Abstract
Type I interferon (IFN) induction is critical for anti-viral and anti-cancer defenses. Proper down-regulation of type I IFN is equally important to avoid deleterious imbalances in the immune response. The cellular FLIP long isoform protein (cFLIPL) controls type I IFN production, but opposing publications show it as either an inhibitor or inducer of type I IFN synthesis. Regardless, the mechanistic basis for cFLIPL regulation is unknown. Because cFLIPL is important in immune cell development and proliferation, and is a target for cancer therapies, it is important to identify how cFLIPL regulates type I IFN production. Data here show that cFLIPL inhibits IRF3, a transcription factor central for IFNβ and ISG expression. This inhibition occurs during virus infection, cellular exposure to poly I:C or TBK1 over-expression. This inhibition is independent of capase-8 activity. cFLIPL binds to IRF3 and disrupts IRF3 interaction with its IFNβ promoter and its co-activator protein (CBP). Mutational analyses reveal that cFLIPL nuclear localization is necessary and sufficient for inhibitory function. This suggests that nuclear cFLIPL prevents IRF3 enhanceosome formation. Unlike other cellular IRF3 inhibitors, cFLIPL did not degrade or dephosphorylate IRF3. Thus, cFLIPL represents a different cellular strategy to inhibit type I IFN production. This new cFLIPL function must be considered to accurately understand how cFLIPL affects immune system development and regulation.
Keywords: IRF3, cFLIP, type I IFN regulation
Introduction
Type I IFNs (IFNα, IFNsβ) are very important cytokines for human health. They are produced in response to virus infection (1). The administration of type I IFN inhibits tumor growth in experimental animals and in some human tumors (2). However, this same immune response must be carefully controlled. For example, increased type I IFN levels cause disease symptoms associated with autoimmunity (3). Thus, there is a cellular regulatory network that precisely modulates type I IFN production to avoid deleterious imbalances in the immune response.
The cellular proteins that control type I IFN production are well-known (1, 4, 5). This includes apical cellular molecules (e.g., MAVS, TLR3) that recognize microbial PAMPs or cancer DNA, and intermediate signaling molecules (e.g. TBK1, IKKε) that directly stimulate IRF3 via phosphorylation. IRF3 then dimerizes, and translocates to the nucleus. IRF3 forms an enhanceosome (6–8), where IRF3 binds to specific recognition sequences in promoters for genes encoding IFNβ and interferon-stimulated genes (ISGs)(6, 7, 9, 10). Importantly, cofactors like CREB-binding protein (CBP) must also be recruited to IRF3 for effective IRF3-controlled transcription (7).
Healthy cells must down-regulate or prevent type I IFN production. Several cellular proteins are known to inhibit IRF3 to this end. These include TRIM26 (11), PPA2 (12), RACK1 (12), FoxO1 (13) and RAUL (14). Most of these proteins bind to and modify the transcriptionally active IRF3 to ultimately induce IRF3 proteosomal degradation. In contrast, PPA2 binds to and dephosphorylates transcriptionally active IRF3 to halt IRF3-driven transcription (12). Regardless, the study of cellular inhibitory mechanisms is critical to rationally control type I IFN to avoid deleterious imbalances in the immune response.
cFLIPL was originally characterized as an anti-apoptosis protein, which has made it an attractive target for cancer therapies (15). cFLIPL also is critical for the development of embryos (16), macrophages (17) and T cells (18), and for lymphocyte proliferation (18, 19). Several publications show that cFLIPL regulates type I IFN production (20–22), which would impact the current thinking about how cFLIPL performs its known functions. However, these reports show opposing effects of cFLIPL, in which cFLIPL induces or inhibit type I IFN production (20–22). Regardless, the molecular mechanism for this cFLIPL function remains unknown. This is an important gap in knowledge if the scientific community expects to understand how to regulate cFLIPL and IRF3 activation to achieve proper immune responses and immune system homeostasis (3, 23).
Type I IFN production is controlled by several transcription factors, including NF-κB, IRF3 and IRF7 and AP1 (5). To ask how cFLIPL inhibits type I IFN production, we used several approaches to stimulate and detect IRF3 activation independent of these other transcription factors. We found that cFLIPL inhibited IRF3-induced gene transcription triggered by virus infection, poly I:C or TBK1 over-expression. When probing events of the IRF3 activation pathway, cFLIPL allowed IRF3 phosphorylation and nuclear translocation. However, cFLIPL prevented IRF3-controlled transcription, IRF3-promoter interactions, and IRF3-CBP interactions. Co-immunoprecipitation analyses showed that cFLIPL interacted with IRF3. Mutational analysis of cFLIPL showed that i) IRF3 interactions and ii) cFLIPL nuclear localization are critical for inhibitory function. Together, these data suggest that cFLIPL prevents the formation of the IRF3 enhanceosome, blocking nascent IRF3 transcriptional action. Unlike other cellular IRF3 inhibitors, cFLIPL did not degrade or dephosphorylate IRF3. This mechanism is distinct from other known cellular IRF3 inhibitors, molecules that act on IRF3 only after IRF3 has initiated transcription.
Materials and Methods
Cell Culture
Human embryonic kidney 293T, mouse embryonic fibroblast (MEFs), the A549 non-small cell lung cancer cell lines cells, HCT116 colorectal carcinoma cells, and AsPC-1 pancreatic adenocarcinoma human cell lines were obtained from the American Type Culture Collection. The 293T-TLR3 cell line stably over-expresses the human TLR3 gene (Dr. R. Tapping, University of Illinois, Urbana, IL). Cells were cultured in appropriate medium supplemented with 10% fetal bovine serum (FBS; Fisher) and 1% Penicillin Streptomycin (Fischer).
Lentivirus infection
Control (LL3.7) and shRNA-expressing (shFLIP) lentiviruses were produced by co-transfecting 293T cells with packaging plasmids pCMV-dR8.2 (Addgene) and pCMV-VSV-G (Addgene) and either control plasmid pLL3.7 (Addgene), or pLL3.7-shFLIP (24). At 24 h post-transfection lentiviruses were isolated from cellular supernatants. Lentiviruses were concentrated with Lent-X Concentrator (ClonTech). A549 cells were infected, and cells were observed for GFP expression as a marker of transduction at 48 h pi. Cellular populations with >80% GFP expression were passaged for use as stably transduced cell lines to create control (LL3.7) or shFLIP-expressing (shFLIP) A549 cell lines.
Plasmids and Transfections
Plasmid pCI was obtained from Promega. Plasmid MC159 encodes the molluscum contagiosum virus MC159L gene (21). Dr. Jeffrey Cohen (National Institutes of Health, Bethesda, MD) provided a plasmid encoding a FLAG epitope tagged human cFLIPL gene (cFLIPL). Dr. Dongwan Yoo (University of Illinois, Urbana, IL) provided a plasmid that encodes the MAVS protein (pMAVS); a plasmid that encodes a constitutively active form of IRF3 (pIRF3CA); a plasmid encoding a GFP-IRF3 fusion protein (pGFP-IRF3)(21). pTBK1 encodes a FLAG epitope-tagged TBK1 protein and was a kind gift from Dr. Siddharth Balachandran (Fox Chase Cancer Center, Philadelphia, PA)(21). The FLAG-tagged CBP plasmid (pCBP) was donated by Dr. David Lebrun (Queens University, Ontario, CA)(25). Plasmids cFLIPR, DED1, and DED2 were kind gifts from Dr. Ingo Schmitz (Helmholtz Center for Infection Research, Braunschweig, DE) (26). A plasmid encoding the 221 residue cFLIPS was a kind gift from Dr. Jae Jung, USC, Los Angeles, CA). It is a FLAG-tagged construct. cFLIPR expresses a FLAG-tagged murine cFLIPR. DED1 encodes a myc-tagged DED1 (aa 1–90) of cFLIPR. DED2 expresses a V5 epitope-tagged DED2 (aa 79–186) of cFLIPR. ΔDED2, ΔDED1, and CLD plasmids were kind gifts from Dr. Gerolama Condorelli (University of Naples, Naples, IT)(27). ΔDED2 expresses human cFLIPL aa 1–93 and aa 178–480, while ΔDED1 expresses human cFLIPL aa 81–480. CLD expresses human cFLIPL residues 178–480. ΔDED2, ΔDED1, and CLD express FLAG epitope tagged proteins.
Plasmid DNA was transfected into cells using TransIT-2020 transfection reagent (Mirus Bio) following manufacturer’s protocol. For experiments in which poly I:C (Invivogen) was a transfected to stimulate IRF3, 1000 ng of poly I:C was transfected using Lipofectamine 2000 (Invitrogen) following manufacturer’s protocol.
Luciferase assays
293T, MEFs, or 293T-TLR3 cells were transfected with pIRF3-luc, a plasmid that contains a firefly luciferase gene that is under the transcriptional control of four copies of the PRDIII promoter sequence specific for IRF3 (21). pRL-null has a sea pansy luciferase gene expressed independent of a promoter and is used to assess transfection efficiency. Cells were transfected in triplicate with 225 ng pIRF3-luc and 25 ng pRL-null, 1000 ng of pCI, MC159, wild-type or mutant cFLIPL-based plasmids and 500 ng pTBK1, or pIRF3CA. Cells were lysed at 24 h post-transfection. When poly I:C was used, 293T-TLR3 cells were transfected as above. At 24 h later, cells were transfected with 1000 ng of poly I:C, and cells were lysed 6 hours later. MVA is an attenuated strain of vaccinia virus (28). Transfected cells were infected with MVA at a multiplicity of infection (MOI) of 5 PFU/cell in serum-free MEM for 1 h, rocking every 15 min. Virus-containing supernatant was removed and replaced with complete medium. At 6 h post-infection, cells were lysed. All cells were lysed in 1X PLB (Promega) and luciferase activity was detected using the Dual Luciferase Reporter Assay System (Promega), and quantified with Clarity Luminescence Microplate Reader (BioTek Instruments). Analysis of firefly and sea pansy luciferase activities was performed as described previously (21). Values are shown as mean ± s.d. The Student’s t-test was used to determine the statistical significance of inhibition of luciferase activity by wild-type and mutant FLIPs versus cells transfected with pCI. Statistically significant inhibition of luciferase activity as compared to untreated, pCI-transfected cells is indicated by asterisks (*P < 0.05, ** P < 0.005). A portion of each lysate was also analyzed for protein expression by immunoblotting.
Quantitative reverse-transcriptase PCR
293T cells or A549 cell lines stably expressing either the shRNA to cflar (shFLIP) or no shRNA (control) were transfected with 1000 ng pCI, pFLAG-cFLIPL, or pMC159 in technical triplicates. In some experiments, 293T cells were co-transfected with 500 ng pCI or pTBK1 for 24 h. Alternatively, 24 h post-transfection, A549 cells were incubated with 2 µg poly I:C for 6 h. Total RNA was extracted using an RNAeasy extraction kit. cDNA was generating using the M-MuLV Reverse Transcriptase (New England BioLabs). Quantitative PCR was performed using a Mastercycler realplex EP (Eppendorf) and SoFast EvaGreen Super Mix (BioRad) per manufacturer’s instructions. The following primers were used: β-actin F (5’-AGTTGCGTTACACCCTTTCT-3’), β-actin R (5’-ACCTTCACCGTTCCAGTTT-3’), isg15 F (5’-CATCTTTGCCAGTACAGGAGCT-3’), isg15 R (5’-ACACCTGGAATTCGTTGCC-3’). Changes in gene expression level were calculated by the 2(−ΔΔC(T)) method. Values obtained from isg15 cDNA levels were divided by β-actin levels for each sample. For normalization, respective β-actin mRNA quantities for each cDNA sample were measured, and then each value was normalized to that of unstimulated, pCI-transfected cells, whose value was set to one. For 293T cells, data are presented as the mean +/− s.d. from three independent experiments. The Student’s t-test was used to determine statistically significant differences in mRNA expression levels. Statistically significant inhibition, as compared to untreated pCI-transfected cells, is indicated by asterisks (*P < 0.05, **P < 0.005). For A549 cells, statistically significant increases in mRNA when cflar is shRNA silenced as compared to cells transduced with empty lentivirus vector, are indicated by asterisks (*P < 0.05), using the Student’s t-test. The data are expressed as mean +/− s.e.m. using data from three independent experiments.
Immunoblotting
For all immunoblots, the protein concentration of lysates was determined by the 660 nm protein assay (Pierce). An equal amount of protein from each lysate was incubated with 5× Non-Reducing Lane Marker (Thermo Scientific) and 5% (vol/vol) 2-mercaptoethanol (Fisher Scientific) and boiled for 5 min, and then electrophoretically separated by SDS-PAGE. Proteins were transferred to polyvinyl difluoride (PVDF) membranes (Millipore). Antibody-antigen reactions were detected by using chemiluminescence reagents (Amersham and Thermo Scientific) and autoradiography. Primary antibodies include: polyclonal rabbit anti-phospho-IRF3 Ser 386 (Millipore), monoclonal rabbit anti-IRF3 (Cell Signaling), or monoclonal mouse anti-IRF3 (ab25950; abcam), monoclonal mouse anti-FLAG (Sigma-Aldrich), monoclonal mouse anti-E3 (Dr. Stuart Isaacs, University of Pennsylvania, Philadelphia), monoclonal mouse anti-TBK1 (Cell Signaling), monoclonal mouse β-actin (Calbiochem), monoclonal mouse anti-myc (Cell Signaling), monoclonal mouse anti-FLIP (7F10; Enzo) or monoclonal rabbit anti-FLIP (D16A8); Cell Signaling monoclonal mouse anti-V5 (Millipore) and polyclonal rabbit anti-MC159 (29). In some cases, the densities of bands were quantified using ImageJ software (30). The values for band intensities for the protein of interest (e.g. IRF3) three independent experiments were averaged and compared to loading controls (e.g. PARP). Values were normalized to unstimulated, pCI-transfected cells. Data are expressed as the mean +/− s.e.m.
Co-immunoprecipitations
293T cells were transfected with 500 ng pTBK1, pCI, pGFP-IRF3, pFLAG-CBP, and 1000 ng cFLIPL, cFLIPS, or CLD. 24 h later, cells were lysed in DED lysis buffer (31). Clarified supernatants were used to assess expression levels of proteins and for co-immunoprecipitations. For the later, lysates were incubated with anti-FLAG, anti-GFP (Santa Cruz), anti-IRF3 or anti-CBP (Cell Signaling) or mouse IgG antibody (Sigma) for 6 h. Protein G-Sepharose beads (Invitrogen) in a 50% slurry were added to each sample and incubation with rotation for 16 h. Beads were collected and washed three times. Pelleted beads were suspended in 2× Laemmli buffer containing 5% 2-mercaptoethanol and boiled for 5 min. Samples were analyzed for the presence of proteins by using immunoblotting.
A549, HCT116 or AsPC-1 cellular monolayers were either untreated or treated with poly I:C (2 micrograms; Sigma) for 6 h. Cells were collected by trypsinization, pelleted by centrifugation, and lysed in DED lysis buffer as above. Clarified cellular lysates were incubated with rabbit anti-IRF3, rabbit anti-FLIP or rabbit IgG antibody (Cell Signaling) for 6 h at 4C. Protein G-Sepharose beads (Invitrogen) in a 50% slurry were added to each sample and incubation with rotation for 16 h. Beads were collected and washed three times. Pelleted beads were suspended in 2× Laemmli buffer containing 5% 2-mercaptoethanol and boiled for 5 min. Samples were analyzed for the presence of proteins by using immunoblotting, probing immunoblots with antibodies to detect endogenous IRF3 (mouse ab25950; abcam) or cFLIPL (mouse 7F10; Enzo).
Cytoplasmic and Nuclear Fractionation Assay
293T cells were transfected with either 1000 ng pCI, cFLIPL, cFLIPS, or CLD and either 500 ng pTBK1 or pCI. At 24 h later, cells were pelleted, and cytoplasmic and nuclear extracts were isolated as described previously (32). Cytoplasmic and nuclear extracts were immunoblotted for tubulin (Abcam), PARP (Santa Cruz), IRF3 (Cell Signaling), or FLAG-tagged cFLIP constructs (anti-FLAG, Sigma). The values for IRF3 band intensities for three independent experiments were determined as described above, using ImageJ software.
Chromatin immunoprecipitation (ChIP)
ChIP assays were performed according to manufacturer’s instructions (Thermo Scientific). Briefly, 293T cells were transfected as described in the co-immunoprecipitation assays for 24 h. Cells were cross-linked with 1% formaldehyde followed by quenching using 125 mM glycine. Cross-linked chromatin and associated proteins were either used as 10% total input or as samples to be immunoprecipitated (IP). Samples were incubated with anti-IRF3 (sc-9082-X; Santa Cruz) pre-conjugated to provided agarose beads. IPs were reverse cross-linked and genetic material was released from the beads by incubation at 65°C. Real-time PCR analyses were performed using the recovered DNA and a Mastercycler realplex EP system (Eppendorf). The primers used for real-time PCR to quantitate the ChIP-enriched DNA were for ifnb1 (33). Relative occupancy values were calculated by determining the apparent IP efficiency (ratios of the final amount of IP DNA as compared to input DNA) and normalized to the level observed at a control region in unstimulated, pCI-transfected cells, which was defined as 1.0. The Student’s T-test was used to determine statistically significant differences in DNA levels. Statistically significant inhibition of IRF3 binding activity as compared to untreated, pCI-transfected cells is indicated by asterisks (*p< 0.05).
Results
cFLIPL inhibits IRF3-controlled transcription of synthetic and natural genes
We published that the long isoform of cFLIP (cFLIPL) inhibits IFNβ gene activation, as measured by an infb1 promoter-controlled luciferase reporter assay (21). The infb1 promoter is controlled by NF-κB, AP-1, IRF3, and IRF7, leaving it unclear as to which transcription factor(s) is targeted by cFLIPL for this inhibition. cFLIP interacts with the IKK complex, an upstream activator of NF-κB (34). However, cFLIPL retains its IFNβ inhibitory function in cells deficient for NF-κB (21), ruling out NF-κB.
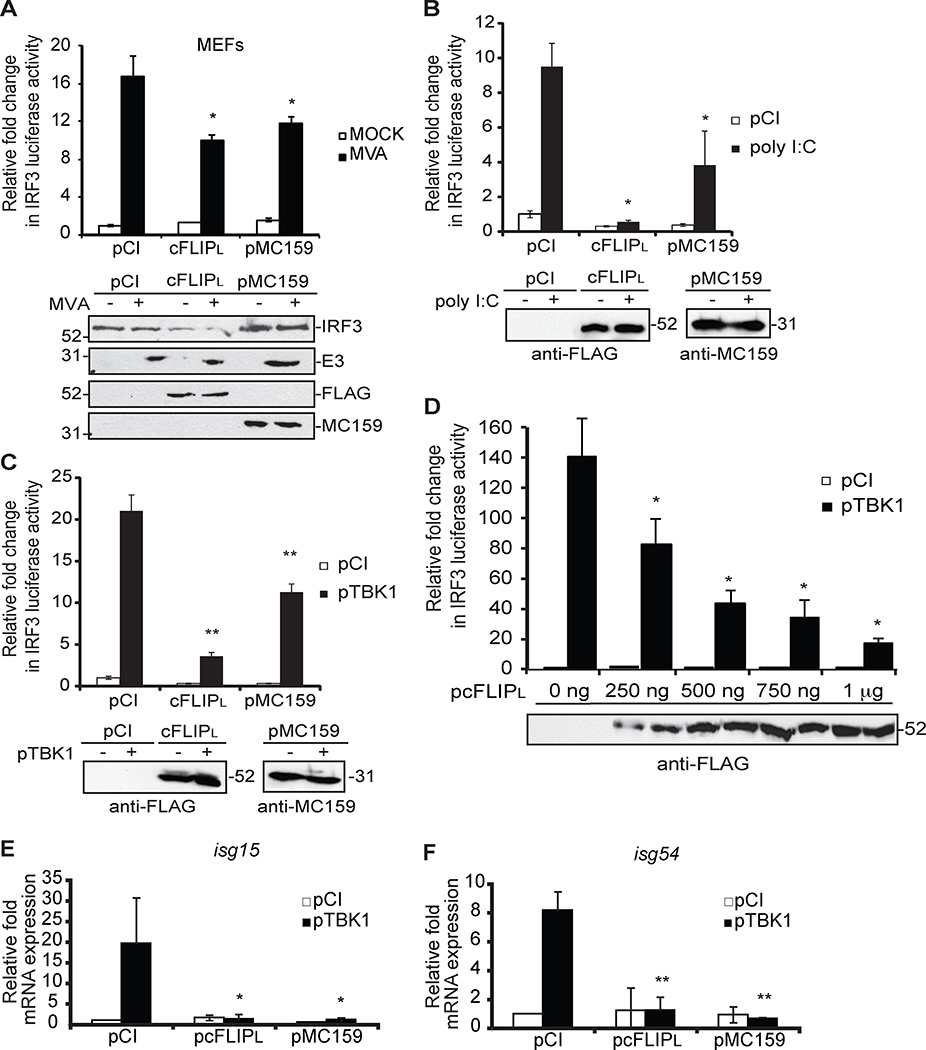
A viral homolog of cFLIPL (the molluscum contagiosum virus (MCV) MC159 protein) inhibits TBK1 activation (21), suggesting that cFLIPL targets IRF3 or IRF7 activation. IRF3 and IRF7 are each stimulated by known and distinct upstream signal transduction pathways (5). However, there are some instances where IRF3 and IRF7 functions overlap, which could complicate the studies with cFLIPL. To overcome this issue, we used an IRF3-controlled firefly luciferase reporter construct, which allows for the measurement of IRF3 activation alone (Fig.1). Several viruses stimulate IRF3 activation during infection (1). For example, MVA is an attenuated vaccinia virus that activates the interferon response in MEFs (35). However, IRF3 activation was blocked in MEFs over-expressing cFLIPL or MC159 as measured by reporter assays (Fig. 1A). MVA-induced IFNβ production is due, in part, to PKR sensing of viral dsRNA (35). We repeated experiments using poly I:C, an IRF3-stimulating agent that stimulates TBK1 activation via RIG-I, MDA-5 and TLR3 (36, 37). Because 293T cells do not express high levels of TLR3, a 293T cell line stably over-expressing TLR3 (293T-TLR3) was used to ensure robust IRF3 signaling upon incubation with poly I:C. The 293T-based cell line was used because of its high transfection efficiency rate and its common use in studying IRF activation pathways (11, 14, 21, 38). cFLIPL, like MC159, also inhibited IRF3-controlled luciferase activity under this condition (Fig. 1B), suggesting cFLIPL targeted a downstream signaling event shared by MVA infection and poly I:C treatment. In support of this, we observed that cFLIPL inhibited IRF3 activation when TBK1 over-expression stimulated IRF3 activation (Fig. 1C), and this inhibition was greater than that observed with MC159 and this inhibition was shown to be dose dependent (Figure 1D). For this set of experiments, regular 293T cell lines that do not over-express TLR3 were used. This suggested that cFLIPL acted on an event occurring at or downstream of TBK1.
Figure 1. cFLIPL inhibits IRF3-controlled luciferase activity.
Luciferase activity in (A) MEF or (B) 293T-TLR3 or (C–D) 293T cells transiently co-transfected for 24 hours with luciferase reporter plasmids (pIRF3-luc, pRL-null) and 1000 ng control plasmid (pCI) or plasmids encoding cFLIPL or MC159. (A) Cells were either mock-infected or infected with MVA (MOI = 5 PFU/cell) or (B) incubated with 1000 ng of poly I:C or (C and D) co-transfected with pTBK1. Cells were lysed at (A and B) 6 h or (C and D) 24 h later. Results are shown as fold-induction of luciferase activity, relative to those of pCI-transfected cells. Immunoblot analysis of lysates also was performed. (E and F) Quantitative RT-PCR (qPCR) analysis of (E) isg15 and (F) isg54 mRNA from 293T cells co-transfected with pTBK1 and 1000 ng control plasmid (pCI) or plasmids encoding cFLIPL or MC159 for 24 h. Results are presented as isg15 and isg54 mRNA relative to β-actin mRNA expression for each sample, and recorded as relative to those of cells transfected with empty vector (pCI). *P < 0.05, compared with cells transfected with empty vector. All experiments were performed at least three times, and data are expressed as the mean +/− s.d.
We evaluated the expression of IRF3-controlled genes as a second independent means to detect cFLIPL action. We examined the transcription of two cellular genes, isg15 and isg54/IFIT2 (Fig. 1E and 1F) that are regulated by IRF3 (39) by using RT-qPCR. In this case, TBK1 over-expression was used to stimulate IRF3, as evidenced by the transcription of isg15 and isg54 (Fig. 1E and 1F). In contrast, cFLIPL expression reduced isg15 and isg54 gene transcription as compared to vector-transfected cells. The transcription of both ISGs was reduced in MC159-expressing cells, and this was expected since MC159 inhibits TBK1 activation (21). Together, data shown in Fig. 1 supported the model that cFLIPL antagonizes the IRF3 activation pathway.
cFLIPL inhibits an event downstream of IRF3 phosphorylation
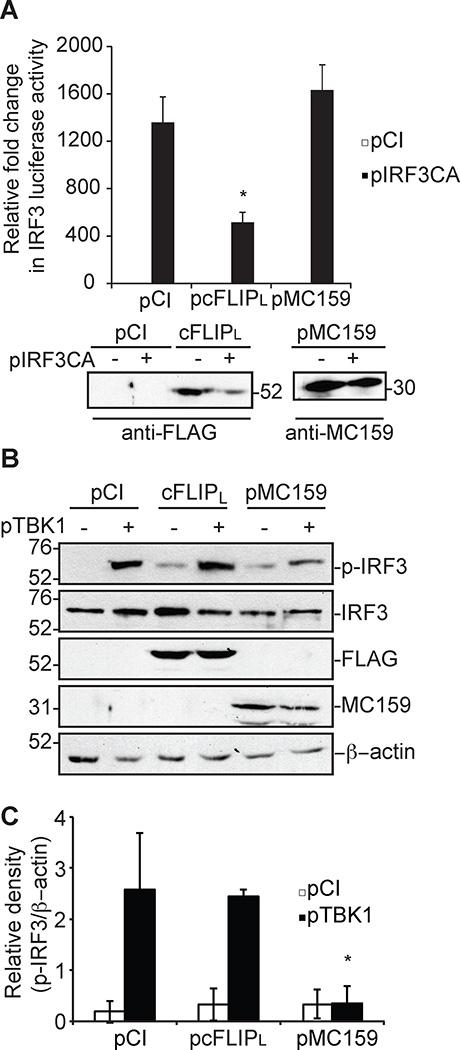
The next goal was to identify the cFLIPL molecular mechanism of inhibition. One obvious possibility was that cFLIPL would act similar to its MC159 homolog, by binding to and inhibiting TBK1 activation (21). However, two pieces of data in Figure 2 showed that cFLIPL used a different mechanism. First, cFLIPL inhibited IRF3 activation triggered by over-expression of a constitutively active phospho-mimetic form of IRF3 (pIRF3CA) (Fig. 2A). MC159 cannot inhibit this because IRF3 activation occurs downstream of TBK1 activation (1, 4, 5). Second, cFLIPL did not prevent IRF3 phosphorylation (Fig. 2B). Fig. 2B shows that MC159 prevented TBK1-induced IRF3 activation, as measured by a decrease in the intensity of the phospho-IRF3-containing band (Fig. 2C). However, there was no observable difference in phospho-IRF3 levels in cFLIPL versus vector-expressing cells (Fig. 2B and 2C).
Figure 2. cFLIPL does not inhibit IRF3 phosphorylation.
Luciferase activity in 293T cells transiently co-transfected for 24 hours with luciferase reporter plasmids, 500 ng pIRF3CA and 1000 ng cFLIPL or MC159. Results are presented as fold-induction of luciferase activity relative to those of cells transfected with empty vector (pCI). * P < 0.05 compared with cells transfected with empty vector. Immunoblot analysis of lysates also was performed. (B) Cells were transfected with 500 ng pTBK1 or pCI and 1000 ng pCI, pCI, cFLIPL or MC159 for 24 h. Protein expression from cellular lysates was evaluated by immunoblotting. (C) The intensity of the phospho-IRF3-containing bands (p-IRF3) from three independent experiments were evaluated using ImageJ software. The density of a phospho-IRF3-containing band was divided by the intensity of the β-actin containing band. The relative fold change in phospho-IRF3 was determined by normalizing the ratio of phospho-IRF3/β-actin to that of pCI-transfected cells, whose value was set to one. Data are expressed at the mean +/− s.e.m.
Several other cellular proteins down-regulate IRF3 activation (11–14). Most target the transcriptionally active form of IRF3, modifying IRF3 such that it is degraded by the cellular proteosomal machinery. Importantly, IRF3 protein levels remained consistent in the presence of cFLIPL (Fig. 2). These data suggest that cFLIPL has a mechanism to inhibit IRF3 that is distinct from other known cellular IRF3 inhibitors and from its viral homologs.
The caspase-like domain (CLD) of cFLIPL is necessary and sufficient to inhibit IRF3 activation
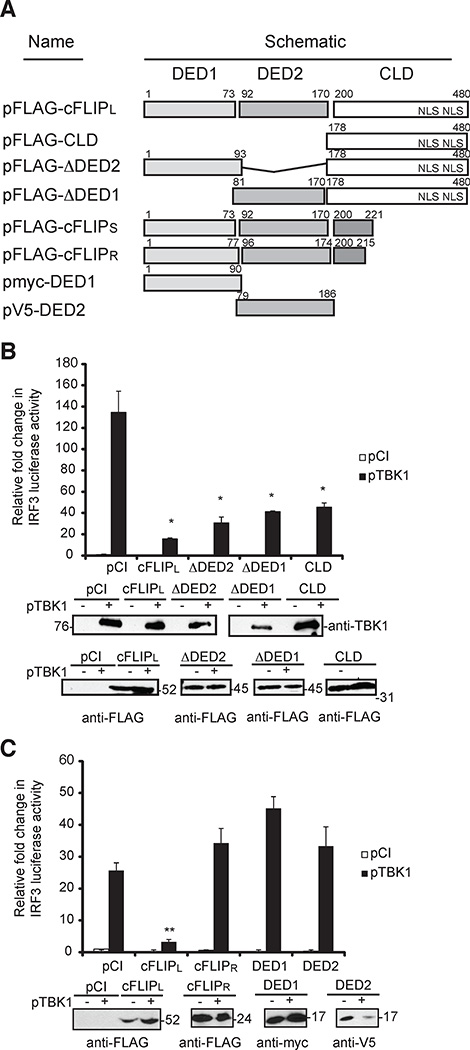
There are three detectable splice variants of cFLIP: cFLIPL (55 kDa), cFLIP short (cFLIPS; 24 kDa), and cFLIP raji (cFLIPR; 24 kDa)(40–42). These are depicted in Figure 3A. Each variant contains two death effector domains (DEDs), which are a death fold motifs present in the cellular apoptotic FADD and procaspase-8 proteins (34, 43). However, cFLIPL alone possesses an additional C-terminal region that is 302 amino acids in length (Fig. 3A). While this region shares some homology to the procaspase-8 caspase domain, the cFLIPL caspase-like domain (CLD) lacks caspase activity because there are several amino acid substitutions in the region required for caspase activity (41). The C-terminal region also possesses nuclear localization sequences (NLS) at residues 435–437 and 472–474 (44).
Figure 3. Analysis of cFLIP constructs for ability to inhibit of IRF3-controlled luciferase activity.
(A) A schematic of cFLIP constructs used for luciferase activity assays. The tandem death effector domains (DEDs) are denoted as DED1 and DED2. The two nuclear localization signals (NLS) in the C-terminal region of cFLIPL are shown. (B and C) Luciferase reporter assays in 293T cells transiently co-transfected with luciferase reporter plasmids (pIRF3-luc and pRL-null), pTBK1 and wild-type or mutant (B) cFLIPL or (C) cFLIPR plasmids. At 24 h later, cells were analyzed for luciferase activities as described in Figure 1. * P < 0.05, compared with cells transfected with empty vector (Student's t-test). Immunoblot analysis of cellular lysates for cFLIP protein expression also was performed. All experiments were performed at least three times.
Luciferase reporter assays were used to identify the cFLIP region that controls IRF3 activation, using TBK1 over-expression to trigger IRF3 activation (Fig. 3). Figure 3B uses mutants in the cFLIPL background. In contrast, mutants DED1 and DED2 were in the cFLIPR background in Fig. 3C. Thus, cFLIPR was used as a control for this set of experiments. Interestingly, Fig. 3B showed that either DED was dispensable for IRF3 activation because mutant cFLIPL that lacked either DED1 (ΔDED1) or DED2 (ΔDED2) still inhibited IRF3 activation. Expression of the CLD-containing C-terminus (CLD; amino acids 178–480) inhibited IRF3 activation, indicating this region was sufficient for the inhibitory phenotype (Fig. 3B). Interestingly, cFLIPR, or constructs expressing only DED1 or DED2 provided no IRF3 inhibitory function (Fig. 3C). Because cFLIPR is 74% similar to cFLIPS (45), cFLIPS is not expected to inhibit IRF3 activation. Indeed, data in Fig. 5 showed that cFLIPS did not inhibit IRF3-induced luciferase activity (Fig. 5B). Thus, the cFLIP DEDs themselves are not sufficient to inhibit IRF3 activity. Together, these data mapped the IRF3 inhibitory region of cFLIPL to residues 222–480. Nearly identical results were observed if over-expression of MAVS (instead of TBK1) was used to trigger IRF3 activation (Supplemental Figs. 1A and 1B).
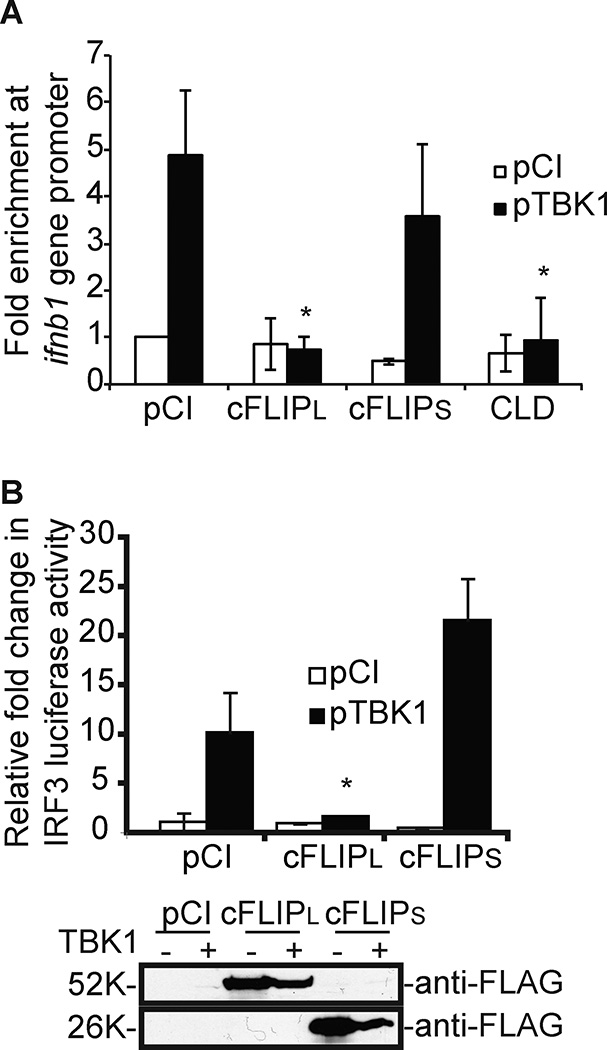
Figure 5. cFLIPL, but not cFLIPS, prevents IRF3 recruitment to the IFNβ promoter.
(A) ChIP assay to detect IRF3 binding to the IFNβ promoter. 293T cells were transfected with 500 ng pTBK1 or pCI and 1000 ng of plasmids encoding cFLIP proteins for 24 h. Results are presented as IFNβ values of immunoprecipitated DNA to total input DNA for each experimental sample. (B) 293T cells were transfected as described in Figure 1C, substituting a plasmid encoding cFLIPS for MC159. For both assays, values were expressed as the mean +/− s.d. Values were normalized to unstimulated, pCI-transfected cells. * P < 0.05, compared with unstimulated cells transfected with empty vector.
cFLIPL does not inhibit IRF3 nuclear translocation and is present in the nucleus
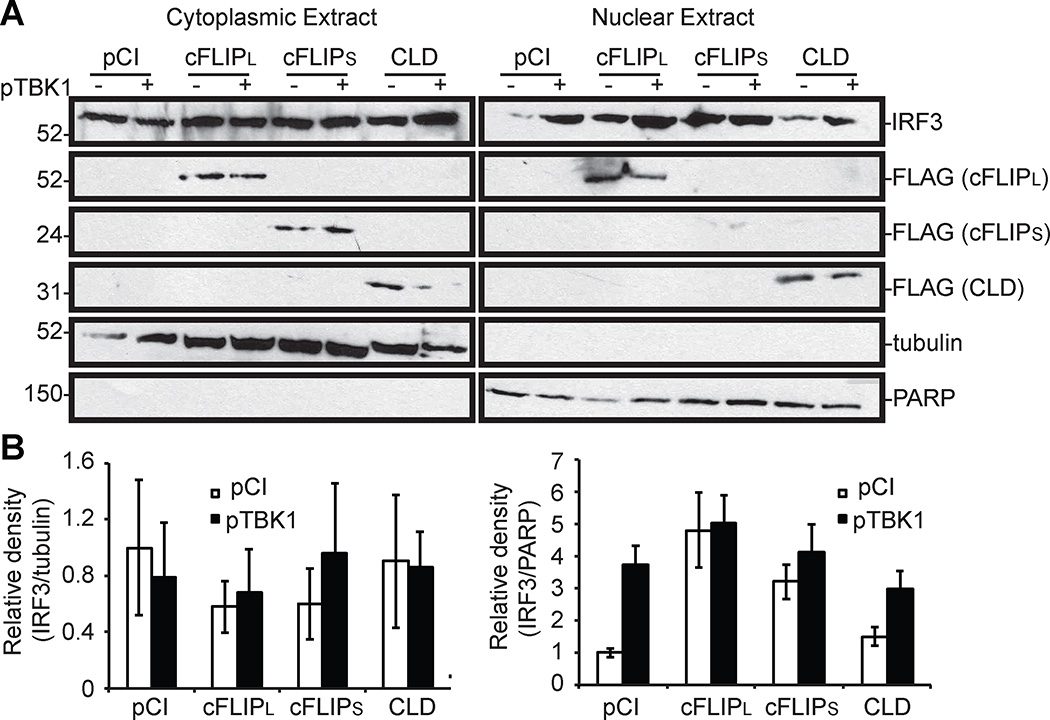
A majority of inactive IRF3 resides in the cytoplasm but the inactive form of IRF3 can cycle between the cytoplasm and nucleus (46). Regardless, the phospho-IRF3 dimer translocates to the nucleus due to an NLS present in IRF3 (7, 9). Thus, a next step was to query if cFLIPL prevented IRF3 nuclear translocation. Cellular fractionation assays were used for this purpose because they are sensitive enough to detect IRF3 nuclear translocation. For example, there was a visible increase in IRF3 protein levels in the nuclear extracts of TBK1 over-expressing cells versus vector-transfected cells (Fig. 4). Note that a small amount of IRF3 was detected in nuclei of unstimulated cells, probably reflecting IRF3 shuttling between the cytoplasm and nucleus (47). Interestingly, a similar trend was observed when cells expressed wild-type cFLIPL: there were no visual differences in nuclearly localized IRF3 in unstimulated versus stimulated cells. This remained true for cells expressing a mutant of cFLIPL (CLD) that inhibited IRF3 activation, and for cells expressing cFLIPS, a protein that did not inhibit IRF3 activation. Similar to Fig. 2B, IRF3 protein levels remained similar under all conditions tested. Thus, cFLIPL does not prevent IRF3 nuclear translocation. It was observed that the nuclear IRF3 protein levels were higher in unstimulated cells that were co-transfected with a cFLIPL- and cFLIPS-expressing cells than in pCI-or CLD-transfected cells. The reasons for this pattern are not clear to us currently, but these data suggest that the DED regions of cFLIP act on other signaling pathways that may indirectly trigger IRF3 nuclear translocation. At least for cFLIPL, this nuclear IRF3 is probably not phosphorylated due to data shown in Figure 2B. When the density of IRF3-containing bands from nuclear extracts was quantified using ImageJ software (Fig. 4B right panel), it was determined that IRF3 nuclear translocation was not significantly inhibited by the presence of cFLIP constructs.
Figure 4. cFLIPL does not inhibit IRF3 nuclear translocation.
(A) 293T cells were co-transfected for 24 h with cFLIP-based plasmids (1000 ng) and pTBK1 (500 ng) or pCI (500 ng). Cells were lysed and cytoplasmic and nuclear proteins were extracted, and samples were probed for the indicated proteins by immunoblotting. β-tubulin and PARP serve as markers of cytoplasmic content and nuclear content, respectively. (B) A graphical representation of the density of IRF3-containing bands in relation to tubulin (cytoplasmic fractions) or PARP (nuclear extracts) from 3 independent experiments. The values for IRF3-, tubulin- or PARP-containing band intensities were determined using ImageJ software, averaged and normalized to untreated, pCI-transfected cells for either cytoplasmic or nuclear extracts separately. Data are expressed as the mean +/− s.e.m.
The localization of cFLIP also was examined in these same extracts to gain a greater understanding of how cFLIPL may inhibit IRF3 activation. cFLIPL is often regarded as a cytoplasmic protein because it interacts with other cytoplasmic proteins including procaspase-8 (34). However, the cFLIPL C-terminus possesses two NLS’s and it has been shown that cFLIPL is present in the nucleus (44). Thus, it was not surprising to detect wild-type cFLIPL or mutant CLD protein in the nucleus and cytoplasm (Fig. 4). The NLS sequences are absent in cFLIPS and this isoform was detected only in the cytoplasm, as would be expected (Fig. 4). Tubulin proteins were detected solely in the cytoplasm and PARP proteins were detected solely in the nuclear extracts, showing the successful separation of cellular compartments.
cFLIPL prevents the IRF3 transcription factor from binding to its target promoter site
Nuclear IRF3 binds to DNA sequences in promoters of its target genes via its DNA binding domain (DBD) (9, 48). A chromatin immunoprecipitation (ChIP) assay was used to examine the extent of IRF3 interactions with the IFNβ promoter in cells expressing wild-type or mutant cFLIP proteins (Fig. 5A). IRF3 was immunoprecipitated from unstimulated and stimulated cells, and IRF3-IFNβ promoter interactions were assessed by using qPCR. As expected, the IFNβ promoter was PCR amplified in TBK1 over-expressing cells, indicating that IRF3 successfully bound to the IFNβ promoter (Fig. 5A). In contrast, IRF3-promoter interactions significantly decreased in cFLIPL-expressing cells (Fig. 5A). Note that both the cFLIPR and cFLIPS proteins have nearly identical phenotypes; neither inhibit IRF3-controlled luciferase activity (Figure 5B and 3C). Finally, the CLD construct prevented IRF3-DNA interactions (Fig. 5A), correlating with its ability to inhibit IRF3-based luciferase activity (Fig. 3).
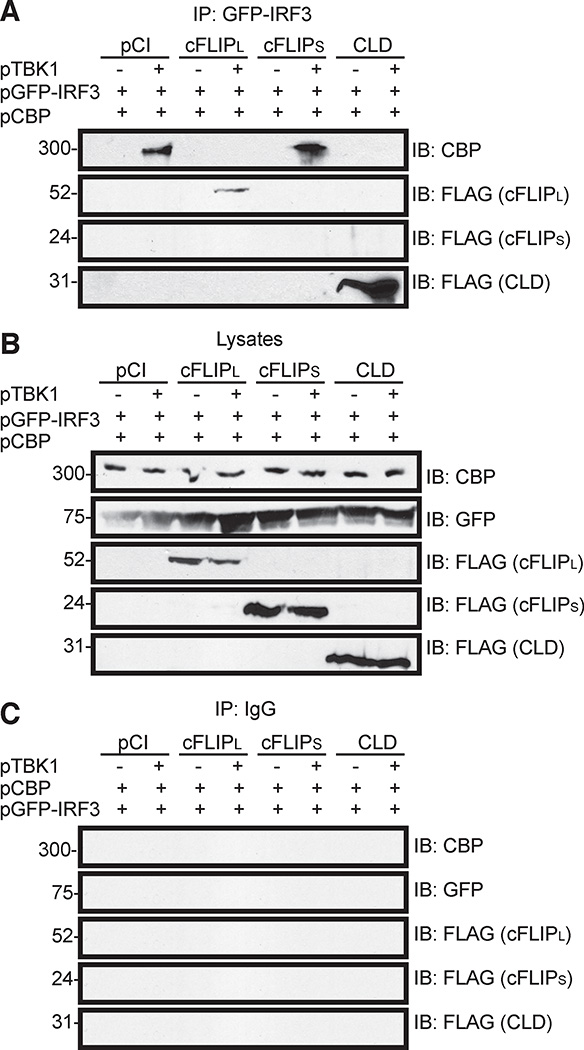
cFLIPL interacts with IRF3 and this interaction correlates with a loss of IRF3-CBP interactions and a loss of IRF3 inhibition
cFLIPL and CLD were the only molecules that inhibited IRF3 activation (Fig. 3), were present in the nucleus (Fig. 4), and prevented IRF3-DNA interactions (Fig. 5). Based on these data, a reasonable hypothesis is that the nuclear-localized cFLIPL interacts with nuclear lRF3 to prevent formation of an IRF3 enhanceosome. This was tested using co-immunoprecipitation assays to detect IRF3-cFLIPL interactions (Fig. 6). Cells were co-transfected to express an IRF3-GFP fusion protein, CBP and wild-type or mutant cFLIPL proteins. In some cases, cells were co-transfected pTBK1 to activate IRF3. As expected, IRF3-CBP interactions were detected under conditions where IRF3 was activated (Fig. 6). However, these interactions were disrupted by cFLIPL. Since this was concomitant with the presence of cFLIPL in the immunoprecipitates (Fig. 6A) it was likely that cFLIPL interacted with IRF3. Similarly, CLD co-immunoprecipitated with IRF3, and IRF3 no longer interacted with CBP in these same reactions. In contrast, no interactions were detected between IRF3 and cFLIPS, and IRF3-CBP interactions were retained in the presence of cFLIPS. All cells expressed similar amounts of IRF3-GFP, CBP or cFLIP (Fig. 6B). Fig. 6C used IgG instead of anti-GFP for immunoprecipitations to show no non-specific binding of our proteins of interest to protein G-sepharose beads.
Figure 6. The CLD of cFLIPL is sufficient and necessary to co-immunoprecipitate with IRF3 and to prevent CBP-IRF3 interactions.
Immunoblot (IB) analysis of (A) GFP-IRF3 fusion protein immunoprecipitated (IP) from 293T cells co-transfected with pGFP-IRF3 (500 ng), pCBP (500 ng) and various combinations of pCI, pTBK1 and FLAG-tagged cFLIP proteins. Anti-GFP was used as the antibody in these IPs. IP samples were probed for the presence of CBP and FLAG-tagged FLIP proteins by immunoblotting (IB). (B) A portion of each lysate prior to immunoprecipitation was set aside as input sample, and immunoblotted to detect protein expression. (C) IB analysis of proteins IP-ed from cellular lysates with non-specific IgG instead of anti-GFP. All experiments were performed at least three times.
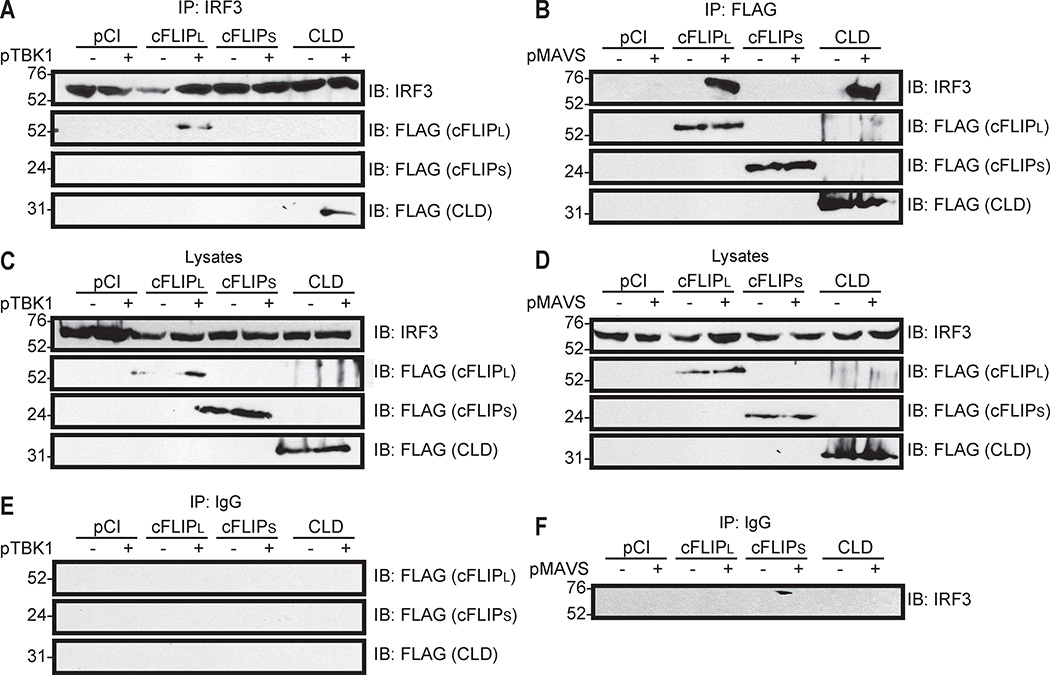
Similar experiments were performed to ask if endogenous IRF3 had the same binding properties as GFP-IRF3 (Fig. 7A). We observed that wild-type cFLIPL and CLD co-immunoprecipitated with endogenous IRF3. As in Fig. 6, these interactions only occurred under conditions known to activate IRF3. We did not detect cFLIPS-IRF3 interactions under any condition tested. A reverse co-immunoprecipitation showed that endogenous IRF3 interacted with the cFLIPL and CLD proteins but not cFLIPS (Fig. 7B). In this case, ectopic MAVS expression was used as an alternative mechanism to trigger IRF3 activation. IRF3 protein expression remained stable regardless of the presence of wild-type or mutant cFLIP constructs (Figs. 7C and 7D). These interactions between IRF3 and cFLIP were specific because neither IRF3 nor cFLIP was immunoprecipitated when a non-specific IgG was used instead of anti-IRF3 (Fig. 7E) or anti-FLAG (Fig. 7F).
Figure 7. Endogenous IRF3 interacts with cFLIPL and the CLD, but not cFLIPS.
Immunoblot (IB) analysis of an immunoprecipitated (IP) endogenous IRF3 from 293T cells co-transfected with plasmids encoding FLAG-tagged cFLIP proteins and various combinations of pCI and pTBK1 or pMAVS. (A) Cellular lysates were IP-ed with (A) anti-IRF3 or (B) anti-FLAG antibodies and IP-ed samples were probed for the presence of IRF3 and FLAG-tagged FLIP proteins by immunoblotting. (C and D) A portion of each lysate prior to immunoprecipitation was set aside as input sample, and immunoblotted to detect protein expression. (E and F) IB analysis of proteins IP-ed with non-specific IgG instead of anti-IRF3 or anti-FLAG. All experiments were performed at least three times.
The role of cFLIPL in preventing IRF3 activation in cancer cell lines
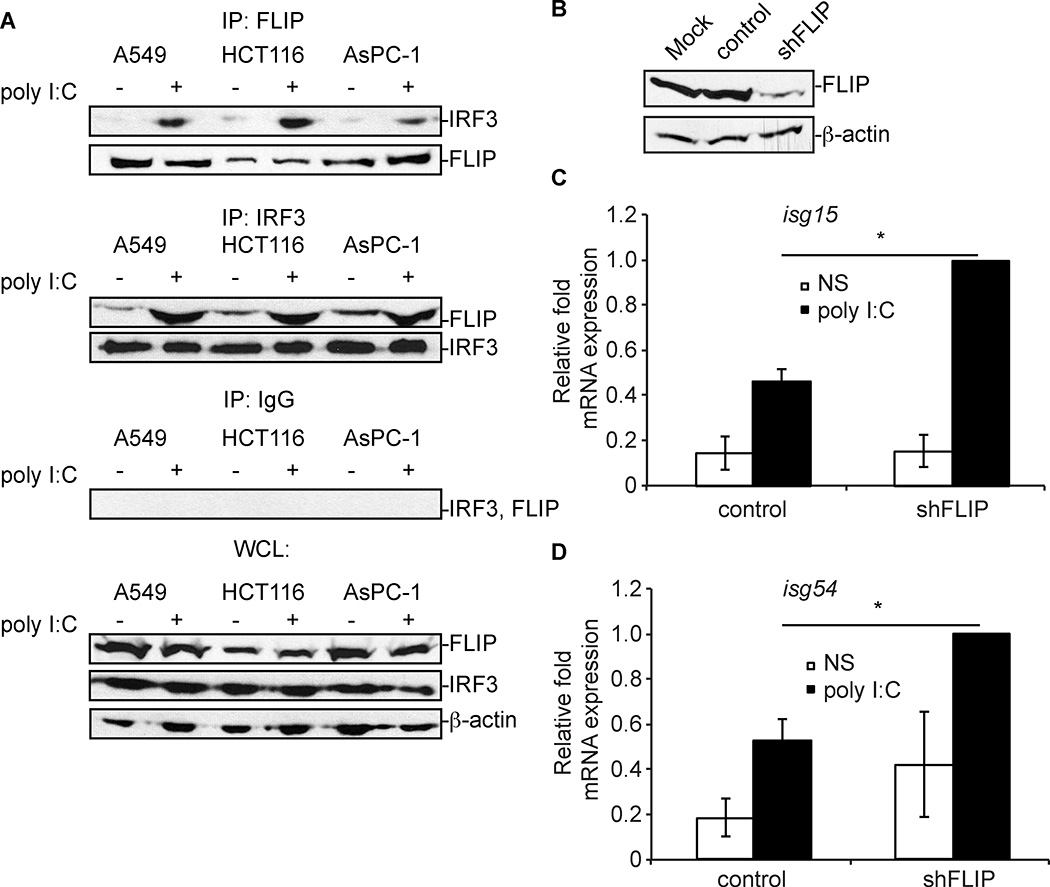
There are many primary cancers, like non-small cell lung cancer (NSCLC), pancreatic cancer, and colorectal cancers, where there is a correlation between a decrease in in ifnb1, and/or isg54 gene transcription (genes controlled by IRF3) and an increase in cflar transcription (the gene that encodes cFLIPL) as identified by using Oncomine (49). These data suggest that cFLIPL may bind to and inhibit IRF3 in tumor cells as a means to prevent IRF3-induced transcription of immune genes. To test this hypothesis, the NSCLC A549 cell line, HCT116 colorectal carcinoma cell line, and the AsPC-1 pancreatic adenocarcinoma cell lines were used. Unlike the 293T cell line used above, A549 cells, HCT116 cells, and AsPC-1 cells express detectable levels of endogenous cFLIPL, allowing us to examine cFLIPL-IRF3 interactions under conditions in which neither protein must be ectopically expressed (50) (51){Liu, 2006 #172}. Figure 8A showed that cFLIPL co-immunoprecipitated with IRF3 in all three of the cell lines examined. This interaction was observed at low levels in unstimulated cells, and greatly increased when cells were stimulated with poly I:C. When examining the lysates from these samples, it was noted that IRF3 and cFLIPL levels were present in similar amounts. The cFLIP-IRF3 interactions were specific because these interactions were not observed if an IgG isotype control antibody was used in co-immunoprecipitations in place of anti-IRF3 or anti-FLIP.
Figure 8. Endogenous cFLIPL-IRF3 interactions and IRF3-controlled isg gene expression in A549 cells.
(A) A549, HCT116, and AsPC-1 cells were unstimulated or stimulated with 2 micrograms poly I:C for 6 h. Clarified cellular lysates were immunoprecipitated with anti-IRF3, anti-FLIP or IgG antibodies. Immunoprecipitates were probed for the presence of endogenous IRF3 and cFLIPL proteins by immunoblotting. A portion of each lysate prior to immunoprecipitation was set aside as input sample, and immunoblotted to detect protein expression. (B) Immunoblot analysis of cFLIP levels in mock-infected A549 cells or A549 cells transduced with a lentivirus expressing (shFLIP) or not expressing (control) shRNA specific for cFLIP (shFLIP). (C and D) Quantitative RT-PCR (qPCR) analysis of isg15 and isg54 mRNA from transduced A549 cells from Figure 8B. Cells were either unstimulated (NS) or incubated with 2 micrograms poly I:C for 6 h. Results are presented as the mean +/− s.e.m. of isg15 and isg54 mRNA relative to β-actin mRNA expression for each sample for three independent experiments, and recorded as relative to those of cells transfected with empty vector (pCI). *P < 0.05, compared with cells transfected with empty vector.
If cFLIPL is indeed responsible for IRF3 inhibition in various cancers, then a lack of cFLIPL would render these cells susceptible to IRF3 activation. To test this hypothesis, we further examined the A549 cell line and stably transduce them with a lentivirus construct that encodes a silencing hairpin RNA specific for cflar (the gene encoding cFLIP; shFLIP). A second set of A549 cells was transduced in parallel with a lentivirus that does not encode shRNA (control) (24). Fig. 8B shows cFLIPL protein levels were decreased only when A549 cells were transduced with a lentivirus containing the shRNA to cflar (shFLIP). In contrast, cFLIPL levels were similar in cells that were not transduced (mock) or transduced with a lentivirus that lacks shRNA (control). When transduced cells were incubated with poly I:C to stimulate IRF3, there was statistically significant increase in the transcription of two genes known to be controlled by IRF3 -- isg15 (Fig. 8C) and isg54 (Fig. 8D) -- when cFLIPL protein levels were decreased versus conditions in which cFLIPL protein levels were unchanged from mock-transduced cells. Thus, IRF3 activation is indeed dampened by cFLIPL in this cell line.
Discussion
The IRF3 transcription factor is very important for the expression of type I IFN and ISGs (2, 5). Data here are the first to report how cFLIPL controls IRF3 activation on a molecular level. In summary, cFLIPL inhibited the transcription of cellular and synthetic genes regulated by promoters containing known IRF3 binding sites. This phenotype was observed in different cell lines and under several conditions that stimulate IRF3. Following the well-known steps of the IRF3 activation pathway, cFLIPL allowed upstream signaling events including IRF3 nuclear translocation. However, cFLIPL inhibited IRF3 from interacting with its DNA promoter and with CBP. This function mapped to the nuclearly localized C-terminal region of cFLIPL. The current model based on these data is that cFLIPL inhibit the formation of an IRF3-based enhanceosome. There are several remaining questions to be asked, including if cFLIPL-IRF3 interactions initially occur in the cytoplasm or in the nucleus. This is currently under investigation in our lab. IRF3 is shuttled from the nucleus to the cytoplasm due to the presence of NLSs and NESs (47). Here, we find that cFLIPL inhibits the action of a dimerized phospho-mimetic form of IRF3 (IRF3CA) (Fig. 2A). cFLIPL also is present in the cytoplasm and nucleus (Fig.4A). This would suggest that cFLIPL preferentially binds to a dimeric IRF3 complex, and may bind to these dimers regardless of whether they are in the cytoplasm or nucleus.
There are other cellular proteins known to inhibit IRF3 function. These proteins target the transcriptionally active form of IRF3. For example, the RTA-associated ubiquitin ligase (RAUL) promotes ubiquitin-mediated degradation of IRF3 (14) as a means of halting IRF3-controlled gene transcription. Protein phosphatase A2 (PPA2) dephosphorylates the activated form of IRF3, which decreases IRF3 transcriptional activity (12). However, neither unmodified nor phospho-IRF3 levels were reduced in the presence of cFLIPL in the system used here. This implies that cFLIPL acts prior to the above-listed cellular inhibitors, and prevents IRF3-controlled transcription from initiating in the first place. As such, cFLIPL may represent a cellular mechanism to prevent low-level or “background” IRF3 activation. Alternatively, cFLIPL may bind to IRF3 to block post-translational modifications (e.g., ubiquitination) of IRF3. In this case, cFLIPL would allow IRF3 to be returned to the cytoplasm for future use. Future studies will examine these possibilities to better understand this new cellular mechanism to down-regulate IRF3 activity.
cFLIPL is a member of the FLIP family, which defined by proteins that possess two tandem DEDs (34, 43). This family includes proteins encoded by the molluscum contagiosum poxvirus (MC159 and MC160) and the Kaposi’s sarcoma herpesvirus (K13). Viruses encode proteins to inhibit IRF3 as a common strategy to fight the anti-viral immune response (4, 52). Both MC159 and MC160 inhibit IRF3 activation, but use mechanisms distinct from cFLIPL. For example, MC159 binds to and inhibits TBK1 activation (21). Moreover, the MC159 DEDs are important for TBK1 binding and subsequent IRF3 inhibition (21). In contrast, the cFLIPL DEDs are dispensable for IRF3 activation in the system used here. The MC159 and cFLIPL DEDs share approximately 30% sequence homology. Therefore, there may be sufficient differences between these DEDs that are responsible for these differences in function. The K13 role in IRF3 regulation remains unclear. K13 induces IFNβ production (53), implying that K13 may activate IRF3. However, this phenotype may be due to the NF-κB activating function of K13 (54). Another approach to examine K13 regulation of IRF3 in an NF-κB-independent manner used p65 −/− MEFs (21). In these conditions, K13 does not stimulate IFNβ activation (as examined by a reporter assay) and weakly inhibits inhibit MAVS-induced IRF3 activation as compared to other FLIPs (21). It is intriguing that these cellular and viral homologs have distinct biochemical and biological properties with regard to IRF3 regulation. These differences give scientists a unique set of reagents to more deeply understand how to manipulate aspects of the IRF3 activation pathway to the benefit of cellular and organismal health.
cFLIPL is a multi-functional protein, regulating events including apoptosis, NF-κB, ERK activation, autophagy, necroptosis and Wnt signaling (23, 55). It also has myriad cellular biding partners including procaspase-8, Ku70, TRAF2 and RIP1 (23). This raised the concern that this newly identified function of cFLIPL is indirect. For example, since caspase-8 cleavage of RIP1 kinase down-regulates IRF3 activation, one worry is that cFLIPL may simply be inhibiting caspase-8 activation to indirectly inhibit the signal transduction pathway leading from MAVS to IRF3 activation (56). Several pieces of data diminish this possibility. Most importantly, we observed that cFLIPL co-immunoprecipitated with IRF3, which suggests that cFLIPL functions directly on the IRF3 signal transduction pathway. Second, in our system, cFLIPL inhibited IRF3 activation in a caspase-8-independent manner (Supplemental Figure 2). Third, we observe a correlation between cFLIPL interactions with IRF3 binding and inhibition of IRF3 activation, when using large deletion mutants of FLIP.
Only a few publications have examined the relationship between cFLIPL and type I IFN production. Two publications report data that complement the findings described here (20, 21). However, Buskiewicz et al. report an opposing function: cFLIPL increases IFNβ production triggered by coxsackievirus B3 (CVB3) infection (22). Potential reasons for these differences are the different stimuli used in those studies. We observed that cFLIPL inhibits IRF3 activation triggered by a DNA virus or by poly I:C (a reagent that uses TLR3 and does not trigger MAVS-induced IRF3 activation). Buskiewicz et al. used an RNA virus for infection, which is known to trigger MAVS-induced IRF3 activation (22). Perhaps the differences we observe reflect different PRR-based signal transduction pathways triggered by RNA versus DNA viruses. Additionally, this phenotype may be cell type specific. For example, we show that caspase-8 is dispensable for IRF3 activation in the 293T cell line used here. However, caspase-8 is required to stimulate IRF3 activation in the cFLIPL-expressing MEF cells (22). The ratio of cFLIPL to procaspase-8 dictates whether cFLIPL has pro- or anti-apoptotic function (57, 58). Thus, another possibility is that the cellular concentrations of cFLIPL dictate its effect on IRF3. Regardless, the continued study of cFLIPL and IRF3 is critical for a deeper understanding of conditions that allow cFLIPL to inhibit or activate type I IFN production.
This study identifies a molecular mechanism for cFLIPL. This finding impacts several scientific disciplines. For cancer, in silico analysis identified a statistically significant relationship between an upregulation of cFLIP gene (cflar) and a down-regulation of the isg54 gene (a gene whose expression is controlled solely by IRF3) in non-small cell lung carcinoma (P = 0.0053), pancreatic carcinomas (P = 0.021) and angioblastic T-cell lymphomas (P = 0.002) and colorectal adenocarcinoma (P = 0.0000000057) as determined by Oncomine (49). This implies that some cancer cells may have an increased level of cFLIPL to inhibit type I IFN production as an immune evasion strategy. In support of this model are our data with the A549 cell line, a model cell line for non-small cell lung carcinomas, shown here (Fig. 8). One prediction is that disruption of cFLIPL inhibitory function in these cancers would increase immune responses, perhaps halting tumorigenesis. Type I IFN production is also associated with several autoimmune diseases (3). Interestingly, IRF3 itself may be responsible for some of the disease observed with systemic lupus erythematosus (59). A next step is to ask if cFLIPL, or a cFLIPL mimetic, can be used as a strategy to diminish the severity or onset of autoimmune diseases. Rapid type I IFN production is a hallmark of acute virus infection (1). Interestingly, type I IFN production also helps establish persistent virus infections (60). Thus, it will be of great interest to infectious disease experts to understand how cFLIPL regulation of IRF3 affects acute versus chronic virus infections and the impact of this on viral pathogenesis.
Supplementary Material
Acknowledgments
The authors thank Drs. Ralph Budd, Andreas Keonig, Inna Buskiewicz and Michelle Arnold for helpful discussion; Drs. Brian Ferguson and Cari Vanderpool for critical review of the manuscript. This manuscript is part of the fulfillment of the Ph.D. requirements for LTG.
Source of funding: University of Illinois and NIH
References
- 1.Hoffmann HH, Schneider WM, Rice CM. Interferons and viruses: an evolutionary arms race of molecular interactions. Trends Immunol. 2015;36:124–138. doi: 10.1016/j.it.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tamura T, Yanai H, Savitsky D, Taniguchi T. The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol. 2008;26:535–584. doi: 10.1146/annurev.immunol.26.021607.090400. [DOI] [PubMed] [Google Scholar]
- 3.Ronnblom L, Eloranta ML. The interferon signature in autoimmune diseases. Curr Opin Rheumatol. 2013;25:248–253. doi: 10.1097/BOR.0b013e32835c7e32. [DOI] [PubMed] [Google Scholar]
- 4.Kumari P, Narayanan S, Kumar H. Herpesviruses: interfering innate immunity by targeting viral sensing and interferon pathways. Rev Med Virol. 2015;25:187–201. doi: 10.1002/rmv.1836. [DOI] [PubMed] [Google Scholar]
- 5.Ikushima H, Negishi H, Taniguchi T. The IRF family transcription factors at the interface of innate and adaptive immune responses. Cold Spring Harb Symp Quant Biol. 2013;78:105–116. doi: 10.1101/sqb.2013.78.020321. [DOI] [PubMed] [Google Scholar]
- 6.Weaver BK, Kumar KP, Reich NC. Interferon regulatory factor 3 and CREB-binding protein/p300 are subunits of double-stranded RNA-activated transcription factor DRAF1. Mol Cell Biol. 1998;18:1359–1368. doi: 10.1128/mcb.18.3.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoneyama M, Suhara W, Fukuhara Y, Fukuda M, Nishida E, Fujita T. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 1998;17:1087–1095. doi: 10.1093/emboj/17.4.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Panne D, McWhirter SM, Maniatis T, Harrison SC. Interferon regulatory factor 3 is regulated by a dual phosphorylation-dependent switch. J Biol Chem. 2007;282:22816–22822. doi: 10.1074/jbc.M703019200. [DOI] [PubMed] [Google Scholar]
- 9.Lin R, Heylbroeck C, Pitha PM, Hiscott J. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol Cell Biol. 1998;18:2986–2996. doi: 10.1128/mcb.18.5.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sato M, Tanaka N, Hata N, Oda E, Taniguchi T. Involvement of the IRF family transcription factor IRF-3 in virus-induced activation of the IFN-beta gene. FEBS Lett. 1998;425:112–116. doi: 10.1016/s0014-5793(98)00210-5. [DOI] [PubMed] [Google Scholar]
- 11.Wang P, Zhao W, Zhao K, Zhang L, Gao C. TRIM26 negatively regulates interferon-beta production and antiviral response through polyubiquitination and degradation of nuclear IRF3. PLoS Pathog. 2015;11:e1004726. doi: 10.1371/journal.ppat.1004726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Long L, Deng Y, Yao F, Guan D, Feng Y, Jiang H, Li X, Hu P, Lu X, Wang H, Li J, Gao X, Xie D. Recruitment of phosphatase PP2A by RACK1 adaptor protein deactivates transcription factor IRF3 and limits type I interferon signaling. Immunity. 2014;40:515–529. doi: 10.1016/j.immuni.2014.01.015. [DOI] [PubMed] [Google Scholar]
- 13.Lei CQ, Zhang Y, Xia T, Jiang LQ, Zhong B, Shu HB. FoxO1 negatively regulates cellular antiviral response by promoting degradation of IRF3. J Biol Chem. 2013;288:12596–12604. doi: 10.1074/jbc.M112.444794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu Y, Hayward GS. The ubiquitin E3 ligase RAUL negatively regulates type i interferon through ubiquitination of the transcription factors IRF7 and IRF3. Immunity. 2010;33:863–877. doi: 10.1016/j.immuni.2010.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shirley S, Micheau O. Targeting c-FLIP in cancer. Cancer Lett. 2013;332:141–150. doi: 10.1016/j.canlet.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 16.Yeh WC, Itie A, Elia AJ, Ng M, Shu HB, Wakeham A, Mirtsos C, Suzuki N, Bonnard M, Goeddel DV, Mak TW. Requirement for Casper (c-FLIP) in regulation of death receptor-induced apoptosis and embryonic development. Immunity. 2000;12:633–642. doi: 10.1016/s1074-7613(00)80214-9. [DOI] [PubMed] [Google Scholar]
- 17.Huang QQ, Perlman H, Huang Z, Birkett R, Kan L, Agrawal H, Misharin A, Gurbuxani S, Crispino JD, Pope RM. FLIP: a novel regulator of macrophage differentiation and granulocyte homeostasis. Blood. 2010;116:4968–4977. doi: 10.1182/blood-2009-11-252841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chau H, Wong V, Chen NJ, Huang HL, Lin WJ, Mirtsos C, Elford AR, Bonnard M, Wakeham A, You-Ten AI, Lemmers B, Salmena L, Pellegrini M, Hakem R, Mak TW, Ohashi P, Yeh WC. Cellular FLICE-inhibitory protein is required for T cell survival and cycling. J Exp Med. 2005;202:405–413. doi: 10.1084/jem.20050118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Budd RC, Yeh WC, Tschopp J. cFLIP regulation of lymphocyte activation and development. Nat Rev Immunol. 2006;6:196–204. doi: 10.1038/nri1787. [DOI] [PubMed] [Google Scholar]
- 20.Handa P, Tupper JC, Jordan KC, Harlan JM. FLIP (Flice-like inhibitory protein) suppresses cytoplasmic double-stranded-RNA-induced apoptosis and NF-kappaB and IRF3-mediated signaling. Cell Commun Signal. 2011;9:16. doi: 10.1186/1478-811X-9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Randall CM, Biswas S, Selen CV, Shisler JL. Inhibition of interferon gene activation by death-effector domain-containing proteins from the molluscum contagiosum virus. Proc Natl Acad Sci U S A. 2014;111:E265–E272. doi: 10.1073/pnas.1314569111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buskiewicz IA, Koenig A, Roberts B, Russell J, Shi C, Lee SH, Jung JU, Huber SA, Budd RC. c-FLIP-Short reduces type I interferon production and increases viremia with coxsackievirus B3. PLoS One. 2014;9:e96156. doi: 10.1371/journal.pone.0096156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Safa AR. Roles of c-FLIP in Apoptosis, Necroptosis, and Autophagy. J Carcinog Mutagen. 2013;(Suppl 6) doi: 10.4172/2157-2518.S6-003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu YH, Kuo WC, Wu YJ, Yang KT, Chen ST, Jiang ST, Gordy C, He YW, Lai MZ. Participation of c-FLIP in NLRP3 and AIM2 inflammasome activation. Cell Death Differ. 2014;21:451–461. doi: 10.1038/cdd.2013.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bayly R, Chuen L, Currie RA, Hyndman BD, Casselman R, Blobel GA, LeBrun DP. E2A-PBX1 interacts directly with the KIX domain of CBP/p300 in the induction of proliferation in primary hematopoietic cells. J Biol Chem. 2004;279:55362–55371. doi: 10.1074/jbc.M408654200. [DOI] [PubMed] [Google Scholar]
- 26.Ueffing N, Keil E, Freund C, Kuhne R, Schulze-Osthoff K, Schmitz I. Mutational analyses of c-FLIPR, the only murine short FLIP isoform, reveal requirements for DISC recruitment. Cell Death Differ. 2008;15:773–782. doi: 10.1038/sj.cdd.4402314. [DOI] [PubMed] [Google Scholar]
- 27.Quintavalle C, Incoronato M, Puca L, Acunzo M, Zanca C, Romano G, Garofalo M, Iaboni M, Croce CM, Condorelli G. c-FLIPL enhances anti-apoptotic Akt functions by modulation of Gsk3beta activity. Cell Death Differ. 2010;17:1908–1916. doi: 10.1038/cdd.2010.65. [DOI] [PubMed] [Google Scholar]
- 28.Antoine G, Scheiflinger F, Dorner F, Falkner FG. The complete genomic sequence of the modified vaccinia Ankara strain: comparison with other orthopoxviruses. Virology. 1998;244:365–396. doi: 10.1006/viro.1998.9123. [DOI] [PubMed] [Google Scholar]
- 29.Shisler JL, Moss B. Molluscum contagiosum virus inhibitors of apoptosis: The MC159 v-FLIP protein blocks Fas-induced activation of procaspases and degradation of the related MC160 protein. Virology. 2001;282:14–25. doi: 10.1006/viro.2001.0834. [DOI] [PubMed] [Google Scholar]
- 30.Abramoff M, Magalhaes P, Ram S. Image Processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
- 31.Randall CM, Jokela JA, Shisler JL. The MC159 protein from the molluscum contagiosum poxvirus inhibits NF-kappaB activation by interacting with the IkappaB kinase complex. J Immunol. 2012;188:2371–2379. doi: 10.4049/jimmunol.1100136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shisler JL, Jin XL. The vaccinia virus K1L gene product inhibits host NF-kappaB activation by preventing IkappaBalpha degradation. J Virol. 2004;78:3553–3560. doi: 10.1128/JVI.78.7.3553-3560.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang M, Liu Y, Wang P, Guan X, He S, Luo S, Li C, Hu K, Jin W, Du T, Yan Y, Zhang Z, Zheng Z, Wang H, Hu Q. HSV-2 immediate-early protein US1 inhibits IFN-beta production by suppressing association of IRF-3 with IFN-beta promoter. J Immunol. 2015;194:3102–3115. doi: 10.4049/jimmunol.1401538. [DOI] [PubMed] [Google Scholar]
- 34.Yu JW, Shi Y. FLIP and the death effector domain family. Oncogene. 2008;27:6216–6227. doi: 10.1038/onc.2008.299. [DOI] [PubMed] [Google Scholar]
- 35.Wolferstatter M, Schweneker M, Spath M, Lukassen S, Klingenberg M, Brinkmann K, Wielert U, Lauterbach H, Hochrein H, Chaplin P, Suter M, Hausmann J. Recombinant modified vaccinia virus Ankara generating excess early double-stranded RNA transiently activates protein kinase R and triggers enhanced innate immune responses. J Virol. 2014;88:14396–14411. doi: 10.1128/JVI.02082-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 37.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, Yamaguchi O, Otsu K, Tsujimura T, Koh CS, Reis e Sousa C, Matsuura Y, Fujita T, Akira S. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 38.Zhou Q, Lavorgna A, Bowman M, Hiscott J, Harhaj EW. Aryl Hydrocarbon Receptor Interacting Protein Targets IRF7 to Suppress Antiviral Signaling and the Induction of Type I Interferon. J Biol Chem. 2015;290:14729–14739. doi: 10.1074/jbc.M114.633065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grandvaux N, Servant MJ, tenOever B, Sen GC, Balachandran S, Barber GN, Lin R, Hiscott J. Transcriptional Profiling of Interferon Regulatory Factor 3 Target Genes: Direct Involvement in the Regulation of Interferon-Stimulated Genes. Journal of Virology. 2002;76:5532–5539. doi: 10.1128/JVI.76.11.5532-5539.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Golks A, Brenner D, Fritsch C, Krammer PH, Lavrik IN. c-FLIPR, a new regulator of death receptor-induced apoptosis. The Journal of biological chemistry. 2005;280:14507–14513. doi: 10.1074/jbc.M414425200. [DOI] [PubMed] [Google Scholar]
- 41.Irmler M, Thome M, Hahne M, Schneider P, Hofmann K, Steiner V, Bodmer JL, Schroter M, Burns K, Mattmann C, Rimoldi D, French LE, Tschopp J. Inhibition of death receptor signals by cellular FLIP. Nature. 1997;388:190–195. doi: 10.1038/40657. [DOI] [PubMed] [Google Scholar]
- 42.Scaffidi C, Schmitz I, Krammer PH, Peter ME. The role of c-FLIP in modulation of CD95-induced apoptosis. J Biol Chem. 1999;274:1541–1548. doi: 10.1074/jbc.274.3.1541. [DOI] [PubMed] [Google Scholar]
- 43.Valmiki MG, Ramos JW. Death effector domain-containing proteins. Cell Mol Life Sci. 2009;66:814–830. doi: 10.1007/s00018-008-8489-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Katayama R, Ishioka T, Takada S, Takada R, Fujita N, Tsuruo T, Naito M. Modulation of Wnt signaling by the nuclear localization of cellular FLIP-L. J Cell Sci. 2010;123:23–28. doi: 10.1242/jcs.058602. [DOI] [PubMed] [Google Scholar]
- 45.Ozturk S, Schleich K, Lavrik IN. Cellular FLICE-like inhibitory proteins (c-FLIPs): fine-tuners of life and death decisions. Exp Cell Res. 2012;318:1324–1331. doi: 10.1016/j.yexcr.2012.01.019. [DOI] [PubMed] [Google Scholar]
- 46.Kumar KP, McBride KM, Weaver BK, Dingwall C, Reich NC. Regulated nuclear-cytoplasmic localization of interferon regulatory factor 3, a subunit of double-stranded RNA-activated factor 1. Mol Cell Biol. 2000;20:4159–4168. doi: 10.1128/mcb.20.11.4159-4168.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu M, Fang T, Li S, Meng K, Guo D. Bipartite Nuclear Localization Signal Controls Nuclear Import and DNA-Binding Activity of IFN Regulatory Factor 3. J Immunol. 2015;195:289–297. doi: 10.4049/jimmunol.1500232. [DOI] [PubMed] [Google Scholar]
- 48.Thornberry NA, Bull HG, Calaycay JR, Chapman KT, Howard AD, Kostura MJ, Miller DK, Molineaux SM, Weidner JR, Aunins J, et al. A novel heterodimeric cysteine protease is required for interleukin-1 beta processing in monocytes. Nature. 1992;356:768–774. doi: 10.1038/356768a0. [DOI] [PubMed] [Google Scholar]
- 49.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang P, Zhang J, Bellail A, Jiang W, Hugh J, Kneteman NM, Hao C. Inhibition of RIP and c-FLIP enhances TRAIL-induced apoptosis in pancreatic cancer cells. Cell Signal. 2007;19:2237–2246. doi: 10.1016/j.cellsig.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 51.Ricci MS, Jin Z, Dews M, Yu D, Thomas-Tikhonenko A, Dicker DT, El-Deiry WS. Direct repression of FLIP expression by c-myc is a major determinant of TRAIL sensitivity. Mol Cell Biol. 2004;24:8541–8555. doi: 10.1128/MCB.24.19.8541-8555.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith GL, Benfield CT, Maluquer de Motes C, Mazzon M, Ember SW, Ferguson BJ, Sumner RP. Vaccinia virus immune evasion: mechanisms, virulence and immunogenicity. J Gen Virol. 2013;94:2367–2392. doi: 10.1099/vir.0.055921-0. [DOI] [PubMed] [Google Scholar]
- 53.Ma Z, Jacobs SR, West JA, Stopford C, Zhang Z, Davis Z, Barber GN, Glaunsinger BA, Dittmer DP, Damania B. Modulation of the cGAS-STING DNA sensing pathway by gammaherpesviruses. Proc Natl Acad Sci U S A. 2015 doi: 10.1073/pnas.1503831112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu L, Eby MT, Rathore N, Sinha SK, Kumar A, Chaudhary PM. The human herpes virus 8-encoded viral FLICE inhibitory protein physically associates with and persistently activates the Ikappa B kinase complex. J Biol Chem. 2002;277:13745–13751. doi: 10.1074/jbc.M110480200. [DOI] [PubMed] [Google Scholar]
- 55.Gong J, Kumar SA, Graham G, Kumar AP. FLIP: molecular switch between apoptosis and necroptosis. Mol Carcinog. 2014;53:675–685. doi: 10.1002/mc.22027. [DOI] [PubMed] [Google Scholar]
- 56.Rajput A, Kovalenko A, Bogdanov K, Yang SH, Kang TB, Kim JC, Du J, Wallach D. RIG-I RNA helicase activation of IRF3 transcription factor is negatively regulated by caspase-8-mediated cleavage of the RIP1 protein. Immunity. 2011;34:340–351. doi: 10.1016/j.immuni.2010.12.018. [DOI] [PubMed] [Google Scholar]
- 57.Fricker N, Beaudouin J, Richter P, Eils R, Krammer PH, Lavrik IN. Model-based dissection of CD95 signaling dynamics reveals both a pro- and antiapoptotic role of c-FLIPL. J Cell Biol. 2010;190:377–389. doi: 10.1083/jcb.201002060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chang DW, Xing Z, Pan Y, Algeciras-Schimnich A, Barnhart BC, Yaish-Ohad S, Peter ME, Yang X. c-FLIP(L) is a dual function regulator for caspase-8 activation and CD95-mediated apoptosis. EMBO J. 2002;21:3704–3714. doi: 10.1093/emboj/cdf356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Santana-de Anda K, Gomez-Martin D, Monsivais-Urenda AE, Salgado-Bustamante M, Gonzalez-Amaro R, Alcocer-Varela J. Interferon regulatory factor 3 as key element of the interferon signature in plasmacytoid dendritic cells from systemic lupus erythematosus patients: novel genetic associations in the Mexican mestizo population. Clin Exp Immunol. 2014;178:428–437. doi: 10.1111/cei.12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Papatriantafyllou M. Infection: the interferon paradox. Nat Rev Immunol. 2013;13:392. doi: 10.1038/nri3461. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.