Abstract
Background
Peripheral nerve injury leads to changes in gene expression in primary sensory neurons of the injured dorsal root ganglia. These changes are believed to be involved in neuropathic pain genesis. Previously, these changes have been identified using gene microarrays or next generation RNA sequencing with poly-A tail selection, but these approaches cannot provide a more thorough analysis of gene expression alterations after nerve injury.
Methods
The present study chose to eliminate mRNA poly-A tail selection and perform strand-specific next generation RNA sequencing to analyze whole transcriptomes in the injured dorsal root ganglia following spinal nerve ligation. Quantitative real-time reverse transcriptase polymerase chain reaction assay was carried out to verify the changes of some differentially expressed RNAs in the injured dorsal root ganglia after spinal nerve ligation.
Results
Our results showed that more than 50 million (M) paired mapped sequences with strand information were yielded in each group (51.87 M–56.12 M in sham vs. 51.08 M–57.99 M in spinal nerve ligation). Six days after spinal nerve ligation, expression levels of 11,163 out of a total of 27,463 identified genes in the injured dorsal root ganglia significantly changed, of which 52.14% were upregulated and 47.86% downregulated. The largest transcriptional changes were observed in protein-coding genes (91.5%) followed by noncoding RNAs. Within 944 differentially expressed noncoding RNAs, the most significant changes were seen in long interspersed noncoding RNAs followed by antisense RNAs, processed transcripts, and pseudogenes. We observed a notable proportion of reads aligning to intronic regions in both groups (44.0% in sham vs. 49.6% in spinal nerve ligation). Using quantitative real-time polymerase chain reaction, we confirmed consistent differential expression of selected genes including Kcna2, Oprm1 as well as lncRNAs Gm21781 and 4732491K20Rik following spinal nerve ligation.
Conclusion
Our findings suggest that next generation RNA sequencing can be used as a promising approach to analyze the changes of whole transcriptomes in dorsal root ganglia following nerve injury and to possibly identify new targets for prevention and treatment of neuropathic pain.
Keywords: Dorsal root ganglion, neuropathic pain, next generation sequencing, transcriptomes
Background
Neuropathic pain resulting from nerve injury is a complex and debilitating public health concern. It is characterized by hyperalgesia, allodynia, and spontaneous pain.1 Opioids provide effective relief of these symptoms, but extended opioid use produces hyperalgesia and tolerance, eliminating their effectiveness in chronic neuropathic pain relief.2 Identifying the mechanisms of peripheral nerve injury-induced neuropathic pain is essential for the discovery of novel treatments and preventative tactics for this disorder. It has been found that peripheral nerve injury produces abnormal ectopic activity at the neuroma of injured sites and within dorsal root ganglia (DRG) neurons.3,4 This abnormal activity, which has been identified as a neuropathic pain contributor, may be related to changes in the expression of noncoding RNAs (ncRNAs), receptors, ion channels, and other intracellular proteins in DRG neurons.5–8 Previous studies used gene microarrays to identify changes in the RNA transcript expression of several annotated genes within the DRG following peripheral nerve injury,5,9,10 but full analysis of the transcriptome and comprehensive gene expression patterns in neuropathic pain is still elusive.
Next-generation RNA sequencing (RNA-seq) is a highly sensitive method of analyzing differential expression of not only mRNAs but also ncRNAs and splice variants. Unlike gene microarrays, RNA-seq is not limited to a subset of known genes for analysis and can map reads to the entire genome of tissues and organs, providing information on possible functions of previously unannotated genes.11 Additionally, by neglecting to select for poly-A tailed mRNA, RNA-seq can detect more ncRNAs, as it has been reported that several ncRNAs lack poly-A tails.12 However, previous studies carried out next-generation RNA-seq with poly-A tail selection in the DRG under neuropathic pain conditions.6,13–15 To this end, the present study chose to perform next-generation RNA-seq with a higher sequencing depth and without mRNA poly-A tail selection to analyze the transcriptome of DRG following spinal nerve ligation (SNL). Compared to previous studies,6,13–15 we identified more splice variants as well as more long ncRNAs (lncRNAs) that displayed significant expression changes in DRG after SNL. Our findings provide novel information that can be used to identify key players in neuropathic pain mechanisms.
Methods
Animal preparation
Eight-week-old male C57BL/6 mice were obtained from Charles River Laboratories (Raleigh, NC) and were housed in the central housing facility under a standard 12 h light/12 h dark cycle at Rutgers New Jersey Medical School, Newark, NJ. Food and water were provided ad libitum. The Animal Care and Use Committee at Rutgers New Jersey Medical School approved all procedures used in this study. Additionally, all procedures are consistent with ethical guidelines produced by the National Institutes of Health and the International Association for the Study of Pain. Every effort was made to minimize the number of animals used and animal suffering. Experimenters were blinded to the treatment conditions of all animals.
L4 SNL-induced neuropathic pain model
For RNA-seq, SNL was performed on the forth lumbar (L4) spinal nerve according to our previous methods.16,17 As several DRG were required for RNA-seq, we carried out bilateral SNL to minimize the amount of animals used. For qPCR verification experiments, unilateral SNL was performed to provide validation that the RNA-seq results are repeatable. Mice were anesthetized with isoflurane and the L4 spinal nerve was exposed through removal of the L6 transverse process. After exposure and isolation of the L4 spinal nerve, a tight ligation with 7–0 silk thread was made and the nerve was transected distal to the ligature. The surgical procedure for the sham group was identical to that of the SNL group, except that the spinal nerves were not transected or ligated.
Behavioral testing
Paw withdrawal responses to mechanical stimuli: Mice were placed in individual Plexiglas chambers elevated on a mesh screen. Calibrated von Frey filament (0.38 g) was used to stimulate the center of each hind paw for 1 second. Each application represented one trial, and 10 trials were completed for each hind paw. The time between stimulation of the left paw and stimulation of the right paw was 5 minutes. The number of times the mouse exhibited a response (withdrawal, flicking, and licking) to the filament was recorded for each set of 10 trials, producing a frequency ([number of paw withdrawals/10 trials] × 100 = % response frequency). This frequency was used as a measurement of mechanical allodynia.
RNA extraction
Bilateral L4 DRG from SNL and sham groups were harvested six days after surgery, immediately treated in RNAlater (Ambion, Austin, TX) and subjected to total RNA extraction. To obtain enough RNA for sequencing, eight L4 DRGs from four mice were pooled together and each group required three biological replicates (n = 12 mice/group). For quantitative real-time PCR assay, four L4 DRGs from the ipsilateral sides of four SNL or sham mice were pooled together and the experiment was repeated three times (n = 12 mice/group). Briefly, total RNA was extracted using the miRNeasy kit with on-column digestion of genomic DNA (QIAGEN, Valencia, CA) according to the manufacturer’s instructions. RNA concentration was measured using the NanoDrop 2000 Spectrophotometer (Thermo Scientific, Wilmington, DE) and Qubit Fluorometric Quantitation (Invitrogen, Carlsbad, CA). Ratios of A260/280 nm were between 1.97 and 2.08. RNA integrity was assessed using RNA Nano chips in an Agilent 2100 Bionalyzer (Agilent technologies, Santa Clara, CA). RNA integrity numbers (RIN) were between 7.5 and 8.4.
RNA sequencing
The above total RNA extracted (1.2 µg/sample) was subjected to rRNA depletion by Ribo-Zero rRNA Removal (Human/Mouse/Rat) Kit (Illumina, San Diego, CA). Strand-specific RNA libraries were prepared using TruSeq Stranded Total RNA Sample Preparation Kit (Illumina) without poly-A selection. All assays were performed according to the manufacturer’s instructions. Sequencing was carried out at the Illumina HiSeq2500 platform High Output Mode, in a 2 × 100 base pairs paired-end configuration, with a total of more than 190 M reads per lane (at least 60 M reads per sample).
Bioinformatics analysis
Six samples were subjected to multiplexing, sequencing, differential gene expression analysis, transcript expression analysis, and ncRNA expression analysis. Briefly, the sequences were quality trimmed using trimmomatic-0.32 (minimal length 50 base pairs, leading and trailing Phred Q 30) first. The resulting sequencing data were then mapped to the musculus genome sequence version GRCm38.72, downloaded from ENSEMBLE. Gene hit counts and reads per kilobase per million (RPKM) mapped reads were calculated for each gene to determine expression levels.11 Comparisons of gene expression between groups were conducted using Student’s t-test. The long ncRNA was identified according to the annotation of biotype. The above RNA-seq analysis, including mapping, read counts, and expression analysis were performed within the CLCbio software environment (CLC genomics Workbench 7.0.2, CLC genomics Server). Mapped reads were visualized on the UCSC browser using bigwig files converted from bam files.
Pathway analysis
For biological pathway categorization, a total of 237 genes were selected based on their significance (false discovery rates, p < 0.05), differential expression (greater than normalized two-fold change following nerve injury), and ability to be categorized using the Panther Classification System database. Using the PANTHER (© 2015, Paul Thomas) system (pantherdb.org), we grouped genes based on biological pathways.
Quantitative real-time RT-PCR
Total RNA was reverse transcribed using the ThermoScript reverse transcriptase (Invitrogen) according to the manufacturer’s instructions with either the oligo (dT) primers or specific RT primers. cDNA was amplified by real-time PCR using the primers listed in Table 1 (Integrated DNA Technologies, Coralville, IA). Each sample was run in triplicate in a 20 μl reaction with 250 nM forward and reverse primers and 10 μl of Advanced Universal SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA). Reactions were performed in a BIO-RAD CFX96 real-time PCR system. The cycle parameters were set as follows: an initial 3-min incubation at 95℃, followed by 40 cycles of 95℃ for 10 s, 60℃ for 30 s, and 72℃ for 30 s. Ratios of ipsilateral-side mRNA levels to contralateral-side mRNA levels were calculated using the ΔCt method (2-ΔΔCt) at a threshold of 0.02. All data were normalized to Tubala (tubulin alpha 1 A gene), which was demonstrated to be stable after SNL in our pilot work.
Table 1.
Primers for RT-qPCR.
| Primer names | Sequences |
|---|---|
| Tuba1a–RT | TTC CAC AGA ATC CAC ACC AA |
| Kcna2-RT | GTC CCC GTC ACA TCT TCT CAC T |
| Gm21781-RT | CGG TGA TAC CAG CAC ACA TC |
| 4732491K20Rik-RT | CAG CAT CCA TGT GAA CCA AC |
| Tuba1a–F | GTG CAT CTC CAT CCA TGT TG |
| Tuba1a–R | GTG GGT TCC AGG TCT ACG AA |
| Kcna2-F | CTG CAA GGG CAA CGT CAC AC |
| Kcna2-R | GGG ACA GTG AGA TGC TTG GC |
| Oprm1-F | TCT TCA CCC TCT GCA CCA TG |
| Oprm1-R | TCT ATG GAC CCC TGC CTG TA |
| Gm21781-F | GAC CTT TGG AAG AGC AGT CG |
| Gm21781-R | CAG CCT GGT CTA CAC AGC AA |
| 4732491K20Rik-F | GGC CAA AAG ATT CCT CCT TC |
| 4732491K20Rik-R | TAG TGC CAG GCT GAC CTT CT |
RT-qPCR: reverse transcriptase quantitative polymerase chain reaction; F: Forward; R: Reverse.
Statistical analysis
All data were collected randomly and expressed as mean ± SEM. The data were statistically analyzed with two-way analysis of variance (ANOVA) or paired Student’s t test. When ANOVA showed a significant difference, pairwise comparison between means were tested by the post hoc Turkey method. p values less than 0.05 were considered statistically significant.
Results
Development of SNL-induced mechanical allodynia in mice
To make sure that all SNL mice used in the sequencing experiments showed pain hypersensitivity, mouse paw withdrawal frequency in response to mechanical stimulation was carried out. Consistent with our previous studies,16 SNL led to mechanical allodynia as demonstrated by a dramatic increase in paw withdrawal frequency for both hind paws on Day 6 post surgery (a time point at which altered gene expression and associated pain-related behavior are well established),16,17 compared to baseline (Figure 1). As expected, sham surgery did not produce a change in paw withdrawal frequency in either hind paw (Figure 1).
Figure 1.
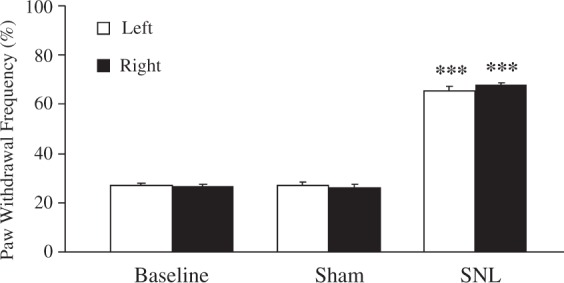
Bilateral L4 spinal nerve ligation produced mechanical allodynia in mice. SNL mice (n = 12) had significantly increased paw withdrawal frequencies in response to a .38 g von Frey hair for both hind paws six days after surgery compared with the sham group (n = 12). The results represent the mean ± SEM; **p < 0.01 compared with the sham group. SNL: spinal nerve ligation.
RNA-seq and genome-wide read mapping in DRGs after nerve injury
Through the use of illumine paired end sequence, 100 base pair long reads were yielded. More than 50 million (M) reads were identified per sample (51.87 M–56.12 M in the sham surgery group vs. 51.08 M–57.99 M in the SNL group; p = 0.68). Reads filtered using Illumina quality control were mapped to the reference mouse genome from ENSEMBLE (GRCm38.72), permitting up to one mismatch when aligning to the genome. The filtered reads that did not map to the genome or that could map to more than one genomic location were excluded from the analysis.
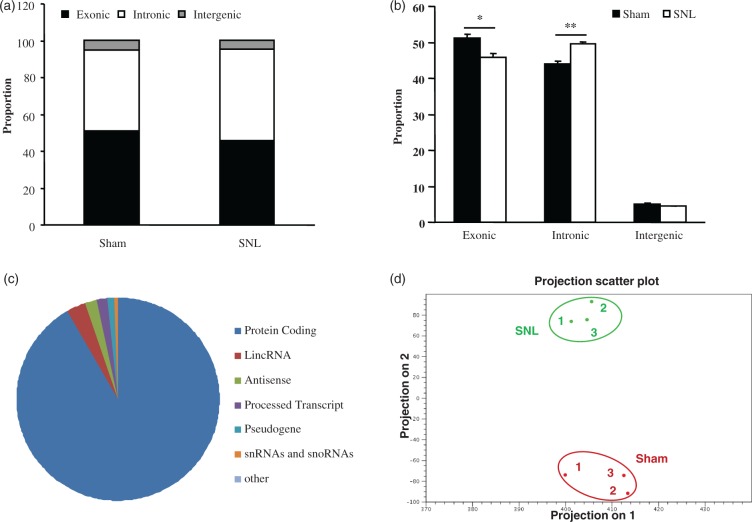
Mapped reads were categorized as exonic, intronic, or intergenic. Figure 2(a) illustrated the proportion of the reads belonging to each category in the sham and SNL groups. As expected, many reads were aligned to exonic regions in both groups (Figure 2(a)). Interestingly, a high proportion of reads also mapped to intronic regions in the sham and SNL groups (44.0% in the sham group and 49.6% in the SNL group; Figure 2(a)). In contrast, only 4.9% and 4.5% of reads mapped to intergenic regions in the sham and SNL groups, respectively (Figure 2(a)). Further analysis showed that SNL significantly decreased the proportion of reads aligning to exonic regions (p < 0.05), increased the proportion of reads aligning to intronic regions (p < 0.01), and did not significantly alter the proportion of reads aligning to intergenic regions, when compared with the sham group (Figure 2(b)).
Figure 2.
Transcriptome profiling in mice fourth lumbar dorsal root ganglia after nerve injury. (a) The mapped proportions of exonic, intronic, and intergenic reads in sham and SNL group. (b) There is a significantly lower proportion of reads that align to exonic regions in SNL samples when compared with sham samples, and the proportion of reads that align to intronic regions is significantly higher in SNL samples compared with sham samples. (c) The distribution of differentially expressed RNAs in the fourth lumbar dorsal root ganglia associated with peripheral nerve injury. (d) PCA was performed using the log2-transformed and quantile-normalized RPKM values. The colors indicate different groups of samples. The results represent the mean ± SEM of three independent experiments. *p < 0.05; **p < 0.01 compared with the sham group.
PCA: Principle component analysis; RPKM: reads per kilobase per million; SNL: spinal nerve ligation.
Finally, differentially expressed RNAs in the DRG associated with peripheral nerve injury were detected. Six days after SNL, the expression levels of 11,163 genes out of a total of 27,463 identified genes were significantly altered in the L4 DRG of SNL mice compared with the sham group (p < 0.05), of which 52.14% were upregulated and 47.86% downregulated. As expected, the largest transcriptional changes were seen in protein-coding RNAs (91.5%) (Figure 2(c)). Of the remaining differentially expressed RNAs, the largest transcriptional changes were observed in long interspersed ncRNAs (lincRNAs) followed in order by antisense RNAs, processed transcripts, pseudogene ncRNAs, snRNAs and snoRNAs, and others (e.g., nonsense-mediated decay) (Figure 2(c)). The variation of these differentially expressed RNAs across the samples from the sham and SNL groups was examined via principal component analysis. This analysis separated the samples into two clusters corresponding to the sham and SNL groups, respectively (Figure 2(d)), indicating that the robust differences in the changes of RNA expression occurred between the sham and SNL groups and not within groups.
Highest differentially expressed RNAs in the injured DRG following peripheral nerve injury
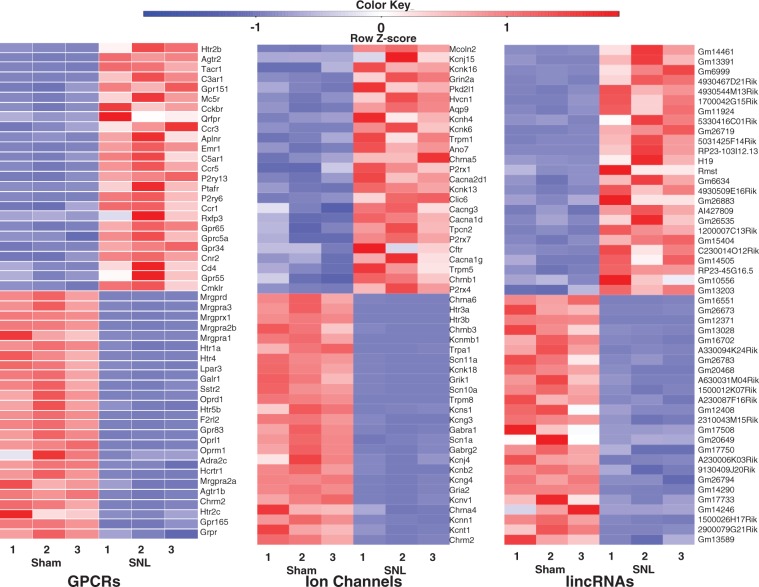
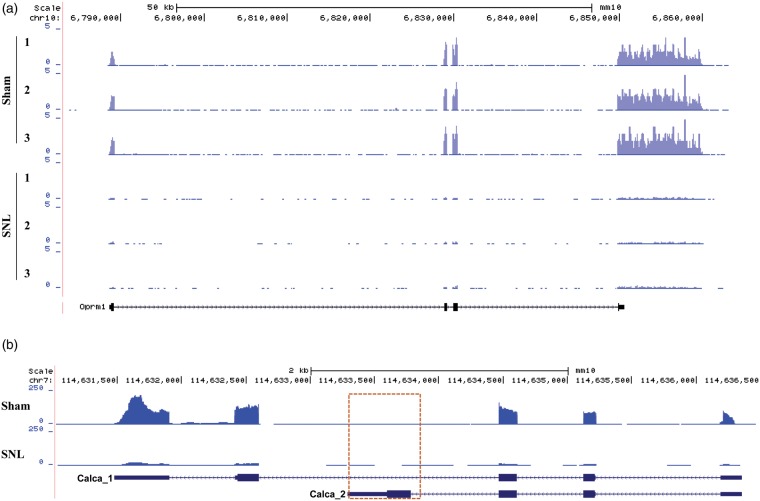
G protein-coupled receptors (GPCRs), ion channels, and lncRNAs play a key role in regulating the transmission of peripheral nociceptive information.18–23 Table 2 depicted expression changes in some of these identified genes that are highly relevant to pain, itch, touch, thermal, and chemical sensitivity. We first analyzed the top 25 significantly (p < 0.05) upregulated and top 25 significantly (p < 0.05) downregulated GPCR genes from the identified 403 GPCR genes in the injured DRG six days after SNL (Figure 3(a); Table 2). Consistent with previous studies, the type-2 angiotensin II receptor,24 substance P receptor,25 and C-C chemokine receptor type 526 increased after nerve injury. Additionally, the RNA-seq exhibited decreases in the mu-type opioid receptor,27 somatostatin receptor type 2,28 and Galanin receptor type 1,29 which was consistent with previous studies. All of the aforementioned GPCRS have known functions in neuropathic pain. The top 25 significantly (p < 0.05) upregulated and top 25 significantly (p < 0.05) downregulated ion channel genes (out of a total of 303 RNA-seq identified ion channels) are depicted in the heat map in Figure 3(b) and Table 2. Interestingly, RNA-seq data showed that genes Kcnk13,30 Chrna5,31 and Cacna2d132 increased following nerve injury, which is consistent with previous studies that found the functional significance of these genes in pain. Also consistent with previous studies, Scn10a, Scn11a,33 Gabra1,34 and Kcns135 decreased after SNL in our RNA-seq results. Our sequencing methods allowed for the identification of more lincRNAs than previous studies, as we neglected to perform poly-A tail selection. The top 25 significantly (p < 0.05) upregulated and top 25 significantly (p < 0.05) downregulated lincRNAs (out of a total of 1,597 RNA-seq identified lincRNAs) were displayed in the heatmap in Figure 3(c). As the discovery of lincRNAs is still at its infancy, the RNA-seq expression analysis of the lincRNAs depicted in Figure 3(c) is novel. Figure 4(a) depicts a representative example of RNA-seq reads for a differentially expressed gene. In this example, the reads mapped to the genomic location corresponding to Oprm1, the mu opioid receptor (MOR) 1 gene, for all samples in each group. MOR has previously been found to be downregulated in the injured DRG after peripheral nerve injury.36 In Figure 4(a), the stacked reads significantly decreased in the SNL samples compared with the sham samples. The reliability in RNA-seq was demonstrated by the repeatable decrease in MOR in all three SNL samples. Our further validation experiments using quantitative real-time reverse transcription polymerase chain reaction (qPCR) analysis confirmed consistent differential expression of selected genes including Kcna2, Oprm1 as well as lncRNAs Gm21781 and 4732491K20Rik (Figure 5).
Table 2.
Changes in some identified genes that are highly relevant to pain, itch, touch, thermal, and chemical sensitivity in the injured DRG after SNL.
| Upregulation |
Downregulation |
||||
|---|---|---|---|---|---|
| Name | Description | Ratio change | Name | Description | Ratio change |
| Agtr2 | Angiotensin II receptor, type 2 | 25.34 | Adra2c | Adrenergic receptor, alpha 2 c | 0.09 |
| Aqp9 | Aquaporin 9 | 4.35 | Asic3 | Acid-sensing (proton-gated) ion channel 3 | 0.39 |
| Atf3 | Activating transcription factor 3 | 9.64 | Atp1a1 | ATPase, Na+/K+ transporting, alpha 1 polypeptide | 0.27 |
| Cacna1d | Calcium channel, voltage-dependent, L type, alpha 1D subunit | 2.30 | Atp1b3 | ATPase, Na+/K+ transporting, beta 3 polypeptide | 0.63 |
| Cacna1g | Calcium channel, voltage-dependent, T type, alpha 1 G subunit | 2.18 | Calca | Calcitonin/calcitonin-related polypeptide, alpha | 0.14 |
| Cacna1h | Calcium channel, voltage-dependent, T type, alpha 1 H subunit | 1.05 (n.s.) | Chrna4 | Cholinergic receptor, nicotinic, alpha polypeptide 4 | 0.12 |
| Cacna2d1 | Calcium channel, voltage-dependent, alpha2/delta subunit 1 | 3.07 | Chrna6 | Cholinergic receptor, nicotinic, alpha polypeptide 6 | 0.01 |
| Cacng3 | Calcium channel, voltage-dependent, gamma subunit 3 | 2.31 | Chrnb3 | Cholinergic receptor, nicotinic, beta polypeptide 3 | 0.02 |
| Cckbr | Cholecystokinin B receptor | 10.00 | Gabbr1 | Gamma-aminobutyric acid (GABA) B receptor, 1 | 0.40 |
| Ccr1 | Chemokine (C-C motif) receptor 1 | 5.67 | Gabra1 | Gamma-aminobutyric acid (GABA) A receptor, alpha 1 | 0.07 |
| Ccr3 | Chemokine (C-C motif) receptor 3 | 8.25 | Gabrg2 | Gamma-aminobutyric acid (GABA) A receptor, subunit gamma 2 | 0.10 |
| Ccr5 | Chemokine (C-C motif) receptor 5 | 7.05 | Galr1 | Galanin receptor 1 | 0.05 |
| Chrna5 | Cholinergic receptor, nicotinic, alpha polypeptide 5 | 3.25 | Gria2 | Glutamate receptor, ionotropic, AMPA2 (alpha 2) | 0.11 |
| Gal | Galanin | 10.06 | Grik1 | Glutamate receptor, ionotropic, kainate 1 | 0.05 |
| Grin2a | Glutamate receptor, ionotropic, N-methyl D-aspartate 2 A | 6.64 | Htr1a | 5-Hydroxytryptamine (serotonin) receptor 1 A | 0.04 |
| Htr2b | 5-Hydroxytryptamine (serotonin) receptor 2B | 35.49 | Htr2c | 5-Hydroxytryptamine (serotonin) receptor 2 C | 0.14 |
| Hvcn1 | Hydrogen voltage-gated channel 1 | 5.90 | Htr3a | 5-Hydroxytryptamine (serotonin) receptor 3a | 0.01 |
| Kcnh4 | Potassium voltage-gated channel, subfamily H (eag-related), member 4 | 4.02 | Htr3b | 5-Hydroxytryptamine (serotonin) receptor 3b | 0.02 |
| Kcnj15 | Potassium inwardly rectifying channel, subfamily J, member 15 | 7.43 | Htr4 | 5-Hydroxytryptamine (serotonin) receptor 4 | 0.04 |
| Kcnk13 | Potassium channel, subfamily K, member 13 | 2.74 | Htr5b | 5-Hydroxytryptamine (serotonin) receptor 5b | 0.08 |
| Kcnk16 | Potassium channel, subfamily K, member 16 | 6.80 | Htr7 | 5-Hydroxytryptamine (serotonin) receptor 7 | 0.44 |
| Kcnk6 | Potassium inwardly rectifying channel, subfamily K, member 6 | 3.84 | Kcnb2 | Potassium voltage gated channel, Sham-related subfamily, member 2 | 0.10 |
| Mcoln2 | Mucolipin 2 | 9.84 | Kcnc2 | Potassium voltage gated channel, Sham-related subfamily, member 2 | 0.78 (n.s.) |
| P2rx1 | Purinergic receptor P2X, ligand-gated ion channel, 1 | 3.13 | Kcng3 | Potassium voltage-gated channel, subfamily G, member 3 | 0.06 |
| P2rx4 | Purinergic receptor P2X, ligand-gated ion channel 4 | 1.97 | Kcng4 | Potassium voltage-gated channel, subfamily G, member 4 | 0.11 |
| P2rx7 | Purinergic receptor P2X, ligand-gated ion channel, 7 | 2.26 | Kcnj4 | Potassium inwardly rectifying channel, subfamily J, member 4 | 0.10 |
| P2ry13 | Purinergic receptor P2Y, G-protein coupled 13 | 6.52 | Kcnk18 | Potassium channel, subfamily K, member 18 | 0.04 |
| P2ry6 | Pyrimidinergic receptor P2Y, G-protein coupled, 6 | 6.07 | Kcnmb1 | Potassium large conductance calcium-activated channel, subfamily M, beta member 1 | 0.03 |
| Pkd2l1 | Polycystic kidney disease 2-like 1 | 6.26 | Kcns1 | Potassium voltage-gated channel, subfamily S, 1 | 0.06 |
| Tacr1 | Tachykinin receptor 1 | 15.43 | Kcnt1 | Potassium channel, subfamily T, member 1 | 0.13 |
| Tpcn2 | Two pore segment channel 2 | 2.28 | Kcnv1 | Potassium channel, subfamily V, member 1 | 0.11 |
| Trpm1 | Transient receptor potential cation channel, subfamily M, member 1 | 3.43 | Mrgpra1 | MAS-related G protein-coupled receptors, member A1 | 0.03 |
| Trpm5 | Transient receptor potential cation channel, subfamily M, member 5 | 2.05 | Mrgpra2a | MAS-related G protein-coupled receptors, member A2A | 0.12 |
| Trpv4 | Transient receptor potential cation channel, subfamily V, member 4 | 1.89 | Mrgpra2b | MAS-related G protein-coupled receptors, member A2B | 0.03 |
| Mrgpra3 | MAS-related G protein-coupled receptors, member A3 | 0.01 | |||
| Mrgprd | MAS-related G protein-coupled receptors, member D | 0.01 | |||
| Mrgprx1 | MAS-related G protein-coupled receptors, member X1 | 0.02 | |||
| Oprd1 | Opioid receptor, delta 1 | 0.08 | |||
| Oprk1 | Opioid receptor, kappa 1 | 0.02 | |||
| Oprl1 | Opioid receptor-like 1 | 0.09 | |||
| Oprm1 | Opioid receptor, mu 1 | 0.09 | |||
| P2ry1 | Purinergic receptor P2Y, G-protein coupled 1 | 0.32 | |||
| Piezo2 | Piezo-type mechanosensitive ion channel component 2 | 0.71 | |||
| Scn10a | Sodium channel, voltage-gated, type X, alpha | 0.05 | |||
| Scn11a | Sodium channel, voltage-gated, type XI, alpha | 0.04 | |||
| Scn1a | Sodium channel, voltage gated, type I alpha subunit | 0.09 | |||
| Sst | Somatostatin | 0.02 | |||
| Sstr2 | Somatostatin receptor 2 | 0.05 | |||
| Trpa1 | Transient receptor potential cation channel, subfamily A, member 1 | 0.04 | |||
| Trpm8 | Transient receptor potential cation channel, subfamily M, member 8 | 0.06 | |||
| Trpv1 | Transient receptor potential cation channel, subfamily V, member 1 | 0.33 | |||
| Vip | Vasoactive intestinal polypeptide | 0.70 (n.s.) | |||
SNL: spinal nerve ligation; DRG: dorsal root ganglia. All ratio changes are significant p < 0.05 except if indicated by “n.s.”—no significant.
Figure 3.
Representative differentially expressed genes in fourth lumbar dorsal root ganglia after nerve injury. The 25 most up- and downregulated G protein-coupled receptors (a), ion channels (b), and long intergenic noncoding RNAs (c) in the fourth lumbar dorsal root ganglia are represented in the form of scaled heat maps created using Z-score values obtained using RNA-seq.
RNA-seq: RNA sequencing.
Figure 4.
Visualized reads mapping to the genomic location of differentially expressed genes and variants. (a) The stacked reads decreased in SNL samples significantly compared with sham samples in the genomic region of the MOR gene. (b) Alternative splicing analysis identified variant Calcal of the calcitonin/calcitonin gene-related polypeptide, alpha gene. The expression of Calca_2, the variant encoding calcitonin, was too low to be detected in DRG. The open box indicates that few reads were aligned to the unique exon region of Calca_2.
SNL: spinal nerve ligation; MOR: mu opioid receptor; DRG: dorsal root ganglia.
Figure 5.
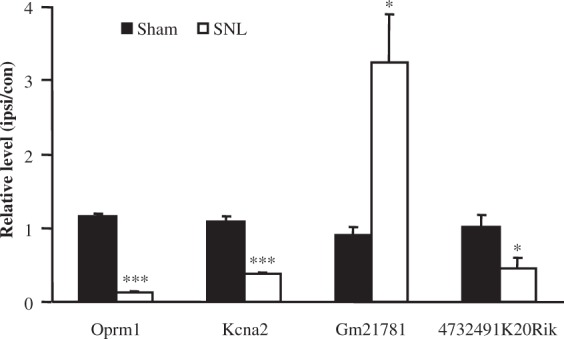
Validation of differentially expressed genes by qPCR. qPCR validated that Gm21781 RNA is significantly upregulated after SNL and Oprm1mRNA, Kcna2 mRNA, and 4732491K20Rik RNA are downregulated. The results represent the mean ± SEM of three independent experiments. *p < 0.05; **p < 0.01 compared with the sham group.
qPCR: quantitative polymerase chain reaction; SNL: spinal nerve ligation.
Alternative splicing variation after SNL
Our alternative splicing analysis identified a large number of specific differentially expressed variants. For example, calcitonin/calcitonin-related polypeptide alpha (Calca) expression levels decreased (−7.14 fold change of original values, p < 0.01) in the SNL group compared with the sham group. Alternative processing of the RNA transcribed from the Calca gene results in the production of calcitonin and another neuropeptide referred to as calcitonin gene-related peptide (CGRP).37 Figure 4(b) depicted the reads for each group for two variants of Calca mRNA. RNA-seq revealed a significant decrease (−6.96 fold, p < 0.01) in variant Calca_1 (ENSMUST00000032906) but not Calca_2 (ENSMUST00000032907) in the SNL group compared with the sham group. Calca_1 encodes the longer mRNA translating to CGRP1, while Calca_2 encodes the shorter mRNA translating to calcitonin polypeptide.37 The expression of Calca_2 is very low in DRG as indicated by the lack of reads in Figure 4(b). This RNA-seq validated decrease in CGRP mRNA in the injured DRG is consistent with previous neuropathic pain studies38,39 and further confirms the reliability of next-generation RNA-seq.
Genes involved in multiple pathways changed following nerve injury
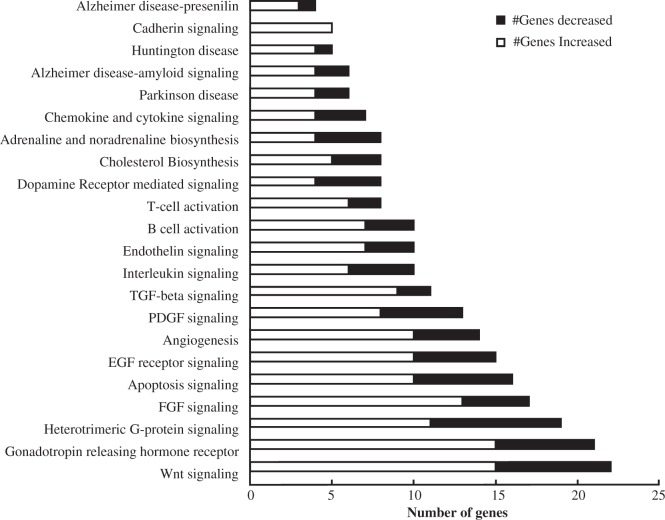
As our RNA-seq analysis resulted in a substantial amount of differentially expressed genes, we grouped genes based on their participation in biological pathways. Genes were selected for gene ontology pathway analysis after meeting the following criteria: the gene (a) has a greater than twofold change in expression (either increased or decreased) following nerve injury, (b) has an FDR p value less than 0.05, and (c) is a coding gene that can be categorized using the PANTHER pathway analysis database. The use of the FDR p value in this analysis further ensures that only the most significantly altered genes are included in the pathway analysis. In total, 237 genes met these criteria and were grouped into pathway categories (Figure 6). Notably, the Wnt signaling pathway, G-protein signaling, and Gonadotropin releasing hormone (GnRH) pathways represented major categories as more than 15 genes in each of these pathways were differentially expressed following nerve injury. Previous studies have identified Wnt Signaling as an important pathway involved in neuropathic pain.40,41 Interestingly, the expression of most of the genes in these pathways increased rather than decreased after nerve injury. This is consistent with previous gene profiling studies which also found more genes to be upregulated rather than downregulated after nerve injury.42,43
Figure 6.
Pathway analyses of the most significant and differentially expressed genes after nerve injury. PANTHER GO analyses groups the most significantly increased (white bars) and decreased (black bars) gene transcripts based on their annotated roles in biological pathways. A total of 237 genes are represented in this pathway analyses, all with greater than a twofold change after nerve injury and a FDR. p < 0.05. FDR: false discovery rate.
Discussion
Nerve injury is associated with gene expression changes within the DRG. Identifying which genes change and even more importantly how they change is a key factor in defining the mechanism of neuropathic pain development and maintenance. RNA-seq assay reveals in-depth and thorough analyses of the transcriptome changes associated with peripheral nerve injury.
Previous studies carrying out RNA-seq after nerve injury used poly-A tail selection and lower sequencing depths, limiting the amount of ncRNAs and differential genes that can be identified.6,15 In the present study, we carried out RNA-seq with no poly-A tail selection and sought to analyze changes in the transcriptome after nerve injury at a greater depth. Additionally, the use of a 100-base pair paired end configuration provides more accurate identification of lncRNAs. However, micro RNAs (miRNAs), due to their size, were overlooked using this configuration. A previous study has focused on identifying miRNAs that change following nerve injury,6 therefore, our focus was on lincRNAs. Strand specificity was maintained using the above kit, which is an important tactic for identifying strand-specific orientation of novel genes and ncRNAs (e.g., antisense ncRNA). Our study provides valuable information on differential gene expression, alternative splicing variation, and ncRNA differential expression after peripheral nerve injury.
Differential gene expression analysis using RNA-seq provided information on not only a select group of genes, as is the case with RNA microarrays,44 but rather the entire mouse transcriptome. This thorough exploration allows for the discovery of new possible pain targets. Figure 4 provides examples of genes that may have a yet unidentified function in pain, as only a few of the top 25 upregulated and downregulated genes have scientifically validated pain-related functions. Further studies into possible pain-related functions of these differentially expressed GPCRS and ion channels could build on our knowledge of pain mechanisms and possible targets for prevention and treatment of neuropathic pain.
In addition to changes in the expression of known genes, ncRNA differential expression could reveal novel epigenetic mechanisms of pain. ncRNAs include both small ncRNAs, such as miRNA, and lncRNAs. While miRNAs involved in pain have been extensively studied,6,45 the roles of lncRNAs in pain are still elusive. LncRNAs include antisense lncRNAs, doubled stranded RNA, and other long RNA species.46 We previously reported an antisense lncRNA, Kcna2 antisense RNA, which contributed to neuropathic pain by specific silencing of Kcna2 (or Kv1.2) expression in DRG.18 However, with the overwhelming amount of lncRNAs identified in the transcriptome, we anticipate that many more lncRNAs with a pain-related function exist. Many of the identified lincRNAs found in Figure 3(c) have no documented function. As these lincRNAs are differentially expressed after peripheral nerve injury, they could be involved in pain mechanisms in some manner. Validation of the expression of a select number of these unannotated ncRNAs reveals true expression and differential expression in the DRG of mice both before and after nerve injury (Figure 5). Further studies into the functions of these lincRNAs could unveil novel neuropathic pain contributors.
The added benefit of using RNA-seq is the ability to identify differential splicing variation. Alternative splicing is the process in which a single gene can result in multiple mRNAs simply due to exon inclusion/exclusion.47 Changes in alternative splicing after nerve injury have been reported12 and alternative splicing variation is thought to contribute to drug efficacy variance in chronic pain patients.48 In our study, we were able to identify a change in the splicing variation of Calca, the gene that encodes for calcitonin or CGRP. RNA-seq showed a decrease in the transcript that translates into CGRP. Interestingly, the Calcal transcript decreased −6.96 fold in relation to the sham group (Figure 4(b)). This is very consistent with the reported −7.14 fold change for the entire Calca gene. This consistency shows that the Calca decrease after SNL may be entirely due to a decrease in the Calca l transcript rather than the Calca_2 transcript. This is expected as calcitonin, the protein translated from the Calca_2 transcript is predominately expressed in the thymus and CGRP and the protein translated from the Calca l transcript is expressed in the DRG.37 The low expression of calcitonin in the DRG could explain the lack of a significant change in Calca_2 transcript expression after SNL. Further analysis of alternative splicing variations after nerve injury could provide insight into how certain genes change in expression in neuropathic pain.
In our study, thousands of genes changed in expression after nerve injury, so grouping these genes based on biological pathways provided an overall view of what may be occurring within the DRG. Our pathway analysis showed that genes involved in GnRH signaling were significantly increased following nerve injury. As males were used for this study, the increase in GnRH signaling would result in testosterone increases in the models. In previous studies of temporomandibular joint (TMJ) pain, gonadal hormones were found to reduce TMJ kappa-mediated antinociception by downregulating the expression of kappa opioid receptors on trigeminal ganglia sensory neurons.49 Also, several previous studies have found the expression of kappa opioid receptors in spinal cord and DRG to vary with the estrous cycle.50–52 An increase in GnRH following nerve injury may lead to reduced kappa opioid expression in DRG which could lead to pain. This evidence of increased GnRH after nerve injury at the transcription level may provide further validation of the role of sex hormones in neuropathic pain.
Conclusion
Much of pain-related research to date has focused on select genes that change after nerve injury, but as novel pain treatments are not readily implemented, it is clear that looking beyond the scope of a select subset of genes is necessary. With this study, we identified change at the transcriptome level of not only coding RNAs but also lncRNAs. Our unique and thorough RNA-seq approach provides insight into alternative splicing, which was overlooked by previous RNA-seq and peripheral nerve injury studies. The RNA-seq data obtained from this study have proven to be repeatable as made evident by previous studies and our own qPCR validation experiments. It is our hope that observations made from this article are pursued further with the goal of identifying novel targets for the prevention and treatment of neuropathic pain.
Author Contributions
SW, BL, PS, BT, AGK, AB, and YXT assisted with experimental design. BL, LL, XM and KM carried out animal surgery, behavioral testing, and tissue collection. SW carried out preparation of RNA samples for sequencing. SW, BL, XM, YC, and PD performed bioinformatics analysis and RT-qPCR confirmation. SW, BL, YH, and YXT assisted with drafting of the manuscript. YXT and YH contributed to the conception and implementation of this study. SW and BML equally contributed to this work.
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by grants from the NIH (NS072206, NS094664, HL117684, and DA033390) and the Rita Allen Foundation.
References
- 1.Latremoliere A, Woolf CJ. Central sensitization: a generator of pain hypersensitivity by central neural plasticity. J Pain 2009; 10: 895–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chu LF, Angst MS, Clark D. Opioid-induced hyperalgesia in humans: molecular mechanisms and clinical considerations. Clin J Pain 2008; 24: 479–496. [DOI] [PubMed] [Google Scholar]
- 3.Chung JM, Chung K. Importance of hyperexcitability of DRG neurons in neuropathic pain. Pain Pract 2002; 2: 87–97. [DOI] [PubMed] [Google Scholar]
- 4.Devor M. Ectopic discharge in Abita afferents as a source of neuropathic pain. Exp Brain Res 2009; 196: 115–128. [DOI] [PubMed] [Google Scholar]
- 5.Coyle DE. Spinal cord transcriptional profile analysis reveals protein trafficking and RNA processing as prominent processes regulated by tactile allodynia. Neuroscience 2007; 144: 144–156. [DOI] [PubMed] [Google Scholar]
- 6.Bali KK, Hackenberg M, Lubin A, et al. Sources of individual variability: miRNAs that predispose to neuropathic pain identified using genome-wide sequencing. Mol Pain 2014; 10: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ishikawa K, Tanaka M, Black JA, et al. Changes in expression of voltage-gated potassium channels in dorsal root ganglion neurons following axotomy. Muscle Nerve 1999; 22: 502–507. [DOI] [PubMed] [Google Scholar]
- 8.Costigan M, Scholz J, Woolf CJ. Neuropathic pain: a maladaptive response of the nervous system to damage. Annu Rev Neurosci 2009; 32: 1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gong Q, Lu Z, Huang Q, et al. Altered microRNAs expression profiling in mice with diabetic neuropathic pain. Biochem Biophys Res Commun 2015; 456: 615–620. [DOI] [PubMed] [Google Scholar]
- 10.Jiang BC, Sun WX, He LN, et al. Identification of lncRNA expression profile in the spinal cord of mice following spinal nerve ligation- induced neuropathic pain. Mol Pain 2015; 11: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mortazavi A, Williams BA, McCue K, et al. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 2008; 5: 621–628. [DOI] [PubMed] [Google Scholar]
- 12.Yi F, Yang F, Liu X, et al. RNA-seq identified a super-long intergenic transcript functioning in adipogenesis. RNA Biol 2013; 10: 991–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perkins JR, Antunes-Martins A, Calvo M, et al. A comparison of RNA-seq and exon arrays for whole genome transcription profiling of the L5 spinal nerve transection model of neuropathic pain in the rat. Mol Pain 2014; 10: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manteniotis S, Lehmann R, Flegel C, et al. Comprehensive RNA-Seq expression analysis of sensory ganglia with a focus on ion channels and GPCRs in trigeminal ganglia. PLoS One 2013; 8: e79523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alvarado S, Tajerian M, Millecamps M, et al. Peripheral nerve injury is accompanied by chronic transcriptome-wide changes in the mouse prefrontal cortex. Mol Pain 2013; 9: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tao YX, Rumbaugh G, Wang GD, et al. Impaired NMDA receptor-mediated postsynaptic function and blunted NMDA receptor-dependent persistent pain in mice lacking postsynaptic density-93 protein. J Neurosci 2003; 23: 6703–6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang W, Petralia RS, Takamiya K, et al. Preserved acute pain and impaired neuropathic pain in mice lacking protein interacting with C Kinase 1. Mol Pain 2011; 7: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao X, Tang Z, Zhang H, et al. A long noncoding RNA contributes to neuropathic pain by silencing Kcna2 in primary afferent neurons. Nat Neurosci 2013; 16: 1024–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caterina MJ, Julius D. The vanilloid receptor: a molecular gateway to the pain pathway. Annu Rev Neurosci 2001; 24: 487–517. [DOI] [PubMed] [Google Scholar]
- 20.Akopian AN, Sivilotti L, Wood JN. A tetrodotoxin-resistant voltage-gated sodium channel expressed by sensory neurons. Nature 1996; 379: 257–262. [DOI] [PubMed] [Google Scholar]
- 21.Wang W, Gu J, Li YQ, et al. Are voltage-gated sodium channels on the dorsal root ganglion involved in the development of neuropathic pain? Mol Pain 2011; 7: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee CY, Perez FM, Wang W, et al. Dynamic temporal and spatial regulation of mu opioid receptor expression in primary afferent neurons following spinal nerve injury. Eur J Pain 2011; 15: 669–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vanderah TW. Delta and kappa opioid receptors as suitable drug targets for pain. Clin J Pain 2010; 26(Suppl 10): S10–S15. [DOI] [PubMed] [Google Scholar]
- 24.Rice ASC, Smith MT. Angiotensin II type 2-receptor: new clinically validated target in the treatment of neuropathic pain. Clin Pharmacol Ther 2015; 97: 128–130. [DOI] [PubMed] [Google Scholar]
- 25.Chen YW, Tzeng JI, Huang PC, et al. Therapeutic ultrasound suppresses neuropathic pain and upregulation of substance P and Neurokinin-1 receptor in rats after peripheral nerve injury. Ultrasound Med Biol 2015; 41: 143–150. [DOI] [PubMed] [Google Scholar]
- 26.Matsushita K, Tozaki-Saitoh H, Kojima C, et al. Chemokine (C-C motif) receptor 5 is an important pathological regulator in the development and maintenance of neuropathic pain. Anesthesiology 2014; 120: 1491–1503. [DOI] [PubMed] [Google Scholar]
- 27.Back SK, Lee J, Hong SK, et al. Loss of spinal mu-opioid receptor is associated with mechanical allodynia in a rat model of peripheral neuropathy. Pain 2006; 123: 117–126. [DOI] [PubMed] [Google Scholar]
- 28.Morell MA, Camprubl-Robles MA, Culler MD, et al. Cortistatin attenuates inflammatory pain via spinal and peripheral actions. Neurobiol Dis 2014; 63: 141–154. [DOI] [PubMed] [Google Scholar]
- 29.Xu X, Yang X, Zhang P, et al. Effects of exogenous galanin on neuropathic pain state and change of galanin and its receptors in DRG and SDH after sciatic nerve-pinch injury in Rat. PLoS One 2012; 7: e37621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marsh B, Acosta C, Djouhri L, et al. Leak K + channel mRNAs in dorsal root ganglia: relation to inflammation and spontaneous pain behaviour. Mol Cell Neurosci 2012; 49: 375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Young T, Wittenauer S, Parker R, et al. Peripheral nerve injury alters spinal nicotinic acetylcholine receptor pharmacology. Eur J Pharmacol 2008; 590: 163–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nieto-Rostro M, Sandhu G, Bauer CS, et al. Altered expression of the voltage-gated calcium channel subunit a25-1: a comparison between two experimental models of epilepsy and a sensory nerve ligation model of neuropathic pain. Neuroscience 2014; 283: 124–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yin R, Liu D, Chhoa M, et al. Voltage-gated sodium channel function and expression in injured and uninjured rat dorsal root ganglia neurons. Int J Neurosci 2016; 126: 182–192. [DOI] [PubMed] [Google Scholar]
- 34.Yang G, Gao J, Jia Y, et al. Oxysophoridine through intrathecal injection induces antinociception and increases the expression of the GABAAa1 receptor in the spinal cord of mice. Planta Med 2012; 78: 874–880. [DOI] [PubMed] [Google Scholar]
- 35.Tsantoulas C, Zhu L, Shaifta Y, et al. Sensory neuron downregulation of the Kv9.1 potassium channel Subunit Mediates Neuropathic Pain following Nerve Injury. J Neurosci 2012; 32: 17502–17513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou XL, Yu LN, Wang Y, et al. Increased methylation of the MOR gene proximal promoter in primary sensory neurons plays a crucial role in the decreased analgesic effect of opioids in neuropathic pain. Mol Pain 2014; 10: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tran Q, Roesser JR. SRp55 is a regulator of Calcitonin/CGRP alternative RNA splicing. Biochemistry 2003; 42: 951–957. [DOI] [PubMed] [Google Scholar]
- 38.McGregor GP, Gibson SJ, Sabate IM, et al. Effect of peripheral nerve section and nerve crush on spinal cord neuropeptides in the rat; increased VIP and PHI in the dorsal horn. Neuroscience 1984; 13: 207–216. [DOI] [PubMed] [Google Scholar]
- 39.Noguchi K, Senba E, Morita Y, et al. aCGRP and PCGRP mRNAs are differentially regulated in the rat spinal cord and dorsal root ganglion. Brain Res Mol Brain Res 1990; 7: 299–304. [DOI] [PubMed] [Google Scholar]
- 40.Kurimoto S, Jung J, Tapadia M, et al. Activation of the Wnt/β catenin signaling cascade after traumatic nerve injury. Neuroscience 2015; 294: 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu Z, Chen Y, Yu J, et al. TCF4 mediates the maintenance of neuropathic pain through Wnt/p catenin signaling following peripheral nerve injury in rats. J Mol Neurosci 2015; 56: 397–408. [DOI] [PubMed] [Google Scholar]
- 42.Li S, Liu Q, Wang Y, et al. Differential gene expression profiling and biological process analysis in proximal nerve segments after sciatic nerve transection. PLoS One 2013; 8: e57000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang N, Li H, Sun Y, et al. Differential gene expression in proximal and distal nerve segments of rats with sciatic nerve injury during Wallerian degeneration. Neural Regen Res 2014; 9: 1186–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 2009; 10: 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lutz BM, Bekker A, Tao YX. Noncoding RNAs: new players in chronic pain. Anesthesiology 2014; 121: 409–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kung JTY, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics 2013; 193: 651–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Amiri S, Movahedin M, Mowla SJ, et al. Differential gene expression and alternative splicing of survivin following mouse sciatic nerve injury. Spinal Cord 2009; 47: 739–744. [DOI] [PubMed] [Google Scholar]
- 48.Jiang YQ, Andrade A, Lipscombe D. Spinal morphine but not ziconotide or gabapentin analgesia is affected by alternative splicing of voltage-gated calcium channel Cav2.2 pre-mRNA. Mol Pain 2013; 9: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clemente-Napimoga JT, Pellegrini-da-Silva A, VcHA Ferreira, et al. Gonadal hormones decrease temporomandibular joint kappa- mediated antinociception through a down-regulation in the expression of kappa opioid receptors in the trigeminal ganglia. Eur J Pharmacol 2009; 617: 41–47. [DOI] [PubMed] [Google Scholar]
- 50.Chang PC, Aicher SA, Drake T. Kappa opioid receptors in rat spinal cord vary across the estrous cycle. Brain Res 2000; 861: 168–172. [DOI] [PubMed] [Google Scholar]
- 51.Maggi R, Ma ZQ, Pimpinelli F, et al. Decrease of the number of opioid receptors and of the responsiveness to morphine during neuronal differentiation induced by 17 beta-estradiol in estrogen receptor-transfected neuroblastoma cells (SK-ER3). Neuroendocrinology 1999; 69: 54–62. [DOI] [PubMed] [Google Scholar]
- 52.Weiland NG, Wise PM. Estrogen and progesterone regulate opiate receptor densities in multiple brain regions. Endocrinology 1990; 126: 804–808. [DOI] [PubMed] [Google Scholar]