Abstract
Mutations of pyrin and mevalonate kinase (MVK) cause distinct interleukin-1β (IL-1β)-mediated autoinflammatory diseases, familial Mediterranean fever (FMF) and hyperimmunoglobulinemia D syndrome (HIDS). Pyrin forms an inflammasome when mutated or in response to bacterial modification of the GTPase RhoA. Here we show that RhoA activates the serine-threonine kinases PKN1 and PKN2 that bind and phosphorylate pyrin. Phosphorylated pyrin binds 14-3-3 proteins, which block the pyrin inflammasome. The binding of 14-3-3 and PKN proteins to FMF-associated mutant pyrin is substantially decreased, and the constitutive IL-1β release from FMF or HIDS patients’ peripheral blood mononuclear cells is attenuated by activating PKN1 and PKN2. Defects in prenylation, seen in HIDS, lead to RhoA inactivation and consequent pyrin inflammasome activation. These data indicate a previously unsuspected fundamental molecular connection between two seemingly distinct autoinflammatory disorders.
Mutations in the genes encoding proteins constituting inflammasomes or regulating inflammasome activation cause interleukin-1β (IL-1β)-mediated autoinflammatory diseases1,2. Familial Mediterranean fever (FMF) and the hyperimmunoglobulinemia D syndrome (HIDS) are two such disorders caused by missense mutations of MEFV and MVK, encoding pyrin and mevalonate kinase (MVK), respectively3,4. Pyrin spontaneously forms an inflammasome dependent on the adaptor protein ASC when mutated5 or in response to bacterial toxins6, and MVK is a key enzyme in the mevalonate pathway, producing isoprenoids7 such as geranylgeranyl pyrophosphate. However, the exact molecular mechanism of pyrin inflammasome activation, as well as the molecular pathology of FMF and HIDS, is unknown.
Previous genetic studies of FMF in Sephardi Jewish families with severe disease indicated a recessive mode of inheritance3,8,9, suggesting that FMF might be caused by loss-of-function mutations in pyrin. However, the availability of genetic testing has led both to the definition of a biochemical phenotype in asymptomatic heterozygotes10 and to the recognition that as many as 30% of patients with clinical FMF have only a single demonstrable mutation in MEFV11–13. Indeed, several cases of apparently dominantly inherited FMF have been reported14,15, while null mutations are extremely rare. Moreover, pyrin knockout mice develop normally and exhibit no overt phenotype5, arguing against a loss-of-function model for FMF. Data from knock-in mice harboring FMF-associated human pyrin mutations strongly support a gain-of-function model with a gene-dosage effect, with the mutant pyrin inducing an ASC-dependent inflammasome and IL-1β-mediated systemic inflammation5.
Certain bacterial toxins that inactivate Rho GTPases induce IL-1β production by the pyrin inflammasome through an indirect mechanism that does not involve an interaction between pyrin and Rho GTPases6. Here we elucidate a pathway by which RhoA activation induces downstream kinases, pyrin phosphorylation, and the resulting inhibitory binding of phosphorylated pyrin by 14-3-3 proteins, and we demonstrate the effects of pyrin and MVK mutations in relieving the tonic inhibition of the pyrin inflammasome. Bacterial sensing by pyrin thus represents the first documented example in which innate immunity in mammals is triggered by an indirect process similar to the ‘guard’ mechanism in plants16,17.
RESULTS
RhoA activity suppresses pyrin inflammasome activation
Bacterial toxins that modify RhoA induce inflammasome formation by wild-type pyrin6. These bacterial toxins mediate the glucosylation, adenylylation, ADP-ribosylation, or deamidation of various residues in the switch-I region of RhoA, inhibiting guanine nucleotide binding and GTPase activity. Taken together with the absence of evidence for direct interaction between pyrin and modified RhoA6, the data suggest that the pyrin inflammasome is activated by the inhibition of downstream RhoA signaling pathways. Hence, we initially explored the correlation between RhoA activity and the pyrin inflammasome by studying bone marrow derived macrophages (BMDMs) in which pyrin had been induced by lipopolysaccharide (LPS)18.
Consistent with the previous report6, the Clostridial TcdB and C3 toxins induced caspase-1 activation and bioactive IL-1β release from LPS-primed BMDMs by a pathway that is pyrin-, ASC-, and caspase-1-dependent, but NLRP3-, NLRC4-, and AIM2-independent (Fig. 1a and Supplementary Fig. 1a). In contrast, C3 toxin-induced IL-1β release was substantially diminished when the BMDMs were treated with calpeptin, an indirect upstream RhoA activator19 or bacterial cytotoxic necrotizing factor (CNF) toxin, a direct RhoA activator that deamidates glutamine-63 of Rho20,21 (Supplementary Fig. 1b–d and Fig. 1b). RhoA activators also reduced IL-1β release from the LPS-primed BMDMs of FMF knock-in mice harboring the V726A mutation (MefvV726A/V726A), in which the pyrin inflammasome is constitutively activated5 (Fig. 1c and Supplementary Fig. 1e). On the other hand, the activation of RhoA had no suppressive effect on the NLRC4 or AIM2 inflammasomes (Fig. 1d and Supplementary Fig. 1f). The NLRP3 inflammasome was also not inhibited by the CNF toxin despite its inhibition by calpeptin (Fig. 1d and Supplementary Fig. 1f).
Figure 1. The pyrin inflammasome is activated by inactivation of RhoA.
(a) BMDMs from wild-type (WT) or Mefv−/− mice were primed with LPS (1 μg/ml) and C3 toxin (0.5 μg/ml) or TcdB (0.5 μg/ml) for 6h. Cell culture supernatants (Sup) and cell lysates (Lys) were analyzed by immunoblotting as indicated. (b,c) BMDMs from (b) WT mice were treated with LPS, C3 toxin, and CNF toxin, or (c) MefvV726A/V726A mice were co-treated with LPS and CNF toxin for 6h and then analyzed for IL-1β release (top) and for activated RhoA levels (bottom), % change of RhoA activity to (b) LPS or (c) UT. ** P ≤ 0.005 (unpaired two-tailed t-test). Data represent the mean ± s.e.m of n = 6 mice. (d) BMDMs from WT mice were co-treated with LPS, the indicated concentration of CNF toxin for 5h, and then ATP (2 mM) for 0.5h, flagellin (0.5 μg/ml with 25 μl/ml DOTAP) for 1h, or dsDNA (1 μg/ml with 2.5 μl/ml Lipofectamine 2000) for 1h and then analyzed for IL-1β release. (e) LPS-primed BMDMs from MefvV726A/V726A mice were treated with the indicated dose of NKH477 for 1h and then analyzed for IL-1β release (top) and for activated RhoA levels (bottom), % change of RhoA activity to LPS. ** P ≤ 0.005 (unpaired two-tailed t-test). Data represent the mean ± s.e.m of n = 8 mice. All immunoblot data shown are representative of at least three independent experiments.
Another line of evidence supporting the inverse relationship between RhoA activation and pyrin inflammasome induction follows from a previous study of the inhibition of the NLRP3 inflammasome by intracellular cAMP, in which cAMP accentuated IL-1β production by peripheral blood mononuclear cells (PBMCs) from FMF patients22. cAMP is known to induce phosphorylation of RhoA at Ser-188 through protein kinase A (PKA), resulting in the translocation of membrane-associated RhoA towards the cytosol23,24. The increase of cAMP synthesis by the adenylate cyclase (ADCY) activator NKH477 (a water-soluble analog of forskolin) potentiated IL-1β release from FMF patients’ PBMCs (Supplementary Fig. 2a), while it moderately inhibited IL-1β release from cryopyrin-associated periodic syndrome (CAPS) patients’ PBMCs or ATP-treated wild-type mouse BMDMs (Supplementary Fig. 2b,c). Indeed, we observed a dose-dependent decrease of RhoA-GTP activity and an increase of IL-1β release from the LPS-primed BMDMs of MefvV726A/V726A mice in response to ADCY activation (Fig. 1e). In sum, the data from both human and murine leukocytes subjected to bacterial toxins and pharmacologic agents support an inverse relationship between RhoA activity and pyrin inflammasome activation.
Colchicine suppresses the pyrin inflammasome
Next, we examined the effect of colchicine on the pyrin inflammasome, since colchicine is a known activator for RhoA and an effective prophylaxis for FMF inflammatory attacks. Within the cytosol, colchicine binds to tubulin and depolymerizes microtubules, resulting in the release of the RhoA activator, guanine nucleotide exchange factor (GEF)-H1, that is inactive when bound to microtubules25. Indeed colchicine activated RhoA or reversed the inhibition of RhoA activity by C3 toxin in LPS-primed BMDMs (Fig. 2a). Consistent with the previous results showing an inverse relationship between RhoA activity and pyrin inflammasome activation, we observed that colchicine inhibited the constitutive IL-1β release from BMDMs of MefvV726A/V726A mice as well as C3 toxin-induced IL-1β release from LPS-primed BMDMs (Fig. 2b,c). In addition, inhibition of IL-1β release from PBMCs of FMF patients was also observed (Fig. 2d). In contrast, colchicine had no consistent dose-dependent inhibitory effect on NLRP3, NLRC4, or AIM2 inflammasome activation (Fig. 2e) or the constitutive IL-1β release from the PBMCs of patients with CAPS (Fig. 2f), although it has been reported that colchicine inhibited NLRP3 inflammasome-induced IL-1β release at higher concentrations26. These data suggest an explanation for the exquisite specificity of colchicine for FMF among the monogenic autoinflammatory diseases.
Figure 2. Colchicine suppresses the pyrin inflammasome.
(a) BMDMs from MefvV726A/V726A or WT mice were treated with the indicated concentration of colchicine without or with C3 toxin, and activated RhoA (GTP-RhoA) was assessed by using a pull-down assay with Rhotekin-RBD beads. (b) LPS-primed BMDMs from MefvV726A/V726A mice were treated with the indicated concentration of colchicine. (c) BMDMs from WT mice were co-treated with LPS, C3 toxin, and indicated concentration of colchicine. (d,f) PBMCs from (d) three FMF (designated patient number 3, 4, and 5) or (f) two CAPS (designated patient number 3 and 4) patients with the indicated mutations in MEFV or NLRP3 were co-treated with LPS and the indicated dose of colchicine for 6h. (e) BMDMs from WT mice were primed with LPS for 3h and then treated with the indicated concentration of colchicine and ATP (2 mM) for 0.5h, flagellin (0.5 μg/ml with 25 μl/ml DOTAP) for 1h, or dsDNA (1 μg/ml with 2.5 μl/ml of Lipofectamine 2000) for 1h. Cell culture supernatants and cell lysates were analyzed by immunoblotting as indicated. Immunoblot data shown are representative of one experiment (d, f) or at least three independent experiments (a–c, e).
RhoA effector kinases suppress pyrin inflammasome activation
Since the pyrin inflammasome is activated by inactivation of RhoA, we hypothesized that a signaling pathway downstream of RhoA may suppress the pyrin inflammasome. A number of proteins have been identified as effectors of RhoA, most notably kinases such as ROCKs and PKNs that belong to the protein kinase C (PKC) superfamily. It is noteworthy that staurosporine, a potent inhibitor of PKC as well as an inducer of cell death, stimulates IL-1β release from LPS-primed macrophages through as yet unknown inflammasome activation27. Here we show that staurosporine-induced IL-1β release is independent of the NLRP3, NLRC4, or AIM2 inflammasomes, but dependent on the pyrin inflammasome (Fig. 3a and Supplementary Fig. 3a), suggesting that the pyrin inflammasome might be activated when a Rho effector kinase is inhibited.
Figure 3. Inhibition of RhoA effector kinases activates the pyrin inflammasome.
(a) LPS-primed BMDMs from WT, Casp1, Asc, Nlrp3, Nlrc4, Aim2, Mefv-deficient mice, or C-terminal pyrin-truncation mice (MefvΔCt/ΔCt) were treated with staurosporine (1 μM) for 1h. (b) BMDMs were transiently transfected with negative control (Ctrl) that has no significant sequence similarity to mouse or human gene sequences, Pkn1, Pkn2, Pkn1 + Pkn2, Rock1, Rock2, Rock1 + Rock2, or Pkn1 + Pkn2 + Rock1 + Rock2 siRNAs and then treated with LPS for 8h. (c) BMDMs from WT or Mefv−/− mice were transiently transfected with Ctrl or Pkn1 + Pkn2 siRNAs and then treated with LPS for 8h. Cell culture supernatants and cell lysates were analyzed by immunoblotting as indicated. All immunoblot data shown are representative of at least three independent experiments.
To find RhoA effector kinases that regulate the pyrin inflammasome, the Rho effector kinase genes encoding PKNs and ROCKs were knocked down in mouse BMDMs by short interfering RNA (siRNA). Knockdown of the Pkn1 or Pkn2 gene in LPS-primed BMDMs induced spontaneous IL-1β release, which was dependent on the pyrin inflammasome, and this effect was accentuated when both Pkn1 and Pkn2 genes were knocked down (Fig. 3b,c). However, the knockdown of Rock1, Rock2, or even double knockdown of Rock1 and Rock2 genes did not induce IL-1β release (Fig. 3b), indicating that Rock1 and Rock2 are dispensable for regulating the pyrin inflammasome.
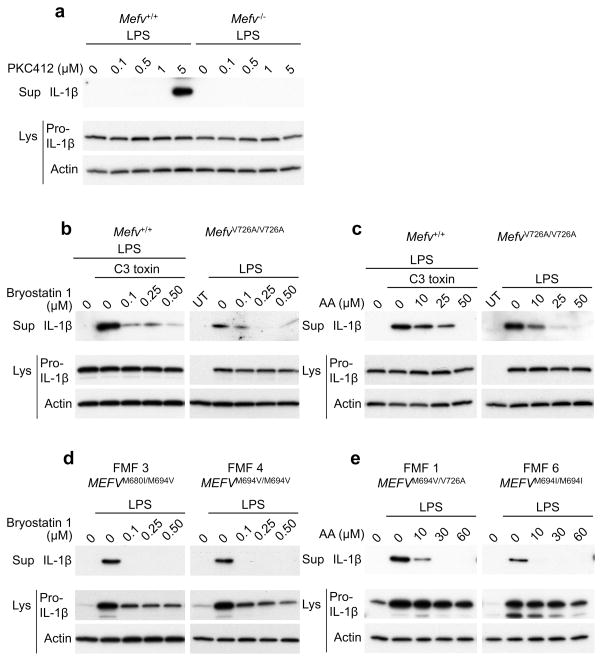
Furthermore, we also observed that the pyrin inflammasome was activated in LPS-primed BMDMs by PKC412, a potent inhibitor of PKNs28 (Fig. 4a). Conversely, IL-1β release from BMDMs treated with C3-toxin or from BMDMs of MefvV726A/V726A mice was markedly inhibited by bryostatin 1, a potent PKC activator (Fig. 4b and Supplementary Fig. 3b). In contrast, NLRP3 inflammasome activation induced by ATP, AIM2 inflammasome activation induced by double-stranded DNA, and NLRC4 inflammasome activation induced by flagellin were not suppressed by bryostatin 1 (Supplementary Fig. 3c). The activation of the pyrin inflammasome was also suppressed by arachidonic acid, a known activator of both PKNs and ROCKs29,30 (Fig. 4c and Supplementary Fig. 3d). The effects of arachidonic acid are not as inflammasome-selective as bryostatin 1, since arachidonic acid suppressed ATP-driven NLRP3 inflammasome activation through its effects on cAMP22,31,32, although it did not suppress the IL-1β release induced by NLRC4 or AIM2 inflammasome activators (Supplementary Fig. 3e). Consistent with the results of the aforementioned siRNA experiments, the constitutive IL-1β release from PBMCs of patients with FMF was substantially decreased by both PKN activators, bryostatin 1 and arachidonic acid (Fig. 4d,e). Taken together, genetic and pharmacologic data from murine and human experimental systems support a role for PKN enzymes in suppressing the pyrin inflammasome.
Figure 4. The RhoA effector kinases, PKNs, suppress pyrin inflammasome activation.
(a) LPS-primed BMDMs from WT or Mefv−/− mice were treated with the indicated dose of PKC412 for 1h. (b,c) BMDMs from WT and FMF-knock-in (FMF-KI) mice were co-treated with LPS and the indicated dose of (b) bryostatin 1 or (c) arachidonic acid (AA) with or without C3 toxin for 6h. (d,e) PBMCs from FMF patients (designated patient number 1, 3, 4, and 6) with the indicated mutations in MEFV were co-treated with LPS and the indicated dose of (d) bryostatin 1 or (e) arachidonic acid for 6h. Cell culture supernatants and cell lysates were analyzed by immunoblotting as indicated. All immunoblot data shown are representative of at least three independent experiments.
PKNs bind to human pyrin and phosphorylate S208 and S242
To understand how PKNs suppress pyrin inflammasome activation, we tested whether pyrin is a substrate of PKNs. In a coimmunoprecipitation assay, we found interaction of endogenous pyrin with PKN1 in lysates of LPS-primed BMDMs (Fig. 5a). The pyrin-PKN1 interaction was decreased by C3 toxin, but the decreased interaction was restored by cotreatment with arachidonic acid or bryostatin 1 (Fig. 5a), indicating that activated PKN1 binds to pyrin and dissociates when PKN1 is inactivated. PKN1 is a serine-threonine protein kinase that has a catalytic kinase domain at its C-terminus and an N-terminal regulatory region where activated RhoA and arachidonic acid bind. Human PKN1 interacts with pyrin through this C-terminal kinase domain (Fig. 5b), suggesting possible phosphorylation of pyrin. Indeed, direct phosphorylation of pyrin by PKN1 and PKN2 was observed from an incubation of the purified N-terminal half of human pyrin with recombinant PKN1 or PKN2 and subsequent phosphoprotein staining or immunoblotting with a phospho-Ser antibody (Fig. 5c).
Figure 5. PKNs bind and phosphorylate pyrin.
(a) Lysates from LPS-primed BMDMs of WT and Mefv−/− mice or lysates from LPS-primed WT BMDMs co-treated with C3 toxin, C3 toxin and arachidonic acid, or C3 toxin and bryostatin 1 were immunoprecipitated with anti-pyrin antibody, and immune complexes were analyzed by immunoblot for PKN1 and pyrin. (b) The lysates of 239T cells transiently expressing human pyrin and full-length or various deleted forms of PKN1 (top) were immunoprecipitated with anti-myc (pyrin) and analyzed by immunoblot using anti-V5 (PKN1) antibody (bottom). (c) Myc&his-tagged N-terminal human pyrin (aa’s 1-330) was expressed and purified by Ni-NTA beads. Purified N-terminal pyrin was incubated with or without recombinant PKN1 or PKN2 and analyzed for phosphorylation by staining with Pro-Q Diamond reagent or immunoblotting with an antibody specific for phospho-Ser followed by Coomassie blue staining and immunoblotting with anti-myc antibody for loading control. All immunoblot and gel staining data shown are representative of at least three independent experiments.
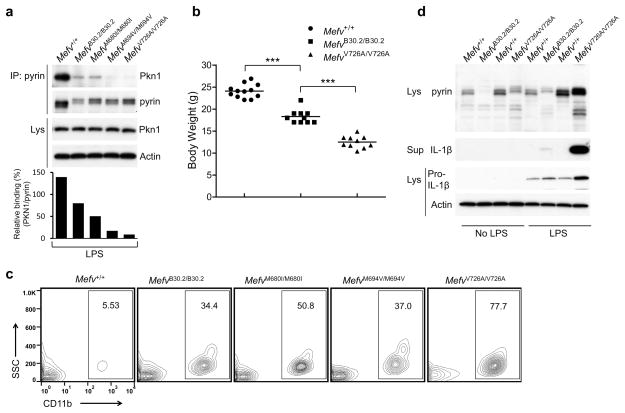
We also examined the binding of PKN1 to pyrin in FMF-knock-in mice. Most FMF-associated mutations are in the C-terminal B30.2 domain of human pyrin, a domain that is not present in mouse pyrin. We found that the binding of PKN1 to the pyrin of FMF-knock-in mice with three frequent FMF-associated B30.2 mutations (MefvM680I/M680I, MefvM694V/M694V, and MefvV726A/V726A) was substantially decreased relative the binding of PKN1 to wild-type (Mefv+/+) mouse pyrin, which lacks a B30.2 orthologous domain (Fig. 6a). The binding of PKN1 to the pyrin of wild-type B30.2 domain knock-in mice (MefvB30.2/B30.2) was also decreased relative its binding to wild-type mouse pyrin, but was not decreased by as much as it was with the pyrin proteins from MefvM694V/M694V or MefvV726A/V726A mice. MefvB30.2/B30.2 mice had been thought to be embryonic lethal in a previous study5, but we have now successfully bred these animals. These mice show spontaneous inflammatory phenotypes but the severity as well as IL-1β release from LPS-primed leukocytes is much diminished compared with MefvV726A/V726A mice that show the most severe inflammation5 (Fig. 6b–d and Supplementary Fig. 4a,b). These findings suggest that the human B30.2 domain has a role in the regulation of PKN1 binding to pyrin.
Figure 6. The binding of PKN1 to the pyrin of FMF-KI mice is substantially decreased relative to wild-type mouse pyrin, which lacks a B30.2 orthologous domain, and KI mice with the WT B30.2 domain of human pyrin have a milder inflammatory phenotype than KI mice with FMF-associated mutations.
(a) BMDMs from WT and FMF-KI mice with WT or the indicated FMF-associated mutant human B30.2 domain were treated with LPS. Lysates were immunoprecipitated with anti-pyrin antibody, and immune complexes were analyzed by immunoblot for PKN1 and pyrin. Densitometry analysis of PKN1 bands, normalized to the pyrin bands in immune complexes, is shown in the histogram below. (b) Body weights of 8-week-old males of Mefv+/+, MefvB30.2/B30.2, and MefvV726A/V726A mice. Each graph point represents one mouse, and means are shown as horizontal bars. *** P ≤ 0.0005 (unpaired two-tailed t-test). (c) Peripheral blood cells from 8-week-old Mefv+/+, MefvB30.2/B30.2, MefvM680I/M680I, MefvM694V/M694V, and MefvV726A/V726A male mice were analyzed for CD11b+ myeloid cells. Numbers indicate percentage of total cells in gates. Data are representative of n = 5 mice (WT and MefvM680I/M680I), n = 7 mice (MefvV726A/V726A), or n = 8 mice (MefvB30.2/B30.2 and MefvM694V/M694V). (d) Bone marrow CD11b+ cells from 8-week-old males of Mefv+/+, MefvB30.2/B30.2, and MefvV726A/V726A mice were treated with/without LPS for 6h. Cell culture supernatants and cell lysates were analyzed by immunoblotting as indicated. All immunoblot data shown are representative of at least three independent experiments.
14-3-3 binds phosphopyrin to inhibit inflammasome activation
Serine residues at 208, 209, and 242 of human pyrin have been reported to be phosphorylated and to mediate the interaction with two isoforms of 14-3-3 proteins33. Here we show that S208 and S242 of human pyrin are phosphorylated by PKN1 or PKN2 (Fig. 7a), and that 14-3-3ε or 14-3-3τ binds to the N-terminal portion of pyrin through serine residues at 208 and 242 but not 209 (Supplementary Fig. 5a,b). Consistent with the C3 toxin-induced attenuation of the PKN1-pyrin interaction (Fig. 5a), the 14-3-3ε-pyrin interaction was substantially decreased by C3 toxin, an effect that was reversed by PKN activators (Fig. 7b).
Figure 7. The pyrin inflammasome is inhibited by phosphorylation and subsequent 14-3-3 binding.
(a) Purified myc- and his-tagged WT N-terminal pyrin (aa’s 1-330) or N-terminal pyrin with S208A and S242A mutations was incubated with recombinant PKN1 or PKN2 and analyzed for phosphorylation by immunoblotting with an antibody specific for phospho-Ser followed by immunoblotting with anti-myc antibody. (b) WT BMDMs were treated with LPS, C3 toxin, and arachidonic acid or bryostatin 1. (c) BMDMs from WT and FMF-KI mice with WT or the indicated mutant human B30.2 domain were treated with LPS. (b, c) Lysates were immunoprecipitated with anti-mouse pyrin antibody, and immune complexes were analyzed by immunoblot for 14-3-3ε. (d) WT or the indicated FMF-associated human mutant pyrin proteins were transiently expressed in 239T cells. (e) Macrophages differentiated from PBMCs of 4 healthy controls and 5 FMF patients were treated with IFN-γ for 16h to induce pyrin expression, and primed with LPS for 6h. (d, e) The lysates were immunoprecipitated with anti-human pyrin antibody, and immune complexes were analyzed by immunoblot for 14-3-3ε. (f) BMDMs from WT and Mefv−/− mice were transfected with Ctrl or 14-3-3ε and 14-3-3τ siRNAs and treated with LPS for 8h. Cell culture supernatants and cell lysates were analyzed by immunoblotting as indicated. (g) U937 cells transiently expressing WT or the indicated mutant pyrin proteins were primed with LPS for 6h. Cell culture supernatants and cell lysates were analyzed by immunoblotting as indicated. All immunoblot data shown are representative at least three independent experiments.
The binding of 14-3-3ε to mouse pyrin from the FMF-knock-in mice was diminished significantly relative to pyrin from wild-type mice (Fig. 7c and Supplementary Fig. 5c), which is also consistent with the PKN1-pyrin interaction (Fig. 6a). In the lysates of BMDMs from MefvV726A/V726A mice, the 14-3-3ε–pyrin interaction was increased by PKN activation with arachidonic acid or bryostatin 1 or by RhoA activation by colchicine (Supplementary Fig. 5d,e). Furthermore, the binding of 14-3-3ε to disease-associated mutant pyrin (M680I, M694V and V726A) was substantially decreased relative to wild-type human pyrin in ectopically expressing 293T cells and macrophages differentiated from the PBMCs of FMF patients (Fig. 7d,e).
Taken together with the inhibition of IL-1β release from BMDMs of MefvV726A/V726A mice (Fig. 4b,c) or PBMCs of FMF patients (Fig. 4d,e) by PKN activation, these findings suggest that the spontaneous activation of the inflammasome in myeloid cells of FMF-knock-in mice or FMF patients might be due to reduced suppression of pyrin activation by 14-3-3 proteins. Consistent with this hypothesis, spontaneous pyrin-dependent IL-1β release was observed in BMDMs upon knockdown of both 14-3-3ε and 14-3-3τ (Fig. 7f). Moreover, ectopic expression of the S208A or S242A pyrin mutants, which do not interact with 14-3-3, induced IL-1β release in U937 cells without upstream pyrin inflammasome activators, while only basal levels of IL-1β were secreted by the non-transfected cells or those expressing wild-type pyrin (Fig. 7g). The binding of 14-3-3 to phosphorylated pyrin thus acts as a molecular switch to turn off pyrin inflammasome activation that would otherwise occur when either 14-3-3 expression or pyrin serine phosphorylation is blocked by bacterial toxins, FMF-associated B30.2 mutations, or pharmacologic agents (Supplementary Figs. 6, 7).
The pyrin inflammasome mediates IL-1β release in HIDS
In addition to the GEF-mediated exchange of GDP with GTP on RhoA, the translocation of RhoA from the cytosol to the plasma membrane is essential for its biological function. Membrane targeting of RhoA is dependent on geranylgeranylation at its C-terminus. Geranylgeranyl pyrophosphate, the substrate of geranylgeranylation, is a product of the mevalonate pathway. HIDS is caused by loss-of-function mutations of mevalonate kinase. The deficiency of mevalonate kinase in HIDS or the pharmacological inhibition of 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA) reductase results in the depletion of geranylgeranyl pyrophosphate, which has been reported to induce IL-1β release in myeloid cells through an as-yet-unknown inflammasome34–36. Since RhoA is inactive without geranylgeranylation and the inactivation of RhoA induces pyrin inflammasome activation, we investigated the role of pyrin in the pathogenesis of HIDS.
Simvastatin, a direct inhibitor of HMG-CoA reductase, induced translocation of RhoA from the membrane into the cytosol and a dose-dependent release of IL-1β from LPS-primed BMDMs (Supplementary Fig. 8a,b). The IL-1β release induced by various statins was dependent on the pyrin inflammasome, but independent of the NLRP3, NLRC4, or AIM2 inflammasomes (Supplementary Fig. 8b–d and Fig. 8a). We also found that the IL-1β release induced by simvastatin was blocked by supplementation with geranylgeranyl pyrophosphate or activation of PKNs with arachidonic acid or bryostatin 1, which suppress the pyrin inflammasome (Fig. 8b,c and Supplementary Fig. 8e–g).
Figure 8. HIDS is caused by a reduced threshold for activation of the pyrin inflammasome.
(a) BMDMs from WT, Casp1, Asc, Nlrp3, Nlrc4, Aim2, Mefv-deficient mice, or MefvΔCt/ΔCt mice were treated with simvastatin (10 μM) for 16h and primed with LPS for 6h. (b) BMDMs from WT mice were treated with simvastatin for 16h and co-treated with LPS and the indicated dose of geranylgeranyl pyrophosphate (GGpp) for 6h. (c) BMDMs from WT mice were co-treated with LPS, simvastatin (10 μM), and the indicated dose of arachidonic acid or bryostatin 1. Cell culture supernatants and cell lysates were analyzed by Immunoblotting as indicated. (d,e) BMDMs from WT mice were treated with LPS, simvastatin, and arachidonic acid or Bryostatin 1. The lysates were immunoprecipitated with anti-pyrin antibody, and immune complexes were analyzed by immunoblot for (d) PKN1 or (e) 14-3-3ε. (f,g) BMDMs from WT mice were treated with simvastatin for 16 h and co-treated with LPS and GGpp for 6h. The lysates were immunoprecipitated with anti-pyrin antibody, and immune complexes were analyzed by immunoblot for (f) PKN1 or (g) 14-3-3ε. (h,i) PBMCs from HIDS patients (designated patient number 1, 2, 3, and 4) with the indicated mutations in MVK were co-treated with LPS and the indicated dose of (h) arachidonic acid or (i) bryostatin 1 for 6h. Cell culture supernatants and cell lysates were analyzed by immunoblotting as indicated. Immunoblot data shown are representative of one experiment (h, i) or at least three independent experiments (a–g).
As seen in cells treated with C3 toxin, simvastatin also significantly decreased the 14-3-3ε-pyrin interaction as well as the PKN1-pyrin interaction in LPS-primed BMDMs, and the decreased interactions were reversed by the activation of PKNs or supplementation with exogenous geranylgeranyl pyrophosphate (Fig. 8 d–g). These data indicate that a blockade of the mevalonate pathway induces pyrin inflammasome activation. Of note, the statin concentrations required to achieve such a blockade are much higher than the blood levels achieved in patients treated with statins.
Consistent with these data, the constitutive IL-1β release from PBMCs of HIDS patients was substantially blocked by the suppression of the pyrin inflammasome with arachidonic acid or bryostatin 1 (Fig. 8h,i). However, unlike FMF, colchicine showed no or little inhibitory effect on pyrin inflammasome activation in PBMCs of HIDS patients (Supplementary Fig. 8h), most likely because colchicine cannot activate RhoA that has not been localized to the cell membrane through geranylgeranylation. These findings indicate an essential role for the pyrin inflammasome in the pathogenesis of HIDS and also provide an ‘experiment of nature’ demonstrating the activation of the pyrin inflammasome by inhibiting the RhoA signaling pathway (Supplementary Fig. 6).
DISCUSSION
Through these studies of RhoA signal transduction, we elucidate how bacterial toxins activate the pyrin inflammasome. The central finding of this study, that the pyrin inflammasome is regulated by RhoA-dependent phosphorylation of pyrin and pyrin’s subsequent interaction with 14-3-3 proteins, also delineates the molecular pathogenesis of FMF and defines a heretofore-unrecognized connection between FMF and the seemingly unrelated autoinflammatory disease, HIDS. The data presented here are consistent with the emerging concept that FMF mutations are gain-of-function with a gene dosage effect5, and the existence of rare FMF mutations at both phosphorylated serine residues of pyrin provides direct evidence for the role of the 14-3-3 proteins in FMF (http://fmf.igh.cnrs.fr/ISSAID/infevers/). This concept is strengthened by the recent description of a new, dominantly-inherited disorder, denoted pyrin-associated autoinflammation and neutrophilic dermatosis (PAAND), caused by the pSer242Arg mutation in pyrin37.
Most of the more common and severe FMF-associated mutations are clustered in the C-terminal B30.2 domain of human pyrin. Perhaps by intramolecular interactions, B30.2 domain mutations are likely to control pyrin phosphorylation by inhibiting the binding of kinases to pyrin. Thus, we hypothesize that FMF-associated mutations in the B30.2 domain block the phosphorylation sites from kinases, such as PKN1, resulting in a lowered threshold for activation of the pyrin inflammasome. In mice, which lack a pyrin B30.2 domain, one would expect that the balance would favor the phosphorylated, inhibited state, unless the mouse has evolved phosphatases that tip the balance toward the dephosphorylated, activated state.
Given the extraordinarily high carrier frequency of FMF-associated MEFV mutations in Mediterranean and Middle Eastern populations, the possibility that heterozygous FMF mutations might confer a selective advantage against one or more pathogenic microbes has long been a topic of intense interest. The findings presented here provide a molecular account of how this may have happened: bacterial toxins that inactivate RhoA have been evolutionarily selected in bacteria because they disable host cell cytoskeletal organization and the numerous downstream host-defense mechanisms such as leukocyte migration and phagocytosis that depend on an intact cytoskeleton. The pyrin inflammasome is likely a host counter-measure that resembles the plant guard-type mechanism, allowing for hosts to defend against a wide range of pathogens by sensing particular virulence-related activities rather than by sensing pathogen-associated molecular patterns (PAMPs) directly17,38.
This hypothesis proposes that the pyrin inflammasome can control a broad spectrum of potential pathogenic infections, perhaps broader than currently appreciated. Normal pyrin exerts its role by nucleating an inflammasome in order to defend against bacteria, such as Clostridium difficile, Burkholderia cenocepacia, and Vibrio cholerae, which utilize toxins to inactivate Rho GTPase, thereby inducing actin depolymerization. The adenylate cyclase toxins secreted from Bordetella pertussis, Bacillus anthracis, Pseudomonas aeruginosa, and Yersinia pestis induce intracellular accumulation of cAMP in the host cell39. While the increase of cAMP suppresses the NLRP3 inflammasome22, it conversely potentiates pyrin inflammasome activation. Thus, pyrin may function as an innate immune ‘guard’ in much the same way that R proteins function in plant antimicrobial defense. The requirement for both a priming step and RhoA inactivation prevents pyrin inflammasome activation triggered by normal cellular processes. However, the end result is a potent mechanism that defends against a major class of pathogens, and the double-edged sword that is genetic variation in this system of defense.
ONLINE METHODS
Reagents
Ultra-pure flagellin (catalogue no. tlrl-pstfla), and ultra-pure LPS (tlrl-pelps) were obtained from InvivoGen. C3 toxin (CT03) and CNF toxin (CN03) were from Cytoskeleton. TcdB toxin (6246-GT) was from R&D Systems. NKH477 (1603), simvastatin (1965), fluvastatin (3309), lovastatin (1530), calpeptin (0448), colchicine (1364), arachidonic acid (2756), bryostatin1 (2383), staurosporine (1285), and PKC412 (2992) were from Tocris Bioscience. Geranylgeranyl pyrophosphate ammonium salt (G6025) was from Sigma.
Mice
Eight- to sixteen-week old male and female mice (Mus musculus) on C57BL/6J background were used in experiments. C57BL/6J mice from The Jackson Laboratory were used for WT control. FMF knock-in (KI) mice harboring an FMF-associated mutant human B30.2 domain (MefvM680I/M680I, MefvM694V/M694V, and MefvV726A/V726A), Mefv-deficient (Mefv−/−), and C-terminal pyrin-truncation mice (MefvΔCt/ΔCt) have been described previously5,18. Wild-type B30.2 domain KI mice (MefvB30.2/B30.2) were generated by homologous recombination of a targeting construct that has been described in our previous study5. Heterozygotes generated from chimeras were crossed with EIIa-Cre transgenic mice (Jackson Laboratory) to remove the neo cassette and backcrossed with C57BL/6J mice at least six generations. ASC-, NLRP3-, and NLRC4-KO mice were a gift from Dr. Vishva M. Dixit (Genentech Inc.). Casp-1-KO mice were from Dr. Richard Flavell (Yale University). AIM2-KO mice were from Dr. Emad Alnemri (Thomas Jefferson University). In each experiment, experimental and control mice were age and sex-matched. Both male and female mice were used according to availability. The investigators were blinded to allocation during experiments and outcome assessment for experiments shown in Fig. 6b and c and Supplementary Fig. 4a and b. All animal studies were performed according to National Institutes of Health guidelines and were approved by the Institutional Animal Care and Use Committee of National Human Genome Research Institute.
Patients
Blood specimens from 4 healthy controls (control 1: 47 years old (Y), male (M); control 2: 53Y, female (F); control 3: 59Y, F; and control 4: 26Y, M), 4 CAPS (CAPS 1: 17Y, F; CAPS 2: 40Y, F; CAPS 3, 6Y, M; and CAPS 4: 46Y, F), 4 HIDS (HIDS 1: 8Y, F; HIDS 2: 12Y, M; HIDS 3: 18Y, M; and HIDS 4: 9Y, M), and 9 FMF (FMF 1: 16Y, M; FMF 2: 46Y, F; FMF 3: 53Y, M; FMF 4: 53Y, M; FMF 5: 34Y, F; FMF 6: 30Y, M; FMF 7: 77Y, F; FMF 8: 29Y, M; and FMF 9: 82Y, M) patients were drawn after obtaining informed consent under a protocol approved by the NIAMS/NIDDK Institutional Review Board.
Cell preparation
BMDMs were obtained by differentiating bone marrow progenitors from the tibia and femur of 6–12 weeks old male or female mice in Iscove’s Modified Dulbecco’s Media (IMDM) containing 20 ng/ml of M-CSF (Peprotech), 10% heat-inactivated fetal bovine serum (FBS, Invitrogen), 1 mM sodium pyruvate, 100 U/ml penicillin, and 100 μg/ml streptomycin (Invitrogen) for 7 days. BMDMs were then replated in 12-well plates one day before experiments. Human PBMCs were isolated by LSM-Lymphocyte Separation Medium (50494, MP Biomedicals) from freshly drawn peripheral venous blood from healthy controls or patients and also differentiated into macrophages with 800 units/ml of GM-CSF (Peprotech) for 7 days. Macrophages were treated with 20 ng/ml of IFN-γ (Peprotech) for 16h and then stimulated with LPS (1 ng/ml) for 6h.
Site-directed mutagenesis
Mutated human pyrin-expressing constructs were generated by site-directed mutagenesis using QuickChange Lightning Kit (210519-5, Agilent Technologies) according to the manufacturer’s instructions. Oligonucleotide primers used to introduce mutations for Ser208Ala were 5′-CGCAGAAACGCCGCCTCCGCGGGGAG-3′ and 5′-CTCCCCGCGGAGGCGGCGTTTCTGCG-3′. For Ser242Ala were 5′-GATGCGACCTAGAGCCCTTGAGGTCAC-3′ and 5′-GTGACCTCAAGGGCTCTAGGTCGCATC-3′.
Inflammasome activation or inhibition
BMDMs (1.0 X 106 cells per well) or PBMCs (3.0 X 106 cells per well) were plated in 12-well plates in RPMI 1640 (Invitrogen) containing 10% FBS and antibiotics and then primed with 1 μg/ml LPS in Opti-MEM (Invitrogen) for 6h. For pyrin inflammasome activation, C3 toxin (0.5 μg/ml) or TcdB toxin (0.5 μg/ml) was added to the medium for 6h. For depletion of cellular mevalonate levels, BMDMs were incubated with RPMI 1640 medium supplemented with simvastatin, fluvastatin, or lovastatin (1 or 10 μM) overnight. Then medium was replaced with Opti-MEM containing LPS (1 μg/ml) and statins for 6h. For AIM2 or NLRC4 inflammasome activation, 1 μg/ml of dsDNA with 2.5 μl/ml of Lipofectamine 2000 (Invitrogen) or 0.5 μg/ml flagellin with 25 μl/ml DOTAP (Roche), respectively, were mixed in Opti-MEM and incubated for 10 min before treatment of the cells. For NLRP3 inflammasome activation, the medium was replaced with RPMI 1640 medium supplemented with ATP (2 mM) and incubated for 30 min. For the inhibition or augmentation of inflammasome activation, LPS-primed BMDMs were treated with arachidonic acid (10–60 μM), NKH477 (10–500 μM), bryostatin 1 (0.1–0.5 μM), cardiolipin (0.1–0.5 μg/ml), CNF toxin (0.5 μg/ml), calpeptin (10–50 μM), colchicine (0.1–104 ng/ml), staurosporine (0.1–2 μM), or PKC412 (0.1–5 μM). After 0.5–6h of treatment, supernatants and cell lysates were collected for immunoblot analysis. None of the reagents in these experiments induced cytotoxicity as confirmed by LDH assay (K311-400, BioVision).
Gene knockdown assay
siRNAs targeting Pkn1, Pkn2, Rock1, Rock2, 14-3-3ε, 14-3-3τ, and negative control siRNA were purchased from Invitrogen. The three siRNAs for Pkn1 knockdown were 5′-AGAUUGACAUCAUCCGCAUTT-3′ (s115927), 5′-GAUCCAGACCUAUAGCAAUTT-3′ (s115925), and 5′-GAGUGGUGGGCUGCAAAAATT-3′ (s203299). The three siRNAs for Pkn2 knockdown were 5′-CGACAUCAAGGAUCGAAUATT-3′ (s99524), 5′-GCAGAACCAGUAUAGCACATT-3′ (s99525), and 5′-GCAACCUCCUGAAAACGGATT-3′ (s99526). The three siRNAs for Rock1 knockdown were 5′-GUACCGAACGGACCCUUAATT-3′ (s73019), 5′-GGUUGGGACUUACAGUAAATT-3′ (s203580), and 5′-GCGAAUGACUUACUUAGGATT-3′ (s73018). The three siRNAs for Rock2 knockdown were 5′-GAGAUUACCUUACGGAAAATT-3′ (s73020), 5′-GCGGAGAUCAUGAAUCGAATT-3′ (s73021), and 5′-GGAUGACAAAGGCGAUGUATT-3′ (s73022). The three siRNAs for 14-3-3ε knockdown were 5′-GGCAAAUGGUUGAAACUGATT-3′ (s76177), 5′-AGGUCUUGCUCUCAACUUUTT-3′ (s76178), and 5′-GUUAAUCUGUUGUGACAUUTT-3′ (s76179). The three siRNAs for 14-3-3τ knockdown were 5′-CAAGGACUAUCGGGAGAAATT-3′ (s76187), 5′-CAACUAAUCCAGAGAGUAATT-3′ (s76186), and 5′-GCAUUGAGCAGAAGACCGATT-3′ (s120785). For siRNA gene knockdown experiments, 50 – 250 pmol siRNA were transfected into BMDMs (1 X 106 cells per well) by electroporation and replated in 12-well plates. After 36h, the siRNA-transfected cells were analyzed for inflammasome activation.
Transfection, immunoprecipitation, immunoblot, and ELISA
A total of 2–5 μg of expression constructs for WT or mutant pyrin proteins were electroporated into U937 (ATCC-CRL-1593.2) cells (5.0 × 106 cells) using a Neon transfection system (Invitrogen) in Tip 100 μl tips at 1500 mV/30 ms/pulse. After transfection, U937 cells were differentiated into macrophages with 400 nM PMA for 48h and then incubated with Opti-MEM containing 1 μg/ml LPS for 6h, then supernatants and cell lysates were collected and analyzed by immunoblot for IL-1β.
PT67 (631510, Clontech) or 293T (ATCC-CRL-3216) cells (4 X 106 cells) were plated in 100 mm2 cell culture dishes one day before transfection. A total of 0.1–10 μg of expression constructs for WT, FMF-associated mutant, or various domain-deleted pyrin proteins were transfected or co-transfected with expression constructs for full-length or various domain-deleted PKN1 proteins with 10–20 μl of Lipofectamine 2000 according to the manufacturer’s instructions. After 36h, the transfected cells were lysed and incubated with anti-myc, anti-mouse pyrin18, or anti-human pyrin antibody40. Lysates from BMDMs or human macrophages were immunoprecipitated with 1 μg/ml anti-mouse pyrin or anti-human pyrin antibody followed by protein A plus agarose (Thermo Scientific). After washing with PBS or PBS containing 0.05–0.1% SDS (for the immunoprecipitation of WT or FMF-associated mutant pyrin proteins), bound proteins were eluted by 2X SDS sample buffer from the beads and analyzed by immunoblot for PKN1 or 14-3-3 proteins. All cell lines were negative for mycoplasma.
Immunoblots were prepared with Novex® Tris-Glycine Gel Systems (Invitrogen) and probed overnight at 4°C with anti-human IL-1β antibody (AF-201-NA); anti-mouse IL-1β antibody (AF-401-NA, R&D Systems); anti-caspase-1 antibody (sc-514), anti-14-3-3ε antibody (sc-23957); anti-14-3-3τ antibody (sc-69720); anti-PKN1 antibody (sc-393344); anti-ROCK1 antibody (sc-365628); anti-ROCK2 antibody (sc-398519); anti-actin antibody (sc-1615); anti-myc antibody (sc-40, Santa Cruz Biotechnology); anti-PKN2 antibody (2612, Cell Signaling); or anti-V5 antibody (Invitrogen).
Secreted IL-1β was also measured in supernatant using commercially available ELISA kit according to the manufacturer’s instructions (MLB00C, R&D Systems).
Flow cytometry analysis
RBC-lysed single-cell suspensions were obtained from peripheral blood and stained with fluorochrome-conjugated CD11b antibodies in PharMingen Stain Buffer (BSA) (BD PharMingen). Cells were analyzed on FACSCaliber flow cytometer (BD Biosciences) and analyzed with FlowJo software (Three Star).
Measurement of active RhoA-GTP levels and differential localization of RhoA
BMDMs (2.0 X 106 cells per well) were plated in 6 well plates and treated with NKH477 (50–500 μM), CNF toxin (0.5–2 μg/ml), or C3 toxin (0.5 μg/ml) and CNF toxin (0.5–2 μg/ml). Activated RhoA (RhoA-GTP) levels were measured by RhoA G-LISA Activation Assay kit (BK121, cytoskeleton) and normalized to total RhoA levels, which were measured by Total RhoA ELISA kit (BK150, Cytoskeleton). RhoA-GTP levels of BMDMs treated with C3 toxin (0.5 μg/ml) and colchicine (102 – 104 ng/ml) were measured by a pull-down assay with Rhotekin-RBD beads using RhoA activation Assay Biochem Kit (BK036, cytoskeleton). Differential localization of RhoA was determined from membrane and cytoplasmic fractions, which were prepared using the Membrane and Cytoplasmic Extraction Buffer (MEB and CEB), respectively (78840, Thermo Scientific). Briefly, BMDMs (3.0 X 106 cells per well) were plated in 6-well plates and treated with simvastatin (10 μM) overnight. Cells were scraped with cold PBS and centrifuged at 500 X g for 3 min. The pellets were resuspended with 100 μl of CEB and incubated on ice for 10 min. After centrifugation at 500 X g for 5 min, supernatants were collected as cytosolic extractions. The pellets were resuspended by 100 μl of MEB by vortexing vigorously and incubated on ice. After 10 min, membrane extractions were collected by centrifugation at 3000 X g for 5 min. The isolated membrane and cytosolic fractions were analyzed by immunoblot using anti-RhoA antibody (sc-418, Santa Cruz Biotechnology), anti-calcium-sensing receptor antibody (ab18200), and anti-glyceraldehyde 3-phosphate dehydrogenase antibody (ab9482, Abcam).
In vitro kinase assay
Myc/his-tagged WT or S208A/S242A mutant N-terminal pyrin (aa’s 1- 330) was overexpressed into 293T cells, and the N-terminal pyrin proteins were purified using His-SpinTrap™ (28-4013-53, GE Healthcare). Each purified N-terminal pyrin protein was incubated with 5 μg of PKN1 (PR7255B) or PKN2 (PR7370A, Thermo Scientific) at 30°C for 0.5h in a kinase buffer (50 mM HEPES pH7.5, 0.01% BRIJ-35, 10 mM MgCl2, 1 mM EGTA, 500 μM ATP and 2.5 mM DTT). The reactants were analyzed by immunoblotting with anti-phospho-Ser antibody (9606, Cell Signaling) or staining with Pro-Q diamond (P33300, Invitrogen). Pro-Q diamond-stained gel was visualized on Molecular Imager FX (Bio-Rad).
Data analysis, statistics and experimental replicates
Statistical analysis was carried out using a nonparametric Mann–Whitney t-test, and the unpaired two-tailed t-test using Prism software (GraphPad). A P value of ≤ 0.05 was considered statistically significant. The number of reproduced experimental repeats is described in the relevant figure legends. The investigators were not blinded to allocation during experiments and outcome assessment, except as noted in Mice section above.
Supplementary Material
Acknowledgments
We thank the patients enrolled in our clinical protocols for providing research specimens; B. Barham, A. Jones, T. Romeo, P. Hoffmann, D. Stone, P. Pinto-Patarroyo, A. Ombrello, and K. Barron for help in caring for patients; I. Aksentijevich for assistance in genotyping patients and for helpful discussions; V. Dixit (Genentech) for providing ASC, NLRP3, and NLRC4 knock-out mice; R. Flavell (Yale University) for providing caspase-1 knock-out mice; E. Alnemri (Thomas Jefferson University) for providing AIM2 knockout mice; and S. Masters and F. Shao for sharing data prior to publication. This work was supported by the Intramural Research Program of the National Human Genome Research Institute.
Footnotes
AUTHOR CONTRIBUTIONS
Y.H.P., D.L.K, and J.J.C. designed the study; Y.H.P., G.W., and J.J.C. performed experiments; Y.H.P., G.W., D.L.K., and J.J.C. analyzed the data; and Y.H.P., D.L.K., and J.J.C. wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Kastner DL, Aksentijevich I, Goldbach-Mansky R. Autoinflammatory disease reloaded: a clinical perspective. Cell. 2010;140:784–790. doi: 10.1016/j.cell.2010.03.002. S0092-8674(10)00238-2 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Masters SL, Simon A, Aksentijevich I, Kastner DL. Horror autoinflammaticus: the molecular pathophysiology of autoinflammatory disease (*) Annu Rev Immunol. 2009;27:621–668. doi: 10.1146/annurev.immunol.25.022106.141627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.International_FMF_Consortium. Ancient missense mutations in a new member of the RoRet gene family are likely to cause familial Mediterranean fever. Cell. 1997;90:797–807. doi: 10.1016/s0092-8674(00)80539-5. [DOI] [PubMed] [Google Scholar]
- 4.Drenth JP, et al. Mutations in the gene encoding mevalonate kinase cause hyper-IgD and periodic fever syndrome. International Hyper-IgD Study Group. Nat Genet. 1999;22:178–181. doi: 10.1038/9696. [DOI] [PubMed] [Google Scholar]
- 5.Chae JJ, et al. Gain-of-function Pyrin mutations induce NLRP3 protein-independent interleukin-1beta activation and severe autoinflammation in mice. Immunity. 2011;34:755–768. doi: 10.1016/j.immuni.2011.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu H, et al. Innate immune sensing of bacterial modifications of Rho GTPases by the Pyrin inflammasome. Nature. 2014;513:237–241. doi: 10.1038/nature13449. [DOI] [PubMed] [Google Scholar]
- 7.Miziorko HM. Enzymes of the mevalonate pathway of isoprenoid biosynthesis. Archives of biochemistry and biophysics. 2011;505:131–143. doi: 10.1016/j.abb.2010.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sohar E, Gafni J, Pras M, Heller H. Familial Mediterranean fever. A survey of 470 cases and review of the literature. Am J Med. 1967;43:227–253. doi: 10.1016/0002-9343(67)90167-2. [DOI] [PubMed] [Google Scholar]
- 9.French_FMF_Consortium. A candidate gene for familial Mediterranean fever. Nat Genet. 1997;17:25–31. doi: 10.1038/ng0997-25. [DOI] [PubMed] [Google Scholar]
- 10.Lachmann HJ, et al. Clinical and subclinical inflammation in patients with familial Mediterranean fever and in heterozygous carriers of MEFV mutations. Rheumatology. 2006;45:746–750. doi: 10.1093/rheumatology/kei279. [DOI] [PubMed] [Google Scholar]
- 11.Booty MG, et al. Familial Mediterranean fever with a single MEFV mutation: where is the second hit? Arthritis Rheum. 2009;60:1851–1861. doi: 10.1002/art.24569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marek-Yagel D, et al. Clinical disease among patients heterozygous for familial Mediterranean fever. Arthritis Rheum. 2009;60:1862–1866. doi: 10.1002/art.24570. [DOI] [PubMed] [Google Scholar]
- 13.Ozen S. Changing concepts in familial Mediterranean fever: is it possible to have an autosomal-recessive disease with only one mutation? Arthritis Rheum. 2009;60:1575–1577. doi: 10.1002/art.24565. [DOI] [PubMed] [Google Scholar]
- 14.Booth DR, et al. The genetic basis of autosomal dominant familial Mediterranean fever. Q J Med. 2000;93:217–221. doi: 10.1093/qjmed/93.4.217. [DOI] [PubMed] [Google Scholar]
- 15.Aldea A, et al. A severe autosomal-dominant periodic inflammatory disorder with renal AA amyloidosis and colchicine resistance associated to the MEFV H478Y variant in a Spanish kindred: an unusual familial Mediterranean fever phenotype or another MEFV-associated periodic inflammatory disorder? American journal of medical genetics. 2004;124A:67–73. doi: 10.1002/ajmg.a.20296. [DOI] [PubMed] [Google Scholar]
- 16.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 17.Mackey D, Holt BF, 3rd, Wiig A, Dangl JL. RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell. 2002;108:743–754. doi: 10.1016/s0092-8674(02)00661-x. [DOI] [PubMed] [Google Scholar]
- 18.Chae JJ, et al. Targeted disruption of pyrin, the FMF protein, causes heightened sensitivity to endotoxin and a defect in macrophage apoptosis. Mol Cell. 2003;11:591–604. doi: 10.1016/s1097-2765(03)00056-x. [DOI] [PubMed] [Google Scholar]
- 19.Schoenwaelder SM, Burridge K. Evidence for a calpeptin-sensitive protein-tyrosine phosphatase upstream of the small GTPase Rho. A novel role for the calpain inhibitor calpeptin in the inhibition of protein-tyrosine phosphatases. The Journal of biological chemistry. 1999;274:14359–14367. doi: 10.1074/jbc.274.20.14359. [DOI] [PubMed] [Google Scholar]
- 20.Schmidt G, et al. Gln 63 of Rho is deamidated by Escherichia coli cytotoxic necrotizing factor-1. Nature. 1997;387:725–729. doi: 10.1038/42735. [DOI] [PubMed] [Google Scholar]
- 21.Flatau G, et al. Toxin-induced activation of the G protein p21 Rho by deamidation of glutamine. Nature. 1997;387:729–733. doi: 10.1038/42743. [DOI] [PubMed] [Google Scholar]
- 22.Lee GS, et al. The calcium-sensing receptor regulates the NLRP3 inflammasome through Ca2+ and cAMP. Nature. 2012;492:123–127. doi: 10.1038/nature11588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lang P, et al. Protein kinase A phosphorylation of RhoA mediates the morphological and functional effects of cyclic AMP in cytotoxic lymphocytes. The EMBO journal. 1996;15:510–519. [PMC free article] [PubMed] [Google Scholar]
- 24.Aburima A, et al. cAMP signaling regulates platelet myosin light chain (MLC) phosphorylation and shape change through targeting the RhoA-Rho kinase-MLC phosphatase signaling pathway. Blood. 2013;122:3533–3545. doi: 10.1182/blood-2013-03-487850. [DOI] [PubMed] [Google Scholar]
- 25.Krendel M, Zenke FT, Bokoch GM. Nucleotide exchange factor GEF-H1 mediates crosstalk between microtubules and the actin cytoskeleton. Nature cell biology. 2002;4:294–301. doi: 10.1038/ncb773. [DOI] [PubMed] [Google Scholar]
- 26.Misawa T, et al. Microtubule-driven spatial arrangement of mitochondria promotes activation of the NLRP3 inflammasome. Nature immunology. 2013;14:454–460. doi: 10.1038/ni.2550. [DOI] [PubMed] [Google Scholar]
- 27.Allam R, et al. Mitochondrial apoptosis is dispensable for NLRP3 inflammasome activation but non-apoptotic caspase-8 is required for inflammasome priming. EMBO reports. 2014;15:982–990. doi: 10.15252/embr.201438463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Falk MD, et al. Enzyme Kinetics and Distinct Modulation of the Protein Kinase N Family of Kinases by Lipid Activators and Small Molecule Inhibitors. Biosci Rep. 2014 doi: 10.1042/BSR20140010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feng J, et al. Rho-associated kinase of chicken gizzard smooth muscle. The Journal of biological chemistry. 1999;274:3744–3752. doi: 10.1074/jbc.274.6.3744. [DOI] [PubMed] [Google Scholar]
- 30.Yoshinaga C, Mukai H, Toshimori M, Miyamoto M, Ono Y. Mutational analysis of the regulatory mechanism of PKN: the regulatory region of PKN contains an arachidonic acid-sensitive autoinhibitory domain. Journal of biochemistry. 1999;126:475–484. doi: 10.1093/oxfordjournals.jbchem.a022476. [DOI] [PubMed] [Google Scholar]
- 31.Regan JW. EP2 and EP4 prostanoid receptor signaling. Life sciences. 2003;74:143–153. doi: 10.1016/j.lfs.2003.09.031. [DOI] [PubMed] [Google Scholar]
- 32.Sokolowska M, et al. Prostaglandin E2 Inhibits NLRP3 Inflammasome Activation through EP4 Receptor and Intracellular Cyclic AMP in Human Macrophages. Journal of immunology. 2015;194:5472–5487. doi: 10.4049/jimmunol.1401343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jeru I, et al. Interaction of pyrin with 14.3.3 in an isoform-specific and phosphorylation-dependent manner regulates its translocation to the nucleus. Arthritis Rheum. 2005;52:1848–1857. doi: 10.1002/art.21050. [DOI] [PubMed] [Google Scholar]
- 34.Mandey SH, Kuijk LM, Frenkel J, Waterham HR. A role for geranylgeranylation in interleukin-1beta secretion. Arthritis and rheumatism. 2006;54:3690–3695. doi: 10.1002/art.22194. [DOI] [PubMed] [Google Scholar]
- 35.Kuijk LM, et al. HMG-CoA reductase inhibition induces IL-1beta release through Rac1/PI3K/PKB-dependent caspase-1 activation. Blood. 2008;112:3563–3573. doi: 10.1182/blood-2008-03-144667. [DOI] [PubMed] [Google Scholar]
- 36.Normand S, et al. Specific increase in caspase-1 activity and secretion of IL-1 family cytokines: a putative link between mevalonate kinase deficiency and inflammation. European cytokine network. 2009;20:101–107. doi: 10.1684/ecn.2009.0163. [DOI] [PubMed] [Google Scholar]
- 37.Masters SM, et al. Familial autoinflammation with neutrophilic dermatosis reveals a regulatory mechanism of pyrin activation. Science Translational Medicine. 2016 doi: 10.1126/scitranslmed.aaf1471. [DOI] [PubMed] [Google Scholar]
- 38.Vance RE. Immunology taught by bacteria. Journal of clinical immunology. 2010;30:507–511. doi: 10.1007/s10875-010-9389-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ahuja N, Kumar P, Bhatnagar R. The adenylate cyclase toxins. Crit Rev Microbiol. 2004;30:187–196. doi: 10.1080/10408410490468795. [DOI] [PubMed] [Google Scholar]
- 40.Chae JJ, et al. The B30.2 domain of pyrin, the familial Mediterranean fever protein, interacts directly with caspase-1 to modulate IL-1beta production. Proc Natl Acad Sci USA. 2006;103:9982–9987. doi: 10.1073/pnas.0602081103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.