Abstract
Cardiolipin is a specific mitochondrial phospholipid that has a high affinity for proteins and that stabilizes the assembly of supercomplexes involved in oxidative phosphorylation. We found that sequestration of cardiolipin in protein complexes is critical to protect it from degradation. The turnover of cardiolipin is slower by almost an order of magnitude than the turnover of other phospholipids. However, in Barth syndrome, cardiolipin is rapidly degraded via the intermediate monolyso-cardiolipin. Treatments that induce supercomplex assembly decrease the turnover of cardiolipin and the concentration of monolyso-cardiolipin whereas dissociation of supercomplexes has the opposite effect. Our data suggest that cardiolipin is uniquely protected from normal lipid turnover by its association with proteins, but in Barth syndrome, where this association is compromised, cardiolipin becomes unstable, which causes the accumulation of monolyso-cardiolipin.
Cardiolipin (CL) is a dimeric phospholipid that occurs in mitochondrial membranes. In those membranes, CL supports a molecular architecture characterized by the lateral segregation of large protein aggregates1, 2. The physical basis of this effect lies in the unique shape of CL and its charge state, which together produce a tendency to cluster and a tendency to form strong non-covalent interactions with proteins3. For example, CL is an integral component of supercomplexes that form from individual complexes of oxidative phosphorylation and it is crucial for their stability4–6.
CL is the primary metabolite affected in Barth syndrome (BTHS)7, a disease caused by mutations in the tafazzin gene8. The gene encodes a phospholipid-lysophospholipid transacylase that shuttles fatty acids between different lipid species9. Although tafazzin can in principle react with all phospholipids, its main substrate in vivo is CL because the acyl chains of CL are under the type of physical constraints that are required to drive transacylations10. In the absence of tafazzin, CL drastically changes its fatty acid composition and is partially replaced by monolyso-cardiolipin (MLCL)11–14. While one can rationalize the altered fatty acid composition, it is more difficult to explain the accumulation of MLCL. It has been proposed that MLCL accumulates because the lack of tafazzin prevents its re-acylation but since transacylations may proceed in either direction one cannot assume a priori that tafazzin will consume MLCL2, 15.
Here we show that BTHS leads to rapid CL degradation and that this is the actual cause of the accumulation of MLCL. Preliminary evidence has suggested that under normal conditions, the turnover of CL is much slower than the turnover of other phospholipids16–18 but no satisfying explanation has been provided for this observation. We demonstrate with stable isotope techniques that the CL turnover is indeed extremely slow and that the compartmentalization of CL within the membrane, specifically the sequestration of CL in protein complexes, is responsible for it. The results are consistent with reports showing that decreased CL concentration rather than altered CL species composition is associated with poor mitochondrial function in the yeast model of BTHS19, 20.
RESULTS
Tafazzin deficiency shortens the half-life of CL
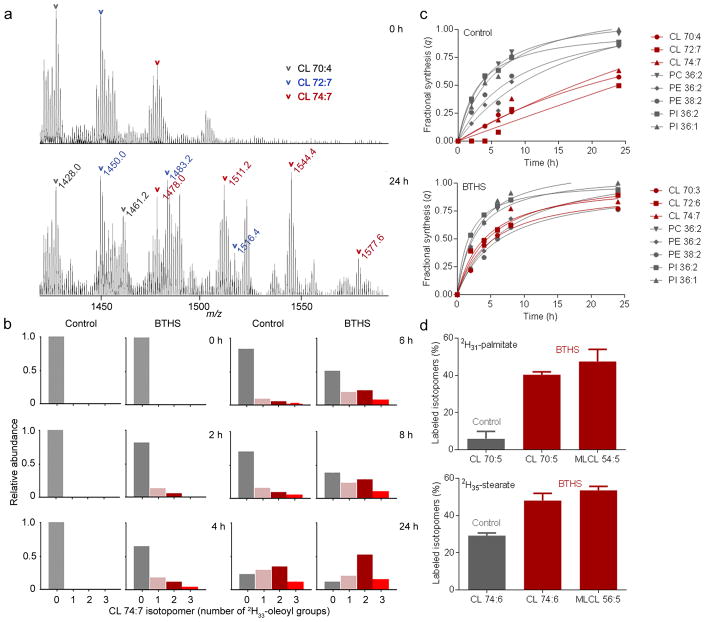
To measure the turnover of CL, we cultured human lymphoblasts with 2H33-oleic acid and analyzed the lipid extracts by mass spectrometry (MS). Heavy isotopomers formed of many lipid species, the number of which depended on the number of oleoyl groups in the molecule; for instance, one heavy isotopomer was found for CL 70:4, two for CL 72:7, and three for CL 74:7 (Fig. 1a) (lipids were abbreviated according to ref. 21). During the incubation, the relative intensity of multi-labeled CL isotopomers increased relative to the intensity of mono-labeled and unlabeled ones. We noted that these changes occurred a lot faster in lymphoblasts from BTHS patients than in controls (Fig. 1b). Previous work has shown that kinetic parameters can be calculated from the intensity pattern of such isotopomers22, 23. Based on the principles of isotopomer spectral analysis, we estimated the fraction of newly synthesized molecules (fractional synthesis) of selected lipid species and found that the fractional synthesis of CL was much lower than the fractional synthesis of diacylglycero-phospholipids. However, the difference between CL and other lipids disappeared in lymphoblasts from BTHS patients (Fig. 1c). BTHS shortened the half-life of CL from 17–26 hours to 4–6 hours but had little effect on phosphatidylinositol (PI), phosphatidylethanolamine (PE), and phosphatidylcholine (PC) (Supplementary Results, Supplementary Table 1). The increased turnover of CL in BTHS lymphoblasts could be verified with other precursors, such as 2H31-palmitic acid and 2H35-stearic acid, although fractional syntheses were not calculated due to the paucity of species with multiple isotopomers (Fig. 1d).
Figure 1. BTHS increases the acyl turnover of CL in human lymphoblasts.
(a) Human lymphoblasts were incubated with 2H33-oleic acid; lipids were analyzed by MS. The CL region of the raw spectra is shown before (0 h) and after (24 h) labeling. Novel isotopomers formed as a result of 2H33-oleoyl incorporation. (b) Intensities of CL 74:7 isotopomers with 0, 1, 2, and 3 2H33-oleoyl groups were determined at different time points. The isotopomer pattern changed faster in BTHS lymphoblasts than in controls. Data are means of duplicate determinations. (c) The fractional syntheses of the indicated lipid species were determined at different time points. The turnover of CL was slower than the turnover of other phospholipids in controls but not in BTHS. Statistical analysis of the data is shown in Supplementary Table 1. (d) Human lymphoblasts were incubated with 2H31-palmitic acid or 2H35-stearic acid for 20 hours. Lipids were analyzed by MS to determine the proportion of labeled isotopomers [labeled/(labeled+unlabeled)]. BTHS caused increased labeling of CL with 2H31-palmitate (P<0.0001) and 2H35-stearate (P<0.002). The proportion of labeled isotopomers was similar in CL and MLCL; the latter was present only in BTHS. Data (means±s.e.m., N=4) were analyzed by t-test.
Next we determined the effect of tafazzin on the lipid turnover of fruit flies. For this experiment, we chose D-[U-13C6]-glucose as precursor, which led to the incorporation of multiple 13C isotopes into glycerol and fatty acid moieties. As a result, the spectrum of PE and other phospholipids became densely populated with novel isotopomers both in the wild type and the tafazzin deletion mutant (ΔTAZ). In contrast, CL was only labeled in ΔTAZ but not in the wild type; in fact, the CL spectra from wild-type flies were completely devoid of novel isotopomers even after feeding D-[U-13C6]-glucose for 9 days (Fig. 2a). When we calculated the fractional synthesis of CL from the intensities of four isotopomers (13C0, 13C3, 13C6, 13C9 ), which in first approximation represented labeling of the glycerol backbone, the computations confirmed that new CL was synthesized in ΔTAZ flies but not in wild type. Similar experiments with human lymphoblasts and mouse embryonic fibroblasts demonstrated a significant increase in the fractional synthesis of CL as a result of tafazzin deletion (Fig. 2b; Supplementary Fig. 1). These data not only confirmed the results obtained with labeled fatty acids but also showed that the turnover of the entire CL molecule, not just the turnover of acyl groups, was elevated in tafazzin mutants from different organisms.
Figure 2. Tafazzin deficiency increases the glycerol turnover of CL in Drosophila and mammalian cells.
(a) Drosophila strains were cultured for 9 days with D-[U-13C6]-glucose or unlabeled glucose as control. Lipids were analyzed by MS and spectra are displayed in centroid mode. 13C was incorporated into PE of the wild-type (WT) and the tafazzin deletion strain (ΔTAZ). 13C was incorporated into CL and MLCL of ΔTAZ but not WT, indicating a very slow CL turnover in the WT. (b) The fractional syntheses of CL and MLCL species were determined from the relative abundances of 13C0, 13C3,13C6 and 13C9 isotopomers following labeling with D-[U-13C6]-glucose for the indicated time periods. Tafazzin deficiency caused an increase in the fractional synthesis of CL species in Drosophila (P<0.04, 3 d; P<0.01, 9 d), mouse fibroblasts (P<0.001), and human lymphoblasts (P<0.0001). The fractional syntheses of CL and MLCL were similar. Data (means±s.e.m., N=3) were analyzed by t-test.
MLCL accumulates in intact mitochondria
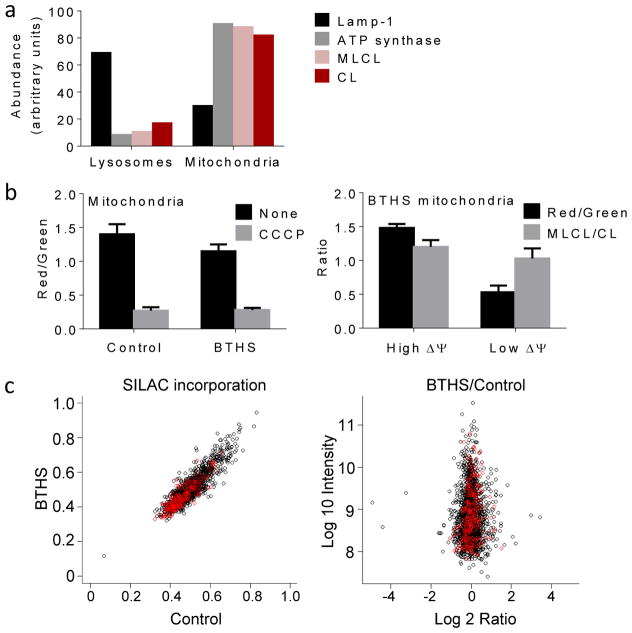
The high turnover of CL in BTHS could be caused by a specific increase in CL synthesis and degradation or by a global increase in mitochondrial biogenesis and mitophagy. If the latter were the case, CL degradation must occur in lysosomes where mitochondria are digested. In order to localize the degradation process, we determined the intracellular distribution of MLCL, an intermediate of the degradation pathway that had the same turnover rate as CL (Figs. 1d, 2b). We found that MLCL was associated with mitochondria rather than with lysosomes of BTHS lymphoblasts, suggesting that at least the first step of CL degradation occurred in mitochondria (Fig. 3a). Next, we asked whether CL degradation was restricted to damaged mitochondria, the proportion of which is increased in tafazzin-deficient organisms19, 24–26. To explore this possibility, we divided isolated BTHS mitochondria with a cell sorter into one population with high and one with low membrane potential. We did not find a difference in their MLCL/CL ratios, which excluded the idea that MLCL accumulated only in damaged mitochondria (Fig. 3b). Finally, we asked whether the turnover of the mitochondrial proteome was accelerated in BTHS lymphoblasts. Using stable isotope labeled amino acids in cell culture (SILAC) combined with mass spectrometric analysis we did not find any difference in the incorporation of label into mitochondrial and non-mitochondrial proteins between BTHS and control (Fig. 3c). We concluded that BTHS increased the turnover of CL but not the turnover of the entire organelle. Importantly, MLCL was present in all mitochondria, not just in those without membrane potential, implying that CL degradation occurred in respiring mitochondria of BTHS lymphoblasts.
Figure 3. MLCL is present in intact mitochondria with normal protein turnover.
(a) Lysosomes and mitochondria were prepared from BTHS lymphoblasts. Marker proteins for lysosomes (Lamp-1) and mitochondria (α-subunit of ATP synthase) were quantified by Western blotting; MLCL and CL were quantified by MS. CL and MLCL were associated with mitochondria. Data are means of duplicate determinations. (b) Mitochondria were isolated from human lymphoblasts and analyzed by flow cytometry after staining with JC-1. The membrane potential, measured by the red/green fluorescence ratio, was slightly lower in BTHS mitochondria than in controls (P<0.03, t-test). CCCP collapsed the membrane potential. Isolated BTHS mitochondria were divided with a cell sorter into two populations one with high and one with low membrane potential (ΔΨ). The populations were analyzed by flow cytometry and MS, which confirmed the difference in membrane potential but did not show any difference in the MLCL/CL ratio. Data are means±s.e.m. (N=3). (c) Human lymphoblasts were cultured with stable isotope labeled amino acids. The level of incorporation of labeled amino acids into mitochondrial (red circles) and non-mitochondrial (black circles) proteins was measured by proteomic analysis. The BTHS/control ratio of incorporated label was measured in 1:1 mixtures of control and BTHS samples, each labeled with different precursors. The turnover of mitochondrial proteins was similar in BTHS and controls.
CL but not MLCL is tightly bound to proteins
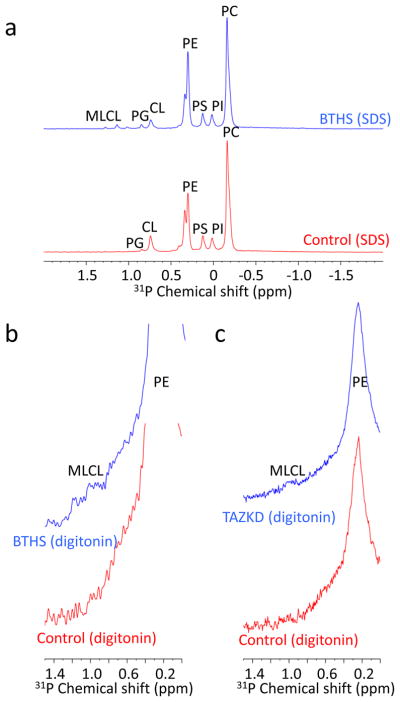
The presence of MLCL in active mitochondria raises the question whether it occupies the same compartment as CL. When we purified mitochondria from BTHS lymphoblasts by Percoll gradient centrifugation, we noticed that a subset of mitochondria floated above the bulk portion. These low-density mitochondria had a slightly higher MLCL/CL ratio than the rest. Furthermore, we noticed that MLCL was solubilized by digitonin more readily than CL (Supplementary Fig. 2a). Because digitonin solubilizes lipids before it solubilizes proteins and a low density in mitochondria may indicate an increased lipid/protein ratio, we hypothesized that CL and MLCL differ in terms of their protein association. To test this hypothesis, we used 31P-NMR spectroscopy. It is known that a large portion of mitochondrial CL is associated with proteins and protein complexes1, 2 and it is also known that this interaction causes line broadening of the 31P-NMR signal of CL, as a result of which CL “disappears” from the spectrum of lipid-protein complexes dissolved with mild detergents27, 28. Consistent with those data, we did not find a detectable 31P-NMR signal of CL in digitonin-solubilized liver mitochondria but did find such signal when mitochondria were solubilized with the strong detergent sodium dodecylsulfate (SDS) (Supplementary Fig. 2b). In SDS-solubilized lymphoblast mitochondria from BTHS patients, both CL and MLCL were detectable (Fig. 4a); however, in the presence of digitonin only MLCL produced a detectable 31P-NMR signal whereas CL remained invisible (Fig. 4b). Essentially the same result was obtained with kidney mitochondria from tafazzin knockdown (TAZKD) mice (Fig. 4c). Quantification of the NMR signals revealed that PC, PE, and MLCL were resolved to nearly the same extent in SDS and in digitonin, whereas CL was resolved only in SDS but not in digitonin (Supplementary Table 2). We concluded from the NMR experiments that CL and MLCL differed in terms of their protein association. This conclusion was corroborated by density gradient centrifugation of n-dodecyl-β-D-maltoside solubilized mitochondria from TAZKD mouse tissues. The MLCL/CL ratio decreased from low to high density, suggesting that CL was associated with proteins but MLCL with free lipids (Supplementary Fig. 2c). Together the data reveal a striking contrast between CL and MLCL, showing that CL was tightly bound to proteins but MLCL was not.
Figure 4. CL but not MLCL is associated with proteins.
(a) Mitochondrial membranes were prepared from human lymphoblasts and solubilized with SDS. 31P-NMR spectra show resonances of all major and some minor phospholipids. CL was detectable in control samples; CL and MLCL were detectable in BTHS samples. (b–c) Mitochondrial membranes, prepared from human lymphoblasts (b) or mouse kidneys (c), were solubilized with digitonin. 31P-NMR spectra show the major phospholipids with broader resonances compared to the SDS experiment. MLCL (chemical shift values of MLCL standard: 1.0, 1.2 ppm) was detectable in tafazzin-deficient samples but CL was not.
Resveratrol protects CL from degradation
When working with lymphoblasts of BTHS patients, we noticed variations in their MLCL/CL ratio in response to the culture conditions. For instance, serum starvation increased the MLCL/CL ratio during exponential growth but had little effect in the stationary phase. Furthermore, the MLCL/CL ratio was higher in the exponential than in the stationary phase but only at low serum concentration (Supplementary Fig. 3). The data suggested that the MLCL/CL ratio is not static but varies with the cellular condition, specifically with the functional state of mitochondria, which is known to be depressed in the logarithmic phase of growth29. Thus, we hypothesized that the MLCL/CL ratio depends on mitochondrial function and therefore might respond to drugs that stimulate mitochondrial biogenesis.
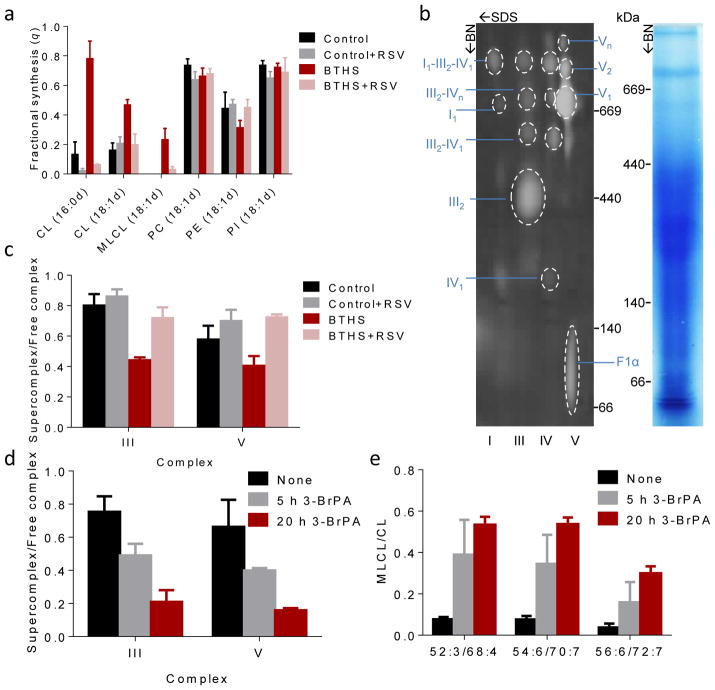
To test this idea, we added two different drugs, bezafibrate and resveratrol, to lymphoblasts from BTHS patients. These drugs activate different transcription programs but their action converges on the PGC-1α pathway, which stimulates the formation of mitochondria and shifts the energy metabolism from a glycolytic to an oxidative state30, 31. Both drugs caused a substantial decrease in the MLCL/CL ratio, far exceeding the effects of growth conditions and strongly suggesting that stimulation of mitochondrial biogenesis reduced the concentration of MLCL in favor of CL (Supplementary Fig. 3). To determine whether the increase in CL was caused by increased synthesis or by reduced degradation, we measured the fractional synthesis of CL with 2H-labeled fatty acids. Resveratrol, the more potent of the two drugs, reduced the fractional synthesis of CL, reversing the elevation of CL turnover in BTHS lymphoblasts. Importantly, the action of resveratrol was restricted to CL and MLCL in BTHS and did not apply to other phospholipids nor did it apply to CL in controls (Fig. 5a). Measurements with D-[U-13C6]-glucose corroborated the inhibitory effect of resveratrol on the turnover of CL and MLCL and showed that the rapid increase in CL was due to the stabilization of pre-existing CL rather than the formation of new CL. Upon withdrawal of resveratrol, an equally rapid transition occurred in the reverse direction, in which case pre-existing CL was degraded faster than new CL could be synthesized (Supplementary Fig. 4). These experiments suggested that resveratrol decreased the turnover of CL by re-distributing it within the membrane. We therefore postulated that intramembranous CL compartmentalization determines CL stability by altering its accessibility to phospholipases.
Figure 5. Supercomplexes protect CL from degradation.
(a) Human lymphoblasts were cultured ±40 μM RSV for 36 hours. The fractional syntheses of lipids were determined by incorporating 2H-labeled palmitic acid (16:0d) or oleic acid (18:1d) for 8 hours. RSV inhibited the turnover of CL in BTHS (P<0.03) but not in controls. RSV had no effect on other phospholipids. (b–c) Lymphoblast mitochondria were solubilized with digitonin, separated by 2D-Blue Native/SDS-PAGE, and analyzed with primary antibodies to NDUFB6 (complex I), UQCRC2 (complex III), MtCO1 (complex IV), and F1α (complex V). The Coomassie-stained lane shows the BN-PAGE separation and the 2-D blot shows individual complexes and supercomplexes with the positions of molecular weight markers. BTHS decreased the proportion of supercomplexes, RSV increased it back to normal (P<0.02). (d–e) Control lymphoblasts were treated with 80 μM 3-bromopyruvate (3-BrPA) for 5 or 20 hours. 3-BrPA decreased the proportion of supercomplexes and increased the MLCL/CL ratio (P<0.03). Data (means±s.e.m., N=3) were analyzed by t-test (a, c) or ANOVA (d, e).
Supercomplexes are essential for CL stability
The question then becomes whether resveratrol, or other treatments that stimulate mitochondrial biogenesis, can change CL compartmentalization. It has been shown that resveratrol promotes the assembly of respiratory supercomplexes31, which could be beneficial for tafazzin-deficient cells, in which the concentration of supercomplexes is low32, 33. Indeed, we confirmed a decreased abundance of large protein assemblies, including respiratory supercomplexes, ATP synthase oligomers, and complexes containing the ADP-ATP carrier in TAZKD mouse tissues and BTHS lymphoblasts (Supplementary Fig. 5a, b) and found that resveratrol increased the abundance of such complexes specifically in BTHS (Fig. 5b, c).
Since supercomplexes contain tightly bound CL5, 6, it is likely that resveratrol not only increases supercomplexes but also the proportion of protein-associated CL. To estimate the amount of supercomplex-trapped CL, we subjected digitonin-solubilized mitochondria to density gradient centrifugation. More than half of the total CL was present in the fraction that contained supercomplexes without preference for any molecular species. In BTHS, less CL was associated with supercomplexes but resveratrol treatment increased that proportion (Supplementary Fig. 6).
Thus, the data suggest that resveratrol promotes the sequestration of CL within large protein complexes, which may be the reason why it protects CL from degradation. To further test this concept, we searched for a way to disassemble such complexes in normal mitochondria. Our search let to 3-bromopyruvate, an alkylating agent that impairs glycolysis and mitochondrial function by inhibiting the activities of several enzymes34, 35. We discovered that although treatment with 3-bromopyruvate caused a slight increase in the abundance of mitochondrial proteins including ATP synthase and respiratory enzymes (Supplementary Fig. 7), the drug induced dissociation of supercomplexes in normal lymphoblasts (Fig. 5d). At the same time, 3-bromopyruvate led to the accumulation of MLCL, but not to the accumulation of other lysophospholipids, directly demonstrating that the disassembly of supercomplexes caused the degradation of CL (Fig. 5e). It had already been reported that 3-bromopyruvate triggered the formation of MLCL in glioblastoma cells36. Together these experiments strongly suggest that the association with supercomplexes protects CL from degradation.
Unsaturation promotes CL-protein association
Previous studies have shown that linoleic acid restores CL levels in fibroblasts37 and cardiomyocytes38 derived from BTHS patients. This effect was thought to result from the scavenging of reactive oxygen species because linoleic acid suppressed the concentration of 4-hydroxynonenal, a lipid oxidation product, and linoleic acid improved the same cellular functions as the antioxidant MitoTempo38. Therefore, we tested the effects of MitoTempo and linoleic acid on the MLCL/CL ratio in BTHS lymphoblasts. Interestingly, we found that MitoTempo had no effect on the MLCL/CL ratio whereas linoleic acid decreased the ratio substantially (Fig. 6a). This suggested that linoleic acid did not act by scavenging free radicals and that CL oxidation was not responsible for the formation of MLCL. Corroborating this conclusion, we found no difference between the average number of double bonds per acyl chain in CL and MLCL from BTHS lymphoblasts and TAZKD mouse tissues (Supplementary Fig. 8a).
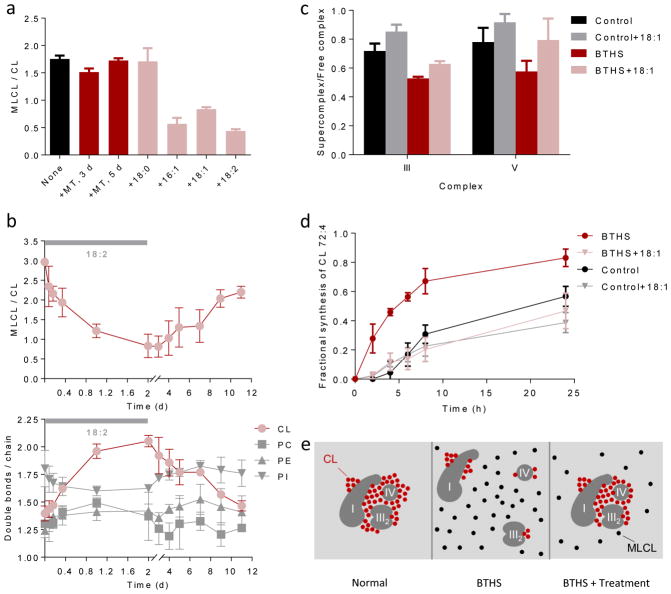
Figure 6. Unsaturated fatty acids stabilize CL.
(a) BTHS lymphoblasts were cultured with 5 μM MitoTempo (MT) for 3 or 5 days or with 0.1 mM exogenous fatty acid (stearic acid, 18:0; palmitoleic acid, 16:1; oleic acid, 18:1; linoleic acid, 18:2) for 4 days. Unsaturated fatty acids decreased the MLCL/CL ratio (P<0.003, t-test). (b) BTHS lymphoblasts were cultured first with and then without linoleic acid. The MLCL/CL ratio inversely correlated with the CL unsaturation. (c–d) Lymphoblasts were cultured ± 18:1 for 4 days followed by measurements of supercomplex abundance and CL turnover with 2H33-oleic acid. BTHS reduced the proportion of supercomplexes and increased the turnover of CL (P<0.03, t-test). 18:1 increased the proportion of supercomplexes and reduced the turnover of CL in BTHS but not in controls (P<0.02, t-test). Half-lives of CL 72:4, estimated by non-linear regression analysis, were 20 hours (control), 30 hours (control+18:1), 5 hours (BTHS), and 26 hours (BTHS+18:1). Data in panels a–d are means±s.e.m. (N=3). (e) The cartoon shows supercomplex I1-III2-IV1 as an example; a similar mechanism is suggested for other complexes. In normal mitochondria, unsaturated CL clusters around protein complexes, which promotes their assembly to supercomplexes and protects CL from degradation. In BTHS, the fatty acid composition of CL is altered. Less CL is associated with proteins and non-associated CL is hydrolyzed to MLCL. Treatments that increase CL unsaturation or force the formation of supercomplexes, increase the proportion of protein-associated CL. As a result, less CL is hydrolyzed to MLCL.
Thus, an alternative explanation had to be sought for the effect of linoleic acid. When we tested other fatty acids, we discovered that they also reduced the MLCL/CL ratio but only if they were unsaturated (Fig. 6a). Since all unsaturated fatty acids increased the double bond density of CL (Supplementary Fig. 8b), we considered the possibility that CL unsaturation itself may control the MLCL/CL ratio. Indeed, we found a strong inverse correlation between CL unsaturation and the MLCL/CL ratio during on-off incubations with linoleic acid, a correlation that existed only for CL but not for other phospholipids (Fig. 6b). Therefore, we hypothesized that high unsaturation in CL promotes the assembly of proteins into large complexes and, as a result, protects CL from degradation. Consistent with that idea, we demonstrated that oleic acid increased the abundance of supercomplexes (Fig. 6c, Supplementary Fig. 5c) and slowed down CL degradation (Fig. 6d), mirroring the effect of resveratrol on BTHS lymphoblasts. In control cells, where CL unsaturation is already high, oleic acid treatment did not alter the CL double bonds (Supplementary Fig. 8b) nor did it affect the abundance of supercomplexes or the CL turnover (Fig. 6c, d). We concluded that unsaturation in CL promotes protein association, which in turn stabilizes CL.
DISCUSSION
We have demonstrated that the lack of tafazzin causes a substantial increase in the turnover of CL. The fractional synthesis of CL, which is an indicator of its turnover rate, equaled the fractional synthesis of MLCL and variations in the fractional synthesis correlated with variations in the MLCL/CL ratio, strongly suggesting that the accumulation of MLCL in tafazzin-deficient cells is a direct consequence of rapid CL degradation. MLCL is an intermediate of this degradation and is itself degraded further, presumably to fatty acids and glycerophosphates. The large elevation in metabolic turnover was reproduced in different experimental models and was a CL-specific phenomenon, neither afflicting other lipids nor mitochondrial proteins. Its ultimate result, the decline in mitochondrial function, may be due to the loss of CL, the increase in MLCL, or both.
Previous studies have already suggested that the turnover of CL is very slow in normal organisms16–18. Here we confirmed and expanded this observation by rigorous kinetic measurements. For example, we demonstrated there was hardly any turnover of CL in fruit flies over the course of an entire week. However, when tafazzin was lacking, the turnover of CL became similar to the turnover of other phospholipids.
In order to maintain a slow turnover, CL must be protected from degradation. Previous studies have shown that CL tightly binds to proteins and protein complexes1, 2 and is entrapped at high concentration in supercomplex assemblies5, 6, to which it also provides structural stability4. Our NMR experiments corroborated that the majority of mitochondrial CL is bound to proteins. Protein binding, in particular the interaction with supercomplexes, segregates CL from the bulk lipid phase and may therefore protect it from degradation. In support of this idea, we found that stimulation of mitochondrial protein synthesis, either by resveratrol, bezafibrate, or stationary growth, partially reversed the elevated MLCL/CL ratio in BTHS lymphoblasts. We also found that supercomplex assembly inversely correlated with the MLCL/CL ratio and the turnover of CL. Importantly, resveratrol did not suppress the capability to degrade CL per se but changed the accessibility of CL to the degrading enzyme. This can be inferred from the fact that the clearance of MLCL continued during resveratrol treatment. Our data suggest that the sequestration of CL in protein complexes is the actual cause of its slow turnover and that it is the loss of this sequestration that makes CL susceptible to degradation in BTHS (Fig. 6e). One would therefore expect CL hydrolysis to occur only in the bulk lipid phase, which is consistent with the localization of MLCL established by NMR. Although our measurements have zoomed in on respiratory supercomplexes and ATP synthase oligomers, CL is associated with many mitochondrial proteins and those may also protect it from degradation. The same protective mechanism may apply to other lipid species that are entrapped together with CL.
Why does CL fail to associate with protein complexes in the absence of tafazzin? In BTHS, CL is more saturated and its species composition is more diverse than in controls2, 15, which may hamper the formation of homogeneous CL clusters around proteins. When we added unsaturated fatty acids to the culture medium, we not only increased the unsaturation and homogeneity of the CL species but also the abundance of supercomplexes. This suggests that unsaturated CL supports protein association better than saturated CL. The underlying mechanism for this difference is not clear at the moment. However, in a previous study with isolated mitochondria, we already observed that CL seemed to dissociate from proteins when its double bonds were reduced by catalytic hydrogenation.39 Hypothetically, the acyl chains of unsaturated CL may adapt better to the various protein surfaces or produce more stable CL clusters within protein complexes. Regardless, we propose that the abnormal CL species composition prevents normal CL-protein assembly in BTHS, which drives CL into the bulk lipid phase where it is degraded to MLCL. Since CL unsaturation drives protein complex formation and protein complex formation increases CL stability, our data suggest a mutual dependence between the stability of CL and the stability of protein complexes (Fig. 6e).
In summary, we propose that CL is protected from degradation in normal cells by its association with proteins but becomes readily degradable when this association is compromised, such as in BTHS. This has two important implications. First, variations in the association-dissociation equilibrium of supercomplexes in response to stress40 may affect the CL concentration in BTHS patients; hypothetically this could provide an explanation for their susceptibility to sudden clinical crises41, 42. Second, drugs that foster supercomplex association may increase the CL concentration in BTHS patients. Since cardiac tissue in vivo may show a different response than lymphoblasts in culture, it remains to be seen whether such drugs lead to functional improvements.
ONLINE METHODS
Cells
Human lymphoblast cell lines were established by Epstein-Barr virus transformation of leukocytes isolated from BTHS patients and control subjects24. Human lymphoblasts were cultured at 37°C in RPMI 1640-PS medium in the presence of 10% fetal bovine serum unless stated otherwise. Mouse embryonic stem cells with and without tafazzin knockout, were obtained from Dr. Zaza Khuchua (Cincinnati Children’s Hospital). The stem cells were differentiated into embryonic fibroblasts in EmbryoMax DMEM containing 15% fetal bovine serum, 0.8% β-mercaptoethanol, 100 IU/mL penicillin, and 0.1 g/L streptomycin.
Animals
Drosophila cultures were maintained on a standard cornmeal-sucrose-yeast medium at 23 °C. The tafazzin gene was inactivated (ΔTAZ) by imprecise excision of a P element from stock y[1]; P{y[+mDint2] w[BR.E.BR] = SUP-P}tafazzin[KG02529]/SM1; ry[506] as described25. The corresponding controls were created by precise excision of the P element from the same stock. All experiments were performed with adult flies (2 weeks old) that were kept in 3-inch vials. Mouse protocols were approved by the Institutional Animal Care and Use Committee of NYU School of Medicine. Colonies were housed under temperature-controlled conditions and a 12-hour light/dark cycle with free access to drinking water and food. Transgenic mice were generated from the C57Bl/6 strain by TaconicArtemis, GmbH (Köln, Germany) under contract from the Barth Syndrome Foundation43, 44. Knock-down of tafazzin (TAZKD) was induced in heterozygous adult mice that contained the H1-tet-On-shRNA promoter by adding doxycycline (2 g/L) to the drinking water. After 6 months of doxycycline treatment, tissues (liver, kidney, heart) were harvested under anesthesia in order to isolate mitochondria. In the TAZKD animals, doxycycline treatment increased the MLCL/CL ratios from zero to 0.2±0.1 (liver), 0.3±0.1 (kidney), and 1.5±0.4 (heart), whereas the ratios remained zero in the doxycycline-treated control animals.
Isolation of mitochondria
Mitochondria were isolated from lymphoblasts as described24. For this purpose, cells were permeabilized by digitonin (0.2 g/L) in chilled isolation buffer (210 mM mannitol, 70 mM sucrose, 1 mM EDTA, 0.5% fatty acid-free bovine serum albumin, 5 mM HEPES, pH 7.2) and disrupted in a tight-fitting glass-Teflon homogenizer. Cell debris and nuclei were removed by centrifugation at 625 g for 5 minutes. Mitochondria were pelleted by centrifugation at 10,000 g for 20 min. They were washed, re-suspended, and pelleted again. For the separation of mitochondria with different densities, the crude pellet was re-suspended in 0.5 mL isolation buffer and placed on top of a medium containing 30% Percoll, 1 mM EGTA, 25 mM Hepes (pH 7.3) and 225 mM mannitol in a centrifugation tube. The tube was spun at 95,000 g for 1 hour after which fractions were collected from the Percoll gradient. Mitochondria were separated from lysosomes using the Organelle Enrichment Kit from the Pierce Protein Biology Product line of Thermo Scientific. This technique also employed Percoll gradient centrifugation.
Mitochondria were also isolated from mouse tissues, including liver, kidney and heart. Tissues were homogenized in chilled isolation buffer using an engine-driven glass-Teflon homogenizer. Debris and nuclei were removed by centrifugation at 750 g for 5 minutes and mitochondria were pelleted from the supernatants by centrifugation at 10,000 g for 10 min. Mitochondria were washed, re-suspended, and pelleted again by the same method.
Density gradient separations of detergent-solubilized mitochondria
Mouse liver and kidney mitochondria were solubilized for 30 min with 2.5 g n-dodecyl-β-D-maltoside per g protein at a protein concentration of 10 g/L. Non-solubilized material was removed by centrifugation and the solubilized mitochondria (0.25 mL) were placed on top of a sucrose gradient consisting of 0.5 mL of 1.5 M sucrose, 1 M sucrose and 0.5 M sucrose respectively in buffer (10 mM KCl, 10 mM Tris, pH 7.4, 0.2 mM n-dodecyl-β-D-maltoside). Tubes were spun at 158,000 g for 20 hours using the TLS-55 swing-out rotor in a Beckman Optima MAX-XP table-top ultracentrifuge. Fractions (0.55 mL) were collected from top to bottom.
Human lymphoblast mitochondria (2 mg protein) were incubated in cold water for 30 min and the membranes were collected by centrifugation (16,000 g, 30 min). Membranes were solubilized in 0.5 mL medium (10 mM Tris, 20 mM KCl, pH 7.4) containing 16 mg digitonin. The sample was stirred on ice for 45 min and subsequently spun at 100,000 g for 30 min in order to remove non-solubilized material. The supernatant was placed on a discontinuous sucrose gradient (7 layers, 0.3 M – 1.5 M sucrose containing 10 mM Tris, pH 7.4, and 0.03% digitonin) in a total volume of 1.4 mL. The gradient was spun at 200,000 g for 20 hrs. Samples were collected from top to bottom.
Digitonin-insoluble mitochondrial membranes
Isolated lymphoblast mitochondria were treated with digitonin in order to collect digitonin-insoluble residues. For this purpose, mitochondria were re-suspended in isolation buffer to a protein concentration of 10 g/L. Aliquots of a 20% digitonin solution were added to adjust digitonin-to-protein ratios ranging from 0 to 10 g/g. Samples were incubated with digitonin for 10 min and subsequently spun at 100,000 g for 1 hour, after which pellets were collected.
Labeling with stable isotopes
Lymphoblasts were cultured in the presence of 2H-labeled fatty acids supplied as albumin complexes at a final concentration of 0.1 mM. Human lymphoblasts and mouse fibroblasts were also grown in glucose-free DMEM supplemented with 4 g/L D-[U-13C6]-glucose. To label flies with 13C isotopes, a 1×1 cm piece of filter paper was soaked in 0.5 M D-[U-13C6]-glucose and placed at the bottom of the culture vial. The flies were incubated up to 9 days; more solution was added from time to time in order to keep the filter paper moist. Five flies were sacrificed per time point. To this end, flies were cooled in a refrigerator until they became immobile and then homogenized in a small aliquot of water.
Lipid analysis
Lipids were extracted into chloroform-methanol45. When internal standards were required, the CL internal standard mixture I, PE 17:0/14:1, and PC 21:0/22:6 from Avanti Polar Lipids were added. The extracts were dried under a stream of nitrogen and re-suspended in 50–200 μL of chloroform-methanol 1:1. MS was performed according to the method of Sun et al.46 Aliquots of the lipid solutions were diluted 1:11 in 2-propanol-acetonitrile (3:2) and then mixed 1:1 with matrix solution containing 20 g/L 9-aminoacridine in 2-propanol-acetonitrile (3:2). One microliter or less was applied onto the target spots and matrix-assisted laser desorption ionization time-of-flight MS was performed with a MALDI Micro MX mass spectrometer (Waters) operated in reflectron mode. The pulse voltage was set to 2000 V, the detector voltage was set to 2200 V, and the TLF delay was set to 700 ns. The nitrogen laser (337 nm) was fired at a rate of 5 Hz and 10 laser shots were acquired per sub-spectrum. For the analysis of PC the instrument was operated in positive ion mode and for all other phospholipids in negative ion mode with a flight tube voltage of 12 kV, a reflectron voltage of 5.2 kV, and a negative anode voltage of 3.5 kV. The instrument was calibrated daily with polyethylene glycol fragments in positive ion mode, or with a reference mixture of LPG 14:0 (m/z = 455.2415), PG 36:2 (m/z = 773.5338), PE 36:2 (m/z = 738.5079), and CL 72:8 (m/z = 1447.9650) in negative ion mode. We typically acquired 100–200 sub-spectra (representing 1000–2000 laser shots) per sample in a mass range from 400 to 2000 Da. Spectra were only acquired if their base-peak-intensity was within 10–95% of the saturation level. Uniform mass adjustment was performed with an internal reference mass. Data were analyzed with the MassLynx 4.1 software. Spectra were displayed in centroid mode after smoothing and baseline subtraction using the processing parameters recommended by the manufacturer.
Isotopomer spectral analysis
Incubation of cell cultures with 2H-labeled fatty acids introduced novel isotopomers into the mass spectra of lipids. For species in which more than one residue can be labeled, several isotopomers exist. It has been demonstrated that the distribution of intensities among those isotopomers carries information about two parameters, (i) the quantity of newly formed species during the observation period and (ii) the concentration of labeled fatty acids in the intracellular precursor pool. Our data analysis followed the principles of isotopomer spectral analysis developed by Kelleher and colleagues23, 47.
As an example, we are demonstrating here the analysis of tetraoleoyl-CL (CL 72:4). During incubation with 2H33-oleic acid, four labeled isotopomers were formed of tetraoleoyl-CL, each having a different number of deuterated chains. Each deuterated chain increased the isotopic mass by 33.20 units, equivalent to the mass difference between 2H33 and 1H33. We transformed the spectral intensities into a normalized vector M=(m0, m1, m2, m3, m4), in which mi was the relative abundance of the isotopomer with i labeled residues (Σmi=1). Because not only newly formed but also pre-existing CL contributed to the spectrum, M can be expressed as the sum of two vectors representing labeled (L) and unlabeled (N) tetraoleoyl-CL:
| (1) |
In equation (1), q is the fraction of newly synthesized tetraoleoyl-CL (fractional synthesis). The spectrum of pre-existing CL (N) was measured at the beginning of the experiment. In contrast, the spectrum of newly synthesized CL (L) cannot be measured but can be calculated from the fraction of labeled fatty acids in the intracellular precursor pool (p) by binomial distribution22, 23. After substituting the binomial term for L, equation (1) becomes:
| (2) |
We estimated p and q by finding the best fit to equation (2) with spectral data obtained before (n0, n1, n2, n3, n4) and after (m0, m1, m2, m3, m4) incubation with 2H33-oleic acid. Numerical fitting was performed in MATLAB using the Levenberg–Marquardt algorithm of the fsolve program in the optimization toolbox.
The same algorithm was applied to other lipid species except for the necessary adjustments in the equation. For molecular species in which three groups can be labeled, we used:
| (3) |
and for molecular species in which two groups can be labeled, we used:
| (4) |
In general, we found the p values of lipid species in good agreement with each other. As already noticed by Kharroubi et al.47, the correlation between p and q was low (R2=0.4399, correlation analysis, N=60), suggesting that the two parameters were relatively independent of each other.
In order to calculate kinetic parameters, q values were obtained at different time points for lipid species with a sufficient number of isotopomers and a good signal-to-noise ratio. Those q values were used to determine the rate constant of synthesis (k) by fitting the equation:
| (5) |
Under steady state conditions, k becomes the turnover rate that relates to the half-life time:
| (6) |
In some instances, we extended our analysis to species with low signal-to-noise ratios and species that contained only a single labeled residue. For those species, we modified the fitting routine to find best-fit q values for fixed p values (obtained from the analysis of dominant species).
Labeling was also performed with D-[U-13C6]-glucose. In this case heavy isotopes entered CL on two separate pathways, 13C2 units were incorporated into the acyl groups and 13C3 units were incorporated into the glycerol groups. The former pathway generated isotopomers of the series 13C2, 13C4, 13C6, 13C8, 13C10 …., whereas the latter generated the isotopomers 13C3, 13C6, and 13C9. Although some contamination with 13C2 units may occur in the 13C6 and 13C9 isotopomers, the 13C3-13C6-13C9 triplet is a good indicator of backbone labeling. In support of this notion, we have previously shown that the 13C9 isotopomer of CL contained heavy carbons primarily in the glycerol groups18. To measure the glycerol turnover of CL, best-fit values for p and q were obtained with equation (3).
2D-Blue Native/SDS PAGE
Mitochondria (0.4 mg protein) were solubilized with digitonin (8 g/g protein) and separated by Blue Native/SDS PAGE as described by Wittig et al.48. The solubilization buffer contained 50 mM NaCl, 50 mM imidazole, 2 mM aminohexanoic acid, and 1 mM EDTA (pH 7.0). Samples were spun at 100,000 x g for 15 min and solubilized proteins were collected from the supernatant. Coomassie blue G-250 was added and the proteins were separated by electrophoresis on a 4–13% gradient Blue Native gel. Gel strips were cut, soaked in 1% SDS for 15 min at 37 °C, and horizontally placed on top of a gel set for 10% SDS-PAGE (10% T, 3% C). After the second electrophoresis, proteins were transferred to a PVDF membrane for Western Blot analysis. PVDF membranes were incubated with primary monoclonal antibodies (1 μg/ml, MitoSciences Inc.) in Odyssey blocking buffer containing 0.01% Tween-20. Fluorescent LiCor GAM-IRDye680 secondary antibodies were used at a dilution of 1:15,000. Proteins were visualized and quantified by the LiCor scanner.
SILAC proteomics
Human lymphoblasts were cultured with stable isotope labeled amino acids. Control cells were cultured in the presence of a medium-labeled mixture containing 2H4-lysine/13C6-arginine (Lys4/Arg6) and BTHS cells were cultured in the presence of a heavy-labeled mixture containing 13C6-15N2-lysine/13C6-15N4-arginine (Lys8/Arg10) according to a protocol previously described49, 50. Cells were harvested after 24 and 32 hours, and each experiment was performed twice. The cell density was measured and aliquots of control and BTHS cells were mixed 1:1. Control cells, BTHS cells, and mixtures were digested separately and peptides were analyzed by LC–MS with two-hour gradients on a self-packed 15 cm C18 column (3 micron beads, Dr. Maisch), using an EASY-nLC 1000 coupled to a Q Exactive hybrid quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific). The raw files were processed using the MaxQuant computational proteomics platform (version 1.2.7.0) for peptide identification (1% FDR) and quantitation51. For visualization of results, the data of each experiment were separated into 2 groups, representing mitochondrial and non-mitochondrial proteins, respectively. The raw MS files have been uploaded to Massive and are accessible through the link ftp://massive.ucsd.edu/MSV000079600/raw/Barth_syndrome/.
Flow cytometry
Freshly isolated lymphoblast mitochondria (protein concentration about 0.5 mg/mL) were incubated with 22.5 μM JC-1 for 10 minutes at room temperature in a medium containing 0.1 mM phosphate, 5 mM glutamate, 5 mM succinate, 0.5 mM MgCl2, 250 mM sucrose and 0.1% bovine serum albumin (pH 7.4). 1 μM CCCP was added if collapse of the mitochondrial membrane potential was desired. Samples were analyzed on a Becton Dickinson FACSCalibur flow cytometry analyzer. JC-1 was excited by a 488nm laser in order to determine mitochondrial membrane potential that correlates with the ratio of red over green fluorescence intensity. JC-1 monomer green fluorescence was detected through a 530/40nm filter. JC-1 aggregate red fluorescence was detected through a 585/42nm filter.
BTHS mitochondria (1–2 mg protein) were sorted into high and low membrane potential populations on a Beckman Coulter MoFlo Legacy cell sorter. On the MoFlo sorter, green and red fluorescence were detected through 520/40nm and 580/30nm filters, respectively. Sorted mitochondria were re-analyzed by flow cytometry to determine the purities of the sorted populations. Lipids were extracted from the two populations and analyzed by MS.
31P NMR
Frozen mitochondria (15–30 mg protein) were thawed, suspended into 40 mL hypotonic buffer (15 mM Tris, pH 7.4), and incubated on ice for 30 minutes in order to release water-soluble phosphorus compounds. Mitochondrial membranes were re-collected by centrifugation at 39,000 g for 15 minutes and re-suspended in a small volume (0.1–0.3 mL) 15 mM Tris. Either digitonin (3–6 g/g protein) or SDS (1.5–2 g/g protein) was added and the samples were transferred into 5 mm diameter NMR tubes. 31P-NMR spectra were recorded on a Bruker AVANCE3 700 MHz NMR spectrometer (31P frequency=283.4 MHz) equipped with a cryoprobe. The cryoprobe had an inner coil capable of tuning to 31P, 13C or 15N frequencies and a 1H-decoupling outer coil. The chemical shift scale was referenced to an external standard of 85% (w/v) phosphoric acid, and broadband proton decoupling was achieved with a WALTZ-16 decoupling type sequence at an applied field of approximately 3,500 Hz. Spectra were recorded at 25 °C over a spectral width of 50 kHz. A 30-degree RF pulse was applied and raw FID data were recorded using 50K data points within an acquisition time of 0.5 s. Each spectrum was acquired using approximately 1000 transients and a recycle delay of 1.0 s. To identify the resonances, 31P NMR measurements were performed on commercial standard lipids in the presence of the same detergents. Small differences in chemical shift were noted between spectra recorded in digitonin and SDS.
Miscellaneous
Protein concentrations were determined by the method of Lowry et al.52. Statistical tests, including Student’s t-test, the paired t-test, analysis of variance (ANOVA), correlation analysis, and non-linear regression analysis were performed with Graph Pad Prism 6. Data were typically reported as mean values and variations estimated as standard error of mean (s.e.m.). The sample size was given for each experiment together with the P value of the statistical hypothesis test.
Supplementary Material
Acknowledgments
This research was supported in part by grant 1R01GM115593 (to MS), the Shared Instrumentation Grant RR027990, and the NINDS core center grant NS050276 (to TAN) from the National Institutes of Health. Funds were also provided from the Barth Syndrome Foundation (to CKLP and RME).
Footnotes
AUTHOR CONTRIBUTIONS
Y.X., C.K.L.P., M.R., and M.S. performed experiments and analyzed data. B.B., K.D., R.M.E., and M.S. designed and performed the NMR experiments. E.H., G. Z., and T.A.N. designed and performed the proteomics analyses. M.S. supervised the entire study and wrote the paper. C.K.L.P., T.A.N., M.R., and R.M.E. revised the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Mileykovskaya E, Dowhan W. Cardiolipin-dependent formation of mitochondrial respiratory supercomplexes. Chem Phys Lipids. 2014;179:42–48. doi: 10.1016/j.chemphyslip.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ren M, Phoon CKL, Schlame M. Metabolism and function of mitochondrial cardiolipin. Progr Lipid Res. 2014;55:1–16. doi: 10.1016/j.plipres.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 3.Lewis RNAH, McElhaney RN. The physicochemical properties of cardiolipin bilayers and cardiolipin-containing lipid membranes. Biochim Biophys Acta. 2009;1788:2069–2079. doi: 10.1016/j.bbamem.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 4.Zhang M, Mileykovskaya E, Dowhan W. Gluing the respiratory chain together. Cardiolipin is required for supercomplex formation in the inner mitochondrial membrane. J Biol Chem. 2002;277:43553–43556. doi: 10.1074/jbc.C200551200. [DOI] [PubMed] [Google Scholar]
- 5.Althoff T, Mills DJ, Popot JL, Kühlbrandt W. Arrangement of electron transport chain components in bovine mitochondrial supercomplex I1III2IV1. EMBO J. 2011;30:4652–4664. doi: 10.1038/emboj.2011.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mileykovskaya E, Penczek PA, Fang J, Mallampalli VKPS, Sparagna GC, Dowhan W. Arrangement of the respiratory chain complexes in Saccharomyces cerevisiae supercomplex III2IV2 revealed by single particle cryo-electron microscopy. J Biol Chem. 2012;287:23095–23103. doi: 10.1074/jbc.M112.367888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barth PG, et al. An X-linked mitochondrial disease affecting cardiac muscle, skeletal muscle and neutrophil leucocytes. J Neurol. 1983;62:327–55. doi: 10.1016/0022-510x(83)90209-5. [DOI] [PubMed] [Google Scholar]
- 8.Bione S, D’Adamo P, Maestrini E, Gedeon AK, Bolhuis PA, Toniolo D. A novel X-linked gene, G4.5. is responsible for Barth syndrome. Nature Genetics. 1996;12:385–389. doi: 10.1038/ng0496-385. [DOI] [PubMed] [Google Scholar]
- 9.Xu Y, Malhotra A, Ren M, Schlame M. The enzymatic function of tafazzin. J Biol Chem. 2006;281:39217–39224. doi: 10.1074/jbc.M606100200. [DOI] [PubMed] [Google Scholar]
- 10.Schlame M, et al. The physical state of lipid substrates provides transacylation specificity for tafazzin. Nature Chemical Biology. 2012;8:862–869. doi: 10.1038/nchembio.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vreken P, et al. Defective remodeling of cardiolipin and phosphatidylglycerol in Barth syndrome. Biochem Biophys Res Comm. 2000;279:378–382. doi: 10.1006/bbrc.2000.3952. [DOI] [PubMed] [Google Scholar]
- 12.Schlame M, et al. Deficiency of tetralinoleoyl-cardiolipin in Barth syndrome. Ann Neurol. 2002;51:634–637. doi: 10.1002/ana.10176. [DOI] [PubMed] [Google Scholar]
- 13.Gu Z, et al. Aberrant cardiolipin metabolism in the yeast taz1 mutant: a model for Barth syndrome. Mol Microbiol. 2004;51:149–158. doi: 10.1046/j.1365-2958.2003.03802.x. [DOI] [PubMed] [Google Scholar]
- 14.Valianpour F, et al. Monolysocardiolipins accumulate in Barth syndrome but do not lead to enhanced apoptosis. J Lipid Res. 2005;46:1182–1195. doi: 10.1194/jlr.M500056-JLR200. [DOI] [PubMed] [Google Scholar]
- 15.Claypool SM, Koehler CM. The complexity of cardiolipin in health and disease. Trends Biochem Sci. 2012;37:32–41. doi: 10.1016/j.tibs.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Landriscina C, Megli FM, Quagliariello E. Turnover of fatty acids in rat liver cardiolipin: comparison with other mitochondrial phospholipids. Lipids. 1976;11:61–66. doi: 10.1007/BF02532585. [DOI] [PubMed] [Google Scholar]
- 17.Wahjudi PN, et al. Turnover of non-essential fatty acids in cardiolipin from the rat heart. J Lipid Res. 2011;52:2226–2233. doi: 10.1194/jlr.M015966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu Y, Schlame M. The turnover of glycerol and acyl moieties of cardiolipin. Chem Phys Lipids. 2014;179:17–24. doi: 10.1016/j.chemphyslip.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 19.Baile MG, et al. Unremodeled and remodeled cardiolipin are functionally indistinguishable in yeast. J Biol Chem. 2014;289:1768–1778. doi: 10.1074/jbc.M113.525733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ye C, et al. Deletion of the cardiolipin-specific phospholipase Cld1 rescues growth and life span defects in the tafazzin mutant: implications for Barth syndrome. J Biol Chem. 2014;289:3114–3125. doi: 10.1074/jbc.M113.529487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liebisch G, et al. Shorthand notation for lipid structures derived from mass spectrometry. J Lipid Res. 2013;54:1523–1530. doi: 10.1194/jlr.M033506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hellerstein MK, et al. Measurement of de novo hepatic lipogenesis in humans using stable isotopes. J Clin Invest. 1991;87:1841–1852. doi: 10.1172/JCI115206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelleher JK, Masterson TM. Model equations for condensation biosynthesis using stable isotopes and radioisotopes. Am J Physiol. 1992;262:E118–E125. doi: 10.1152/ajpendo.1992.262.1.E118. [DOI] [PubMed] [Google Scholar]
- 24.Xu Y, Sutachan JJ, Plesken H, Kelley RI, Schlame M. Characterization of lymphoblast mitochondria from patients with Barth syndrome. Lab Invest. 2005;85:823–830. doi: 10.1038/labinvest.3700274. [DOI] [PubMed] [Google Scholar]
- 25.Xu Y, et al. A Drosophila model of Barth syndrome. Proc Natl Acad Sci USA. 2006;103:11584–11588. doi: 10.1073/pnas.0603242103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dudek J, et al. Cardiolipin deficiency affects respiratory chain function and organization in an induced pluripotent stem cell model of Barth syndrome. Stem Cell Res. 2013;11:806–819. doi: 10.1016/j.scr.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 27.Beyer K, Klingenberg M. ADP/ATP carrier protein from beef heart mitochondria has high amounts of tightly bound cardiolipin, as revealed by 31P nuclear magnetic resonance. Biochemistry. 1985;24:3821–3826. doi: 10.1021/bi00336a001. [DOI] [PubMed] [Google Scholar]
- 28.Eble KS, Coleman WB, Hantgan RR, Cunningham CC. Tightly associated cardiolipin in the bovine heart mitochondrial ATP synthase as analyzed by 31P nuclear magnetic resonance spectroscopy. J Biol Chem. 1990;265:19434–19440. [PubMed] [Google Scholar]
- 29.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metabol. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 31.Hofer A, et al. Defining the action spectrum of potential PGC-1α activators on a mitochondrial and cellular level in vivo. Human Mol Genet. 2014;23:2400–2415. doi: 10.1093/hmg/ddt631. [DOI] [PubMed] [Google Scholar]
- 32.Brandner K, et al. Taz1, an outer mitochondrial membrane protein, affects stability and assembly of inner membrane protein complexes: Implications for Barth Syndrome. Mol Biol Cell. 2005;16:5202–5214. doi: 10.1091/mbc.E05-03-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McKenzie M, Lazarou M, Thorburn DR, Ryan MT. Mitochondrial respiratory chain supercomplexes are destabilized in Barth syndrome patients. J Mol Biol. 2006;361:462–469. doi: 10.1016/j.jmb.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 34.Pereira Da Silva AP, et al. Inhibition of energy-producing pathways of HepG2 cells by 3-bromopyruvate. Biochem J. 2009;417:717–726. doi: 10.1042/BJ20080805. [DOI] [PubMed] [Google Scholar]
- 35.Macchioni L, Davidescu M, Roberti R, Corazzi L. The energy blockers 3-bromopyruvate and lonidamine: effects on bioenergetics of brain mitochondria. J Bioenerg Biomembr. 2014;46:389–394. doi: 10.1007/s10863-014-9577-5. [DOI] [PubMed] [Google Scholar]
- 36.Davidescu M, et al. Bromopyruvate mediates autophagy and cardiolipin degradation to monolyso-cardiolipin in GL15 glioblastoma cells. J Bioenerg Biomembr. 2012;44:51–60. doi: 10.1007/s10863-012-9411-x. [DOI] [PubMed] [Google Scholar]
- 37.Valianpour F, et al. Linoleic acid supplementation of Barth syndrome fibroblasts restores cardiolipin levels: Implications for treatment. J Lipid Res. 2003;44:560–566. doi: 10.1194/jlr.M200217-JLR200. [DOI] [PubMed] [Google Scholar]
- 38.Wang G, et al. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nature Medicine. 2014;20:616–623. doi: 10.1038/nm.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schlame M, Horvath L, Vigh L. Relationship between lipid saturation and lipid-protein interaction in liver mitochondria modified by catalytic hydrogenation with reference to cardiolipin molecular species. Biochem J. 1990;265:79–85. doi: 10.1042/bj2650079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Acin-Perez R, Enriquez JA. The function of the respiratory supercomplexes: The plasticity model. Biochim Biophys Acta. 2014;1837:444–450. doi: 10.1016/j.bbabio.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 41.Spencer CT, et al. Cardiac and clinical phenotype in Barth syndrome. Pediatrics. 2006;118:e337. doi: 10.1542/peds.2005-2667. [DOI] [PubMed] [Google Scholar]
- 42.Clarke SL, et al. Barth syndrome. Orphanet J Rare Dis. 2013;8:23. doi: 10.1186/1750-1172-8-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soustek MS, et al. Characterization of a transgenic short hairpin RNA-induced murine model of tafazzin deficiency. Human Gene Therapy. 2011;22:865–871. doi: 10.1089/hum.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Acehan D, et al. Cardiac and skeletal muscle defects in a mouse model of human Barth syndrome. J Biol Chem. 2011;286:899–908. doi: 10.1074/jbc.M110.171439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bligh EF, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 46.Sun G, et al. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometric analysis of cellular glycerophospholipids enabled by multiplexed solvent dependent analyte-matrix interactions. Anal Chem. 2008;80:7576–7585. doi: 10.1021/ac801200w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kharroubi AT, Masterson TM, Aldaghlas TA, Kennedy KA, Kelleher JK. Isotopomer spectral analysis of triglyceride fatty acid synthesis in 3T3-L1 cells. Am J Physiol. 1992;263:E667–E675. doi: 10.1152/ajpendo.1992.263.4.E667. [DOI] [PubMed] [Google Scholar]
- 48.Wittig I, Braun HP, Schägger H. Blue native PAGE. Nature Protocols. 2006;1:418–428. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
- 49.Selbach M, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 50.Zhang G, Deinhardt K, Chao MV, Neubert TA. Study of neurotrophin 3 signaling in primary cultured neurons using multiplex stable isotope labeling with amino acids in cell culture. J Proteome Res. 2011;10:2546–2554. doi: 10.1021/pr200016n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature Biotechnology. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 52.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.