutsenko S. Atp7b−/− mice as a model for studies of Wilson's disease. Biochem Soc Trans. 2008;36:1233–8. doi: 10.1042/BST0361233. [DOI] [PubMed] [Google Scholar]
- 26.Buiakova OI, et al. Null mutation of the murine ATP7B (Wilson disease) gene results in intracellular copper accumulation and late-onset hepatic nodular transformation. Hum Mol Genet. 1999;8:1665–71. doi: 10.1093/hmg/8.9.1665. [DOI] [PubMed] [Google Scholar]
- 27.Gerbasi V, Lutsenko S, Lewis EJ. A mutation in the ATP7B copper transporter causes reduced dopamine beta-hydroxylase and norepinephrine in mouse adrenal. Neurochem Res. 2003;28:867–73. doi: 10.1023/a:1023219308890. [DOI] [PubMed] [Google Scholar]
- 28.Aigner E, et al. A role for low hepatic copper concentrations in nonalcoholic fatty liver disease. Am J Gastroenterol. 2010;105:1978–85. doi: 10.1038/ajg.2010.170. [DOI] [PubMed] [Google Scholar]
- 29.Kawamura M, et al. Hormone-sensitive lipase in differentiated 3T3-L1 cells and its activation by cyclic AMP-dependent protein kinase. Proc Natl Acad Sci U S A. 1981;78:732–6. doi: 10.1073/pnas.78.2.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim BE, et al. Cardiac copper deficiency activates a systemic signaling mechanism that communicates with the copper acquisition and storage organs. Cell Metab. 2010;11:353–63. doi: 10.1016/j.cmet.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duncan RE, Ahmadian M, Jaworski K, Sarkadi-Nagy E, Sul HS. Regulation of lipolysis in adipocytes. Annu Rev Nutr. 2007;27:79–101. doi: 10.1146/annurev.nutr.27.061406.093734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chijiwa T, et al. Inhibition of forskolin-induced neurite outgrowth and protein phosphorylation by a newly synthesized selective inhibitor of cyclic AMP-dependent protein kinase, N-[2-(p-bromocinnamylamino)ethyl]-5-isoquinolinesulfonamide (H-89), of PC12D pheochromocytoma cells. J Biol Chem. 1990;265:5267–72. [PubMed] [Google Scholar]
- 33.Greenberg AS, et al. Stimulation of lipolysis and hormone-sensitive lipase via the extracellular signal-regulated kinase pathway. J Biol Chem. 2001;276:45456–61. doi: 10.1074/jbc.M104436200. [DOI] [PubMed] [Google Scholar]
- 34.Xue B, Greenberg AG, Kraemer FB, Zemel MB. Mechanism of intracellular calcium ([Ca2+]i) inhibition of lipolysis in human adipocytes. FASEB J. 2001;15:2527–9. doi: 10.1096/fj.01-0278fje. [DOI] [PubMed] [Google Scholar]
- 35.Zhou L, et al. Berberine attenuates cAMP-induced lipolysis via reducing the inhibition of phosphodiesterase in 3T3-L1 adipocytes. Biochim Biophys Acta. 2011;1812:527–35. doi: 10.1016/j.bbadis.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 36.Zhou D, et al. CD36 level and trafficking are determinants of lipolysis in adipocytes. FASEB J. 2012;26:4733–42. doi: 10.1096/fj.12-206862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Snyder PB, Esselstyn JM, Loughney K, Wolda SL, Florio VA. The role of cyclic nucleotide phosphodiesterases in the regulation of adipocyte lipolysis. J Lipid Res. 2005;46:494–503. doi: 10.1194/jlr.M400362-JLR200. [DOI] [PubMed] [Google Scholar]
- 38.Ahmad F, et al. Insulin-induced formation of macromolecular complexes involved in activation of cyclic nucleotide phosphodiesterase 3B (PDE3B) and its interaction with PKB. Biochem J. 2007;404:257–68. doi: 10.1042/BJ20060960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Varnerin JP, et al. Expression, refolding, and purification of recombinant human phosphodiesterase 3B: definition of the N-terminus of the catalytic core. Protein Expr Purif. 2004;35:225–36. doi: 10.1016/j.pep.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 40.McMillin DR. The origin of the intense absorption in azurin. Bioinorg Chem. 1978;8:179–84. doi: 10.1016/s0006-3061(00)80243-4. [DOI] [PubMed] [Google Scholar]
- 41.Pountney DL, Schauwecker I, Zarn J, Vasák M. Formation of mammalian Cu8-metallothionein in vitro: evidence for the existence of two Cu(I)4-thiolate clusters. Biochemistry. 1994;33:9699–705. doi: 10.1021/bi00198a040. [DOI] [PubMed] [Google Scholar]
- 42.Angeletti B, et al. BACE1 cytoplasmic domain interacts with the copper chaperone for superoxide dismutase-1 and binds copper. J Biol Chem. 2005;280:17930–7. doi: 10.1074/jbc.M412034200. [DOI] [PubMed] [Google Scholar]
- 43.Omburo GA, Brickus T, Ghazaleh FA, Colman RW. Divalent metal cation requirement and possible classification of cGMP-inhibited phosphodiesterase as a metallohydrolase. Arch Biochem Biophys. 1995;323:1–5. doi: 10.1006/abbi.1995.0001. [DOI] [PubMed] [Google Scholar]
- 44.Scapin G, et al. Crystal structure of human phosphodiesterase 3B: atomic basis for substrate and inhibitor specificity. Biochemistry. 2004;43:6091–100. doi: 10.1021/bi049868i. [DOI] [PubMed] [Google Scholar]
- 45.Degerman E, Belfrage P, Manganiello VC. Structure, localization, and regulation of cGMP-inhibited phosphodiesterase (PDE3) J Biol Chem. 1997;272:6823–6. doi: 10.1074/jbc.272.11.6823. [DOI] [PubMed] [Google Scholar]
- 46.Hung SH, et al. New insights from the structure-function analysis of the catalytic region of human platelet phosphodiesterase 3A: a role for the unique 44-amino acid insert. J Biol Chem. 2006;281:29236–44. doi: 10.1074/jbc.M606558200. [DOI] [PubMed] [Google Scholar]
- 47.Tang KM, Jang EK, Haslam RJ. Expression and mutagenesis of the catalytic domain of cGMP-inhibited phosphodiesterase (PDE3) cloned from human platelets. Biochem J. 1997;323:217–24. doi: 10.1042/bj3230217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ovadia H, et al. Increased adipocyte S-nitrosylation targets anti-lipolytic action of insulin: relevance to adipose tissue dysfunction in obesity. J Biol Chem. 2011;286:30433–43. doi: 10.1074/jbc.M111.235945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maurice DH, et al. Advances in targeting cyclic nucleotide phosphodiesterases. Nat Rev Drug Discov. 2014;13:290–314. doi: 10.1038/nrd4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chung YW, et al. Targeted disruption of PDE3B, but not PDE3A, protects murine heart from ischemia/reperfusion injury. Proc Natl Acad Sci U S A. 2015;112:E2253–62. doi: 10.1073/pnas.1416230112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gray NE, et al. Angiopoietin-like 4 (Angptl4) protein is a physiological mediator of intracellular lipolysis in murine adipocytes. J Biol Chem. 2012;287:8444–56. doi: 10.1074/jbc.M111.294124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stephens JM, Lee J, Pilch PF. Tumor necrosis factor-alpha-induced insulin resistance in 3T3-L1 adipocytes is accompanied by a loss of insulin receptor substrate-1 and GLUT4 expression without a loss of insulin receptor-mediated signal transduction. J Biol Chem. 1997;272:971–6. doi: 10.1074/jbc.272.2.971. [DOI] [PubMed] [Google Scholar]
- 53.Hong-Hermesdorf A, et al. Subcellular metal imaging identifies dynamic sites of Cu accumulation in Chlamydomonas. Nat Chem Biol. 2014;10:1034–42. doi: 10.1038/nchembio.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kenan Y, Murata T, Shakur Y, Degerman E, Manganiello VC. Functions of the N-terminal region of cyclic nucleotide phosphodiesterase 3 (PDE 3) isoforms. J Biol Chem. 2000;275:12331–8. doi: 10.1074/jbc.275.16.12331. [DOI] [PubMed] [Google Scholar]
- 55.Cotruvo JA, Stubbe J. NrdI, a flavodoxin involved in maintenance of the diferric-tyrosyl radical cofactor in Escherichia coli class Ib ribonucleotide reductase. Proc Natl Acad Sci U S A. 2008;105:14383–8. doi: 10.1073/pnas.0807348105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leonard WR, Romine JL, Meyers AI. A rapid and efficient synthesis of chiral 2-hydro-2-oxazolines. Journal of Organic Chemistry. 1991;56:1961–63. [Google Scholar]
- 57.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–5. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 58.Cobine PA, et al. Stoichiometry of complex formation between Copper(I) and the N-terminal domain of the Menkes protein. Biochemistry. 2000;39:6857–63. doi: 10.1021/bi000015+. [DOI] [PubMed] [Google Scholar]
- 59.Xiao Z, et al. Unification of the copper(I) binding affinities of the metallo-chaperones Atx1, Atox1, and related proteins: detection probes and affinity standards. J Biol Chem. 2011;286:11047–55. doi: 10.1074/jbc.M110.213074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dworkin M, Keller KH. Solubility and diffusion coefficient of adenosine 3':5'-monophosphate. J Biol Chem. 1977;252:864–5. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.