Abstract
Dyneins are large microtubule motor proteins that convert ATP energy to mechanical power. High-resolution crystal structures of ADP-bound cytoplasmic dynein have revealed the organization of the motor domain, comprising the AAA+ ring, the linker, the stalk/strut and the C sequence. Recently, the ADP.vanadate-bound structure, which is similar to the ATP hydrolysis transition state, revealed how the structure of dynein changes upon ATP binding. Although both the ADP- and ATP-bound state structures have been resolved, the dynamic properties at the atomic level remain unclear. In this work, we built two models named ‘the ADP model’ and ‘the ATP model’, where ADP and ATP are bound to AAA1 in the AAA+ ring, respectively, to observe the initial procedure of the structural change from the unprimed to the primed state. We performed 200-ns molecular dynamics simulations for both models and compared their structures and dynamics. The motions of the stalk, consisting of a long coiled coil with a microtubule-binding domain, significantly differed between the two models. The elastic properties of the stalk were analyzed and compared with the experimental results.
Keywords: ADP and ATP, dynein, elastic properties, molecular dynamics simulation
Introduction
Dyneins are large microtubule motor proteins that play important roles in various biological movements along microtubules (Gibbons and Rowe, 1965; Paschal and Vallee, 1987). Their motor activity is generated by the energy conversion gained from ATP hydrolysis (Neuwald et al., 1999). Cytoplasmic dynein is responsible for cell division, cell migration and other basic cellular functions (Karki and Holzbaur, 1999). Axonemal dynein plays a role in the motion of cilia and flagella (Gibbons, 1981). Dyneins exist as a complex of heavy, intermediate and light chains. The core part of the heavy chain that performs the major functions for motor activities is called the motor domain. The motor domain contains an N-terminal linker, central ring-shaped AAA+ modules (AAA1–AAA6) (Neuwald et al., 1999) and a stalk extending from the AAA+ ring. The linker is considered to function as a mechanical element for generating the power stroke (Burgess et al., 2003). The three AAA+ modules, AAA1, AAA3 and AAA4, perform the ATP hydrolysis (Kon et al., 2012). The stalk consists of a long coiled coil with an ATP-dependent microtubule-binding domain (MTBD) at its tip (Gee et al., 1997).
Electron microscopy and FRET studies have clarified the existence of primed (pre-power stroke) and unprimed (post-power stroke) states, depending on the nucleotide state in the AAA1 module (Burgess et al., 2003; Kon et al., 2005; Roberts et al., 2009). The nucleotide state changes following the so-called mechanochemical cycle, from the apo-state to the ATP-bound state, ADP.Pi state, and ADP-bound state, and finally returns to the original apo-state. The apo- and ADP-bound states correspond to the unprimed state, while the ATP-bound and ADP.Pi states are the primed state. The linker moves during the mechanochemical cycle, and its tip is close to AAA4 and AAA2 in the unprimed and primed states, respectively.
X-ray crystallographic studies have revealed the structure of the stalk including the MTBD (Carter et al., 2008; Nishikawa et al., 2014) and the organization of the AAA+ ring, the linker, the stalk, the strut (or buttress) and the C sequence in the motor domain (Carter et al., 2011; Kon et al., 2011, 2012; Schmidt et al., 2012). The AAA+ ring is formed by the tandem linkage of the AAA1–AAA6 modules, from the N-terminal. The stalk and the strut, which both have a coiled-coil structural motif, extend from AAA4 and AAA5, respectively. The strut contacts the lower basis part of the stalk. The C sequence is linked to AAA6 at the C-terminal end. High-resolution structures of the motor domain of wild-type cytoplasmic dynein and its truncated MTBD mutant (ΔMTBD), which are both in the unprimed states, revealed the interface between the AAA+ ring and the linker, and the junctional structures between the ring and the stalk (Kon et al., 2012). The nucleotide binding sites contain four ADP molecules in AAA1–AAA4, while no nucleotides are bound to AAA5 and AAA6. A mutational analysis demonstrated that the nucleotide binding site of AAA1 is essential for the dynein motility (Kon et al., 2004). Recently, a high-resolution structure of dynein in the primed state, with ADP.vanadate being bound to AAA1 to mimic the ATP hydrolysis transition state, was reported (Schmidt et al., 2015). The structures of the linker and the stalk are quite different between the primed and unprimed states. In the primed state, one of the α-helices of the stalk contains a kink, resulting in the sliding of the kinked helix relative to the other helix. Thus, while the linker forms a straight structure in the unprimed state, it is bent in the primed state. Although the structures of the two states have been resolved, the dynamic properties of the states at the atomic level remain unclear.
We previously introduced the zero-dipole summation (ZD) method (Fukuda et al., 2011) to treat electrostatic interactions for molecular dynamics (MD) simulation. The ZD method takes into account the neutralities of charges (zero-monopole) and dipoles (zero-dipole) in a truncated subset. Recently, the ZD method has been expanded to the zero-multipole method (Fukuda, 2013; Fukuda et al., 2014; Wang et al., 2016). The ZD method has reproduced structures and dynamics properties similar to those obtained by the particle mesh Ewald method (Essmann et al., 1995), which is one of the most accurate methods, in a pure TIP3P water system (Fukuda et al., 2012), a membrane protein system (Kamiya et al., 2013) and a DNA–ion–water system (Arakawa et al., 2013). We have performed MD simulations on the stalk fragment of the dynein motor domain using the ZD method, which revealed the function-related motion of the stalk (Nishikawa et al., 2014). Since the ZD method is free from the evaluation of the reciprocal Fourier part required in the particle mesh Ewald method, an order N scheme is implied for a large system by computing only local interactions. In fact, by using the ZD method, effective scalability has been attained from the EGFR system consisting of ∼40 000 atoms to the dynein motor domain system consisting of one million atoms (Mashimo et al., 2013).
In this work, we built two models, named the ADP and ATP models, where ADP and ATP ligands are bound to AAA1, respectively. As the ADP model, the X-ray crystal structure of the dynein from Dictyostelium discoideum was used, in which four ADP molecules are bound to AAA1–AAA4. In the ATP model, the ADP molecule in AAA1, which is essential for dynein function, was substituted with ATP. We executed an MD simulation starting from the dynein structure of the unprimed state, to investigate the dynamic properties of both models. The computational system consists of the dynein motor domain, nucleotides, explicit water molecules and ions (size of the systems: approximately one million atoms). To execute the MD simulation of such a large system, we used our program, psygene-G (Mashimo et al., 2013), which utilizes GPGPUs for acceleration of the non-bonded interactions, including the electrostatic computation by the ZD method. We performed 200-ns MD simulations for both the ADP and ATP models, and compared their structures and dynamics. The motions of the stalks in the two models were quite different. The elastic properties of the stalks were analyzed and compared with the experimental results.
Materials and methods
Computational system
Our target, cytoplasmic dynein, exists as a homodimer in vivo, but the monomer is the minimal functional unit (Roberts et al., 2013; Schiavo et al., 2013). We built a system consisting of a dynein monomer to reduce the computational costs. Two different models were prepared for MD simulations.
The ADP model, in which all four AAA+ modules, AAA1–AAA4, bind ADPs, was built from the high-resolution structures of D. discoideum dynein (Kon et al., 2012). The wild-type and ΔMTBD mutant structures (PDB IDs: 3 vkh and 3 vkg, respectively) were obtained from the Protein Data Bank (Berman et al., 2007). Since the resolution of the ΔMTBD mutant structure (2.8 Å) is higher than that of the wild-type structure (3.8 Å) and the ΔMTBD mutant retains the ATPase activity, the structure of the ΔMTBD mutant including nucleotides was chosen as the initial model for the current MD simulations. The missing MTBD domain, residues Ala3372–Lys3495, was modeled from the wild-type structure. In order to include the effects of the missing charged residues, we built the atomic models for the other residues with electron densities that were not observed in the crystal structures, listed in Table I, by the spanner modeling server (Lis et al., 2011) (http://sysimm.ifrec.osaka-u.ac.jp/spanner/). The N- and C-termini were capped by acetyl and N-methyl groups, respectively. After the modeling, the system consists of 3320 residues (Glu1410–Asp4729), four ADP molecules and one magnesium ion (Fig. 1A). A rectangular water box with dimensions of 167 × 333 × 180 Å3 was placed around dynein. Sodium and chloride ions were added to neutralize the system and to achieve a physiological concentration (100 mM). The final system consists of 980 265 atoms (53 443 dynein atoms, 4 ADP molecules, 1 magnesium ion, 574 sodium ions, 528 chloride ions and 308 521 water molecules).
Table I.
Domain names and residue numbers of D. discoideum dynein
| Domain name | Residue number (initial) | Residue number (final) | Missing residuesa |
|---|---|---|---|
| Linker | 1410 | 1935 | 1425–1449: 1620–1624 |
| AAA1 | 1936 | 2229 | 2061–2063: 2170–2175 |
| AAA2 | 2230 | 2631 | 2454–2487: 2631 |
| AAA3 | 2632 | 2948 | 2632–2633 |
| AAA4 (N-terminal end) | 2949 | 3262 | 2958–2961: 2981–2986: 3042–3045: 3157–3160: 3212–3215 |
| Stalk (N-terminal helix) | 3263 | 3371 | |
| Stalk (MTBD) | 3372 | 3486 | |
| Stalk (C-terminal helix) | 3487 | 3596 | 3492–3495 |
| AAA4 (C-terminal end) | 3597 | 3638 | |
| AAA5 (N-terminal end) | 3639 | 3806 | 3699–3703: 3725–3758 |
| Strut | 3807 | 3885 | |
| AAA5 (C-terminal end) | 3886 | 4114 | 4114 |
| AAA6 | 4115 | 4411 | 4115 |
| C-sequence | 4412 | 4729 | 4440–4493: 4522–4550: 4582–4588: 4614–4634: 4674–4676 |
aMissing residues of each domain in the crystal structure are shown.
Fig. 1.
(A) Structure of dynein motor domain. The linker, AAA1–AAA6 and C-sequence are colored purple, blue, cyan, green, yellow, orange, red and black, respectively. Domain names are shown in the figure. (B) Definitions of L and r for the calculation of the elastic properties of the stalk. The principal axis of the moment of inertia of the initial structure of MD is depicted by the solid line with arrow. Domain names of the coiled coil (CC1 and CC2), the MTBD and residue numbers are shown in the figure.
The ATP model, in which ATP is bound to AAA1 and the structure of dynein is exactly the same as the ADP model, was built from the ADP model by adding the PO3 group to the terminal oxygen atom of the ADP molecule. Then, the chloride ion and water molecule nearest to the PO3 group of the ATP were removed. In total, 980 265 atoms (53 443 dynein atoms, 1 ATP molecule, 3 ADP molecules, 1 magnesium ion, 574 sodium ions, 527 chloride ions and 308 520 water molecules) were in the rectangular box.
The force field parameters for dynein, the other ions and water molecules were derived from our AMBER-based hybrid force field (Kamiya et al., 2005), the halide monovalent ion parameters (Joung and Cheatham, 2008) and the flexible TIP3P water model (Jorgensen et al., 1983), respectively, and the parameters for the nucleotides and the magnesium ion originated from the General AMBER Force Field (Wang et al., 2004). The partial atomic charges of ADP and ATP were taken from the AMBER parameter database (http://www.pharmacy.manchester.ac.uk/bryce/amber).
MD simulations
All of the energy minimization calculations and MD simulations were performed by the psygene-G program (Mashimo et al., 2013). The simulation protocols for the ADP and ATP models were the same. Conjugated gradient energy minimization was executed with positional restraints on the heavy atoms of dynein, nucleotides and the magnesium ion, using a force constant of 10 kcal/mol Å2, followed by minimization without these restraints. The system was equilibrated for 100 ps by adopting Berendsen's NPT algorithm (Berendsen et al., 1984), with the temperature set at 300 K, pressure under 1 bar and a time step of 0.5 fs. Here, the positions of the heavy atoms of dynein, nucleotides and the magnesium ion were restrained with a force constant of 10 kcal/mol Å2. Electrostatic interactions were treated by the ZD method, with a cutoff distance of 12 Å and a damping factor α of 0.00 Å−1. The cutoff distance of the van der Waals interactions was set to 12 Å. After the NPT simulations, the cell sizes of the ADP and ATP models were 166.2 × 329.4 × 176.1 and 165.9 × 333.7 × 175.0 Å3, respectively. An NVT heating run from 0 to 300 K for 1 ns was performed with a time step of 1.0 fs. The SHAKE algorithm (Ryckaert et al., 1977) was applied to constrain the covalent bonds between heavy and hydrogen atoms. Finally, the NVT production run at 300 K was performed for 200 ns with SHAKE and a time step of 1.0 fs for each model. The electrostatic and van der Waals treatments were the same as those of the NPT simulation. The structure trajectory was stored every 10 000 MD steps (10 ps). All of the productive runs were performed using the HA-PACS supercomputer system at Tsukuba University (http://www.ccs.tsukuba.ac.jp/eng/research-activities/projects/ha-pacs/). It took ∼12 h to execute a 1-ns MD simulation in parallel with 64 CPUs and 64 GPGPUs.
Analysis of elastic properties
The stalk of dynein consists of a long coiled coil (∼150 Å) and the MTBD. In the coiled coil, two α-helices are aligned with anti-parallel geometry (see Fig. 1B). The elastic properties of the coiled coil were analyzed by the same method used for the analysis of myosin II (Adamovic et al., 2008). Here, the method is briefly summarized below. The elastic properties of the coiled coil are divided into the stretching stiffness (i.e. spring constant) (ks) and the bending stiffness (kb), which are given by the following equations:
| (1) |
| (2) |
Here, k, T, ΔL, Δr and < > are the Boltzmann constant, temperature (= 300 K), fluctuations of the stalk in the parallel (stretching) direction, those in the perpendicular (bending) direction and the time average over the MD trajectory, respectively.
In the current study, the length of L was calculated as the average distance between selected pairs of Cα atoms (Val3267–Gln3358, Ile3271–Glu3354, Leu3504–Val3593 and Ala3508–Val3589 in Fig. 1B), which are close to the N- and C-termini of the coiled coil. The first principal axis of the moment of inertia for the stalk (solid line with arrow in Fig. 1B) was calculated from the X-ray structure (Kon et al., 2012). The atomic structures obtained during the MD trajectory were superimposed on those of the X-ray structure, and all of the heavy atoms other than the stalk were used for the superimposition. The value r was then calculated as the minimum distance from selected Cα atoms to the principal axis, using the Cα atoms of Glu3354, Gln3358, Leu3504 and Ala3508 close to the MTBD (Fig. 1B).
Since there is currently no experimental data available on the stretching stiffness of the stalk, we calculated the apparent stretching modulus (E), for comparison with that of another protein (i.e. myosin II), according to the following equation:
| (3) |
Here, <L> is the average distance over the MD trajectory.
Fortunately, the bending stiffness of the stalk has been obtained from the analysis of the electron micrograph data (Burgess et al., 2003) and is 0.14 pN/nm in the ADP–vanadate configuration, and 0.47 pN/nm in the apo-state (Lindemann and Hunt, 2003). Here, the ADP–vanadate state is a model for the ADP.Pi state, which corresponds to the primed state, while the apo-state corresponds to the unprimed state. Therefore, it is possible for us to compare the bending stiffness of the ATP and ADP models from our simulation data with those of the ADP–vanadate and apo-states from the experimental data, respectively.
Results and discussion
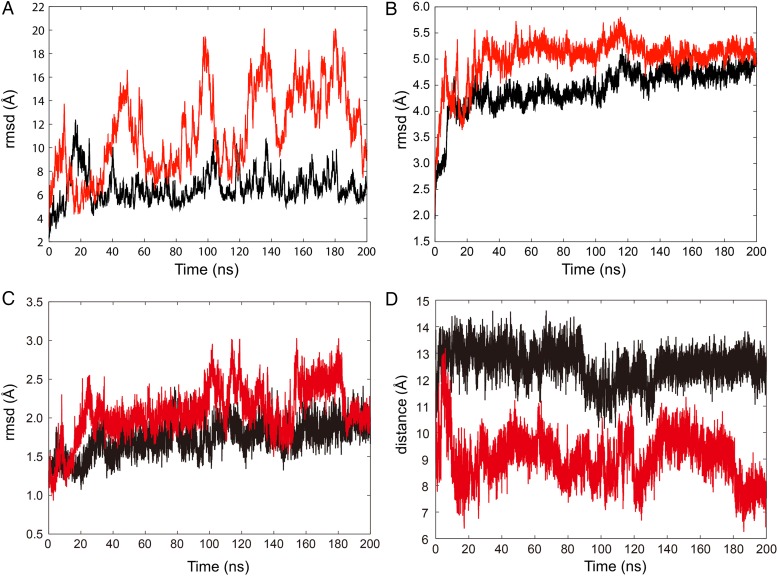
To compare the thermodynamic stability between the ADP and ATP models, we analyzed the root-mean-square deviations (rmsds) of the backbone atoms along the 200-ns MD trajectory (Fig. 2). The rmsds of all backbone atoms for both models are shown in Fig. 2A. The rmsd of the ATP model fluctuated much more broadly between 4 and 20 Å than that of the ADP model. In contrast, the trajectories of the rmsds of the backbone atoms other than the stalk were similar in both models, where each of them reached a plateau within 40 ns, followed by fluctuations between 4.0 and 5.0 Å in the ADP model and 4.5 and 5.5 Å in the ATP model (Fig. 2B). The result suggests that the stalk in the ATP model fluctuated broadly, even after the system was equilibrated. The dynamics of the AAA1 module itself were similar in the both ADP and ATP models from Fig. 2C. However, the distances between the phosphate group of ATP bound to the AAA1 module and Arg2410 in the AAA2 module were much shorter in the ATP model than the corresponding distances in the unprimed ADP-bound model along the MD trajectories (Fig. 2D).
Fig. 2.
The rmsds of (A) all backbone atoms and (B) backbone atoms other than the stalk, and (C) backbone atoms in AAA1 along the MD trajectory. (D) The distance between Pβ of ADP or ATP in AAA1 and Cζ of Arg2410 in AAA2 along the MD trajectory. Black and red lines represent the rmsd or the distance of the ADP and ATP models, respectively.
Next, to determine which residues moved during the initial stage of the trajectory (i.e. <40 ns), the rmsd of each residue with respect to the initial structure was calculated. In the AAA2 module, the modeled residues, which form the loop structure as shown in the missing residues column in Table I (i.e. 2454–2487), were flexible in both models (Fig. 3). The stalk of the ATP model moved in a broader conformational space than that of the ADP model at the beginning (<10 ns). The linker, strut and C-sequence of the ATP model began to deviate from the X-ray structure at 2, 27 and 8 ns, respectively (Fig. 3B), while those of the ADP model remained stable with similar structures to the initial ones (Fig. 3A). The magnitudes of these movements in the ATP model were, however, limited to the local area in comparison with those of the stalk.
Fig. 3.
The rmsd of each residue in (A) the ADP model and (B) the ATP model along the initial stage of the trajectory. Domain names are shown with the colored bar. The rmsd value corresponds to the color in the contour bar.
The snapshot structures during the MD simulations, taken every 40 ns, are depicted in Fig. 4. While the stalk and the MTBD underwent significantly large motions in random directions in the ATP model, they slightly fluctuated around the initial structure in the ADP model. In the ATP model, the boundary region between the stalk and the MTBD, which formed a helical structure before the MD, adopted disordered conformations during the MD. Therefore, the ATP in AAA1 appeared to contribute to the large movement of the stalk in the ATP model. No other significant structural changes occurred in either model, and no dissociation of the nucleotides was observed. In order to confirm these characteristic dynamic properties, we performed further MD simulations, in which the ADP model was switched to the ATP model and vice versa, at the middle of the MD trajectory. As shown in Supplementary Fig. S1A and B, the dynamic properties changed from the ATP to the ADP model, and the ADP to the ATP model, respectively. It is evident that although the dynamic feature did not change after 44 ns switching from the ATP to the ADP model in Supplementary Fig. S1A, it dramatically changed in Supplementary Fig. S1B after switching from the ADP to the ATP model at 40 ns. Namely, the current study could simulate the initial procedure from the unprimed to the primed state in a reproducible manner.
Fig. 4.
Structures during MD taken every 40 ns from (A) the ADP model and (B) the ATP model. Structures at 40, 80, 120, 160 and 200 ns are colored blue, green, cyan, red and purple, respectively. The backbone atoms other than the stalk are superimposed with respect to the initial structure.
In Fig. 5, the cross-correlation matrix elements are shown in the fluctuations of the unprimed ADP model (upper triangle) and those in the ATP model (lower triangle). Because the amplitudes of the structural fluctuation of the ATP model are much larger than those of the ADP model as shown in Figs 2–4, the absolute values of the cross-correlation matrix elements of the ATP model are also significantly larger than those of the ADP model. In addition, when the matrix elements between the two models are precisely compared, the following strong correlations are found:
Strong negative correlations between N-terminal linker region and AAA1, and between the linker and C-sequence plus strut, as indicated by dotted circles a and b, respectively.
Positive correlation between AAA1 and AAA2 by c.
Strong negative and positive correlations between stalk (CC1, CC2 and MTBD) and both AAA2 and AAA3 by d.
Strong negative correlation between strut and both AAA2 and AAA3 by e.
Negative correlation between strut and stalk by f.
Positive correlations between C-sequence and AAA1 plus AAA2 by g.
Strong positive correlation between C-sequence and strut by h.
Fig. 5.
The cross-correlation matrix elements in the structural fluctuations of the ADP model (upper triangle) and those in the ATP model (lower triangle) during 200 ns MD simulations. Red and blue colors indicate positive and negative correlations, respectively, as indicated in the right bar. The dotted circles are area, where the ATP model gave different dynamic features from those in the ADP model (see text).
These correlations seem to well support the mechanism underlying the motor actions of dynein proposed by Kon et al. (2012).
In fact, the correlation identified by a dotted circle c in Fig. 5 could correspond to the interaction between the bound ATP to AAA1 and the Arg2410 in AAA2 (see Fig. 2D). In order to dissect the interaction between the bound nucleotide, ADP or ATP, and the side chain of Arg2410 in AAA2, the shortest distances between the atom pairs of a negatively charged phosphate oxygen atom in ADP or ATP and a positively charged hydrogen atom in the guanidinium group of Arg2410 side chain are plotted during the MD simulations in Supplementary Fig. S2A. In the ADP model, the shortest distances between an ADP phosphate oxygen and the side chain of Arg2410 along the MD trajectory were large, separating the pairs far around 8–10 Å as shown in a snapshot at 200 ns (Supplementary Fig. S2B). On the contrary, in the ATP model, the shortest distances became very close, sometimes forming a salt bridge between the oxygen atom of ATP γ-phosphate and the Hη atom of Arg2410, as in a snapshot near 200 ns (Supplementary Fig. S2D). This salt bridge is similar to what was observed for the arginine finger of AAA2L of human cytoplasmic dynein-2 in the primed state with the bound ADP.vanadate to AAA1 (Schmidt et al., 2015). However, the current salt bridge is not permanent, but it fluctuates along the MD simulation shown by red lines in Supplementary Fig. S2A. In fact, in a snapshot at 150 ns (Supplementary Fig. S2C), the shortest length was too distant to form a salt bridge. This phenomenon is so characteristic observed only in the ATP model, that it may cause the larger dynamics of the ATP model than those of the stable ADP model.
The correlations d, e and f may correspond to the secondary propagation path of the structural change from AAA2 and AAA3 to stalk, strut and MTBD. The correlations g and h suggest the role of C-sequence, which relays the main structural change derived by ATP hydrolysis in AAA1 to MTBD through strut. Finally, the large negative correlations of the N-terminal linker with AAA1, AAA2, strut and C-sequence may support a model that the linker swing motion for the power stroke is induced by the ATPase activity (Kon et al., 2012).
We calculated the Φ- and Ψ-angles of the stalk along the trajectory, to analyze the structure of the stalk in detail. The Φ- and Ψ-angles are shown in Supplementary Fig. S3A and B, respectively, in the ADP model along the trajectory. Most Φ- and Ψ-angles of the N-terminal helix, called the CC1 helix, were within the α-helical values (−125° < Φ < −30°, −75° < Ψ < −10°), except for Glu3370 located near MTBD (bottom panels in Supplementary Fig. S3A and B). While the Φ- and Ψ-angles of the C-terminal helix, called the CC2 helix, deviated from the standard α-helical values in five residues (Arg3493, Lys3495, Leu3497, Arg3498 and Glu3499) near MTBD and in Thr3587 near AAA4, they were within the α-helical values in the middle region between AAA4 and MTBD (top panels in Supplementary Fig. S3A and B). These results indicate that the overall region of the CC1 helix and the middle region of the CC2 helix were sufficiently stable during the entire simulation. In contrast, the Φ- and Ψ-angles of the ATP model deviated from the standard α-helical values, not only in the terminal regions but also in the middle region (Supplementary Fig. S4A and B). The Φ- and Ψ-angles of 20 residues (Leu3278, Gly3288, Arg3311, Leu3313, Met3327, Gln3329, Gln3332, Ile3355, Ala3356, Val3357, Gln3358, Ala3362, Tyr3363, Ala3364, Asp3365, Leu3366, Glu3367, Lys3368, Ala3369 and Glu3370) in the CC1 helix (bottom panels in Supplementary Fig. S4A and B) and of 17 residues (Asp3492, Arg3493, Ile3494, Lys3495, Pro3496, Glu3500, Gln3503, Ser3528, Ala3530, Tyr3532, Arg3541, Thr3543, Ile3546, Thr3548, Ser3551, Val3553 and Thr3587) in the CC2 helix (top panels in Supplementary Fig. S4A and B) varied widely. These 37 residues, especially two residues (Asp3365 and Asp3492) located near the MTBD, deviated largely and formed an extended structure (−180° < Φ < −125°, 100° < Ψ < 180° and −180° < Ψ < −160°). Since Asp3492 is one of the missing residues in the ΔMTBD mutant structure (Table I), this residue was unstable during the MD simulation.
The stretching elastic property of the stalk has not been reported yet, to the best of our knowledge. We estimated the stretching stiffness and modulus, using the MD trajectories after equilibration was attained (40–200 ns). The length of the stalk (L), calculated by the distances between the four pairs of Cα atoms, is shown in Fig. 6. The changes in the length of the CC1 helix (Val3267–Gln3358 and Ile3271–Glu3354) and of the CC2 helix (Leu3504–Val3593 and Ala3508–Val3589) correlated well in each model, suggesting that these helices collectively change their structures in the direction parallel to the principal axis. The length of the stalk L in the ATP model (Fig. 6B) fluctuated more broadly than that in the ADP model (Fig. 6A). The initial lengths, average lengths, average deviations and stretching stiffnesses are listed in Table II. The average length is similar between the two models, but the deviation is significantly different. The stretching stiffness estimated from the average stiffness of the four pairs is 160.9 pN/nm in the ADP model and 35.4 pN/nm in the ATP model. Namely, the stalk of the ADP model is 4.5 times stiffer than that of the ATP model.
Fig. 6.
The length of the stalk (L) calculated by the distances between four pairs of Cα atoms in (A) the ADP model and (B) the ATP model. The lengths of Val3267–Gln3358, Ile3271–Glu3354, Leu3504–Val3593 and Ala3508–Val3589 are shown by red, green, blue and purple lines, respectively.
Table II.
Stretching property of the stalk obtained from MD simulation
| Model | Residue pair | Linia (Å) | Laveb (Å) | <(L–Lave)2>c (Å2) | Stiffness (pN/nm) | Apparent stretching modulus (pN) |
|---|---|---|---|---|---|---|
| ADP model | Val3267–Gln3358 | 134.0 | 132.9 | 3.0 | 138.5 | 1855.9 |
| Ile3271–Glu3354 | 121.8 | 121.5 | 2.6 | 162.4 | 1978.0 | |
| Leu3504–Val3593 | 131.6 | 129.7 | 3.4 | 120.9 | 1591.0 | |
| Ala3508–Val3589 | 120.6 | 119.6 | 1.9 | 221.6 | 2672.5 | |
| ATP model | Val3267–Gln3358 | 134.0 | 130.1 | 12.8 | 32.3 | 432.8 |
| Ile3271–Glu3354 | 121.8 | 118.8 | 9.9 | 41.7 | 507.9 | |
| Leu3504–Val3593 | 131.6 | 124.8 | 14.0 | 29.7 | 390.9 | |
| Ala3508–Val3589 | 120.6 | 115.8 | 10.9 | 37.9 | 457.1 |
aLini is the length of the stalk of the initial structure.
bLave is the average length over from 40 to 200 ns trajectories.
c<(L–Lave)2> is the average deviation of the length over from 40 to 200 ns.
As there are no reports on the stretching stiffness of the stalk, we calculated the apparent stretching modulus of the stalk, by comparing it with another protein that has a similar coiled-coil structure. The apparent stretching moduli of the stalk, calculated from the current average stretching stiffness, are 2024 pN in the ADP model and 447 pN in the ATP model. These stretching moduli are less stiff than that of the myosin II s2 subdomain, 3448 pN (Adamovic et al., 2008).
As shown in the Materials and Methods section, the bending stiffnesses of the stalk corresponding to the ADP and ATP models have been experimentally obtained (Lindemann and Hunt, 2003). We estimated the bending stiffness from the MD data, for comparison with the experimental value. The distance from the principal axis of the moment of inertia (r) to each of the four atoms is shown in Fig. 7. The changes in the distance from the principal axis to the atoms in the CC1 helix (Glu3354 and Gln3358) and the CC2 helix (Leu3504 and Ala3508) clearly correlated in both models, indicating that these helices moved collectively in the direction perpendicular to the principal axis. Although the magnitude of the distance (∼50 Å) is similar in both models, the distance in the case of the ATP model (Fig. 7B) vibrated much more frequently than that of the ADP model (Fig. 7A). The initial distances from the principal axis of moment of inertia, average distances, average deviations and bending stiffnesses are listed in Table III. The average distance in the ATP model was approximately twice that in the ADP model, and the deviation was also significantly different.
Fig. 7.
The distance from the principal axis of the moment of inertia (r) to each of four Cα atoms in (A) the ADP model and (B) the ATP model. The distances to Gln3358, Glu3354, Leu3504 and Ala3508 are shown by red, green, blue and purple lines, respectively.
Table III.
Bending property of the stalk obtained from MD simulation
| Model | Residue | rinia (Å) | raveb (Å) | <(r–rave)2>c (Å2) | Stiffness (pN/nm) |
|---|---|---|---|---|---|
| ADP model | Gln3358 | 6.9 | 16.4 | 70.6 | 5.9 |
| Glu3354 | 9.2 | 16.3 | 60.6 | 6.8 | |
| Leu3504 | 0.7 | 12.9 | 54.0 | 7.7 | |
| Ala3508 | 2.5 | 10.8 | 40.5 | 10.2 | |
| ATP model | Gln3358 | 6.9 | 29.4 | 134.3 | 3.1 |
| Glu3354 | 9.2 | 26.9 | 119.6 | 3.5 | |
| Leu3504 | 0.7 | 28.7 | 141.5 | 2.9 | |
| Ala3508 | 2.5 | 24.3 | 117.9 | 3.5 |
arini is the distance from the principal axis (see Fig. 1B) of the initial structure.
brave is the average distance over from 40 to 200 ns trajectories.
c<(r–rave)2> is the average deviation of the distance over from 40 to 200 ns.
The bending stiffnesses estimated from the average over the stiffness of the four pairs were 7.6 pN/nm in the ADP model and 3.2 pN/nm in the ATP model, indicating that the stalk of the ADP model was 2.4 times stiffer than that of the ATP model. The experimental bending stiffness was 0.14 pN/nm in the ADP–vanadate configuration and 0.47 pN/nm in the apo-state (Lindemann and Hunt, 2003), which correspond to the ATP and ADP models, respectively. The ratios of the stiffnesses between the ADP and ATP models were 2.4 in our simulation and 3.4 in the experiment, and these values agree well with each other. Thus, the tendency of the stalk to become more flexible in the presence of ATP at AAA1 seemed to be reproduced by our simulation.
The absolute value of the bending stiffness in our simulation was, however, ∼10 times higher than the experimental value. The reason may be due to the fact that the current simulation time was too short to capture any large-scale structural changes. Since the computational system consisting of about one million atoms is very big, a simulation with a duration longer than 1 ms should be necessary to see large-scale motion. In fact, the sliding of the helix of the stalk and the structural change of the linker from the straight to bent conformation (Schmidt et al., 2015), upon binding the ATP molecule in AAA1, were not observed during our simulation. Further simulation studies should be required. However, the simulation and analysis of the stiffness in the present work will surely help us to understand the initial procedure of the structural change from the unprimed to primed state during the mechanochemical cycle of dynein.
Conclusion
To clarify the structural changes in dynein from the unprimed state with ADP bound in AAA1 to the initial transition state with ATP bound in AAA1, 200-ns MD simulations of the dynein motor domain for both models were executed, starting from the structure of the unprimed state. The stalk of the ATP model was more flexible than that of the ADP model, based on analyses of the rmsds and the backbone torsion angles. The stretching and bending stiffness of the stalk were analyzed using the trajectory. The stretching stiffness, which has not been previously reported, was estimated to be 160.9 pN/nm in the ADP model and 35.4 pN/nm in the ATP model. The stretching modulus derived from those values has a more flexible nature than that of the similar coiled-coil structure in the myosin II s2 subdomain. The bending stiffness was estimated to be 7.6 pN/nm in the ADP model and 3.2 pN/nm in the ATP model, and these values qualitatively agree with the experimental values. The simulation and analysis of the stiffness in the present work should help to clarify the structure–function relationships during the mechanochemical cycle of dynein.
Supplementary data
Funding
This work was supported by a Grant-in-Aid for Scientific Research C (25440070) from the Japan Society for the Promotion of Science (JSPS) to N.K. H.N. was supported by a Grant-in-Aid for Scientific Research on Innovative Areas, Grant Number 24118008 from JSPS. G.K. was supported by a Grant-in-Aid for Scientific Research B (26291014) from JSPS, and T.K. was supported by a Grant-in-Aid for Scientific Research B (26291034) from JSPS. This research was partially supported by the Platform Project for Supporting in Drug Discovery and Life Science Research (Platform for Drug Discovery, Informatics, and Structural Life Science) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) and the Japan Agency for Medical Research and Development (AMED). This work was performed in part under the Cooperative Research Program of Institute for Protein Research, Osaka University, CR-15-05 to N.K.
Supplementary Material
References
- Adamovic I., Mijailovich S.M., Karplus M. (2008) Biophys. J., 94, 3779–3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakawa T., Kamiya N., Nakamura H., Fukuda I. (2013) Plos One, 8, e76606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen H.J.C., Postma J.P.M., Vangunsteren W.F., Dinola A., Haak J.R. (1984) J. Chem. Phys., 81, 3684–3690. [Google Scholar]
- Berman H., Henrick K., Nakamura H., Markley J.L. (2007) Nucleic Acids Res., 35, D301–D303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S.A., Walker M.L., Sakakibara H., Knight P.J., Oiwa K. (2003) Nature, 421, 715–718. [DOI] [PubMed] [Google Scholar]
- Carter A.P., Cho C., Jin L., Vale R.D. (2011) Science, 331, 1159–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter A.P., Garbarino J.E., Wilson-Kubalek E.M., Shipley W.E., Cho C., Milligan R.A., Vale R.D., Gibbons I.R. (2008) Science, 322, 1691–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essmann U., Perera L., Berkowitz M.L., Darden T., Lee H., Pedersen L.G. (1995) J. Chem. Phys., 103, 8577–8593. [Google Scholar]
- Fukuda I. (2013) J. Chem. Phys., 139, 174107. [DOI] [PubMed] [Google Scholar]
- Fukuda I., Kamiya N., Nakamura H. (2014) J. Chem. Phys., 140, 194307. [DOI] [PubMed] [Google Scholar]
- Fukuda I., Kamiya N., Yonezawa Y., Nakamura H. (2012) J. Chem. Phys., 137, 054314. [DOI] [PubMed] [Google Scholar]
- Fukuda I., Yonezawa Y., Nakamura H. (2011) J. Chem. Phys., 134, 164107. [DOI] [PubMed] [Google Scholar]
- Gee M.A., Heuser J.E., Vallee R.B. (1997) Nature, 390, 636–639. [DOI] [PubMed] [Google Scholar]
- Gibbons I.R. (1981) J. Cell Biol., 91, S107–S124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons I.R., Rowe A.J. (1965) Science, 149, 424–426. [DOI] [PubMed] [Google Scholar]
- Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. (1983) J. Chem. Phys., 79, 926–935. [Google Scholar]
- Joung I.S., Cheatham T.E. (2008) J. Phys. Chem. B, 112, 9020–9041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya N., Fukuda I., Nakamura H. (2013) Chem. Phys. Lett., 568, 26–32. [Google Scholar]
- Kamiya N., Watanabe Y.S., Ono S., Higo J. (2005) Chem. Phys. Lett., 401, 312–317. [Google Scholar]
- Karki S., Holzbaur E.L.F. (1999) Curr. Opin. Cell Biol., 11, 45–53. [DOI] [PubMed] [Google Scholar]
- Kon T., Mogami T., Ohkura R., Nishiura M., Sutoh K. (2005) Nat. Struct. Mol. Biol., 12, 513–519. [DOI] [PubMed] [Google Scholar]
- Kon T., Nishiura M., Ohkura R., Toyoshima Y.Y., Sutoh K. (2004) Biochemistry, 43, 11266–11274. [DOI] [PubMed] [Google Scholar]
- Kon T., Oyama T., Shimo-Kon R., Imamula K., Shima T., Sutoh K., Kurisu G. (2012) Nature, 484, 345–350. [DOI] [PubMed] [Google Scholar]
- Kon T., Sutoh K., Kurisu G. (2011) Nat. Struct. Mol. Biol., 18, 638–642. [DOI] [PubMed] [Google Scholar]
- Lindemann C.B., Hunt A.J. (2003) Cell Motil. Cytoskel., 56, 237–244. [DOI] [PubMed] [Google Scholar]
- Lis M., Kim T., Sarmiento J.J. et al. (2011) Immunome Res., 7, 1–8. [Google Scholar]
- Mashimo T., Fukunishi Y., Kamiya N., Takano Y., Fukuda I., Nakamura H. (2013) J. Chem. Theory Comput., 9, 5599–5609. [DOI] [PubMed] [Google Scholar]
- Neuwald A.F., Aravind L., Spouge J.L., Koonin E.V. (1999) Genome Res., 9, 27–43. [PubMed] [Google Scholar]
- Nishikawa Y., Oyama T., Kamiya N., Kon T., Toyoshima Y.Y., Nakamura H., Kurisu G. (2014) J. Mol. Biol., 426, 3232–3245. [DOI] [PubMed] [Google Scholar]
- Paschal B.M., Vallee R.B. (1987) Nature, 330, 181–183. [DOI] [PubMed] [Google Scholar]
- Roberts A.J., Kon T., Knight P.J., Sutoh K., Burgess S.A. (2013) Nat. Rev. Mol. Cell. Bio., 14, 713–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A.J., Numata N., Walker M.L. et al. (2009) Cell, 136, 485–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryckaert J.P., Ciccotti G., Berendsen H.J.C. (1977) J. Comput. Phys., 23, 327–341. [Google Scholar]
- Schiavo G., Greensmith L., Hafezparast M., Fisher E.M.C. (2013) Trends Neurosci., 36, 641–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H., Gleave E.S., Carter A.P. (2012) Nat. Struct. Mol. Biol., 19, 492–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H., Zalyte R., Urnavicius L., Carter A.P. (2015) Nature, 518, 435–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Nakamura H., Fukuda I. (2016) J. Chem. Phys., 144, 114503. [DOI] [PubMed] [Google Scholar]
- Wang J.M., Wolf R.M., Caldwell J.W., Kollman P.A., Case D.A. (2004) J. Comput. Chem., 25, 1157–1174. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.