Abstract
Introduction
Pulmonary complications due to infection and Idiopathic Pneumonia Syndrome (IPS), a non-infectious lung injury in the hematopoietic stem cell transplant (HSCT) recipients, are frequent causes of transplant-related mortality and morbidity. Our objective was to characterize the global bronchoalveolar lavage fluid (BALF) protein expression of IPS to identify proteins and pathways that differentiate IPS from infectious lung injury following HSCT.
Methods
We studied 30 BALF samples from patients who developed lung injury within 180 days of HSCT, or cellular therapy transfusion (natural killer cell transfusion). Adult subjects were classified as IPS or infectious lung injury by the criteria outlined in the 2011 ATS statement. BALF was depleted of hemoglobin and 14 high abundance proteins, treated with trypsin and labeled with iTRAQ® 8-plex reagent for 2D capillary LC and data dependent peptide tandem MS on an Orbitrap Velos system in HCD mode. Protein identification employed a target-decoy strategy using ProteinPilot within Galaxy P. The relative protein abundance was determined with reference to a global internal standard consisting of pooled BALF from patients with respiratory failure and no history of HSCT. A variance weighted t-test controlling for a false discovery rate of ≤ 5% was used to identify proteins that showed differential expression between IPS and infectious lung injury. The biological relevance of these proteins was determined by using Gene Ontology (GO) enrichment analysis and Ingenuity Pathway Analysis.
Results
We characterized 12 IPS and 18 infectious lung injury BALF samples. In the five iTRAQ LC-MS/MS experiments 845, 735, 532, 615 and 594 proteins were identified for a total of 1125 unique proteins and 368 common proteins across all five LC-MS/MS experiments. When comparing IPS to infectious lung injury, 96 proteins were differentially expressed. Gene Ontology enrichment analysis showed that these proteins participate in biological processes involved in the development of lung injury after HSCT. These include acute phase response signaling, complement system, coagulation system, LXR / RXR, and FXR/RXR modulation. We identified two canonical pathways modulated by TNF-α, FXR/RXR activation and IL2 signaling in macrophages. The proteins also mapped to blood coagulation, fibrinolysis, and wound healing; processes that participate in organ repair. Cell movement was identified as significantly overrepresented by proteins with differential expression between IPS and infection.
Conclusion
The BALF protein expression in IPS differed significantly from infectious lung injury in HSCT recipients. These differences provide insights into mechanisms that are activated in lung injury in HSCT recipients and suggest potential therapeutic targets to augment lung repair.
Keywords: IPS, BAL, Proteomics, Bioinformatics, Pathways
INTRODUCTION
Hematopoietic stem cell transplant (HSCT) is a potentially curative treatment for otherwise fatal hematologic and lymphoid malignancies. Pulmonary complications have been reported in 40% to 60% of HSCT recipients, and may contribute to death in over one-third of these cases [1-4]. The advent of non-myeloablative and reduced intensity conditioning regimens have decreased the duration of neutropenia and lowered the incidence of infectious complications though the mortality from these complications remains high [5]. Idiopathic Pneumonia Syndrome (IPS) is a non-infectious lung injury found in HSCT recipients. In 2011, the American Thoracic Society (ATS) updated the definition of IPS to include widespread alveolar injury in the absence of infection and the absence of cardiac, renal or iatrogenic etiology [6]. Historically, the cumulative incidence of IPS has been reported to be 3-15% in the first 120 days after HSCT. In a more recent study, the cumulative incidence of IPS has been shown to be lower following nonmyeloablative conditioning (2.2%) versus myeloablative conditioning (8.4%). Once established however it was associated with a high mortality [7].
IPS is less common than infectious lung injury but has a poorer prognosis with a mortality rate of 60-80% [6]. IPS occurs in up to 15% of allogeneic HSCT recipients after myeloablative conditioning [7-11]. Although it is less frequent with autologous HSCT [12, 13], the mortality of IPS remains high at approximately 80% [7, 11, 14]. The median time to development of IPS has been reported to be 19 days after allogeneic (range 4-106 days) and 63 days after autologous HSCT (range 7-336); the period when the risk of infectious complications is also high. Escalated immunosuppression with high dose steroids [15] or TNF blockade [11, 16, 17] are current treatments for IPS. As these agents may worsen infectious lung injury, it is critical to differentiate infection from IPS.
The goal of this study was to apply state-of-the-art protein expression profiling tools in cases of lung injury following HSCT or cellular therapy infusion to identify the proteins and biological processes that differentiate IPS from infectious lung injury. We performed comprehensive label-based quantitative protein profiling of BALF in patients undergoing HSCT. Our hypothesis was that subsets of proteins expressed in the BALF represent the biological processes responsible for lung injury and recovery. To identify these processes we performed Gene Ontology enrichment analysis and pathway analysis on proteins that showed differences in abundance in IPS compared to infectious lung injury.
METHODS
Study population
The University of Minnesota Institutional Review Board (IRB) Human Subjects Committee approved this study. Subjects were recruited from the Adult Blood and Marrow Transplant unit at the University of Minnesota Medical Center from November 2009 to June 2013. We included subjects within 180 days of HSCT who underwent a clinically indicated bronchoscopy for whom excess BALF was available. This period represents a high risk period for lung injury and has been used in a prior interventional study of IPS [16]. There was no exclusion based on underlying hematological disease, donor source, the degree of HLA match and conditioning regimen [16, 17]. Demographic and transplant characteristics of the study population are presented in Table 1.
Table 1.
Demographic and transplant characteristics of study subjects
| IPS | Infection | p-value | |
|---|---|---|---|
| Recipient age (mean ± SD) | 47 ± 15 | 49 ± 16 | 0.727 |
| Gender (M/F) and % | 3/9 (25 / 75%) | 6/10 (37.5 / 62.5%) | 0.48 |
| Diagnosis | 0.61 | ||
| Acute Leukemia (AML/ALL) | 7 | 12 | |
| Lymphoma (HD/NHL) | 2 | 2 | |
| MDS/ Myeloproliferative disorder (MDS,myelofibrosis, PV) | 2 | 2 | |
| Chronic Leukemia (CLL) | 1 | 0 | |
| Type of transplant | 0.92 | ||
| Autologous | 1 | 1 | |
| Allogeneic | 10 | 13 | |
| Haploidentical NK cell infusion | 1 | 2 | |
| Donor Type | 0.66 | ||
| Unrelated donor | 8 | 13 | |
| Autologous | 1 | 1 | |
| Haploidentical | 3 | 2 | |
| Graft type | |||
| DUCBT | 5 | 13 | 0.067 |
| PBSCT | 5 | 1 | |
| Marrow | 1 | 0 | |
| Haploidentical NK cell infusion | 1 | 2 | |
| HLA match | 0.003 | ||
| Matched | 3 | 0 | |
| Mismatched | 5 | 16 | |
| Haploidentical | 3 | 0 | |
| Conditioning | 0.13 | ||
| Myeloablative | 3 | 9 | |
| Reduced intensity conditioning | 9 | 7 | |
| Recipient / donor CMV status | 0.079 | ||
| Positive / Negative | 3 | 11 | |
| Positive / Positive | 3 | 0 | |
| Negative / Negative | 4 | 4 | |
| Negative / Positive | 1 | 0 | |
| Positive (autologous transplant) | 1 | 1 | |
chi-square test for all except age.
Amongst the study subjects, 2 underwent autologous HSCT, 23 allogeneic HSCT, and three patients were enrolled in a clinical trial of haploidentical natural-killer cell infusion for refractory acute myeloid leukemia. Twelve patients underwent myeloablative conditioning, and 16 underwent reduced intensity conditioning before HSCT. Graft sources included bone marrow (1), peripheral blood stem cells (6), umbilical cord blood (18) and natural killer cell (3). All BALF samples were collected from clinically indicated bronchoscopies performed using a standard protocol [18, 19] with excess, cell-free supernatant used for these studies. The diagnosis of IPS or infectious lung injury was made using the criteria established by the American Thoracic Society [6] (Table 2). The patients had evidence of widespread alveolar injury, abnormal respiratory physiology and no evidence of pulmonary edema or volume overload or acute kidney injury as the primary cause of respiratory dysfunction. A diagnosis of IPS requires the absence of bacterial, viral, mycobacterial or fungal infection assessed by cultures, cytology or PCR-based studies. A diagnosis of infectious lung injury was made based on evidence of a pathogenic organism. In this group, two subjects had isolation of fungal organisms only, three had viral organisms only, six had bacterial organisms only and the rest of the subjects had a polymicrobial infection (isolation of bacteria with either fungal and/ or viral organisms). This study was conceived and the protein profiling completed prior to a recent publication where advanced genetic methodologies demonstrate viral pathogens in IPS [20].
Table 2.
IPS diagnostic criteria
| 1. Presence of widespread alveolar injury |
| a. CT of CXR evidence of bilateral of multilobar infiltrates |
| b. Abnormal respiratory physiology based on room air O2 saturation of < 93% or need for supplemental oxygen to keep O2 > 93 |
| 2. Absence of lower respiratory tract infection assessed by sputum or BAL fluid negative for pathogenic bacterial and nonbacterial organism1 |
| a. Gram stain, fungal stain, AFB stain |
| b. Bacterial2, fungal and viral (RSV CMV,adenovirus, parainfluenza, influenza A and B, rhinovirus). |
| c. Viral PCR studies for CMV, HSV, VZV, HHV-6 |
| d. Pneumocystis jeroveci assay by cytology |
| 3.Absence of cardiac dysfunction (echo, pro BNP), acute kidney injury and fluid overload |
mixed oral flora and rare Candida or penicillium did not rule out IPS.
Any positive culture as quantitative bacterial cultures not always done.
A global internal standard was used to compare relative protein abundance across multiple isobaric tagging for relative and absolute quantification- two dimensional liquid chromatography tandem mass spectrometry (iTRAQ 2DLC-MS/MS) experiments. This consisted of pooled BALF from 27 cases (‘mastermix”) with respiratory failure without previous HSCT and who did not meet the criterion for acute respiratory distress syndrome (except in one case with ARDS).
Sample preparation
Cell and debris free BALF supernatant was stored at −80°C within 60 minutes of collection. BALF samples containing at least 1.2 mg of proteins were processed for LC-MS/MS from individual patients employing a protocol previously published with minor modifications [18]. BALF was concentrated and desalted using Amicon 3-MWCO filters. Hemoglobin depletion was performed with Hemoglobind (BioTech Support Group LLC, Monmouth Junction, NJ) per the manufacturer's instructions. Subsequent processing was similar to our prior study with immunoaffinity depletion of high abundance proteins to reduce the dynamic range and appropriate buffer exchanges for labeling with iTRAQ reagent [18].
iTRAQ labeling and 2D LC-MS/MS
After depletion the remaining medium and low abundant BALF proteins were digested with trypsin-gold (Promega cat#V5280), dried and suspended in 0.1- 0.2% formic acid (pH < 3) and MCX cation exchange (Oasis MCX Cartridge, Waters, Milford, MA, Cat no 186000254) performed to remove SDS. Proteins were eluted and labeled with eight-plex iTRAQ reagent per the manufacturer's (AB Sciex, Framingham, MA) instructions as described previously [18, 21]. The relative protein abundance was determined with reference to the pooled “mastermix” across all iTRAQ LC-MS/MS runs. In each LC-MS/MS experiment, two iTRAQ reporter ion channels were labeled with the mastermix to assure the accuracy of fold change measurements while the remaining six channels contained study samples. Thus, to characterize 30 samples in this study, we performed five separate iTRAQ LC-MS/MS experiments. The labeling strategy for the 30 BALF samples studied is outlined in Supplemental Table S1. To prevent reporter ion signal (channel) bias the mastermix, IPS, and infectious lung injury samples were randomly placed in different iTRAQ reporter ion channels in each experiment.
Each iTRAQ-labeled peptide mixture was purified with an MCX cartridge before off-line peptide separation in the first dimension and if needed C18 stage tipping. Peptides were separated offline into 15 different peptide-containing fractions collected in 2-minute intervals on a C18 Gemini column (Phenomenex, Torrance, CA) at pH 10. Peptide fractions were concentrated, purified by the Stage Tip procedure [22] with Empore SDB-RPS extraction disks [mixed mode strong cation exchange and reversed phase], 3M (St. Paul MN), and separated in the 2nd dimension by C18 reversed phase capillary LC with a nano LC system (Eksigent, Dublin, CA) online with MS. Data-dependent acquisition was performed on an Orbitrap Velos system with HCD (higher-energy collision induced dissociation) activation for peptide tandem MS. LC and MS experimental details were previously reported, with the exception that the activation time was 20 msec [18, 23].
Database search for protein identification
.RAW files obtained directly from the Orbitrap Velos mass spectrometer were imported into GalaxyP ( https://usegalaxyp.org/ for public instance) for further processing (as described in emz.umn.edu/ppingp). Within GalaxyP, all .RAW files together (using a multifile format that has been recently changed to dataset collection) were converted to mzml format using msconvert and then into ProteinPilot compatible Mascot Generic Format (MGF) files with preselected iTRAQ reporter ions. The MGF files were searched against the target-decoy version of Human UniProt database along with contaminant protein sequences (84,838 target sequences in total; July 2014) using ProteinPilot version 4.5. Following search parameters were used: Sample Type: iTRAQ 8-plex (peptide labeled); Cys-alkylation: MMTS; Instrument: Orbi MS, Orbi MS/MS; run quant; bias correction on; search focus on biological modifications and amino-acid substitutions; thorough search and with a detected protein threshold (Unused Protscore (Conf)): 10%. The ProteinPilot searches and subsequent generation of Proteomics System Performance Evaluation Pipeline Software (PSPEP) –FDR reports and protein and peptide level summaries were generated within Galaxy-P as previously described [18, 24]. The mass spectrometry data have been deposited to the ProteomeXchange Consortium [25] via the PRIDE partner repository with the dataset identifier PXD002437
The results of multiple iTRAQ LC-MS/MS experiments were aligned to compare protein-level quantitative data using Protein Alignment Template vs. 2.00p (AB Sciex) [26]. The Protein Alignment Template provides the overlap of proteins across multiple iTRAQ LC-MS/MS experiments by matching proteins in a ‘reference master list’ to the ‘test list’ of proteins identified in each iTRAQ LC-MS/MS run. For this alignment, we created a master reference list by performing a database search using .RAW files from two iTRAQ LC-MS/MS experiments i.e. iTRAQ experiment 1 and 5. To ensure that the proteins in this list are of high ID quality, we used a local FDR ≤ 5% as a threshold for the master reference protein list. As per the recommendation of the Protein Alignment Template, for assembling of feature table with quantitative values, the threshold of ≤ 5% global FDR was used for ‘test list’ consisting of individual sets of the five-iTRAQ LC-MS/MS experiments. The Protein Alignment Template resulted in aligning of the ratios, p-values and error factors of the proteins across replicate experiments by using accession numbers of isoforms within protein summary and UniProt database.
Statistical analysis
Identification of differentially expressed proteins between IPS and infectious lung injury
We performed inverse variance weighted ratio test to account for the peptide level variance in fold changes measured for each protein across multiple iTRAQ runs. An inverse variance weighted t-test was performed to identify the proteins that were differentially expressed between two groups (IPS vs. infection). This model allows for comparison of averaged protein ratios across the five-iTRAQ LC-MS/MS runs. However, to avoid overfitting, we did not compare the protein ratios in the patients with repeat BALF sampling, as they were not analyzed on the same iTRAQ LC-MS/MS run. We controlled for multiple comparisons by using FDR [27] as done previously [18]. An R script employed for this statistical analysis is included in the supplementary material as R script.
Computational Analysis
To gain insight into the biological significance of the differentially expressed proteins, we used the Database for Annotation, Visualization, and Integrated Discovery (DAVID, http://david.abcc.ncifcrf.gov) [28] as previously reported [18, 21]. DAVID generates an enrichment score for a group of genes indicating annotation term member associations in a given experiment. We used the ‘highest stringency’ Functional Annotation Clustering algorithm and focused on clusters with an enrichment score ≥ 1.3 because it is equivalent to a non-log scale p-value of 0.05 (carried out on 02/11/2016). To confirm the findings observed in DAVID we also performed functional analysis using Ingenuity Pathway Analysis (IPA® QIAGEN, Redwood City www.quiagen.com/ingenuty Build 366632M, Version 26127183, carried out on 2/12/2016). We focused on the protein subsets represented in canonical pathways, upstream regulators and ‘molecular and cellular functions’. In IPA, the significance of the association between the differentially expressed proteins and the canonical pathway was measured in 2 ways: 1) a ratio of the number of BALF proteins that mapped to the pathway divided by the total number of genes assigned to that canonical pathway as it provides information about the depth of association; 2) a Benjamini and Hochberg corrected p-value (obtained using the right-tailed Fisher Exact Test) determining the probability that the association between the BALF proteins and the signaling canonical pathway/ biological function was explained by chance alone. All pathways with an FDR ≤ 0.05 are reported (equivalent to –log (B-H )p-value > 1.3). Canonical pathways over-represented in allogeneic HSCT were compared to all HSCT (allogeneic and autologous) recipients utilizing IPA comparison analysis algorithm.
Identifying the universe of BALF proteins
The Protein Alignment Template uses a reference master list, which introduces a certain degree of arbitrariness into the result. Thus, to match corresponding proteins across runs, we also implemented our own approach that seeks to overcome this limitation. This method uses the ProteinPilot output files that characterize each protein regarding the peptides that support its identification and matches proteins across runs when they share peptides. FDR filtering, as described previously, was performed before analysis to eliminate those proteins with less reliable identification. More specifically, our approach creates a graph, where each protein from any run is a node in the graph. Links are established between two nodes (proteins) for those nodes that share at least one peptide. We eliminated proteins that have links to protein within the same run or more than two proteins within another run. This resulted in the elimination of a small number of proteins (~8). Each connected component of the graph was interpreted as representing a single protein and was used to align the ratios and P-values of the proteins across replicate experiments.
RESULTS
Characteristics of study participants
In this study, we characterized BALF from HSCT subjects collected between November 2009 and June 2013. Twelve samples were classified as IPS and 18 BALF samples were classified as infectious lung injury. Two cases of IPS met the criterion for diffuse alveolar hemorrhage suggesting vascular injury and the remaining patients in the IPS groups had findings consistent with injury to the pulmonary parenchyma. The median time from transplantation to BALF sample collection did not differ between IPS (median 33 days, range 15-138 days) and infectious lung injury (median 51 days, range 8-179 days). The indication and type of transplantation are shown in Table 1 and the subject characteristics are shown in Table 3. Between the two groups, there was no difference in the age, time after HSCT when BALF samples were collected, the total BALF leukocyte count and the percent of BALF neutrophils, lymphocytes, and monocytes. Six cases had a diagnosis of GVHD at the time of BALF collection (IPS=1, infectious lung injury =5) and were on corticosteroids with prednisone equivalent dose ranging from 25-112.5mg/day.
Table 3.
Clinical characteristics of the study participants
| IPS | Infectious Lung Injury | P-value | |
|---|---|---|---|
| Number of BAL Samples | 12 | 18 | |
| Time from transplant to bronchoscopy (days) | 60 ± 39 | 51 ± 50 | 0.60 |
| S Creatinine (mg/dl) on day of bronchoscopy | 1.2 ± 0.6 | 1.4 ± 0.8 | 0.47 |
| BALF leucocytes (per μl) | 314 ± 337 | 203 ± 173 | 0.24 |
| BALF Neutrophils (%) | 29 ± 35 | 20 ± 30 | 0.47 |
| BALF lymphocytes (%) | 8.7 ± 13 | 4.1 ± 6.9 | 0.23 |
| BAL monocytes (%) | 39 ± 43 | 59 ± 43 | 0.55 |
Protein identified by database searching
Most of the BALF samples in HSCT recipients appeared blood tinged; therefore for the first iTRAQ LC-MS/MS experiment we tested whether removal of hemoglobin (in addition to high abundance proteins) would improve the depth of coverage. Hemoglobin removal improved protein identification to 845 proteins at 1% global FDR compared to 496 proteins with high abundance protein depletion alone. Subsequent iTRAQ LC-MS/MS experiments were performed with hemoglobin removal followed by high abundance protein depletion.
Table 4 shows the ProteinPilot PSPEP FDR summary with the number of spectra, peptides and the proteins identified at ≤ 1% global FDR for the five-iTRAQ experiments. The total numbers of proteins identified in each of the five different iTRAQ LC-MS/MS experiments were 845, 735, 532, 615 and 594 respectively. The ProteinPilot summary report of the proteins identified is included in the supplemental information Table S2. A total of 1125 unique proteins were identified in the five separate experiments (Table S3) of which 368 proteins were present in all five LC-MS/MS experiments.
Table 4.
PSPEP protein summary report for number of spectra, peptides and proteins identified at ≤ 1% global FDR
| Spectra | Peptides | Proteins | |
|---|---|---|---|
| iTRAQ1 | 21933 | 8635 | 845 |
| iTRAQ2 | 24691 | 9466 | 735 |
| iTRAQ3 | 21826 | 5842 | 532 |
| iTRAQ 4 | 17893 | 7285 | 615 |
| iTRAQ 5 | 16789 | 6515 | 594 |
Proteins differentially expressed in IPS
The 793 proteins identified in the master reference list at ≤ 5% local FDR were aligned to the proteins from individual iTRAQ LC-MS/MS experiment (Table S4, Reference list alighed to sets tab). Of these 793 proteins, quantitative information was present to perform inverse variance weighted t-test on 509 proteins and 111 proteins were differentially expressed when all patients (autologous and allogeneic HSCT) with IPS were compared to infectious lung injury, controlling for an FDR ≤ 5% (Table S5, FDR BMT tab). We manually removed the high abundance proteins that should have been eliminated by the immune-depletion column along with misidentified or contaminant proteins from the list of 111 proteins resulting in 96 differentially expressed proteins (Table S5, 5% FDR (96) tab). Proteins representing the greatest difference in abundance between IPS and infectious lung injury are shown in Table 5.
Table 5.
Selected proteins with highest difference in abundance in BALF of cases with IPS compared to infectious lung injury.
| Uniprot Accession Number | Protein Name | Mean variance weighted fold change IPS: mastermix | Mean Variance weighted Fold change Infection: marstermix | FDR (Comparing fold change in IPS vs infectious lung injury) | Fold Change IPS: Infection |
|---|---|---|---|---|---|
| Proteins that are high in IPS compared to infectious lung injury (ten proteins with highest differential expression) | |||||
| tr|F6SYF8|F6SYF8_HUMAN | Dickkopf-related protein 3 | 8.87 | 2.71 | 8.98E-03 | 3.27 |
| sp|P24158|PRTN3_HUMAN | Myeloblastin | 1.09 | 0.34 | 5.27E-03 | 3.25 |
| sp|Q9BTY2|FUCO2 HUMAN | Plasma alpha-L-fucosidase | 4.10 | 1.53 | 8.05E-03 | 2.69 |
| sp|O95497|VNN1_HUMAN | Pantetheinase | 4.97 | 1.87 | 1.07E-02 | 2.66 |
| sp|P14151|LYAM1_HUMAN | L-selectin | 2.33 | 0.89 | 1.17E-03 | 2.63 |
| sp|P13598|ICAM2_HUMAN | Intercellular adhesion molecule 2 | 2.63 | 1.25 | 3.64E-02 | 2.10 |
| sp|P22352|GPX3_HUMAN | Glutathione peroxidase 3 | 0.96 | 0.47 | 8.41E-06 | 2.06 |
| sp|P48637|GSHB_HUMAN | Glutathione synthetase | 2.15 | 1.10 | 1.59E-03 | 1.96 |
| sp|Q6UX71|PXDC2 HUMAN | Plexin domain-containing protein 2 | 4.16 | 2.21 | 2.66E-02 | 1.89 |
| sp|P01033|TIMP1_HUMAN | Metalloproteinase inhibitor 1 | 0.69 | 0.37 | 2.49E-04 | 1.85 |
| Proteins that are high in infectious lung injury compared to IPS (ten proteins with highest differential expression) | |||||
| tr|H0Y7A7|H0Y7A7_HUMAN | Calmodulin (Fragment) | 0.51 | 0.90 | 1.42E-03 | 0.57 |
| sp|P07108-5|ACBP_HUMAN | Isoform 5 of Acyl-CoA-binding protein | 1.02 | 1.78 | 2.10E-02 | 0.57 |
| sp|P61626|LYSC_HUMAN | Lysozyme C | 0.27 | 0.47 | 2.57E-05 | 0.57 |
| sp|P01011|AACT_HUMAN | Alpha-1-antichymotrypsin | 0.68 | 1.22 | 3.31E-06 | 0.56 |
| sp|Q8WUM4|PDC6I_HUMAN | Programmed cell death 6-interacting protein | 1.12 | 2.01 | 2.49E-04 | 0.56 |
| sp|Q16651|PRSS8_HUMAN | Prostasin | 1.80 | 3.23 | 8.98E-03 | 0.56 |
| tr|X6R8F3|X6R8F3_HUMAN | Neutrophil gelatinase-associated lipocalin | 0.30 | 0.54 | 1.30E-03 | 0.56 |
| sp|P15311|EZRI_HUMAN | Ezrin | 0.83 | 1.49 | 2.56E-08 | 0.55 |
| sp|P05109|S10A8_HUMAN | Protein S100-A8 SV=1 | 0.15 | 0.28 | 6.90E-04 | 0.54 |
| sp|P08118|MSMB_HUMAN | Beta-microseminoprotein | 0.81 | 1.87 | 3.10E-03 | 0.43 |
In this study, two subjects had autologous HSCT one each in the infection and IPS groups. Since lung injury in autologous HSCT recipients is felt to be different than that in allogeneic HSCT, we analyzed our data after removing the two autologous HSCT cases. In the allogeneic HSCT recipients, we identified 127 differentially expressed proteins controlling for an FDR ≤ 5%. Eighty-five proteins differentially expressed in the allogeneic HSCT overlapped with the differentially expressed proteins identified in the analysis performed on all HSCT recipients (allogeneic and autologous).
Biological processes represented by differentially expressed proteins
To gain insight into biological processes that differ in IPS and infection, we performed Functional Annotation Clustering analysis on the differentially expressed proteins employing ‘highest stringency’ algorithm in DAVID. Fifteen biological modules (functional annotation clusters with an enrichment score > 1.3) were identified (Table S6). Our analysis revealed proteins that mapped to processes involved in inflammation mediated by lymphocytes, complement activation and B cell-mediated adaptive immune response (clusters 1, 5 and 14). Other functional modules that were identified included those involved in blood coagulation/wound healing (cluster 3), cell migration (cluster 6,11), and apoptosis (cluster 4,15). There were modules involved in cholesterol / triglyceride metabolism (cluster 7, 12), lipid / cholesterol transport (cluster 8, 9) and oxidative stress / cellular ion homeostasis (cluster 2,10). We also identified one module involved in regulation of endocytosis (cluster 13).
To understand the relative impact of changes in protein levels in the context of well-characterized pathways and to independently test the validity of the findings observed using DAVID, we performed IPA core analysis; focussing mainly on molecular and cellular functions, canonical pathways and the upstream regulators. Several of the GO terms that were over- represented by the differentially expressed proteins in DAVID were also identified by IPA core analysis. The top five molecular and cellular functions represented by the differentially expressed proteins included cellular movement, cell-to-cell signaling and interaction, cell death and survival, free radical scavenging and lipid metabolism.
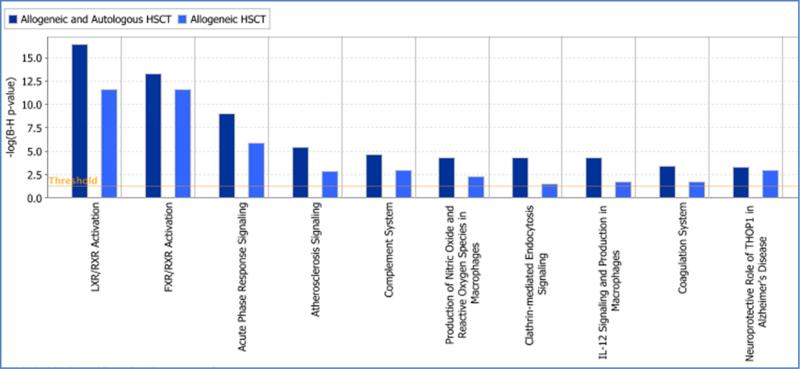
Controlling for an FDR ≤ 5% (equivalent to –log (B-H p-value) > 1.3), we identified 17 IPA canonical pathways that were represented by differentially expressed proteins (Table 6). These included processes identified by GO enrichment analysis such as acute phase response signaling (APRS), complement system, coagulation system, intrinsic and extrinsic prothrombin activation, production of nitric oxide and reactive oxygen species in macrophages, granulocyte adhesion and diapedesis (leukocyte migration) and glutathione redox reactions. Additionally, pathways in the IPA knowledge base that were affected included clathrin-mediated endocytotic signaling, IL-12 signaling and production in macrophages. Additionally, we performed IPA core analysis on the differentially expressed proteins identified in allogeneic HSCT. Though there were differences in the level of significance of enrichment (Table S7), the canonical pathways overrepresented in allogeneic HSCT patients overlapped with those identified in all HSCT recipients (Figure 1).
Table 6.
Ingenuity Pathway Analysis Canonical pathways represented by proteins that are differentially expressed between IPS and infectious lung injury.
| Ingenuity Canonical Pathways | −log(B-H p-value) | Ratio$ | z-score | Molecules |
|---|---|---|---|---|
| LXR/RXR Activation | 1.64E01 | 1.32E-01 | 1.500 | KNG1, APOE, TTR, HPX, APOA4, C3, APOH, A1BG, PON1, LYZ, ITIH4, S100A8, GC, CLU, APOD, AGT |
| FXR/RXR Activation | 1.33E01 | 1.11E-01 | NaN | KNG1, APOE, TTR, HPX, APOA4, C3, APOH, A1BG, PON1, ITIH4, GC, CLU, APOD, AGT |
| Acute Phase Response Signaling | 9.03E00 | 7.1E-02 | −0.378 | PLG, TTR, HPX, ITIH3, C3, APOH, ITIH4, SERPINA3, HRG, F2, AGT, C5 |
| Atherosclerosis Signaling | 5.38E00 | 6.45E-02 | NaN | APOE, PON1, LYZ, APOA4, S100A8, CLU, PRDX6, APOD |
| Complement System | 4.63E00 | 1.35E-01 | 0.447 | CD55, C3, CFI, CFH, C5 |
| Production of Nitric Oxide and Reactive Oxygen Species in Macrophages | 4.32E00 | 4.44E-02 | 0.000 | APOE, PON1, LYZ, APOA4, S100A8, CLU, SIRPA, APOD |
| Clathrin-mediated Endocytosis Signaling | 4.3E00 | 4.32E-02 | NaN | APOE, PON1, LYZ, APOA4, S100A8, F2, CLU, APOD |
| IL-12 Signaling and Production in Macrophages | 4.27E00 | 5.26E-02 | NaN | APOE, PON1, LYZ, APOA4, S100A8, CLU, APOD |
| Coagulation System | 3.44E00 | 1.14E-01 | 0.000 | KNG1, PLG, PROS1, F2 |
| Neuroprotective Role of THOP1 in Alzheimer's Disease | 3.25E00 | 1E-01 | NaN | KNG1, PLG, SERPINA3, AGT |
| Intrinsic Prothrombin Activation Pathway | 2.32E00 | 1.03E-01 | NaN | KNG1, PROS1, F2 |
| Granulocyte Adhesion and Diapedesis | 1.73E00 | 2.82E-02 | NaN | SELL, ICAM2, EZR, MSN, C5 |
| Agranulocyte Adhesion and Diapedesis | 1.64E00 | 2.65E-02 | NaN | SELL, ICAM2, EZR, MSN, C5 |
| γ-glutamyl Cycle | 1.6E00 | 1.33E-01 | NaN | GSS, ANPEP |
| Phagosome maturation | 1.6E00 | 3.33E-02 | NaN | CALR, LAMP2, PRDX1, PRDX6 |
| Extrinsic Prothrombin Activation Pathway | 1.6E00 | 1.25E-01 | NaN | PROS1, F2 |
| Glutathione Redox Reactions I | 1.47E00 | 1.05E-01 | NaN | GPX3, PRDX6 |
Ratio is the number of genes in a given pathway divided by the total number genes in that pathway in a reference gene set
Figure 1. Canonical pathways overrepresented by proteins differentially expressed in infectious lung injury compared to IPS.
Pathways over-represented in allogeneic HSCT recipients (light blue bars) compared to all (allogeneic and autologous) HSCT recipients (dark blue bars) are shown. The y-axis is the level of significance of enrichment represented as the negative log of Benjamini and Hochberg corrected p-value where 1.3 is equivalent to a non-log p-value of 0.05.
Upstream regulator analysis in IPA demonstrated that nitrofurantoin, PD98059 (2-(2-amino-3-methoxyphenyl)-4H-1-benzopyran-4-one, an ERK /MAPK regulator) lipopolysaccharide (LPS), FOXA2 (forkhead box A2) and tretinoin were the top five upstream regulators of the differentially expressed proteins between IPS and infectious lung injury. TNF was also identified as an upstream regulator of twenty-five of the differentially expressed proteins by this analysis.
DISCUSSION
This is the first study that characterizes the BALF protein profile by mass spectrometry in HSCT recipients. In this study, we show broad coverage of the BALF proteome with the identification of 1125 proteins. We also identified differentially expressed proteins that provide insights into the biological processes involved in the development of lung injury after HSCT. IPS continues to have a high mortality and rapid diagnosis and novel insights into the biology of the disease process will advance our goal of improving the outcomes of HSCT recipients with lung injury.
Four of the five most significant canonical pathways that we identified in our BALF study were previously reported to be relevant in IPS from mass spectrometry studies performed on blood samples comparing IPS to subjects with no lung damage following HSCT [29]. The pathways identified in blood include acute phase reactants (APRS), complement system, LXR / RXR activation and FXR/RXR activation. As BALF is the most proximate fluid to the site of injury, it is not surprising that BALF proteins are annotated to pathways that previously were implicated in the development of IPS. This also supports the appropriateness of BALF as a biofluid for discovery studies in lung injury as systemic changes are identified both in the BALF and blood. A positive z-score in IPA predicts that the LXR/RXR is activated in IPS compared to infectious lung injury.
TNF- α plays a major role in the development of IPS and etanercept has been an effective agent in IPS [11, 15, 17]. However, the beneficial response to TNF blockade is not universal as a recent study reported that patients with IPS already receiving corticosteroids did not have therapeutic benefit with etanercept [16]. In our study, although we did not identify TNF- α at the threshold for protein identification, there were indicators of TNF-α involvement in IPS after HSCT. In the upstream regulator analysis, , LPS, a powerful stimulator of TNF-α was identified as the top regulator of the proteins that are differentially expressed between IPS and infectious lung injury. We also identified two canonical pathways that are targets for TNF-α blockade. These pathways were Farsenoid X Receptor (FXR) / Retinoid X Receptor (RXR) activation and IL12 signaling and production in macrophages. FXR is a nuclear receptor that In the liver and intestinal epithelia plays a crucial role in lipoprotein and bile acid metabolism [30]. Increasing literature supports the role of FXR in inflammation mediated by NF-κB in the liver [31] and intestinal epithelium [32]. In the lung, FXR recently was found to be expressed on the pulmonary endothelial cells [33, 34] and it protects against LPS induced lung injury, possibly via attenuation of P-selectin mediated neutrophil recruitment [34]. Interestingly, FXR ligand attenuated TNF-α induced up-regulation of P-selectin in a pure culture of endothelial cells, suggesting its role in TNF- α induced inflammation. This provides a potential mechanism by which selected patients with IPS respond to TNF-α inhibition. Our study identified L-selectin to be more abundant in IPS though we did not identify P-selectin. These findings will need further exploration.
We also found that differentially expressed proteins mapped to IL-12 signaling in macrophages. IL -12 belongs to a diverse family of cytokines that have both pro-inflammatory and anti-inflammatory roles. In macrophages, IL-12 favors the differentiation of Th1 cells stimulating the adaptive immune response. Also, IL-12 activates MAPK- mediated induction of tumor necrosis factor suggesting that it may be a link between innate and adaptive immune responses (IPA knowledgebase). Our study did not provide adequate information to conclude if this pathway is activated or inhibited and warrants further investigation.
Several biological pathways were identified that could provide mechanistic insights into the lung injury-repair after HSCT. Cell migration / cell movement was identified by both the IPA disease and function algorithm and also by DAVID functional annotation clustering. Alveolar epithelial cell motility is an understudied and likely important mechanism contributing to the re-epithelialization of the alveolus. Ezirin-Radixin- Moesin (ERM) family of proteins could participate in this process and we identified ezrin and moesin in HSCT related lung injury. ERM proteins co-localize in cell matrix adhesion sites, filopodia, and membrane protrusions [35]. ERMs function by binding to and organizing the actin cytoskeleton and in turn, stabilize adherens junctions that are involved in cell migration [36-38]. ERM protein activation requires c-terminal phosphorylation involving the small GTP-binding Rho [39]. Upon activation, the n-terminus of the ERM protein interacts with trans-membrane proteins such as CD 44, with the subsequent stabilization of the actin cytoskeleton or activating signaling molecules. These interactions during lung injury and repair will need better characterization but offer potential therapeutic targets. In the IPA canonical pathway analysis, differentially expressed proteins mapped to RhoGDI signaling and signaling by Rho family of GTPases but this did not reach statistical significance using B and H corrected p-values. This, however, could provide a mechanism by which an injured lung may repair itself. These proteins annotated to RhoGDI signaling included cytoplasmic proteins: beta actin, ezrin, moesin and guanine nucleotide binding protein (all low in IPS) and the membrane proteins: CD44 and cadherin 1 (high in IPS). Rho GDI belongs to a family of small GTPases comprised of Cdc 42, Rac and Rho that regulate several critical processes including cell proliferation, apoptosis, cell differentiation and cell motility, which are important in lung injury-repair cycle. Rho GDI signaling may be involved in regulation of cell migration by regulation of ezrin-radixin-moesin (ERM) proteins.
Limitations of this study include the subjects that were selected based on the availability of excess BALF. We do no have information on patients with lung injury who either did not have excess BALF or who did not undergo a bronchoscopy. We elected to deplete high abundance proteins to enrich medium and low abundance proteins. This is an important and controversial issue in proteomic studies, especially when protein quantification is performed. Depletion of high abundance protein is crucial in data dependent MS acquisition where only selected MS1 precursor ions are isolated for fragmentation and contribute to missing values. As high abundance protein can preclude quantification of less abundant MS1 precursor ions, analyzing undepleted BALF could severely limit the proteome coverage. The downside of depletion of higher abundance protein is that other proteins might be co-depleted. It also increases the amount of sample handling needed before MS data acquisition. In this study, despite using high-resolution MS platforms, the overlap of the proteins identified in the five-iTRAQ runs was partial i.e. there were missing data points across the five iTRAQ LC-MS/MS experiments. For this reason, we cannot compare the patients who were included in the study twice but were no analyzed in the same TRAQ LC-MS/MS run. We expect that with improvement in MS platforms, the depth and overlap in protein identified across multiple LC-MS/MS experiments will improve. More recent methodologies using data-independent acquisition (such as SWATH-MS), also will directly address the limitation of MS platforms. Another limitation of this study is the small number and heterogeneity of study subjects. A larger study with an adequate size of subgroups will allow identifying differences according to the graft source, relatedness and conditioning regimen. Despite these limitations, our study provides proof of concept for future work in this area. We also acknowledge that serum proteins likely contaminated the BALF proteome to some extent in our study. Though this could be addressed by direct comparison of comprehensive protein expression changes in serum and BALF, BALF analysis is a critical first step that provides a framework for future studies of lung dysfunction in HSCT recipients.
CONCLUSIONS
In this study where we characterize the BALF proteome in subjects with lung injury following HSCT, we found differences in the BALF proteome that distinguish IPS from infectious lung injury. These differences likely reflect the underlying mechanisms implicated in the development of lung damage and the repair response that is activated. Though somewhat speculative, we also identify possible mechanisms that could explain the variable response to TNF blockade in IPS. Studies in a larger validation cohort of subjects will likely provide valuable insights into disease biology and also identify novel therapeutic targets for intervention.
Supplementary Material
HIGHLIGHTS.
We characterize the bronchoalveolar lavage fluid protein expression in hematopoietic stem cell transplant recipients with lung injury.
Protein abundance of several proteins differs in subjects with IPS compared to infectious lung injury.
These differences could provide insights into the biological processes that are activated in IPS compared to infectious lung injury.
Proteins that are differentially expressed could be systematically studied to identify a panel to rapidly differentiate IPS from infectious lung injury.
ACKNOWLEDGEMENTS
We greatly appreciate assistance by personnel at Center for Mass Spectrometry and Proteomics (CMSP) – especially Todd Markowski for sample preparation. GalaxyP is maintained by the Minnesota Supercomputing Institute at University of Minnesota and the infrastructure is supported by NSF grant 1147079. This project was supported by University of Minnesota Foundation Biomedical Research Grant (UMF 4158-9216-13, Bhargava PI) and University of Minnesota CTSI KL2 Scholars Program (KL2 RR0333182, Bhargava).
ABBREVIATIONS
- AFB-
Acid-fast bacilli
- ALL
Acute lymphoblastic leukemia
- AML
Acute Myeloid Leukemia
- BALF
Bronchoalveolar lavage fluid
- CLL
Chronic lymphocytic leukemia
- CMV
Cytomegalo virus
- CT
Computed tomography
- CXR-
Chest X Ray
- DAVID
Database for Annotation, Visualization, and Integrated Discovery
- DUCBT
Double umbilical cord blood transplant
- FDR
False Discovery Rate
- HSCT
Hematopoietic stem cell transplant
- HSV
Herpes simplex virus
- iTRAQ
Isobaric tagging for relative and absolute quantification
- IPS
Idiopathic pneumonia syndrome
- LC
Liquid chromatography
- MCX
mixed-mode cationic exchange
- MDS
Myelodysplastic Syndrome
- MGF
Mascot Generic Format
- MMTS
Methyl methanethiosulfonate
- MS
Mass spectrometer
- PBSCT
Peripheral blood stem cell transplant
- PCR
Polymerase chain reaction
- PSPEP
Proteomics System Performance Evaluation Pipeline
- PV
Polycytehemia vera
- RSV
respiratory syncytial virus
- SDS
sodium dodecyl sulfate
- TNF
Tumor necrosis factor
- UCB
Umbilical cord blood transplant
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Soubani AO, Miller KB, Hassoun PM. Pulmonary complications of bone marrow transplantation. Chest. 1996;109:1066–77. doi: 10.1378/chest.109.4.1066. [DOI] [PubMed] [Google Scholar]
- 2.Griese M, Rampf U, Hofmann D, Fuhrer M, Reinhardt D, Bender-Gotze C. Pulmonary complications after bone marrow transplantation in children: twenty-four years of experience in a single pediatric center. Pediatr Pulmonol. 2000;30:393–401. doi: 10.1002/1099-0496(200011)30:5<393::aid-ppul5>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 3.Roychowdhury M, Pambuccian SE, Aslan DL, Jessurun J, Rose AG, Manivel JC, et al. Pulmonary complications after bone marrow transplantation: an autopsy study from a large transplantation center. Archives of pathology & laboratory medicine. 2005;129:366–71. doi: 10.5858/2005-129-366-PCABMT. [DOI] [PubMed] [Google Scholar]
- 4.Kaya Z, Weiner DJ, Yilmaz D, Rowan J, Goyal RK. Lung function, pulmonary complications, and mortality after allogeneic blood and marrow transplantation in children. Biol Blood Marrow Transplant. 2009;15:817–26. doi: 10.1016/j.bbmt.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 5.Nusair S, Breuer R, Shapira MY, Berkman N, Or R. Low incidence of pulmonary complications following nonmyeloablative stem cell transplantation. The European respiratory journal. 2004;23:440–5. doi: 10.1183/09031936.04.00053004. [DOI] [PubMed] [Google Scholar]
- 6.Panoskaltsis-Mortari A, Griese M, Madtes DK, Belperio JA, Haddad IY, Folz RJ, et al. An official American Thoracic Society research statement: noninfectious lung injury after hematopoietic stem cell transplantation: idiopathic pneumonia syndrome. American journal of respiratory and critical care medicine. 2011;183:1262–79. doi: 10.1164/rccm.2007-413ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fukuda T, Hackman RC, Guthrie KA, Sandmaier BM, Boeckh M, Maris MB, et al. Risks and outcomes of idiopathic pneumonia syndrome after nonmyeloablative and conventional conditioning regimens for allogeneic hematopoietic stem cell transplantation. Blood. 2003;102:2777–85. doi: 10.1182/blood-2003-05-1597. [DOI] [PubMed] [Google Scholar]
- 8.Huisman C, van der Straaten HM, Canninga-van Dijk MR, Fijnheer R, Verdonck LF. Pulmonary complications after T-cell-depleted allogeneic stem cell transplantation: low incidence and strong association with acute graft-versus-host disease. Bone Marrow Transplant. 2006;38:561–6. doi: 10.1038/sj.bmt.1705484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keates-Baleeiro J, Moore P, Koyama T, Manes B, Calder C, Frangoul H. Incidence and outcome of idiopathic pneumonia syndrome in pediatric stem cell transplant recipients. Bone Marrow Transplant. 2006;38:285–9. doi: 10.1038/sj.bmt.1705436. [DOI] [PubMed] [Google Scholar]
- 10.Patriarca F, Skert C, Bonifazi F, Sperotto A, Fili C, Stanzani M, et al. Effect on survival of the development of late-onset non-infectious pulmonary complications after stem cell transplantation. Haematologica. 2006;91:1268–72. [PubMed] [Google Scholar]
- 11.Yanik G, Hellerstedt B, Custer J, Hutchinson R, Kwon D, Ferrara JL, et al. Etanercept (Enbrel) administration for idiopathic pneumonia syndrome after allogeneic hematopoietic stem cell transplantation. Biol Blood Marrow Transplant. 2002;8:395–400. doi: 10.1053/bbmt.2002.v8.pm12171486. [DOI] [PubMed] [Google Scholar]
- 12.Afessa B, Abdulai RM, Kremers WK, Hogan WJ, Litzow MR, Peters SG. Risk factors and outcome of pulmonary complications after autologous hematopoietic stem cell transplant. Chest. 2012;141:442–50. doi: 10.1378/chest.10-2889. [DOI] [PubMed] [Google Scholar]
- 13.Wong R, Rondon G, Saliba RM, Shannon VR, Giralt SA, Champlin RE, et al. Idiopathic pneumonia syndrome after high-dose chemotherapy and autologous hematopoietic stem cell transplantation for high-risk breast cancer. Bone Marrow Transplant. 2003;31:1157–63. doi: 10.1038/sj.bmt.1704141. [DOI] [PubMed] [Google Scholar]
- 14.Crawford SW, Hackman RC. Clinical course of idiopathic pneumonia after bone marrow transplantation. Am Rev Respir Dis. 1993;147:1393–400. doi: 10.1164/ajrccm/147.6_Pt_1.1393. [DOI] [PubMed] [Google Scholar]
- 15.Tizon R, Frey N, Heitjan DF, Tan KS, Goldstein SC, Hexner EO, et al. High-dose corticosteroids with or without etanercept for the treatment of idiopathic pneumonia syndrome after allo-SCT. Bone Marrow Transplant. 2012;47:1332–7. doi: 10.1038/bmt.2011.260. [DOI] [PubMed] [Google Scholar]
- 16.Yanik GA, Horowitz MM, Weisdorf DJ, Logan BR, Ho VT, Soiffer RJ, et al. Randomized, double-blind, placebo-controlled trial of soluble tumor necrosis factor receptor: enbrel (etanercept) for the treatment of idiopathic pneumonia syndrome after allogeneic stem cell transplantation: blood and marrow transplant clinical trials network protocol. Biol Blood Marrow Transplant. 2014;20:858–64. doi: 10.1016/j.bbmt.2014.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yanik GA, Grupp SA, Pulsipher MA, Levine JE, Schultz KR, Wall DA, et al. TNF receptor inhibitor therapy for the treatment of children with idiopathic pneumonia syndrome (IPS). A joint Pediatric Blood and Marrow Transplant Consortium (PBMTC) and Children's Oncology Group (COG) study (ASCT0521). Biol Blood Marrow Transplant. 2014 doi: 10.1016/j.bbmt.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhargava M, Becker TL, Viken KJ, Jagtap PD, Dey S, Steinbach MS, et al. Proteomic profiles in acute respiratory distress syndrome differentiates survivors from non-survivors. PloS one. 2014;9:e109713. doi: 10.1371/journal.pone.0109713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Wroblewski M, Hertz MI, Wendt CH, Cervenka TM, Nelsestuen GL. Analysis of chronic lung transplant rejection by MALDI-TOF profiles of bronchoalveolar lavage fluid. Proteomics. 2006;6:1001–10. doi: 10.1002/pmic.200500105. [DOI] [PubMed] [Google Scholar]
- 20.Seo S, Renaud C, Kuypers JM, Chiu CY, Huang ML, Samayoa E, et al. Idiopathic pneumonia syndrome after hematopoietic cell transplantation: evidence of occult infectious etiologies. Blood. 2015;125:3789–97. doi: 10.1182/blood-2014-12-617035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bhargava M, Dey S, Becker T, Steinbach M, Wu B, Lee SM, et al. Protein expression profile of rat type two alveolar epithelial cells during hyperoxic stress and recovery. Am J Physiol Lung Cell Mol Physiol. 2013;305:L604–14. doi: 10.1152/ajplung.00079.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem. 2003;75:663–70. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 23.Lin-Moshier Y, Sebastian PJ, Higgins L, Sampson ND, Hewitt JE, Marchant JS. Re-evaluation of the role of calcium homeostasis endoplasmic reticulum protein (CHERP) in cellular calcium signaling. The Journal of biological chemistry. 2013;288:355–67. doi: 10.1074/jbc.M112.405761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Timothy Griffin JC, Johnson James, de Jong Ebbing. American Society of Mass Spectrometry and Allied Topiecs. Minneapolis, USA: 2013. Getiria Onsongo and Pratik Jagtap. Galaxy-P: Transforming MS-based proteomic informatics via innovative workflow development, dissemination, standardization and transparency. [Google Scholar]
- 25.Vizcaino JA, Deutsch EW, Wang R, Csordas A, Reisinger F, Rios D, et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nature biotechnology. 2014;32:223–6. doi: 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Champion MM, Williams EA, Kennedy GM, Champion PA. Direct detection of bacterial protein secretion using whole colony proteomics. Mol Cell Proteomics. 2012;11:596–604. doi: 10.1074/mcp.M112.017533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Y B. Discovering the False Discovery Rate. J R Statist Soc B. 2010;72:405–16. [Google Scholar]
- 28.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 29.Schlatzer DM, Dazard JE, Ewing RM, Ilchenko S, Tomcheko SE, Eid S, et al. Human biomarker discovery and predictive models for disease progression for idiopathic pneumonia syndrome following allogeneic stem cell transplantation. Mol Cell Proteomics. 2012;11:M111 015479. doi: 10.1074/mcp.M111.015479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shaik FB, Prasad DV, Narala VR. Role of farnesoid X receptor in inflammation and resolution. Inflammation research : official journal of the European Histamine Research Society [et al] 2014 doi: 10.1007/s00011-014-0780-y. [DOI] [PubMed] [Google Scholar]
- 31.Wang YD, Chen WD, Wang M, Yu D, Forman BM, Huang W. Farnesoid X receptor antagonizes nuclear factor kappaB in hepatic inflammatory response. Hepatology. 2008;48:1632–43. doi: 10.1002/hep.22519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vavassori P, Mencarelli A, Renga B, Distrutti E, Fiorucci S. The bile acid receptor FXR is a modulator of intestinal innate immunity. J Immunol. 2009;183:6251–61. doi: 10.4049/jimmunol.0803978. [DOI] [PubMed] [Google Scholar]
- 33.Zhang L, Li T, Yu D, Forman BM, Huang W. FXR protects lung from lipopolysaccharide-induced acute injury. Molecular endocrinology. 2012;26:27–36. doi: 10.1210/me.2011-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He F, Li J, Mu Y, Kuruba R, Ma Z, Wilson A, et al. Downregulation of endothelin-1 by farnesoid X receptor in vascular endothelial cells. Circulation research. 2006;98:192–9. doi: 10.1161/01.RES.0000200400.55539.85. [DOI] [PubMed] [Google Scholar]
- 35.Berryman M, Franck Z, Bretscher A. Ezrin is concentrated in the apical microvilli of a wide variety of epithelial cells whereas moesin is found primarily in endothelial cells. Journal of cell science. 1993;105(Pt 4):1025–43. doi: 10.1242/jcs.105.4.1025. [DOI] [PubMed] [Google Scholar]
- 36.Niggli V, Rossy J. Ezrin/radixin/moesin: versatile controllers of signaling molecules and of the cortical cytoskeleton. Int J Biochem Cell Biol. 2008;40:344–9. doi: 10.1016/j.biocel.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 37.Arpin M, Chirivino D, Naba A, Zwaenepoel I. Emerging role for ERM proteins in cell adhesion and migration. Cell Adh Migr. 2011;5:199–206. doi: 10.4161/cam.5.2.15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okayama T, Kikuchi S, Ochiai T, Ikoma H, Kubota T, Ichikawa D, et al. Attenuated response to liver injury in moesin-deficient mice: impaired stellate cell migration and decreased fibrosis. Biochim Biophys Acta. 2008;1782:542–8. doi: 10.1016/j.bbadis.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 39.Kahsai AW, Zhu S, Fenteany G. G protein-coupled receptor kinase 2 activates radixin, regulating membrane protrusion and motility in epithelial cells. Biochim Biophys Acta. 2009 doi: 10.1016/j.bbamcr.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.