Abstract
Fragile X syndrome (FXS), a trinucleotide repeat disorder, is the most common heritable form of cognitive impairment. Since the discovery of the FMR1 gene in 1991, great strides have been made in the field of molecular diagnosis for FXS. Cytogenetic analysis, which was the method of diagnosis in the early 1990, was replaced by Southern blot and PCR analysis albeit with some limitations. In the past few years many PCR-based methodologies, able to amplify large full mutation expanded alleles, with or without methylation, have been proposed. Reviewed here are the advantages, disadvantages and limitations of the most recent developments in the field of FXS diagnosis.
Introduction
Fragile X syndrome (FXS) (OMIM 300624) is an X-linked disorder and the leading cause of intellectual disabilities (ID). Individuals with FXS can suffer from an array of behavioral, cognitive, neurologic, and physical problems of varying degree, seizure, and macroorchidism (1); approximately 60% can present with autism making FXS the leading monogenic cause of autism spectrum disorders (2–4). In addition to cognitive impairment and typical physical features, individuals with FXS are at increased risk for maladaptive behaviors or symptoms, including social anxiety and withdrawal, social deficits, abnormalities in communication, unusual responses to sensory stimuli, stereotypic behavior, impulsivity, hyperactivity, aggression, and self-injurious behaviors.
FXS is caused by a CGG trinucleotide expansion, greater than 200 repeats, in the 5’UTR of the fragile X mental retardation 1 gene (FMR1), which leads to methylation and transcriptional silencing of the gene with consequent lack of the encoded product, FMRP (5, 6). FMRP is an RNA binding protein that acts as a translational repressor of many mRNA targets at the synapse, thus, it plays a key role in synaptic maturation, plasticity and function (7, 8).
Smaller premutation expansions (55–200 CGG repeats) also can lead to a constellation of neurodevelopmental problems, including autism spectrum disorder, attention deficit hyperactivity disorders and seizures (9–11). In addition, individual carriers of a premutation allele are at risk for primary ovarian insufficiency and for the late-adult-onset neurodegenerative disorder, fragile X-associated tremor/ataxia syndrome (FXTAS) (12). The broad spectrum of clinical involvement (referred to as FMR1-associated disorders) is caused by a completely distinct mechanism from that leading to FXS, one involving CGG-repeat-induced toxicity due to elevated FMR1 mRNA levels (13, 14). Interestingly, subjects carrying an allele with a CGG repeat number within the intermediate range (45–54 CGG repeats) have been reported to show similar phenotypes to those observed in premutation carriers, including neurological and cognitive signs and endocrinal problems (15–19) in addition to showing an altered molecular phenotype including elevated FMR1mRNA and low FMRP expression levels (20–23).
The significant clinical overlap between FXS and the premutation disorders has recently led to the introduction of the term fragile X spectrum disorder (FXSD) (24) to indicate the continuity of clinical involvement expanded alleles including even those in the gray zone (45 to 54 CGG repeats) and extending throughout the premutation and into the full mutation range.
FMR1 allele categories and prevalence
The CGG repeat is highly polymorphic and according to the American College of Medical Genetics guidelines (25, 26) can be divided into four distinct FMR1 allelic categories: a) normal alleles harboring between 6 and 44 CGG repeats, with the most common alleles in the general population being 29 and 30 CGG repeats; b) intermediate or gray zone alleles between 45 and 54 CGG repeats; c) premutation alleles harboring between 55 and 200 CGG repeats and d) full mutation alleles with greater than 200 CGG repeats. At least 40% of individuals with a full mutation can present with mosaicism (27), which can be differentiated in size mosaicism and methylation mosaicism. In the FXS field size mosaicism is defined as having both premutation and full mutation alleles, not simply multiple full mutation or premutation alleles. Individuals with methylation mosaicism have cells with unmethylated alleles, which can span the entire CGG size range, and cells with methylated alleles, usually in the full mutation range.
Approximately 1 in 5000 males and 1 in 2500 – 8000 females have FXS (reviewed in 28). The prevalence of premutation alleles varies across populations ranging from 110 to 250 in females and from 260 and 810 in males (28, 29). The prevalence of intermediate alleles has been determined in many studies and varies between 1:22–66 in females and 1:42–112 in males. The wide variations in prevalence observed in different studies likely depend on the CGG repeat range adopted for each category in any given study and possibly on ethnicity (28).
The molecular diagnosis of fragile X syndrome
Recent advances in the field of molecular diagnosis of FXS have been made since the discovery of the FMR1 gene in 1991 (30–33). Prior to the identification of the FMR1 gene, culturing cells in a folate-deficient medium followed by cytogenetic analysis was the method of choice for the diagnosis for FXS. However, this approach, while assessing for the presence of “fragile sites” (visualized as discontinuity of staining in the region of the gene) on the long arm of the X chromosome, proved to be difficult (34) as the fragile site was often seen only in small percent of cells. This was not as much as of a problem in males, where the fragile site could generally be seen in at least 10% of cells, but rather in female, where the mutation often could not be visualized.
Thus, research for approximately the past 20 years has focused on the acquisition of more sensitive, mostly PCR-based, molecular techniques and several approaches have been developed, although with different limitations mainly driven by the intractability of expanded CGG repeats. Indeed, currently the diagnosis of FXS is generally based on the measurement of the CGG repeat size and the assessment of the methylation status of the FMR1 gene, using mainly PCR-based approaches.
The gold standard DNA methodologies for the diagnosis of FXS use a combination of polymerase chain reaction (PCR), particularly useful for CGG sizing within the premutation range and, Southern blot analysis for sizing larger alleles and for determining their methylation status (Figure 1). However, it is worth noting that, because these methodologies only test for expansion of the CGG repeat, any individual with FXS caused by a deletion or by a missense mutation, which may include all or portions of the FMR1 gene, will not be detected using this test (35). Although FXS is caused in most of the cases, by the CGG expansion, other mutations, even if not very frequent, can lead to FXS. Many of these cases have indeed been described and many small, gross deletions, splicing, missense, nonsense mutations have been identified and are reported in the Human Gene Mutation Database (http://www.hgmd.cf.ac.uk/ac/gene.php?gene=FMR1) (36). Because Southern blot and PCR analysis do not identify these mutations, their prevalence is not known.
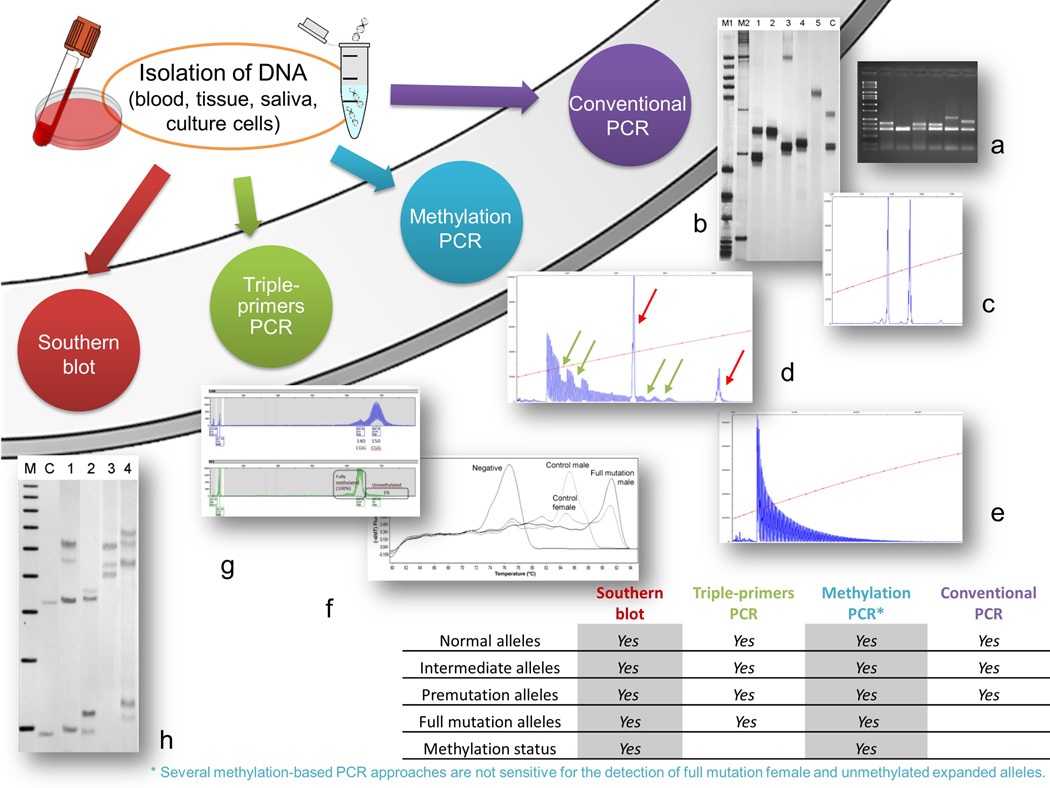
Figure 1.
Diagram showing the different methodologies utilized for the diagnosis of FXS. Genomic DNA (gDNA) can be isolated from whole blood, tissue, saliva or culture cells. Isolated gDNA can be amplified by PCR or digested with methylation sensitive restriction enzymes for Southern blot analysis. Conventional PCR using primers that flank the CGG repeat can amplify FMR1 alleles containing CGG repeat usually up the lower premutation range. The alleles can be visualized either on an agarose gel (a), on an acrylamide gel (b) or by capillary electrophoresis (CE) (c). CE provides accurate CGG sizing particularly if a CGG size marker is utilized. Triple primer PCR (d= premutation female; e= full mutation male) utilizes two FMR1 specific primers that flank the CGG repeat as well as a third primer that is complementary to the CGG repeat element (CGG primer). The PCR produces both full-length gene-specific FMR1 amplicons (red arrows in d) as well as triplet repeat-specific products visualized on CE as a series of peaks (d, e). Two AGG interruptions within the CGG repeat are visible, both within the normal and the premutation allele (green arrows in d) from a woman premutation carrier. Methylation status (including measure of the percent of methylation for any given allele, percent of cells carrying a methylated allele or measure of the activation ration in females) can be assessed by various mPCR strategies (g, mPCR-CE,(47); f, MM-RTPCR, modified from (69)) or by Southern blot analysis (h). Southern blot analysis (h) is performed using genomic DNA digested with the restriction enzymes EcoRI and NruI and separated on agarose gel. The DNA is transferred to a nylon membrane and then hybridized with the dig labeled specific FMR1 probe, StB12.3 (46). M= marker; C= normal control female; lane 1= full mutation female; lane 2= premutation female; lane 3= full mutation male; lane 4= mosaic male. The table in the right lower corner shows the information that can be obtained by the various PCR-methodologies. Specifically, the detection of normal, intermediate, premutation and full mutation alleles, can be obtained using any of the indicated approaches except that full mutation alleles are not amplifiable using conventional PCR. Southern blot can detect alleles from the normal throughout the full mutation range; however, sizing is obtained only for full mutation alleles.
Southern blot analysis is a costly and time consuming procedure that requires larger amount of high molecular weight genomic DNA (37, 38) than does PCR but allows the unbiased detection of larger alleles, difficult to achieve by PCR and, importantly, also provides information on allele methylation. However, because only a limited number of samples can be run simultaneously, a high throughput cannot be achieved with this method. As alternative to Southern Blot analysis, several PCR-based diagnostic strategies have been proposed in the past several years for the identification of FMR1 gene CGG repeat expansions. Conventional PCR-based methodologies do not reliably achieve CGG amplification as this is particularly problematic due to the high CG content and to the tendency to form undenaturable secondary structures; thus, the amplification is mostly limited to alleles spanning from the normal to the lower premutation range.
The combinations of polymerases and co-solvents, such as DMSO and betaine (37, 39–43) are generally used to overcome this issue. Most of these approaches rely on the use of 7-deaza-2’-GTP, which greatly reduces the detection of stained PCR products with ethidium bromide (37) and thus requires an additional detection method (such as silver staining or hybridization with a radioactive or chemiluminescent probe). The use of betaine instead of DMSO to reduce secondary structures, or the Expand Long Template PCR system, have been independently proposed in the analysis of repeat expansions, including CGG, often in the context of fluorescent PCR-based assays (41–43).
The Expand Long Template PCR system in conjunction with the osmolyte betaine proposed by Saluto and collaborators (44) can amplify CGG repeat expansions ranging from the normal throughout the premutation range in both genders. A robust FMR1 gene–specific PCR technology has also demonstrated to be able to accurately identify expanded FMR1 alleles including very large full mutation alleles and present at low abundance. This approach was validated using a series of samples previously evaluated by the combination of polymerase chain reaction (PCR) and Southern blot analysis producing a strong correlation with the results obtained with Southern blot analysis (45). However, these assays are not always able to resolve the apparent homozygosity in females (two normal identical FMR1 alleles vs one normal allele and one unamplified full mutation allele). This obstacle was overcome by the development of the hybrid PCR primer (so named CGG linker primer) that by annealing randomly within the CGG repeat region, creates multiple PCR-amplified products visualized as a smear on agarose gel (46) or as a series of peaks on a capillary electrophoresis (CE) (47) (Figure 1). The CGG linker PCR-based approach which amplifies large FMR1 expanded alleles (46), has been further developed as triplet repeat-primed PCR (TR-PCR) method and has been utilized by several laboratories to replace the use of Southern Blot analysis (45, 47–51) (Figure 1). These studies, and others (51, 52) have validated this PCR approach by providing comparable results to the combination of PCR/Southern blot methodologies. The TR-PCR assay utilizes two primers targeting sequences outside the CGG repeat region and, the CGG primer, which anneals within the CGG region. Thus multiple PCR products are amplified and visualized as serial or stutter peaks on CE (Figure 1). TR-PCR is capable of identifying all expanded alleles from the normal throughout the full mutation range, of distinguishing normal from full mutation females and it provides CGG sizing information.
Importantly, the use of the CGG primer has allowed one to determine the number and location of the AGG interruptions (Figure 1) which when present within the CGG repeat locus, stabilizes the gene during transmission (53), presumably by decreasing the risk of DNA polymerase slippage during DNA replication (54). As premutation alleles are very unstable and tend to expand during transmission, women who carry premutation alleles are at risk of having a child with FXS. Their mode of transmission, which explains the Sherman paradox (55), is such that the risk of expansion for any given premutation allele increases with the number of CGG repeats and also with the presence of AGG interruption (55, 56). Knowledge of the number and position of the AGG interruptions within a premutation allele has permitted the development of a model for estimating the risk of expansion to a full mutation given the total CGG length, number of AGG interruptions and maternal age (56, 57). The AGG interruptions also facilitate assessment of the magnitude of the intergenerational allele expansion from the normal to the premutation range (56–59). This information can be very useful in genetic counseling for premutation carriers as it allows assessment of the risk of expansion in their offspring. Indeed the information gained from AGG analysis can help in reproductive decision-making and options for premutation carriers.
Methylation status
Although several of the current “CGG sizing” methodologies can amplify alleles throughout the full mutation range (44, 45, 47, 49, 50, 60) they cannot determine methylation status, the epigenetic modification leading to FXS. This is of relevance for the diagnosis of FXS as the degree of methylation has been shown to be associated with the degree of intellectual disabilities and/or of the clinical involvement (38, 61–63).
Methylation specific PCR approaches using bisulfite modification of the CGG repeat sequence are based on the conversion of unmethylated cytosine into uracil residues, with methylated cytosine remaining resistant to this modification (64). When amplified and sequenced, this “modified DNA” can provide information about methylation at specific CpG sites within the amplified DNA sequence (65). Although the use of PCR-based amplification coupled with bisulfite modification, while attractive for subsequent long PCR amplification, it could fail unpredictably due to the well-known degradation of DNA during the bisulfite conversion process (66). Nevertheless, the PCR/bisulfite is an approach that has been proposed for the detection of fragile X syndrome (67), for methylation analysis of FXS (68, 69) and successfully used to establish the prevalence of full mutations, in a very large sample size of males from the general population (70).
Additional PCR-based methods for the detection of methylation in the FMR1 gene have been developed, mainly based on the use of methylation sensitive restriction enzymes, followed by PCR and visualization of the amplified products by CE (71–73). Specifically, multiplex ligation-dependent probe amplification techniques (MS-MLPA) have been proposed to distinguish premutation from full mutation alleles; however, these methods are limited to males and therefore less useful for females with an expanded allele (72). More recently, Chen et al. (71) have developed a high resolution methylation PCR able to assess allele-specific methylation states in both genders and even for low abundance alleles, observed in some mosaic males that go undetected by Southern blot analysis. However, although the percent of methylation for each allele can be easily determined, the assessment of the total methylation levels appears to be more complex. A novel and alternative PCR approach in which the bisulfite treatment is followed by a melting curve analysis was recently proposed and based on the melting temperatures of different PCR amplified products (Fig. 1). Coupled to either a methylation specific (MCA; (74)) or a multiplex methylation specific real time PCR (MM-RTPCR) (69), it can differentiate between different PCR amplicons as they melt at specific temperatures and it allows the quantification of the methylation status. In a more recent study, Lim GX, et al. (75) validated this approach as screening tool for the identification of expanded FMR1 alleles, showing high analytic specificity and sensitivity. Since there is no need to analyze the amplified products by gel electrophoresis, this approach has the advantage of reducing the risk of contamination and may be more cost effective of those methodologies that require more post-PCR analysis and more expensive equipments/reagents such as capillary electrophoresis. However, the TP-PCR with MCA approach has the limitation of being very laborious, not being able to distinguish between premutation and full mutation alleles and proven to be not as sensitive for the detection of full mutation females. Lastly and very importantly it does not provide CGG repeat size and FMR1 methylation status (75). However, it may represent a valid screening tool to use in a large population screening studies where the initial step flags for the presence of an expanded allele (>55 CGG repeats).
Finally, a matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS) test can assess the methylation status in large segments of the FMR1 gene from a number of different tissues (76). This approach interrogates two regions within the FMR1 gene, namely fragile X related epigenetic 1 (FREE1) and 2 (FREE2) and was proposed as unique tool for both FXS diagnosis and for population screening for FXS (76, 77). These regions, located within the FMR1 promoter (FREE1) and within exon1/intron1 (FREE2) are methylated in FXS but unmethylated in typical developing controls or in those carrying small-expanded alleles. The methylation status of these regions associates with the methylation in the FMR1 promoter region in subjects with FXS, with FMRP expression (76), and with an increased risk for clinical involvement in premutation carriers including comorbid dysexecutive and social anxiety symptoms, verbal impairment and working memory in premutation women (78–80). As the other bisulfite based approach, the MALDI-TOF MS possesses the limitation that females with a full mutation or carriers cannot be easily distinguished from normal females.
Interestingly, in a recent report, a potential approach based on the use of the single-molecule real-time (SMRT) sequencing proved, for the first time, to be suited for long, repetitive DNA sequences, including CGG expanded repeat alleles (81). The study reported on the sequencing of an expanded FMR1 allele of 750 CGG-repeat, therefore in the full mutation range, demonstrating that sequencing long expanded CG rich regions is feasible. Indeed, until recently, conventional DNA sequencing technologies have been unable to sequence repeat-expansion alleles; thus, the real-time SMRT sequencing could represent a potential future tool for the diagnosis of FXS and future studies could also lead to further development providing information on differential methylation, particularly in mosaic individuals. Moreover, this could eventually permit genotyping in a high-throughput mode required for the screening large populations for CGG repeat alleles.
Population screening
The development of these PCR-based methodologies has also given impetus to several large population efforts. Population screening for FXS, including pre-conception, prenatal, newborn, carriers and high risk screening, has been proposed for the identification of expanded alleles in the general population, to rule out FXS or the presence of a premutation alleles in populations at risk for FMR1 associated disorders (29, 70, 82, 83) but has been prevented, for many years, by the lack of a rapid and cost effective test. As mentioned above, accurate diagnostic DNA testing has been available for a long time and traditionally included a combination of conventional PCR and Southern Blot analysis. However, because it is an expensive methodology and requires large amount of template DNA, this combined approach is not suitable for large population screening. The development of novel PCR based approaches, particularly the ones based on the use of the triplet-primed PCR, of the melting curve analysis or of the fragile X-related epigenetic FREE2 FMR1 methylation, has changed this scenario. These methodologies, although can present with some of the issues discussed above, are able to flag the presence of an expanded allele, are rapid, can be cost-effective and importantly they work with small amount of DNA as the one provided by blood spots cards (28, 70), 75, 77) satisfying therefore, the requirement for a valid and effective screening tool for expanded alleles of the FMR1 gene in a large sample size.
FXS is not included in the Recommended Uniform Screening Panel (RUSP) (84) although patient advocates have generally favorable opinions about earlier identification and feel that early identification of children with FXS could certainly benefit them as well as the families. Their reasoning includes access to services, avoid the diagnostic odyssey, and providing access to participate in behavioral and pharmacological intervention studies. Offering an expanded newborn screening panel under a research protocol with opt-in informed consent, to which families have a choice to participate, could represent a possible alternative. This will lead to a gain of knowledge about the true prevalence of expanded FMR1 alleles and will allow to determining potential patterns of symptom at earlier stages (85, 86). However, the presence of FMR1 expanded alleles is not clinically limited to FXS but it extends to premutation carriers, who can develop a number of medical, cognitive, and emotional problems. Specifically carrier females are at risk for Fragile X-associated Primary Ovarian Insufficiency (FXPOI) (87) and both males and females are at risk for Fragile X-associated Tremor Ataxia Syndrome (FXTAS) (88). In addition, several premutation carriers can present a risk for autism spectrum disorders, seizures, learning disabilities, anxiety, depression attention and visual perceptual deficits (89–92). The identification of premutation carriers and family members at risk for these conditions has led to a controversial debate on when it is the right timing for the early identification of expanded FMR1 alleles. Prenatal and preconception screening will mainly identify premutation carriers. The issues raised by offering carriers screening in pregnant and not pregnant women from the general population have been discussed in a number of studies (93–96) and focused on the significance of obtaining reproductive options and family planning and indicated the importance of ensuring informed decisions and wide accessibility and facilitation of decision-making. Recommendations should perhaps include FMR1 screening of general population to all preconception and prenatal women regardless of their family history, which is cost effective when considering the costs associated with raising a child with FXS
Carrier screening represents a controversial issue because the identification of a carrier status can inform about personal health risks particularly related to late-onset neurological problems, FXTAS, anxiety, depression, or other medical conditions documented in premutation carriers (12). However, knowing of carrier status could encourage changes and preventive lifestyle measures that can potentially reduce the risk of the medical problems observed in premutation carriers. To this regard, more studies are warranted to inform on policy and appropriate approaches and for better understanding of the potential consequences/benefits of offering carrier screening for FXS.
Expert commentary
In the past several years many methodologies, mainly PCR-based, have been validated and proposed for the diagnosis of FXS. For many of them the specificity and the sensitivity have been demonstrated and the limitations stated. Currently, many laboratories have abandoned the use of the traditional Southern Blot approach and used PCR as first screening step coupled by methylation PCR assay (or Southern blot analysis) if an individual with an expanded allele is identified. However, because the limitations presented by the various PCR approaches recently developed, including failing in determining CGG sizing in some cases or the methylation status in other, Southern blot analysis coupled with PCR is still considered the gold standard methodology for the diagnosis of FXS.
Future studies may provide further development of such approaches and yield information of epigenetic modification of the FMR1 gene. More importantly, they may provide the tools to refine the sensitivity and lead to the development of an approach that fulfill the need for reliable diagnosis of FXS throughout the spectrum of the various mutations and, for the FXS screening of large populations.
Five year view
The field of fragile X diagnosis has greatly evolved since the discovery of the FMR1 gene in 1991. For the past 7–10 years a number of PCR based methodologies have proposed for their capability to reliably amplify expanded FMR1 alleles. Although several issues remain to be solved, some of these approaches have provided compelling alternatives to the gold standard DNA methodologies for the diagnosis of FXS, which use a combination of PCR and Southern blot analysis.
It is likely that the advent of rapid DNA sequencing methodologies, which has undoubtedly and greatly accelerated biological discoveries, will allow in the next future to obtain a complete genetic and epigenetic characterization of the FMR1 gene, as well as of other repeat-expansion containing genes. It is also likely that the further development of the sequencing technologies used to assess individuals with intellectual and behavior disabilities will provide the means for detecting non-CGG repeat FMR1 mutations.
Key issues.
The gold standard DNA methodologies for the diagnosis of FXS use a combination of PCR and Southern blot analysis.
Several PCR based methodologies have been developed for the detection of FMR1 expanded alleles.
Limitations of these methodologies include higher costs, inability to define the methylation status of the expanded FMR1 alleles, to detect expanded alleles in both genders, to identify expanded alleles only if methylated or failing in providing a precise CGG allele size.
Several of these approaches are able to identify premutation carriers, which has led to a controversial debate on when it is the appropriate timing for early identification of the FMR1 mutations
Acknowledgments
The author was supported by NICHD grant HD02274.
Footnotes
Financial & competing interests disclosure
The author has no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
References
Papers of special note have been highlighted as:
* of interest
- 1.Hagerman RJ, Des-Portes V, Gasparini F, et al. Translating molecular advances in fragile X syndrome into therapy: a review. J Clin Psychiatry. 2014;75(4):e294–e307. doi: 10.4088/JCP.13r08714. [DOI] [PubMed] [Google Scholar]
- 2.Bailey A, Luthert P, Dean A, et al. A clinicopathological study of autism. Brain. 1998;121(Pt 5):889–905. doi: 10.1093/brain/121.5.889. [DOI] [PubMed] [Google Scholar]
- 3.Harris SW, Hessl D, Goodlin-Jones B, et al. Autism profiles of males with fragile X syndrome. American Journal of Mental Retardation. 2008;113(6):427–438. doi: 10.1352/2008.113:427-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rogers SJ, Wehner DE, Hagerman R. The behavioral phenotype in fragile X: symptoms of autism in very young children with fragile X syndrome, idiopathic autism, and other developmental disorders. J Dev Behav Pediatr. 2001;22(6):409–417. doi: 10.1097/00004703-200112000-00008. [DOI] [PubMed] [Google Scholar]
- 5.Pieretti M, Zhang FP, Fu YH, et al. Absence of expression of the FMR-1 gene in fragile X syndrome. Cell. 1991;66(4):817–822. doi: 10.1016/0092-8674(91)90125-i. [DOI] [PubMed] [Google Scholar]
- 6.Sutcliffe JS, Nelson DL, Zhang F, et al. DNA methylation represses FMR-1 transcription in fragile X syndrome. Hum Mol Genet. 1992;1(6):397–400. doi: 10.1093/hmg/1.6.397. [DOI] [PubMed] [Google Scholar]
- 7.Darnell JC, Klann E. The translation of translational control by FMRP: therapeutic targets for FXS. Nat Neurosci. 2013;16(11):1530–1536. doi: 10.1038/nn.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pasciuto E, Bagni C. SnapShot: FMRP mRNA targets and diseases. Cell. 2014;158(6):1446–1446. e1441. doi: 10.1016/j.cell.2014.08.035. [DOI] [PubMed] [Google Scholar]
- 9.Chonchaiya W, Au J, Schneider A, et al. Increased prevalence of seizures in boys who were probands with the FMR1 premutation and co-morbid autism spectrum disorder. Hum Genet. 2012;131(4):581–589. doi: 10.1007/s00439-011-1106-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clifford S, Dissanayake C, Bui QM, et al. Autism spectrum phenotype in males and females with fragile X full mutation and premutation. J Autism Dev Disord. 2007;37(4):738–747. doi: 10.1007/s10803-006-0205-z. [DOI] [PubMed] [Google Scholar]
- 11.Farzin F, Perry H, Hessl D, et al. Autism spectrum disorders and attention-deficit/hyperactivity disorder in boys with the fragile X premutation. J Dev Behav Pediatr. 2006;27(2 Suppl):S137–S144. doi: 10.1097/00004703-200604002-00012. [DOI] [PubMed] [Google Scholar]
- 12. Hagerman R, Hagerman P. Advances in clinical and molecular understanding of the FMR1 premutation and fragile X-associated tremor/ataxia syndrome. Lancet Neurol. 2013;12(8):786–798. doi: 10.1016/S1474-4422(13)70125-X. * This paper is a review on the broad spectrum of clinical involvements obserevd in premutation carriers.
- 13.Tassone F, Hagerman RJ, Taylor AK, et al. Elevated levels of FMR1 mRNA in carrier males: a new mechanism of involvement in the fragile-X syndrome. Am J Hum Genet. 2000;66(1):6–15. doi: 10.1086/302720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hagerman RJ, Hagerman PJ. Fragile X syndrome: a model of gene-brain-behavior relationships. Molecular Genetics and Metabolism. 2001;74:89–97. doi: 10.1006/mgme.2001.3225. [DOI] [PubMed] [Google Scholar]
- 15.Bodega B, Bione S, Dalpra L, et al. Influence of intermediate and uninterrupted FMR1 CGG expansions in premature ovarian failure manifestation. Hum Reprod. 2006;21(4):952–957. doi: 10.1093/humrep/dei432. [DOI] [PubMed] [Google Scholar]
- 16.Bretherick KL, Fluker MR, Robinson WP. FMR1 repeat sizes in the gray zone and high end of the normal range are associated with premature ovarian failure. Hum Genet. 2005;117(4):376–382. doi: 10.1007/s00439-005-1326-8. [DOI] [PubMed] [Google Scholar]
- 17.Hall D, Tassone F, Klepitskaya O, et al. Fragile X–associated tremor ataxia syndrome in FMR1 gray zone allele carriers. Mov Disord. 2012;27(2):297–301. doi: 10.1002/mds.24021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Y, Winarni T, Zhang L, et al. Fragile X-associated tremor/ataxia syndrome (FXTAS) in grey zone carriers. Clin Genet. 2013;84(1):74–77. doi: 10.1111/cge.12026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loesch DZ, Churchyard A, Brotchie P, et al. Evidence for, a spectrum of, neurological involvement in carriers of the fragile X pre-mutation: FXTAS and beyond. Clin Genet. 2005;67(5):412–417. doi: 10.1111/j.1399-0004.2005.00425.x. [DOI] [PubMed] [Google Scholar]
- 20.Kenneson A, Zhang F, Hagedorn CH, et al. Reduced FMRP and increased FMR1 transcription is proportionally associated with CGG repeat number in intermediate-length and premutation carriers. Hum Mol Genet. 2001;10(14):1449–1454. doi: 10.1093/hmg/10.14.1449. [DOI] [PubMed] [Google Scholar]
- 21.Loesch DZ, Bui QM, Huggins RM, et al. Transcript levels of the intermediate size or grey zone fragile X mental retardation 1 alleles are raised, and correlate with the number of CGG repeats. J Med Genet. 2007;44(3):200–204. doi: 10.1136/jmg.2006.043950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loesch DZ, Godler DE, Khaniani M, et al. Linking the FMR1 alleles with small CGG expansions with neurodevelopmental disorders: preliminary data suggest an involvement of epigenetic mechanisms. Am J Med Genet A. 2009;149A(10):2306–2310. doi: 10.1002/ajmg.a.32990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sellier C, Usdin K, Pastori C, et al. The multiple molecular facets of fragile X-associated tremor/ataxia syndrome. J Neurodev Disord. 2014;6(1):23. doi: 10.1186/1866-1955-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lozano R, Rosero CA, Hagerman RJ. Fragile X spectrum disorders. Intractable Rare Dis Res. 2014;3(4):134–146. doi: 10.5582/irdr.2014.01022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maddalena A, Richards CS, McGinniss MJ, et al. Technical standards and guidelines for fragile X: the first of a series of disease-specific supplements to the Standards and Guidelines for Clinical Genetics Laboratories of the American College of Medical Genetics. Quality Assurance Subcommittee of the Laboratory Practice Committee. Genet Med. 2001;3(3):200–205. doi: 10.1097/00125817-200105000-00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sherman S, Pletcher BA, Driscoll DA. Fragile X syndrome: Diagnostic and carrier testing. Genet Med. 2005;7(8):584–587. doi: 10.1097/01.GIM.0000182468.22666.dd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nolin SL, Glicksman A, Houck GE, Jr, et al. Mosaicism in fragile X affected males. Am J Med Genet. 1994;51(4):509–512. doi: 10.1002/ajmg.1320510444. [DOI] [PubMed] [Google Scholar]
- 28. Tassone F, Iong KP, Tong TH, et al. FMR1 CGG allele size and prevalence ascertained through newborn screening in the United States. Genome Med. 2012;4(12):100. doi: 10.1186/gm401. * This paper provides for the first time the prevalence of intermediate and premutation alleles in males and female newborns in US
- 29.Maenner MJ, Baker MW, Broman KW, et al. FMR1 CGG expansions: prevalence and sex ratios. Am J Med Genet B Neuropsychiatr Genet. 2013;162B(5):466–473. doi: 10.1002/ajmg.b.32176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kremer EJ, Pritchard M, Lynch M, et al. Mapping of DNA instability at the fragile X to a trinucleotide repeat sequence p(CCG)n. Science. 1991;252(5013):1711–1714. doi: 10.1126/science.1675488. [DOI] [PubMed] [Google Scholar]
- 31.Oberle I, Rousseau F, Heitz D, et al. Instability of a 550-base pair DNA segment and abnormal methylation in fragile X syndrome. Science. 1991;252(5010):1097–1102. doi: 10.1126/science.252.5009.1097. [DOI] [PubMed] [Google Scholar]
- 32.Verkerk AJ, Pieretti M, Sutcliffe JS, et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65(5):905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- 33.Yu S, Mulley J, Loesch D, et al. Fragile-X syndrome: unique genetics of the heritable unstable element. Am J Hum Genet. 1992;50(5):968–980. [PMC free article] [PubMed] [Google Scholar]
- 34.Sutherland GR, Baker E, Fratini A. Excess thymidine induces folate sensitive fragile sites. American Journal of Medical Genetics. 1985;22(2):433–443. doi: 10.1002/ajmg.1320220234. [DOI] [PubMed] [Google Scholar]
- 35.Wells RD. Mutation spectra in fragile X syndrome induced by deletions of CGG*CCG repeats. J Biol Chem. 2009;284(12):7407–7411. doi: 10.1074/jbc.R800024200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Handt M, Epplen A, Hoffjan S, et al. Point mutation frequency in the FMR1 gene as revealed by fragile X syndrome screening. Mol Cell Probes. 2014;28(5–6):279–283. doi: 10.1016/j.mcp.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 37.Brown WT, Nolin S, Houck G, Jr, et al. Prenatal diagnosis and carrier screening for fragile X by PCR. Am J Med Genet. 1996;64(1):191–195. doi: 10.1002/(SICI)1096-8628(19960712)64:1<191::AID-AJMG34>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 38.Snow K, Doud LK, Hagerman R, et al. Analysis of a CGG sequence at the FMR-1 locus in fragile X families and in the general population. Am J Hum Genet. 1993;53(6):1217–1228. [PMC free article] [PubMed] [Google Scholar]
- 39.O'Connell CD, Atha DH, Jakupciak JP, et al. Standardization of PCR amplification for fragile X trinucleotide repeat measurements. Clin Genet. 2002;61(1):13–20. doi: 10.1034/j.1399-0004.2002.610103.x. [DOI] [PubMed] [Google Scholar]
- 40.Haddad LA, Mingroni-Netto RC, Vianna-Morgante AM, et al. A PCR-based test suitable for screening for fragile X syndrome among mentally retarded males. Hum Genet. 1996;97(6):808–812. doi: 10.1007/BF02346194. [DOI] [PubMed] [Google Scholar]
- 41.Houdayer C, Lemonnier A, Gerard M, et al. Improved fluorescent PCR-based assay for sizing CGG repeats at the FRAXA locus. Clin Chem Lab Med. 1999;37(4):397–402. doi: 10.1515/CCLM.1999.065. [DOI] [PubMed] [Google Scholar]
- 42.Hamdan H, Tynan JA, Fenwick RA, et al. Automated Detection of Trinucleotide Repeats in Fragile X Syndrome. Mol Diagn. 1997;2(4):259–269. doi: 10.1054/MODI00200259. [DOI] [PubMed] [Google Scholar]
- 43.Hecimovic S, Vlasic J, Barisic L, et al. A simple and rapid analysis of triplet repeat diseases by expand long PCR. Clin Chem Lab Med. 2001;39(12):1259–1262. doi: 10.1515/CCLM.2001.202. [DOI] [PubMed] [Google Scholar]
- 44.Saluto A, Brussino A, Tassone F, et al. An enhanced polymerase chain reaction assay to detect pre- and full mutation alleles of the fragile X mental retardation 1 gene. J Mol Diagn. 2005;7(5):605–612. doi: 10.1016/S1525-1578(10)60594-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Filipovic-Sadic S, Sah S, Chen L, et al. A Novel FMR1 PCR Method for the Routine Detection of Low-Abundance Expanded Alleles and Full Mutations in Fragile X Syndrome. Clin Chem. 2010;56(3):399–408. doi: 10.1373/clinchem.2009.136101. * This paper describes a novel PCR technology able to accurately identify FMR1 expanded alleles throughout the spectrum of CGG sizes in both males and females potentially replacing the need of Southern blot analysis.
- 46. Tassone F, Pan R, Amiri K, et al. A rapid polymerase chain reaction-based screening method for identification of all expanded alleles of the fragile X (FMR1) gene in newborn and high-risk populations. J Mol Diagn. 2008;10(1):43–49. doi: 10.2353/jmoldx.2008.070073. * This paper describes for the first time a novel approach based on the use of a CGG primer able to amplify FMR1 expanded alleles from the normal to the full mutation range in both males and females on DNA isolated also from blood spot cards.
- 47.Chen L, Hadd A, Sah S, et al. An information-rich CGG repeat primed PCR that detects the full range of fragile X expanded alleles and minimizes the need for southern blot analysis. J Mol Diagn. 2010;12(5):589–600. doi: 10.2353/jmoldx.2010.090227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hantash FM, Goos DG, Tsao D, et al. Qualitative assessment of FMR1 (CGG)n triplet repeat status in normal, intermediate, premutation, full mutation, and mosaic carriers in both sexes: implications for fragile X syndrome carrier and newborn screening. Genet Med. 2010;12(3):162–173. doi: 10.1097/GIM.0b013e3181d0d40e. [DOI] [PubMed] [Google Scholar]
- 49.Lyon E, Laver T, Yu P, et al. A simple, high-throughput assay for Fragile X expanded alleles using triple repeat primed PCR and capillary electrophoresis. J Mol Diagn. 2010;12(4):505–511. doi: 10.2353/jmoldx.2010.090229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Strom CM, Huang D, Li Y, et al. Development of a novel, accurate, automated, rapid, high-throughput technique suitable for population-based carrier screening for Fragile X syndrome. Genet Med. 2007;9(4):199–207. doi: 10.1097/gim.0b013e31803d3ac9. [DOI] [PubMed] [Google Scholar]
- 51.Nahhas F, Monroe T, Prior T, et al. Evaluation of the human fragile X mental retardation 1 polymerase chain reaction reagents to amplify the FMR1 gene: testing in a clinical diagnostic laboratory. Genet Test Mol Biomarkers. 2012;16(3):187–192. doi: 10.1089/gtmb.2011.0128. [DOI] [PubMed] [Google Scholar]
- 52.Curtis-Cioffi KMC, Rodrigueiro DA, Rodrigues VC, et al. Comparison between the polymerase chain reaction-based screening and the Southern blot methods for identification of fragile X syndrome. Genet Test Mol Biomarkers. 2012;16(11):1303–1308. doi: 10.1089/gtmb.2012.0158. [DOI] [PubMed] [Google Scholar]
- 53.Eichler EE, Holden JJ, Popovich BW, et al. Length of uninterrupted CGG repeats determines instability in the FMR1 gene. Nat Genet. 1994;8(1):88–94. doi: 10.1038/ng0994-88. [DOI] [PubMed] [Google Scholar]
- 54.Gacy AM, Goellner G, Juranic N, et al. Trinucleotide repeats that expand in human disease form hairpin structures in vitro. Cell. 1995;81(4):533–540. doi: 10.1016/0092-8674(95)90074-8. [DOI] [PubMed] [Google Scholar]
- 55.Fu YH, Kuhl DP, Pizzuti A, et al. Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell. 1991;67(6):1047–1058. doi: 10.1016/0092-8674(91)90283-5. [DOI] [PubMed] [Google Scholar]
- 56.Yrigollen CM, Durbin-Johnson B, Gane L, et al. AGG interruptions within the maternal FMR1 gene reduce the risk of offspring with fragile X syndrome. Genet Med. 2012;14(8):729–736. doi: 10.1038/gim.2012.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yrigollen CM, Martorell L, Durbin-Johnson B, et al. AGG interruptions and maternal age affect FMR1 CGG repeat allele stability during transmission. J Neurodev Disord. 2014;6(1):24. doi: 10.1186/1866-1955-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nolin SL, Glicksman A, Ersalesi N, et al. Fragile X full mutation expansions are inhibited by one or more AGG interruptions in premutation carriers. Genet Med. 2015;17(5):358–364. doi: 10.1038/gim.2014.106. [DOI] [PubMed] [Google Scholar]
- 59.Nolin SL, Sah S, Glicksman A, et al. Fragile X AGG analysis provides new risk predictions for 45–69 repeat alleles. Am J Med Genet A. 2013;161(4):771–778. doi: 10.1002/ajmg.a.35833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hantash FM. Fragile X syndrome: is now the time for population screening? MLO Med Lab Obs. 2010;42(5):20, 22. [PubMed] [Google Scholar]
- 61.Hagerman RJ, Hull CE, Safanda JF, et al. High functioning fragile X males: demonstration of an unmethylated fully expanded FMR-1 mutation associated with protein expression. Am J Med Genet. 1994;51(4):298–308. doi: 10.1002/ajmg.1320510404. [DOI] [PubMed] [Google Scholar]
- 62.McConkie-Rosell A, Lachiewicz AM, Spiridigliozzi GA, et al. Evidence that methylation of the FMR-I locus is responsible for variable phenotypic expression of the fragile X syndrome. Am J Hum Genet. 1993;53(4):800–809. [PMC free article] [PubMed] [Google Scholar]
- 63.Pretto D, Yrigollen CM, Tang HT, et al. Clinical and molecular implications of mosaicism in FMR1 full mutations. Front Genet. 2014;5:318. doi: 10.3389/fgene.2014.00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Clark SJ, Harrison J, Paul CL, et al. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22(15):2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Laird PW. Principles and challenges of genomewide DNA methylation analysis. Nat Rev Genet. 2010;11(3):191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- 66.Grunau C, Clark SJ, Rosenthal A. Bisulfite genomic sequencing: systematic investigation of critical experimental parameters. Nucleic Acids Res. 2001;29(13):E65–E65. doi: 10.1093/nar/29.13.e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rosales-Reynoso M, Vilatela E, Ojeda R, et al. PCR approach for detection of Fragile X syndrome and Huntington disease based on modified DNA: limits and utility. Genet Test. 2007;11(2):153–159. doi: 10.1089/gte.2006.0508. [DOI] [PubMed] [Google Scholar]
- 68.Das S, Kubota T, Song M, et al. Methylation analysis of the fragile X syndrome by PCR. Genet Test. 1997;1(3):151–155. doi: 10.1089/gte.1997.1.151. [DOI] [PubMed] [Google Scholar]
- 69.Elias MH, Ankathil R, Salmi AR, et al. A new method for FMR1 gene methylation screening by multiplex methylation-specific real-time polymerase chain reaction. Genet Test Mol Biomarkers. 2011;15(6):387–393. doi: 10.1089/gtmb.2010.0191. [DOI] [PubMed] [Google Scholar]
- 70. Coffee B, Keith K, Albizua I, et al. Incidence of fragile X syndrome by newborn screening for methylated FMR1 DNA. Am J Hum Genet. 2009;85(4):503–514. doi: 10.1016/j.ajhg.2009.09.007. * This paper represents the largest male newborn screening study in US and it provides the prevalence of full mutation in males form the general population.
- 71.Chen L, Hadd AG, Sah S, et al. High-resolution methylation polymerase chain reaction for fragile X analysis: evidence for novel FMR1 methylation patterns undetected in Southern blot analyses. Genet Med. 2011;13(6):528–538. doi: 10.1097/GIM.0b013e31820a780f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nygren AO, Lens SI, Carvalho R. Methylation-specific multiplex ligation-dependent probe amplification enables a rapid and reliable distinction between male FMR1 premutation and full-mutation alleles. J Mol Diagn. 2008;10(6):496–501. doi: 10.2353/jmoldx.2008.080053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhou Y, Law HY, Boehm CD, et al. Robust fragile X (CGG)n genotype classification using a methylation specific triple PCR assay. J Med Genet. 2004;41(4):e45. doi: 10.1136/jmg.2003.012716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dahl C, Gronskov K, Larsen LA, et al. A homogeneous assay for analysis of FMR1 promoter methylation in patients with fragile X syndrome. Clin Chem. 2007;53(4):790–793. doi: 10.1373/clinchem.2006.080762. [DOI] [PubMed] [Google Scholar]
- 75.Lim GX, Loo YL, Mundhofir FE, et al. Validation of a Commercially Available Screening Tool for the Rapid Identification of CGG Trinucleotide Repeat Expansions in FMR1. The Journal of Molecular Diagnostics. 2015;17(3):302–314. doi: 10.1016/j.jmoldx.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 76.Godler DE, Tassone F, Loesch DZ, et al. Methylation of novel markers of fragile X alleles is inversely correlated with FMRP expression and FMR1 activation ratio. Hum Mol Genet. 2010;19(8):1618–1632. doi: 10.1093/hmg/ddq037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Inaba Y, Schwartz CE, Bui QM, et al. Early detection of fragile X syndrome: applications of a novel approach for improved quantitative methylation analysis in venous blood and newborn blood spots. Clinical chemistry. 2014;60(7):963–973. doi: 10.1373/clinchem.2013.217331. [DOI] [PubMed] [Google Scholar]
- 78.Cornish KM, Kraan CM, Bui QM, et al. Novel methylation markers of the dysexecutive-psychiatric phenotype in FMR1 premutation women. Neurology. 2015;84(16):1631–1638. doi: 10.1212/WNL.0000000000001496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Godler DE, Slater HR, Bui QM, et al. Fragile X mental retardation 1 (FMR1) intron 1 methylation in blood predicts verbal cognitive impairment in female carriers of expanded FMR1 alleles: evidence from a pilot study. Clin Chem. 2012;58(3):590–598. doi: 10.1373/clinchem.2011.177626. [DOI] [PubMed] [Google Scholar]
- 80.Shelton AL, Cornish KM, Godler DE, et al. Delineation of the working memory profile in female FMR1 premutation carriers: the effect of cognitive load on ocular motor responses. Behav Brain Res. 2015;282:194–200. doi: 10.1016/j.bbr.2015.01.011. [DOI] [PubMed] [Google Scholar]
- 81. Loomis EW, Eid JS, Peluso P, et al. Sequencing the unsequenceable: expanded CGG-repeat alleles of the fragile X gene. Genome Res. 2013;23(1):121–128. doi: 10.1101/gr.141705.112. * This paper demonstrates the ability of single-molecule, real-time (SMRT) sequencing to sequence full mutation FMR1 alleles.
- 82.Musci TJ, Caughey AB. Cost-effectiveness analysis of prenatal population-based fragile X carrier screening. Am J Obstet Gynecol. 2005;192(6):1905–1912. doi: 10.1016/j.ajog.2005.02.052. discussion 1912-1905. [DOI] [PubMed] [Google Scholar]
- 83.Song FJ, Barton P, Sleightholme V, et al. Screening for fragile X syndrome: a literature review and modelling study. Health Technol Assess. 2003;7(16):1–106. doi: 10.3310/hta7160. [DOI] [PubMed] [Google Scholar]
- 84.Kemper AR, Green NS, Calonge N, et al. Decision-making process for conditions nominated to the recommended uniform screening panel: statement of the US Department of Health and Human Services Secretary's Advisory Committee on Heritable Disorders in Newborns and Children. Genet Med. 2014;16(2):183–187. doi: 10.1038/gim.2013.98. [DOI] [PubMed] [Google Scholar]
- 85.Bailey DB, Jr, Gehtland L. Newborn screening: evolving challenges in an era of rapid discovery. JAMA. 2015;313(15):1511–1512. doi: 10.1001/jama.2014.17488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tassone F. Newborn screening for fragile X syndrome. JAMA Neurol. 2014;71(3):355–359. doi: 10.1001/jamaneurol.2013.4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sherman SL, Curnow EC, Easley CA, et al. Use of model systems to understand the etiology of fragile X-associated primary ovarian insufficiency (FXPOI) J Neurodev Disord. 2014;6(1):26. doi: 10.1186/1866-1955-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hagerman P, Hagerman R. Fragile X-associated tremor/ataxia syndrome. Ann N Y Acad Sci. 2015;1338:58–70. doi: 10.1111/nyas.12693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gallego PK, Burris JL, Rivera SM. Visual motion processing deficits in infants with the fragile X premutation. J Neurodev Disord. 2014;6:29. doi: 10.1186/1866-1955-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Grigsby J, Cornish K, Hocking D, et al. The cognitive neuropsychological phenotype of carriers of the FMR1 premutation. J Neurodev Disord. 2014;6:28. doi: 10.1186/1866-1955-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Roberts JE, Tonnsen BL, McCary LM, et al. Trajectory and predictors of depression and anxiety disorders in mothers with the FMR1 premutation. Biological psychiatry. 2015 doi: 10.1016/j.biopsych.2015.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wheeler AC, Bailey DB, Jr, Berry-Kravis E, et al. Associated features in females with an FMR1 premutation. J Neurodev Disord. 2014;6:30. doi: 10.1186/1866-1955-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ames AG, Jaques A, Ukoumunne OC, et al. Development of a fragile X syndrome (FXS) knowledge scale: towards a modified multidimensional measure of informed choice for FXS population carrier screening. Health Expectations. 2015;18(1):69–80. doi: 10.1111/hex.12009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Anido A, Carlson LM, Taft L, et al. Women's attitudes toward testing for fragile X carrier status: a qualitative analysis. J Genet Couns. 2005;14(4):295–306. doi: 10.1007/s10897-005-1159-6. [DOI] [PubMed] [Google Scholar]
- 95.Archibald AD, Jaques AM, Wake S, et al. “It's something I need to consider”: Decisions about carrier screening for fragile X syndrome in a population of non-pregnant women. American Journal of Medical Genetics Part A. 2009;149(12):2731–2738. doi: 10.1002/ajmg.a.33122. [DOI] [PubMed] [Google Scholar]
- 96.Martyn M, Anderson V, Archibald A, et al. Offering fragile X syndrome carrier screening: a prospective mixed-methods observational study comparing carrier screening of pregnant and non-pregnant women in the general population. BMJ open. 2013;3(9):e003660. doi: 10.1136/bmjopen-2013-003660. [DOI] [PMC free article] [PubMed] [Google Scholar]