Abstract
Males are XY and females are XX in most mammalian species. Other species such as birds have a different sex chromosome make-up: ZZ in males and ZW in females. In both types of organisms one of the sex chromosomes, Y or W, has degenerated due to lack of recombination with its respective homolog X or Z. Since autosomes are present in two copies in diploid organisms the heterogametic sex has become a natural "aneuploid" with haploinsufficiency for X- or Z-linked genes. Specific mechanisms have evolved to restore a balance between critical gene products throughout the genome and between males and females. Some of these mechanisms were co-opted from and/or added to compensatory processes that alleviate autosomal aneuploidy. Surprisingly, several modes of dosage compensation have evolved. In this review we will consider the evidence for dosage compensation and the molecular mechanisms implicated.
Keywords: Dosage compensation, X chromosome, aneuploidy, sex chromosome evolution, sex differences
Introduction
The adaptive advantages of recombination favor sexual reproduction [1], which is often accompanied by differentiation of sex chromosomes. In mammals, males are XY and females XX, while in birds, males are ZZ and females ZW. These systems evolved because sex is genetically determined [2, 3]. Other vertebrates such as reptiles rely on temperature-sensitive systems for sex determination. Muller hypothesized that differentiation of the sex chromosomes would inevitably arise from lack of recombination due to the appearance of a sex-determining gene on the Y or W chromosome [4]. Ohno expanded these ideas by proposing the concept of ancestral sex chromosomes (protosex chromosomes) that progressively evolved to the present-day sex chromosomes by degeneration of the Y or W. [5].
Sex chromosomes have evolved independently multiple times: for example, mammalian and avian sex chromosomes derive from different ancestral autosomes. It has been proposed that some chromosomes may be better suited to become sex chromosomes based on their gene content [3, 6]. Both Muller and Ohno predicted that sex chromosome divergence would lead to dosage compensation of the natural type of aneuploidy caused by degeneration of one chromosome in the heterogametic sex. Indeed, a variety of dosage compensation mechanisms that regulate the sex chromosomes have evolved, resulting in a dazzling array of systems throughout the plant and animal kingdoms. The evolution of vertebrate sex chromosomes and of dosage compensation were recently comprehensively reviewed by us and by others [3, 7–9]. In addition, X chromosome inactivation, one of the main form of X regulation in mammals is discussed in detail by others in this issue.
Here, we summarize salient features of dosage compensation of sex-linked and autosomal genes with a focus on molecular mechanisms of dosage regulation. Two major types of sex chromosome dosage compensation, often confounded in the literature, can be recognized; one type balances gene expression throughout the genome by changing the relative expression of X-linked or Z-linked genes versus autosomal genes or vice versa, and the other equalizes sex-linked gene expression between homogametic and heterogametic sexes. The former type of dosage regulation is critical to maintain fitness. Finally, a narrower definition of dosage compensation has been proposed as representing evolutionary adaptive changes in expression of ancestral autosomal genes that evolved into sex-linked genes [10]. Such definition is necessarily based on a restricted number of conserved genes in different species.
Dosage regulation of the sex chromosomes can be viewed as either global, i.e. employing mechanisms that modify most - but not all - genes on an entire chromosome, or local, i.e. acting on individual genes. This distinction is somewhat fluid as the number of dosage-compensated genes on a given sex chromosome varies between tissues, and also depends on methods of analysis. Intuitively, not all genes need to be regulated by either type of dosage compensation mentioned above. Indeed, balanced expression throughout the genome may be critical only for dosage-sensitive genes implicated in protein complexes and functional networks, but may not apply to dosage-insensitive genes unless they are swept in a global regulatory system. Deleterious effects of copy-number changes may be subtle at the individual gene level, but cumulative effects of large chromosomal imbalance are often severe and yet to be fully understood. Conversely, patchy dosage compensation may be advantageous and selected for in terms of sex-specific traits important in male/female conflicts. This is particularly relevant for testis- or ovary-specific genes abundant on the sex chromosomes.
Differentiation of the sex chromosomes
Dosage compensation of sex-linked genes should be considered in light of sex chromosome evolution. One of the best studied systems in which progressive evolutionary steps have been deciphered is represented by the human sex chromosomes that have evolved for about 300 million years [11–14]. Based on DNA sequence analyses of genes retained on the human sex chromosomes it is apparent that the Y underwent large inversions that progressively prevented large regions from undergoing X/Y recombination. This may have been helped by a gradual spread of regions with reduced recombination [15]. Detailed sequence analyses led to the definition of six evolutionary strata on the X chromosome, each containing genes that diverged from their Y paralogs for a similar length of time [13]. Evolutionary strata have also been found in other mammals and in birds [11, 14, 16].
The Y chromosome in mammals and the W chromosome in birds are gene-poor, having lost many functional genes [17]. It is estimated that the human Y retains about 3% of the genes originally located on the proto-sex chromosomes, whereas the X retains about 98% of genes [14]. The human Y is rich in palindromic duplicated sequences that may help retention of specific Y-linked genes important in male fertility, but also facilitate deletions and gene loss [18]. Such deletions are often associated with male infertility [19, 20], in which case they would not be transmitted, thus preserving some integrity of the Y chromosome.
When comparing sex chromosomes in divergent mammalian species such as marsupials (metatherian) it is evident that the eutherian sex chromosomes acquired a large piece of chromosome that is autosomal in marsupials [21]. Graves proposed that successive cycles of addition and attrition have shaped the sex chromosomes [3]. Small regions have also been added or deleted more recently to the eutherian sex chromosomes, reshaping the pseudoautosomal region (PAR) but rarely changing the content of the rest of the X [22, 23]. Altogether, the X chromosome in eutherian mammals is highly conserved, probably because it is subjected to complex mechanisms of dosage regulation [5].
Specialized gene content of the sex chromosomes
When studying dosage regulation of the sex chromosomes one must consider their gene content. For example, male-biased genes often expressed in testis are abundant on the Y chromosome, a location predicted to be favorable to the accumulation of sexually antagonistic genes with a male benefit [24]. Interestingly, the X chromosome is also highly enriched in male-biased genes [25]. Hemizygosity in males favors the accumulation of male-advantageous mutations at both X and Y locations. In addition to being enriched in male-biased genes the X chromosome is also enriched in femalebiased genes expressed in ovary [26]. Of special interest is the accumulation of brain expressed genes on the X chromosome, possibly a by-product of sexual reproduction [27–29].
Some of the male-biased genes located on the sex chromosomes were recently and independently acquired in different clades [30]. The convergent evolution of the bird Z and mammalian X chromosomes, both of which demonstrate massive enrichment in multi-copy genes expressed in testis, shows striking similarities between the two types of heterogametic systems [31]. The mouse Y represents an extreme case of specialization with accumulation of hundreds of copies of fertility genes. In this case, both Y- and X-linked paralogs are amplified, suggesting meiotic driver and suppressor pairs with cycles of amplification in response to interchromosomal X/Y conflict [32].
The mammalian sex chromosomes also carry genes essential for survival of human embryos. Over 95% of 45,X embryos die during development and those that survive are often mosaic for a normal XX or XY line [33]. A subset of highly conserved dosage-sensitive X/Y paralogs with essential functions are the main candidates in the context of embryo survival [11, 14]. These genes usually escape X inactivation in females and thus are bi-allelically expressed in both sexes [34–36]. Note that even though many of the X/Y paralogs retain apparently similar functions their sequence has in some cases diverged, suggesting that the Y-linked paralog may be acquiring a male-specific function. An early example of concerted divergence between X/Y paralogs and acquisition of X inactivation is represented by the gene pairs ZFX/ZFY and Zfx/Zfy1-2 in human and mouse. In human, which would represent the more primitive condition, both ZFX and ZFY are ubiquitously expressed and ZFX escapes X inactivation, ensuring similar expression between sexes. In mouse, Zfy1-2 have acquired a testis-specific function, while the ubiquitously expressed Zfx is subject to X inactivation [37–39].
The specialized gene content of the sex chromosomes results in phenotypic sex differences manifested in health and disease susceptibility. Specific mouse breeding schemes including the four-core genotype to generate sex-reversed animals have helped sort out the roles of X- and Y-linked genes versus those of sex hormones [40, 41]. The complicated dosage regulation of the sex chromosomes, for example escape from X inactivation, may have evolved in part to enhance such sex-specific differences [7] (see sex differences below).
Dosage compensation responses to aneuploidy
The heterogametic sex would have had to survive a natural form of aneuploidy. How do organisms respond to any aneuploidy, whether autosomal or sex-linked? Aneuploidy causes significant phenotypic abnormalities and loss of fitness [42]. Duplications are generally better tolerated than deletions. In Drosophila melanogaster, deletions that affect 1% of the genome reduce viability [43], but of course, this very much depends on the gene content of the deleted region (see dosage-sensitive genes below). Usually, the larger the deletion the more lethal it is, indicating a clear cumulative effect. The effects of deletions and duplications in mammals are less well-studied than in model organisms such as yeast and fly, and no systematic series of deletions have been generated yet. Thus, most information comes from spontaneously occurring copy-number changes in human. Overall, copy-number changes of ≥1Mb are associated with abnormal phenotypes and rare in normal individuals [44]. Monosomy for a whole human chromosome is lethal but trisomy is better tolerated. However, even trisomy for the smallest human chromosome, trisomy 21, causes widespread gene dysregulation [45]. Interestingly, not all genes located on chromosome 21 show the expected 1.5 expression increase, indicating dampening. Furthermore, the expression and chromatin features of genes located elsewhere in the genome is also altered [46]. In human trisomy 8 and 21, differentially methylated regions are found not only along the trisomic chromosome but also genome-wide, suggesting that DNA methylation plays important roles in compensatory adjustments of gene expression [47, 48].
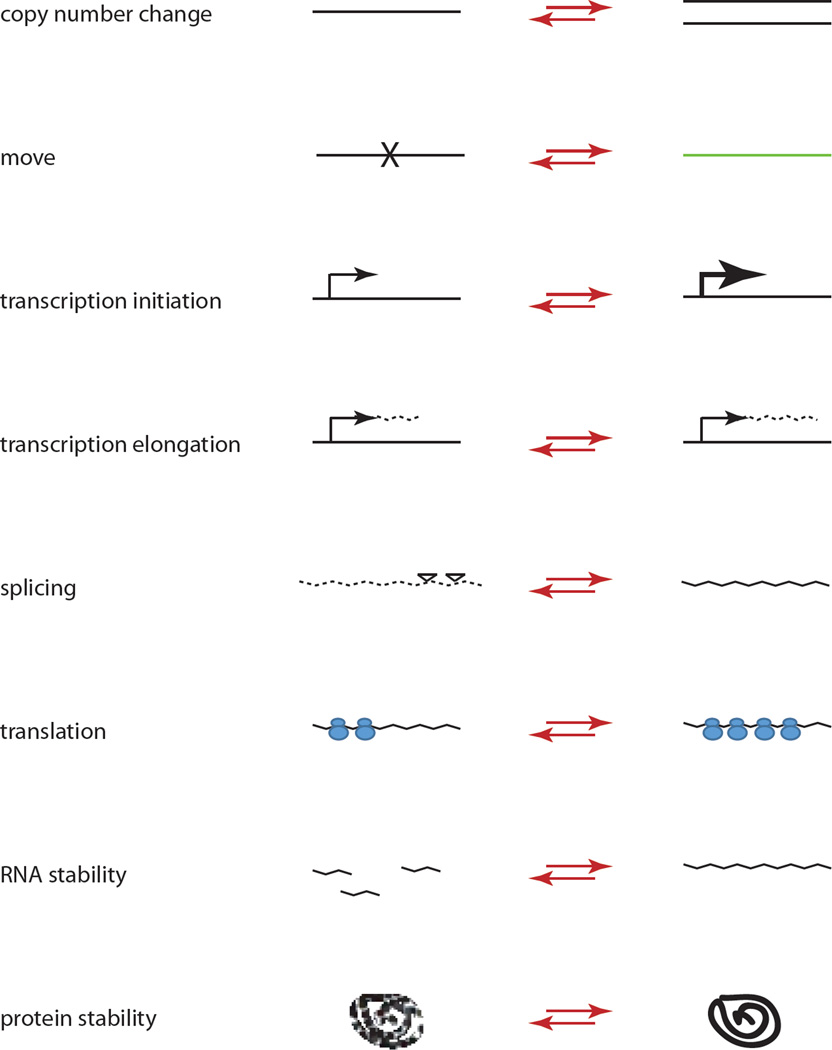
While the expression of many genes varies in direct proportion to their dose, expression of some genes is compensated, presumably to reduce deleterious dosage imbalance [9, 49, 50]. Oliver defined three types of compensatory responses: (1) buffering or passive absorption of gene dose perturbation by inherent system properties, (2) feedback or gene-specific sensing and adjustment of levels, which can result in overexpression, and (3) feedforward responses representing systems that apply only to special cases, for example the male X chromosome in Drosophila [51]. In a diploid Drosophila cell line with multiple copy-number changes, monosomic regions show 0.75 expression and trisomic regions 1.33, relative to a value of 1.0 in disomic regions [52]. By comparing multiple cell lines it is evident that even for the same deletion compensatory responses may differ [52]. Such compensation of aneuploidy has also been observed in yeast aneuploids in which different mechanisms adjust mRNA abundance, splicing, stability, or translation in response to gene amplification [53]. Alterations and potential adjustments in levels of long non-coding RNAs (lncRNA) and miRNAs are less well documented. Other mechanisms operate at the protein level: abnormal protein levels and macromolecular interactions such as folding, aggregation, and interactions can be alleviated by induction of proteolysis [42, 54]. Figure 1 summarizes various types of adjustments in response to dosage imbalance.
Figure 1. Types of mechanisms that can modulate dosage responses to imbalance.
From top: 1. Copy-number adjustments occur, for example, in the case of multi-copy ribosomal genes or mouse sex-linked genes engaged in a meiotic conflict; 2. Move to a different genomic location occurs in the case of genes lost from the mammalian Y but translocated to the X or to an autosome to preserve function; 3. Increased initiation of transcription can represent a feedback mechanism or a feedforward mechanism, as in mammalian and Drosophila X upregulation; 4. Increased elongation of transcription represents a feedforward mechanism in Drosophila X upregulation; 5. Increased efficiency of splicing have been observed in aneuploid yeast; 6. Increased number of ribosomes on RNA to enhance translation occurs in mammalian X upregulation; 7. Increased RNA stability has been associated with mammalian X upregulation; 8. Adjustment at the protein level, for example, by changing stability has been reported in Drosophila aneuploidy. Note that all types of adjustments can work in reverse to decrease gene/protein products, for example to adjust genome-wide expression in response to a deficiency (inverse effects). The mechanisms included here have been implicated in dosage responses, but this does not exclude other mechanisms known to change gene expression (e.g. miRNA, lncRNA) and protein production. Epigenetic mechanisms including chromatin and nuclear organization associated with enhancement or repression of gene expression are not included in this figure. See text for further details and references.
In addition to compensatory mechanisms that control gene expression and amounts of protein products other systems may rely on adjustments in copy number for the restoration of a balanced genome, for example in the case of multi-copy ribosomal genes that encode the 5S and 45S rRNA in human cells (Fig. 1). Although the multi-copy arrays are on different human chromosomes they display rapid and concerted copy number adjustment to fulfill their stoichiometric requirements [55]. Rapid co-variation of rDNA gene copy number has been observed not only between siblings but also in response to environment changes, underlying the need to efficiently maintain a balance between ribosomal components. Correlated copy number changes have also been observed in Drosophila cell lines [52]. Interestingly, of 142 protein-protein interaction networks identified, 84 had a greater than 90% co-occurrence of copy number changes in the same direction.
Therapeutic interventions may ultimately be developed to artificially correct deleterious effects of aneuploidy. In the case of trisomy 21 amelioration of neural cell growth was obtained by insertion of a highly expressed copy of the lncRNA XIST within one of the trisomic chromosome 21 in a cell line. This approach took advantage of the property of XIST, which is essential for the onset of X inactivation, in cis-induction of chromosome-wide gene silencing [56]. The timing of intervention in terms of dosage compensation may be critical. This conundrum is exemplified in a recent report of a zebrafish with a mutation in an extracellular matrix protein, which shows no phenotypes, whereas a morpholino-induced knockdown causes severely abnormal phenotypes [57]. This observation is attributed to adaptive compensatory mechanisms that upregulate other extracellular proteins during development, whereas the rapid effects of the knockdown fail to be compensated.
Finally, it is important to point out that aneuploidy can be neutral or advantageous. Adaptive aneuploidy has been reported in yeast exposed to oxidative stress [58]. In human, Grompe’s group has reported frequent aneuploidy together with genomic deletions in hepatocytes [59]. Frequent copy-number changes have also been observed in human neuronal cells, but it is unclear whether these have any advantage [60]. Overall, somatic mosaicism for copy-number changes is widespread and much work needs to be done to determine whether this is simply tolerated or selected for. Thus, there may be a subtle equilibrium between maintenance of dosage by compensation systems and selection of specific imbalances advantageous to specific cell types. Clearly, copy-number changes in cancer cells such as amplification of oncogenes or deletion of tumor suppressor genes provide growth advantage to these cells [61]. In contrast, germ cells would be expected to maintain an intact genome with sporadic copy-number changes strongly selected against or compensated during development.
Allele-specific expression
Some genes are normally expressed from a single allele. Do these genes get compensated? While a majority of these genes are X-linked in mammals and will be discussed below, others are autosomal. One category of such autosomal genes is represented by imprinted genes. It should be noted that there are interesting parallels between mechanisms that silence imprinted genes and those that silence genes regulated by X inactivation [62], both types of genes being regulated by cis-acting lncRNAs, histone modifications, DNA methylation, and specific nuclear positioning [63]. One study has addressed the question of dosage compensation of imprinted genes by measuring expression of 59 such genes in mice using expression arrays and quantitative RT-PCR [64]. While inherently limited due to the paucity of imprinted genes this study concludes that imprinted genes are partially upregulated. Increased expression could help alleviate deleterious effects due to transcriptional noise at mono-allelically expressed genes.
A large number of non-imprinted autosomal genes show consistent heritable allelic biases in terms of chromatin structure and levels of gene expression in human [65]. Such allelic biases reflect close proximity of enhancers and promoters, or strong interactions at longer distances [65]. Pervasive allelic imbalance has been reported in mouse crosses where 80% genes show cis-regulatory variation [66], and in humans in relation to specific haplotypes [67]. The role of allelic differences in causing subtle deficiency or overexpression is not well understood and it is unclear whether specific compensatory mechanisms adjust allelic expression. Random allelic silencing of autosomal genes was initially illustrated in clonal cell lines [68], but subsequent studies in single cells have confirmed this phenomenon [69]. There are again parallels between molecular signatures of randomly silenced autosomal and X-linked alleles. For example, SMCHD1, a protein implicated in X inactivation in part for the establishment of DNA methylation, plays a role in mono-allelic repression of autosomal genes, e.g. protocadherin genes [70, 71]. Common epigenetic features such as H3K27me3 also decorate both silenced X-linked genes and mono-allelically expressed autosomal genes [70–73].
Dosage compensation between sex chromosomes and autosomes
Balanced expression between X/Z-linked and autosomal genes can be attained by increasing X/Z expression or by decreasing autosomal expression in the heterogametic sex. X upregulation is best documented in Drosophila males to increase expression of a large portion of genes [74]. In organisms where dosage adjustment is sex-specific - as in Drosophila - there would be no need for adjustment in the homogametic sex. However, in systems in which X/Z expression is increased relative to autosomal expression in both sexes, there would be a need to reduce expression in the homogametic sex. This is achieved by X inactivation in mammals, or X repression in Caenorhabditis elegans. These repressive systems have been extensively reviewed [9, 75]. Here, we will limit our discussion to the balance in the heterogametic sex.
Ratios of gene expression between autosomes and sex chromosomes could result from modulations of either sex-linked or autosomal genes [76]. However, it is inherently difficult to measure absolute amounts of transcripts in cells. Global studies of gene expression make specific assumptions about normalization to the total transcriptome that may not always be correct and lead to aberrant conclusions when comparing samples [77, 78]. Co-isolation of RNA and genomic DNA together with counting the number of cells from which the nucleic acids are extracted are helpful, as well as spiking reactions with exogenous RNA. Almost no studies of dosage compensation include such controls. We performed one study in human triploid lines where correlated changes in X expression and number of active X chromosomes, but no detectable autosomal gene expression changes, were observed [79].
As measured by X:autosome expression ratios mammalian X upregulation is controversial and more studies are needed [80]. Initial expression microarray studies supported the existence of increased expression of X-linked versus autosomal genes in multiple mammalian species and tissues [81, 82]. Subsequent RNA-seq studies carried out in multiple somatic tissues either disproved or confirmed X upregulation [83, 84]. Presence of X upregulation is based on finding average X:autosome expression ratios between 0.8 and 1.0, while absence of X upregulation is based on finding low (around 0.5) median X:autosome ratios. Clearly, X upregulation only operates on expressed genes. As we have shown, low median X:autosome ratios reported by some [83] are obtained in somatic tissues when all X-linked genes whose expression distribution comprises a long tail of non-expressed testis-specific genes, are included. When testis-specific genes are excluded, X:autosome expression ratios increase [84, 85]. Furthermore, both the depth of sequencing and the methodologies used to analyze RNA-seq data can lead to low expression medians [86]. A recent analysis in multiple mouse strains confirmed similar expression between X-linked and autosomal genes [66]. However, analyses of protein levels suggest absence of X upregulation [87].
One study that supports X upregulation addresses expression changes during ES cell differentiation when X inactivation takes place [88]. Undifferentiated male and female ES cells show X:autosome expression ratios of 1.4–1.6. After 2–3 weeks of differentiating male ES cells X upregulation results in equal X-linked and autosomal expression throughout the genome. In differentiating female ES cells gene-by-gene silencing on one allele results in a progressive decrease in X expression to achieve a balanced expression [88]. Another study done in haploid cells shows that a high proportion (35%) of genes upregulated (in comparison to diploid cells) are X-linked [89]. In bovine blastocysts there is also evidence of increased X expression in females [90]. However, a recent study in human and mouse mature oocytes reports low X:autosome expression ratios [91].
One way championed by Kaessman to evaluate the existence of X upregulation is to compare expression of X-linked genes in mammals to that of “ancestral” genes in chicken [10]. This study concludes that there is no or very little evolutionary upregulation of X-linked genes in mammals. The authors propose that genome balance is maintained in eutherians by an overall expression reduction in a subset of autosomal genes to compensate for a decrease in X-linked expression. X inactivation then would serve to reduce X expression in females to compensate for the inverse effect on autosomes. While this comparative study may address evolutionary aspects of X expression it remains unclear whether contemporary mammals can be directly compared to contemporary birds with different physiology. In addition, the number of genes that can be compared is restricted and it has not been shown that haploinsufficiency of the genes in question would be harmless if deleted in chicken. Indeed, it would be interesting to produce monosomy for chicken chromosomes 1 and 4 to evaluate the consequences of such large chromosomal defects.
Surprisingly, marsupial but not monotreme X-linked genes were fully upregulated compared to ancestral genes in chicken [10, 92]. It will be informative to determine whether the autosomal genes with reduced expression in eutherians have increased expression in marsupials. Birds only have partial dosage compensation, leading to sex differences in levels of Z-linked gene products [93]. This was confirmed in multiple somatic tissues in chicken where the Z:autosome ratios for protein and RNA were 1.0 and 0.8 in males, and 0.8 and 0.6 in females, respectively [94, 95]. A potentially useful avenue of research into evolution of dosage regulation would be to compare X- or Z-linked ohnologs to their autosomal copy. Ohnologs are genes that result from evolutionary whole genome duplications hypothesized by Ohno [96]. A subset of ohnologs are thought to have been retained because of balance constraints. In human, persistent ohnologs are refractory to copy-number changes and cause abnormal phenotypes [97].
Species with newly evolved sex chromosomes have been tested to determine whether dosage compensation can rapidly evolve. In sticklebacks recently evolved sex chromosomes allowed the definition of two main strata along the X, with the older strata showing evidence of upregulation in males and overexpression in females at least for a subset of genes [98]. Complete dosage compensation by upregulation of the male X chromosome evolved independently in Drosophila melanogaster and in a mosquito Anopheles stephensi [99], while in another insect, Heliconius butterfly, there is only partial dosage compensation [100]. In a plant (Silene latifola) with newly evolved sex chromosomes no global dosage compensation between sexes was detected in one study [101], while two others showed increased X expression [102, 103], further demonstrating the difficulty of reaching a consensus. In C. elegans an initial study that showed absence of X upregulation [83] was subsequently shown to be flawed because it did not take into account the progressive accumulation of germ cells in which the X chromosome is silenced [84]. A more recent study confirmed balanced expression of X-linked and autosomal genes in C. elegans, but argued against evolutionary X upregulation by measuring ancestral gene expression in another species [104].
We and others have proposed that mammalian X upregulation would probably operate gene-by-gene once the Y-linked homolog is lost by recruiting a number of different molecular mechanisms of gene regulation (see below and Fig. 1) [7, 84]. Expression of multiple genes in large X regions could also have been concomitantly adjusted if several adjacent Y-linked genes degenerated together [105]. In this case deleterious effects of haploinsufficiency would likely have been cumulative, in a manner analogous to loss of a whole chromosome or a large copy-number change. Upregulation is expected to only regulate expressed genes [7, 106], and to especially target dosage-sensitive genes that need to remain in stoichiometric equilibrium in large macromolecular complexes [107]. A study of dosage-sensitive genes defined as those included in large protein complexes is consistent with upregulation of X-linked genes, with no downregulation of autosomal genes [108].
A recent study has proposed an alternative novel mechanism to compensate for loss of a Y paralog, which is to move a copy of that gene to another genomic location (Fig. 1) [109]. This rescue of dosage-sensitive Y-linked genes by translocation to the X or to an autosome was initially discovered by studying the spiny rat where the Y chromosome has disappeared altogether [110, 111]. This rescue mechanism was subsequently shown to have happened multiple times in mammalian lineages [109].
Molecular evidence of X upregulation
Precise adjustment of X-linked gene expression requires a combination of systems for enhancement and suppression of expression. In Drosophila, upregulation of the male X chromosome is achieved by recruitment of the MSL complex to increase levels of H4K16 acetylation and open chromatin, which results in increased transcription initiation and elongation (Fig. 1) [74]. Recruitment of the MSL complex to the male X is mediated by a 2–4 fold enrichment in specific binding motifs. Heterochromatin elements counteract chromatin unfolding to achieve doubling in gene expression [112]. An alternative hypothesis proposed by Birchler who examined MSL mutants is that the X:autosome balance in Drosophila depends genome-wide adjustments of gene expression. This is the so-called inverse-hypothesis [113, 114]. In C. elegans, X upregulation is associated with visible de-condensation of the active X together with increased levels of H4K16 acetylation [115].
In mammals, genes on the active X chromosome are distinguishable from autosomal genes in several respects. We and others have shown an average of 30% enrichment in RNA polymerase II at the 5’end of expressed X-linked genes compared to autosomal genes, suggesting increased initiation of transcription (Fig. 1) [84, 106, 116]. In addition, a subset of X-linked genes is sensitive to MOF, the acetyltransferase that modifies H4K16 [106]. Reduced RNA decay at X-linked versus autosomal transcripts is also evident and X-linked transcripts have a longer half-life (Fig. 1) [117]. A new analysis further confirms increased mRNA stability and also demonstrates a greater number of ribosomes on X-linked versus autosomal genes, indicating a higher rate of translation (Fig. 1) [118]. Manipulation of mRNA stability causes a greater decrease in X than autosomal expression, but the increased stability of X-linked transcripts does not appear to be related to poly-A tail length, GC and GC3 contents, or any detected 3’UTR sequence features [118]. Control of ribosome translocation for polypeptide elongation, which is also influenced by relative abundance of tRNAs, is another potential regulatory mechanism that has not been fully evaluated in terms of X chromosome regulation [119].
Thus, several mechanisms have been demonstrated that increase transcription, RNA stability, and translation, for X-linked genes (Fig. 1). Additional studies are needed to fully characterize these processes, for example in terms of regulatory motifs to help understand targeting of molecular changes to the X chromosome. In contrast to mammals, Drosophila X-linked genes have fewer ribosomes and lower translational rates than autosomal genes in males [120]. It may be that mammals employ compensatory mechanisms that act on RNA, while Drosophila mechanisms are more specifically targeted to the rate of transcription. In mammals, both ancestral genes defined as those located on the X in marsupials and acquired genes that are autosomal in marsupials have similar characteristics, suggesting that the molecular changes observed rapidly occurred during differentiation of the sex chromosomes [7].
An emerging feature of dosage regulation of chromosomes concerns their nuclear position and structure. In mammals, the heterochromatic inactive X is often located near the nuclear membrane or the nucleolus, possibly helped by specific elements including the lncRNA Firre [121]. In addition, the inactive X forms a bipartite structure with two superdomains of condensation separated by a hinge that binds the lncRNA DXZ4/Dxz4 in both human and mouse [122, 123]. Genes that escape X inactivation tend to be located at the periphery of the condensed structure, suggesting that location is important in terms of expression [122, 124]. Factors involved in location of the active X, which is often located near the nuclear membrane in mammals, remain to be determined. In Drosophila and C. elegans, nuclear pore complexes associate with the active X chromosome, consistent with a specific nuclear location [125, 126].
Sex differences due to dosage
In mammals, gene expression in somatic tissues is fairly similar between sexes except in sex organs. Indeed, the Y chromosome contains few genes except for those implicated in male fertility, and X inactivation effectively silences most genes on one allele in females, except for genes that escape X inactivation [127, 128]. Escape from X inactivation may enhance sex-specific differences via female bias. Such effects can be indirect, as shown for the reproduction-related Hox genes Rhox6/9 expressed highly in ovary and regulated by the histone demethylase KDM6A encoded by a female-biased escape gene [129]. Sexual dimorphisms in gene expression arise in early development prior to hormonal influence [130], for example, in male and female ES cells also shown to differ in epigenetic features such as DNA methylation and histone modifications [129, 131]. In addition, the testis-determining gene Sry acts as a repressor in somatic tissues and plays a role in sexual dimorphisms throughout the genome [132]. X-linked imprinted genes could also lead to sexual dimorphisms since males only inherit a maternal X, and females both a maternal and paternal X [133]. An interesting hypothesis is that X inactivation evolved not to reduce overexpression in females, but rather as a type of imprinting mechanism to silence growth-inhibiting genes in embryos [134].
Both the inactive X and the Y chromosomes are heterochromatic and this structural feature is probably important to consider in terms of sexual dimorphisms in the epigenetic landscape of the genome in males and females. These large heterochromatic structures represent sinks that sequester specific factors otherwise important for epigenetic modifications. Since the Y chromosome is smaller than the inactive X such effects may differ between sexes [135]. In cases of abnormal sex chromosome complements sink effects are multiplied and could influence the entire genome by altering the overall nuclear structure.
In contrast to mammals, birds have extensive sex-bias in gene expression due to incomplete dosage compensation [10]. This is also the case for snakes and some fish (reviewed in [3]). In birds, Z-linked genes have about 30% higher expression in males than in females, and thus may play important roles in sexual dimorphisms. The male-tofemale expression ratio ranges from 1 to 2, suggesting a continuum in the level of piecemeal compensation. Protein analyses are consistent with a sex ratio of 1.32, similar to that obtained by RNA-seq (1.29) [94].
Partial rather than complete dosage compensation between the sexes is common in insects other than Drosophila, as well as in other organisms, leading to the suggestion that efficient mechanisms to equalize X or Z expression between males and females is in fact rare [136]. Overall, this type of dosage compensation appears to be more frequent in XX/XY than in ZZ/ZW systems, which may be due to stronger selection on X-linked genes in males or to faster Y chromosome decay due to a greater number of cell divisions in spermatogenesis than oogenesis [137]. Alternatively, there may be an insufficient number of organisms examined so far to draw a meaningful conclusion about the prevalence of sex bias due to sex-linked genes.
Dosage-sensitive genes
In human, dosage-sensitive genes have been identified as those causing disease if mutated, deleted, or present in supernumerary copies [138]. A comprehensive list of DNA sequences intolerant to alterations was compiled by considering coding as well as non-coding regions to include regulatory features [139]. Gene expression analysis using expression microarrays in a very large number of tumors has shown that many genes are dosage-sensitive, with their expression usually correlated to copy number changes [140]. Furthermore, abundantly expressed genes are more dosage-sensitive. Conversely, some genes are apparently dosage-insensitive and heterozygous deletions do not cause anomalies.
Copy-number aberrations of the human X chromosome are frequently associated with abnormal transcript levels, which lead to abnormal phenotypes in males, while females are often protected by mosaicism or skewing of X inactivation. Numerous examples of such effects have been reported, suggesting a greater abundance of dosage-sensitive genes on the X chromosome than previously suspected. One interesting example is a 0.3Mb copy-number (2–5 copies) change at band Xq38 [141]. The expression level of genes (including GDI1, a gene associated with intellectual disability) in the region was proportional to the number of copies, as was the severity of phenotypes in males and the degree of skewing in females.
Two important studies published in 2014 have shown that a conserved set of genes with paralogs on the X/Y chromosomes in mammals and on the Z/W chromosomes in birds have persisted in many species [11, 31]. In mammals, persistence of such genes in multiple species indicate that despite widespread Y degeneration, the Y copy could not get lost because of dosage-sensitivity. Interestingly, these intolerant genes are all engaged in critical functions such as transcription, translation, chromatin structure, splicing and ubiquitination [11, 31]. The X paralogs usually escape X inactivation, thus preserving bi-allelic expression in both sexes. Consistent with being dosage-sensitive genes escape genes are under greater purifying selection than genes subject to X inactivation [142, 143].
Thus, dosage of the broadly expressed X/Y genes is important and preserved. The mouse has fewer of these genes compared to human, which may explain the difference in severity between the near-lethal Turner syndrome versus X0 mice that have a much milder phenotype [127]. Women with a single X chromosome are often mosaic, suggesting that the presence of two copies of the X/Y dosage-sensitive genes at least in some tissues may be necessary for survival [33]. Two of the X paralogs of preserved X/Y gene pairs are KDM5C and KDM6A both included in the list of intolerant genes known to cause disease if present in an abnormal number of copies [139]. Note that for X/Y gene pairs Y-linked expression is often lower than X-linked expression, consistent with X upregulation and suggesting Y decay or evolution of new male function.
Are dosage-sensitive genes excluded from the X compared to autosomes? Chen and Oliver measured the effects of X-linked and autosomal deletions in terms of gene expression alterations in Drosophila and concluded that the X chromosome is neither more robust nor sensitive to dosage change [144]. They measured a 1.1 response to these deletions, which suggests that even with the contribution of the MSL complex estimated to be around 1.4 there must be an additional, yet to be discovered, mechanism to reach a two-fold adjustment.
Emerging view
Conflicting statements about the existence of dosage compensation in haploid chromosomes such as the sex chromosomes in part result from unclear definition of the term “dosage compensation”. Any adjustment of dosage, partial or global, chromosome-specific or genome-wide, indicates some form of dosage regulation. For sex-linked genes such regulation is in part co-opted from responses to any form of autosomal aneuploidy, with added mechanisms to help restore genome balance. Findings of molecular differences between sex chromosomes and autosomes are a strong indicator that organisms respond to haploinsufficiency due to differentiation of the sex chromosomes. The overall picture that is emerging about any form of so-called dosage compensation of the sex chromosomes is that not all genes are dosage compensated in an organism and that mechanisms widely differ between species. There is clear a continuum between small and large adaptive changes in gene expression. Additional studies are needed to understand the importance of this regulation in relation to sex differences.
Acknowledgments
This work was supported by grants from the National Institutes of Health (GM113943, GM046883, DK107979). I thank X. Deng for helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hoekstra RF. Evolutionary biology: why sex is good. Nature. 2005;434:571–573. doi: 10.1038/434571a. [DOI] [PubMed] [Google Scholar]
- 2.Graves JA. Sex chromosome specialization and degeneration in mammals. Cell. 2006;124:901–914. doi: 10.1016/j.cell.2006.02.024. [DOI] [PubMed] [Google Scholar]
- 3.Graves JA. Evolution of vertebrate sex chromosomes and dosage compensation. Nat Rev Genet. 2016;17:33–46. doi: 10.1038/nrg.2015.2. [DOI] [PubMed] [Google Scholar]
- 4.Muller HJ. A gene for the fourth chromosome of Drosophila. J Exp Zool. 1914;17:325–336. [Google Scholar]
- 5.Ohno S. Sex Chromosomes and Sex Linked Genes. Berlin: Springer Verlag; 1967. [Google Scholar]
- 6.O'Meally D, Ezaz T, Georges A, Sarre SD, Graves JA. Are some chromosomes particularly good at sex? Insights from amniotes. Chromosome research : an international journal on the molecular, supramolecular and evolutionary aspects of chromosome biology. 2012;20:7–19. doi: 10.1007/s10577-011-9266-8. [DOI] [PubMed] [Google Scholar]
- 7.Deng X, Berletch JB, Nguyen DK, Disteche CM. X chromosome regulation: diverse patterns in development, tissues and disease. Nat Rev Genet. 2014;15:367–378. doi: 10.1038/nrg3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Disteche CM. Dosage compensation of the sex chromosomes. Annu Rev Genet. 2012;46:537–560. doi: 10.1146/annurev-genet-110711-155454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ercan S. Mechanisms of x chromosome dosage compensation. Journal of genomics. 2015;3:1–19. doi: 10.7150/jgen.10404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Julien P, Brawand D, Soumillon M, Necsulea A, Liechti A, Schutz F, et al. Mechanisms and evolutionary patterns of mammalian and avian dosage compensation. PLoS biology. 2012;10:e1001328. doi: 10.1371/journal.pbio.1001328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cortez D, Marin R, Toledo-Flores D, Froidevaux L, Liechti A, Waters PD, et al. Origins and functional evolution of Y chromosomes across mammals. Nature. 2014;508:488–493. doi: 10.1038/nature13151. [DOI] [PubMed] [Google Scholar]
- 12.Lahn BT, Page DC. Four evolutionary strata on the human X chromosome. Science. 1999;286:877–879. doi: 10.1126/science.286.5441.964. [DOI] [PubMed] [Google Scholar]
- 13.Ross MT, Grafham DV, Coffey AJ, Scherer S, McLay K, Muzny D, et al. The DNA sequence of the human X chromosome. Nature. 2005;434:325–337. doi: 10.1038/nature03440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bellott DW, Hughes JF, Skaletsky H, Brown LG, Pyntikova T, Cho TJ, et al. Mammalian Y chromosomes retain widely expressed dosage-sensitive regulators. Nature. 2014;508:494–499. doi: 10.1038/nature13206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marais G, Galtier N. Sex chromosomes: how X–Y recombination stops. Current biology : CB. 2003;13:R641–R643. doi: 10.1016/s0960-9822(03)00570-0. [DOI] [PubMed] [Google Scholar]
- 16.Sandstedt SA, Tucker PK. Evolutionary strata on the mouse X chromosome correspond to strata on the human X chromosome. Genome Res. 2004;14:267–272. doi: 10.1101/gr.1796204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Charlesworth B, Charlesworth D. The degeneration of Y chromosomes. Philos Trans R Soc Lond B Biol Sci. 2000;355:1563–1572. doi: 10.1098/rstb.2000.0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rozen S, Skaletsky H, Marszalek JD, Minx PJ, Cordum HS, Waterston RH, et al. Abundant gene conversion between arms of palindromes in human and ape Y chromosomes. Nature. 2003;423:873–876. doi: 10.1038/nature01723. [DOI] [PubMed] [Google Scholar]
- 19.Silber SJ, Disteche CM. Y Chromosome Infertility. In: Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH, et al., editors. GeneReviews(R) Seattle (WA): 2012. [Google Scholar]
- 20.Repping S, Skaletsky H, Lange J, Silber S, Van Der Veen F, Oates RD, et al. Recombination between palindromes P5 and P1 on the human Y chromosome causes massive deletions and spermatogenic failure. Am J Hum Genet. 2002;71:906–922. doi: 10.1086/342928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Graves JAM. The origin and function of the mammalian Y chromosome and Y-borne genes- -an evolving understanding. Bioessays. 1995;17:311–320. doi: 10.1002/bies.950170407. [DOI] [PubMed] [Google Scholar]
- 22.Disteche CM, Brannan CI, Larsen A, Adler DA, Schorderet DF, Gearing D, et al. The human pseudoautosomal GM-CSF receptor alpha subunit gene is autosomal in mouse. Nat Genet. 1992;1:333–336. doi: 10.1038/ng0892-333. [DOI] [PubMed] [Google Scholar]
- 23.Rugarli EI, Adler DA, Borsani G, Tsuchiya K, Franco B, Hauge X, et al. Different chromosomal localization of the Clcn4 gene in Mus spretus and C57BL/6J mice. Nat Genet. 1995;10:466–471. doi: 10.1038/ng0895-466. [DOI] [PubMed] [Google Scholar]
- 24.Rice WR. Evolution of the Y sex chromosome in animals: Y chromosomes evolve through the degeneration of autosomes. BioScience. 1996;46:331–343. [Google Scholar]
- 25.Vicoso B, Charlesworth B. Evolution on the X chromosome: unusual patterns and processes. Nat Rev Genet. 2006;7:645–653. doi: 10.1038/nrg1914. [DOI] [PubMed] [Google Scholar]
- 26.Khil PP, Smirnova NA, Romanienko PJ, Camerini-Otero RD. The mouse X chromosome is enriched for sex-biased genes not subject to selection by meiotic sex chromosome inactivation. Nat Genet. 2004;36:642–646. doi: 10.1038/ng1368. [DOI] [PubMed] [Google Scholar]
- 27.Zechner U, Wilda M, Kehrer-Sawatzki H, Vogel W, Fundele R, Hameister H. A high density of X-linked genes for general cognitive ability: a run-away process shaping human evolution? Trends Genet. 2001;17:697–701. doi: 10.1016/s0168-9525(01)02446-5. [DOI] [PubMed] [Google Scholar]
- 28.Nguyen DK, Disteche CM. High expression of the mammalian X chromosome in brain. Brain Res. 2006b;1126:46–49. doi: 10.1016/j.brainres.2006.08.053. [DOI] [PubMed] [Google Scholar]
- 29.Skuse DH. X-linked genes and mental functioning. Hum Mol Genet. 2005;14(Spec No 1):R27–R32. doi: 10.1093/hmg/ddi112. [DOI] [PubMed] [Google Scholar]
- 30.Mueller JL, Mahadevaiah SK, Park PJ, Warburton PE, Page DC, Turner JM. The mouse X chromosome is enriched for multicopy testis genes showing postmeiotic expression. Nat Genet. 2008;40:794–799. doi: 10.1038/ng.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bellott DW, Skaletsky H, Pyntikova T, Mardis ER, Graves T, Kremitzki C, et al. Convergent evolution of chicken Z and human X chromosomes by expansion and gene acquisition. Nature. 2010;466:612–616. doi: 10.1038/nature09172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soh YQ, Alfoldi J, Pyntikova T, Brown LG, Graves T, Minx PJ, et al. Sequencing the mouse Y chromosome reveals convergent gene acquisition and amplification on both sex chromosomes. Cell. 2014;159:800–813. doi: 10.1016/j.cell.2014.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hassold T, Benham F, Leppert M. Cytogenetic and molecular analysis of sex-chromosome monosomy. Am J Hum Genet. 1988;42:534–541. [PMC free article] [PubMed] [Google Scholar]
- 34.Jegalian K, Page DC. A proposed path by which genes common to mammalian X and Y chromosomes evolve to become X inactivated. Nature. 1998;394:776–780. doi: 10.1038/29522. [DOI] [PubMed] [Google Scholar]
- 35.Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- 36.Yang F, Babak T, Shendure J, Disteche CM. Global survey of escape from X inactivation by RNA-sequencing in mouse. Genome Res. 2010;20:614–622. doi: 10.1101/gr.103200.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koopman P, Gubbay J, Collignon J, Lovell-Badge R. Zfy gene expression patterns are not compatible with a primary role in mouse sex determination. Nature. 1989;342:940–942. doi: 10.1038/342940a0. [DOI] [PubMed] [Google Scholar]
- 38.Adler DA, Bressler SL, Chapman VM, Page DC, Disteche CM. Inactivation of the Zfx gene on the mouse X chromosome. Proc Natl Acad Sci U S A. 1991;88:4592–4595. doi: 10.1073/pnas.88.11.4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schneider-Gadicke A, Beer-Romero P, Brown LG, Nussbaum R, Page DC. ZFX has a gene structure similar to ZFY, the putative human sex determinant, and escapes X inactivation. Cell. 1989;57:1247–1258. doi: 10.1016/0092-8674(89)90061-5. [DOI] [PubMed] [Google Scholar]
- 40.Arnold AP. Mouse models for evaluating sex chromosome effects that cause sex differences in non-gonadal tissues. J Neuroendocrinol. 2009;21:377–386. doi: 10.1111/j.1365-2826.2009.01831.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arnold AP. The end of gonad-centric sex determination in mammals. Trends Genet. 2012;28:55–61. doi: 10.1016/j.tig.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Veitia RA, Potier MC. Gene dosage imbalances: action, reaction, and models. Trends in biochemical sciences. 2015;40:309–317. doi: 10.1016/j.tibs.2015.03.011. [DOI] [PubMed] [Google Scholar]
- 43.Chen ZX, Golovnina K, Sultana H, Kumar S, Oliver B. Transcriptional effects of gene dose reduction. Biology of sex differences. 2014;5:5. doi: 10.1186/2042-6410-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Girirajan S, Campbell CD, Eichler EE. Human copy number variation and complex genetic disease. Annu Rev Genet. 2011;45:203–226. doi: 10.1146/annurev-genet-102209-163544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.FitzPatrick DR, Ramsay J, McGill NI, Shade M, Carothers AD, Hastie ND. Transcriptome analysis of human autosomal trisomy. Hum Mol Genet. 2002;11:3249–3256. doi: 10.1093/hmg/11.26.3249. [DOI] [PubMed] [Google Scholar]
- 46.Letourneau A, Santoni FA, Bonilla X, Sailani MR, Gonzalez D, Kind J, et al. Domains of genome-wide gene expression dysregulation in Down's syndrome. Nature. 2014;508:345–350. doi: 10.1038/nature13200. [DOI] [PubMed] [Google Scholar]
- 47.Davidsson J, Veerla S, Johansson B. Constitutional trisomy 8 mosaicism as a model for epigenetic studies of aneuploidy. Epigenetics Chromatin. 2013;6:18. doi: 10.1186/1756-8935-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jin S, Lee YK, Lim YC, Zheng Z, Lin XM, Ng DP, et al. Global DNA hypermethylation in down syndrome placenta. PLoS Genet. 2013;9:e1003515. doi: 10.1371/journal.pgen.1003515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Y, Oliver B. Dosage compensation goes global. Curr Opin Genet Dev. 2007;17:113–120. doi: 10.1016/j.gde.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 50.Birchler JA. Reflections on studies of gene expression in aneuploids. Biochem J. 2010;426:119–123. doi: 10.1042/BJ20091617. [DOI] [PubMed] [Google Scholar]
- 51.Zhang Y, Malone JH, Powell SK, Periwal V, Spana E, Macalpine DM, et al. Expression in aneuploid Drosophila S2 cells. PLoS biology. 2010;8:e1000320. doi: 10.1371/journal.pbio.1000320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee H, McManus CJ, Cho DY, Eaton M, Renda F, Somma MP, et al. DNA copy number evolution in Drosophila cell lines. Genome Biol. 2014;15:R70. doi: 10.1186/gb-2014-15-8-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hose J, Yong CM, Sardi M, Wang Z, Newton MA, Gasch AP. Dosage compensation can buffer copy-number variation in wild yeast. eLife. 2015;4 doi: 10.7554/eLife.05462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lundberg LE, Figueiredo ML, Stenberg P, Larsson J. Buffering and proteolysis are induced by segmental monosomy in Drosophila melanogaster. Nucleic Acids Res. 2012;40:5926–5937. doi: 10.1093/nar/gks245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gibbons JG, Branco AT, Godinho SA, Yu S, Lemos B. Concerted copy number variation balances ribosomal DNA dosage in human and mouse genomes. Proc Natl Acad Sci U S A. 2015;112:2485–2490. doi: 10.1073/pnas.1416878112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jiang J, Jing Y, Cost GJ, Chiang JC, Kolpa HJ, Cotton AM, et al. Translating dosage compensation to trisomy 21. Nature. 2013;500:296–300. doi: 10.1038/nature12394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rossi A, Kontarakis Z, Gerri C, Nolte H, Holper S, Kruger M, et al. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature. 2015;524:230–233. doi: 10.1038/nature14580. [DOI] [PubMed] [Google Scholar]
- 58.Kaya A, Gerashchenko MV, Seim I, Labarre J, Toledano MB, Gladyshev VN. Adaptive aneuploidy protects against thiol peroxidase deficiency by increasing respiration via key mitochondrial proteins. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:10685–10690. doi: 10.1073/pnas.1505315112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Duncan AW, Hanlon Newell AE, Bi W, Finegold MJ, Olson SB, Beaud AL, et al. Aneuploidy as a mechanism for stress-induced liver adaptation. The Journal of clinical investigation. 2012;122:3307–3315. doi: 10.1172/JCI64026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fischer HG, Morawski M, Bruckner MK, Mittag A, Tarnok A, Arendt T. Changes in neuronal DNA content variation in the human brain during aging. Aging cell. 2012;11:628–633. doi: 10.1111/j.1474-9726.2012.00826.x. [DOI] [PubMed] [Google Scholar]
- 61.Torres EM, Williams BR, Amon A. Aneuploidy: cells losing their balance. Genetics. 2008;179:737–746. doi: 10.1534/genetics.108.090878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bartolomei MS, Ferguson-Smith AC. Mammalian genomic imprinting. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a002592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fedoriw AM, Calabrese JM, Mu W, Yee D, Magnuson T. Differentiation-driven nucleolar association of the mouse imprinted Kcnq1 locus. G3 (Bethesda) 2012;2:1521–1528. doi: 10.1534/g3.112.004226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zaitoun I, Downs KM, Rosa GJ, Khatib H. Upregulation of imprinted genes in mice: an insight into the intensity of gene expression and the evolution of genomic imprinting. Epigenetics. 2010;5:149–158. doi: 10.4161/epi.5.2.11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Leung D, Jung I, Rajagopal N, Schmitt A, Selvaraj S, Lee AY, et al. Integrative analysis of haplotype-resolved epigenomes across human tissues. Nature. 2015;518:350–354. doi: 10.1038/nature14217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Crowley JJ, Zhabotynsky V, Sun W, Huang S, Pakatci IK, Kim Y, et al. Analyses of allele-specific gene expression in highly divergent mouse crosses identifies pervasive allelic imbalance. Nat Genet. 2015;47:353–360. doi: 10.1038/ng.3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cowles CR, Hirschhorn JN, Altshuler D, Lander ES. Detection of regulatory variation in mouse genes. Nat Genet. 2002;32:432–437. doi: 10.1038/ng992. [DOI] [PubMed] [Google Scholar]
- 68.Gimelbrant A, Hutchinson JN, Thompson BR, Chess A. Widespread monoallelic expression on human autosomes. Science. 2007;318:1136–1140. doi: 10.1126/science.1148910. [DOI] [PubMed] [Google Scholar]
- 69.Deng Q, Ramskold D, Reinius B, Sandberg R. Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science. 2014;343:193–196. doi: 10.1126/science.1245316. [DOI] [PubMed] [Google Scholar]
- 70.Blewitt ME, Gendrel AV, Pang Z, Sparrow DB, Whitelaw N, Craig JM, et al. SmcHD1, containing a structural-maintenance-of-chromosomes hinge domain, has a critical role in X inactivation. Nat Genet. 2008;40:663–669. doi: 10.1038/ng.142. [DOI] [PubMed] [Google Scholar]
- 71.Mould AW, Pang Z, Pakusch M, Tonks ID, Stark M, Carrie D, et al. Smchd1 regulates a subset of autosomal genes subject to monoallelic expression in addition to being critical for X inactivation. Epigenetics Chromatin. 2013;6:19. doi: 10.1186/1756-8935-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nag A, Vigneau S, Savova V, Zwemer LM, Gimelbrant AA. Chromatin Signature Identifies Monoallelic Gene Expression Across Mammalian Cell Types. G3 (Bethesda) 2015;5:1713–1720. doi: 10.1534/g3.115.018853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Marks H, Chow JC, Denissov S, Francoijs KJ, Brockdorff N, Heard E, et al. High-resolution analysis of epigenetic changes associated with X inactivation. Genome Res. 2009;19:1361–1373. doi: 10.1101/gr.092643.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lucchesi JC, Kuroda MI. Dosage compensation in Drosophila. Cold Spring Harb Perspect Biol. 2015;7 doi: 10.1101/cshperspect.a019398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Veitia RA, Veyrunes F, Bottani S, Birchler JA. X chromosome inactivation and active X upregulation in therian mammals: facts, questions, and hypotheses. Journal of molecular cell biology. 2015;7:2–11. doi: 10.1093/jmcb/mjv001. [DOI] [PubMed] [Google Scholar]
- 76.Birchler JA. Facts and artifacts in studies of gene expression in aneuploids and sex chromosomes. Chromosoma. 2014;123:459–469. doi: 10.1007/s00412-014-0478-5. [DOI] [PubMed] [Google Scholar]
- 77.Coate JE, Doyle JJ. Variation in transcriptome size: are we getting the message? Chromosoma. 2015;124:27–43. doi: 10.1007/s00412-014-0496-3. [DOI] [PubMed] [Google Scholar]
- 78.Loven J, Orlando DA, Sigova AA, Lin CY, Rahl PB, Burge CB, et al. Revisiting global gene expression analysis. Cell. 2012;151:476–482. doi: 10.1016/j.cell.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Deng X, Nguyen DK, Hansen RS, Van Dyke DL, Gartler SM, Disteche CM. Dosage regulation of the active X chromosome in human triploid cells. PLoS Genet. 2009;5:e1000751. doi: 10.1371/journal.pgen.1000751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pessia E, Engelstadter J, Marais GA. The evolution of X chromosome inactivation in mammals: the demise of Ohno's hypothesis? Cellular and molecular life sciences : CMLS. 2014;71:1383–1394. doi: 10.1007/s00018-013-1499-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gupta V, Parisi M, Sturgill D, Nuttall R, Doctolero M, Dudko OK, et al. Global analysis of X-chromosome dosage compensation. J Biol. 2006;5:3. doi: 10.1186/jbiol30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nguyen DK, Disteche CM. Dosage compensation of the active X chromosome in mammals. Nat Genet. 2006a;38:47–53. doi: 10.1038/ng1705. [DOI] [PubMed] [Google Scholar]
- 83.Xiong Y, Chen X, Chen Z, Wang X, Shi S, Wang X, et al. RNA sequencing shows no dosage compensation of the active X-chromosome. Nat Genet. 2010;42:1043–1047. doi: 10.1038/ng.711. [DOI] [PubMed] [Google Scholar]
- 84.Deng X, Hiatt JB, Nguyen DK, Ercan S, Sturgill D, Hillier LW, et al. Evidence for compensatory upregulation of expressed X-linked genes in mammals, Caenorhabditis elegans and Drosophila melanogaster. Nat Genet. 2011;43:1179–1185. doi: 10.1038/ng.948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kharchenko PV, Xi R, Park PJ. Evidence for dosage compensation between the X chromosome and autosomes in mammals. Nat Genet. 2011;43:1167–1169. doi: 10.1038/ng.991. author reply 71-2. [DOI] [PubMed] [Google Scholar]
- 86.Jue NK, Murphy MB, Kasowitz SD, Qureshi SM, Obergfell CJ, Elsisi S, et al. Determination of dosage compensation of the mammalian X chromosome by RNA-seq is dependent on analytical approach. BMC Genomics. 2013;14:150. doi: 10.1186/1471-2164-14-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen X, Zhang J. No X-chromosome dosage compensation in human proteomes. Molecular biology and evolution. 2015;32:1456–1460. doi: 10.1093/molbev/msv036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lin H, Gupta V, Vermilyea MD, Falciani F, Lee JT, O'Neill LP, et al. Dosage compensation in the mouse balances up-regulation and silencing of X-linked genes. PLoS biology. 2007;5:e326. doi: 10.1371/journal.pbio.0050326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Leeb M, Wutz A. Derivation of haploid embryonic stem cells from mouse embryos. Nature. 2011;479:131–134. doi: 10.1038/nature10448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Itoh Y, Arnold AP. X chromosome regulation of autosomal gene expression in bovine blastocysts. Chromosoma. 2014;123:481–489. doi: 10.1007/s00412-014-0461-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fukuda A, Tanino M, Matoba R, Umezawa A, Akutsu H. Imbalance between the expression dosages of X-chromosome and autosomal genes in mammalian oocytes. Scientific reports. 2015;5:14101. doi: 10.1038/srep14101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Whitworth DJ, Pask AJ. The X factor: X chromosome dosage compensation in the evolutionarily divergent monotremes and marsupials. Seminars in cell & developmental biology. 2016 doi: 10.1016/j.semcdb.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 93.Itoh Y, Melamed E, Yang X, Kampf K, Wang S, Yehya N, et al. Dosage compensation is less effective in birds than in mammals. J Biol. 2007;6:2. doi: 10.1186/jbiol53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Uebbing S, Konzer A, Xu L, Backstrom N, Brunstrom B, Bergquist J, et al. Quantitative Mass Spectrometry Reveals Partial Translational Regulation for Dosage Compensation in Chicken. Molecular biology and evolution. 2015;32:2716–2725. doi: 10.1093/molbev/msv147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Uebbing S, Kunstner A, Makinen H, Ellegren H. Transcriptome sequencing reveals the character of incomplete dosage compensation across multiple tissues in flycatchers. Genome biology and evolution. 2013;5:1555–1566. doi: 10.1093/gbe/evt114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ohno S, Wolf U, Atkin NB. Evolution from fish to mammals by gene duplication. Hereditas. 1968;59:169–187. doi: 10.1111/j.1601-5223.1968.tb02169.x. [DOI] [PubMed] [Google Scholar]
- 97.Makino T, McLysaght A. Ohnologs in the human genome are dosage balanced and frequently associated with disease. Proc Natl Acad Sci U S A. 2010;107:9270–9274. doi: 10.1073/pnas.0914697107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Schultheiss R, Viitaniemi HM, Leder EH. Spatial dynamics of evolving dosage compensation in a young sex chromosome system. Genome biology and evolution. 2015;7:581–590. doi: 10.1093/gbe/evv013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jiang X, Biedler JK, Qi Y, Hall AB, Tu Z. Complete Dosage Compensation in Anopheles stephensi and the Evolution of Sex-Biased Genes in Mosquitoes. Genome biology and evolution. 2015;7:1914–1924. doi: 10.1093/gbe/evv115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Walters JR, Hardcastle TJ, Jiggins CD. Sex Chromosome Dosage Compensation in Heliconius Butterflies: Global yet Still Incomplete? Genome biology and evolution. 2015;7:2545–2559. doi: 10.1093/gbe/evv156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bergero R, Qiu S, Charlesworth D. Gene loss from a plant sex chromosome system. Current biology : CB. 2015;25:1234–1240. doi: 10.1016/j.cub.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 102.Muyle A, Zemp N, Deschamps C, Mousset S, Widmer A, Marais GA. Rapid de novo evolution of X chromosome dosage compensation in Silene latifolia, a plant with young sex chromosomes. PLoS biology. 2012;10:e1001308. doi: 10.1371/journal.pbio.1001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Papadopulos AS, Chester M, Ridout K, Filatov DA. Rapid Y degeneration and dosage compensation in plant sex chromosomes. Proc Natl Acad Sci U S A. 2015;112:13021–13026. doi: 10.1073/pnas.1508454112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Albritton SE, Kranz AL, Rao P, Kramer M, Dieterich C, Ercan S. Sex-biased gene expression and evolution of the x chromosome in nematodes. Genetics. 2014;197:865–883. doi: 10.1534/genetics.114.163311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Vicoso B, Bachtrog D. Progress and prospects toward our understanding of the evolution of dosage compensation. Chromosome research : an international journal on the molecular, supramolecular and evolutionary aspects of chromosome biology. 2009;17:585–602. doi: 10.1007/s10577-009-9053-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Deng X, Berletch JB, Ma W, Nguyen DK, Hiatt JB, Noble WS, et al. Mammalian X upregulation is associated with enhanced transcription initiation, RNA half-life, and MOF-mediated H4K16 acetylation. Dev Cell. 2013;25:55–68. doi: 10.1016/j.devcel.2013.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Veitia RA, Birchler JA. Dominance and gene dosage balance in health and disease: why levels matter. J Pathol. 2010;220:174–185. doi: 10.1002/path.2623. [DOI] [PubMed] [Google Scholar]
- 108.Pessia E, Makino T, Bailly-Bechet M, McLysaght A, Marais GA. Mammalian X chromosome inactivation evolved as a dosage-compensation mechanism for dosage-sensitive genes on the X chromosome. Proc Natl Acad Sci U S A. 2012;109:5346–5351. doi: 10.1073/pnas.1116763109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hughes JF, Skaletsky H, Koutseva N, Pyntikova T, Page DC. Sex chromosome-to-autosome transposition events counter Y-chromosome gene loss in mammals. Genome Biol. 2015;16:104. doi: 10.1186/s13059-015-0667-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Arakawa Y, Nishida-Umehara C, Matsuda Y, Sutou S, Suzuki H. X-chromosomal localization of mammalian Y-linked genes in two XO species of the Ryukyu spiny rat. Cytogenet Genome Res. 2002;99:303–309. doi: 10.1159/000071608. [DOI] [PubMed] [Google Scholar]
- 111.Kuroiwa A, Ishiguchi Y, Yamada F, Shintaro A, Matsuda Y. The process of a Y-loss event in an XO/XO mammal, the Ryukyu spiny rat. Chromosoma. 2010;119:519–526. doi: 10.1007/s00412-010-0275-8. [DOI] [PubMed] [Google Scholar]
- 112.Prestel M, Feller C, Becker PB. Dosage compensation and the global re-balancing of aneuploid genomes. Genome Biol. 2010;11:216. doi: 10.1186/gb-2010-11-8-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bhadra MP, Bhadra U, Kundu J, Birchler JA. Gene expression analysis of the function of the male-specific lethal complex in Drosophila. Genetics. 2005;169:2061–2074. doi: 10.1534/genetics.104.036020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Birchler JA, Veitia RA. The Gene Balance Hypothesis: dosage effects in plants. Methods in molecular biology. 2014;1112:25–32. doi: 10.1007/978-1-62703-773-0_2. [DOI] [PubMed] [Google Scholar]
- 115.Lau AC, Csankovszki G. Balancing up and downregulation of the C. elegans X chromosomes. Current opinion in genetics & development. 2015;31:50–56. doi: 10.1016/j.gde.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yildirim E, Sadreyev RI, Pinter SF, Lee JT. X-chromosome hyperactivation in mammals via nonlinear relationships between chromatin states and transcription. Nat Struct Mol Biol. 2011;19:56–61. doi: 10.1038/nsmb.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yin S, Deng W, Zheng H, Zhang Z, Hu L, Kong X. Evidence that the nonsense-mediated mRNA decay pathway participates in X chromosome dosage compensation in mammals. Biochem Biophys Res Commun. 2009a;383:378–382. doi: 10.1016/j.bbrc.2009.04.021. [DOI] [PubMed] [Google Scholar]
- 118.Faucillion ML, Larsson J. Increased expression of X-linked genes in mammals is associated with a higher stability of transcripts and an increased ribosome density. Genome biology and evolution. 2015;7:1039–1052. doi: 10.1093/gbe/evv054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Richter JD, Coller J. Pausing on Polyribosomes: Make Way for Elongation in Translational Control. Cell. 2015;163:292–300. doi: 10.1016/j.cell.2015.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang Z, Presgraves DC. Drosophila X-linked Genes Have Lower Translation Rates than Autosomal Genes. Molecular biology and evolution. 2015 doi: 10.1093/molbev/msv227. [DOI] [PubMed] [Google Scholar]
- 121.Yang F, Deng X, Ma W, Berletch JB, Rabaia N, Wei G, et al. The lncRNA Firre anchors the inactive X chromosome to the nucleolus by binding CTCF and maintains H3K27me3 methylation. Genome Biology. 2015:1–17. doi: 10.1186/s13059-015-0618-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Deng X, Ma W, Ramani V, Hill A, FY, Ay F, et al. Bipartite structure of the inactive mouse X chromosome. Genome Biology. 2015;16:152. doi: 10.1186/s13059-015-0728-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Heard E, Bickmore W. The ins and outs of gene regulation and chromosome territory organisation. Curr Opin Cell Biol. 2007;19:311–316. doi: 10.1016/j.ceb.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 125.Sharma R, Meister P. Linking dosage compensation and X chromosome nuclear organization in C. elegans. Nucleus. 2015;6:266–272. doi: 10.1080/19491034.2015.1059546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Vaquerizas JM, Suyama R, Kind J, Miura K, Luscombe NM, Akhtar A. Nuclear pore proteins nup153 and megator define transcriptionally active regions in the Drosophila genome. PLoS Genet. 2010;6:e1000846. doi: 10.1371/journal.pgen.1000846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Berletch JB, Yang F, Xu J, Carrel L, Disteche CM. Genes that escape from X inactivation. Hum Genet. 2011;130:237–245. doi: 10.1007/s00439-011-1011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Peeters SB, Cotton AM, Brown CJ. Variable escape from X-chromosome inactivation: identifying factors that tip the scales towards expression. BioEssays : news and reviews in molecular, cellular and developmental biology. 2014;36:746–756. doi: 10.1002/bies.201400032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Berletch JB, Deng X, Nguyen DK, Disteche CM. Female bias in Rhox6 and 9 regulation by the histone demethylase KDM6A. PLoS Genet. 2013;9:e1003489. doi: 10.1371/journal.pgen.1003489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Schulz EG, Heard E. Role and control of X chromosome dosage in mammalian development. Curr Opin Genet Dev. 2013;23:109–115. doi: 10.1016/j.gde.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 131.Zvetkova I, Apedaile A, Ramsahoye B, Mermoud JE, Crompton LA, John R, et al. Global hypomethylation of the genome in XX embryonic stem cells. Nat Genet. 2005;37:1274–1279. doi: 10.1038/ng1663. [DOI] [PubMed] [Google Scholar]
- 132.Wijchers PJ, Yandim C, Panousopoulou E, Ahmad M, Harker N, Saveliev A, et al. Sexual dimorphism in Mammalian autosomal gene regulation is determined not only by Sry but by sex chromosome complement as well. Dev Cell. 2010;19:477–484. doi: 10.1016/j.devcel.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 133.Raefski AS, O'Neill MJ. Identification of a cluster of X-linked imprinted genes in mice. Nat Genet. 2005;37:620–624. doi: 10.1038/ng1567. [DOI] [PubMed] [Google Scholar]
- 134.Haig D. Self-imposed silence: parental antagonism and the evolution of X-chromosome inactivation. Evolution; international journal of organic evolution. 2006;60:440–447. [PubMed] [Google Scholar]
- 135.Wijchers PJ, Festenstein RJ. Epigenetic regulation of autosomal gene expression by sex chromosomes. Trends Genet. 2011;27:132–140. doi: 10.1016/j.tig.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 136.Mank JE. Sex chromosome dosage compensation: definitely not for everyone. Trends Genet. 2013;29:677–683. doi: 10.1016/j.tig.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 137.Bachtrog D, Kirkpatrick M, Mank JE, McDaniel SF, Pires JC, Rice W, et al. Are all sex chromosomes created equal? Trends Genet. 2011;27:350–357. doi: 10.1016/j.tig.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 138.Petrovski S, Wang Q, Heinzen EL, Allen AS, Goldstein DB. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 2013;9:e1003709. doi: 10.1371/journal.pgen.1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Petrovski S, Gussow AB, Wang Q, Halvorsen M, Han Y, Weir WH, et al. The Intolerance of Regulatory Sequence to Genetic Variation Predicts Gene Dosage Sensitivity. PLoS Genet. 2015;11:e1005492. doi: 10.1371/journal.pgen.1005492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Fehrmann RS, Karjalainen JM, Krajewska M, Westra HJ, Maloney D, Simeonov A, et al. Gene expression analysis identifies global gene dosage sensitivity in cancer. Nat Genet. 2015;47:115–125. doi: 10.1038/ng.3173. [DOI] [PubMed] [Google Scholar]
- 141.Vandewalle J, Van Esch H, Govaerts K, Verbeeck J, Zweier C, Madrigal I, et al. Dosage-dependent severity of the phenotype in patients with mental retardation due to a recurrent copy-number gain at Xq28 mediated by an unusual recombination. Am J Hum Genet. 2009;85:809–822. doi: 10.1016/j.ajhg.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Park C, Carrel L, Makova KD. Strong purifying selection at genes escaping X chromosome inactivation. Molecular biology and evolution. 2010;27:2446–2450. doi: 10.1093/molbev/msq143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Slavney A, Arbiza L, Clark AG, Keinan A. Strong Constraint on Human Genes Escaping X-Inactivation Is Modulated by their Expression Level and Breadth in Both Sexes. Molecular biology and evolution. 2015 doi: 10.1093/molbev/msv225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Chen ZX, Oliver B. X Chromosome and Autosome Dosage Responses in Drosophila melanogaster Heads. G3. 2015;5:1057–1063. doi: 10.1534/g3.115.017632. [DOI] [PMC free article] [PubMed] [Google Scholar]