Abstract
Most of the known regulatory mechanisms that curb inflammatory gene expression target pre-transcription initiation steps and evidence for regulation of inflammatory gene expression post initiation remains scarce. Here we show that transcription repressor hairy and enhancer of split 1 (Hes1) suppresses production of CXCL1, a chemokine crucial for recruiting neutrophils. Hes1 negatively regulates neutrophil recruitment in vivo in a manner that is dependent on macrophage-produced CXCL1 and attenuates severity of inflammatory arthritis. Mechanistically, inhibition of Cxcl1 expression by Hes1 does not involve modification of transcription initiation. Instead, Hes1 inhibits signal-induced recruitment of positive transcription elongation complex P-TEFb, thereby preventing phosphorylation of RNA polymerase II on serine-2 and productive elongation. Thus, our results identify Hes1 as a homeostatic suppressor of inflammatory responses which exerts its suppressive function by regulating transcription elongation.
Introduction
Cytokines and chemokines recruit and activate specialized effector cells to sites of inflammation1. However, excessive production of inflammatory mediators leads to immune hyper-activation and tissue damage and contributes to pathogenesis of inflammatory and autoimmune disorders such as rheumatoid arthritis (RA)2. Therefore, expression of inflammatory mediators must be precisely controlled to avoid inappropriate inflammation and tissue damage. Many negative regulatory mechanisms have been described to curb inflammatory mediator production at multiple levels3. In particular the complex nature of transcription makes it suitable for precise and selective regulation essential for mounting inflammatory responses most appropriate to given environmental cues4. Transcription of inflammatory genes can be negatively regulated via direct inhibition or epigenetic modifications to close chromatin structures5. Indeed, most of the described mechanisms of inflammatory gene regulation occur at or prior to transcriptional initiation by modulating RNA polymerase II (Pol II) recruitment to transcription start sites (TSS)6. However, advancements in investigation of the transcription cycle facilitated by high-throughput sequencing technology strongly argue that regulation at the post-initiation stage is extensive in scope and highly conserved across species from Drosophila to mammals7,8.
Transcription elongation is a stepwise process during which Pol II ultimately synthesizes the full length RNA transcript. During early elongation, Pol II escapes the promoter, transcribes a short RNA transcript and pauses at ∼50 nucleotides downstream of the transcription start site. Pausing can be subsequently released by the positive transcription elongation factor b (P-TEFb) that phosphorylates the regulatory C-terminal domain (CTD) of Pol II and facilitates productive elongation7,9. Regulation of transcription elongation in the immune system has not been widely appreciated yet accumulating evidence suggests that such regulation is critical for fine tuning expression of a subset of key inflammatory mediators10-12.
Transcription repressor hairy and enhancer of split 1 (Hes1) belongs to a family of basic helix-loop-helix (bHLH) DNA binding proteins and plays key roles in the development of multiple organs and cell types13. As a result, mice globally deficient in the Hes1 gene are not viable and display multiple developmental defects14. Recent studies reveal that expression of Hes1 can be modulated by innate and inflammatory signals15-17 and Hes1, in turn, negatively regulates macrophage TLR responses15, expanding the role of Hes1 in immune regulation beyond developmental processes18 and suggesting potential involvement of Hes1 in autoimmune and inflammatory disorders such as RA and systemic lupus erythematosus (SLE)19-22. However, the molecular mechanisms, transcription targets, and physiological significance of Hes1-mediated regulation of inflammation remain largely unknown.
Here, we evaluated the role of Hes1 on gene regulation in primary macrophages and in inflammatory conditions in vivo. We show that Hes1 restrains inflammation and especially neutrophil-mediated responses by controlling production of macrophage-derived chemokines. The inhibitory effects of Hes1 are highly restricted to a small subset of genes in the macrophage inflammatory transcriptome. Finally, we used one such Hes1-sensitive gene (Cxcl1) to assess the mechanistic basis of the non-conventional function of the Hes1 repressor.
Results
Hes1 and Hey1 regulate the inflammatory transcriptome
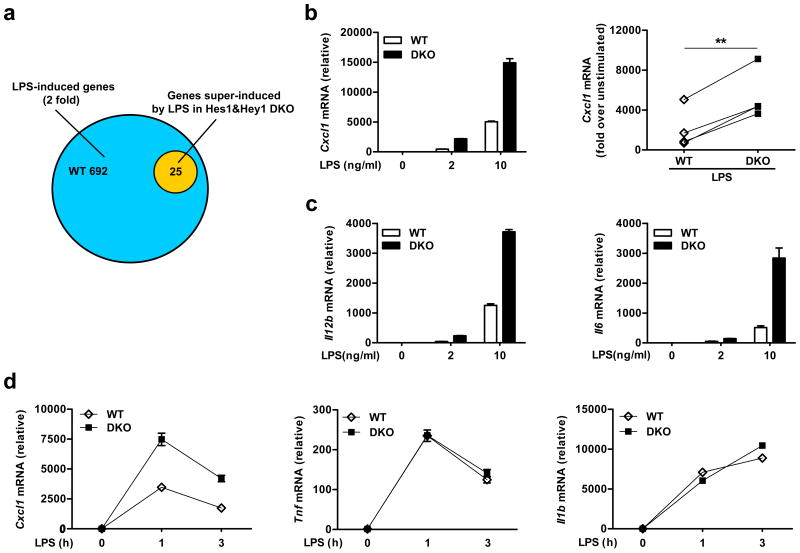
Inflammatory stimuli such as TLR ligands consistently induce expression of two Hes family members Hes1 and Hey1 in macrophages15. To evaluate the effects of these two factors on inflammatory responses while circumventing their potential functional redundancy, we generated mice lacking both Hes1 and Hey1. Mice with global deletion of Hes1 are not viable14 whereas Hey1-deificent mice are phenotypically normal23. Therefore, we generated mice with the genotype of Hey1–/–Hes1fl/flMx1-Cre. These mice were phenotypically indistinguishable from WT littermate controls after induction of deletion and did not display gross abnormalities up to 20 weeks of age. To identify Hes1- and Hey1-regulated inflammatory genes, we generated bone marrow-derived macrophages (BMDMs) from these mice and confirmed efficient deletion in Hes1 and Hey1-deficeint BMDMs (Supplementary Fig. 1a). Microarray analysis of lipopolysaccharide (LPS)-treated BMDMs showed that out of hundreds of genes induced by LPS (>2 fold) in wild-type cells, 25 genes were super-induced in Hes1 and Hey1-deficient BMDMs compared to wild-type (Fig. 1a and Supplementary Fig. 1b), suggesting that Hes1 and Hey1 inhibited inflammatory gene expression in a highly selective manner. Moreover, Hes1 and Hey1 regulated expression of a small subset of LPS-suppressed genes (Supplementary Fig. 1c). Super-induced genes encoded several key immune and inflammatory effectors including CXCL1, IL-12p40, and IL-6 (Supplementary Fig. 1b). Super-induction of Cxcl1, Il12b and Il6 in Hes1 and Hey1-deficient BMDMs was confirmed by quantitative real-time PCR (qPCR) in multiple independent experiments (Fig. 1b,c). We chose to focus on Cxcl1 as its regulation by Hes1 and Hey1 was among the most striking and reliable. Super-induction of Cxcl1 was also observed in Hes1 and Hey1-deficient BMDMs in response to other TLR ligands such as Pam3Cys, a TLR2 ligand, and R848, a TLR7 ligand (Supplementary Fig. 1d), demonstrating that Hes1 and Hey1-mediated suppression of Cxcl1 is not specific to TLR4. In contrast to the regulation of Cxcl1, Hes1 and Hey1 deficiency did not affect expression of two genes encoding prototypical pro-inflammatory cytokines TNF and IL-1β (Fig. 1d). Taken together, these data indicated that Hes1 and Hey1 functioned as selective regulators of the macrophage inflammatory transcriptome.
Figure 1.
Hes1 and Hey1 inhibit expression of a subset of inflammatory genes. (a) Pie graph showing the numbers of LPS-induced genes in wild-type (Hey1+/+Hes1fl/fl; blue) BMDMs (wild-type, WT) and super-induced genes in Hes1 and Hey1-deficient (Hey1–/–Hes1fl/flMx1-Cre; orange) BMDMs (DKO) stimulated for 3 h with LPS (2 ng/ml). (b, c) Quantification of mRNA expression of Cxcl1 (b, left panel), Il12b and Il6 (c) in WT and DKO BMDMs stimulated for 3h with LPS at the indicated doses. (d) Quantification of mRNA expression of Cxcl1, Tnf, and Il1b in WT and DKO BMDMs stimulated with LPS (10 ng/ml) for the indicated periods. Representative data of four (b, left panel) or three (c,d) independent experiments are shown as means and s.d. of technical triplicate determinants. Cumulative data (LPS, 10 ng/ml LPS) of Cxcl1 induction (fold change over unstimulated condition) in WT and DKO BMDMs from four independent experiments are shown in b, right panel. **P<0.01, Student's paired t-test.
Hes1, but not Hey1, suppresses Cxcl1 expression
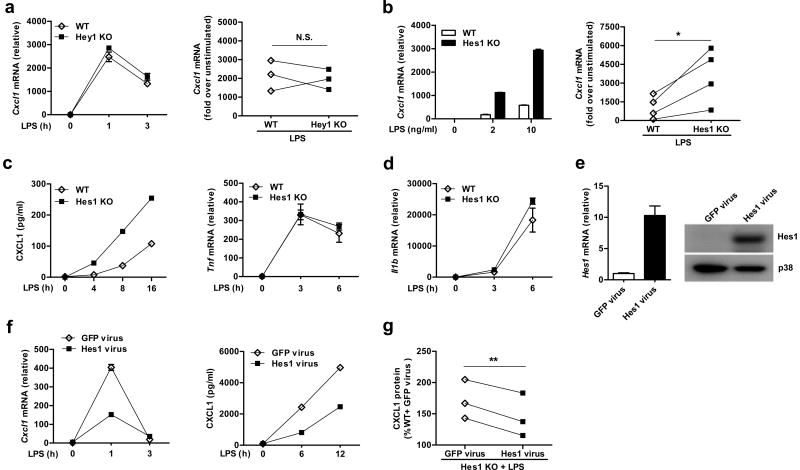
Given the gene regulation patterns in Hes1 and Hey1-deficient BMDMs, we next asked whether both Hes1 and Hey1 contributed to suppression of Cxcl1 expression. The results from multiple experiments showed that Cxcl1 induction was comparable in WT and Hey1–/– KO BMDMs (Fig. 2a), even in response to a relatively high dose (100 ng/ml) of LPS stimulation (Supplementary Fig. 2a), indicating that Hey1 deficiency alone is not sufficient to potentiate Cxcl1 expression. To investigate whether Hes1 suppresses Cxcl1 expression, we generated Hes1 inducible knock-out mice by crossing Hes1fl/fl animals with Mx1-Cre animals (Hes1fl/flMx1-Cre). Efficient deletion of Hes1 was verified in BMDMs at both mRNA and protein levels (Supplementary Fig. 2b,c). Upon stimulation, Cxcl1 was super-induced in Hes1-deficient BMDMs in a pattern similar to that observed in Hes1 and Hey1-deficient cells (Fig. 2b). CXCL1 protein was also increased in supernatants of Hes1-deficient BMDMs (Fig. 2c). Similar to observations made in Hes1 and Hey1-deficient BMDMs, Hes1 single deficiency did not affect expression of Tnf and Il1b (Fig. 2d) and Hes1-mediated suppression of Cxcl1 was not specific to TLR4 signaling (Supplementary Fig. 2d). In contrast to an apparent lack of a role of Hey1 in Cxcl1 regulation, deletion of either Hey1 or Hes1 led to super-induction of Il12b (Supplementary Fig. 2e), suggesting that Il12b expression was inhibited by Hes and Hey family members via mechanisms distinct from those regulating Cxcl1.
Figure 2.
Hes1 but not Hey1 suppresses Cxcl1 expression. (a,b) Quantification of Cxcl1 mRNA expression in Hey1+/+ (WT) and Hey1–/–(Hey1 KO) BMDMs stimulated with LPS for the indicated time (a) or in Hes1+/+Mx1-Cre (WT) and Hes1fl/flMx1-Cre (Hes1 KO) BMDMs stimulated with LPS for 3 h (b). (c,d) ELISA for CXCL1 protein levels (c) or quantification of Tnf and Il1b mRNA expression (d) in WT and Hes1 KO BMDMs stimulated with LPS for indicated times. (e) Quantification of Hes1 mRNA expression (left panel) and immunoblot analysis of Hes1 protein (right panel) in BMDMs transduced with GFP or Hes1 retrovirus. p38 served as a loading control. (f) Quantification of Cxcl1 mRNA expression (left panel) and ELISA of CXCL1 protein levels (right panel) in WT BMDMs transduced with GFP or Hes1 retrovirus and subsequently stimulated with LPS for the indicated periods. (g) ELISA for CXCL1 protein levels in WT and Hes1 KO BMDMs transduced with GFP or Hes1 retrovirus and subsequently stimulated with LPS (10 ng/ml) for 6 h. Representative data from three (a, left panel), four (b, left panel) or two independent experiments (c-e,f) are shown and data are mean and s.d. of technical triplicate determinants (a,b,e, left panels & d,f). Cumulative data from three (a, left panel, g) or four (b, left panel) independent experiments are shown as Cxcl1 induction in a paired manner (left panels of a,b) or percentages of CXCL1 levels in LPS-stimulated WT BMDMs transduced with GFP virus (g). *P<0.05, **P<0.01, Student's paired t-test.
To rule out the possibility that our observations are linked to a specific deletion strategy, we generated two more mouse lines, namely the Hes1fl/flCre-ERT2 mice in which gene deletion was induced by tamoxifen treatment and myeloid-specific Hes1-deficient mice in which gene deletion was constitutive (Supplementary Fig. 3a,b,d). Compared to wild-type controls, super-induction of Cxcl1 was also observed in macrophages derived from animals with inducible Cre-ERT2 or myeloid-specific Hes1-deficient animals (Supplementary Fig. 3c,e), indicating that Hes1-mediated gene inhibition is a robust strain-independent phenomenon. As Mx1-Cre consistently achieved high deletion efficiency in macrophages, we proceeded with Hes1fl/flMx1-Cre animals in most of the following experiments. To further corroborate these observations in a gain-of-function system, Hes1 was over-expressed in wild-type BMDMs by retroviral transduction (Fig. 2e). Over-expression of Hes1 decreased Cxcl1 mRNA and protein production (Fig. 2f), showing that exogenously introduced Hes1 was sufficient to mediate the inhibition. The suppressive effects of Hes1 on Cxcl1 expression were lost upon deleting or mutating Hes1 key functional domains13,24 (Supplementary Fig. 4a-c). In addition, Hes1 reconstitution in Hes1-deficient BMDMs inhibited CXCL1 production (Fig. 2g). Taken together, these loss-of-function and gain-of-function studies support a critical regulatory role for Hes1 in macrophages as a selective suppressor of inflammatory gene expression.
Hes1 inhibits neutrophil recruitment in vivo
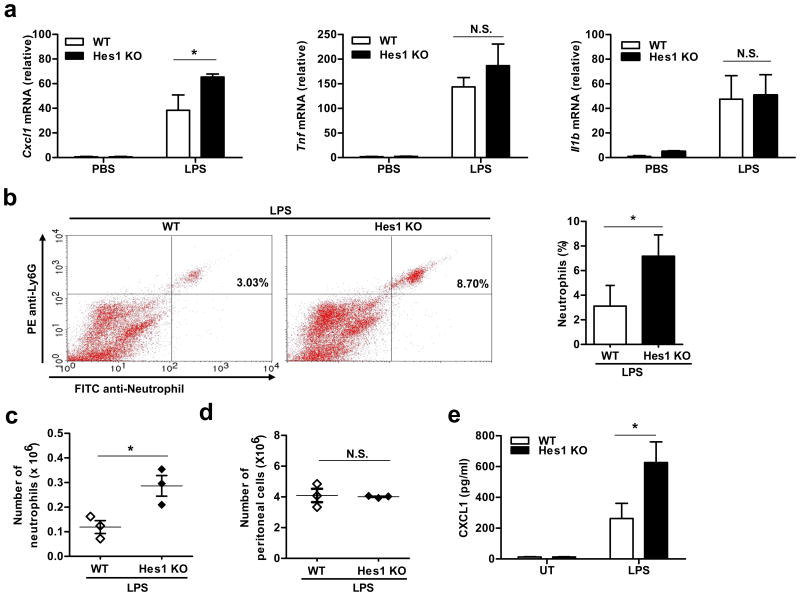
Having established that Hes1 negatively regulated Cxcl1 expression in vitro in cultured BMDMs, we wished to determine whether Hes1-mediated regulation also occurred in vivo. In an LPS-induced peritonitis model, Hes1 deficiency in the hematopoietic compartment promoted expression of Cxcl1 in peritoneal exudates without significantly altering Tnf and Il1b mRNA levels (Fig. 3a). Given the importance of CXCL1 in control of neutrophil recruitment during inflammatory responses25, we next examined whether Hes1 deficiency affected neutrophil recruitment in vivo. In the peritonitis model, Hes1 deficiency resulted in increased percentage and number of neutrophils in the peritoneum without affecting the total number of peritoneal cells and composition of other peritoneal populations (Fig. 3b-d and Supplementary Fig. 5a-c), indicating that Hes1 specifically suppressed neutrophil recruitment in vivo. The increased recruitment of neutrophils seen in Hes1–/– animals was not due to intrinsically enhanced chemotactic capacity of neutrophils as Hes1-deficient and wild-type neutrophils displayed similar motility in response to CXCL1 (Supplementary Fig. 5d-f). Instead, macrophages in the peritoneal cavity of Hes1–/– mice produced significantly higher CXCL1 than those in wild-type animals (Fig. 3e), suggesting that Hes1 acted primarily to inhibit chemokine production by macrophages and thereby indirectly reduced neutrophil trafficking. This notion was further supported by the findings that adoptive transfer of Hes1-deficient macrophages into wild-type hosts led to increased recruitment of wild-type neutrophils (Supplementary Fig. 5g). These data establish Hes1 as a selective suppressor of Cxcl1 expression in vitro and in vivo with a non-redundant role in controlling neutrophil responses during inflammatory conditions.
Figure 3.
Hes1 negatively regulates Cxcl1 expression and neutrophil recruitment in vivo. (a) qPCR analysis of the indicated gene expression in peritoneal cells from WT and Hes1 KO bone marrow chimeric mice. Peritoneal cells were harvested 4 h after intraperitoneal injection of PBS or LPS (100 ng/mouse). Results from three independent experiments are shown as mean and s.d. (n = 3). (b) FACS analysis of neutrophil population (Ly-6G and 7/4 double positive) in peritoneal exudates of WT and Hes1 KO mice treated with LPS as in (a) Representative flow cytometry distribution is shown in the left panel and percentage of neutrophils from three independent experiments are quantified as mean and s.d. in the right panel (n=3). (c,d) Cumulative data from three independent experiments show total neutrophil numbers (c) and total peritoneal cell numbers (d) from WT and Hes1 KO mice treated as in (b). (e) ELISA for Cxcl1 protein levels in peritoneal macrophages from WT (Hes1+/+Cre-ERT2) and Hes1 KO (Hes1fl/flCre-ERT2) mice were stimulated with 100 ng/ml of LPS for 12 h. Data from three independent experiments are shown as mean and s.d. (n=3). *P<0.05 (paired Student's t-test).
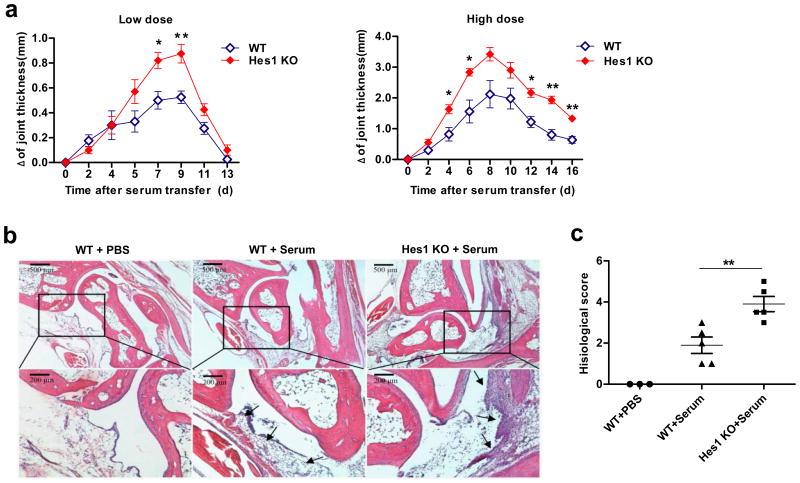
Neutrophils are required for development of inflammatory arthritic symptoms in several experimental animal models including K/B×N-serum transfer-induced arthritis25,26. We examined whether Hes1 deficiency regulated disease severity in serum-induced arthritis by monitoring joint swelling, a hallmark of acute inflammatory responses. Joint swelling was more severe in Hes1-deficient mice at multiple time points following both low and high dose of serum administrations (Fig. 4a), suggesting that Hes1 deficiency exacerbated disease severity. Consistent with the joint measurement results, histological analysis of ankle joints showed heightened signs of inflammation and histological scores in Hes1-deficient mice compared to wild-type controls (Fig. 4b-c). Collectively, these data showed that the presence of Hes1 attenuated development of inflammatory arthritis and suggested that Hes1 functions as an endogenous brake of neutrophil-mediated tissue inflammation in a complex disease setting.
Figure 4.
Hes1 deficiency exacerbates inflammatory arthritis. (a) Kinetics of joint swelling induced by K/B×N serum transfer with low dose (100 μl) and high dose (150 μl) in WT and Hes1 KO chimeric mice at the indicated days. Total joint swelling is shown as Δ of joint thickness. Data are shown as mean ± SEM. (low dose, n=4 in each group; high dose, WT: n=5, Hes1 KO: n=3). *P<0.05, **P<0.01 (Student's t test). (b) Histological features of ankle joints of mice examined on day 3 after serum transfer (high dose). H&E-stained images of low-resolution (4×, upper panels) and higher-resolution (10×, lower panels) of selected areas from corresponding sections (boxed area) are shown. Infiltration of inflammatory cells after serum transfer is shown with arrows. (c) Histological scores of ankle joints examined on day 3 after serum transfer (high dose). Each symbol indicates one mouse and score of each mouse is average of two ankles (n=3-5 per group). **P<0.01 (Student's t test).
Hes1 represses Cxcl1 transcription
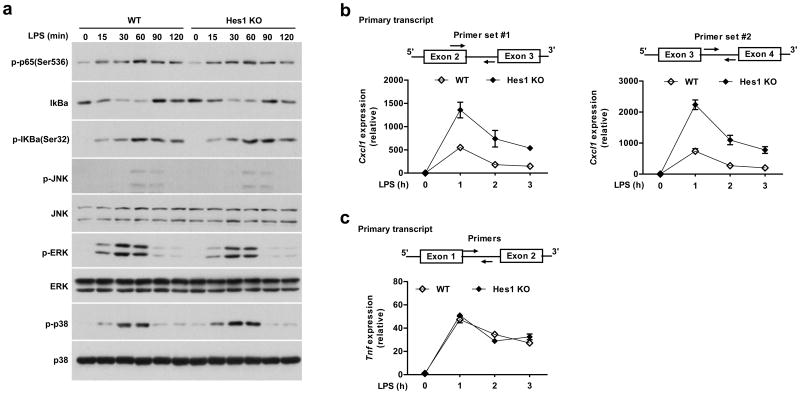
Next we sought to investigate the mechanisms by which Hes1 suppressed Cxcl1 expression. It is well established that TLR-induced activation of nuclear factor-kappa B (NF-κB) and mitogen-activated protein kinase (MAPK) signaling cascades contribute to inflammatory gene activation27. We found that Hes1 deficiency did not affect TLR4-induced activation of NF-κB or of MAPKs p38, Jnk, and Erk (Fig. 5a), indicating that Hes1 did not inhibit Cxcl1 expression by altering TLR-induced canonical signaling events. Next, to assess whether Hes1 suppressed Cxcl1 gene transcription, Cxcl1 primary transcripts were detected by using intronic PCR primers. Similar to Cxcl1 steady-state mRNA levels (Fig. 2b,c), levels of Cxcl1 primary transcripts were elevated in Hes1-deficient BMDMs compared with those in wild-type cells (Fig. 5b), implying that Hes1-mediated suppression of Cxcl1 expression is at the transcriptional level. In contrast, Hes1 deficiency did not affect levels of Tnf primary transcripts (Fig. 5c).
Figure 5.
Hes1 represses Cxcl1 transcription without altering NF-κB and MAPK activation. (a) Immunoblot analysis of the indicated proteins in whole cell lysates of LPS-treated BMDMs from WT and Hes1 KO paired littermates. (b,c) Levels of primary transcripts of Cxcl1 (b) and Tnf (c) in BMDMs from WT and Hes1 KO littermates were quantitated by qPCR. Data are shown as mean and s.d. of triplicate determinants.
A previously characterized mode of Hes1's action is via binding to the conserved DNA sequences such as N-box and E-box near gene promoters13. Analysis of the Cxcl1 core promoter region revealed two putative E-box sequences (Supplementary Fig. 6a). We thus investigated whether Hes1 suppressed Cxcl1 transcription via acting through these putative E-boxes using luciferase assays with a Cxcl1 reporter construct containing the E box sites. Over-expression of Hes1 did not inhibit Cxcl1 promoter-driven luciferase activities in RAW264.7 cells under either basal or stimulated conditions (Supplementary Fig. 6b), indicating that the presence of E-box sequences was not sufficient for Hes1-mediated suppression of Cxcl1 transcription in this system.
Hes1 suppresses Cxcl1 gene transcription elongation
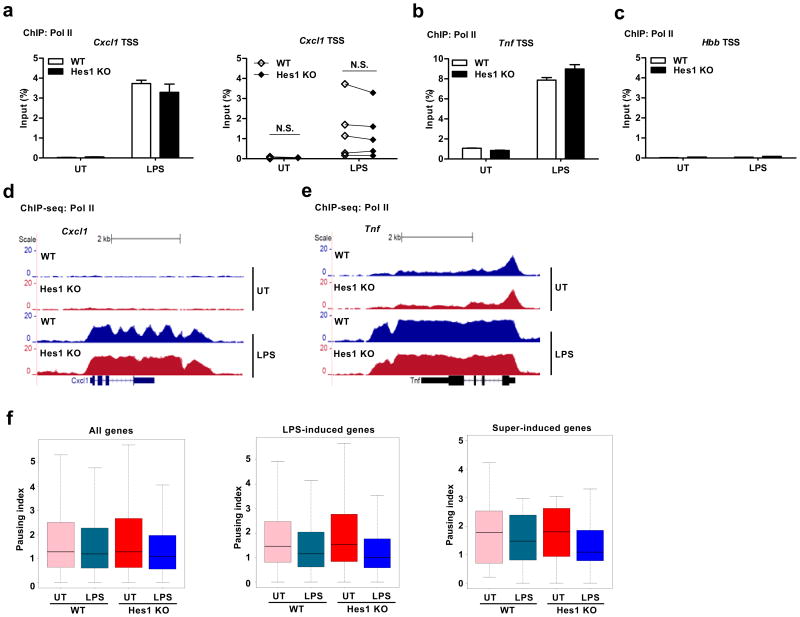
Given that Hes1 did not act upon either upstream signaling pathways or promoter response elements to suppress Cxcl1 gene transcription, we considered other plausible mechanisms of gene regulation. Previous studies have demonstrated that in macrophages, Cxcl1 is a primary response gene whose locus adopts an open chromatin configuration at baseline4 and therefore, epigenetic mechanisms that target chromatin structures are unlikely to be involved in gene regulation. Thus, we examined whether Hes1 targeted specific steps of the transcription cycle to suppress Cxcl1 expression. First, we tested whether Hes1 suppressed Cxcl1 gene transcription initiation by using chromatin immunoprecipitation (ChIP) assays to assess binding of Pol II to the TSS region. While minimal Pol II occupancy (< 0.05% of input) was observed near the Cxcl1 TSS region at the basal level in resting BMDMs, occupancy of Pol II dramatically increased upon LPS stimulation (Fig. 6a), indicating that Cxcl1 fits the criteria of a non-paused gene that displays low levels of promoter-bound Pol II prior to activation and robustly recruits Pol II for gene induction. In contrast, substantial Pol II occupancy was detected at baseline near the Tnf TSS region (Fig. 6b), an observation consistent with the fact that Tnf has been reported to be a paused gene10. Lack of Pol II occupancy at the silent hemoglobin beta (Hbb) locus served as a negative control (Fig. 6c). Levels of Pol II occupancy near the Tnf TSS region did not differ between wild-type and Hes1-deficient BMDMs after LPS stimulation (Fig. 6b) as predicted from the failure of Hes1 to regulate Tnf gene transcription (Fig. 5c). Hes1 deficiency also did not affect Pol II occupancy near the Cxcl1 TSS region (Fig. 6a) despite the fact that Cxcl1 gene transcription was markedly enhanced in Hes1-deficient cells (Fig. 5b), suggesting that Hes1 did not regulate Cxcl1 transcription initiation and may instead target a post-initiation step(s). In contrast to Cxcl1, Hes1 deficiency correlated with increased recruitment of Pol II to the TSS regions of Il6 and Il12b genes (Supplementary Fig. 6c), indicating that Hes1 inhibited Il6 and Il12b transcription initiation. This result was consistent with our earlier data that Hes1 acts on promoter elements to repress these genes15 and with the observation that these gene were sensitive to suppression by either Hes1 or Hey1 (Supplementary Fig. 2e), pointing to a mechanism of inhibition likely distinct from that operating at Cxcl1.
Figure 6.
Hes1 attenuates Pol II occupancy at the Cxcl1 gene body region. (a-c) Pol II occupancy on the TSS region of Cxcl1 (a), Tnf (b) and Hbb (c) in Hes1+/+Mx1-Cre (WT) and Hes1fl/flMx1-Cre (Hes1 KO) BMDMs untreated (UT) or stimulated with LPS (10 ng/ml) for 1 h. Occupancy at the Hbb locus served as a negative control (c). (d,e) Pol II ChIP followed by deep sequencing (ChIP-seq) shows Pol II distribution along the Cxcl1 (d) and Tnf (e) gene tracks in WT and Hes1 KO BMDMs untreated or stimulated with LPS (10 ng/ml) for 1 h. (f) Pausing indices computed from Pol II distribution of all genes and two gene subsets (LPS-induced genes in WT and super-induced genes as in Fig. 1a) in untreated or LPS stimulated WT and Hes1 KO BMDMs. Data shown are representative results (a-c, mean and s.d. of technical triplicate determinants) from one of five (a, left panel) or two (b-e) independent experiments, or cumulative data from five independent experiments (a, right panel).
To gain further insight into post-initiation regulation, we assessed by ChIP-seq genome wide distribution of Pol II before or after LPS stimulation. Pol II occupancy was undetectable throughout the Cxcl1 gene locus under resting conditions in both wild-type and Hes1-deificient BMDMs (Fig. 6d, upper two panels), which was consistent with the results of the ChIP assays and confirmed that Cxcl1 was indeed a non-paused gene. Upon LPS stimulation, Pol II occupancy on Cxcl1 gene locus drastically increased near the TSS and throughout the gene body in wild-type BMDMs (Fig. 6d, panel 3). In line with the ChIP data (Fig. 6a), Pol II recruitment near the Cxcl1 TSS did not significantly differ between wild-type and Hes1-deficient BMDMs (Fig. 6d, lower two panels). Pol II occupancy throughout the gene body was markedly increased in Hes1-deficient cells compared to wild-type controls (Fig. 6d), which implied that Hes1 deficiency increased the amounts of Pol II molecules available to transcribe the Cxcl1 gene potentially accounting for its enhanced expression in Hes1-deficient cells. In contrast to the Cxcl1 gene locus, substantial basal Pol II occupancy near the Tnf TSS was observed under unstimulated conditions (Fig. 6e, upper two panels), consistent with previous studies establishing Tnf as a paused gene10. Upon LPS treatment, Pol II occupancy in the Tnf gene body also drastically increased but showed similar patterns in wild-type and Hes1-deficient BMDMs (Fig. 6e, lower two panels), suggesting that Hes1 did not affect transcriptional elongation of this gene. Next, based on whole genome Pol II distribution patterns (Supplementary Fig. 7a,b), we quantitatively analyzed whether Hes1 deficiency altered transcription elongation by calculating pausing indices. Pausing index for a given gene is the ratio of Pol II occupancy near the TSS region versus Pol II occupancy in the gene body region and thus negatively correlates with productive elongation. Hes1 deficiency did not dramatically change global pausing indices in untreated or LPS-stimulated conditions (Fig. 6f, left panel). For LPS-induced genes, LPS stimulation led to reduced pausing indices (Fig. 6f, middle panel), supporting the notion that LPS activates gene expression in part by promoting transcription elongation. For the group of super-induced genes (Fig. 1a), Hes1 deficiency further lowered pausing indices under the LPS-stimulated conditions (Fig. 6f, right panel), suggesting that Hes1 deficiency promotes transcription elongation at select gene loci. Taken together, these data suggested that Hes1 suppressed Cxcl1 transcription elongation and did so in a gene-specific manner.
To identify genes that like Cxcl1 were the targets of Hes1-mediated suppression of transcriptional elongation, we performed bioinformatic analysis of the ChIP-seq data sets to calculate Pol II density throughout the gene body regions. By comparing Pol II occupancy between LPS-stimulated wild-type and Hes1–/– cells, we found that Hes1 deficiency resulted in enhanced Pol II occupancy in gene body regions of 9 genes (Supplementary Fig. 7c). Notably, these 9 genes included Cxcl1 and Dclbd2, which were among super-induced genes in DKO macrophages in the microarray analysis (Fig. 1a and Supplementary Fig. 1b). Therefore, the genome-wide ChIP-seq analysis corroborated our microarray data and helped reveal the underlying mechanisms for Hes1-mediated selective gene regulation.
Hes1 targets P-TEFb to suppress productive elongation
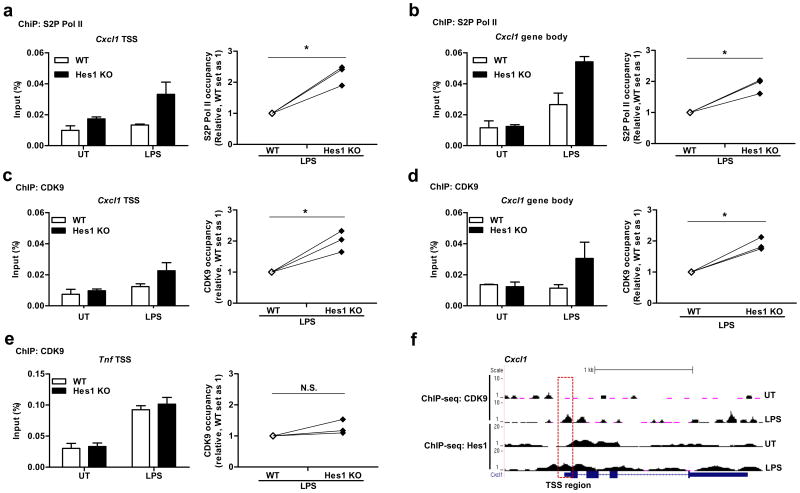
Next, we investigated mechanisms by which Hes1 inhibited Pol II-mediated elongation. The Pol II CTD contains a series of highly conserved heptapeptide repeats (YS2TPS5PS7) with three serine (S) residues, and S2 phosphorylation within these repeats marks elongation-competent Pol II9. To examine the possibility that Hes1 suppressed productive elongation by affecting S2 phosphorylation, we examined S2P Pol II levels at the Cxcl1 gene locus by ChIP. Hes1 deficiency significantly enhanced S2P Pol II occupancy of the Cxcl1 gene locus at both the TSS and gene body regions (Fig. 7a,b), suggesting that Hes1 may suppress Cxcl1 productive Pol II elongation by attenuating S2 phosphorylation.
Figure 7.
Hes1 inhibits Cxcl1 productive elongation by suppressing recruitment of the P-TEFb complex. ChIP assays showing the occupancy of S2P Pol II (a-b, left panels) and CDK9 (c-e, left panels) at the TSS regions and gene body regions of Cxcl1 (a-d) and Tnf (e) in Hes1+/+Mx1-Cre (WT) and Hes1fl/flMx1-Cre (Hes1 KO) BMDMs untreated or stimulated with LPS (10 ng/ml) for 1 h. Cumulative results from three independent experiments were shown for S2P Pol II (a-b, right panels) and CDK9 (c-e, right panels) on the Cxcl1 and Tnf gene loci under LPS-stimulated conditions and relative occupancy in WT cells was set to 1. (f) CDK9 ChIP-seq and Hes1 ChIP-seq gene tracks show CDK9 (upper) and Hes1 (lower) distribution along the Cxcl1 locus in BMDMs untreated or stimulated with LPS (10 ng/ml) for 1 h. Red box, TSS region of Cxcl1. *P<0.05 (Student's t-test).
S2 phosphorylation of the Pol II CTD is carried out by the P-TEFb heterodimeric complex comprised of cyclin T1 and cyclin-dependent kinase 9 (CDK9) subunits9. During the transcription cycle, recruitment of P-TEFb to gene loci to phosphorylate Pol II is a prerequisite for releasing Pol II into productive elongation7. We therefore investigated whether Hes1 inhibited S2P Pol II occupancy by targeting recruitment of P-TEFb. ChIP showed increased CDK9 occupancy on the Cxcl1 gene locus in Hes1-deficient BMDMs compared to wild-type (Fig. 7c,d) in a manner that closely resembled the regulation of S2P Pol II (Fig. 7a,b). In contrast, Hes1 deficiency did not significantly affect CDK9 recruitment to the Tnf gene locus in the presence or absence of LPS (Fig. 7e), suggesting that regulation of CDK9 occupancy by Hes1 is gene-specific. To gain further insight into the potential mechanisms of Hes1-imposed regulation, we carried out genome-wide analysis of CDK9 and Hes1 distribution by ChIP-seq in primary macrophages. The results show that Hes1 co-localized with CDK9 near the Cxcl1 TSS region in LPS-activated BMDMs (Fig. 7f), suggesting that Hes1 may regulate transcription of specific genes by acting at CDK9 recruitment sites. As expected, a CDK inhibitor flavopiridol suppressed Cxcl1 mRNA levels in both wild-type and Hes1–/– BMDMs (Supplementary Fig. 8a). Importantly, Hes1 deficiency did not globally alter cellular CDK9 protein levels (Supplementary Fig. 8b).
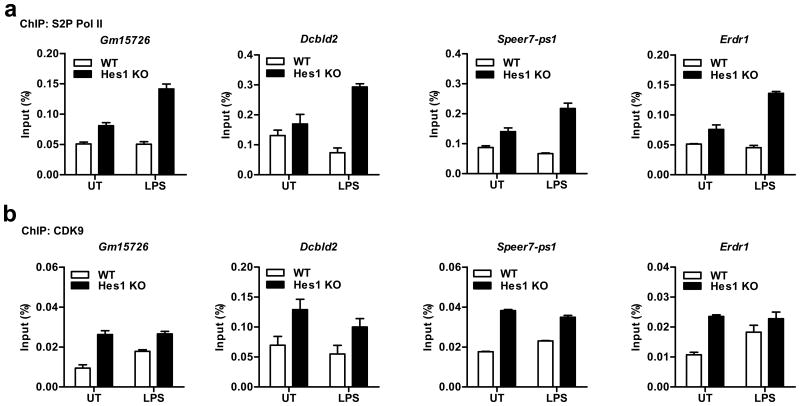
In addition to Cxcl1, enhanced S2P Pol II occupancy in Hes1–/– relative to wild-type BMDMs (Fig. 8a) was also observed on loci of several other genes including Gm15726 and Dcbld2 that harbored more Pol II in their gene body regions as revealed by Pol II ChIP-seq and computational analysis (Supplementary Fig. 7c). Similar to Cxcl1, these genes also displayed higher, and even LPS-independent occupancy by CDK9 in Hes1–/– BMDMs (Fig. 8b), thus confirming the robustness of our bioinformatic analysis for identifying genes that were targets for Hes1-mediated regulation. In summary, these data demonstrate that Hes1 suppressed productive transcription of Cxc1l by regulating P-TEFb recruitment and subsequent S2 phosphorylation of Pol II (Supplementary Fig. 8c).
Figure 8.
Hes1-mediated suppression of P-TEFb is not restricted to the Cxcl1 gene. (a,b) ChIP assays showing occupancy of S2P Pol II (a) and CDK9 (b) on the gene body regions of Gm15726, Dcbld2, Speer7-ps1, and Erdr1 in Hes1+/+Mx1-Cre (WT) and Hes1fl/flMx1-Cre (Hes1 KO) BMDMs stimulated with LPS (10 ng/ml) for 1 h. Representative data are shown as means ± s.d. of technical triplicate determinants from two independent experiments (a,b).
Discussion
Attenuation of production of inflammatory mediators by endogenous inhibitory factors has been regarded as an important mechanism by which body restrains inflammation and maintains immune homeostasis. Yet, most of these inhibitory events converge at the transcription initiation step and result in diminished assembly of the pre-initiation complex signified by decreased Pol II occupancy near the target gene TSS28. Here, we found that transcription repressor Hes1 selectively inhibits the expression of a subset of inflammatory mediators including CXCL1, a chemokine crucial for recruitment of neutrophils to sites of inflammation. Interestingly, events such as TLR-induced signal transduction, activation of effector transcription factors responsible for Cxcl1 expression, or its transcription initiation were unaffected by Hes1 deletion. Instead, Hes1 inhibits Cxcl1 transcription through attenuation of Pol II-mediated productive elongation by antagonizing the recruitment and, possibly, activity of the Pol II CTD kinase, P-TEFb – a tonic inhibition of elongation, which is relieved in Hes1-deficient macrophages.
Transcriptional elongation can be divided into two functional stages: early elongation and productive (late) elongation7. During early elongation, Pol II synthesizes short RNA transcripts and then, on many genes, pauses or stalls approximately 25-60 nucleotides downstream from TSS. Promoter-proximal pausing of Pol II has been postulated to represent a critical and rate-limiting step in transcription regulation for approximately half of all active Drosophila and mammalian genes7. In fact, several studies including ours revealed that transcription of several key immune genes such as Tnf are positively and negatively regulated at the early elongation step10-12. In contrast, the Cxcl1 gene locus in resting macrophages lacks appreciable Pol II occupancy, consistent with Pol II recruitment and subsequent initiation being rate-limiting for gene activation. Nonetheless, the inhibitory effects of Hes1 affect predominantly elongation and the efficiency of late processive elongation rather than pause-release appears to be attenuated. It is tempting to speculate that for the class of Cxcl1-like “non-paused” genes, targeting late elongation represents an effective way to regulate transcription rate beyond the point of initiation. Interestingly, Hes1 appears to employ different mechanisms to regulate different genes. Il6 and Il12b, for example, are also negatively regulated by Hes1 yet inhibition occurs at the transcription initiation step, and could result from Hes1-mediated repression via promoter elements such as N box or E box15.
Compared to the wealth of knowledge on the regulation of promoter-proximal pausing, little is known about mechanisms that control late elongation. We found that Hes1 broadly inhibits CDK9 recruitment to the Cxcl1 locus thereby attenuating Pol II CTD S2 phosphorylation. It remains unclear how Hes1 attenuates P-TEFb occupancy in a gene-specific manner. P-TEFb is recruited to its target genes by transcription factors such as NF-κB and c-Myc7,29, but given that most genes require P-TEFb for activation, clearly, additional regulators can direct this kinase to promoters. Indeed, in our system, Hes1 depletion had no effect on the NF-κB p65 occupancy at the Cxcl1 gene locus (data not shown), suggesting that Hes1 acts upon as-yet-undetermined factors to inhibit CDK9 recruitment. In fact, our ChIP-seq analysis for CDK9 and Hes1 revealed Hes1 recruitment to the CDK9 sites near the Cxcl1 locus (also detected by CDK9 ChIP-seq in BMDMs under slightly different stimulation conditions30). Co-localization of Hes1 and CDK9 suggests that Hes1 may physically hinder excessive CDK9 binding serving to constrain inflammatory response while still preserving signal-dependent transcription activation.
Transcription regulation at the post-initiation steps has been long appreciated in non-mammalian organisms such as Drosophila and more recently in mammalian systems31. Although we are only beginning to appreciate the impact of elongation control in inflammation and immunity10-12, the data emerging from recent genome-wide studies32-35 make it extremely likely that such mode of regulation is broadly applicable to the immune system and may serve to fine-tune expression of key immune effector molecules. The newly described role of Hes1 in this study provides such an example whereby targeting transcription elongation of a chemokine gene yielded important (patho)physiological consequences and affected inflammatory responses in vivo. In our study, the presence of Hes1 under homeostatic conditions is permissive for robust production of cytokines critically involved in host defense against pathogens, e.g., TNF and IL-1β, while limiting CXCL1 production and paracrine signaling, thereby restraining neutrophil trafficking to inflammatory sites and tissue damage. It is plausible that dysregulation of Hes1 expression or function may contribute to heightened inflammation in human diseases such as asthma by promoting neutrophil recruitment. Conversely, enhancing Hes1-mediated repressive pathway may represent a novel therapeutic approach for selectively curbing excessive inflammation while preserving other aspects of immunity.
Online Methods
Mice
The experiments using mice were approved by the Institutional Animal Care and Use Committees at Tsinghua University and the Hospital for Special Surgery. C57/BL6/J mice were purchased from Jackson Laboratory. Hes1fl/fl mice were originally obtained from R. Kageyama36. Mice with an inducible deletion of Hes1 (Hes1fl/flMx1-Cre) or (Hes1fl/flCre-ERT2) were generated by crossing Hes1fl/fl animals to animals with a Mx1 promoter-driven Cre transgene or Cre-ERT2 mice on the C57/BL6/J background. Gender-matched littermates with genotype of Hes1fl/flMx1-Cre and Hes1+/+Mx1-Cre or Hes1fl/flCre-ERT2 and Hes1+/+Cre-ERT2 were used for experiments. Mice with a myeloid-specific deletion of Hes1 (Hes1fl/flLyz2-Cre) were generated by crossing Hes1fl/fl animals to Lyz2-Cre mice. Littermates with genotype of Hes1fl/flLyz2-Cre and Hes1+/+Lyz2-Cre were used for experiments. Hes1 and Hey1-deficient mice were obtained by crossing Hes1fl/flMx1-Cre mice with Hey1–/– mice. Littermates with genotype of Hey1+/+Hes1fl/fl and Hey1–/–Hes1fl/flMx1-Cre were used for experiments. For Hes1fl/flMx1-Cre mice and Hey1–/–Hes1fl/flMx1-Cre mice, deletion of Hes1 was induced by intraperitoneal (i.p.) injection with 200 μg/mouse of Poly (I:C) for three times in five days. For Hes1fl/flCre-ERT2 mice, deletion of Hes1 was induced by i.p. injection with 2 mg/mouse of tamoxifen (dissolved in corn oil) for three times in three days. For all inducible deletion of Hes1, wild-type littermates were also injected with Poly (I:C) or tamoxifen with the same dosage. Mice were used for experiments two weeks later. We previously have comprehensively evaluated the potential effects of poly (I:C) in wild-type mice and observed no measurable impact of prior poly (I:C) injections on basal or LPS-induced cytokine production in vitro and in vivo or on chromatin state and Pol II occupancy37. For in vitro experiments involving Hes1-deficient macrophages, cells were derived from Hes1fl/flMx1-Cre animals where consistent and strong deletion of Hes1 (approximately 80%) was observed and the conditional deletion was controlled for expression of Cre and genetic background. For Hey1-deficient mice, littermates with genotype of Hey1+/+ and Hey1–/– were used for experiments. Experiments on mice were performed at 8-12 weeks of age with gender matched littermates. Bone marrow chimeras were generated as previously described15. Briefly, 6-week-age recipient C57/BL6/J mice were subjected to irradiation at a dose of 875 cGy, followed by intravenous injection of 1 × 106 donor bone marrow cells from the Hes1-deficient mice with the genotype Hes1fl/flMx1-Cre or the littermate controls with the genotype Hes1+/+Mx1-Cre. Chimeric mice were used for experiments 6 weeks after the initial bone marrow transfer.
Cell culture and reagents
Murine BMDMs were obtained as previously described38 and maintained in DMEM supplemented with 10% FBS and 10% L929 cell supernatant as conditioned medium providing macrophage colony stimulating factor. Cell culture grade LPS (Escherichia coli 0111:B4) was purchased from Sigma-Aldrich, Pam3Cys was purchased from EMC Microcollections and R848 was obtained from Invivogen.
Microarray analysis
Total RNA were extracted with an RNeasy Mini kit (Qiagen) and RNA quality was determined by Bioanalyer (Agilent). aRNAs for microarray were amplified by using a MessageAmp™ Premier RNA Amplification Kit according to the manufacturer's protocol (Ambion). Fragmented aRNAs (10 μg per sample) were then processed for hybridization to GeneChip Mouse Genome 430 2.0 according to the manufacturer's instruction (Affymetrix). Genespring Software v11.0 (Agilent) was used to analyze the microarray data and 2 fold induction was set as cut-off. Heat maps were generated by gplots software package in R (version 3.3.2). In the heat maps, genes were ranked according to the order of numeric identifiers of microarray probes.
Cloning and expression of Hes1 mutants
The full-length murine Hes1 cDNA with an N-terminal FLAG tag was cloned into the pEasy-blunt cloning vector (Transgene). Key domains of Hes1 were identified as previously described13,24. A dominant-negative Hes1 (dnHes1) was generated by mutating E43, K44, and R47 in the basic region to A. Hes1(ΔHLH) and Hes1(ΔWPRW) deletion mutants were generated by deleting HLH domain (amino acids 48-92) and the last 6 amino acids of Hes1 respectively. All Hes1 mutants were amplified by PfuUltra II Fusion HS DNA Polymerase (Stratagene) with pEASY-blut-Flag-Hes1 as a template (20 ng), following by DpnI (NEB) digestion. The primers used to construct these mutants were listed in Supplemental Table 1. The cDNA fragments of Flag-Hes1 mutants were then cut from pEASY-blunt vectors with BamHI and XhoI, and were ligated with pMx-puro retroviral vectors. All clones were sequenced and expression was confirmed by immunoblotting transiently transfected HEK-293T cell lysates with an anti-FLAG antibody (Sigma).
Retroviral transduction
Retroviral transduction was performed as previously described38. Briefly, 3×106 Plat-E cells were seeded into 100 cm plates and were cultured for 24 hours. Cells were then transfected with 17 μg retroviral vectors of pMx-Puro-GFP,pMx-Puro-Flag-Hes1 or pMx-Puro-Flag-Hes1 mutants by using Fugene HD (Promega) reagents. 48 hours after transfection, viral supernatants were collected and filtered and 5 ml of viral supernatant was used to transduce 5×106 BMDMs in presence of 8 μg/ml of polybrene (Sigma). 24 hours post viral infection, BMDMs were selected by puromycin (2 μg/ml) for 3 days and then were used for experiments.
Reverse transcription and qPCR
RNA was extracted from whole cell lysates with an RNeasy Mini Kit (Qiagen) and was reversely transcribed to cDNA with a First Strand cDNA Synthesis Kit (Fermentas). qPCR was performed in triplicate determinants with an ABI7500 thermal cycler. Primary transcripts were measured with primers that amplify either exon-intron junctions or intronic sequences. Threshold cycle numbers were normalized to triplicate samples amplified with primers specific for glyceraldehyde-3-phosphate dehydrogenase (Gapdh). Primer sequences are listed in the Supplementary Table 1.
Enzyme-linked immunosorbent assay (ELISA)
Chemokine secretion was quantified by a murine CXCL1/KC ELISA Kit (R&D System) according to the manufacturers' instructions.
Isolation of neutrophils
Murine neutrophils were isolated from bone marrow as described previously39. Briefly, bone marrow cells were flushed from femur and tibia using a 25-G needle syringe filled with RPMI 1640 supplemented with 10% FBS and 2 mM EDTA, followed by filtration through a 100 μm cell strainer into 50 ml Falcon tubes. Cells were spun down, red blood cells were removed with ACK lysing buffer, and remaining cells were resuspended in 1 ml of ice-cold sterile PBS. Neutrophils were then separated by density gradient centrifugation for 30 min at 2,000 rpm at RT without brake with 3 ml of Histopaque 1119 (Sigma) in a 15-ml conical tube overlaid with 3 ml of Histopaque 1077 (Sigma). Cell viability and purity were determined by FACS analysis.
In vitro transwell cell migration assay
Cell chemotaxis was determined using a transwell system (Corning). Transwell inserts with 8 μm polycarbonate membrane were used. Neutrophil suspensions were prepared in RPM1640 medium supplemented with 10% FBS. For chemotaxis assay, 2 × 105 neutrophils in 200 μl culture medium were added into transwell inserts placed in a 24-well plate containing 500 μl medium or medium with 100 ng/ml murine CXCL1 (Peprotech) in the outside compartment. The migration system was incubated for 1 h in cell culture incubator. The number of cells in the outside compartment was determined by direct cell counting using a hemocytometer. The chemotaxis index for each sample was calculated by the ratio of number of migrated cells in Cxcl1 conditions versus number of migrated cells in medium only conditions (representing spontaneous cell migration)40.
Isolation of resident peritoneal macrophages
Resident peritoneal macrophages were prepared as described elsewhere41. Briefly, peritoneal exudate cells were washed out with ice cold PBS containing 2 mM EDTA. After washing twice with PBS, peritoneal cells were resuspended in DMEM supplement with 10% FBS. The cells were then allowed to adhere for overnight in petri dishes at 37 °C. Non-adherent cells are removed by gently washing three times with warm PBS. The adherent cells were used as peritoneal macrophages.
LPS-induced peritonitis
Peritonitis was induced by intraperitoneal injection of 100 ng LPS/mouse in 500 μl PBS as previously described42 with minor modifications. 4 hours post injection, mice were euthanized by carbon dioxide exposure and peritoneal cavities were washed with 5 ml PBS containing 5 mM EDTA. Numbers of total peritoneal cells were counted by hemocytometer.
Adoptive transfer of macrophages
Macrophage adoptive transfer was performed as described elsewhere with minor modification43. Briefly, BMDMs were untreated or activated with LPS (10 ng/ml) for 30 min. The cells were washed three times with PBS and cell numbers were adjusted to 1×106 cells/ml in sterile PBS. 500 microliters suspension of BMDMs (0.5×106 cells) were injected i.p. into WT C57/BL6/J mice. After 4 h, peritoneal cells were isolated for flow cytometric analysis.
Flow cytometry
Peritoneal cells were stained with an FITC-conjugated anti-neutrophil antibody (7/4, 1:200, Abcam) and PE or PerCP-cy5.5-conjugated anti-Ly-6G antibody (1A8,1:400, BD Biosciences) for gating neutrophils as previously described42. APC anti-mouse F4/80 (BM8, 1:400, Biolegend), Alexa Flour 700 anti-mouse CD45R/B220 (RA3-6B2,1:200, Biolegend) and PE anti-mouse Ly6C (AL21, 1:400, BD Biosciences) together with PE/Cy7 anti-mouse CD11b (M1/70, 1:400, BD Biosciences) were used to stain macrophages, B cells and monocytes respectively. Cells were washed three times and analyzed on FACSCalibur or FACSAira II flow cytometer (BD Biosciences) using CellQuest or Flowjo softwares (BD Biosciences).
K/B×N serum-induced arthritis
K/B×N serum pools were prepared as described44. Arthritis was induced by i.p. injection of total volume of 100 μl (low dose) or 150 μl (high dose) K/B×N serum per mouse. WT and Hes1 KO bone marrow chimeric mice were given serum twice at day 0 and day 2 respectively. Physical measurement of wrist and ankle thickness was performed using a metric vernier caliper (Bel-Art Products). For each animal, the total joint thickness was calculated as the sum of the measurements of all wrists and ankles. Change of total joint thickness (Δ of joint thickness) was calculated by the formula: Δ of joint thickness = Measured joint thickness - Initial joint thickness.
Histopathology
Joint histology was performed as previously described45. Briefly, paws were fixed overnight in 10% neutral buffered formalin and decalcified in 10% EDTA (pH 7.4) until the bones were pliable, trimmed, and embedded in paraffin. Sections were cut into 5 μm and stained with hematoxylin and eosin (H&E). Images were acquired on a Nikon Eclipse 50i microscope using NIS-Elements imaging software. For histopathological analysis, the degree of inflammation was scored as described previously46. Briefly, the histological score was determined as following: 0, normal; 1, minimal; 2, mild; 3, moderate; 4, marked; and 5, severe.
Immunoblotting analysis
Whole cell lysates and nuclear extracts were prepared as described previously38. For immunoblotting analysis, lysates were separated by 10% SDS-PAGE and transferred to a PVDF membrane (Millipore) for probing with specific antibodies. Antibodies against p38 (sc-535), Hes1 (sc-25392), CDK9 (sc-8338), and TBP (sc-204) were purchased from Santa Cruz Biotechnology. All the other antibodies were obtained from Cell Signaling Technology.
Dual-luciferase reporter assay
Murine Cxcl1 reporter plasmid containing promoter sequences from positions −701 to +30 (pGL3-Basic-KC701) was a gift from K.F. Roby47. RAW 264.7 cells were co-transfected in duplicate with the Cxcl1 reporter plasmid and an expression plasmid (pCMV6-XL4-Hes1) encoding human HES1 or a control vector (pCMV6-XL4) using Lipofectamine LTX reagent (Invitrogen). The renilla luciferase reporter gene (pRL-TK, Promega) was used as an internal control. 24 hours post transfection, cells were stimulated with LPS (100 ng/ml) for 4 hours and cell lysates were prepared and analyzed using Dual-Luciferase Report Assay System (Promega).
Chromatin immunoprecipitation (ChIP) assay and ChIP-seq
For Pol II, S2P Pol II and CDK9 ChIP assay, BMDMs from wild type or Hes1fl/flMx1-Cre mice were used for ChIP assay. For Hes1 ChIP assay, BMDMs transduced with pMx-Hes1 viral particles were used. Cells were left untreated or stimulated with LPS (10 ng/ml) for 1 h. 15∼20×106 cells in each condition were fixed by 1% methanol-free formaldehyde (Thermo Scientific) for 5 min at room temperature followed by quenching with 125 mM glycine for another 5 min. Cells were then lysed in 1% SDS lysis buffer. Chromatin DNAs were sheared to an average size of 300 bp by using a Bioruptor UCD-400 (Diagenode). For immunoprecipitations, the following antibodies were used: Pol II (sc-9001x, Santa Cruz); Pol II-S2P (ab5095, Abcam), CDK9 (sc-8338x, Santa Cruz) and Hes1 (sc-25392, Santa Cruz). After purification, immunoprecipitated DNAs were analyzed by qPCR with corresponding primers (sequences are listed in Supplementary Table 1) and relative occupancies were normalized to input DNA. For ChIP-seq, 10 ng DNAs of each sample were ligated with adaptors and 100∼300 bp DNA fragments were purified to prepare DNA libraries. The DNA libraries were generated using Illumina ChIP-seq Sample Prep Kit following the manufacturer's instructions. ChIP libraries were sequenced with the 50 bp single end option using an Illumina HiSeq 2000 Sequencer at the Weill Cornell Medical College Epigenetic Core Facility or at BGI, China per manufacturer's recommended protocol. FASTQ and aligned files were generated using CASAVA 1.8.2. The aligned reads were mapped to mouse reference sequence (GRCm38/mm10) and generated in BAM format using default parameters and clonal reads were removed from further analysis. More than 60 million nonclonal mapped tags were obtained for each condition in each experiment for analysis. The WIG files displayed in UCSC genome browser were generated using the bioinformatic program ChIPseeqer48.
Bioinformatic analysis
To identify genes with differential Pol II occupancy on gene body regions, the read counts from the Pol II ChIP-seq data in specialized genome region were compared. A custom R script was used to modify the mouse annotated gtf file, which was downloaded from Ensembl Biomart Database (http://uswest.ensembl.org/biomart/). Gene body region was defined as from +100 bp to 3′ end. With these edited gene feature annotation files, we ran the featureCounts read quantification program within the Subread software package49 on the computation cluster to process the read summarization from the Pol II ChIP-seq read alignment files (input file format: Bam, alignment reference Genome: UCSC Mus musculus mm10). DESeq program (http://www-huber.embl.de/users/anders/DESeq/) was used for statistical analysis of the Pol II differentially binding genes (DBGs) for each gene features between WT + LPS condition and Hes1 KO + LPS condition. Each condition contained two biological replicates. DBGs were screened out according to the difference of Pol II binding amount fold change and significant p-value (padj < 0.05), which was adjusted for FDR due to multiple testing procedures to control type I error. Heat maps of Pol II distribution and pausing indices were generated as previously described35. Briefly, pausing indices were calculated as the ratio of promoter window density to gene body density as read pairs/kb. For display of the data as heat maps, counts of same-strand read pairs intersecting eighty 50 nt bins (from -1500 to +2500 nt relative to each TSS) were determined and visualized using an internally developed computational program.
Statistics
P values were calculated with a two-tailed paired or unpaired Student's t-test. P values of 0.05 or less were considered significant.
Supplementary Material
Acknowledgments
We thank R. Kageyama (Koyto University) for Hes1flox/flox mice; K. F. Roby (University of Kansas Medical Center) for Cxcl1 reporter plasmids; L. B. Ivashkiv for helpful discussion; and S. Chakravarty, K. Au, S. Smith, and J. Zhu for technical assistance. We also thank C. Tang, J. Lu, Z. Zhang, X. Yang, and J. Zhang (Peking University) for constructive advice on bioinformatic analysis. This research was supported by Ministry of Science and Technology of China National Key Research Project 2015CB943201 (X.H.), National Natural Science Foundation of China Young Investigator Award 81422019 (X.H.), funds from Tsinghua-Peking Center for Life Sciences (X.H.), Institute for Immunology at Tsinghua University (X.H.), and The David Rosensweig HSS Genomics Center (M.C., I.R.), and grants from Rheumatology Research Foundation (I.R.), the U.S. Department of Defense (I.R.), and the National Institutes of Health (I.R., B.Z.).
Footnotes
Accession codes. All genomic data described herein are deposited in Gene Expression Omnibus under accession number GSE73484, GSE73568, and GSE77334.
Author Contributions: Y.S. designed research, performed experiments, analyzed data, and wrote the manuscript; M.C. performed Pol II ChIP-seq experiments; T.H. analyzed Pol II ChIP-seq data and wrote methods for bioinformatic analysis; F.N. constructed retroviral plasmids, performed retroviral related experiments, and prepared mice for experiments; L.Y. performed CDK9 and Hes1 ChIP-seq experiments; L.K. performed FACS experiments and analysis; Bi.Z. contributed to bioinformatics analysis; C.J. prepared mice for experiments; Y.Q. contributed to bioinformatics analysis; Ba.Z. provided K/B×N serum and advice on experiments; M.G. provided Hey1–/– KO mice; I.R. provided advice on experiments and wrote the manuscript; X.H. conceptualized the project, designed research, supervised experiments, and wrote the manuscript.
Competing Financial Interests: The authors declare no competing financial interests.
References
- 1.Griffith JW, Sokol CL, Luster AD. Chemokines and chemokine receptors: positioning cells for host defense and immunity. Annu Rev Immunol. 2014;32:659–702. doi: 10.1146/annurev-immunol-032713-120145. [DOI] [PubMed] [Google Scholar]
- 2.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 3.Kondo T, Kawai T, Akira S. Dissecting negative regulation of Toll-like receptor signaling. Trends in immunology. 2012;33:449–458. doi: 10.1016/j.it.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 4.Smale ST. Selective transcription in response to an inflammatory stimulus. Cell. 2010;140:833–844. doi: 10.1016/j.cell.2010.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ivashkiv LB. Epigenetic regulation of macrophage polarization and function. Trends in immunology. 2013;34:216–223. doi: 10.1016/j.it.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rogatsky I, Adelman K. Preparing the first responders: building the inflammatory transcriptome from the ground up. Mol Cell. 2014;54:245–254. doi: 10.1016/j.molcel.2014.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwak H, Lis JT. Control of transcriptional elongation. Annu Rev Genet. 2013;47:483–508. doi: 10.1146/annurev-genet-110711-155440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nature reviews Genetics. 2012;13:720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou Q, Li T, Price DH. RNA polymerase II elongation control. Annu Rev Biochem. 2012;81:119–143. doi: 10.1146/annurev-biochem-052610-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adelman K, et al. Immediate mediators of the inflammatory response are poised for gene activation through RNA polymerase II stalling. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:18207–18212. doi: 10.1073/pnas.0910177106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138:129–145. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gupte R, Muse GW, Chinenov Y, Adelman K, Rogatsky I. Glucocorticoid receptor represses proinflammatory genes at distinct steps of the transcription cycle. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:14616–14621. doi: 10.1073/pnas.1309898110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kobayashi T, Kageyama R. Expression dynamics and functions of Hes factors in development and diseases. Curr Top Dev Biol. 2014;110:263–283. doi: 10.1016/B978-0-12-405943-6.00007-5. [DOI] [PubMed] [Google Scholar]
- 14.Ishibashi M, et al. Targeted disruption of mammalian hairy and Enhancer of split homolog-1 (HES-1) leads to up-regulation of neural helix-loop-helix factors, premature neurogenesis, and severe neural tube defects. Genes Dev. 1995;9:3136–3148. doi: 10.1101/gad.9.24.3136. [DOI] [PubMed] [Google Scholar]
- 15.Hu X, et al. Integrated regulation of Toll-like receptor responses by Notch and interferon-gamma pathways. Immunity. 2008;29:691–703. doi: 10.1016/j.immuni.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Su X, et al. Interferon-gamma regulates cellular metabolism and mRNA translation to potentiate macrophage activation. Nat Immunol. 2015;16:838–849. doi: 10.1038/ni.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alvarez Y, et al. Notch- and transducin-like enhancer of split (TLE)-dependent histone deacetylation explain interleukin 12 (IL-12) p70 inhibition by zymosan. J Biol Chem. 2011;286:16583–16595. doi: 10.1074/jbc.M111.222158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shang Y, Smith S, Hu X. Role of Notch signaling in regulating innate immunity and inflammation in health and disease. Protein Cell. 2016;7:159–174. doi: 10.1007/s13238-016-0250-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang W, Xu W, Xiong S. Blockade of Notch1 signaling alleviates murine lupus via blunting macrophage activation and M2b polarization. J Immunol. 2010;184:6465–6478. doi: 10.4049/jimmunol.0904016. [DOI] [PubMed] [Google Scholar]
- 20.Sodsai P, Hirankarn N, Avihingsanon Y, Palaga T. Defects in Notch1 upregulation upon activation of T Cells from patients with systemic lupus erythematosus are related to lupus disease activity. Lupus. 2008;17:645–653. doi: 10.1177/0961203308089406. [DOI] [PubMed] [Google Scholar]
- 21.Antoniv TT, Ivashkiv LB. Dysregulation of interleukin-10-dependent gene expression in rheumatoid arthritis synovial macrophages. Arthritis Rheum. 2006;54:2711–2721. doi: 10.1002/art.22055. [DOI] [PubMed] [Google Scholar]
- 22.Teachey DT, et al. Targeting Notch signaling in autoimmune and lymphoproliferative disease. Blood. 2008;111:705–714. doi: 10.1182/blood-2007-05-087353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fischer A, Schumacher N, Maier M, Sendtner M, Gessler M. The Notch target genes Hey1 and Hey2 are required for embryonic vascular development. Genes Dev. 2004;18:901–911. doi: 10.1101/gad.291004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strom A, Castella P, Rockwood J, Wagner J, Caudy M. Mediation of NGF signaling by post-translational inhibition of HES-1, a basic helix-loop-helix repressor of neuronal differentiation. Genes Dev. 1997;11:3168–3181. doi: 10.1101/gad.11.23.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kolaczkowska E, Kubes P. Neutrophil recruitment and function in health and inflammation. Nature reviews Immunology. 2013;13:159–175. doi: 10.1038/nri3399. [DOI] [PubMed] [Google Scholar]
- 26.Wipke BT, Allen PM. Essential role of neutrophils in the initiation and progression of a murine model of rheumatoid arthritis. J Immunol. 2001;167:1601–1608. doi: 10.4049/jimmunol.167.3.1601. [DOI] [PubMed] [Google Scholar]
- 27.Richmond A. Nf-kappa B, chemokine gene transcription and tumour growth. Nature reviews Immunology. 2002;2:664–674. doi: 10.1038/nri887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stender JD, Glass CK. Epigenomic control of the innate immune response. Curr Opin Pharmacol. 2013;13:582–587. doi: 10.1016/j.coph.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barboric M, Nissen RM, Kanazawa S, Jabrane-Ferrat N, Peterlin BM. NF-kappaB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol Cell. 2001;8:327–337. doi: 10.1016/s1097-2765(01)00314-8. [DOI] [PubMed] [Google Scholar]
- 30.Nicodeme E, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119–1123. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Min IM, et al. Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells. Genes Dev. 2011;25:742–754. doi: 10.1101/gad.2005511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jonkers I, Kwak H, Lis JT. Genome-wide dynamics of Pol II elongation and its interplay with promoter proximal pausing, chromatin, and exons. Elife. 2014;3:e02407. doi: 10.7554/eLife.02407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saponaro M, et al. RECQL5 controls transcript elongation and suppresses genome instability associated with transcription stress. Cell. 2014;157:1037–1049. doi: 10.1016/j.cell.2014.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nilson KA, et al. THZ1 Reveals Roles for Cdk7 in Co-transcriptional Capping and Pausing. Mol Cell. 2015 doi: 10.1016/j.molcel.2015.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams LH, et al. Pausing of RNA polymerase II regulates mammalian developmental potential through control of signaling networks. Mol Cell. 2015;58:311–322. doi: 10.1016/j.molcel.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Imayoshi I, Shimogori T, Ohtsuka T, Kageyama R. Hes genes and neurogenin regulate non-neural versus neural fate specification in the dorsal telencephalic midline. Development. 2008;135:2531–2541. doi: 10.1242/dev.021535. [DOI] [PubMed] [Google Scholar]
- 37.Chinenov Y, et al. Role of transcriptional coregulator GRIP1 in the anti-inflammatory actions of glucocorticoids. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:11776–11781. doi: 10.1073/pnas.1206059109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu H, et al. Notch-RBP-J signaling regulates the transcription factor IRF8 to promote inflammatory macrophage polarization. Nat Immunol. 2012;13:642–650. doi: 10.1038/ni.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swamydas M, Lionakis MS. Isolation, purification and labeling of mouse bone marrow neutrophils for functional studies and adoptive transfer experiments. J Vis Exp. 2013:e50586. doi: 10.3791/50586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lima TF, et al. Warifteine, an alkaloid purified from Cissampelos sympodialis, inhibits neutrophil migration in vitro and in vivo. J Immunol Res. 2014;2014:752923. doi: 10.1155/2014/752923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang X, Goncalves R, Mosser DM. The isolation and characterization of murine macrophages. Curr Protoc Immunol Chapter 14. Unit. 2008;14:11. doi: 10.1002/0471142735.im1401s83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Filippo K, Henderson RB, Laschinger M, Hogg N. Neutrophil chemokines KC and macrophage-inflammatory protein-2 are newly synthesized by tissue macrophages using distinct TLR signaling pathways. J Immunol. 2008;180:4308–4315. doi: 10.4049/jimmunol.180.6.4308. [DOI] [PubMed] [Google Scholar]
- 43.Wongchana W, Lawlor RG, Osborne BA, Palaga T. Impact of Notch1 Deletion in Macrophages on Proinflammatory Cytokine Production and the Outcome of Experimental Autoimmune Encephalomyelitis. J Immunol. 2015;195:5337–5346. doi: 10.4049/jimmunol.1401770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Monach PA, Mathis D, Benoist C. The K/BxN arthritis model. Curr Protoc Immunol Chapter 15. Unit. 2008;15:22. doi: 10.1002/0471142735.im1522s81. [DOI] [PubMed] [Google Scholar]
- 45.Li S, et al. RBP-J imposes a requirement for ITAM-mediated costimulation of osteoclastogenesis. J Clin Invest. 2014;124:5057–5073. doi: 10.1172/JCI71882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sadik CD, Kim ND, Iwakura Y, Luster AD. Neutrophils orchestrate their own recruitment in murine arthritis through C5aR and FcgammaR signaling. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E3177–3185. doi: 10.1073/pnas.1213797109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Son DS, Roby KF. Interleukin-1alpha-induced chemokines in mouse granulosa cells: impact on keratinocyte chemoattractant chemokine, a CXC subfamily. Molecular endocrinology. 2006;20:2999–3013. doi: 10.1210/me.2006-0001. [DOI] [PubMed] [Google Scholar]
- 48.Giannopoulou EG, Elemento O. An integrated ChIP-seq analysis platform with customizable workflows. BMC bioinformatics. 2011;12:277. doi: 10.1186/1471-2105-12-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.