Abstract
A range of current candidate AIDS vaccine regimens are focused on generating protective HIV neutralizing antibody responses. Many of these efforts rely on the rhesus macaque animal model. Understanding how protective antibody responses develop and how to increase their efficacy are both major knowledge gaps. Germinal centers (GC) are the engines of antibody affinity maturation. GC T follicular helper (GC Tfh) CD4 T cells are required for GCs. Studying vaccine-specific GC Tfh cells after protein immunizations has been challenging, as antigen-specific GC Tfh cells are difficult to identify by conventional intracellular cytokine staining (ICS). Cytokine production by GC Tfh cells may be intrinsically limited in comparison to other T helper effector cells, as the biological role of a GC Tfh cell is to provide help to individual B cells within the GC, rather than secreting large amounts of cytokines bathing a tissue. To test this idea, we developed a cytokine-independent method to identify antigen-specific GC Tfh cells. RNAseq was performed using TCR stimulated GC Tfh cells to identify candidate markers. Validation experiments determined CD25 (IL2Rα) and OX40 to be highly upregulated activation induced markers (AIM) on the surface of GC Tfh cells after stimulation. In comparison to ICS, the AIM assay identified > 10-fold more antigen-specific GC Tfh cells in HIV Env protein immunized macaques (BG505 SOSIP). CD4 T cells in blood were also studied. In sum, AIM demonstrates that antigen-specific GC Tfh cells are intrinsically stingy producers of cytokines, which is likely an essential part of their biological function.
Keywords: CD4 T cells, vaccines, rhesus macaques, antigen-specific, Tfh
INTRODUCTION
The vast majority of vaccines are effective by inducing protective antibody responses (1). The induction of high quality antibody responses is strongly dependent on the germinal center response. A number of current HIV vaccine candidates being tested in macaques aim to generate protective antibody responses against HIV/SHIV, with broadly neutralizing antibodies (bnAbs) being the most lofty goal (2). HIV bnAbs are extensively somatically mutated Abs by affinity maturation, implying a central role of germinal centers in bnAb development (3, 4). Germinal centers are sites of intricate B cell and T cell interaction necessary for the development of antibody affinity maturation and affinity matured memory B cells (5). For vaccine development, a greater understanding of T follicular helper (Tfh) CD4 T cells and germinal center biology is needed (6–10). In the germinal center, GC B cells bind antigen in proportion to the affinity of each B cell receptor for that antigen and present it to GC Tfh cells. A GC Tfh cell then provides ‘help’ in the form of survival, proliferation, and/or differentiation signals to instruct a GC B cell (8, 11). In this way, higher affinity B cells are selected by the GC Tfh cells to drive affinity maturation to the antigen.
Tfh help to B cells is antigen-specific. However, most analysis of GC Tfh cells is not done in an antigen-specific manner (12–17). In both humans and non-human primates, it is extremely difficult to quantify antigen-specific GC Tfh cells (18–20). GC Tfh cells generally do not express Th1, Th2, or Th17 cytokines (21–23). GC Tfh cell production of IL-21, IL-4, CXCL13, and CD40L are important mitogenic and differentiation factors provided to GC B cells. Each of these factors are induced in GC Tfh cells at the mRNA level upon non-specific stimulation (e.g. PMA/ionomycin), but robust cytokine protein expression after antigen stimulation of GC Tfh cells has generally not been observed. Approximately, 0.1% or fewer of GC Tfh cells are generally identified as specific for the antigen of interest (19, 20). The difficulty in identifying GC Tfh cells by measuring cytokine production may be a consequence of germinal center biology. To create competition among GC B cells, GC Tfh cells must discriminately provide ‘help’ signals only to GC B cells in a one-on-one manner. It is this direct competition between the GC B cells for GC Tfh cell help which drives the rapid evolution of affinity maturation in germinal centers. This biology is unlike other effector T helper subsets, such as Th1 or Th17 cells, which are frequently tasked to produce large amounts of cytokines such as IFNγ or IL-17 to recruit pro-inflammatory cells over long distances. Therefore, we considered that perhaps GC Tfh cells are intrinsically stingy cytokine producers.
MATERIALS AND METHODS
Macaque immunization
Macaques were immunized subcutaneously with HIV BG505 SOSIP.v5.2 [a version of BG505 SOSIP.664 (2) that has been stabilized (24) and then further modified (Steven W. de Taeye and R.W.S., manuscript in preparation)], or SIVE660 gp140 Env protein in Iscomatrix or MPL (monophosphoryl lipid A) and R848 encapsulated PLGA [poly(lactic-co-glycolic acid)] nanoparticles (25). A full description of the results of both vaccine trials will be published elsewhere (C.H.-D. and S.C., manuscript in preparation; S.K. and B.P., manuscript in preparation). All rhesus macaque study procedures were performed in accordance with Emory School of Medicine Institutional Animal Care and Use Committee approved protocols.
Macaque lymph node, spleen, and blood processing
Previously cryopreserved macaque LN, spleen or PBMC were used. Excisional LN biopsies of inguinal LNs were conducted three weeks post immunization. Splenic tissue was obtained at necropsy. LN or spleen were ground through 70 um strainers and washed with PBS. Samples were centrifuged and treated with Ammonium-Chloride-Potassium (ACK) lysing buffer (Lonza), if needed, and washed with R10. Blood was collected in EDTA tubes for PBMC and plasma isolation by gradient centrifugation.
Human tonsil tissue
Previously cryopreserved human tonsil were used. Non-identifiable discarded tonsil tissue was obtained from Rady Children’s Hospital, San Diego. Tonsil tissue was processed as previously described (21). Informed, written consent was obtained from all human study participants before enrollment in the human studies listed above and approved by the La Jolla Institute Internal Review Board, the Institutional Review Board at Rady Children’s Hospital, and the UCSD Institutional Review Board.
Flow cytometry
GC Tfh cells were characterized by flow cytometry as previously described in human tonsil (21) or macaque lymph node (26). Cells were acquired on a BD Fortessa Analyzer using FACSDivaTM software and analyzed with FlowJo v9.9. See Supplemental Table 1 for antibody panels. A BD FACSAria was used for the cell sorting experiment in Figure 2.
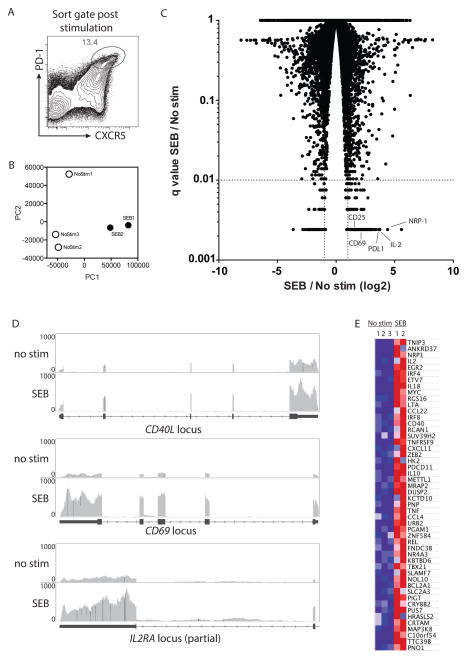
Figure 2. RNAseq of stimulated GC Tfh cells to identify candidate activation induced markers.
A. Lymph node cells from Env protein + adjuvant immunized macaques were either left unstimulated or stimulated with SEB for 6 hours without golgi or vesicle transport inhibitors. B. PCA analysis of unstimulated and SEB stimulated samples. C. Volcano plot showing fold change of stimulated vs unstimulated genes with associated q values. Genes with 2-fold change and q < 0.01 were considered positive. Example genes of interest are labeled. D. RNAseq tracks of unstimulated and SEB stimulated samples at gene loci of interest (from Integrative Genomics Viewer). E. The top 50 upregulated genes in SEB stimulated vs unstimulated samples.
RNA sequencing
Total draining LN cells from immunized macaques were either left unstimulated with no exogenous antigen (n=3) or stimulated with 1 μg/ml SEB (Staphylococcal Enterotoxin B, n=2) at 37°C for 6 hours. 2×104 to 5×104 CXCR5hiPD-1hi CD4+ GC Tfh cells were sorted from each sample. Total RNA was purified using miRNAeasy Mini Kit (Qiagen). Standard quality control steps were included to determine total RNA quality using Agilent 2100 Bioanalyzer (RNA integrity number (RIN) >8.5; Eukaryote Total RNA Pico Kit, Agilent, USA) and quantity using a nanoliter spectrophotometer (Nanodrop, Thermofisher, USA). For each sample, 500 pg of total RNA was prepared into mRNA libraries. According to manufacturer’s instructions Clontech Ultralow HV Kit was used for cDNA synthesis, followed by Illumina NexteraXT DNA Kit for library preparation. The resulting libraries were deep sequenced using the Illumina HiSeq1000 system. Each sample was split into 2, sequenced, and then pooled to obtain between 20 to 23 millions single-end reads per library. Reads were aligned to the Norgren rhesus reference (UNMC_v7.6). RPKM values were filtered by setting a cutoff value of 0.5 and a heat map generated with GenePattern (Broad Institute). Fold change was calculated for the SEB stimulated group value over the unstimulated group value and “q” significance values (p values corrected for multiple corrections) were determined.
Activation Induced Marker (AIM) assay
After thawing, cells were treated with DNase I (100 μg/ml, Stem Cell Technologies) for 15 minutes at 37°C and then rested at 37°C for 3 hours. After resting, the cells were separated into three groups of 1×106 cells each: no exogenous stimulation (“—”), antigen stimulation (5 μg/ml BG505 Env protein and 5 μg/ml of each 15mer peptide of a peptide pool spanning BG505 Env), or SEB (1 μg/ml) and incubated for 18 hours at 37° C. AIM-V serum-free media (GibcoTM by ThermoFisher Scientific) was used for all steps above. Following stimulation, the cells were stained for 1 hour: CD4 (L200 or OKT-4), CD5RA (5H9), OX40 (L106), CD20 (2H7), PD-1 (EH12.2H7), CD25 (BC96), CXCR5 (MU5UBEE) and APCe780 fixable viability dye (see Supplemental Table 1). For PBMC samples, CD14 (61D3) and CD16 (eBioCB16) were included to gate out monocytes. For staining of human tonsilar cells, PD-L1 (29E.2A3), CD83 (HB15e), and CD304 (12C2) were also used. The cells were then washed, fixed with 1% formaldehyde, and acquired the same day. The antigen-specific CD4 T cell frequency was determined by subtracting the frequency of CD25+OX40+ cells in the no exogenous stimulation (“—”) condition from the Env antigen stimulation condition. Samples from animals receiving only adjuvant were used to set the baseline response. A two-fold increase over the average response from the ‘adjuvant only’ animals was considered positive.
ICS Assay
Frozen cells were thawed, washed with AIM-V serum-free media, and treated with DNase for 15 minutes at 37°C. The cells were then rested in AIM-V media at 37°C for 3 hours. After resting, the cells were separated into three groups of 1×106 cells each: no exogenous stimulation (“—”), antigen stimulation (5 μg/ml Env protein plus 5 μg/ml of each 15mer peptide of an overlapping peptide pool spanning BG505 Env; each peptide overlapped by 10 amino acids), or SEB (1 μg/ml) and incubated for a total of 6 hours at 37°C. The experiments shown in Figure S1 used a consensus Clade C Env peptide pool for stimulation (NIH Aids Reagents Program). The final 4 hours of incubation were in the presence of 2 μg/ml Brefeldin A (Sigma). As a positive control, cells were stimulated with 25 ng/mL PMA and 1 μg/mL ionomycin in the presence of brefeldin A for 4 hours. Previous experiments determined that extending the incubation time from a total of 6 to 18 hours did not substantially improve antigen-specific CD4 T cell detection by ICS. Following stimulation, the cells were stained for surface markers CXCR5 (MU5UBEE), CD4 (L200 or OKT-4), CD20 (2H7), CD45RA (5H9), PD-1 (EH12.2H7), and APCe780 fixable viability dye for 30 minutes at 4°C (see Supplemental Table 1). The cells were fixed with 1% formaldehyde for 20 minutes at 4°C, permeabilized with 0.5% saponin (Sigma, S7900) buffer, and stained intracellularly with CD40L (24–31), IL2 (MQ1-17H12), TNF (both MAb11), IL21 (3A3-N2), and IFNγ (45.B3) for 30 minutes. The cells were then washed, fixed with 1% formaldehyde, and acquired the same day.
Statistics
The two tailed Mann-Whitney test was used for evaluating differences among groups. The Wilcoxon test was used to evaluate differences between time points for the same individuals. The Friedmen test, considered a repeated measure non-parametric one-way ANOVA test, was used to analyze the data in Figure 7D. Prism 5.0 (GraphPad) was used for all statistical analyses.
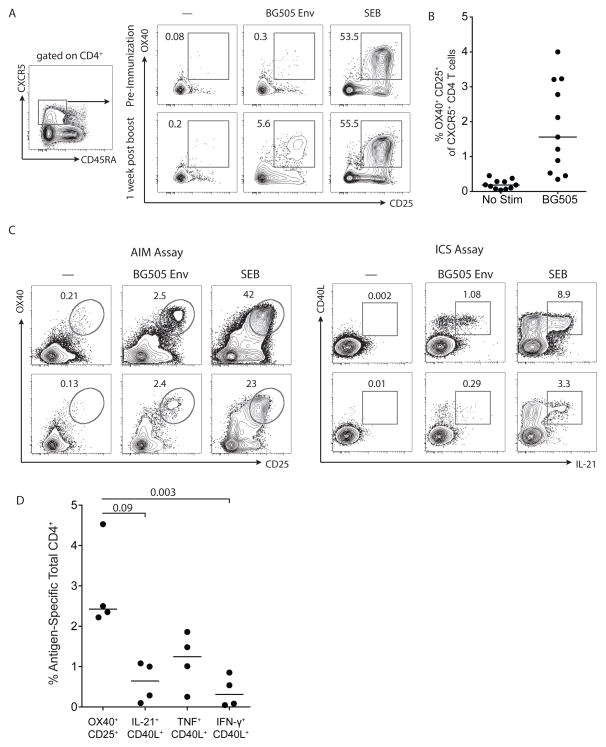
Figure 7. AIM assay detection of immunogen-specific CD4 T cells in blood.
A. BG505 Env-specific responses detected with the AIM assay among CXCR5+ CD4 T cells from PBMC of immunized animals. PBMC from a pre-immunization time point (data from one representative animal of 3) and 1 week after the 4th immunization time point are shown (one representative animal of 11). B. Frequency of BG505 Env-specific responses in the AIM assay by CXCR5+ CD4 T cells from macaque PBMC one week after the 4th immunization. C. PBMC obtained from BG505 Env-immunized rhesus macaques at five weeks after the final immunization were split into two groups for AIM assay and ICS assay comparisons. Assays were compared by evaluation of a condition in which no exogenous antigen was added (marked as “—”), an immunogen stimulation condition (incubation with BG505 Env whole protein and peptides, marked as “BG505 Env”), and a SEB stimulation condition. The cells in the AIM assay were stimulated for 24 hours; cells in the ICS assay were stimulated for 6 hours. Plots are gated on total CD4+ T cells and compare the AIM assay markers OX40+CD25+ to ICS markers CD40L+IL-21+. D. Quantitation of antigen-specific total CD4+ T cells from macaque PBMC identified by the AIM assay (OX40+CD25+) versus IL-21+CD40L+, TNF+ CD40L+, or IFNγ+ CD40L+ by ICS assay (n=4).
RESULTS
Few antigen specific GC Tfh cells are detected by intracellular cytokine staining
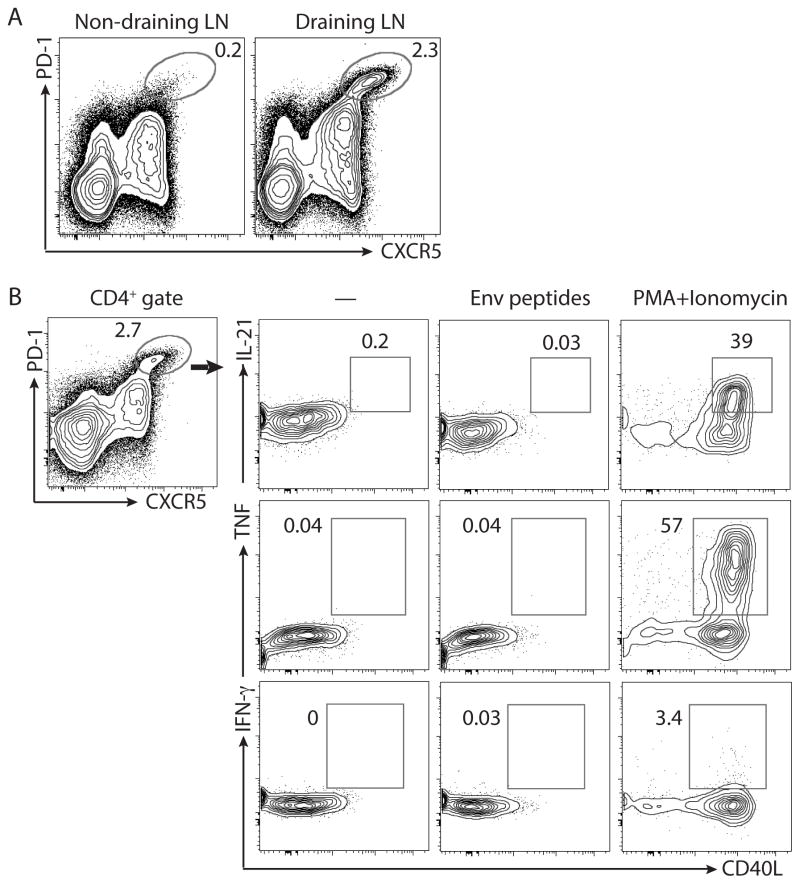
Germinal center Tfh cells, together with GC B cells, are induced in draining secondary lymphoid organs after exposure to foreign antigen. After rhesus macaques were immunized with SIVE660 gp140 protein and adjuvant, GC Tfh cells were found at greater frequencies in the draining LN in comparison to the non-draining LN (Figure 1A). Proportional increases in GC B cells were observed as well (manuscript in preparation). The large increase in GC Tfh cells in the draining LN was consistent with the likely scenario that the GC Tfh cells were specific to the gp140 immunogen. However, conventional ICS assay failed to identify the GC Tfh cells as gp140-specific. 0.1% or fewer of the GC Tfh cells were identifiable as immunogen-specific by ICS, using different combinations of CD40L and TNF, IFNγ, or IL-21 flow cytometry panels (Figure 1B). GC Tfh cells were capable of expressing CD40L, IL-21, and TNF when stimulated with PMA and Ionomycin (Figure 1B). Since GC Tfh cells help GC B cells in an antigen-specific manner and the vast majority of the GC Tfh cells induced by immunization were not identified as antigen-specific by the cytokines analyzed in the ICS assay, we hypothesized that the ICS assay was failing to identify immunogen-specific GC Tfh cells.
Figure 1. Failure of ICS assay to detect antigen-specific GC Tfh cells.
A. Lymph node cells from SIVE660 gp140 Env protein + adjuvant [PLGA (MPL and R848)] immunized macaques 1 week after the 3rd immunization. Cells are gated on total CD4+ T cells. PD-1hiCXCR5hi GC Tfh cell are shown in the oval gate. Representative data of 1 of 8 animals are shown. B. Cytokine expression by GC Tfh cells in an ICS assay. Cells incubated with no exogenous antigen added are marked by “—”. Alternatively, cells were incubated with consensus clade C ENV peptides or PMA+Ionomycin. Representative data from 1 of 8 animals are shown.
Identification of candidate markers to detect antigen-specific GC Tfh cells
GC Tfh cells provide signals to specific GC B cells in one-on-one interactions. Lack of large scale cytokine synthesis by stimulated GC Tfh cells was unlikely to be due to lack of T cell activation, as GC Tfh cells exhibit rapid TCR signaling upon cognate interactions with B cells in GCs (27, 28). Therefore, we took a global approach to identify gene expression changes induced in GC Tfh cells upon TCR stimulation. Cells from draining lymph nodes of immunized macaques were stimulated with SEB or left non-stimulated for 6 hours and then GC Tfh cells were isolated by multiparameter fluorescence cell sorting (Figure 2A). RNAseq was performed on the isolated cells and gene expression of non-stimulated and stimulated GC Tfh cell populations were compared (Figure 2B). From a total of 11,733 mapped genes, we identified 288 upregulated and 174 downregulated genes in stimulated versus unstimulated GC Tfh cells (Figure 2C). Mapped RNAseq traces of CD40L, CD69, and CD25 before and after stimulation are shown as examples of genes induced in GC Tfh cells by TCR stimulation (Figure 2D). In sum, GC Tfh cells respond to TCR stimulation with extensive gene expression changes.
TCR induced genes in antigen-specific GC Tfh cells
Among upregulated genes, we observed TCR stimulation dependent increases in known activation induced genes CD69 and CD40L, supporting the potential identification of GC Tfh activation markers useful for detecting antigen-specific GC Tfh cells. Candidate targets (shown in Table I) were selected based on a combination of four criteria: 1) highest fold change between stimulated and unstimulated conditions (Figure 2E), 2) high absolute expression by RPKM (Reads Per Kilobase Million) after stimulation, 3) secreted or surface molecule, and 4) availability of a monoclonal antibody for flow cytometry analysis.
Table I.
Macaque GC Tfh cell RNAseq targets of interest.
| Cytokines/Chemokines | ||
|---|---|---|
| Tracking ID | SEB/nostim | RPKM |
| IL2 | 14.2 | 173 |
| LTA | 7.8 | 1343 |
| IL10 | 5.9 | 32 |
| TNF | 5.8 | 150 |
| IFNG | 3.8 | 43 |
| IL21 | 2.9 | 129 |
| CCL22 | 7.8 | 277 |
| CCL4 | 6.0 | 587 |
| Surface markers | ||
|---|---|---|
| Tracking ID | SEB/nostim | RPKM |
| NRP1 | 14.7 | 27 |
| CD274 | 11.1 | 97 |
| TNFRSF9 | 6.9 | 634 |
| CD69 | 4.6 | 475 |
| ICAM1 | 3.4 | 62 |
| CD83 | 3.4 | 208 |
| IL2RA | 3.1 | 696 |
| CTLA4 | 2.6 | 377 |
| TNFSF8 | 2.6 | 415 |
| CD40LG | 2.5 | 628 |
| CD200 | 2.4 | 567 |
RNA of multiple cytokine genes were upregulated more than 2.0-fold in GC Tfh cells after TCR stimulation, including TNF, IL-2, IFNγ, and IL-21 (Table I). Compared to TNF, IFNγ was minimally expressed after PMA/ionomycin (Figure 1B). Of the few IFNγ+ cells within the GC Tfh gate (CXCR5hi PD-1hi), most were closer in phenotype to a mantle Tfh (CXCR5int PD-1int) phenotype, indicating that these IFNγ+ cells are not representative of GC Tfh cells (data not shown). While differential expression of IL-2 was detected by RNA-seq, IL-2 protein production came from 2% of SEB stimulated GC Tfh cells (Figure S1). Since cytokine genes differentially expressed by RNAseq were not reliable identifiers of TCR stimulated GC Tfh cells, we investigated whether TCR activation-induced surface molecules may be more reliable identifiers of antigen-specific GC Tfh cells.
For many differentially expressed genes identified by RNAseq, antibodies suitable for NHP flow cytometry were not available for validation of protein expression. Therefore, initial validation screening was done analyzing human GC Tfh cells from tonsils, a lymphoid tissue rich in germinal centers and GC Tfh cells. CD69 was identified in the RNAseq as one of the top upregulated genes, and is widely used as a T cell activation marker. Effector CD4 T cells (CXCR5−) in the tonsil are CD69lo and markedly upregulated the protein after TCR stimulation. However, GC Tfh cells already expressed high levels of CD69 prior to stimulation, and little change in CD69 expression at the protein level was detected after stimulation (Figure 3A), precluding its use as an activation marker to identify antigen-specific GC Tfh cells in vitro. CD200, an immunoregulatory molecule used to identify mouse, human, and macaque GC Tfh cells (12, 29), was excluded for the same reason (Figure 3B). These surface molecules already present on GC Tfh cells were not sufficiently up-regulated after activation to accurately distinguish antigen-specific GC Tfh cells.
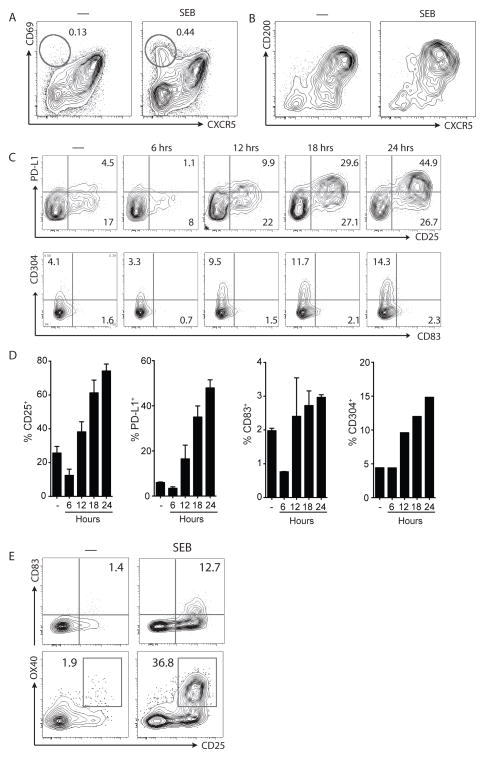
Figure 3. Characterization of candidate GC Tfh cell activation markers.
A. Expression of CD69 on human tonsil CD4 T cells left unstimulated (marked by “—”) or stimulated with SEB for 6 hours. A population of CXCR5− cells upregulates CD69 after SEB stimulation (oval gates). B. Expression of CD200 on human tonsil CD4 T cells, unstimulated “—“ or stimulated with SEB for 24 hours. C. Kinetics of PD-L1, CD25, CD304, and CD83 surface expression on human tonsil GC Tfh cells following stimulation with SEB over the course of 24 hours. The unstimulated “—“ condition was analyzed after 24 hours of incubation; it is not an ex vivo analysis. D. Frequency of single positive CD25-, PD-L1-, CD83-, and CD304-expressing cells in C. Data are from 2 samples, except for CD304 (n=1). E. CD83, OX40, and CD25 expression on GC Tfh cell-gated rhesus macaque spleen or LN cells left unstimulated (marked by “—”) or stimulated with SEB for 24 hours. Data are from 2 samples.
Surprisingly, we observed up-regulation of the IL2α receptor, CD25, on GC Tfh cells after TCR stimulation (q < 0.005, Figure 2C). IL-2 is an inhibitor of murine Tfh differentiation, and CD25 is minimally expressed on differentiating Tfh cells ex vivo (30–33). Surface expression of CD25 protein on GC Tfh cells activated in vitro was minimal at 6 hours after stimulation, but showed large increases at ≥18 hours (Figure 3C). At 18 hours post stimulation, a robust 2 log increase in MFI was observed with ~60% of the GC Tfh cells expressing CD25 (Figure 3C and D). CD25 protein expression was also up-regulated on CXCR5int PD-1int follicular mantle Tfh (mTfh) and CXCR5− effector CD4 T cells from both lymphoid tissue and PBMC, with similar kinetics (Figure S2). In summary, CD25 was validated as an in vitro marker of GC Tfh cell activation.
Additional proteins potentially responsive to GC Tfh cell TCR stimulation were examined. PD-L1 was one such candidate (11.1-fold increase, q < 0.005; Fig 2C, Table I). As GC Tfh cells are high expressers of PD-1, expression of the ligand PD-L1 by T cells after stimulations was unexpected. PD-L1 expression by GC Tfh cells progressively increases to ~35% after 18 hours of stimulation, with a 1 log MFI increase (Figure 3C and D). PD-L1 was co-expressed with CD25 on activated GC Tfh cells (Figure 3C).
More heterogeneous increases in CD83+, a Siglec binding protein, and NRP-1+ (CD304), a Tfh associated gene (34), were observed on GC Tfh cells after TCR activation (Figure 3C and 3D). Few cells co-expressed CD83 and NRP-1, while virtually all CD83+ or NRP-1+ positive cells co-expressed CD25 (data not shown). A separate study of human GC Tfh cell activation revealed OX40 as an additional candidate marker (35). OX40 was not identified as a candidate molecule in the macaque RNAseq, possibly due to the relatively short 6 hr stimulation used (36, 37).
The most promising candidate markers were then reassessed with rhesus macaque GC Tfh cells from immunized animals. Detectable increases in the expression of CD25, CD83, and OX40 were observed after rhesus GC Tfh cell stimulation, although CD83 MFI increases were limited (Figure 3F). No increase was detected for PD-L1 and CD304 on rhesus GC Tfh cells post stimulation (data not shown). Lack of PD-L1 detection on activated GC Tfh cells was likely due to poor cross-reactivity of available anti-PD-L1 mAb to rhesus macaque PD-L1, as minimal PD-L1 was detectable on any cell type (data not shown). Using CD25 and CD83 as activation markers, we were able to identify a population of HIV Env-specific GC Tfh cells from the draining LN of immunized macaques in preliminary experiments (data not shown). However, the most robust and reproducible detection of TCR stimulated GC Tfh cells was observed for OX40 and CD25. Thus, utilizing OX40 and CD25 co-expression may function as an activation induced marker (AIM) technique to detect antigen-specific GC Tfh cells in NHPs in a cytokine-independent manner.
Comparison of AIM and conventional ICS assays in NHP
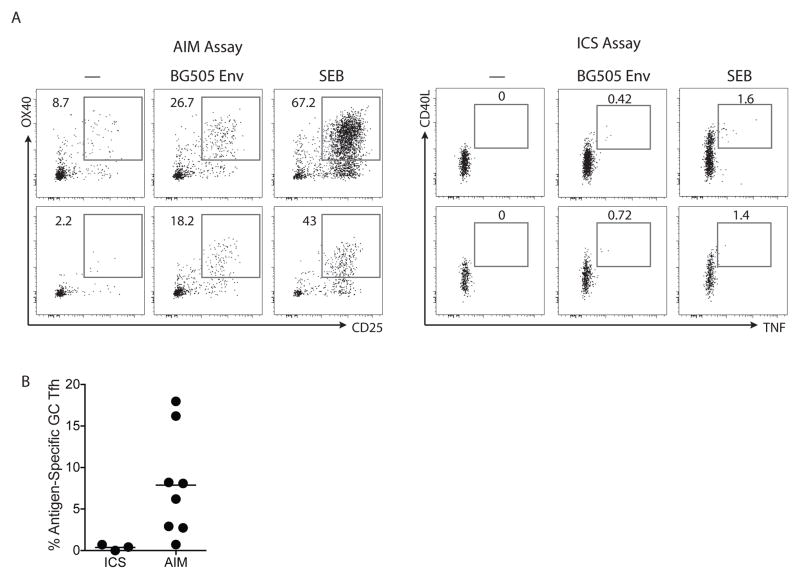
The AIM technique was then assessed for detection of antigen-specific GC Tfh cells. Eight LN samples were tested from a new cohort of rhesus macaques immunized with BG505 SOSIP HIV Env trimers. By AIM assay, robust populations of Env-specific GC Tfh cells were detected in response to BG505 Env stimulation (CD25+OX40+, Figure 4A). SEB stimulation was used as a positive control (Figure 4A). By ICS, CD40L+IFNaγ+ BG505 Env-specific GC Tfh cells were undetectable (Figure S3A). A small population of antigen-specific GC Tfh cells were detectable as CD40L+TNF+ (Figure 4A and 4B). The magnitude of the GC Tfh cell responses detected using AIM were >10-fold higher than the responses detected by conventional ICS (Figure 4B). Thus, the majority of antigen-specific GC Tfh cells in protein immunized macaques do not make sufficient cytokine to be detectable based on conventional cytokine staining.
Figure 4. Comparison of ICS and AIM assays for detection of immunogen-specific GC Tfh cells.
A. BG505 Env-immunized rhesus macaque lymph node cells were split into two groups for the AIM and ICS assays. Assays were compared by evaluation of no exogenous antigen added condition (marked as “—”), an BG505 Env stimulation condition (marked as “BG505 Env”), and a SEB stimulation condition. The “BG505 Env” stimulation condition consisted of both whole Env protein and pooled, overlapping BG505 peptides, which was previously determined to be superior to either stimulation alone. The cells in the AIM assay were stimulated for 24 hours; cells in the ICS assay were stimulated for a total of 6 hours. Plots are gated on live, CD20− CD4+ PD-1hi CXCR5hi GC Tfh cells. B. Quantitation of antigen-specific GC Tfh cells by ICS (n=3) and AIM (n=8) assays. Potential background signals (quantified from the “—” condition) may be true antigen-specific cells responding to antigen already present in the LN preparation from the immunization in vivo, but here were subtracted from BG505 Env responses to be conservative. Raw data are shown in Figure S3B.
In some LN samples, low frequencies of CD25+OX40+ GC Tfh cells were also detected in the absence of exogenous antigen (‘—‘) condition (Figure 4A). Given that ongoing GCs depend on antigen, these GC Tfh cells may be antigen-specific cells responding to antigen residually present in the LN preparation. Consistent with this conclusion, such signals in the absence of exogenous antigen were not observed when peripheral blood cells were used, wherein no antigen is present (see below). Nevertheless, as a conservative calculation of antigen-specific GC Tfh cells, the frequency of AIM+ cells in the absence of exogenous antigen was subtracted as potentially background signal. Raw results are shown in Supplementary Figure 3B.
Antigen-specific GC Tfh cells in the primary draining lymph node
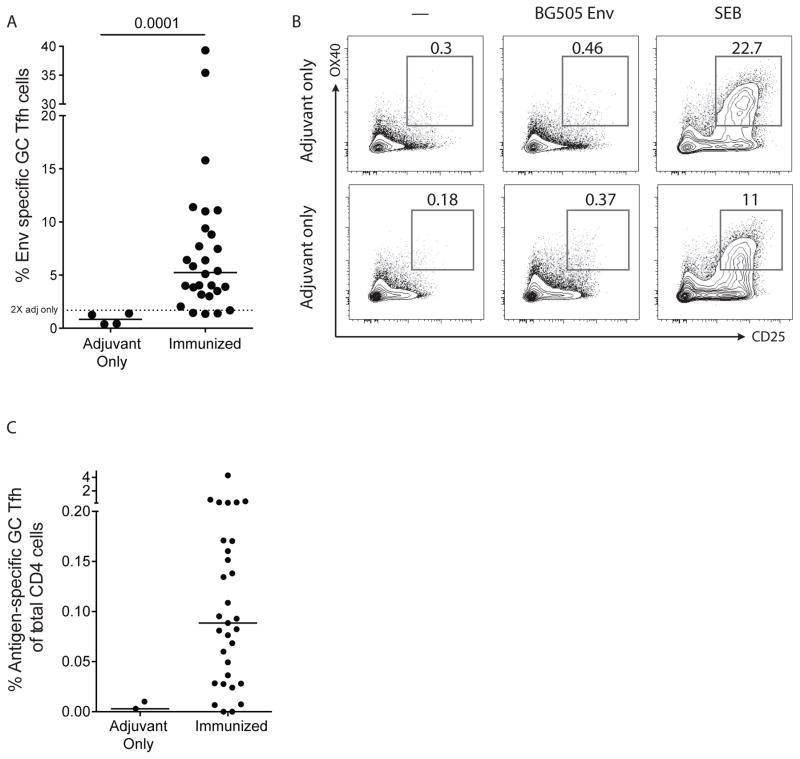
We next quantified the BG505 Env-specific GC Tfh cell response in 28 primary draining lymph nodes of macaques immunized with BG505 SOSIP Env. AIM+ GC Tfh cell responses were detected in 24 of 28 LNs, based on the criterion that positive responses are two-fold greater than the average response of ‘adjuvant only’ animals. The frequency of Env-specific cells ranged from 1% to 15% of GC Tfh cells (Figure 5A). To test the specificity of the AIM assay, we then tested samples from the draining lymph nodes of macaques immunized with adjuvant only (BG505 Env protein was not included in the immunization). As expected, few GC Tfh cells were generated in these animals (data not shown). In the adjuvant-only immunized animals, the few GC Tfh cells and the total LN CD4 T cell population showed virtually no AIM response to BG505 Env above the control ‘no exogenous antigen’ condition (Figure 5A and 5B). Therefore, the frequency of CD25+OX40+ GC Tfh cells among total CD4+ T cells was dramatically higher in immunized versus adjuvant only immunized animals (Figure 5C). In sum, AIM is both a highly sensitive and specific assay to detect antigen-specific GC Tfh cells in rhesus macaques.
Figure 5. Quantitation of immunogen-specific GC Tfh cells in a cohort of BG505 Env-immunized macaques.
A. Frequency of AIM+ (OX40+CD25+) BG505 Env-specific GC Tfh cells from the draining LNs of either adjuvant-only immunized or adjuvant + BG505 Env protein-immunized macaques. Dotted line indicates two times the average response from LN of ‘adjuvant only’ animals. B. Lack of the AIM responses for BG505 Env in total CD4+ cells in draining lymph nodes from adjuvant-only immunized rhesus macaque LNs. C. Frequency of AIM+ (OX40+CD25+) BG505 Env-specific GC Tfh cells from the draining LNs of either adjuvant-only immunized or adjuvant + BG505 Env protein-immunized macaques in panel A shown as a percent of total CD4 T cells. Potential background signals (“—” condition) were subtracted from BG505 Env responses.
Detection of non-Tfh CD4 T cells using the AIM assay
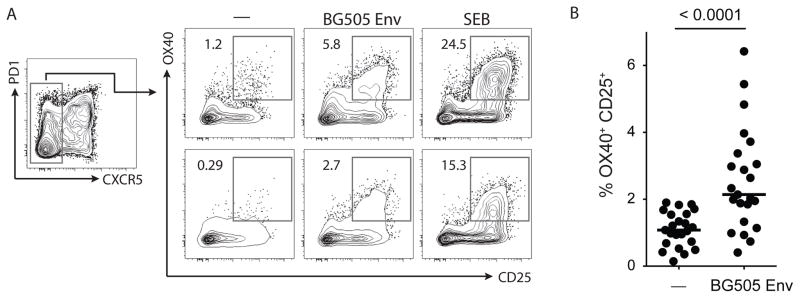
To expand the potential utility of the AIM assay, we investigated its ability to identify total non-Tfh (CXCR5−) CD4 T cells responses in draining lymph nodes after immunization, putatively including Th1, Th2, and Th17 cells. BG505 Env-specific CXCR5− CD4 T cells were detected in BG505 Env-immunized animals (Figure 6A and 6B). In some samples, the control ‘no exogenous antigen’ condition showed a population of AIM+ cells, which again was likely due to the presence of antigen in the LN from the immunization. Thus, detection of antigen-specific CD4 T cells among both CXCR5+ and CXCR5− CD4 T cells indicates that AIM can be used as a cytokine-independent method to identify NHP antigen-specific CD4 T cells irrespective of T helper differentiation state.
Figure 6. AIM assay detection of immunogen-specific non-Tfh cells.
A. BG505 Env-specific responses with the AIM assay among non-Tfh (CXCR5−) CD4 T cells from LN cells of immunized animals. B. Frequency of the AIM+ (OX40+CD25+) BG505 Env-specific non-Tfh CD4 T cells as in A. Potential background signals (“—” condition) may be true antigen-specific cells responding to antigen already present in the LN preparation from the immunization in vivo. Data are from 23 inguinal LN samples.
Detection of antigen-specific CXCR5+ CD4 T cells in peripheral blood
GC Tfh cells and GC B cells do not circulate in blood. However, there is a population of CXCR5+ CD4 T cells present in peripheral blood. These blood CXCR5+ CD4 T cells can be broadly divided into two groups: resting memory Tfh cells and activated CXCR5+ cells induced upon immune activation (38–41). While memory Tfh cells express low or intermediate levels of PD-1 and low levels of ICOS, the activated CXCR5+ cells are identifiable by high expression of both PD-1 and ICOS. This population of activated CXCR5+ cells is likely heterogeneous, consisting of recently activated Tfh cells coming from or transiting to the LN, and recently activated cells only transiently expressing CXCR5 (8, 39, 42).
We assessed whether antigen-specific CXCR5+ CD4 T cells in the blood could be detected in immunized macaques using AIM. No CXCR5+ CD4 T cell responses were detected to BG505 Env using baseline blood samples taken before immunization (Figure 7A). AIM identified antigen-specific CXCR5+ CD4 T cells in blood, one week after a booster immunization (Figure 7A and B). In a group of 11 immunized animals, antigen-specific responses were detectable in the majority of animals (81%), with a range of 0.2 to 4% of CXCR5+ CD4 T cells identified as Env-specific (Figure 7B). Total CD4+ T cells BG505 Env-specific responses were also quantified using the AIM assay (Figure S3C and D). The AIM assay outperformed ICS in a head-to-head comparison for detection of antigen specific CD4 T cells in PBMC (Figure 7C and 7D). Therefore, in addition to the detection of antigen-specific GC Tfh and non-GC Tfh CD4 T cell in lymphoid tissue, AIM can also be used to detect CXCR5+ and total CD4+ antigen-specific CD4 T cells in blood.
DISCUSSION
Tfh cells are recognized as critical contributors to immune responses generated in the settings of immunization, infectious disease, allergy, and autoimmunity. However, the means to specifically identify and quantify antigen-specific GC Tfh cells has been elusive. The non-human primate experimental animal model is an essential tool for understanding primate immunology, and HIV vaccine research depends heavily on the rhesus macaque model (10, 43). Previous NHP studies have generally had difficulty detecting antigen-specific GC Tfh cell responses after immunization or infection when using a standard ICS assay and quantifying antigen-specific cell by cytokine production. Here we show that antigen-specific GC Tfh cells are ‘stingy’ cytokine producing cells, likely focusing small amounts of cytokines and other help signals to individual GC B cells. Therefore, we have developed a cytokine-independent AIM assay to identify and quantify antigen-specific GC Tfh cells by upregulation of CD25 and OX40.
The AIM assay detects far more antigen specific GC Tfh cells than conventional ICS in macaques immunized with HIV Env. This indicates that after immunization, antigen-specific GC Tfh cells are far more abundant than previously shown and that this is likely due to meager cytokine production by GC Tfh cells after TCR stimulation. Even the AIM assay is likely to underestimate the true frequency of antigen-specific GC Tfh cells, considering the numerous negative regulators of TCR activation expressed by GC Tfh cells. PMA and Ionomycin stimulation, by-passing the TCR, induces large frequencies of cytokine production by GC Tfh cells. This suggests that cytokine production by GC Tfh cells is more restrained in response to TCR signaling than other T cell populations. To identify antigen-specific GC Tfh cells, a more sensitive assay was needed.
The AIM methodology can be used to monitor vaccine-specific responses from both GC Tfh cells and other CD4 T cell populations. Importantly, human tonsil sorting experiments showed that CXCR5 and PD-1 expression profiles were maintained on GC Tfh cells and mTfh cells (35). The AIM assay can also be used to identify antigen-specific CXCR5+ cells circulating in blood. Subsets of blood CXCR5+ CD4 T cells have been associated with autoimmune antibodies (41), anti-influenza Ab responses after vaccination (40), and the ability of rare HIV+ individuals to generate broadly neutralizing antibodies against HIV (39). Memory CXCR5+ Tfh cells found in circulating blood are resting cells. When assayed in vitro, these resting memory cells are much more capable of producing detectable quantities of cytokines than GC Tfh cells isolated from ongoing germinal centers. Indeed, cytokine production was observed to a greater extent by macaque memory CD4 T cells in blood, 6 weeks after the final vaccination. Nevertheless, the AIM assay still identified higher frequencies of antigen specific circulating memory Tfh cells than conventional ICS. Thus, we have confirmed that upregulation of CD25 and OX40 can be used to quantify antigen specific CD4 T cells in NHP blood (44), and that this method can be extended to also quantify blood CXCR5+ Tfh cells. Furthermore, the AIM assay can also be used to study human blood samples, as CD25+OX40+ antigen-specific CD4 T cells have been identified against numerous antigens in human blood (45). Pairing immunogen-specific detection of Tfh cells in the blood with quantitation of plasma CXCL13, a biomarker of lymphoid tissue germinal center activity (26), could potential greatly aid in the evaluation and understanding of vaccine induced antibody responses in macaques and humans.
Upregulation of CD25 and OX40 encompassed the broadest set of antigen-specific NHP GC Tfh cells in comparison to other markers, such as PD-L1, CD83, and NRP-1. Nevertheless, the diversity of upregulated surface molecules after stimulation suggests varying functional abilities of different subsets of GC Tfh cells. Using surface markers to identify antigen specific GC Tfh cells has the advantages of a live cell assay, in that these cells can be FACS sorted for further downstream analyses, such as RNAseq or TCR sequencing. Antigen-specific GC Tfh cell subsets expressing various surface markers may have distinct functions that can now be investigated. IL-2 signaling can disturb human Tfh cells (46) presumably through CD25 signaling. OX40-OX40L interactions have been associated with lupus in humans (47). PD-L1 expression on GC Tfh cells was unexpected, but it has been observed on activated CD4 T cells in blood from HIV+ individuals (48). In the GC, given the high concentration of GC Tfh cells in the GC light zone, PD-L1 expression by GC Tfh cells after TCR stimulation may contribute in trans to the highly-constrained proliferative phenotype of GC Tfh cells. Subsetting antigen-specific GC Tfh cells with these and other markers could yield important insights into potential GC Tfh cell diversity and how to best to direct the GC response during immunization regimen.
In summary, the AIM assay is an excellent method for detecting antigen-specific GC Tfh cells in non-human primates, which will provide a valuable means of monitoring the immune responses responsible for generating protective an antibody responses by vaccination.
Supplementary Material
Acknowledgments
We would like to thank Steven Bosinger, Greg K. Tharp, and Nirav Patel (Yerkes National Primate Center) for assistance with rhesus macaque RNAseq experiment, and Jason Greenbaum and LJI bioinformatics for additional RNAseq analysis. We thank Devin Sok and Dennis Burton (Scripps Research Institute) for assistance with cell sorting.
Footnotes
This work was supported by grants from NIAID, the Gates Foundation (CAVD), the European Research Council (ERC), and the Scripps CHAVI-ID. This work was funded in part by P51 RR000165/OD011132 to the Yerkes National Primate Research Center, and the Emory Center for AIDS Research, NIH Grant # P30-AI-504.
The authors declare no competing interests.
References
- 1.Plotkin SA. Vaccines: correlates of vaccine-induced immunity. CLIN INFECT DIS. 2008;47:401–409. doi: 10.1086/589862. [DOI] [PubMed] [Google Scholar]
- 2.Sanders RW, van Gils MJ, Derking R, Sok D, Ketas TJ, Burger JA, Ozorowski G, Cupo A, Simonich C, Goo L, Arendt H, Kim HJ, Lee JH, Pugach P, Williams M, Debnath G, Moldt B, van Breemen MJ, Isik G, Medina-Ramírez M, Back JW, Koff WC, Julien JP, Rakasz EG, Seaman MS, Guttman M, Lee KK, Klasse PJ, Labranche C, Schief WR, Wilson IA, Overbaugh J, Burton DR, Ward AB, Montefiori DC, Dean H, Moore JP. HIV-1 VACCINES. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science. 2015;349:aac4223–aac4223. doi: 10.1126/science.aac4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haynes BF, Moody MA, Alam M, Bonsignori M, Verkoczy L, Ferrari G, Gao F, Tomaras GD, Liao HX, Kelsoe G. Progress in HIV-1 vaccine development. J Allergy Clin Immunol. 2014;134:3–10. doi: 10.1016/j.jaci.2014.04.025. quiz 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burton DR, Mascola JR. Antibody responses to envelope glycoproteins in HIV-1 infection. Nat Immunol. 2015;16:571–576. doi: 10.1038/ni.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Victora GD, Nussenzweig MC. Germinal centers. Annu Rev Immunol. 2012;30:429–457. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- 6.Burton DR, Ahmed R, Barouch DH, Butera ST, Crotty S, Godzik A, Kaufmann DE, McElrath MJ, Nussenzweig MC, Pulendran B, Scanlan CN, Schief WR, Silvestri G, Streeck H, Walker BD, Walker LM, Ward AB, Wilson IA, Wyatt R. A Blueprint for HIV Vaccine Discovery. Cell Host and Microbe. 2012;12:396–407. doi: 10.1016/j.chom.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Victora GD, Wilson PC. Germinal center selection and the antibody response to influenza. Cell. 2015;163:545–548. doi: 10.1016/j.cell.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crotty S. T follicular helper cell differentiation, function, and roles in disease. Immunity. 2014;41:529–542. doi: 10.1016/j.immuni.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chahroudi A, Silvestri G. HIV and Tfh Cells: Circulating New Ideas to Identify and Protect. Immunity. 2016;44:16–18. doi: 10.1016/j.immuni.2015.12.016. [DOI] [PubMed] [Google Scholar]
- 10.Hessell AJ, Haigwood NL. Animal models in HIV-1 protection and therapy. Curr Opin HIV AIDS. 2015;10:170–176. doi: 10.1097/COH.0000000000000152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crotty S. A brief history of T cell help to B cells. Nature Reviews Immunology. 2015;15:185–189. doi: 10.1038/nri3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fukazawa Y, Park H, Cameron MJ, Lefebvre F, Lum R, Coombes N, Mahyari E, Hagen SI, Bae JY, MDR, Swanson T, Legasse AW, Sylwester A, Hansen SG, Smith AT, Stafova P, Shoemaker R, Li Y, Oswald K, Axthelm MK, McDermott A, Ferrari G, Montefiori DC, Edlefsen PT, Piatak M, Lifson JD, Sékaly RP, Picker LJ. Lymph node T cell responses predict the efficacy of live attenuated SIV vaccines. Nat Med. 2012;18:1673–1681. doi: 10.1038/nm.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chowdhury A, Del Rio PME, Tharp GK, Trible RP, Amara RR, Chahroudi A, Reyes-Teran G, Bosinger SE, Silvestri G. Decreased T Follicular Regulatory Cell/T Follicular Helper Cell (TFH) in Simian Immunodeficiency Virus-Infected Rhesus Macaques May Contribute to Accumulation of TFH in Chronic Infection. The Journal of Immunology. 2015;195:3237–3247. doi: 10.4049/jimmunol.1402701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petrovas C, Yamamoto T, Gerner MY, Boswell KL, Wloka K, Smith EC, Ambrozak DR, Sandler NG, Timmer KJ, Sun X, Pan L, Poholek A, Rao SS, Brenchley JM, Alam SM, Tomaras GD, Roederer M, Douek DC, Seder RA, Germain RN, Haddad EK, Koup RA. CD4 T follicular helper cell dynamics during SIV infection. J Clin Invest. 2012;122:3281–3294. doi: 10.1172/JCI63039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hong JJ, Amancha PK, Rogers KA, Courtney CL, Havenar-Daughton C, Crotty S, Ansari AA, Villinger F. Early lymphoid responses and germinal center formation correlate with lower viral load set points and better prognosis of simian immunodeficiency virus infection. The Journal of Immunology. 2014;193:797–806. doi: 10.4049/jimmunol.1400749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miles B, Miller SM, Folkvord JM, Kimball A, Chamanian M, Meditz AL, Arends T, McCarter MD, Levy DN, Rakasz EG, Skinner PJ, Connick E. Follicular regulatory T cells impair follicular T helper cells in HIV and SIV infection. Nature Communications. 2015;6:1–16. doi: 10.1038/ncomms9608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mylvaganam GH, Velu V, Hong JJ, Sadagopal S, Kwa S, Basu R, Lawson B, Villinger F, Amara RR. Diminished viral control during simian immunodeficiency virus infection is associated with aberrant PD-1hi CD4 T cell enrichment in the lymphoid follicles of the rectal mucosa. The Journal of Immunology. 2014;193:4527–4536. doi: 10.4049/jimmunol.1401222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindqvist M, van Lunzen J, Soghoian DZ, Kuhl BD, Ranasinghe S, Kranias G, Flanders MD, Cutler S, Yudanin N, Muller MI, Davis I, Farber D, Hartjen P, Haag F, Alter G, Schulze zur Wiesch J, Streeck H. Expansion of HIV-specific T follicular helper cells in chronic HIV infection. J Clin Invest. 2012;122:3271–3280. doi: 10.1172/JCI64314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamamoto T, Lynch RM, Gautam R, Matus-Nicodemos R, Schmidt SD, Boswell KL, Darko S, Wong P, Sheng Z, Petrovas C, McDermott AB, Seder RA, Keele BF, Shapiro L, Douek DC, Nishimura Y, Mascola JR, Martin MA, Koup RA. Quality and quantity of TFH cells are critical for broad antibody development in SHIVAD8 infection. Science Translational Medicine. 2015;7:298ra120–298ra120. doi: 10.1126/scitranslmed.aab3964. [DOI] [PubMed] [Google Scholar]
- 20.Vargas-Inchaustegui DA, Demers A, Shaw JM, Kang G, Ball D, Tuero I, Musich T, Mohanram V, Demberg T, Karpova TS, Li Q, Robert-Guroff M. Vaccine Induction of Lymph Node-Resident Simian Immunodeficiency Virus Env-Specific T Follicular Helper Cells in Rhesus Macaques. The Journal of Immunology. 2016 doi: 10.4049/jimmunol.1502137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kroenke MA, Eto D, Locci M, Cho M, Davidson T, Haddad EK, Crotty S. Bcl6 and Maf cooperate to instruct human follicular helper CD4 T cell differentiation. The Journal of Immunology. 2012;188:3734–3744. doi: 10.4049/jimmunol.1103246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma CS, Suryani S, Avery DT, Chan A, Nanan R, Santner-Nanan B, Deenick EK, Tangye SG. Early commitment of naive human CD4+; T cells to the T follicular helper (TFH) cell lineage is induced by IL-12. Immunology and Cell Biology. 2009;87:590–600. doi: 10.1038/icb.2009.64. [DOI] [PubMed] [Google Scholar]
- 23.Yu D, Rao S, Tsai LM, Lee SK, He Y, Sutcliffe EL, Srivastava M, Linterman M, Zheng L, Simpson N, Ellyard JI, Parish IA, Ma CS, Li QJJ, Parish CR, Mackay CR, Vinuesa CG. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 2009;31:457–468. doi: 10.1016/j.immuni.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 24.de Taeye SW, Ozorowski G, Torrents de la Peña A, Guttman M, Julien J-P, van den Kerkhof TLGM, Burger JA, Pritchard LK, Pugach P, Yasmeen A, Crampton J, Hu J, Bontjer I, Torres JL, Arendt H, DeStefano J, Koff WC, Schuitemaker H, Eggink D, Berkhout B, Dean H, Labranche C, Crotty S, Crispin M, Montefiori DC, Klasse PJ, Lee KK, Moore JP, Wilson IA, Ward AB, Sanders RW. Immunogenicity of Stabilized HIV-1 Envelope Trimers with Reduced Exposure of Non-neutralizing Epitopes. Cell. 2015;163:1702–1715. doi: 10.1016/j.cell.2015.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kasturi SP, Skountzou I, Albrecht RA, Koutsonanos D, Hua T, Nakaya HI, Ravindran R, Stewart S, Alam M, Kwissa M, Villinger F, Murthy N, Steel J, Jacob J, Hogan RJ, García-Sastre A, Compans R, Pulendran B. Programming the magnitude and persistence of antibody responses with innate immunity. Nature. 2011;470:543–547. doi: 10.1038/nature09737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Havenar-Daughton C. CXCL13 is a Plasma Biomarker of Germinal Center Activity. Proceedings of the National Academy of Sciences. 2016 doi: 10.1073/pnas.1520112113. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shulman Z, Gitlin AD, Weinstein JS, Lainez B, Esplugues E, Flavell RA, Craft JE, Nussenzweig MC. Dynamic signaling by T follicular helper cells during germinal center B cell selection. Science. 2014;345:1058–1062. doi: 10.1126/science.1257861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu D, Xu H, Shih C, Wan Z, Ma X, Ma W, Luo D, Qi H. T-B-cell entanglement and ICOSL-driven feed-forward regulation of germinal centre reaction. Nature. 2015;517:214–218. doi: 10.1038/nature13803. [DOI] [PubMed] [Google Scholar]
- 29.Crotty S. Follicular helper CD4 T cells (TFH) Annu Rev Immunol. 2011;29:621–663. doi: 10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- 30.Johnston RJ, Choi YS, Diamond JA, Yang JA, Crotty S. STAT5 is a potent negative regulator of TFH cell differentiation. Journal of Experimental Medicine. 2012;209:243–250. doi: 10.1084/jem.20111174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choi YS, Kageyama R, Eto D, Escobar TC, Johnston RJ, Monticelli L, Lao C, Crotty S. ICOS Receptor Instructs T Follicular Helper Cell versus Effector Cell Differentiation via Induction of the Transcriptional Repressor Bcl6. Immunity. 2011;34:932–946. doi: 10.1016/j.immuni.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pepper M, Pagán AJ, Igyártó BZ, Taylor JJ, Jenkins MK. Opposing Signals from the Bcl6 Transcription Factor and the Interleukin-2 Receptor Generate T Helper 1 Central and Effector Memory Cells. Immunity. 2011:1–13. doi: 10.1016/j.immuni.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ballesteros-Tato AE, León B, Graf BA, Moquin A, Adams PS, Lund FE, Randall TD. Interleukin-2 inhibits germinal center formation by limiting T follicular helper cell differentiation. Immunity. 2012;36:847–856. doi: 10.1016/j.immuni.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Renand A, Milpied P, Rossignol J, Bruneau J, Lemonnier F, Dussiot M, Coulon S, Hermine O. Neuropilin-1 Expression Characterizes T Follicular Helper (Tfh) Cells Activated during B Cell Differentiation in Human Secondary Lymphoid Organs. PLoS ONE. 2013;8:e85589. doi: 10.1371/journal.pone.0085589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dan J, Crotty S. A cytokine-independent approach to identify antigen- specific human germinal center Tfh cells and rare antigen- specific CD4. 2016:1–38. doi: 10.4049/jimmunol.1600318. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soroosh P, Ine S, Sugamura K, Ishii N. OX40-OX40 ligand interaction through T cell-T cell contact contributes to CD4 T cell longevity. J Immunol. 2006;176:5975–5987. doi: 10.4049/jimmunol.176.10.5975. [DOI] [PubMed] [Google Scholar]
- 37.Croft M, So T, Duan W, Soroosh P. The significance of OX40 and OX40L to T-cell biology and immune disease. Immunol Rev. 2009;229:173–191. doi: 10.1111/j.1600-065X.2009.00766.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmitt N, Ueno H. Blood Tfh cells come with colors. Immunity. 2013;39:629–630. doi: 10.1016/j.immuni.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Locci M, Havenar-Daughton C, Landais E, Wu J, Kroenke MA, Arlehamn CL, Su LF, Cubas R, Davis MM, Sette A, Haddad EK, Poignard P, Crotty S International AIDS Vaccine Initiative Protocol C Principal Investigators. Human circulating PD-1+CXCR3-CXCR5+ memory Tfh cells are highly functional and correlate with broadly neutralizing HIV antibody responses. Immunity. 2013;39:758–769. doi: 10.1016/j.immuni.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bentebibel SE, Lopez S, Obermoser G, Schmitt N, Mueller C, Harrod C, Flano E, Mejias A, Albrecht RA, Blankenship D, Xu H, Pascual V, Banchereau J, Garcia-Sastre A, Palucka AK, Ramilo O, Ueno H. Induction of ICOS+CXCR3+CXCR5+ TH Cells Correlates with Antibody Responses to Influenza Vaccination. Science Translational Medicine. 2013;5:176ra32–176ra32. doi: 10.1126/scitranslmed.3005191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morita R, Schmitt N, Bentebibel S-E, Ranganathan R, Bourdery L, Zurawski G, Foucat E, Dullaers M, Oh S, Sabzghabaei N, Lavecchio EM, Punaro M, Pascual V, Banchereau J, Ueno H. Human Blood CXCR5+CD4+ T Cells Are Counterparts of T Follicular Cells and Contain Specific Subsets that Differentially Support Antibody Secretion. Immunity. 2011:1–14. doi: 10.1016/j.immuni.2010.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He J, Tsai LM, Leong YA, Hu X, Ma CS, Chevalier N, Sun X, Vandenberg K, Rockman S, Ding Y, Zhu L, Wei W, Wang C, Karnowski A, Belz GT, Ghali JR, Cook MC, Riminton DS, Veillette A, Schwartzberg PL, Mackay F, Brink R, Tangye SG, Vinuesa CG, Mackay CR, Li Z, Yu D. Circulating Precursor CCR7loPD-1hi CXCR5+ CD4+ T Cells Indicate Tfh Cell Activity and Promote Antibody Responses upon Antigen Reexposure. Immunity. 2013;39:770–781. doi: 10.1016/j.immuni.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 43.Shedlock DJ, Silvestri G, Weiner DB. Monkeying around with HIV vaccines: using rhesus macaques to define “gatekeepers” for clinical trials. Nature Reviews Immunology. 2009;9:717–728. doi: 10.1038/nri2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Rose R, Mason RD, Loh L, Peut V, Smith MZ, Fernandez CS, Alcantara S, Amarasena T, Reece J, Seddiki N, Kelleher AD, Zaunders J, Kent SJ. Safety, immunogenicity and efficacy of peptide-pulsed cellular immunotherapy in macaques. J Med Primatol. 2008;37(Suppl 2):69–78. doi: 10.1111/j.1600-0684.2008.00329.x. [DOI] [PubMed] [Google Scholar]
- 45.Zaunders JJ, Munier ML, Seddiki N, Pett S, Ip S, Bailey M, Xu Y, Brown K, Dyer WB, Kim M, De Rose R, Kent SJ, Jiang L, Breit SN, Emery S, Cunningham AL, Cooper DA, Kelleher AD. High levels of human antigen-specific CD4+ T cells in peripheral blood revealed by stimulated coexpression of CD25 and CD134 (OX40) The Journal of Immunology. 2009;183:2827–2836. doi: 10.4049/jimmunol.0803548. [DOI] [PubMed] [Google Scholar]
- 46.Cubas R, van Grevenynghe J, Wills S, Kardava L, Santich BH, Buckner CM, Muir R, Tardif V, Nichols C, Procopio F, He Z, Metcalf T, Ghneim K, Locci M, Ancuta P, Routy JP, Trautmann L, Li Y, McDermott AB, Koup RA, Petrovas C, Migueles SA, Connors M, Tomaras GD, Moir S, Crotty S, Haddad EK. Reversible Reprogramming of Circulating Memory T Follicular Helper Cell Function during Chronic HIV Infection. The Journal of Immunology. 2015;195:5625–5636. doi: 10.4049/jimmunol.1501524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jacquemin C, Schmitt N, Contin-Bordes C, Liu Y, Narayanan P, Seneschal J, Maurouard T, Dougall D, Davizon ES, Dumortier H, Douchet I, Raffray L, Richez C, Lazaro E, Duffau P, Truchetet ME, Khoryati L, Mercié P, Couzi L, Merville P, Schaeverbeke T, Viallard JF, Pellegrin JL, Moreau JF, Muller S, Zurawski S, Coffman RL, Pascual V, Ueno H, Blanco P. OX40 Ligand Contributes to Human Lupus Pathogenesis by Promoting T Follicular Helper Response. Immunity. 2015;42:1159–1170. doi: 10.1016/j.immuni.2015.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rosignoli G, Lim CH, Bower M, Gotch F, Imami N. Programmed death (PD)-1 molecule and its ligand PD-L1 distribution among memory CD4 and CD8 T cell subsets in human immunodeficiency virus-1-infected individuals. Clin Exp Immunol. 2009;157:90–97. doi: 10.1111/j.1365-2249.2009.03960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.