SUMMARY
Follicular helper T (TFH) cells are CD4+ T cells specialized in helping B cells and are associated both with protective antibody responses and autoimmune diseases. The promise of targeting TFH cells therapeutically has been limited by fragmentary understanding of extrinsic signals regulating human TFH cell differentiation. A screen of a human protein library identified activin A as new regulator of TFH cell differentiation. Activin A orchestrated expression of multiple TFH-associated genes, independently or in concert with additional signals. TFH programming by activin A was antagonized by the cytokine IL-2. Activin A’s capacity to drive TFH cell differentiation in vitro was conserved for non-human primates but not mice. Finally, activin A-induced TFH programming was dependent on SMAD2 and SMAD3 signaling and blocked by pharmacological inhibitors.
Antibody responses are a powerful arm of the adaptive immune system to prevent and fight pathogen infections. In order to produce antibodies with high affinity and pathogen neutralization potential, naïve B cells are activated and divide in response to cognate antigen and subsequently undergo rounds of somatic hypermutation and selection in germinal centers (GC), which are specialized microanatomical sites in secondary lymphoid organs. This process of affinity maturation is tightly regulated by follicular helper T (TFH) cells1–3, a distinct subset of CD4+ T cells specialized in helping B cells. TFH cells provide signals for selection, survival and proliferation of GC B cells via cytokine production and costimulatory molecule expression4. TFH cell help to B cells has been found to be a limiting factor for determining the magnitude of germinal center reactions and the mutation rate of GC B cells1, 5.
The distinctive features of TFH cells are acquired during a multi-stage, multi-factorial differentiation process that takes place in secondary lymphoid organs1, 6, 7. Upon priming of naïve CD4+ T cells by DCs or other myeloid antigen presenting cells (APCs), activated antigen-specific CD4+ T cells may upregulate the chemokine receptor CXCR5 and become early TFH cell precursors. The activated CXCR5+ CD4+ T cells migrate towards the B cell follicles upon downregulation of chemokine receptor CCR7. At this early stage of differentiation, the CXCR5intCCR7lo TFH cells can be found at the border between T cell area and B cell follicles (T cell-B cell border) and in B cell follicles. Here, TFH cells interact with cognate B cells, which deliver stimuli to TFH cells to complete their differentiation. Fully polarized GC TFH cells are located within GCs and highly express CXCR5 (CXCR5hi) and the TFH lineage defining transcription factor Bcl61, 6. GC TFH cells are functionally mature B cell helpers, expressing high levels of canonical functional mediators (interleukin 21 (IL-21), CXCL13, CD40L, ICOS, SLAMF1, interleukin 4 (IL-4)), along with a series of inhibitory receptors (PD-1, BTLA, and others), which restrain TFH cells from excessive proliferation1, 4.
Multiple signaling pathways cooperate to imprint the TFH differentiation program. Over the past decade, roles for the costimulatory receptor ICOS, interleukin 6 (IL-6), IL-21, interleukin 2 (IL-2), SLAM family receptors, and other molecules were shown to regulate mouse TFH cell differentiation1, 6. However, few studies identified regulators of human TFH biology. Interestingly, some data suggest evolutionary divergences between the signaling pathways shaping human and mouse TFH cell differentiation. IL-6 is the dominant inducer of murine IL-21 production and an important early regulator of TFH differentiation in mice8–11, but it has minimal effects on human CD4+ T cell IL-21 induction and in vitro TFH cell generation12. In contrast, interleukin 12 (IL-12) is a potent regulator of IL-21 production and TFH cell differentiation in humans13, 14, but it has only a minor effect on mouse CD4+ T cell IL-21 expression8, 15. Thus, while extremely valuable for studies of evolutionarily shared pathways, mice might represent a limited model for identification of regulators of human TFH cell differentiation. In light of those apparent differences, we developed an in vitro screen of a large human recombinant protein collection to uncover novel regulators of human TFH cell differentiation.
RESULTS
Screen for novel regulators of human TFH differentiation
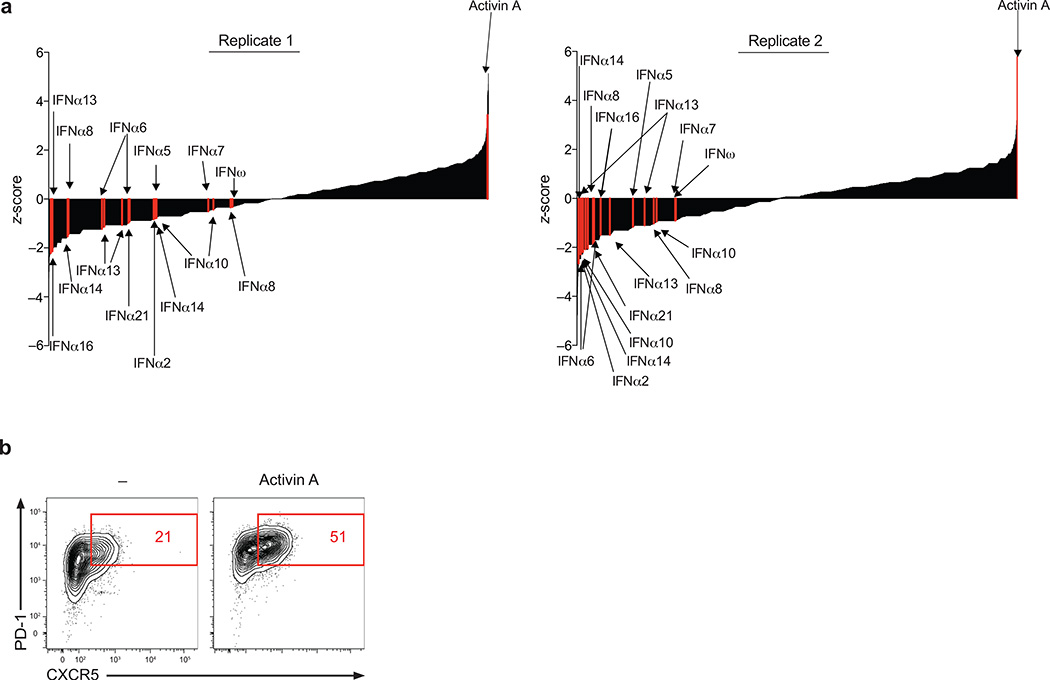
To discover novel regulators of human TFH cell differentiation, we performed an unbiased high-throughput screen of a human extracellular proteome library consisting of over 2000 human proteins predicted or known to be cytokines, chemokines, morphogens, costimulatory receptors, or single pass transmembrane molecules16. Each unique protein in this proteome, or ‘secretomics’, library was produced as a secreted recombinant molecule and tested for its capacity to modulate the differentiation of activated naïve CD4+ T cells into TFH cells in vitro (Supplementary Fig. 1a and Supplementary Fig. 1b). Briefly, activated human naïve CD4+ T cells were cultured for 5d in the presence of the human secretome library in duplicate. Expression of the TFH cell signature markers CXCR5 and PD-1 were measured by flow cytometry, in an automated fashion. The primary screen revealed multiple recombinant proteins that either induced or inhibited CXCR5 and PD-1 expression, as identified by PD-1+CXCR5+ cell % (Fig. 1a) or CXCR5+ cell % (Supplementary Fig. 1c) ranked by z-score. Eleven members of the type I interferon cytokine family, known inhibitors of mouse17 and human12 TFH cell differentiation, were identified in the primary screen as suppressors of CXCR5 and PD-1 expression (Fig. 1a). Activin A, the inhibin beta A (INHBA) homodimer, emerged as the most potent inducer of CXCR5 and PD-1 (Fig. 1a). We then confirmed the primary screen results, showing that activin A induces CXCR5 and PD-1 expression (Fig. 1b).
Figure 1. High throughput screening identifies activin A as a potent regulator of human TFH cell differentiation.
(a) Primary screen results. Enrichment of PD-1+CXCR5+ cell induction is reported as z-score for each recombinant protein from 8 × 384 well plates. Activin A and 11 type I interferon family proteins are shown in red. (b) Expression of CXCR5 and PD-1 from a screen repeat experiment re-testing activin A from secretomics library for its capacity to induce TFH-like differentiation. The data are representative of 2 independent replicates.
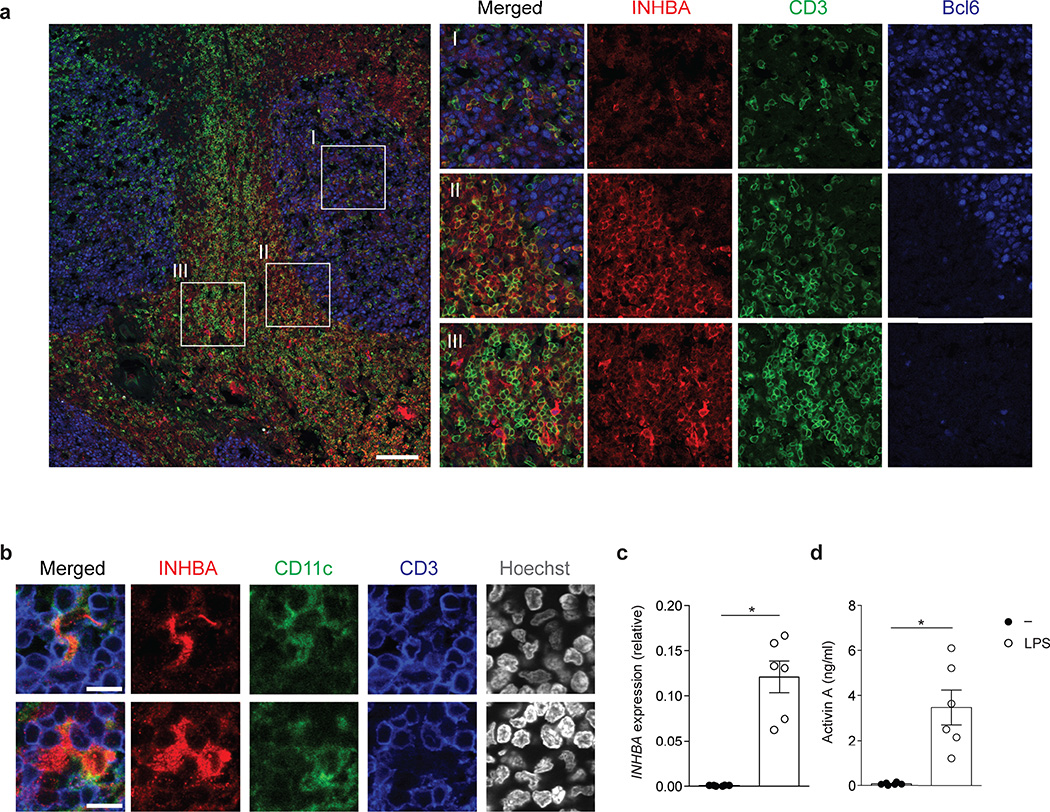
Activin A belongs to the family of activins-inhibins, a group of five distinct dimeric cytokines that can arise from three monomer subunits, inhibin alpha, beta A, beta B18. Activin A is a pleiotropic cytokine involved in multiple vital biological processes, including maintenance of pluripotent stem cells19–23. However, the role of activin A in the immune system is not fully understood, with only a small number of studies providing evidence for involvement of activin A in T cell biology22, 24–26. We evaluated the presence of activin A in human tonsils, a lymphoid tissue enriched in TFH cells and GCs where TFH cell differentiation physiologically occurs. INHBA protein was found in T cell zones, including the T cell-B cell borders, of all tonsils analyzed (Fig. 2a and Supplementary Fig. 2a). INHBA+ T cells were observed. The strongest INHBA production was in CD3− cells with myeloid cell morphology, consistent with the known ability of monocytes, dendritic cells (DCs) and macrophages to produce activin A27–29. CD11c staining revealed both CD11c+ and CD11c− cells expressing INHBA in T cell areas (Fig. 2b and Supplementary Fig. 2b). Furthermore, blood monocytes upregulated INHBA expression within 4h following LPS stimulation, and secreted activin A protein (Fig. 2c,d), confirming that human APCs are capable of activin A production.
Figure 2. INHBA is present in sites relevant for TFH cell differentiation and can be produced by myeloid cells.
(a) Confocal microscopy of INHBA expression in human tonsils stained with anti-INHBA (red) anti-Bcl6 (blue) and anti-CD3 (green). An overlay from one donor representative of six is shown on the left panel. Enlarged images on the right show representative INHBA expression in (I) a germinal center, (II) the T cell-B cell border and (III) T cell areas. Scale bar=100µm.(b) Confocal microscopy of INHBA expression in tonsil CD11c+ DCs. Tonsil sections were stained with anti-INHBA (red) anti-CD11c (green), anti-CD3 (blue) and counterstained with Hoechst. An overlay from one donor representative of two is shown on the left panel. The images were enlarged from a larger section depicted in Supplementary Fig 2b. Scale bar=10µm.(c) INHBA expression relative to GAPDH in purified monocytes cultured with and without E.Coli LPS (100ng/ml) for 4 h. The relative expression is shown as 2^-ΔCt. (d) Quantification of activin A secretion from purified monocytes cultured with and without E. coli LPS (100ng/ml) for 48h. Data (c,d) are combined from 2 independent experiments (n=6). Each symbol indicates an individual sample. * P < 0.05 (two-tailed Wilcoxon matched-pairs signed ranked test). (c,d; error bars represent mean and s.e.m.)
The role of activin A in in vitro TFH cell differentiation was then validated using primary naïve CD4+ T cells from numerous human donors (Fig. 3a–c) and activin A from multiple commercial vendors (data not shown). Furthermore, serum-free medium was used throughout these validation experiments to rule out possible indirect effects of undefined serum components. In stringent serum free conditions, activin A induced both PD-1 and CXCR5 expression on activated naïve CD4+ T cells in a dose dependent fashion (Fig. 3a,b and Supplementary Fig. 3a), demonstrating a direct effect of activin A on human TFH cell differentiation. Overall, these data indicate that a screen of human proteins enables the identification of factors that can function as early regulators of human TFH cell differentiation, with activin A identified as the top hit in the screen.
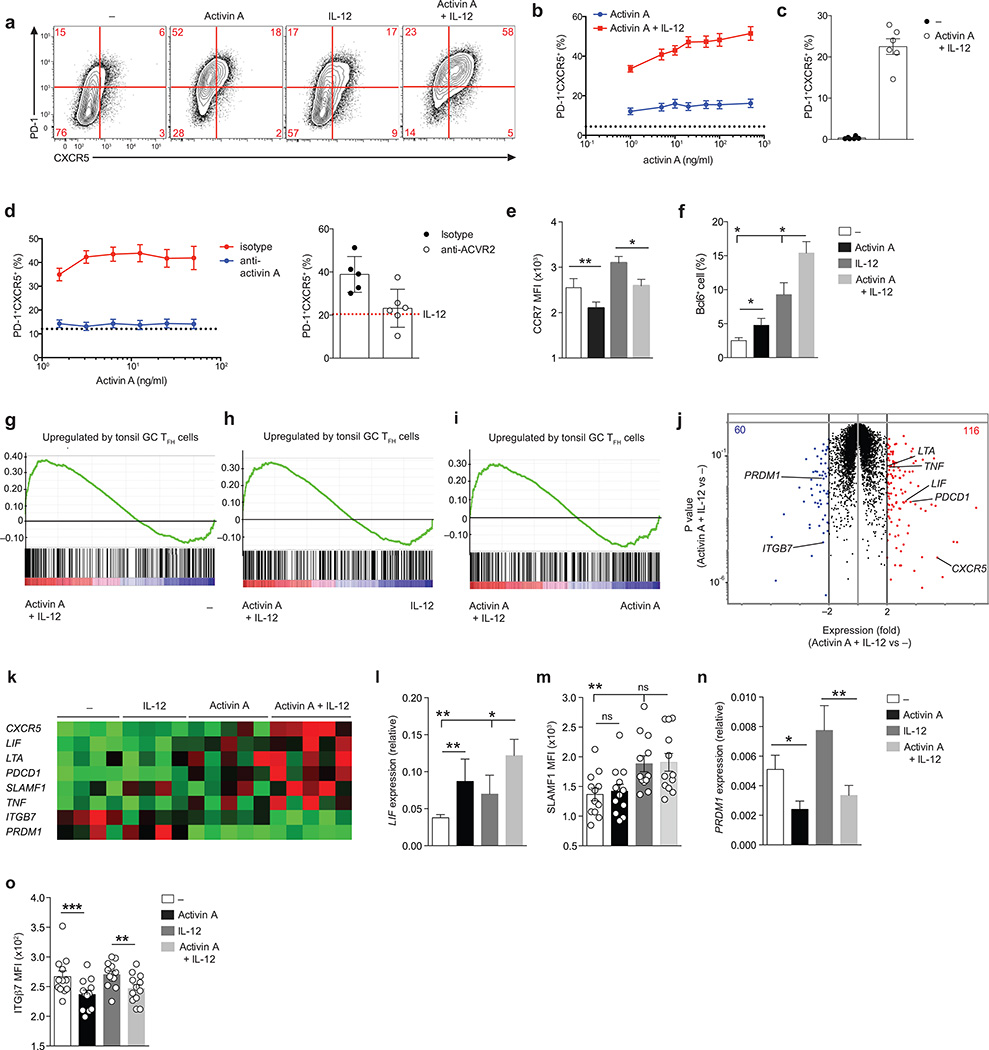
Figure 3. Activin A synergizes with IL-12 and molds the human TFH gene program.
(a) Flow cytometry of naïve CD4+ T cells stimulated by anti-CD3/CD28 beads for 5 d, in the presence of commercial human recombinant activin A, with or without IL-12. Cells stimulated with beads only (−) were used as control. Numbers in quadrants indicate percentage of cells in the outlined areas. (b) Frequency of PD-1+CXCR5+ cells as in (a). The dotted line shows the average basal induction of PD-1+CXCR5+ cells induced by beads only (−) from 13 donors. (c) Frequency of PD-1+CXCR5+ cells following 10 d of in vitro differentiation as in (a). (d) Left: frequency of PD-1+CXCR5+ cells from bead-activated naïve CD4+ T cells cultured in vitro with different doses of activin A combined with a fixed amount of IL-12 for 5 d in the presence of anti-activin A (block) or isotype control antibody (isotype). Dotted lines indicate the average percentages of PD-1+CXCR5+ cells induced by IL-12 with isotype control antibody. Right: frequency of PD-1+CXCR5+ cells from bead-activated naïve CD4+ T cells differentiated with activin A and IL-12, or IL-12 only (red dotted line, average value), in the presence of pan anti-ACVR2 or isotype control antibody. (e) Mean fluorescence intensity (MFI) of CCR7 on cells cultured as in (a). (f) Frequency of Bcl6 intranuclear expression on cells differentiated as in (a). (g-i) Enrichment of tonsil GC TFH signature genes in genes upregulated by activin A + IL-12 cultured cells in comparison to beads only (−) (g), IL-12 (h) or activin A only (i) stimulated cells. (j) RNAseq gene expression of cells differentiated as in (a) for 3d. Red: activin A + IL-12 versus – fold change (FC) > 2; blue: activin A + IL-12 versus – FC < −2). (k) RNAseq gene expression of selected TFH signature genes from (j). (l) LIF expression relative to ACTB in cells cultured as in (j)(m) MFI of SLAMF1 expression on cells cultured as in (j)(n) PRDM1 expression relative to ACTB in cells cultured as in (j) (o) MFI of ITGβ7 expression on cells cultured as in (j).Data in (b-f) and (l-o) data are from 2 or more independent experiments (n=6 or higher). * P < 0.05, ** P < 0.01 and *** P < 0.001 (two-tailed Wilcoxon matched-pairs signed ranked test). In (c), (e, f) and (l-o) bars show mean and s.e.m..
Activin A activity is modulated by IL-12 family members
IL-12 is the strongest identified IL-21 inducer for human CD4+ T cells13, 14. IL-12 can also modulate CXCR5 expression30. Therefore, we asked whether IL-12 might act in synergy with activin A. Activin A regulation of CXCR5 was enhanced by IL-12 (Fig. 3a,b). Expression of CXCR5 and PD-1 was retained by cells stimulated with activin A in combination with IL-12 for 10d of in vitro culture (Fig. 3c). Two additional IL-12 cytokine family members, IL-23 and IL-35, were also evaluated. IL-23 had a small activity in combination with activin A, while IL-35 had no relevant effect on CXCR5 and PD-1 expression in serum-free conditions (Supplementary Fig. 3c,d).
Treatment of activated CD4+ T cells with an activin A neutralizing monoclonal antibody abolished differentiation of PD-1+CXCR5+ cells at a range of activin A concentrations (Fig. 3d and Supplementary Fig. 3b). Activin A primarily binds and signals through the type I receptor ACVR1B (ALK4), in complex with the activin A receptor type IIA (ACVR2A) or IIB (ACVR2B)31, 32. A monoclonal antibody against both human Type II activin A receptors (‘pan anti-ACVR2’) resulted in a severe decrease of PD-1 and CXCR5 induction driven by activin A (Fig. 3d). Thus, activin A acts directly on activated human CD4+ T cells and regulates PD-1 and CXCR5 expression in a Type II activin A receptor-specific manner.
Instruction of a human TFH gene program by activin A
Since activin A promoted expression of the canonical TFH markers PD-1 and CXCR5, we asked if activin A, alone or in combination with IL-12, likewise regulated expression of additional molecules known to be important for TFH cell biology. Activin A was sufficient to decrease CCR7 expression on activated CD4+ T cells (Fig. 3e). Moreover, a fraction of CD4+ T cells differentiated with activin A plus IL-12 increased expression of Bcl6 (Fig. 3f and Supplementary Fig. 4a,b). Next, we ascertained the global transcriptomic profile of the in vitro differentiated TFH-like cells by RNA sequencing (RNA-seq). A comparison of gene expression profiles of in vitro cultured CD4+ T cells and bona fide human GC TFH cells revealed that cells differentiated in presence of activin A or activin A plus IL-12 were imprinted with bona fide GC TFH signature genes (activin A+IL-12 versus “−“: FDRq=0.015, Fig. 3g–i). Activin A contributed more than IL-12 to driving expression of GC TFH associated genes (activin A+IL-12 versus IL-12: p=0.046; activin A+IL-12 versus activin A: p=0.14, Fig. 3h,i). Activin A plus IL-12 resulted in selective upregulation of 116 genes versus cells differentiated with beads only (Fig. 3j and Supplementary Table 1), including several TFH-associated genes such as SLAMF1, LIF, LTA, and TNF (Fig. 3k and Supplementary Fig. 5), in addition to CXCR5 and PDCD1 (PD-1). Activin A emerged as the major factor regulating LIF, LTA, PDCD1, and TNF genes (Fig. 3k). Differential LIF and SLAMF1 expression were confirmed (Fig. 3l,m).
In addition to the upregulated genes, a set of 60 genes was suppressed by activin A plus IL-12 (Fig. 3j and Supplementary Table 2). Activin A caused significant downregulation of PRDM1, which encodes the transcription factor Blimp-1 (Fig. 3k,n). Blimp-1 is a potent and well-established Bcl6 antagonist that is expressed at low levels in bona fide TFH cells1. ITGB7, which encodes integrin β7 and is expressed at low levels on human GC TFH cells (Supplementary Fig. 5), was downregulated in the presence of activin A (Fig. 3o). Altogether, gene expression analysis indicated that activin A is sufficient to regulate expression of multiple molecules important for TFH cell biology, including genes involved in migration, differentiation, and proliferation.
Activin A regulation of TFH cell function
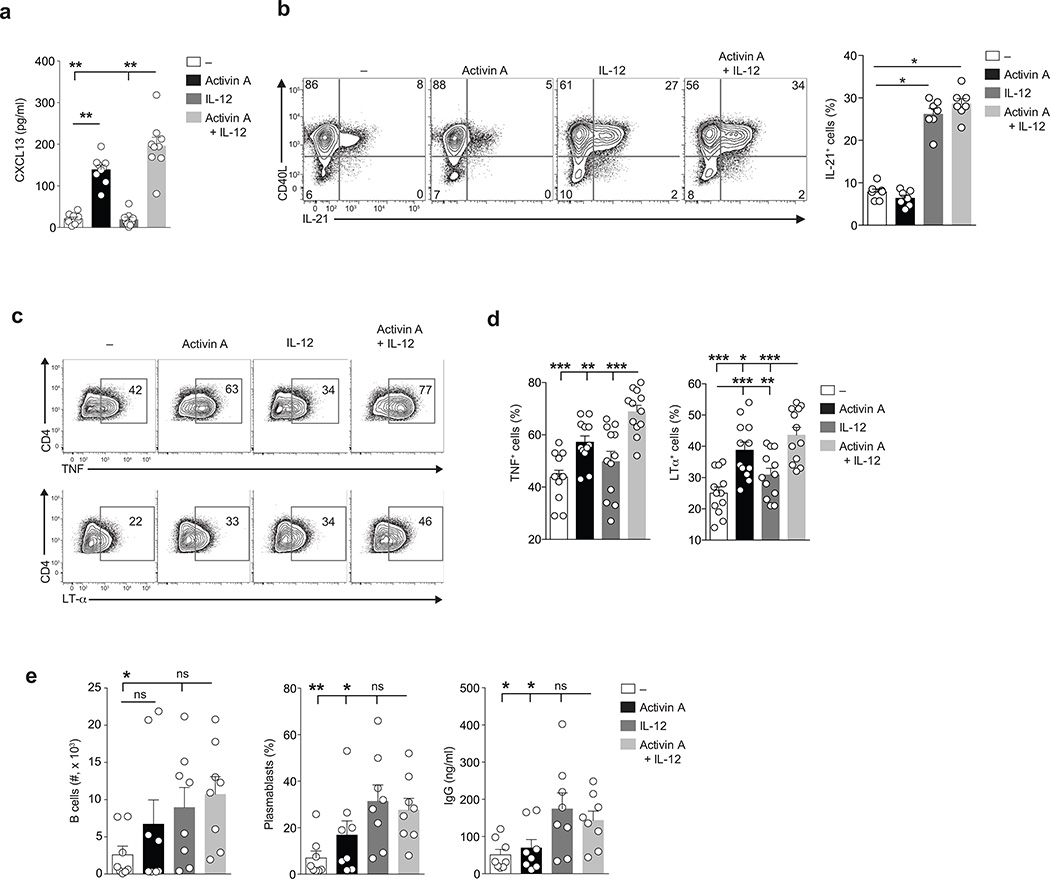
TFH cells are defined by their specialized function as B cell helpers. To investigate whether activin A can modulate TFH cell function, we evaluated production of canonical TFH lymphokines by the in vitro differentiated cells. CXCL13, the chemokine ligand of CXCR5, exhibits cytokine-type activity on B lymphocytes33 and is one of the defining factors secreted by human TFH cells. CXCL13 is constitutively produced in large quantities by human GC TFH cells and is selectively expressed by circulating memory TFH cells upon restimulation34–36. Activin A selectively induced CXCL13 expression by CD4+ T cells in vitro (Fig. 4a).
Figure 4. CD4+ T cells differentiated with activin A acquire functional signature molecules of TFH cells.
(a) Production of CXCL13 by naïve CD4+ T cells stimulated by plate bound anti-CD3 and hrICOS-L chimera, in the presence of human recombinant activin A, with or without IL-12 for 5 d. (b) Flow cytometry of CD40L and IL-21 intracellular expression on naive CD4+ T cells stimulated stimulated by anti-CD3/CD28 beads for 5 d, in the presence of activin A, with or without IL-12 and re-stimulated on day 5 with PMA/Ionomycin. Numbers in quadrants indicate percentage of cells in the outlined areas. (c) Flow cytometry of TNFα and LTα intracellular expression on cells differentiated as in (b) for 3 d and stimulated on day 3 with PMA/Ionomycin. Numbers in quadrants indicate percentage of cells in the outlined areas. (d) Frequency of cells in (c). (e) Absolute numbers of live CD19+ B cells (left plot), frequency of plasmablasts (CD19+CD20loCD38+ cells, center plot), and secreted IgG generated from coculture of memory B cells with autologous naïve CD4+ T cells differentiated as in (b) for 3d in the presence of SEB. In all (a-e) data are from 3 or more independent experiments (n=7 or more), and bars are mean and s.e.m.. * P < 0.05, ** P < 0.01 and *** P < 0.001. (a-d: two-tailed Wilcoxon matched-pairs signed ranked test; e: one-tailed Wilcoxon matched-pairs signed ranked test).
IL-21 is a cytokine highly produced by GC TFH cells and circulating memory TFH cells36, 37. IL-21 regulates GC B cell survival and plasma cell differentiation4, 38. As expected, CD4+ T cells cultured in vitro with IL-12 produced IL-21 after a short re-stimulation with PMA plus ionomycin (Fig. 4b). Moreover, cells cultured with activin A plus IL-12 retained full IL-21 production potential (Fig. 4b). Activin A cultured CD4+ T cells expressed elevated amounts of TNF and LTα proteins upon restimulation (Fig. 4c,d), consistent with the RNA-seq data. Finally, we evaluated the capacity of activin A differentiated cells to provide help to B cells. CD4+ T cells differentiated with activin A plus IL-12 were functionally competent B cell helpers, capable of supporting B cell proliferation and survival, plasmablast differentiation, and IgG production (Fig. 4e).
Overall, our data indicate that activin A, in combination with IL-12, promotes the generation of TFH-like cells that express high levels of CXCR5 and PD-1, modulate expression of Bcl6, PRDM1, CCR7, ITGB7, and SLAMF1, and display TFH functional properties, including B cell helper activity and production of CXCL13, IL-21, TNF, and LTα
Activin A-TGF-βrelationship in TFH differentiation
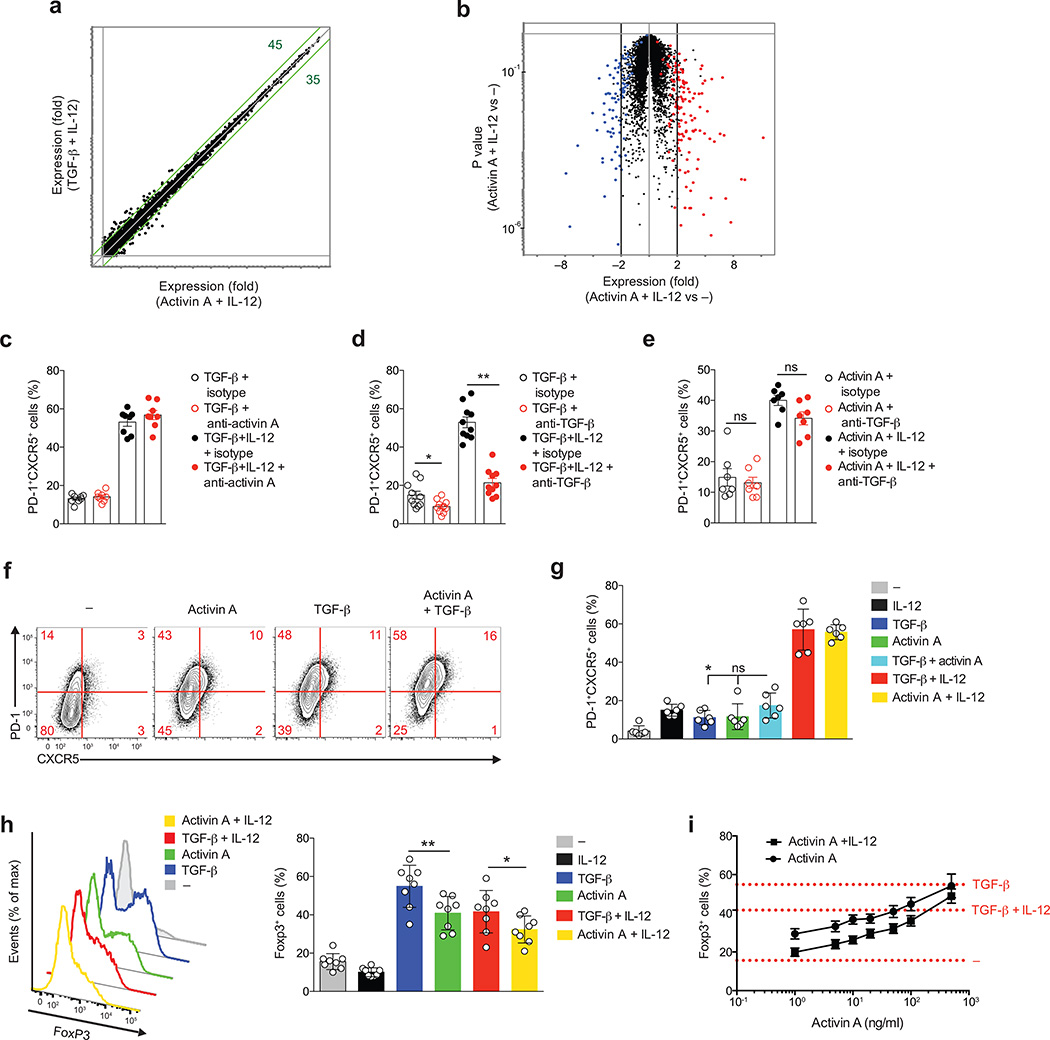
Transforming growth factor beta (TGF-β is a pleiotropic cytokine involved in the differentiation of multiple CD4+ T cell subsets in a context dependent fashion39. A recent study showed that TGF-β in conjunction with IL-12 and other cytokines, played a role in human TFH cell differentiation12. While activin A and TGF-β use independent receptors, both cytokines can trigger SMAD2/3 signaling pathways downstream of their respective receptors31, 32, 39. We utilized stringent serum-free conditions and found that TGF-β synergized with IL-12 to induce CXCR5 and PD-1 protein expression in activated naïve CD4+ T cells cultured in vitro for 5d (Supplementary Fig. 6a). While a TFH gene signature was present in RNA-seq of the serum-free cultures, a TH17 gene signature12 was not observed (data not shown). TGF-β and activin A displayed a similar capacity to regulate the differentiation of PD-1+CXCR5+ and Bcl6+ cells (Supplementary Fig. 6b) when combined with IL-12. A comparison of activin A plus IL-12 versus TGF-β plus IL-12 revealed a high degree of similarity (Fig. 5a,b). “Volcano” plot analysis indicated that the majority of the genes selectively regulated by activin A plus IL-12 in comparison to beads only controls followed the same trend as the genes regulated by TGF-β plus IL-12 versus beads only controls(Fig. 5b).
Figure 5. Activin A and TGF-β act independently from each other to drive in vitro TFH cell differentiation.
(a) RNA-seq gene expression on naive CD4+ T cells stimulated by anti-CD3/CD28 beads for 3 d, in the presence of activin A + IL-12 or TGF-β + IL-12.(b) RNA-seq gene expression on cells from (a). Red: TGF-β + IL-12 versus –, fold change (FC) > 2), blue: TGF-β + IL-12 versus –, FC < −2. (c) Frequency of PD-1+CXCR5+ naïve CD4+ T cells stimulated by anti-CD3/CD28 beads for 5 d with TGF-β, alone or with IL-12, in the presence of anti-activin A or isotype control antibody. (d-e) Frequency of PD-1+CXCR5+ naïve CD4+ T cells stimulated by anti-CD3/CD28 beads for 5 d with TGF-β and TGF-β + IL-12 (d) or activin A and activin A+IL-12 (e), in the presence of anti- TGFβ or isotype control antibody. (f) Flow cytometry of naïve CD4+ T cells stimulated by anti-CD3/CD28 beads for 5 d, in the presence of activin A, with or without TGF-β. Cells stimulated with beads only (−) were used as control. Numbers in quadrants indicate percentage of cells in the outlined areas. (g) Quantification of (f). (h-i) Expression of FoxP3 on naïve CD4+ T cells differentiated in vitro for 5d with different cytokine combinations. In (c-i) data are cumulative from 3 or more experiments (n=6 or more). * P < 0.05, ** P < 0.01 and *** P <0.001 (Two-tailed Wilcoxon matched-pairs signed ranked test).
Activated CD4+ T cells are capable of secreting activin A following TCR stimulation26. Because TGF-β mirrored activin A in driving human TFH cell differentiation, we explored the possibility that activin A accounted for the TGF-β-induced phenotype in vitro. However, activin A blockade did not alter the frequency of PD-1+CXCR5+ cells induced by TGF-β (Fig. 5c), indicating that activin A is not responsible for TGF-β induced TFH cell differentiation. Conversely, anti-TGF-β did not have any relevant effect on activin A–mediated TFH cell differentiation, suggesting that TGF-β is not responsible for activin A-induced TFH cell differentiation (Fig. 5d,e). Next, we evaluated if these two cytokines could induce PD-1 and CXCR5 expression in an additive or synergistic fashion. Neither PD-1 nor CXCR5 expression changed when activin A and TGF-β were combined in vitro (Fig. 5f,g). Despite driving highly similar gene expression, activin A and TGF-β significantly differed in their ability to induce Foxp3 expression, with activin A being less potent than TGF-β in driving the generation of Foxp3+ cells (Fig. 5h,i). Differential induction of Foxp3 may be important in vivo, since Foxp3 is the lineage defining transcription factor of regulatory T (Treg) cells. Thus, TGF-β may preferentially bias cells toward a Treg phenotype and activin A may bias cells toward a TFH phenotype. In summary, activin A and TGF-β do not account for each other’s ability to regulate TFH biology, and TGF-β preferentially induces Foxp3.
IL-2 modulation of activin A-mediated TFH cell differentiation
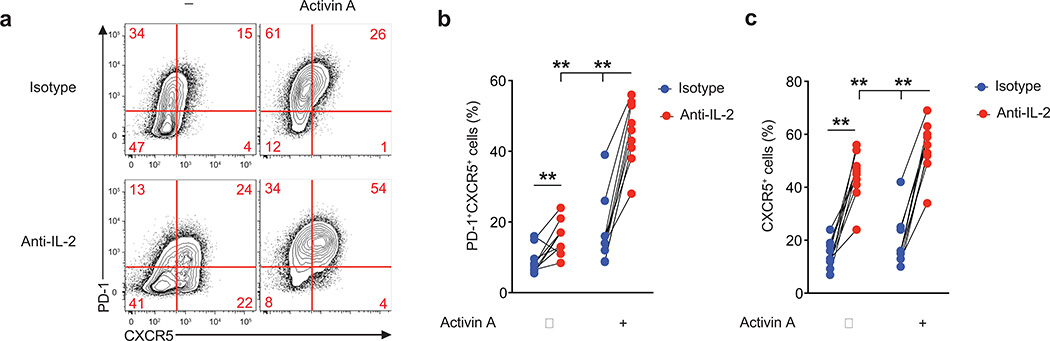
We noted that PRDM1 was expressed by activated human CD4+ T cells, and that PRDM1 expression was lower in activin A-stimulated T cells. Blimp-1 is one of the strongest negative regulators of TFH differentiation, through its abilities to antagonize Bcl6 and directly suppress CXCR5 expression1, 40. IL-2 potently induces Blimp-1 in mouse CD4+ T cells40–42. We considered that reduced Blimp-1 expression may be occurring via suppression of CD25 expression, but RNA-seq data revealed no difference in IL2RA in the presence of activin A (data not shown). We hypothesized that CD4+ T cell secreted IL-2 may be inhibiting activin A induction of TFH differentiation. Therefore, we evaluated the effect of IL-2 blockade on activin A treated CD4+ T cells. In the presence of activin A alone, differentiation of PD-1+CXCR5+ cells was remarkably increased when IL-2 was neutralized (Fig. 6a–c). IL-2 blockade also modestly potentiated the expression of PD-1 and CXCR5 in cells cultured with activin A plus IL-12 (Supplementary Fig. 7). Thus, IL-2 is a potent inhibitor of human TFH cell differentiation, and activin A can directly induce human TFH-associated gene expression in the absence of any additional cytokines.
Figure 6. IL-2 antagonizes activin A driven TFH cell differentiation.
(a-c) Flow cytometry of naïve CD4+ T cells stimulated by anti-CD3/CD28 beads for 5d with activin A, or beads only (−), in the presence of anti-IL-2, or isotype control antibody. Numbers in quadrants indicate percentage of cells in the outlined areas. Data are cumulative from 3 independent experiments (n=10). ** p<0.01 (Two-tailed Wilcoxon matched-pairs signed ranked test).
Evolutionary requirement for activin A in TFH differentiation
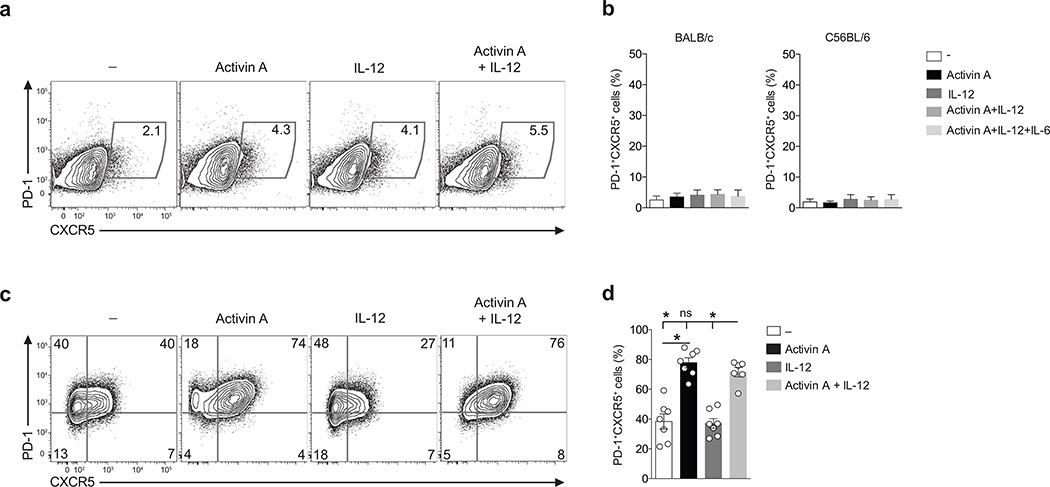
To test the functional conservation of activin A in TFH differentiation, we activated murine naïve CD4+ T cells in the presence of activin A alone or combined with IL-12. The activin A amino acid sequence is 100% conserved between human and mouse. Strikingly, activin A did not have any effect on PD-1 and CXCR5 expression by mouse CD4+ T cells (Fig. 7a,b). This was true for both C57BL/6 and BALB/c mouse strains (Fig. 7a,b). The presence of IL-6, a known regulator of early murine in vivo TFH cell differentiation, in combination with activin A did not promote in vitro mouse TFH cell differentiation (Fig. 7b).
Figure 7. The role of activin A in TFH cell differentiation is conserved for non-human primate, but not mouse CD4+ T cells.
(a) Flow cytometry of naïve CD4+ T cells from BALB/c or C57BL/6 mice cultured in vitro for 5 d with IL-12, activin A, activin A and IL12 or medium only (−), in the presence of plate-bound anti-CD3 and anti-CD28. Numbers in gates indicate percentages of PD-1+ CXCR5+ cells.(b) Frequency of cells in (a). (c) Flow cytometry of naïve CD4+ T cells from rhesus macaque activated by plate bound anti-CD3 and anti-CD28 in the presence of activin A, with or without IL-12, or medium only (−) for 5d. Numbers in quadrants indicate percentage of cells in the outlined areas.(d) Frequency of cells in (c). In (a-d) data are from 4 or more independent experiments (n=4 or more). Bars are mean and s.e.m. * p<0.05 (Two-tailed Wilcoxon matched-pairs signed ranked test).
The lack of any measurable effect of activin A on TFH-associated gene expression by mouse CD4+ T cells prompted us to investigate whether the role of activin A in TFH differentiation was unique to humans. Thus, we evaluated the capacity of activin A to drive in vitro TFH cell differentiation by a species in a different taxonomical family, macaca mulatta (rhesus macaques) of the non-human primate family Cercopithecidae. Potent induction of PD-1+CXCR5+ cells was observed by activated m. mulatta naïve CD4+ T cells in response to activin A (Fig. 7c,d). These data indicate that the TFH-like differentiation mediated by activin A in vitro is not preserved in mice, but is conserved between non-primate species and humans. This finding suggests the existence of a possible evolutionary divergence in activin A-mediated TFH cell differentiation.
SMAD2/3 pathway in activin A-induced TFH differentiation
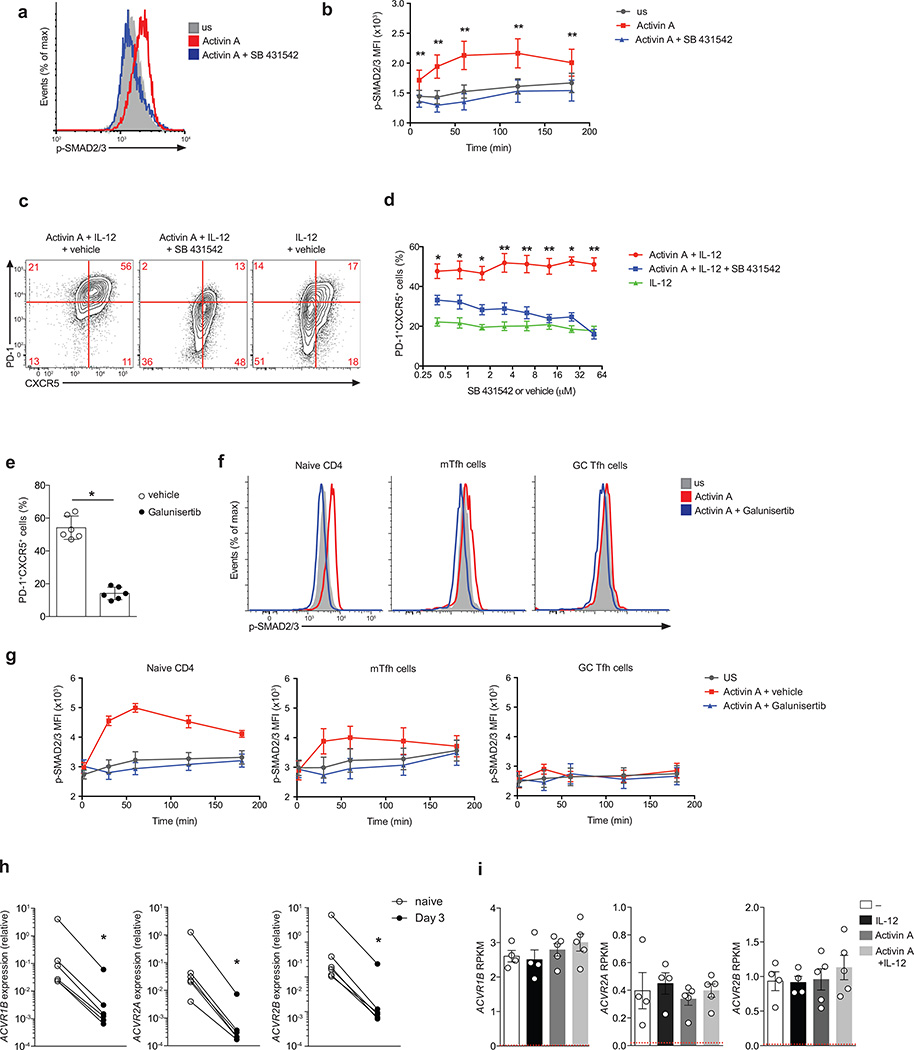
SMAD2/3 is the canonical signaling pathway activated downstream of activin A binding to type IIA and/or IIB receptors and type I receptor ACVR1B (ALK4)31, 32. Phosphorylation of SMAD2/3 occurred in naïve human CD4+ T cells in response to activin A (Fig. 8a,b). Potential SMAD-independent pathways downstream of activin A were not observed (Supplementary Fig. 8). Therefore, we hypothesized that SMAD2/3 activation is a central signaling pathway in activin A mediated induction of TFH-associated gene expression. A pharmacological inhibitor of ALK4, SB431542, known to suppress SMAD2/3 activation but not the SMAD-independent pathways43, resulted in complete inhibition of activin A-mediated SMAD2/3 phosphorylation (Fig. 8a,b). Furthermore, ALK4 inhibition by SB431542 blocked the differentiation of PD-1+CXCR5+ cells in activin A containing culture conditions (Fig. 8c,d). Galunisertib44 is an independent small molecule ALK4 inhibitor currently in clinical trials. Galunisertib potently suppressed PD-1+CXCR5+ cell generation in response to activin A (Fig. 8e and Supplementary Fig. 8c). Altogether, these data point to SMAD2/3 as an important signaling pathway downstream of activin A in the regulation of human TFH cell differentiation.
Figure 8. Activin A activity is mediated by an ALK4-SMAD2/3 pathway.
(a-b) Flow cytometry of phosphorylated-SMAD2/3 (p-SMAD) by human naïve CD4+ T cells following stimulation for 60 min (a) or up to 180 min (b) with activin A (red), activin A + SB 431542 (blue) and in unstimulated cells (US, grey). (c) Flow cytometry of human naïve CD4+ T cells differentiated for 5 d with activin A + IL-12 or IL-12 only in the presence of SB 431542 (50µM) or vehicle (DMSO). Numbers in quadrants indicate percentage of cells in the outlined areas.(d) Frequency of PD-1+CXCR5+ on cells differentiated as in (c) in the presence of different doses of SB 431542 (50µM) or vehicle (DMSO). (e) Frequency of PD-1+CXCR5+ on cells naïve CD4+ T cells differentiated for 5 d with activin A + IL-12 in the presence of Galunisertib or vehicle (DMSO). (f-g) Flow cytometry of phosphorylated-SMAD2/3 (p-SMAD) by tonsillar naïve CD4+ T cells (CD4+CD45RO−), mTFH (CD4+CD45RO+PD-1intCXCR5int) and GC TFH cells (CD4+CD45RO+PD-1hiCXCR5hi) following stimulation for 60 min (f) or up to 180 min (g) with activin A (red), activin A + Galunisertib (blue) and in unstimulated cells (US, grey). (h) Type I (ACVR1B) and Type II (ACVR2A and ACVR2B) activin A receptor expression relative to GAPDH on naïve CD4+ T cells ex vivo (naïve), or upon 3 d following in vitro anti-CD/CD28 bead stimulation. (i) RNA-seq gene expression of ACVR1B, ACVR2A and ACVR2B on naïve CD4+ T cells stimulated with antiCD3/CD28 beads for 3 d with activin A, with or without IL-12, IL-12 and beads only (−). NGFR average expression by cells differentiated with beads only (−) is shown in red. Bars are mean and s.e.m.. In (c-h) data are cumulative from 2 or more experiments (n=6 or more). * P < 0.05, ** P < 0.01 and *** P <0.001 (Two-tailed Wilcoxon matched-pairs signed ranked test).
Activin A is expressed in human lymphoid tissue in both the T cell area and the T cell-B cell border area. We observed a gradient in activin A induction of phosphorylated SMAD2/3 in tonsillar CD4+ T cells, with naïve CD4+ T cells having the highest responsiveness to activin A (Fig. 8f,g). PD-1intCXCR5int CD4+ T cells, a heterogeneous population of TFH-associated cells located at the T cell-B cell border and in the follicular mantle (mantle TFH, mTFH), also induced SMAD phosphorylation in response to activin A (Fig. 8f,g), although to a lesser extent than naïve CD4+ T cells. This reduced responsiveness might at least partially be explained by a trend of lower expression of the activin A Type I and II receptors in differentiated TFH cells in comparison to naïve CD4+ T cells (Supplementary Fig. 8d). The tonsillar expression pattern of activin A receptors is consistent with the finding that naïve CD4+ T cells from blood expressed ACVR1B, ACVR2A and ACVR2B, but that following TCR stimulation the activin A receptors were down regulated (Fig. 8h). The presence of activin A did not influence the reduction in Type I and II activin A receptors driven by TCR triggering (Fig. 8i). In summary, activin A is expressed by myeloid cells in areas where TFH cell differentiation is initiated, and CD4+ T cells in those locations are responsive to activin A signaling, consistent with an in vivo role for activin A as an early regulator of human TFH cell differentiation.
DISCUSSION
This work establishes a role for activin A in human TFH differentiation. Proper localization is a key feature of TFH biology. Activin A acts by dampening CCR7 and ITGβ7 while fostering CXCR5 expression. Besides regulating localization, activin A also modulates the expression of TFH functional regulators. Activin A promotes the production of CXCL13, TNF, LIF, and LTα. TNF can act as co-stimulatory signal to mediate B cell activation and Ig production38. The roles of LIF and LTα in B cell help have not been formally addressed. However, B cell hyperplasia and polyclonal hypergammaglobulemia are found in mice overexpressing LIF45. In T cell-B cell co-cultures, the B cell helper activity of the in vitro differentiated TFH cells was driven by IL-12. This assay has major limitations in that it mostly relies on CD40L (comparably expressed by all activated CD4+ T cells in vitro) and IL-21. Therefore, TFH cell features regulated by activin A vital for B cell helper function in vivo, such as CXCL13 and cell migration (CXCR5, CCR7) are not emphasized in the available assay.
One important open question is whether activin A and TGF-β physiological distribution indicates which of these two cytokines is more relevant for Tfh differentiation in vivo. There are many means by which TGF-β in vivo bioavailability is regulated, including the fact that TGF-β is physiologically activated in an extremely localized fashion by integrin expressing cells39. Activin A bioavailability is regulated differently31, 32. Therefore, it is not currently possible to predict if activin A or TGF-β is more dominant at inducing Tfh differentiation in a particular in vivo context. Clearly activin A is present in human lymphoid tissue and is induced in myeloid cells by inflammatory TLR ligands. It is still controversial as to whether acquisition of Foxp3 in vitro by human CD4+ T cells confers them with the capacity to suppress immune responses46. Nevertheless, lower Foxp3 induction by activin A in comparison to TGF-β appears to be a favorable event when the ultimate goal is the induction induce human Tfh cells.
Activin A alone was sufficient for the regulation of many Tfh signature molecules, particularly in the absence of high IL-2. Nevertheless, integration of activin A and IL-12 mediated signals was required to acquire a more complete phenotype, including high IL-21 expression. These findings support a working model in which activin A is at the center of a cytokine network orchestrating human Tfh cell differentiation. The cytokine network includes not only additional agonistic players such as IL-12, but also antagonistic signals such as IL-2. We envision the cytokine network driving Tfh cell differentiation to be more complex than what has been described so far.
A significant limitation in the usage of rodent models results from the existence of evolutionary divergence in pathways shaping TFH biology. Fully mature mouse and human TFH cells express a virtually identical set of signature molecules. Despite the phenotypical similarity, a growing body of evidence suggested that there are signaling pathways crucial for human, but not mouse, TFH cell differentiation. Three recent studies reported different outcomes on mouse TFH cell differentiation when TGF-β signaling was impaired, with one study finding that TGF-β is a positive regulator of TFH biology by virtue of its ability to insulate CD4+ T cells from IL-2 signaling and suppress Th1 differentiation47. A second study, however, found no effect on TFH cells when TGF-β signaling was blocked48. Finally, a third study indicated that TGF-β behaved as a TFH cell inhibitor49. Additional studies are therefore required to clarify TGF-β’s role in TFH biology in mice. While we could not find any relevant effect of activin A on mouse CD4+ T cells in vitro, activin A potently induced human and non-human primate TFH differentiation. These data suggest the existence of a potential evolutionary divergence in the role that activin A plays in TFH cell biology. Alternatively, the inability of activin A to induce mouse CD4+ T cell TFH differentiation in vitro may reflect an unknown inhibitor or cofactor.
It is highly desirable to foster TFH cells in vivo for vaccines, when the generation of a protective vaccine relies on the production of neutralizing antibodies1. Memory TFH cells and recently activated TFH-related cells can be found in blood1, 7, 35–37, 50. Highly functional memory TFH cells were the strongest immunological correlate for HIV broadly neutralizing antibody development among HIV+ donors36. In contrast, dysregulated antibody responses may lead to auto-antibody mediated diseases3, 7 and memory TFH cells are perturbed in rheumatoid arthritis, systemic lupus erythematous and juvenile dermatomyositis3, 7, 37. In these contexts, hampering TFH cells in a targeted fashion represents a promising therapeutic approach. An appealing candidate could be Galunisertib44, which is currently being tested in clinical trials.
In conclusion, our study revealed a powerful approach for identification of novel regulators of human TFH cell differentiation, which led to the discovery of activin A as potent regulator of human TFH cell differentiation.
METHODS
Human samples
Leukapheresis sample from a healthy donor was obtained from AllCells, Inc., after a prescreening of multiple donors. Fresh whole blood samples from healthy donors were obtained from the La Jolla Institute for Allergy and Immunology (LJI) in-house normal blood donor program (NBDP). Informed consent was obtained from all donors. Fresh human tonsils were obtained from the National Disease Resource Interchange. Tonsil preparation was previously described34. All the protocols were approved by LJI institutional review board. Peripheral blood mononuclear cells (PBMCs) were isolated from both leukapheresis and whole blood samples by density-gradient centrifugation using Histopaque-1077 (Sigma-Aldrich).
Mouse samples
C57BL/6J and BALB/cJ mice were from Jackson Laboratory. Mice were housed in specific pathogen–free conditions in accordance with the Institutional Animal Care and Use Guidelines of the La Jolla Institute.
Non-human primate samples
All rhesus macaque study procedures were performed in accordance with Emory School of Medicine Institutional Animal Care and Use Committee approved protocols. Previously cryopreserved macaque PBMCs were used.
Flow cytometry and cell sorting
For surface staining, primary-staining panels used for phenotypic analysis or for cell sorting are described in supplemental antibody table (Supplementary Table 3). For the intranuclear staining: cells were treated with the Foxp3 Fixation/Permeabilization kit (eBioscience) and stained in Permeabilization buffer (eBioscience). For cytokine staining: day 3 or day 5 in vitro-differentiated cells (specified in the main text or in figure legends) cells were stimulated for 5 h at 37°C with Phorbol 12-myristate 13-acetate (PMA, 2 5ng/ml) and Ionomycin (1 µg/ml) in the presence of Brefeldin A (5 µg/ml). Cells were fixed with PBS 2% FBS, 1% PFA and permeabilized with PBS 2% FBS, 0.5% Saponin (from quillaja bark, Sigma-Aldrich). Samples were analyzed on a LSR Fortessa Cell Analyzer (BD, Bioscience). Flow cytometry data were analyzed with FlowJo (Tree Star). For cell sorting, cells were sorted using sorted using a FACS Aria (BD, Bioscience).
Screening of ‘secretomics’ recombinant proteins
Naïve CD4+ T cells were isolated from leukapheresis-derived mononuclear cells via CliniMACS negative selection (Miltenyi Biotec). The ‘secretomics’ collection was previously described16. An expanded secretomics set (PhaseII), composed of 2688 unique proteins (total 1772 genes) was tested. Naïve CD4+ T cells (1.25 × 104 cells/well) were cultured with secretomics proteins in the presence of Dynabeads Human T-Activator CD3/CD28 (0.5 µl/well, Life Technologies), recombinant human IL-7 (4 ng/ml, Peprotech), and anti-human-TGF-β blocking antibody (100 µg/ml, clone 1D11.16.8, BioXcell) in RPMI medium (Cellgro), supplemented with 10% fetal bovine serum (Omega Scientific), GlutaMAX (Gibco) and Penicillin/Streptomycin (Gibco). Induction of CXCR5 and PD-1 was assessed after 5 d of in vitro culture using automated FACS acquisition via HTS on LSR Fortessa (BD Biosciences). Flow cytometry panel is listed in Supplementary Table 3. For each 384-well plate, mean and standard deviation of PD-1+CXCR5+ or CXCR5+ cell frequency induced by each individual recombinant proteins was determined and used to calculate the z-score (value indicating how many standard deviations an element is from the mean). Z-scores of replicate plates were analyzed independently.
In vitro CD4+ T cell differentiation
Unless differently specified, naïve CD4+ T cells were enriched from PBMCs by magnetic bead negative selection with the EasySep™ Naïve CD4+ T Cell Isolation Kit (STEMCELL Technologies). Purity (CD4+CD45RA+) was 90% or higher. Cells (7.5 × 104 cells/well) were activated by Dynabeads Human T-Activator CD3/CD28 (2 µl/well, Life Technologies) and cultured with recombinant human/mouse/rat activin A (50 ng/ml or 100 ng/ml), recombinant human IL-12 (5 ng/ml), recombinant human TGF-β (1 ng/ml), recombinant human IL-23 (10 ng/ml) or recombinant human IL-35 (10 ng/ml) in the presence of recombinant human IL-7 (4 ng/ml) in AIM-V medium (Life Technologies). All cytokines were from R&D System and Peprotech. The 14 kDa mature human beta A chain of human activin A shares 100% amino acid sequence identity with bovine, feline, mouse, porcine, and rat beta A. Phenotype was quantified after 3 or 5 d, as specified in the Figure legends. Similar results were obtained when using Treg cell-depleted purified naïve CD4+ T cells (Supplementary Fig. 2a), in which cells were sorted from total PBMCs by flow cytometry as CD19−CD8−CD14−CD16−CD4+CD45RO−CD25− cells. For experiments quantifying supernatant CXCL13 concentration, cells were activated by plate-bound anti-human CD3 (5 µg/ml, BD) and ICOS-L (5 µg/ml, recombinant human B7-H2, R&D). CXCL13 concentrations were determined via human CXCL13 DuoSet ELISA (R&D) on supernatants harvested after 5 d of in vitro culture. For blocking experiments, cells were cultured with anti-activin A (5 µg/ml, clone 69403, R&D), pan anti-activin A Type II receptor (ACVR2) (100µg/ml, Novartis), anti-TGF-β (100µg/ml, clone 1D11.16.8, BioXcell), or equivalent concentration of mouse IgG1 isotype antibody (BioXcell). For blockade of IL-2, cells were cultured with anti-IL-2 (10µg/ml, clone 5334, R&D) or mouse IgG1 isotype antibody (BioXcell). For SMAD2/3 inhibition experiments, cells were cultured with different concentrations of SB 431542 (Tocris Bioscience), Galunisertib (LY2157299, Selleck Chemicals) or vehicle (DMSO) for 5 d in AIM-V supplemented with 2% heat-inactivated FBS or in RPMI-1640 (Corning) supplemented with 10% heat-inactivated FBS, 1% GlutaMax, 1% PenStrep.
Microscopy
Tonsils were fixed for 4 h in freshly diluted 4% paraformaldehyde in PBS (Electron Microscopy Sciences, EMS Diasum), then washed 3 times in PBS and equilibrated in 30% w/v sucrose solution in PBS for 24 h. Tissues were briefly washed in PBS, frozen in OCT and stored at −80 °C. Cryostat sections were cut at 12 µm thickness, and air-dried for 30 min. Sections were rehydrated for 10 min in PBS and non-specific binding sites were saturated with 5% normal donkey serum (Jackson Immunoresearch) in the presence of 0.3% Triton X-100 in PBS for 1 h. Tissues were incubated with rabbit polyclonal anti-INHBA (HPA020031, Sigma-Aldrich), mouse anti-CD3−FITC (UCHT1, Tonbo Biosciences) and mouse anti-Bcl6–Alexa Fluor 647 (K112-91, BD Biosciences) overnight at 4 °C. Following washing, sections were reacted with donkey anti-rabbit–Alexa Fluor 568 and goat anti-FITC–Alexa Fluor 488 secondary antibodies (Invitrogen) for 1 h at 23 °C. For CD11c staining, slides were first incubated with anti-INHBA and mouse anti-CD11c (3.9, BD), washed and reacted with donkey anti-rabbit–Alexa Fluor 568 and goat anti-mouse–DyLight 649 secondary antibodies (Biolegend). Following brief fixation slides were stained with anti-CD3 as described above. Slides were counterstained with 10 µg/ml Hoechst for 10 min, washed and mounted in Prolong Gold antifade reagent (Invitrogen) with a cover slip and examined with AxioScan Z1 slide scanner equipped in 20x/0.8 NA air objective using appropriate filter sets. High resolution imaging of selected regions was done on a FluoView FV10i confocal microscope (Olympus) using 60x/1.35 NA oil objective. To reduce blurriness epifluorescence images were processed with an extended depth of field algorithm (ZEN2, Zeiss). To improve feature visibility images were processed by contrast stretching using identical procedures (ZEN2 and Olympus FluoView software).
A separate anti-INHBA from Abcam (ab56057) was used for staining validation and comparable results were obtained. In addition, two independent rabbit IgG (R&D and EMD Millipore) and mouse IgG1 (Biolegend) were used as negative controls. Equipment and settings used to acquire microscopy data are described in Supplementary Table 4.
Monocyte stimulation
Monocytes were isolated by human peripheral blood mononuclear cells by EasySep™ human CD14-positive selection kit (STEMCELL Technologies). Purity, measured as percentage of CD14+ cells by flow cytometry, was 90% or higher. For qPCR analysis of INHBA expression, 106 CD14+ cells were cultured in 96-well plates, with or without Escherichia coli-derived LPS (100 ng/ml, Invivogen) for 4 h at 37°C and then frozen in RLT buffer (Qiagen). For quantitation of activin A secretion by ELISA, 106 CD14+ cells were cultured in 48-well plates, with or without E. coli-derived LPS (100 ng/ml, Invivogen) for 48 h at 37°C. After 48 h, supernatant was collected and then frozen for subsequent measure of secreted activin A. Supernatants were diluited 1:20 and activin A was measured by Human/Mouse/Rat activin A Quantikine ELISA kit (R&D) following manufacturer instructions.
T–B co-culture
Naïve CD4+ T cells were differentiated with activin A and/or IL-12. At day 3, live CD4+ T cells were sorted by flow cytometry. Fresh autologous B cells were enriched from PBMCs by CD19 magnetic-bead positive selection (Miltenyi Biotec). Memory B cells were then sorted by flow cytometry as CD3−CD14−CD4−CD19+IgD−CD27+ cells as previously described36 and cultured (4 × 105 cells/well) with the in vitro differentiated T cells (2.5 × 103 cells/well) in the presence of staphylococcal enterotoxin B (SEB, 0.25 ng/ml, Toxin Technology) in AIM-V medium. B cell counts and plasmablast frequencies were measured after 7 d by flow cytometry as previously described36. Ig concentrations in the supernatants were determined by ELISA.
Mouse CD4+ T cell differentiation
Naïve CD4+ T cells were isolated from spleen using the Naïve CD4+ T cell isolation kit (Stem Cell). Purity was 92% or higher. Naïve CD4+ T cells (2 × 105 cells/well) were cultured for 3 d with recombinant human/mouse/rat activin A (50 ng/ml, R&D Systems), recombinant mouse IL-12 (10 ng/ml, Peprotech), and/or recombinant mouse IL-6 (20 ng/ml, Peprotech) in the presence of plate-bound anti-mouse-CD3 mAb (8 µg/ml, clone 145-2C11, BioXcell) and anti-mouse-CD28 (8 µg/ml, clone 37.51, BioXcell), in RPMI medium, supplemented with 10% fetal bovine serum, GlutaMAX, penicillin/streptomycin, and 2.5 µM β-mercaptoethanol. After 3 d, cells were removed from stimuli and further cultured for 2 additional d in IL-2 (50 U/ml,) and the same cytokine combination used at day 0. Phenotype was quantified by flow cytometry (Supplementary Table 3) at day 5 of the in vitro culture.
Non-human primate (NHP) CD4+ T differentiation
CD4+ T cells from rhesus macaque PBMCs were enriched using the Non-Human Primate CD4+ T Cell Isolation Kit (Miltenyi Biotec). Naïve CD4+ T cells were isolated by flow cytometry as CD4+CD45RA+CD95−CD28+CCR7+ cells. Naïve CD4+ T cells (7.5 × 104 cells/well) were activated by plate-bound anti-human-CD3 and anti-human-CD28 (both at 5 µg/ml, BD) and cultured with recombinant human/mouse/rat activin A (50 ng/ml) and/or recombinant human IL-12 (5 ng/ml) for 5 d. Phenotype was assessed by flow cytometry, following the staining panel in Supplementary Table 3.
RNA sequencing
Differentiation of naïve CD4+ T cells with activin A and/or IL-12 was described above. Naïve CD4+ T cells from 4 or more donors were cultured with the following conditions: beads only (−), IL-12, activin A (100 ng/ml), activin A (100 ng/ml) + IL-12, TGF-β, TGF-β + IL-12. After 3 d of in vitro culture, live CD4+ T cells were sorted by flow cytometry. Total RNA was purified using miRNAeasy Mini kit (Qiagen). Standard quality control steps were included to determine total RNA quality using Agilent Bioanalyzer (RNA integrity number (RIN) > 8.5; Agilent RNA 6000 Nano Kit) and quantity using a nanoliter spectrophotometer (Nanodrop, Thermofisher). For every sample, 500 ng of purified total RNA was prepared into mRNA libraries, according to manufacturer’s instructions, using the Truseq Stranded mRNA Library Prep Kit (Illumina, RS-122-2103). The resulting libraries were deep sequenced, using the Illumina HiSeq2500 system in rapid run mode, to obtain between 6–7 × 106 of 50-bp length single-end reads per library (Supplementary Table 5).
Analysis of RNA-seq data
The single-end reads that passed Illumina filters were filtered for reads aligning to tRNA, rRNA, adapter sequences, and spike-in controls. The reads were then aligned to UCSC hg19 reference genome using TopHat (v 1.4.1). DUST scores were calculated with PRINSEQ Lite (v 0.20.3) and low-complexity reads (DUST > 4) were removed from the BAM files. The alignment results were parsed via the SAMtools to generate SAM files. Read counts to each genomic feature were obtained with the htseq-count program (v 0.6.0) using the “union” option. After removing absent features (zero counts in all samples), the raw counts were converted to RPKM value followed by quantile normalization via R library ‘aroma.light’. Then the quantile normalized RPKM values were filtered by setting a cutoff value of 1 and analyzed with the Multiplot module in GenePattern suite (http://www.broadinstitute.org/cancer/software/genepattern/) to generate the Volcano, Heatmap and Expression plots. Meanwhile the filtered raw counts were imported to R/Bioconductor package DESeq2 (v 3.1) to normalize counts and identify differentially expressed genes among conditions.
Gene Set Enrichment analysis (GSEA)
Gene Set Enrichment Analysis (v 2.2.0; http://www.broadinstitute.org/gsea/index.jsp) was used to evaluate if a pre-defined tonsil GC-TFH gene set showed statistically significant enrichment between two phenotypes when cells are given a particular stimulus. The tonsil GC TFH gene set (Supplementary Table 6) was generated from previously published microarray data36 and defined by genes with more than two-fold higher expression in GC-TFH cells (CD4+CD45RO+PD-1hiCXCR5hi) compared to non-TFH cells (CD4+CD45RO+PD-1−CXCR5−). For RNA-seq profiles used as the query list, genes were ranked from the most up-regulated to the most down-regulated in cells stimulated with beads versus activin A + IL-12, IL-12 versus activin A + IL-12, and activin A versus activin A + IL-12 on the basis of the DESeq2 analysis results. The signal-to-noise metric was used for ranking the genes in the RNA-seq expression datasets and 105 cycles of permutations on the phenotype labels were performed to determine the normalized enrichment score (NES).
Quantitative Real-Time PCR
RNA was isolated by QIAGEN RNeasy spin columns and reverse-transcribed into cDNA using Superscript II Reverse Transcriptase (Invitrogen). Quantitative real-time PCR of ACTB, LIF, and PRDM1 was performed using the following primers: ACTB forward, 5′-ACCTTCTACAATGAGCTGCG-3′, ACTB reverse, 5′- CCTGGATAGCAACGTACATGG-3′; LIF forward, 5′-ATACGCCACCCATGTCAC-3′, LIF reverse, 5′- CCACATAGCTTGTCCAGGTTG-3′; and PRDM1 forward, 5′-TGTGGTATTGTCGGGACTTTG-3′, PRDM1 reverse 5′-CTTTGGGACATTCTTTGGGCCTG-3′. The following primers were used to quantify the expression of ACVR1B (ALK4), ACVR2A, ACVR2B and INHBA: GAPDH forward, 5′-ACATCGCTCAGACACCATG-3′, GAPDH reverse 5′-TGTAGTTGAGGTCAATGAAGGG-3′, ACVR1B forward, 5′-GGAAGCAGAGATATACCAGACG-3′, ACVR1B reverse, 5′-AGATAATCAAACAGGGACCCG-3′, ACVR2A forward, 5′-CCTGGAATGAAGCATGAGAAC-3′, ACVR2A reverse, 5′-TTCCAAGAGACCACATTAGCC-3′, ACVR2B forward, 5′-GAGATCTTCAGCACACCTGG-3′, ACVR2B reverse, 5′-GATGTTCCCCTTGAGGTAATCC-3′, INHBA forward, 5′-ACGGGTATGTGGAGATAGAGG-3′, and INHBA reverse, 5′-TGGAAATCTCGAAGTGCAGC-3′. Real-time PCR was set up with Applied Biosystem SYBR Green Master Mix.
Analysis of phosphorylated SMAD2/3
Fresh PBMCs and tonsil mononuclear cells were serum-starved overnight in AIM-V medium. The following day, the cells (10^6 cells/condition) were stimulated for 10, 30, 60, 120 or 180 min with activin A (400ng/ml) + vehicle (DMSO), activin A + SB 431542 (10µM, Tocris Bioscience), Galunisertib (LY2157299, 10 µg/ml, Selleck Chemicals) or vehicle only in AIM-V medium. Following stimulation, the cells were fixed in BD Phosflow™ Fix Buffer I (BD Biosciences), then permeabilized using BD Phosflow™ Perm Buffer III (BD Biosciences) following manufacture’s instruction. The staining panel is provided in Supplementary Table 3.
Statistical Analysis
All the statistical analyses, unless differently specified, were done with two-tailed Wilcoxon matched-pairs signed ranked test, which does not assume Gaussian distribution of the data, but it allows the direct comparison of populations within donors. Independent experiments were repeated several times with controls. Prism 6.0 (GraphPad) was used for all analysis.
Supplementary Material
Acknowledgments
This work was supported by NIH grant UM1-AI100663 (S.C.), internal La Jolla Institute funding (S.C.) and internal GNF funding (A.M.). We thank the Protein Sciences Group at GNF for protein production. The content of GNF secretomics collection is proprietary. Access to the collection may be considered for further research collaboration agreements on a case-by-case basis. The NHP PBMCs samples were kindly provided by G. Silvestri (Emory University). We thank the sequencing core at La Jolla Institute for RNA-seq data generation. We acknowledge the National Disease Resource Interchange for providing tonsil samples.
Footnotes
Accession codes
GEO: RNA-seq datasets, GSE78276.
AUTHOR CONTRIBUTIONS
M.L. and S.C. conceived and designed the experiments; M.L. and J.W. performed experiments and analyzed data; F.A. performed SMAD-related experiments; Z.M. generated and analyzed immunofluorescence data; C.D. contributed to the screen optimization; A.T.M. provided the secretomics library; M.L. and S.C. wrote the paper; S.C. supervised the study.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interest.
REFERENCES
- 1.Crotty S. T Follicular Helper Cell Differentiation, Function, and Roles in Disease. Immunity. 2014;41:529–542. doi: 10.1016/j.immuni.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Victora GD, Nussenzweig MC. Germinal centers. Annu Rev Immunol. 2012;30:429–457. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- 3.Craft JE. Follicular helper T cells in immunity and systemic autoimmunity. Nat Rev Rheumatol. 2012:1–11. doi: 10.1038/nrrheum.2012.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crotty S. A brief history of T cell help to B cells. Nat. Rev. Immunol. 2015;15:185–189. doi: 10.1038/nri3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gitlin AD, Shulman Z, Nussenzweig MC. Clonal selection in the germinal centre by regulated proliferation and hypermutation. Nature. 2014;509:637–640. doi: 10.1038/nature13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vinuesa CG, Cyster JG. How T cells earn the follicular rite of passage. Immunity. 2011;35:671–680. doi: 10.1016/j.immuni.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 7.Ueno H, Banchereau J, Vinuesa CG. Pathophysiology of T follicular helper cells in humans and mice. Nat Immunol. 2015;16:142–152. doi: 10.1038/ni.3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suto A, et al. Development and characterization of IL-21-producing CD4+ T cells. J Exp Med. 2008;205:1369–1379. doi: 10.1084/jem.20072057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nurieva RI, et al. Generation of T follicular helper cells is mediated by interleukin-21 but independent of T helper 1, 2, or 17 cell lineages. Immunity. 2008;29:138–149. doi: 10.1016/j.immuni.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eto D, et al. IL-21 and IL-6 are critical for different aspects of B cell immunity and redundantly induce optimal follicular helper CD4 T cell (Tfh) differentiation. PLoS ONE. 2011;6:e17739. doi: 10.1371/journal.pone.0017739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi YS, Eto D, Yang JA, Lao C, Crotty S. Cutting edge: STAT1 is required for IL-6-mediated Bcl6 induction for early follicular helper cell differentiation. J Immunol. 2013;190:3049–3053. doi: 10.4049/jimmunol.1203032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmitt N, et al. The cytokine TGF-β co-opts signaling via STAT3-STAT4 to promote the differentiation of human TFH cells. Nat Immunol. 2014 doi: 10.1038/ni.2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma CS, et al. Early commitment of naïve human CD4(+) T cells to the T follicular helper (T(FH)) cell lineage is induced by IL-12. Immunol Cell Biol. 2009;87:590–600. doi: 10.1038/icb.2009.64. [DOI] [PubMed] [Google Scholar]
- 14.Schmitt N, et al. Human dendritic cells induce the differentiation of interleukin-21-producing T follicular helper-like cells through interleukin-12. Immunity. 2009;31:158–169. doi: 10.1016/j.immuni.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakayamada S, et al. Early Th1 cell differentiation is marked by a Tfh cell-like transition. Immunity. 2011;35:919–931. doi: 10.1016/j.immuni.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonzalez R, et al. Screening the mammalian extracellular proteome for regulators of embryonic human stem cell pluripotency. Proc Natl Acad Sci USA. 2010;107:3552–3557. doi: 10.1073/pnas.0914019107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ray JP, et al. Transcription factor STAT3 and type I interferons are corepressive insulators for differentiation of follicular helper and T helper 1 cells. Immunity. 2014;40:367–377. doi: 10.1016/j.immuni.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gold E, Risbridger G. Activins and activin antagonists in the prostate and prostate cancer. Mol. Cell. Endocrinol. 2012;359:107–112. doi: 10.1016/j.mce.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 19.Muttukrishna S, Tannetta D, Groome N, Sargent I. Activin and follistatin in female reproduction. Mol. Cell. Endocrinol. 2004;225:45–56. doi: 10.1016/j.mce.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 20.Munz B, et al. The roles of activins in repair processes of the skin and the brain. Mol. Cell. Endocrinol. 2001;180:169–177. doi: 10.1016/s0303-7207(01)00514-7. [DOI] [PubMed] [Google Scholar]
- 21.Phillips DJ, de Kretser DM, Hedger MP. Activin and related proteins in inflammation: not just interested bystanders. Cytokine Growth Factor Rev. 2009;20:153–164. doi: 10.1016/j.cytogfr.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 22.Aleman-Muench GR, Soldevila G. When versatility matters: activins/inhibins as key regulators of immunity. Immunol Cell Biol. 2012;90:137–148. doi: 10.1038/icb.2011.32. [DOI] [PubMed] [Google Scholar]
- 23.Dalton S. Signaling networks in human pluripotent stem cells. Curr. Opin. Cell Biol. 2013;25:241–246. doi: 10.1016/j.ceb.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones CP, Gregory LG, Causton B, Campbell GA, Lloyd CM. Activin A and TGF-β promote T(H)9 cell-mediated pulmonary allergic pathology. J. Allergy Clin. Immunol. 2012;129:1000.e3–1010.e3. doi: 10.1016/j.jaci.2011.12.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huber S, et al. Activin a promotes the TGF-beta-induced conversion of CD4+CD25− T cells into Foxp3+ induced regulatory T cells. J Immunol. 2009;182:4633–4640. doi: 10.4049/jimmunol.0803143. [DOI] [PubMed] [Google Scholar]
- 26.Ogawa K, Funaba M, Chen Y, Tsujimoto M. Activin A functions as a Th2 cytokine in the promotion of the alternative activation of macrophages. J Immunol. 2006;177:6787–6794. doi: 10.4049/jimmunol.177.10.6787. [DOI] [PubMed] [Google Scholar]
- 27.Robson NC, et al. Activin-A: a novel dendritic cell-derived cytokine that potently attenuates CD40 ligand-specific cytokine and chemokine production. Blood. 2008;111:2733–2743. doi: 10.1182/blood-2007-03-080994. [DOI] [PubMed] [Google Scholar]
- 28.Erämaa M, Hurme M, Stenman UH, Ritvos O. Activin A/erythroid differentiation factor is induced during human monocyte activation. J Exp Med. 1992;176:1449–1452. doi: 10.1084/jem.176.5.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ogawa K, Funaba M, Mathews LS, Mizutani T. Activin A stimulates type IV collagenase (matrix metalloproteinase-2) production in mouse peritoneal macrophages. J Immunol. 2000;165:2997–3003. doi: 10.4049/jimmunol.165.6.2997. [DOI] [PubMed] [Google Scholar]
- 30.Schmitt N, et al. IL-12 receptor β1 deficiency alters in vivo T follicular helper cell response in humans. Blood. 2013;121:3375–3385. doi: 10.1182/blood-2012-08-448902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abe Y, Minegishi T, Leung PCK. Activin receptor signaling. Growth Factors. 2004;22:105–110. doi: 10.1080/08977190410001704688. [DOI] [PubMed] [Google Scholar]
- 32.Tsuchida K, et al. Activin signaling as an emerging target for therapeutic interventions. Cell Commun. Signal. 2009;7:15. doi: 10.1186/1478-811X-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sáez de Guinoa J, Barrio L, Mellado M, Carrasco YR. CXCL13/CXCR5 signaling enhances BCR-triggered B-cell activation by shaping cell dynamics. Blood. 2011;118:1560–1569. doi: 10.1182/blood-2011-01-332106. [DOI] [PubMed] [Google Scholar]
- 34.Kroenke MA, et al. Bcl6 and Maf Cooperate To Instruct Human Follicular Helper CD4 T Cell Differentiation. J Immunol. 2012;188:3734–3744. doi: 10.4049/jimmunol.1103246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chevalier N, et al. CXCR5 Expressing Human Central Memory CD4 T Cells and Their Relevance for Humoral Immune Responses. J Immunol. 2011;186:5556–5568. doi: 10.4049/jimmunol.1002828. [DOI] [PubMed] [Google Scholar]
- 36.Locci M, et al. Human circulating PD-1+CXCR3-CXCR5+ memory Tfh cells are highly functional and correlate with broadly neutralizing HIV antibody responses. Immunity. 2013;39:758–769. doi: 10.1016/j.immuni.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morita R, et al. Human blood CXCR5(+)CD4(+) T cells are counterparts of T follicular cells and contain specific subsets that differentially support antibody secretion. Immunity. 2011;34:108–121. doi: 10.1016/j.immuni.2010.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moens L, Tangye SG. Cytokine-Mediated Regulation of Plasma Cell Generation: IL-21 Takes Center Stage. Front Immunol. 2014;5:65. doi: 10.3389/fimmu.2014.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Travis MA, Sheppard D. TGF-β Activation and Function in Immunity. Annu Rev Immunol. 2013 doi: 10.1146/annurev-immunol-032713-120257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oestreich KJ, Mohn SE, Weinmann AS. Molecular mechanisms that control the expression and activity of Bcl-6 in TH1 cells to regulate flexibility with a TFH-like gene profile. Nat Immunol. 2012;13:405–411. doi: 10.1038/ni.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnston RJ, Choi YS, Diamond JA, Yang JA, Crotty S. STAT5 is a potent negative regulator of TFH cell differentiation. J Exp Med. 2012;209:243–250. doi: 10.1084/jem.20111174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ray JP, et al. The Interleukin-2-mTORc1 Kinase Axis Defines the Signaling, Differentiation, and Metabolism of T Helper 1 and Follicular B Helper T Cells. Immunity. 2015 doi: 10.1016/j.immuni.2015.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Inman GJ, et al. SB-431542 is a potent and specific inhibitor of transforming growth factor-beta superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol. Pharmacol. 2002;62:65–74. doi: 10.1124/mol.62.1.65. [DOI] [PubMed] [Google Scholar]
- 44.Herbertz S, et al. Clinical development of galunisertib (LY2157299 monohydrate), a small molecule inhibitor of transforming growth factor-beta signaling pathway. Drug Des Devel Ther. 2015;9:4479–4499. doi: 10.2147/DDDT.S86621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shen MM, et al. Expression of LIF in transgenic mice results in altered thymic epithelium and apparent interconversion of thymic and lymph node morphologies. EMBO J. 1994;13:1375–1385. doi: 10.1002/j.1460-2075.1994.tb06391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tran DQ, Ramsey H, Shevach EM. Induction of FOXP3 expression in naive human CD4+FOXP3 T cells by T-cell receptor stimulation is transforming growth factor-beta dependent but does not confer a regulatory phenotype. Blood. 2007;110:2983–2990. doi: 10.1182/blood-2007-06-094656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marshall HD, et al. The tumor growth factor beta signaling pathway is critical for the formation of CD4 T follicular helper cells and isotype-switched antibody responses in the lung mucosa. Elife. 2015;4 doi: 10.7554/eLife.04851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.León B, Bradley JE, Lund FE, Randall TD, Ballesteros-Tato A. FoxP3+ regulatory T cells promote influenza-specific Tfh responses by controlling IL-2 availability. Nat Comms. 2014;5:3495. doi: 10.1038/ncomms4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McCarron MJ, Marie JC. TGF-β prevents T follicular helper cell accumulation and B cell autoreactivity. J. Clin. Invest. 2014;124:4375–4386. doi: 10.1172/JCI76179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bentebibel S-E, et al. Induction of ICOS+CXCR3+CXCR5+ TH Cells Correlates with Antibody Responses to Influenza Vaccination. Sci Transl Med. 2013;5:176ra32–176ra32. doi: 10.1126/scitranslmed.3005191. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.