Abstract
The enzyme UDP-glucose dehydrogenase (UGDH) catalyzes the reaction of UDP-glucose to UDP-glucuronate through two successive NAD+-dependent oxidation steps. Human UGDH apoprotein purifies as a mixture of dimeric and hexameric species. Addition of substrate and cofactor stabilizes the oligomeric state to primarily the hexameric form. To determine if the dynamic conformations of hUGDH are required for catalytic activity, we used site-specific unnatural amino acid incorporation to facilitate crosslinking of monomeric subunits into predominantly obligate oligomeric species. Optimal crosslinking was achieved by encoding p-benzoyl-L-phenylalanine at position 458, normally a glutamine located within the dimer-dimer interface, and exposing to long wavelength UV in the presence of substrate and cofactor. Hexameric complexes were purified by gel filtration chromatography and found to contain significant fractions of dimer and trimer (approximately 50%) along with another 10% tetramer and higher molecular mass species. Activity of the crosslinked enzyme was reduced by almost 60% relative to the uncrosslinked UGDH mutant, and UV exposure had no effect on activity of the wildtype enzyme. These results support a model for catalysis in which the ability to dissociate the dimer-dimer interface is as important for maximal enzyme function as has been previously shown for the formation of the hexamer.
Graphical abstract
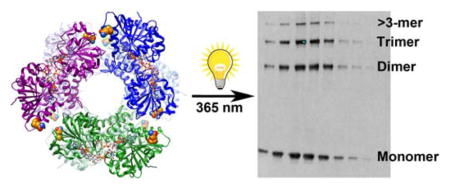
UDP-glucose dehydrogenase (UGDH) is a unique enzyme that produces UDP-glucuronate, which is a precursor for the synthesis of extracellular matrix polysaccharides, such as hyaluronan, chondroitin sulfate, and heparan sulfate 1. UDP-glucuronate is also decarboxylated by UDP-xylose synthase to form UDP-xylose, which can then be used as a sugar donor for synthesis of glycoproteins in the Golgi or for initiation of heparan sulfate proteoglycan synthesis. UDP-glucuronate also acts as a substrate for glucuronosyltransferase enzymes that function in detoxifying xenobiotic compounds in the liver as well as inactivating lipophilic androgens such as dihydrotestosterone in the prostate 2. Human UGDH is expressed primarily in liver, kidney, prostate, and cardiac tissues 3. Loss of function mutations caused by UGDH polymorphisms found in the human population result in congenital cardiac valve defects due to a reduction in enzymatic activity during morphogenesis 4. Characterization of these thermally unstable mutations revealed that the quaternary structure of two mutants, R141C and E416D, was destabilized such that the equilibrium between dimeric and hexameric species was shifted to favor the dimeric species.
The monomeric subunit of hUGDH is a 57-kDa, 494 amino acid protein with orthologues in murine, bovine, streptococcal, and plant systems. All catalyze two successive NAD+-dependent oxidations of UDP-α-D-glucose to form UDP-α-D-glucuronic acid (scheme 1) 3, 5–8. Interestingly, bacterial UGDH is catalytically active as a homodimer and does not form higher-order oligomeric assemblies, while eukaryotic homologues of UGDH exist primarily as a homohexameric species, or trimer of dimers 9, 10. Studies of a similar enzyme, GDP-mannose dehydrogenase, suggest higher-order quaternary assembly is not required for catalytic activity, but provides the ability for allosteric regulation and cooperativity between adjacent subunits with respect to substrate utilization 11, 12. We previously generated an obligatory homodimeric point mutant of hUGDH, T325D, in which interactions at the dimer-dimer interface were disrupted to show that the enzyme retained only ≈20% of wild-type activity in this state and lacked the ability to contribute significant UDP-glucuronate to downstream products 13. However, an obligate hexamer caused by a single amino acid deletion, Δ132, also showed no detectable activity 13, 14. Moreover, crystal structure determination and sedimentation velocity characterization have shown that an endogenous inhibitor of UGDH, UDP-xylose, functions by binding within the UDP-glucose active site and altering subunit association to yield an inactivated hexamer 15, 16. These findings suggest quaternary assembly and the dynamic fluctuation at the interfaces between subunits of the enzyme is significant in achieving optimal enzymatic activity.
Scheme 1.
In the current study, we sought to modify quaternary structure by covalently crosslinking subunits through site-specific photoreactive unnatural amino acid incorporation. We screened several positions at the interface between dimeric units for their ability to support quaternary structure when substituted, while also efficiently crosslinking hUGDH subunits in higher order oligomers upon exposure to long wavelength UV light. By mutagenizing the coding sequence for each residue of interest to an amber stop codon (TAG), and with the assistance of engineered pairs of tRNAs and aminoacyl tRNA synthetases, we introduced a photoreactive unnatural amino acid at the amber codon 17–19. The resultant protein species were used to test the requirement for dynamic dissociation at the subunit interfaces for hUGDH catalytic activity. Photocrosslinking of hUGDH was most efficient when performed in the holocomplex containing NAD+ and UDP-glucose. The substrate and cofactor were freely diffusible in the photocrosslinked species, but crosslinking resulted in complete loss of activity. These results suggest that dissociation of the dimer-dimer interface during dynamic reorganization of hydrogen bonding is an essential component of the catalytic mechanism.
Experimental Procedures
Reagents and materials
All reagents were purchased from Fisher Scientific except where otherwise indicated. p-azido-L-phenylalanine (AzF) and p-benzoyl-L-phenylalanine (Bpa) were from Chem-Impex International, Inc. (Wood Dale, IL) Trypsin for limited proteolysis, UDP-glucose, and NAD+ were obtained from Sigma Chemical Company.
Generation and Purification of UGDH Unnatural Amino Acid Point Mutations
The coding sequence of human wildtype (WT) UGDH was codon optimized for expression in E. coli, purchased as a synthetic gene insert from GeneArt (Thermo Fisher), and subcloned into the pET28a vector (EMD Millipore). Selection of positions for unnatural amino acid substitution was facilitated by molecular graphics analysis in the UCSF Chimera package 20. Amber mutants of human UGDH were generated from the codon optimized E. coli expression construct, WT-UGDH pET28a, using PCR mutagenesis with appropriate primers. Sequences were verified by Eurofins MWG Operon (Huntsville, AL). E. coli strain BL21(DE3) was co-transformed with each UGDH amber mutant and the AzF- or Bpa-specific tRNA synthetase encoded by pEVOL plasmids 21. Cultures were grown in 2xYT media containing 34 mg/L chloramphenicol and 50 mg/L kanamycin at 37°C. At OD600 of 0.6–0.8, AzF or Bpa was added from a 1M stock to a final concentration of 1mM, along with 0.2% arabinose to induce expression of the tRNA synthetases. Cultures were adjusted to 30°C and then IPTG was added to a final concentration of 0.5 mM to induce protein expression, following which cells were maintained at 18°C overnight. The cells were harvested by centrifugation and lysed by sonication 4, 13. All hUGDH AzF and Bpa mutants were expressed in the soluble fraction. The enzymes were purified by affinity chromatography using a HisTrap FF column (GE Healthcare) according to the manufacturer’s protocol. Average protein yield was 10–15 mg/L of culture following purification. Proteins were dialyzed against 20 mM Tris-HCl, pH 7.4, containing 1 mM DTT, then concentrated, flash frozen in liquid nitrogen, and stored at −80°C.
Proteolytic Sensitivity of UGDH
Purified recombinant WT and unnatural amino acid substituted mutants of UGDH (10 μg protein) were digested with 10 ng of trypsin in 1X PBS, pH 7.4, for 2.5 h in the absence or presence of 5 mM NADH and/or 1 mM UDP-glucose. Samples were analyzed by reducing SDS-PAGE stained with Gel Code Blue (Pierce Chemical Company).
Crosslinking of Mutant UGDH
To determine optimal crosslinking conditions, solutions of purified recombinant UGDH AzF or Bpa mutants were prepared in apo form, or with 5 mM NAD+, 1 mM UDP-glucose, or both. These samples were exposed to long wavelength UV light (365 nm) for 1 h. Control samples were prepared identically, but with no exposure to UV light. As an additional control, WT UGDH without the amber mutation was exposed to the same conditions as the UGDH AzF and Bpa mutants, with and without UV exposure. Aliquots were removed at 0, 5, 15, 30, 45, and 60 min for analysis by SDS-PAGE to confirm specificity and timing of crosslinking.
Crosslinked Species Quantification
SDS-PAGE gels from the crosslinking procedure were either stained with Gel Code Blue or analyzed by western blot probed with anti-UGDH rabbit polyclonal antibody. For western blots, indicated fractions were transferred to PVDF membranes and blocked with Pierce Superblock reagent before incubation with anti-UGDH primary at a dilution of 1:1000. Membranes were washed and proteins detected by secondary incubation with IRDye 800 conjugated anti-rabbit IgG (Rockland, Gilberstville, PA, 1:5000). Images were captured in the 800-channel of the Odyssey Near Infrared Imager and analyzed using the Image Studio Lite program (LI-COR Biosciences). We quantified the fluorescence intensity of bands representing monomeric and oligomeric UGDH. By dividing the total of each oligomeric species by the total intensity of all species, we obtained a normalized percentage of crosslinking efficiency. Dynamic range of the instrument was determined by similar analysis of a standard curve consisting of 12 concentrations of purified wild-type UGDH spanning 0.1 ng to 10 μg. All values for western imaging were analyzed in the linear range of the standard curve by adjusting the instrument settings appropriately and loading equal volumes from each sample such that the highest protein concentration per fraction loaded was ≤10 μg.
Purification of Multimeric Crosslinked Species
All crosslinked and non-crosslinked samples from WT and WT-Q458Bpa were separated using size exclusion chromatography. After UV exposure, all samples were dialyzed against 50 mM Tris pH 8.5 with 1 mM DTT for 1 h at room temperature and centrifuged to remove precipitants. Each sample was loaded on a Superdex 300 10/200 GL gel filtration column (GE Healthcare) and separated by FPLC in 0.1 M Tris, pH 8.5 at a flow rate of 0.5 mL/min. Fractions were collected and analyzed by SDS-PAGE to determine oligomeric state of the sample. The fractions representative of the hexameric peak were pooled and dialyzed against 20 mM Tris pH 7.4 and 1 mM DTT for 1 h at room temperature prior to activity measurements.
Enzymatic Activity Assay
Activity of UGDH WT and Q458Bpa samples was determined by monitoring the change in absorbance at 340 nm that accompanies the reduction of NAD+ to NADH. For standard screening of enzymatic activity, the assay was performed for 5 min at room temperature in 0.1 M sodium phosphate buffer, pH 7.4, containing 2 μM enzyme, 1 mM UDP-glucose, and 5 mM NAD+. Change in absorbance versus time was recorded for a 0–1 min interval. For each condition, enzymatic activity was measured in triplicate on three individually prepared and crosslinked biological replicates.
Results
UGDH oligomeric association can be altered by covalent crosslinking of monomeric subunits
To covalently alter the oligomeric state of purified UGDH in solution, we designed, expressed, and purified amber mutant species of hUGDH with an N-terminal His6-tag fusion. The amber mutations were selected to replace residues we expected to interact across adjacent subunits, but that were not critical contacts needed to maintain the interface and would not perturb catalytic sites, based on crystallographic data for wild-type UGDH in a ternary complex (PDB code 2Q3E) 10. Orthogonal tRNA and aminoacyl-tRNA synthetase pairs for AzF and Bpa incorporation at amber codons were previously generated and reported 17, 18. We tested both AzF and Bpa for incorporation in our amber mutants and ultimately chose Bpa based on previous studies that showed Bpa is the least reactive in ambient light compared to other photoactive unnatural amino acids, is chemically stable, and has efficient crosslinking capabilities 17, 22.
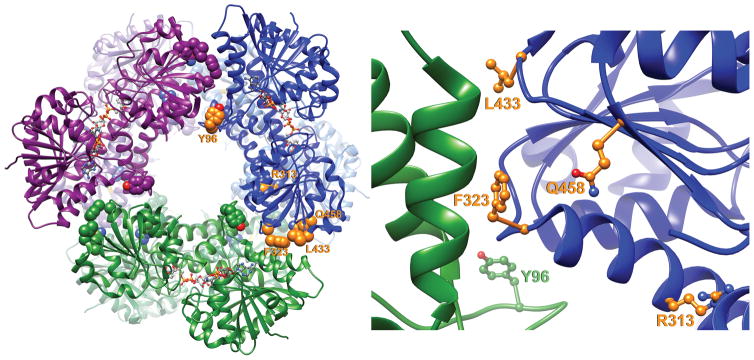
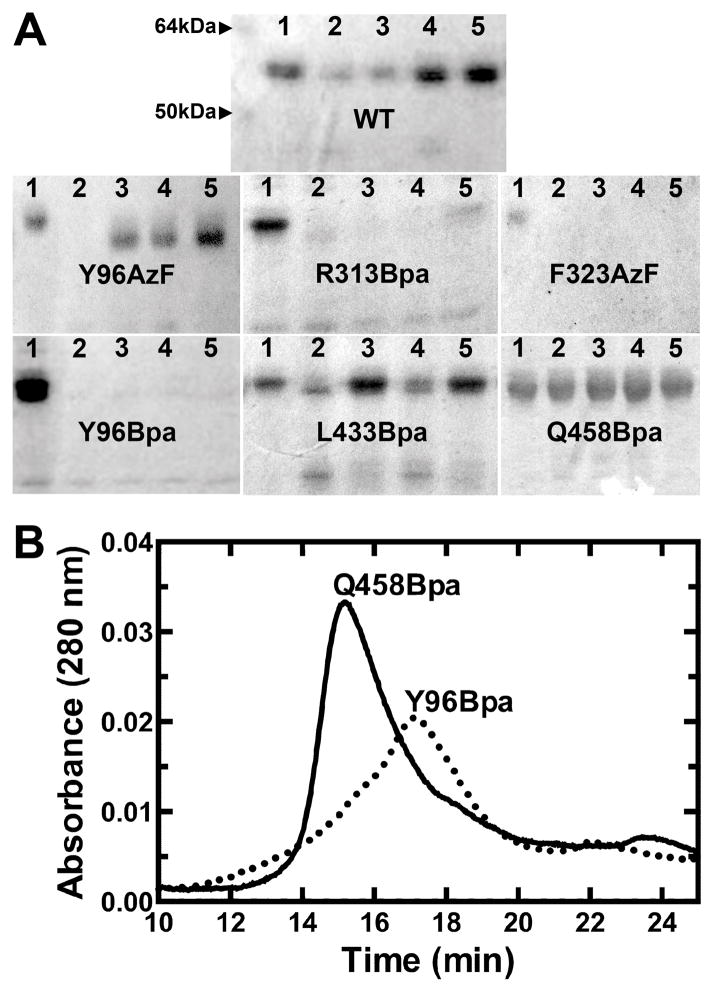
We selected six different positions to incorporate the amber codon, Y96, R313, F323, R393, L433, and Q458, due to their locations within the dimer-dimer interface (Fig 1). Our goal was to minimally perturb intrinsic properties of the enzyme with the substitution in the absence of crosslinking, so that if crosslinking affected activity, we could attribute the effects specifically to the restraint of conformational flexibility. We first screened each mutant for its ability to form a hexamer without crosslinking, using limited trypsin proteolysis as we previously described 13. As expected, the wild-type UGDH enzyme (WT) was relatively stable to proteolysis in the apoenzyme form but was further stabilized by addition of NADH cofactor, UDP-glucose substrate, or a combination of the two to form a ternary enzyme complex (Fig 2A). In contrast, substituting AzF or Bpa for tyrosine at position 96 compromised the ability of the apoenzyme to spontaneously adopt the hexamer conformation, as evidenced by complete proteolytic susceptibility of Y96Bpa and only partially protected degradation of Y96AzF. Similar substitutions for arginine at position 313 and phenylalanine at position 323 also resulted in destabilized protein. Only L433Bpa and Q458Bpa were capable of adopting wild-type quaternary structures that protected against degradation either in apo, binary, or ternary holo complexes. To confirm that proteolytic sensitivity of the mutants corresponded with quaternary structure as reported in the wild-type enzyme, we analyzed Y96Bpa and Q458Bpa apoenzymes by size exclusion chromatography. As predicted by limited proteolysis, UGDH Q458Bpa eluted in the peak corresponding to the hexamer (≈360 kDa), while Y96Bpa eluted as the dimeric (≈120 kDa) species (Fig 2B).
Figure 1. Model of the UGDH hexamer indicating locations of unnatural amino acid substitution.
The human UGDH ternary crystal structure is shown in ribbon representation (pdb 2Q3E). Individual dimeric units are colored blue, green and purple, with single subunits of each dimer colored light and dark in the ribbon diagram. Side chains are colored orange as space filling (A) or ball and stick representations (B) of Y96, R313, F323, L433, and Q458. Panel B is a zoom of one interface to illustrate the locations of each amino acid.
Figure 2. Competence of UGDH mutants for stable hexameric assembly.
UGDH amber codon mutants were expressed in E. coli with either AzF or Bpa and screened for quaternary structure integrity by limited proteolysis (A). Proteins were briefly trypsinized in the presence or absence of substrate and/or cofactor and analyzed by SDS-PAGE. In each panel, lanes are numbered as follows: 1, apoprotein undigested; 2, apoprotein digested; 3, NADH added; 4, UDP-glucose added; 5, NADH and UDP-glucose added. (B) Two mutants were further analyzed by size exclusion chromatography to confirm quaternary structure assembly. Chromatogram traces are shown for Y96Bpa and Q458Bpa, which are dimeric and hexameric, respectively.
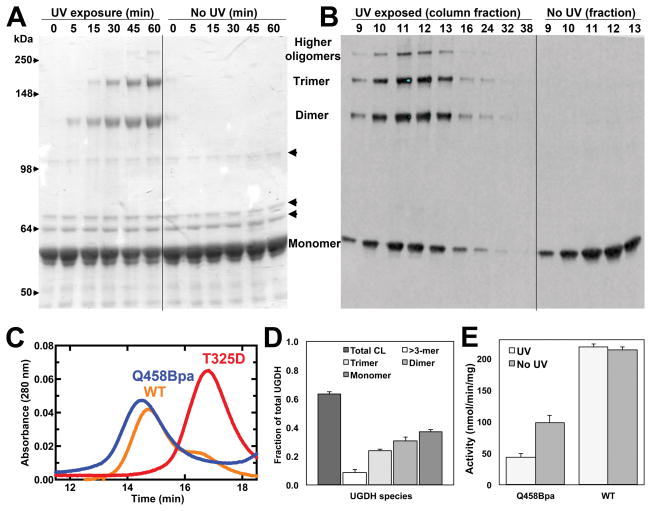
Next, we optimized conditions for efficient crosslinking, using both UGDH L433Bpa and Q458Bpa. We exposed proteins to long wavelength UV (365 nm) for periods of 5 min to 4 h, sampling a pH range from 6–9 in the presence and absence of reducing agents, and then analyzed by SDS-PAGE and western blot. Crosslinking was defined by the appearance of new bands in molecular weight multiples of ≈60 kDa. UGDH L433Bpa formed covalent linkages upon UV exposure, but the crosslinked bands were only detectable by western analyses (not shown), whereas UGDH Q458Bpa crosslinking was clearly visible in a Coomassie blue stained gel (Fig 3A). Three minor non-specific protein bands were also present (indicated by arrowheads on the right side of the gel); however, these bands co-purify in small amounts with the wild-type enzyme as well and do not appear in western blots probed for UGDH. Therefore, we pursued further optimization with the Q458Bpa mutant, testing the effect of substrate and/or cofactor addition, in conjunction with a time course. In comparing UGDH apoenzyme to the holoenzyme ternary complex with UDP-glucose and NAD+, the ternary complex exhibited significantly increased crosslinking efficiency (Fig 3A). Although either UDP-glucose or NAD+ alone promoted dimer and trimer formation (not shown), subunits crosslinked most efficiently in a solution of 5 mM NAD+ and 1 mM UDP-glucose. In time trials, we found crosslinking reached a plateau after 60 min. Longer exposure did not increase the percentage of crosslinked species.
Figure 3. Initial crosslinking and enzymatic activity of UGDH Q458Bpa.
UGDH Q458Bpa apoprotein (A) or holoenzyme (H) complexes containing both UDP-glucose substrate and NAD+ cofactor were incubated in the presence or absence of UV light exposure. Crosslinked species were analyzed by SDS-PAGE to confirm the presence of higher order oligomers (A). Arrowheads on the right indicate non-specific bands that co-purify with UGDH. Fractional crosslinking was quantified by densitometry (B) and compared to the activity of the enzyme in standard assay conditions with saturating substrate and cofactor (C).
Covalent crosslinking reduces activity of UGDH
To determine if covalently binding subunits and consequently changing the oligomeric state of the enzyme in solution affects enzyme activity, we crosslinked Q458Bpa mutant subunits, semi-quantitatively estimated fractional crosslinking efficiency by gel densitometry (Fig 3B), and measured enzyme activity in this mixture. There was a clear negative correlation between crosslinking and activity, where the fraction of unaltered protein in the solution was approximately equivalent to the fraction of residual activity.
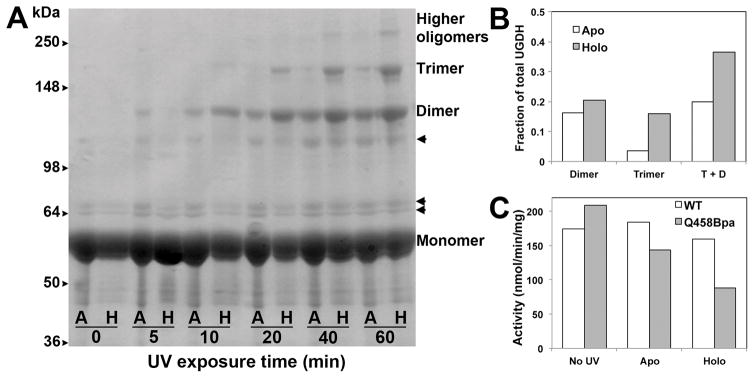
To examine this more analytically, we next purified the crosslinked oligomeric species, and compared kinetic activity of the purified crosslinked species to that of non-crosslinked Q458Bpa and wild-type UGDH. Using the optimized crosslinking conditions, we exposed Q458Bpa and wild-type UGDH to UV light for 1 h, removing aliquots at varying time points to evaluate crosslinking. A separate aliquot of UGDH Q458Bpa and wild-type UGDH was exposed to similar conditions, including the same concentrations of NAD+ and UDP-glucose, but this aliquot was not UV irradiated. The non-crosslinked UGDH Q458Bpa, and the wild-type enzyme both exposed and not exposed to UV light were used as controls to ensure that any change in kinetic activity was due to crosslinking of enzyme interfaces, and not to UV exposure or the unnatural amino acid incorporation. We dialyzed all samples and separated by size exclusion chromatography. Using dimeric (UGDH T325A13) and hexameric (UGDH wild-type) standards, we pooled fractions that corresponded to the hexamer peak, with minimal expected overlap from the dimers as indicated by the elution profiles (Fig 4C). This peak proved to have the greatest amount of oligomeric species as assessed by SDS-PAGE, western blot analysis, and Coomassie blue stain (Fig 4A and 4B). It is important to note that there was a single elution peak for UGDH Q458Bpa following crosslinking, and that even though the peak constituted the hexameric form of UGDH, crosslinked dimer, trimer, and higher oligomeric species were present on the gel as well as the monomer.
Figure 4. Crosslinked hexameric UGDH Q458Bpa is catalytically inactive.
A) Purified UGDH Q458Bpa was incubated with or without photoactivation by UV light exposure and analyzed by SDS-PAGE. B) Cross-linked UGDH Q458Bpa was fractionated by size exclusion chromatography. Arrowheads on the right of the gel and the left of the western indicate non-specific bands that co-purify with UGDH. Fractions corresponding to the elution of the single hexameric peak were analyzed by western blot probed for UGDH and compared to hexameric fractions of similarly purified non-crosslinked UGDH. C) The elution profile for crosslinked Q458Bpa (blue trace) is superimposed with sequentially collected profiles for the wild-type enzyme (hexamer-dimer, orange trace) and the previously characterized T325D obligate dimeric mutant (red). Pooled fractions 9–13 correspond to the 12–15 minute range on the elution profile. D) Fractional crosslinking was determined by densitometric analysis of the western blot in (B). D) Enzymatic activity of crosslinked and non-crosslinked UGDH Q458Bpa was compared in standard assay conditions with saturating substrate and cofactor.
We tested three separate preparations of UGDH Q458Bpa, which were similar in final concentrations and initial kinetic activities. The total crosslinking efficiency (total CL) was consistently ≈63% across Q458Bpa protein preparations (Fig 4D). As observed in the unpurified solution, the specific activity of the purified hexameric enzyme was ≈58% reduced in the crosslinked species compared to non-crosslinked Q458Bpa (Fig 4E). The wild-type control was exposed to the same conditions as the Q458Bpa mutants, but in contrast, no changes were observed in the enzymatic activities.
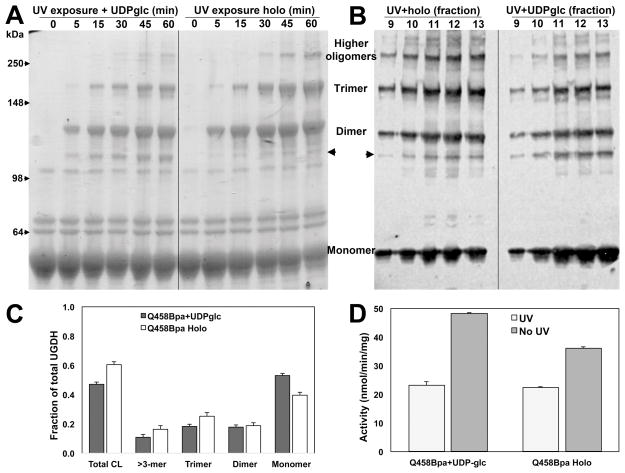
Cofactor and substrate can associate and dissociate from the crosslinked UGDH complex
To confirm that neither cofactor or substrate were covalently crosslinked and/or trapped in the active site of the UGDH Q458Bpa crosslinked enzyme, we performed photoactivation in the presence of substrate alone and compared the gel filtration purified hexamer to similarly treated holoenzyme. Time dependence of crosslinking and fractional crosslinking were analyzed by SDS-PAGE and western blot (Figure 5A and 5B). Cross-linking efficiency was reduced in the absence of NAD+ cofactor (Fig 5C), but significant loss of activity was nonetheless observed (>50%), comparable to that of the holoenzyme (Fig 5D). An additional band appeared at ≈110 kDa (indicated by the arrowheads on the right in 5A and left in 5B), which intensified with UV irradiation time and was detected by the UGDH antibody. This band was visible to an equal extent in both the UDP-glc only and the holoenzyme conditions, and was a minor contribution to the overall crosslinked fraction (<2%), so we did not consider it to have a specific impact on activity. One possibility is that this band represented a cyclized dimer, which we would also expect to lack activity. Overall, these results suggest the cofactor is not trapped by crosslinking, either covalently or non-covalently, and the loss of enzyme activity is not attributable to bound NAD+ in the complex.
Figure 5. Fractional crosslinking is less effective in the absence of NAD+ cofactor but enzymatic activity is equally diminished.
A) UGDH Q458Bpa was incubated in the presence or absence of UV exposure either in its binary complex with UDP-glucose substrate alone or as a ternary complex with UDP-glucose and NAD+. Aliquots were removed at the indicated times for analysis by SDS-PAGE. Arrowheads indicate a new band resulting from crosslinking that does not directly align with the predicted dimer. B) Crosslinked UGDH Q458Bpa binary and ternary complexes were fractionated by size exclusion chromatography. Fractions corresponding to the elution of each single hexameric peak were compared by western blot probed for UGDH. C) Fractional crosslinking was determined by densitometric analysis of the western blot in (B). D) Fractions were pooled and dialyzed prior to comparison of enzymatic activity of crosslinked binary and ternary UGDH Q458Bpa complexes. Standard assay conditions with saturating substrate and cofactor were used.
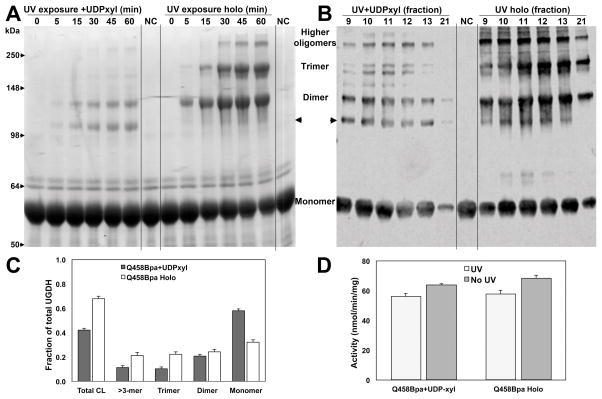
Crosslinking UGDH in an alternative conformation precluding closure of its interfaces preserves activity
Finally, we tested the effect of cross-linking in the presence of UDP-xylose, which is a competitive inhibitor of UGDH that binds tightly at the UDP-glucose binding site and induces a structural shift in the conformation of the hexamer 16. As observed for UDP-glucose alone, cross-linking efficiency in the presence of UDP-xylose alone was only ≈40% relative to >60% for the holoenzyme (Fig 6A and 6B). Similarly, the additional band at ≈110 kDa was also present (indicated by arrowheads on the right of the gel in 6A and the left in 6B), but to a larger extent in reactions with UDP-xylose alone, where it represented approximately half of the total crosslinked species. Following size exclusion purification and dialysis, activity of both the binary and ternary complexes crosslinked in the presence of UDP-xylose was significantly higher than that of the complexes using UDP-glucose, consistent with the trapping of an altered conformation of the UGDH hexamer by covalent crosslinking (Fig 6C and 6D). The activity of this catalytically impaired complex, as expected, was only modestly affected by crosslinking (<10% activity loss). Overall, these results suggest that the hexameric conformation of the enzyme requires significant potential for dynamic motion during catalysis to sustain maximal activity. Specifically, the conformational flexibility at the dimer-dimer interfaces is a critical component of UGDH catalytic competence.
Figure 6. Loss of activity in crosslinked UGDH Q458Bpa does not result from trapping substrate in the active site.
A) UGDH Q458Bpa was incubated with and without UV exposure either in a binary complex with UDP-xylose, a competitive inhibitor of UGDH, or as a ternary complex with UDP-xylose and NAD+. Aliquots were removed at the indicated times for analysis by SDS-PAGE. Arrowheads indicate a new band resulting from crosslinking that does not directly align with the predicted dimer. B) Cross-linked UGDH Q458Bpa binary and ternary complexes were fractionated by size exclusion chromatography. Fractions corresponding to the elution of each single hexameric peak were compared by western blot probed for UGDH. C) Fractional crosslinking was determined by densitometric analysis of the western blot in (B). D) Fractions were pooled and dialyzed prior to comparison of enzymatic activity of crosslinked binary and ternary UGDH Q458Bpa complexes. Standard assay conditions with saturating substrate and cofactor were used.
Discussion
Human UGDH spontaneously assembles and functions in a homohexamer, which is stabilized by the presence of its substrate and cofactor. Previous mutagenesis studies have found that activity is reduced by preventing hexamer formation, but can also be nearly eliminated by a point deletion that prevents hexamer dissociation 13–16. Thus, the functional state of UGDH higher oligomeric assemblies has not been explicitly defined. In the current study, our goal was to address the requirement for hexamer disassembly by trapping the enzyme in an obligatory hexameric conformation while minimally impairing its stability or intrinsic activity. We chose to use photoreactive unnatural amino acid incorporation so we could experimentally control the location and the timing of stable oligomeric crosslinking. This is a unique approach and an application for unnatural amino acid substitution that has not been previously reported. Though several of the sites we attempted to substitute were not predicted to perturb UGDH structural or functional properties to a significant extent, all but the UGDH Q458Bpa mutant formed unstable or inactive species that were not suitable for quantitative analysis. However, UGDH Q458Bpa adopted the hexamer conformation and retained 60–80% of wild-type activity in the absence of UV exposure, while crosslinking efficiently and specifically into the expected dimer and trimer when exposed to UV for 1 h. Covalently linked oligomeric species exhibited significantly reduced catalytic activity relative to the uncrosslinked, crosslinked apoenzyme, and wildtype UGDH controls, consistent with a critical role for both dimer-dimer association and dissociation in UGDH kinetic activity. This apparent need for conformational flexibility at the dimer-dimer interface likely confers allosteric regulatory control in vivo.
The addition of substrate and cofactor increased the efficiency of UGDH crosslinking. We expected this outcome on the basis of previous studies showing that the hexamer predominates in solutions that contain saturating substrate and cofactor 4, 13, 16. Moreover, comparison of the crystal structures of the UGDH apoenzyme and the ternary complex suggests the potential for more extensive contacts at the dimer-dimer interface in the vicinity of the Q458 substitution 10. The time course for crosslinking is consistent with those of previous studies using Bpa as a photocrosslinker in other systems 17, 21, 22. Though we achieved a significant fraction of covalent linkage between subunits, sufficient to purify and proceed with activity measurements, we were not able to achieve complete crosslinking. This may be a physical consequence of hysteresis in the homohexameric species, which results from the stable adoption of an altered conformation in the product inhibited state that could preclude sufficient contact for crosslinking at every interface of a single complex 15, 23. Additional support for this interpretation comes from the analysis of the crosslinking reactions with UDP-xylose added, in which new species of crosslinked proteins are observed that suggest an altered conformation. The hysteretic conformation of UGDH has significantly greater apparent affinity for UDP-xylose (evidenced as a lower Ki), and the crystal structure of UGDH with bound UDP-xylose reveals different exposure and solvent accessibility of interface contacts 15, 23.
Size exclusion chromatography was used to separate the hexameric fraction of UGDH Q458Bpa following crosslinking, so we could determine the resultant subunit composition and the specific activity associated with the conformational restraint. In addition to crosslinked trimer species, we also found molecular masses corresponding to dimer, monomer, and minor amounts of higher oligomeric species. The non-hexameric species present in the hexameric fractions probably indicate a mixture of associated crosslinked and non-crosslinked subunits in the hexameric forms, which then dissociate to yield the observed components. We saw ≈55% average decrease in catalytic activity among the three replicates, while crosslinking efficiency was greater than 60%. The difference in decreased activity versus crosslinking efficiency may again stem from the possibility of higher oligomeric species forming between both crosslinked and non-crosslinked subunits. For example, in a crosslinked trimer, three non-crosslinked monomers could potentially associate with the crosslinked trimer to form a hexameric structure that may retain sufficient flexibility to support catalytic activity.
An obvious concern about reduced activity upon crosslinking is that the additional components of the mixture, in this case NAD+ and UDP-glucose or the NADH and UDP-glucuronate products, could be covalently linked to the enzyme or structurally entrapped within the overall crosslinked hexamer as a consequence of reduced active site flexibility. We addressed this by crosslinking the enzyme in the presence of UDP-xylose, which is a competitive inhibitor of UGDH that binds at the UDP-glucose site. Formation of binary and ternary complexes is associated with a quaternary structural change that leaves several of the active sites more open to solvent and conformationally dynamic on an individual subunit level than the UDP-glucose binary and ternary complexes. These differences were demonstrated effectively by crystallographic and sedimentation velocity experiments 15, 23. When we UV-irradiated UGDH Q458Bpa with saturating amounts of UDP-xylose, purified the hexameric species, and dialyzed any residual inhibitor, the enzyme was crosslinked to an equivalent extent but retained comparable activity to the uncrosslinked control. The crosslinked bands were similar in fractional composition to those of the UDP-glucose and the ternary complexes, with the addition of a new band we speculate is the cyclized dimer.
The above results distinctly show that the substrate and cofactor are not locked into the complex by crosslinking but do not preclude the possibility that some portion of the enzyme is locked into an unproductive conformation that is not a hexamer and is still capable of associating in hexamers. Though one additional possibility for inactive subunits could be intrasubunit crosslinks, the placement of the unnatural amino acid substitution makes such crosslinks unlikely without significant conformational changes that we have typically seen associated with increased sensitivity to limited proteolysis. Moreover, intrasubunit crosslinking would be expected to produce a shifted monomer band in the western analysis, for which we saw no evidence. In contrast, we did observe such bands in analysis of the UGDH Y96AzF mutant (not shown). We also expect that intrasubunit crosslinking would be incompatible with hexamer formation and that such subunits would therefore not be represented among those present in the hexameric fractions isolated from the gel filtration column prior to analysis.
In summary, it appears that dynamic oligomeric structure has a significant role in hUGDH activity and subsequent formation of UDP-glucuronate. Understanding the relevance of oligomeric structure in UGDH activity may have a potential role in modulating UGDH activity for targeted intervention in prostate cancer. Due to the apparent implications of UGDH in pathogenesis of prostate cancer, further investigation of quaternary state modification may prove worthwhile in developing a more thorough understanding of UGDH function in disease.
Acknowledgments
Funding information:
This work was supported by NIH R01 CA165574 (MAS) and NIH R21 CA185993 (MAS, JJB, and JG).
ABBREVIATIONS
- UGDH
UDP-glucose dehydrogenase
- UDP
uridine-5′-diphosphate
- UDP-glc
UDP-glucose
- UDP-glcA
UDP-glucuronate
- NAD
nicotinamide adenine dinucleotide
- SDS-PAGE
SDS-polyacrylamide gel electrophoresis
- IPTG
isopropyl-β-D-thiogalactopyranoside
- DTT
dithiothreitol
- FPLC
fast performance liquid chromatography
- PDB
protein database
- AzF
p-azido-L-phenylalanine
- Bpa
p-benzoyl-L-phenylalanine
References
- 1.Roden L, Koerner T, Olson C, Schwartz NB. Mechanisms of chain initiation in the biosynthesis of connective tissue polysaccharides. Federation proceedings. 1985;44:373–380. [PubMed] [Google Scholar]
- 2.Tukey RH, Strassburg CP. Human UDP-glucuronosyltransferases: metabolism, expression, and disease. Annu Rev Pharmacol Toxicol. 2000;40:581–616. doi: 10.1146/annurev.pharmtox.40.1.581. [DOI] [PubMed] [Google Scholar]
- 3.Spicer AP, Kaback LA, Smith TJ, Seldin MF. Molecular cloning and characterization of the human and mouse UDP-glucose dehydrogenase genes. J Biol Chem. 1998;273:25117–25124. doi: 10.1074/jbc.273.39.25117. [DOI] [PubMed] [Google Scholar]
- 4.Hyde AS, Farmer EL, Easley KE, van Lammeren K, Christoffels VM, Barycki JJ, Bakkers J, Simpson MA. UDP-glucose dehydrogenase polymorphisms from patients with congenital heart valve defects disrupt enzyme stability and quaternary assembly. J Biol Chem. 2012;287:32708–32716. doi: 10.1074/jbc.M112.395202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell RE, Sala RF, van de Rijn I, Tanner ME. Properties and kinetic analysis of UDP-glucose dehydrogenase from group A streptococci. Irreversible inhibition by UDP-chloroacetol. J Biol Chem. 1997;272:3416–3422. doi: 10.1074/jbc.272.6.3416. [DOI] [PubMed] [Google Scholar]
- 6.Nelsestuen GL, Kirkwood S. The mechanism of action of uridine diphosphoglucose dehydrogenase. Uridine diphosphohexodialdoses as intermediates. J Biol Chem. 1971;246:3824–3834. [PubMed] [Google Scholar]
- 7.Schiller JG, Bowser AM, Feingold DS. Studies on the mechanism of action of udp-D-glucose dehydrogenase from beef liver II. Carbohydr Res. 1972;25:403–410. doi: 10.1016/s0008-6215(00)81651-1. [DOI] [PubMed] [Google Scholar]
- 8.Stewart DC, Copeland L. Uridine 5′-Diphosphate-Glucose Dehydrogenase from Soybean Nodules. Plant Physiol. 1998;116:349–355. [Google Scholar]
- 9.Campbell RE, Mosimann SC, van De Rijn I, Tanner ME, Strynadka NC. The first structure of UDP-glucose dehydrogenase reveals the catalytic residues necessary for the two-fold oxidation. Biochemistry. 2000;39:7012–7023. [PubMed] [Google Scholar]
- 10.Egger S, Chaikuad A, Kavanagh KL, Oppermann U, Nidetzky B. Structure and mechanism of human UDP-glucose 6-dehydrogenase. J Biol Chem. 2011;286:23877–23887. doi: 10.1074/jbc.M111.234682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Naught LE, Gilbert S, Imhoff R, Snook C, Beamer L, Tipton P. Allosterism and cooperativity in Pseudomonas aeruginosa GDP-mannose dehydrogenase. Biochemistry. 2002;41:9637–9645. doi: 10.1021/bi025862m. [DOI] [PubMed] [Google Scholar]
- 12.Snook CF, Tipton PA, Beamer LJ. Crystal structure of GDP-mannose dehydrogenase: a key enzyme of alginate biosynthesis in P aeruginosa. Biochemistry. 2003;42:4658–4668. doi: 10.1021/bi027328k. [DOI] [PubMed] [Google Scholar]
- 13.Hyde AS, Thelen AM, Barycki JJ, Simpson MA. UDP-glucose dehydrogenase activity and optimal downstream cellular function require dynamic reorganization at the dimer-dimer subunit interfaces. J Biol Chem. 2013;288:35049–35057. doi: 10.1074/jbc.M113.519090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sennett NC, Kadirvelraj R, Wood ZA. Conformational flexibility in the allosteric regulation of human UDP-alpha-D-glucose 6-dehydrogenase. Biochemistry. 2011;50:9651–9663. doi: 10.1021/bi201381e. [DOI] [PubMed] [Google Scholar]
- 15.Kadirvelraj R, Sennett NC, Custer GS, Phillips RS, Wood ZA. Hysteresis and negative cooperativity in human UDP-glucose dehydrogenase. Biochemistry. 2013;52:1456–1465. doi: 10.1021/bi301593c. [DOI] [PubMed] [Google Scholar]
- 16.Sennett NC, Kadirvelraj R, Wood ZA. Cofactor binding triggers a molecular switch to allosterically activate human UDP-alpha-D-glucose 6-dehydrogenase. Biochemistry. 2012;51:9364–9374. doi: 10.1021/bi301067w. [DOI] [PubMed] [Google Scholar]
- 17.Chin JW, Martin AB, King DS, Wang L, Schultz PG. Addition of a photocrosslinking amino acid to the genetic code of Escherichiacoli. Proc Natl Acad Sci U S A. 2002;99:11020–11024. doi: 10.1073/pnas.172226299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chin JW, Santoro SW, Martin AB, King DS, Wang L, Schultz PG. Addition of p-azido-L-phenylalanine to the genetic code of Escherichia coli. J Am Chem Soc. 2002;124:9026–9027. doi: 10.1021/ja027007w. [DOI] [PubMed] [Google Scholar]
- 19.Liu CC, Mack AV, Brustad EM, Mills JH, Groff D, Smider VV, Schultz PG. Evolution of proteins with genetically encoded “chemical warheads”. J Am Chem Soc. 2009;131:9616–9617. doi: 10.1021/ja902985e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. Journal of computational chemistry. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 21.Young TS, Ahmad I, Yin JA, Schultz PG. An enhanced system for unnatural amino acid mutagenesis in E coli. J Mol Biol. 2010;395:361–374. doi: 10.1016/j.jmb.2009.10.030. [DOI] [PubMed] [Google Scholar]
- 22.Farrell IS, Toroney R, Hazen JL, Mehl RA, Chin JW. Photo-cross-linking interacting proteins with a genetically encoded benzophenone. Nature methods. 2005;2:377–384. doi: 10.1038/nmeth0505-377. [DOI] [PubMed] [Google Scholar]
- 23.Kadirvelraj R, Custer GS, Keul ND, Sennett NC, Sidlo AM, Walsh RM, Jr, Wood ZA. Hysteresis in human UDP-glucose dehydrogenase is due to a restrained hexameric structure that favors feedback inhibition. Biochemistry. 2014;53:8043–8051. doi: 10.1021/bi500594x. [DOI] [PubMed] [Google Scholar]