Abstract
The mammalian target of rapamycin complex 1 (mTORC1) regulates several biological processes, although the key downstream mechanisms responsible for these effects are poorly defined. Using mice with deletion of eukaryotic translation initiation factor 4E-binding protein 2 (4E-BP2), we determine that this downstream target is a major regulator of glucose homeostasis and β-cell mass, proliferation, and survival by increasing insulin receptor substrate 2 (IRS2) levels and identify a novel feedback mechanism by which mTORC1 signaling increases IRS2 levels. In this feedback loop, we show that 4E-BP2 deletion induces translation of the adaptor protein SH2B1 and promotes the formation of a complex with IRS2 and Janus kinase 2, preventing IRS2 ubiquitination. The changes in IRS2 levels result in increases in cell cycle progression, cell survival, and β-cell mass by increasing Akt signaling and reducing p27 levels. Importantly, 4E-BP2 deletion confers resistance to cytokine treatment in vitro. Our data identify SH2B1 as a major regulator of IRS2 stability, demonstrate a novel feedback mechanism linking mTORC1 signaling with IRS2, and identify 4E-BP2 as a major regulator of proliferation and survival of β-cells.
Introduction
Type 2 diabetes is characterized by insufficient β-cell expansion in conditions of obesity-induced insulin resistance. Recent data suggest that the nutrient environment in states of overnutrition and obesity could play a role in the adaptation of β-cells to insulin resistance. How the nutrient environment modulates the β-cell response during adaptation to diabetogenic conditions is not completely understood.
The mammalian target of rapamycin complex 1 (mTORC1) signaling pathway integrates signals from growth factors and nutrients signals to regulate cell size and proliferation (1–3). In β-cells, mTORC1 activity is increased during conditions of insulin resistance (4). Modulation of mTORC1 function by genetic or pharmacologic manipulation highlights the role of this pathway in the regulation of β-cell mass (4–9). Genetic models with activation of mTORC1 in β-cells exhibit β-cell mass expansion caused by increases in both proliferation and cell size (4–9). mTORC1 controls growth (cell size) and proliferation (cell number) by modulating protein translation through phosphorylation of 4E-binding proteins (4E-BPs) and the ribosomal protein S6 kinases (10–13). However, how mTORC1, acting upon 4E-BPs and S6K, modulates β-cell mass and function is unclear.
The members of the 4E-BP family include three paralogs (4E-BP1, -2, and -3) that have variable expression in different tissues. Phosphorylation of 4E-BPs by mTORC1 prevents the repression of eIF4E, resulting in augmented translation of highly cap-dependent mRNAs (14). Although the three 4E-BPs have some degree of functional redundancy (14,15), there also seems to be some tissue-specific differences (16). Most current knowledge about the role of these proteins is based on experiments using 4E-BP1–deficient cells or mice (14,17–19). Growth factors, amino acids, glucose, and insulin induce phosphorylation of 4E-BP1 in islets and insulinoma cells, and deletion of the Eif4ebp1 gene increases susceptibility to endoplasmic reticulum stress–mediated apoptosis (20–23). Little is known about 4E-BP2, but this protein is highly expressed in the brain and is required for learning, memory, and autism (24,25). The importance of the different 4E-BPs and the function of each in the regulation of β-cell proliferation, size, survival, mass, and function has not been clearly defined.
We previously explored the role of S6K in pancreatic β-cells by transgenic overexpression of a constitutively active isoform (26). These studies revealed that S6K activation recapitulates the cell size but not the proliferative phenotype of models with activated mTORC1 signaling. The current study describes the role of 4E-BP2 and the interaction with S6K in β-cells using mice with global genetic deletion of Eif4ebp2. We demonstrate that 4E-BP2, and not 4E-BP1, plays a major role in the regulation of β-cell mass. Mice deficient for 4E-BP2 exhibited increased β-cell mass caused by enhanced proliferation and survival. These beneficial effects of 4E-BP2 deletion resulted from increased insulin receptor substrate 2 (IRS2) levels in β-cells resulting from enhanced synthesis of SH2B1, stabilization of a Janus kinase 2 (Jak2)/SH2B1/IRS2 complex, and enhanced Jak2 activity. This work provides evidence for a novel feedback mechanism by which mTORC1 signaling regulates IRS2 levels, proliferation, and survival in pancreatic β-cells.
Research Design and Methods
Animals and Treatments
Mice with targeted deletion of Eif4ebp1 and Eif4ebp2 have been previously described (14,27). Male mice on a C57BL/6J background were used for these experiments. All procedures were performed in accordance with the University Committee on Use and Care of Animals at the University of Michigan.
Cell Culture
MIN6 cells were cultured in DMEM supplemented with 10% FBS, glutamine, and antibiotics. Stable MIN6 knockdown cell lines were generated by infecting MIN6 cells with lentiviral particles containing a short hairpin RNA targeting 4ebp2 or control.
For protein stability studies, cells were harvested after treatment with cycloheximide (CHX) (12.5 μg/mL; Sigma-Aldrich) for various periods of time. The cells were lysed and sonicated as described elsewhere (28). Cytokine treatment was performed by treating islets with human interleukin-1β (50 U/mL), recombinant rat interferon-γ (1,000 U/mL), and recombinant rat tumor necrosis factor-α (1,000 U/mL). These concentrations were based on the results of previously published studies (29).
Metabolic Studies
Blood glucose concentrations were determined using an AlphaTrak glucose meter (Abbott Laboratories). Glucose tolerance tests were performed on animals fasted overnight by intraperitoneally injecting glucose (2 mg/kg), as previously described (30,31). Plasma insulin concentrations were determined using a Mouse Insulin ELISA kit (ALPCO). For an insulin tolerance test, animals fasted for 6 h received an intraperitoneal injection of either saline or human insulin (0.5 units/kg; Novolin; Novo Nordisk). Fasting glucose and insulin were measured after an overnight fast.
Islets Studies
Islets were isolated by collagenase digestion and insulin secretion, as previously described (32). Secreted insulin was then measured using an ELISA and normalized to DNA content.
Islets were perifused using batches of 20 islets per 70-μL chamber. The flow rate was 1.0 mL/min. From time 0 to 60 min, the islets were perifused under basal conditions (Krebs-Ringer bicarbonate buffer and 2.8 mmol/L glucose). At 62 min, islets were exposed to Krebs-Ringer bicarbonate buffer, 2.8 mmol/L glucose, and KCl (30 mmol/L) for 20 min.
A group of 10 islets were lysed and the samples were sonicated for 15 s. Aliquots were then frozen for the subsequent analysis of insulin content.
AG490 (a Jak2 inhibitor) and 4E1RCat (inhibitor of a cap-dependent translation) were purchased from Calbiochem.
Immunofluorescence and Morphometry
Formalin-fixed pancreatic tissues were embedded and sectioned as previously described (28). Sections were incubated overnight at 4°C with the primary antibodies described in Supplementary Table 1, followed by fluorophore-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories). Mounting media containing DAPI (Vector Laboratories) was added to coverslips. β-Cell mass assessment, proliferation, and TUNEL assay (ApopTag Red In Situ Apoptosis Detection Kit; Chemicon) were performed as previously described (28).
Western Blotting and Immunoprecipitation
Lysed islets in lysis buffer containing a protease inhibitor cocktail (Roche Diagnostics) were boiled for 5 min and then loaded and electrophoresed on 10–12% SDS-PAGE. Antibodies used for immunoblotting are listed in Supplementary Table 1. Protein-band densitometry was determined using National Institutes of Health ImageJ software version 1.49d (33) (freely available at https://imagej.nih.gov/ij/) and normalized to tubulin/actin in the same membrane. Immunoprecipitation experiments were carried out using 300 μg of total protein for each sample, with 2 μg of IRS2 antibody and 50 μL of protein A agarose beads (Sigma-Aldrich). All immunoblotting images were developed using an Immun-Star Chemiluminescent Kit (Bio-Rad Laboratories). For pulse experiments, cells were pulse-labeled with [35S]-methionine for 2 h followed by immunoprecipitation with IRS2 or SH2B1 antibodies.
Quantitative Real-Time PCR
Total RNA was isolated using RNeasy (Qiagen). cDNA was synthesized using random hexamers and was reverse transcribed using Superscript II (Invitrogen), according to the manufacturer’s protocol. Real-time PCR was performed on an ABI 7000 sequence detection system using Taq-man gene expression assays (Applied Biosystems). Primers were purchased from Applied Biosystems.
Polyribosomal Profiling and Gradient Fraction Quantitative PCR
Polyribosomal profiles from sh4ebp2 and control cells were analyzed using a sucrose gradient, as described previously (34). Briefly, 5 × 106 MIN6 cells washed in cold PBS containing CHX followed by harvesting on lysis buffer; the remainder of the lysate was then subjected to separation on a 10% to 40% sucrose gradient containing CHX for 2 h at 270,000g. A piston gradient fractionator (BioComp Instruments) was used to fractionate the sample and measure RNAA254 with an in-line ultraviolet monitor. Gradients were collected in ten 1-mL fractions. Fractions 1–5 were combined as the monoribosome-associated mRNA pool, and fractions 6–10 were combined as the polyribosome-associated RNA pool. Total RNA was isolated from the fractions and subjected to reversed transcription and quantitative RT-PCR.
Adenoviral Infection
MIN6 cells were infected with SH2B1β or control adenovirus expressing β-galactosidase gene (β-gal) (3.8 × 1010 viral particles in 4 mL growth medium per plate) for 4 h. A cell extract was prepared and used for immunoblotting analysis.
Data Analysis
Data were analyzed using the Student t test or ANOVA followed by post hoc analysis, where appropriate. In some experiments, a Student paired t test was used. Results were considered statistically significant when the P value was less than 0.05.
Results
4E-BP2–Deficient Mice Exhibit Improved Glucose Tolerance by Enhanced β-Cell Mass
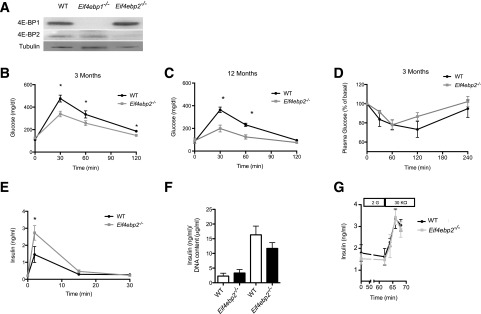
To begin to elucidate the role of 4E-BPs in β-cells, we first determined the expression levels of 4E-BP1 and 4E-BP2 in islet lysates from wild-type, Eif4ebp1−/−, and Eif4ebp2−/− mice. Both are expressed in islets, and deletion of either does not result in a compensatory increase in the level of the other protein (Fig. 1A). Body weight of Eif4ebp1−/− and Eif4ebp2−/− mice was comparable to that of wild-type controls (Supplementary Fig. 1A). As previously described, examination of glucose homeostasis in Eif4ebp1−/− demonstrated improved glucose clearance (Supplementary Fig. 1B–G), and this phenotype resulted from enhanced insulin sensitivity (14). Examination of glucose tolerance at 3 and 12 months of age in Eif4ebp2−/− mice revealed improved glucose clearance at 30, 60, and 120 min after glucose injection (Fig. 1B and C). In contrast to Eif4ebp1−/− mice (14), no alterations in insulin sensitivity were observed in Eif4ebp2−/− mice at 3 months old (Fig. 1D). Glucose-stimulated insulin secretion showed that Eif4ebp2−/− mice exhibited enhanced insulin concentrations in response to glucose at 2 min (Fig. 1E). These experiments suggest that, in contrast to Eif4ebp1−/− mice, 4E-BP2 deletion results in improved glucose tolerance as a result of increased insulin secretion rather than changes in insulin sensitivity.
Figure 1.
Eif4ebp2-deficient mice exhibit improved glucose tolerance and β-cell mass expansion. A: Western blotting for 4E-BP1 and 4E-BP2 in wild-type (WT), Eif4ebp1−/−, and Eif4ebp2−/− mice. B and C: Glucose tolerance test (2 g/kg) in wild-type (black circles) and Eif4ebp2−/− mice (gray squares) at 3 and 12 months of age. D: Insulin tolerance test in wild-type and Eif4ebp2−/− mice. Mice were fasted for 6 h before insulin tolerance tests. E: Glucose-stimulated insulin secretion in the wild-type and Eif4ebp2−/− mice. F: Glucose-stimulated insulin secretion determined by static incubation of isolated islets. G: Glucose-induced insulin secretion from isolated islets was measured in perifusion experiments at 2 mmol/L (basal) or with stimulation of KCl (30 mmol/L). The perfusate was collected at 3-min intervals, and insulin concentrations were determined using ELISA. Data are shown as mean ± SEM (n = 7 mice per group). *P < 0.05.
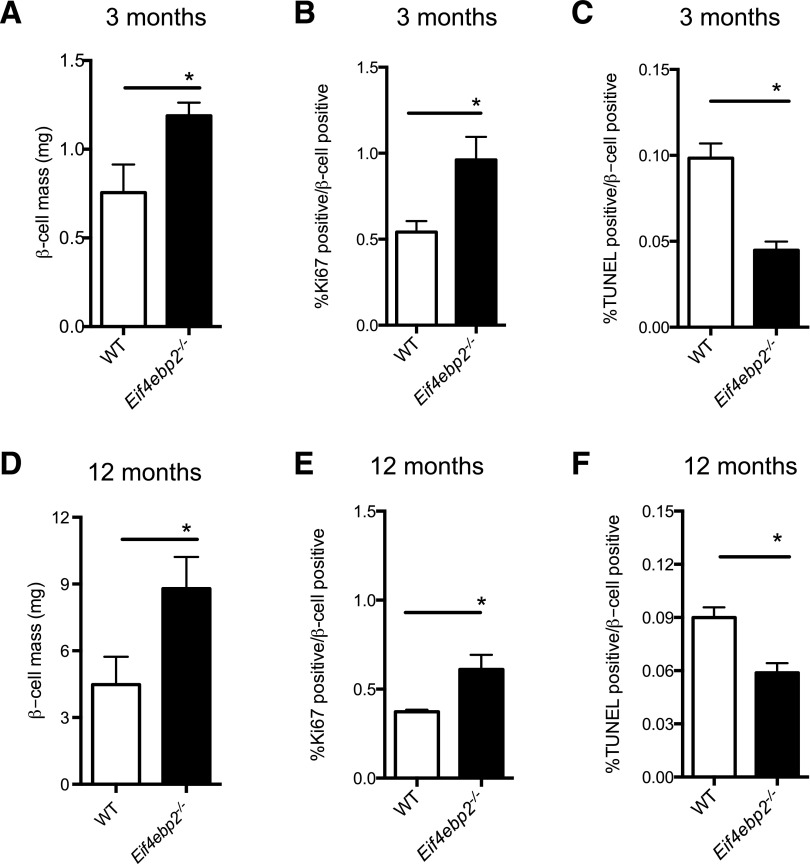
To begin to elucidate the changes in insulin secretory responses observed in vivo, we statically incubated isolated islets, causing glucose-stimulated insulin secretion. Insulin secretion in response to glucose was comparable between Eif4ebp2−/− and wild-type islets (Fig. 1F), and maximum islet secretory responses to KCl in a perifusion system demonstrated no differences between Eif4ebp2−/− and wild-type islets (Fig. 1G). The normal secretory responses suggested that the increased glucose-stimulated insulin secretion observed in vivo was most likely a result of changes in β-cell mass. Indeed, assessment of islet morphology showed that Eif4ebp2−/− mice exhibited increased β-cell mass at 3 and 12 months of age (Fig. 2A and D and Supplementary Fig. 2G). β-Cell proliferation was increased and β-cell apoptosis, measured using a TUNEL assay, was decreased in Eif4ebp2−/− mice compared with wild-type mice at 3 and 12 months of age (Fig. 2B, C, E, and F). The average size of the β-cells was not different between the groups at 3 or 12 months of age (data not shown). Compared with Eif4ebp2−/− mice, islet morphometry was completely normal in Eif4ebp1−/− mice (Supplementary Fig. 2A–F). Assessment of proliferation by Ki67 staining in glucagon- and somatostatin-positive cells from Eif4ebp1−/−, Eif4ebp2−/−, and control mice showed no differences at 3 months of age (Supplementary Fig. 2H). Taken together, these results suggest that improved glucose homeostasis in Eif4ebp2−/− mice is caused by increases in β-cell mass, proliferation, and survival. By contrast, improved glucose control in Eif4ebp1−/− mice results from enhanced insulin sensitivity.
Figure 2.
Eif4ebp2-deficient mice exhibit increased β-cell mass. A and D: Assessment of β-cell mass at 3 and 12 months of age in wild-type (WT; white bars) and Eif4ebp2−/− (black bars) mice. B and E: Proliferative index in sections stained for Ki67 and insulin. C and F: TUNEL assay in insulin-stained sections (apoptotic rate). Data are shown as mean ± SEM (n = 5 mice per group). *P < 0.05.
Increased β-Cell Proliferation in Eif4ebp2−/− Mice Is Associated With Reduced Levels and Stability of p27
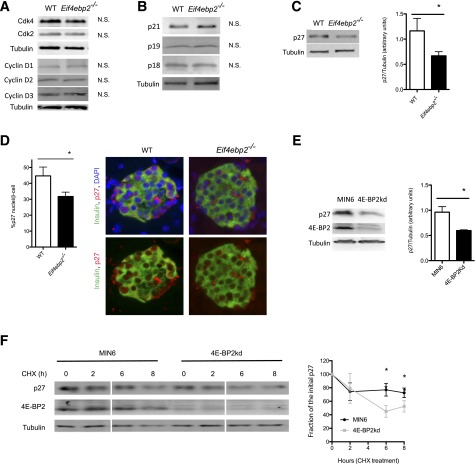
To determine how loss of 4E-BP2 induces proliferation in β-cells, we assessed cell cycle components responsible for progression from G1 to S. Protein levels for cyclins D1, D2, and D3, as well as Cdk2 and Cdk4, were similar in islets from Eif4ebp2−/− and wild-type mice (Fig. 3A). Assessment demonstrated no differences in cell cycle inhibitors, including p21, p19, and p18 (Fig. 3B). By contrast, levels of p27, an important inhibitor of β-cell proliferation, were lower in Eif4ebp2−/− mice (Fig. 3C). In addition, p27 immunostaining revealed that the number of β-cells with nuclear p27 was decreased in Eif4ebp2−/− mice (Fig. 3D). To investigate the mechanisms for the reduction of p27 levels by loss of 4E-BP2, we silenced 4E-BP2 in MIN6 cells (4E-BP2kd). In agreement with the results from Eif4ebp2−/− islets, 4E-BP2kd cells also showed decreased p27 levels (Fig. 3E). Evaluation of p27 mRNA levels revealed similar p27 expression between 4E-BP2kd and control cells, suggesting that the alterations were posttranscriptional (data not shown). Assessment of protein stability using CHX showed that the stability of p27 was decreased in 4E-BP2kd cells (Fig. 3F). These studies indicate that a loss of 4E-BP2 function induces β-cell proliferation, at least in part, by decreasing p27 stability in 4E-BP2kd cells.
Figure 3.
Decreased level and stability of p27 is related to increased proliferation in Eif4ebp2-deficient mice. A: Assessment of cell cycle components. Immunoblotting for Cdk4, Cdk2, and cyclins D1, D2, and D3 using islet lysates from wild-type (WT) and Eif4ebp2−/− mice. N.S., not significant. B: Assessment of cell cycle inhibitors. Immunoblotting for p21, p19, and p18 using islet lysates from WT and Eif4ebp2−/− mice. N.S., not significant. C: Immunoblotting and densitometric analysis for p27 in islets lysate from WT and Eif4ebp2−/− mice. D: Quantification (left) and staining (right) of nuclear p27 in β-cells from WT and Eif4ebp2−/− mice. E: Immunoblotting (left) and densitometric analysis (right) for p27 in 4E-BP2kd and control cells. F: p27 protein stability assessed by immunoblotting (left) and quantification of p27 and tubulin in 4E-BP2kd and control cells cultured with 12.5 μg/mL CHX for 0, 2, 6, and 8 h (right). (Samples were run in the same gel but, for consistency, appear spliced at the time points in Fig. 7A). Data are shown as mean ± SEM (n = 4 mice per group). *P < 0.05.
β-Cells From Eif4ebp2−/− Mice Exhibit Increased IRS2, but Not IRS1, Protein Levels
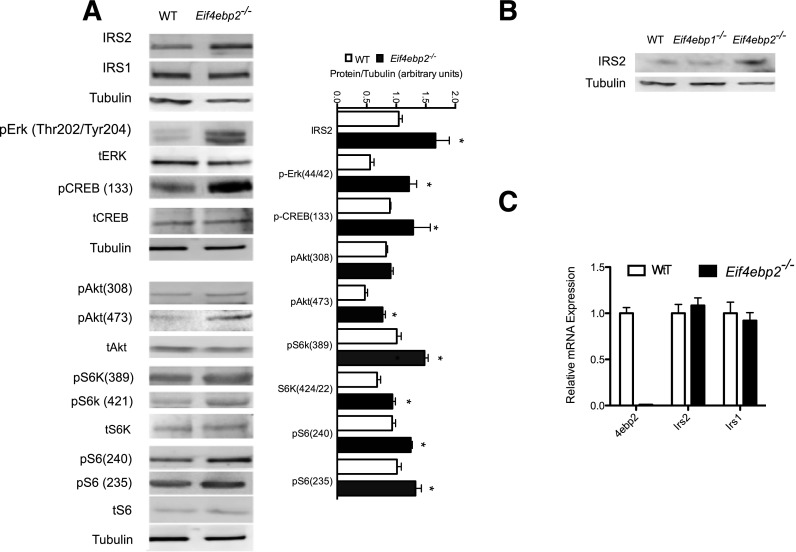
To determine the molecular mechanisms responsible for the increased β-cell mass, proliferation, and survival in Eif4ebp2−/− mice, we assessed signaling pathways involved in growth responses. Evaluation of proximal growth factor signaling components demonstrated that IRS2, but not IRS1, levels were increased in β-cells from Eif4ebp2−/− mice (Fig. 4A). Next, we assessed pathways downstream of IRS2 and observed increased phosphorylation of the mitogen-activated protein kinase Erk1/2 (Thr202/Tyr204) and its downstream targets, S6 (Ser235) and CREB (Ser133), in islets from Eif4ebp2−/− mice. In addition, AKT showed increased phosphorylation at Ser473, but not Thr308. The changes in AKT phosphorylation were accompanied by phosphorylation of the mTORC1 targets, S6 kinase and S6 protein (Ser240) in islets from Eif4ebp2−/− mice (Fig. 4A). The increase in IRS2 levels was not observed in islets from Eif4ebp1−/− mice (Fig. 4B). To determine whether the increase in IRS2 protein resulted from enhanced transcription of Irs2 mRNA, we performed quantitative RT-PCR on isolated islets. Expression of Irs2 and Irs1 were not different between Eif4ebp2−/− and wild-type mice (Fig. 4C), indicating that the changes in IRS2 levels are posttranscriptional at the level of protein synthesis or stability.
Figure 4.
IRS2 levels are increased and downstream pathways are activated in Eif4ebp2-deficient islets. A: Immunoblotting (left) and quantification (right) of IRS2, IRS1, phospho-Erk, phospho-Creb, phospho-Akt, and mTORC1 targets (pS6k and pS6) using islet lysates from wild-type (WT; white bars) and Eif4ebp2−/− (black bars) mice. B: Immunoblotting for IRS2 and tubulin using islet lysates from WT, Eif4ebp1−/−, and Eif4ebp2−/− mice. C: Assessment of 4ebp2, Irs2, and Irs1 mRNA levels in islets lysates from WT and Eif4ebp2−/− mice using TaqMan RT-PCR. Data are shown as mean ± SEM (n = 4 mice per group). *P < 0.05.
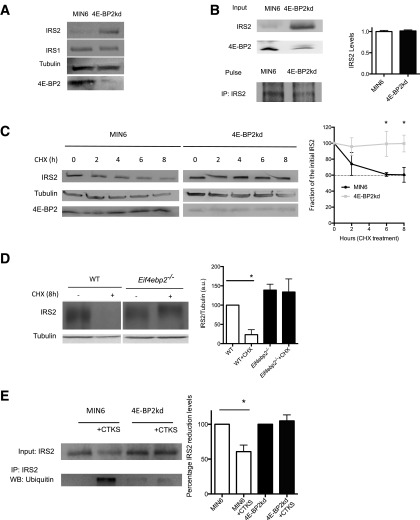
Loss of 4E-BP2 Function Induces IRS2 Levels by Augmenting Protein Stability
We next determined the mechanisms by which loss of 4E-BP2 regulates IRS2 levels. To this end, we assessed the contribution of protein synthesis and/or stability to the regulation of IRS2 levels by loss of 4E-BP2 using metabolic labeling and CHX experiments in 4E-BP2kd cells. In agreement with the results from Eif4ebp2−/− islets, 4E-BP2kd cells also showed increased IRS2, but not IRS1, protein levels (Fig. 5A). 4E-BP2kd and control cells given a pulse of radioactive methionine had similar levels of radiolabeled IRS2, demonstrating that IRS2 synthesis was unaltered (Fig. 5B). Moreover, assessment of IRS2 phosphorylation by a decreased electrophoretic mobility of IRS2 protein immunoblot analysis was performed on an 8% polyacrylamide gel analysis as described elsewhere (35). Treatment with alkaline phosphatase demonstrated that IRS2 was phosphorylated in 4E-BP2kd and control cells. We observed increased IRS2 levels in 4E-BP2kd cells, but no upward mobility shift in IRS2 protein migration between MIN6 or 4E-BP2kd cells was detected, suggesting that there were no alterations in IRS2 phosphorylation in 4E-BP2kd cells (Supplementary Fig. 3A). 4E-BP2kd and control cells were treated with CHX to determine whether the stability of IRS2 is altered by 4E-BP2 deficiency (Fig. 5C). In control cells, IRS2 levels were reduced by 50% after 8 h of CHX treatment (Fig. 5C). By contrast, IRS2 levels were maintained during the 8-h treatment with CHX in 4E-BP2kd cells (Fig. 5C). More important, IRS2 stability was also enhanced in islets from Eif4ebp2−/− mice when compared with islets from controls, confirming the results obtained in cell lines (Fig. 5D). Previous studies have shown that proinflammatory cytokines promote IRS1 and IRS2 ubiquitination and subsequent degradation in multiple cell types, providing a good model to study IRS2 stability (36). Therefore, we studied the responses to proinflammatory cytokines to assess the mechanisms responsible for enhanced IRS2 stability after loss of 4E-BP2. For these experiments, we examined the ubiquitinated IRS2 in lysates from MIN6 cells and 4E-BP2kd cells treated with a cocktail of proinflammatory cytokines (interferon-γ, tumor necrosis factor-α, interleukin-1β) for 24 h. As expected, treatment of MIN6 cells with proinflammatory cytokines resulted in marked ubiquitination and degradation of IRS2 levels (Fig. 5E). By contrast, treatment with cytokines failed to induce IRS2 ubiquitination, and levels were maintained in 4E-BP2kd cells (Fig. 5E). These experiments suggest that loss of 4E-BP2 enhances IRS2 stability by inhibiting ubiquitin-mediated degradation after cytokine treatment.
Figure 5.
IRS2 stability in Eif4ebp2-deficient islets and 4E-BP2 knockdown cells. A: Immunoblotting for IRS2 and IRS1 in 4E-BP2kd and control cells. B: Pulse-chased cells with IRS2 antibody immunoblotting (left) and quantification (right). C: IRS2 protein stability assessed by immunoblotting (left) and quantification (right) for IRS2 and tubulin in 4E-BP2kd (gray squares) and control cells (black circles) cultured with 12.5 μg/mL CHX for 0, 2, 4, 6, and 8 h. D: IRS2 protein stability assessed by immunoblotting for IRS2 and tubulin in Eif4ebp2−/− and wild-type (WT) islets cultured with 12.5 μg/mL CHX for 0 and 8 h (samples were run in the same gel but appear spliced to show WT data to the left of Eif4ebp2−/−). E: Immunoprecipitation (IP) and quantification for IRS2 in MIN6 and 4E-BP2kd cells and Western blotting (WB) for ubiquitin in cells treated with cytokines or not treated. Data are shown as mean ± SEM (n = 4 mice per group). *P < 0.05.
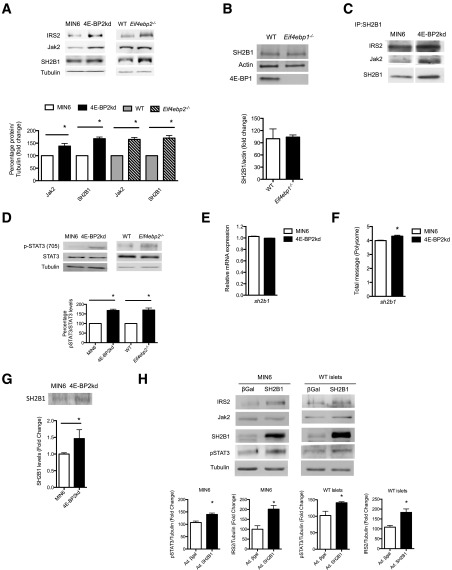
SH2B1 Synthesis and Jak2 Activity Regulate IRS2 Levels in β-Cells With Loss of 4E-BP2
The responses to cytokines suggest that the differences in ubiquitin-dependent degradation of IRS2 could be mediated by activation of Jak2 signaling. The adaptor protein SH2B1 binds and recruits IRS2 not only to growth factor receptors but also to cytokine receptors, forming a complex to enhance Jak2 activity (37). We hypothesized that the increased IRS2 stability resulted from the binding of IRS2 to SH2B1 and recruitment to form a complex with Jak2. To explore this possibility, we first assessed levels of different components of the complex. Basal Jak2 and SH2B1 levels were higher in the 4E-BP2kd cells and in islets from Eif4ebp2−/− mice when compared with controls (Fig. 6A), but not in islets from Eif4ebp1−/− mice (Fig. 6B). Immunoprecipitation of SH2B1 in 4E-BP2kd and control cells showed that this adaptor forms a complex with IRS2 and Jak2 in 4E-BP2kd cells but is only bound to IRS2 in MIN6 cells without 4E-BP2 (Fig. 6C). The formation of the Jak2/SH2B1/IRS2 complex resulted in activation of Stat3 (Ser705) (Fig. 6D). Because SH2B1 is the adaptor protein that recruits IRS2 to the complex, we postulated that increased levels of this adaptor could provide a link between 4E-BP2/eIF4E and increased IRS2 levels. Assessment of Sh2b1 at the mRNA levels demonstrated that Sh2b1 transcription was not different between MIN6 and 4E-BP2kd cells (Fig. 6E), suggesting that the changes in SH2B1 were posttranscriptional at the level of protein synthesis or stability. 4E-BP2 deletion releases eIF4E and favors the interaction with eIF4G, resulting in enhanced cap-dependent translation of a subset of mRNAs with highly structured 5′ untranslated region (UTR) (38). mRNAs with a complex secondary structure in the 5′ UTR are characterized by high guanine cytosine content and thermodynamically stable structures (low ΔG) (39). Indeed, the SH2B1 5′ UTR is richer in guanine cytosine (67.9%) than average (actin) and exhibits a lower free energy (ΔG: −281.50 kcal/mol), indicating that this 5′ UTR contains complex secondary structures and could be favorably translated by eIF4E (40) (Supplementary Fig. 4A). To assess whether SH2B1 is favorably translated in the absence of 4E-BP2, we performed polysomal fractionation and determined sh2b1 mRNA levels in polyribosomal fractions from 4E-BP2 knockdown and control MIN6 cells. The polyribosome profile showed a shift from the monosome to the polysome fraction in 4E-BP2kd cells (data not shown), and sh2b1 mRNA levels were significantly increased in the polyribosomal fractions from 4E-BP2kd cells, indicating that a decrease in 4E-BP2 levels enhances sh2b1 translation (Fig. 6F). 4E-BP2kd cells exhibited higher levels of radiolabeled SH2B1 following a pulse of radioactive methionine, demonstrating that SH2B1 synthesis was enhanced (Fig. 6G). In addition, adenoviral overexpression of SH2B1 in MIN6 cells and islets seemed to be sufficient to increase IRS2 protein stability but not Jak2 levels (Fig. 6H).
Figure 6.
Stability of IRS2 depends of Jak2 activation. A: Immunoblotting for IRS2, Jak2, and SH2B1 in MIN6 cells, 4E-BP2kd cells, and wild-type (WT) and Eif4ebp2−/− islets, and quantification of Jak2 and SH2B1. B: Immunoblotting for SH2B1 in isolated islets from wild-type and Eif4ebp1−/− mice. C: Immunoprecipitation (IP) for SHB1 and Western blotting for IRS2, Jak2, and SH2B1 (samples were run in the same gel but appear spliced to exclude data on AG490 + CHX treatment shown in Fig. 7B). D: Immunoblotting (top) and quantification (bottom) for Stat3 phosphorylated at Tyr705, total Stat3, and tubulin in cells and islets. E: Assessment of sh2b1 mRNA levels in MIN6 and 4E-BP2kd cell lysates using TaqMan RT-PCR. F: Assessment of sh2b1 mRNA levels in polyribosomal fractions from MIN6 and 4E-BP2kd cells. G: Pulse-chased cells with SH2B1 antibody immunoblotting (top) and quantification (bottom). H: Immunoblotting (top) and quantification (bottom) for IRS2, Jak2, SH2B1, phospho-STAT3, and tubulin in MIN6 or WT islets infected with SH2B1 or control adenovirus (Ad.) expressing β-gal. Data are shown as mean ± SEM (n = 4 mice per group). *P < 0.05.
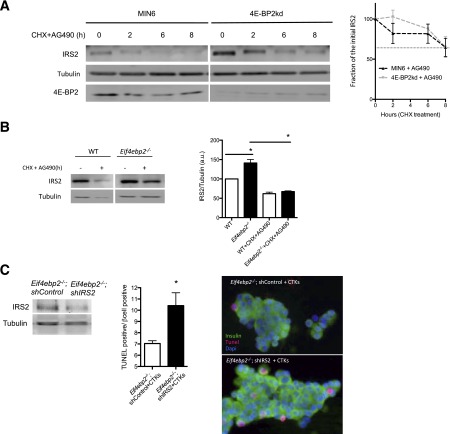
Jak2 Activity Regulates IRS2 Stability in Cells With Loss of 4E-BP2
We then tested the contribution of Jak2 activity to the regulation of IRS2 levels by assessing IRS2 stability in the presence of a Jak2 inhibitor. Inhibition of Jak2 activity by AG490 treatment induced IRS2 ubiquitination in 4E-BP2kd cells treated with proinflammatory cytokines or the vehicle, suggesting that Jak2 activity plays an important role in regulating IRS2 ubiquitination (Supplementary Fig. 5A). We then assessed the effect of Jak2 inhibition on IRS2 stability in 4E-BP2kd and Eif4ebp2−/− islets. IRS2 levels in MIN6 cells treated with AG490 decreased by 50% in the first 8 h, which was similar to that observed in cells that were not treated with the Jak2 inhibitor (Fig. 7A vs. Fig. 5C). By contrast, the IRS2 stability observed in 4E-BP2kd cells was completely lost upon Jak2 inhibition (Fig. 7A vs. Fig. 5C). More important, inhibition of Jak2 signaling decreased IRS2 in Eif4ebp2−/− islets to a level similar to that observed in controls (Fig. 7B). In addition, silencing of IRS2 in dispersed islets from Eif4ebp2−/− mice was sufficient to enhance cytokine-induced apoptosis by 50% (Fig. 7C). Taken together, these results demonstrate that increased levels of SH2B1 and the activation of the Jak2 pathway are responsible for the increased stability of IRS2 and survival in cells with loss of 4E-BP2.
Figure 7.
Stability of IRS2 depends on Jak2 activation. A: IRS2 protein stability assessed by immunoblotting (left) and quantification (right) for IRS2 and tubulin in 4E-BP2kd (gray line) and control cells (black line) cultured with 12.5 μg/mL CHX and AG490 for 0, 2, 6, and 8 h. B: IRS2 protein stability assessed by immunoblotting (left) and quantification (right) for IRS2 and tubulin in Eif4ebp2−/− and wild-type (WT) islets cultured with 12.5 μg/mL CHX and AG490 for 0 and 8 h (samples were run in the same gel but appear spliced to show WT data to the left of Eif4ebp2−/−). C: Immunoblotting for IRS2 and tubulin in dispersed islets from Eif4ebp2−/− (control or with IRS2 silenced) (left). Apoptotic rate (middle) and images (right) in Eif4ebp2−/− dispersed cells (control or with IRS2 silenced) treated with cytokines for 24 h. Data are shown as mean ± SEM (n = 4 mice per group). *P < 0.05. a.u., arbitrary units.
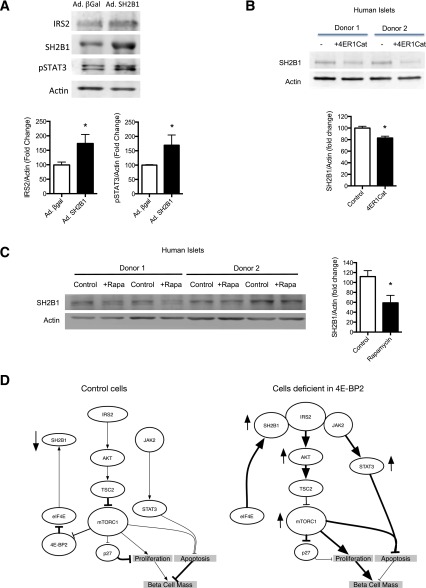
4E-BP2 Regulates SH2B1/IRS2 Levels in Human Islets
To demonstrate the role of SH2B1 in the stability of IRS2 in human islets, we overexpressed SH2B1 by adenoviral infection of human islets. IRS2 levels and Jak2 signaling, measured by phosphorylation of Stat3, were increased in human islets overexpressing SH2B1 (Fig. 8A). To assess the effect of the mTORC1/4E-BP axis on SH2B1 levels using pharmacologic inhibitors, we used 4ER1Cat (inhibitor of cap-dependent translation) and rapamycin (mTORC1 inhibitor). Human islets treated for 24 h with 4ER1Cat, a cap-dependent translation inhibitor that prevents eIF4E–eIF4G interaction, showed a decrease in SH2B1 protein levels, suggesting that SH2B1 levels are regulated by cap-dependent translation (Fig. 8B). We then assessed the effect of short-term inhibition of mTORC1, the upstream regulator of 4E-BP signaling, by rapamycin treatment of human islets. Rapamycin treatment for 48 h was sufficient to decrease SH2B1 levels (Fig. 8C). Taken together, these data demonstrate that the inhibition of mTORC1/4E-BP2 decreased SH2B1 levels and could be detrimental to the proliferation and survival of β-cells.
Figure 8.
Increased SH2B1 stabilizes IRS2 and inhibition of eIF4E reduces SH2B1 levels in human islets. A: Immunoblotting (top) and quantification (bottom) for IRS2, SH2B1, phospho-STAT3 (pSTAT3), and actin in human islets infected with SH2B1 or control adenovirus (Ad.) expressing β-gal. B: Immunoblotting (top) and quantification (bottom) for SH2B1 and actin in human islets treated with or without 4ER1Cat for 24 h. C: Immunoblotting (left) and quantification (right) for SH2B1 and actin in human islets treated with or without rapamycin for 24 h. D: Schematics showing the Akt/TSC2/mTORC1/4E-BP2/IRS2 axis in control cells (left) and cells deficient in 4E-BP2 (right). Data are shown as mean ± SEM (n = 4 mice per group). *P < 0.05.
Discussion
This study describes the contribution of 4E-BPs to the regulation of β-cell mass, growth, and proliferation. We identify a novel role of 4E-BP2/eIF4E in the modulation of glucose homeostasis by regulating β-cell mass. In addition, we determine that 4E-BP2/eIF4E regulates β-cell proliferation and survival by a novel positive feedback mechanism to increase IRS2 stability and levels (Fig. 8D). This novel mechanism results from upregulation of the adaptor protein SH2B1 and activation of Jak2 signaling (Fig. 8D). These studies provide the first evidence for a unique regulatory pathway downstream of mTORC1 that is responsible for β-cell proliferation and survival; they also demonstrate a link between translational control and insulin sensitivity in β-cells.
Our previous work showed that activation of mTORC1 signaling in mice with conditional deletion of TSC2 results in β-cell mass expansion caused by increases in both proliferation and cell size (6). This work demonstrates that the 4E-BP2/eIF4E axis relates proliferative and survival signals downstream of mTORC1. Assessment of the signaling mechanisms in islets from Eif4ebp2−/− mice demonstrated that the β-cell phenotype in these mice resulted from increased levels of IRS2, a critical molecule for β-cells (41). The activation of IRS2/Akt signaling ultimately results in β-cell cycle progression by reduction in the levels and nuclear localization of p27 (Fig. 8D). The role of the IRS2/p27 axis in the modulation of β-cell cycle progression has been well-established previously (42). The elevated levels of IRS2 and reduction in p27 observed in Eif4ebp2−/− mice and 4E-BP2kd cells are in marked contrast to the changes observed in transgenic mice overexpressing constitutively active S6K and suggest that overactivation of S6K signaling and a loss of 4E-BP2 converge to regulate IRS2 and p27 levels, thereby modulating β-cell mass expansion. In addition, deletion of 4E-BP2 recapitulates the β-cell proliferation and expansion observed in mice with conditional deletion of TSC2 in β-cells (6).
These studies demonstrate that the positive impact of the loss of 4E-BP2 in β-cell proliferation is mediated, to a great extent, by the induction of IRS2 levels. Examination of mRNA suggested that the elevation in IRS2 levels after the loss of 4E-BP2 was posttranscriptional. Given the major role of 4E-BP2 in the regulation of translation of specific mRNAs, we reasoned that IRS2 protein synthesis was enhanced. However, we found that a loss of 4E-BP2 increased IRS2 levels by enhancing stability rather than synthesis. These findings are consistent with a model in which increased synthesis of the adaptor protein SH2B1 plays a central role in the regulation of IRS2 ubiquitination, stability, and levels in cells with a loss of 4E-BP2 by forming a complex with IRS2 and Jak2 and activating Jak2 signaling (Figs. 6C and 8D) under basal conditions (Figs. 5E and 8D) or after cytokine treatment (Fig. 5E and Supplementary Fig. 5A). The importance of the SH2B1/Jak2/IRS2 complex in Jak2 activation has also been reported in fibroblasts (43,44). SH2B family members couple upstream activators of multiple receptor tyrosine kinases to downstream effectors by forming multiprotein complexes with different partners; they enhance the catalytic activity of its bound enzymes. How SH2B members bind other partners to regulate signaling is not completely understood, but there is evidence that this process could be regulated by phosphorylation in a stimulus-specific manner (reviewed in refs. 45 and 46). Our studies showed that an increase in SH2B levels seemed to be sufficient to drive the formation of the Jak2/SH2B1/IRS2 complex and basal Jak2 activity. The importance of SH2B1 in β-cells has been demonstrated by the inhibition of compensatory β-cell expansion in mice with pancreas-specific deletion of SH2B1 (47). Finally, this work demonstrates that the mTORC1/4E-BP2/SH2B axis seems to exist in human islets and suggests that this mechanism could regulate IRS2 levels in human β-cells.
These experiments demonstrate that a loss of 4E-BP2, but not 4E-BP1, plays a major role in the regulation of β-cell mass by driving cell cycle progression and survival. One potential limitation of these studies is the use of global knockouts and the potential of systemic effects on the regulation of IRS2. Although possible, this is less likely as Eif4ebp2−/− mice exhibited normal insulin sensitivity and the findings related to IRS2 levels and proliferation in islets were validated in ex vivo experiments using isolated islets and MIN6 cells (data on not shown for MIN6 proliferation). Transplanting islets from wild-type into Eif4ebp2−/− mice could assess this possibility. The role of 4E-BP2 loss on proliferation and survival is in marked contrast to the deleterious role of the loss of 4E-BP1 in the response to endoplasmic reticulum stress (48). Our results also confirm that there are major differences between 4E-BP1– and 4E-BP2–deficient mice with regard to the regulation of insulin sensitivity and suggest that improvement in glucose homeostasis in Eif4ebp1−/− mice is mainly modulated at the insulin sensitivity level (14). This, combined with normal β-cell mass in these mice, led us to conclude that loss of 4E-BP1 has a minor effect on β-cells. These major differences between these two translational regulators are intriguing, as the cellular functions of 4E-BP1 and 4E-BP2 were believed to be redundant. It is possible that different expressions of 4E-BP1 and 4E-BP2 in tissue explain these differences. However, recent data demonstrate a major role of 4E-BP2 in the nervous system, not only regulating learning and memory but also clinical implications in autism (49). Our results suggest that, in the β-cell, enhanced SH2B1 synthesis is one distinction between translational responses regulated by 4E-BP2 versus 4E-BP1.
In summary, these studies indicate a novel mechanism about the pathways responsible for β-cell mass and function induced by signals downstream of mTORC1. These studies suggest that mTORC1 regulates β-cell mass by regulating two processes: cell growth and function by activating mTORC1/S6K1, and cell cycle progression by activating mTORC1/4E-BP2. In addition, this study revealed a second feedback loop downstream of mTORC1 signaling and suggests that both S6K and 4E-BP2 converge on IRS2 and p27 to regulate β-cell expansion. These findings provide a better understanding of how nutrients and growth factors regulate β-cell mass expansion and the critical components involved, an important step for designing novel strategies for the treatment and cure of diabetes.
Article Information
Acknowledgments. The authors thank Drs. Masayuki Hatanaka (Yamaguchi University Graduate School of Medicine, Ube, Yamaguchi, Japan) and Raghavendra G. Mirmira (Indiana University School of Medicine, Indianapolis, IN) for assistance with polyribosomal profile experiments.
Funding. This work was supported by National Institutes of Health (grant nos. RO1-DK073716 and DK084236) and JDRF (grant no. 17-2013-416 to E.B.M.). The authors acknowledge support from the Morphology and Image Analysis Core and the Molecular Biology and DNA Sequencing Core of the National Institutes of Health–funded Diabetes Research Center (NIH P60-DK20572).
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. M.B.-R., J.O.S., M.J.-P., R.B., A.S.B., and M.L. performed the experiments and analyzed results. M.B.-R. and E.B.-M. wrote the article and designed the experiments. A.Y., L.R., and N.S. generated mice and/or reagents. All authors contributed to discussion and reviewed and edited the manuscript. E.B.-M. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db15-1443/-/DC1.
M.B.-R. and E.B.-M. are currently affiliated with the Miami VA Healthcare System and the Division of Endocrinology, Metabolism and Diabetes, Department of Medicine, University of Miami Miller School of Medicine, Miami, FL.
References
- 1.Shimobayashi M, Hall MN. Making new contacts: the mTOR network in metabolism and signalling crosstalk. Nat Rev Mol Cell Biol 2014;15:155–162 [DOI] [PubMed] [Google Scholar]
- 2.Newgard CB. Interplay between lipids and branched-chain amino acids in development of insulin resistance. Cell Metab 2012;15:606–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Efeyan A, Comb WC, Sabatini DM. Nutrient-sensing mechanisms and pathways. Nature 2015;517:302–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shigeyama Y, Kobayashi T, Kido Y, et al. Biphasic response of pancreatic beta-cell mass to ablation of tuberous sclerosis complex 2 in mice. Mol Cell Biol 2008;28:2971–2979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balcazar N, Sathyamurthy A, Elghazi L, et al. mTORC1 activation regulates beta-cell mass and proliferation by modulation of cyclin D2 synthesis and stability. J Biol Chem 2009;284:7832–7842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rachdi L, Balcazar N, Osorio-Duque F, et al. Disruption of Tsc2 in pancreatic beta cells induces beta cell mass expansion and improved glucose tolerance in a TORC1-dependent manner. Proc Natl Acad Sci U S A 2008;105:9250–9255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fraenkel M, Ketzinel-Gilad M, Ariav Y, et al. mTOR inhibition by rapamycin prevents beta-cell adaptation to hyperglycemia and exacerbates the metabolic state in type 2 diabetes. Diabetes 2008;57:945–957 [DOI] [PubMed] [Google Scholar]
- 8.Hamada S, Hara K, Hamada T, et al. Upregulation of the mammalian target of rapamycin complex 1 pathway by Ras homolog enriched in brain in pancreatic beta-cells leads to increased beta-cell mass and prevention of hyperglycemia. Diabetes 2009;58:1321–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu H, Remedi MS, Pappan KL, et al. Both glycogen synthase kinase-3 (GSK-3) and mammalian target of rapamycin (mTOR) pathways contribute to DNA synthesis, cell cycle progression and proliferation in human islets. Diabetes 2009;58:663–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell 2007;12:9–22 [DOI] [PubMed] [Google Scholar]
- 11.Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev 2004;18:1926–1945 [DOI] [PubMed] [Google Scholar]
- 12.Harris TE, Lawrence JC Jr. TOR signaling. Sci STKE 2003;2003:re15. [DOI] [PubMed] [Google Scholar]
- 13.Fingar DC, Salama S, Tsou C, Harlow E, Blenis J. Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev 2002;16:1472–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsukiyama-Kohara K, Poulin F, Kohara M, et al. Adipose tissue reduction in mice lacking the translational inhibitor 4E-BP1. Nat Med 2001;7:1128–1132 [DOI] [PubMed] [Google Scholar]
- 15.Poulin F, Gingras AC, Olsen H, Chevalier S, Sonenberg N. 4E-BP3, a new member of the eukaryotic initiation factor 4E-binding protein family. J Biol Chem 1998;273:14002–14007 [DOI] [PubMed] [Google Scholar]
- 16.Tomoo K, Matsushita Y, Fujisaki H, et al. Structural basis for mRNA cap-binding regulation of eukaryotic initiation factor 4E by 4E-binding protein, studied by spectroscopic, X-ray crystal structural, and molecular dynamics simulation methods. Biochim Biophys Acta 2005;1753:191–208 [DOI] [PubMed] [Google Scholar]
- 17.Gingras AC, Sonenberg N. Adenovirus infection inactivates the translational inhibitors 4E-BP1 and 4E-BP2. Virology 1997;237:182–186 [DOI] [PubMed] [Google Scholar]
- 18.Le Bacquer O, Petroulakis E, Paglialunga S, et al. Elevated sensitivity to diet-induced obesity and insulin resistance in mice lacking 4E-BP1 and 4E-BP2. J Clin Invest 2007;117:387–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olson KE, Booth GC, Poulin F, Sonenberg N, Beretta L. Impaired myelopoiesis in mice lacking the repressors of translation initiation, 4E-BP1 and 4E-BP2. Immunology 2009;128(Suppl.):e376–e384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu G, Kwon G, Marshall CA, Lin TA, Lawrence JC Jr, McDaniel ML. Branched-chain amino acids are essential in the regulation of PHAS-I and p70 S6 kinase by pancreatic beta-cells. A possible role in protein translation and mitogenic signaling. J Biol Chem 1998;273:28178–28184 [DOI] [PubMed] [Google Scholar]
- 21.Xu G, Marshall CA, Lin TA, et al. Insulin mediates glucose-stimulated phosphorylation of PHAS-I by pancreatic beta cells. An insulin-receptor mechanism for autoregulation of protein synthesis by translation. J Biol Chem 1998;273:4485–4491 [DOI] [PubMed] [Google Scholar]
- 22.Xu G, Kwon G, Cruz WS, Marshall CA, McDaniel ML. Metabolic regulation by leucine of translation initiation through the mTOR-signaling pathway by pancreatic beta-cells. Diabetes 2001;50:353–360 [DOI] [PubMed] [Google Scholar]
- 23.Kwon G, Marshall CA, Pappan KL, Remedi MS, McDaniel ML. Signaling elements involved in the metabolic regulation of mTOR by nutrients, incretins, and growth factors in islets. Diabetes 2004;53(Suppl. 3):S225–S232 [DOI] [PubMed] [Google Scholar]
- 24.Bidinosti M, Ran I, Sanchez-Carbente MR, et al. Postnatal deamidation of 4E-BP2 in brain enhances its association with raptor and alters kinetics of excitatory synaptic transmission. Mol Cell 2010;37:797–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bachmann-Gagescu R, Mefford HC, Cowan C, et al. Recurrent 200-kb deletions of 16p11.2 that include the SH2B1 gene are associated with developmental delay and obesity. Genet Med 2010;12:641–647 [DOI] [PubMed] [Google Scholar]
- 26.Elghazi L, Balcazar N, Blandino-Rosano M, et al. Decreased IRS signaling impairs beta-cell cycle progression and survival in transgenic mice overexpressing S6K in beta-cells. Diabetes 2010;59:2390–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Banko JL, Poulin F, Hou L, DeMaria CT, Sonenberg N, Klann E. The translation repressor 4E-BP2 is critical for eIF4F complex formation, synaptic plasticity, and memory in the hippocampus. J Neurosci 2005;25:9581–9590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elghazi L, Balcazar N, Blandino-Rosano M, et al. Decreased IRS signaling impairs beta-cell cycle progression and survival in transgenic mice overexpressing S6K in beta-cells. Diabetes 2010;59:2390–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blandino-Rosano M, Perez-Arana G, Mellado-Gil JM, Segundo C, Aguilar-Diosdado M. Anti-proliferative effect of pro-inflammatory cytokines in cultured beta cells is associated with extracellular signal-regulated kinase 1/2 pathway inhibition: protective role of glucagon-like peptide-1. J Mol Endocrinol 2008;41:35–44 [DOI] [PubMed] [Google Scholar]
- 30.Bernal-Mizrachi E, Wen W, Stahlhut S, Welling CM, Permutt MA. Islet beta cell expression of constitutively active Akt1/PKB alpha induces striking hypertrophy, hyperplasia, and hyperinsulinemia. J Clin Invest 2001;108:1631–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fatrai S, Elghazi L, Balcazar N, et al. Akt induces beta-cell proliferation by regulating cyclin D1, cyclin D2, and p21 levels and cyclin-dependent kinase-4 activity. Diabetes 2006;55:318–325 [DOI] [PubMed] [Google Scholar]
- 32.Bernal-Mizrachi E, Fatrai S, Johnson JD, et al. Defective insulin secretion and increased susceptibility to experimental diabetes are induced by reduced Akt activity in pancreatic islet beta cells. J Clin Invest 2004;114:928–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 2012;9:671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tersey SA, Nishiki Y, Templin AT, et al. Islet β-cell endoplasmic reticulum stress precedes the onset of type 1 diabetes in the nonobese diabetic mouse model. Diabetes 2012;61:818–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Briaud I, Dickson LM, Lingohr MK, McCuaig JF, Lawrence JC, Rhodes CJ. Insulin receptor substrate-2 proteasomal degradation mediated by a mammalian target of rapamycin (mTOR)-induced negative feedback down-regulates protein kinase B-mediated signaling pathway in beta-cells. J Biol Chem 2005;280:2282–2293 [DOI] [PubMed] [Google Scholar]
- 36.Rui L, Fisher TL, Thomas J, White MF. Regulation of insulin/insulin-like growth factor-1 signaling by proteasome-mediated degradation of insulin receptor substrate-2. J Biol Chem 2001;276:40362–40367 [DOI] [PubMed] [Google Scholar]
- 37.Morris DL, Cho KW, Zhou Y, Rui L. SH2B1 enhances insulin sensitivity by both stimulating the insulin receptor and inhibiting tyrosine dephosphorylation of insulin receptor substrate proteins. Diabetes 2009;58:2039–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem 1999;68:913–963 [DOI] [PubMed] [Google Scholar]
- 39.Koromilas AE, Lazaris-Karatzas A, Sonenberg N. mRNAs containing extensive secondary structure in their 5′ non-coding region translate efficiently in cells overexpressing initiation factor eIF-4E. EMBO J 1992;11:4153–4158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davuluri RV, Suzuki Y, Sugano S, Zhang MQ. CART classification of human 5′ UTR sequences. Genome Res 2000;10:1807–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.White MF. Regulating insulin signaling and beta-cell function through IRS proteins. Can J Physiol Pharmacol 2006;84:725–737 [DOI] [PubMed] [Google Scholar]
- 42.Uchida T, Nakamura T, Hashimoto N, et al. Deletion of Cdkn1b ameliorates hyperglycemia by maintaining compensatory hyperinsulinemia in diabetic mice. Nat Med 2005;11:175–182 [DOI] [PubMed] [Google Scholar]
- 43.Duan C, Li M, Rui L. SH2-B promotes insulin receptor substrate 1 (IRS1)- and IRS2-mediated activation of the phosphatidylinositol 3-kinase pathway in response to leptin. J Biol Chem 2004;279:43684–43691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Z, Zhou Y, Carter-Su C, Myers MG Jr, Rui L. SH2B1 enhances leptin signaling by both Janus kinase 2 Tyr813 phosphorylation-dependent and -independent mechanisms. Mol Endocrinol 2007;21:2270–2281 [DOI] [PubMed] [Google Scholar]
- 45.Maures TJ, Kurzer JH, Carter-Su C. SH2B1 (SH2-B) and JAK2: a multifunctional adaptor protein and kinase made for each other. Trends Endocrinol Metab 2007;18:38–45 [DOI] [PubMed] [Google Scholar]
- 46.Rui L. SH2B1 regulation of energy balance, body weight, and glucose metabolism. World J Diabetes 2014;5:511–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen Z, Morris DL, Jiang L, Liu Y, Rui L. SH2B1 in β-cells regulates glucose metabolism by promoting β-cell survival and islet expansion. Diabetes 2014;63:585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamaguchi S, Ishihara H, Yamada T, et al. ATF4-mediated induction of 4E-BP1 contributes to pancreatic beta cell survival under endoplasmic reticulum stress. Cell Metab 2008;7:269–276 [DOI] [PubMed] [Google Scholar]
- 49.Gkogkas CG, Khoutorsky A, Ran I, et al. Autism-related deficits via dysregulated eIF4E-dependent translational control. Nature 2013;493:371–377 [DOI] [PMC free article] [PubMed] [Google Scholar]