Abstract
Purpose
Accumulation of bisretinoids as lipofuscin in retinal pigment epithelial (RPE) cells is implicated in the pathogenesis of some blinding diseases including age-related macular degeneration (AMD). To identify genes whose expression may change under conditions of bisretinoid accumulation, we investigated the differential gene expression in RPE cells that had accumulated the lipofuscin fluorophore A2E and were exposed to blue light (430 nm).
Methods
A2E-laden RPE cells were exposed to blue light (A2E/430 nm) at various time intervals. Cell death was quantified using Dead Red staining, and RNA levels for the entire genome was determined using DNA microarrays (Affymetrix GeneChip Human Genome 2.0 Plus). Array results for selected genes were confirmed by real-time reverse-transcriptase polymerase chain reaction.
Results
Principal component analysis revealed that the A2E-laden RPE cells irradiated with blue light were clearly distinguishable from the control samples. We found differential regulation of genes belonging to the following functional groups: transcription factors, stress response, apoptosis and immune response. Among the last mentioned were downregulation of four genes that coded for proteins that have an inhibitory effect on the complement cascade: (complement factor H, complement factor H-related 1, complement factor I and vitronectin) and of two belonging to the classical pathway (complement component 1, s subcomponent and complement component 1, r subcomponent).
Conclusion
This study demonstrates that blue light irradiation of A2E-laden RPE cells can alter the transcription of genes belonging to different functional pathways including stress response, apoptosis and the immune response. We suggest that these molecules may be associated to the pathogenesis of AMD and can potentially serve as future therapeutic targets.
Keywords: A2E, age-related macular degeneration, apoptosis, complement cascade, gene expression, retinal pigment epithelial cells
Introduction
Bisretinoid adducts accumulate in retinal pigm’ent epithelial (RPE) cells with age and constitute the autofluorescent lipofuscin of the cells. The lipofuscin deposits of RPE are primarily derivatives of vitamin A that originate in photoreceptor cells and are deposited in RPE as components of phagocytized photoreceptor outer segments. RPE lipofuscin can be monitored in vivo as fundus autofluorescence and may be associated with retinal degenerative diseases such as best macular dystrophy, recessive Stargardt disease and age-related macular degeneration (AMD; Sparrow & Boulton 2005; Sparrow 2007). Numerous fluorophores have been detected in RPE lipofuscin such as the pyridinium bis-retinoid A2E and isoA2E (Eldred & Lasky 1993; Parish et al. 1998), oxidized derivatives of A2E, conjugates of the all-trans-retinal dimer and trans-retinal dimer-phosphatidylethanolamine (atRAL dimer-PE; Kim et al. 2007; Sparrow 2007) and A-aldehyde dihydropyridine-phosphatidylethanolamine (A2-DHP-PE; Wu et al. 2009). Of these, the best characterized is A2E, an amphiphilic molecule that contains a quaternary amine nitrogen and that forms from two molecules of all-trans-retinal and one of phosphatidylethanolamine; the latter is a component of photoreceptor outer segment membranes (Liu et al. 2000).
A2E is photoreactive, and blue light exposure of intracellular accumulated A2E leads to singlet oxygen production. The formed singlet oxygen adds to the carbon–carbon-double bonds of the retinoid derived side-arms of the A2E molecule resulting in formation of oxygen-containing moieties (Ben-Shabat et al. 2002; Sparrow et al. 2002; Jang et al. 2005). The oxygen-containing groups in photooxidized A2E include epoxides that rearrange to furanoid structures and cyclic peroxides, which are highly reactive and therefore suspected to be cytotoxic (Ben-Shabat et al. 2002; Dillon et al. 2004; Jang et al. 2005). It is well established that exposure of cultured A2E-laden RPE cells to blue light (430 nm) results in cell death (Schutt et al. 2000; Sparrow et al. 2000; Davies et al. 2001; Sparrow & Cai 2001; Westlund et al. 2009). This process has been shown to be dependent on the formation of reactive oxygen species (Sparrow et al. 2002). Of particular importance is the finding that A2E-laden RPE cells irradiated at 430 nm as well as photooxidized forms of A2E tested in a noncellular assay can activate the complement system (Zhou et al. 2006, 2009). Studies indicate that complement activation by this mechanism may be dependent on the alternative pathway (Zhou et al. 2009).
Age-related macular degeneration is considered to be a multifactorial disease with many contributing factors and mechanisms working together in a complicated interplay. Different elements, for instance, complement dysregulation and oxidative mechanisms are suspected to play a role in the pathogenesis (Beatty et al. 2000; Holz et al. 2001; Anderson et al. 2002; Hageman et al. 2005).
The role of photooxidized A2E in retinal disorders is far from fully elucidated. Using microarrays, we sought to clarify how intracellular A2E-accumulation and exposure to blue light affects gene expression. Studying changes in gene expression can provide important information about pathways that might be implicated in RPE cell death and provide clues to other cellular processes that might be influenced by photooxidative process in RPE.
Methods
A2E accumulation and irradiation
Cells were treated as previously described (Sparrow et al. 1999, 2000, 2002). In short, A2E was synthesized from all-trans-retinal and ethanolamine (2:1; Parish et al. 1998). A human adult RPE cell line (ARPE-19; American Type Culture Collection, Manassas, VA, USA) lacking endogenous A2E (Sparrow et al. 1999) was grown to confluence in 35-mm dishes in cell culture medium consisting of Dulbecco’s Modified Eagle Medium (DME; Gibco, Gaithersburg, MD, USA) with 10% heat-inactivated fetal calf serum (Hyclone, Logan, UT, USA), 2 mM glutamine (Gibco), 0.1 mM minimum essential medium nonessential amino acids solution (Gibco) and gentamicin sulphate (10 μg/ml). Subsequently, cells were incubated with synthesized A2E in cell culture media at a concentration of 10 μM to allow A2E accumulation; the latter was confirmed by light microscopy.
Culture medium was replaced with Dulbecco’s phosphate-buffered saline containing calcium, magnesium (Gibco) and 5.5 mM glucose, and cells grown for a minimum of 4 weeks were irradiated with blue light (430 ± 30 nm 1 mW/cm2, 6, 10, 15 or 20 min exposure) followed by 6 hr incubation. These wavelengths are consistent with the excitation spectrum of A2E. (Parish et al. 1998; Sparrow et al. 2000) The time intervals were chosen to ensure both sublethal and lethal exposure to blue light of the A2E-laden RPE cells. Controls included untreated, A2E-free RPE cells, A2E-free RPE cells irradiated for 20 min and RPE cells that had accumulated A2E but were not exposed to blue light. The experiment was repeated in triplicates. The three experiments were performed on subsequent passages of the ARPE-19 cells and therefore in the same passage intervals.
Cell viability assays
Cell viability was quantified 8 hr after light exposure by labelling nuclei of nonviable cells with the membrane impermeant dye Dead Red (Molecular Probes, Eugene, OR, USA) and the nuclei of all cells with 4′,6′-diamino-2-phenylindole (DAPI), as previously described (Sparrow et al. 2002).
Preparation of total RNA
Total RNA was extracted using NucleoSpin® RNA II (Macherey-Nagel GmbH, Düren, Germany) according to the manufacturer’s protocol.
Affymetrix GeneChip analysis
The genome-wide Human Genome U133 Plus 2.0 Array (Affymetrix, Santa Clara, CA: http://www.affymetrix.com) representing more than 54 000 probe sets translating into approximately 47 000 transcripts and 38 500 well-characterized genes were used. Processing of the Affymetrix GeneChips® were performed by the RH Microarray Centre at Rigshospitalet (Copenhagen, Denmark), following the guidelines from Affymetrix.
Data analysis
Summation and normalization of Probe set expression measures were carried out using the Robust Multichip Average (RMA) method of background correction, quantile normalization and summarization of signal intensity (Irizarry et al. 2003). Calculations were made using the statistical software environment R (http://www.R-project.org). Principal component analysis (PCA), functional annotation for biological processes and identification of overrepresentation of potential transcription factor binding sites were calculated using R functions developed by the authors as previously described (Hansen et al. 2012). In brief, the 2.5% of genes contributing the most to the first principal component were identified and overrepresentation of gene ontology (GO) terms for biological processes and of predicted transcription factor binding sites were calculated using Fisher’s exact test for proportions followed by correction for multiple testing using the Bonferroni procedure. Student’s t-test was used to compare RPE cell cultures analysed at different time points. Data were pooled for groups where no statistically significance was observed. Microarray data are reported according to the MIAME statement (Brazma et al. 2001).
Quantitative reverse-transcriptase polymerase chain reaction
Several differentially regulated genes with relation to the immune system or cell death were selected and quantitative reverse-transcriptase polymerase chain reaction (qRT-PCR) was performed for these candidate genes to validate the results obtained from the microarrays. cDNA was produced from 2 μg of each original RNA sample using the ‘Revertaid First Strand Synthesis’ kit (Fermentas, Burlington, ON, Canada).
For the qPCR reaction, a Brilliant SYBR Green QPCR Mastermix was used according to the manufacturer’s instructions (Stratagene; AH Diagnostics, Aarhus, Denmark). The reactions were run on the Stratagene Mx3000P. Reactions were performed in triplicates plus a control without reverse-transcriptase and a control without template. Table 1 shows primer sequences used. To account for differences in the amount of total RNA added to each reaction, the housekeeping gene GAPDH was used as internal amplification control. Melting curves were routinely checked to rule out the amplification of unrelated fragments during qRT-PCR. Pfaffl’s method (Pfaffl 2001) was used to calculate the relative gene expression, and mean and Standard Error of Mean (SEM) of the relative quantity of target mRNA were calculated for all groups (n = 3). Means for all experimental groups were compared with untreated controls by one-way ANOVA and the Newman–Keul Multiple Comparison test (Prism; GraphPad Software, San Diego, CA, USA). Values were considered statistical significantly different when p < 0.05.
Table 1.
Primers for qRT-PCR.
| Gene symbol | Forward (5′–3′) | Reverse (5′–3′) |
|---|---|---|
| CFI | CGAACACCTCCAACATGAAG | TCTCCACCAGATCCTCCTGA |
| CFH | CCAGATGCATCCGTGTCA | TCGCTTTTTCTTTTAAGGCA |
| C3 | ACTGTGCTGACCCCTGCC | TCACGATCCCAGCCAACA |
| HSPA6 | TGCAAGAGGAAAGCCTTAGGGACA | ACAGATTTGCTCCAGCTCCCTCTT |
| HSPA1A | GGGCCTTTCCAAGATTGCTGTT | TCATCTCTGCATGTAGAAACCGGA |
| HSPA1B | TGCTTCAGCTCTTTGCTGCTTCAC | ACTCGTACAGAAGGTGGCAGTGTT |
| ATF3 | AGAGTCGGAGAAGCTGGAAAGTGT | AATACACGTGGGCCGATGAAGGTT |
| ATF4 | CAACAACAGCAAGGAGGATGCCTT | TGTCATCCAACGTGGTCAGAAGGT |
| DITT3 | TGGAACCTGAGGAGAGAGTGTTCA | TGTCCCGAAGGAGAAAGGCAATGA |
| CFLAR | AGAGTGCTGATGGCAGAGATTGGT | TCTCCAACTCAACCACAAGGTCCA |
| HMOX1 | GGGCCAGCAACAAAGTGCAAGATT | TCGCCACCAGAAAGCTGAGTGTAA |
| CCL2 | GCCTCCAGCATGAAAGTCTC | AGGTGACTGGGGCATTGAT |
| GAPDH | AGCCTCAAGATCATCAGCAATGCC | TGTGGTCATGAGTCCTTCCACGAT |
CFH = complement factor H; CFI = complement factor I; C3, complement component 3; HSPA6, heat shock 70 kDa protein 6; HSPA1A, heat shock 70 kDa protein 1A; HSPA1B, heat shock 70 kDa protein 1B; ATF3, activating transcription factor 3; ATF4, activating transcription factor 4; DITT3, DNA-damage-inducible transcript 3; CFLAR, CASP8 and FAAD-like apoptosis regulator; HMOX1, heme oxygenase 1; CCL2, C-C motif chemokine ligand 2; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Results
Assessment of irradiation-induced cell death
Retinal pigment epithelial cells that had accumulated A2E were exposed to blue light (A2E/430 nm) for 6, 10, 15 and 20 min. Cell death was measured after 8 hr by colabelling all cells with DAPI and nuclei of dead cells with a membrane impermeant dye. In A2E-containing RPE cells irradiated for 6 or 10 min, <1% of the cells were nonviable. The rate of cell death increased to 5.1% (±3.4%, p > 0.05) and 16.3% (±3.6%, p < 0.001) for A2E-laden RPE cells irradiated for 15 and 20 min, respectively. Cell death did not occur in any of the control groups (Fig. 1A).
Fig. 1.
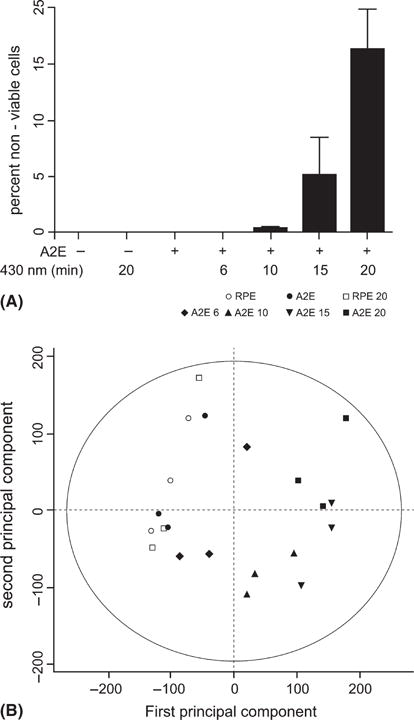
Cell death and changes in the gene expression profile induced by blue light exposure of A2E-laden retinal pigment epithelial (RPE) cells were more pronounced with longer exposure time. A2E-laden RPE cells were exposed to blue light for 6, 10, 15 and 20 min. Untreated A2E-free RPE cells, A2E-free RPE cells irradiated for 20 min and A2E-laden RPE cells that had not been exposed to blue light served as controls. The experiment was repeated three times. Cell death was quantified, and RNA was extracted for microarray analysis in each experiment. (A) Per cent of nonviable cells was determined by labelling nuclei in death cells with death red staining and all nuclei with DAPI. Mean ± SEM of three experiments, *p < 0.001, + presence of condition + indicates presence of condition. (B) The gene expression data from the microarrays were analysed by principal component analysis (PCA). Two principal components were extracted, and the projections of the samples were plotted in a two-dimensional score plot using the first and second principal component axes. The first PCA dimension holds most of the variation (21%) in the model. (RPE) refers to A2E-free RPE cells and (RPE + A2E) to A2E-laden RPE cells. The duration of blue light exposure is indicated in minutes (6, 10, 15, 20). (○) RPE, (●) RPE + A2E (□) RPE 20, (◆) RPE + A2E 6, (▲) RPE + A2E 10 (▼) RPE + A2E 15, (■) RPE + A2E 20.
Principal component analysis revealed changes in the gene expression pattern for blue light irradiated A2E-laden RPE cells
It is the hypothesis that many genes will change their expression in a coordinated way in the illuminated A2E-laden RPE cells. The goal of the bioinformatics analysis is to simplify this complex response of the many covarying genes to identify patterns in the data and to interpret the response in a biologically meaningful way. One bioinformatics strategy to achieve this goal is to use principal component analysis (PCA). The PCA combines genes that are up- and downregulated almost identically into a single variable, a principal component, which represents the gene expression pattern of such covarying genes. The remaining genes are then analysed for other gene expression patterns that could be represented by another new common variable, which would constitute a second principal component. This allows the most important patterns of the gene expression to be visualized in a two-dimensional plot, where each component represents a large group of covarying genes with similar expression patterns. In this plot, samples that are plotted close to each other are more related with respect to gene expression than samples that are plotted further apart.
PCA carried out on the gene expression data presented here, revealed that the first principal component explained 21%, and the second component explained 10% of the variation. A two-dimensional plot using these first two components as axes demonstrated differential distribution of the samples (Fig. 1B). Samples from the irradiated A2E-laden RPE cells were scattered mainly in the positive direction on the axis for the first principal component (Fig. 1B). The longer duration of blue light exposure the more the samples deviated from untreated cells, with the A2E-laden RPE cells irradiated for 20 min deviating the most. In contrast, irradiation of A2E-free RPE cells for 20 min and A2E accumulation did not show changes in gene expression, and the samples from these two groups and the samples from the untreated RPE cells were scattered close to each other and mainly towards the negative values on the axes of the first principal component (Fig. 1B).
Subsequently, the genes contributing the most to the definition of the first principal component were identified and subjected to functional annotation analysis for overrepresentation of terms for biological processes defined by the GO consortium. This functional analysis showed that the positive direction of the first axis represents processes related to programmed cell death, GO term: 12 501, p = 1.89e-05, protein kinase cascade, GO term: 7243, p = 3.23e-04, response to stress, GO term: 6950, p = 3.73e-05, and response to unfolded protein, GO term: 6986, p = 3.70e-07.
Transcription factors
Gene expression is tightly controlled by transcription factors that bind to transcription factor binding sites, which are regulatory elements in the promoter regions of the genes. The coordination of gene expression can happen via induction of different signalling transduction pathways that converge on transcription factors that regulate subsets of genes sharing same sequence for transcription factor binding sites. Thus, analysis of transcriptional regulators can be useful to understand how large sets of genes can be controlled by a small number of upstream signalling molecules.
To identify possible regulatory mechanisms and early events initiating pathological processes, we analysed the genes comprising the first dimension of the PCA for overrepresentation of potential transcription factor binding sites. The corresponding transcription factors are shown in Table 2. Among these where transcription factors belonging to the ATF/CREB family. This family of transcription factors has been implicated in the regulation of a wide range of biological functions including regulation of cell growth, apoptosis and innate immune system (Persengiev & Green 2003; Matthew et al. 2009).
Table 2.
Predicted transcription factor binding sites.
| PWM | PWM length | TF | p-value | Bonferroni |
|---|---|---|---|---|
| M00800 | 16 | AP-2 | 1.8E-09 | 1.5E-06 |
| M00470 | 9 | AP-2gamma | 1.3E-05 | 1.1E-02 |
| M00017 | 14 | ATF | 9.5E-14 | 8.1E-11 |
| M00338 | 12 | ATF | 1.7E-12 | 1.5E-09 |
| M00179 | 12 | ATF2 | 3.7E-13 | 3.1E-10 |
| M00513 | 14 | ATF3 | 8.4E-08 | 7.1E-05 |
| M00514 | 12 | ATF4 | 2.7E-08 | 2.3E-05 |
| M00483 | 8 | ATF6 | 2.1E-16 | 1.8E-13 |
| M01175 | 9 | CKROX | 3.6E-08 | 3.0E-05 |
| M00178 | 12 | CREB | 1.2E-14 | 1.1E-11 |
| M00803 | 6 | E2F | 2.1E-28 | 1.8E-25 |
| M00940 | 10 | E2F-1 | 1.7E-06 | 1.5E-03 |
| M00938 | 16 | E2F-1 | 2.5E-05 | 2.1E-02 |
| M00736 | 8 | E2F-1:DP-1 | 2.4E-07 | 2.0E-04 |
| M00737 | 8 | E2F-1:DP-2 | 2.4E-07 | 2.0E-04 |
| M00738 | 8 | E2F-4:DP-1 | 1.9E-05 | 1.6E-02 |
| M00694 | 10 | E4F1 | 2.4E-13 | 2.0E-10 |
| M00695 | 7 | ETF | 1.6E-31 | 1.3E-28 |
| M00182 | 12 | GBP | 1.3E-05 | 1.1E-02 |
| M01072 | 15 | HIC1 | 4.7E-08 | 4.0E-05 |
| M01160 | 5 | Kid3 | 3.4E-19 | 2.9E-16 |
| M00982 | 14 | KROX | 7.5E-06 | 6.3E-03 |
| M00649 | 8 | MAZ | 2.0E-06 | 1.7E-03 |
| M00652 | 10 | Nrf-1 | 1.6E-06 | 1.3E-03 |
| M00931 | 10 | Sp1 | 1.1E-05 | 9.1E-03 |
| M00706 | 9 | TFII-I | 4.5E-08 | 3.8E-05 |
| M00121 | 14 | USF | 5.3E-05 | 4.4E-02 |
| M00187 | 10 | USF | 2.0E-06 | 1.7E-03 |
| M01118 | 9 | WT1 | 1.5E-12 | 1.3E-09 |
| M00793 | 9 | YY1 | 7.4E-06 | 6.2E-03 |
| M01035 | 11 | YY1 | 1.5E-05 | 1.3E-02 |
PWM = position weight matrices; TF = transcription factor.
Exposure of A2E-laden RPE cells to blue light resulted in differential gene expression in genes belonging to various functional groups
Expression values from the three control groups (A2E-free RPE cells, A2E-free RPE cells irradiated for 20 min and A2E-laden RPE cells that had not been exposed to blue light) were compared using ANOVA. This analysis revealed that blue light exposure of the A2E-free RPE cells and A2E accumulation alone did not exert a statistical significant effect upon gene expression. We compared the A2E-laden cells irradiated for 6 and 10 min with a t-test and found no statistically significant difference. The same were true when comparing the A2E-laden cells irradiated for 15 and 20 min. Therefore, the data from A2E-free RPE cells, A2E-free RPE cells irradiated for 20 min and the nonirradiated A2E-laden RPE cells were pooled as a single control group (control). The A2E-laden RPE cells irradiated for 6 and 10 min were pooled to a new group termed ‘A2E/430 nm short duration’ (A2E/430 nm-S), and A2E-laden RPE cells irradiated for 15 and 20 min were pooled as a group referred to as ‘A2E/430-nm long duration’ (A2E/430 nm-L).
Differences between the control group and the A2E/430 nm-S and A2E/430 nm-L groups were analysed with a t-test, and the set of p-values was used to calculate the false discovery rate (FDR; Benjamini & Hochberg 1995), which was chosen to be 1%. This revealed that 97 probe sets – corresponding to 75 genes – were differentially regulated in the sublethal irradiated A2E-laden RPE cells as compared to controls (fold change ≥2 or ≤0.5, p-value ≤ 0.01, FDR = 1%, minimum difference between expression values were set to ≥50). The GO-terms were used to extract differentially regulated genes involved in apoptosis, stress response and in the immune response (Table 3). In addition, differentially regulated genes with biological functions that might have relevance in the pathogenesis of AMD are also shown in Table 3. As some of the genes do not show significantly change in expression levels in both A2E/430 nm-S and A2E/430 nm-L, the fold change for A2E/430 nm-L are also shown in Table 3. The genes SQSTM1 (pro-apoptotic, response to stress, and immune response), and the gene TRIB3 (pro-apoptotic and response to stress), RRAGC (pro-apoptotic) and ASNS (anti-apoptotic) were found to be upregulated. In contrast, DHRS3 that is involved in visual perception and retinol metabolic processes was found to be downregulated.
Table 3.
Differentially regulated genes in the A2E/430 nm-S group.
| Gene symbol | Gene name | Fold change
|
p-value adjusted* | |
|---|---|---|---|---|
| A2E/430 nm-S | A2E/430 nm-L | |||
| Immune response | ||||
| CEBPG | CCAAT/enhancer binding protein (C/EBP), gamma | 4.8 | 5.5 | 1.1E-02 |
| SQSTM1 | Sequestosome 1 | 2.7 | 4.2 | 6.5E-03 |
| XBP1 | X-box binding protein 1 | 2.2 | 2.4 | 5.3E-03 |
| GPI | Glucose phosphate isomerase | 0.5 | 0.5 | 5.7E-03 |
| Anti-apoptotic | ||||
| ASNS | Asparagine synthetase | 4.0 | 3.5 | 9.9E-03 |
| NUAK2 | NUAK family, SNF1-like kinase, 2 | 0.5 | 0.4 | 1.1E-02 |
| Pro-apoptotic | ||||
| TRIB3 | Tribbles homolog 3 (Drosophila) | 10.9 | 7.4 | 5.5E-03 |
| SQSTM1 | Sequestosome 1 | 2.7 | 4.2 | 6.5E-03 |
| RRAGC | Ras-related GTP binding C | 2.0 | 2.7 | 7.0E-03 |
| Response to stress | ||||
| TRIB3 | Tribbles homolog 3 (Drosophila) | 10.9 | 7.4 | 5.5E-03 |
| SQSTM1 | Sequestosome 1 | 2.7 | 4.2 | 6.5E-03 |
| Positive regulation of proliferation | ||||
| HES1 | Hairy and enhancer of split 1, (Drosophila) | 0.3 | 0.2 | 1.3E-02 |
| EDN1 | Endothelin 1 | 0.2 | 0.2 | 6.4E-03 |
| Negative regulation of proliferation | ||||
| CDC6 | Cell division cycle 6 homolog (S. cerevisiae) | 2.7 | 2.3 | 5.5E-03 |
| DAB2 | Disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila) | 2.1 | 1.8 | 1.1E-02 |
| Cell cycle | ||||
| E2F7 | E2F transcription factor 7 | 7.8 | 12.6 | 5.3E-03 |
| ASNS | Asparagine synthetase | 4.0 | 3.5 | 9.9E-03 |
| CDC6 | Cell division cycle 6 homolog (S. cerevisiae) | 2.7 | 2.3 | 5.5E-03 |
| RASSF1 | Ras association (RalGDS/AF-6) domain family member 1 | 2.4 | 2.8 | 2.9E-03 |
| FBXO5 | F-box protein 5 | 2.0 | 2.0 | 1.3E-02 |
| KANK1 | KN motif and ankyrin repeat domains 1 | 0.5 | 0.6 | 6.5E-03 |
| Autophagy | ||||
| WIPI1 | WD repeat domain, phosphoinositide interacting 1 | 3.4 | 3.2 | 6.6E-03 |
| Visual perseption | ||||
| DHRS3 | Dehydrogenase/reductase (SDR family) member 3 | 0.4 | 0.3 | 9.6E-03 |
Adjusted p-value to false discovery rate according to the methods of (Benjamini & Hochberg 1995).
Comparing the control group with the A2E/430 nm-L group showed that 1584 probe sets – corresponding to 1131 genes – were differentially regulated in the lethal irradiated A2E-laden RPE cells as compared to controls (fold change ≥2 or ≤0.5, p-value ≤ 0.01, FDR = 1%, minimum difference between expression values were set to ≥50). GO-terms were used to search specifically for genes involved in apoptosis, stress response and in the immune response (Appendix S1). Corresponding results for A2E/430 nm-S are included in Appendix S1, to illustrate a possible trend for an expression of a gene, even when the expression is only significantly different in the A2E/430 nm-L group. We found that 35 pro-apoptotic genes were upregulated and 20 were downregulated in the A2E/430 nm-L group. We also found upregulation of 25 anti-apoptotic genes and downregulation of three anti-apoptotic genes.
Heat-shock proteins (HSPs) are molecular chaperons that inhibit protein aggregation, facilitate optimal folding of newly synthesized proteins and help maintain the stability of protein–protein interactions within the cell. Genes coding for HSPs can be upregulated in response to unfolded protein or stress, and HSPs are also recognized to exert cytoprotective and both anti-inflammatory and pro-inflammatory effects depending on the cell type, stimulus and cellular location. Various members of the heat-shock protein (HSP) family were upregulated in the A2E/430 nm-L group; heat-shock 70 kDa protein six HSP70B’ (HSPA6), heat-shock 70 kDa protein 1A (HSPA1A), heat-shock 70 kDa protein 1B (HSPA1B), DnaJ Hsp40 homolog, subfamily B, member 1, 2 and 4 (DNAJB1, DNAJB2 and DNAJB4) and heat-shock 60 kDa protein 1 (HSPD1).
We found that various genes related to the immune system were upregulated in the A2E/430 nm-L group (Appendix S1). Among these were Interleukin 8 (IL8), Interleukin 6 (IL6) and the receptor for IL6 (IL6R). Transcription of both IL8 and IL6 are positively regulated by the transcription factor DDIT3 that were found to be upregulated as well. Further, IL6 has been shown to release IL1R antagonist (IL1RN) in the liver. We found an upregulation of IL1RN in response to A2E/430 nm-L.
Studies implicate the complement cascade as playing an important role in development of AMD (Hageman et al. 2005), and we have previously demonstrated that irradiated A2E-laden RPE cells and photooxidized compounds of A2E can activate the complement cascade (Zhou et al. 2006, 2009). In the present study, we found statistically significant downregulation of six complement-related genes in A2E-laden RPE cells irradiated with blue light. Four of these genes encoded proteins that have an inhibitory effect on the complement cascade: complement factor H (CFH), complement factor H-related 1 (CFHR1), complement factor I (CFI) and vitronectin (VTN) and two belonging to the classical pathway: complement component 1, s subcomponent (C1S) and complement component 1, r subcomponent (C1R).
Validation of microarray results using quantitative RT-PCR
Selected genes found by t-test to be significantly up or downregulated were further investigated by qRT-PCR. The results can be seen in Fig. 2. All genes were found to be regulated in the same direction with both microarray technology and qRT-PCR, and fold changes were found to be significant. From Fig. 2, it is visualized that the longer irradiation duration of the A2E-laden RPE cells, the more pronounced is the change in mRNA levels for the differential regulated genes we tested with qRT-PCR.
Fig. 2.
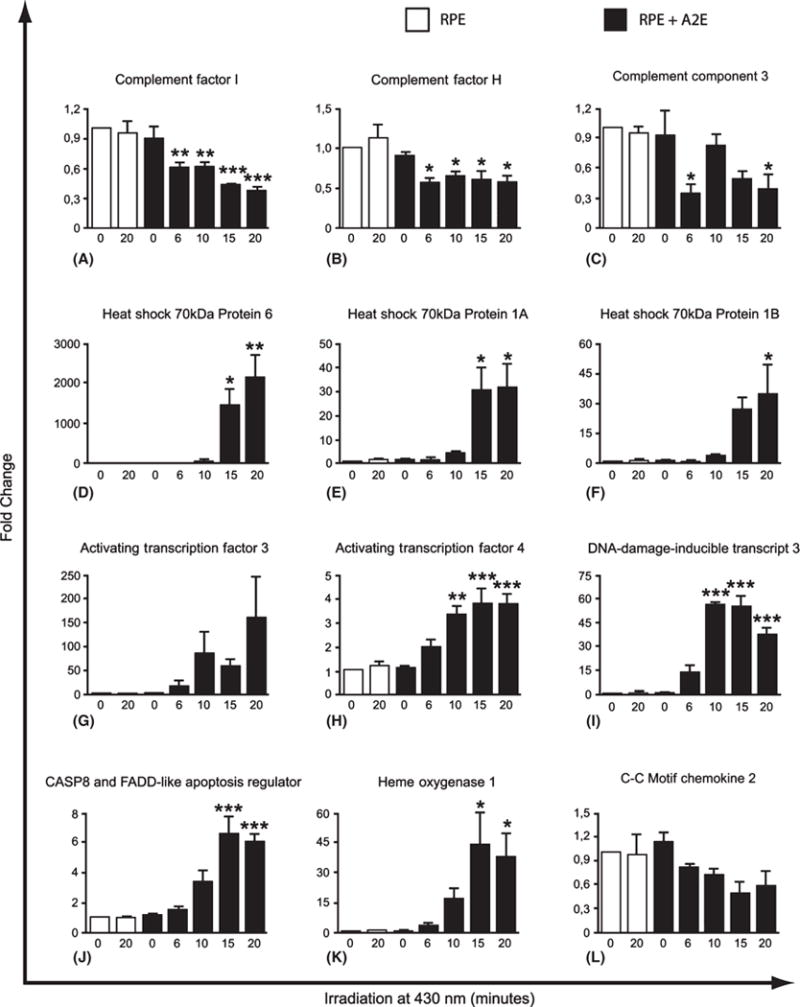
Quantitative reverse-transcriptase PCR analysis of 12 selected genes confirmed the expression patterns observed in the Affymetrix microarrays. Retinal pigment epithelial (RPE) cells that had accumulated A2E (A2E) were irradiated at 430 nm for 6, 10, 15 and 20 min. RNA was extracted after 6-hr incubation and expression of selected genes was measured by quantitative RT-PCR to validate the microarray data. Controls were A2E-free RPE cells (A2E-free) that were not irradiated or were irradiated for 20 min and cells that had accumulated A2E but were not irradiated (A2E). The RNA level of GAPDH was used as internal amplification control, and Pfaffl method was used to calculate the relative gene expression. (A) Complement Factor I; (B) Complement Factor H; (C) C3; (D) Heat-shock 70 kDa Protein 6; (E) Heat-shock 70 kDa Protein 1A; (F) Heat-shock 70 kDa Protein 1B; (G) Activating Transcription Factor 3; (H) Activating Transcription Factor 4; (I) DNA-damage-inducible transcript 3; (J) CASP8 and FAAD-like apoptosis regulator; (K) Heme Oxygenase 1; (L) CCL2. Mean ± SEM of three experiments. *p < 0.05, **p < 0.01 and ***p < 0.001.
Discussion
Using microarrays, we investigated the early changes in gene expression induced by blue light exposure of A2E-laden RPE cells. We used the well-characterized human RPE cell line ARPE-19, because it is devoid of melanin and endogenous A2E (Sparrow et al. 1999). The absence of these pigments is essential to eliminate problems due to natural variations in the concentrations of these pigments, which could confound studies on light damage. However, we are aware that one should be cautious about extrapolation from the in vitro to the in vivo situation.
The study presented here indicates that there were no significant differences in gene expression between untreated RPE cells and A2E-free RPE cells irradiated for 20 min and A2E-laden RPE cells that had not been exposed to blue light. Thus, there were no effects due to either blue light or A2E accumulation alone. Effects were only observed when the combination of blue light and A2E accumulation were present. The PCA analysis of the microarray data clearly demonstrated that the samples from A2E-laden RPE cells exposed to blue light were distinguished from the controls. The longer the samples were exposed to blue light the more they deviated from the controls. However, it is denoted that some degree of variation are observed among samples sharing the same experimental conditions. The samples are three individual replicates for each experimental condition, and the observed variation is ascribed to the variance expected in biological material, due to the fact that microarrays are very sensitive to small changes.
According to the functional analysis performed with the GO-terms, there were no unambiguous signalling pathways or functional group that could account for the observed differences. This indicates that A2E and blue light might cause changes in various cellular processes besides apoptosis.
Less than 1% cell death was detected in the A2E/430 nm-S group, and RNA expression was altered for 75 genes. This might be an important observation because it indicates that pathological changes could emerge with exposure to a small amount of blue light and before RPE cell death occur.
When grouping the differentially upregulated genes according to functionality, we found that several of these genes and transcription factors were involved in response to stress, programmed cell death and immune response. Both anti- and pro-apoptotic-related genes and transcription factors were upregulated in response to A2E/430 nm-S and A2E/430 nm-L. This might seem inconsistent but could indicate that a balance between pro- and anti-apoptotic stimuli is of importance for the cell fate in response to A2E/430 nm-S and A2E/430 nm-L.
In the A2E/430 nm-L group, there were from 2% up to 23% cell death. The fact that only a relatively small fraction of the cells were dead and that changes in RNA levels were detected in the A2E/430 nm-S group indicates that part of the observed changes in gene expression could originate from damaged – but not yet dead cells.
Polymorphisms in several genes coding for complement factors have been shown to influence the risk of developing AMD (Gold et al. 2006). Especially, CFH (Hageman et al. 2005) has been considered to be a key element. CFH and CFI inhibit the activity of the alternative pathway in the complement cascade, and it has been speculated that lack of inhibition of the complement system might result in bystander damage of the RPE cells. The microarray analysis demonstrates that the gene expression of six genes belonging to the complement system was suppressed in A2E-laden RPE cells irradiated with lethal dose of blue light. Further validation performed with the more accurate method qRT-PCR indicates that a sublethal dose of blue light also is sufficient to induce downregulation of CFH and CFI. We speculate that a reduced production of these inhibitors of the alternative pathway could cause the cells to be less resilient to complement attack. This might reinforce the inherited susceptibility to develop AMD due to certain polymorphisms in genes coding for complement.
In a murine model of recessive Stargardt macular degeneration, the Abca4−/− mouse, RPE lipofuscin formation is accentuated (Weng et al. 1999; Radu et al. 2011). It has recently been reported that the mRNA encoding the complement regulatory proteins CD46, CD55, CD59 and CFH are downregulated in RPE cells of Abca4−/− mice when compared to wild-type mice. Concurrently, in the Abca4−/− mice, lipid peroxidation products are increased and a number of genes related to oxidative stress (SOD1, GSTM, CAT1 and HO1) are upregulated (Radu et al. 2011). These observations are partly in agreement with our findings of downregulation of CFH (twofold) and upregulation of HMOX1 (>5-fold) in RPE cells that have accumulated A2E and are exposed to blue light. However, Radu et al. (2011) also reported upregulation of complement regulatory genes in human foetal RPE cells after feeding these cells with photoreceptor outer segments from mice deficient in ABCA4 transporter. The difference between the in vivo and in vitro observations is not clear. Our in vitro data did not show any differential gene regulation of ARPE-19 cells in response to accumulation of A2E alone, but only after exposure to blue light of A2E loaded RPE cells.
Oxidative stress has been suspected to play an important role in retinal degenerative diseases such as AMD, and studies have been performed to clarify the mechanisms of oxidative stress injury to the RPE cells. Strunnikova et al. (2004) examined differential gene expression of RPE cells exposed to a sublethal dose of oxidative stress and found upregulation of several genes involved in antioxidation, protection, detoxification and reparation. Of these genes, we also found that MGST1, DNAJB1, DNAJA1 and HSP70 were upregulated in the A2E/430 nm-L group. In contrast, TXNRD1, PRDX1, FTH1 were differentially regulated in response to nonlethal oxidative injury but showed no significant difference due to A2E/430 nm-S or A2E/430 nm-L. Further, RNA levels were increased for the extracellular matrix-related genes CYR61 and FN1 in cells stimulated with oxidative stress as well as in A2E/430 nm-L treated cells. On the other hand, Strunnikova et al. found upregulation of the metalloproteinases MMP15 and MMP3, whereas our results demonstrated a downregulation of MMP11, MMP24 and MMP28. Matrix metalloproteinases (MMPs) are thought to play a major role in degradation of extracellular matrix proteins. One might speculate that the downregulation of the MMPs observed in RPE cells stressed with photooxidation could compromise the ability to degrade extracellular proteins. This could eventually lead to enhanced accumulation of extracellular materials. The differences in gene expression of MMPs observed in RPE cells challenged with oxidative stress as compared to photooxidative stress emphasizes that the molecular consequences of the two forms of stress might be different.
Exposure of lipofuscin to short wavelength light generates reactive oxygen intermediates (ROI). This process not only leads to the production of ROI, singlet oxygen also reacts with the A2E molecule with new, highly reactive moieties resulting from this photooxidation. The comparisons with the study of Strunnikova et al. suggest that there might be some similarity in the cellular response to ROI and to A2E/430 nm-L. However, there are also differences on the gene expression level, and therefore, it is likely that the production of ROI by A2E/430 nm-L accounts for some of the effects and that the photooxidation and photodegradation of A2E (Wu et al. 2010) accounts for a large part as well.
In conclusion, the data presented here demonstrate that intracellular A2E accumulation accompanied with exposure to blue light – in a sublethal as well as a lethal dose – causes differential expression of genes belonging to various functional categories, including response to stress, cell death and the immune response. These data raise the possibility that A2E/430 nm conditions can perturb RPE cells and cause changes in gene expression in the absence of cell death. These questions will have to be investigated further.
Furthermore, gene expression and presence of for example transcription factors may differ substantially in for instance primary RPE cells and in vivo as compared to the ARPE-19 cell line. Therefore, additional gene expressions studies in RPE cells from the Abca4−/− mouse could serve as an informative in vivo model to explore similarities and discrepancies between findings in vitro versus in vivo.
Supplementary Material
Appendix S1. Differentially regulated genes in the A2Eã 430 nm-l group.
Acknowledgments
This work was supported by Kristine Petrea Marius Claus og Erik Feldthusens Fond, Generalkonsul Friederich Bøhn og datter Else Bøhm’s Fond, Carl og Ellen Hertz’ Legat til Dansk Læge- og Naturvidenskab, Grosserer L.F. Foghts Fond, Fabrikant Einar Willumsens Mindelegat, Mogens Svarre Mogensens Fond, Frimodt-Heineke Fonden and The Danish Eye Health Society to BVDB and by National Institutes of Health grant EY12951 to JRS.
Footnotes
Supporting Information
Additional Supporting Information may be found in the online version of this article:
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer-reviewed and may be re-organized for online delivery, but are not copy-edited. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
References
- Anderson DH, Mullins RF, Hageman GS, Johnson LV. A role for local inflammation in the formation of drusen in the aging eye. Am J Ophthalmol. 2002;134:411–431. doi: 10.1016/s0002-9394(02)01624-0. [DOI] [PubMed] [Google Scholar]
- Beatty S, Koh H, Phil M, Henson D, Boulton M. The role of oxidative stress in the pathogenesis of age-related macular degeneration. Surv Ophthalmol. 2000;45:115–134. doi: 10.1016/s0039-6257(00)00140-5. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- Ben-Shabat S, Itagaki Y, Jockusch S, et al. Formation of a nonaoxirane from A2E, a lipofuscin fluorophore related to macular degeneration, and evidence of singlet oxygen involvement. Angew Chem Int Ed Engl. 2002;41:814–817. doi: 10.1002/1521-3773(20020301)41:5<814::aid-anie814>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Brazma A, Hingamp P, Quackenbush J, et al. Minimum information about a microarray experiment (MIAME)-towards standards for microarray data. Nat Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- Davies S, Elliott MH, Floor E, et al. Photocytotoxicity of lipofuscin in human retinal pigment epithelial cells. Free Radical Biol Med. 2001;31:256–265. doi: 10.1016/s0891-5849(01)00582-2. [DOI] [PubMed] [Google Scholar]
- Dillon J, Wang Z, Avalle LB, Gaillard ER. The photochemical oxidation of A2E results in the formation of a 5,8,5′,8′-bis-furanoid oxide. Exp Eye Res. 2004;79:537–542. doi: 10.1016/j.exer.2004.06.024. [DOI] [PubMed] [Google Scholar]
- Eldred GE, Lasky MR. Retinal age pigments generated by self-assembling lysosomotropic detergents. Nature. 1993;361:724–726. doi: 10.1038/361724a0. [DOI] [PubMed] [Google Scholar]
- Gold B, Merriam JE, Zernant J, et al. Variation in factor B (BF) and complement component 2 (C2) genes is associated with age-related macular degeneration. Nat Genet. 2006;38:458–462. doi: 10.1038/ng1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hageman GS, Anderson DH, Johnson LV, et al. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc Natl Acad Sci USA. 2005;102:7227–7232. doi: 10.1073/pnas.0501536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Gerds TA, Nielsen OH, et al. pcaGoPromoter – an R package for biological and regulatory interpretation of principal components in genome-wide gene expression data. PLoS ONE. 2012;7:e32394. doi: 10.1371/journal.pone.0032394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holz FG, Bellman C, Staudt S, et al. Fundus autofluorescence and development of geographic atrophy in age-related macular degeneration. Invest Ophthalmol Vis Sci. 2001;42:1051–1056. [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, et al. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang YP, Matsuda H, Itagaki Y, et al. Characterization of peroxy-A2E and furan-A2E photooxidation products and detection in human and mouse retinal pigment epithelial cell lipofuscin. J Biol Chem. 2005;280:39732–39739. doi: 10.1074/jbc.M504933200. [DOI] [PubMed] [Google Scholar]
- Kim SR, Jang YP, Jockusch S, et al. The all-trans-retinal dimer series of lipofuscin pigments in retinal pigment epithelial cells in a recessive Stargardt disease model. Proc Natl Acad Sci USA. 2007;104:19273–19278. doi: 10.1073/pnas.0708714104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Itagaki Y, Ben-Shabat S, et al. The biosynthesis of A2E, a fluorophore of aging retina, involves the formation of the precursor, A2-PE, in the photoreceptor outer segment membrane. J Biol Chem. 2000;275:29354–29360. doi: 10.1074/jbc.M910191199. [DOI] [PubMed] [Google Scholar]
- Matthew T, Dakang X, Williams BRG. ATF3 transcription factor and its emerging roles in immunity and cancer. J Mol Med. 2009;87:1053–1060. doi: 10.1007/s00109-009-0520-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parish CA, Hashimoto M, Nakanishi K, et al. Isolation and one-step preparation of A2E and iso-A2E, fluorophores from human retinal pigment epithelium. Proc Natl Acad Sci USA. 1998;95:14609–14613. doi: 10.1073/pnas.95.25.14609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persengiev SP, Green MR. The role of ATF/CREB family members in cell growth, survival and apoptosis. Apoptosis. 2003;8:225–228. doi: 10.1023/a:1023633704132. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radu RA, Hu J, Yuan Q, et al. Complement system dysregulation and inflammation in the retinal pigment epithelium of a mouse model for Stargardt macular degeneration. J Biol Chem. 2011;286:18593–18601. doi: 10.1074/jbc.M110.191866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutt F, Davies S, Kopitz J, et al. Photodamage to human RPE cells by A2E, a retinoid component of lipofuscin. Invest Ophthalmol Vis Sci. 2000;41:2303–2308. [PubMed] [Google Scholar]
- Sparrow JR. RPE lipofuscin: formation, properties and relevance to retinal degeneration. Retinal Degenerations. 2007:213–236. [Google Scholar]
- Sparrow JR, Boulton M. RPE lipofuscin and its role in retinal pathobiology. Exp Eye Res. 2005;80:595–606. doi: 10.1016/j.exer.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Sparrow JR, Cai B. Blue light-induced apoptosis of A2E-containing RPE: involvement of caspase-3 and protection by Bcl-2. Invest Ophthalmol Vis Sci. 2001;42:1356–1362. [PubMed] [Google Scholar]
- Sparrow JR, Parish CA, Hashimoto M, Nakanishi K. A2E, a lipofuscin fluorophore, in human retinal pigmented epithelial cells in culture. Invest Ophthalmol Vis Sci. 1999;40:2988–2995. [PubMed] [Google Scholar]
- Sparrow JR, Nakanishi K, Parish CA. The lipofuscin fluorophore A2E mediates blue light-induced damage to retinal pigmented epithelial cells. Invest Ophthalmol Vis Sci. 2000;41:1981–1989. [PubMed] [Google Scholar]
- Sparrow JR, Zhou J, Ben-Shabat S, et al. Involvement of oxidative mechanisms in blue-light-induced damage to A2E-laden RPE. Invest Ophthalmol Vis Sci. 2002;43:1222–1227. [PubMed] [Google Scholar]
- Strunnikova N, Zhang C, Teichberg D, et al. Survival of retinal pigment epithelium after exposure to prolonged oxidative injury: a detailed gene expression and cellular analysis. Invest Ophthalmol Vis Sci. 2004;45:3767–3777. doi: 10.1167/iovs.04-0311. [DOI] [PubMed] [Google Scholar]
- Weng J, Mata NL, Azarian SM, et al. Insights into the function of Rim protein in photoreceptors and etiology of Stargardt’s disease from the phenotype in abcr knockout mice. Cell. 1999;98:13–23. doi: 10.1016/S0092-8674(00)80602-9. [DOI] [PubMed] [Google Scholar]
- Westlund BS, Cai B, Zhou J, Sparrow JR. Involvement of c-Abl, p53 and the MAP kinase JNK in the cell death program initiated in A2E-laden ARPE-19 cells by exposure to blue light. Apoptosis. 2009;14:31–41. doi: 10.1007/s10495-008-0285-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Fishkin NE, Pande A, et al. Novel lipofuscin bisretinoids prominent in human retina and in a model of recessive stargardt disease. J Biol Chem. 2009;284:20155–20166. doi: 10.1074/jbc.M109.021345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Yanase E, Feng X, et al. Structural characterization of bisretinoid A2E photocleavage products and implications for age-related macular degeneration. Proc Natl Acad Sci USA. 2010;107:7275–7280. doi: 10.1073/pnas.0913112107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Jang YP, Kim SR, Sparrow JR. Complement activation by photooxidation products of A2E, a lipofuscin constituent of the retinal pigment epithelium. Proc Natl Acad Sci USA. 2006;103:16182–16187. doi: 10.1073/pnas.0604255103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Kim SR, Westlund BS, Sparrow JR. Complement activation by bisretinoid constituents of RPE lipofuscin. Invest Ophthalmol Vis Sci. 2009;50:2–1399. doi: 10.1167/iovs.08-2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1. Differentially regulated genes in the A2Eã 430 nm-l group.


