Abstract
Background
Alterations in the gut microbial community composition maybe influential in neurological disease. Microbial community profiles were compared between early onset pediatric multiple sclerosis (MS) and control children similar for age and sex.
Methods
Children ≤18 years old within two years of MS onset, or controls without autoimmune disorders attending a University of California, San Francisco, USA pediatric clinic were examined for fecal bacterial community composition and predicted function by 16S ribosomal RNA sequencing and Phylogenetic Reconstruction of Unobserved States (PICRUST) analysis. Associations between subject characteristics and the microbiota, including beta diversity and taxa abundance were identified using non-parametric tests, permutational multivariate analysis of variance and negative binomial regression.
Results
Eighteen relapsing-remitting MS cases and 17 controls (mean age 13 years; range 4-18) were studied. Cases had a short disease duration (mean=11 months; range 2-24) and half were immunomodulatory (IMD) drug naïve. While overall gut bacterial beta diversity was not significantly related to MS status, IMD exposure was (Canberra, p<0.02). However relative to controls, MS cases had a significant enrichment in relative abundance for members of the Desulfovibrionaceae (Bilophila, Desulfovibrio and Christensenellaceae) and depletion in Lachnospiraceae and Ruminococcaceae (all p and q<0.000005). Microbial genes predicted as enriched in MS vs. controls included those involved in glutathione metabolism (Mann-Whitney, p=0.017); findings that were consistent regardless of IMD exposure.
Conclusions
In recent onset pediatric MS, perturbations in the gut microbiome composition were observed, in parallel with predicted enrichment of metabolic pathways associated with neurodegeneration. Findings were suggestive of a pro-inflammatory milieu.
Keywords: pediatric multiple sclerosis, peadiatric multiple sclerosis, gut microbiota, gut microbiome, 16S rRNA, case-control study, risk factors, immunomodulatory drugs
Background
Alterations in the gut microbiota may be influential in disease, including neurological and autoimmune conditions.[1, 2] Humans comprise of 10 times more microbial than human cells, of which >90% reside in the gastrointestinal tract.[3, 4] The role of these microbial communities (the gut microbiota) includes synthesis of vitamins, modulation of the immune system and resistance to infection.[5] Multiple sclerosis (MS) is considered an autoimmune disease, with both genetic and early life environmental exposures (including infections), implicated as risk factors. Recent studies suggest a role for the gut microbiota in MS, with most evidence derived from experimental animal models. Emerging work reported perturbation in the adult MS gut microbiota, relative to healthy controls.[6, 7] While of major interest, adults with MS have had a lifetime of exposures, with potential for disease trigger(s) or modifiers to have occurred years previously or accumulated over decades. Pediatric MS, while relatively rare, represents a unique opportunity to examine disease processes after few years of life have accrued, narrowing the search for potential disease triggers. We conducted a pilot study to explore the gut microbiota in early onset pediatric MS, compared to controls of similar age and sex. We also explored the potential influence of other characteristics such as immunomodulatory drug exposure to guide future studies.
Methods
Children ≤18 years old attending a University of California, San Francisco (UCSF) pediatric clinic (e.g. a general or MS specific clinic) were invited to participate in an environmental risk factor study; those providing a stool sample formed the current study cohort. MS cases were within 2 years of symptom onset. Use of immunomodulatory drugs (IMDs) or systemic corticosteroids were permissible, although prior cytotoxic immunosuppressant exposure was not. In the absence of consensus on the most appropriate controls for microbiota studies, we selected controls of similar age and sex, free from autoimmune disorders (conditions such as asthma and eczema were allowed), with neither parent diagnosed with MS nor a related disorder. Neither cases nor controls were exposed to systemic antibiotics in the two months before stool collection.
Demographic, lifestyle and clinical characteristics
The children's characteristics were obtained from the physician and research coordinator (JH) clinic visits (using standardized forms and chart abstraction), as well as questionnaires which could be completed at home (summarized in eTable 1). For the current study, we included: sex; age; race (white, non-white); ethnicity (Hispanic, non-Hispanic), co-morbidity (present, absent), body mass index (healthy or underweight, overweight or obese [<85th,≥85th percentile]), fat-fibre diet groups (high fat-low fibre, based on recent diet[8] [yes, no]); early life diet (breastfed <12, ≥12 months); antibiotics in the first year of life (yes, no); attendance at day-care (yes, no); mode of delivery (vaginal, caesarean section); prior close contact with animals (direct physical contact [ever, never]; dog or cat slept in the house on a regular basis [yes, no]). MS specific clinical characteristics included: age at MS symptom onset, disability (Expanded Disability Status Scale Score, EDSS); relapses pre-stool sample (including number, severity and location); use of immunomodulatory drugs (IMDs) and systemic corticosteroids (in the previous 2 months).
Stool sample collection, DNA extraction and 16S rRNA profiling
The parent was asked to collect the child's first stool of the day and to ship on ice (via FedEx®) to UCSF where it was stored at −80°C before processing. DNA was extracted from approximately 25mg of stool using the PowerSoil® DNA Isolation Kit (MO BIO Laboratories, Inc, Carlsbad, California, USA) according to the manufacturer's recommendations. The V4 hypervariable region of the bacterial 16S rRNA gene was amplified in triplicate as previously described,[9] then combined, purified and pooled in equimolar concentrations. Sequencing was performed via the Illumina MiSeq platform using a 251×151 base pair run. The 16S rRNA reads were clustered at ≥ 97% similarity into operational taxonomic units (OTUs), and the resulting OTU table was singly rarefied to 201,546 reads per sample. OTUs were assigned taxonomy using the Greengenes database and analysis was performed using QIIME (Quantitative Insights Into Microbial Ecology).[10, 11]
Alpha and beta diversity
Alpha diversity was expressed as evenness, richness and Faith's phylogenic diversity metric, using the vegan and picante packages [R]. Beta diversity was calculated using a Canberra distance matrix, a non-phylogenetic, equal-weight measure where each OTU affects the distance value equally.
Predicted metagenome
Metagenomic predictions were made using PICRUSt[12] (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States) and summarized as KEGG (Kyoto Encyclopaedia of Genes and Genomes) pathways.
Statistical analyses
Epidemiological characteristics were presented descriptively. The gut microbiota metrics were primarily compared between cases and controls and secondly by IMD exposure. Additional exploratory comparisons were performed for the diversity metrics only, including characteristics selected a priori as either potential confounders or if a reasonable (e.g. ≥10% difference in proportions) between cases and controls existed (sex, age, race, ethnicity fat/fibre groups, breastfeeding, IMD exposure and for the MS cases only, EDSS, corticosteroid use or history of a recent relapse).[13] Alpha diversity was compared between groups using non-parametric tests (Mann-Whitney or Kruskal-Wallis), beta diversity was explored using principal coordinate analyses (PCoA) and permutational multivariate analysis of variance (PERMANOVA). At the taxon level, the relative abundance of individual OTUs for the primary (cases and controls) and secondary (IMD naïve vs. IMD exposed cases vs. controls) comparisons were explored using generalized linear models (GLM, negative binomial regression), with findings expressed as rate ratios (RR), and 95% confidence intervals (95%CI), along with p and q values (false discovery rate adjusted p-values; <0.05 was considered significant). All other comparisons were made without adjustment. Phylum-level and KEGG Pathway abundance were explored using non-parametric tests. Statistical analyses were performed using R (Version 3.0.2)[14] and the Statistical Package for the Social Sciences (SPSS for Windows, Ver. 22.0. Armonk, NY: IBM Corp. 2013).
Secondary platform – DNA microarray
In the absence of a validation cohort, we opted for an additional, secondary platform to corroborate the 16S rRNA sequencing data, using the G3 PhyloChip™ (Second Genome, Inc., CA), see Appendix #1, online.
The University of California San Francisco Institutional Review Board approved the study.
Results
Between Nov/2011-Nov/2013, 18 MS (10 girls, 8 boys) and 17 controls (9 girls, 7 boys) fulfilled study criteria. The mean age at stool collection was 13.0 years (SD=3.82; range 4-18). Although controls averaged one year older than cases and more self-identified as white (77% controls, 50% cases), the proportion of Hispanics was relatively similar (44% cases, 35% controls) and no characteristic (Table 1) differed significantly between cases and controls (all p>0.1, not shown).
Table 1.
Characteristics of the pediatric multiple sclerosis (MS) cases and controls
| Characteristic, n (%) unless stated otherwise | MS cases, n=18 | All controls, n=17 |
|---|---|---|
| Sex: Girl | 10 (55.6%) | 9 (52.9%) |
| Boy | 8 (44.4%) | 8 (47.1%) |
| Age at stool sample collection, years: mean (SD; range) | 12.5 years (SD=4.44; 4-17) | 13.5 years (SD=3.08; 9-18) |
| Age at stool sample collection: ≤12 years old | 6 (33.3%) | 8 (47.1%) |
| >12 years old | 12 (66.7%) | 9 (52.9%) |
| Race: White | 9 (50.0%) | 13 (76.5%) |
| Non-white | 9 (50.0%) | 4 (23.5%) |
| Ethnicity: Hispanic | 8 (44.4%) | 6 (35.3%) |
| Non- Hispanic | 10 (55.6%) | 11 (64.7%) |
| Co-morbid condition [1]: present | 7 (38.9%) | 5 (29.4%) |
| Absent | 11 (61.1%) | 12 (70.6%) |
| Diet, lifestyle and very early life exposures | ||
| Mode of delivery: vaginal birth | 16 (88.9%) | 15 (88.2%) |
| Caesarean section | 2 (11.1%) | 1 (5.9%) |
| Unknown | 0 | 1 (5.9%) |
| Breastfed: <12 months | 13 (72.2%) | 8 (47.1%) |
| ≥12 months | 5 (27.8%) | 9 (52.9%) |
| Antibiotic exposure in 1st year of life: yes | 5 (27.8%) | 6 (35.3%) |
| No | 12 (66.7%) | 10 (58.8%) |
| Unknown | 1 (5.6%) | 1 (5.9%) |
| Attended day-care (‘preschool’): yes | 12 (66.7%) | 12 (70.6%) |
| No | 4 (22.2%) | 4 (23.5%) |
| Unknown | 2 (11.1%) | 1 (5.9%) |
| BMI [2]: crude mean (SD; range) | 22.2 (SD:5.66; range:15.1-35.3) | 22.8 (SD:7.10; range:15.0-43.9) [missing=1] |
| Healthy or underweight(<85th percentile) | 12 (66.7%) | 11 (64.7%) |
| Overweight or obese (≥85th percentile) | 6 (33.3%) | 5 (29.4%) [missing=1] |
| Fat/fibre diet groups [3]: High fat, and low or very fibre – yes | 5 (27.8%) | 5 (29.4%) |
| No | 13 (72.2%) | 12 (70.6%) |
| Animal exposure | ||
| Direct physical contact with animals: yes | 12 (66.7%) | 12 (70.6%) |
| No | 5 (27.8%) | 3 (17.6%) |
| Unknown | 1 (5.6%) | 2 (11.8%) |
| Animal contact – dog or cat slept in house on regular basis: yes | 8 (44.4%) | 8 (47.1%) |
| No | 8 (44.4%) | 7 (41.2%) |
| Unknown | 2 (11.1%) | 2 (11.8%) |
Key: SD=standard deviation
Comorbid conditions were collected pre-stool sample (but were not necessarily present pre-MS onset): for cases: headache (n=1); atopic dermatitis/eczema (n=1); long-term constipation (n=1); history of shingles (n=1); history of seizures (n=1); reactive airways disease and headache (n=1); scoliosis (n=1). For controls: kyphosis (n=1); Raynaud phenomenon (n=1); asthma (n=2); pancreatitis (n=1)
Body Mass Index (BMI)=height(kg)/weight(m)sq, with age and sex-specific percentiles (see eTable 1). One individual (case) was below the 5th percentile; 3/18 cases and 3/17 controls were ≥95th percentile (obese)
Recent diet derived from the Block Kids Food Screener (NutritionQuest©), recorded over one week (see eTable 1 for further details)
For cases, the mean disease duration at stool collection (from MS symptom onset) was 10.6 months (SD=6.34; range 2-23), the median EDSS at enrolment was 2.0 (range 0-4.0), half were exposed to an IMD for MS (5 glatiramer acetate, 3 beta-interferon, 1 natalizumab) and 6 (33%) to a systemic corticosteroid (in the 2 months pre-stool sample). All met McDonald criteria and had relapsing-remitting MS, see Table 2, and online eTables 2, 3 for additional characteristics.
Table 2.
Multiple sclerosis (MS) specific clinical characteristics of the children (cases)
| Characteristic, n (%) unless stated otherwise | N=18 |
|---|---|
| Age at MS symptom onset, years: mean (SD; range) | 12.1 years (SD=4.66; 4-17) |
| Disease duration at stool collection [1], months: mean (SD; range) | 10.6 months (SD=6.45; 2.3-23.1) |
| Annualized relapse rate pre-stool sample [excl. onset attack], mean (SD; range) [2] | 0.86 (SD=0.914; range: 0-2.9) |
| Time since last relapse (in relation to stool sample; onset attack considered): days: mean (SD; range) | 183.3 days (SD=135.74; 4 to 489 days) |
| Time since last relapse (in relation to stool sample; onset attack considered): <90 days | 6 (33.3%) |
| 90-<200 days | 6 (33.3%) |
| 200+days | 6 (33.3%) |
| Disability level - EDSS at enrolment, median (range) | 2.0 (0-4.0) |
| 0-<2.0 | 7 |
| 2.0-<3.0 | 8 |
| 3.0+ | 3 |
| Immunomodulatory drug exposure status [3]: IMD naïve | 9 (50.0%) |
| IMD exposed | 9 (50.0%) |
| Corticosteroids – systemic [4]: No | 12 (60.0%) |
| Yes | 6 (33.3%) |
Key: excl.=excluding; SD=standard deviation; EDSS=Expanded Disability Status Scale score; IMD=immunomodulatory drug
disease duration: time from symptom onset to stool collection
relapse rate = relapse count/disease duration; note however that while a major strength of the study, the disease duration was low; i.e. those cases with the shortest disease durations pre-stool sample can influence the apparent relapse rates as follows: a very short disease duration implies less opportunity for a relapse to occur, therefore often the relapse rate was ‘zero.’ However, if a relapse did occur, the resulting relapse rate can appear high (inflated) because of the small denominator (disease duration)
‘IMD naïve’ indicates never exposed pre-stool sample. ‘IMD exposed’ indicates ever exposed pre-stool sample; all were also currently exposed (at the time of stool collection) as follows: beta-interferon (n=3); glatiramer acetate (n=5); natalizumab (n=1). No child had switched or stopped an IMD (although one child had previously been exposed to plasma exchange before taking glatiramer acetate).
within previous 2 months
Gut microbiota profiling
All samples underwent 16S rRNA sequencing, though one control sample produced insufficient read depth and was excluded from these analyses. The majority of samples produced a 16S rRNA PhyloChip profile (3 control samples produced insufficient 16S rRNA amplicon for analyses by this secondary profiling technique).
Alpha and beta diversity
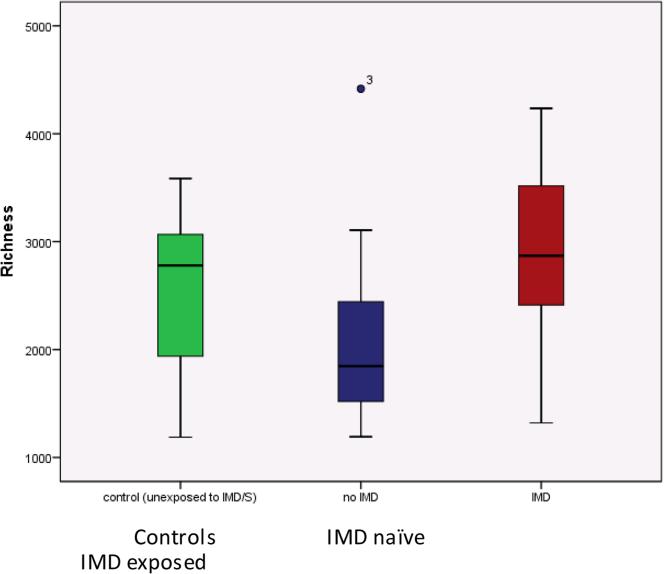
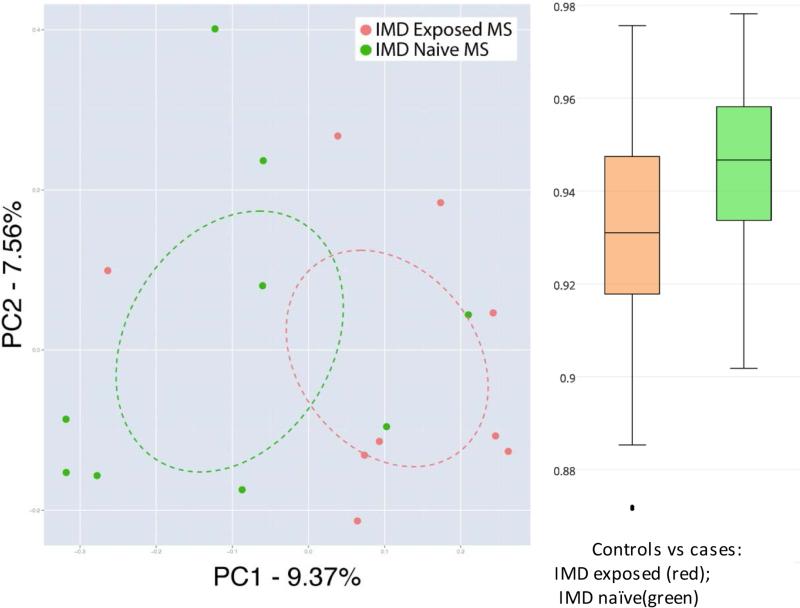
Gut microbiota alpha diversity did not significantly differ between cases and controls (all p>0.2 for the diversity metrics, eTable 4A-B, online). While there was a trend for greater alpha diversity in the IMD exposed cases>controls>IMD naïve cases, no comparison reached significance (all p>0.05, see Figure 1, Panel A and eTable 4A-B, online). However, IMD exposure was identified as a significant factor that explained 6.8% of the variation in gut microbiota beta diversity (Canberra distance matrix; IMD exposed vs. naïve cases vs controls, p=0.012, PERMANOVA). When only MS cases were considered in this analysis, IMD exposure explained a slightly larger proportion (7.1 %) of the observed compositional variation (p=0.016, Figure 1, Panel B and eTable 4B), indicating that IMD exposure was associated with differences in gut microbiota composition. This was supported by an (ad-hoc) comparison of compositional distance from controls – IMD exposed cases exhibited gut microbiota that were more similar to that of control subjects when compared to IMD-naïve patients (Figure 1, Panel C, Bonferroni-corrected 2-sample t-test p<0.001). Hispanics exhibited lower community evenness compared to non-Hispanics (p=0.040), however no other measured variables reached significance (all p>0.1, data not shown).
Figure 1 (Panel A, B, C). Alpha and beta-diversity of the gut microbiota community composition for controls and cases (IMD naïve and exposed).
Panel A: Alpha diversity (richness): controls vs. IMD naïve and exposed cases. The box plot depicts findings; there were no significant differences between groups (or overall across groups), all p>0.05 (Mann-Whitney or Kruskal-Wallis). All alpha diversity data are shown in eTable 4A, online
Key for Figure 1
Panel A, C: Box plots represent median (black horizontal line), upper and lower quartiles (edge of boxes), upper and lower extremes (whiskers). Outliers represented by a single data point.
Panel B: Each dot represents one child with MS. Red=IMD exposed child (n=9); Green=IMD naïve child (n=9); all exposed cases were currently on IMD treatment at the time of the stool sample, the naïve cases had never been exposed to an IMD. The controls are not depicted.
PC1 and PC2 represent the top two principal coordinates that captured most diversity (generated using the Canberra dissimilarity matrices), the percentages represent the fraction of the diversity captured by each coordinate shown.
Ellipses=95% confidence intervals (via SE of weighted mean axes values)
IMD exposure was associated with differences in beta diversity, explaining 7.1% of the gut microbiota variation (Canberra metric, IMD exposed vs naïve cases, p=0.016, Adonis)
Panel B (left): The immunomodulatory drug [IMD] exposure status of the children with MS was associated with gross perturbations in the overall microbial community composition: A principal coordinate analyses (PCoA) plot indicating the similarity and difference's in the gut microbiota composition of the IMD exposed and IMD naïve MS cases
Panel C (right): IMD-exposed subjects are compositionally more similar to the controls (less distant, as measured by Canberra) than the IMD-naïve cases (Bonferroni-corrected 2-sample t-test p<0.001). The box plot depicts the Canberra compositional distance of cases (IMD exposed and naïve) from controls
Taxonomic characteristics of cases and controls
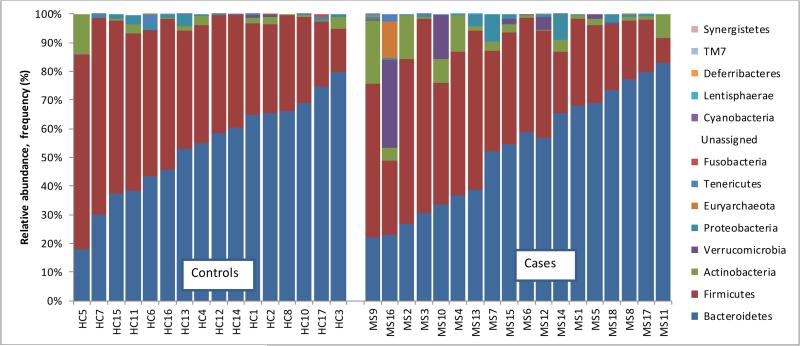
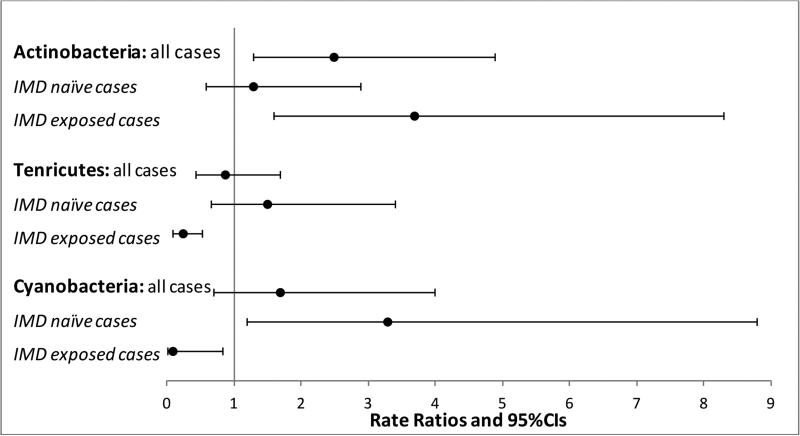
From the 16S rRNA sequencing data, a total of 25,134 OTUs were identified. These were primarily bacterial (>99%) with the remainder belonging to the Archaea. In total, 14 phyla, 24 classes, 37 orders, and 61 families were identified. Twelve phyla were common to cases and controls, and 2 found exclusively in cases (Synergistetes and Lentisphaerae). As previously described in other US populations,[15] Bacteroidetes and Firmicutes dominated the gut microbiome of study participants (Figure 2A). Cases had 2.5 times the abundance of Actinobacteria compared to controls (95%CI:1.3-4.9), p=0.008 (Figure 2B). While no other phylum reached significance (cases vs. controls), IMD status influenced findings; the rate ratio for Actinobacteria was attenuated for the IMD naïve cases (decreasing to 1.3;95%CI:0.58-2.9, p=0.527 vs controls), but remained significantly elevated in the IMD exposed cases (RR=3.7;95%CI:1.6-8.3, p=0.002), see Figure 2B. IMD-based differences in two other phyla (Tenericutes and Cyanobacteria) were also observed, see Figure 2B, suggesting that even at the phylum level IMD exposure significantly influences microbiota composition.
Figures 2A-B. Phylum-level findings (relative abundance) for cases and controls [A] and by immunomodulatory drug exposure [B].
Figure 2A: Phylum-level findings: Bacteriodetes and Firmicutes dominated the gut microbiota of children with and without MS
Key: Each bar represents one child; HC=health controls; MS=multiple sclerosis.
Y axis depicts relative abundance, reported as a proportion for each child, relative to all cases and controls.
Figure 2B: Phylum-level differences were observed for MS cases (n=18) vs controls (n=16), but were influenced by the immunomodulatory drug [IMD] exposure status of the cases.
Key: Controls formed the reference group. Rate ratios derived from the negative binomial regression; CI=confidence intervals. IMD=immunomodulatory drug (n=9 MS cases were naïve, 9 were exposed [5 glatiramer acetate, 3 beta-interferon, 1 natalizumab]). Only phylum shown where a significant difference was observed (p<0.05) in at least one comparison. No adjustments made for multiple comparisons.
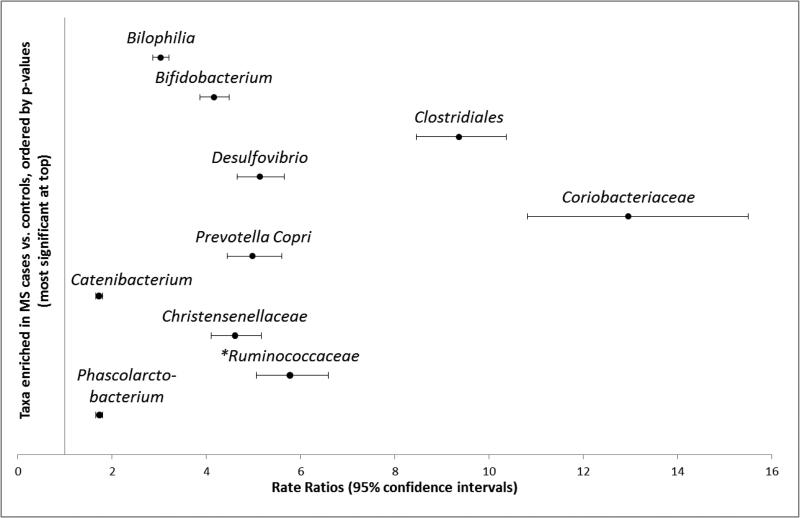
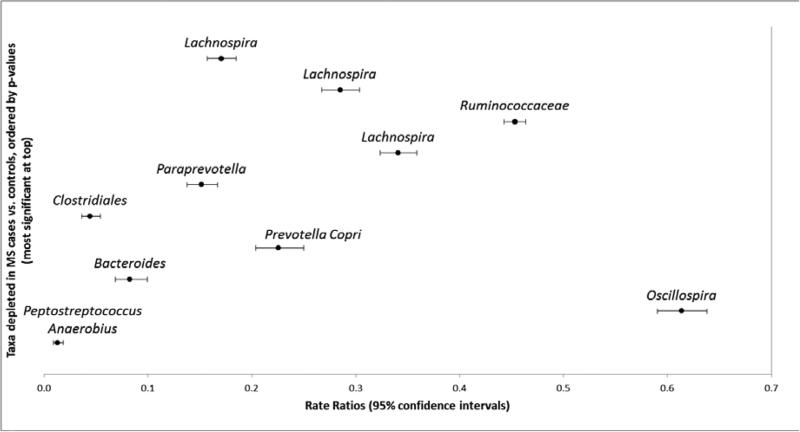
At the individual OTU level, 160 of the 25,134 taxa (0.6%) were significantly enriched and 163 taxa (0.6%) depleted in MS cases compared to controls (all p, q<0.05, negative binomial regression, eTables 5A,B, online). At the order level, 57% (91/160) of the enriched and 49% (79/163) of depleted taxa belonged to Clostridiales (Firmicutes), with the second most common being Bacteroidales (Bacteriodetes), representing 26% (41/160) and 45% (74/163), respectively. Although these dominated, a range of additional taxa were found to be significantly altered in relative abundance across these groups (eTables 5A,B). Some of the most significant (i.e. lowest p and q values) are depicted in Figure 3A, B and summarized in context of the wider microbiota literature in eTable 6. For example, MS cases exhibited 3.0 times the relative abundance of Bilophilia (order Desulfovibrionales) compared to controls (95%CI:2.9-3.2). Members of the Bifidobacterium, Desulfovibrio, and Christensenellaceae family were similarly enriched (RR>4), among others (Figure 3A). There was additionally evidence for decreased relative abundances of members of the Clostridiales order (e.g. the butyrate producing. Lachnospiraceae and Ruminococcaceae). A comparison of the absolute differences in genera abundance for cases and controls included enrichment for Akkermansia, Parabacteroides, Methanobrevibacter (Archaea), and Bacteroides and depletion for members of the orders Bacteroidales and Clostridiales (genus unknown); see eTables 5A-B for all findings. The direction of findings, and those in Figure 3A-B were similar, regardless of IMD exposure (aside from one taxon belonging to the Ruminococcaceae family), see Figure 3A). The influence of IMD exposure status on the primary comparisons (cases vs. controls) are depicted in eTables 5A,B, with the original rate ratios for these secondary comparisons in eTables 7A-D. Briefly, 45 of the 160 taxa enriched in MS relative to controls were depleted in IMD naïve cases and 40/160 were depleted in IMD exposed cases (all relative to controls; most being from the Clostridiales order, n=28 and 25, respectively). Fewer (7) of the 163 depleted taxa were similarly affected (eTable5B).
Figure 3 A-B.
Rate ratios and 95% confidence intervals depicting taxa-level [OTU] differences between MS cases and controls in the gut microbial communities (relative abundance).
Ten most significantly enriched taxa shown in Fig. 3A (top), and ten depleted in Fig. 3B (bottom), selected and ordered based on the lowest p-values, from eTables5A,B; all p and q values <0.000005.
Key for Figure 3A, B: All findings are shown in eTable 5A,B (online only), including full taxonomic nomenclature; the lowest available shown here (typically family or genus). In total, of the 25,134 taxa identified, 160 were significantly enriched in MS cases; 163 were depleted, relative to controls. The most (statistically) significant findings are depicted here for illustrative purposes, representing, OTU# (Fig 3A, from top to bottom: 359872, 365385, 2740950 4453773, 4397092,196296, 4480861, 176318, 4476065, 3138798; Fig 3B: 193191, 4457427 4409280, 176269, 181432, 146564, 518820, New.ReferenceOTU477, 4468466, 4473664).
Example interpretation of results: Children with MS had 3.0 times the abundance of Bilophila (95%CI: 2.9-3.2) than control children.
All depicted findings were consistent, regardless of IMD exposure, except for: *Ruminococcaceae which while significantly enriched for all cases vs controls (Figure3A) and for IMD exposed children [RR= 10.9; 95%CI:9.4-12.3], relative to controls], this taxa was significantly depleted for the IMD naïve children [RR= 0.76; 95%CI:0.61-0.95]
See eTable 6 (online only) for a summary of the taxa depicted in Figure 3A, B in relation to the secondary platform (DNA microarray) wider gut microbiota literature
The rate ratios and 95% confidence intervals are derived from negative binomial regression models which may not meet the default convergence limits
Validation of findings
The G3 phylogenetic microarray data broadly concurred with the main findings, in that cases and controls did not significantly differ in alpha diversity indices (all p>0.1, data not shown). Additional results, including comparisons with findings from the primary platform, are provided in Table 3, eTable 6 and Appendix #1.
Table 3.
Current study findings in context of the emerging studies in multiple sclerosis (MS) or animal models of MS
| Authors, year(s), location | Study design‡ or target population | Platform | Main finding(s) | Comparison with current study findings |
|---|---|---|---|---|
| Peer-reviewed publications | ||||
| Miyake et al. 2015. Japan (Tokyo / Kanagawa Prefecture) [6, 7] | Case-control; 20 RRMS cases [9 were IMD exposeda]; National Center of Neurology and Psychiatry Hospital, Tokyo; 40 controls from Azabu University, Kanagawa Prefecture | 16S rRNA; Roche 454 sequencing | 1.Alpha diversity did not differ between cases and controlsa; beta-diversity differed [IMD status not stated] 2.Genus level: Faecalibacterium, Prevotella, and Anaerostipes less abundant in MS cases vs controls; p<0.05 (Welch's test) 3.Species level: Lower abundance of clostridial species [Clostridia clusters XIVa and IV] & several Bacteroidetes. |
1.Alpha diversity similar for cases & controls, p>0.05b 2. Lower abundance: cFaecalibacterium (genus), in MS vs. controls 3. Lower abundances of several dClostridiales (order) and eBacteroidaceae (family) in MS vs controls |
| Cantarel et al. Mowry, 2015. San Francisco, California, USA[6] | Vitamin D intervention; pre-intervention (baseline): 4 RRMS [2 GA, 2 IMD naïve], 8 healthy controls; all white, non-hispanic women with low serum vitamin D | 16S rRNA Phylochip (akin to DNA microarray) | 1.Beta-diversity similar for cases & controls, p=0.74f; but differed for IMD-exposed vs unexposed cases, p=0.007f 2. Lower number of OTU identified as: Faecalibacterium (genus), Bacteroidaceae (family) MS vs. controls |
1.Beta-diversity similar for cases & controls, p=0.22g, but differed for IMD-exposed vs unexposed cases, p=0.016g; 2. Lower abundance: cFaecalibacterium (genus); eBacteroidaceae (family) in MS vs. controls |
| Kasper et al., [multiple papers/years] Dartmouth, New Hampshire, USA[29] | Animal model (EAE); in vitro studies in MS | Multiple | Findings suggest gut colonization of Bacteroides fragilis can protect against EAE | hBacteroides fragilis was significantly depleted in MS cases vs controls (RR: 0.039; 95%CI: 0.014-0.106). Consistent findings in IMD exposed or naïve cases, eTables 5, 7 |
| Rumah et al. 2013, New York, USA[41] | Case-control: 30 relapsing-onset MS patients (disease duration not stated); 31 controls. Cases recruited via MS clinic, not stated for controls.k | Clostridium perfringens (type A) present in i7/30 (23%) MS patients vs 16/31 (52%) controls | Sequencing: unknown [several Clostridiaceae (family) identified, genus and species level could not be classified in all instances; no clostridium (genus) identified]. DNA microarray: 10 Clostridium perfringens detected. Abundance (fluorescence) did not differ between cases & controls, all p>0.08. No suggestion that taxa was less prevalent in casesj (OTU# [no.cases/no.controls]: 72305 [6/4]; 71846 [17/13]; 72764 [10/9]; 72181 [17/12]; 72505 [5/2]; 72179 [5/2]; 72587 [18/12]; 72718 [5/2]; 72823 [10/5]; 72153 [14/7]). |
|
| Conference abstracts | ||||
| Jangi et al., 2014, Boston, Massachusetts USA.[23] | Case-control: 53 MS clinic patients (22 IMD naïve, 31 IMD exposed [GA or IFNB]; 44 healthy controls from lPhenoGenetic project | Roche 454 sequencing | 1.Increase in Archaea (Methanobrevibacteriaceae, Methanobrevibacter [family, genus]) in MS vs. healthy controls (p <0.00001). 2. Decreased: Butyricimonas (genus) in IMD naïve MS vs. controls |
1.Increase in Archaea (φMethanobrevibacter[genus]) in MS vs controls (p<0.000005), see eTable 5A; 2.Butyricimonas (genus) depleted in MS cases [all cases or IMD naïve] vs. controls (p<0.000005), see eTables 5, 7 |
| Baranzini et al. 2014, New York and San Francisco, California, USA[22] | Not specified (in the published abstract) | Significant enrichment of Enterobacteriaceae (family) were identified when comparing female patients to sex-matched controls | 5 members of Enterobacteriaceae (family) significantly enriched in MS cases (boys and girls) vs controls (OTU# 3232397; 3101394; NR106; 114510; 814442, all p<0.0003), see eTable 5A | |
Key: IMD=immunomodulatory drug; GA=glatiramer acetate; IFNB=beta-interferon; RR=rate ratio (from negative binomial regression); EAE = experimental autoimmune encephalomyelitis; OTU=Operational Taxonomic Unit
no.cases/no.controls = number cases / number controls with fluorescence on chip (>0); no formal statistical comparisons performed for presence vs absence of taxa. Total possible cases=18; controls=14
all study results reported here were based on cross-sectional findings. While two studies[6] included a longitudinal, i.e. repeated sampling component (e.g., Cantarel et al., 2015[6] included a vitamin D supplementation intervention), although the cross-sectional or baseline comparisons (pre-intervention) were reported here to allow comparisons with the current study.
Φpossible confounder was constipation; while gut transit time was not measured, comorbidity information was collected; the one child (a case) was reported as having ‘long-term constipation.’ This child also had high abundances of Methanobrevibacter.
9/20 (45%) were exposed to beta-interferon (derived from Table S1). [6, 7] No formal comparisons for IMD exposed / unexposed individuals reported. No p-value given; authors stated ‘the Shannon index, a metric for evaluating bacterial diversity, was not significantly different between the samples obtained from HC40 (3.39 ± 0.29) and MS20 subjects (3.29 ± 0.46).’ [6, 7]
p-values derived from non-parametric tests. See Figure 1 and in eTable 4A, online. All p>0.05 for all comparisons with diversity measured as richness, evenness or Faiths: all cases vs. controls; IMD naïve cases vs IMD exposed cases vs controls; IMD naïve cases vs IMD exposed cases; IMD naïve cases vs controls (p>0.05, Mann-Whitney or Kruskal-Wallis)
Faecalibacterium (genus): all were prausnitzii (species), OTU# 181422; 365717; 180572), all p<0.0002; q<0.013). No members of this genus were identified as significantly increased in MS.
Clostridiales (order) 79 of 163 identified OTU which were significantly depleted in MS cases vs. controls were from the Clostridiales order; species unknown, all p and q<0.05, see eTable 5B
Bacteroidaceae (family) 44 identified OTU were significantly reduced in MS cases vs. controls (and 17 were increased), all were Bacteroides (genus), all p and q<0.05, see eTable 5B
p-values derived from the Adonis test, using the Unifrac metric to assess beta-diversity. Additional (post-baseline) IMD exposed cases were included for this comparison, increasing numbers to 5 IMD exposed (all were glatiramer acetate) vs 2 IMD naïve MS cases
p-values derived from the Adonis test, using the Canberra metric to assess beta diversity, see eTable 4B
Bacteroides fragilis (New.CleanUp.ReferenceOTU66109) – the only OTU identified at this species level in eTable 5B
Numbers were calculated based on the %'s reported by the authors; exact numbers [numerators] were not stated in the paper
No formal statistical analyses performed in these subgroups
No subjects had been exposed to cytotoxic or immunosuppressive drugs, or antibiotics in the previous 6 months. Not stated if immunomodulatory drugs had been used.
PhenoGenetic project based at the Brigham and Women's Hospital, Boston
Predicted metagenome
From the 16S rRNA sequencing data, 271 KEGG pathways were identified; 27 were predicted as significantly enriched in MS and 1 depleted, relative to controls (p <0.05). IMD exposure affected findings (eTable 8, online). Compared to controls, pathways predicted as enriched in the microbiome of MS patients included those involved in glutathione metabolism (p=0.017) and lipopolysaccharide biosynthesis (IMD naïve cases only, p=0.008), see online eFigure 1, Panel A. IMD exposure was associated with enrichment of several pathways, including those related to immune responses and folate biosynthesis (see eFigure 1, Panel B).
Discussion
We observed perturbations in the gut microbiota of children very early in their MS disease course relative to children without MS. These perturbations were not evident at the beta diversity level, but rather were characterized by discrete taxonomic enrichments and depletions.
This suggests that recent MS onset may be associated with more subtle changes in a limited number of microbial taxa, rather than large shifts in the community composition.
In contrast, immunomodulatory drug (IMD) exposure was associated with a substantial portion of microbiota compositional variation. Microbial taxa alterations associated with MS which remained significant irrespective of IMD exposure, included an increase in relative abundance of members of the Desulfovibrio, Bifidobacterium genera and members of the recently identified and highly heritable Christensenellaceae family.[16] There was additionally evidence for decreased relative abundances of members of the Clostridiales order (e.g. the butyrate producing. Lachnospiraceae and Ruminococcaceae). Combined, these data indicate that early dysregulation in MS involves an increase in pro-inflammatory and a decrease in anti-inflammatory gut microbiota milieu.[17, 18] The suggestion of a heritable taxon contributing to MS in a disease with known (host) familial and genetic contribution deserves further investigation. [19] [19]
We were unable to find any studies exploring the potential association between the gut microbiota and pediatric MS or with individuals sampled so close to MS onset. Remarkably however, patterns are emerging from the sparse literature, despite different platforms and approaches (summarised in Table 3). One USA-based pilot study compared 7 relapsing-remitting MS and 8 healthy control women (all were non-Hispanic whites).[6] A second study from Japan. [6, 7] included 20 relapsing-remitting MS adults and 40 controls. Similar to our findings, no overall differences in the gut microbiota composition between cases and controls (beta-diversity in the former study[6] and alpha-diversity in the latter). Both studies identified discrete taxon-level differences as characteristic of cases.[6, 7] In addition, IMD exposure was also related to microbiota compositional variation,[6] concurring with our findings. While 9/20 individuals in the Japanese study were IMD exposed, not formal comparisons by IMD status were reported. [6, 7]
Common to both USA based studies was the significant depletion of Faecalibacterium and Bacteroidaceae members in MS cases (independent of IMD treatment). Faecalibacterium prausnitzii (the species identified as depleted in MS cases in our study), has been shown to exert anti-inflammatory effects and its presence is considered a ‘sensor and marker of health.’[20] Its depletion has also been observed in children newly diagnosed with the autoimmune condition Crohn's disease.[20, 21]
Additional gut microbiota studies in MS have appeared as conference proceedings only23, 24 or primarily studied in animal models of MS. [22, 23][22, 23] Briefly, significant enrichment of the pro-inflammatory Enterobacteriaceae family have been reported in MS, relative to controls (in women only);23 a finding we also observed, regardless of sex. [20, 22, 24, 25] [20, 22, 24, 25] Depletion of the Butyricimonas genus and enrichment for the Archaea Methanobrevibacter in 53 adults with MS as compared to 44 controls also concurred with our observations.[23] Although consistent with the broader literature, this Archaea was not ubiquitous; relatively few individuals (cases or controls) had detectable levels in their gut (eTable 5A). This may be due to a lack of detection related to the depth of sequencing, or because this genus is a marker of established disease in adults. However, overgrowth of the methane-producing Methanobrevibacter has recently been attributed to underlying constipation.[26] These slow-growing microbes thrive in environments with low water availability; methane further slows gut transit, promoting their self-survival.[26] Future studies should consider assessing gut transit to help disentangle whether these differences in Archaea are directly related to MS or are a consequence of comorbidity, such as constipation.
Bacteroides fragilis was also significantly depleted in our MS cases compared to controls, with findings consistent regardless of IMD exposure. Similar reductions are observed in early rheumatoid arthritis (n=51), relative to fibromyalgia (n=50), all prior to initiation of specific drug treatments).[27, 28] Gut colonization of Bacteroides fragilis protects against experimental autoimmune encephalomyelitis (EAE) via polysaccharide A production and induction of IL-10 from CD4+ Foxp3+ regulatory T cells in the gut.[29] Additionally, this species has been shown to ameliorate neurodevelopmental issues in a mouse model, indicating a gut-microbiota-brain connection.[30]
Exposure to an IMD influenced some findings, but not all; suggestive of a partial, but not full resolution of gut perturbation in addition to possible IMD-specific perturbations. Longitudinal studies are needed to establish the nature and direction of these associations. We did observe putative functional differences congruent with the IMDs known pharmacodynamics, including predicted enrichment of microbial genes encoding immunomodulatory pathways,[31] involving CD8+, CD4+ and B-cell maturation and differentiation.[31] In addition, the MS gut microbiota was predicted as enriched for genes encoding glutathione metabolism[32] and lipopolysaccaride biosynthesis.[33] While the latter appeared ‘normalized’ for the children exposed to an IMD, glutathione remained elevated, regardless of IMD exposure status (all relative to controls). Glutathione is an important agent of the endogenous antioxidant defence system; loss of the oxidant/antioxidant balance has been implicated in MS neurodegeneration, and glutathione has been pursued as a therapeutic target and potential in vivo (brain) biomarker.[34] However, monitoring or targeting glutathione in the CNS is challenging. The potential for gut microbiota involvement in glutathione regulation - perhaps via metabotropic glutamate gut receptors - is intriguing and deserves further exploration.[35] Lipopolysaccharides can induce and worsen EAE,[36] perhaps via bystander activation, supporting an antigen-independent link between infection and autoimmunity.[36] We also observed a number of infectious disease-related pathways upregulated in MS (eTable 8). A better understanding of these alterations is needed, especially if future ventures include interventions (e.g. diet or pre/probiotics) targeted at ‘normalizing’ the functional capacity of the gut microbiota in MS.
Others have explored the role of Fungi in MS, specifically Candida spp., via blood serum levels from 80 prevalent MS patients (local MS association members) relative to 240 age and sex matched blood donors.[37] The MS participants had a higher odds of serum antigen presence (vs absence) as measured by slot-blot analyses to: Candida parapsilosis (OR=7.3;95%CI:3.2-16.6,p<0.001) and Candida glabrata (OR=3.0;95%CI:1.5-6.1,p=0.002).[37] Authors indicated that findings were similar after excluding the MS participants (50%) exposed to an IMD or immunosuppressant or an extended MS disease duration (>10 years).[37] Although the original sources of the Candida spp. were unknown (e.g. skin, nails, mouth, gut or vagina[38]), it raises the possibility that other microbiota, such as Candida spp, could be associated with MS.[37]
Strengths of our study include a well phenotyped group of children sampled very close to MS onset, and diagnosed using internationally recognized criteria by MS specialist pediatric neurologists. We accessed a well-validated microbiota pipeline.[39] In the absence of a validation cohort we accessed a secondary platform (DNA microarray) and included an inferred functional metagenome assessment. Detailed demographic and lifestyle information indicated similarities between cases and controls; controls were also recruited from the same catchment area as cases. Pediatric MS represents a unique opportunity to explore the gut microbiota after relatively few years of life or disease have accrued. However, its relative rarity was reflected by our small numbers, and combined with the cross-sectional design precluded a rigorous assessment of potential confounders. Future studies need to carefully consider those identified here, including ethnicity and ideally enrol IMD naïve individuals. There was also considerable variation (from <10% to 100%) in the proportion of children with any given taxon in their gut. The contribution of low or high penetrance microbes which all differed significantly in abundance between cases and controls requires further investigation.
What triggers MS is unknown. Whether changes in the gut microbiota community play a role in precipitating or enabling the disease or in facilitating a persistent underlying immune dysregulation in MS is intriguing. We found that even in the very early stages of MS, and independent of IMD exposure, taxon level differences in the gut microbiota were observed. These perturbations suggested a shift towards a pro-inflammatory state concur with observations in other autoimmune diseases, including traditionally ‘gut unrelated’ conditions, such as rheumatoid arthritis.[40] We also observed similarities between emerging MS studies; whether these indicate a ‘gut signature’ of MS or of broader autoimmune disease remains to be determined, as does the relationship with IMDs. A better understanding of the gut microbiota's role in MS may identify novel drug targets, preventative measures and pathways to improved health.
Supplementary Material
Acknowledgements
Gratitude s extended to the families and children for participation in this study.
Shirley Bi for conducting relevant literature reviews and valuable administrative tasks related to compiling the manuscript.
Tom Duggan for facilitating in the preparation of Tables and graphics
Funding Statement: This work was supported by the National MS Society RG4861A3/1 (PI Waubant), National Institutes of Health NS071463 (PI Waubant), The Race to Erase MS (PI Waubant) and the Canada Research Chair program (PI Tremlett). The funding source(s) had no role in the study design, collection, analysis or interpretation of the data, or in the decision to submit the article for publication.
Footnotes
Statistical analyses were performed by Helen Tremlett (University of British Columbia), Douglas Fadrosh (University of California, San Francisco), Ali Faruqi (University of California, San Francisco), Feng Zhu (University of British Columbia)
Contributorship statement: The corresponding author (HT) takes responsibility for the integrity of the data and the accuracy of the data analysis and had full access to the data. DF performed the gut microbiota analyses. HT, DF, AF, FZ performed the statistical analyses. EW, JH and JG facilitated sample and data collection.SR facilitated electronic linkage and quality assurance of data from environmental risk factors parent study. All authors were involved in the current study design, contributed to interpretation of data. SL and EW designed the original study and obtained funding. DF and AF drafted the microbiota methods. HT drafted the remaining manuscript. All authors revised the manuscript and approved of the final version to be published.
Competing Interests Statements:
Helen Tremlett is funded by the Canada Research Chair program. She has received research support from the National Multiple Sclerosis Society, the Canadian Institutes of Health Research, and the UK MS Trust; speaker honoraria and/or travel expenses to attend conferences from the Consortium of MS Centres (2013), the National MS Society (2012, 2014), Bayer Pharmaceuticals (2010), Teva Pharmaceuticals (2011), ECTRIMS (2011, 2012, 2013, 2014), UK MS Trust (2011), the Chesapeake Health Education Program, US Veterans Affairs (2012), Novartis Canada (2012), Biogen Idec (2014), American Academy of Neurology (2013, 2014, 2015). Unless otherwise stated, all speaker honoraria were either donated to an MS charity or to an unrestricted grant for use by her research group.
Douglas Fadrosh has no disclosures
Ali Faruqi has no disclosures
Feng Zhu has no disclosures
Janace Hart has no disclosures
Shelly Roalstad has no disclosures
Jennifer Graves is funded by the Race to Erase MS and the National MS Society
Susan Lynch is funded by the NIH, Sloan Foundation, Cystic Fibrosis Foundation, Broad Foundation, Jannsen Pharmaceuticals, Gilead and Pfizer. She has recently or currently acts as an ad hoc consultant for Janssen Pharmaceuticals, Regeneron, Boston Consulting Group, Theravance and Novartis. She volunteers as a members of the Scientific Advisory Board of Second Genome and has received honoraria for lectures from American Thoracic Society, American Academy of Allergy Asthma and Immunology, Georgia Regents University, Alta Bates and Kaiser Permanente. She holds four patents and has received royalties for IP licensed by KaloBIos Inc.
Emmanuelle Waubant is funded by the NIH, the NMSS, and the Race to Erase MS. She has received honorarium for one educational lecture from Genentech. She volunteers on an advisory board for a Novartis trial. She has received honorarium or travel support from ACTRIMS, ECTRIMS, and AAN. She has been an ad hoc consultant for Actelion, Roche and Novartis
The US Network of Pediatric MS Centers (principal and co-principal investigators listed in alphabetical order): Greg Aaen1, Anita Belman2, Leslie Benson3, T. Charles Casper4, Tanuja Chitnis3, Mark Gorman3, Yolanda Harris8, Lauren Krupp2, Tim E Lotze6, Jayne Ness8, Cody Olsen4, Moses Rodriguez5, John Rose4, Timothy C Simmons4, Jan-Mendelt Tillema5, Bianca Weinstock-Guttman9
1. Loma Linda University, Loma Linda, CA, United States
2. Stony Brook University, Stony Brook, NY, United States
3. Harvard University, Cambridge, MA, United States
4. University of Utah, Salt Lake City, UT, United States
5. Mayo Clinic, Rochester, MN, United States
6. Baylor College of Medicine, Houston, TX, United States
7. University of California, San Francisco, San Francisco, CA, United States
8. University of Alabama, Birmingham, AL, United States
9. State University of New York at Buffalo, Buffalo, NY, United States
eFigure 1: Functionality of the gut microbiota community: examples of pathways differing between cases and controls, and by immunomodulatory drug [IMD] exposure status
eTable 1: Summary of the demographic, lifestyle and clinical characteristics collected for cases and controls and derived variables
eTable 2: Additional characteristics of the pediatric multiple sclerosis (MS) cases and controls (all controls, n=17; controls with sequencing (n=16) and/or chip data (n=14))
eTable 3: Additional clinical characteristics of the pediatric MS cases: onset symptoms and attack severity
eTable 4A: The alpha diversity metrics for cases and controls, and by immunomodulatory drug (IMD) exposure
eTable 4B: Beta diversity: the Canberra metric for cases and controls and by immunomodulatory drug (IMD) exposure
eTable 5A, B: All taxa-level differences in the gut microbial communities: cases compared to controls for individual OTUs (p and q values <0.05).
Uploaded as separate files
eTable 6: Summary of the taxa depicted in Figure 3A, B in relation to the secondary platform (DNA microarray) and the wider gut microbiota literature
eTable 7A-D: All taxa-level differences in the gut microbial communities: cases compared to controls, by immunomodulatory drug (IMD) exposure for individual OTUs (p and q values <0.05).
Uploaded as separate files
eTable 8: Enriched and depleted metabolic pathways: predicted function of the gut microbiota community for cases and controls, and by immunomodulatory drug (IMD)
Uploaded as separate files
Appendix #1: Secondary platform – DNA microarray
References
- 1.Wang Y, Kasper LH. The role of microbiome in central nervous system disorders. Brain Behav Immun. 2014;38:1–12. doi: 10.1016/j.bbi.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berer K, Krishnamoorthy G. Microbial view of central nervous system autoimmunity. FEBS Lett. 2014;588(22):4207–13. doi: 10.1016/j.febslet.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 3.Savage DC. Microbial Ecology of the Gastrointestinal Tract. Annu Rev Microbiol. 1977;31:107–33. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- 4.Knight R. Follow your gut: the enormous impact of tiny microbes. Simon & Schuster; New York, NY: 2015. [Google Scholar]
- 5.Siezen RJ, Kleerebezem M. The human gut microbiome: are we our enterotypes? Micro biotech. 2011;4(5):550–3. doi: 10.1111/j.1751-7915.2011.00290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cantarel BL, Waubant E, Chehoud C, Kuczynski J, DeSantis TZ, Warrington J, et al. Gut microbiota in multiple sclerosis: possible influence of immunomodulators. J Invest Med. 2015;63(5):729–34. doi: 10.1097/JIM.0000000000000192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miyake S, Kim S, Suda W, Oshima K, Nakamura M, Matsuoka T, et al. Dysbiosis in the Gut Microbiota of Patients with Multiple Sclerosis, with a Striking Depletion of Species Belonging to Clostridia XIVa and IV Clusters. PLoS One. 2015;10(9):e0137429. doi: 10.1371/journal.pone.0137429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hunsberger M, O'Malley J, Block T, Norris JC. Relative validation of Block Kids Food Screener for dietary assessment in children and adolescents. Maternal & Child nutrition. 2015;11(2):260–70. doi: 10.1111/j.1740-8709.2012.00446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, et al. Ultra- high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME. 2012;6(8):1621–4. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nature Methods. 2010;7(5):335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faruqi AA. Microbiome Analysis using Workflow of QIIME. GitHub; 2015. [June/2015]. [cited 2015]. Available from: https://github.com/alifar76/MAWQ. [Google Scholar]
- 12.Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nature Bio. 2013;31(9):814–21. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foxman B, Martin ET. Use of the Microbiome in the Practice of Epidemiology: A Primer on -Omic Technologies. Am J Epidemiol. 2015;182(1):1–8. doi: 10.1093/aje/kwv102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.R Core Team . R: A language and environment for statistical computing. 3.0.2. ed. R Foundation for Statistical Computing; Vienna, Austria: 2013. [Google Scholar]
- 15.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: Human gut microbes associated with obesity. Nature. 2006;444:1022–3. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 16.Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, et al. Human genetics shape the gut microbiome. Cell. 2014;159(4):789–99. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Segain J, de la Bletiere DR, Bourreille A, Leray V, Gervois N, Rosales C, et al. Butyrate inhibits inflammatory responses through NFκB inhibition: implications for Crohn's disease. Gut. 2000;47(3):397–403. doi: 10.1136/gut.47.3.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang W, Chen L, Zhou R, Wang X, Song L, Huang S, et al. Increased Proportions of Bifidobacterium and the Lactobacillus Group and Loss of Butyrate-Producing Bacteria in Inflammatory Bowel Disease. J Clin Microbiol. 2014;52(2):398–406. doi: 10.1128/JCM.01500-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.International Multiple Sclerosis Genetics Consortium. Beecham AH, Patsopoulos NA, Xifara DK, Davis MF, Kemppinen A, et al. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat Genet. 2013;45(11):1353–60. doi: 10.1038/ng.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gevers D, Kugathasan S, Denson LA, Vazquez-Baeza Y, Van Treuren W, Ren B, et al. The treatment-naive microbiome in new-onset Crohn's disease. Cell host & microbe. 2014;15(3):382–92. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105(43):16731–6. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baranzini SE, Katz-Sand I, Mazmanian SK, Becosme Y, London J, Farber R, et al. The MS Microbiome Consortium (MSMC): an academic multi-disciplinary collaborative effort to elucidate the role of the gut microbiota in MS Mult Scler. 2014;20(S1):285–496. [Google Scholar]
- 23.Jhangi S, Gandhi R, Glanz B, Cook S, Nejad P, Ward D, et al. Increased Archaea Species and Changes with Therapy in Gut Microbiome of Multiple Sclerosis Subjects (S24.001). Neurol. 2014;82(10):S24.001.AAN. [Google Scholar]
- 24.Nagalingam NA, Lynch SV. Role of the microbiota in inflammatory bowel diseases. Inflamm Bowel Dis. 2012;18(5):968–84. doi: 10.1002/ibd.21866. [DOI] [PubMed] [Google Scholar]
- 25.Touchefeu Y, Montassier E, Nieman K, Gastinne T, Potel G, Bruley des Varannes S, et al. Systematic review: the role of the gut microbiota in chemotherapy- or radiation- induced gastrointestinal mucositis - current evidence and potential clinical applications. Aliment Pharmacol Ther. 2014;40(5):409–21. doi: 10.1111/apt.12878. [DOI] [PubMed] [Google Scholar]
- 26.Vandeputte D, Falony G, Vieira-Silva S, Tito RY, Joossens M, Raes J. Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates. Gut. 2015 doi: 10.1136/gutjnl-2015-309618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Surana NK, Kasper DL. The yin yang of bacterial polysaccharides: lessons learned from B. fragilis PSA. Immunol Rev. 2012;245(1):13–26. doi: 10.1111/j.1600-065X.2011.01075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vaahtovuo J, Munukka E, Korkeamäki M, Luukkainen R, Toivanen P. Fecal microbiota in early rheumatoid arthritis. J Rheumatol. 2008;35(8):1500–5. [PubMed] [Google Scholar]
- 29.Mielcarz DW, Kasper LH. The gut microbiome in multiple sclerosis. Cur Treat Options Neurol. 2015;17(4):344. doi: 10.1007/s11940-015-0344-7. [DOI] [PubMed] [Google Scholar]
- 30.Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155(7):1451–63. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trown PW, Kramer MJ, Dennin RA, Jr., Connell EV, Palleroni AV, Quesada J, et al. Antibodies to human leucocyte interferons in cancer patients. Lancet. 1983;1(8316):81–4. doi: 10.1016/s0140-6736(83)91737-3. [DOI] [PubMed] [Google Scholar]
- 32.Glutathione metabolism - Reference pathway. KEGG; 2014. [December 15, 2014]. Available from: http://www.genome.jp/kegg/pathway/map/map00480.html. [Google Scholar]
- 33.Lipopolysaccharide biosynthesis - Reference pathway. KEGG; 2015. [December 15, 2014]. Available from: http://www.genome.jp/kegg bin/show_pathway?map=map00540&show_description=show. [Google Scholar]
- 34.Carvalho AN, Lim JL, Nijland PG, Witte ME, Van Horssen J. Glutathione in multiple sclerosis: more than just an antioxidant? Mult Scler. 2014;20(11):1425–31. doi: 10.1177/1352458514533400. [DOI] [PubMed] [Google Scholar]
- 35.Julio-Pieper M, Flor PJ, Dinan TG, Cryan JF. Exciting times beyond the brain: metabotropic glutamate receptors in peripheral and non-neural tissues. Pharmacolog Rev. 2011;63(1):35–58. doi: 10.1124/pr.110.004036. [DOI] [PubMed] [Google Scholar]
- 36.Nogai A, Siffrin V, Bonhagen K, Pfueller CF, Hohnstein T, Volkmer-Engert R, et al. Lipopolysaccharide Injection Induces Relapses of Experimental Autoimmune Encephalomyelitis in Nontransgenic Mice via Bystander Activation of Autoreactive CD4+ Cells. J Immunol. 2005;175(2):959–66. doi: 10.4049/jimmunol.175.2.959. [DOI] [PubMed] [Google Scholar]
- 37.Benito-Leon J, Pisa D, Alonso R, Calleja P, Diaz-Sanchez M, Carrasco L. Association between multiple sclerosis and Candida species: evidence from a case-control study. Eur J Clin Microbiol Infect Dis. 2010;29(9):1139–45. doi: 10.1007/s10096-010-0979-y. [DOI] [PubMed] [Google Scholar]
- 38.Hube B. From commensal to pathogen: stage- and tissue-specific gene expression of Candida albicans. Curr Opin Microbiol. 2004;7(4):336–41. doi: 10.1016/j.mib.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 39.Panzer AR, Lynch SV. Influence and effect of the human microbiome in allergy and asthma. Current opinion in rheumatology. 2015;27(4):373–80. doi: 10.1097/BOR.0000000000000191. [DOI] [PubMed] [Google Scholar]
- 40.Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife. 2013;2:e01202. doi: 10.7554/eLife.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.