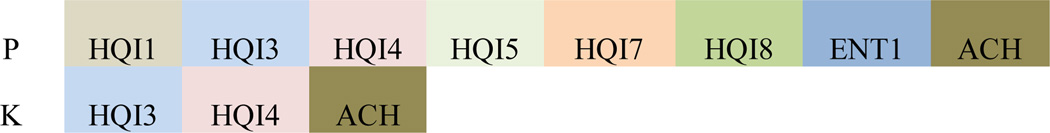Abstract
Protein hydroxylation is an emerging posttranslational modification involved in both normal cellular processes and a growing number of pathological states, including several cancers. Protein hydroxylation is mediated by members of the hydroxylase family of enzymes, which catalyze the conversion of an alkyne group at select lysine or proline residues on their target substrates to a hydroxyl. Traditionally, hydroxylation has been identified using expensive and time-consuming experimental methods, such as tandem mass spectrometry. Therefore, to facilitate identification of putative hydroxylation sites and to complement existing experimental approaches, computational methods designed to predict the hydroxylation sites in protein sequences have recently been developed. Building on these efforts, we have developed a new method, termed RF-Hydroxysite, that uses random forest to identify putative hydroxylysine and hydroxyproline residues in proteins using only the primary amino acid sequence as input. RF-Hydroxysite integrates features previously shown to contribute to hydroxylation site prediction with several new features that we found to augment the performance remarkably. These include features that capture physicochemical, structural, sequence-order and evolutionary information from the protein sequences. The features used in the final model were selected based on their contribution to the prediction. Physicochemical information was found to contribute the most to the model. The present study also sheds light on the contribution of evolutionary, sequence order, and protein disordered region information to hydroxylation site prediction. The web server for RF-Hydroxysite is available online at http://bcb.ncat.edu/RF_hydroxy/.
1. Introduction
Though it was first identified over fifty years ago as an essential component of collagen fibres, protein hydroxylation has only recently begun to emerge as an important posttranslational modification (PTM) involved in the etiology of a variety of diseases, including breast, stomach and lung cancers [1]. Protein hydroxylation is mediated by a family of approximately 70 hydroxylase enzymes that utilize the cofactors oxygen, Fe(II), ascorbate and 2-oxoglutarate to catalyze the conversion of specific lysine and proline residues on their target proteins to hydroxylysine (HyK) and hydroxyproline (HyP), respectively [2]. The hydroxylated residues are essential elements of collagen and connective tissues, as well as precursors for subsequent PTMs, such as glycosylation. As a consequence, hydroxylation plays an important role in key physiological processes, such as tissue stability, molecular assembly, metabolism and oxygen-dependent regulation of hypoxia.
In order to gain a better understanding of the role of hydroxylation in normal cellular physiology and disease, it is important to identify hydroxylation sites in cellular proteins. This information can also help inform the development of pharmacological interventions for diseases associated with the dysregulation of protein hydroxylation. Experimental identification of hydroxylation sites is typically labor-intensive and time consuming, requiring expensive instrumentation, such as tandem mass spectrometers, and a high degree of technical expertise. Moreover, due to the large number of peptide fragments analyzed during a given experiment, standard approaches, such as shotgun mass spectrometry, may miss low abundance hydroxylation sites. Recently, three studies focused on the development of computational methods for hydroxylation site prediction have been reported. In the first study, Hu et al. combined support vector machines (SVMs) with position-specific scoring matrices (PSSM) and physicochemical properties to predict sites of hydroxylation in sliding windows of size of 9 [3]. The accuracies achieved using this approach were 76% and 82.1% for HyP and HyK, respectively. Importantly, this study was one of the first to demonstrate that evolutionary and physicochemical information can contribute to the prediction of hydroxylation sites. However, no web server or standalone software is currently available for this method. The second method, termed iHyd-PseAAC [4], incorporated dipeptide position-specific propensity into amino acid composition (PseAAC) for a sequence window of size 15 and used discriminant analysis for training and prediction. The reported average accuracy of iHyd-PseAAC was 79.5% and 83.34% for HyP and HyK, respectively. Most recently, Shi and colleagues developed PredHydroxy [5], which uses SVM to integrate position weight amino acid composition (PWAAC) information with 8 high-quality amino acid physicochemical property indices (HQI). Using this approach, the authors observed accuracies of 84.51% and 83.33% for HyP and HyK, respectively.
Though these studies have laid a solid foundation for hydroxylation site prediction, there is still room for improvement with respect to accuracy, efficiency and overall performance. Here, we describe a new method based on random forest (RF) that combines information about physicochemical, structural, evolutionary and sequence-order features to accurately predict sites of protein hydroxylation using the primary amino acid sequence as input. This method, which we term RF-Hydroxysite, benefits from the integration of several features, such as average cumulative hydrophobicity and position-specific entropy, that have not previously been used in hydroxysite prediction. Aside from their impact on hydroxysite prediction, these features may also offer important insights into the biochemical parameters underlying substrate recognition and subsequent hydroxylation by hydroxylase family members.
2. Material and methods
A. Sequences and sequence preparation
The protein sequences used in this study, which are the same as those used during the development of PredHydroxy, were extracted from UniProtKB /Swiss-Prot database (version 2014_1). The sequences correspond to known, experimentally-verified hydroxylation sites for both lysine (34 sequences) and proline (265 sequences). Similarly, sequences that contain lysine and proline residues that have been shown not to be hydroxylated under physiological conditions were downloaded from the UniProt database. These non-hydroxylated sites served as negative controls during method development and evaluation. To minimize the possibility that some negative sites may be reported as positive sites in the future, Rvp-net [6] was used to filter any negative site with absolute surface area more than 40%. To eliminate redundancies in the datasets, the sequences that share more than 40% sequence identity were removed using the CD-HIT standalone program [7] from both datasets. From the remaining unique sequences, windows of length 7, 9, 11, 13, 15, 17, and 19 amino acid residues were prepared with positive or negative lysine/proline residues in the center of the window. The various windows lengths were then used to identify the one that yielded the best performance of the model. The number of windows with positive hydroxylated sites used in this study is 97 for lysine and 719 for proline. The same numbers of windows with non-hydroxylated lysine and proline residues were selected randomly as unbiased negative controls to balance the positive windows. Finally, before training was initiated, 10% of the dataset was drawn randomly and put aside to serve as an independent sample for testing.
B. Sequence features
Protein sequences are represented with 11 feature types reflecting evolutionary, physiochemical, sequential, structural, and functional information. The features include position weight amino acid composition (PWAA), high-quality physicochemical property indices (HQI), type I entropy (ENT1), type I relative entropy (RE1), type I information gain (IG1), overlapping properties (OP), average cumulative hydrophobicity (ACH), protein disordered region features (PDR), type II entropy (ENT2), type II relative entropy (RE2), and type II information gain (IG2).
1-Position weight amino acid composition
Position weight amino acid composition (PWAA) is used to extract position information about amino acid residues surrounding potential hydroxylation sites in a protein sequence fragment [5]. There are 20 PSWAA features, with each one representing the position weight of an amino aai =(A, C, D, E, F, G, H, I, K, L, M, N, P,Q, R, S, T, V,W, Y) where i denotes the position index in the amino acid list. In a protein sequence window of length 2n+1, in which the potential hydroxylation site is in the center at position n+1, the surrounding amino acid position weights are given by
| (1) |
where n denotes the number of downstream or upstream residues from the center of the window, j denotes the position of the amino acid relative to the center (−n ≤ j ≤ n), and xij =1 if the amino acid aai is found in position j of the sequence window or xij = 0 otherwise.
2-High quality physicochemical indices
High quality physicochemical indices (HQI) as described in PredHydroxy [5] are the indices of eight physicochemical properties that represent the eight group identified by clustering the 544 correlated amino acid properties in the AAindex1 database [8]. The properties are propensity (BLAM930101) [9] (HQI1), information value for accessibility (BIOV880101) [10] (HQI2), normalized frequency of alpha-helix (MAXF760101) [11] (HQI3), volumes including the crystallographic waters (TSAJ990101) [12] (HQI4), amino acid composition of MEM of multi-spanning proteins (NAKH920108) [13] (HQI5), composition of amino acids in intracellular proteins (CEDJ970104) [14] (HQI6), conformational preference for all beta-strand (LIFS790101) [15] (HQI7), and optimized relative partition energies (MIYS990104) [16] (HQI8). HQI features are extracted by representing the amino acid residues surrounding the hydroxylation site by a property of a corresponding index. The number of HQI features for a sequence window of size n is 8×(n−1).
3-Type I entropy
Type I entropy (ENT1) is calculated using probabilities of the individual amino acids in the window to generate one numeric feature. It is calculated as
| (2) |
where pi is the probability of an amino acid i=(A, C, D, E, F, G, H, I, K, L, M, N, P,Q, R, S, T, V,W, Y) in the sequence and it is computed as the total number of amino acids, i, divided by the length of the window, assuming that the probability of any amino acid that does not exist in the window is zero. Entropy ranges between zero, where only one type of residue in the entire sequence is found, and 3.17, where all types of amino acids have equal occurrence in the window.
4-Type I relative entropy
Type I relative entropy (RE1) of the distribution pi of an amino acid and its random distribution, p0, is calculated as
| (3) |
where p0=1/n, the probability that all amino acids have equal occurrence in the window of size n. RE is always non-negative and becomes zero if and only if pi = p0. Like entropy, the relative entropy is represented by one feature for each window. We again assumed that the probability of any amino acid that does not exist in the window is zero.
5-Type I Information gain
Type I information gain (IG1) is computed by subtracting RE1 from H1. It measures the transformation of information in a sequence fragment influenced by a grouping factor.
| (4) |
6-Overlapping properties
Overlapping properties (OP) captures information from common physicochemical properties shared by the amino acids in a protein fragment [17, 18]. The amino acids were classified based on ten physicochemical properties: polar (NQSDECTK-RHYW), positive (KHR), negative (DE), charged (KHRDE), hydrophobic (AGCTIVLKHFWYM), aliphatic (IVL), aromatic (FYWH), small (PNDTCAGSV), tiny (ASGC), and proline (P). An amino acid may fall into more than one group (i.e., be overlapping). Each amino acid was encoded with 10-bit, where each bit in the code represents a group, respectively. The position of the bit is set to 1 if the amino acid belongs to the corresponding group and 0 if it does not. For example, histidine (H) is encoded with 1101101000, which indicates that it belongs to polar, positive, charged, hydrophobic, and aromatic groups. The number of features extracted with this method is n×10 where n is the window size [18].
7-Average cumulative hydrophobicity
The average cumulative hydrophobicity (ACH) quantifies the tendency of the amino acids that surround the hydroxylation sites in a protein fragment to interact with solvents. The Eisenberg hydrophobicity scales [19] were used, where:
A: 0.62, C: 0.29, D: −0.90, E: −0.74, F: 1.19, G: 0.48, H: −0.40, I: 1.38, K: −1.50, L: 1.06, M: 0.64, N: −0.78, P: 0.12, Q: −0.85, R: −2.53, S: −0.18, T: −0.05, V: 1.08, W: 0.81, Y: 0.26
The number of ACH features depends on the size of the window. For a window of size 9, the ACH is computed by averaging the cumulative hydrophobicity indices of the amino acids around the putative hydroxylation site for the sub-windows of sizes 3, 5, 7 and 9, respectively, where K/P is always in the centre of the window. For example, to calculate ACH for the sequence KAGVPHED, we need first to create the sub-windows AGVPHED, GVPHE, and VPH. Then we can calculate the feature of each window as:
| (5) |
where n is the sub-window size and Pi is hydrophobicity index for the amino acid in the position i in the window. For this example the number of features is four.
8-Protein disordered region
Previous studies suggested that many PTMs take place in disordered regions of the protein, where enzymes and solvent interact easily with the residues [20]. The protein disordered region information was extracted for the residues surrounding the hydroxylation sites using DISOPRED [21], which is standalone software for the prediction of protein disorder. The software assigns an amino acid 1 if it is disordered and 0 otherwise.
9-Type II entropy, relative entropy and information gain
Type II entropy (ENT2), relative entropy (RE2), and information gain (IG2) are computed using the above entropy, relative entropy, and information gain equations but the probabilities are substituted with the position-specific weighted observed percentages (WOP) [22], which are obtained by aligning each sequence to related homologous protein sequences in the NCBI non-redundant protein database (nr). The alignment is performed with the NCBI executable position specific iterative basic local alignment sequence tool (psi-blast) [23], which uses BLOSUM62 [24] as a scoring matrix for the initial alignment. Then, a position specific scoring matrix (PSSM) and WOP are generated iteratively and each time the new PSSM is used as a scoring matrix for a new alignment until the convergence or the stopping criterion is reached. The WOP generated from the last psi-blast alignment is used to calculate H2, RE2, and IG2. These three feature types provide evolutionary information for each training sequence by reflecting the conservatism of the amino acid residues surrounding the hydroxylation sites.
C. Model learning and testing
In this study, random forest (RF) [30], which is a popular tree-based ensemble machine learning technique, was used to construct a model using the features extracted from the benchmark sequences to predict lysine and proline hydroxylation sites in a protein sequences. The RF is a combination of a number of decision trees. Each tree is constructed with a bootstrap sample from the training dataset. A tree is composed of a root node, internal nodes, and terminal nodes. Each internal node represents a subset of the training data split based on a decision function of the best discriminatory feature. An internal node may further split into two nodes. The splitting features are selected based on feature importance. The terminal nodes represent the classified dataset. Put simply, training a model with an RF algorithm is the process of finding the tree structures and decision rules from the training data. Unknown sequence windows are classified by each tree in the forest whether they are positive hydroxylation sites or negative sites by traversing each tree starting from the root node down to terminal nodes where the path is determined according to the outcome of the splitting function at each node. The final classification is based on the general agreement of most decision trees rather than only one. Scikit-learn, a Python package for machine learning, was used to implement RF algorithm [25]. The RF parameters were chosen to optimize the performance of the model. These parameters included the number of trees (100) [26], the depth (i.e., until node purity is achieved) and the number of features. Though the number of features varies depending on the window size, in each case the maximum number of features for a given window was considered (Sup-B). For comparison, four other ensemble learning methods (AdaBoost, Bagging, Gradient Boosting, and Extra-Trees Classifier) were also tested and analyzed. The results for the other four ensemble methods, which demonstrate the robustness of the selected features, are provided in supplemental information (Sup-A).
The models were rigorously tested with both jackknife cross validation and an independent test set. In the jackknife test, an instance of the dataset is left out for testing and the remaining instances are used for training one at a time until all instances in the dataset are tested without being in the training dataset. For the independent test, a sample of 10% of the sequences was selected randomly from both positive and negative windows and left out for testing the model while the remaining 90% of the sequences were used to construct the model. Furthermore, entire protein sequences of experimentally verified positive hydroxylation sites were selected randomly as independent sequences and were removed from the training data. These sequences then were tested with RF-Hydroxysite to compare the numbers and positions of the experimentally verified sites in each sequence to those that were predicted by our model.
The testing results from both jackknife cross validation and the independent test were evaluated for accuracy, specificity, sensitivity, precision, F1-score, Matthew’s correlation coefficient (MCC), and area under the receiver operating characteristic curve (AUC). These parameters are defined below:
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
| (6) |
where TP is the true positive rate, TN is the rate of true negative rate, FP is the false positive rate and FN is the false negative rate.
3. Results and discussion
To develop a robust computational tool that is able to identify putative hydroxylation sites using only the primary amino acid sequence as input, we examined a diverse set of sequence features using a random forest (RF)-based approach. To this end, we first evaluated sequence windows of lengths 7, 9, 11, 13, 15, 17, and 19 residues for both training and testing with RF. The number of trees in the RF was chosen to be 100 based on a previous study [26]. The features extracted from the sequences represented information pertaining to sequence-order, physicochemical properties, protein structure and evolutionary relationships. To capture this information, we integrated a variety of features, including position weight amino acid composition (PWAA), high quality indices (HQI1–HQI8), type I entropies (H1, RE1, and IG1), overlapping properties (OP), average cumulative hydrophobicity (ACH), protein disordered regions (PDR) and type II entropies (H2, RE2, and IG2), into a sequence-window represented with a numerical feature vector.
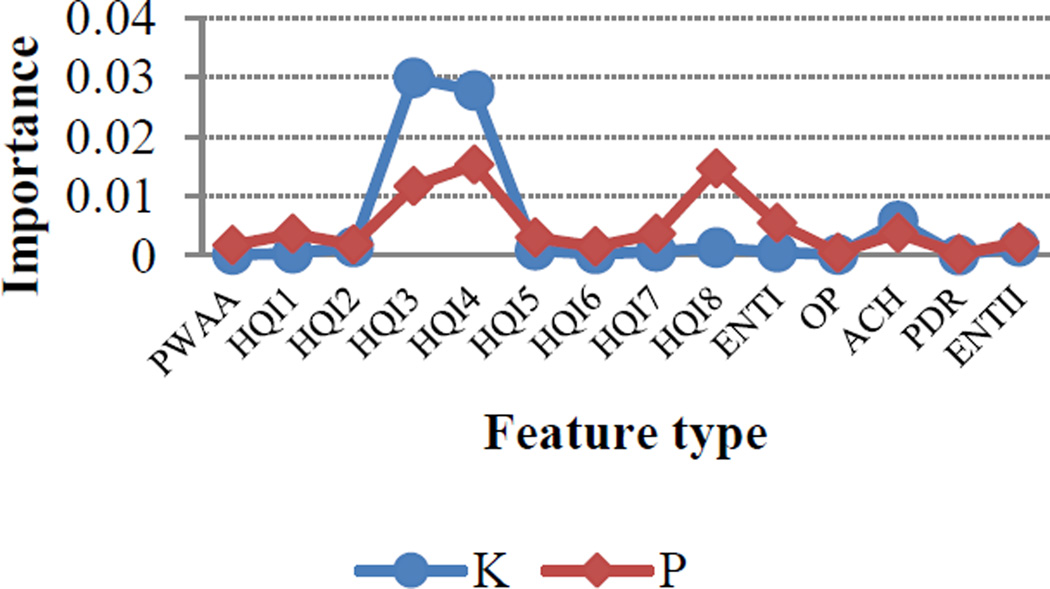
One of the primary advantages of using an RF-based approach was that it allowed us to evaluate the relative impact of each feature on method performance. To identify the feature types that contributed most significantly to the performance, we implemented Gini feature index to quantify the relative importance of each feature. Although all feature types contributed to model improvement to some degree, some features had a greater impact than others. For instance, Figure 1 shows that HQI3 (normalized frequency of alpha-helix) and HQI4 (protein volumes, including the crystallographic waters) had a strong impact for both HyK and HyP residues.
Figure 1.
The importance scale of feature types for hydroxylysine (blue) and hydroxyproline (red)
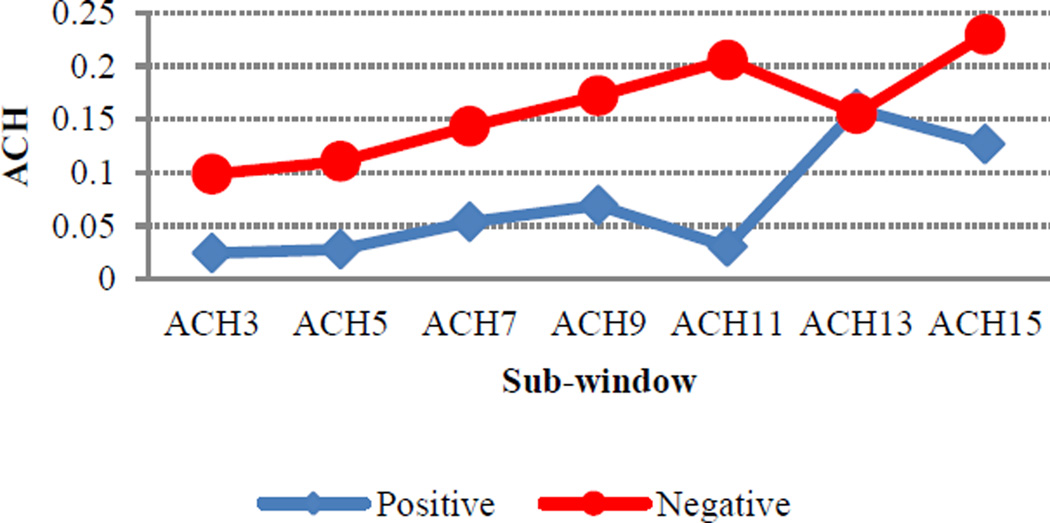
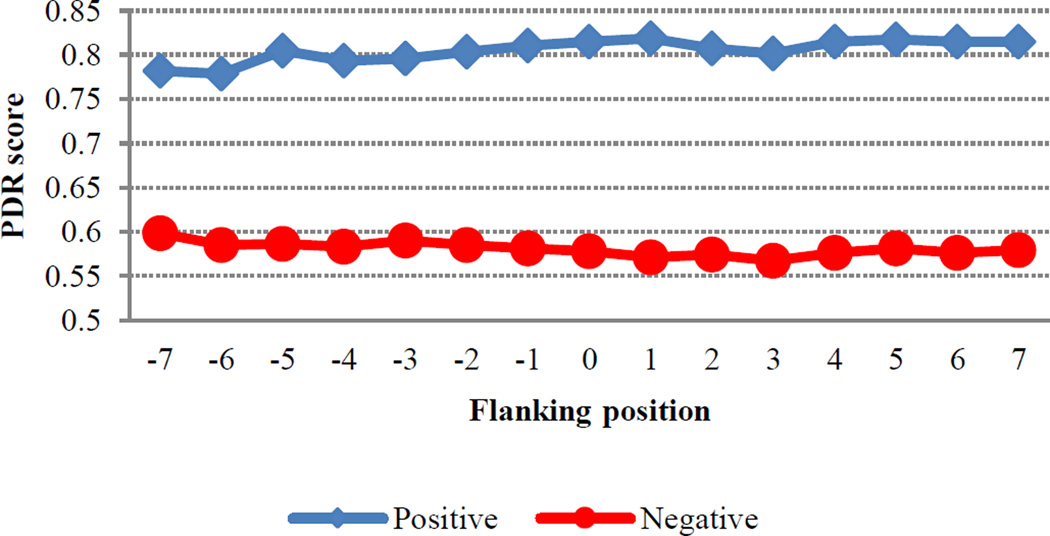
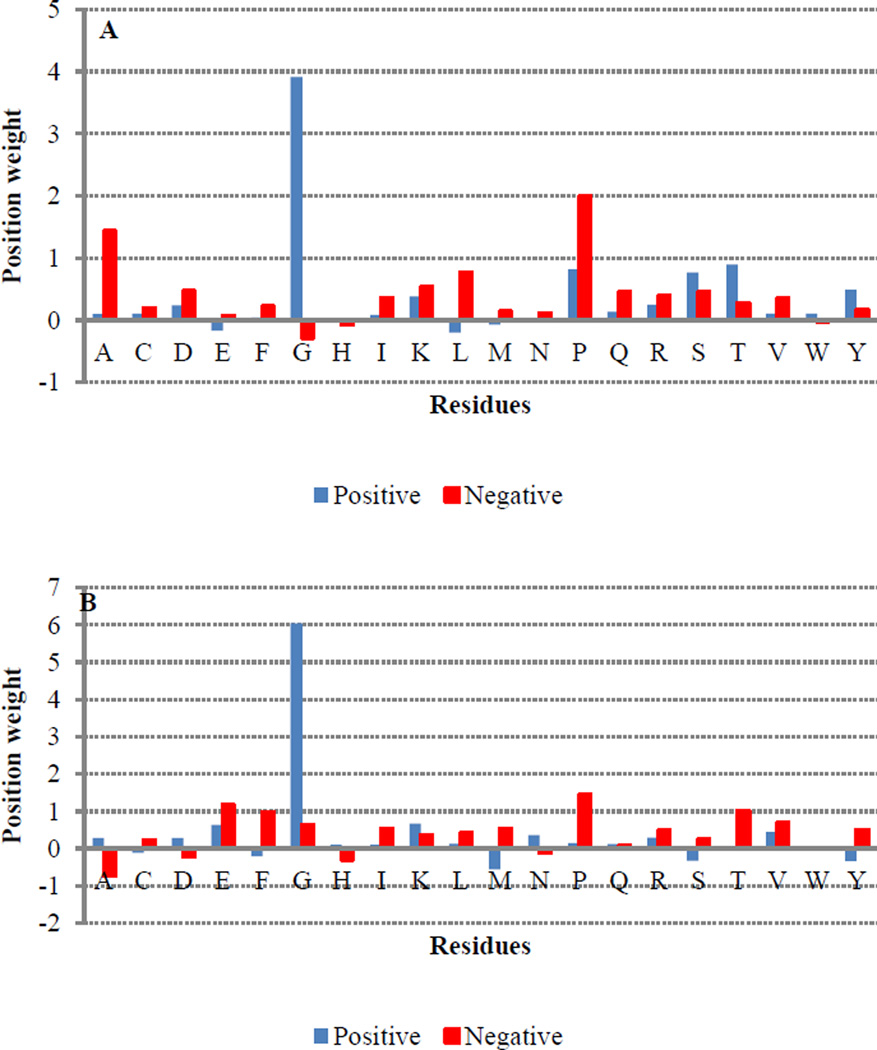
Likewise, ACH also had a substantial impact for both residues, with more hydrophilic environments being favored for positive sites. This is perhaps not surprising since sites of hydroxylation are expected to be found near the protein surface. However, it is important to note that, though the influence of ACH is quite pronounced for sequence windows less than 11, its impact decreases dramatically at window sizes larger than 13 (Figure 2). Interestingly, HQI8 (optimized relative partition energies) appeared to be highly important for prediction of HyP residues but had little impact on the prediction of HyK residues. While exploring the feature profiles, we also noticed that the sequences surrounding positive hydroxylation sites exhibited consistently higher protein disorder region (PDR) scores than those surrounding negative sites (Figure 3). The difference between positive and negative sites is particularly pronounced at positions +1, +2 and +3, suggesting that a high degree of flexibility may be required immediately C-terminal to the site of hydroxylation. This is consistent with the PWAA feature profiles, which show that glycine exhibits the heaviest position-weight amongst the upstream flanking residues for both HyP and HyK (Figure 4). Glycine is the smallest amino acid and is frequently found in disordered protein regions [27] where flexibility is required. Interestingly, other residues also associated with disordered regions, such as proline, were found more often in the negative set than the positive set for both residues.
Figure 2.
Average cumulative hydrophobicity for hydroxyproline flanking regions where x-axis represents the sub-windows of size 3, 5, 7, 9, 11, 13, and 15 and y-axis the average of ACH for positive and negative hydroxyproline windows.
Figure 3.
Protein disordered region (PDR) scores for hydroxyproline flanking position, where y-axis shows the average of PDR scores. Higher PDR scores correspond to more disordered regions.
Figure 4.
The average of position weight amino acid composition in the flanking regions for positive and negative proline (A) and lysine (B) hydroxylation sites
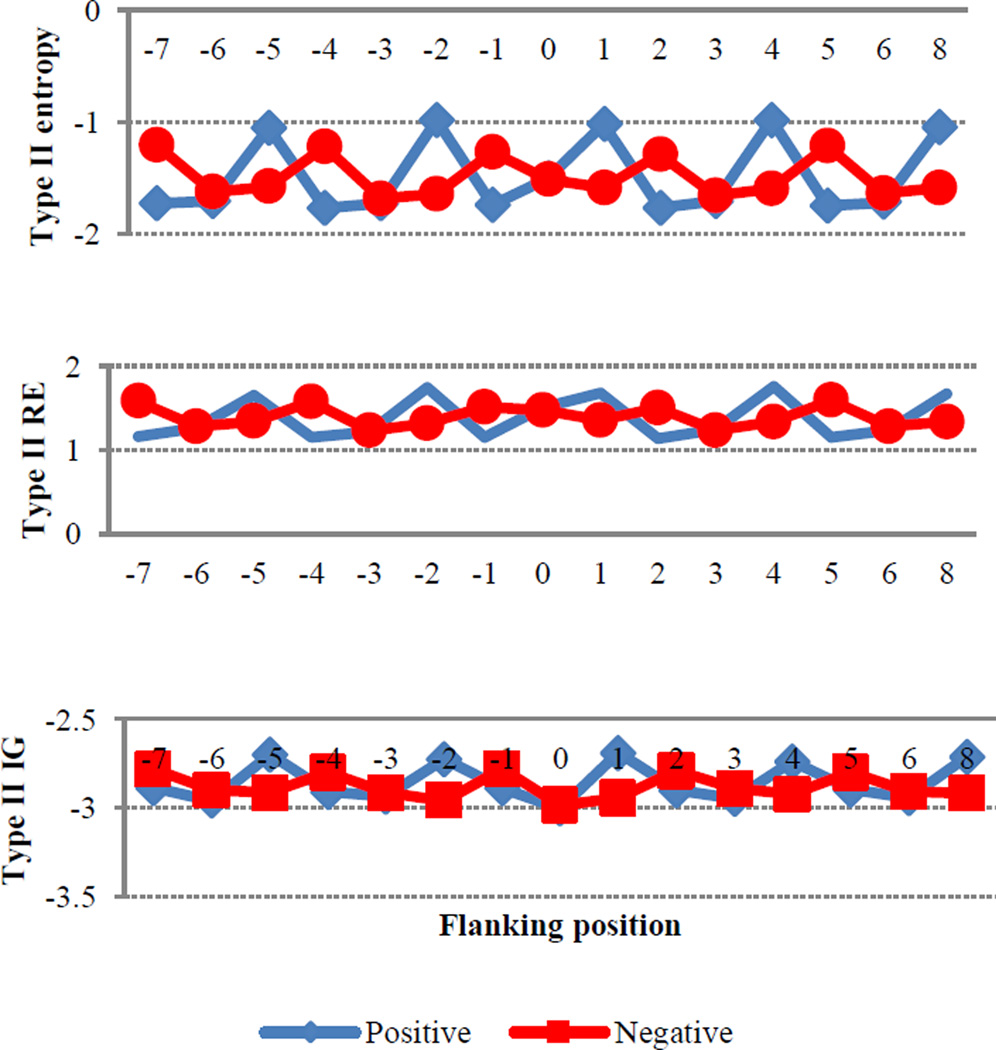
In addition to information about disorder, the type II entropies (EN2, RE2, and IG2) also contain evolutionary information that may reveal the conservation of a function. Since hydroxylation sites have important implications with regard to protein function, we hypothesized that some degree of conservation in the flanking regions of both HyP and HyK would be observed. As can be seen in Figure 5, differences in the evolutionary information between positive and negative HyP sites fluctuate in a reciprocal manner. The flanking positions of HyK show a similar pattern (please see Sup-C Figure C-7 in supplemental information). The biological significance of this observation requires further investigation.
Figure 5.
The average of type II entropies for positive and negative hydroxyproline flanking positions
In addition to profiling the ensemble dataset, we also evaluated a randomly selected collection of individual sequences (Sup-C). To facilitate comparison between positive and negative sets, the window size for all analyses was arbitrarily set at 15. Interestingly, several features exhibited distinct patterns, both between the positive and negative datasets and within a given dataset. For instance, using HQI-1 for hydroxyproline, nearly all of the sequences in the negative dataset were characterized by relatively flat feature profiles. In contrast, the positive hydroxyproline sequences appeared to cluster into two distinct groups: one in which the feature profiles remained flat and another that was characterized by an oscillatory pattern throughout the window. Likewise, Shannon Entropy exhibited distinct patterns between the positive and negative hydroxyproline sequences. However, in this case, nearly all of the negative sites exhibited an oscillatory pattern while the majority of the regions surrounding positive sites exhibited uniformly flat profiles (albeit with one subset clustered around 0 AU and another set clustered around −2.0 AU). It will be interesting to see whether these patterns correlate with different hydroxylases and/or hydroxylase subfamilies.
Together, these analyses allowed us to identify the most important features for method development. Though the contribution of all eleven feature types and their overall impact on the performance of the model is evident, only the top contributors were selected in the final model for each hydroxylation site (Table 1). These features were selected based on the level of their average feature importance. This was done in order to develop a model that is simultaneously non-complicated and efficient. Figure 6 shows the feature order in the final hydroxyproline (P) and hydroxylysine (K) models. Likewise, the lengths of the feature vectors, which were dependent on the window-size, are provided in Supplemental Figure B (Sup-B). Table 2A and 2B show the evaluation metrics for the RF-based models before and after feature selection, respectively. The evaluation metrics of the other ensemble learning methods are also included in Supplemental Figure A (Sup-A).
Table 1.
The list of the features used to develop the models. Only the checked features were selected for the final hydroxyproline and hydroxylysine model while the crossed features were omitted. The shaded features belong to the HQI feature type.
| Features | P | K |
|---|---|---|
| Position weight amino acid (PWAA) | ✖ | ✖ |
| Propensity (HQI1) | ✓ | ✖ |
| Solven accessibility (HQI2) | ✖ | ✖ |
| Alpha-helix frequency (HQI3) | ✓ | ✓ |
| Crystallographic waters (HQI4) | ✓ | ✓ |
| Amino acid composition of MEM (HQI5) | ✓ | ✖ |
| Composition of AA in intracellular (HQI6) | ✖ | ✖ |
| Conformational preference (HQI7) | ✓ | ✖ |
| Partition energies (HQI8) | ✓ | ✖ |
| Type I entropy (ENTI) | ✓ | ✖ |
| Overlapping properties (OP) | ✖ | ✖ |
| Average cumulative hydrophobicity (ACH) | ✓ | ✓ |
| Protein disordered region (PDR) | ✖ | ✖ |
| ENTII (Type II entropy) | ✖ | ✖ |
Figure 6.
Feature order in the final hydroxyproline and hydroxylysine models.
Table 2.
Method performance across different window sizes before (A) and after (B) feature selection.
| Win-size | Residue | Accu | Prec | Sens | Spec | F1sc | MCC |
|---|---|---|---|---|---|---|---|
| 7 | K | 0.98 | 1.00 | 0.95 | 1.00 | 0.98 | 0.95 |
| P | 0.94 | 0.96 | 0.91 | 0.97 | 0.94 | 0.89 | |
| 9 | K | 0.97 | 0.95 | 0.99 | 0.96 | 0.97 | 0.94 |
| P | 0.94 | 0.92 | 0.97 | 0.91 | 0.95 | 0.89 | |
| 11 | K | 0.98 | 0.95 | 1.00 | 0.96 | 0.98 | 0.95 |
| P | 0.94 | 0.92 | 0.97 | 0.91 | 0.94 | 0.88 | |
| 13 | K | 0.98 | 0.95 | 1.00 | 0.96 | 0.98 | 0.95 |
| P | 0.94 | 0.92 | 0.96 | 0.91 | 0.94 | 0.87 | |
| 15 | K | 0.97 | 0.95 | 0.99 | 0.96 | 0.97 | 0.94 |
| P | 0.95 | 0.96 | 0.93 | 0.96 | 0.94 | 0.89 | |
| 17 | K | 0.97 | 1.00 | 0.94 | 1.00 | 0.97 | 0.94 |
| P | 0.94 | 0.96 | 0.92 | 0.96 | 0.94 | 0.89 | |
| 19 | K | 0.98 | 0.95 | 1.00 | 0.96 | 0.98 | 0.95 |
| P | 0.94 | 0.96 | 0.92 | 0.97 | 0.94 | 0.89 | |
| A | |||||||
| Win-size | Residue | Accu | Prec | Sens | Spec | F1sc | MCC |
|---|---|---|---|---|---|---|---|
| 7 | K | 0.97 | 1.00 | 0.93 | 1.00 | 0.97 | 0.93 |
| P | 0.94 | 0.96 | 0.92 | 0.96 | 0.94 | 0.88 | |
| 9 | K | 0.97 | 0.94 | 0.99 | 0.94 | 0.96 | 0.93 |
| P | 0.94 | 0.92 | 0.98 | 0.90 | 0.94 | 0.88 | |
| 11 | K | 0.96 | 0.93 | 0.99 | 0.93 | 0.96 | 0.92 |
| P | 0.94 | 0.92 | 0.96 | 0.91 | 0.94 | 0.87 | |
| 13 | K | 0.97 | 0.94 | 0.99 | 0.94 | 0.96 | 0.93 |
| P | 0.93 | 0.92 | 0.96 | 0.91 | 0.94 | 0.87 | |
| 15 | K | 0.96 | 0.94 | 0.98 | 0.94 | 0.96 | 0.92 |
| P | 0.94 | 0.96 | 0.92 | 0.97 | 0.94 | 0.89 | |
| 17 | K | 0.96 | 0.99 | 0.93 | 0.99 | 0.96 | 0.92 |
| P | 0.95 | 0.97 | 0.92 | 0.97 | 0.94 | 0.90 | |
| 19 | K | 0.96 | 0.94 | 0.99 | 0.94 | 0.96 | 0.93 |
| P | 0.94 | 0.96 | 0.91 | 0.96 | 0.93 | 0.88 | |
| B | |||||||
By testing several lengths of sequence windows, we expected that information about the hydroxylation sites would concentrate or fade in a particular range of flanking depths. However, to our surprise, after comparing the results from various window lengths, we found that the results of all window sizes are similar to one another, with window size 7 showing marginally better performance across most metrics. Therefore, in the final model, which we termed RF-Hydroxysite, we utilized a window size of seven. However, it is important to note that users can choose from any of the window sizes in the web-based interface (http://bcb.ncat.edu/RF_hydroxy/).
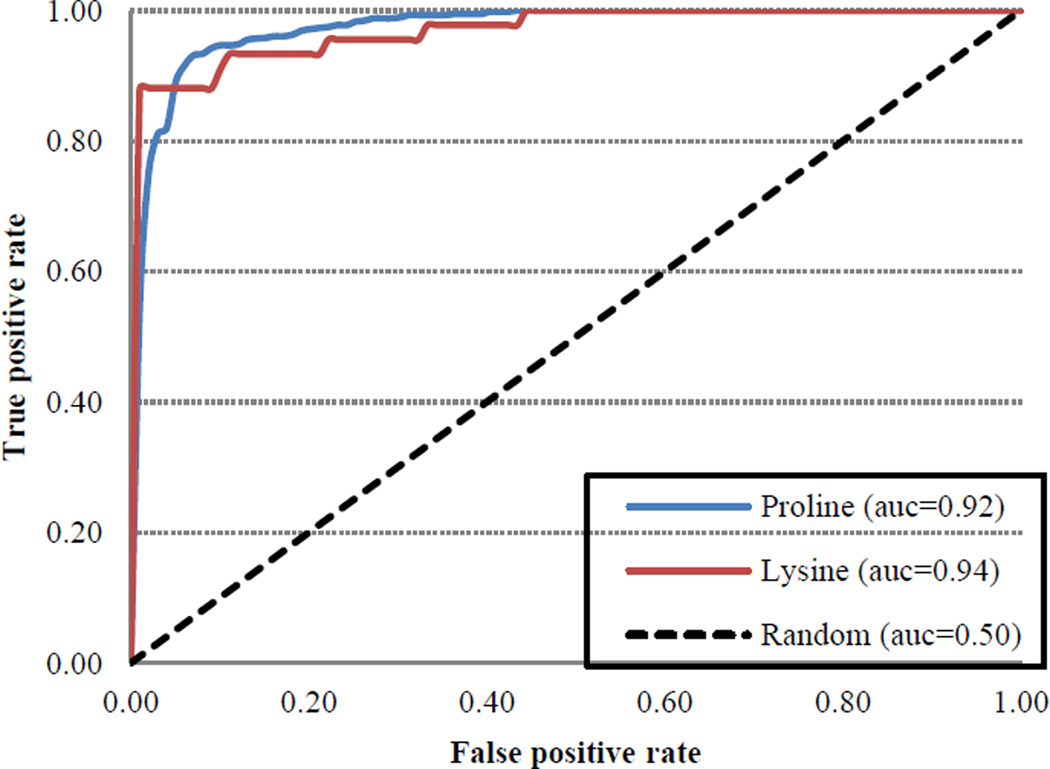
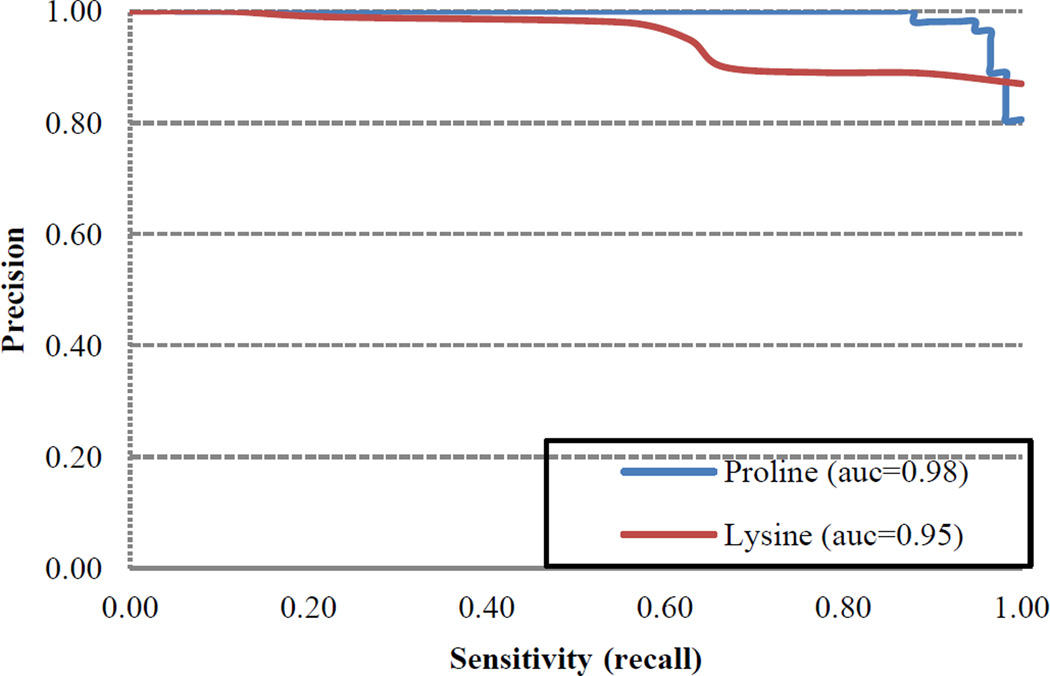
Jackknife cross validation reflected robust performance with respect to all metrics for both HyP and HyK (Table 2B, Figures 7–8). For instance, RF-Hydroxysite was characterized by high true positive (TP) and true negative (TN) rates coupled with low false positive (FP) and false negative (FN) rates. As a consequence, RF-Hydroxysite was both highly precise, exhibiting precision scores of 98.9% for HyK and 96.9% for HyP, and highly sensitive, exhibiting sensitivity scores of 93.8% and 92.0% for HyK and HyP, respectively. On the other hand, the specificity evaluates the ability of the method to predict negative hydroxylation sites. Owing to high TN and low FP rates, RF-Hydroxysite exhibited specificity scores of 98.9% for HyK and 97.4% for HyP. Likewise, RF-Hydroxysite performed well with respect to composite scores, such as accuracy, which reflects the ability of the method to predict positive and negative hydroxylation sites correctly. Indeed, the accuracy of our method is 96.3% and 94.8% for HyK and HyP, respectively. Similarly, the F1-scores, which combine both precision and sensitivity, were 96.3% for HyK and 94.4% for HyP. Finally, the Matthew’s correlation coefficient (MCC), which quantifies the quality of binary prediction, can be used as a surrogate for overall method performance. Accordingly, an MCC score of 1.0 denotes perfection, 0 denotes poor quality (i.e., the method performance is no better than random prediction) and −1 denotes total disagreement between the prediction and observation. Our method exhibited MCC scores of 0.927 for HyK and 0.897 for HyP. The relatively small trade-off between true positive rate (TPR) and false positive rate (FPR) is apparent in the receiver-operating characteristic (ROC) curve (Figure 7). Likewise, as can be seen in Figure 8, the precision-recall (PR) curve suggests a small trade-off between prediction and sensitivity. Indeed, the large area under both curves reflects excellent model performance (AUCROC > 0.92 and AUCPR > 0.95). Moreover, similar results were obtained using an independent test set (Table 3). Taken together, these results suggest that there is strong agreement between the prediction and observation, indicating that the quality of our method is high.
Figure 7.
Receiver operating characteristics curve (ROC) of RF-based model for the prediction of hydroxylation sites
Figure 8.
Precision-recall (PR) curve of RF-based model for the prediction of hydroxylation sites
Table 3.
The evaluation metrics of models of the seven window-sizes based on independent samples
| Size | Resid. | Accu | Prec | Sens | Spec | F1sc | MCC | AUC |
|---|---|---|---|---|---|---|---|---|
| 7 | K | 0.95 | 1.00 | 0.89 | 0.89 | 0.94 | 0.90 | 0.94 |
| P | 0.95 | 1.00 | 0.90 | 0.90 | 0.95 | 0.90 | 0.95 | |
| 9 | K | 0.95 | 0.88 | 1.00 | 1.00 | 0.93 | 0.90 | 0.96 |
| P | 0.95 | 0.93 | 0.95 | 0.95 | 0.94 | 0.89 | 0.95 | |
| 11 | K | 0.95 | 0.88 | 1.00 | 1.00 | 0.93 | 0.90 | 0.96 |
| P | 0.95 | 0.92 | 0.97 | 0.97 | 0.94 | 0.89 | 0.95 | |
| 13 | K | 0.95 | 0.88 | 1.00 | 1.00 | 0.93 | 0.90 | 0.96 |
| P | 0.95 | 0.92 | 0.96 | 0.96 | 0.94 | 0.90 | 0.95 | |
| 15 | K | 0.95 | 0.88 | 1.00 | 1.00 | 0.93 | 0.90 | 0.96 |
| P | 0.91 | 0.93 | 0.90 | 0.90 | 0.91 | 0.81 | 0.91 | |
| 17 | K | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| P | 0.94 | 0.97 | 0.91 | 0.91 | 0.94 | 0.88 | 0.94 | |
| 19 | K | 0.95 | 0.88 | 1.00 | 1.00 | 0.93 | 0.90 | 0.96 |
| P | 0.96 | 0.97 | 0.95 | 0.95 | 0.96 | 0.92 | 0.96 | |
To see how our method performed relative to existing hydroxylation site prediction methods, we compared RF-Hydroxysite to iHyd-PseAAC and PredHydroxy, the most popular hydroxysite methods developed to date. As can be seen in Tables 4 and 5, in side-by-side comparisons using both jackknife cross-validation and an independent test set, our method performed as well or better than the existing methods in each of the metrics, suggesting that the features introduced during the development of RF-Hydroxysite positively impact method performance. This was also evident when the features were used to train other machine learning methods, namely Adaptive Boosting, Bagging, Gradient Boosting and Extra-Trees Classifier (Sup-A). Together, these results suggest that the selected features are highly robust. Indeed, the results of testing the entire set of independent sequences showed that RF-Hydroxysite was able to successfully predict 100% of the experimentally verified hydroxylysine sites and 97.83% of the hydroxyproline sites (Sup-D).
Table 4.
Comparison between PredHydroxy and our method. The results were based on jackknife cross validation
| Metrics | PredHydroxy | RF-Hydroxysite | ||
|---|---|---|---|---|
| P | K | P | K | |
| Accuracy | 0.85 | 0.83 | 0.95 | 0.95 |
| Sensitivity | 0.84 | 0.84 | 0.90 | 0.89 |
| Specificity | 0.85 | 0.82 | 0.90 | 0.89 |
| MCC | 0.69 | 0.67 | 0.90 | 0.90 |
Table 5.
Side-by-side comparing RF-Hydroxysite with iHyd-PseAAC and PredHydroxy using an independent dataset
| Method | Residue | Accu | Prec | Sens | Spec | F1sc | MCC |
|---|---|---|---|---|---|---|---|
| iHyd-PSeAAc | P | 0.84 | 0.91 | 0.74 | 0.93 | 0.82 | 0.68 |
| PredHydroxy | 0.94 | 0.97 | 0.91 | 0.97 | 0.94 | 0.89 | |
| RF-Hydroxysite | 0.96 | 0.96 | 0.97 | 0.96 | 0.96 | 0.93 | |
| iHyd-PSeAAc | K | 0.95 | 1.00 | 0.90 | 1.00 | 0.95 | 0.90 |
| PredHydroxy | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | |
| RF-Hydroxysite | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | |
Conclusions
In this study, we describe the development of RF-Hydroxysite, a new method for identification of putative hydroxylation sites in a protein given only the primary amino acid sequence as input. The features used to develop this new method capture physicochemical (HQIs, OP, and ACH), sequence-order (PWAA, ENT1, RE1, and IG1), structural (PDR), and evolutionary (ENT2, RE2, and IG2) information from protein sequences. The relative importance of each feature type was evaluated by averaging the Gini importance indices across the features in the group, allowing us to identify those feature types that most strongly impacted the fidelity of hydroxylation site identification for proline and lysine residues. For instance, for HyK, the most decisive features were HQI3, HQI4, and ACH (derived from physicochemical information) followed by ENT2, RE2, and IG2 (derived from evolutionary information). Meanwhile, for HyP, the most decisive features were HQI1, HQI3, HQI4, HQI7, HQI8, and ACH (physicochemical) and ENT2, RE2, and IG2 (evolutionary).
The physicochemical information and evolutionary information are important for both types of hydroxylation sites, which may suggest that the biochemical process is physicochemical in nature but that evolutionary factors serve to preserve the biological functions that rely on hydroxylation, thereby reinforcing the features. The finding that ACH features are important may indicate that the flanking regions of the hydroxylation sites are highly hydrophilic in nature and that there are clear distinctions between the local environment of positive sites and that of negative sites, which tend to be found in more hydrophobic contexts. With regard to other features, such as PWAA and OP, although they showed clear patterns, these differences were not large enough to create clear distinctions between positive and negative sites. Therefore, their roles in prediction were limited and they were ultimately omitted from the final model.
The method was evaluated using both jackknife cross validation (Table 2B) and an independent test set (Table 3). Both evaluation methods suggest that RF-Hydroxysite performs as well or better than other existing hydroxylation site prediction methods. Importantly, its high accuracy and specificity suggest that RF-Hydroxysite has the power to annotate potential hydroxylation sites within a protein with high confidence. Importantly, model building based on the selected features using four other ensemble learning methods (AdaBoost, Bagging, Gradient Boosting and the Extra Trees Classifier) showed no significant difference from that of RF-based models. This strongly supports the robustness of the features selected as determining factors for hydroxylation site prediction. Though subsequent experimental validation will be necessary to verify putative hydroxylation sites, accurate prediction will allow for targeted analysis that will complement global identification methods, such as shotgun tandem MS and protein microarray-based approaches (28–29). To promote its use, RF-Hydroxysite is freely available online as bioinformatics tool at http://bcb.ncat.edu/RF_hydroxy/.
Supplementary Material
Acknowledgments
DBKC is partly supported by a start-up grant from the Department of Computational Science & Engineering at North Carolina A&T State University. DBKC and RHN are also partly supported by the National Science Foundation under cooperative agreement No. DBI-0939454. RHN is supported by 1SC2GM113784-01 from the NIH and NSF grant 1038160. Some of the computational works were performed on the resources provided by NSF MRI Grant 1429464.
References
- 1.Bradlow HL, et al. 16 alpha-hydroxylation of estradiol: a possible risk marker for breast cancer. Ann N Y Acad Sci. 1986;464:138–151. doi: 10.1111/j.1749-6632.1986.tb16001.x. [DOI] [PubMed] [Google Scholar]
- 2.Ploumakis A, Coleman ML. OH, the Places You'll Go! Hydroxylation, Gene Expression, and Cancer. Mol Cell. 2015;58(5):729–741. doi: 10.1016/j.molcel.2015.05.026. [DOI] [PubMed] [Google Scholar]
- 3.Hu LL, et al. Prediction and Analysis of Protein Hydroxyproline and Hydroxylysine. PLoS One. 2010;5(12) doi: 10.1371/journal.pone.0015917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu Y, et al. iHyd-PseAAC: predicting hydroxyproline and hydroxylysine in proteins by incorporating dipeptide position-specific propensity into pseudo amino acid composition. Int J Mol Sci. 2014;15(5):7594–7610. doi: 10.3390/ijms15057594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi SP, et al. PredHydroxy: computational prediction of protein hydroxylation site locations based on the primary structure. Mol Biosyst. 2015;11(3):819–825. doi: 10.1039/c4mb00646a. [DOI] [PubMed] [Google Scholar]
- 6.Ahmad S, Gromiha MM, Sarai A. RVP-net: online prediction of real valued accessible surface area of proteins from single sequences. Bioinformatics. 2003;19(14):1849–1851. doi: 10.1093/bioinformatics/btg249. [DOI] [PubMed] [Google Scholar]
- 7.Huang Y, et al. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics. 2010;26(5):680–682. doi: 10.1093/bioinformatics/btq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saha I, et al. Fuzzy clustering of physicochemical and biochemical properties of amino acids. Amino Acids. 2012;43(2):583–594. doi: 10.1007/s00726-011-1106-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blaber M, Zhang XJ, Matthews BW. Structural basis of amino acid alpha helix propensity. Science. 1993;260(5114):1637–1640. doi: 10.1126/science.8503008. [DOI] [PubMed] [Google Scholar]
- 10.Biou V, et al. Secondary structure prediction: combination of three different methods. Protein Eng. 1988;2(3):185–191. doi: 10.1093/protein/2.3.185. [DOI] [PubMed] [Google Scholar]
- 11.Maxfield FR, Scheraga HA. Status of empirical methods for the prediction of protein backbone topography. Biochemistry. 1976;15(23):5138–5153. doi: 10.1021/bi00668a030. [DOI] [PubMed] [Google Scholar]
- 12.Tsai J, et al. The packing density in proteins: standard radii and volumes. J Mol Biol. 1999;290(1):253–266. doi: 10.1006/jmbi.1999.2829. [DOI] [PubMed] [Google Scholar]
- 13.Nakashima H, Nishikawa K. The amino acid composition is different between the cytoplasmic and extracellular sides in membrane proteins. FEBS Lett. 1992;303(2–3):141–146. doi: 10.1016/0014-5793(92)80506-c. [DOI] [PubMed] [Google Scholar]
- 14.Cedano J, et al. Relation between amino acid composition and cellular location of proteins. J Mol Biol. 1997;266(3):594–600. doi: 10.1006/jmbi.1996.0804. [DOI] [PubMed] [Google Scholar]
- 15.Lifson S, Sander C. Antiparallel and parallel beta-strands differ in amino acid residue preferences. Nature. 1979;282(5734):109–111. doi: 10.1038/282109a0. [DOI] [PubMed] [Google Scholar]
- 16.Miyazawa S, Jernigan RL. Self-consistent estimation of inter-residue protein contact energies based on an equilibrium mixture approximation of residues. Proteins. 1999;34(1):49–68. doi: 10.1002/(sici)1097-0134(19990101)34:1<49::aid-prot5>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 17.Dou Y, Yao B, Zhang C. PhosphoSVM: prediction of phosphorylation sites by integrating various protein sequence attributes with a support vector machine. Amino Acids. 2014;46(6):1459–1469. doi: 10.1007/s00726-014-1711-5. [DOI] [PubMed] [Google Scholar]
- 18.Dou Y, et al. Prediction of catalytic residues based on an overlapping amino acid classification. Amino acids. 2010;39(5):1353–1361. doi: 10.1007/s00726-010-0587-2. [DOI] [PubMed] [Google Scholar]
- 19.Eisenberg D, et al. Faraday Symposia of the Chemical Society. Royal Society of Chemistry; 1982. Hydrophobic moments and protein structure. [Google Scholar]
- 20.Iakoucheva LM, et al. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004;32(3):1037–1049. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ward JJ, et al. The DISOPRED server for the prediction of protein disorder. Bioinformatics. 2004;20(13):2138–2139. doi: 10.1093/bioinformatics/bth195. [DOI] [PubMed] [Google Scholar]
- 22.Stormo GD, et al. Use of the 'Perceptron' algorithm to distinguish translational initiation sites in E. coli. Nucleic Acids Res. 1982;10(9):2997–3011. doi: 10.1093/nar/10.9.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci U S A. 1992;89(22):10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pedregosa F, et al. Scikit-learn: Machine learning in Python. The Journal of Machine Learning Research. 2011;12:2825–2830. [Google Scholar]
- 26.Ismail HD, et al. Computational Advances in Bio and Medical Sciences (ICCABS), 2015 IEEE 5th International Conference on. IEEE; 2015. Phosphorylation sites prediction using Random Forest. [Google Scholar]
- 27.Brown CJ, Johnson AK, Daughdrill GW. Comparing Models of Evolution for Ordered and Disordered Proteins. Molecular Biology and Evolution. 2010;27(3):609–621. doi: 10.1093/molbev/msp277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newman RH, Zhang J, Zhu H. Toward a systems-level view of dynamic phosphorylation networks. Front Genet. 2014;5:263. doi: 10.3389/fgene.2014.00263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu S, Xie Z, Qian J, Blackshaw S, Zhu H. Functional protein microarray technology. Wiley Interdiscip Rev Syst Biol Med. 2011;3(3):255–268. doi: 10.1002/wsbm.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.