Abstract
Human polynucleotide phosphorylase (hPNPaseold-35) is an evolutionary conserved RNA processing enzyme with expanding roles in regulating cellular physiology. hPNPaseold-35 was cloned using an innovative “overlapping pathway screening” strategy designed to identify genes coordinately regulated during the processes of cellular differentiation and senescence. Although hPNPaseold-35 structurally and biochemically resembles PNPase of other species, overexpression and inhibition studies reveal that hPNPaseold-35 has evolved to serve more specialized and diversified functions in humans. Targeting specific mRNA or non-coding small microRNA (miRNA), hPNPaseold-35 modulates gene expression that in turn plays a pivotal role in regulating normal physiological and pathological processes. In these contexts, targeted overexpression of hPNPaseold-35 represents a novel strategy to selectively downregulate RNA expression and consequently intervene in a variety of pathophysiological conditions.
Keywords: hPNPaseold-35, Senescence, RNA degradation, c-myc, miRNA
1. Introduction
RNA degradation and/turnover are major processes controlling RNA levels and are important regulators of physiological and pathological processes (Parker and Song, 2004). Labile messenger RNAs to more stable non-coding RNAs (mostly rRNA and tRNA, but also the expanding class of small regulatory RNAs) are eventually degraded by a complex process involving the simultaneous or sequential interplay of multiple proteins. Many of these proteins are evolutionary conserved extending from prokaryotes to higher mammals and serving comparable functions. In eukaryotic cells, a variety of ribonucleases (RNases) act cooperatively, initially shortening the 3′ poly (A) tail of an mRNA by deadenylases followed by the removal of the 5′ cap structure by a decapping enzyme, which enables the degradation of the transcript by a 5′→3′ exoribonuclease. Alternatively, the mRNA might be degraded from the 3′ end by the cytoplasmic exosome, a multiprotein complex of diverse 3′→5′ exoribonucleases. Polynucleotide phosphorylase (PNPase) is a 3′→5′ exoribonuclease that uses the phosphorolytic mechanism to degrade RNA (Sarkar et al., 2006; Mohanty et al., 2000; Yehudai et al., 2001). It is conserved evolutionarily and is expressed in different species including bacteria, plants, worms, flies, mice and humans. Our group cloned the human homolog of polynucleotide phosphorylase (hPNPaseold-35) in the unique contexts of differentiation and senescence, using an “overlapping pathway screening” scheme. We documented that hPNPaseold-35 plays a key role in regulating both of these fundamental physiological processes (Leszczyniecka et al., 2002, 2003, 2004; Sarkar et al., 2003, 2004, 2005, 2006, 2007; Sarkar & Fisher, 2006). We presently review recent advances in our understanding of hPNPaseold-35-mediated RNA degradation, in particular its ability to target different classes of RNAs.
2. Cloning, expression and localization of human polynucleotide phosphorylase (hPNPaseold-35)
hPNPaseold-35 was cloned by using an overlapping pathway screening (OPS) approach (Leszczyniecka et al., 2002) during a screen for genes up-regulated in the process of terminal cellular differentiation and senescence. Although terminal cell differentiation and cellular senescence represent two discrete phenomena, these processes have several common characteristics. Both are distinguished by irreversible growth arrest associated with marked inhibition of DNA synthesis, inhibition of telomerase activity, and modulation of discrete programs of gene expression, especially up-regulation of cyclin-dependent kinase inhibitors (CDKI) (Campisi, 1992; Fisher et al. 1985, 1986). Combined treatment of metastatic human melanoma cells HO-1 with recombinant human fibroblast interferon (IFN)-β and the protein kinase C activator mezerein (MEZ) induces irreversible growth arrest accompanied by morphological, biochemical, antigenic and gene expression changes culminating in a state of ‘terminal differentiation (Fisher and Grant, 1985; Fisher et al., 1985, 1986; Guarini et al., 1989, 1992; Jiang et al., 1993, 1995). Screening of a temporal cDNA library generated from terminally differentiated HO-1 melanoma cells with cDNAs from senescent progeriod fibroblasts identified 75 genes, termed old-1 to -75, that were upregulated during both terminal differentiation and senescence. Sequence analysis of one particular clone, old-35, confirmed its identity to the PNPase gene, resulting in the gene being renamed hPNPaseold-35 (Leszczyniecka et al., 2002). The hPNPaseold-35 gene consists of 28 exons and 27 introns spanning 54 kb in chromosome 2p15–2p16.1 (Leszczyniecka et al., 2003). Of interest, this unstable genomic region is prone to cytogenetic alterations in human cancers and in various genetic disorders (Kirschner et al., 1999) such as B-cell lymphoma (Fukuhara et al., 2006), type I hereditary nonpolyposis colorectal cancer, familial male precocious puberty, Carney complex, Doyne’s honeycomb retinal dystrophy and DYX-3, a form of familial dyslexia (Kirschner et al., 1999). hPNPaseold-35 mRNA expression could be detected in all normal tissues analyzed with the highest expression being detected in heart and brain (Leszczyniecka et al., 2002). However, to date, there is no evidence linking expression and function of hPNPase to any of the aforementioned pathological processes.
In bacteria, PNPase autogenously regulates its expression by promoting the decay of PNPase mRNA by binding to the 5′-untranslated leader region of an RNase III-processed form of this transcript (Jarrige et al., 2001). To date the only known regulators of hPNPaseold-35 transcription are type I interferon (IFN-α and IFN-β) in both normal and cancer cells with diverse backgrounds irrespective of their p53 and Rb status (Leszczyniecka et al., 2002). Double-stranded RNA and poly(I) poly(C ), a known inducer of IFN-α and IFN-β, also stimulate hPNPaseold-35 expression while IFN-γ and TNF-α have minimal or no effect, respectively. hPNPaseold-35 is an early IFN response gene and its induction depends on the Janus-activated kinase ((JAK)/STAT (signal transducers and activators of transcription) signal transduction pathways. Analysis of the hPNPaseold-35 promoter identified an IFN-stimulated response element (ISRE) that showed increased binding of ISGF3 complex upon IFN-β treatment (Leszczyniecka et al., 2003). Mutation in this site abolished IFN-β induction of the promoter indicating that hPNPaseold-35 is regulated at the level of transcription. In addition to the ISRE, the hPNPaseold-35 promoter contains additional putative regulatory protein-binding sites, including a site for E2F transcription factor 3 (E2F3), a transcriptional repressor that is responsible for gene silencing during the G1 to S phase transition (Gewartowski et al. 2006).
Studies using cellular fractionation and/or immunofluorescence showed that endogenous hPNPaseold-35 and overexpressed C-terminal myc- tagged hPNPaseold-35 localize only to the mitochondria (Chen et al., 2006; Piwowarski et al., 2003). hPNPaseold-35 has a typical mitochondrial localization signal (MTS) at the NH2-terminal and it is imported into the mitochondria by i-AAA (ATPases associated with several diverse cellular activities) protease Yme1, localized into mitochondrial intermembrane space (IMS) and maintains mitochondrial homeostasis (Chen et al., 2006). However, our studies document that overexpressed C-terminal HA-tagged hPNPaseold-35 localizes both in cytosol and mitochondria (Sarkar et al., 2005) indicating that hPNPaseold-35 might reside within or outside mitochondria. In these contexts, the targets and the consequences of hPNPaseold-35 expression in different cellular compartments may be distinct and diverse, thereby expanding the repertoire of activities of this interesting enzyme.
3. RNA degradation machinery: PNPase and Exosome
Ribonucleases (RNase) are enzymes that are master regulators of stability and decay of RNA (Deutscher et al., 1993a, 1993b; Deutscher and Li, 2001). Depending upon their degradative properties, RNases are divided into 2 functional classes, endoribonucleases that cleave RNA molecules internally and exoribonucleases that act at the end of RNA chains (Deutscher et al, 1993b). RNA decay pathways in two of the most comprehensively studied model systems, the prokaroyte Escherichia coli and the eukaryote Saccharomyces cerevisiae, are different. In eukaryotes exoribonucleases can degrade RNA both at 5′ to 3′ and 3′ to 5′ directions (Deutscher and Li, 2001), whereas in prokaryotes RNA degradation takes place only in the 3′ to 5′direction. However, an interesting aspect of RNA decay in both prokaryotes and eukaryotes is the presence of multiprotein complexes known as the degradosome and the exosome, respectively.
In E.coli, PNPase is associated with the endonuclease RNase E, RNA helicase and the glycolytic enzyme enolase to form the degradosome and executes its processive 3′ to 5′ phosphorolysis or degradation of RNA species upon endonucleolytic cleavage by RNase E. PNPase acts as an integral component of degradosome like RNA-helicase and enolase. In contrast, in yeast, PNPase is absent and the exosome, a complex of multiple exoribonuclease performs RNA degradation in both the cytoplasm and the nucleus (Buttner et al. 2006; Mitchel et al. 1997; Hoof et al. 1999; Raijmakers et al. 2004). However, PNPase has been identified in higher animals such as mouse, rat and human indicating that the exosome and PNPase might serve specialized functions in these species (Leszczyniecka et al., 2002, 2003, 2004; Raijmakers et al., 2002).
In all species, PNPase contains five motifs that are conspicuously preserved through evolution extending from prokaryotes and plants to mammals (Leszczyniecka et al., 2004; Almeida et al., 2008). Two conserved catalytic RNase PH regions, a small domain of ~250 a.a. residues related to the Escherichia coli RNase PH enzyme and involved primarily in the 3′ processing of transfer RNA (tRNA) precursors, are present at the N-terminus of hPNPaseOld-35 (Leszczyniecka M et al. 2004). These RNase PH domains are separated by an α-helix that is unique to PNPase (Symmons et al., 2002). The RNA binding property of hPNPaseOld-35 is conferred by two C-terminal RNA binding domains, KH and S1 (Symmons et al., 2000, 2002; Leszczyniecka et al., 2002, 2004; Raijmakers et al., 2002). In addition to the characteristic 5 motifs, plant PNPase contains an N-terminal target peptide allowing translocation to chloroplast and the mammalian PNPase contains an N-terminal mitochondrial localization signal facilitating its subcellular localization in mitochondria (Piwowarski et al., 2003, Yehudai-Resheff et al., 2003 and Sarkar et al., 2005). The catalytic activity of PNPase enzyme in bacteria is located mainly in the second RNase PH domain (Jarrige et al., 2002) while in spinach chloroplasts, both RNase PH domains have equal polyadenylation and exoribonuclease activity. Deletion and mutation analysis have identified the critical regions of bacterial PNPase activity. Deletion of the S1 or KH RNA binding domain reduces the enzymatic activity by 50- or 19-fold, respectively, while deletion of both S1 and KH domains results in 1% of the enzymatic activity (Stickney et al., 2005). These deletions do not interfere with interaction with RNase E to form the degradosome. Site-directed mutagenesis reveals that the catalytic center of PNPase is located in the second RNase PH domain around the binding site for tungstate, a phosphate analogue (Jarrige et al., 2002). Interestingly, a G454D mutation in the second RNase PH domain displays defective RNA binding and impairs the autogenous regulation of PNPase mRNA indicating that in addition to S1 and KH domains, the catalytic domains are also capable of binding to RNA (Regonesi et al., 2004).
The alignment of the proteins in eukaryotic exosome clearly points to a structural similarity with bacterial PNPase. The three-dimensional structure of the PNPase from the bacterium Streptomyces antibioticus has revealed that the enzyme is a ring (doughnut)- shape formed by a homotrimeric complex, with the hexameric PH-domains surrounding a central channel that can accommodate a single-stranded RNA molecule (Symmons et al., 2002). Similarly, the same core structure is formed in the exosome: 6 RNase PH homologues comprise the core while 3 additional exosome subunits, which contain S1 RNA-binding domains, are positioned on the outer surface of the ring (Mitchell and Tollervey, 2000; Raijmakers et al., 2002; Symmons et al., 2002).
4. Specialized functions of hPNPaseold-35: RNA degradation
In eukaryotes, the poly(A) tail of mRNA first becomes deadenylated by 3 different enzyme complexes, the first composed mainly of Ccr4p and Pop2p, the second containing Pan2p and Pan3p and the third is a poly(A)-specific exonuclease called PARN (poly(A) ribonucleases) (Parker and Song, 2004). Following deadenylation, a decapping enzyme consisting of Dcp1p and Dcp2p removes the 5′ cap structure thus facilitating degradation by the 5′, 3′ exoribonuclease Xrn1p (Tucker and Parker, 2000). Alternatively, after deadenylation, mRNA can be degraded in a 3′ to 5′ direction by the exosome complex and the oligonucleotide cap structure is hydrolyzed by the decapping enzyme DcpS (Wang and Kiledjian, 2001). In addition, mRNA can also be degraded by endoribonucleases and aberrant mRNA may be degraded by specialized pathways such as nonsense-mediated decay (NMD) or nonstop decay (NSD) that degrade mRNA containing a premature stop codon or lacking a stop codon, respectively (Dodson and Shapiro, 2002; Frischmeyer et al., 2002 and Cao & Parker, 2003).
PNPase, as a phopsphorylase, it incorporates Pi and ADP in degradation and polymerization process, respectively (Littauer and Grunberg, 1999). Optimal degradation activity depends on the concentration of Pi and it varies from species to species (Portnoy et al., 2008). The human PNPase is active in much lower concentration of Pi compared with bacterial PNPase (Portnoy et al., 2008). The specificity of the enzyme for the polymerization reaction is, like that of the E.coli. PNPase, is high for ADP, with much less activity for other NDPs and no activity for ATP/NTPs. More interestingly, hPNPase displays no preferential activity for polyadenylated RNA like bacterial or chloroplast PNPase (Portnoy et al., 2008).
4.1. c-myc mRNA is the target of hPNPaseold-35
In the cytoplasm, adenoviral mediated overexpression of hPNPaseold-35 could directly degrade c-myc mRNA by virtue of its 3′→5′ exoribonuclease property and this degradation is specific for c-myc as compared with other mRNAs such as c-jun, GAPDH or GADD 34 (Sarkar D et al., 2003). It is still not clear what confers the specificity of hPNPaseold-35 for c-myc mRNA. There might be a specific sequence in c-myc mRNA that allows hPNPaseold-35 binding and degradation. In E. coli, PNPase degrades a family of cold shock proteins (CSP) that do not show any sequence similarity (Yamanaka and Inouye, 2001). Considering this observation, the secondary structure of the mRNA rather than its primary sequence might be the determining factor for specificity of hPNPaseold-35 binding and degradation of c-myc. The presence of either of the RNase PH domains was sufficient for degradation of c-myc mRNA and induction of morphological, biochemical and gene expression changes by hPNPaseold-35 (Sarkar et al., 2005) (described in next section of this review). While in bacteria, presence of the KH and the S1 RNA binding domains are required for functional activity of PNPase, hPNPaseold-35 still retained its functional activity upon removal of KH and S1 domains (Sarkar et al., 2005; Stickney et al., 2005).
4.2. hPNPase might be involved in degradation of RNA in mammalian mitochondria
In mammals, mitochondrial RNA (mtRNA) degradation is not well defined as no RNA degrading complex has been identified. The current view is largely based on our understanding of the E.coli RNA degradosome and yeast mitochondrial exosome. Similar to cytoplasmic mRNAs, mtRNAs also require long poly (A) tails for recruitment of poly (A)-binding proteins for maintenance of stability (Temperley et al., 2003). Human mitochondria-specific poly (A) polymerase (PAP) has been shown to synthesize mtRNA poly(A) tails. Knocking down PAP by siRNA decreases the length of poly (A) tail of mitochondrial mRNA and decreases their steady state levels (Nagaike et al., 2005). The mitochondrial membrane potential (Δψ) and oxygen consumption also decreased in PAP siRNA treated cells. In contrast, knocking down of hPNPase showed significantly extended poly(A) tails of mtRNA although it did not affect the steady-state levels of these mRNAs or their translational products (Nagaike et al., 2005; Temperley et al., 2003). In these studies, only the levels of full-length mtRNAs were quantified without considering the truncated and polyadenylated mtRNAs that normally occur in human mitochodria (Slomovic et al., 2005). In normal mammalian mitochondria, truncated and polyadenylated transcripts do not accumulate and are rapidly degraded (See et al., 1972). Recent studies suggest that knocking down hSUV3 (human Suppressor of Var1 3) helicase leads to an accumulation of shortened polyadenylated mtRNA species (Khidr et al., 2008). hSUV3 makes a heteropentameric complex with hPNPase at a 2:3 molar ratio in a coordinated manner to degrade dsRNA substrates (Wang et al., 2009). Considering this recent finding it is hypothesized that the hSUV3-hPNPase complex might be involved in removing truncated RNA species through hPNPase-mediated degradation. Further experimentation is required to validate this possibility.
4.3. Modulation of miRNAs by hPNPaseold-35
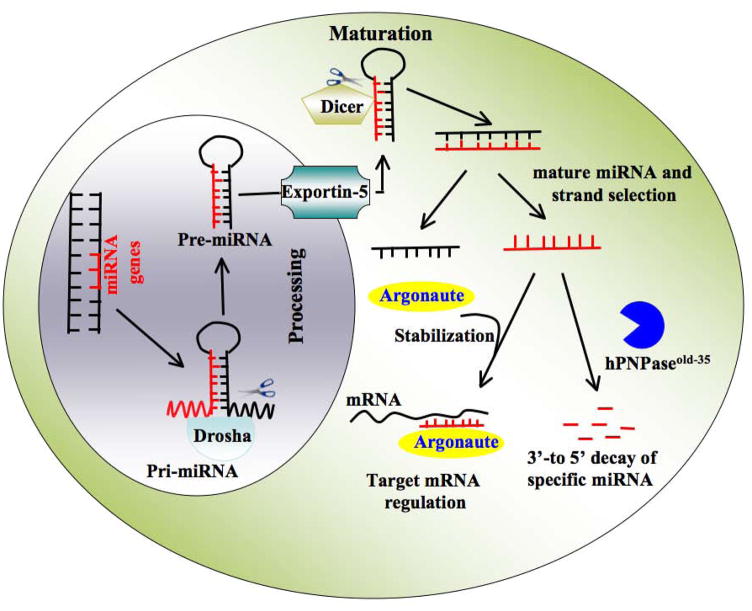
MicroRNAs (miRNAs), a subset of non-coding RNAs, are 22–25-nt long endogenously-initiated short RNA molecules that regulate gene expression at the post-transcriptional level and play important roles in a multiplicity of biological functions, including cell differentiation, tumorigenesis, apoptosis and metabolism (Ambros, 2004). An individual miRNA is able to control the expression of more than one target mRNA and each mRNA may be regulated by multiple miRNAs. The interaction between miRNA and mRNA are usually restricted to the “seed”- 6 to 8-nt at the 5′ region of miRNA and partially complementary sites in the 3′ UTRs of target mRNAs, resulting in either translational repression or target degradation (Kim, 2005). miRNAs are initially transcribed principally by either RNA polymerase II or RNA polymerase III as long primary transcripts (pri-miRNAs), which are further processed by the nuclear RNase Drosha and cytoplasmic RNase Dicer to produce precursor miRNAs (pre-miRNAs) and mature miRNAs, respectively (Calin and Croce, 2006) (Figure 1). In principle, miRNA abundance could be controlled by developmental and tissue specific signaling (Landgraf, 2009). The steady state levels of miRNAs, crucial for its profound impact on a wide array of biological processes (Filipowicz et al., 2008; Tsuchiya et al., 2006) are presumably regulated by the opposing activities of miRNA biogenesis and degradation.
Figure 1.
Schematic model of miRNA biogenesis and stability. After synthesis by RNA polymerase II, primary transcripts of (pri) miRNA are recognized by Drosha, which excises the hairpin precursor and released precursor (pre) miRNA. From nucleus, exportin five delivers the miRNA precursor to Dicer and its RNA binding partner in the cytoplasm for final processing to the mature 22-nt miRNAs. One strand is selected for stable association with Argonaute, where it serves as a guide to target and regulate specific mRNAs. By executing exonuclease activity hPNPaseold-35 specifically degrades mature miRNAs. However, their substrate recognition mechanism is unknown.
In the biogenesis process, miRNAs might be regulated both transcriptionally and post-transcriptionally. Numerous Pol II-associated transcription factors such as myogenin and MYOD1 are involved in transcriptional control of miR-1 and miR-133 genes during myogenesis (Chen JF et al., 2006; Rao et al., 2006). Some miRNAs are under the control of tumor suppressive p53 (reviewed by He et al., 2007) or the oncogenic transcription factor c-myc (Chang et al., 2008; He et al., 2005). Epigenetic control also contributes to miRNA gene regulation (Bueno et al., 2008). Several miRNAs expressions are also regulated at the post-transcriptional level. The primary transcript of let-7 (pri-let-7) is expressed in both undifferentiated and differentiated ES cells, whereas mature let-7 is detected only in differentiated cells, indicating that let-7a might be posttranscriptionally controlled (Suh et al., 2004; Thomson et al., 2006; Wulczyn et al., 2007). Excluding this activity, turnover of miRNA also might contribute in maintaining its level, however, this remains a largely unexplored area. In C.elegans (Kennedy et al., 2006) an exoribonuclease ERI1 (also known as THEX1) was previously shown to degrade siRNAs. Another exoribonuclease named small RNA degrading nuclease (SDN) proteins were reported to affect the stability of miRNAs in plants (Ramachandran and Chen, 2008). Very recently we demonstrated a role of hPNPaseold-35 in human miRNAs degradation (Das et al. 2010). This novel way of regulating miRNA levels is being actively explored by our research group.
4.3.1. hPNPaseold-35 downregulates specific miRNAs
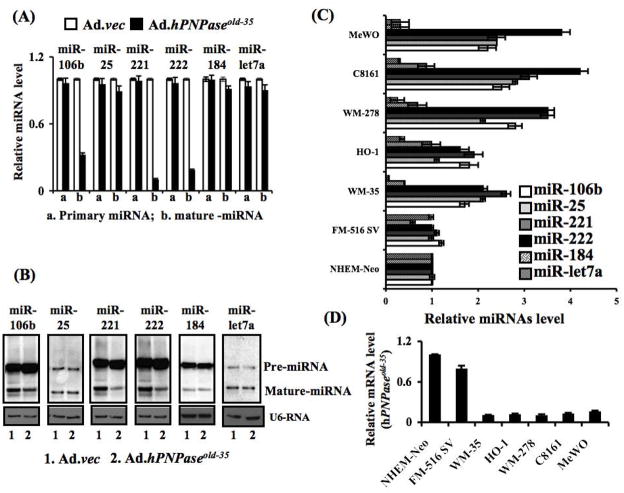
miRNA microarray analysis between Ad.hPNPaseold-35- and Ad.vec-infected human melanoma cells identified specific miRNAs differentially regulated by hPNPaseold-35, which have been subsequently validated by qPCR and Northern Blot analyses. Of interest, robust downregulation of several miRNAs (e.g., miR-221, miR-222, miR-106b) by hPNPaseold-35 was observed while a number of miRNAs were not affected (miR-184, miR-let7a) suggesting specificity and selectivity of this enzyme-mediated miRNA downregulation. Moreover, an inverse correlation between these miRNAs and hPNPaseold-35 in primary melanocytes and different melanoma cell lines support the biological relevance of this protein in regulating specific miRNAs (Figure 2)
Figure 2. hPNPaseold-35 target miRNAs.
HO-1 cells were either infected with Ad.vec or Ad.hPNPaseold-35 at a m.o.i. of 5000 vp/cells for three days and subjected to miRNA microarrays and potential target miRNAs miR-106b, miR-25, miR-221, miR-222, miR-let7a and miR-184 were validated for differential expression by using primary- and mature miRNA-specific Taqman® probes with qPCR. (B) Northern blotting was performed to detect mature miRNAs and its precursor species by using specific probes. Expression of GAPDH, miR-RNU44 and U6 RNA were used to normalize the pri- and mature miRNA in qPCR and Northern blotting data, respectively. (C & D) The basal level of different miRNAs (C) and mRNA for hPNPaseold-35 (D) were evaluated by qPCR in different cell lines, including Normal human epidermal melanocytes (NHEM), Normal immortal human melanocytes FM-516-SV (referred to as FM-516), radial growth phase primary melanoma WM-35, vertical growth phase primary melanoma WM-278 and metastatic melanoma HO-1, C8161.9, and MeWo. (Taken from Das et al., PNAS, 2010)
4.3.2. Post-transcriptional modification of miRNA biogenesis by hPNPaseold-35
miRNA genes are initially transcribed as long primary transcripts (pri-miRNAs), which are further processed to produce precursor miRNAs (pre-miRNAs) and mature miRNAs (Figure 1) (Calin and Croce, 2008). Ad.hPNPaseold-35 did not show any effect on pri-miRNA and pre-miRNA. Only mature miRNAs were downregulated by hPNPaseold-35. hPNPaseold-35 is an exoribonulease and in vivo immunopricipitation assays clearly demonstrate specific binding of mature miRNAs to hPNPaseold-35, a pre-requisite step for enzymatic activity indicating that miRNAs undergo enzymatic degradation.
4.3.3. hPNPaseold-35 preferentially degrades miR-221
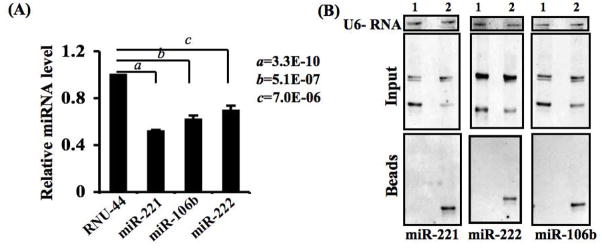
Although the overexpression of hPNPaseold-35 selectively downregulates specific miRNAs, the preferences to miRNAs varied significantly. In melanoma cells, degradation of miR-221 by hPNPaseold-35 was more profound compared to other miRNAs tested. This preferential activity might be attributed to high binding affinity of hPNPaseold-35 to the mature miR-221, a hypothesis that needs to be tested (Figure 3).
Figure 3. hPNPaseold-35 preferentially degrades miR-221.
(A) Comparative degradation between miR-221, miR-222 and miR-106b at a 2 h time point using in vitro degradation assays. P value was calculated using student’s t-test by comparing the specific miRNA normalized by miR-RNU-44. The data represent mean ± S.D. of two independent experiments each done in triplicate. (B) A direct interaction between targeted miRNAs with in vitro translated hPNPaseold-35 was confirmed by immunoprecipitation and Northern blotting.
4.3.4. Role of human polynucleotide phosphorylaseold-35 mediated miRNA degradation in interferon-β-mediated growth inhibition
hPNPaseold-35 is an IFN-β-inducible early response gene and treating a panel of melanoma cells with IFN-β also downregulates hPNPaseold-35-target miRNAs. This effect was abrogated by knocking down hPNPaseold-35, thus establishing functional and mechanistic links between IFN-β-mediated hPNPaseold-35 induction and miRNA downregulation. IFN-β is a potent growth inhibitor and one of the growth inhibiting mechanisms in melanoma is the induction of hPNPaseold-35, which in turn raises the level of p27Kip1, a cyclin-dependent kinase inhibitor protein, by degrading c-myc mRNA (Sarkar et al., 2006). However, forced degradation of c-myc by siRNA did not completely block IFN-β-mediated growth arrest indicating the existence of additional pathways potentially regulating this effect (Barnes and Karin, 2008; Obaya et al., 1999; O’Hagan et al., 2000). Since miR-221 directly targets p27Kip1 mRNA (Fornari et al., 2008), IFN-β mediated hPNPaseold-35-dependent miR-221 downregulation might also be involved in inducing growth arrest.
5. Other biological functions of human polynucleotide phosphorylaseold-35 protein
5.1 hPNPaseold-35 and cellular senescence
Senescence is a state of irreversible growth arrest induced spontaneously in primary cells after a finite number of population doublings (replicative senescence) or induced by endogenous and exogenous acute and chronic stress signals (stress- or aberrant-signaling-induced senescence (Hayflick, 1976; Serrano et al., 2004). hPNPaseold-35 was first described as an induced gene during terminal differentiation and cellular senescence two end-stage processes sharing several overlapping features including irreversible growth arrest, marked inhibition of DNA synthesis and modulation of telomerase activity (Sarkar et al., 2006). Overexpression of full length hPNPaseold-35 or either one of its RNase PH domains in human melanoma cells and melanocytes induces distinctive changes associated with senescent phenotypes (Sarker et al., 2003, 2005) including induction of SA-β-gal activity, cell cycle arrest in G1 phase with inhibition of DNA synthesis followed by induction of apoptosis, and inhibition of telomerase activity. In total, these findings indicate that hPNPaseold-35 might play an essential role in senescence- and differentiation-associated growth inhibition. Mechanistically, hPNPaseold-35-induced senescence is associated with an increase in p27kip1 and a decrease in p21CIP1/WAF-/MDA-6. The increase of p27kip1 is most likely secondary to the decrease in c-myc that controls p27kip1 expression by multiple mechanisms (Barnes and Karin, 2008; Obaya et al., 1999; O’Hagan et al., 2000). However, it is also a consequence of downregulation of miR-221 that specifically targets the mRNA of p27kip1. Since oxidative stress mediates cellular senescence (Colavitti et al., 2005; PAssos et al., 2006) the induction of reactive oxygen species (ROS) following upregulation of hPNPaseold-35 could also be involved in induction of the senescent phenotype. Further studies are necessary to investigate the role of the interactive network of c-myc, miR-221, p27kip1 and ROS in regulating the senescence process.
5.2. Human polynucleotide phosphorylaseold-35 and aging-associated inflammation
Aging represents a conundrum in the evolution of higher organisms that is associated with progressive degenerative diseases the underlying pathology of which is often chronic inflammation (Kiecolt-Glaser et al., 2003). Oxidative stress plays an important role in induction of chronic inflammation. Reactive oxygen species (ROS) generated by mitochondria induce oxidative stress in DNA, protein and lipid and this activity is more prominent in tissues from aged individuals or aged experimental animals than their young counterparts (Finkel et al., 2000; Harman, 1957; Chen, 2000; Hagen, 1997). A prominent mechanism by which ROS modulates diverse intracellular molecular processes is by turning on the expression of proinflammatory cytokines (Baldwin, 1996) and through regulating the activity of NF-κB (Schreck et al., 1992). hPNPaseold-35 is localized in mitochondria and induces ROS that subsequently leads to NF-κB activation, which is inhibited by anti-oxidant N-acetyl-L-cysteine (NAC). Activation of NF-κB leads to increased production of pro-inflammatory cytokines such as IL-6, IL-8, RANTES and MMP-3, which could also be inhibited by treatment with NAC suggesting the involvement of hPNPaseold-35 in producing pathological changes associated with aging by generating pro-inflammatory cytokines via ROS and NF-κB (Sarkar et al., 2004; Sarkar & Fisher, 2006)
5.3. Overexpression of hPNPaseold-35 induced growth inhibition in different cancer cells and its molecular mechanism
Based on cell line studies it is observed that slow and sustained overexpression of hPNPaseold-35 induces growth arrest ultimately culminating in apoptosis, whereas rapid overexpression of hPNPaseold-35 directly promotes apoptosis without cell cycle changes. These observations imply that inhibition of cell cycle progression and induction of apoptosis by hPNPaseold-35 involves multiple intracellular targets and signaling pathways. In the context of cell cycle, hPNPaseold-35 overexpression induces growth arrest at both G1/S and G2/M phase depending on the cell types, although the molecular mechanism is quite similar (Sarkar et al., 2004; Maerken et al., 2009; Chan et al., 2008 ). In all cases, c-myc is downregulated by hPNPaseold-35 accompanied with upregulation of CDKI, p21CIP1/WAF-1/MDA-6 and p27KIP1 that play essential roles in cell cycle arrest either in the G1 or G2/M phase. Apoptosis-inducing activity of hPNPaseold-35 is mediated by activation of double-stranded RNA–dependent protein kinase (PKR) (Sarkar et al., 2007). Activation of PKR by hPNPaseold-35 precedes phosphorylation of eukaryotic initiation factor-2A and induction of growth arrest and DNA damage-inducible gene 153 (GADD153) that culminates in the shutdown of protein synthesis and apoptosis. Activation of PKR by hPNPaseold-35 also initiates down-regulation of the anti-apoptotic protein Bcl-xL. All of these studies elucidate a novel pathway by which an evolutionary conserved RNA-metabolizing enzyme, hPNPaseold-35, regulates cell growth and viability.
5.4. Role of hPNPaseold-35 in maintaining mitochondrial homeostasis
Based on the localization in the mitochondrial intermembrane space (IMS), hPNPaseold-35 is predicted to play a major role in mitochondrial bioenergetics. Evidence for maintaining homeostasis comes from both loss-of-function and gain-of-function studies, although contradictory results also indicate that knocking down hPNPaseold-35 does not affect mitochondrial morphology or the rate of oxygen consumption (Nagaike T et al., 2005). Knocking down of hPNPaseold-35 reduced mitochondrial ΔΨ and enzymatic activities of couple respiratory complexes compared with control cells resulting in mitochondrial dysfunction such as lactate accumulation and reduction of steady state ATP levels (Chen et al., 2006, 2007). Concurrently, overexpression results in increased reactive oxygen species accumulation over time confirming a role for hPNPaseold-35 in mitochondrial homeostasis (Sarkar et al., 2004). However, it is not understood how mitochondrial homeostasis is precisely regulated by hPNPaseold-35, since no known substrates are located in the IMS. Accordingly, one must also consider potential non-enzymatic functions of hPNPaseold-35 in maintenance of mitochondrial homeostasis. It is possible that hPNPaseold-35 affects oxidative phosphorylation directly by impacting on components of the respiratory complexes or indirectly through interference in mitochondrial fusion. Consistent with this possibility, a very recent study (Wang et al. 2010) showed that components of the Electron Transport Chains (ETC) were reduced at both RNA and protein levels in PNPase RNAi transfected HEK293 cells as compared to controls. A similar result was also observed in liver mitochondria from a liver-specific PNPase knockout mouse model suggesting that the decrease in functional ETC complexes was responsible for decreased respiration. Furthermore, the ultrastructure of liver mitochondria from liver-specific knockout mice displayed disordered circular and smooth inner membrane criste, similar to mitochondria having impaired components of oxidative phosphorylation pathways. Citrate synthase activity, routinely used as a marker of aerobic capacity, also decreased in the liver of PNPase knockout mice compared with the wild type mice establishing a pivotal role of PNPase in mitochondrial morphogenesis and respiration in vivo. Additionally, a number of studies indicate that hPNPaseold-35 potentially maintains high-fidelity translation by sequestering oxidative stress-damaged RNA (Hayakawa et al., 2006; Wu et al., 2008) supporting potential exoribonuclease-independent activity of this enzyme in maintaining normal mitochondrial function.
5.5. Role of PNPase in translocation of RNA in mitochondria
While the molecular mechanism of mitochondrial import of nuclear-encoded proteins is well defined, little is known about the factors that control mitochondrial RNA import. Wang and colleagues (Wang et al. 2010) have now demonstrated a direct involvement of PNPase in regulating specific cytosolic RNA import into the mitochondrial matrix. This function of PNPase is independent of its RNA processing function, as inactivation of RNA processing by mutation did not affect RNA import. Mammalian RNase P localizes in mitochondria and functions in the processing of tRNAs during the maturation of mitochondrial transcripts (Chang and Clayton, 1987). RNase P is encoded in the nucleus and PNPase is directly involved in translocation of this enzyme. However, the exact mechanism by which PNPase augments RNA import is not yet known. It is hypothesized that PNPase imports RNAs from the cytosol into the IMS and then passes this RNA to another protein or complex that facilitates passage of the RNAs through the IM into the matrix. Interestingly PNPase showed specificity towards import of cytosolic RNAs based on their secondary structure. Consistent with in vitro findings, mitochondrial RNA import was severely compromised in the liver mitochondria from liver-specific PNPase (pnpt1) knock out mice (Wang et al. 2010).
6. Summary
The gradual unraveling of the multifaceted functions of hPNPaseold-35 illustrates its prominent role in regulating diverse physiological and pathological processes (Figure 4). Since, hPNPaseold-35 displays substrate specificity, either mRNA or miRNA, the elucidation of how it elicits such specificity will significantly improve our understanding of its RNA-processing functions. The preferential degradative effects on specific miRNAs might be applicable to selectively downregulate oncogenic miRNA in different malignancies and thus might be amenable for developing a novel potential anti-cancer therapy. miRNAs have recently been shown to be involved in developing drug resistance and thus combinatorial therapy of hPNPaseold-35 and appropriate therapeutic drug might be an effective approach for combating cancer. The lack of expression in different cancer cells while the ubiquitious expression in multiple organ/tissues raises the relevant question whether hPNPaseold-35 serves any specialized function in specific organs? Animal models with either conditional overexpression or knockdown of PNPase expression will be extremely valuable in answering these questions. Importantly, these models will facilitate studies on senescence and IFN action, including its anti-viral properties and role in inflammatory disease processes as well as maintaining mitochondrial homeostasis. Strategies using natural compounds and/or small molecules to block the enzymatic functions of hPNPaseold-35 might provide effective therapeutic benefit for inflammation or even provide a means of retarding the aging process and enhancing longevity.
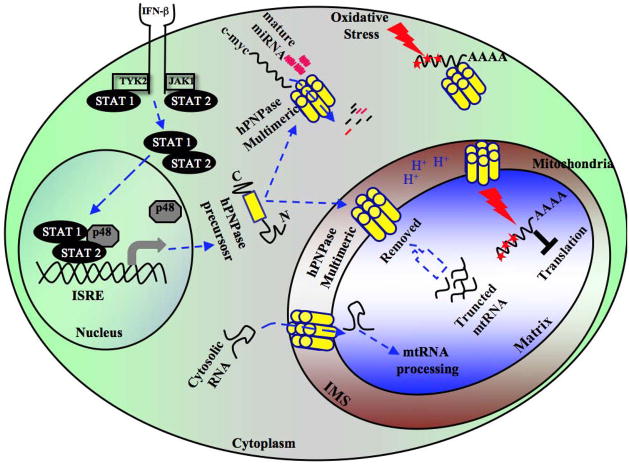
Figure 4. Schematic representation of the diversified functions of hPNPaseold-35.
Upon Type I IFNs binding with its corresponding receptor, hPNPaseold-35 transcription is promoted through activation of the JAK/STAT pathway. Once transcribed, hPNPaseold-35 is either imported into the mitochondria IMS or mobilized into the cytosol by unknown mechanism(s). In the cytoplasm, by executing exoribonuclease activity it specifically targets mRNA/miRNAs thereby significantly affecting various biological functions, e.g., induction of cell cycle arrest by targeting c-myc. In addition to maintaining mitochondrial homeostasis, hPNPaseold-35 actively participates to block the translation of oxidative stress-mediated damaged RNA by sequestering it. In addition, although not evident, hPNPaseold-35 might also be involved in removal of truncated mtRNA from mitochondria.
Overall, delineation of the nuances of how hPNPaseold-35 induces its diverse effects in different organisms offers significant potential to comprehend the evolutionary relevance of this molecule and this will help delineate its quintessential role in both normal physiology and in disease pathology.
Acknowledgments
The present study was supported in part by NIH grants R01 CA097318, R01 CA127641, CA134721, and P01 CA104177, the Samuel Waxman Cancer Research Foundation and the National Foundation for Cancer Research (PBF) and NIH grant R01 CA134721 (DS). DS is the Harrison Endowed Scholar in Cancer Research. PBF holds the Thelma Newmeyer Corman Chair in Cancer Research and is a SWCRF Investigator.
Footnotes
Conflict of Interest
The authors declare no conflicts of interest.
References
- Allmang C, Petfalski E, Podtelejnikov A, Mann M, Tollervey D, Mitchell P. The yeast exosome and human PM-Scl are related complexes of 3′→5′ exonucleases. Genes Dev. 1999;13:2148–2158. doi: 10.1101/gad.13.16.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida FC, DeSalle R, Leszczyniecka M, Fisher PB. Examining ancient inter-domain horizontal gene transfer. Evolutionary Bioinformatics. 2008;4:109–119. [PMC free article] [PubMed] [Google Scholar]
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Baldwin AS. The NF-kappa B and I kappa B proteins: new discoveries and insights. Annu Rev Immunol. 1996;14:649–683. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- Barnes PJ, Karin M. Nuclear factor-kappaB: A pivotal transcription factor in chronic inflammatory diseases. N Engl J Med. 1997;336:1–10. doi: 10.1056/NEJM199704103361506. [DOI] [PubMed] [Google Scholar]
- Bueno MJ, Pérez de Castro I, Gómez de Cedrón M, Santos J, Calin GA, Cigudosa JC, et al. Genetic and epigenetic silencing of microRNA-203 enhances ABL1 and BCR-ABL1. Cancer Cell. 2008;13:496–506. doi: 10.1016/j.ccr.2008.04.018. [DOI] [PubMed] [Google Scholar]
- Buttner K, Wenig K, Hopfner KP. The exosome: a macromolecular cage for controlled RNA degradation. Mol Microbiol. 2006;61:1372–1379. doi: 10.1111/j.1365-2958.2006.05331.x. [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006;66:7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2008;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Campisi J. Gene expression in quiescent and senescent fibroblasts. Ann N Y Acad Sci. 1992;663:195–201. doi: 10.1111/j.1749-6632.1992.tb38663.x. [DOI] [PubMed] [Google Scholar]
- Cao D, Parker R. Computational modeling and experimental analysis of nonsense-mediated decay in yeast. Cell. 2003;113:533–545. doi: 10.1016/s0092-8674(03)00353-2. [DOI] [PubMed] [Google Scholar]
- Chang DD, Clayton DA. A mammalian mitochondrial RNA processing activity contains nucleus-encoded RNA. Science. 1987;235:1178–1184. doi: 10.1126/science.2434997. [DOI] [PubMed] [Google Scholar]
- Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, et al. Widespread microRNA repression by Myc contributes to tumorigenesis. Nature Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HW, Koehler CM, Teitell MA. Human polynucleotide phosphorylase: location matters. Trends Cell Biol. 2006;17:600–608. doi: 10.1016/j.tcb.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Chen HW, Rainey RN, Balatoni CE, Dawson DW, Troke JJ, Wasiak S, et al. Mammalian polynucleotide phosphorylase is an intermembrane space RNase that maintains mitochondrial homeostasis. Mol Cell Biol. 2006;26:8475–8487. doi: 10.1128/MCB.01002-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nature Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan I, Lebedeva IV, Su ZZ, Sarkar D, Valerie K, Fisher PB. Progression elevated gene-3 promoter (PEG-Prom) confers cancer cell selectivity to human polynucleotide phosphorylase (hPNPase(old-35))-mediated growth suppression. J Cell Physiol. 2008;215:401–409. doi: 10.1002/jcp.21320. [DOI] [PubMed] [Google Scholar]
- Chen QM. Replicative senescence and oxidant-induced premature senescence. Beyond the control of cell cycle checkpoints. Ann NY Acad Sci. 2000;908:111–125. doi: 10.1111/j.1749-6632.2000.tb06640.x. [DOI] [PubMed] [Google Scholar]
- Colavitti R, Finkel T. Reactive oxygen species as mediators of cellular senescence. IUBMB Life. 2005;57:277–281. doi: 10.1080/15216540500091890. [DOI] [PubMed] [Google Scholar]
- Das SK, Sokhi UK, Bhutia SK, Azab B, Su ZZ, Sarkar D, et al. Human polynucleotide phosphorylase selectively and preferentially degrades microRNA-221 in human melanoma cells. Proc Natl Acad Sci U S A. 2010;107:11948–11953. doi: 10.1073/pnas.0914143107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher MP, Li Z. Exoribonucleases and their multiple roles in RNA metabolism. Prog Nucleic Acid Res Mol Biol. 2001;66:67–105. doi: 10.1016/s0079-6603(00)66027-0. [DOI] [PubMed] [Google Scholar]
- Deutscher MP. Promiscuous exoribonucleases of Escherichia coli. J Bacteriol. 1993;175:4577–4583. doi: 10.1128/jb.175.15.4577-4583.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher MP. Ribonuclease multiplicity, diversity, and complexity. J Biol Chem. 1993;268:13011–13014. [PubMed] [Google Scholar]
- Dodson RE, Shapiro DJ. Regulation of pathways of mRNA destabilization and stabilization. Prog Nucleic Acid Res Mol Biol. 2002;72:129–164. doi: 10.1016/s0079-6603(02)72069-2. [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya S, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- Fisher PB, Grant S. Effects of interferon on differentiation of normal and tumor cells. Pharmacol Ther. 1985;27:143–66. doi: 10.1016/0163-7258(85)90067-1. [DOI] [PubMed] [Google Scholar]
- Fisher PB, Hermo H, Jr, Solowey WE, Dietrich MC, Edwalds GM, Weinstein IB, et al. Effect of recombinant human fibroblast interferonand mezerein on growth, differentiation, immune interferon binding and tumor associated antigen expression in human melanoma cells. Anticancer Res. 1986;6:765–774. [PubMed] [Google Scholar]
- Fisher PB, Prignoli DR, Hermo H, Jr, Weinstein IB, Pestka S. Effects of combined treatment with interferon and mezerein on melanogenesis and growth in human melanoma cells. J Interferon Res. 1985;5:11–22. doi: 10.1089/jir.1985.5.11. [DOI] [PubMed] [Google Scholar]
- Fornari F, Gramantieri L, Ferracin M, Veronese A, Sabbioni S, Calin GA, et al. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene. 2008;27:5651–5661. doi: 10.1038/onc.2008.178. [DOI] [PubMed] [Google Scholar]
- Frischmeyer PA, van Hoof A, O’Donnell K, Guerrerio AL, Parker R, Dietz HC. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science. 2002;295:2258–2261. doi: 10.1126/science.1067338. [DOI] [PubMed] [Google Scholar]
- Gewartowski K, Tomecki R, Muchowski L, Dmochow Ska A, Dzwonek A, Malecki M, et al. Up-regulation of human PNPase mRNA by beta-interferon has no effect on protein level in melanoma cell lines. Acta Biochim Pol. 2006;53:179–188. [PubMed] [Google Scholar]
- Guarini L, Graham GM, Jiang H, Ferrone S, Zucker S, Fisher PB. Modulation of the antigenic phenotype of human melanoma cells by differentiation-inducing and growth suppressing agents. Pigment Cell Res. 1992;2:123–131. doi: 10.1111/j.1600-0749.1990.tb00361.x. [DOI] [PubMed] [Google Scholar]
- Guarini L, Temponi M, Edwalds GM, Vita JR, Fisher PB, Ferrone S. In vitro differentiation and antigenic changes in human melanoma cell lines. Cancer Immunol Immunother. 1989;30:262–268. doi: 10.1007/BF01744892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen TM, Yowe DL, Bartholomew JC, Wehr CM, Do KL, Park JY, et al. Mitochondrial decay in hepatocytes from old rats: membrane potential declines, heterogeneity and oxidants increase. Proc Natl Acad Sci U S A. 1997;94:3064–3069. doi: 10.1073/pnas.94.7.3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harman D. Aging: a theory based on free radical and radiation chemistry. J Gerontol. 1957;2:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- Hayflick L. The cell biology of human aging. N Engl J Med. 1976;295:1302–1308. doi: 10.1056/NEJM197612022952308. [DOI] [PubMed] [Google Scholar]
- He L, He X, Lowe SW, Hannon GJ. microRNAs join the p53 network — another piece in the tumour suppression puzzle. Nature Rev Cancer. 2007;7:819–822. doi: 10.1038/nrc2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoof A, Parker R. The exosome: a proteasome for RNA? Cell. 1999;99:347–350. doi: 10.1016/s0092-8674(00)81520-2. [DOI] [PubMed] [Google Scholar]
- Hayakawa H, Sekiguchi M. Human polynucleotide phosphorylase protein in response to oxidative stress. Biochemistry. 2006;45:6749–6755. doi: 10.1021/bi052585l. [DOI] [PubMed] [Google Scholar]
- Jarrige AC, Mathy N, Portier C. PNPase autocontrols its expression by degrading a double-stranded structure in the pnp mRNA leader. EMBO J. 2001;20:6845–6855. doi: 10.1093/emboj/20.23.6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrige A, Brechemier-Baey D, Mathy N, Duche O, Portier C. Mutational analysis of polynucleotide phosphorylase from Escherichia coli. J Mol Biol. 2002;321:397–409. doi: 10.1016/s0022-2836(02)00645-9. [DOI] [PubMed] [Google Scholar]
- Jiang H, Su ZZ, Boyd J, Fisher PB. Gene expression changes associated with reversible growth suppression and the induction of terminal differentiation in human melanoma cells. Mol Cell Differ. 1993;1:41–66. [Google Scholar]
- Jiang H, Lin J, Young SM, Goldstein NI, Waxman S, Davila V, et al. Cell cycle gene expression and E2F transcription factor complexes in human melanoma cells induced to terminally differentiate. Oncogene. 1995;11:1179–1189. [PubMed] [Google Scholar]
- Kennedy S, Wang D, Ruvkun GA. Conserved siRNA degrading RNase negatively regulates RNA interference in C. elegans. Nature. 2004;427:645–649. doi: 10.1038/nature02302. [DOI] [PubMed] [Google Scholar]
- Khidr L, Wu G, Davila A, Procaccio V, Wallace D, Lee WH. Role of SUV3 helicase in maintaining mitochondrial homeostasis in human cells. J Biol Chem. 2008;283:27064–27073. doi: 10.1074/jbc.M802991200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiecolt-Glaser JK, Preacher KJ, MacCallum RC, Atkinson C, Malarkey WB, Glaser R. Chronic stress and age-related increases in the proinflammatory cytokine IL-6. Proc Natl Acad Sci U S A. 2003;100:9090–9095. doi: 10.1073/pnas.1531903100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim V. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Kirschner LS, Taymans SE, Pack S, Pak E, Pike BL, Chandrasekharappa SC, et al. Genomic mapping of chromosomal region 2p15-p21 (D2S378-D2S391): integration of Genemap’98 within a framework of yeast and bacterial artificial chromosomes. Genomics. 1999;62:21–33. doi: 10.1006/geno.1999.5957. [DOI] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leszczyniecka M, Kang DC, Sarkar D, Su ZZ, Holmes M, Valerie K, et al. Identification and cloning of human polynucleotide phosphorylase, hPNPaseold-35, in the context of terminal differentiation and cellular senescence. Proc Natl Acad Sci U S A. 2002;99:16636–16641. doi: 10.1073/pnas.252643699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leszczyniecka M, Su ZZ, Kang DC, Sarkar D, Fisher PB. Expression regulation and genomic organization of human polynucleotide phosphorylase, hPNPaseold-35, a type I interferon inducible early response gene. Gene. 2003;316:143–156. doi: 10.1016/s0378-1119(03)00752-2. [DOI] [PubMed] [Google Scholar]
- Leszczyniecka M, DeSalle R, Kang DC, Fisher PB. The origin of polynucleotide phosphorylase domains. Mol Phylogenet Evol. 2004;31:123–130. doi: 10.1016/j.ympev.2003.07.012. [DOI] [PubMed] [Google Scholar]
- Littauer UZ, Grunberg-Manago M. Polynucleotide phosphorylase. In: Creighton TE, editor. Encyclopedia of Molecular Biology. Vol. 3. 1999. p. 1911. [Google Scholar]
- Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- Mitchell P, Tollervey D. Musing on the structural organization of the exosome complex. Nat Struct Biol. 2000;7:843–846. doi: 10.1038/82817. [DOI] [PubMed] [Google Scholar]
- Mohanty BK, Kushner SR. Polynucleotide phosphorylase functions both as a 3′ right-arrow 5′ exonuclease and a poly(A) polymerase in Escherichia coli. Proc Natl Acad Sci U S A. 2000;97:11966–11971. doi: 10.1073/pnas.220295997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaike T, Suzuki T, Katoh T, Ueda T. Human mitochondrial mRNA are stabilized with polyadenylation regulated by mitochondria-specific poly (A) polymerase and polynucleotide phosphorylase. J Biol Chem. 2005;280:19721–19727. doi: 10.1074/jbc.M500804200. [DOI] [PubMed] [Google Scholar]
- Obaya AJ, Mateyak MK, Sedivy JM. Mysterious liaisons: the relationship between c-Myc and the cell cycle. Oncogene. 1999;18:2934–2941. doi: 10.1038/sj.onc.1202749. [DOI] [PubMed] [Google Scholar]
- O’Hagan RC, Ohh M, David G, de Alboran IM, Alt FW, Kaelin WG, Jr, et al. Myc-enhanced expression of Cul1 promotes ubiquitin-dependent proteolysis and cell cycle progression. Genes Dev. 2000;14:2185–2191. doi: 10.1101/gad.827200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nat Struct Mol Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- Passos JF, von Zglinicki T, Saretzki G. Mitochondrial dysfunction and cell senescence: cause or consequence? Rejuvenation Res. 2006;9:64–68. doi: 10.1089/rej.2006.9.64. [DOI] [PubMed] [Google Scholar]
- Piwowarski J, Grzechnik P, Dziembowski A, Dmochowska A, Minczuk M, Stepien PP. Human polynucleotide phosphorylase, hPNPase, is localized in mitochondria. J Mol Biol. 2003;329:853–857. doi: 10.1016/s0022-2836(03)00528-x. [DOI] [PubMed] [Google Scholar]
- Portnoy V, Evguenieva-Hackenberg E, Klein F, Walter P, Lorentzen E, Klug G, et al. RNA polyadenylation in Archaea: not observed in Haloferax while the exosome polyadenylates RNA in Sulfolobus. EMBO Rep. 2005;6:1188–1193. doi: 10.1038/sj.embor.7400571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran V, Chen X. Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science. 2008;321:1490–1492. doi: 10.1126/science.1163728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raijmakers R, Egberts WV, van Venrooij WJ, Pruijn GJ. Protein-protein interactions between human exosome components support the assembly of RNase PH-type subunits into a six-membered PNPase-like ring. J Mol Biol. 2002;323:653–663. doi: 10.1016/s0022-2836(02)00947-6. [DOI] [PubMed] [Google Scholar]
- Raijmakers R, Schilders G, Pruijn GJ. The exosome, a molecular machine for controlled RNA degradation in both nucleus and cytoplasm. Eur J Cell Biol. 2004;83:175–183. doi: 10.1078/0171-9335-00385. [DOI] [PubMed] [Google Scholar]
- Rao PK, Kumar RM, Farkhondeh M, Baskerville S, Lodish HF. Myogenic factors that regulateexpression of muscle-specific microRNAs. Proc Natl Acad Sci USA. 2006;103:8721–8726. doi: 10.1073/pnas.0602831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regonesi ME, Briani F, Ghetta A, Zangrossi S, Ghisotti D, Tortora P, et al. A mutation in polynucleotide phosphorylase from Escherichia coli impairing RNA binding and degradosome stability. Nucleic Acids Res. 2004;32:1006–1017. doi: 10.1093/nar/gkh268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar D, Fisher PB. Molecular mechanisms of aging-associated inflammation. Cancer Lett. 2006;236:13–23. doi: 10.1016/j.canlet.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Sarkar D, Leszczyniecka M, Kang DC, Lebedeva IV, Valerie K, Dhar S, et al. Downregulation of Myc as a potential target for growth arrest induced by human polynucleotide phosphorylase (hPNPaseold-35) in human melanoma cells. J Biol Chem. 2003;278:24542–24551. doi: 10.1074/jbc.M302421200. [DOI] [PubMed] [Google Scholar]
- Sarkar D, Lebedeva IV, Emdad L, Kang DC, Baldwin AS, Jr, Fisher PB. Human polynucleotide phosphorylase (hPNPaseold-35): a potential link between aging and inflammation. Cancer Res. 2004;64:7473–7478. doi: 10.1158/0008-5472.CAN-04-1772. [DOI] [PubMed] [Google Scholar]
- Sarkar D, Park ES, Emdad L, Randolph A, Valerie K, Fisher PB. Defining the domains of human polynucleotide phosphorylase (hPNPaseOLD-35) mediating cellular senescence. Mol Cell Biol. 2005;25:7333–7343. doi: 10.1128/MCB.25.16.7333-7343.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar D, Park ES, Fisher PB. Defining the mechanism by which IFN-beta downregulates c-myc expression in human melanoma cells: pivotal role for human polynucleotide phosphorylase (hPNPaseold-35) Cell Death Differ. 2006;13:1541–1553. doi: 10.1038/sj.cdd.4401829. [DOI] [PubMed] [Google Scholar]
- Sarkar D, Park ES, Barber GN, Fisher PB. Activation of double-stranded RNA dependent protein kinase, a new pathway by which human polynucleotide phosphorylase (hPNPase(old-35)) induces apoptosis. Cancer Res. 2007;67:7948–7953. doi: 10.1158/0008-5472.CAN-07-0872. [DOI] [PubMed] [Google Scholar]
- Schreck R, Albermann K, Baeuerle PA. Nuclear factor kappa B: an oxidative stress-responsive transcription factor of eukaryotic cells. Free Radic Res Commun. 1992;17:221–237. doi: 10.3109/10715769209079515. [DOI] [PubMed] [Google Scholar]
- See YP, Fitt PS. Partial purification and properties of rat liver mitochondrial polynucleotide phosphorylase. Biochem J. 1972;130:343–353. doi: 10.1042/bj1300343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M, Blasco MA. Putting the stress on senescence. Curr Opin Cell Biol. 2001;13:748–753. doi: 10.1016/s0955-0674(00)00278-7. [DOI] [PubMed] [Google Scholar]
- Slomovic S, Slomovic S, Laufer D, Geiger D, Schuster G. Polyadenylation and degradation of human mitochondrial RNA: the prokaryotic past leaves its mark. Mol Cell Biol. 2005;25:6427–6435. doi: 10.1128/MCB.25.15.6427-6435.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stickney LM, Hankins JS, Miao X, Mackie GA. Function of the conserved S1 and KH domains in polynucleotide phosphorylase. J Bacteriol. 2005;187:7214–7221. doi: 10.1128/JB.187.21.7214-7221.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh MR, Lee Y, Kim JY, Kim SK, Moon SH, Lee JY, et al. Human embryonic stem cells express a unique set of microRNAs. Dev Biol. 2004;270:488–498. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- Symmons MF, Jones GH, Luisi BF. A duplicated fold is the structural basis for polynucleotide phosphorylase catalytic activity, processivity, and regulation. Structure Fold Des. 2000;8:1215–1226. doi: 10.1016/s0969-2126(00)00521-9. [DOI] [PubMed] [Google Scholar]
- Temperley RJ, Seneca SH, Tonska K, Bartnik E, Bindoff LA, Lightowlers RN, et al. Investigation of a pathogenic mtDNA microdeletion reveals a translation-dependent deadenylation decay pathway in human mitochondria. Hum Mol Genet. 2003;12:2341–2348. doi: 10.1093/hmg/ddg238. [DOI] [PubMed] [Google Scholar]
- Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya S, Okuno Y, Tsujimoto G. Micro-RNA: Biogenetic and functional mechanisms and involvements in cell differentiation and cancer. J Pharmacol Sci. 2006;101:267–270. doi: 10.1254/jphs.cpj06013x. [DOI] [PubMed] [Google Scholar]
- Tucker M, Parker R. Mechanisms and control of mRNA decapping in Saccharomyces cerevisiae. Annu Rev Biochem. 2000;69:571–595. doi: 10.1146/annurev.biochem.69.1.571. [DOI] [PubMed] [Google Scholar]
- Wang G, Chen HW, Oktay Y, Zhang J, Allen EL, Smith GM, et al. PNPase regulates RNA import into mitochondria. Cell. 2010;142:456–467. doi: 10.1016/j.cell.2010.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Kiledjian M. Functional link between the mammalian exosome and mRNA decapping. Cell. 2001;107:751–762. doi: 10.1016/s0092-8674(01)00592-x. [DOI] [PubMed] [Google Scholar]
- Wang DD, Shu Z, Lieser SA, Chen PL, Lee WH. Human mitochondrial SUV3 and polynucleotide phosphorylase form a 330-kDa heteropentamer to cooperatively degrade double-stranded RNA with a 3′-to-5′ directionality. J Biol Chem. 2009;284:20812–20821. doi: 10.1074/jbc.M109.009605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Li Z. Human polynucleotide phosphorylase reduces oxidative RNA damage and protects HeLa cell against oxidative stress. Biochem Biophys Res Commun. 2008;372:288–92. doi: 10.1016/j.bbrc.2008.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulczyn FG, Smirnova L, Rybak A, Brandt C, Kwidzinski E, Ninnemann O, et al. Post-transcriptional regulation of the let-7 microRNA during neural cell specification. FASEB J. 2007;21:415–426. doi: 10.1096/fj.06-6130com. [DOI] [PubMed] [Google Scholar]
- Yamanaka K, Inouye M. Selective mRNA degradation by polynucleotide phosphorylase in cold shock adaptation in Escherichia coli. J Bacteriol. 2001;183:2808–2816. doi: 10.1128/JB.183.9.2808-2816.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehudai-Resheff S, Hirsh M, Schuster G. Polynucleotide phosphorylase functions as both an exonuclease and a poly (A) polymerase in spinach chloroplasts. Mol Cell Biol. 2001;21:5408–5416. doi: 10.1128/MCB.21.16.5408-5416.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehudai-Resheff S, Portnoy V, Yogev S, Adir N, Schuster G. Domain analysis of the chloroplast polynucleotide phosphorylase reveals discrete functions in RNA degradation, polyadenylation, and sequence homology with exosome proteins. Plant Cell. 2003;15:2003–2019. doi: 10.1105/tpc.013326. [DOI] [PMC free article] [PubMed] [Google Scholar]