Abstract
Background
Dorsal root ganglion neuron-derived immortal cell lines including ND7/23 and F-11 cells have been used extensively as in vitro model systems of native peripheral sensory neurons. However, while it is clear that some sensory neuron-specific receptors and ion channels are present in these cell lines, a systematic comparison of the molecular targets expressed by these cell lines with those expressed in intact peripheral neurons is lacking.
Results
In this study, we examined the expression of RNA transcripts in the human neuroblastoma-derived cell line, SH-SY5Y, and two dorsal root ganglion hybridoma cell lines, F-11 and ND7/23, using Illumina next-generation sequencing, and compared the results with native whole murine dorsal root ganglions. The gene expression profiles of these three cell lines did not resemble any specific defined dorsal root ganglion subclass. The cell lines lacked many markers for nociceptive sensory neurons, such as the Transient receptor potential V1 gene, but expressed markers for both myelinated and unmyelinated neurons. Global gene ontology analysis on whole dorsal root ganglions and cell lines showed similar enrichment of biological process terms across all samples.
Conclusions
This paper provides insights into the receptor repertoire expressed in common dorsal root ganglion neuron-derived cell lines compared with whole murine dorsal root ganglions, and illustrates the limits and potentials of these cell lines as tools for neuropharmacological exploration.
Keywords: Dorsal root ganglions, SH-SY5Y, F-11, ND7/23, transcriptomic analysis, molecular expression
Background
Peripheral sensory neurons enable humans to sense the world around us. Sensory nerve endings innervating the skin, muscle, and viscera are responsible for detecting external stimuli such as touch, vibration, temperature, and pain, which are then relayed to the spinal cord and the central nervous system. Peripheral sensory neurons are a heterogeneous group of neurons that can be classified according to axonal transmission speed, degrees of myelination, and functional characteristics.1 The cell bodies of these pseudo-unipolar neurons are located in the dorsal root ganglia (DRG) and are used extensively to study mammalian sensory processes, peripheral mechanisms of pain, and the effect of drugs on the peripheral nervous system. In recent years, significant advances have been made in unravelling the molecular and pharmacological basis of the diverse functional properties defining subsets of peripheral sensory neurons, as well as the G-protein coupled receptors (GPCRs) and ion channels contributing to sensory neuron physiology and pathophysiology. For example, the transient receptor potential (TRP) channels including TRPV1, TRPA1, and TRPM8 are activated by temperature and reactive chemicals, while GPCRs such as protease-activated receptors, histamine receptors, and bradykinin receptors are activated by inflammatory mediators. While primary DRG neurons are used extensively to study the pharmacological mechanisms involved in sensory neuron functions such as pain and itch, this approach has several drawbacks. The isolation and culture of DRG neurons is time consuming, highly technical, and requires the use of animals. Importantly, not all neurons survive this process.2 Removal of DRG neurons from the in vivo environment and axotomy of the peripheral processes also leads to adaptive changes such as down-regulation of the potassium channel subunit KV9.1, overexpression of the nerve growth factor receptor TrkA, and increased neuronal excitability.3–6 In addition, as DRG neurons are heterogeneous, only a subset of cells is likely to express the target of interest and a substantial proportion of cultured cells include non-neuronal cell types such as fibroblasts and glial cells. Accordingly, researchers have turned to DRG-derived immortal cell lines as substitutes for intact DRG neurons when investigating endogenous receptors. Common candidates are the DRG-hybridoma cell lines F-11 and ND7/23, which were derived from embryonic or neonatal rat DRG neurons by fusion with the mouse neuroblastoma cell line N18Tg27,8 and display neuron-like properties including excitable membranes and sensory neuronal cell surface markers. Similarly, we have recently shown that the neuroblastoma cell line SH-SY5Y, which is commonly used as a model of adrenergic or dopaminergic neurons, exhibits traits of peripheral sensory neurons such as functional expression of the sensory neuron-specific sodium channel isoform NaV1.7.9 However, although these cell lines are often considered suitable models to study mechanisms of nociceptor activation and individual investigations of specific protein families have been conducted,9–11 a systematic analysis of the expression profile of these cell lines in comparison with DRG neurons is lacking.
In this study, we conducted RNA-Seq analysis with particular emphasis on ion channel and GPCR expression in DRG neuron-derived cell lines including the human neuroblastoma cells SH-SY5Y,12,13 the mouse neuroblastoma/rat embryonic DRG neuron hybrid cell line F-11,8 and the mouse neuroblastoma/rat neonatal DRG neuron hybrid cell line ND7/23.7 Data from cell lines were then compared to RNA-Seq analysis conducted on whole murine DRG. Functional expression of selected targets was confirmed by Ca2+ imaging. While a number of sensory neuron-specific targets including NaV1.7 were expressed in all cell lines, we also found several discrepancies, and none of the cell lines displayed an expression profile consistent with specific subclasses of sensory neurons. This stresses the need for further exploration on the functional receptors present on neuronal cell lines used as model DRG neurons, and highlights the importance for cautious interpretation of results obtained using SH-SY5Y, F-11, and ND7/23 cells.
Methods
Animals
Male wild-type C57BL/6 mice age six to eight weeks and weighing 20–23 g were used in the study. Ethical approval for experiments involving animal tissues was obtained from the University of Queensland animal ethics committee. Experiments were conducted in accordance with the Animal Care and Protection Regulation Qld (2012), the Australian Code of Practice for the Care and Use of Animals for Scientific Purposes, 8th edition (2013), and the International Association for the Study of Pain Guidelines for the Use of Animals in Research. All animals were obtained from University of Queensland Biological Resources (Brisbane, Australia).
Chemicals
The following compounds were obtained from Life Technologies (Mulgrave, Victoria, Australia): L-glutamine, Dulbecco’s Modified Eagle Medium (DMEM), Ham’s F12 media, Roswell Park Memorial Institute medium, 0.25% trypsin/EDTA, foetal bovine serum (FBS), and phosphate buffered saline (PBS). The following compounds were obtained from Sigma-Aldrich (Castle Hill, New South Wales, Australia): HAT media supplement Hybri-Max™, capsaicin, collagenase, protease, menthol, and allyl isothiocyanate (AITC). Oxytocin and vasopressin were a kind gift from Professor Paul Alewood (Institute for Molecular Bioscience, The University of Queensland). Calcium 4 No Wash dye was purchased from Molecular Devices (Sunnyvale, CA, USA).
Cell culture
Human neuroblastoma SH-SY5Y cells (The European Collection of Cell Cultures) were grown in Roswell Park Memorial Institute medium with 15% FBS and 2 mM L-glutamine and kept at 37℃/5% CO2; 0.25% trypsin/EDTA was used to passage the cells every three to five days at a 1:5 dilution ratio or when approximately 90% confluent.
Mouse neuroblastoma/rat embryonic DRG neuron hybrid F-11 cells (The European Collection of Cell Cultures) were grown in Ham’s F12 media with 10% FBS, 100 mM hypoxanthine, 0.4 mM aminopterin, and 16 mM thymidine (HAT media supplement Hybri-Max™) and kept at 37℃/5% CO2; 0.25% trypsin/EDTA was used to passage the cells every two to three days at a 1:5 dilution ratio or when approximately 80% confluent.
Mouse neuroblastoma/rat DRG neuron hybrid ND7/23 cells (The European Collection of Cell Cultures) were grown in DMEM with 10% FBS, 2 mM L-glutamine, pyridoxine, and 110 mg/ml sodium pyruvate and kept at 37℃/5% CO2; 0.25% trypsin/EDTA was used to passage the cells every three to five days at a 1:5 dilution ratio or when approximately 90% confluent.
Sample extraction and preparation
Mice were euthanised with carbon dioxide followed by cervical dislocation. The spine was immediately removed, the dorsal side of the vertebra bones removed, and the tissue immersed in ice-cold DMEM. The spinal cord was removed, and thoracic and lumbar DRGs (T6-L6 inclusive) were dissected with forceps and placed in DMEM, followed by trimming of excess myelin. Dissected DRGs were then placed into fresh DMEM with 1 mg/mL collagenase and 0.5 mg/mL protease, and incubated at 37℃/5% CO2 for 45 min to allow for dissociation. The dissociated cells were centrifuged at 20,000 r/min for 3 min and excess DMEM removed. The cell pellet was then washed in PBS, centrifuged briefly, and excess PBS removed. Whole DRG samples from two mice were pooled together for RNA extraction.
All cell lines were allowed to reach ∼90% confluence in a T75 flask in culture before harvesting. Cells were harvested by washing with PBS, followed by trypsinisation with 0.25% trypsin/EDTA. The appropriate cell culture media were added into the flask and the cell suspension was centrifuged at 14,000 r/min for 5 min to form a pellet. Excess media was removed and the cell pellet processed for RNA extraction.
RNA-seq, bioinformatics analysis, and data analysis
Cells were brought up from frozen stocks and allowed to recover for at least three to five passages. RNA was extracted from SH-SY5Y cells at passage 29, from F-11 cells at passage 9, and from ND7/23 cells at passage 28 using the RNeasy Mini Kit (Qiagen, Melbourne, Australia) according to the manufacturer’s instructions, including on-column DNase digestion. The RNA Integrity Number (RIN) was above 8.5 for all samples. Library construction and bioinformatics analysis were conducted by the Australian Genome Research Facility. RNA-Seq was conducted on an Illumina Hiseq-2000 machine as a 100-nucleotide single-end run, and primary.fastq data was generated with the CASAVA 1.8.2 pipeline (Illumina). Reads were screened for the presence of any adaptor or overrepresented sequences, ambiguous characters were clipped where required, and sequence bias was corrected. Alignment was performed with Tophat with default analysis parameters in single-end mode. The number of mismatches were restricted to no more than two bases. Reads from SH-SY5Y cells were mapped against the human genome (hg19), DRG reads were mapped against the mouse genome (mm9), and F-11 and ND7/23 reads were mapped against both the mouse genome (mm9) and rat genome (rn5). Transcripts assembly was performed by Cufflinks with the reference annotation-based assembly option14 using the UCSC annotation, and multi-reads correction was used. Gene counts were then compiled using the HTSeq software.15 Heatmaps were generated using R with the gplots, pheatmap, and RColorBrewer libraries.
Accession number and gene ontology analysis
Raw bioinformatics data in the form of .fastq files and count tables generated with HTSeq are both available at the Gene Expression Omnibus database under the accession number GSE75811. Gene ontology (GO) analysis was conducted with the PANTHER database16 with the Gene List Analysis tool.17 Genes with >100 reads in each sample were compiled and submitted to Gene List Analysis and analysed according to Biological Processes (BP) terms. Genes from whole DRGs were assigned according to the Mus musculus genome, the SH-SY5Y sample was assigned to the Homo sapiens genome and F-11 and ND7/23 samples were assigned to both Mus musculus and Rattus norvegicus genomes each.
Transfection
HEK293 (American Tissue Culture Collection, Manassas, VA, USA) were routinely maintained in DMEM containing 10% FBS, 2 mM L-glutamine, pyridoxine, and 110 mg/ml sodium pyruvate. Cells were split every three to six days in a ratio of 1:5 using 0.25% trypsin/EDTA. Cells were plated on T75 tissue culture flasks (Nunc) 24 h prior to transfection, and transfected with plasmid DNA of rTRPV1 (D. Julius, Department of Physiology, University of California, Berkeley, CA, USA), mTRPA1 (A. Patapoutian, The Scripps Research Institute, La Jolla, CA, USA), rTRPM8 (K. Zimmermann, Department of Anesthesiology, Friedrich-Alexander-University, Erlangen-Nuremberg, Erlangen, Germany), hV1A, or hOTR (OriGene Technologies, Rockville, MD, USA) using Lipofectamine 2000 according to the manufacturer's instructions. Twenty-four hours after transfection, cells were plated on 384-well plates and used for Ca2+ experiments 24 h after plating.
Fluorescent Ca2+ assays
Cells were prepared as previously reported.9 Briefly, cells were seeded onto 384-well black-walled imaging plates (Corning, MA, USA) 48 h prior to calcium imaging experiments and incubated at 37℃/5% CO2. SH-SY5Y cells were placed onto uncoated plates (Corning, Cat No #3542), while F-11 and ND7/23 cells were placed onto Cell-bind (Corning, Cat no #3683) plates. Cells were seeded to ensure ∼90% confluency on the day of the experiment.
Cells were loaded 30 min prior to imaging with Calcium 4 No-Wash dye in physiological salt solution (PSS; NaCl 140 mM, glucose 11.5 mM, KCl 5.9 mM, MgCl2 1.4 mM, NaH2PO4 1.2 mM, NaHCO3 5 mM, CaCl2 1.8 mM, HEPES 10 mM), and kept at 37℃/5% CO2. Ca2+ responses were measured using a FLIPRTETRA (Molecular Devices, Sunnyvale, CA, USA) fluorescent plate reader with excitation at 470–495 nM and emission at 515–575 nM. Camera gain and intensity were adjusted for each plate to yield a minimum of 1000–1500 arbitrary fluorescence units baseline fluorescence. Prior to addition of agonists, 10 baseline fluorescence readings were taken, followed by fluorescent readings every 0.5 s for 150 s. Data was analysed using Screenworks 3.2 and FLIPR™ data were plotted using GraphPad Prism™ software (Version 6.00).
Results
All samples had 84% or more reads aligned to the respective reference genomes (Table 1). The whole DRG sample included heterogenous populations of cells including neurons and non-neuronal cells, as exemplified by the presence of the non-neuronal glial cell marker vimentin (VIM).18 All cell samples expressed the neuronal and glial marker N-Cam, the neuronal marker F-3, and the mouse neuronal marker Thy-1, which was particularly abundant in the whole DRG sample. As data normalisation relies upon housekeeping genes expressed in similar levels across all samples, we examined the expression of various housekeeping genes due to the recognised issue of differential expression levels for standard housekeeping genes19 and found TUBA1A to be one of the only candidates expressed at a similar global level (0.02–0.08% of all aligned reads) across all samples in our data. In contrast, other popular housekeeping genes commonly used in data normalisation, such as GAPDH and β-actin, were not expressed with similar intensity across all our samples. For instance, 64 reads aligned to GAPDH in the DRG sample, while 12,475 reads aligned to GAPDH in the SH-SY5Y sample. We thus normalised all reads to the highly expressed α-tubulin gene, with the intensities of gene expression presented as normalised gene ratios.
Table 1.
Read alignments for all RNA samples.
| Mouse DRGs (mm9) | SH-SY5Y (hg19) | F-11 (mm9) | F-11 (rn5) | ND7/23 (mm9) | ND7/23 (rn5) | |
|---|---|---|---|---|---|---|
| Total reads | 35723774 | 31364133 | 29496604 | 13580565 | 30759267 | 12005128 |
| Aligned reads | 32480739 (90.92%) | 27118145 (86.46%) | 26346262 (89.32%) | 11513489 (84.78%) | 27667233 (89.95%) | 10337475 (86.11%) |
| Non-matching reads | 2217836 (6.83%) | 2407972 (7.68%) | 2346986 (7.96%) | 1987513 (14.63%) | 2225020 (7.23%) | 1612465 (13.43%) |
| Multiple matching reads | 1025199 (3.15%) | 1838016 (5.86%) | 803356 (2.72%) | 47147 (0.59%) | 867014 (2.82%) | 42507 (0.46%) |
DRG: dorsal root ganglion.
The two hybrid cell lines examined, F-11 and ND7/23, are known to contain both rat and mouse chromosomes (F-11)8 and express rat surface markers after fusion with mouse neuroblastoma cells (ND7/23).7 It was not clear which genes are transcribed from the rat genome and which are from the mouse genome in these cell lines. We therefore chose to align F-11 and ND7/23 reads twice, once against the mouse genome and once against the rat. We found that qualitatively, expression profiles largely mirrored each other between mouse and rat alignments in these two cell lines, varying predominantly in intensity.
Neuronal markers expressed by neuronal cell lines
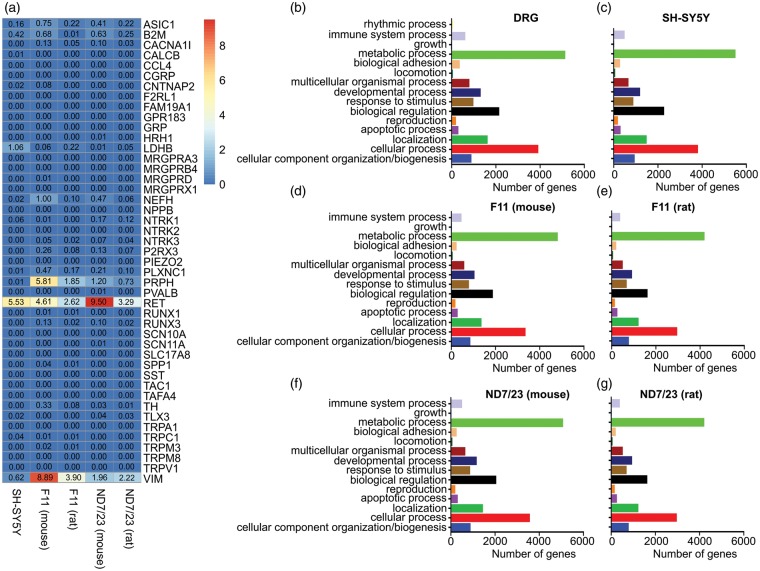
We initially analysed the cell line expression data against a wide range of known markers of specific neuronal types (Figure 1(a)). Some of these include the peptidergic nociceptor marker TrkA (NTRK1), the mechanosensor marker PIEZO2/FAM38B, the myelinated neuronal marker CNTNAP2 (contactin associated protein-like 2), and voltage-gated calcium channel (CaV) subtype 3.2 that was recently identified to be uniquely expressed on unmyelinated low-threshold mechanoreceptors (C-LTMR)20 (CACNA1H). Out of the 43 neuronal markers investigated, transcripts of nine were found in all cell lines. These nine genes included the nociceptor markers voltage-gated sodium channel (NaV) subtype 1.9 (SCN11A) and acid-sensing ion channel 1 (ASIC1); TLX3, a transcription factor that regulates the expression of several sensory channels and receptors implicated in sensing pain, itch, and temperature21; PLXNC1, a marker for unmyelinated nonpeptidergic nociceptors and thermosensors; RET, a marker for myelinated and unmyelinated neurons;22 the C-LTMR marker tyrosine hydroxylase (TH);23 RUNX3, a transcription factor that regulates the development of proprioceptive DRG neurons24; the unmyelinated C-fibre marker peripherin (PRPH); and LDHB, a marker of myelinated neurons22 (Figure 1(a)). However, the co-expression of some of these markers in individual cell lines was surprising given their expression profiles do not overlap significantly in subtypes of DRG neurons. Specifically, some markers are known to be selectively expressed in unmyelinated C-fibre neurons (such as PRPH and TH), while others are indicative of myelinated A-fibres (such as LDHB) and would thus not be expected to be present in a single sensory neuron type. Many sensory neuron genes were also absent across all cell line samples, including nociceptive markers such as TRPV1,25 TRPA1,26 calcitonin gene-related peptide (CGRP),27 and NaV1.8 (SCN10A); mechanoreceptor markers such as PIEZO228 and VGLUT3/SLC17A829; and markers for neurons responsible for itch such as PAR2 (F2RL1).30 This suggests that while the cell lines are known to express receptors found on DRG neurons, they do not mimic the entire repertoire of proteins expressed by functional in vivo DRG neuron classes.
Figure 1.
Neuronal markers and gene ontology analysis on RNA samples. The SH-SY5Y, F-11, and ND7/23 cell lines do not fit into existing categories of peripheral neurons, yet gene ontology analysis demonstrates that the cell lines transcriptomes retain similar ratios of enriched biological processes compared to whole DRGs. (a) Expression of markers commonly used to identify specific peripheral neuron sub-populations, microglia, and non-neuronal cells by the SH-SY5Y, F-11, and ND7/23 cell lines. (b) Gene ontology analysis of whole DRGs. (c) Gene ontology analysis of SH-SY5Y cells. (d) Gene ontology analysis of F-11 cells (aligned to the mouse genome). (e) Gene ontology analysis of F-11 cells (aligned to the rat genome). (f) Gene ontology analysis of ND7/23 cells (aligned to the mouse genome). (g) Gene ontology analysis of ND7/23 cells (aligned to the rat genome).
In SH-SY5Y cells, the most enriched marker was RET, the glial cell line-derived neurotrophic factor (GDNF) receptor tyrosine kinase, consistent with previous reports that these cells respond to GDNF with increased neurite outgrowth.31,32 SH-SY5Y cells expressed markers of A-fibres, including LDHB, CNTNAP2, and neurofilament heavy polypeptide (NEFH). However, TrkA (NTRK1), a C-fibre marker, was also expressed; while the A-fibre marker TrkC (NTRK3) was completely absent. SH-SY5Y also lacked the nociceptor markers TRPV1, MAS-related GPR member D (MRGPRD), the peptidergic neuron markers CGRP, substance P (TAC1), the itch receptor marker somatostatin, the mechanoreceptor marker TAFA4/FAM19A4, and the proprioceptor marker parvalbumin (PVALB). Overall, SH-SY5Y cells expressed a range of genes associated with different subtypes of sensory neurons and could not be classified according to known DRG neuron subclasses, including those defined by Usoskin et al.,22 Li et al.,33 or simpler categorisation based on expression of established markers relating to functional aspects, such as CGRP, IB4, and NF200. Although further classification of sensory neurons was recently reported by Li et al.,33 DRG classification criteria based on transcriptional markers remain relatively broad and as yet do not unambiguously encompass the possibly 17 or more different fibre types that have been proposed based on responsiveness to mechanical and thermal stimuli as well as conduction velocity and rate of adaptation.34 Thus, while these cells express genes – as well as their respective protein products9,10 – known to be involved in key sensory neuron functions, they should not be regarded as a model for DRG neurons.
F-11 and ND7/23 cells were both created from a fusion of mouse neuroblastoma cells and rat embryonic or neonatal DRG neurons, respectively.7,8 Accordingly, the expression profile of neuronal markers was similar in both cell lines, and included a number of markers of A-fibre mechanosensors and proprioceptors, including LDHB, NEFH, and TrkC (Figure 1(a)). As with SH-SY5Y cells, markers for C-fibres were also present in both cell lines, including P2RX3, a purinergic ion channel expressed in C-fibre non-peptidergic neurons; plexin C1 (PLXNC1); TH; PRPH; RET; and CaV3.3 (CACNA1I). ND7/23 cells expressed comparatively higher levels of TLX3, SCN11A, TrkC, and TrkA than F-11 cells, while ASIC1, PLXNC1, TH, PRPH, NEFH, CNTNAP2, LDHB, CACNA1H, and CACNA1I had reduced expression in ND7/23 cells compared with F-11 cells. TrkB was expressed at very low levels in both cell lines. The C-fibre marker TrkA was enriched approximately 20-fold in ND7/23 cells compared with F-11 cells, while TH was elevated approximately 10-fold in F-11 cells compared to ND7/23 cells. This established that, similar to SH-SY5Y cells, both F-11 and ND7/23 cells expressed a range of cellular markers that are usually found in distinct sensory neuron subtypes and a specific neuronal origin can thus not be definitely assigned.
All samples showed a similar distribution of genes expressed according to BP terms following global GO analysis. The BP term with the most genes associated was ‘metabolic process’ across all samples, followed by the terms ‘cellular process’ and ‘biological regulation’. Fourteen different BP terms were found to be present in all samples, with whole DRGs being the only sample that contained one extra term, that of ‘rhythmic process’ (Figure 1(b) to (g)). F-11 and ND7/23 samples exhibited reduced numbers of genes associated with each BP term when analysed against the rat genome; however, the distribution of associations across different terms remains very similar across the mouse and rat genomes for both samples.
Ion channel and GPCR expression by neuronal cell lines
We next analysed the expression of ion channels and GPCRs in whole DRGs and F-11, ND7/23, and SH-SY5Y cells. Ion channels have an important role in regulating neuronal excitability and firing rate, making them promising novel therapeutic targets in the treatment of pain. Pharmacological modulators of certain families of ion channels, including voltage-gated calcium channels (CaV), voltage-gated potassium channels (KV), and voltage-gated sodium channels (NaV), are already used as drugs in the clinic to treat neuropathic pain.35,36 Similarly, many GPCRs initiate intracellular changes after activation from external chemicals, making this family of receptors ideal pharmacological targets. A number of GPCRs are already effective and established analgesic targets, such as the opioid receptors, while others have recognised roles in inflammation and nociception, such as bradykinin and protease-activated receptors.37–39 We assessed expression of ion channels and GPCRs with putative therapeutic importance based on classification obtained from IUPHAR (http://www.guidetopharmacology.org).
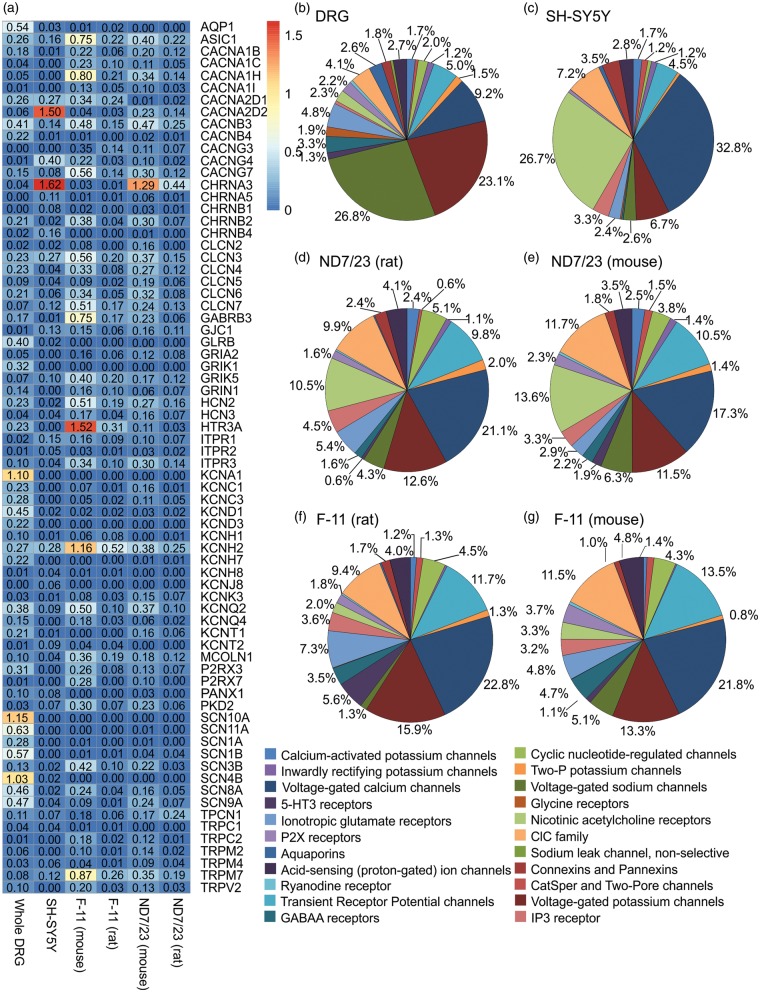
There were 261 ion channel genes detected in whole DRGs (≥1 read aligned to the gene), 217 in SH-SY5Y cells, 223 in F-11 (mouse), 201 in F-11 (rat), 214 in ND7/23 (mouse), and 189 genes in ND7/23 (rat). Twenty-one families of ion channels were expressed by all samples, including several considered as therapeutic targets for pain such as ASIC receptors, GABAA receptors, and voltage-gated sodium, potassium, and calcium channels (Figure 2(a)). Whole DRGs had the highest number of ion channel transcripts among the samples, with 494,237 reads aligned to ion channel genes. The level of ion channel expression was also at least three times higher in whole DRGs (1.42% of total aligned reads) than in the cell lines examined; 0.53% of total aligned reads in SH-SY5Y cells consisted of ion channel reads, while ion channel reads made up 0.43% of total aligned reads in F-11 (mouse), 0.48% of total aligned reads in F-11 (rat), 0.42% of total aligned reads in ND7/23 (mouse), and 0.45% of total aligned reads in ND7/23 (rat). A total of 58 ion channels were found to be uniquely expressed in whole DRGs, while eight uniquely expressed ion channel genes were found in SH-SY5Y, three in F-11, and four in ND7/23 (Table 2).
Figure 2.
Whole DRGs and cell lines have unique expression patterns of ion channels families. Sections of the pie chart without an annotated percentage constituted ≤0.5% of total ion channel expression in the sample. (a) Compilation of the ion channel genes with the highest expression levels for each sample. (b) Distribution of ion channel expression in whole DRGs. (c) Distribution of ion channel expression in SH-SY5Y cells. (d) Distribution of ion channel expression in ND7/23 cells (aligned to the rat genome). (e) Distribution of ion channel expression in ND7/23 cells (aligned to the mouse genome). (f) Distribution of ion channel expression in F-11 cells (aligned to the rat genome). (g) Distribution of ion channel expression in F-11 cells (aligned to the mouse genome).
Table 2.
Ion channels and GPCRs uniquely expressed in whole DRGs and cell lines.
| Whole DRGs | SH-SY5Y | F-11 | ND7/23 | |
|---|---|---|---|---|
| Uniquely expressed | ANO1 | AQP10 | CATSPER2 | KCNK6 |
| ion channels | AQP4 | AQP3 | GJB3 | KCNN3 |
| ASIC2 | CHRND | MCOLN3 | SCNN1A | |
| CACNA1D | GABRQ | TRPM5 | ||
| CACNA1E | GJC2 | |||
| CACNA2D3 | KCNJ8 | |||
| CHRNA6 | KCNQ1 | |||
| CHRNB3 | SCNN1D | |||
| GABRA1 | ||||
| GABRA2 | ||||
| GABRA3 | ||||
| GABRA5 | ||||
| GABRB2 | ||||
| GABRG1 | ||||
| GABRG2 | ||||
| GABRG3 | ||||
| GJB1 | ||||
| GJC3 | ||||
| GJD2 | ||||
| GRIA1 | ||||
| GRID2 | ||||
| GRIK1 | ||||
| GRIK4 | ||||
| GRIN3A | ||||
| HCN1 | ||||
| HVCN1 | ||||
| KCNA1 | ||||
| KCNA2 | ||||
| KCNB2 | ||||
| KCNC2 | ||||
| KCND3 | ||||
| KCNG2 | ||||
| KCNG4 | ||||
| KCNH5 | ||||
| KCNJ16 | ||||
| KCNJ3 | ||||
| KCNJ9 | ||||
| KCNK10 | ||||
| KCNK13 | ||||
| KCNK18 | ||||
| KCNK4 | ||||
| KCNQ3 | ||||
| KCNS1 | ||||
| KCNU1 | ||||
| KCNV1 | ||||
| NALCN | ||||
| P2RX5 | ||||
| RYR3 | ||||
| SCN10A | ||||
| SCN1A | ||||
| SCN4A | ||||
| SCN2B | ||||
| TRPA1 | ||||
| TRPC4 | ||||
| TRPC6 | ||||
| TRPM3 | ||||
| TRPM8 | ||||
| TRPV1 | ||||
| Uniquely expressed | ADRA1D | APLNR | GPBAR1 | AGTR2 |
| GPCRs | ADRA2A | GPR63 | DRD2 | |
| ADRB2 | GPR98 | DRD4 | ||
| AGTR1B | HTR2B | GIPR | ||
| CCKAR | LTB4R1 | GPR20 | ||
| CHRM2 | NPFFR2 | GPR4 | ||
| CYSLTR2 | PTGER2 | HRH3 | ||
| DARC | PTH2R | SSTR5 | ||
| ELTD1 | TAS1R2 | |||
| GABBR2 | VIPR2 | |||
| GALR1 | ||||
| GPR126 | ||||
| GPR139 | ||||
| GPR149 | ||||
| GPR179 | ||||
| GPR26 | ||||
| GPR35 | ||||
| GPR37L1 | ||||
| GPR61 | ||||
| GPR75 | ||||
| GPR83 | ||||
| GRM4 | ||||
| GRM8 | ||||
| HCRTR1 | ||||
| HCRTR2 | ||||
| HRH1 | ||||
| HTR1A | ||||
| HTR1B | ||||
| HTR1D | ||||
| HTR1F | ||||
| HTR2A | ||||
| HTR4 | ||||
| HTR5A | ||||
| HTR7 | ||||
| LPAR5 | ||||
| MCHR1 | ||||
| MRGPRD | ||||
| MRGPRX1 | ||||
| NPY1R | ||||
| OPRK1 | ||||
| OXTR | ||||
| P2RY1 | ||||
| P2RY12 | ||||
| PTGDR | ||||
| PTGER3 | ||||
| S1PR1 |
GPCR: G-protein coupled receptor; DRG: dorsal root ganglion. A gene is considered to be uniquely expressed if >100 reads are detected only in one sample.
The most striking difference in the distribution of ion channel transcripts between DRGs and cell lines lies in the percentage of voltage-gated sodium, potassium, and calcium channels. Together, NaV, KV, and CaV consisted of 59% of all reads aligned to ion channels in whole DRG, a trend that was reflected in the three cell lines. NaV, KV, and CaV constituted 43% of all ion channel transcripts in SH-SY5Y cells, 40% in F-11 cells (mouse and rat), 35% in ND7/23 cells (mouse), and 38% in ND7/23 cells (rat) counts. However, much of the CaV expression in the cell lines was contributed by CaV auxiliary units rather than the channel-forming CaV α units, and this was especially true for SH-SY5Y cells. SH-SY5Y cells also abundantly expressed cholinergic nicotinic receptors, amounting to 27% of all ion channel transcripts. In contrast, cholinergic nicotinic receptors consisted of 15% or less ion channel transcripts in all other samples (whole DRG 2%, F-11 mouse 3%, F-11 rat 2%, ND7/23 mouse 14%, and ND7/23 rat 11%) (Figure 2(b) to (g)).
A compilation of the 30 ion channels with the highest expression in each sample revealed significant differences as well as similarities between the cell lines and compared with whole DRGs (Figure 2(a)). The ion channels with the highest expression in whole DRGs were NaV1.8 (SCN10A), KV1.1 (KCNA1), and NaV β4 subunit (SCN4B). Other ion channels highly expressed in whole DRGs, but expressed at very low levels in the cell lines, included NaV1.9, aquaporin 1, the NaV β1 subunit, KV4.1 (KCND1), and glycine receptor β unit (GLRB). In addition, transcripts for NaV1.1 (SCN1A), ionotropic glutamate receptor 1 (GRIK1), and KV4.3 (KCND3) were found at lower levels in DRGs but were virtually absent in the three cell lines. Transcripts for SCN8A (NaV1.6) and SCN9A (NaV1.7), channels responsible for signal transduction and known to have important roles in pain, were present in all cell lines as well as being highly expressed in whole DRGs. F-11 and ND7/23 cells in particularly expressed NaV1.6 and NaV1.7 at higher levels than SH-SY5Y cells, with F-11 cells expressing more NaV1.6 than ND7/23, and ND7/23 cells expressing more NaV1.7 than F-11 cells.
The nicotinic cholinergic receptor α3 (CHRNA3) was the ion channel with the highest expression in SH-SY5Y and ND7/23 cells, but few transcripts were found in whole DRGs and F-11 cells. The second-most enriched ion channel in SH-SY5Y cells, the CaVα2δ subunit 2 (CACNA2D2), also had very little expression in the other samples. Other ion channels highly expressed in SH-SY5Y cells included CaV auxiliary units such as CACNG4 and CACNA2D1, and voltage-gated chloride channels such as CLCN3 (Figure 2(a)).
While the overall gene expression profiles of F-11 and ND7/23 cells were similar, there was notable disparity in ion channel expression. F-11 cells expressed a large amount of transcripts for KV11.1 (KCNH2, otherwise known as Human ether-a-go-go-related gene (hERG)) and 5-HT receptor 3 a (HTR3A), genes that were expressed at much lower level in ND7/23 cells (Figure 2(a)). In contrast, CHRNA3 was the most highly expressed ion channel transcript in ND7/23 cells, but was only expressed at low levels in F-11 cells. Apart from these exceptions, many ion channel genes where expressed at similar levels in F-11 and ND7/23 cells. Several of these, including ion channels with recognised roles in nociception such as ASIC1, CaV3.2 (CACNA1H), and KV7.2 (KCNQ2) were highly expressed in F-11 and ND7/23 cells as well as in whole DRGs and SH-SY5Y cells (Figure 2(a)). Similarly, P2RX3, CaV3.3 (CACNA1I), TRPV2, and CaV2.2 (CACNA1B) were enriched in F-11 and ND7/23 cells to a lesser extent compared with DRGs and SH-SY5Y cells. For all of these ion channels, transcript numbers were higher in F-11 cells compared with ND7/23 cells. In contrast, TRPM2, CLCN2, KCNK3, CLCN5, GRIA2, TPCN1, KCNC3, KCNC1, and KCNT1 were expressed at higher levels in ND7/23 cells than F-11 cells.
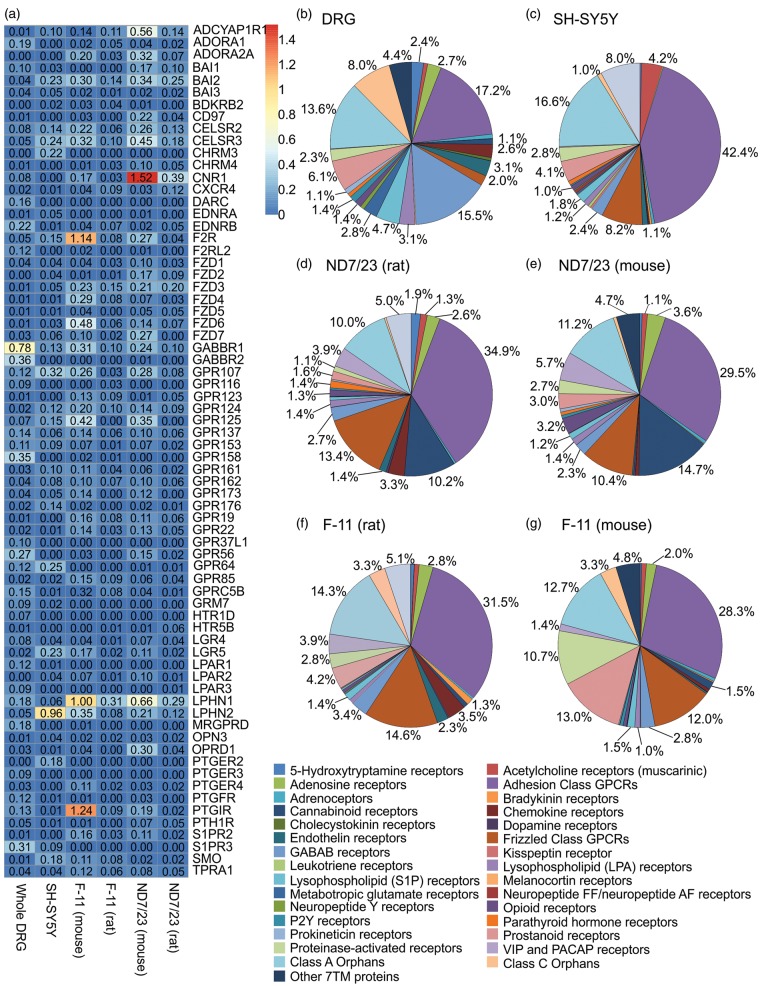
A total of 300 different GPCR genes from the IUPHAR list were expressed in whole DRGs, 218 genes in SH-SY5Y cells, 215 in F-11 (mouse), 188 in F-11 (rat), 225 in ND7/23 (mouse), and 187 in ND7/23 cells (rat) (Figure 3(a) to (g)). Whole DRGs contained the most GPCR transcripts, with 185,707 reads aligned to GPCR genes accounting for 0.54% of all aligned reads. SH-SY5Y cells had the second-most abundant GPCR expression (0.38% of all aligned reads), followed by ND7/23 mouse (0.35%), ND7/23 rat (0.33%), and F-11 rat and mouse (both 0.26%). Similar to the trend seen for ion channels, whole DRGs had both the highest GPCR expression level and the highest number of uniquely expressed GPCRs, with 46 GPCRs uniquely expressed in whole DRGs. SH-SY5Y cells expressed 8 unique GPCRs, F-11 cells expressed 1, and ND7/23 cells expressed 10 (Table 2).
Figure 3.
Whole DRGs and cell lines have unique expression patterns of GPCRs. Sections of the pie chart without an annotated percentage constituted ≤0.5% of total ion channel expression in the sample. (a) Compilation of the GPCR genes with the highest expression levels for each sample. (b) Distribution of GPCR expression in whole DRGs. (c) Distribution of GPCR expression in SH-SY5Y cells. (d) Distribution of GPCR expression in ND7/23 cells (aligned to the rat genome). (e) Distribution of GPCR expression in ND7/23 cells (aligned to the mouse genome). (f) Distribution of GPCR expression in F-11 cells (aligned to the rat genome). (g) Distribution of GPCR expression in F-11 cells (aligned to the mouse genome).
A total of 45 families of GPCRs were expressed by all four samples. Among these, the adhesion class GPCR family had the highest expression levels in all samples, constituting 42% of all GPCR transcripts from SH-SY5Y cells, 35% in ND7/23 cells (rat), 32% in F-11 cells (rat), 28.3% in ND7/23 (mouse), 27.5% in F-11 (mouse), and 17% in whole DRGs. The GPCR family with the second highest expression in whole DRGs were the GABAB receptors GABBR1 and GABBR2 (16% of all GPCR reads), a trend that was not observed in the cell lines where GABAB receptors only consisted of 3% of GPCR reads in F-11 and ND7/23 (rat) and 2% in ND7/23 (mouse) and SH-SY5Y cells. However, expression of approximately equivalent transcript numbers for both the R1 and R2 subunit of the GABAB receptor was only observed in DRG neurons, suggesting that functional GABAB receptors do not form in the neuronal cell lines (Figure 3(b)).
Other GPCRs with high expression in whole DRGs include sphingosine-1-phosphate receptor 3 and the orphan GPCRs GPR158 and GPR56. These genes, together with the prostaglandin F receptor (PTGFR), par3 (F2RL2), 5-HT receptor 1D (HTR1D), prostaglandin E receptor 3 (PTGER3), and adenosine A1 receptor (ADORA1), were expressed at very low levels in neuronal cell lines. Two exceptions in this group include the orphan GPCRs with unknown function, GPR137, and GPR153, which were expressed in all samples (Figure 3(a)).
The GPCR with the highest level of expression in SH-SY5Y cells was latrophilin 2 (LPHN2). Latrophilin 2 is an adhesion GPCR that plays a role in exocytosis and is closed related to latrophilin 1 (LPHN1), the receptor for latrotoxin, the active peptide in black widow spider venom.40 Interestingly, latrophilin 1 was the GPCR with the highest expression in F-11 cells and the second highest expressed in ND7/23 cells, and was also present in whole DRGs. Many genes enriched in SH-SY5Y cells, such as smoothened and par1 (F2R), were also expressed in other samples. However, a small group of genes, including the muscarinic acetylcholine receptor M3 (CHRM3) and prostaglandin E2 receptor (PTGER2), were enriched in SH-SY5Y cells but largely absent in other samples. Overall, GPCR expression patterns were similar for all neuronal cell lines, but distinct from that of whole DRGs (Figure 3(a)).
In F-11 cells, the prostaglandin I2 receptor (PTGIR), par1 (F2R), and LPHN1 were the most highly expressed GPCRs. In ND7/23 cells, the GPCRs with the highest expression were the cannabinoid receptor 1, LPHN1, and pituitary adenylate cyclase-activating polypeptide type I receptor. The majority of GPCR genes were expressed at similar levels in F-11 and ND7/23 cells. GPCRs enriched in F-11 and ND7/23 tended to be expressed at very low levels in whole DRGs, whereas a group of GPCRs highly expressed in DRGs, including CELSR3 and BAI2, were enriched in SH-SY5Y cells. Of note, GPR123 and PTGER4 were enriched exclusively in F-11 cells, and CD97 and FZD2 were enriched only in ND7/23 cells. When aligned to the mouse genomes, the top 30 GPCRs expressed for F-11 and ND7/23 cells resembled each other very closely. Interestingly, F-11 cells expressed mouse but not rat transcripts of PTGIR, while the same was the case for ADCYAP1R in ND7/23 cells (Figure 3(a)).
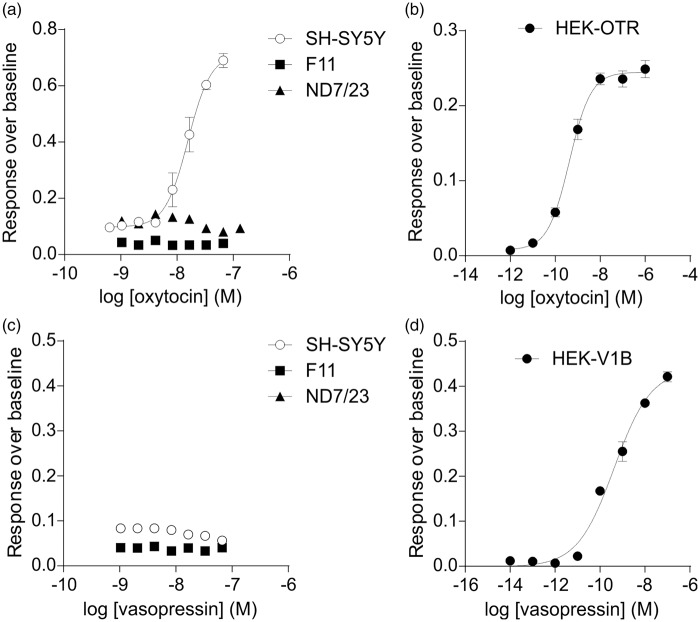
Interestingly, we noted expression of both arginine vasopressin receptor 1A (AVPR1A) and oxytocin receptors (OXTR) in DRGs, consistent with an emerging functional role of these GPCRs in pain, where AVPRA1 has been linked to strain-dependent pain sensitivity to formalin and capsaicin41 and oxytocin activation ameliorates visceral pain in particular.42 Consistent with a specific role of these GPCRs in pain, both vasopressin receptor 1B and vasopressin receptor 2 were expressed at very low level or absent in DRGs. In contrast, all three cell lines expressed OXTRs, but vasopressin receptor 1A and 1B were absent. We thus sought to verify this gene expression profile using functional Ca2+ imaging. The OXTR was expressed in SH-SY5Y cells, absent in F-11 cells, and only aligned to three reads in ND7/23 cells. Accordingly, only SH-SY5Y responded to oxytocin at concentrations that elicit Ca2+ responses in HEK293 cells transfected with the OXTR, confirming the presence of functional OXTRs in SH-SY5Y cells and its absence in F-11 and ND7/23 cells (Figure 4(a) and (b)). Moreover, consistent with our transcriptomic results, vasopressin did not elicit an increase in intracellular Ca2+ in any of the cell lines tested, albeit robust Ca2+ responses were elicited in HEK293 cells transiently transfected with AVPR1A (Figure 4(c) and (d)).
Figure 4.
Calcium response to oxytocin and vasopressin addition in DRG neurons and cell lines. Functional oxytocin receptors are expressed in SH-SY5Y cells, but not in F-11 or ND7/23 cells. Functional vasopressin receptors are absent from all three cell lines. (a) Calcium influx responses of SH-SY5Y, F-11, and ND7/23 cell lines upon exposure to oxytocin. (b) Calcium response of HEK293 cells transfected with oxytocin receptor (OXTR) upon exposure to oxytocin. (c) Calcium influx responses of SH-SY5Y, F-11, and ND7/23 cell lines upon exposure to vasopressin. (d) Calcium response of HEK293 cells transfected with the vasopressin V1B receptor (AVPR1B) upon exposure to vasopressin.
The gene expression profiles observed in this study are supported by several studies assessing expression of isolated ion channels and GPCRs in sensory neurons, SH-SY5Y neuroblastoma, as well as F-11 and ND7/23 cells. Specifically, the expression profile of CaV transcripts in SH-SY5Y cells was entirely consistent with that previously reported,11 with CaV2.1, CaV2.3, CaV3.2, and CaV1.1 being completely absent in SH-SY5Y cells. CaV3.1 had the largest number of reads, followed by CaV2.2 and CaV1.3. SH-SY5Y cells also expressed several CaV auxilliary units at high level, a trend that was also observed in F-11 and ND7/23 cell lines. In contrast, the order of expression of CaV alpha units in DRGs was observed as CaV2.2 > CaV2.1 > CaV3.2 > CaV1.2 > CaV3.3 > CaV1.3 > CaV2.3 > CaV3.1 > CaV1.1 > CaV1.4, with all alpha isoforms being present (supplementary data). This was mirrored by the F-11 and the ND7/23 cells, both of which expressed all CaV isoforms with a similar pattern. F-11 and ND7/23 expressed most CaV3.2 out of all CaV alpha subunits, closely followed by CaV2.2, CaV1.2, and CaV3.3.
Similarly, expression of KCNQ2 and KCNQ3, the genes encoding KV7.2 and KV7.3, the K+ channel isoforms which underlie the M-current, were consistent with previous results reporting presence of this channel in DRGs,43 F-11 cells,44 and SH-SY5Y cells;45 although curiously, KCNQ3 was absent in ND7/23 cells. Interestingly, KV1.1, a key KV channel regulating excitability in DRG neurons, was found to be virtually absent in all cell lines, where the KV11 subtype family was expressed most abundantly (55.06% of all KV transcripts in SH-SY5Y, 58.56% in F-11, and 38.30% in ND7/23), and KV1 family expression was at least 30-fold lower in all cell line samples compared to whole DRGs. KV11.1 (KCNH2), commonly known as the hERG, was the most abundant KV subtype channel in all three cell line samples. These findings are consistent with previous studies reporting absence of KV1.1, and endogenous expression of hERG, in these cell lines.46–48
The expression profile of TRP channel transcripts was similar across all cell lines. TRPA1 and TRPV1, the TRP channels receptive to the noxious stimuli mustard oil and capsaicin, respectively, were largely missing from the three cell lines. Very small amounts of TRPV1 were present in the three cell lines and TRPA1 was completely absent. TRPM8, the menthol and cold receptor, was also expressed at very low levels in all cell lines as previously reported. The recently identified cold-sensitive channel TRPC549 was revealed to be present in whole DRGs and in ND7/23 cells, with very low expression in SH-SY5Y and F-11 cells. TRPM3, another novel channel suggested to be a potential nociceptor and thermoceptor,50 was present in whole DRGs, F-11 cells, and ND7/23 cells, with very low expression levels in SH-SY5Y cells (supplementary data).
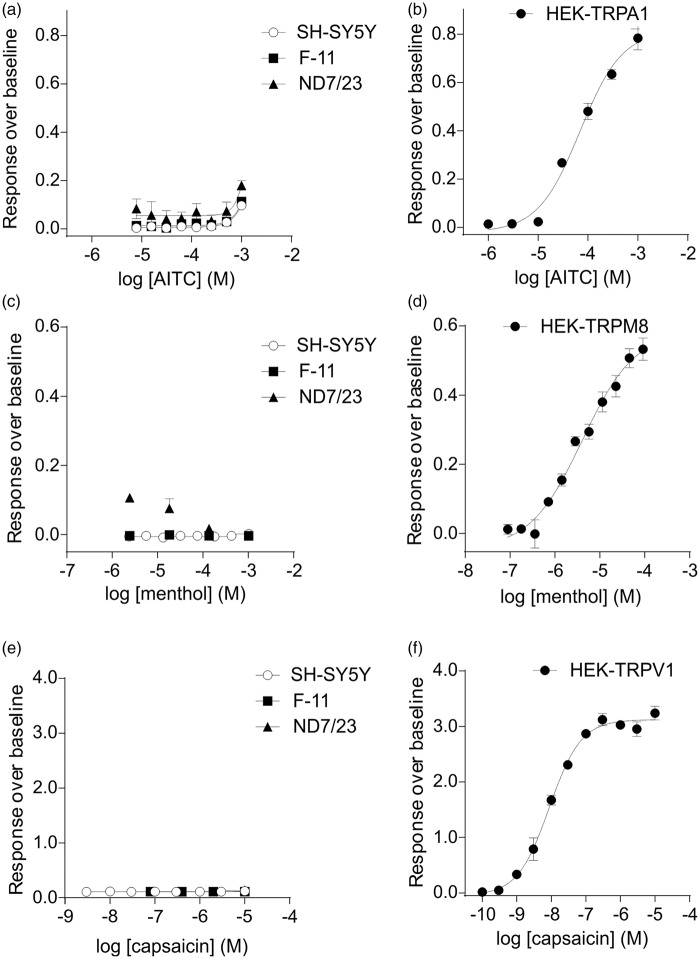
These findings were confirmed with fluorescent intracellular calcium measurements after the application of selective TRP channel agonists (Figure 5). The TRPA1 activator, AITC, did not elicit a response from any of the cell lines at concentrations despite activating transfected HEK293 cells transiently expressing rat TRPA1 (Figure 5(a) and (b)). The application of the TRPM8 agonist menthol also elicited minimal responses from any of the three cell lines, while robust Ca2+ responses were observed in transfected HEK293 cells (Figure 5(c) and (D)). Similarly, capsaicin, known to activate rat, mouse, and human TRPV1,51–53 elicited no responses in the cell lines, but robustly activated TRPV1-transfected HEK293 cells (Figure 5(e) and (F)). The relatively lack of expression of these key thermosensitive TRP channels suggests that none of the three cell line examined should be used as model nociceptive or thermosensitive neurons.
Figure 5.
Calcium responses to TRP activators for cell lines and transfected HEK293 cells. Calcium responses to the application of selective TRP activators for cell lines and HEK293 cells transiently expressing TRPA1, TRPM8, or TRPV1. None of the cell lines responded to AITC, menthol, or capsaicin, confirming the absence of TRPA1, TRPM8, and TRPV1 expression in the three cell lines examined. (a) Calcium responses of the cell lines to AITC. (b) Calcium responses of HEK293 cells transiently expressing rat TRPA1 channels to AITC. (c) Calcium responses of the cell lines to menthol. (d) Calcium responses of HEK293 cells transiently expressing rat TRPM8 channels to menthol. (e) Calcium responses of the cell lines to capsaicin. (f) Calcium responses of HEK293 cells transiently expressing rat TRPV1 channels to capsaicin.
Discussion
Immortalised neuronal cell lines as models of primary sensory neurons represent a comfortable compromise for the investigation of the neuropharmacological mechanisms underlying pain. These neuroblastoma-derived cell lines often express molecular targets consistent with those found in sensory neurons, bypassing the need for excision and primary culture of tissue while – presumably – retaining endogenous protein expression profiles. Indeed, one advantage of using immortalised neuronal cell lines over heterologously transfected cells is the co-expression of relevant auxillary subunits as well as interacting proteins, and the constellation of channels and receptors that would form the “signallosome” in a native neuron, providing a more holistic insight to nociceptive pharmacology and signalling. However, these properties are often based on assumptions about the gene expression profile of such cell lines, and the lack of comprehensive analyses of receptor expression in these cell lines limits their applicability.
We thus set out to assess the gene expression profile of the model sensory neuron cell lines ND7/23 and F-11 as well as the neuroblastoma cell line SH-SY5Y, which has previously been reported to express genes and ion channels normally restricted to peripheral sensory neurons such as NaV1.7.10 Accordingly, when compared with the gene expression profile of whole DRG, we found expression of many individual genes known to be expressed in, or play an important role in the function of, sensory neurons. These include RET, TH, PRPH, and LDHB. However, we also found many sensory neuron-enriched genes that were absent from the cell lines, such as TRPV1 and SCN10A. Moreover, given that whole DRGs and all three cell lines derive from peripheral neurons, the absence of central nervous system microglial markers such as CCL4 and GPR18354 in the neuronal cell lines was expected.
Sensory neurons are a heterogeneous group of neurons with distinct functional characteristics and physiological functions. Since cell lines can be considered monoclonal and originate from a single cell colony, the absence of genes enriched in specific subgroups of neurons could be expected – it is plausible that the gene expression profile of ND7/23 and F-11 cells would resemble only one subset of the many different types of sensory neurons. Indeed, both ND7/23 and F-11 cells are often considered a model for small nociceptive neurons that give rise to unmyelinated C-fibres. However, the precise type of sensory neuron that these cell lines may have arisen from, or that they most closely resemble, is difficult to assess currently, not least because the precise number of individual neuronal subtypes found in DRG are unclear. Based on functional characteristics of the fibres innervating target organs including skin, at least 16 subclasses of purely sensory neurons should exist.34 In recent years, significant efforts have been made towards identifying the gene expression profile of these individual neuronal classes, as it is becoming increasingly clear that it is the constellation of ion channels, GPCRs, auxiliary subunits, and interacting proteins expressed in a given neuron that defines its functional properties. For example, cold sensitivity in trigeminal neurons is defined by co-expression of KV1.1/1.2 channels with TRPM8,55 while in sensory neurons innervating the skin, a similar functional relationship was observed for TRPM8 and KV7.2/7.3 channels.43 There is also existing evidence towards the presence of multiple neuron types within F-11 cell lines,56 indicating that while neuron-derived cell lines are supposedly clones of the parent cell line, a myriad of neuronal subtypes exists within one population.
Based on a comprehensive study assessing the gene expression profiles of sensory neurons using single-cell RNA sequencing, Usoskin et al.22 defined four basic sensory neuron subtypes, characterised by expression of TAC1, NTRK1, and CALCA (peptidergic cluster); MRGPRD and P2RX3 (non-peptidergic cluster); TH (tyrosine hydroxylase containing cluster); NEFH and PVALB (neurofilament-containing cluster); as well as 11 subgroups thereof by correlating both old and novel markers to physiological function in peripheral nerves. Li et al.33 also examined the types of neurons present in DRGs in a separate study, and classified mouse DRG neurons into 10 types and 14 subordinate subtypes based on molecular markers, functional annotations, and transcriptional patterns. The resulting clusters matched the classification by Usoskin et al. to some extent, but also introduced additional transcriptional markers defining subgroups of sensory neurons. Importantly, the gene expression profile of F-11, ND7/23, and SH-SY5Y cells resembled none of these subgroup classifications closely, with all cell lines expressing markers from several different clusters as well as markers of non-neuronal cells including B2M and VIM.22 Indeed, F-11 cells expressed high levels of P2RX3, TH, and NEFH, thus simultaneously incorporating markers of non-peptidergic, TH-containing, and neurofilament-expressing neurons based on the classification proposed by Usoskin et al.; while ND7/23 cells expressed high levels of the peptidergic neuron marker NTRK1, the non-peptidergic marker P2RX3, TH, and the neurofilament-expressing marker NEFH. The corresponding gene modules based on the classifications by Li et al. also belonged to separate clusters, including black (TH), turquoise (P2RX3, NTRK1), and blue (NEFH).33
Similar discrepancies were also observed when comparing the gene expression profile of the cell lines to the transcriptome of neurons expressing NaV1.8, which are typically considered nociceptive and expressed high levels of ANO3, TRPA1, and CHRNA6,57 none of which were present in the immortalised neuronal cell lines examined. However, these discrepancies are perhaps less surprising given none of the cell lines expressed more than one read of NaV1.8 transcripts in the entire sample. Indeed, our results showed that allocation of neuronal cell lines to any single meaningful subtype of sensory neuron is hardly possible, as they expressed a mixture of cellular markers that are typically found in non-overlapping cell populations, including markers for myelinated neurons as well as for unmyelinated neurons. Some examples include the presence of TrkA, TrkB, and TrkC in all cell line samples, suggesting a mixed heritage of the cell lines. Thermosensitive proteins responsible for temperature sensing such as TRPV1, TRPA1, and TRPM8 were all virtually absent in the cell lines, making them unlikely to have been derived from thermosensors. The complete lack of PIEZO2 also indicate that the cell lines are not to be used to investigate stretch-sensitive neurons in their native state.58 Another subclasses we can rule out are subsets of pruriceptors due to the lack of PAR2 and somatostatin in the cell lines.
Global GO analysis of all samples according to BP terms revealed similar functional distribution of genes, suggesting that these neuronal cell lines maintain broad functional similarities with DRG neurons despite the apparent poor fit into the existing framework of neuron classification. It is possible that despite the lack of expression of certain gene markers, other genes with similar roles are expressed through compensatory mechanisms to maintain functional similarity with in vivo peripheral neurons.
While the present study assessed gene expression profiles in undifferentiated ND7/23, F-11, and SH-SY5Y cells, all three cell lines have the capacity and plasticity to develop into mature, terminally differentiated neurons upon exposure to various reagents such as retinoic acid, cAMP derivatives, or forskolin.45,59 These agents can be used to drive the cells towards various phenotypes away from their native form. This is certainly observed in SH-SY5Y cells, which can express different receptors based on the method used to induce differentiation (for a brief review see Kovalevich and Langford60). It is thus possible that differential cells may resemble certain neuronal subtypes more closely, and a transcriptomic analysis on differentiated cells could provide additional insight into processes involved in neuronal maturation.
A drawback of transcriptomic studies lies in difficulties reconciling gene expression with expression of functional proteins. However, we have confirmed the presence of N-methyl-D-aspartate receptors, HCN2 channels, NaV1.7 (SCN9A), CaV2.2, and opioid receptors in all cell lines, consistent with previous findings,9,10 and we additionally confirmed the observed pattern of expression for selected targets using a pharmacological approach where sufficiently selective agonists were readily available.
Conclusions
To conclude, we present the first comprehensive transcriptomic analysis of the gene expression profile in F-11 and ND7/23 cell lines in comparison with whole mouse DRGs and the human neuroblastoma-derived cell line SH-SY5Y. We found that all cell lines expressed a multitude of receptors and ion channels with relevance to nociceptive signalling. However, although the cell lines investigated express molecular targets found in native DRGs, they are not representative of specific subclasses of peripheral sensory neurons and do not fit into existing peripheral neuron categories. Thus, these findings support the continued use of immortal cell lines with neuronal origins in pain research by defining their gene expression profiles relative to native DRG neurons.
Acknowledgement
The authors would also like to thank the IMB Sequencing Facility for their assistance in data analysis.
Declaration of conflicting interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This research was supported by an ARC Future Fellowship to IV, a UQ postgraduate research scholarship to KY, and a Ramaciotti Foundation Establishment Grant (IV).
Supplementary Material
The online data supplements are available at http://mpx.sagepub.com/supplemental.
References
- 1.Fang X, McMullan S, Lawson SN, et al. Electrophysiological differences between nociceptive and non-nociceptive dorsal root ganglion neurones in the rat in vivo. J Physiol (Camb) 2005; 565: 927–943. doi:10.1113/jphysiol.2005.086199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lindsay RM. Nerve growth factors (NGF, BDNF) enhance axonal regeneration but are not required for survival of adult sensory neurons. J Neurosci 1988; 8: 2394–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zheng J-H, Walters ET, Song X-J. Dissociation of dorsal root ganglion neurons induces hyperexcitability that is maintained by increased responsiveness to cAMP and cGMP. J Neurophysiol (Bethesda) 2007; 97: 15–25. doi:10.1152/jn.00559.2006. [DOI] [PubMed] [Google Scholar]
- 4.Huang Z-J, Li H-C, Cowan AA, et al. Chronic compression or acute dissociation of dorsal root ganglion induces cAMP-dependent neuronal hyperexcitability through activation of PAR2. Pain 2012; 153: 1426–1437. doi:10.1016/j.pain.2012.03.025. [DOI] [PubMed] [Google Scholar]
- 5.Genc B, Ulupınar E, Erzurumlu RS. Differential Trk expression in explant and dissociated trigeminal ganglion cell cultures. J Neurobiol 2005; 64: 145–156. doi:10.1002/neu.20134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsantoulas C, Zhu L, Shaifta Y, et al. Sensory neuron downregulation of the Kv9.1 potassium channel subunit mediates neuropathic pain following nerve injury. J Neurosci 2012; 32: 17502–17513. doi:10.1523/JNEUROSCI.3561-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wood JN, Bevan SJ, Coote PR, et al. Novel cell lines display properties of nociceptive sensory neurons. Proc Biol Sci 1990; 241: 187–194. [DOI] [PubMed] [Google Scholar]
- 8.Platika D, Boulos MH, Baizer L, et al. Neuronal traits of clonal cell lines derived by fusion of dorsal root ganglia neurons with neuroblastoma cells. Proc Natl Acad Sci USA 1985; 82: 3499–3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vetter I, Lewis RJ. Characterization of endogenous calcium responses in neuronal cell lines. Biochem Pharmacol 2010; 79: 908–920. doi:10.1016/j.bcp.2009.10.020. [DOI] [PubMed] [Google Scholar]
- 10.Vetter I, Mozar CA, Durek T, et al. Characterisation of Nav types endogenously expressed in human SH-SY5Y neuroblastoma cells. Biochem Pharmacol 2012; 83: 1562–1571. doi:10.1016/j.bcp.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 11.Sousa SR, Vetter I, Ragnarsson L, et al. Expression and pharmacology of endogenous Cav channels in SH-SY5Y human neuroblastoma cells. PLoS One 2013; 8: e5923 doi:10.1371/journal.pone.0059293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Biedler JL, Helson L, Spengler BA. Morphology and growth, tumorigenicity, and cytogenetics of human neuroblastoma cells in continuous culture. Cancer Res 1973; 33: 2643–2652. [PubMed] [Google Scholar]
- 13.Biedler JL, Roffler-Tarlov S, Schachner M, et al. Multiple neurotransmitter synthesis by human neuroblastoma cell lines and clones. Cancer Res 1978; 38: 3751–3757. [PubMed] [Google Scholar]
- 14.Roberts A, Pimentel H, Trapnell C, et al. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics (Oxf) 2011; 27: 2325–2329. doi:10.1093/bioinformatics/btr355. [DOI] [PubMed] [Google Scholar]
- 15.Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics (Oxf) 2015; 31: 166–169. doi:10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mi H, Muruganujan A, Thomas PD. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res 2012; 41: D377–D386. doi:10.1093/nar/gks1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mi H, Muruganujan A, Casagrande JT, et al. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc 2013; 8: 1551–1566. doi:10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chiu F-C, Norton WT, Fields KL. The cytoskeleton of primary astrocytes in culture contains actin, glial fibrillary acidic protein, and the fibroblast-type filament protein, vimentin. J Neurochem 1981; 37: 147–155. [DOI] [PubMed] [Google Scholar]
- 19.Gubern C, Hurtado O, Rodríguez R, et al. Validation of housekeeping genes for quantitative real-time PCR in in-vivo and in-vitro models of cerebral ischaemia. BMC Mol Biol 2009; 10: 57 doi:10.1186/1471-2199-10-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reynders A, Mantilleri A, Malapert P, et al. Transcriptional profiling of cutaneous MRGPRD free nerve endings and C-LTMRs. Cell Reports 2015; 10: 1007–1019. doi:10.1016/j.celrep.2015.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopes C, Liu Z, Xu Y, et al. Tlx3 and Runx1 Act in combination to coordinate the development of a cohort of nociceptors, thermoceptors, and pruriceptors. J Neurosci 2012; 32: 9706–9715. doi:10.1523/JNEUROSCI.1109-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Usoskin D, Furlan A, Islam S, et al. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat Neurosci 2015; 18: 145–153. doi:10.1038/nn.3881. [DOI] [PubMed] [Google Scholar]
- 23.Li L, Rutlin M, Abraira VE, et al. The functional organization of cutaneous low-threshold mechanosensory neurons. Cell 2011; 147: 1615–1627. doi:10.1016/j.cell.2011.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inoue K-i, Ozaki S, Shiga T, et al. Runx3 controls the axonal projection of proprioceptive dorsal root ganglion neurons. Nat Neurosci 2002; 5: 946–954. doi:10.1038/nn925. [DOI] [PubMed] [Google Scholar]
- 25.Caterina MJ, Leffler A, Malmberg AB, et al. Impaired nociception and pain sensation in mice lacking the capsaicin receptor. Science 2000; 288: 306–313. [DOI] [PubMed] [Google Scholar]
- 26.Hoffmann T, Kistner K, Miermeister F, et al. TRPA1 and TRPV1 are differentially involved in heat nociception of mice. Eur J Pain 2013; 17: 1472–1482. doi:10.1002/j.1532-2149.2013.00331.x. [DOI] [PubMed] [Google Scholar]
- 27.Ju G, Hokfelt T, Brodin E, et al. Primary sensory neurons of the rat showing calcitonin gene-related peptide immunoreactivity and their relation to substance P-, somatostatin-, galanin-, vasoactive intestinal polypeptide- and cholecystokinin-immunoreactive ganglion cells. Cell Tissue Res 1987; 247: 417–431. [DOI] [PubMed] [Google Scholar]
- 28.Woo S-H, Ranade S, Weyer AD, et al. Piezo2 is required for Merkel-cell mechanotransduction. Nature (Lond) 2014; 509: 622–628. doi:10.1038/nature13251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gras C, Herzog E, Bellenchi GC, et al. A third vesicular glutamate transporter expressed by cholinergic and serotoninergic neurons. J Neurosci 2002; 22: 5442–5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Akiyama T, Carstens MI, Carstens E. Excitation of mouse superficial dorsal horn neurons by histamine and/or PAR-2 agonist: potential role in itch. J Neurophysiol (Bethesda) 2009; 102: 2176–2183. doi:10.1152/jn.00463.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamada S, Nomura T, Uebersax L, et al. Retinoic acid induces functional c-Ret tyrosine kinase in human neuroblastoma. Neuroreport 2006; 18: 359–363. [DOI] [PubMed] [Google Scholar]
- 32.Park S, Lee J. Proteome profile changes in SH-SY5y neuronal cells after treatment with neurotrophic factors. J Cell Biochem Suppl 2011; 112: 3845–3855. doi:10.1002/jcb.23316. [DOI] [PubMed] [Google Scholar]
- 33.Li C-L, Li K-C, Wu D, et al. Somatosensory neuron types identified by high-coverage single-cell RNA-sequencing and functional heterogeneity. Cell Res 2016; 26: 83–102. doi:10.1038/cr.2015.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zimmermann K, Hein A, Hager U, et al. Phenotyping sensory nerve endings in vitro in the mouse. Nat Protoc 2009; 4: 174–196. doi:10.1038/nprot.2008.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ocana M, Cendan CM, Cobos EJ, et al. Potassium channels and pain: present realities and future opportunities. Eur J Pharmacol 2004; 500: 203–219. doi:10.1016/j.ejphar.2004.07.026. [DOI] [PubMed] [Google Scholar]
- 36.Wallace MS. Ziconotide: a new nonopioid intrathecal analgesic for the treatment of chronic pain. Expert Rev Neurother 2006; 6: 1423–1428. doi:10.1586/14737175.6.10.1423. [DOI] [PubMed] [Google Scholar]
- 37.Blaes N, Girolami J-P. Targeting the ‘Janus face’ of the B2-bradykinin receptor. Expert Opin Ther Targets 2013; 17: 1145–1166. doi:10.1517/14728222.2013.827664. [DOI] [PubMed] [Google Scholar]
- 38.Grastilleur S, Mouledous L, Bedel J, et al. Role of kinin B2 receptors in opioid-induced hyperalgesia in inflammatory pain in mice. Biol Chem Hoppe Seyler 2013; 394: 361–368. doi:10.1515/hsz-2012-0305. [DOI] [PubMed] [Google Scholar]
- 39.Paterson KJ, Zambreanu L, Bennett DLH, et al. Characterisation and mechanisms of bradykinin-evoked pain in man using iontophoresis. Pain 2013; 154: 782–792. doi:10.1016/j.pain.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bittner MA. α-Latrotoxin and its receptors CIRL (latrophilin) and neurexin 1α mediate effects on secretion through multiple mechanisms. Biochimie (Paris) 2000; 82: 447–452. [DOI] [PubMed] [Google Scholar]
- 41.Mogil JS, Sorge RE, LaCroix-Fralish ML, et al. Pain sensitivity and vasopressin analgesia are mediated by a gene-sex-environment interaction. Nat Neurosci 2011; 14: 1569–1573. doi:10.1038/nn.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.ADd Araujo, Mobli M, Castro J, et al. Selenoether oxytocin analogues have analgesic properties in a mouse model of chronic abdominal pain. Nat Commun 2014; 5: 3165 doi:510.1038/ncomms4165. [DOI] [PubMed] [Google Scholar]
- 43.Vetter I, Hein A, Sattler S, et al. Amplified cold transduction in native nociceptors by M-channel inhibition. J Neurosci 2013; 33: 16627–16641. doi:10.1523/JNEUROSCI.1473-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jow F, He L, Kramer A, et al. Validation of DRG-Like F11 cells for evaluation of KCNQ/M-channel modulators. Assay Drug Dev Technol 2006; 4: 49–56. [DOI] [PubMed] [Google Scholar]
- 45.Wickenden AD, Krajewski JL, London B, et al. N-(6-Chloro-pyridin-3-yl)-3,4-difluoro-benzamide (ICA-27243): a novel, selective KCNQ2/Q3 potassium channel activator. Mol Pharmacol 2008; 73: 977–986. doi:10.1124/mol.107.043216. [DOI] [PubMed] [Google Scholar]
- 46.Zhao J, Wei X-L, Jia Y-S, et al. Silencing of herg gene by shRNA inhibits SH-SY5Y cell growth in vitro and in vivo. Eur J Pharmacol 2008; 579: 50–57. doi:10.1016/j.ejphar.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 47.Faravelli L, Arcangeli A, Olivotto M, et al. A HERG-like K+ channel in rat F-Il DRG cell line: pharmacological identification and biophysical characterization. J Physiol (Camb) 1996; 496: 13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Friederich P, Benzenberg D, Trellakis S, et al. Interaction of volatile anesthetics with human Kv channels in relation to clinical concentrations. Anesthesiology (Hagerst) 2001; 95: 954–958. [DOI] [PubMed] [Google Scholar]
- 49.Zimmermann K, Lennerz JK, Hein A, et al. Transient receptor potential cation channel, subfamily C, member 5 (TRPC5) is a cold-transducer in the peripheral nervous system. Proc Natl Acad Sci USA 2011; 108: 18114–18119. doi:10.1073/pnas.1115387108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vriens J, Owsianik G, Hofmann T, et al. TRPM3 is a nociceptor channel involved in the detection of noxious heat. Neuron 2011; 70: 482–494. doi:10.1016/j.neuron.2011.02.051. [DOI] [PubMed] [Google Scholar]
- 51.Jerman JC, Brough SJ, Prinjha R, et al. Characterization using FLIPR of rat vanilloid receptor (rVR1) pharmacology. Br J Pharmacol 2000; 130: 916–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Correll CC, Phelps PT, Anthes JC, et al. Cloning and pharmacological characterization of mouse TRPV1. Neurosci Lett 2004; 370: 55–60. doi:10.1016/j.neulet.2004.07.058. [DOI] [PubMed] [Google Scholar]
- 53.Witte DG, Cassar SC, Masters JN, et al. Use of a fluorescent imaging plate reader–based calcium assay to assess pharmacological differences between the human and rat vanilloid receptor. J Biomol Screen 2002; 7: 466–475. doi:10.1177/108705702237679. [DOI] [PubMed] [Google Scholar]
- 54.Darmanis S, Sloan SA, Zhang Y, et al. A survey of human brain transcriptome diversity at the single cell level. Proc Natl Acad Sci USA 2015; 112: 7285–7290. doi:10.1073/pnas.1507125112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Madrid R, Edl Pena, Donovan-Rodriguez T, et al. Variable threshold of trigeminal cold-thermosensitive neurons is determined by a balance between TRPM8 and Kv1 potassium channels. J Neurosci 2009; 29: 3120–3131. doi:10.1523/JNEUROSCI.4778-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boland LM, Dingledine R. Expression of sensory neuron antigens by a dorsal root ganglion cell line, F-11. Dev Brain Res 1990; 51: 259–266. [DOI] [PubMed] [Google Scholar]
- 57.Thakur M, Crow M, Richards N, et al. Defining the nociceptor transcriptome. Front Mol Neurosci 2014; 7: 87 doi:10.3389/fnmol.2014.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ranade SS, Woo S-H, Dubin AE, et al. Piezo2 is themajor transducer of mechanical forces for touch sensation inmice. Nature (Lond) 2014; 516: 121–125. doi:10.1038/nature13980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chiesa N, Rosati B, Arcangeli A, et al. A novel role for HERG K+ channels: spike-frequency adaptation. J Physiol (Camb) 1997; 501: 313–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kovalevich J, Langford D. Considerations for the use of SH - SY5Y neuroblastoma cells in neurobiology. In: Amini S, White MK. (eds). Neuronal cell culture: methods and protocols. Methods in molecular biology, New York: Humana Press, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]