Abstract
The renal cell carcinomas, clear cell, papillary, and chromophobe, have recently undergone an unmatched genomic characterization by The Cancer Genome Atlas (TCGA). This analysis has revealed new insights into each of these malignancies, and underscores the unique biology of clear cell, papillary, and chromophobe renal cell carcinoma. Themes that have emerged include distinct mechanisms of metabolic dysregulation and common mutations in chromatin modifier genes. Importantly, the papillary renal cell carcinoma classification encompasses a heterogeneous group of diseases, each with highly distinct genetic and molecular features. In conclusion, this review summarizes renal cell carcinomas which represent a diverse set of malignancies, each with novel biological programs that define new paradigms for cancer biology.
Keywords: Renal cell carcinoma, Clear cell renal cell carcinoma, Papillary renal cell carcinoma, Chromophobe renal cell carcinoma, TCGA
INTRODUCTION
Cancers of the kidney have long fascinated physicians with their highly divergent phenotypes and patterns of behavior (1). Kidney tumors are conventionally grouped based upon their anatomical origin and cell type into four general categories: renal cell carcinomas (RCC), which arise from the renal cortex epithelial cell compartment; collecting duct carcinomas and renal medullary carcinomas, both of which arise from the renal medulla; and papillary urothelial carcinomas, which arise from the transitional epithelium lining the renal pelvis and ureter (2). These classifications roughly follow the anatomical groupings of the nephron and the route of passage toward the urinary bladder, with papillary urothelial carcinomas recognized as sharing more pathological and histological characteristics with transitional cell carcinoma of the urinary bladder and ureter than other types of kidney cancer (3).
The renal cell carcinoma classification is yet further broken down into three main pathological subtypes defined by their histological and morphological characteristics: clear cell renal cell carcinoma (ccRCC), papillary renal cell carcinoma (pRCC), and chromophobe renal cell carcinoma (chRCC). Of these, ccRCC is by far the most common, comprising roughly 70% of all renal cortical tumors. Next most common is pRCC, making up about 10–15% of RCC cases, followed by chRCC, which is a rare carcinoma that accounts for only about 5% of RCC (4). Marked differences in risk for development of metastatic disease and tumor aggressiveness are exhibited between all of these different classes of RCC tumors.
Although rare, there are yet additional cases of RCC that do not fit into these broad categories. These include renal medullary carcinoma and translocation carcinoma. It is anticipated that a revised World Health Organization pathological classification recognizing even more rare and largely low-risk entities such as clear cell tubulopapillary RCC and multilocular cystic RCC will be published in 2016 (5, 6). There are also a handful of related benign tumors that occur in the kidney, including angiomyolipoma, which shares some common features with ccRCC, and renal oncocytoma, which is genetically and histologically similar to chRCC, although with key differences (7).
One major challenge that has stymied the field has been classifying these tumors for therapeutic considerations. Modern clinical trials now dictate the histology of cases that are included, but until even a few years ago it was commonplace for all RCC histologies to be lumped together in clinical studies. The result is that today, the FDA approved treatments applied to the renal cell carcinomas are identical for what we now know are highly disparate tumor types. However, thanks to the highly forward thinking of The Cancer Genome Atlas (TCGA) project, the three major renal cell carcinoma subtypes have each been examined independently in TCGA genomic profiling efforts. Had all RCCs been considered as a single group, the numbers of tumors for the important but rarer entities of chRCC and the type II pRCC (pRCC-II) would have been insufficient to draw new conclusions about these clinically distinct and meaningful diseases. Thus, in this review, we compare and discuss the unique findings observed by the TCGA for tumors from each of the three primary RCC classifications, as a more complete understanding of the biology and signaling of these different types of RCC is a major step forward in determining the best way to treat each of these distinct tumor types.
1. CLEAR CELL RENAL CELL CARCINOMA (ccRCC or KIRC)
1.1 Background
The most commonly encountered malignancy in the cortex of the kidney is ccRCC (also referred to as KIRC in the TCGA studies), making up roughly 70% of renal cell carcinomas. This tumor type is well known to overlap significantly with the ccRCC occurring in the setting of von Hippel-Lindau (VHL) disease, and indeed, mutation of the VHL gene which causes VHL disease is observed in up to 90% of sporadic cases of this cancer (8). These tumors are characterized by a histology pattern of cleared cytoplasm with an acentric nucleus, and cells that are organized into small, tight vascular bundles. While patients with ccRCC usually present with non-metastatic disease, ~25% of patients have metastases at initial presentation, and another 30% of individuals with ccRCC will eventually develop distant metastases.
Historically, renal cell carcinomas, and ccRCC specifically, display inherent resistance to conventional cytotoxic chemotherapies. As a result, despite the lack of actionable tumor cell intrinsic targets, the field has sought to take advantage of the unique biology imparted on this cancer as a result of VHL mutation: namely, VEGF-promoted angiogenesis (9, 10). Anti-angiogenic agents targeting VEGF or the endothelial cell VEGF receptor (VEGFR) have been approved for the treatment of ccRCC (11–14) and, due to the dominance of this histologic subtype, these agents have been given broad approval for use against all of the renal cell carcinomas. When used to treat ccRCC, although rarely curative, these agents have been shown to significantly reduce metastatic growth and enhance progression free survival (11–13) and overall survival as determined by implicit meta-analysis (15). The benefit of these drugs as monotherapy in this setting, in contrast to many other tumors, is clearly tied to the central biological feature of enhanced vascularity (16) driven by the deregulation of hypoxia inducible factors (HIFs) that is inherent in VHL-mutated ccRCC.
1.2 TCGA Analysis of ccRCC
Even though ccRCC is the most studied of the renal cell carcinomas, the large set of over 400 ccRCC tumors examined by TCGA provided additional insight into the genetic and molecular makeup of this disease (17). As with each of the types of renal cell carcinomas detailed below, the TCGA applied a multiplatform analysis consisting of copy number analysis, whole exome sequencing, RNA sequencing (RNAseq), miRNA sequencing (miRNAseq), methylation analysis, and proteomic analysis (RPPA). In addition, selected cases also underwent whole genome sequencing (WGS). All samples were annotated with demographic, outcome, and treatment information.
1.2.1 ccRCC genomics and emerging biomarkers
The copy number analysis of ccRCC samples revealed a strong signature of 3p deletion and 5q gains, with other modifications largely scattered across the genome. Some alterations of interest included frequent deletions of CDKN2A (p16/INK4A tumor suppressors) and RB loci, as well as MYC amplification. In general, the number of focal and arm level alterations was much smaller on a per-tumor basis than that commonly observed in other types of cancer (Fig. 1). WES revealed common VHL mutations, as expected, which when coupled with promoter hypermethylation, indicated VHL inactivation occurred in over 60% of cases. The next most commonly mutated genes were all chromatin modifiers: PBRM1 (BAF180), SETD2, KDM5C (JARID1C), and BAP1. These proteins all play diverse roles in chromatin maintenance, ranging from Swi/Snf nucleosome repositioning (PBRM1) to histone modification such as deubiquitination (BAP1), methylation (SETD2) and demethylation (KDM5C). Intriguingly, three of these genes (PBRM1, SETD2, and BAP1) reside in the commonly deleted 3p region, which includes the VHL gene. 3p loss occurs in > 95% of ccRCC, and this clustering of genes may account for some of the unusual aspects of this tumor type, and is explored in greater detail in a recent review (18). Subsequent studies have suggested that mutations in BAP1 may identify a subgroup of tumors with a higher malignant potential (19, 20). Another pathway recurrently mutated in the clear cell RCC tumors were genes in the PI3K/AKT/mTOR signaling pathway. Alterations in one or more components of this pathway were present in approximately 28% of TCGA ccRCC samples. Recently, study of aggressive sarcomatoid renal cell carcinomas found an association between mTOR pathway activation and increased proliferation, and tumors with mutations in this pathway are, in some instances, more sensitive to mTOR inhibitors (21).
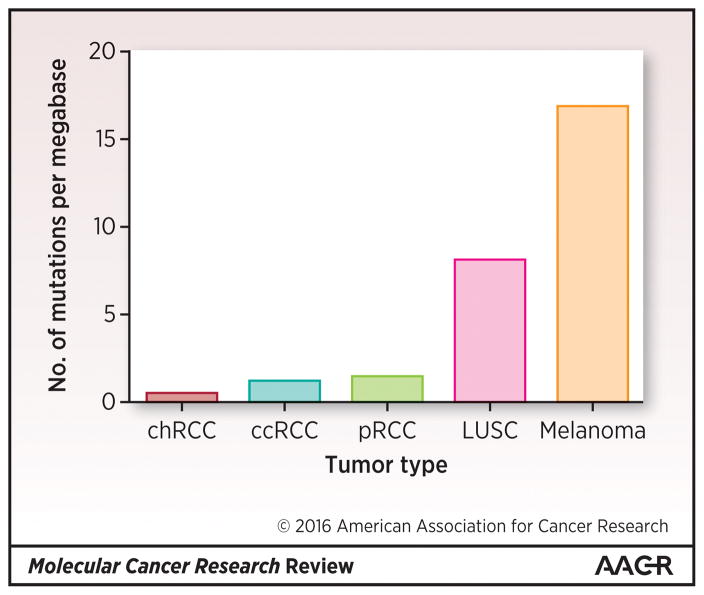
Figure 1. Mutational Burden Across Renal Cell Carcinoma Spectrum in Comparison with Other Solid Tumors.
The renal cell carcinoma TCGA projects identified lower mutational burdens relative to TCGA projects in classic, carcinogen-associated solid tumors such as squamous lung cancer and melanoma.
While sequencing analysis of ccRCC did not reveal distinct genetic subgroupings, gene expression and microRNA analyses did identify four unique subgroups (m1–m4 and mi1-mi4, respectively), including predicted mRNA targets for the miRNA subgroups. With the emergence of mRNA-based prognostic and predictive biomarker panels in breast and colon cancer (22, 23) as well as lymphoma (24), expression-based subgroups are becoming more widely accepted for diagnostic use. It is now clear that gene expression biomarkers also may be useful for the classification of ccRCC. Interestingly, the gene expression clusters identified by the TCGA overlap clearly with the previously reported subtypes of ccRCC known as ccA (corresponding to m1) and ccB (corresponding to m2 and m3) (25). Likewise, a distinct overall survival advantage was associated with the m1 subtype, as was previously observed for the corresponding ccA tumors (25). Thus, continued study of these subgroups as potential biomarkers remains an area of active investigation.
1.2.2 Metabolic features contribute to the overall outcome of ccRCC
Dysregulation of metabolic pathways is a common feature of ccRCC, owing to the upregulation of numerous key enzymes of glycolysis downstream of HIF transcriptional activation (26, 27). Examination of the HIF family members has demonstrated that changes in the glycolytic genes are largely driven by a HIF-1α specific transcription (28), which has both direct and indirect influences on metabolism (29). Similar changes in the expression of metabolic pathway genes have also been observed in a HIF1α-mutant mouse model of renal cell carcinoma (30). The interactions between HIF and metabolism are complex and bidirectional, with recent studies suggesting that expression of one glycolytic enzyme, fructose-1,6-bisphosphatase, has tumor suppressive activity via direct repression of HIF functionality, while loss of this enzyme is associated with the progression of disease (31).
Surprisingly, through the integrated analysis of hundreds of tumors in the TCGA study, it was revealed that the upregulation of genes involved in glycolysis and fatty acid synthesis were associated with a significantly worse survival outcome in ccRCC patients, whereas genes involved in Krebs cycle or AMPK signaling were associated with an overall better survival outcome. These findings suggest that there are either inherent metabolic differences that may influence the progression of ccRCC tumors, or that a metabolic switch occurs favoring HIF targeted glycolytic gene expression and higher rates of glycolysis during tumor progression, the latter of which has been suggested by studies showing fructose bisphosphatase 1 downregulation, and commensurate increase in glucose uptake in tumor samples displaying loss of this enzyme and patterns of poor risk gene expression (31, 32). Although the mechanisms by which these metabolic differences contribute to tumor progression have not yet been elucidated, these findings may help to explain some of the heterogeneity that is observed between ccRCC cases.
1.2.3 Linking chromatin modifier gene mutations to the progression of ccRCC
As indicated above, after VHL, the most commonly mutated genes in ccRCC, PBRM1, SETD2, KDM5C, and BAP1, are all involved in regulation of chromatin. Overall, methylation patterns in ccRCC tumors in the TCGA favored hypermethylation when compared with normal kidney. In addition, the extent of hypermethylated promoter sites increased with both stage and grade, suggesting that hypermethylation was a feature associated with progression. In contrast, the SETD2 mutant tumors displayed a unique signature pattern of global DNA hypomethylation at non-promoter regions that distinguished them from other tumors. The implications of this remain uncertain, although recent studies have demonstrated that SETD2 mutations alter chromatin accessibility (33) and that expanded methylation coordinated with changes in histone methylation across the genome (34). As mutations in these chromatin-regulators were associated with altered expression patterns in a large number of other genes, chromatin remodeling appears to play a key role in the progression of clear cell RCC.
1.3 Summary of TCGA analysis of ccRCC
Analysis of over 400 clear cell RCC tumors by the TCGA revealed that the primary genetic changes contributing to this tumor type include those underlying cellular oxygen sensing, including VHL and its signaling pathways, and those involved in the maintenance of chromatin states, specifically PBRM1, SETD2, KDM5C, and BAP1. Importantly, a metabolic shift in ccRCC appears to be associated with disease progression, as more aggressive tumors demonstrated up-regulation of genes involved in glycolysis and fatty acid synthesis, and down regulation of genes involved in Krebs (TCA) cycle and AMPK pathway signaling. Likewise, overall changes in promoter hypermethylation were correlated with higher grade tumors. These findings, combined with the clustering of tumors into distinct subsets using gene expression data, have provided a foundation for future identification of subtype- and pathway-specific diagnostics and treatments.
2. PAPILLARY RENAL CELL CARCINOMA (pRCC or KIRP)
2.1. Background
pRCC is the second most common histologic subtype of renal cell carcinoma, representing 10–15% of cases (4). Based on histology, pRCC can be further divided approximately 1:1 into type I (pRCC-I) and type II (pRCC-II) tumors (35, 36). pRCC-I tumors feature small, basophilic cells forming distinct papillae whereas pRCC-II tumors exhibit large, eosinophilic cells with pseudostratification (35). The pRCC-II tumors are larger, more likely to metastasize, and have an inferior prognosis (37).
The clinical care of pRCC remains largely uninformed by the tumor’s biology, especially among metastatic patients. These patients are typically treated with VEGFR-directed therapies developed predominantly for ccRCC, a biologically and genetically divergent disease. Outcomes for pRCC patients treated with these medications are, predictably, inferior relative to ccRCC (38). If clinical care is to be advanced for these patients, a more complete understanding of the molecular biology of these tumors is needed.
2.2. TCGA Analysis of pRCC
Prior to the TCGA report on pRCC, no single large study had systematically examined the sporadic form of this disease. The TCGA examined 161 papillary RCC tumors, which after expert pathology review were classified as pRCC-I (75), pRCC-II (60), and pRCC not otherwise specified (NOS) (26). A number of significantly mutated genes were identified in these tumors. Of particular note were alterations in several genes previously known to be commonly mutated in other cancers. These included NF1, involved in the Hippo signaling pathway, and SMARCB1, PBRM1, SETD2, KDM6A, and BAP1, all involved in chromatin modification pathways. TFE3 and TFEB gene fusions were also found to occur frequently in pRCC, resulting in higher levels of expression of their transcriptional targets.
Similar to prior gene expression studies (39), the multiple molecular platforms used in the TCGA, including those evaluating somatic copy number (SCNA), miRNA and mRNA expression, were able to confirm the divergent biology of pRCC-I and pRCC-II tumors. In addition, correlation with clinical data demonstrated a higher stage and poorer survival for the pRCC-II patients, as observed in previous studies (37). Moreover, strategies integrating these molecular platforms (40, 41) not only reinforced the pRCC-I vs. pRCC-II classification schema, but further resolved pRCC-II into three distinct subtypes: pRCC-IIa, pRCC-IIb, and CIMP (CpG Island Methylator Phenotype). Thus, pRCC represents at least four molecularly distinct subtypes (Table 1).
Table 1. Clinical and Molecular Distinctions among the Papillary RCC Molecular Subtypes.
161 papillary RCC tumor samples were collected with clinical correlative data, subtyped as pRCC-I or -II during central pathology review, and molecularly profiled. MET activation corresponds to a diverse set of molecular perturbations including trisomy 7, promoter and exon mutations, RNA overexpression, and others. NRF2/ARE activation includes NQO1 overexpression. CDKN2A inactivation includes focal deletion, promoter methylation, and mutations. SETD2 was significantly mutated in type 2 papillary tumors and enriched within the pRCC-IIb subtype. SWI/SNF pathway activation as determined by HOTNET analysis was significant across all tumors but enriched within the pRCC-IIb subtype. The CIMP (CpG island methylator phenotype) subtype was unique in terms of poor survival, young age of onset, extensive DNA hypermethylation, and fumarate hydratase mutations as well as evidence of hypoxia and Warburg-like metabolism.
| pRCC-I | pRCC-IIa | pRCC-IIb | pRCC-IIc | |
|---|---|---|---|---|
| Survival | Good | Good | Intermediate | Poor |
| MET activation | ++++ | − | − | − |
| NRF2/ARE activation | − | + | + | +++ |
| CDKN2A/RB alterations | −/+ | ++ | ++ | +++ |
| SETD2 mutation | − | − | ++ | − |
| SWI/SNF alteration | + | + | ++ | + |
| DNA hypermethylation | − | − | + | ++++ |
| FH mutation | − | − | − | ++++ |
2.2.1. Papillary RCC Type I
Tumors classified as pRCC-I demonstrated a lower clinical stage and better survival relative to pRCC-II tumors. Consistent with prior observations (42, 43), pRCC-I tumors were enriched for MET mutations: of the 14 observed somatic MET mutations, only one was in a pRCC-II tumor. In addition to somatic tumor mutations, several of the samples contained a previously described germline MET mutation reported in hereditary papillary renal cell carcinoma (HPRCC) (44)). Other molecular mechanisms of MET activation were also observed in the pRCC-I patients. For example, about 10% of pRCC-I patients contained a novel MET RNA transcript lacking exons 1 and 2. As these exons encode the HGF ligand binding domain of the MET-encoded HGF receptor, the transcribed protein is hypothesized to exhibit ligand-independent activation. Furthermore, four tumors that underwent whole genome sequencing were observed to have MET promoter mutations that were predicted to be functional (45). In addition to MET mutations, pRCC-I tumors were frequently observed to have chromosomal gains of the MET-encoding chromosome 7. Thus, trisomy 7 may also contribute to the overall increase in MET mRNA expression and HGF receptor activation observed in pRCC-I relative to pRCC-II tumors. When considering these various molecular events, the role of MET as a driver in pRCC-I tumors was reinforced.
2.2.2. Papillary RCC Type II
As described previously, pRCC type I and II tumors are distinct in terms of histology, clinical stage, and patient survival. As such, it is not surprising that several molecular features emerged that distinguished pRCC-II tumors from pRCC-I. For example, alterations in CDKN2A, which encodes p16 (INK4A), were more common amongst pRCC-II samples, resulting in proliferation-associated increases in expression of phosphorylated Rb and cell cycle genes. In addition, as seen for ccRCC, mutations in the chromosome modifier genes SETD2, BAP1, and PBRM1 were also more commonly observed in pRCC-II tumors.
When examining differential gene expression between pRCC-I and pRCC-II tumors, one of the most divergent gene groups corresponded to the NRF2/antioxidant response element (ARE) pathway. This observation is consistent with reports that have documented mutations in NRF2/ARE pathway genes in pRCC-II tumors (46). NFE2L2, the canonical NRF2 gene, is a transcription factor that, when stable, triggers a cellular response that is protective against oxidative and electrophilic stresses. While initially thought to act as a tumor suppressor by inhibiting carcinogenesis, recent reports implicate this pathway as oncogenic. Indeed, inactivating mutations in genes involved in NFE2L2 degradation, such as KEAP1 and CUL3, as well NFE2L2 mutations that render it resistant to degradation, have been observed in cancers and are associated with poor outcomes (47–49). Thus, the NRF2/ARE pathway has been suggested to be oncogenic, conferring protection against the stress incurred by rapid proliferation and chemotherapy. Expression of NQO1, one of the chief transcriptional targets of NFE2L2, was observed to be significantly higher in pRCC-II tumors in the TCGA study, under scoring the fact that the NRF2/ARE pathway seems to play a particularly important role in these tumors.
As previously mentioned, TCGA analysis of the pRCC tumors further subdivided the pRCC-II tumors into three additional subtypes: pRCC-IIa, pRCC-IIb, and CIMP. These subgroups were distinguished from one another based upon combined differences in histology, genetic alterations, gene expression and methylation patterns, and clinical outcomes.
2.2.2.1 Papillary RCC type IIa and IIb
Among the pRCC-II tumors, patients with pRCC-IIa tumors trend towards the best survival while those with pRCC-IIb tumors having intermediate survival. Similarly, patients with pRCC-IIa tumors tend to have more clinical stage I and II disease, while the pRCC-IIb patients had more clinical stage III and IV disease. In addition, the pRCC-IIb tumors harbored the majority of the SETD2 mutations identified among all the pRCC samples.
2.2.2.2 Papillary RCC type II-CIMP
The pRCC-II CIMP subtype is of particular interest as it is associated with the extremes in terms of age of onset, prognosis, and several oncogenic pathways. The CIMP subgroup in the TCGA analysis was relatively small (9 tumors or 6% of all pRCC), and was identified primarily from the DNA methylation data as having significant, genome-wide hypermethylation. The CIMP patients were the youngest of the pRCC-II subtypes with a median age of 42 years and had the worst overall survival. The previously described NRF2/ARE pathway was highly dysregulated in these tumors, as was the CDKN2A/RB tumor suppressor pathway. In addition, the CIMP tumors demonstrated marked metabolic dysregulation highlighted by the presence of either somatic or germline mutations in the gene fumarate hydratase (FH) in 6 of the 9 CIMP tumors and low FH expression in all CIMP tumors. Overall, the CIMP tumors demonstrated a metabolic shift toward increased glycolysis, with the upregulation of hypoxia-related and glycolytic pathway genes, accompanied by lower expression of Krebs (TCA) cycle and AMPK complex genes.
2.3. Summary of TCGA analysis of papillary RCC
The powerful and integrated analysis of papillary RCC performed by the TCGA provided important insights into this disease’s genetic basis. This study not only validated previous disease classifications and their genetic drivers, but also identified new differences between tumor types that may prove valuable in treating this disease.
Known biologic distinctions, such as pRCC-I versus pRCC-II, were validated and described in greater molecular detail than was previously possible. MET was validated as a likely oncogenic driver in many pRCC-I tumors, but other oncogenic pathways were also identified as playing an important role in pRCC-I tumorigenesis. For example, while the NRF2/ARE pathway is activated more frequently in pRCC-II, a small minority of pRCC-I tumors also demonstrated high levels of NRF2/ARE target gene expression. The more aggressive pRCC-II tumors are a heterogeneous group that were further resolved through the multiplatform TCGA analysis into pRCC-IIa, pRCC-IIb, and CIMP. Collectively, these tumors exhibit activated NRF2/ARE as well as inactivation of the tumor suppressor CDKN2A. The CIMP tumors were identified as a group with especially poor prognosis and early onset, characterized by genome-wide hypermethylation, FH mutations or low expression, and a shift to a Warburg-like metabolism.
With the knowledge gained, investigators are now presented with the challenge of designing logical strategies to therapeutically target the diverse biology and signaling patterns exhibited by these tumor subtypes, and to select for patients most likely to benefit from these therapies.
3. CHROMOPHOBE RENAL CELL CARCINOMA (chRCC or KICH)
3.1 Background
chRCC is traditionally an indolent disease, with tumors characterized by cells with mildly granular to pale and finely reticular cytoplasm with central clearing and irregular nuclear borders. The disease shares many histologic features of the benign condition oncocytoma, but unlike oncocytoma, does carry risk for metastasis, and can transform with sarcomatoid features, rendering it highly aggressive and lethal. Only about 5% of RCC cases are classified as chRCC, making it the kidney cancer subgroup occurring at the lowest frequency amongst those included in TCGA studies. Interestingly, however, this normally rare cancer is more common than pRCC among young women with non-clear cell histology tumors. This raises questions about how gender differences, such as hormonal factors or specific pathway dysregulation, could be involved in the development of chRCC (50).
The inclusion of chRCC in the TCGA marked the first commitment to mapping the integrated genome of rare tumor types (51). Although the number of deaths annually attributed to chRCC is undoubtedly low, the argument for interrogating this rare disease was two-fold: 1) to reveal important features of chRCC, so that these patients could benefit from appropriate treatment advances, and 2) to allow a rare and unusual, but homogeneous, set of tumors to inform new aspects of tumor biology which may be relevant for other more common diseases.
3.2 Mutational burden and profiles
One of the most characteristic features of chRCC is monosomy of many chromosomes. Originally described in 1992 (52, 53), chromophobe tumors show almost complete uniformity in the wholesale loss of chromosomes 1, 2, 6, 10, 13, 17 and often 21. This signature was confirmed by the TCGA analysis, which also demonstrated that other minor copy number changes are generally absent in this tumor type. Such a fingerprint of whole chromosome loss, in the absence of other evidence of genomic instability, is unprecedented. Unfortunately, the genomic error or process that allows for this pattern of chromosomal mis-segregation to occur remains unknown.
3.2.1 The TP53 axis and PTEN pathway
Tumor suppressor gene alterations are prominent in chRCC. Unlike either ccRCC or pRCC tumors, the most commonly mutated gene in chRCC is TP53. As anticipated, TP53 mutations are largely inactivating and, combined with chromosome 17 deletion, results in loss of function of this important tumor suppressor. Thus, complete inactivation of p53 signaling is likely a major driving event in chRCC tumorigenesis. This is an important feature to consider, as it distinguishes these tumors from all other forms of RCC, and aligns them more closely (in genetic terms) with breast and ovarian cancers. Similarly, the next most commonly mutated gene in chRCC is PTEN. Again, pairing these mutations with the near ubiquitous loss of chromosome 10 results in complete loss of function of this tumor suppressor, which acts as a brake on the PI3K signaling pathway. The expected activation of mTOR signaling down-stream from PI3K has been previously observed in small studies, and provides additional validation for the expanded use of mTOR inhibitors in this disease (21, 54). Together, the frequent loss of these tumor suppressors have important therapeutic implications as the current standard practice focuses on anti-angiogenic therapies (55).
3.2.2 TERT fusions
A highly unique finding in the chRCC TCGA evaluation arose from the extended analysis of whole genomes in this set of tumors. Whole-genome sequencing identified a number of genomic rearrangements in the TERT promoter region. This finding was also coupled with the observation that these same tumors displayed elevated TERT gene expression, suggesting a functional role for these gene fusions, and selection for these events in tumor progression. Mutations of the TERT promoter were also identified, as had been previously described in melanoma (56, 57), though tumors harboring these mutations had less robust surges in TERT gene expression levels. Collectively, genomic alterations leading to increased TERT expression represents a novel mechanism of RCC tumor promotion. High expression of TERT and the occurrence of these gene fusions was also associated with regional kataegis, a pattern of highly localized substitutions observed in a subset of chRCC tumors. Tumors displaying kataegis also had a mutation pattern consistent with APOBEC cytidine deaminase-mediated mutagenesis (58) This new finding remains an incompletely understood set of events guiding the mutational remodeling of the chRCC genome, but which is being increasingly observed in other cancers (58).
3.3 chRCC: A distinct metabolic disease
3.3.1 Mutations in the electron transport chain
Another special feature of the chRCC TCGA dataset was the inclusion of mitochondrial gene sequencing. chRCCs and oncocytomas have previously been reported to harbor mitochondrial gene mutations, but the frequency of these events, or the association with other features of chRCC, were unknown due to the rarity of this cancer. This analysis revealed a surprisingly high rate of mutations in genes encoding proteins involved in electron transport chain complex I. In particular, mutations in MT-ND5, a key component of this large complex, dominated the mutation landscape. No mutations were observed in other metabolic regulatory elements, such as glucose transport or the Krebs cycle. The association between gene expression of glycolytic enzymes and clinical outcome was not observed as it had been for ccRCC. However, the overall favorable outcome of chRCC tumors in this cohort limited analyses linked with survival based outcomes.
3.3.2 Contributions to the eosinophilic variant subtype
A tight correlation was observed between mitochondrial gene mutations and the eosinophilic phenotype as identified by the TCGA expert pathology group members. An independent set of tumors was examined for eosinophilic histology, and measured for mitochondrial mass, clearly tying this phenotype to an accumulation of mitochondrial density. It remains uncertain whether these mutations promote a setting in which mitochondrial electron transport is hindered to such an extent that compensatory mitochondrial function (and mitogenesis) is needed to allow survival of these cells, or if the accumulation of mitochondria represents an alternate metabolic program fueling the growth of these cells. A recent assessment of the metabolic blockade in these tumors favors the former explanation (59).
3.4 Summary of TCGA analysis of chRCC
Overall, TCGA analysis of this unique and rare tumor type identified several interesting facets of tumor biology that render it highly distinct from the more common RCCs, and also sheds light on the range of tumor biological features that exist to drive cancer. The major findings in this cancer are 1) a highly stochastic copy number profile, indicative of a cellular genomic event that results in aneuploidy and massive elimination of chromosomal material; 2) a program of TP53 and PTEN mutations more aligned with breast and ovarian tumors than the classical tumors of the kidney cortex; 3) high frequency gene fusions involving the TERT promoter that are associated with increased gene expression, and presumably contribute to the self-renewing phenotype of these cells as well as kataegis, and an APOBEC-type mutational spectrum in a subset of tumors; and 4) a unique phenotype of mitochondrial perturbation resulting from inactivating mutations in key members of the electron transport chain. Although these tumors may be finding compensatory mechanisms of mitochondrial over-duplication, the selection for these events suggests a growth advantage associated with alternative metabolic fuel utilization and/or resource generation.
4. COMPARATIVE FEATURES OF THE RENAL CELL CARCINOMAS
The comprehensive molecular profiling of the major RCC subtypes achieved by TCGA allows for more extensive analysis and comparison of the biology across the RCC spectrum. Such an analysis was previously difficult and highlights both similarities and important distinctions among these related but distinct diseases (Fig. 2).
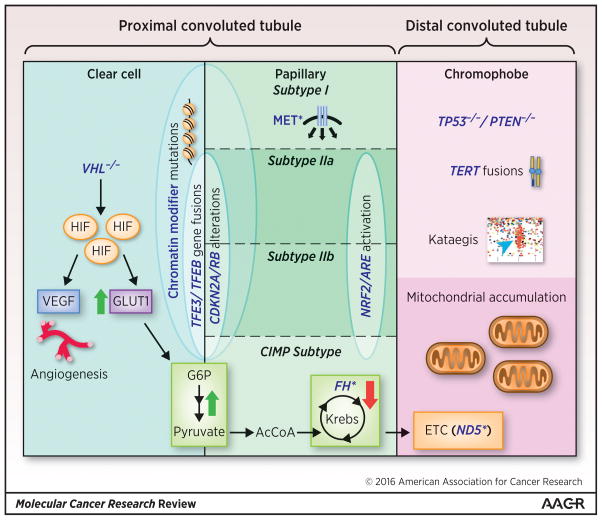
Figure 2. Molecular Comparison of Clear Cell, Papillary, and Chromophobe Renal Cell Carcinoma.
The molecular annotation of renal cell carcinoma subtypes enhances our ability to compare these tumors at the molecular level. Clear cell and papillary RCC exhibit gene expression profiles most similar to the proximal convoluted tubule while chromophobe is most similar to the distal convoluted tubule. The loss of VHL with resultant HIF stabilization is unique to clear cell RCC. However, CDKN2A/RB alterations and TFE3/TFEB gene fusions were identified in both clear cell and papillary type 2 tumors. Mutations (*) of MET were specific to papillary type 1. The chromophobe RCC tumors included TERT fusions and overexpression, kataegis, homozygous loss of TP53 and PTEN, and an eosinophilic subtype characterized by mitochondrial accumulation and mutations in electron transport chain (ETC) genes (most notably MT-ND5). Across the spectrum, significant metabolic reprogramming was observed. Both clear cell and papillary RCC featured increased glycolysis and decreased oxidative phosphorylation, including the Krebs cycle, as measured by gene and protein expression. The CIMP subtype of type 2 papillary RCC was unique in that it featured mutated and/or decreased expression of the Krebs cycle enzyme FH as well as dramatic Warburg effect. Conversely, the chromophobe RCC tumors demonstrate upregulation of the Krebs cycle and electron transport chain as well as mitochondrial accumulation.
4.1 Using comparative features to reveal the origin of RCCs
Macroscopically, the kidney is divided into two distinct regions: the outer cortex and the inner medulla. Microscopically, each kidney consists of about a million complex, multicellular units called nephrons. The portion of the nephron that dips deepest into the medulla (the loop of Henle) divides the nephron into the proximal segment (including the proximal convoluted tubule) and the distal segment (including the distal convoluted tubule and collecting duct) (60). Cells of the nephron are morphologically distinct in different regions with unique gene expression patterns (61) reflecting their often non-overlapping and specialized physiological roles. Indeed, distinct kidney cancers may arise from cells of these distinct portions of the nephron. At the level of gene expression, pRCC and ccRCC seem to be most similar to the proximal nephron as opposed to chRCC, which appears most similar to the distal nephron (51). These findings suggest that some portion of the biologic divergences among kidney cancer subtypes likely stems from their unique sites of origin within the nephron.
4.2 Mutation spectrum
The mutation rate in RCC tumors is generally low. For example, WES in the pRCC dataset revealed an average of 1.45 non-silent mutations per megabase pair (MBP) (62), comparable to the rate of 1.1 mutations/MBP observed in the ccRCC (17) and significantly higher than the 0.4 mutations/MBP seen in most of the chRCC tumors (51) (Fig. 1). Despite these differences among subtypes, overall, RCC tumors seem to have a significantly lower mutation rate than classic mutagen-associated cancers such as lung squamous and melanoma (8 and 17 mutations/MBP, respectively) (63, 64). This is important to note when considering the roughly 20% response rate in RCC to checkpoint inhibitor immunotherapy (65) and emerging evidence supporting a mutational burden association with response to checkpoint inhibition in other cancers (66).
Among the significantly mutated genes identified in each RCC subtype were those anticipated from the genetic syndromes known to predispose individuals to RCC, including the enrichment of VHL mutations in ccRCC, MET mutations in pRCC-I, and FH mutations in pRCC-II. However, mutations in genes previously not associated with specific subtypes of RCC also emerged. While NFE2L2 mutations were observed in some ccRCC tumors, they did not reach statistical significance as they did in pRCC tumors. No mutations in the NRF2 pathway were identified in the chRCC tumors. Thus, mutations in NRF2/ARE genes may be most important in pRCC, especially type II. Similarly, the relatively high rate of TP53 mutations in chRCC was not shared by ccRCC or pRCC tumors. In contrast, other pathways were mutated across all three subtypes. For example, mutations in mTOR pathway genes, including those shown to correlate with robust mTOR-inhibitor response in RCC (67), were seen in all three subtypes (clear cell 14%, papillary 7%, and chromophobe 14%). Other examples of mutated pathways across the subtypes are described in greater detail below.
4.3 The common theme of chromatin modifier mutations
ccRCC tumors had frequent mutations in the chromatin modifier pathway (35%) as did pRCC-I and -II (35% and 38%, respectively). chRCC, however, consistent with its overall relatively low mutational burden, had far less (3%). Similarly, components of the chromatin-remodeling complex SWI/SNF were frequently mutated in both papillary subtypes (20% and 27%, respectively), a frequency intermediate between clear cell RCC (43%) and chromophobe RCC (3%). Interestingly, in an unbiased, network-driven analysis that evaluates significantly mutated genes, as well as less frequently mutated genes (HotNet analysis), SWI/SNF emerged as a significant pathway in pRCC and ccRCC. How mutations in chromatin modifier mutations impact growth, metastasis, and drug sensitivity of these tumors remains an intense area of research.
4.4 CDKN2A/RB pathway
The TCGA analysis also identified alterations in CDKN2A as an oncogenic pathway of importance across the RCC spectrum. A variety of mechanisms were identified that could inactivate CDKN2A, including mutations in the gene, focal deletions in 9p21, and hypermethylation of the CDKN2A promoter. Among the 23 pRCC where these alterations were observed, most were the aggressive pRCC-II tumors. Among these CDKN2A-altered tumors, consistent with loss of function of the p16/INK4A tumor suppressor, RB phosphorylation and expression of cell cycle genes were significantly higher and survival was decreased. Among the ccRCC tumors, 9p21 deletions were enriched among the mRNA cluster referred to as “m3”. This m3 cluster, contained within the poor prognostic “ccB” subtype referenced in other ccRCC literature, had poor survival (17, 68, 69). Thus, in both papillary and clear cell RCC, tumors with CDKN2A alterations correlate with aggressive subtypes. While no mutations were observed, 4 of the chRCC tumors displayed epigenetic silencing of CDKN2A, therefore strategies to target CDKN2A biology may prove useful across the kidney cancer spectrum.
4.5 RCC is a metabolic disease—in many different ways
The kidneys are large organs with a very high blood flow (~400 mL/min per 100 g of tissue relative to ~80 mL/min for the heart) (60). This blood flow is in excess of what is needed for their metabolic needs and instead facilitates their role in regulating water and electrolyte balance. This regulation, especially sodium reabsorption, requires extensive energy as reflected by the high oxygen consumption by the kidneys, second only to the heart. Given this high metabolic activity of the native organ, it is perhaps not surprising to find metabolic reprogramming to be a common phenomenon across RCC subtypes. For example, fumarate hydratase (FH) is a Krebs cycle enzyme as well as the gene responsible for the inherited cancer predisposition syndrome of hereditary leiomyomatosis and renal cell cancer (HLRCC), including pRCC-II (70). The accumulation of fumarate in FH-mutated RCC may cause multiple oncogenic sequelae, including activation of the NRF2/ARE pathway (71–73) as well as HIF stabilization with upregulation of hypoxia-related genes (74, 75). These, and possibly other mechanisms, likely contribute to the dramatic metabolic shift towards Warburg-like metabolism seen in the pRCC-II tumors (especially the CIMP subtype). Similar metabolic trends were seen in the ccRCC tumors driven by loss of VHL function and subsequent HIF stabilization. In both of these groups, high expression of genes and proteins involved in glycolysis, pentose-phosphate pathway, and fatty acid synthesis, and low expression of those involved in Krebs cycle and AMPK signaling, correlated with poor survival. This is in stark contrast to the chromophobe tumors, where evidence of enhanced cellular respiration including increased expression of Krebs cycle and oxidative phosphorylation genes was observed. Thus, while the type of metabolic defects differ among the subtypes, metabolic reprogramming continues to emerge as a core principle in RCC biology.
4.6 Translocation RCC
A molecular event once thought to be unique to pRCC-II tumors are the cytogenetic translocations referred to as TFE3 and TFEB fusions. TFE3 is a transcription factor from the MiT family located on Xp11.2. Tumors with TFE3 fusions are rare and referred to as translocation RCC. They represent one third of the ~25 pediatric RCC cases diagnosed in the United States per year (41) and are recognized by the WHO as a distinct RCC subtype (2), but are rarely seen in adults (76). How TFE3 fusions promote oncogenesis in RCC is incompletely understood, though TFE3 is known to regulate several oncogenic pathways involved in cell growth and metabolism, including the mTOR and TGFβ signaling pathways, MET, and AMPK (77). While the disease often follows an indolent clinical course in pediatric patients (78), it can be very aggressive and metastasize early, especially in adults. In one retrospective analysis of adults with translocation RCC receiving anti-angiogenesis therapies, the median overall survival was a mere 14.3 months (79). Interestingly, the true estimate of translocation RCC incidence in adults may be underestimated. Among the TCGA samples, in addition to seven TFE3/TFEB translocations identified in pRCC, five were observed in ccRCC cases. Thus, translocation RCC may be histologically indistinguishable from other RCC types, and identifiable only at the genetic level. This finding suggests that it is important to be alert to the possibility that translocation tumors may present with different phenotypical characteristics..
CONCLUSIONS
The three major projects in renal cell carcinoma conducted by TCGA have revealed new insights into this heretofore enigmatic disease. The details summarized above are available in the three published TCGA index papers (17, 51, 62). These findings provide a foundation of genetic and molecular evidence, which combined with histological and morphological data, demonstrate that subtypes of the RCCs are quite biologically distinct. Collectively, this information will be useful for improving both diagnosis and treatment of RCC patients.
Acknowledgments
This project could not have been done without the massive effort of the TCGA. WKR received support from K24CA172355. SMH received an ASCO YIA award to support this effort.
Footnotes
The authors declare no conflict of interest.
References
- 1.Campbell SC, Rini BI. Editorial comment. The Journal of urology. 2009;182(6):2599–600. doi: 10.1016/j.juro.2009.08.194. [DOI] [PubMed] [Google Scholar]
- 2.Eble JN, Sauter G, Epstein J. Tumours of the Genitourinary and Male Genital Organs. Lyon: IARC Press; 2004. WHO Classification of Tumours. [Google Scholar]
- 3.Solomon JP, Hansel DE. Morphologic and Molecular Characteristics of Bladder Cancer. Surgical pathology clinics. 2015;8(4):663–76. doi: 10.1016/j.path.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 4.Patard JJ, Leray E, Rioux-Leclercq N, Cindolo L, Ficarra V, Zisman A, et al. Prognostic value of histologic subtypes in renal cell carcinoma: a multicenter experience. J Clin Oncol. 2005;23(12):2763–71. doi: 10.1200/JCO.2005.07.055. [DOI] [PubMed] [Google Scholar]
- 5.Aydin H, Chen L, Cheng L, Vaziri S, He H, Ganapathi R, et al. Clear cell tubulopapillary renal cell carcinoma: a study of 36 distinctive low-grade epithelial tumors of the kidney. The American journal of surgical pathology. 2010;34(11):1608–21. doi: 10.1097/PAS.0b013e3181f2ee0b. [DOI] [PubMed] [Google Scholar]
- 6.Williamson SR, Halat S, Eble JN, Grignon DJ, Lopez-Beltran A, Montironi R, et al. Multilocular cystic renal cell carcinoma: similarities and differences in immunoprofile compared with clear cell renal cell carcinoma. The American journal of surgical pathology. 2012;36(10):1425–33. doi: 10.1097/PAS.0b013e31825b37f0. [DOI] [PubMed] [Google Scholar]
- 7.Tan MH, Wong CF, Tan HL, Yang XJ, Ditlev J, Matsuda D, et al. Genomic expression and single-nucleotide polymorphism profiling discriminates chromophobe renal cell carcinoma and oncocytoma. BMC cancer. 2010;10:196. doi: 10.1186/1471-2407-10-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nickerson ML, Jaeger E, Shi Y, Durocher JA, Mahurkar S, Zaridze D, et al. Improved identification of von Hippel-Lindau gene alterations in clear cell renal tumors. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14(15):4726–34. doi: 10.1158/1078-0432.CCR-07-4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen C, Kaelin WG., Jr The VHL/HIF axis in clear cell renal carcinoma. Seminars in cancer biology. 2013;23(1):18–25. doi: 10.1016/j.semcancer.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choueiri TK, Fay AP, Gagnon R, Lin Y, Bahamon B, Brown V, et al. The role of aberrant VHL/HIF pathway elements in predicting clinical outcome to pazopanib therapy in patients with metastatic clear-cell renal cell carcinoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19(18):5218–26. doi: 10.1158/1078-0432.CCR-13-0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Motzer RJ, Hutson TE, Cella D, Reeves J, Hawkins R, Guo J, et al. Pazopanib versus sunitinib in metastatic renal-cell carcinoma. The New England journal of medicine. 2013;369(8):722–31. doi: 10.1056/NEJMoa1303989. [DOI] [PubMed] [Google Scholar]
- 12.Motzer RJ, Hutson TE, Tomczak P, Michaelson MD, Bukowski RM, Rixe O, et al. Sunitinib versus interferon alfa in metastatic renal-cell carcinoma. The New England journal of medicine. 2007;356(2):115–24. doi: 10.1056/NEJMoa065044. [DOI] [PubMed] [Google Scholar]
- 13.Escudier B, Szczylik C, Hutson TE, Demkow T, Staehler M, Rolland F, et al. Randomized phase II trial of first-line treatment with sorafenib versus interferon Alfa-2a in patients with metastatic renal cell carcinoma. J Clin Oncol. 2009;27(8):1280–9. doi: 10.1200/JCO.2008.19.3342. [DOI] [PubMed] [Google Scholar]
- 14.Patard JJ, Pignot G, Escudier B, Eisen T, Bex A, Sternberg C, et al. ICUD-EAU International Consultation on Kidney Cancer 2010: treatment of metastatic disease. European urology. 2011;60(4):684–90. doi: 10.1016/j.eururo.2011.06.017. [DOI] [PubMed] [Google Scholar]
- 15.Heng DY, Xie W, Regan MM, Warren MA, Golshayan AR, Sahi C, et al. Prognostic factors for overall survival in patients with metastatic renal cell carcinoma treated with vascular endothelial growth factor-targeted agents: results from a large, multicenter study. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2009;27(34):5794–9. doi: 10.1200/JCO.2008.21.4809. [DOI] [PubMed] [Google Scholar]
- 16.Kim WY, Kaelin WG. Role of VHL gene mutation in human cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2004;22(24):4991–5004. doi: 10.1200/JCO.2004.05.061. [DOI] [PubMed] [Google Scholar]
- 17.Cancer Genome Atlas Research N. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499(7456):43–9. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hakimi AA, Chen YB, Wren J, Gonen M, Abdel-Wahab O, Heguy A, et al. Clinical and pathologic impact of select chromatin-modulating tumor suppressors in clear cell renal cell carcinoma. European urology. 2013;63(5):848–54. doi: 10.1016/j.eururo.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hakimi AA, Ostrovnaya I, Reva B, Schultz N, Chen YB, Gonen M, et al. Adverse outcomes in clear cell renal cell carcinoma with mutations of 3p21 epigenetic regulators BAP1 and SETD2: a report by MSKCC and the KIRC TCGA research network. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19(12):3259–67. doi: 10.1158/1078-0432.CCR-12-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brugarolas J. Molecular genetics of clear-cell renal cell carcinoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2014;32(18):1968–76. doi: 10.1200/JCO.2012.45.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Voss MH, Bastos DA, Karlo CA, Ajeti A, Hakimi AA, Feldman DR, et al. Treatment outcome with mTOR inhibitors for metastatic renal cell carcinoma with nonclear and sarcomatoid histologies. Annals of oncology : official journal of the European Society for Medical Oncology/ESMO. 2014;25(3):663–8. doi: 10.1093/annonc/mdt578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaklamani V. A genetic signature can predict prognosis and response to therapy in breast cancer: Oncotype DX. Expert review of molecular diagnostics. 2006;6(6):803–9. doi: 10.1586/14737159.6.6.803. [DOI] [PubMed] [Google Scholar]
- 23.Webber EM, Lin JS, Evelyn PW. Oncotype DX tumor gene expression profiling in stage II colon cancer. Application: prognostic, risk prediction. PLoS currents. 2010;2 doi: 10.1371/currents.RRN1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goy A, Stewart J, Barkoh BA, Remache YK, Katz R, Sneige N, et al. The feasibility of gene expression profiling generated in fine-needle aspiration specimens from patients with follicular lymphoma and diffuse large B-cell lymphoma. Cancer. 2006;108(1):10–20. doi: 10.1002/cncr.21500. [DOI] [PubMed] [Google Scholar]
- 25.Brooks SA, Brannon AR, Parker JS, Fisher JC, Sen O, Kattan MW, et al. ClearCode34: A prognostic risk predictor for localized clear cell renal cell carcinoma. European urology. 2014;66(1):77–84. doi: 10.1016/j.eururo.2014.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masoud GN, Li W. HIF-1alpha pathway: role, regulation and intervention for cancer therapy. Acta pharmaceutica Sinica B. 2015;5(5):378–89. doi: 10.1016/j.apsb.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mucaj V, Shay JE, Simon MC. Effects of hypoxia and HIFs on cancer metabolism. International journal of hematology. 2012;95(5):464–70. doi: 10.1007/s12185-012-1070-5. [DOI] [PubMed] [Google Scholar]
- 28.Hu CJ, Wang LY, Chodosh LA, Keith B, Simon MC. Differential roles of hypoxia-inducible factor 1alpha (HIF-1alpha) and HIF-2alpha in hypoxic gene regulation. Molecular and cellular biology. 2003;23(24):9361–74. doi: 10.1128/MCB.23.24.9361-9374.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lum JJ, Bui T, Gruber M, Gordan JD, DeBerardinis RJ, Covello KL, et al. The transcription factor HIF-1alpha plays a critical role in the growth factor-dependent regulation of both aerobic and anaerobic glycolysis. Genes & development. 2007;21(9):1037–49. doi: 10.1101/gad.1529107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fu L, Minton DR, Zhang T, Nanus DM, Gudas LJ. Genome-Wide Profiling of TRACK Kidneys Shows Similarity to the Human ccRCC Transcriptome. Molecular cancer research : MCR. 2015;13(5):870–8. doi: 10.1158/1541-7786.MCR-14-0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li B, Qiu B, Lee DS, Walton ZE, Ochocki JD, Mathew LK, et al. Fructose-1,6-bisphosphatase opposes renal carcinoma progression. Nature. 2014;513(7517):251–5. doi: 10.1038/nature13557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brooks SA, Khandani A, Fielding J, Lin W, Sills T, Lee Y, et al. Alternate metabolic programs define regional variation of relevant biological features in renal cell carcinoma progression. Clinical cancer research : an official journal of the American Association for Cancer Research. 2016 doi: 10.1158/1078-0432.CCR-15-2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simon JM, Hacker KE, Singh D, Brannon AR, Parker JS, Weiser M, et al. Variation in chromatin accessibility in human kidney cancer links H3K36 methyltransferase loss with widespread RNA processing defects. Genome research. 2014;24(2):241–50. doi: 10.1101/gr.158253.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tiedemann RL, Hlady RA, Hanavan PD, Lake DF, Tibes R, Lee JH, et al. Dynamic reprogramming of DNA methylation in SETD2-deregulated renal cell carcinoma. Oncotarget. 2015 doi: 10.18632/oncotarget.6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Delahunt B, Eble JN. Papillary renal cell carcinoma: a clinicopathologic and immunohistochemical study of 105 tumors. Mod Pathol. 1997;10(6):537–44. [PubMed] [Google Scholar]
- 36.Pignot G, Elie C, Conquy S, Vieillefond A, Flam T, Zerbib M, et al. Survival analysis of 130 patients with papillary renal cell carcinoma: prognostic utility of type 1 and type 2 subclassification. Urology. 2007;69(2):230–5. doi: 10.1016/j.urology.2006.09.052. [DOI] [PubMed] [Google Scholar]
- 37.Klatte T, Pantuck AJ, Said JW, Seligson DB, Rao NP, LaRochelle JC, et al. Cytogenetic and molecular tumor profiling for type 1 and type 2 papillary renal cell carcinoma. Clin Cancer Res. 2009;15(4):1162–9. doi: 10.1158/1078-0432.CCR-08-1229. [DOI] [PubMed] [Google Scholar]
- 38.Ravaud A, Oudard S, De Fromont M, Chevreau C, Gravis G, Zanetta S, et al. First-line treatment with sunitinib for type 1 and type 2 locally advanced or metastatic papillary renal cell carcinoma: a phase II study (SUPAP) by the French Genitourinary Group (GETUG)dagger. Annals of oncology : official journal of the European Society for Medical Oncology/ESMO. 2015 doi: 10.1093/annonc/mdv149. [DOI] [PubMed] [Google Scholar]
- 39.Yang XJ, Tan MH, Kim HL, Ditlev JA, Betten MW, Png CE, et al. A molecular classification of papillary renal cell carcinoma. Cancer Res. 2005;65(13):5628–37. doi: 10.1158/0008-5472.CAN-05-0533. [DOI] [PubMed] [Google Scholar]
- 40.Hoadley KA, Yau C, Wolf DM, Cherniack AD, Tamborero D, Ng S, et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell. 2014;158(4):929–44. doi: 10.1016/j.cell.2014.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Durinck S, Stawiski EW, Pavia-Jimenez A, Modrusan Z, Kapur P, Jaiswal BS, et al. Spectrum of diverse genomic alterations define non-clear cell renal carcinoma subtypes. Nat Genet. 2015;47(1):13–21. doi: 10.1038/ng.3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmidt L, Junker K, Nakaigawa N, Kinjerski T, Weirich G, Miller M, et al. Novel mutations of the MET proto-oncogene in papillary renal carcinomas. Oncogene. 1999;18(14):2343–50. doi: 10.1038/sj.onc.1202547. [DOI] [PubMed] [Google Scholar]
- 44.Schmidt L, Duh FM, Chen F, Kishida T, Glenn G, Choyke P, et al. Germline and somatic mutations in the tyrosine kinase domain of the MET proto-oncogene in papillary renal carcinomas. Nature genetics. 1997;16(1):68–73. doi: 10.1038/ng0597-68. [DOI] [PubMed] [Google Scholar]
- 45.Khurana E, Fu Y, Colonna V, Mu XJ, Kang HM, Lappalainen T, et al. Integrative annotation of variants from 1092 humans: application to cancer genomics. Science. 2013;342(6154):1235587. doi: 10.1126/science.1235587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ooi A, Dykema K, Ansari A, Petillo D, Snider J, Kahnoski R, et al. CUL3 and NRF2 mutations confer an NRF2 activation phenotype in a sporadic form of papillary renal cell carcinoma. Cancer Res. 2013;73(7):2044–51. doi: 10.1158/0008-5472.CAN-12-3227. [DOI] [PubMed] [Google Scholar]
- 47.Singh A, Misra V, Thimmulappa RK, Lee H, Ames S, Hoque MO, et al. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS medicine. 2006;3(10):e420. doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shibata T, Ohta T, Tong KI, Kokubu A, Odogawa R, Tsuta K, et al. Cancer related mutations in NRF2 impair its recognition by Keap1-Cul3 E3 ligase and promote malignancy. Proc Natl Acad Sci U S A. 2008;105(36):13568–73. doi: 10.1073/pnas.0806268105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Solis LM, Behrens C, Dong W, Suraokar M, Ozburn NC, Moran CA, et al. Nrf2 and Keap1 abnormalities in non-small cell lung carcinoma and association with clinicopathologic features. Clin Cancer Res. 2010;16(14):3743–53. doi: 10.1158/1078-0432.CCR-09-3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Daugherty M, Blakely S, Shapiro O, Vourganti S, Mollapour M, Bratslavsky G. Chromophobe RCC is the most common non-clear RCC in young women: results from the SEER database. The Journal of urology. 2015 doi: 10.1016/j.juro.2015.10.177. [DOI] [PubMed] [Google Scholar]
- 51.Davis CF, Ricketts CJ, Wang M, Yang L, Cherniack AD, Shen H, et al. The somatic genomic landscape of chromophobe renal cell carcinoma. Cancer Cell. 2014;26(3):319–30. doi: 10.1016/j.ccr.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kovacs A, Kovacs G. Low chromosome number in chromophobe renal cell carcinomas. Genes, chromosomes & cancer. 1992;4(3):267–8. doi: 10.1002/gcc.2870040313. [DOI] [PubMed] [Google Scholar]
- 53.Kovacs A, Storkel S, Thoenes W, Kovacs G. Mitochondrial and chromosomal DNA alterations in human chromophobe renal cell carcinomas. The Journal of pathology. 1992;167(3):273–7. doi: 10.1002/path.1711670303. [DOI] [PubMed] [Google Scholar]
- 54.Chen J, Huang D, Rubera I, Futami K, Wang P, Zickert P, et al. Disruption of tubular Flcn expression as a mouse model for renal tumor induction. Kidney international. 2015;88(5):1057–69. doi: 10.1038/ki.2015.177. [DOI] [PubMed] [Google Scholar]
- 55.Tannir NM, Jonasch E, Albiges L, Altinmakas E, Ng CS, Matin SF, et al. Everolimus Versus Sunitinib Prospective Evaluation in Metastatic Non-Clear Cell Renal Cell Carcinoma (ESPN): A Randomized Multicenter Phase 2 Trial. European urology. 2015 doi: 10.1016/j.eururo.2015.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Horn S, Figl A, Rachakonda PS, Fischer C, Sucker A, Gast A, et al. TERT promoter mutations in familial and sporadic melanoma. Science (New York, NY) 2013;339(6122):959–61. doi: 10.1126/science.1230062. [DOI] [PubMed] [Google Scholar]
- 57.Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin L, Garraway LA. Highly recurrent TERT promoter mutations in human melanoma. Science (New York, NY) 2013;339(6122):957–9. doi: 10.1126/science.1229259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–21. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Joshi S, Tolkunov D, Aviv H, Hakimi AA, Yao M, Hsieh JJ, et al. The Genomic Landscape of Renal Oncocytoma Identifies a Metabolic Barrier to Tumorigenesis. Cell reports. 2015;13(9):1895–908. doi: 10.1016/j.celrep.2015.10.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Michael J, editor. Fundamentals of Medical Physiology. 1. New York, NY: Thieme; 2011. [Google Scholar]
- 61.Cheval L, Pierrat F, Rajerison R, Piquemal D, Doucet A. Of mice and men: divergence of gene expression patterns in kidney. PLoS One. 2012;7(10):e46876. doi: 10.1371/journal.pone.0046876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Linehan WM, Spellman PT, Ricketts CJ, Creighton CJ, Fei SS, Davis C, et al. Comprehensive Molecular Characterization of Papillary Renal-Cell Carcinoma. The New England journal of medicine. 2016;374(2):135–45. doi: 10.1056/NEJMoa1505917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489(7417):519–25. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Genomic Classification of Cutaneous Melanoma. Cell. 2015;161(7):1681–96. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Motzer RJ, Escudier B, McDermott DF, George S, Hammers HJ, Srinivas S, et al. Nivolumab versus Everolimus in Advanced Renal-Cell Carcinoma. The New England journal of medicine. 2015;373(19):1803–13. doi: 10.1056/NEJMoa1510665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science (New York, NY) 2015;348(6230):124–8. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Voss MH, Hakimi AA, Pham CG, Brannon AR, Chen YB, Cunha LF, et al. Tumor genetic analyses of patients with metastatic renal cell carcinoma and extended benefit from mTOR inhibitor therapy. Clin Cancer Res. 2014;20(7):1955–64. doi: 10.1158/1078-0432.CCR-13-2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brannon AR, Reddy A, Seiler M, Arreola A, Moore DT, Pruthi RS, et al. Molecular Stratification of Clear Cell Renal Cell Carcinoma by Consensus Clustering Reveals Distinct Subtypes and Survival Patterns. Genes Cancer. 2010;1(2):152–63. doi: 10.1177/1947601909359929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brannon AR, Haake SM, Hacker KE, Pruthi RS, Wallen EM, Nielsen ME, et al. Meta-analysis of clear cell renal cell carcinoma gene expression defines a variant subgroup and identifies gender influences on tumor biology. European urology. 2012;61(2):258–68. doi: 10.1016/j.eururo.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Toro JR, Nickerson ML, Wei MH, Warren MB, Glenn GM, Turner ML, et al. Mutations in the fumarate hydratase gene cause hereditary leiomyomatosis and renal cell cancer in families in North America. American journal of human genetics. 2003;73(1):95–106. doi: 10.1086/376435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kinch L, Grishin NV, Brugarolas J. Succination of Keap1 and activation of Nrf2-dependent antioxidant pathways in FH-deficient papillary renal cell carcinoma type 2. Cancer Cell. 2011;20(4):418–20. doi: 10.1016/j.ccr.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ooi A, Wong JC, Petillo D, Roossien D, Perrier-Trudova V, Whitten D, et al. An antioxidant response phenotype shared between hereditary and sporadic type 2 papillary renal cell carcinoma. Cancer Cell. 2011;20(4):511–23. doi: 10.1016/j.ccr.2011.08.024. [DOI] [PubMed] [Google Scholar]
- 73.Adam J, Hatipoglu E, O’Flaherty L, Ternette N, Sahgal N, Lockstone H, et al. Renal cyst formation in Fh1-deficient mice is independent of the Hif/Phd pathway: roles for fumarate in KEAP1 succination and Nrf2 signaling. Cancer Cell. 2011;20(4):524–37. doi: 10.1016/j.ccr.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Isaacs JS, Jung YJ, Mole DR, Lee S, Torres-Cabala C, Chung YL, et al. HIF overexpression correlates with biallelic loss of fumarate hydratase in renal cancer: novel role of fumarate in regulation of HIF stability. Cancer Cell. 2005;8(2):143–53. doi: 10.1016/j.ccr.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 75.O’Flaherty L, Adam J, Heather LC, Zhdanov AV, Chung YL, Miranda MX, et al. Dysregulation of hypoxia pathways in fumarate hydratase-deficient cells is independent of defective mitochondrial metabolism. Human molecular genetics. 2010;19(19):3844–51. doi: 10.1093/hmg/ddq305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Argani P, Olgac S, Tickoo SK, Goldfischer M, Moch H, Chan DY, et al. Xp11 translocation renal cell carcinoma in adults: expanded clinical, pathologic, and genetic spectrum. The American journal of surgical pathology. 2007;31(8):1149–60. doi: 10.1097/PAS.0b013e318031ffff. [DOI] [PubMed] [Google Scholar]
- 77.Kauffman EC, Ricketts CJ, Rais-Bahrami S, Yang Y, Merino MJ, Bottaro DP, et al. Molecular genetics and cellular features of TFE3 and TFEB fusion kidney cancers. Nature reviews Urology. 2014;11(8):465–75. doi: 10.1038/nrurol.2014.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ramphal R, Pappo A, Zielenska M, Grant R, Ngan BY. Pediatric renal cell carcinoma: clinical, pathologic, and molecular abnormalities associated with the members of the mit transcription factor family. American journal of clinical pathology. 2006;126(3):349–64. doi: 10.1309/98YE9E442AR7LX2X. [DOI] [PubMed] [Google Scholar]
- 79.Choueiri TK, Lim ZD, Hirsch MS, Tamboli P, Jonasch E, McDermott DF, et al. Vascular endothelial growth factor-targeted therapy for the treatment of adult metastatic Xp11.2 translocation renal cell carcinoma. Cancer. 2010;116(22):5219–25. doi: 10.1002/cncr.25512. [DOI] [PMC free article] [PubMed] [Google Scholar]