Abstract
Several members of the matrix metalloproteinase (MMP) family control a range of immune processes, such as leukocyte influx and chemokine activity. Stromelysin-2 (MMP10) is expressed by macrophages in numerous tissues after injury; however, little is known of its function. Here, we report that MMP10 is expressed by macrophages in human lungs from patients with cystic fibrosis and induced in mouse macrophages in response to Pseudomonas aeruginosa infection both in vivo and by isolated resident alveolar and bone marrow-derived macrophages (BMDM). Our data indicates that macrophage MMP10 serves a beneficial function in response to acute infection. Whereas wildtype mice survived infection with minimal morbidity, 50% of Mmp10−/− mice died and all showed sustained weight loss (morbidity). Although bacterial clearance and neutrophil influx did not differ between genotypes, macrophage numbers were about 3-fold greater in infected Mmp10−/− lungs than in wildtypes. Adoptive transfer of wildtype BMDM normalized infection-induced morbidity in Mmp10−/− recipients to wildtype levels demonstrating that the protective effect of MMP10 was due to its production by macrophages. Both in vivo and in cultured alveolar macrophages and BMDM, expression of several M1 macrophage markers was elevated, while M2 markers were reduced in Mmp10−/− tissue and cells. Global gene expression analysis revealed that infection-mediated transcriptional changes persisted in Mmp10−/− BMDM long after they were down-regulated in wildtype cells. These results indicate that MMP10 serves a beneficial role in response to acute infection by moderating the pro-inflammatory response of resident and infiltrating macrophages.
Introduction
Macrophages are critical effector cells of the immune system and play essential, yet distinct roles in both promoting and resolving inflammation and in facilitating tissue repair and contributing to its destruction (1). That a single cell type can serve opposing functions may seem counterintuitive, but dramatic phenotypic changes occur when macrophages respond to local stimuli (1–4). Based on patterns of gene and protein expression and function, macrophages are commonly classified as classically activated (M1) or alternatively activated (M2) cells, as well as a variety of M2 subtypes (1, 4, 5). M1 macrophages are induced by infection and pro-inflammatory TH1 cytokines, are effective at killing bacteria, and release pro-inflammatory cytokines, such as IL-1β, IL-12, and TNFα. M2 macrophages are induced by the TH2 cytokines IL-4 and IL-13 and other factors, release anti-inflammatory factors, such as IL-10, are weakly microbicidal, and promote repair. We recognize, however, that dividing macrophages into M1 and M2 classes oversimplifies the complex continuum of functional and reversible states in which these cells exist (6–9).
Several proteins influence macrophage behavior, including some members of the matrix metalloproteinase (MMP) gene family. For example, MMP12 and MMP28, both macrophage products, either promote or restrict macrophage influx into lung (10, 11), and MMP8 promotes M2 polarization (12). In addition, we reported that MMP28 and TIMP3, an MMP inhibitor, moderate M1 activation of macrophages in models of lung infection and fibrosis (13, 14). As their name implies, MMPs are thought to degrade extracellular matrix proteins, a function that is indeed performed by some members (15–17). However, matrix degradation is neither the shared nor predominant function of these enzymes. Rather, individual MMPs have been shown to regulate specific immune processes, such as leukocyte influx and activation, antimicrobial activity, and restoration of barrier function, typically by processing of a range of non-matrix protein substrates (18–25).
MMP10 (stromelysin-2) is not expressed in developing or normal adult tissues, including lung (26, 27). However, in both human conditions and mouse models, MMP10 is induced in response to injury, infection, or transformation in essentially all tissues (28–33). In a meta-analysis of gene array experiments involving numerous different host-pathogen interactions, MMP10 was identified as a common host response gene (34). The widespread expression of MMP10 among tissues suggests that this proteinase serves critical roles in the host response to environmental insults. As we show here, MMP10 serves a protective role in acute infection by moderating the pro-inflammatory activity of macrophages.
Recently, we reported that macrophage MMP10 promotes the ability of M2 macrophages to clear scar tissues in normal skin wounds by controlling the expression of collagenolytic MMPs, particularly MMP13 (35). However, because sterile excision wounds are not associated with a profound inflammatory response, the stimulus (i.e., a clean wound) may not have been sufficiently robust to reveal other MMP10-dependent roles in macrophage inflammation. Thus, for the studies reported here, we challenged Mmp10−/− mouse lungs with Pseudomonas aeruginosa, an opportunistic pathogen and common cause of hospital-acquired infections and pneumonia (36) and the major pathogen in lungs of patients with cystic fibrosis (CF) (37). Compared to wildtype mice, we found that Mmp10−/− mice were much more susceptible to airway infection and that MMP10 influenced activation of resident and infiltrated macrophages by curbing M1 polarization and P. aeruginosa-induced transcriptional changes. Our findings indicate that MMP10 is a critical cell autonomous mediator controlling macrophage activation.
Materials and Methods
Animals
Mmp10−/− mice and wildtype littermates (C57BL/6J, male and female, 8–16 wk old) were used for these studies. All procedures were approved by the Office of Animal Welfare at the University of Washington and the IACUC at Cedars-Sinai. Mmp10−/− mice were generated by targeting exons 3–5 that code the catalytic domain as described (38). Mmp10−/− mice breed and appear normal, with similar litter sizes. We also generated Mmp10−/− mice (Mmp10tm1Lex/Mmucd) with embryonic stem cells from the Mutant Mouse Resource & Research Center (MMRRC; https://www.mmrrc.org/catalog/sds.php?mmrrc_id=11737). These cells were originally generated by Lexicon Pharmaceuticals (The Woodlands, TX), who used a targeting strategy distinct from ours.
Infection Model
Mice were challenged with live P. aeruginosa strain K either by aerosolization in a whole animal nebulizer chamber for 30 min to produce a deposition of bacteria between 105 to 106 cfu/left lung or by oropharyngeal aspiration as described (11, 39). Briefly, bacteria were grown in LB overnight, washed twice, and suspended in sterile PBS. For oropharyngeal aspiration, bacteria were quantified by OD600 and suspended at 2 × 108 cells/ml, and 50 µl was instilled per mouse. After exposure by either method, the lungs of 3–5 mice were immediately harvested and homogenized to determine 0-h bacterial counts (CFU). CFU were determined by plating serial dilutions of left lungs or spleens homogenized in 1 ml PBS on LB agar plates. Duplicate plates of each dilution were incubated 24–48 h at 37°C and counted using a Quebec Colony Counter (Reichert Technologies, Depew NY).
Sample Collection and Analyses
The left lung hilum was ligated, and the left lung and spleen were removed for determination of bacterial load. Bronchoalveolar lavage (BAL) was collected by flushing the right lung via the trachea 4 times with 0.5 ml saline. The recovered lavage (typically 1.7 ml) was centrifuged at 400×g, 4°C for 10 min. Supernatants were collected and frozen at −70°C for later analysis. Total protein in BAL was quantified using the Pierce BCA Protein Assay Kit (Thermo Scientific, Rockford IL) and IgM by ELISA (Bethyl Laboratories, Montgomery TX). Pelleted cells were resuspended in DMEM with 10% FBS, counted in a hemocytometer, and a 100-µl aliquot was cytospun onto slides and differentially stained with LeukoStat (Fisher Scientific, Pittsburgh PA). At least 300 cells were counted per lavage. Lungs from separate mice were used for RNA isolation or flow cytometry (below).
Immunostaining and In Situ Hybridization
De-identified human lung specimens were obtained with approval of the University of Washington Institutional Review Board. Sections (5 µm) were stained with antibodies specific for human MMP10 (Abcam, Cambridge MA) as described (40). Mouse lungs were perfused-fixed with PBS-buffered formalin via the trachea under 25-cm pressure for 5 min, incubated in fixative at room temperature for 48 h, dehydrated through graded ethanol, and embedded in paraffin. Deparaffinized sections (5 µm) underwent heat-induced (95°C, 30 min) antigen retrieval in pH 7.0 citrate. Endogenous peroxidase activity was quenched with 3% H2O2 for 10 min. Sections were incubated with 4% normal rabbit serum for 1 h then overnight at 4°C with anti-Mac2 antibody (1:500 dilution; clone M3/38.1.2.8 HL.2, ATCC, Manassas VA). Bound antibody was detected using a Vectastain Elite ABC (Rat IgG) kit (Vector Laboratories, Burlingame CA) and 3,3′-diaminobenzidine peroxidase substrate and counterstained with hematoxylin. For combined in situ hybridization/immunostaining, deparaffinized rehydrated sections were treated with 10 µg/ml proteinase K in RNase-free PBS for 20 min at ambient temperature, incubated with 0.1M triethanolamine in RNase-free water, washed in RNase-free PBS, then probed overnight at 55°C with either a 570-bp sense or antisense digoxigenin-labeled RNA probe targeting bases 1050–1620 of murine Mmp10. Probes were labeled using DIG RNA Labeling Mix and SP6/T7 DIG Labeling Kit (Roche Diagnostics, Indianapolis IN). After hybridization, slides were washed 3 times in 2× SSC/50% formamide, for 15 min at 50°C, once in 2× SSC at ambient temperature for 20 min, and once in 0.5× SSC for 30 min then developed with anti-digoxigenin-alkaline phosphatase using NBT/BCIP as substrate. Slides were washed with PBS and images were captured using an Olympus BX51 with DP70 digital camera (Olympus America, Center Valley, PA). Sections were then processed for immunostaining with anti-Mac-2 as described above minus antigen retrieval and counterstaining.
Flow Cytometry
Whole lungs were perfused with 10 ml ice-cold PBS through the left ventricle and then harvested, minced into tiny pieces using a razor blade, and digested in RPMI containing 2.5 mg/ml Collagenase Type IV (Gibco) for 45 min at 37°C with shaking. The digestion was stopped with addition of FBS to 10%. The lung cell suspension was applied to a 70-µm cell strainer and washed with cold RPMI. The sample was subjected to RBC lysis (ACK Lysing Buffer; Thermo-Fisher), followed by washing in RPMI and cell counting using a Bio-Rad Cell Counter. Lung cells were stained with fluorophore-conjugated antibodies and analyzed using a BD LSRFortessa or sorted using a BD FACSARIA III (BD Biosciences, San Jose CA). Cell populations in the lungs were identified using the following combinations of cell surface markers: alveolar macrophages, F4/80+ CD11c+ SiglecF+; interstitial macrophages, F4/80+ CD11b+ CD11c+ MHCII+ SiglecF−; Ly6C+ monocytes/macrophages, F4/80+ Ly6C+ CD11b+ CD11c+ MHCII− SiglecF−, and neutrophils, F4/80− Ly6G+ CD11b+. Sorted cells were kept at 4°C during sorting, centrifuged, and immediately processed for RNA isolation as described below. SiglecF antibody was from BD Biosciences; all others were from eBioscience (San Diego CA).
Macrophage Culture, Transfer, and Treatments
Bone marrow-derived macrophages (BMDM) and alveolar macrophages were isolated and cultured as described (11, 39). To generate BMDM, marrow cells were differentiated in CSF-1-containing medium for 7 d (referred to as M0 macrophages). To generate M1-biased macrophages, M0 BMDM (1.5 × 106 per well in 6-well plates) were stimulated with 100 ng/ml E. coli 0111:B4 lipopolysaccharide (LPS) for 24 h or with live P. aeruginosa strain K at a multiplicity of infection of 5 bacteria per macrophage for 1 h before washing away bacteria and then culturing in medium with 75 µg/ml gentamicin to kill live extracellular bacteria. For M2 polarized macrophages, BMDM were stimulated with 10 ng/ml each of IL-4 and IL-13 for 48 h. For adoptive transfer, recipient mice were given 2 × 106 wildtype or Mmp10−/− M0 BMDM in 100 µl PBS via retro-orbital 24 h post infection. For the microarray experiments, M0 BMDM cells were exposed to P. aeruginosa strain K as described above. The cells were then cultured in medium containing gentamicin, 100 IU/ml penicillin, and 100 µg/ml streptomycin, and RNA was collected 6 h or 24 h later. For alveolar macrophages, cells in BAL (98% macrophages) from 4 groups of 5 naïve wildtype mice/group were pooled, and 2 × 105 cells were cultured in 24-well plates overnight in RPMI containing 10% FBS and penicillin/streptomycin before stimulating with LPS or bacteria. Phagocytosis assay is described in the supplement.
mRNA Assays
Total RNA was isolated from lungs or macrophages and RT-PCR was performed and quantified as described (41). Unlabeled PCR primers and TaqMan probes (FAM dye-labeled) were used to detect mRNAs for Nos2, Ifng, Ccl2, Ccl3, Ccl5, Il6, Tnf, Ccr2, Arg1, Il10, Mrc1, Clec10a, Retnla, Mmp3, 7, 8, 10, 12, and 28, and Hprt (Applied Biosystems, Foster City CA). The data are expressed as relative quantification, which is the fold change and calculated as 2−ΔΔCt.
Cytokine/Chemokine Assays
Lung samples were homogenized in 2 ml lysis buffer: 150 mM NaCl, 15 mM Tris (pH 7.2), 1 mM MgCl2, 1 mM CaCl2, 1% Triton X-100, and Complete Mini Protease Inhibitor Cocktail (Roche Molecular Biochemicals, Mannheim, Germany). Homogenates were centrifuged at 10,000×g for 15 min at 4°C, and the supernatants were collected. Multiplex reagents were from R&D Systems (Minneapolis MN) including the mouse Fluorokine MAP kit and analytes for IFNγ, IL-1β, IL-6, IL-10, IL-12, IL-17, TNFα, CXCL1, CXCL2, and CCL2.
Microarray Analysis
Total RNA from BMDM was purified using RNeasy Mini-kits (Qiagen, Hilden, Germany). All RNA samples were of high quality as assessed by an Agilent 2100 Bioanalyzer, with RIN ≥ 9.8. Each RNA sample was converted to cDNA, labeled, and hybridized to GeneChip Mouse Gene 2.0 ST arrays (Affymetrix, Santa Clara CA) following the manufacturer’s protocols. The whole genome GeneChip Mouse Gene 2.0 ST is comprised of 35,240 transcripts including 26,515 unique genes. Hybridized arrays were scanned with an Affymetrix Gene Chip 3000 scanner, and image generation and feature extraction were performed using Affymetrix GeneChip Operating Software. All arrays passed the manufacturer’s quality specifications with respect to background and percent present call rates. Gene expression levels were estimated from probe intensities using the robust multi-array analysis method with quantile normalization and background adjustment (42). Differential gene expression between wildtype and Mmp10-null samples at each time point was determined using a Bayesian implementation of the parametric t-test (43) and multiple hypothesis testing was controlled using false discovery rate analysis (q-value) (44). A q-value < 0.01 was used to identify significant differential gene expression. Functional enrichment analysis of differentially expressed genes was performed with WebGestalt program (45) based on multiple publicly available resources including Gene Ontology and Kyoto Encyclopedia of Genes and Genomes. Enrichment p-values were determined using Fisher’s exact test and corrected for multiple testing using Benjamini-Hochberg’s method (46) with a significance adjusted p-value < 0.01. Minimal Information About Microarray Experiments (MIAME)-compliant data have been submitted to the NIH-GEO (accession no. GSE78175; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=epkvmssmvhmrlmf&acc=GSE78175).
Statistics
Statistical analysis was performed using 2-way ANOVA with Bonferroni post-test where appropriate. Data are presented as mean ± SEM, with p ≤ 0.05 considered statistically significant. Mortality data was analyzed by Kaplan-Meier log rank test. Prism 5 software (GraphPad Software, La Jolla CA) was used for all statistical analyses.
Results
MMP10 is Expressed in Infected Lungs
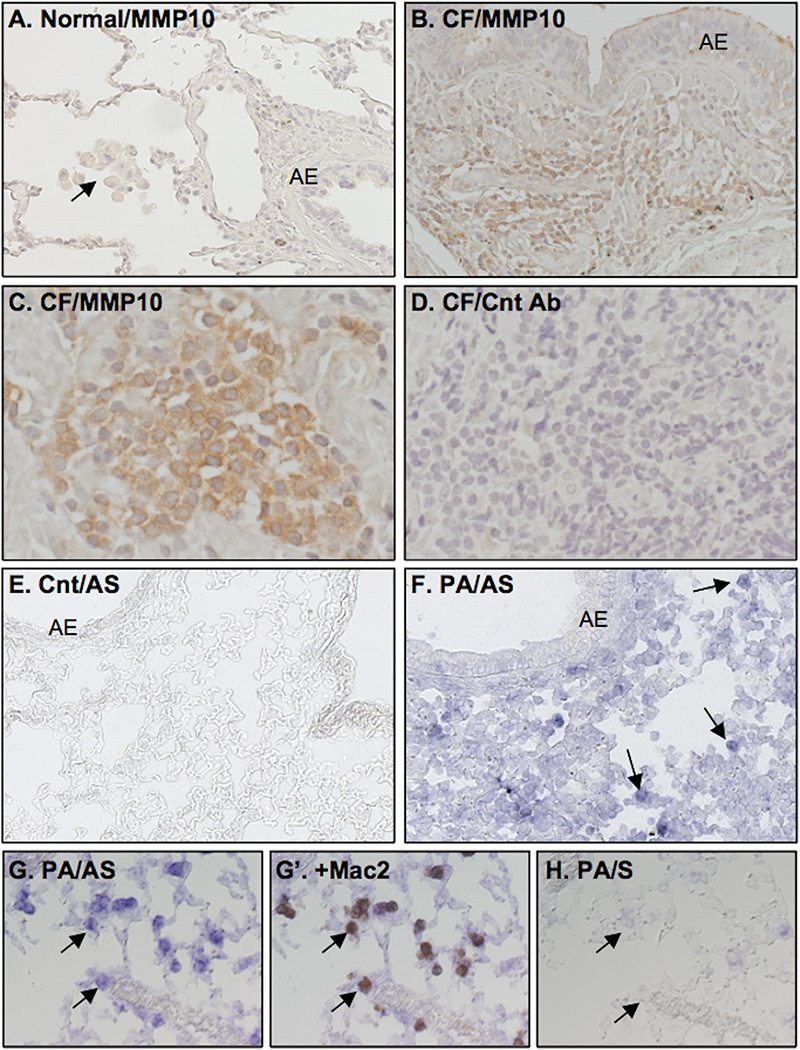
We stained de-identified samples of CF lungs – all colonized with P. aeruginosa – from transplant recipients (n = 5) and normal human lungs (n = 3) for MMP10 protein using a human-specific antibody. In all CF samples, signal for MMP10 was seen in most mononuclear cells (Fig. 1B,C). The large, pale nuclei of these cells and their accumulation within interstitial areas are characteristic features of macrophages. No signal for MMP10 was detected in normal human lung, including resident alveolar macrophages (Fig. 1A).
Figure 1. MMP10 is expressed in the lungs of patients with cystic fibrosis (CF) and frzom mice infected with P. aeruginosa.
(A–C) Sections of normal or CF human lungs were stained for MMP10 protein. (A) MMP10 was not detected in alveolar macrophages (arrow), airway epithelium (AE), or elsewhere in normal human lung. (B) MMP10 protein is expressed strongly in mononuclear cells; weak signal was seen in some AE. (C) Higher magnification shows a nest of mononuclear cells with strong MMP10 expression. (D) No specific signal was seen in CF sections processed with pre-immune serum (Cnt Ab). (E–H) Wildtype mice were infected with P. aeruginosa (PA) by oropharyngeal aspiration and controls (Cnt) received an equal volume of saline. Lungs were harvested 72 h later, and sections were probed for Mmp10 mRNA by hybridization with digoxigenin anti-sense (AS) or sense (S) RNA probes. Images were collected, and sections were stained with Mac-2 antibody. (E) No signal for Mmp10 mRNA was seen in lung sections from control mice. (F) Signal for Mmp10 mRNA was detected in mononuclear cells (arrows) in infected lungs, whereas weak signal was observed in AE cells. Higher magnification images show Mmp10 RNA expression before (G) and after co-staining with Mac-2 antibody (G’). (H) The section serial to that in G was hybridized with sense RNA and showed no signal.
We used in situ hybridization coupled with immunostaining to identify the cell sources of Mmp10 mRNA expression in mouse lung. Similar to human lung, we detected no signal for Mmp10 mRNA in naïve mouse lungs (Fig. 1E). However, at 72 h post-infection with P. aeruginosa, we detected prominent signal for Mmp10 mRNA (Fig. 1F) in cells that stained positive for Mac-2, a pan-macrophage marker (11), demonstrating that all cells strongly positive for Mmp10 mRNA were macrophages (Fig. 1G, G’). No specific signal was seen in sections processed with sense RNA probe (Fig. 1H). These findings demonstrate that MMP10 is not expressed in healthy lung but is induced by macrophages in human and murine lung in response to bacterial infection.
MMP10 Protects against Lethality Induced by Lung Infection
To determine the role of MMP10 in the host response to infection, we exposed wildtype and Mmp10−/− mice to live P. aeruginosa by aerosolization or oropharyngeal aspiration. Unchallenged Mmp10−/− mice develop normally and reveal no overt phenotype in any tissue (33, 35, 38). Consistent with our in situ hybridization data (Fig. 1E), Mmp10 mRNA levels were very low (average Ct = 38) in naïve wildtype lung, indicating that this transcript was not expressed by any resident cell, including alveolar and interstitial macrophages in unchallenged lung. In response to P. aeruginosa infection, expression of Mmp10 mRNA was induced in wildtype lungs, peaked 3 d after infection, and remained elevated at 7 d post infection (Fig. 2A). The pattern and extent of MMP10 expression were equivalent between the two infection protocols and mirrored the 0–24 h induction response we reported in mice infected by nasal inoculation with P. aeruginosa strain PA51673, CF clinical isolate (38). In contrast, expression of other MMPs increased modestly (MMP3, MMP12), if at all (MMP28), or transiently (MMP7, MMP8) (Fig. S1A). MMP8, which is produced by many cell types and is particularly abundant in neutrophils, quickly increased then fell (Fig. S1B), likely reflecting the rapid influx and efflux of neutrophils in this model (11). Thus, among MMPs, MMP10 revealed a potent and sustained response to infection.
Figure 2. MMP10 is induced in P. aeruginosa-infected mouse lung and protects against the lethal effects of lung infection.
Wildtype and Mmp10−/− mice were infected with P. aeruginosa by oropharyngeal aspiration (A,C), or by aerosolization (B, D, E). (A) Wildtype mice were sacrificed at 0 (uninfected), 1, 2, 3, 4, and 7 d post-infection (n = 5 mice/time point). RNA was isolated and processed for RT-PCR for Mmp10 mRNA. Data are presented as relative quantification (RQ). The number above the 3-d time point is the average Ct value for those samples. For the 0-d samples, the Ct > 35. (B) Survival was significantly reduced in Mmp10−/− post infection (n = 11 wildtype, 10 Mmp10−/− mice; Kaplan-Meier log rank test, Χ2 = 5.775, p = 0.0163). (C) Surviving Mmp10−/− (n = 15) mice had delayed recovery of body weight post-infection compared to wildtype mice (n = 13). *p ≤ 0.0288 by 2-way paired ANOVA between genotypes. (D) Clearance of bacteria from lungs did not differ between genotypes. (E) Septicemia as measured by live bacteria counts in spleen also did not differ between genotypes.
Whereas infection with P. aeruginosa led to no mortality in wildtype mice, half of Mmp10−/− mice had died by 48 h post P. aeruginosa exposure (Fig. 2B). Among the survivors, Mmp10−/− mice were more susceptible to infection than wildtype mice as manifested by a reproducible and significant delay in their ability to regain body weight (Fig. 2C). Lethality and delayed weight recovery in Mmp10−/− mice coincided with the marked upswing in Mmp10 expression in wildtype mice (Fig. 2A). In addition, we observed greater mortality (Fig. S1C) and delayed weight gain (data not shown) in Mmp10tm1Lex/Mmucd generated with embryonic stem cells in which the Mmp10 locus was targeted by a gene-trap approach. Thus, the phenotypes we observed in our Mmp10−/− mice were not due to an off-target artifact. These data indicate that MMP10 serves a critical and protective role in the host response to infection.
Lethality and morbidity in Mmp10−/− mice were not due to impaired bacterial clearance. By 48 h, both wildtype and Mmp10−/− mice had effectively cleared the infection (Fig. 2D). In addition, bacteremia, as measured by the recovery of live bacteria from spleen (Fig. 2E) and spleen weights (data not shown) before and during infection did not differ between genotypes. Furthermore, infection-induced increase in lung permeability, a marker of acute lung injury, also did not differ between genotypes (Fig. S1D). We did not see any indication (edema, hemorrhage, inflammation) that other tissues (liver, intestines) were involved (data not shown).
Whereas mortality of Mmp10−/− mice was more pronounced when P. aeruginosa was administered by aerosolization (≥ 50%; Fig. 2B, S1C) compared to oropharyngeal aspiration (≤ 14%; n > 90 mice/genotype from several experiments; data not shown), morbidity as gauged by degree of weight loss and delay in weight recovery was similar by either method of P. aeruginosa infection. Thus, to minimize a potential survival bias, our subsequent in vivo data were obtained with mice infected by oropharyngeal aspiration.
Increased Macrophage Influx in Lungs of Mmp10−/− Mice
Because lethality in Mmp10−/− mice was not associated with impaired bacterial clearance, we predicted that Mmp10−/− mice died due to altered or excessive inflammation. We examined BAL and lung sections at various times post infection. Consistent with the similar rate of bacterial clearance (Fig. 2D), neutrophils – which are critical for clearance of P. aeruginosa – were the predominant cell in BAL post infection (>95% of total cells), and their numbers did not differ between genotypes (Fig. 3B). Lymphocyte numbers, which increased in response to infection, also did not differ significantly between genotypes (data not shown). In contrast, we detected 3-fold more macrophages in BAL (Fig. 3C) at 48 h post-infection, coincident with the upswing in MMP10 expression (Fig. 2A), and even more in tissue sections from Mmp10−/− mice (Fig. 3E). At 4 and 24 h post-infection, before the significant increase in MMP10 expression, the number of macrophages in BAL did not differ between genotypes (Fig. 3C). Flow cytometry of CD45+ cells from non-lavaged lung demonstrated that numbers of alveolar (F4/80+ CD11c+ SiglecF+) and interstitial (F4/80+ CD11b+ CD11c+ MHCII+ SiglecF−) macrophages did not differ between naïve wildtype and Mmp10−/− mice and that the influx of macrophages into lungs of both genotypes was predictably due to infiltrated cells (F4/80+ Ly6C+ CD11b+ CD11c+ MHCII− SiglecF−; data not shown). Similarly, we observed a 2-fold increase in BAL macrophages in Mmp10tm1Lex/Mmucd mice post-infection (data not shown).
Figure 3. Increased macrophage influx in Mmp10−/− lungs.
Mmp10−/− and wildtype mice (n = 6–8/genotype/time point) were infected with P. aeruginosa, and BAL and lung tissue were collected 4 and 48 h later. In BAL samples, total cell (A) and neutrophil (B) counts did not differ between genotypes. (C) Macrophage numbers in BAL did not differ at 4 h post-infection, but at 48 h, about 3-fold more macrophages were detected in Mmp10−/− BAL compared to wildtype. *p = 0.0045 by unpaired t-test. (D) Sections of wildtype (WT) and Mmp10−/− lungs collected at 48 h post-infection were stained for macrophages with Mac-2 antibody.
The number and differential of circulating leukocytes did not differ between wildtype and Mmp10−/− mice (data not shown). In addition, we reported no difference in the ability of wildtype and Mmp10−/− macrophages to migrate toward serum or other chemotactic stimuli (35) indicating that MMP10 does not affect the migratory machinery of macrophages. We also assessed the expression of various chemokines and cytokines that could affect macrophage influx or that have pro-inflammatory activity. We found no differences in the levels for TNFα, CXCL1/2, IL-6, or IL-17 in BAL or lung homogenates (data not shown). In contrast, we found elevated expression of CCL2/MCP1, a potent macrophage chemokine (47, 48), in Mmp10−/− lungs (see Fig. S3A), a finding that was mirrored in isolated macrophages (below).
MMP10 Expression by Macrophages
Macrophages that influx into the lung – or any tissue – in response to infection or injury are distinct from the resident population (49, 50). We found that both resident and recruited macrophages expressed MMP10 at fairly similar levels post-infection (data not shown). We then assessed if macrophages in culture respond as these cells did in vivo. Consistent with our in vivo data, Mmp10 mRNA was not expressed by alveolar macrophages (Fig. 4B) or bone marrow (Fig. 4C) freshly isolated from naïve wildtype mice. Upon CSF-1-mediated differentiation into macrophages (M0), MMP10 was induced in bone marrow-derived macrophages (BMDM) (Fig. S2A, Fig. 4C). MMP10 was not induced by marrow cells cultured with GM-CSF (Fig. S2A). As GM-CSF promotes differentiation into dendritic cells (5), these negative data indicate further that macrophages are the source of MMP10. Expression of MMP10 by both alveolar macrophages and BMDM was stimulated with LPS and even more so by exposure to live P. aeruginosa (Fig. 4B,C) reaching levels that were quite similar between cell types and comparable to the levels detected for Mmp10 mRNA in lung in response to airway infection (see Fig. 2A). Together, these findings indicate that both resident alveolar and infiltrated macrophages express similar levels of MMP10 in response to infection.
Figure 4. MMP10 is expressed by both resident and infiltrated macrophages.
(A) Alveolar macrophages were isolated from BAL of naïve wildtype mice, cultured for 24 h, and exposed to 100 ng/ml LPS for 24 h or to live P. aeruginosa (Pa) at an MOI 5:1 for 1 h then washed and refed as described under Methods. RNA was collected 24 h later, and Mmp10 expression was determined by RT-PCR. The numbers above the bars are the average Ct values. (n = cells from 4 mice/condition) (B) RNA was isolated from fresh wildtype bone marrow (d0), from cells following differentiation to macrophages in culture for 7 d (M0), and from M0 macrophages exposure to LPS or P. aeruginosa. (n = marrow from 3 mice/condition)
Rescue of Mmp10−/− Phenotype
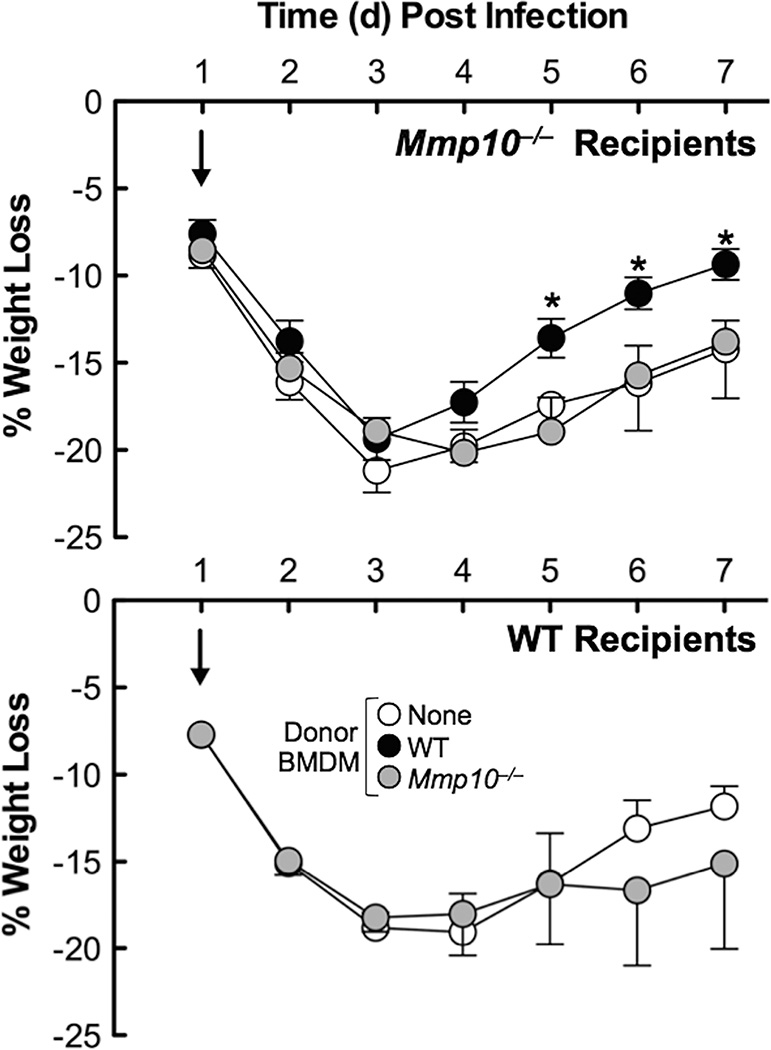
We performed adoptive transfer experiments to determine if wildtype macrophages could ameliorate morbidity in infected Mmp10−/− mice. Indeed, adoptive transfer of wildtype BMDM 24 h after infection with P. aeruginosa resulted in significantly more rapid recovery of weight loss – used as a surrogate for morbidity – comparable to that seen in wildtype mice (Fig. 5). Mmp10−/− BMDM did not affect weight loss in either wildtype or Mmp10−/− recipients (Fig. 5). These findings demonstrate that macrophages are the key cells mediating the MMP10-dependent phenotypes in response to P. aeruginosa lung infection.
Figure 5. Adoptive transfer of wildtype macrophages improves body weight recovery.
Wildtype and Mmp10−/− mice (n = 6–7 mice/group) were exposed to P. aeruginosa by oropharyngeal aspiration and 24 h later were injected retro-orbitally with vehicle (100 µl saline) or with 2 × 106 wildtype or Mmp10−/− BMDM. Mice were weighed and monitored daily for 7 d post infection. *p ≤ 0.03 between genotypes by unpaired t-test.
MMP10 Regulates Macrophage Activation
We assessed if MMP10 influences the state of macrophage activation by measuring the in vivo expression of M1 and M2 markers (5). Compared to levels in wildtype mice, we found elevated expression of mRNAs for the M1 markers IFNγ, iNOS, and CCL2 (Fig. S3A) and reduced expression levels of M2 markers arginase-1 and IL-10 in infected Mmp10−/− lungs (Fig. S3C). In contrast, IL-12a, an M1 marker, was reduced in Mmp10−/− lungs (Fig. S3A), and protein levels of TNFα and IL-1β did not differ between genotypes (Fig. S3B). In addition, as we found in a skin wound model (35), expression of the M2 marker resistin-like α (Retnla, FIZZ1) was not affected by MMP10 (Fig. S3D). Overall, the in vivo phenotypes we saw in Mmp10−/− mice, as summarized in Table I, support the idea that MMP10 mitigates the pro-inflammatory activity of macrophages.
Table I.
Summary of In Vivo Phenotypes
| Process/Factors | Response in Mmp10−/− Mice Relative to Wildtypes |
|
|---|---|---|
| Neutrophil Influx | No Δ | |
| Bacterial Clearance | No Δ | |
| Lethality, Morbidity | ⬆ | |
| Macrophage Influx | ⬆ | |
| M1 Markers | iNOS, IFNγ, CCL21 | ⬆ |
| IL-12a | ⬇ | |
| CCL31, IL-1β2, TNFα2 | No Δ | |
| M2 Markers | IL-10, Arg1 | ⬇ |
| Retnla | No Δ | |
Not necessarily a consensus M1 marker.
Assessed by protein levels in BAL (Fig. 2S)
No Δ: no significance difference between genotypes.
We next assessed if MMP10 plays a cell autonomous role in regulating macrophage polarization. We treated wildtype and Mmp10−/− M0 BMDM with live P. aeruginosa to stimulate M1 differentiation or with IL-4 and IL-13 for M2 differentiation and assessed expression of various markers. Expression of MMP10 by M0 macrophages was stimulated by M1 agonists (Fig. 4C, S2B) but was not further elevated by M2 agonists (Fig. S2B), consistent with findings in human monocytes (51). In M0 and M2-polarized BMDM, we saw no difference in the expression of M1 and M2 markers between wildtype and Mmp10−/− BMDM (Table II, Fig. S4), with one notable exception. IFNγ expression was elevated about 10-fold in Mmp10−/− M0 BMDM compared to wildtype M0 cells (Fig. S4A), suggesting that in the absence of MMP10 macrophages are primed toward a pro-inflammatory phenotype.
Table II.
Relative Expression of Macrophage Activation Markers
| Pathway | Factor | Response in Mmp10−/− Macrophages1 | ||
|---|---|---|---|---|
| M02 | M1 Activated3 | M2 Activated4 | ||
|
M1 Markers or Function |
IFNγ | ⬆ | ⬆ | ⬆ |
| iNOS, CCL5 | nd | ⬆ | nd | |
| CCL2 | No Δ or nd | ⬆ | No Δ | |
| Phagocytosis | ⬆ | |||
| IL-1β, IL-12a | No Δ or nd | ⬇ | No Δ | |
| IL-6, TNFα | No Δ | No Δ | No Δ | |
| M2 Markers | Arg1, CD206, IL-10 | No Δ | ⬇ | No Δ |
| CCR2, Retnla | No Δ | No Δ | No Δ | |
Relative to expression levels in wildtype BMDM. Arrow indicates about a 2-fold or greater difference between genotypes (P < 0.01).
No Δ: no difference between genotypes.
nd: not detected in either genotype.
BMDM 7 d in culture after marrow harvest.
BMDM were exposed to live P. aeruginosa for 1 h, and RNA was isolated 24 h later.
BMDM were treated with IL-4 and IL-13 for 24 h.
We did observe significant MMP10-dependent effects in M1-polarized macrophages (Table II). In particular, the induction of CCL2 in response to P. aeruginosa was markedly greater in Mmp10−/− alveolar macrophages and BMDM compared to wildtype cells (Fig. 6). In addition, expression of mRNAs for the M1 markers iNOS, IFNγ, and CCL5 and the ability to phagocytize E. coli particles – an M1 property – were significantly elevated in M1-polarized Mmp10−/− macrophages (Fig. S4A). Expression of IL-6 did not differ between genotypes (Table I, Fig. S4B). Furthermore, expression of the M2 markers arginase-1 and IL-10 was stimulated in P. aeruginosa-treated BMDM, a response that was significantly diminished in Mmp10−/− BMDM (Fig. S4C). CD206 (mannose receptor) was down-regulated in M1-polarized wildtype BMDM and significantly more so in Mmp10−/− BMDM (Table I, Fig. S4C). The levels of other M2 markers, including CCR2 (Fig. S4D) and resistin-like α (35), did not differ between M1-polarized wildtype and Mmp10−/− BMDM. Collectively, these data indicate that MMP10 functions to moderate several characteristic responses of M1 macrophage activation.
Figure 6. Elevated expression of CCL2 by Mmp10−/− macrophages.
(A) Alveolar macrophages (n = cells from 4 mice/genotype) and (B) BMDM (n = marrow from 8 mice/genotype) from naïve wildtype and Mmp10−/− mice were infected with P. aeruginosa, and expression of CCL2 was assessed by RT-PCR. *p < 0.05 between genotypes by unpaired t-test.
MMP10 Moderates Bacteria-induced Transcriptional Changes in Macrophages
To gain insight into potential mechanisms by which MMP10 influences the macrophage response to bacterial infection, we performed global gene expression analysis. Wildtype and Mmp10−/− BMDM were exposed to P. aeruginosa for 1 h, and cells were harvested at 6 h and 24 h later to assess acute and persistent gene expression changes. We identified many genes differentially expressed (q < 0.01) compared to untreated controls at 6 h after infection in both wildtype and Mmp10−/− BMDM. As expected, P. aeruginosa infection caused a robust transcriptional response in both wildtype (12,672 genes) and Mmp10−/− (15,164 genes) BMDM. The vast majority of differentially expressed genes (n = 10,286) were common between wildtype and Mmp10−/− BMDM (Fig. 7A). Moreover, the direction and pattern of differential expression of the common genes were similar in wildtype and Mmp10−/− cells, indicating that P. aeruginosa infection was the dominant cause of early transcriptional changes (Fig. 7B). Functional enrichment analysis of the smaller subsets of genes that were differentially expressed only in wildtype or Mmp10−/− at 6 h did not reveal coherent phenotypes (data not shown).
Figure 7. Global gene expression analysis of wildtype and Mmp10−/− BMDM in response to P. aeruginosa exposure.
BMDM from wildtype and Mmp10−/− mice were exposed to P. aeruginosa for 1 h at MOI 5:1. Infection medium was removed, and cells were cultured in medium containing antibiotics until cells were harvested for RNA collection 6 or 24 h later. (A) Bacteria exposure induced significant transcriptional changes in both wildtype and Mmp10−/− BMDM 6 h post challenge. (B) Heatmap of the genes commonly differentially expressed in both wildtype and Mmp10−/− BMDM shows similar patterns of up- and down-regulation 6 h post-infection. (C) Of the genes that were commonly differentially expressed at 6 h, many genes remained significantly changed at 24 h post infection exclusively in Mmp10−/− BMDM. (D) Functional analysis of genes exclusively up-regulated in Mmp10−/− at 24 h reveals significant enrichment in immune and inflammatory signaling pathways.
Because gene expression is a temporally dynamic process, we postulated that the role of Mmp10 in regulating transcriptional programs induced by bacterial challenge may be revealed by examining a later time point. Thus, we focused on the large set of common differentially expressed genes at 6 h and assessed if their levels remained significantly altered at 24 h post-infection. Of the 10,286 common genes, 5,609 remained differentially expressed 24 h after infection (Fig. 7C). Notably, we observed that the majority (3,981) of genes that remained differentially expressed at the 24-h time point were unique to Mmp10−/− BMDM, implying that MMP10 plays a distinct role in moderating the post-infection transcriptional response. Of the 3,981 genes that remained differentially expressed only in Mmp10−/− BMDM, 1,386 were up-regulated and 2,505 were down-regulated at 24 h. We applied functional analysis using WebGestalt to identify biological processes enriched within the subset of persistently up-regulated genes (Fig. 7D). We found that the absence of Mmp10 in P. aeruginosa-exposed BMDM was associated with persistent activation of a wide repertoire of pathways mapping to immunity, inflammation, apoptosis, and cell signaling. In contrast, functional analysis of the persistently down-regulated genes showed enrichment of processes involved in metabolism, transcription, and translation.
Discussion
Here, we report on the expression and function of macrophage MMP10 in the host response to infection. Our findings indicate that MMP10 is not expressed in healthy, unchallenged human and mouse lung but is markedly induced in both resident and infiltrated macrophages following infection with P. aeruginosa. With a combination of in vivo and primary macrophage models, we demonstrate that MMP10 protects mice from morbidity and mortality induced by acute P. aeruginosa lung infection. By assessing the expression of various markers and overall gene expression, our findings indicate that MMP10 functions to drive the activation of macrophages from proinflammatory (M1-like) cells into immunosuppressive (M2-like) cells. Our findings complement observations we reported with others showing greater mortality and delayed weight gain in Mmp10−/− mice following acute liver injury (52) and more macrophages biased towards M1 activation and more tissue injury in Mmp10−/− mice subjected to acute colon injury (33). Furthermore, Mmp10−/− mice are much more susceptible to the lethal effects of acute lung injury caused by bleomycin toxicity than are wildtype animals (unpublished observations). Thus, across organ systems, MMP10 appears to play a beneficial role in mitigating the deleterious responses to injury and infection by promoting the conversion of pro-inflammatory macrophages toward immunosuppressive cells. Our findings support the general concept that in response to acute injury or infection macrophage activation evolves from potentially detrimental M1-based cells to suppressive M2-biased macrophages (6, 39, 50).
Although more macrophages emigrated into Mmp10−/− lungs in response to infection, we found no difference in the ability of wildtype and Mmp10−/− macrophages to migrate toward serum or wound-tissue homogenate (35) indicating that MMP10 does not affect the migratory machinery of macrophages. With others (53), we reported that Mmp10−/− macrophages have an impaired ability to migrate over fibronectin and to invade into Matrigel. Although these findings are seemingly opposed to our in vivo observations, there are two important caveats with these in vitro studies. First, it is not clear if macrophages migrate over fibronectin on their way into and through the interstitial space. Although fibronectin would be present in the area, it appears that other matrix components, particularly versican and hyaluronan (54), serve as the interstitial substrata that macrophages follow as they migrate through tissue (55, 56). Second, Matrigel is a highly dense material that does not mirror the more porous architecture of interstitial matrix (57, 58). In fact, Matrigel does not contain interstitial matrix components; it is comprised of misassembled basement membrane components (57). Thus, unlike how they move through the interstitium in vivo, macrophages may require proteases to burrow through a thick plug of Matrigel in a culture dish. As we reported that MMP10 activates matrix-remodeling programs in macrophages (35, 59), Mmp10−/− cells may not possess the enzymes needed to invade through the artificial setting of Matrigel. We conclude that MMP10 does not affect macrophage migration per se in physiologic settings.
An open question is if increased morbidity and mortality in Mmp10−/− animals was due to more macrophage numbers, to a bias toward M1 activation, or a combination of these two processes. We propose that altered activation of both resident and infiltrated macrophages towards an M1, pro-inflammatory state led to worse outcomes for infected Mmp10−/− mice and this deleterious effect was augmented by the presence of more M1-biased cells. Consistent with this conclusion, Johnston et al. reported that the macrophage phenotypes begins to evolve toward an M2 state about 48 h after lung infection with P. aeruginosa (39), and D’Alessio et al. recently demonstrated that therapeutically promoting macrophage differentiation toward M2 cells (via instillation of IL-4) markedly reduces lethality and morbidity in response to acute P. aeruginosa infection (60). In Mmp10−/− mice and macrophages, we found increased expression levels of M1 markers and decreased levels of M2 markers (Table I, II), such as the increased and prolonged expression of CCL2 (MCP-1), which was also seen in injured Mmp10−/− colons (33). CCL2, a potent macrophage chemokine, is expressed predominantly by resident lung macrophages (47, 48), and over-expression of this factor may account for the excess macrophage inflammation seen in Mmp10−/− mice. The diminished expression of IL-10 and Arg-1, both M2 markers, in infected Mmp10−/− lungs and BMDM provides further evidence that MMP10 broadly affects macrophage polarity to moderate inflammation.
Neutrophil influx, which accounted for about 95% of the cells recovered from BAL, and the levels of CXCL1 and CXCL2, critical neutrophil chemokines, did not differ between wildtype and Mmp10−/− mice in response to infection. Similarly, Koller et al. found no difference in neutrophil numbers between wildtype and Mmp10−/− mice over the first 14 days post colon injury (33). However, they did find that elevated neutrophil counts persisted in Mmp10−/− mice at 21 days post-injury, at which time their numbers had dropped in wildtype colons. Although sustained neutrophil influx may have been secondary to worse injury of Mmp10−/− colons, it is also possible that a bias toward M1 macrophages maintained neutrophil influx that, in turn, led to more tissue damage. In our lung infection model, the lack of difference in neutrophil numbers and indices of acute lung injury between genotypes indicates that neutrophils did not contribute to worse outcomes in Mmp10−/− mice. A potential caveat to this conclusion is if MMP10 influences the state of neutrophil activation. For example, in other studies, we found that MMP7 shedding of syndecan-1/CXCL1 complexes from injured lung epithelium permits both neutrophil migration through the mucosal barrier and their subsequent activation (20, 61). Without this processing mechanism, Mmp7−/− mice are markedly protected from the lethal effects of neutrophil-mediated lethality in response to acute lung injury (20). However, we propose that the similar rate of bacterial clearance in wildtype and Mmp10−/− mice does not support an MMP10-dependent role in neutrophil activation.
While MMP10 can also be expressed by injured/infected epithelium, including in lung (38 and unpublished observations), we do not think that the enzyme produced by these cells contributed to the key phenotypes and responses we report here. Indeed, adoptive transfer of wildtype macrophages into Mmp10−/− recipients reduced post-infection morbidity to levels seen in wildtypes. Similarly, using bone marrow transplantation, Koller et al. concluded that the lack of MMP10 in macrophages led to worse tissue damage in a model of acute colon injury (33). Furthermore, the macrophage polarization changes we observed in vivo (Table I) were largely duplicated with isolated macrophages in culture (Table II). These findings support the conclusion that MMP10 is a cell autonomous regulator of macrophage activation. In contrast, the function of epithelial MMP10 remains unanswered. In skin wound models, we found no re-epithelialization phenotype in Mmp10−/− mice (35) or in mice over-expressing active MMP-10 in basal keratinocytes (32).
We recently contributed to a multi-center genome-wide association study that identified MMP10 as a candidate gene for chronic obstructive pulmonary disease (59). Using a model of chronic (6 mo) exposure to cigarette smoke, we found that Mmp10−/− mice are fully resistant to the development of emphysema. In addition, MMP10 is produced by macrophages from human smokers with emphysema (62) and is one of two genes whose expression correlates with reduced lung function (i.e., FEV1) in smokers (63). These findings indicate that macrophage MMP10 contributes to disease progression in emphysema, which is seemingly opposed to the protective role for this MMP in acute infection we reported here. However, there are important differences between these models, especially with respect to macrophage biology. Macrophages that function early in inflammation are functionally distinct from those that function late in inflammation or in a persistent inflammatory response (4, 6, 50, 64, 65), and this concept has been demonstrated in models of liver (66), vascular (67), cardiac (68), and kidney disease (69). In addition, whereas acute infection and injury bias toward an M1 phenotype (4), cigarette smoke promotes expansion of M2 macrophages (70). Macrophages are considered to be the destructive cell in emphysema (71, 72), and our studies in skin wounds indicate that MMP10 promotes expression of matrix-degrading proteinases in M2 macrophages (35). Thus, in an acute setting, MMP10 is beneficial by moderating the pro-inflammatory activity of M1-biased macrophages and by stimulating the ability of M2-biased macrophages to remodel scar tissue. But in a chronic setting, MMP10-driven matrix remodeling could be excessive and detrimental, as suggested in our smoke-exposure studies. Still, an important and shared conclusion among these models is that MMP10 functions to control macrophage behavior.
Significant differences in expression of M1 and M2 markers were only observed between infected (M1-biased) wildtype and Mmp10−/− macrophages but not between M0 or M2 cells (Table II). Consistent with these data, transcriptomic analysis did not reveal significant enrichment of any inflammation-related pathways in genes differentially expressed between M0 wildtype and Mmp10−/− BMDM (data not shown). Thus, the effects of MMP10, at least in terms of polarization, are more significant in M1-activated macrophages. However, MMP10 does shape some functional properties of M2-biased macrophages, such as their ability to clear collagen (35). The P. aeruginosa-induced transcriptional changes between wildtype and Mmp10−/− BMDM were quite similar at 6 h post infection but diverged significantly at 24 h. Nearly 4,000 of the genes that were differentially expressed in both genotypes at 6 h remained significantly changed only in Mmp10−/− BMDM at 24 h. These data indicate that MMP10 functions to broadly moderate or quench the temporal transcriptional response induced by bacteria in infiltrating macrophages. The genes that remained persistently up-regulated only in Mmp10−/− macrophages at 24 h were significantly enriched for inflammation and immune-related pathways, consistent with our observations in the infected Mmp10−/− lung.
In summary, our data demonstrate a novel role for MMP10 in moderating lung inflammation by altering the balance of M1/M2 transcripts and broadly suppressing immune-related pathways in macrophages. Importantly, the patterns of MMP10 expression are conserved between human and mouse lung and macrophages. The critical functions of MMP10 in lung inflammation are largely attributable to infiltrated macrophages and may therefore be relevant in other organs and inflammatory conditions. Given the importance of macrophage polarization in the initiation and resolution of inflammation, further study of MMP10 function is warranted in other disease models, especially those involving bacterial infection. Though beyond the scope of this study, identification of the physiologic substrates cleaved by MMP10 will be essential to determine the mechanisms by which MMP10 regulates macrophage polarization. Schlage et al. have identified multiple MMP10 substrates in primary mouse fibroblasts (73), but we predict that macrophage-derived MMP10 may cleave a limited and distinct set of substrates, some of which may only be expressed in an inflamed microenvironment. Determining the relevant MMP10 cleavage events in the lung is a significant challenge but a priority for our future work.
Supplementary Material
Acknowledgments
We thank Drs. Ann Chen and Gail Deutsch for help acquiring human lung specimens, Ying Wang for maintaining mouse strains, Dr. Elaine Raines for providing Mac-2 antibody, and Fred Farin, Theo Bammler, and Jesse Thai for assistance with microarray experimental design and processing.
This work was supported by NIH grants HL089455 (WCP), HL098067 (WCP), HL093022 (JKM), and HL116514 (AMM).
Abbreviations used in this paper
- MMP
matrix metalloproteinase
- BMDM
bone marrow-derived macrophage
- BAL
bronchoalveolar lavage
- iNOS
nitric oxide synthase 2
- CF
cystic fibrosis
References
- 1.Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nat Rev Immunol. 2005;5:953–964. doi: 10.1038/nri1733. [DOI] [PubMed] [Google Scholar]
- 2.Mantovani A, Sica A, Locati M. Macrophage polarization comes of age. Immunity. 2005;23:344–346. doi: 10.1016/j.immuni.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 3.Gordon S. Alternative activation of macrophages. Nat Rev Immunol. 2003;3:23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- 4.Benoit M, Desnues B, Mege JL. Macrophage polarization in bacterial infections. J Immunol. 2008;181:3733–3739. doi: 10.4049/jimmunol.181.6.3733. [DOI] [PubMed] [Google Scholar]
- 5.Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, Goerdt S, Gordon S, Hamilton JA, Ivashkiv LB, Lawrence T, Locati M, Mantovani A, Martinez FO, Mege JL, Mosser DM, Natoli G, Saeij JP, Schultze JL, Shirey KA, Sica A, Suttles J, Udalova I, van Ginderachter JA, Vogel SN, Wynn TA. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41:14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lichtnekert J, Kawakami T, Parks WC, Duffield JS. Changes in macrophage phenotype as the immune response evolves. Curr Opin Pharmacol. 2013;13:555–564. doi: 10.1016/j.coph.2013.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kratz M, Coats BR, Hisert KB, Hagman D, Mutskov V, Peris E, Schoenfelt KQ, Kuzma JN, Larson I, Billing PS, Landerholm RW, Crouthamel M, Gozal D, Hwang S, Singh PK, Becker L. Metabolic dysfunction drives a mechanistically distinct proinflammatory phenotype in adipose tissue macrophages. Cell Metab. 2014;20:614–625. doi: 10.1016/j.cmet.2014.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Becker L, Gharib SA, Irwin AD, Wijsman E, Vaisar T, Oram JF, Heinecke JW. A macrophage sterol-responsive network linked to atherogenesis. Cell Metab. 2010;11:125–135. doi: 10.1016/j.cmet.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Becker L, Liu NC, Averill MM, Yuan W, Pamir N, Peng Y, Irwin AD, Fu X, Bornfeldt KE, Heinecke JW. Unique proteomic signatures distinguish macrophages and dendritic cells. PLoS One. 2012;7:e33297. doi: 10.1371/journal.pone.0033297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hautamaki RD, Kobayashi DK, Senior RM, Shapiro SD. Requirement for macrophage elastase for cigarette smoke-induced emphysema. Science. 1997;277:2002–2004. doi: 10.1126/science.277.5334.2002. [DOI] [PubMed] [Google Scholar]
- 11.Manicone AM, Birkland TP, Lin M, Betsuyaku T, van Rooijen N, Lohi J, Keski-Oja J, Wang Y, Skerrett SJ, Parks WC. Epilysin (MMP-28) restrains early macrophage recruitment in Pseudomonas aeruginosa pneumonia. J Immunol. 2009;182:3866–3876. doi: 10.4049/jimmunol.0713949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wen G, Zhang C, Chen Q, Luong le A, Mustafa A, Ye S, Xiao Q. A novel role of matrix metalloproteinase-8 in macrophage differentiation and polarization. J Biol Chem. 2015;290:19158–19172. doi: 10.1074/jbc.M114.634022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gharib SA, Johnston LK, Huizar I, Birkland TP, Hanson J, Wang Y, Parks WC, Manicone AM. MMP28 promotes macrophage polarization toward M2 cells and augments pulmonary fibrosis. J Leukoc Biol. 2014;95:9–18. doi: 10.1189/jlb.1112587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gill SE, Gharib SA, Bench EM, Sussman SW, Wang RT, Rims C, Birkland TP, Wang Y, Manicone AM, McGuire JK, Parks WC. Tissue inhibitor of metalloproteinases-3 moderates the proinflammatory status of macrophages. Am J Respir Cell Mol Biol. 2013;49:768–777. doi: 10.1165/rcmb.2012-0377OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Apte SS, Parks WC. Metalloproteinases: A parade of functions in matrix biology and an outlook for the future. Matrix Biol. 2015;44–46:1–6. doi: 10.1016/j.matbio.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 16.Houghton AM. Matrix metalloproteinases in destructive lung disease. Matrix Biol. 2015;44–46:164–174. doi: 10.1016/j.matbio.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 17.Rohani MG, Parks WC. Matrix remodeling by MMPs during wound repair. Matrix Biol. 2015;44–46:113–121. doi: 10.1016/j.matbio.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 18.Wilson CL, Ouellette AJ, Satchell DP, Ayabe T, López-Boado YS, Stratman JL, Hultgren SJ, Matrisian LM, Parks WC. Regulation of intestinal a-defensin activation by the metalloproteinase matrilysin in innate host defense. Science. 1999;286:113–117. doi: 10.1126/science.286.5437.113. [DOI] [PubMed] [Google Scholar]
- 19.McQuibban GA, Gong JH, Tam EM, McCulloch CA, Clark-Lewis I, Overall CM. Inflammation dampened by gelatinase A cleavage of monocyte chemoattractant protein-3. Science. 2000;289:1202–1206. doi: 10.1126/science.289.5482.1202. [DOI] [PubMed] [Google Scholar]
- 20.Li Q, Park PW, Wilson CL, Parks WC. Matrilysin shedding of syndecan-1 regulates chemokine mobilization and transepithelial efflux of neutrophils in acute lung injury. Cell. 2002;111:635–646. doi: 10.1016/s0092-8674(02)01079-6. [DOI] [PubMed] [Google Scholar]
- 21.Parks WC, Wilson CL, Lopez-Boado YS. Matrix metalloproteinases as modulators of inflammation and innate immunity. Nat Rev Immunol. 2004;4:617–629. doi: 10.1038/nri1418. [DOI] [PubMed] [Google Scholar]
- 22.Cauwe B, Van den Steen PE, Opdenakker G. The biochemical, biological, and pathological kaleidoscope of cell surface substrates processed by matrix metalloproteinases. Crit. Rev. Biochem. Mol. Biol. 2007;42:113–185. doi: 10.1080/10409230701340019. [DOI] [PubMed] [Google Scholar]
- 23.Greenlee KJ, Werb Z, Kheradmand F. Matrix metalloproteinases in lung: multiple, multifarious, and multifaceted. Physiol Rev. 2007;87:69–98. doi: 10.1152/physrev.00022.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Lint P, Libert C. Chemokine and cytokine processing by matrix metalloproteinases and its effect on leukocyte migration and inflammation. J Leukoc Biol. 2007;82:1375–1381. doi: 10.1189/jlb.0607338. [DOI] [PubMed] [Google Scholar]
- 25.Chen P, Abacherli LE, Nadler ST, Wang Y, Li Q, Parks WC. MMP7 shedding of syndecan-1 facilitates re-epithelialization by affecting alpha(2)beta(1) integrin activation. PLoS ONE. 2009;4:e6565. doi: 10.1371/journal.pone.0006565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nuttall RK, Sampieri CL, Pennington CJ, Gill SE, Schultz GA, Edwards DR. Expression analysis of the entire MMP and TIMP gene families during mouse tissue development. FEBS Lett. 2004;563:129–134. doi: 10.1016/S0014-5793(04)00281-9. [DOI] [PubMed] [Google Scholar]
- 27.Madlener M, Werner S. cDNA cloning and expression of the gene encoding murine stromelysin-2 (MMP-10) Gene. 1997;202:75–81. doi: 10.1016/s0378-1119(97)00456-3. [DOI] [PubMed] [Google Scholar]
- 28.Vaalamo M, Karjalainen-Lindsberg ML, Puolakkainen P, Kere J, Saarialho-Kere U. Distinct expression profiles of stromelysin-2 (MMP-10), collagenase-3 (MMP-13), macrophage metalloelastase (MMP-12), and tissue inhibitor of metalloproteinases-3 (TIMP-3) in intestinal ulcerations. Am J Pathol. 1998;152:1005–1014. [PMC free article] [PubMed] [Google Scholar]
- 29.Saghizadeh M, Brown DJ, Castellon R, Chwa M, Huang GH, Ljubimova JY, Rosenberg S, Spirin KS, Stolitenko RB, Adachi W, Kinoshita S, Murphy G, Windsor LJ, Kenney MC, Ljubimov AV. Overexpression of matrix metalloproteinase-10 and matrix metalloproteinase-3 in human diabetic corneas: a possible mechanism of basement membrane and integrin alterations. Am J Pathol. 2001;158:723–734. doi: 10.1016/S0002-9440(10)64015-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gill JH, Kirwan IG, Seargent JM, Martin SW, Tijani S, Anikin VA, Mearns AJ, Bibby MC, Anthoney A, Loadman PM. MMP-10 is overexpressed, proteolytically active, and a potential target for therapeutic intervention in human lung carcinomas. Neoplasia. 2004;6:777–785. doi: 10.1593/neo.04283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bodey B, Bodey B, Jr, Siegel SE, Kaiser HE. Immunocytochemical detection of MMP-3 and-10 expression in hepatocellular carcinomas. Anticancer Res. 2000;20:4585–4590. [PubMed] [Google Scholar]
- 32.Krampert M, Bloch W, Sasaki T, Bugnon P, Rulicke T, Wolf E, Aumailley M, Parks WC, Werner S. Activities of the matrix metalloproteinase stromelysin-2 (MMP-10) in matrix degradation and keratinocyte organization in wounded skin. Mol Biol Cell. 2004;15:5242–5254. doi: 10.1091/mbc.E04-02-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koller FL, Dozier EA, Nam KT, Swee M, Birkland TP, Parks WC, Fingleton B. Lack of MMP10 exacerbates experimental colitis and promotes development of inflammation-associated colonic dysplasia. Lab Invest. 2012;92:1749–1759. doi: 10.1038/labinvest.2012.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jenner RG, Young RA. Insights into host responses against pathogens from transcriptional profiling. Nat Rev Microbiol. 2005;3:281–294. doi: 10.1038/nrmicro1126. [DOI] [PubMed] [Google Scholar]
- 35.Rohani MG, McMahan RS, Razumova MV, Hertz AL, Cieslewicz M, Pun SH, Regnier M, Wang Y, Birkland TP, Parks WC. MMP-10 regulates collagenolytic activity of alternatively activated resident macrophages. J Invest Dermatol. 2015;135:2377–2384. doi: 10.1038/jid.2015.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richards MJ, Edwards JR, Culver DH, Gaynes RP. Nosocomial infections in medical intensive care units in the United States. National Nosocomial Infections Surveillance System. Crit Care Med. 1999;27:887–892. doi: 10.1097/00003246-199905000-00020. [DOI] [PubMed] [Google Scholar]
- 37.Gibson RL, Burns JL, Ramsey BW. Pathophysiology and management of pulmonary infections in cystic fibrosis. Am J Respir Crit Care Med. 2003;168:918–951. doi: 10.1164/rccm.200304-505SO. [DOI] [PubMed] [Google Scholar]
- 38.Kassim SY, Gharib SA, Mecham BH, Birkland TP, Parks WC, McGuire JK. Individual matrix metalloproteinases control distinct transcriptional responses in airway epithelial cells infected with Pseudomonas aeruginosa. Infect Immun. 2007;75:5640–5650. doi: 10.1128/IAI.00799-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnston LK, Rims CR, Gill SE, McGuire JK, Manicone AM. Pulmonary macrophage subpopulations in the induction and resolution of acute lung injury. Am J Respir Cell Mol Biol. 2012;47:417–426. doi: 10.1165/rcmb.2012-0090OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saarialho-Kere UK, Kovacs SO, Pentland AP, Parks WC, Welgus HG. Distinct populations of keratinocytes express stromelysin-1 and-2 in chronic wounds. J Clin. Invest. 1994;94:79–88. doi: 10.1172/JCI117351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swee M, Wilson CL, Wang Y, McGuire JK, Parks WC. Matrix metalloproteinase-7 (matrilysin) controls neutrophil egress by generating chemokine gradients. J Leukoc Biol. 2008;83:1404–1412. doi: 10.1189/jlb.0108016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 43.Kayala MA, Baldi P. Cyber-T web server: differential analysis of high-throughput data. Nucleic Acids Res. 2012;40:W553–559. doi: 10.1093/nar/gks420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Storey JD, Tibshirani R. Statistical methods for identifying differentially expressed genes in DNA microarrays. Methods Mol Biol. 2003;224:149–157. doi: 10.1385/1-59259-364-X:149. [DOI] [PubMed] [Google Scholar]
- 45.Wang J, Duncan D, Shi Z, Zhang B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 2013;41:W77–W83. doi: 10.1093/nar/gkt439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19:368–375. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- 47.Belperio JA, Keane MP, Burdick MD, Lynch JP, 3rd, Xue YY, Berlin A, Ross DJ, Kunkel SL, Charo IF, Strieter RM. Critical role for the chemokine MCP-1/CCR2 in the pathogenesis of bronchiolitis obliterans syndrome. J Clin Invest. 2001;108:547–556. doi: 10.1172/JCI12214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sakanashi Y, Takeya M, Yoshimura T, Feng L, Morioka T, Takahashi K. Kinetics of macrophage subpopulations and expression of monocyte chemoattractant protein-1 (MCP-1) in bleomycin-induced lung injury of rats studied by a novel monoclonal antibody against rat MCP-1. J Leukoc Biol. 1994;56:741–750. doi: 10.1002/jlb.56.6.741. [DOI] [PubMed] [Google Scholar]
- 49.Laskin DL, Weinberger B, Laskin JD. Functional heterogeneity in liver and lung macrophages. J Leukoc Biol. 2001;70:163–170. [PubMed] [Google Scholar]
- 50.Aggarwal NR, King LS, D'Alessio FR. Diverse macrophage populations mediate acute lung inflammation and resolution. Am J Physiol Lung Cell Mol Physiol. 2014;306:L709–725. doi: 10.1152/ajplung.00341.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roch T, Akymenko O, Kruger A, Jung F, Ma N, Lendlein A. Expression pattern analysis and activity determination of matrix metalloproteinase derived from human macrophage subsets. Clin Hemorheol Microcirc. 2014;58:147–158. doi: 10.3233/CH-141885. [DOI] [PubMed] [Google Scholar]
- 52.Garcia-Irigoyen O, Carotti S, Latasa MU, Uriarte I, Fernandez-Barrena MG, Elizalde M, Urtasun R, Vespasiani-Gentilucci U, Morini S, Banales JM, Parks WC, Rodriguez JA, Orbe J, Prieto J, Paramo JA, Berasain C, Avila MA. Matrix metalloproteinase-10 expression is induced during hepatic injury and plays a fundamental role in liver tissue repair. Liver Int. 2014;34:e257–e270. doi: 10.1111/liv.12337. [DOI] [PubMed] [Google Scholar]
- 53.Murray MY, Birkland TP, Howe JD, Rowan AD, Fidock M, Parks WC, Gavrilovic J. Macrophage migration and invasion is regulated by MMP10 expression. PLoS One. 2013;8:e63555. doi: 10.1371/journal.pone.0063555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chang MY, Tanino Y, Vidova V, Kinsella MG, Chan CK, Johnson PY, Wight TN, Frevert CW. A rapid increase in macrophage-derived versican and hyaluronan in infectious lung disease. Matrix Biol. 2014;34:1–12. doi: 10.1016/j.matbio.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de La Motte CA, Hascall VC, Calabro A, Yen-Lieberman B, Strong SA. Mononuclear leukocytes preferentially bind via CD44 to hyaluronan on human intestinal mucosal smooth muscle cells after virus infection or treatment with poly(I.C) J Biol Chem. 1999;274:30747–30755. doi: 10.1074/jbc.274.43.30747. [DOI] [PubMed] [Google Scholar]
- 56.de la Motte CA, Hascall VC, Drazba J, Bandyopadhyay SK, Strong SA. Mononuclear leukocytes bind to specific hyaluronan structures on colon mucosal smooth muscle cells treated with polyinosinic acid:polycytidylic acid: inter-alpha-trypsin inhibitor is crucial to structure and function. Am J Pathol. 2003;163:121–133. doi: 10.1016/s0002-9440(10)63636-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rowe RG, Weiss SJ. Breaching the basement membrane: who, when and how? Trends in cell biology. 2008;18:560–574. doi: 10.1016/j.tcb.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 58.Sabeh F, Shimizu-Hirota R, Weiss SJ. Protease-dependent versus - independent cancer cell invasion programs: three-dimensional amoeboid movement revisited. J Cell Biol. 2009;185:11–19. doi: 10.1083/jcb.200807195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gharib SA, Loth DW, Soler Artigas M, Birkland TP, Wilk JB, Wain LV, Brody JA, Obeidat M, Hancock DB, Tang W, Rawal R, Boezen HM, Imboden M, Huffman JE, Lahousse L, Alves AC, Manichaikul A, Hui J, Morrison AC, Ramasamy A, Smith AV, Gudnason V, Surakka I, Vitart V, Evans DM, Strachan DP, Deary IJ, Hofman A, Glaser S, Wilson JF, North KE, Zhao JH, Heckbert SR, Jarvis DL, Probst-Hensch N, Schulz H, Barr RG, Jarvelin MR, O'Connor GT, Kahonen M, Cassano PA, Hysi PG, Dupuis J, Hayward C, Psaty BM, Hall IP, Parks WC, Tobin MD, London SJ, Consortium C, SpiroMeta C. Integrative pathway genomics of lung function and airflow obstruction. Hum Mol Genet. 2015;24:6836–6848. doi: 10.1093/hmg/ddv378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.D'Alessio FR, Craig JM, Singer BD, Files DC, Mock JR, Garibaldi BT, Fallica J, Tripathi A, Mandke P, Gans JH, Limjunyawong N, Sidhaye VK, Heller NM, Mitzner W, King LS, Aggarwal NR. Enhanced resolution of experimental ARDS through IL-4-mediated lung macrophage reprogramming. Am J Physiol Lung Cell Mol Physiol. 2016 doi: 10.1152/ajplung.00419.2015. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gill SE, Nadler ST, Li Q, Frevert CW, Park PW, Chen P, Parks WC. Shedding of syndecan-1/CXCL1 complexes by MMP7 functions as an epithelial checkpoint of neutrophil activation. Am J Respir Cell Mol Biol. 2016 doi: 10.1165/rcmb.2015-0193OC. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kaner R, Santiago F, Crystal R. Up-regulation of alveolar macrophage matrix metalloproteinases in HIV1+ smokers with early emphysema. J Leukoc Biol. 2009;86:913–922. doi: 10.1189/jlb.0408240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gosselink JV, Hayashi S, Elliott WM, Xing L, Chan B, Yang L, Wright C, Sin D, Pare PD, Pierce JA, Pierce RA, Patterson A, Cooper J, Hogg JC. Differential expression of tissue repair genes in the pathogenesis of COPD. Am J Respir Crit Care Med. 2010;181:1329–1335. doi: 10.1164/rccm.200812-1902OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Porcheray F, Viaud S, Rimaniol AC, Leone C, Samah B, Dereuddre-Bosquet N, Dormont D, Gras G. Macrophage activation switching: an asset for the resolution of inflammation. Clin Exp Immunol. 2005;142:481–489. doi: 10.1111/j.1365-2249.2005.02934.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stout RD, Jiang C, Matta B, Tietzel I, Watkins SK, Suttles J. Macrophages sequentially change their functional phenotype in response to changes in microenvironmental influences. J Immunol. 2005;175:342–349. doi: 10.4049/jimmunol.175.1.342. [DOI] [PubMed] [Google Scholar]
- 66.Duffield JS, Forbes SJ, Constandinou CM, Clay S, Partolina M, Vuthoori S, Wu S, Lang R, Iredale JP. Selective depletion of macrophages reveals distinct, opposing roles during liver injury and repair. J Clin Invest. 2005;115:56–65. doi: 10.1172/JCI22675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tabas I, Bornfeldt KE. Macrophage phenotype and function in different stages of atherosclerosis. Circ Res. 2016;118:653–667. doi: 10.1161/CIRCRESAHA.115.306256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nahrendorf M, Swirski FK, Aikawa E, Stangenberg L, Wurdinger T, Figueiredo JL, Libby P, Weissleder R, Pittet MJ. The healing myocardium sequentially mobilizes two monocyte subsets with divergent and complementary functions. J Exp Med. 2007;204:3037–3047. doi: 10.1084/jem.20070885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ricardo SD, van Goor H, Eddy AA. Macrophage diversity in renal injury and repair. J Clin Invest. 2008;118:3522–3530. doi: 10.1172/JCI36150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shaykhiev R, Krause A, Salit J, Strulovici-Barel Y, Harvey B-G, O'Connor TP, Crystal RG. Smoking-dependent reprogramming of alveolar macrophage polarization: implication for pathogenesis of chronic obstructive pulmonary disease. J Immunol. 2009;183:2867–2883. doi: 10.4049/jimmunol.0900473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shapiro SD. The macrophage in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 1999;160:S29–S32. doi: 10.1164/ajrccm.160.supplement_1.9. [DOI] [PubMed] [Google Scholar]
- 72.Tetley TD. Macrophages and the pathogenesis of COPD. Chest. 2002;121:156S–159S. doi: 10.1378/chest.121.5_suppl.156s. [DOI] [PubMed] [Google Scholar]
- 73.Schlage P, Egli FE, Nanni P, Wang LW, Kizhakkedathu JN, Apte SS, auf dem Keller U. Time-resolved analysis of the matrix metalloproteinase 10 substrate degradome. Mol Cell Proteomics. 2014;13:580–593. doi: 10.1074/mcp.M113.035139. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.