Abstract
ENU mutagenesis is a powerful method for generating novel lines of mice that are informative with respect to both fundamental biological processes and human disease. Rapid developments in genomic technology have made the task of identifying causal mutations by positional cloning remarkably efficient. One limitation of this approach remains the mutation frequency achievable using standard treatment protocols, which currently generate approximately 1–2 sequence changes per megabase when optimized. In this study we used two strategies to attempt to increase the number of mutations induced by ENU treatment. One approach employed mice carrying a mutation in the DNA repair enzyme Msh6. The second strategy involved injection of ENU to successive generations of mice. To evaluate the number of ENU-induced mutations, single mice or pooled samples were analyzed using whole exome sequencing. The results showed that there is considerable variability in the induced mutation frequency using these approaches, but an overall increase in ENU-induced variants from one generation to another was observed. The analysis of the mice deficient for Msh6 also showed an increase in the ENU-induced variants compared to the wild-type ENU-treated mice. However, in both cases the increase in ENU-induced mutation frequency was modest.
Introduction
Forward genetic screens using N-ethyl-N-nitrosurea (ENU) as a chemical mutagen have revealed a wide spectrum of biological and disease processes [1]. ENU causes DNA damage by transferring a methyl or ethyl group to the oxygen and nitrogen atoms of nucleotide bases (reviewed in [2] and [3]). The resulting base adducts tend to mispair during semi-conservative replication. If this is not corrected, the following round of replication will convert the mismatch to a point mutation. This method creates random mutations throughout the genome, which are potentially more representative of the mutations responsible for human disease than null or conditional mutations generated by genome targeting.
While mutagenesis screens have been successful in creating a wide range of phenotypes for disease modeling, the efficiency of this approach is still constrained by the number of mutations that can be induced in a single organism. The frequency of ENU-induced mutations is affected by treatment regimens and dosage, but generally ranges from 1–2 mutations/Mb of genomic DNA [4–8]. In a strain such as C57BL/6J, each mutated gamete would carry approximately 3000–6000 mutations, of which around 30–60 mutations would be in coding regions. A major limitation is the dose of ENU that can be administered, as a ceiling is reached such that treatment results in sterility or lethality [9, 10]. As a consequence, ENU screens frequently entail treatment of a large number of animals in order to obtain a mutant phenotype of interest.
A number of different strategies have previously been tested in a bid to increase the number of mutations per animal without decreasing fertility. One involves manipulating DNA mismatch repair (MMR), which is part of the repair mechanism that prevents alkylation damage in cells and protects them from naturally occurring mutations [3, 11, 12]. This machinery corrects small replication errors such as insertion/deletion loops (IDLs) and base-base mismatches. There are five MMR genes in mammals that produce three different heterodimers. The mismatch-bound MutS heterodimer recognizes the replication errors and exists in two varieties: MutSα and MutSβ. MutSα consists of the MMR proteins MSH2 and MSH6, and recognizes single base pair mismatches and small IDLs. MutSβ is a heterodimer of MSH2 and MSH3 and mediates recognition of larger IDLs. Subsequent recognition of the mismatch by MutSα or MutSβ will induce the recruitment of multiple molecules of MutLα. MutLα is the predominant form of the MutL family functioning in MMR, consisting of MLH1 and PMS2 [13].The recruitment of MutLα will activate its endonuclease activity [14] together with the exonuclease EXO1, causing DNA excision to initiate at an upstream nick. The gap created by this excision is repaired by the combined action of proliferating cell nuclear antigen (PCNA), replication factor C (RFC) and DNA polymerase III that functions as its processivity factor. The remaining nick is then sealed by DNA ligase [15].
This mechanism not only recognizes and repairs natural occurring mutations, but has also been shown to be involved in mutations induced by ENU. Claij et al. [16] found that mouse embryonic stem cells lacking one of the components of the MMR, Msh2, had a strongly increased mutation frequency when treated with ENU as compared to wild-type. A number of studies have attempted to use the MMR mechanism to increase the number of mutations induced by ENU treatment in vivo. Specifically studies were done using Msh6 mutant zebrafish [17] or rats [18] with different outcomes ranging from no significant increase in zebrafish to a ~2.5 fold increase in ENU-induced variant number in rats. In this study we describe a similar strategy using mice.
An alternative approach to increase ENU-induced mutations would be to use treatment of serial generations, in which ENU injections are administered to the progeny of treated animals. In this report we describe the consequences of both the use of serial injection and the use of an MMR-deficient mouse line. Both methods resulted in increased numbers of mutations detected by exome sequence analysis. However, the maximal increase obtained by either method compared to a single injection was less than 1.8-fold, suggesting these strategies have limited benefit.
Experimental Methods
Animals and mutant mouse generation
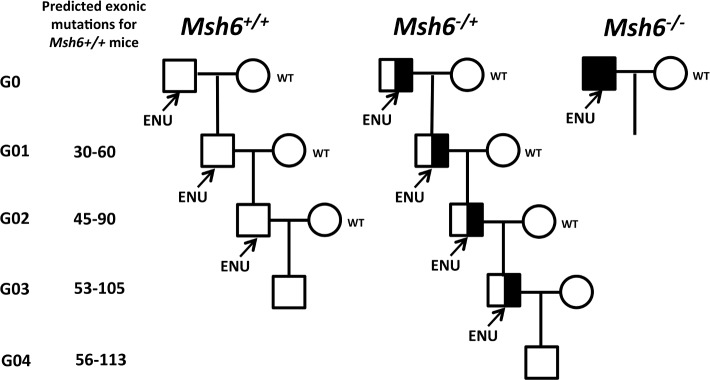
All animals were bred and maintained under specific pathogen-free conditions at McLaughlin Research Institute’s AALAC International accredited Animal Resource Center. All protocols were reviewed and approved by the Institutional Animal Care and Use Committee. The Msh6 mutant line Msh6tm1Rak [19] was kindly provided by Dr. Winfred Edelmann. Wild-type (Msh6+/+), Msh6+/-, and Msh6-/- mice were treated with ENU as previously described (reviewed in [20]). Briefly, adult male animals (Generation 0 (G0)) were treated with three weekly intraperitoneal injections of 90mg of ENU per kilogram of body weight. Treated males were individually housed with ICR females to test sterility and the time to recovery of fertility. After fertility was recovered, the animals were mated with C57BL/6J (B6) females to generate G01 offspring (Fig 1, Table 1). The G01, G02, and, from the Msh6+/- cohort, G03 progeny males were treated with the same ENU regimen. Females from the different generations (G01, G02, G03, and G04) and mutation cohorts were sequenced. Given the various breeding patterns (see S1 Fig for an example), the relatedness of mice in the different cohorts was not identical.
Fig 1. Breeding scheme for analysis of serial ENU-treatment in wild-type and MMR defective mice.
G01 males carry a random set of de novo point mutations induced by ENU treatment of wild-type or MMR-mutant G0 mice. G02, G03 and G04 carry mutations that were induced in their respective parents, as well as those inherited from previous generations. Each generation treated with ENU will on average have 3000–6000 new ENU-induced variants genome wide. After ENU treatment each male is crossed with a wild-type female; their progeny will inherit newly induced mutations and 50% of those of the parent. The mutations are sampled by exome analysis; the expected number of ascertained mutations (E) is shown. The ENU treatment was performed on three successive generations for the Msh6+/+ mice and four generation for the Msh6+/- mice. Msh6-/- mice did not tolerate ENU treatment.
Table 1. Fertility after ENU treatment.
| Genotype x dose1 | |||||
|---|---|---|---|---|---|
| Generation | Msh6+/+ x 90 | Msh6-/+ x 90 | Msh6-/- x 90 | Msh6-/- x 75 | |
| G01 | # males injected | 17 | 31 | 8 | 4 |
| # recovering fertility2 | 8 | 14 | 0 | 0 | |
| total # of progeny | 142 | 198 | 0 | 0 | |
| G02 | # males injected | 18 | 30 | ||
| # recovering fertility | 11 | 22 | |||
| total # of progeny | 101 | 140 | |||
| G03 | # males injected | 21 | 22 | ||
| # recovering fertility | 16 | 17 | |||
| total # of progeny | 118 | 248 | |||
| G04 | # males injected | 15 | |||
| # recovering fertility | 9 | ||||
| total # of progeny | 117 | ||||
1Dose is indicated as mg of ENU injected per Kg bodyweight x 3 injections.
2Fertility is measured by presence at least of one litter within 10 weeks after the last injection.
Mice treated with ENU are at risk for developing tumors and were observed every work day for signs of distress, including wasting, hairloss, inactivity, mass formation, etc. Mice identified as possibly having a tumor were euthanized using carbon dioxide narcosis, followed by cervical dislocation and bilateral thoracotomy, consistent with the recommendations of the American Veterinary Medical Association (AVMA) Guidelines on Euthanasia. No mice were found dead. As mice were euthanized immediately upon showing signs of illness, no other methods were taken to ameliorate suffering.
DNA extraction
We isolated DNA from liver by phenol/chloroform extraction or using a Qiagen DNeasy Blood and Tissue Kit (Qiagen, Santa Clarita, CA USA). Individuals or pools of 6 to 10 samples of extracted DNA for each generation treated with ENU were then sent for library preparation and whole exome sequencing at Centrillion Genomics Technology (CA, USA).
Sequencing data analysis
We mapped paired end sequencing data to the reference genome mm10 using BWA-MEM (Burrows-Wheeler alignment tool)[21] employing default parameters. We used the Genome Analysis Toolkit (GATK)[22] to realign reads around known indels, and recalibrate quality scores to reduce sequencing artifacts. Picard (Broad Institute: http://broadinstitute.github.io/picard/) was used to identify duplicate reads. SNPs were identified using SAMtools (mpileup, bcftools)[23]. SNPs were annotated with ANNOVAR [24] employing refGene as the gene model. To control for differences in the amount of sequence per sample we identified all regions covered by either 20 reads (individual mice) or 100 reads (pooled mice) using generated Python scripts and the Python version of BEDTools [25] (Pybedtools) [26]. We applied additional filtering to identify ENU induced SNPs with the highest confidence. SNPs in publically available databases (Sanger Mouse exomes, dbSNP) or in-house controls, located within a repeat region, or with low likelihood of being real (<Q30) were removed. Sequence data have been deposited in the SRA repository with the accession number SRP074832. Statistical analyses were done using two-tailed T-tests assuming unequal variances.
Results
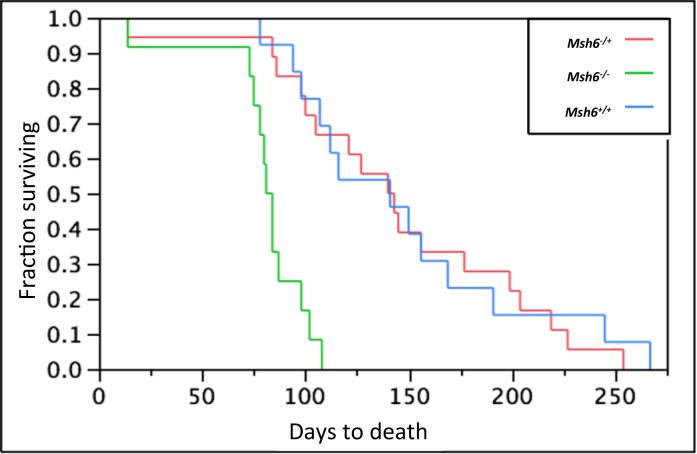
ENU treatment was done on 3 serial generations of B6 wild-type and 4 serial generations of B6 Msh6+/- mice (Fig 1). We initially also included Msh6-/- mice, but they did not tolerate ENU treatment (Fig 2) even at a reduced dose (Table 1), as they developed thymic tumors with enlarged lymph nodes and spleens (data not shown). Wild-type and Msh6+/- mice tolerated the ENU administration and more than half of injected mice recovered fertility to allow the production of offspring (Table 1). Serial ENU administration to the progeny of the ENU-treated animals had no cumulative effect on fecundity as after each round of injection there was no decrease in the number of animals that recovered fertility and produced progeny (Table 1).
Fig 2. Effect of ENU treatment on survival.
Msh6+/- males show survival comparable to wild-type males after treatment with the standard concentration of ENU (90mg/kg x 3). Survival of ENU-treated Msh6-/-males is much reduced. Death is expressed in days.
The efficiency of the ENU treatment for inducing mutations has generally been reported to range from 1–2 mutations/Mb. Half of the autosomal mutations present in G01 mice will be transmitted to the next generation. Therefore, in the second treated generation there should be a 1.5 fold increase in mutation frequency relative to the first treated generation and in the third generation there should be a 1.75 fold increase. We analyzed the number of variants present in each of 3 individual mice from 3 generations of wild-type or Msh6+/- mice using whole exome sequencing. This strategy was chosen as it enabled us to sample the genome with high confidence and reproducibility, allowing reliable comparison of the cohorts. On average there was an increase in the number of variants present in the next generation for both backgrounds of mice, albeit less than expected (Table 2). Notably, individual mice from the same cohort had very different variant numbers (Table 2 and S1 Fig). Given this variability and the small number of samples per cohort, the differences in mean variant number do not reach statistical significance.
Table 2. Variant analysis for individual mice.
| Genotype and Generation | variant for each individual | average | ratio1 | ratio H/W2 | ||
|---|---|---|---|---|---|---|
| wild-type G01 | 86 | 60 | 72 | 73 | 1.0 | |
| wild-type G02 | 103 | 73 | 54 | 77 | 1.1 | |
| wild-type G03 | 110 | 118 | 53 | 94 | 1.3 | |
| Msh6+/- G01 | 70 | 77 | 49 | 65 | 1.0 | 0.9 |
| Msh6+/-G02 | 135 | 106 | 57 | 99 | 1.5 | 1.3 |
| Msh6+/- G03 | 108 | 56 | 124 | 96 | 1.5 | 1.0 |
1Ratio: the increase of variant present compared to the first ENU injection (G01)
2Ratio H/W: the increase of variant present in Msh6+/- compared to the same wild-type generation.
To obtain a more robust representation of the ENU-induced mutation frequency, we analyzed a pool of 10 mice for each cohort (except for the Msh6+/- G04 pool, which was 6 mice). While this has the virtue of including more samples, coverage of each sample was on average 10X (compared to 20X for the individual analysis), so the number of mutations per exome ascertained was smaller. However, this sample depth still enabled comparison among cohorts. The variant number in each pool increased after each generation as previously observed, except for the fourth generation of ENU-treated Msh6+/- mice (Table 3). However, also as previously observed, the increase in mutation frequency was lower than predicted.
Table 3. Variant analysis for pooled mice.
| Genotype and Generation | variants | ratio1 | ratio H/W2 |
|---|---|---|---|
| wild-type G01 | 186 | 1.00 | |
| wild-type G02 | 259 | 1.39 | |
| wild-type G03 | 295 | 1.59 | |
| Msh6+/- G01 | 256 | 1.00 | 1.38 |
| Msh6+/-G02 | 313 | 1.22 | 1.21 |
| Msh6+/- G03 | 325 | 1.27 | 1.10 |
| Msh6+/- G04 | 311 | 1.21 |
1Ratio: the increase of variant present compared to the first ENU injection (G01)
2Ratio H/W: the increase of variant present in Msh6+/- compared to the same wild-type generation.
An aim of this experiment was to test whether treating mice at least partially deficient in Msh6 would increase the mutagenesis frequency. In the individually analyzed mice, only one of the 3 generations showed an increase of mutation frequency in the Msh6+/- group, and this was small (Table 2). Again, the variability within a cohort was problematic. The analysis of pools of mice revealed a marked increase in the variant number for the Msh6+/- mice compared to the wild-type mice (Table 3). The mutation frequency was further increased for the second (G02) and third generations (G03); however, the ratio of mutations compared to the wild-type cohort decreased for each successive generation.
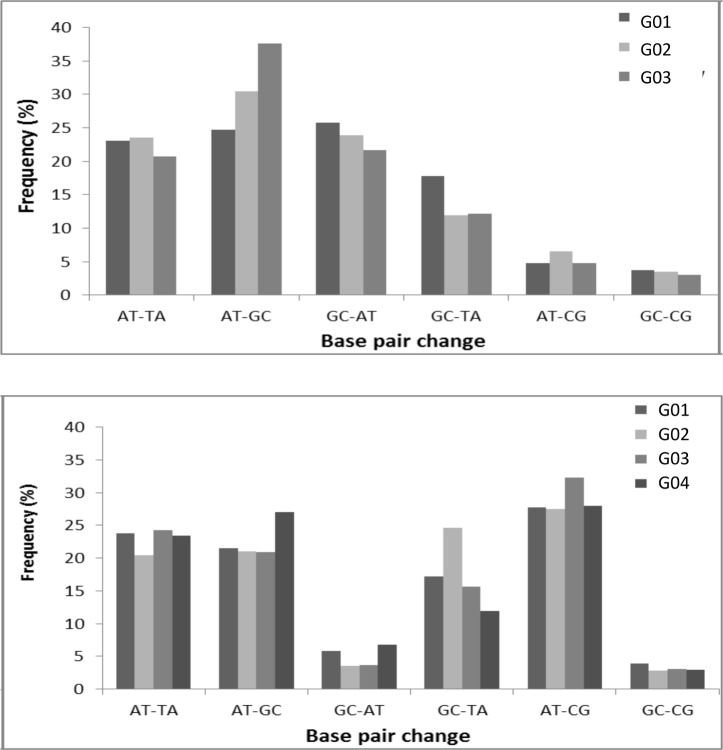
Examination of the type of mutations produced by the ENU treatment revealed a different spectrum in the Msh6+/-mice compared to wild-type. The most frequent base-pair alterations induced by ENU affected A-T base pairs, with A-T to T-A and A-T to G-C substitutions accounting for almost fifty percent of the mutations for both the wild-type and Msh6+/- ENU treated mice (Fig 3), which is consistent with previous results [27]. However, G-C to A-T transitions were much reduced in the Msh6+/-mice compared to wild-type mice, while A-T to C-G transitions were elevated (Fig 3). Overall, wild-type mice have a similar fraction of A-T and G-C modifications, in contrast to the Msh6+/- mice that had an average of 75% A-T modifications compared to 25% G-C modifications (p = 0.015, Table 4).
Fig 3. ENU-induced mutation spectrum.
(a) Frequencies of mutations in successive generation of Msh6+/+ mice in the pooled samples. (b) Frequencies of mutations in the successive generation of Msh6+/- mice in the pooled samples.
Table 4. Distribution of ENU-induced mutations in A-T vs. G-C base-pairs.
| %A-T | %G-C | Avg %A-T | Avg %G-C | |
|---|---|---|---|---|
| wild-type G01 | 53 | 47 | ||
| wild-type G02 | 61 | 39 | 59 | 41 |
| wild-type G03 | 63 | 37 | ||
| Msh6+/- G01 | 73 | 27 | ||
| Msh6+/-G02 | 69 | 31 | ||
| Msh6+/- G03 | 78 | 22 | 75 | 25 |
| Msh6+/- G04 | 78 | 22 |
Discussion
In this study we demonstrate that serial administration of ENU to successive generations of mice increases the ENU-induced mutation frequency in both wild-type and Msh6+/-mice. Our initial analysis of 3 individual mice in each of 3 generations supported this observation; however, we found considerable variation among individuals. We therefore analyzed pooled DNA obtained from up to 10 different mice from each generation, which confirmed this observation.
The increase we observed was less than expected based on Mendelian inheritance, which could be explained in a number of ways. For example, we used stringent parameters for whole exome sequencing analysis in order to increase the likelihood that we identified de novo ENU-induced variants; however, these may have excluded some true variants These parameters could be modified but the filtering of false positives would be less efficient. Another possibility is that we are not accounting for loss due to lethality; however, given that even null mutations of important developmental genes are generally well tolerated in mice when heterozygous, this is unlikely to be an important factor. Of note, this is the case for individual mutant loci; it is possible that an accumulation of heterozygous mutants could affect viability or fertility.
We and others have hypothesized that a deficiency in the repair system that detects or corrects DNA mismatches would result in an increased ENU-induced mutation frequency. This is based on the biochemistry of the mutagen ENU, which can transfer its ethyl group to oxygen or nitrogen radicals present in DNA; this results in lesions that can cause mispairing during replication and eventually give rise to a single base pair substitution[28, 29]. Three other studies addressed this question in ES cells and mice [16], zebrafish [17] and rats [18].
In an Msh2 null ES cell line, the background mutation rate (assayed by analysis of loss of HPRT activity) was elevated at least two orders of magnitude compared to wild-type cells, and the mutation rate after ENU treatment was further increased 2–3 fold [16]. In an Msh2-deficient line that had 10% of wild-type activity, the mutation rate in untreated cells was close to background, and was increased 7–10 fold after ENU treatment. While this would seem promising for a strategy of mutagenesis in an MMR deficient mouse, it is difficult to compare the dose regimen of ENU treatments for in vitro and in vivo studies, and it is possible the effective dose given in these studies would not be tolerated in vivo. Indeed, in our hands Msh6-/- mice did not tolerate even low doses of ENU.
In Msh6-/- rats, treatment with ¾ of a dose used in wild-type was modestly successful, elevating the mutation rate to 0.8 mutations/Mb, a 1.5-fold increase compared to the mutation rate in Msh6-proficient animals [18]. Msh6+/- rats were not tested. In contrast, in zebrafish there were no differences found for the mutation frequency for ENU-treated wild-type, Msh6+/- and Msh6-/- lines [17]. The authors speculate that these results may indicate that the maximum mutation load for zebrafish has been reached with the currently used, highly optimized ENU mutagenesis protocol; it is notable that the mutation rate obtained of 6–7 mutations /Mb is 3–6 fold greater than that generally obtained in ENU-treated mice. Alternatively, the authors suggest that the MMR system in the zebrafish germ line may be saturated very rapidly, thereby having a limited effect on high-dose ENU mutagenesis.
In our analysis we see a small increase in mutation frequency in treated Msh6+/- mice compared to wild-type for both individually analyzed and pooled cohorts. In the latter, which likely represents a more representative sample, after 3 serial treatments 325 variants were detected in 6 Mb obtained from 10 mice, for a mutation rate of 5.4 mutations/Mb per mouse. This is 1.1-fold the rate in an equivalent wild-type cohort, and 1.75-fold the frequency found in a wild-type cohort after a single treatment.
Our results also demonstrate that the common mutation both in wild-type and Msh6+/- background were alterations of A-T base pairs, with Msh6+/- having a higher ratio of A-T transitions. These results are consistent with what was observed in the Msh6-deficient rats and the Msh2-deficient mice [16, 18] where they also observed a decrease of A-T to G-C transitions. This phenomenon was associated with the fact that ethylated bases have been shown to direct misincorporation of bases when DNA polymerases succeed in trans-lesion synthesis [30, 31]. Bypass of O2-ethylthymineT subsequently induced A-T to T-A transversions, whereas O4-ethylthymine causes A-T to G-C transitions [30, 31]. These DNA adducts contribute to the toxicity and mutagenicity of ENU and are speculated to be the target of MMR activity [16]. It has been suggested that MutSα preferentially recognizes the O2-ethylthymineT lesion [18], which is consistent with our observation that transversions are elevated in the Msh6+/- mutants at the expense of transitions.
There are a number of possible explanations for why the strategies we tested failed to substantially elevate the mutation frequency induced by ENU. One possibility for the limited effect of reducing Msh6 gene dosage is that 50% of enzyme was sufficient for normal DNA repair. This is clearly not the case, as the spectrum of mutations in the Msh6+/- cohort is different than in wild-type, indicating that this reduction has biological consequences. It also suggests the possibility that more comprehensive testing in homozygous Msh6-/- mice might identify an ENU dose that is compatible with survival but is still mutagenic. However, it should also be noted that there are other DNA damage repair pathways that perform partially redundant functions with MMR. For example, the nucleotide excision repair pathway has been shown to mediate ENU-induced mutation repair in flies [32]. One could imagine mutagenizing mice carrying multiple DNA repair defect mutations, although the husbandry task could potentially outweigh the benefit.
The results also suggest the possibility that our protocols do not succeed because we reached a ceiling on the level of mutations that mice can tolerate. While intriguing, this seems unlikely for a number of reasons. Firstly, one would require treating multiple additional generations to insure that the mutation frequency has truly peaked. Secondly, the mutations generated in our strategy are heterozygous (as we outcross at every generation), and these are generally well-tolerated in mice, even when the homozygous phenotype is lethal. Lastly, we see no obvious effect on fertility, fecundity, gross morphology, or general viability, which one might expect if there were additive effects of multiple mutations. Determining what the upper limit of heterozygous mutational load might be in a mammalian system would be interesting, but we do not think this study conclusively addresses this.
The wealth of studies using ENU mutagenesis for a wide variety of phenotypic analyses demonstrates unequivocally that this approach is efficient, that these mutations can be readily mapped and that they can ultimately be identified using well-established approaches for positional cloning [33]. However, given the development of powerful new methods for generating targeted mutations [34,35,36], it is necessary to examine the utility of an approach that requires positional cloning for mutation discovery. In this regard, the expedience of a genotype-driven approach must be balanced against that of a phenotype-driven method, in which one may obtain unbiased insight into the genetic basis of a trait. Many studies employing forward genetic analysis have provided unexpected insights and have identified mutations in unannotated genes whose function were otherwise unknown.
Importantly, the mutations induced by ENU often have functional consequences that are more informative than the null mutations that are commonly generated by genomic targeting. ENU induced mutations often create hypomorphic alleles, due either to “leaky” splice-site mutations or due to missense mutations that do not completely abolish gene function. Missense mutations induced by ENU can also result in dominant “gain-of-function” effects, which can provide insight into protein structure-function and roles in specific developmental pathways that are not revealed by characterization of null mutants.
In summary, the two strategies that we have described in this study do increase the ENU mutagenesis efficiency. However, the increase is small, and can arguably be equaled by simply expanding a screen that employs a standard treatment for a single generation. It is possible that the small increase reflects a biological limit on the number of heterozygous mutations that are tolerated in mice, but this would require additional analysis.
Supporting Information
The G0 through G04 generations are shown. Msh6+/- heterozygotes are shown as half-filled symbols. Mice used for DNA sample preparation are shown in red. For the total pool mice were obtained from multiple pedigrees, and relatedness of those used was not identical across generations.
(TIFF)
G01, G02, and G03 samples of W (Msh+/+) and H (Msh+/-) mice are shown.
(TIFF)
Data Availability
Sequence data have been deposited in the SRA repository with the accession number SRP074832.
Funding Statement
This work was supported by NIH grants R01NS41997, R01MH081187 and R01HD36404. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Herron BJ, Lu W, Rao C, Liu S, Peters H, Bronson RT, et al. Efficient generation and mapping of recessive developmental mutations using ENU mutagenesis. Nat Genet. 2002;30(2):185–9. [DOI] [PubMed] [Google Scholar]
- 2.Noveroske JK, Weber JS, Justice MJ. The mutagenic action of N-ethyl-N-nitrosourea in the mouse. Mamm Genome. 2000;11(7):478–83. [DOI] [PubMed] [Google Scholar]
- 3.Wyatt MD, Pittman DL. Methylating agents and DNA repair responses: Methylated bases and sources of strand breaks. Chem Res Toxicol. 2006;19:1580–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arnold CN, Barnes MJ, Berger M, Blasius AL, Brandl K, Croker B, et al. ENU-induced phenovariance in mice: inferences from 587 mutations. BMC Res Notes. 2012;5:577 Epub 2012/10/26. 10.1186/1756-0500-5-577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nguyen N, Judd LM, Kalantzis A, Whittle B, Giraud AS, van Driel IR. Random mutagenesis of the mouse genome: a strategy for discovering gene function and the molecular basis of disease. American journal of physiology Gastrointestinal and liver physiology. 2011;300(1):G1–11. 10.1152/ajpgi.00343.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andrews TD, Whittle B, Field MA, Balakishnan B, Zhang Y, Shao Y, et al. Massively parallel sequencing of the mouse exome to accurately identify rare, induced mutations: an immediate source for thousands of new mouse models. Open Biol. 2012;2(5):120061 Epub 2012/06/23. 10.1098/rsob.120061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Justice MJ, Carpenter DA, Favor J, Neuhauser-Klaus A, Hrabe de Angelis M, Soewarto D, et al. Effects of ENU dosage on mouse strains. Mamm Genome. 2000;11(7):484–8. [DOI] [PubMed] [Google Scholar]
- 8.Beutler B, Du X, Xia Y. Precis on forward genetics in mice. Nat Immunol. 2007;8(7):659–64. [DOI] [PubMed] [Google Scholar]
- 9.Augustin M, Sedlmeier R, Peters T, Huffstadt U, Kochmann E, Simon D, et al. Efficient and fast targeted production of murine models based on ENU mutagenesis. Mammalian genome: official journal of the International Mammalian Genome Society. 2005;16(6):405–13. [DOI] [PubMed] [Google Scholar]
- 10.Sakuraba Y, Sezutsu H, Takahasi KR, Tsuchihashi K, Ichikawa R, Fujimoto N, et al. Molecular characterization of ENU mouse mutagenesis and archives. Biochemical and biophysical research communications. 2005;336(2):609–16. [DOI] [PubMed] [Google Scholar]
- 11.Fishel R. Mismatch Repair. The Journal of biological chemistry. 2015;290(44):26395–403. 10.1074/jbc.R115.660142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spies M, Fishel R. Mismatch repair during homologous and homeologous recombination. Cold Spring Harb Perspect Biol. 2015;7(3):a022657 10.1101/cshperspect.a022657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiricny J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol. 2006;7(5):335–46. [DOI] [PubMed] [Google Scholar]
- 14.Kadyrov FA, Dzantiev L, Constantin N, Modrich P. Endonucleolytic function of MutLalpha in human mismatch repair. Cell. 2006;126(2):297–308. [DOI] [PubMed] [Google Scholar]
- 15.Li GM. New insights and challenges in mismatch repair: getting over the chromatin hurdle. DNA repair. 2014;19:48–54. 10.1016/j.dnarep.2014.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Claij N, van der Wal A, Dekker M, Jansen L, te Riele H. DNA mismatch repair deficiency stimulates N-ethyl-N-nitrosourea-induced mutagenesis and lymphomagenesis. Cancer research. 2003;63(9):2062–6. [PubMed] [Google Scholar]
- 17.Feitsma H, de Bruijn E, van de Belt J, Nijman IJ, Cuppen E. Mismatch repair deficiency does not enhance ENU mutagenesis in the zebrafish germ line. Mutagenesis. 2008;23(4):325–9. 10.1093/mutage/gen019 [DOI] [PubMed] [Google Scholar]
- 18.van Boxtel R, Toonen PW, Verheul M, van Roekel HS, Nijman IJ, Guryev V, et al. Improved generation of rat gene knockouts by target-selected mutagenesis in mismatch repair-deficient animals. BMC genomics. 2008;9:460 10.1186/1471-2164-9-460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edelmann W, Yang K, Umar A, Heyer J, Lau K, Fan K, et al. Mutation in the mismatch repair gene Msh6 causes cancer susceptibility. Cell. 1997;91(4):467–77. [DOI] [PubMed] [Google Scholar]
- 20.Stottmann R, Beier D. ENU Mutagenesis in the Mouse. Current protocols in mouse biology. 2014;4(2):25–35. 10.1002/9780470942390.mo140029 [DOI] [PubMed] [Google Scholar]
- 21.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–60. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome research. 2010;20(9):1297–303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–9. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic acids research. 2010;38(16):e164 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–2. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dale RK, Pedersen BS, Quinlan AR. Pybedtools: a flexible Python library for manipulating genomic datasets and annotations. Bioinformatics. 2011;27(24):3423–4. 10.1093/bioinformatics/btr539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takahasi KR, Sakuraba Y, Gondo Y. Mutational pattern and frequency of induced nucleotide changes in mouse ENU mutagenesis. BMC molecular biology. 2007;8:52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Justice MJ, Noveroske JK, Weber JS, Zheng B, Bradley A. Mouse ENU mutagenesis. Hum Mol Genet. 1999;8(10):1955–63. [DOI] [PubMed] [Google Scholar]
- 29.Shibuya T, Murota T, Horiya N, Matsuda H, Hara T. The induction of recessive mutations in mouse primordial germ cells with N-ethyl-N-nitrosourea. Mutat Res. 1993;290(2):273–80. [DOI] [PubMed] [Google Scholar]
- 30.Klein JC, Bleeker MJ, Lutgerink JT, van Dijk WJ, Brugghe HF, van den Elst H, et al. Use of shuttle vectors to study the molecular processing of defined carcinogen-induced DNA damage: mutagenicity of single O4-ethylthymine adducts in HeLa cells. Nucleic acids research. 1990;18(14):4131–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhanot OS, Grevatt PC, Donahue JM, Gabrielides CN, Solomon JJ. In vitro DNA replication implicates O2-ethyldeoxythymidine in transversion mutagenesis by ethylating agents. Nucleic acids research. 1992;20(3):587–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tosal L, Comendador MA, Sierra LM. In vivo repair of ENU-induced oxygen alkylation damage by the nucleotide excision repair mechanism in Drosophila melanogaster. Mol Genet Genomics. 2001; 265(2):327–35. [DOI] [PubMed] [Google Scholar]
- 33.Stottmann RW, Beier DR. 2010. Using ENU mutagenesis for phenotype-driven analysis of the mouse. Methods Enzymol 477: 329–348. 10.1016/S0076-6879(10)77017-8 [DOI] [PubMed] [Google Scholar]
- 34.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA et al. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. 2013. RNA-guided human genome engineering via Cas9. Science 339: 823–826. 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R. 2013. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154: 1370–1379. 10.1016/j.cell.2013.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The G0 through G04 generations are shown. Msh6+/- heterozygotes are shown as half-filled symbols. Mice used for DNA sample preparation are shown in red. For the total pool mice were obtained from multiple pedigrees, and relatedness of those used was not identical across generations.
(TIFF)
G01, G02, and G03 samples of W (Msh+/+) and H (Msh+/-) mice are shown.
(TIFF)
Data Availability Statement
Sequence data have been deposited in the SRA repository with the accession number SRP074832.