Abstract
The cells that stimulate positive selection express different specialized proteasome β-subunits than all other cells, including those involved in negative selection. Mice that lack all four specialized proteasome β-subunits, and therefore express only constitutive proteasomes in all cells, had a profound defect in the generation of CD8+ T cells. While a defect in positive selection would reflect an inability to generate the appropriate positively selecting peptides, a block at negative selection would point to the potential need to switch peptides between positive and negative selection to avoid the two processes often cancelling each other out. We found that the block in T cell development occurred around the checkpoints of positive and, surprisingly, also negative selection.
During T cell development, immature thymocytes rearrange their T cell receptor (TCR) genes to generate receptors of random specificities. The immune system must then positively select T cells with useful receptors, match the cells' receptor specificities with function through the process of lineage commitment, and eliminate dangerous autoreactive clones by negative selection1. Each of these key developmental check points is driven by signaling through the TCR, via interactions with self-peptide-MHC complexes1,2.
The majority of MHC class I-presented self-peptides are generated when proteasomes, proteolytic particles present in the nucleus and cytosol of all nucleated mammalian cells, degrade cellular proteins3. Proteasomes contain six active sites that are formed by three different beta subunits – two each of β1, β2, and β5 4 to form thymoproteasomes (β1i, β2i and β5t), constitutive proteasome (β1c, β2c and β5c), immunoproteasomes (β1i, β2i and β5i), or mixed proteasomes (particles with various combinations of the c and i subunits)5,6 Because the active sites have different specificities/catalytic properties and allosterically influence one another 7, proteasomes with different active site subunit combinations cleave substrates differently and thereby produce different peptides, and where examined, thymoproteasomes generate both unique (∼30%) and commonly (∼70%) presented peptides 6,8,9. Thymoproteasomes are expressed exclusively in cortical thymic epithelial cells (cTECs), and therefore some of the peptides presented on the cTECs that drive positive selection will be different from the peptides presented anywhere else in the body, including on the medullary thymic epithelial cells (mTECs) and thymic DCs that drive negative selection 5,6.
Psmb11−/− mice that lacked the β5t thymoproteasome subunit had a partial but substantial reduction in the development of mature CD8+ T cells5,10. These findings resonated with earlier studies that found that only a minority of exogenous antigenic or splenic peptides added to B2m−/− or Tap1−/− (MHC class I-peptide-deficient) fetal thymic organ cultures (FTOCs) could stimulate the maturation of CD8+ T cells (reviewed in1). Using these cultures it was found that variants of antigenic epitopes that had been modified to become antagonists or weak agonists/antagonists for mature T cells (altered peptide ligands), promoted CD8+ T cell maturation, whereas the native peptide sequence did not11. This led to the idea that specialized epitopes might be needed to drive positive selection. Given these findings it was logical that thymoproteasomes might help generate such special peptides, and some experiments suggested that the developmental block in Psmb11−/−thymi occurred before negative selection (reviewed in 12). Accordingly, the favored model for why CD8+ T cell development was decreased in β5t-deficient mice was impaired positive selection due to a loss of the special peptides needed for positive selection 13. An alternate and disfavored model was that thymoproteasome-derived peptides allowed cells to survive negative selection because they were uniquely displayed by cTECs and would not be present at negative selection. Therefore positive and negative selection would not cancel each other14.
The present study was initiated to examine the role in T cell development of the specialized proteasomes that contain β5t, β1i, β2i and/or β5i and the potential importance of switching presented peptides between positive and negative selection. To investigate these issues we analyzed T cell development in animals that expressed only constitutive proteasomes in all cells, and that would therefore largely present the same peptide-MHC class I complexes during positive and negative selection.
Results
Specialized proteasomes required for CD8+ T cell development
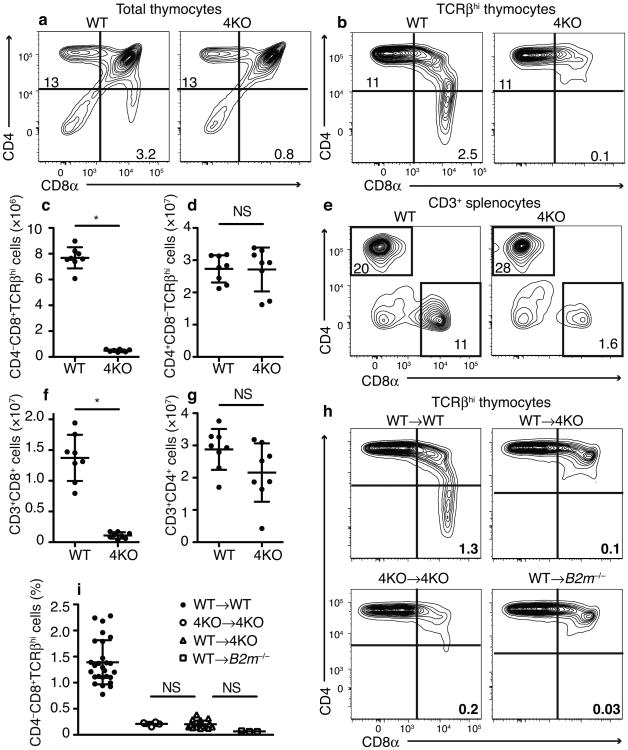
To investigate the role of specialized proteasomes (i.e. thymoproteasomes, immunoproteasomes and mixed proteasomes) in T cell development, we generated Psmb8−/− Psmb9−/− Psmb10−/− Psmb11−/− (4KO) mice. These animals, which contain only constitutive proteasomes, were viable, healthy, and bred normally. Their thymi were of normal size (Supplementary Fig 1a). Compared to C57BL6/J wild-type mice, there were normal numbers of CD4–CD8– double negative (DN) and CD4+CD8+ double positive (DP) thymocytes (Supplementary Fig 1b,c,d). In contrast, there was a >90% loss of mature TCRβhiCD4–CD8αβ+ single positive thymocytes (Fig 1a-c, Supplementary Fig 1f,g). Similarly the spleens of 4KO mice had a ≥90% reduction in CD8+ T lymphocytes (Fig 1e,f). This block in polyclonal T cell development in the 4KO mice was substantially more than was observed in the β5t-deficient animals and immunoproteasome-deficient mice, which had reductions in CD8+ single-positive (CD8SP) cells of ∼75% and 50%, respectively5,8. Thus the analysis of the 4KO mice revealed specialized proteasomes are required for the development of the vast majority of mature CD8+ T cells.
Fig 1.
Severe defect in mature polyclonal CD8+ T cells in 4KO mice. Flow cytometric plots of (a) total and (b) TCRβhi thymocytes in wild-type and 4KO mice. Numbers on gates indicate percent of total thymocytes. Quantitation of total number of TCRβhi (c) CD4SP and (d) CD8SP thymocytes. (e) Flow cytometric plots of CD3+ splenocytes in wild-type and 4KO mice. Numbers on gates indicate percent of total splenocytes. Quantitation of total number of CD3+ (f) CD8+ or (g) CD4+ splenocytes. (n= 8 mice each, male and female, combined from 2 experiments.) (h) Flow cytometric plots of TCRβhi thymocytes in indicated BM→host chimeras. Numbers on gates indicate percent of total thymocytes. (i) Frequency of CD8SP TCRβhi thymocytes in the indicated BM→host chimeras. (n = 27 WT→WT, 4 4KO →4KO, 21 WT→4KO, 3 WT→B2m−/−, male and female, combined from 5 experiments). *P< 0.001 in c,f (Student's t-test) NS, not significant in d,g (Student's t-test) and i (One-way ANOVA with Dunnett's multiple comparison post-test)., c,d,f,g,and i (mean±SD)
The CD8SP cells that developed in the 4KO thymus expressed all of the TCR Vβ chains (2, 3, 4, 5.1 or 5.3,6, 7, 8.1 or 8.2, 8.3, 9, 10a, 11, 12, 13, 14) we analyzed, although the frequency of some of these Vβs was modestly different than in wild-type mice (Supplementary Fig 2). Interestingly, the CD8SP cells in 4KO mice expressed significantly lower amounts of CD5 (Supplementary Fig 1h,i), which suggests that their TCRs have been selected on lower affinity peptide-MHC I complexes15. This altered CD5 expression is almost certainly due to a change in presented peptides because it was only seen on CD8+ T cells. This finding suggests that thymoproteasomes are important for the positive selection of higher affinity TCRs and/or that in the absence of thymoproteasomes lower affinity TCRs are better able to survive positive and negative selection on the exact same peptides (i.e. those made by constitutive proteasomes).
In contrast to the block in development of mature CD8+ T cells in the 4KO mice, these animals generated normal numbers of mature CD4+ single-positive (CD4SP) thymocytes and there was no significant reduction in CD4+ lymphocytes in the spleen (Fig. 1a,b,d,e,g). These results indicated that the block in T development in the 4KO mice was selective for the CD8 pathway. Because CD4+ and CD8+ T cells develop in parallel in the same thymic microenvironments and rely on the same thymic stromal cells, the finding that CD4+ T cell development was intact strongly suggests that there was no general defect in the function of the thymic cellular elements, such as cTECs.
To determine the contribution of thymic stromal cells to the defect in CD8+ T cell development in 4KO animals, we created bone marrow (BM) chimeras. We transferred wild-type BM into irradiated 4KO hosts, and found that the frequency of CD8+ T cells in the thymus of these mice was profoundly reduced, down to the numbers that were observed in 4KO animals that received 4KO BM, and similar to the numbers found in B2m−/− (MHC I-deficient) animals that received wild-type BM (Fig 1h,i). Therefore, specialized proteasomes were required in thymic stromal cells for development of most CD8+ T cells. These results indicate that the defect in 4KO animals was not simply due to a cell intrinsic defect in T cells lacking specialized proteasomes.
Because altered MHC class I expression can affect CD8+ T cell development, we next investigated MHC class I expression on TECs that lacked specialized proteasomes (Supplementary Fig 3). We found that H-2Kb and H-2Db expression on 4KO CD45– CD249+CD326+ cTECs was approximately half of that in wild-type mice (Supplementary Fig 3d). We found a similar reduction in MHC class I molecules on 4KO CD45–CD326+UEA1+ mTECs (Supplementary Fig 3e). The 50% reduction of MHC class I expression was similar to what we observed in dendritic cells and lymphocytes from Psmb8−/− Psmb9−/− Psmb10−/− (immunoproteasome-deficient) 3KO mice (which, like 4KO hematopoietic cells, only have constitutive proteasomes)8. To test whether a 50% reduction in MHC class I expression could account for the block in CD8+ T cell development observed in the 4KO mice, we used the heterozygous progeny of H-2Kb−/−H-2Db−/− crossed with wild-type mice. Due to the single copy of the MHC class I genes, these animals had a similar 50% reduction in MHC class I expression on both hematopoietic cells and TECs, but their development of CD8+ T cells was not reduced (Supplementary Fig 3).
Taken together, our data indicate that specialized proteasome subunits were needed in non-hematopoietic cellular elements to support the development of the vast majority of CD8+ T cells, and this key role was almost certainly due to their production of the MHC I-presented peptides that are required for the selection the CD8+ T cells.
Positive selection of monoclonal and polyclonal thymocytes
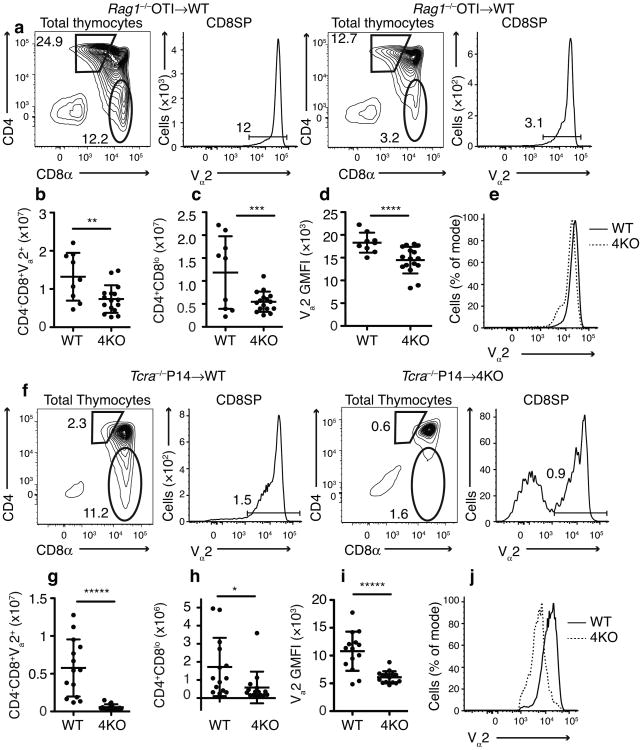
We examined how the loss of specialized proteasomes affected the positive selection of TCR transgenic cells in chimeric mice. We observed a marked reduction in the development of mature CD8SP T cells from OT-I BM (which has transgenic expression of an ovalbumin-specific, H-2Kb–specific TCR) that developed in 4KO compared to wild-type thymi (Fig 2a,b). This result was markedly different from what was seen in Psmb11−/− thymi, where there was no significant reduction in OT-I CD8SP cells13,16. There was also a marked reduction in the number of CD4+CD8lo OT-1 thymocytes that developed in the 4KO thymus (Fig 2a,c). Moreover, in the 4KO mice, the CD4+CD8lo transgenic thymocytes that did develop failed to upregulate expression of the TCR to wild-type amounts (Fig 2d,e). Because the CD4+CD8lo phenotype indicated that the thymocytes had been positively selected, it showed that in the absence of specialized proteasomes, positive selection of OT-1 TCR transgenic T cells was defective. These findings are consistent with the recent finding that the thymoproteasome could generate, at least in fibroblasts9, peptides that were known to positively select OT-1 cells17. If these peptides are actually the bone fide ones that positively select OT-1 in vivo, which is unknown, then presumably constitutive proteasomes cannot generate them in sufficient amounts. We found similar results for the development of mature CD8SP T cells from P14 BM (which has transgenic expression of lymphocytic choriomeningitis virus glycoprotein epitope gp33-specific, H-2Db-specific TCR) in the 4KO thymus (Fig 2f-j).
Figure 2.
Defect in positive selection of TCR transgenic CD8+ T cells in 4KO thymi. Gating of (a) Rag1−/− OT-I → WT or 4KO BM chimeras and (f) Tcra−/− P14 → wild type or 4KO BM chimeras. Number of (b) CD4-CD8+Vα2hi or (c) CD4+CD8lo thymocytes, (d) Vα2 GMFI on CD4+CD8lo thymocytes, and (e) histograms of Vα2 on CD4+CD8lo thymocytes in mice receiving Rag1−/− OT-I BM (n=9 wild-type, 16 4KO, male and female, combined from 2 experiments). Number of (g) CD4–CD8+Vα2hi or (h) CD4+CD8lo thymocytes, (i) Vα2 GMFI on CD4+CD8loVα2+ thymocytes, and (j) histograms of Vα2 on CD4+CD8loVα2+ thymocytes in mice receiving Tcra–/P14 BM (n=14 wild-type, 15 4KO, male and female, combined from 2 experiments). *P <0.0245 **P <0.0068 ***P <0.0056 ****P <0.0025 *****P <0.0001 in b-d, g and h (Student's t-test mean±SD)
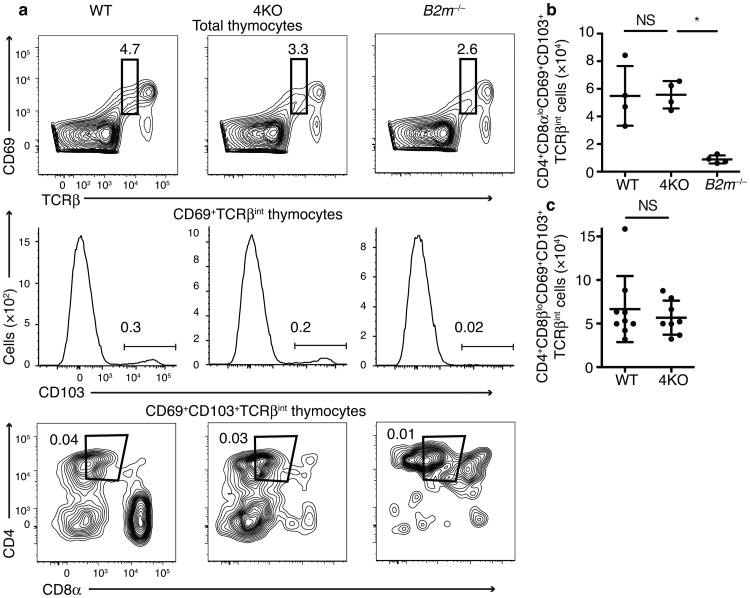
We also examined how the loss of specialized proteasome subunits affected the development of polyclonal T cells. As noted above, the decreased expression of CD5 on polyclonal CD8SP cells in 4KOs suggested that there could be defective positive selection of these cells in the absence of specialized proteasomes, at least for the small subset of these cells that successfully transited all of the developmental checkpoints. We sought to further examine this issue for the bulk developing cells. DP cells that have received positive selection signals upregulate CD6918,19 along with the TCR and downregulate CD8 expression. The CD69+TCRβint CD4+CD8lo thymocyte population includes cells that will develop into both CD4SP and CD8SP cells 2. To distinguish between these two populations, we analyzed the cell surface marker CD103, an integrin expressed on CD8 lineage cells in the thymus 20,21. Surprisingly, we found that the CD4+CD8αβloCD69+CD103+TCRβint population was not reduced in 4KO animals (Fig 3a,b,c). To verify that the development of these CD103+CD8lo cells was MHC class I-dependent, we examined their development in MHC class I-deficient (B2m−/−) mice, and found that their numbers were markedly reduced in the B2m−/− mice compared to wild-type or 4KO animals.
Fig 3.
CD8 lineage cells express CD69 and TCRbeta in 4KO mice. Flow cytometric plots (a) and quantitation (b) of CD69+TCRβintCD103+CD4+CD8αlo thymocytes in wild-type, 4KO, or B2m−/− mice (n=4 mice each group, male and female, combined from 2 experiments). (c) Quantitation of CD69+TCRβintCD103+CD4+CD8βlo thymocytes in wild-type or 4KO mice (n=9 wild-type, 8 4KO mice, male and female, combined from 2 experiments) (Numbers on gates indicate percent of total thymocytes.) *P<0.05 and NS, not significant in b (One-way ANOVA with Dunnett's multiple comparison post-test). NS in c (Student's t-test) b and c (mean±SD)
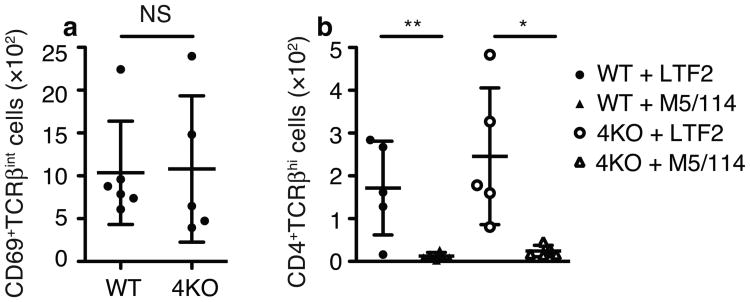
We also wanted to rule out the possibility that in the absence of specialized proteasome-derived peptides in the 4KO thymus, MHC class II-presented peptides might somehow be aberrantly positively selecting CD8+ T cells. Given the close proximity of MHC class II genes to Psmb8 and Psmb9 on chromosome 17, it is impractical to breed homozygous MHC class II-deficient 4KO mice for such an analysis. Therefore we blocked TCR-MHC II interactions during development by adding anti-MHC class II antibodies to FTOC22. CD4+ T cell development was blocked by anti-MHC class II treatment, as expected (Fig 4 and Supplementary Fig 4). This served as a positive control that MHC class II molecules were indeed blocked by addition of the exogenous antibody. Since there were no CD4 lineage cells being selected we analyzed the development of the MHC class I-selected CD8+ T cells by staining with anti-CD69 and anti-TCRβ. Consistent with what we found in intact 4KO animals, similar numbers of CD8 lineage thymocytes were positively selected in both wild-type and 4KO even when MHC class II molecules were blocked with antibody (Fig 4 and Supplementary Fig 4).
Fig 4.
4KO thymocytes receive positive selection signals independent of MHC II. (a) Number of CD69+TCRβint thymocytes in FTOC in the presence of 500μg/ml M5/114. (b) Number of CD4+CD8–TCRβhi thymocytes in the presence of 500μg/ml control (LTF-2) or anti-MHC class II (M5/114) mAbs. (5 WT + LTF-2 lobes, 6 WT + M5/114 lobes, 5 4KO + LTF-2 lobes, 5 4KO + M5/114 lobes, from one representative experiment. Combined data from 5 experiments with 20 WT + LTF-2 lobes, 21 WT + M5/114 lobes, 21 4KO + LTF-2 lobes, 17 4KO + M5/114 lobes are in Supplementary Fig 4). NS, not significant in a (Student's t-test). *P=0.0149 **P=0.006 in b (Student's t-test) a and b (mean±SD)
Taken together, these data indicate that in the absence of specialized proteasomes, approximately normal numbers of polyclonal CD8+ T cells had phenotypic evidence of having undergone positive selection, whereas the development TCR transgenic T cells was impaired at this first selection checkpoint.
Polyclonal 4KO CD8+ thymocytes undergo lineage commitment
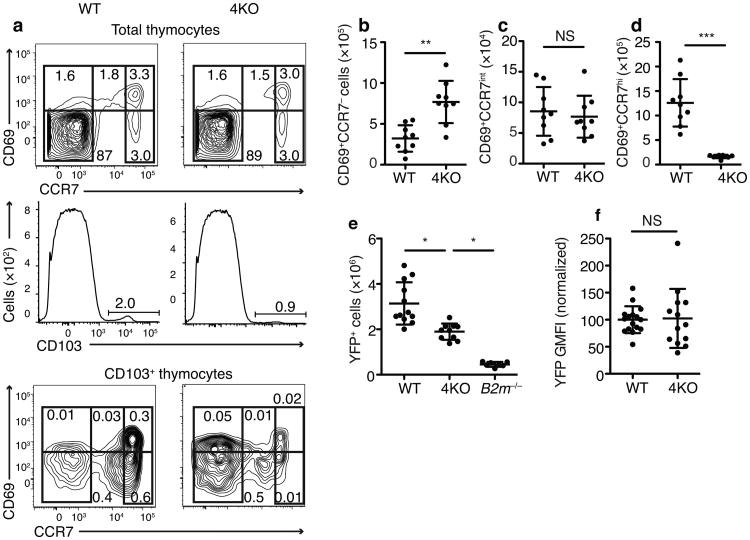
Following positive selection, CD69+ thymocytes upregulate expression of CCR7, a chemokine receptor that enables the thymocytes to migrate to the thymic medulla23. We found that CCR7intCD69+CD103+ cells were present in similar numbers in both wild-type and 4KO thymi (Fig 5a-d). This suggested that polyclonal CD8+ T cells in the 4KO thymus were not arresting during positive selection but had already successfully transited through this developmental checkpoint.
Fig 5.
Expression of CCR7 and markers of CD8 lineage commitment on 4KO thymocytes Flow cytometric plots (a) and quantitation of (b) CD103+CD69+CCR7–, (c) CD103+CD69+CCR7int, or (d) CD103+CD69+CCR7hi thymocytes. (n=9 mice per group, male and female, combined from 3 experiments.) (e) Number of YFP+ thymocytes in Runx3dYFP/+→WT, Runx3dYFP/+→4KO, or Runx3dYFP/+→B2m−/− chimeras and (f) YFP GMFI on CD69+TCRβintCD103+ cells. Mean of wild-type BM recipient results set to 100% in each experiment. (n=12 wild-type, 10 4KO, 7 B2m−/− combined from 2 experiments). Numbers on gates indicate percent of total thymocytes (a). NS, not significant, **P=0.0005 ***P<0.0001 in b-d, and f, (Student's t-test) or *P<0.05 in e, (One-way ANOVA with Dunnett's multiple comparison post-test). b-f (mean±SD).
To further explore this issue, we investigated whether CD8+ T cells in the 4KO thymus were progressing through lineage commitment (a key event that occurs after positive selection2) by analyzing the expression of Runx3d, a transcription factor that commits thymocytes to the CD8 lineage both phenotypically and functionally 20,24. Recent evidence showed that, in cells that required the CD8 co-receptor for TCR signaling, the developmental downregulation of CD8 expression interrupted TCR signaling, which in conjunction with thymic cytokines, drove the expression of Runx3d and subsequent commitment of these cells to the CD8 lineage 25. We transferred Runx3dYFP/+ BM into wild-type, 4KO, and B2m−/− hosts and analyzed the number of YFP+ (Runx3d+) cells that developed in the three strains. Substantial numbers of Runx3d+ thymocytes developed in 4KO mice, significantly more than in B2m−/− mice (Fig 5e). The total number of Runx3d+ cells was lower in 4KO mice compared to wild-type controls, presumably because mature CD8SP (CD4–CD8+Runx3d+) T cells developed in wild-type but not 4KO thymi. Consistent with this, the Runx3d expression on CD69+CD103+TCRβint cells in 4KO mice was not reduced relative to wild-type mice (Fig 5f).
Altogether these results suggest that polyclonal CD8+ T cells completed positive selection on MHC class I molecules in 4KO thymi and then expressed medullary-homing chemokine receptors and successfully underwent lineage commitment.
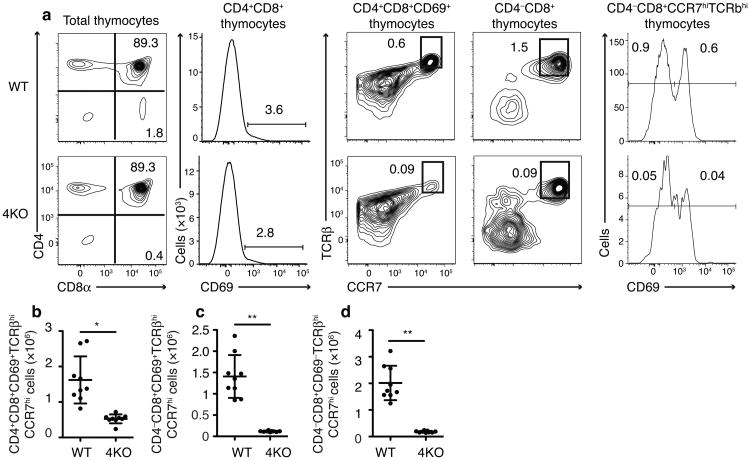
Block in CD8+ thymocyte development at negative selection
As positively selected CD8 lineage cells (CD4+CD8loCD69+ CD103+TCRβint) progress in development they upregulate CD8 and TCR expression to become CD4+CD8+ CD69+TCRβhiCCR7hi cells. This population of cells is committed to the CD8 lineage20,26 and subjected to negative selection27. Interestingly, we found CD4+CD8+CD69+TCRβhiCCR7hi cell numbers were significantly decreased (by 68%) in 4KO mice (Fig 6a,b). All TCRβhiCCR7hi CD8SP cell numbers (normally found in the medulla)28 including both immature CD69+ cells and mature CD69– cells, were decreased by >90% (Fig 6a,c,d, Supplementary Fig 5). Therefore a block in CD8+ T cell development in the absence of specialized proteasome subunits appeared to be occurring post-positive selection and coincident with the last TCR-MHC class I checkpoint — negative selection.
Fig 6.
4KO thymocytes are lost during negative selection. (a) Flow cytometric plots and quantitation of (b) CD4+CD8+CD69+TCRβhiCCR7hi, (c) CD4–CD8+CCR7hiCD69+TCRβhi, and (d) CD4–CD8+CCR7hiCD69–TCRβhi thymocytes in wild type and 4KO mice (n=9 mice each group, male and female, combined from 3 experiments). *P=0.0002 **P<0.0001 (Student's t-test) in b-d (mean±SD)
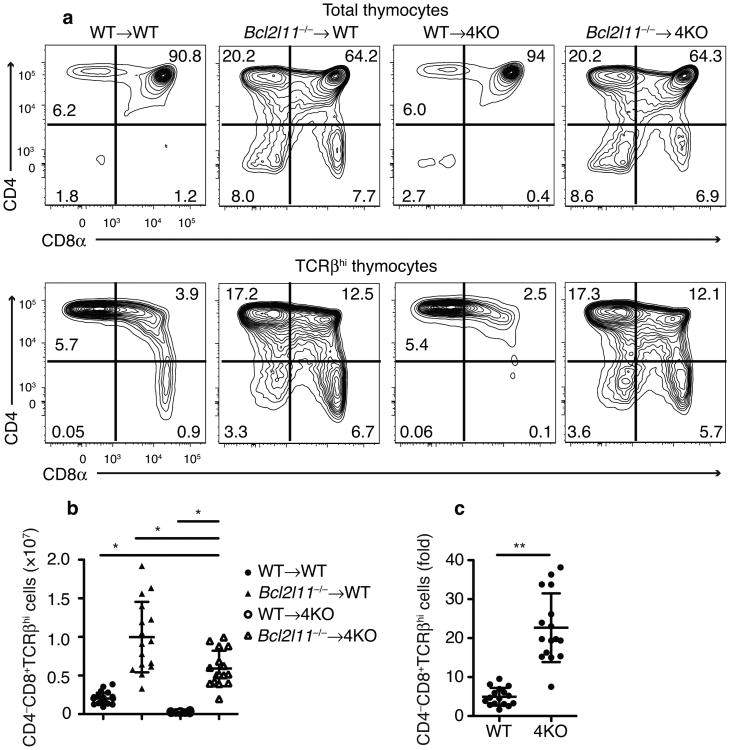
We sought to further test this idea by analyzing whether impairing negative selection in 4KO mice would rescue CD8+ T cell development. Therefore, we reconstituted 4KO mice with BM from Bcl2l11−/− (Bim deficient) mice, a strain with a substantial defect in negative selection (though cells that fail to undergo positive selection still die normally) 27,29. We found that the development of mature CD8+ T cells in the thymi of Bcl2l11−/− → 4KO mice was rescued by 2.94 fold over the numbers of these cells observed in WT→WT mice (Fig 7a,b). The loss of Bim in 4KO chimeras did not increase CD8+ T cells numbers to that observed in Bim-deficient wild-type chimeras (∼40% less), but this was not surprising because some T cells may be negatively selected by a Bim-independent pathway30 perhaps involving other pro-apoptotic factors, such as Puma that contribute to negative selection 31 and should also disproportionally eliminate more cells in the 4KO versus wild-type thymus. Importantly, however, there was a 22.7±8.8-fold increase in mature CD8+ thymocytes (CD4–CD8+TCRβhi) in Bcl2l11−/− →4KO chimeras compared to only a 5.0±2.3 -fold increase in Bcl2l11−/− → WT mice (mean±SD, p<0.0001, Fig 7c). In contrast, the changes in DP and CD4SP numbers were much more modest; the fold increase in CD4SP cells in the Bcl2l11−/− → 4KO was actually less than in the Bcl2l11−/−→WT mice (Supplementary Fig 6).
Fig 7.
Rescue of CD8 development in 4KO thymus by Bcl2l11−/−thymocytes. (a) Flow cytometric plots and (b) quantitation of CD4–CD8+ TCRβhi thymocytes in indicated BM → host chimeras. (c) Fold increase in CD4-CD8+TCRβhi thymocytes from mice receiving Bcl2l11−/− vs. wild-type BM. (n=22 WT→WT,16 Bcl211−/−→WT,16 WT→4KO, 16 Bcl211−/−→4KO; male and female, combined from 5 experiments) Numbers on gates indicate percent of total thymocytes. *P<0.05 in b, (One-way ANOVA with Dunnett's multiple comparison post-test). **P=0.0005 in c, (Student's t-test) b and c (mean±SD)
These findings were surprising because previous studies of less complete KOs of specialized proteasomes (mice deficient in β5t only), concluded that the block in CD8+ T cell development was at an earlier stage than negative selection5,10; however in our opinion, these earlier findings do not definitively demonstrate that this is indeed the case. In one study, it was found that a transplant of MHC class I-deficient (B2m−/−) BM into irradiated Psmb11−/− mice did not rescue CD8+ T cell development13. It was suggested that if the problem in these mice was due to excessive negative selection, then the loss of MHC class I on cells of hematopoietic origin should have rescued thymocytes from negative selection; the fact that this did not occur led to the suggestion that the defect in these mice must be at an earlier stage. However, this is not a definitive because B2m−/− hematopoietic cells also don't rescue CD8SP cells (i.e. increase their numbers) in a wild-type thymus. This may be because B2m−/− hematopoietic cells can also present some peptides32, or because non-bone marrow-derived cells express MHC class I and can either drive negative selection directly or transfer MHC class I complexes to hematopoietic cells (cross dressing)33. Therefore this model still had MHC class I molecules that could participate in negative selection. In other experiments it was found that CCR7-deficiency (which prevents thymocytes from migrating to the medulla, where negative selection on mTECs occurs) did not rescue CD8+ T cell development13. However, negative selection also occurs in the thymic cortex27,34 and therefore the block in development could still be caused by negative selection. In yet another study it was found that Bcl2l11−/− BM rescued the development of some CD8+ T cells, but it was concluded that the magnitude of the rescue was smaller than the authors expected for a defect at negative selection10. However, in this study Bcl2l11−/− BM caused a proportionally greater increase in CD8SP cells in the Psmb11−/− compared to the wild-type thymi and in our current study this increase was even greater than observed in the 4KO thymi.
Altogether our findings that the block in CD8+ thymocyte development mapped to negative selection and that CD8+ T cell development could be rescued using Bim-deficient cells, suggests that the majority of CD8+ T cell that were positively selected in 4KO mice were failing to pass the negative selection checkpoint (Supplemental Fig 7).
Discussion
Here we show that specialized proteasome subunits are more essential for CD8+ T cell development than previously appreciated from studies of the β5t-subunit deficient mice. Moreover, while impaired CD8+ T cell development in Psmb11−/− mouse was attributed solely to a defect in positive selection, analysis of 4KO mice revealed an additional major block occurring around negative selection.
A block at negative selection is consistent with a peptide-switching model wherein most developing CD8+ T cells need to be selected on different peptides at positive and negative selection 14,35. In this model, the uniquely presented peptides (∼30%) generated by thymoproteasomes allow the system to select host MHC class I-restricted TCRs on cTECs. Negative selection then occurs on peptides produced by constitutive, immuno, and/or mixed proteasomes. These are the peptides that are presented in the periphery, and therefore it is essential to eliminate autoreactive T cells that recognize them. In the 4KO thymus such peptide-switching cannot occur and consequently both selection steps take place on peptides generated by constitutive proteasomes. While some unique peptides could come from proteins that are only expressed in cTECs, these unique proteins are rare36 and without proteasome-switching seem to at most only be able to select the small number of CD8+ T cells found in 4KO mice. An analogous peptide-switching mechanism may also operate in the selection of CD4 T cells37,38. A previous study found that a switch between β1i,β2i,β5i proteasomes in cTECs and β1i,β2i,β5c proteasomes elsewhere in the thymus resulted in a reduction of ∼75% in CD8SP cells (similar to Psmb11−/−)10. However, there may be little peptide switching in this system because the difference in antigen presentation by hematopoietic APCs that contain β1i,β2i,β5c vs β1i,β2i,β5i proteasomes is relatively minor 39. The uncertainty about how much peptide switching occurs in the partial β-subunit KOs is a limitation of these models.
Selection of thymocytes has been thought to be via a differential-affinity mechanism14 wherein positive selection is triggered by a lower TCR affinity than negative selection. Such a mechanism is not mutually exclusive with the peptide-switching model, but peptide-switching offers the advantage that it could in theory generate a larger TCR repertoire than differential-affinity. This is because the window of permissible affinities could be much larger for the peptide-switching mechanism; higher affinity TCRs that are positively selected on unique thymoproteasome-derived peptides would not subsequently encounter these same peptides at negative selection. The survival of higher affinity TCRs would also increase the functionality of the T cell repertoire because such TCRs are the most effective ones for immunity to pathogens40. On the other hand, in FTOCs positive selection on some high affinity agonist peptides results in T cells with abnormal phenotype and impaired function41. However, it is not clear whether this effect is seen with all high affinity peptides and/or when such presentation is restricted to cTECs. In fact, functional high affinity T cells are clearly positively selected in vivo because they cause autoimmunity when negative selection is blocked31,42,43. It might be argued that the selection of high affinity TCRs for unique thymoproteasome-generated peptides would put cTECs at risk of attack from mature T cells. However, cTECs lack the costimulatory molecules needed to activate T cells44,45 and even more importantly, mature T cells do not traffic into the thymic cortex46.
The peptide-switching model can also explain why the development of TCR transgenic cells in the 4KO thymus was blocked at positive selection. Transgenic TCRs that were originally positively selected on unique thymoproteasome peptides (in a wild type thymus) would not encounter their positively-selecting peptides on 4KO cTECs. Polyclonal T cells do not suffer the same fate because this population begins with a random TCR repertoire.
The peptide-switching model raises a number of unanswered questions. Why don't higher affinity peptides on cTECs delete CD8+ T cells? This may be because negative selection in the thymic cortex occurs on dendritic cells and not cTECs34. Another question is how T cells that have been selected by unique thymoproteasome-generated peptides are maintained in the periphery. It has been suggested that the peptides that are required to sustain CD8+ T cells in the periphery are the same ones that the thymocytes were positively selected on47,48. However, an essential role for positively selecting peptides is not established for CD8+ T cells homeostasis and if this is required, then presumably T cells selected by unique thymoproteasome-peptides are sustained in the periphery on low affinity, cross-reactive peptides.
Our observations suggest that evolution of the thymo- and immuno- proteasome subunits may have been essential for the development of the CD8 arm of adaptive immunity. This may explain why these subunits arose in phylogeny at the same point as the RAG recombination enzymes and the TCR genes that needed to be selected49
Analysis of 4KO mice revealed that the absence of specialized proteasome subunits in non-hematopoietic cells disrupted both positive and negative selection. These findings support the idea that peptide-switching between positive and negative selection is important to establish a broad TCR repertoire.
On-line Methods
Mice
All mouse strains were bred and maintained in specific pathogen–free conditions in the animal facilities at the University of Massachusetts Medical School. All experiments involving live animals were approved by and performed in accordance with guidelines set forth by the University of Massachusetts Medical School Department of Animal Medicine and the Institutional Animal Care and Use Committee. Psmb8−/−Psmb9−/− Psmb10−/−Psmb11−/− immunoproteasome/thymoproteasome deficient mice were created by breeding Psmb8−/−Psmb9−/−Psmb10−/− mice 8 to Psmb11−/− mice 5. All wild-type animals were C57BL/6J (Jackson Labs). B6;129P2-Runx3tm1Litt (Runx3dYFP/YFP 20) mice were from NYU SOM, New York, NY. B6.SJL-Ptprca Pep3b/BoyJ (CD45.1, Jackson Labs) C57BL/6-, B6.129P2-H2-Kbtm1 H2-Dbtm1 (H-2Kb−/− H-2Db−/−, Taconic), B6.129S7-Rag1tm1Mom Tg(TcraTcrb)1100Mjb (Rag1−/− OT-I, Taconic), B6.Cg-Tcratm1Mom Tg(TcrLCMV)327Sdz (Tcra−/− P14, Taconic) were purchased from the indicated vendor and/or bred in the UMMS animal facility. Male and female mice were used in approximately equal numbers. All mice were between 6 and 12 weeks old, except BM chimeras, as described below.
Bone Marrow Chimeras
BM recipients (at least 8 weeks old) were irradiated with Cesium 137, in 2 rounds of 550 rad each, 5 h apart, using a Gammacell 40 irradiator (Atomic Energy of Canada). One day later, BM cells (isolated as previously described 8) were injected retroorbitally. All mice received Sulfamethoxazole and Trimethoprim, Oral Suspension, USP 200 mg/40 mg per 5 mL in their drinking water for 6 weeks while engraftment took place.
Cell preparation and flow cytometry analysis
Mice were sacrificed and spleen, inguinal lymph nodes and/ or thymus were harvested. In the case of spleens, single cell suspensions were isolated, erythrocytes were lysed using ACK buffer. In the case of thymus, the tissue was disrupted and passed through a 70μm cell strainer (Costar). In thymic dissections, care was taken to exclude lymph nodes; the normal numbers of thymic CD4SP cells and absence of increased numbers of B cells (Supplementary Fig 1) in the 4KO thymi verified that thymus preparations were not contaminated with lymph nodes. To analyze TECs, the tissue that remained in the strainer was digested using the Miltenyii Splenocyte isolation kit and the GentleMACS dissociator (using program m_spleen_02 before incubation at 37° and m_spleen_01 after). For TEC staining, 1:400 biotin-anti-CD249 (eBioscience) or 1:100 biotin-UEA-1 (Vector Labs) was added to the staining panel, followed by 1:400 V450 Streptavidin (BD Bioscience). Cells were stained for 20 minutes at 4 degrees using the antibodies and dilutions in Supplementary Table 1. For CCR7 staining, cells were first stained as described above, then incubated at 37deg for 30 min, with 1μg biotin-anti-CD197 per 2,500,000 cells, followed by 1:200 Brilliant Violet 421 Streptavidin (BD Bioscience). Flow Cytometry was performed on a FACS Caliber or LSR II (Becton Dickenson), and data was compensated and analyzed using FlowJo(TreeStar). We used biexponential transformations (which only affects the visualization of the data but does not exclude data points) to plot the compensated data. Cell numbers for gated populations were calculated by multiplying total thymocyte number (counted by hemocytometer) by the frequency of that population in the flow cytometry sample.
Fetal Thymic Organ Cultures
E16-E17 embryos from timed pregnant mice were cultured in Costar transwell plates, essentially as previously described50 500μg M5/114 (anti-MHC II) or control LTF-2 mAbs (both from Bio X Cell) was added to the culture media for the 5 d of culture.
Statistical analysis
Statistical analysis of data (SD, t tests, One- and Two-way ANOVA, as indicated in figure legends) was performed using GraphPad Prism version 5.0a for Mac OSX, GraphPad Software, San Diego California USA, www.graphpad.com. We used a pre-established criterion by which thymi with more than 5× the percentage of CD8SP cells compared to comparable samples (indicating contamination with mature lymphocytes) were excluded. Samples size was based on past experience, and were sufficient to show significant differences. Animals were not randomly assigned. No investigator blinding was done.
Supplementary Material
Acknowledgments
We thank D. Littman (NYU SOM, New York, NY) for the Runx3dYFP/YFP mice and A. Singer and E. Huseby for helpful discussions. This work was supported by National Institutes of Health Grants AI20248 and AI110374 (to K.L.R.) and T32CA130807-02 (to E.Z.K.). Kenneth Rock is a member of the UMass DERC (DK32520). Core resources supported by the Diabetes Endocrinology Research Center grant DK32520 were also used. This work was supported in part by JSPS KAKENHI (Grant Numbers 21000012 to K.T. and 25221102 to S.M.), Japan.
Footnotes
Author contribution: E.Z.K. designed and did experiments, analyzed data and wrote the paper. S.M. and K.T. generated the β5t-defidient mice and discussed results and conclusions. K.L.R. designed experiments, supervised the experiments and wrote the paper.
Competing Financial Interests Statement: The authors declare no competing financial interests.
References
- 1.Starr TK, Jameson SC, Hogquist KA. Positive and negative selection of T cells. Annu Rev Immunol. 2003;21:139–176. doi: 10.1146/annurev.immunol.21.120601.141107. [DOI] [PubMed] [Google Scholar]
- 2.Singer A, Adoro S, Park JH. Lineage fate and intense debate: myths, models and mechanisms of CD4- versus CD8-lineage choice. Nat Rev Immunol. 2008;8:788–801. doi: 10.1038/nri2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rock KL, et al. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 4.Coux O, Tanaka K, Goldberg AL. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 5.Murata S, et al. Regulation of CD8+ T cell development by thymus-specific proteasomes. Science. 2007;316:1349–1353. doi: 10.1126/science.1141915. [DOI] [PubMed] [Google Scholar]
- 6.Guillaume B, et al. Two abundant proteasome subtypes that uniquely process some antigens presented by HLA class I molecules. Proc Natl Acad Sci U S A. 2010;107:18599–18604. doi: 10.1073/pnas.1009778107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kisselev AF, Akopian TN, Castillo V, Goldberg AL. Proteasome active sites allosterically regulate each other, suggesting a cyclical bite-chew mechanism for protein breakdown. Mol Cell. 1999;4:395–402. doi: 10.1016/s1097-2765(00)80341-x. [DOI] [PubMed] [Google Scholar]
- 8.Kincaid EZ, et al. Mice completely lacking immunoproteasomes show major changes in antigen presentation. Nat Immunol. 2012;13:129–135. doi: 10.1038/ni.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sasaki K, et al. Thymoproteasomes produce unique peptide motifs for positive selection of CD8(+) T cells. Nat Commun. 2015;6:7484. doi: 10.1038/ncomms8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xing Y, Jameson SC, Hogquist KA. Thymoproteasome subunit-β5T generates peptide-MHC complexes specialized for positive selection. Proc Natl Acad Sci U S A. 2013;110:6979–6984. doi: 10.1073/pnas.1222244110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hogquist KA, Jameson SC, Bevan MJ. Strong agonist ligands for the T cell receptor do not mediate positive selection of functional CD8+ T cells. Immunity. 1995;3:79–86. doi: 10.1016/1074-7613(95)90160-4. [DOI] [PubMed] [Google Scholar]
- 12.Klein L, Kyewski B, Allen PM, Hogquist KA. Positive and negative selection of the T cell repertoire: what thymocytes see (and don't see) Nat Rev Immunol. 2014;14:377–391. doi: 10.1038/nri3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nitta T, et al. Thymoproteasome shapes immunocompetent repertoire of CD8+ T cells. Immunity. 2010;32:29–40. doi: 10.1016/j.immuni.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 14.Klein L, Hinterberger M, Wirnsberger G, Kyewski B. Antigen presentation in the thymus for positive selection and central tolerance induction. Nat Rev Immunol. 2009;9:833–844. doi: 10.1038/nri2669. [DOI] [PubMed] [Google Scholar]
- 15.Azzam HS, et al. CD5 expression is developmentally regulated by T cell receptor (TCR) signals and TCR avidity. J Exp Med. 1998;188:2301–2311. doi: 10.1084/jem.188.12.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takada K, et al. TCR affinity for thymoproteasome-dependent positively selecting peptides conditions antigen responsiveness in CD8(+) T cells. Nat Immunol. 2015;16:1069–1076. doi: 10.1038/ni.3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hogquist KA, et al. Identification of a naturally occurring ligand for thymic positive selection. Immunity. 1997;6:389–399. doi: 10.1016/s1074-7613(00)80282-4. [DOI] [PubMed] [Google Scholar]
- 18.Yamashita I, Nagata T, Tada T, Nakayama T. CD69 cell surface expression identifies developing thymocytes which audition for T cell antigen receptor-mediated positive selection. Int Immunol. 1993;5:1139–1150. doi: 10.1093/intimm/5.9.1139. [DOI] [PubMed] [Google Scholar]
- 19.Davey GM, et al. Preselection thymocytes are more sensitive to T cell receptor stimulation than mature T cells. J Exp Med. 1998;188:1867–1874. doi: 10.1084/jem.188.10.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Egawa T, Littman DR. ThPOK acts late in specification of the helper T cell lineage and suppresses Runx-mediated commitment to the cytotoxic T cell lineage. Nat Immunol. 2008;9:1131–1139. doi: 10.1038/ni.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grueter B, et al. Runx3 regulates integrin alpha E/CD103 and CD4 expression during development of CD4-/CD8+ T cells. J Immunol Baltim Md 1950. 2005;175:1694–1705. doi: 10.4049/jimmunol.175.3.1694. [DOI] [PubMed] [Google Scholar]
- 22.Takeuchi Y, Habu S, Okumura K, Suzuki G. Cyclosporin A and anti-Ia antibody cause a maturation defect of CD4+8- cells in organ-cultured fetal thymus. Immunology. 1989;66:362–367. [PMC free article] [PubMed] [Google Scholar]
- 23.Ueno T, et al. CCR7 signals are essential for cortex-medulla migration of developing thymocytes. J Exp Med. 2004;200:493–505. doi: 10.1084/jem.20040643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Egawa T, Tillman RE, Naoe Y, Taniuchi I, Littman DR. The role of the Runx transcription factors in thymocyte differentiation and in homeostasis of naive T cells. J Exp Med. 2007;204:1945–1957. doi: 10.1084/jem.20070133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adoro S, et al. Coreceptor gene imprinting governs thymocyte lineage fate. EMBO J. 2012;31:366–377. doi: 10.1038/emboj.2011.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merkenschlager M, et al. Centromeric repositioning of coreceptor loci predicts their stable silencing and the CD4/CD8 lineage choice. J Exp Med. 2004;200:1437–1444. doi: 10.1084/jem.20041127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stritesky GL, et al. Murine thymic selection quantified using a unique method to capture deleted T cells. Proc Natl Acad Sci U S A. 2013;110:4679–4684. doi: 10.1073/pnas.1217532110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takahama Y. Journey through the thymus: stromal guides for T-cell development and selection. Nat Rev Immunol. 2006;6:127–135. doi: 10.1038/nri1781. [DOI] [PubMed] [Google Scholar]
- 29.Bouillet P, et al. BH3-only Bcl-2 family member Bim is required for apoptosis of autoreactive thymocytes. Nature. 2002;415:922–926. doi: 10.1038/415922a. [DOI] [PubMed] [Google Scholar]
- 30.Suen AYW, Baldwin TA. Proapoptotic protein Bim is differentially required during thymic clonal deletion to ubiquitous versus tissue-restricted antigens. Proc Natl Acad Sci U S A. 2012;109:893–898. doi: 10.1073/pnas.1114834109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gray DHD, et al. The BH3-only proteins Bim and Puma cooperate to impose deletional tolerance of organ-specific antigens. Immunity. 2012;37:451–462. doi: 10.1016/j.immuni.2012.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lehmann-Grube F, Dralle H, Utermöhlen O, Löhler J. MHC class I molecule-restricted presentation of viral antigen in beta 2-microglobulin-deficient mice. J Immunol Baltim Md 1950. 1994;153:595–603. [PubMed] [Google Scholar]
- 33.Dolan BP, Gibbs KD, Ostrand-Rosenberg S. Dendritic cells cross-dressed with peptide MHC class I complexes prime CD8+ T cells. J Immunol Baltim Md 1950. 2006;177:6018–6024. doi: 10.4049/jimmunol.177.9.6018. [DOI] [PubMed] [Google Scholar]
- 34.McCaughtry TM, Baldwin TA, Wilken MS, Hogquist KA. Clonal deletion of thymocytes can occur in the cortex with no involvement of the medulla. J Exp Med. 2008;205:2575–2584. doi: 10.1084/jem.20080866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marrack P, Kappler J. The T cell receptor. Science. 1987;238:1073–1079. doi: 10.1126/science.3317824. [DOI] [PubMed] [Google Scholar]
- 36.Derbinski J, et al. Promiscuous gene expression in thymic epithelial cells is regulated at multiple levels. J Exp Med. 2005;202:33–45. doi: 10.1084/jem.20050471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Honey K, Rudensky AY. Lysosomal cysteine proteases regulate antigen presentation. Nat Rev Immunol. 2003;3:472–482. doi: 10.1038/nri1110. [DOI] [PubMed] [Google Scholar]
- 38.Gommeaux J, et al. Thymus-specific serine protease regulates positive selection of a subset of CD4+ thymocytes. Eur J Immunol. 2009;39:956–964. doi: 10.1002/eji.200839175. [DOI] [PubMed] [Google Scholar]
- 39.Nussbaum AK, Rodriguez-Carreno MP, Benning N, Botten J, Whitton JL. Immunoproteasome-deficient mice mount largely normal CD8+ T cell responses to lymphocytic choriomeningitis virus infection and DNA vaccination. J Immunol Baltim Md 1950. 2005;175:1153–1160. doi: 10.4049/jimmunol.175.2.1153. [DOI] [PubMed] [Google Scholar]
- 40.Mandl JN, Monteiro JP, Vrisekoop N, Germain RN. T cell-positive selection uses self-ligand binding strength to optimize repertoire recognition of foreign antigens. Immunity. 2013;38:263–274. doi: 10.1016/j.immuni.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baldwin TA, Hogquist KA, Jameson SC. The fourth way? Harnessing aggressive tendencies in the thymus. J Immunol Baltim Md 1950. 2004;173:6515–6520. doi: 10.4049/jimmunol.173.11.6515. [DOI] [PubMed] [Google Scholar]
- 42.Kurobe H, et al. CCR7-dependent cortex-to-medulla migration of positively selected thymocytes is essential for establishing central tolerance. Immunity. 2006;24:165–177. doi: 10.1016/j.immuni.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 43.Anderson MS, et al. Projection of an immunological self shadow within the thymus by the aire protein. Science. 2002;298:1395–1401. doi: 10.1126/science.1075958. [DOI] [PubMed] [Google Scholar]
- 44.Reiser H, Schneeberger EE. The costimulatory molecule B7 is expressed in the medullary region of the murine thymus. Immunology. 1994;81:532–537. [PMC free article] [PubMed] [Google Scholar]
- 45.Jenkinson EJ, Anderson G, Moore NC, Smith CA, Owen JJ. Positive selection by purified MHC class II+ thymic epithelial cells in vitro: costimulatory signals mediated by B7 are not involved. Dev Immunol. 1994;3:265–271. doi: 10.1155/1994/75434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sprent J, Surh CD. Re-entry of mature T cells to the thymus: an epiphenomenon? Immunol Cell Biol. 2009;87:46–49. doi: 10.1038/icb.2008.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ernst B, Lee DS, Chang JM, Sprent J, Surh CD. The peptide ligands mediating positive selection in the thymus control T cell survival and homeostatic proliferation in the periphery. Immunity. 1999;11:173–181. doi: 10.1016/s1074-7613(00)80092-8. [DOI] [PubMed] [Google Scholar]
- 48.Goldrath AW, Bevan MJ. Low-affinity ligands for the TCR drive proliferation of mature CD8+ T cells in lymphopenic hosts. Immunity. 1999;11:183–190. doi: 10.1016/s1074-7613(00)80093-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ohta Y, McKinney EC, Criscitiello MF, Flajnik MF. Proteasome, transporter associated with antigen processing, and class I genes in the nurse shark Ginglymostoma cirratum: evidence for a stable class I region and MHC haplotype lineages. J Immunol Baltim Md 1950. 2002;168:771–781. doi: 10.4049/jimmunol.168.2.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only references
- 50.Ramsdell F. In: Current Protocols in Immunology. Coligan JE, Bierer BE, Margulies DH, Shevach EM, Strober W, editors. John Wiley & Sons, Inc; 2001. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.