Abstract
Structure-based protein design tests our understanding of the minimal determinants of protein structure and function. Previous studies have demonstrated that placing zinc binding amino acids (His, Glu, Asp or Cys) near each other in a folded protein in an arrangement predicted to be tetrahedral is often sufficient to achieve binding to zinc. However, few designs have been characterized with high-resolution structures. Here, we use X-ray crystallography, binding studies and mutation analysis to evaluate three alternative strategies for designing zinc binding sites with the molecular modeling program Rosetta. While several of the designs were observed to bind zinc, crystal structures of two designs reveal binding configurations that differ from the design model. In both cases, the modeling did not accurately capture the presence or absence of second-shell hydrogen bonds critical in determining binding-site structure. Efforts to more explicitly design second-shell hydrogen bonds were largely unsuccessful as evidenced by mutation analysis and low expression of proteins engineered with extensive primary and secondary networks. Our results suggest that improved methods for designing interaction networks will be needed for creating metal binding sites with high accuracy.
Keywords: metalloproteins, protein design, Rosetta, zinc binding
Introduction
Over 30% of nonredundant protein chains in the PDB contain bound metal ions. These ions are often key to metalloproteins' functions including stability and folding (Tainer et al., 1992; O'Brien et al., 2015), intracellular signaling (Burgoyne, 2007) and catalysis (Andreini et al., 2008).
Due to their prevalence and the range of functions that they can provide, metal binding sites are appealing targets for protein design. Previous studies have designed metal binding sites for functions including allosteric control of an enzyme (Browner et al., 1994; Dwyer et al., 2003), heavy-metal sequestration (Eskandari et al., 2013; Zhou et al., 2014; Plegaria et al., 2015) and catalysis (Der et al., 2012a; Zastrow et al., 2012). Since metal ions have only one possible conformation to place in any given position and orientation, they may also be a good starting point for computational design of ligand binding proteins. Furthermore, the coordination geometry of many metals has been extensively studied (Tainer et al., 1992; Karlin and Zhu, 1997; Auld, 2001).
Zinc ions are among the most common in metalloproteins, occurring in 58% of known human metalloproteins, and serve a wide range of functions (Azia et al., 2015). Many zinc binding sites provide structural stability, such as the sites that give superoxide dismutase (Nedd et al., 2014) and zinc finger proteins (Matthews and Sunde, 2002) their active conformations. Zinc binding can also induce oligomerization (Derewenda et al., 1989) and conformational changes that lead to signaling (Chen et al., 2014). About half of the eukaryotic zinc binding proteins are enzymes, where zinc often acts as a catalytic cofactor. Ten percent of enzyme-catalyzed reactions, including all six classes of enzyme, are thought to require zinc for at least one of their mechanisms (Andreini and Bertini, 2012).
While structural zinc sites are most often buried and are coordinated by four protein ligands, predominately histidines and cysteines, catalytic sites are often solvent accessible and have only three coordinating residues with one site occupied by a water molecule. These sites are predominately coordinated by histidine, aspartate and glutamate residues, and cysteines are relatively uncommon (Auld, 2001; Andreini and Bertini, 2012). First-shell residues are often stabilized by hydrogen bonds with second-shell residues, most commonly Asp/Glu for catalytic histidines, backbone carbonyl groups for structural histidines and backbone amide groups for Cys, Asp and Glu (Dudev et al., 2003; Lin and Lim, 2004). These interactions have been shown to contribute to both metal affinity and, in the case of zinc enzymes, catalytic activity (Kiefer et al., 1995; Marino and Regan, 1999).
While efforts at metalloprotein design frequently yield metal binding, there are few examples of designs that have been structurally characterized and have structures that closely match their designed models. Such precision will be key for certain functional applications of metalloprotein design, most notably catalysis. Studies which directly graft a native protein's metal binding site onto a different scaffold (Müller and Skerra, 1994) or insert a binding motif into a surface-exposed region (Eskandari et al., 2013) have been successful in achieving metal binding but lack both models and crystal structures of designed proteins. Other labs have designed sites rationally with no computational scoring. The Pecoraro lab has designed binding sites for zinc and mercury in a trimeric peptide (Zastrow et al., 2012) and for zinc in a single-chain three-helix bundle (Plegaria et al., 2015); however, both studies lack structural models for their designs, so their precision cannot be determined.
Computational metalloprotein designs provide structural models that can be used to evaluate the precision of designs when compared with a crystal structure; however, many of these studies do not acquire high-resolution structures of the designed proteins. The Regan group has produced several successful zinc binding protein designs, including designs containing second-shell interactions, but crystal structures for those designs are not available (Regan and Clarke, 1990; Klemba and Regan, 1995; Klemba et al., 1995; Marino and Regan, 1999). Designs from the Hellinga group have likewise lacked structural studies (Hellinga et al., 1991; Dwyer et al., 2003). Perhaps more informative are examples in which structural information is obtained but shows discrepancies with the design models. For instance, designed zinc binding sites from the Tezcan lab were found to bind residues that were not anticipated and did not bind some of the designed residues (Salgado et al., 2010). Similarly, the crystal structure of Zhou et al.'s uranyl binding protein does not demonstrate coordination by two of the designed residues (although this may be an artifact of crystallization conditions) (Zhou et al., 2014), and the initial design model for the zinc binding site of MID1 (Der et al., 2012b) includes a fourth histidine that was not found to coordinate the metal in the crystal structure.
Even designs that would typically be considered successful, i.e. those which specifically bind the intended metal with the intended residues and whose structures have a low RMSD to the design model, often show slight differences in metal coordination that could make such goals a considerable challenge. Notably, Mills et al. reported using the RosettaMatch algorithm (Zanghellini et al., 2006) with negative design states and a non-natural amino acid that forms a bidentate interaction with zinc to design a binding site that only deviates by 0.9 Å RMSD in the crystal structure from the designed model at the binding site (Mills et al., 2013). Even this structure, however, is missing one predicted water molecule, and one ligand is bidentate instead of monodentate as predicted. Even slight differences such as these could have functional consequences. Coordinating water molecules often participate in catalysis, particularly for zinc binding sites, and any change in coordination will likely alter the electrostatic properties of the metal ion, which would affect its ability to perform catalysis. Therefore, only structures with all of the coordinating residues in the correct rotamers and with the same basic coordination geometry can truly be considered successful.
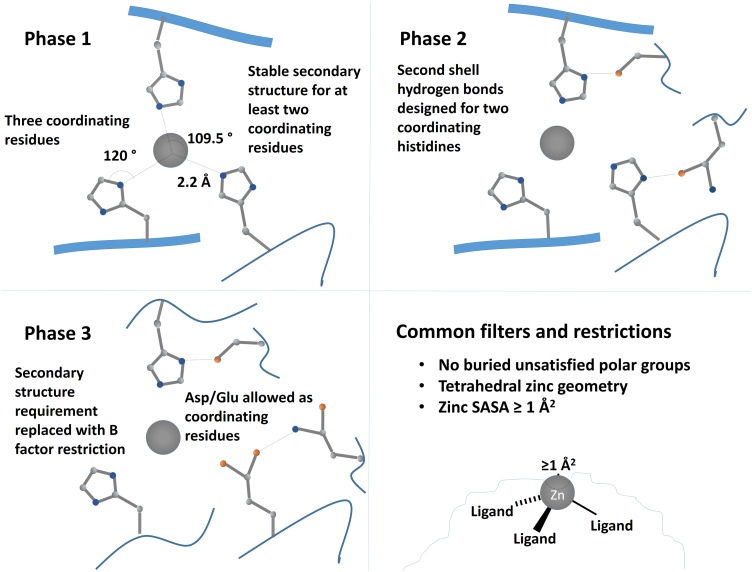
Here, we describe our approach for designing zinc binding proteins in native protein scaffolds. Briefly, binding sites with three liganding residues were first built in binding pockets from a library of scaffold proteins (Rothlisberger et al., 2008) using the RosettaMatch algorithm (Zanghellini et al., 2006) subject to geometric constraints. Residues within 10 Å of the site were designed using Rosetta (Leaver-Fay et al., 2011) to improve the site's stability, and the resulting designs were filtered based on site geometry and other factors described in more detail below. Three separate design approaches (called phases) were performed with additional features being taken into account in each (Fig. 1). The outcomes of these results give further insight into the challenges and factors that must be considered when designing metalloproteins.
Fig. 1.
Schematic of design strategies for first, second and third rounds of design. In the first set of designs (Phase 1), tetrahedral zinc binding sites were designed with three coordinating residues and one open coordination site with at least two of the ligands on stable secondary-structure elements. In Phase 2, we designed two second-shell hydrogen bonding residues in addition to three coordinating histidines. To increase the diversity of designed sites, Phase 3 allowed one of the coordinating residues to be an aspartate or glutamate and removed the secondary-structure requirement for ligand residues, instead requiring all ligand residues to have Cα B factor values of 30 or greater.
Materials and methods
Computational methods
For all designed zinc binding proteins, initial zinc sites were constructed using the RosettaMatch protocol (Zanghellini et al., 2006). Briefly, we first obtained starting scaffolds by limiting a set of 85 enzyme scaffolds from previous studies to the 55 smallest scaffolds (<340 residues) (Jiang et al., 2008; Rothlisberger et al., 2008). These 55 scaffolds are listed in Supplementary Table SI. The open-source software Fpocket (Le Guilloux et al., 2009) was then used to identify residue positions in potential binding pockets within each of these scaffolds. For each of these sets of residues, a search was conducted to determine if a zinc binding site satisfying a provided set of geometric constraints could be formed from residues within that set; these constraints varied across the three design phases. Each of these constraints specified the geometry of binding site residues relative to a potential ligand (in this case, a zinc ion). For all possible residue positions within a candidate binding pocket, all common rotamers of potential coordinating residues were placed, and the resulting zinc ion was stored in a six-dimensional hash of its position and orientation. After this search had been conducted for all constraints at all positions, the hash was searched for bins that contain hits for all of the specified constraints, indicating that the rotamers composing those hits could bind a ligand with the position and orientation specified by that bin. If these rotamers did not clash with one another or with the protein backbone, then the set of hits was output as a potential binding site (a ‘match’).
In each phase, after these binding sites had been constructed, the geometry of the zinc binding site was scored (Der et al., 2012b) such that a score of <2.0 is (on average) within 1 SD of the typical distances, angles and dihedral angles for a zinc binding site. Designs were then refined using backbone minimization and design of surrounding residues as additional filters were applied in each phase as described below, and a final set of designs was selected for testing by visual inspection.
Phase 1
In the first design phase, designed sites were required to contain three coordinating residues in a tetrahedral arrangement; these sites included three-histidine (HHH) sites; sites with two histidines and one aspartate (HHD), glutamate (HHE) or cysteine (HHC); and sites with one histidine and two cysteines (HCC). In each of these cases, instead of a simple zinc ion, a zinc ion with a bound histidine was used as the test ligand to ensure that the designed binding site would have one open coordination site. The ideal distance between the nitrogen and zinc atoms was set to 2.10 Å with a 0.15 Å SD. The ideal angle about the zinc ion was set to 109.5° (tetrahedral coordination), and the ideal angle about the coordinating nitrogen was set to 120°. The zinc was further constrained to be planar with the histidine ring with a 120° torsion about the bond between the zinc ion and the histidine nitrogen; rather than using a second coordinating residue to determine this torsion, it was measured with respect to the theoretical location of a ligand if the coordination environment were perfectly tetrahedral. Designs with zinc geometry scores <2.0 (within 1 SD of a typical zinc coordination) were selected for refinement.
To refine the resulting models, we performed 10 runs of the Rosetta enzyme design protocol EnzDes (Richter et al., 2011) per site. EnzDes holds the designed coordinating residues fixed while allowing backbone minimization of the remainder of the scaffold and designing and repacking residues within specified distances of the zinc binding site. Both the design and repacking shells specify two cutoff distances; residues that have alpha carbons closer than the first cutoff to the ligand or with alpha carbons closer than the second cutoff with side chains oriented toward the ligand are included in that shell. The two design cutoffs were set to 6 and 8 Å, and the two repacking cutoffs were set to 10 and 12 Å. Designs containing no clashes were then filtered to remove sites containing rare rotamers of coordinating residues based on Rosetta's rotamer energy term (Shapovalov and Dunbrack, 2011) and to remove designs containing buried unsatisfied polar atoms. An atom was considered buried if it was inaccessible to a 1.2 Å probe; all such polar atoms were required to have at least one hydrogen bond partner.
Phase 2
The second design phase required that each zinc binding site contain three coordinating histidines, in this case using a free zinc ion as the ligand. However, this set of designs further required that two of the three coordinating histidines form hydrogen bonds with either the carbonyl oxygen or a side chain oxygen from a neighboring residue. These hydrogen bonding residues were again placed using the RosettaMatch protocol as secondary matches. Briefly, matches were initially identified using the coordinating residues as described above. For each of these matches, possible rotamers of secondary match residues were built off at each position; if that residue position could form an interaction with the primary match residues that satisfies the match constraint, then it was stored. All possible sets of second-shell interactions for a given primary match were output as separate candidate binding sites. The ideal hydrogen bond distance between the donor and acceptor was set to 2.80 Å, and the angles about the donor atoms were set to 109° for sp3-hybridized atoms and 120° for sp2-hybridized atoms.
Designs were then refined using the following protocol: All residues within 10 Å of the binding site (excluding the residues placed during the RosettaMatch protocol) were allowed to change rotamers and identities. Since we had already made up to five mutations due to the inclusion of second-shell residues in this phase, we wanted to minimize the number of mutations made to prevent destabilizing the native folds of the scaffolds. A 10-Å cutoff allowed changes in residues which could potentially interact or clash with the new binding site while maintaining the native sequence in more distal regions. Five rounds of gradient-based minimization using Rosetta's score12 score function and ‘dfpmin’ minimizer were then performed on the torsion angles of both the backbone and side chains of these residues. In each case, sites were then filtered such that the total zinc geometry score was <4.0 (indicating binding site geometry within 2 SDs of the mean), all liganding atoms were <3.1 Å from the zinc ion and were within 30° of the proper tetrahedral angles, the total score for the protein was <0 Rosetta energy units (REU) to avoid structures containing clashes, the RMSD of the design to the starting scaffold was <2 Å, the RMSD of the match residues was <1 Å and the solvent accessible surface area of the zinc ion was at least 1 Å.
After refinement, sites were further filtered on the geometry of the hydrogen bonds placed by the RosettaMatch protocol. All donor–acceptor distances were required to be <3.7 Å, and the Rosetta scores for each of these hydrogen bonds were calculated and required to be <−1 REU. All match residues were required to be in favorable rotamers (Rosetta rotamer scores of <6 REU), and the zinc geometry score was required to be <2.5. Designs were further filtered to prohibit buried unsatisfied polar atoms within the designed portion of the protein, to limit the total number of mutations to seven, to have at least two of the three coordinating residues on stable secondary-structure elements and to have no more than one glutamate or glutamine as a hydrogen bond donor. Designs were chosen from the best-scoring 50% of the structures that passed all of these filters.
Phase 3
Rather than requiring all three coordinating residues to be histidines, the third design phase allowed one coordinating aspartate or glutamate in the binding site. Distances, angles and dihedrals about the zinc ion remained the same as for a coordinating histidine, and the zinc ion was required to be in the plane of the carboxylate group. Constraints were also added for possible hydrogen bond donors for these residues; again, ideal hydrogen bond distances were set to 2.80 Å, and hydrogen bond angles remained 109° for sp3-hybridized atoms and 120° for sp2-hybridized atoms.
Designs were then refined and filtered as in Phase 2 with the following exceptions: The filter on the secondary structure of coordinating residues was replaced with a filter that required the B factor of each coordinating residue's alpha carbon to be <30.0, and the filter limiting the number of glutamates and glutamines in the binding site was replaced with a filter that limited the total number of lysines, arginines, glutamates and glutamines in the binding site to two. The maximum number of mutations allowed was increased to nine. During visual inspection, designs from different scaffold types were evaluated independently so that at least one design from each representative scaffold type was chosen.
Experimental methods
Protein expression and purification
DNA sequences for designed proteins were ordered as gBlocks gene fragments from Integrated DNA Technologies (IDT) optimized for expression in Escherichia coli with a C-terminal stop codon. For all designs, an N-terminal BamH1 restriction site and a C-terminal Sal1 restriction site were added to each design, and the designed proteins were inserted into the pQE-80L vector with an N-terminal 6-His tag, an MBP fusion and a TEV cleavage site. Phase 3 designs were inserted into the same vector with no MBP fusion (pQE-80L with an N-terminal 6-His tag and TEV cleavage site). Plasmids were transformed into BL21 star cells (Phases 1 and 2) or BL21 cells (Phase 3) for expression. Cells were initially grown at 37°C in 1.5 L LB broth containing 67 µg/ml ampicillin to an OD of 0.6–0.8. Expression was then induced with 0.33 mM IPTG. Phase 1 designs were expressed at 30°C for 5 h, and Phase 2 and 3 designs were expressed at 18°C for 18 h.
After expression, cultures were centrifuged at 12 000 rpm for 20 min to remove the growth media. Cell pellets were resuspended in lysis buffer (10% glycerol, 20 mM Tris pH 8.0, 100 mM NaCl, 0.5 mM PMSF, 0.5 mM DTT, 1× leupeptin, 1× pepstatin, 1× bestatin) and lysed by sonication. Two units each of DNase and RNase A were added to the lysates, and lysates were incubated at room temperature for 15 min to remove nucleic acids. The lysates were then cleared by centrifugation at 15 000 rpm for 30 min. Designed proteins were purified from cleared lysates by immobilized-metal affinity chromatography (IMAC) with a 5 ml Ni-NTA HisTrap HP column (GE Healthcare). Columns were equilibrated with 20 mM Tris pH 8.0, 100 mM NaCl, 25 mM imidazole (IMAC wash buffer) before and after loading the lysate, and proteins were eluted with 20 mM Tris pH 8.0, 100 mM NaCl and 500 mM imidazole (IMAC elution buffer).
After elution, samples were treated with 5 mM EDTA to chelate excess nickel ions and 0.05 mg/ml TEV protease to remove the polyhistidine tag and/or MBP fusion. Samples were cleaved overnight while being dialyzed into 20 mM Tris pH 8.0, 100 mM NaCl with stirring. To remove the cleaved MBP and/or polyhistidine tags, the samples were again purified by IMAC as before but were collected in the flowthrough and wash steps. Samples were again treated with 5 mM EDTA and were concentrated to <2 ml for size exclusion chromatography on a Superdex-75 column (GE Healthcare, HiLoad 16/60 prep grade); during this process, they were exchanged either into 100 mM ammonium acetate pH 7.0 (crystallography buffer) or 100 mM ammonium acetate, 100 mM NaCl, pH 7.0 (sample buffer). Fractions containing the purified protein were identified both by the fractions' absorbance at 280 nm and by SDS-PAGE, and pure fractions were combined for subsequent experiments. Protein concentrations were determined by their absorbance at 280 nm using molar extinction coefficients calculated from their sequences (Gasteiger et al., 2005).
Production of point mutants
Point mutations of all coordinating and second-shell residues in Hinge2 were produced using a three-step PCR method. Both forward and reverse primers were ordered containing each point mutation, and the first and second halves of the gene were amplified separately using these primers and the appropriate cloning primers. These fragments were combined in a final PCR reaction to produce the full-length mutant gene. All proteins were cloned and expressed as previously described for Phase 3 designs, and gene sequences were verified by sequencing.
Circular dichroism
All circular dichroism spectra and thermal melts were collected on a JASCO J-815 CD spectrometer. Cell temperatures were controlled by a JASCO Peltier device and water bath. CD spectra were measured in sample buffer described above. ZE2 spectra were collected with 5 mM protein; ZE2 thermal denaturation curves and the spectra and denaturation curves of Hinge2 and its point mutants were collected with 15 mM protein. Spectra were measured from 190 to 250 nm, and thermal denaturation was measured by monitoring circular dichroism at 220 nm as the temperature was increased from 20 to 95°C at 3°C/min. Thermal denaturation was measured in the presence and absence of 30 mM zinc sulfate.
Isothermal titration calorimetry
Zinc binding affinities were measured by isothermal titration calorimetry (ITC) on a MicroCal Auto-iTC200 instrument in UNC's Macromolecular Interactions Facility. All experiments were run at 20°C with 20 2-µl injections of zinc into 200 µl protein in sample buffer. ZE2 affinity measurements were carried out with a cell concentration of 40 µM ZE2 and a syringe concentration of 800 µM ZnSO4; affinity measurements for Hinge2 and its mutants were performed at ∼50 µM protein and 1 mM ZnSO4.
X-Ray crystallography
For crystallization, ZE2 was exchanged into crystallization buffer as described above and concentrated to 19 mg/ml. An equimolar concentration of zinc sulfate was added prior to crystallization. Protein crystals were initially grown in 0.1 mM succinate pH 6.0 and 22% PEG 3350 in a 96-well format at 20°C. These crystals were combined 1:1 with mother liquor from the same well and were used as seed stock for crystal seeding in a 24-well format under the same conditions. Crystals in this format grew within 3–4 days. Cells from this screen were cryoprotected in a 1:1 solution of 50% glycerol and mother liquor before being stored in liquid nitrogen.
Spelter was stored in 10 mM ammonium acetate pH 7.0 prior to crystallization at a 1:1:1 molar ratio with zinc sulfate and ubiquitin at 15 mg/ml total protein concentration. Protein crystals were grown in 0.22 M sodium iodide and 26% PEG 3350 at 4°C. Crystals took over 2 weeks to appear and were stored in liquid nitrogen prior to data collection.
All diffraction data were collected at the Advanced Photon Source (APS), Argonne National Laboratory (ANL), Argonne, IL. Data were initially processed using the program HKL2000 (Otwinowski and Minor, 1997). Molecular replacement into the starting scaffold protein was performed using Phaser (McCoy et al., 2007). The structure was refined by alternating manual refinement using Coot (Emsley et al., 2010) and anisotropic refinement using Refmac (Murshudov et al., 2011).
Results and discussion
We searched potential ligand binding pockets in 55 monomeric scaffold proteins for three-histidine zinc binding sites using the RosettaMatch application (Zanghellini et al., 2006). During the initial search, few three-histidine (HHH) sites were found; therefore, the search was expanded to include HHD, HHE, HHC and HCC sites. After filtering sites based on their zinc geometry score (described in the Materials and Methods section), we identified 500 potential designs (108 HHH sites, 193 HHD, 102 HHC and 98 HCC).
To resolve clashes with side chains of surrounding residues and identify mutations that would stabilize our designs, we used the EnzDes Rosetta protocol. This application allows small backbone movements to the protein scaffold and repacks and designs side chains within user-specified distances of the binding site. In this protocol, we designed residues which had alpha carbons within 6 Å of the zinc ion or with alpha carbons within 8 Å of zinc with side chains oriented toward the zinc ion; likewise, the two repacking shells were set to 10 and 12 Å, respectively. Although it could potentially design hydrogen bonds to the coordinating histidines, we found that it very rarely did so; instead, its main function was to remove clashes with neighboring residues. We performed 10 runs of this protocol for each of the designed zinc sites, and these results were then filtered to remove any models containing buried unsatisfied polar atoms. The remaining sites were also filtered to remove designs with rare rotamers for zinc-coordinating residues. We then used visual inspection of the remaining designs to select three (ZE1, ZE2 and ZE3) with zinc binding sites in deep but accessible pockets. For these designs, we also reverted any mutations that were not close to the zinc coordination sphere.
All of our three selected designs were in α/β scaffolds; two of these (ZE2 and ZE3) contained zinc binding sites in the beta barrel of TIM barrel scaffolds (Table I), while ZE1's zinc binding site sits within a somewhat shallower cleft. ZE1 and ZE2 both contained three-histidine zinc binding sites, whereas the ZE3 binding site consisted of two histidines and one cysteine.
Table I.
Summary of zinc binding site designs and experimental results
| Name | Scaffold | Scaffold type | Mutations | Zinc binding residues | Second-shell hydrogen bond residues | Experimental outcome | Zinc binding affinity | |
|---|---|---|---|---|---|---|---|---|
| Phase 1 | ZE1 | 4fua | Hydrolase-like (α/β) | G37A | H92, H94, H155 | N/A | Destabilized by zinc | ND |
| ZE2 | 1a53 | TIM barrel (α/β) | K109A, E158G, N179H, R181A, L183H, E209A, S210H | H179, H183, H210 | N/A | Soluble, binds zinc | 1.4 µM | |
| ZE3 | 1dl3 | TIM barrel (α/β) | L130G, E158H, M79C, R181S, E209H | H7, C7, H79 | N/A | Soluble aggregates | ND | |
| Phase 2 | 1473 | 1icm | Lipocalin-like (Beta barrel) | L36S, E51H, R56H, V60H, Y70N | H51, H56, H60 | S36, N70 | Soluble aggregates | ND |
| 225 | 1q7f | 6-bladed beta propeller | A32N, R79H, T120H, V163S, V164S, I207H | H79, H120, H207 | N32, S164 | Did not express | ND | |
| 255 | 1m4w | Jelly roll (Beta sandwich) | Y78H, W80H, E87H, Y89N, D169G | H78, H80, H87 | N89, S126 | Soluble aggregates | ND | |
| 339 | 1f5j | Jelly roll (Beta sandwich) | Y81H, W83H, E90H, Y92T, S130T | H81, H83, H90 | D92, T130 | Soluble aggregates | ND | |
| 548 | 1lbm | TIM barrel (α/β) | K5H, A25G, F55H, V57H, Q81I, I101E, L124I, A165N, D167G | H5, H55, H57 | E10, N165 | Soluble aggregates | ND | |
| 1032 | 1suu | 6-bladed beta propeller | V186T, K187H, D239H, I241T, V287A, S288H, V289A | H187, H239, H288 | T24, T186 | Aggregated after cleavage | ND | |
| 289 | 2h13 | 6-bladed beta propeller | K21R, D61H, C103H, N105D, F232H | H61, H103, H232 | S60, D105 | Soluble aggregates | ND | |
| Phase 3 | Alpha1 | 1ovk | Lysozyme-like (α+β) | T21Q, N101A, Q105E, M106D, W118H, T122H, R125G | E105, H138, H142 | N21, D106 | Did not express | ND |
| AlphaBeta1 | 6cpa | Hydrolase-like (α/β) | R71A, E72H, R127H, E163Q, H196D, E270H, F279H | H72, H127, H279 | Q163, D196 | Did not express | ND | |
| TIM1 | 1igs | TIM barrel (α/β) | W7E, S55H, P56A, S57G, F87N, R181E, L186H | H55, E181, H186 | E7, S210 | Low expression, bound zinc | ∼50 µM | |
| TIM2 | 1tml | 7-stranded TIM barrel (α/β) | F42E, N46T, Q53H, L57K | E42, H44, H53 | T46, K57 | Did not express | ND | |
| Hinge1 | 1abe | Periplasmic binding protein (α/β) | W16F, D88H, M107Q, T146H, R150M, M203S, N204H, T207Q | H88, H146, H204 | E107, D231 | Expressed, bound zinc | 6.5 µM | |
| Hinge2 | 1gca | Periplasmic binding protein (α/β) | F16D, N91G, T110H, D154E, N256H, Q261E, Y295C | H110, H256, E261 | D16, E154 | Expressed, bound zinc | 1.1 µM | |
| Beta1 | 1cbs | Lipocalin-like (Beta barrel) | F15H, L18S, L19A, A32H, A36S, V76E | H15, H32, E76 | S18, S36 | Did not express | ND | |
| Beta2 | 1cbs | Lipocalin-like (Beta barrel) | F15A, L18E, A32D, A36H, T56N, V76H | D32, H36, H76 | E18, N56 | Did not express | ND | |
| Beta3 | 1ifc | Lipocalin-like (Beta barrel) | L36S, E51H, F55S, R56H, I58E, V60H | H51, H56, H60 | S36, S55 | Soluble aggregates | ND | |
| Beta4 | 1cbs | Lipocalin-like (Beta barrel) | F15A, L18Q, A32D, A36H, T56N, V76H | D32, H36, H76 | E18, N56 | Did not express | ND | |
| Pocket1 | 1f5j | Jelly roll (Beta sandwich) | N43E, L45D, Q128R, E180H, Y182H | E43, H180, H182 | D45, R128 | Soluble aggregates | ND | |
| Pocket2 | 1m4w | Jelly roll (Beta sandwich) | Y78H, W80H,Y89D, I127D, D169G | H78, H80, E87 | D89, D127 | Soluble aggregates | ND |
All three of the tested designs were solubly expressed in E. coli as described in the Materials and Methods section. When the three designs were purified by size exclusion, ZE1 and ZE2 eluted at the predicted monomeric sizes; however, ZE3 formed soluble aggregates which eluted in the void volume and was thus excluded from further analysis. ZE1 and ZE2 were initially tested for zinc binding by determining their change in thermal stability as determined by circular dichroism in the presence and absence of saturating zinc. ZE1 was found to denature at a lower temperature in the presence of zinc, indicating that partial unfolding may be required for binding; therefore, it was excluded from additional studies. ZE2 showed a 2.4°C increase in its transition temperature in the presence of zinc, indicating that it successfully binds zinc in the folded state (Fig. 2E).
Fig. 2.
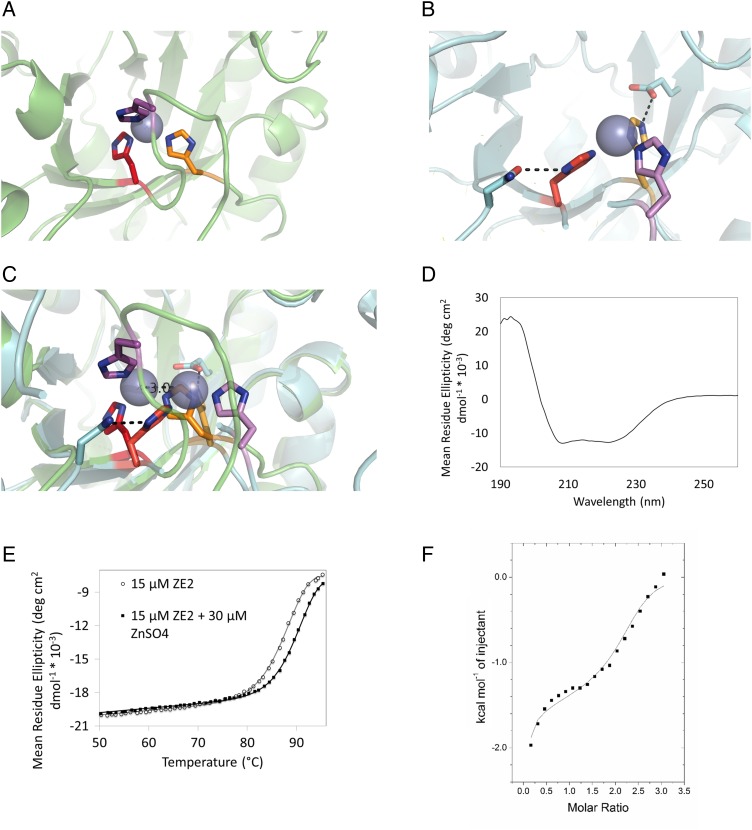
(A) Predicted structure of ZE2 zinc binding site. (B) Crystal structure of ZE2 zinc binding site. (C) Crystal structure of ZE2 overlaid with the design model. Although the predicted residues do form the zinc binding site, the histidines (along with the zinc ion) have shifted their positions to form stabilizing hydrogen bonds with surrounding residues. H210 (bottom center) has changed rotamers to form a hydrogen bond with the side chain of E50; H179 (left) has changed rotamers to form a hydrogen bond with N160 and is coordinating the zinc ion with the epsilon nitrogen instead of the delta nitrogen; and the loop containing H183 (top center, far right) has moved over 9 Å to accommodate the new position of the binding site. (D) ZE2 is well folded in the absence of zinc. (E) ZE2 shows increased thermal stability in the presence of saturating zinc, which increases the thermal transition temperature from 90.2 to 92.4°C. (F) ZE2 binds zinc in the designed site with 1.4 µM affinity as measured by ITC. A second nonspecific binding event (KD = 92 µM) is also observed by ITC, but only the first site appears in the crystal structure.
Given this result, we next determined the affinity of ZE2 for zinc using ITC (Fig. 2F). Our data indicate that ZE2 binds zinc in an exothermic reaction with an affinity of 1.4 µM. However, we also observed a second nonspecific binding event (KD ≈ 90 µM) in this experiment. Since only one zinc ion was identified in the crystal structure of ZE2 (described below), it is unclear where this second binding event occurs; ZE2 contains several patches of acidic residues which could transiently bind zinc ions. However, due to the low affinity of this interaction, it is unlikely to affect the results of our other assays.
The crystal structure of ZE2 was solved to 1.4 Å resolution using molecular replacement into the native PDB scaffold (PDB ID 1a53). We found that all of the designed zinc binding residues do participate in the binding site (Fig. 2A and B); however, two of the coordinating histidines are in different rotamers than were predicted in the design model, allowing them to form stabilizing hydrogen bonds to two second-shell residues (Fig. 2B). H210 has changed rotamers so that its protonated ε nitrogen hydrogen bonds with the side chain of E50, and the δ nitrogen H179 hydrogen bonds with the side chain oxygen of N160. Both of these residues are also in a different tautomeric state than we predicted; H210 and H179 bind zinc with their δ and ε nitrogen, respectively, instead of the ε and δ nitrogens as predicted. This adjustment has repositioned the binding site such that the zinc ion is 3 Å from its predicted location. To accommodate this motion, the flexible loop containing the third coordinating histidine (H183) has shifted to complete the new binding site, placing its alpha carbon 9 Å from its predicted location.
In our model, all of ZE2's designed histidines were predicted to be solvent-exposed with no hydrogen bonds to other protein residues; however, in native proteins, such a configuration is very rare (Dudev et al., 2003). We reasoned that explicit design of hydrogen bond partners for these residues would stabilize the binding site and increase the likelihood that our predicted structures would be accurate. Given this result, we decided to incorporate these second-shell residues into our next round of designs.
Our second design phase followed a similar procedure to the first; however, we added secondary match constraints to build hydrogen bond acceptors for two of the three coordinating histidines, and the design models were further filtered on the geometry and energy of the resulting hydrogen bonds. This procedure generated 144 722 potential binding sites in 54 scaffolds, of which 7302 in 52 scaffolds passed our initial filters for zinc coordination and hydrogen bond geometry. The increased number of initial hits reflects the increase in our sample space; the same set of primary residues often has multiple potential sets of second-shell residues. It quickly became apparent that this set of constraints strongly favored scaffolds that were mostly β sheets; these designs were common in our initial output and became further enriched as we further filtered the designs as described in the Materials and Methods section. Briefly, these filters, included restrictions on the predicted change in protein stability, the number of mutations made, the rotamers and identities of second-shell residues, buried unsatisfied polar groups, the geometries of both the zinc coordination and the designed hydrogen bonds and the secondary structure of first- and second-shell residues. The filters for rotamer energies and stable secondary structure were especially restrictive in this respect; while most filters eliminated <15% of our prospective binding pockets, these removed 23 and 42% of pockets, respectively. They also removed a large proportion of potential designs, particularly within helical scaffolds (51 and 59%, respectively); the only filter that eliminated a larger proportion of designs was our initial filter for buried unsatisfied polar groups, which eliminated 64% of potential models. Only one design with any appreciable alpha helical character was selected for testing (Table I).
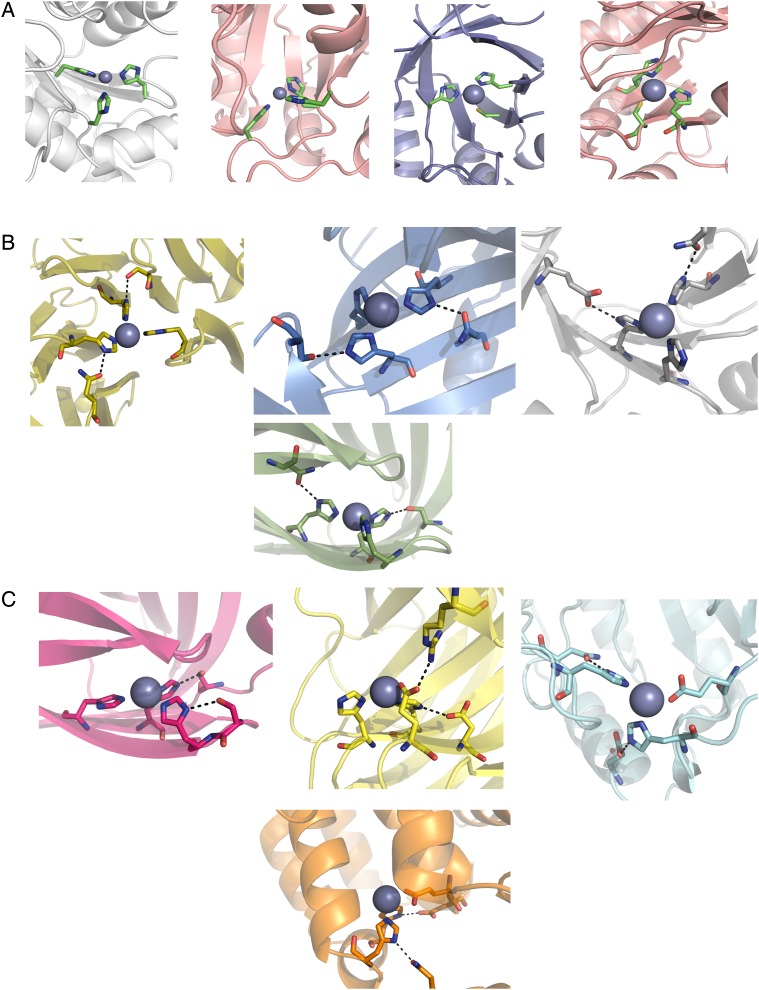
Despite being common in nature (Dudev et al., 2003), we saw very few designed zinc binding sites with second-shell hydrogen bonds to backbone carbonyl oxygens. This result may be due to the fact that RosettaMatch is intended for side chain design and does not allow backbone flexibility; since the backbone torsion angles of these residues, even in flexible loop regions, are not allowed to change, we are far less likely to find matches that include these atoms. The majority of second-shell residues in our initial matches were aspartate, asparagine, glutamate and glutamine; however, due to our restrictions on both the number of second-shell glutamates/glutamines and buried unsatisfied polar groups in the second shell (since these groups introduce an extra polar atom requiring a hydrogen bond partner), serine and threonine hydrogen bond partners were slightly enriched during the filtering process. We also favored hydrogen bonds to serine and threonine during our manual screen of designs. Figure 3B shows examples of designed zinc binding sites chosen for experimental testing.
Fig. 3.
Sample models of designed zinc binding sites showing coordinating residues and hydrogen bonding residues. (A) Phase 1 sites include three histidines or cysteines with no designed second-shell residues. (B) Phase 2 sites add two second-shell hydrogen bonding residues. (C) Phase 3 sites introduce liganding Asp/Glu and relaxed secondary-structure requirements to increase site variability.
Six of the seven Phase 2 designs tested were solubly expressed as fusions to maltose binding protein (MBP). However, all of these six designs either formed soluble aggregates as fusion proteins or aggregated immediately upon MBP cleavage (Table I). When expressed with no MBP fusion, only designs 255 and 339 were detected in the soluble fraction of bacterial lysates, and they continued to form soluble aggregates; these problems persisted with low expression temperatures and exogenous zinc.
We reasoned that the mutations we made in these designs, many of which were on β strands, may have disrupted β sheet assembly and prevented them from folding properly. Although most of these designs did score slightly worse in the absence of zinc than their starting scaffolds, two of the proteins that formed soluble aggregates (548 and 289) were predicted to be stabilized. However, 48% of the mutations made in these designs converted a hydrophobic or aromatic residue to a polar or charged residue (Table I). We reasoned that if we lifted our requirement on binding site secondary structure, we could potentially avoid making these destabilizing mutations to the protein core and we may also increase our scaffold diversity to include more proteins with helical secondary structure. We also reasoned that increasing the diversity of our potential binding sites by allowing alternative coordinating residues would give us a larger set of designs to select from that passed our selection criteria and allow us to be more stringent in our filter for protein stability. Therefore, in our next design round, we allowed acidic coordinating residues and lifted our requirement that the majority of ligand residues be placed on stable secondary-structure elements.
While our efforts to increase diversity in our designs did give us a varied pool of starting structures, their scaffold diversity quickly deteriorated during filtering. The initial RosettaMatch run generated 2 868 308 potential binding sites in 54 scaffolds. After applying initial filters for zinc site geometry and eliminating models containing clashes, we produced 95 318 starting designs in 47 scaffold proteins across all scaffold types. After applying all filters, however, 12 of these scaffolds were eliminated, 11 of which were mixed α/β scaffolds. Of the 549 remaining designs, 448 of them were in mostly-beta scaffolds (lipocailin, jelly roll or beta propeller folds). To ensure that designs with helical character were still represented in our test set, we chose 1–3 designs from each scaffold type excluding beta propellers, which were considerably larger than our other scaffolds. The selected designs are summarized in Table I, and examples of binding sites from selected designs are shown in Fig. 3C.
Of the 12 selected designs expressed with N-terminal polyhistidine tags, 6 failed to express and 3 formed soluble aggregates similar to those seen in Phase 2 (Table I). Notably, all of the proteins in β sheet scaffolds (lipocailin or jelly roll) either failed to express or formed soluble aggregates. The three remaining designs, which did express successfully, were all in α/β scaffolds with binding sites primarily located on loops rather than on stable secondary-structure elements. Using ITC, we determined that these three designs all bind zinc with the 1:1 stoichiometry that we predicted; however, all of their affinities were substantially lower than those typically found in naturally occurring zinc binding proteins (Table I). TIM1 was found to have a very low zinc affinity (>30 µM) and expressed at very low levels, whereas the two designs in periplasmic binding protein scaffolds (Hinge1 and Hinge2) bound somewhat more tightly (KD = 6.5 and 1.1 µM, respectively). Hinge2 was also found to be stabilized in the presence of zinc (Fig. 4B), whereas both TIM1 and Hinge1 showed no change in stability. Since it had the highest zinc affinity of the three, we focused on Hinge2 for further characterization.
Fig. 4.
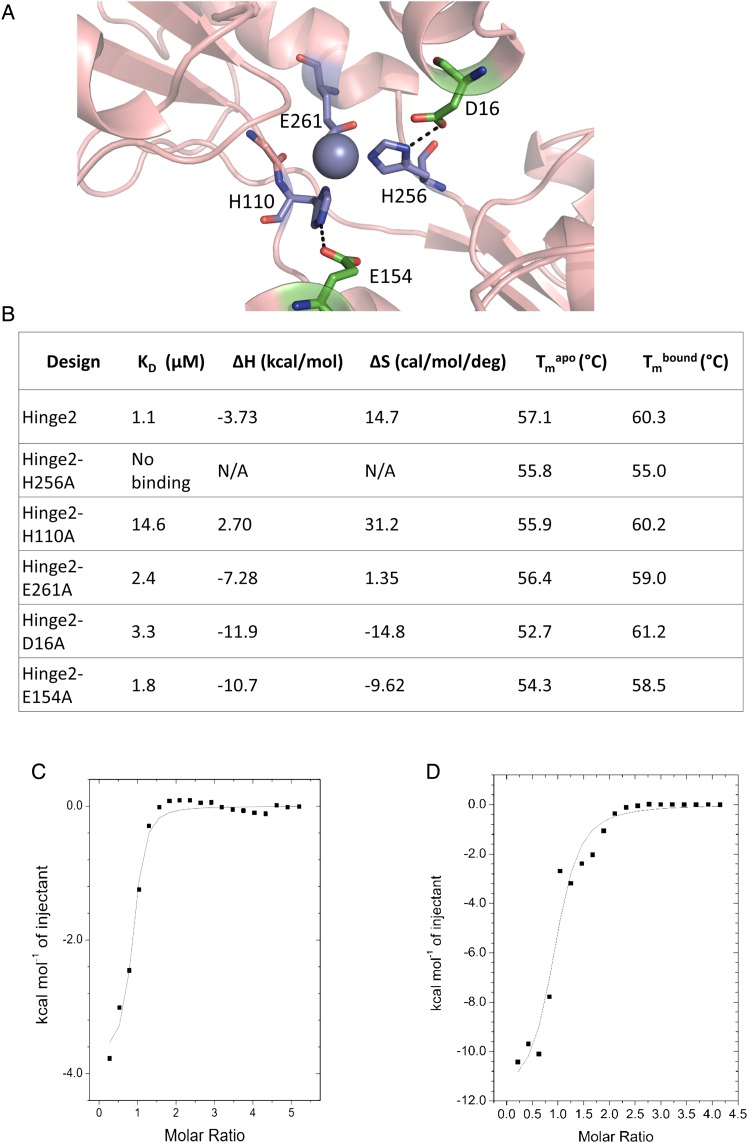
(A) Design model for Hinge2. The predicted binding site is composed of two histidines and one aspartate. Stabilizing hydrogen bonds were designed for two of the residues. (B) Comparison of zinc binding measurements for Hinge2 and point mutants of its three zinc-coordinating residues (H256A, H110A, E261A) and two hydrogen bond partners for those residues (D16A, E154A). All mutants show decreased affinity for zinc. The H256A mutation abolishes zinc binding. N/A, not applicable. (C and D) Sample ITC data for Hinge2 (C) and its D16A mutant (D).
We next determined the zinc affinities of alanine point mutations of the three coordinating and two second-shell residues of Hinge2 by ITC (Fig. 4B–D). Mutation of H256 leads to a complete elimination of zinc affinity as evidenced by both a lack of binding on ITC and a lack of thermal stabilization in the presence of zinc. Mutation of H110 leads to a substantial (∼10-fold) decrease in zinc affinity; however, the other three mutations lead to little change in affinity (∼1.6- to 3-fold decrease) (Fig. 4B). Overall, the data suggest that H256 and H110 participate in zinc binding; however, the remaining three point mutants (E261A, D16A and E154A) show little change in zinc affinity. Therefore, we cannot conclusively say that the site is formed as predicted.
The flexibility of the protein backbone at the zinc binding site may have contributed to differences in its metal coordination. Natural zinc binding sites favor ligand residues that are stabilized on secondary-structure elements, especially beta strands (Vallee and Auld, 1990; Mccall et al., 2000). Unfortunately, in our experience, protein scaffolds are less able to tolerate mutations in these positions, making many of the most promising metalloprotein designs difficult to express and purify.
In this case, the scaffold chosen for our designs is also known to undergo conformational changes upon ligand binding; binding of the natural ligand stabilizes proteins of this family in the closed conformation, which we used in our design models (Dwyer et al., 2003; Narunsky et al., 2015). Although our zinc binding site is near the original ligand binding site, we did not explicitly model the open conformation of our scaffold; therefore, the binding site likely takes on multiple conformations, and some of these alternate conformations may also be able to coordinate zinc in unexpected ways.
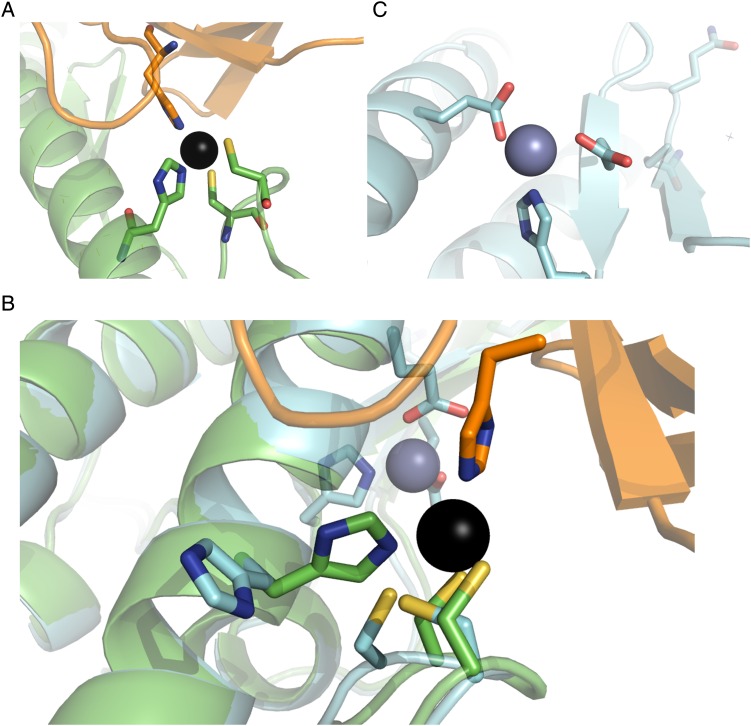
Unfortunately, possibly due to the issues discussed above, we have not yet been able to obtain a crystal structure of Hinge2 to confirm the geometry of its binding site. Our experiences with other designs have demonstrated that biophysical characterization will not necessarily reveal the geometry of a zinc binding site. In a 2013 study, Der et al. describe Spelter, a designed protein which was intended to form a zinc binding site at an interface with ubiquitin (Der et al., 2013). However, when we attempted to co-crystallize Spelter and ubiquitin, the resulting crystal structure did not contain ubiquitin. Instead, molecular replacement with two copies of Spelter's starting scaffold (PDB ID 2D4X) yielded a crystal structure with 1.7 Å resolution. As the designed zinc binding site occurs at the interface between Spelter and ubiquitin, this was a strong indication that the metal binding site was most likely not forming as we originally predicted. Indeed, despite extensive validation, including testing zinc binding to a variant reverting all mutations except the residues in the designed binding site to wild type, the crystal structure of Spelter does not show zinc binding at the designed site; the histidine (H192) and one of the cysteines (C137) predicted to participate in zinc binding have changed rotamers, and the loop containing the two cysteines (C135 and C137) has changed conformation resulting in a 2 Å movement in the alpha carbon of C135. Instead, a zinc ion is bound at a nearby site composed of two wild-type residues (H125 and E200) and one mutation (V122E) that was made for stability (Fig. 5). As in the case of ZE2, this difference may be partially explained by the lack of designed second-shell interactions in the zinc binding site. Cysteines in natural structural zinc binding sites typically interact with main-chain amide groups (Supplementary Fig. S2B); these interactions are not present in Spelter's designed zinc binding site (Supplementary Fig. S2A). In the crystal structure of Spelter, these cysteines have changed rotamers so that their side chains interact with neighboring backbone amide groups. H192 has likewise changed rotamers to form pi-stacking interactions with a nearby tryptophan.
Fig. 5.
Crystal structure of Spelter (Der et al., 2013) compared with design model. (A) The modeled zinc binding site includes two cysteines and a histidine from Spelter in addition to a residue from ubiquitin. (B) Overlay of design model and crystal structure. The designed zinc binding site (foreground) does not form. (C) An additional unmodeled zinc binding site was formed due to a mutation (V122E) made to increase stability during the design process.
Conclusion
Although several studies, including this one, have succeeded in designing sites that bind the desired metal, our results indicate that further advancements in computational metal binding site design methods will be necessary to design these sites with the precision necessary to achieve native-like affinity, specificity and function.
To solve this problem, it will be necessary to more carefully consider other factors in metal coordination that were not accounted for in this or previous studies. For instance, we did not directly take into account the electrostatic environment of the designed zinc binding sites, which can affect both their affinities and their functions. For instance, sites containing negatively charged residues stabilize the positive charge on zinc ions; while this increases their affinity for zinc, it simultaneously decreases the ion's catalytic activity by reducing its ability to activate water molecules (Dudev and Lim, 2008). Furthermore, we did not explicitly design a stabilizing hydrogen bond partner for such a water molecule; naturally occurring zinc enzymes such as carbonic anhydrase II typically have such an interaction (Christianson and Fierke, 1996). Although we did test to ensure that our designs did not contain buried unsatisfied polar atoms, we did not explicitly design additional interactions; the presence of a larger hydrogen bond network may improve the success of this approach.
Although we did perform backbone minimization during the refinement of our designs, backbone flexibility was not allowed during the initial search for binding sites and was limited throughout the protocol. Therefore, our search space was somewhat limited; in particular, it was difficult for us to find potential second-shell hydrogen bonds to backbone atoms, and we could not detect potential conformational changes caused by our mutations (such as those seen in the crystal structure of ZE2). Future techniques that take into account these effects on both the immediate environment of the site and the scaffold as a whole will be necessary to consistently achieve accurate design of these sites.
Supplementary data
Funding
This work was funded by National Science Foundation grant CBET-1403663 and National Institutes of Health grant GM117968 to B.K.
Supplementary Material
Acknowledgments
We thank Michael Miley, Mischa Machias, Ashutosh Tripathy and Samantha Piszkiewicz for help with crystallization, data analysis and biophysical characterization of designed proteins.
References
- Andreini C., Bertini I. (2012) J. Inorg. Biochem., 111, 150–156. [DOI] [PubMed] [Google Scholar]
- Andreini C., Bertini I., Cavallaro G., Holliday G.L., Thornton J.M. (2008) J. Biol. Inorg. Chem., 13, 1205–1218. [DOI] [PubMed] [Google Scholar]
- Auld D.S. (2001) Biometals, 14, 271–313. [DOI] [PubMed] [Google Scholar]
- Azia A., Levy R., Unger R., Edelman M., Sobolev V. (2015) Proteins Struct. Funct. Bioinforma., 83, 931–939. [DOI] [PubMed] [Google Scholar]
- Browner M.F., Hackos D., Fletterick R. (1994) Nat. Struct. Mol. Biol., 1, 327–333. [DOI] [PubMed] [Google Scholar]
- Burgoyne R.D. (2007) Nat. Rev. Neurosci., 8, 182–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S.H., Chen L., Russell D.H. (2014) J. Am. Chem. Soc., 136, 9499–9508. [DOI] [PubMed] [Google Scholar]
- Christianson D.W., Fierke C.A. (1996) Acc. Chem. Res., 29, 331–339. [Google Scholar]
- Der B.S., Edwards D.R., Kuhlman B. (2012a) Biochemistry, 51, 3933–3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Der B.S., Jha R.K., Lewis S.M., Thompson P.M., Guntas G., Kuhlman B. (2013) Proteins Struct. Funct. Bioinforma., 81, 1245–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Der B.S., Machius M., Miley M.J., Mills J.L., Szyperski T., Kuhlman B. (2012b) J. Am. Chem. Soc., 134, 375–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derewenda U., Derewenda Z., Dodson G.G., Hubbard R.E., Korber F. (1989) Br. Med. Bull., 45, 4–18. [DOI] [PubMed] [Google Scholar]
- Dudev T., Lim C. (2008) Annu. Rev. Biophys., 37, 97–116. [DOI] [PubMed] [Google Scholar]
- Dudev T., Lin Y., Dudev M., Lim C. (2003) J. Am. Chem. Soc., 125, 3168–3180. [DOI] [PubMed] [Google Scholar]
- Dwyer M.A., Looger L.L., Hellinga H.W. (2003) Proc. Natl. Acad. Sci. USA., 100, 11255–11260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. (2010) Acta Crystallogr. Sect. D Biol. Crystallogr., 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskandari V., Yakhchali B., Sadeghi M., Karkhane A.A. (2013) Biotechnol. Appl. Biochem., 60, 564–572. [DOI] [PubMed] [Google Scholar]
- Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M.R., Appel R.D., Bairoch A. (2005) In Walkered J.M. (eds), The Proteomics Protocols Handbook. Humana Press, Totowa, NJ, pp. 571–607. [Google Scholar]
- Hellinga H.W., Caradonna J.P., Richards F.M. (1991) J. Mol. Biol., 222, 787–803. [DOI] [PubMed] [Google Scholar]
- Jiang L., Althoff E.A., Clemente F.R. et al. (2008) Science, 319, 1387–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin S., Zhu Z.Y. (1997) Proc. Natl. Acad. Sci. USA., 94, 14231–14236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer L.L., Paterno S.A., Fierke C.A. (1995) J. Am. Chem. Soc., 117, 6831–6837. [Google Scholar]
- Klemba M., Gardner K.H., Marino S., Clarke N.D., Regan L. (1995) Nat. Struct. Biol., 2, 368–373. [DOI] [PubMed] [Google Scholar]
- Klemba M., Regan L. (1995) Biochemistry, 34, 10094–10100. [DOI] [PubMed] [Google Scholar]
- Le Guilloux V., Schmidtke P., Tuffery P. (2009) BMC Bioinformatics, 10, 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaver-Fay A., et al. (2011) Methods Enzymol., 487, 545–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.L., Lim C. (2004) J. Am. Chem. Soc., 126, 2602–2612. [DOI] [PubMed] [Google Scholar]
- Marino S.F., Regan L. (1999) Chem. Biol., 6, 649–655. [DOI] [PubMed] [Google Scholar]
- Matthews J.M., Sunde M. (2002) IUMB Life, 54, 351–355. [DOI] [PubMed] [Google Scholar]
- Mccall K.A., Huang C.chin, Fierke C.A. (2000) J. Nutr., 130, 1437S–1446S. [DOI] [PubMed] [Google Scholar]
- McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. (2007) J. Appl. Crystallogr., 40, 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills J.H., et al. (2013) J. Am. Chem. Soc., 135, 13393–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller H.N., Skerra A. (1994) Biochemistry, 33, 14126–14135. [DOI] [PubMed] [Google Scholar]
- Murshudov G.N., Skubák P., Lebedev A.A., Pannu N.S., Steiner R.A., Nicholls R.A., Winn M.D., Long F., Vagin A.A. (2011) Acta Crystallogr. Sect. D Biol. Crystallogr., 67, 355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narunsky A., Nepomnyachiy S., Ashkenazy H., Kolodny R., Ben-Tal N. (2015) Structure, 23, 2162–2170. [DOI] [PubMed] [Google Scholar]
- Nedd S., Redler R.L., Proctor E.A., Dokholyan N.V., Alexandrova A.N. (2014) J. Mol. Biol., 426, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien D.P., et al. (2015) Sci. Rep., 5, 14223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. (1997) Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Plegaria J.S., Dzul S.P., Zuiderweg E.R.P., Stemmler T.L., Pecoraro V.L. (2015) Biochemistry, 54, 2858–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan L., Clarke N.D. (1990) Biochemistry, 29, 10878–10883. [DOI] [PubMed] [Google Scholar]
- Richter F., Leaver-Fay A., Khare S.D., Bjelic S., Baker D. (2011) PLoS One, 6, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothlisberger D., et al. (2008) Nature, 453, 190–195. [DOI] [PubMed] [Google Scholar]
- Salgado E.N., Radford R.J., Tezcan F.A. (2010) Acc. Chem. Res., 43, 661–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapovalov M.V., Dunbrack R.L. (2011) Structure, 19, 844–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tainer J.A., Roberts V.A., Getzoff E.D. (1992) Curr. Opin. Biotechnol., 3, 378–387. [DOI] [PubMed] [Google Scholar]
- Vallee B.L., Auld D.S. (1990) Biochemistry, 29, 5647–5659. [DOI] [PubMed] [Google Scholar]
- Zanghellini A., Jiang L., Wollacott A.M., Cheng G., Meiler J., Althoff E.A., Rothlisberger D., Baker D. (2006) Protein Sci., 15, 2785–2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zastrow M.L., Peacock A.F.A., Stuckey J.A., Pecoraro V.L. (2012) Nat. Chem., 4, 118–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., et al. (2014) Nat. Chem., 6, 236–241. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.