Abstract
An allelic variant of protein tyrosine phosphatase nonreceptor type 22 (PTPN22), PTPN22R620W, is strongly associated with type 1 diabetes (T1D) in humans and increases the risk of T1D by two- to fourfold. The NOD mouse is a spontaneous T1D model that shares with humans many genetic pathways contributing to T1D. We hypothesized that the introduction of the murine orthologous Ptpn22R619W mutation to the NOD genome would enhance the spontaneous development of T1D. We microinjected CRISPR-Cas9 and a homology-directed repair template into NOD single-cell zygotes to introduce the Ptpn22R619W mutation to its endogenous locus. The resulting Ptpn22R619W mice showed increased insulin autoantibodies and earlier onset and higher penetrance of T1D. This is the first report demonstrating enhanced T1D in a mouse modeling human PTPN22R620W and the utility of CRISPR-Cas9 for direct genetic alternation of NOD mice.
Introduction
Protein tyrosine phosphatase nonreceptor type 22 (PTPN22) is expressed in essentially all hematopoietic cell types and serves as a multifunctional molecular switch that fine-tunes T-cell, B-cell, and myeloid cell signaling and plays important roles in autoimmunity and infections (1). A minor allelic form, PTPN22R620W, increases type 1 diabetes (T1D) by two- to fourfold in humans (1,2). Studies of how this allele contributes to the etiopathology of T1D would be greatly assisted by the availability of a murine model in which spontaneous T1D is enhanced by this allele, as it is in humans.
The development of T1D results from the impairment of multiple immune tolerance mechanisms (3). Presumably due to this multigenic nature of T1D, previously constructed murine models in which human PTPN22R620W or its murine ortholog PTPN22R619W was introduced into nondiabetes-prone mice showed no spontaneous development of T1D (4–6). The NOD mouse is a spontaneous T1D model that shares with humans many interacting genetic pathways (3). We hypothesized that if we placed the murine ortholog of the R620W allele in its endogenous locus under physiological regulation, it would interact with other susceptibility alleles on the NOD background to increase T1D incidence and provide a spontaneous T1D model to study the mechanisms by which the disease-associated variant affects immune tolerance.
Research Design and Methods
Generation of Ptpn22R619W and Ptpn22 Knockout Mice Using a CRISPR-Cas9 System
Ptpn22R619W knock-in and Ptpn22 knockout (KO) mice were generated using CRISPR-Cas9 technology as previously reported (7). Four nucleotides were substituted in exon 14 of Ptpn22 to replace arginine (R) 619 to tryptophan (W) and to insert a BspEI restriction site. Homologous recombination was achieved by the coinjection of Cas9 mRNA transcript, single-guide RNA (sgRNA) for Ptpn22, and a homology-directed repair (HDR) template. The HDR template Ptpn22-R619W-HDR1 is a 200-nucleotide-long single-stranded DNA (ssDNA) oligo generated by Ultramer technology (Integrated DNA Technologies). The sgRNA target site was selected and the sgRNA synthesis performed as described previously (7). Briefly, a double-stranded DNA (dsDNA) template was generated by annealing Ptpn22-E14–60-sgRNA-F oligo with sgRNA common primer. A fill-in reaction produced dsDNA that was subsequently amplified by PCR, using T7–19 and sgRNA-R primers, and then purified using QIAquick PCR Purification Kit (Qiagen). dsDNA templates were subsequently used for in vitro transcription using the MEGAshortscript T7 Kit and enriched using the MEGAclear Kit (Invitrogen). Cas9 mRNA was in vitro transcribed from PmeI-lineralized and column-purified pcDNA3.3topo-T7-hCas9 plasmid using mMESSAGE mMACHINE T7 Ultra Kit (Life Technologies). pcDNA3.3topo-T7-hCas9 was created by adding a T7 promoter in the XbaI site of plasmid hCas9 (Addgene 41815), encoding the human codon optimized Cas9 nuclease. Cas9 mRNA transcripts were purified using the MEGAclear kit (Life Technologies) All oligo and primer sequences are shown in Supplementary Table 1.
Microinjection Into One-Cell Mouse Embryos
Embryos were generated by mating superovulated 12-week-old NOD/shiLtJ females to NOD/shiLtJ males (The Jackson Laboratory). ssDNA, sgRNA, and Cas9 mRNA were mixed in water (130 ng/μL, 50 ng/μL, and 100 ng/μL, respectively) and coinjected into cytoplasm using standard micromanipulation equipment (Narishige). Injected embryos were surgically transplanted into oviducts of pseudopregnant CD1 recipient mice.
Breeding, Genotyping, Cloning, and Sequencing
To minimize the risk of off-target effects caused by CRISPR-Cas9, founder M3 and F6 mice were backcrossed with NOD wild-type (WT) mice for two generations and selected solely for the presence of Ptpn22R619W allele or KO allele to generate N2 mice, which were then intercrossed to generate N2F1 mice that were used for this study. DNA extraction of mouse tail snips (BioPioneer) was used for PCR genotyping using forward primer Ptpn22F1 and reverse primer Ptpn22R1. PCR products were analyzed in one of the three ways: 1) electrophoresis of restriction enzyme BspEI digests, 2) sequencing (Eton Bioscience) of treated PCR products (ExoSAP-IT; Affymetrix), and 3) sequencing of PCR products cloned into vector pJET1.2 (CloneJET; Thermo Scientific).
Measurement of Insulin Autoantibodies
Insulin autoantibodies (IAAs) were measured with a currently standard 96-well plate micro-IAA radiobinding assay at the Barbara Davis Center Autoantibody/HLA Service Center, as previously described (8). The result was expressed as an index and an upper limit of normal range (index 0.010) was established as 99th percentile of nonautoimmune-prone mice.
Reverse Transcription PCR and Western Blot Analysis
mRNAs were extracted from ∼5 × 106 cells from each sample by RNeasy Mini Kit (Qiagen). A total of 500 ng of mRNA was used to perform reverse transcription (RT) using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). A total of 300 ng cDNA was then used for PCR reactions using primers Ptpn22F2 and Ptpn22R2. Primer sequences are shown in Supplementary Table 1. Western blot analysis was performed as described previously (9). PTPN22 protein was detected using P2 antibody against PTPN22 amino acids 670-740 (5,10), a region conserved between R and W alleles. Actin was detected using β-actin antibody #4967 (Cell Signaling Technology).
Assessment of Diabetes in Mice
Experimental procedures were carried out according to the National Institutes of Health Guide for the Care and Use of Laboratory Animals and approved by The Scripps Research Institute Animal Care and Use Committee. Mice were monitored weekly for nonfasting blood glucose via retro-orbital bleeding and measurement with blood glucose test strips and TRUEresult meters (TRUEtest). Mice having nonfasting blood glucose ≥250 mg/dL for two consecutive tests were classified as diabetic and euthanized thereafter.
Statistical Analyses
Kaplan-Meier survival curves were plotted for T1D incidence. The P values were calculated using log-rank test. To analyze the measurements of IAAs, the P value was calculated using Mann-Whitney U test, without assuming normal distribution of the data.
Results
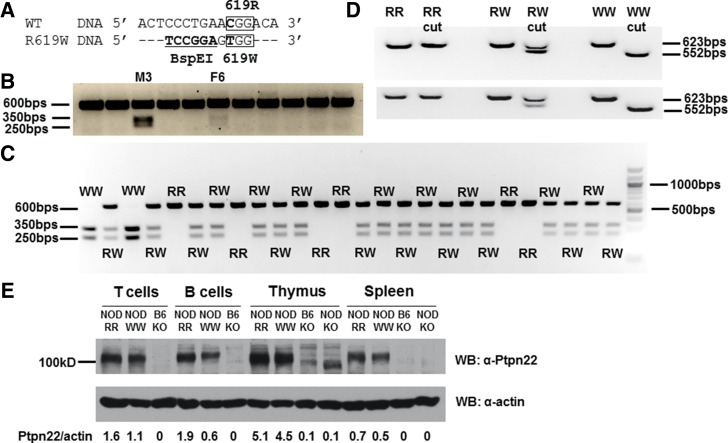
In order to directly modify genes on a pure NOD genetic background, we used the novel CRISPR-Cas9 technology coupled with a microinjection of single-cell mouse zygotes (11–14). We designed and microinjected a three-component CRISPR system (7) consisting of Cas9 mRNA, sgRNA, and ssDNA repair template into NOD zygotes to introduce a single amino-acid mutation R619W in the endogenous Ptpn22 gene (Fig. 1A). We also introduced synonymous mutations to create a BspEI restriction site to facilitate genotyping (Fig. 1A). This injection attempt led to the birth of 11 mice, 2 of which were found to contain Ptpn22R619W as indicated by restriction digestion (Fig. 1B). These two mice, male #3 (M3) and female #6 (F6), were subsequently used as founders of two independent strains and were backcrossed with WT NOD mice for two generations to produce N2 descendants that were intercrossed to generate N2F1 mice used in this study. This breeding strategy further reduced the risk of potential off-target effects caused by CRISPR-Cas9. Sequencing of M3 mouse genomic PCR confirmed all four nucleotide substitutions (Supplementary Fig. 1A). The F6 mouse was found to be a mosaic mouse (13,14) that had not only WT and R619W alleles but also a KO allele resulting from a random addition of 13 nucleotides incorporated by a nonhomologous end-joining mechanism (Supplementary Fig. 1B–D). Both mutant alleles and the KO allele were bred to homozygosity in the N2F1 generation to produce three new lines on the NOD background (Fig. 1C and Supplementary Figs. 2 and 3).
Figure 1.
Ptpn22R619W design, expression, and germline transmission. A: WT and R619W Ptpn22 DNA sense strains are aligned. C-to-T transition results in R619W mutation. Synonymous nucleotide substitutions create a BspEI restriction site. B: Genomic PCR generated a 600 bps fragment from all CRISPR mice. Only mutant (W) PCR product could be cut into 350 and 250 bps. M3 and F6 carried the mutation. C: RR, RW, and WW mice were identified in M3 strain N2F1 descendants. D: Traditional RT-PCR of mRNA derived from thymus (top) and spleen (bottom) of RR, RW, and WW mice. E: Expression of PTPN22 proteins in T and B cells, thymus, and spleen was analyzed by Western blot (WB). NOD Ptpn22 RR, WW, KO strains were analyzed, and B6 Ptpn22 KO strain was included as a negative control.
Traditional RT-PCR followed by BspEI digestion verified the expression of Ptpn22 mRNA from either R allele or W allele in both thymocytes and splenocytes of homozygous WT (RR), heterozygous mutant (RW), and homozygous mutant (WW) mice (Fig. 1D), as only the W allele had a restriction site. However, KO mice did not show any detectable Ptpn22 mRNAs that could be PCR amplified (Supplementary Fig. 4). Using a previously published polyclonal antibody that binds to a conserved PTPN22 protein region, we detected the expression of the W form of PTPN22 protein in thymocytes, splenocytes, and purified T and B cells (Fig. 1E). NOD KO spleen showed no expression of PTPN22, while B6 KO strain (10) was used as a negative control (Fig. 1E). Considering the absence of Ptpn22 mRNA (Supplementary Fig. 4) and the fact that no PTPN22 protein was observed in the NOD KO spleen (Fig. 1E), the bands observed in the NOD KO thymus (Fig. 1E) were likely due to the other mouse proteins.
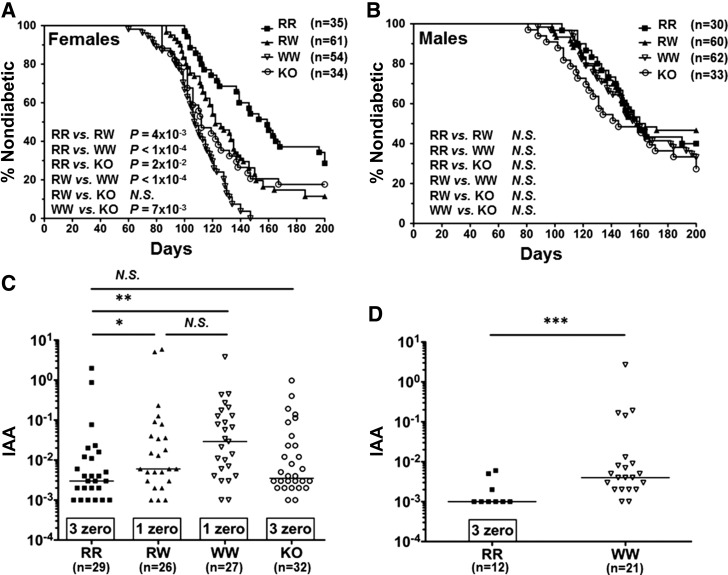
Having validated Ptpn22R619W and KO alleles at the DNA, mRNA, and protein levels, we used these mutants to test the effects of modified PTPN22 alleles on spontaneous T1D development. Ptpn22R619W variant was found to be associated with an earlier onset of T1D and an increased penetrance in RW females (Fig. 2A). WW females showed even earlier disease onset and further increased penetrance (Fig. 2A). Ptpn22 KO females showed a diabetogenic phenotype that is significantly more severe than RR, less severe than WW, and yet comparable to RW females in terms of disease onset and penetrance (Fig. 2A).
Figure 2.
Ptpn22R619W increases diabetes incidence and IAAs. A and B: Kaplan-Meier survival curves were plotted showing percentage of nondiabetic animals over time. A: Survival curves of NOD RR, RW, WW, and KO females are shown. B: Survival curves of RR, RW, WW, and KO males are shown. C: IAAs in the sera of 8-week-old RR, RW, WW, and KO females are shown. D: IAAs in the sera of 4-week-old RR and WW females are shown. *P < 0.05; **P < 10−3; ***P < 10−4; N.S., P > 0.05.
No significant difference of T1D incidence between M3 and F6 strains was observed, in either males or females (Supplementary Fig. 5A–D). Female NOD mice generally develop T1D sooner and with greater penetrance than males (15). We found that the diabetogenic effects of Ptpn22R619W did not penetrate the NOD mice sex barrier (15) and that RW, WW, or KO males did not show significantly increased T1D incidence compared with WT males (Fig. 2B).
It has recently been reported that PTPN22R620W human carriers with diabetes exhibited elevated IAAs (16,17) relative to RR homozygous individuals. We investigated whether our mutant mice also exhibited an insulin antibody phenotype resembling human carriers and discovered that 8-week-old RW and WW females exhibited significantly increased serum IAA levels compared with RR females (Fig. 2C). This significant increase was detectable as early as 4 weeks of age in WW females (Fig. 2D). KO females did not exhibit increased serum IAAs (Fig. 2C).
Discussion
In the past 20 years, gene targeting of NOD mice was generally carried out in the 129/Sv mouse–derived embryonic stem cells (ESCs) and then required backcrossing with NOD mice for at least 10 generations (18). There were few studies reporting generation and use of germline-competent NOD ESCs suitable for gene targeting (19,20). To circumvent the necessity to modify NOD ESCs and test germline transmission, a study reported the use of zinc-finger nuclease coupled with embryo microinjection to modify genes on a pure NOD background (21). The emerging CRISRP-Cas9 technology coupled with embryo microinjection or in vitro fertilization has demonstrated the relative ease of design and high efficiency of mouse genome modification in a single step (13,14,22).
To our knowledge, this is the first time the murine ortholog of the autoimmune-associated PTPN22 allele has been introduced into NOD mice using CRISPR-Cas9 technology to successfully produce a disease phenotype that closely mimics the effects of the allele in humans (1). The association of W variant with an earlier onset of T1D and an increased penetrance in heterozygous mice (Fig. 2A) suggests strong effect of Ptpn22R619W allele in heterozygosity, which closely resembles the diabetogenic effects of PTPN22R620W on human heterozygous carriers (1). In fact, PTPN22R620W is most commonly carried in heterozygosity in humans (1,2). The association of W variant homozygosity with significantly earlier disease onset and further enhanced penetrance (Fig. 2A) suggests a gene dosage effect, which was also consistent with the effect of PTPN22R620W observed in human homozygous carriers (1,2). Therefore, our newly generated NOD Ptpn22R619W strain is an excellent physiological model that will provide mechanistic insights into how the human PTPN22R620W allele contributes to T1D. We should be able to target other genes using the same techniques to generate physiological models closely resembling human diseases and to test various human disease-associated polymorphisms in mice that were genetically difficult to modify in the past.
Converging lines of evidence pointed to insulin as a primary antigen in the diabetogenic process (23). It was reported that 129/B6 mice expressing homozygous Ptpn22R619W alleles showed increased IAAs, but no spontaneous diabetes was observed, presumably due to the multigenic nature of T1D requiring the presence of multiple T1D-specific susceptible alleles in the genome (5). As PTPN22R620W human carriers with diabetes exhibited elevated IAAs (16,17), we tested whether Ptpn22R619W mutant NOD mice showed increased IAAs. Indeed, as early as 4 weeks of age, WW mice showed significantly increased prevalence and elevated titer of IAAs (Fig. 2D), suggesting an early loss of tolerance to insulin, which was not observed in WT mice (8). The increased prevalence and elevated titer of IAAs could also be detected in RW and WW mice at 8 weeks of age (Fig. 2C), a time when such autoantibodies peak in WT mice (8). Interestingly, Ptpn22 KO mice did not show increased prevalence and elevated titer of IAAs (Fig. 2C) and did not exhibit as severe an enhancement of T1D as seen in WW mice, consistent with the hypothesis that the W variant is not simply a loss-of-function variant (1,24,25).
Article Information
Acknowledgments. The authors thank Linda S. Wicker, of University of Cambridge; Nunzio Bottini and Stephanie Stanford, of La Jolla Institute for Allergy and Immunology; and Bruce E. Torbett, David Nemazee, Kerri Mowen, and Dwight Kono, of The Scripps Research Institute, for their helpful discussions.
Funding. Research was supported by a grant from the National Institute of Allergy and Infectious Diseases (R21AI119353) of the National Institutes of Health. X.L. is supported by a graduate student fellowship from The Scripps Research Institute Kellogg School of Science and Technology.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. X.L. designed the study, performed experiments, analyzed data, made the figures, and wrote and edited the manuscript. S.P. and S.G. designed the study, performed experiments, and edited the manuscript. S.R., C.J.M., K.M., A.R.R., G.M., and L.J. performed experiments. Y.D.D. contributed mice. K.S. contributed reagents. S.K. and D.R.G. designed the study. L.Y. designed the study, contributed reagents, and performed experiments. L.A.S. designed the study, analyzed data, supervised the research, and edited the manuscript. L.A.S. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db16-0061/-/DC1.
References
- 1.Bottini N, Peterson EJ. Tyrosine phosphatase PTPN22: multifunctional regulator of immune signaling, development, and disease. Annu Rev Immunol 2014;32:83–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Todd JA. Etiology of type 1 diabetes. Immunity 2010;32:457–467 [DOI] [PubMed] [Google Scholar]
- 3.Ridgway WM, Peterson LB, Todd JA, et al. Gene-gene interactions in the NOD mouse model of type 1 diabetes. Adv Immunol 2008;100:151–175 [DOI] [PubMed] [Google Scholar]
- 4.Zhang J, Zahir N, Jiang Q, et al. The autoimmune disease-associated PTPN22 variant promotes calpain-mediated Lyp/Pep degradation associated with lymphocyte and dendritic cell hyperresponsiveness. Nat Genet 2011;43:902–907 [DOI] [PubMed] [Google Scholar]
- 5.Dai X, James RG, Habib T, et al. A disease-associated PTPN22 variant promotes systemic autoimmunity in murine models. J Clin Invest 2013;123:2024–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y, Shaked I, Stanford SM, et al. The autoimmunity-associated gene PTPN22 potentiates toll-like receptor-driven, type 1 interferon-dependent immunity. Immunity 2013;39:111–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pelletier S, Gingras S, Green DR. Mouse genome engineering via CRISPR-Cas9 for study of immune function. Immunity 2015;42:18–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu L, Robles DT, Abiru N, et al. Early expression of antiinsulin autoantibodies of humans and the NOD mouse: evidence for early determination of subsequent diabetes. Proc Natl Acad Sci U S A 2000;97:1701–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu G, Casas J, Rigaud S, et al. Themis sets the signal threshold for positive and negative selection in T-cell development. Nature 2013;504:441–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hasegawa K, Martin F, Huang G, Tumas D, Diehl L, Chan AC. PEST domain-enriched tyrosine phosphatase (PEP) regulation of effector/memory T cells. Science 2004;303:685–689 [DOI] [PubMed] [Google Scholar]
- 11.Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013;339:819–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mali P, Yang L, Esvelt KM, et al. RNA-guided human genome engineering via Cas9. Science 2013;339:823–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang H, Yang H, Shivalila CS, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 2013;153:910–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 2013;154:1370–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pozzilli P, Signore A, Williams AJ, Beales PE. NOD mouse colonies around the world--recent facts and figures. Immunol Today 1993;14:193–196 [DOI] [PubMed] [Google Scholar]
- 16.Lempainen J, Laine AP, Hammais A, et al. Non-HLA gene effects on the disease process of type 1 diabetes: From HLA susceptibility to overt disease. J Autoimmun 2015;61:45–53 [DOI] [PubMed] [Google Scholar]
- 17.Pihoker C, Gilliam LK, Hampe CS, Lernmark A. Autoantibodies in diabetes. Diabetes 2005;54(Suppl. 2):S52–S61 [DOI] [PubMed] [Google Scholar]
- 18.Markel P, Shu P, Ebeling C, et al. Theoretical and empirical issues for marker-assisted breeding of congenic mouse strains. Nat Genet 1997;17:280–284 [DOI] [PubMed] [Google Scholar]
- 19.Nichols J, Jones K, Phillips JM, et al. Validated germline-competent embryonic stem cell lines from nonobese diabetic mice. Nat Med 2009;15:814–818 [DOI] [PubMed] [Google Scholar]
- 20.Jakubczik F, Jones K, Nichols J, Mansfield W, Cooke A, Holmes N. A SNP in the immunoregulatory molecule CTLA-4 controls mRNA splicing in vivo but does not alter diabetes susceptibility in the NOD mouse. Diabetes 2016;65:120–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen YG, Forsberg MH, Khaja S, Ciecko AE, Hessner MJ, Geurts AM. Gene targeting in NOD mouse embryos using zinc-finger nucleases. Diabetes 2014;63:68–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li F, Cowley DO, Banner D, Holle E, Zhang L, Su L. Efficient genetic manipulation of the NOD-Rag1-/-IL2RgammaC-null mouse by combining in vitro fertilization and CRISPR/Cas9 technology. Sci Rep 2014;4:5290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Unanue ER. Antigen presentation in the autoimmune diabetes of the NOD mouse. Annu Rev Immunol 2014;32:579–608 [DOI] [PubMed] [Google Scholar]
- 24.Vang T, Congia M, Macis MD, et al. Autoimmune-associated lymphoid tyrosine phosphatase is a gain-of-function variant. Nat Genet 2005;37:1317–1319 [DOI] [PubMed] [Google Scholar]
- 25.Zheng P, Kissler S. PTPN22 silencing in the NOD model indicates the type 1 diabetes-associated allele is not a loss-of-function variant. Diabetes 2013;62:896–904 [DOI] [PMC free article] [PubMed] [Google Scholar]