Abstract
Proper organ patterning depends on a tight coordination between cell proliferation and differentiation. The patterning of Drosophila retina occurs both very fast and with high precision. This process is driven by the dynamic changes in signaling activity of the conserved Hedgehog (Hh) pathway, which coordinates cell fate determination, cell cycle and tissue morphogenesis. Here we show that during Drosophila retinogenesis, the retinal determination gene dachshund (dac) is not only a target of the Hh signaling pathway, but is also a modulator of its activity. Using developmental genetics techniques, we demonstrate that dac enhances Hh signaling by promoting the accumulation of the Gli transcription factor Cubitus interruptus (Ci) parallel to or downstream of fused. In the absence of dac, all Hh-mediated events associated to the morphogenetic furrow are delayed. One of the consequences is that, posterior to the furrow, dac- cells cannot activate a Roadkill-Cullin3 negative feedback loop that attenuates Hh signaling and which is necessary for retinal cells to continue normal differentiation. Therefore, dac is part of an essential positive feedback loop in the Hh pathway, guaranteeing the speed and the accuracy of Drosophila retinogenesis.
Author Summary
Molecules of the Hedgehog (Hh) family are involved in the control of many developmental processes in both vertebrates and invertebrates. One of these processes is the formation of the retina in the fruitfly Drosophila. Here, Hh orchestrates a differentiation wave that allows the fast and precise differentiation of the fly retina, by controlling cell cycle, fate and morphogenesis. In this work we identify the gene dachshund (dac) as necessary to potentiate Hh signaling. In its absence, all Hh-dependent processes are delayed and retinal differentiation is severely impaired. Using genetic analysis, we find that dac, a nuclear factor that can bind DNA, is required for the stabilization of the nuclear transducer of the Hh signal, the Gli transcription factor Ci. dac expression is activated by Hh signaling and therefore is a key element in a positive feedback loop within the Hh signaling pathway that ensures a fast and robust differentiation of the retina. The vertebrate dac homologues, the DACH1 and 2 genes, are also important developmental regulators and cancer genes and a potential link between DACH genes and the Hh pathway in vertebrates awaits investigation.
Introduction
Temporal and spatial coordination between cell proliferation and differentiation is essential for proper organ patterning. A way to ensure this coordination is through the use of regulatory signaling pathways that control both processes. Among those is the Hedgehog (Hh) signaling pathway that regulates organ growth and patterning in embryos and tissue homeostasis in adults, both in vertebrates and invertebrates [1–4]. Not surprisingly, mutations in components of the Hh signaling pathway cause a number of human disorders, including congenital abnormalities and cancer [2–4].
One of the processes in which Hh signaling plays an essential role is the patterning of the retina in vertebrates and invertebrates [5–7]. In Drosophila, Hh is responsible for organizing a moving signaling wave that patterns the primordium of the fly eye during the last larval stage (L3). The processes under Hh signaling control have been extensively studied and summarized in what follows. The front of the differentiation wave is marked by a straight indentation of the eye epithelium, called morphogenetic furrow (MF), that runs across the dorsoventral axis of the eye primordium, or “eye imaginal disc” [8–10]. hh, initially expressed along the posterior margin of the eye disc [11] and later by the differentiating photoreceptors (PRs), activates the expression of the BMP2 decapentaplegic (dpp) within the MF [12]. Hh instructs undifferentiated proliferating progenitor cells to synchronously undergo mitosis (First Mitotic Wave, FMW) and then stop temporarily their cell cycle in G1 phase through Dpp, which acts long range [13–16]. At a shorter range, Hh initiates the expression of the proneural gene atonal (ato) [17–23] and stabilizes the G1 state by activating the expression of the p21/p27 Cdk inhibitor homologue dacapo (dap) [24–27]. In addition, together with Dpp, Hh induces coordinated cell shape changes responsible for MF formation [18,19,23,28–31] by promoting the apical constriction, apical-basal contraction and basal nuclei migration of cells (Fig 1A). These cellular changes are mediated, at least in part, through the contraction of the acto-myosin cytoskeleton [32,33]. Immediately behind the MF, Ato expression is restricted to evenly spaced cells, which become the ommatidial founder photoreceptors (PR8s). Then, PR8s induce neuronal differentiation of the adjacent precursor cells. Precursor cells that did not start their differentiation program immediately after the MF suffer one last round of mitosis, the Second Mitotic Wave (SMW) [10]. Therefore, Hh secreted by differentiating PR cells drives the anterior propagation of the MF and its associated differentiation wave, while regulating the SMW locally [25,34–36]. Thus, the MF coincides spatially with the onset of differentiation. Interestingly, the MF state is transient: while anterior precursor cells are recruited to enter the MF state, the newly differentiating PRs and cells at the SMW exit this “furrowed” state.
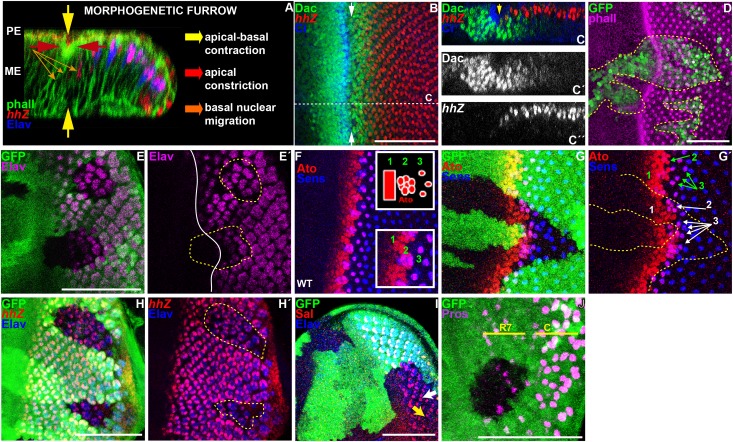
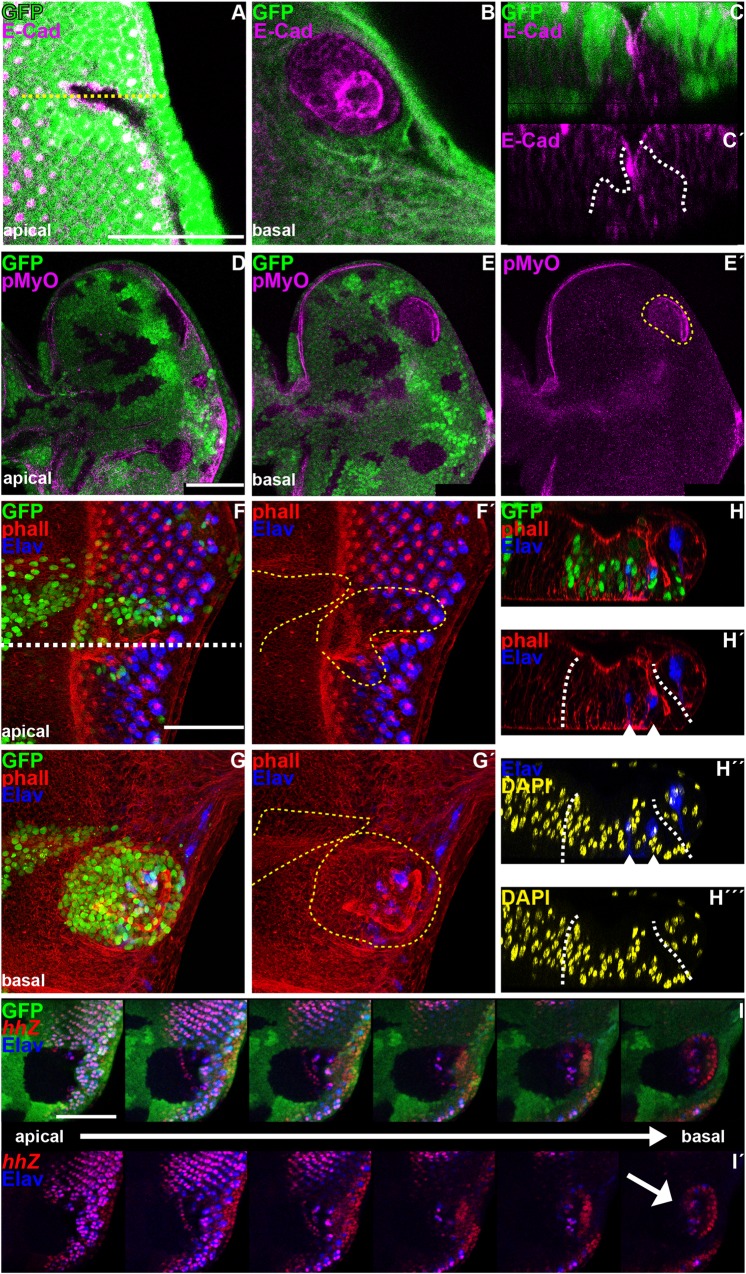
Fig 1. Loss of dac function affects the dynamics of the MF and proper retinogenesis.
All panels show L3 eye imaginal discs. (A) Cross-section through a wild type disc with anterior to the left and apical side up, revealing that the formation of the MF depends on the coordination of three different cell processes: apical constriction (red arrow), apical-basal contraction (yellow arrow), seen by phalloidin staining, which outlines cell shape (green) and nuclear basal migration (orange arrow), labeled with anti-Elav (blue), which marks PR nuclei and anti-β-Galactosidase to detect hhZ (red). (B,D-J) Standard confocal sections with anterior to the left and dorsal up on this and all subsequent eye disc panels. (C-C´´) Cross section through the wild type disc in B (dashed white line). (B,C-C´´,F) wild type discs stained with (B,C-C´´) anti-Dac (green in B,C and white in C´), anti-β-Galactosidase to detect hhZ (red in B,C and white in C´´) and anti-Ci (blue in B,C) or (F) anti-Ato (red) and anti-Sens (blue). Note that Dac is expressed at high levels in a stripe that comprises the MF (arrows in B,C). (F) Ato shows three distinct expression pattern: ahead of the MF, all cells express Ato (1), posterior to the MF, Ato is restricted to a group of cells that start to co-express Sens (2). More posterior, its expression is only maintained in R8 cell until the 2nd row of cells (3), while Sens expression persists onwards [21,62,88,93,94]. (D-E´ and G-J) dac3 clones marked by the (D) presence or (E-E´ and ´G-J) absence of GFP (green) and outlined by yellow dashed lines in D,E´,G´ and H´. Discs are stained with (D) phalloidin (magenta) or (E-E´) anti-Elav (magenta) or (G-G´) anti-Ato (red) and anti-Sens (blue) or (H-H´) anti-Elav (blue) and anti-β-Galactosidase (red) to reveal hhZ or (I) anti-Elav (blue) and anti-Sal (red) or (J) anti-Pros (magenta), R7 indicates PR7 and C indicates cone cells. Plain white line in E’ marks the MF. Note that in dac mutant clones, the refinement of Ato expression is atypical: its expression is not restricted to the groups of cells that start to co-express Sens (2) and is not singled out properly in R8 cells (3) (1G´, compare green with white annotations). dac mutant ommatidia show irregular number of cells: with only one (white arrow in I) or 2 Sal-expressing PR (yellow arrow in I) and fewer PRs (Elav positive cells, E-E´, H-H´). Scale bars represent 50μm.
The coordinated action of Hh has been shown to rely on dynamic changes of its signaling activity. In flies, Hh signaling regulates the post-translational proteolytic processing of the Gli-family transcription factor Cubitus interruptus (Ci). Hh binding to the receptor Patched (Ptc) relieves the inhibition exerted by unbound Ptc on the transducer Smoothened (Smo), and thus promotes the activation of the Fused (Fu) kinase [1–3]. In turn, activated Fu promotes the conversion of the full-length form of Ci (CiFL) to Ci activator (CiA) form [37]. As a result, CiFL is no longer phosphorylated by Protein Kinase A (PKA) [38–40] and other kinases. Otherwise, CiFL phosphorylation leads to the generation of the Ci repressor form (CiR) through partial CiFL degradation by the F-box-containing protein E3 ubiquitin ligase complex (SCFSlim-Cullin1). The relative amount of both CiR and CiA determines the transcriptional status of Hh-target genes, such as ptc, dpp and engrailed (en) [41–44].
In the developing Drosophila eye, while low levels of Hh signaling promotes cell shape changes associated with MF formation and concomitant dpp expression, high levels are required for the re-entry of the precursor cells in the cell cycle at the SMW and for the activation of roadkill (rdx) expression [45–48]. Rdx targets CiFL to full degradation through the recruitment of the Cullin3 (Cul3)-based E3 ligase complex [45,46]. Thus, posterior to the MF, high levels of Hh signaling attenuate its own activity by Rdx:Cul3 complex, allowing retinogenesis to occur properly [45,46]. Therefore, mutations that affect MF progression could be additional components of the machinery that regulates Hh signaling intensity and dynamics. Mutations in the retinal determination gene dachshund (dac) affect MF movement, without blocking differentiation [49]. dac expression depends on Hh signaling itself [25,50]. It localizes to all nuclei straddling the CiFL-expressing domain, from the progenitor domain to the SMW, where differentiating PR cells start expressing hh (Fig 1B and 1C–1C´´). Therefore, high Dac levels coincide with the major neuronal differentiation and morphogenetic processes controlled by Hh signaling. Altogether, these results indicate that dac exhibits the traits required for being a candidate modulator of Hh signaling intensity. Here, we show that indeed dac potentiates Hh signaling in the MF by promoting CiFL accumulation and CiA activity downstream or in parallel of Fu. Our observations argue that this mechanism is absolutely required to promote proper retinogenesis by controlling the timing of MF formation, accurate specification of the founder PR cell and to trigger the Rdx-dependent negative feedback, which turns Hh signaling off posterior to the MF. Thus, Hh signaling potentiation by Dac allows the fast building up of signaling that is required for the swift processes associated with the moving retinal differentiation wave in Drosophila.
Results
dac controls the timing of MF formation and proper retinogenesis
To investigate a role of dac as a candidate modulator of Hh signaling, we reexamined the consequence of removing dac on MF-associated processes. Consistent with previous observations [49], all GFP-marked dac- clones larger than 6 cells straddling this region showed a delay in MF formation (Fig 1D, S1A and S1A´ Fig; n = 53). Accordingly, the onset of PR differentiation, detected by labeling with an antibody against the neuronal marker Elav, was also retarded in these clones (Fig 1E and 1E´, S1D and S1D´ Fig). This retardation was associated with a delay in the onset of ato expression and with an aberrant spacing of Ato-positive PR8 cells (Fig 1, compare 1G and 1G´ with 1F). These defects were not specific of any ommatidial cell type: in dac- clones posterior to the MF, we detected expression of the hh-Z enhancer trap, which marks PR2-5 cells (Fig 1H and 1H´), of the PR3/4 marker Spalt (Sal) (Fig 1I) and of the PR7 marker Prospero (Pros) (Fig 1J). However, the density of ommatidia (Fig 1E and 1E´, 1H and 1H´, S1D and S1D´ Fig) and the proper number of cell types per ommatidium were affected by the loss of dac function. For instance, some ommatidia only contained one Sal-expressing cell instead of two (yellow arrow in Fig 1I).
Concomitant with the delayed MF and differentiation onset, the SMW was also retarded and became asynchronous. Posterior to the SMW, dac- clones showed persistent reentry into the cell cycle, as detected by elevated expression of the G2/M CyclinB (CycB) (S2A and S2A´ Fig), increased number of cycling cells (S2B and S2B´, S2C and S2C´ Fig), as well as maintenance of the expression of the G1/S cyclin CyclinE (CycE) and loss of dap (S2D and S2D´ and S2E Fig). Taken together, we conclude that dac is required for three essential roles played by Hh signaling: MF movement, regulation of the cell cycle and proper retinogenesis.
Dac potentiates Hh signaling
Downregulating the function of the Hh-signal transducer smo (smo3 clones) also caused a delay in MF movement (seen by E-Cadherin (E-Cad) higher signal intensity) and affected apical constriction of cells within the MF (S1B and S1B´ Fig). In addition, the density of ommatidia was reduced in smo- clones (S1E and S1E´ Fig). Thus, smo- and dac-mutant clones shared similar phenotypes (Fig 1D, 1E and 1E´ and S2A and S2B´, S2D and S2E´ Fig). In agreement with dac and smo being part of the same signaling pathway, dac synergized with smo in MF formation and PRs differentiation. All smo, dac double-mutant cells failed to undergo the cell shape changes associated with the MF (S1C and S1C´ Fig, n = 33 discs) and to differentiate PRs (S1F and S1F´ Fig, n = 20 discs). As dac was expressed in cells that accumulated CiFL at high levels in the MF and at low levels posterior to the MF where Ci promotes the transcription of the rdxZ reporter (Fig 1B–1C´´ and 2A–2B´´´), we next analyzed if dac affected Hh signaling. Strikingly, 67% of discs containing GFP-marked dac- clones straddling the MF showed reduced CiFL levels (Fig 2C–2F; n = 24 discs) and lower transcription of dpp, monitored by the transcriptional reporter dppZ (Fig 2G–2J; 64% of discs, n = 14). In addition, all dac- clones failed to trigger high levels of a lacZ enhancer trap insertion in the rdx gene (rdxZ) posterior to the MF (Fig 2K–2N; n = 9 discs). All these results indicate that dac is required for a full activation of the Hh signaling pathway.
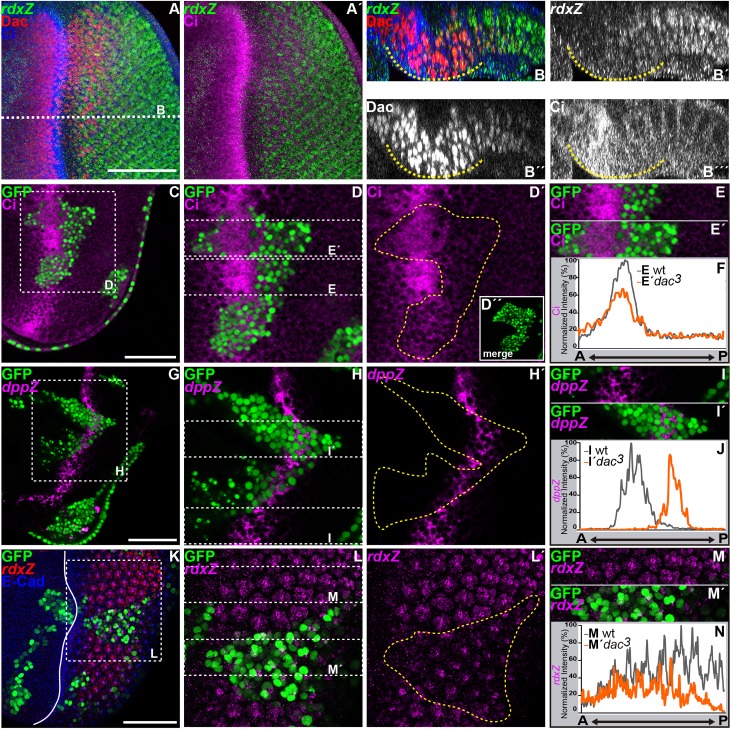
Fig 2. Loss of dac reduces CiFL and dppZ levels in the MF and fails to potentiate rdxZ posterior to the MF.
All panels show L3 eye imaginal discs, except F,J,N. (A-B´´´) Standard confocal section (A-A´) and cross-section (B´´´) through the dashed white line in B of a wild type disc stained with (A-B´) anti-β-Galactosidase to detect rdxZ (green in A-A´, B and white in B´), anti-Dac (red in A,B and white in B´´) and anti-Ci (blue in A,B; magenta in A´ and white in B´´´). Dashed yellow lines in B-B´´´ outline the domain where Dac is expressed at higher levels. (C-E´, G-I´, K-M´) dac3 clones marked by the presence of GFP (green in C,D,D´´,E-E´,G,H,I-I´,K,L,M-M´) and outlined by yellow dashed lines in D´,H´ and L´. Discs are stained with (C-E´) anti-Ci (magenta), (G-I´,K-M´) anti-β-Galactosidase to reveal (G-I´) dppZ (magenta) or (K-M ´) rdxZ (red in K and magenta in L-M´) and (K) anti-E-Cad (blue in K). Dashed white squares in C,G,K delimit the area shown in D-D´´, H-H´, L-L´, respectively. D´´ shows a merge of GFP signal relative to D. (F,J,N) Profiles of the intensity signals along the anterior-posterior (AP) axis of wild type (grey line) or dac3 mutant clones (orange line) for (F) Ci (at the levels of E or E´, respectively), or (J) dppZ (at the levels of I or I´, respectively) or (N) rdxZ (at the levels of M or M´, respectively). E-E´, I-I´ and M-M´ are outlined by dashed white rectangles in D,H,L. Plain white line in K’ indicates the MF. Scale bars represent 50μm.
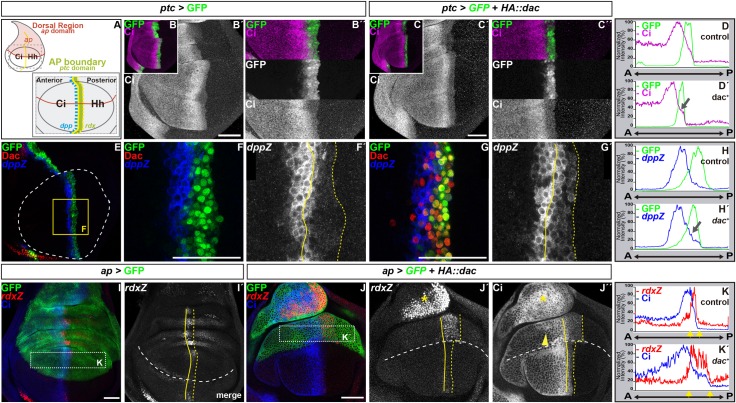
To determine if Dac is sufficient to potentiate Hh signaling, we analyzed the effect of overexpressing dac on Hh signaling activity in the wing imaginal disc, where endogenous Dac protein is expressed only in a few restricted patches [49]. In this tissue, Hh produced in the posterior (P) compartment signals to the anterior (A) compartment (Fig 3A). Thus, cells along the AP boundary compartment receive maximal Hh signaling, leading to the activation of rdx expression and consequently signaling attenuation through Rdx:Cul3-mediated CiFL degradation [45,46]. At these signaling levels, immediately adjacent to the P compartment, ptc expression is induced [41–44]. Next to this domain and further away from the AP boundary dpp is expressed [41–44]. Therefore, if dac potentiates Hh signaling, we expected that its expression along the AP boundary should enhance signaling levels and allow dpp transcription. Although overexpressing dac (HA::dac) along the AP boundary compartment using a ptc-Gal4 driver (Fig 3A) promoted CiFL accumulation (Fig 3, compare 3C–3C´´ with 3B–3B´´ and 3D´ with 3D) and increased expression of dppZ (Fig 3, compare 3G and 3G´ with 3E, 3F and 3F´ and 3H´ with 3H), these effects were relatively modest. Overactivation of Hh signaling would also be expected to potentiate rdx transcription, which would limit CiFL accumulation and Hh signaling activity. In agreement with this, overexpressing HA::dac in the dorsal compartment using the apterous-Gal4 (ap-Gal4) driver (Fig 3A) upregulated rdxZ expression in 85% of discs analyzed (n = 20) and drastically extended the CiFL-expression domain in the dorsal wing disc cells closed to the AP boundary in 82% of cases (n = 17) (Fig 3, compare 3J–J´´ with 3I and 3I´ and 3K´ with 3K). Taken together, we conclude that dac is necessary and sufficient to potentiate Hh signaling activity by promoting CiFL accumulation and the activation of Hh target genes.
Fig 3. Expressing dac ectopically in the wing disc is sufficient to enhance Hh signaling.
(A) Schematics of the whole wing disc (upper) and of the distal wing disc (lower) showing the expression domain of ap and of Hh pathway components. ap and hh are expressed in the dorsal (red) and posterior domain, respectively. Hh signals to the anterior domain leading to CiFL accumulation and activation of rdx and ptc in the anterior-posterior (AP) compartment boundary (green) and of dpp (blue). (B-C´´,E-G´, I-J´´) Standard confocal sections of L3 wing imaginal discs with anterior to the left and dorsal up. (B-C´´,E-G´) ptc-Gal4 driving the expression of UAS-GFP (B-B´´,E-F´) or UAS-GFP (C-C´´,G-G´) and UAS-HA::dac. (I-J´´) ap-Gal4 driving the expression of (I-I´) UAS-GFP or (J-J´´) UAS-GFP and UAS-HA::dac. Discs are stained with (B-C´´) anti-Ci (magenta in B,C, upper panel in B´´ and C´´; white in B´,C´, bottom panel in B´´ and C´´) or (E-G´) anti-Dac (red in E,F,G) and anti-β-Galactosidase to reveal dppZ (blue in E,F,G and white in F´,G´) or (I-J´´) anti-Ci (blue in I,J and white in J´´) and anti-β-Galactosidase to reveal rdxZ (red in I-J and white in I´,J´). B´´ and C´´ are magnification of B and C, respectively. (D-D´) Profiles of the GFP (green lines) or Ci (magenta lines) intensity signals across the AP axis in B´´ and C´´, respectively. (H-H´) Profiles of the GFP (green lines) and dppZ (blue lines) intensity signals across the AP axis in F and G, respectively. The arrows in D´ and H´ highlight the accumulation of Ci and dppZ in the ptc>dac-expressing domain, respectively. (K-K´) Profiles of the Ci (blue lines) and rdxZ (red lines) intensity signals across the AP axis in the dorsal areal delimited by dashed line in I and J, respectively. Yellow arrows in K and K´ indicate the position of the AP compartment boundary. The plain and dashed yellow lines in F´,G´,I´ and J´-J´´ delimited the AP compartment boundaries. The white dashed line in E outlines the distal wing blade. F-F´ is a magnification of the yellow square in E. The white dashed line in I´,J´-J´´ indicate the dorsal-ventral boundaries. The yellow arrowhead in J´´ indicates the accumulation of Ci in the dorsal, anterior blade. The yellow asterisks in J´,J´´ indicate the upregulation of rdxZ (J´) and the accumulation of Ci (J´´) in the proximal dorsal domain. Scale bars represent 50μm.
dac is required for Hh signaling downregulation and for ‘furrow state’ exit
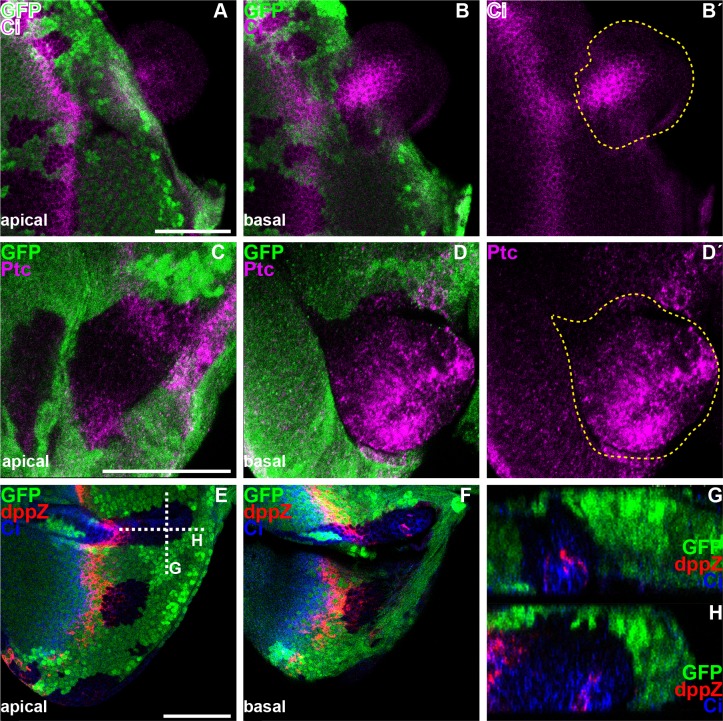
The Hh pathway has built-in a negative feedback that serves to attenuate signaling following maximal activation. This feedback rests on the activation of rdx by high signal levels. Once expressed, Rdx drives a Cul3-dependent CiFL degradation posterior to the MF thus allowing the exit from the furrow state [45,46]. We therefore tested if the loss of dac function induces the persistence of Hh signaling posterior to the MF. Indeed, some dac- clones located in internal region of the disc primordium showed ectopic expression of CiFL (Fig 4B and 4B´) and of the Hh-target genes Ptc (Fig 4D and 4D´) and dppZ (Fig 4F–4H). Strikingly, all these clones dropped basally (Fig 4A, 4C and 4E). As sustained exposure to Hh signaling promotes MyOII-dependent cell ingression and groove formation [32,33], we analyzed the shape of dac- clones posterior to the MF using the apical marker E-Cad. We confirmed that the disappearance of dac- cells from the apical surface (Fig 5A) did not result from a loss of cell polarity, as dac- cells maintained E-Cad expression apically (Fig 5B). However, cross section through the eye disc showed that dac- clones ingressed within the epithelium, forming grooves (Fig 5C and 5C´). In addition, these clones accumulated activated MyOII, detected by phospho-Myosin Light Chain (pMLC) antibody at the apical surface of ingressed clones (Fig 5D, 5E and 5E´). These drastic changes in cell shape were associated with the presence of PR nuclei, expressing Elav and hhZ that were still localized close to the apical cell surface but appeared on basal focal planes compared to control GFP-positive neighboring nuclei (Fig 5F and 5F´ to 5I and 5I´). dac- clones spanning the disc margin that delayed MF progression and PRs differentiation also contained higher Ptc levels (S3A and S3A´´ Fig). However, those in which MF initiation and retinogenesis were compromised [49] showed reduced Ptc expression (S3B and S3B´ Fig). Thus, dac is required for the swift dynamic changes in Hh signaling associated to the passing MF. In its absence, Hh target genes activation and Hh-regulated processes suffer a general delay. Interestingly, one of the consequences is that Hh signaling persists for longer, as its attenuation mediated by rdx is also delayed.
Fig 4. Loss of dac function leads to the persistence of Hh target genes expression posterior to the MF.
All panels show L3 eye imaginal discs, containing dac3 clones marked by the absence of GFP (green in A,B,C,D,E-H) and outlined by yellow dashed lines in B’ and D’. A and B-B´ or C and D-D´ or E and F are apical (A,C,E) or basal (B-B´, D-D´,F) standard confocal sections of the same discs. (G-H) Cross-section through the disc epithelium at the level of the white dashed lines in E. Discs are stained with (A-B´) anti-Ci (magenta) or (C-D´) anti-Ptc (magenta) or (E-G´) anti-Ci (blue) and anti-β-Galactosidase (red) to reveal dppZ. Note that dac mutant tissues appear to drop out from the disc epithelium. Scale bars represent 50μm.
Fig 5. Loss of dac maintains a ‘furrow state’ posterior to the MF.
All panels show L3 eye imaginal discs, containing dac3 clones marked by the (A-E´ and I-I´) absence or (F-H´´´) presence of GFP (green in A, B, C, D, E, F, H, G and I) and outlined by dashed lines in C´, E’, F´ and H´-H´´. A and B or D and E-E´ or F-F´ and G-G´ are apical (A,D,F-F´) or basal (B, E-E´,G-G´) standard confocal sections of the same discs. I and I´ show subsequent standard confocal sections from apical to basal. (C-C´,H-H´´´) Cross-sections through the disc epithelium at the level of the yellow dashed lines in A or F, respectively. Discs are stained with (A-C´) anti-E-Cad (magenta) or (D-E´) anti-pMLC (magenta) or (F-H´´´) anti-Elav (blue) and phalloidin (red in F,F´,G,G´,H and H´) and DAPI (yellow in H´´ and H´´´) or (I-I´) anti-Elav (blue) and anti-β-Galactosidase (red) to reveal hhZ. Arrows in H´,H´´ and I´ indicate the basal localization of PRs in dac- clones. A minimum of 24 clones in internal region of the disc primordium was observed to maintain a “furrow state” posterior to the MF. Scale bars represent 50μm.
dac is required downstream of fu to promote Ci accumulation
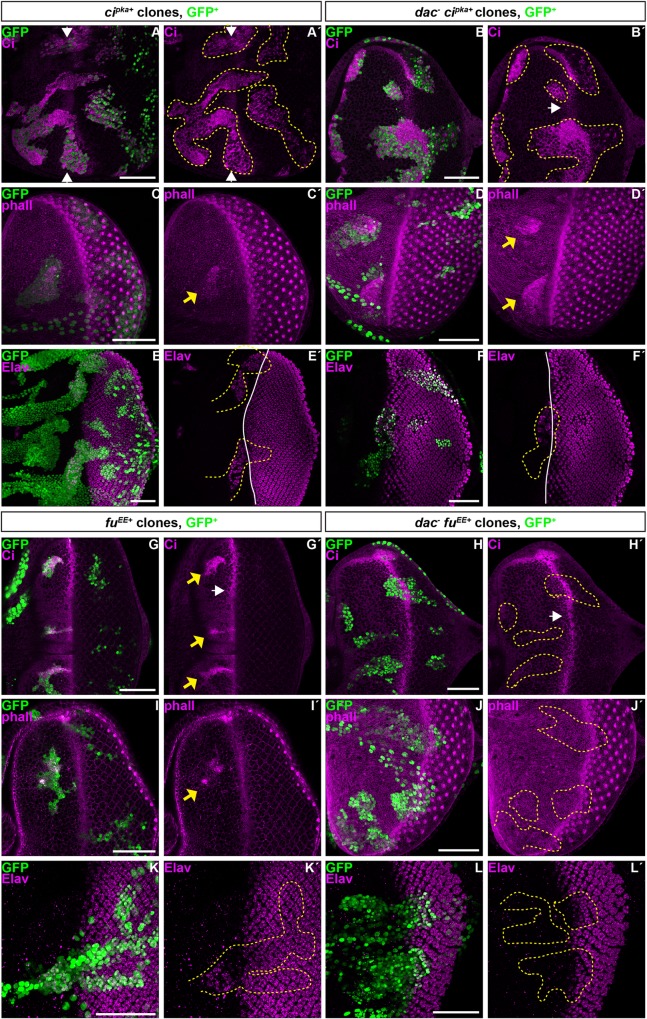
To understand how dac promotes CiFL accumulation, we analyzed the requirement for dac to transduce Hh signaling in cells expressing constitutive active forms of Hh pathway components. We first expressed in clones a form of ci insensitive to phosphorylation by PKA (cipka+). Consistent with previous observations, cipka+ clones promoted precocious differentiation anterior to the MF, where cells express dac endogenously [20,51–56]. All cipka+ clones accumulated CiFL (Fig 6A and 6A´; n = 7 discs), while 89% displayed an enrichment of F-actin, reminiscent to the apical cell constriction in the MF (Fig 6C and 6C´; n = 27 discs) and 62% formed ectopic PRs (Fig 6E and 6E´; n = 8 discs). However, posterior to the MF, where CiFL degradation is independent of PKA [57], Cipka+ accumulation was reduced when compared to clones located in the MF and anterior to the MF (Fig 6A and 6A´). Thus, Cipka+ may suffer degradation in this domain. Removing dac function did not affect the ability of clones expressing ectopic cipka+ to accumulate CiFL (Fig 6B and 6B´; 100% of discs, n = 10), F-actin (Fig 6D and 6D´; 95% of discs, n = 22) or differentiate ectopic PRs (Fig 6F and 6F´; 22% of discs, n = 12) anterior to the MF. Therefore, dac acts upstream of CiFL.
Fig 6. Removing Dac function suppresses the activity of FuEE, but not of Cipka.
All panels show standard confocal sections of L3 eye imaginal discs, containing clones labeled with GFP (green in A,B,C,D,E,F,G,H,I,J,K,L) and outlined by dashed lines in A´,B´,E´,F´,H´,J´,K´,L´ and containing clones (A-A´,C-C´,E-E´) expressing UAS-cipka and (B-B´,D-D´,F-F´) mutant for dac3 or (G-G´,I-I´,K-K´) expressing UAS-fuEE and (H-H´,J-J´,L-L´) mutant for dac3. Discs are stained with (A-B´,G-H´) anti-Ci (magenta) or (C-D´,I-J´) phalloidin (magenta) to outline cell shape or (E-F´,K-L´) anti-Elav (magenta)White arrows in B´,G´ and H´ indicate CiFL accumulation in wild type cells in the MF. Yellow arrows in G´ indicate CiFL accumulation anterior to the MF. Yellow arrows in C´, D´ and I´ highlight increased levels of phalloidin staining, reminiscent to apical contraction of the MF. Arrows in A-A´ and solid lines in E´ and F´ indicate MF position. Scale bars represent 50μm.
We next investigated if dac was required for the activity of the upstream Ci regulator Fused (Fu). Overexpressing a constitutive active form of fu (fuEE+) anterior to the MF also triggered CiFL accumulation in 93% of discs with clones (Fig 6G and 6G´, n = 27 discs), an enrichment of F-actin or E-Cad, reminiscent to the apical cell constriction in the MF in 85% of cases (Fig 6I and 6I´; n = 13 discs), and ectopic PR differentiation in 11% of discs with clones (Fig 6K and 6K´; n = 27 discs). In contrast, when these clones were also mutant for dac, the accumulation of CiFL (Fig 6H and 6H´; 36% of discs, n = 11) and of F-actin (Fig 6J and 6J´; 41% of discs, n = 27) was severely reduced. In addition, these clones were no longer able to differentiate ectopic PRs (Fig 6L and 6L´, n = 6). Further, overexpressing fuEE+ did not rescue the MF delay of dac-mutant tissue (Fig 6H and 6H´ and 6J and 6J´)). Thus, dac potentiates Hh signaling by promoting CiFL accumulation downstream of, or parallel to fu.
dac encodes a nuclear protein, which has been shown to bind double-stranded nucleic acids [58] and to activate transcription of a reporter gene in yeast [59]. We therefore tested, using a genome-wide ChIP-seq approach, the possibility that Dac regulated the expression of some of the components along the Hh signaling pathway (see Materials and Methods). We analyzed specifically ChIP peaks in distal regions (i.e. excluding those falling in 5’UTRs and 1kb upstream of transcription start sites). Within the ChIP peaks, we identified a set of 352 distal regions in which we found significantly enriched an A-rich motif. This motif is similar to the DNA binding motif identified previously for the human DACH by protein structure, in-silico and ChIP-seq analyses (S4 Fig, [60]). This fact indicated that the set of 352 ChIP peaks were likely directly bound by Dac. However, among the nearby genes (S4 Fig), we could not identify any of the major components of the Hh signaling pathway (including hh itself plus smo, PKA, CKI, ci, cullin1, Slimb, skp-1, and cul3). This result suggested that the effect of dac on the activity of the Hh pathway was unlikely to be mediated by the transcriptional regulation of these signaling components. To test experimentally this point, we generated dac- clones and asked whether dac loss affected the expression of the hhZ and ciZ enhancer traps, which serve as transcriptional reporters. Although dac mutant clones located in internal region of the eye primordium appeared to contain reduced hhZ expression (Fig 1H and 1H´), this effect likely resulted from a reduction in the number of PR cells per ommatidia in dac- mutant tissue, as PR hhZ levels were not different than in wild type cells. Similarly, in clones straddling the MF, the absence of dac function delayed hhZ expression, but appeared normally as PR cells gained ELAV signal (S5A and S5A´´´ Fig). In addition, dac was neither necessary nor sufficient to control ci transcription, as ciZ expression was not affected in dac mutant in the eye discs (S5B and S5C´ Fig) or in wing discs overexpressing dac using the ptc-Gal4 driver (S5 Fig, compare S5H to S5J with S5D to S5F and S5K with S5G). We conclude that dac does not promote CiFL accumulation downstream of, or parallel to fu by affecting hh or ci expression and, most likely, neither affecting the transcription of other major pathway components.
Discussion
The differentiation of the retina in Drosophila occurs both very fast and with high precision: every 2 hours, one new column of assembled ommatidia is added to the developing eye [61]. This phenomenon requires the coordination of gene expression, cell cycle and tissue morphogenesis. All these processes depend critically on the dynamics of Hh signaling. In this report, we show that Dac is an essential element in this dynamics. Dac potentiates Hh signaling in the MF, upstream of Ci and downstream of, or in parallel to Fu. By doing so, dac ensures proper retinogenesis by controlling the timing of MF formation, the accuracy of cell cycle control, the tissue changes associated to the MF, the correct specification of the founder PR8 cell and the attenuation of Hh signaling posterior to the MF, which allows the progression of the differentiation wave.
dac potentiates Hh signaling
Our observations demonstrate that dac is required to strengthen Hh signaling. First, the absence of dac function recapitulates phenotypically a reduction of Hh signaling. Removing smo or dac function delays MF progression (Figs 1 and 2, S1 Fig, [23,28–31,49]) and the re-entry in S-phase in the SMW (S2 Fig; [25]). In addition, loss of dac (Fig 1) or smo [23,29,31] results in reduced ato expression ahead of the MF and affects the restriction of ato, first in proneural clusters and then in single PR8 posterior to the MF. Furthermore, neither loss of dac (Fig 1), nor smo [31,35] affects ommatidial cell fate. The cause of the reduced number of ommatidia and variable number of cells per ommatidium in dac mutant clones (Fig 1) is not totally clear. It could result from the effects in ato expression, including its abnormal spacing posterior to the MF and the singling of the ommatidial founder PR8 [62–67]; it could also arise from alterations in cell cycle control, as we detect persistent cell cycling posterior to the MF, which may affect cell recruitment into the ommatidium [25,36]; or from a combination of both. Second, smo and dac synergize to promote MF progression and PR differentiation (S1 Fig). Accordingly, removing one copy of hh enhances the dac mutant eye phenotype and fully suppresses PRs differentiation of dac mutant clones located in internal regions of the disc primordium [49]. Third, dac mutant clones show reduced levels of CiFL and lower expression of the Hh target gene dpp in the MF and rdx posterior to the MF (Fig 2). In addition, ptc expression, another Hh target, is reduced in marginal dac mutant clones that fail to differentiate photoreceptor cells (S3 Fig). Conversely, expressing dac ectopically in the wing disc is sufficient to enhance CiFL accumulation and rdx expression, and to induce dpp in the domain immediately adjacent to the compartment boundary (Fig 3).
How does dac regulate Hh signaling?
Our observations argue that dac potentiates Hh signaling by promoting CiFL accumulation downstream of, or in parallel to Fu. First, loss of dac reduces CiFL levels in the MF (Fig 2), while expressing dac ectopically in the wing disc has the opposite effect (Fig 3). Second, an activated form of Fu (FuEE+), which inhibits Ci processing (into CiR) and promotes CiFL/CiA accumulation [68], requires dac function for this accumulation of CiFL and to induce MF-like features and precocious PR differentiation anterior to the MF. In contrast, removing dac function has no major effect on the ability of a PKA-insensitive form of Ci (Cipka+) to trigger features associated to ectopic Hh signaling in this domain (Fig 6). The precise molecular mechanism by which Dac affects this step along the Hh signaling pathway is unknown at the moment. dac encodes a nuclear protein, which has been shown to bind double-stranded nucleic acids [58] and to activate transcription of a reporter gene in yeast [59]. Our functional and ChIP-seq experiments indicate that hh and ci are not direct transcriptional targets of Dac. In addition, the ChIP-seq data suggest that neither are any of the major pathway components of the Hh pathway (S4 Fig)–although without further studies this possibility cannot be ruled out completely. This would shift the control of the Hh signaling activity to other Dac targets, which would exert this control directly or indirectly. Another, not mutually exclusive possibility is that Dac collaborates with CiA in enhancing the expression of CiA-dependent target genes, such as rdx or ptc (Figs 2 and 3). Although we have previously shown that Dac inhibits the transcriptional ability of the Homothorax/Yorkie (Hth/Yki) complex [69], Dac may act as an activator or repressor depending on the cellular context [70]. Therefore, Dac may contribute to the transcriptional activity of CiA promoting the expression of target genes responsible for proneural fate acquisition and differentiation. In agreement with this hypothesis, using the bioinformatic tool Clover [71], with standard parameters, we also found, besides the Dac motif, the Gli/Ci consensus binding motif significantly enriched (p<0.001) in the distal Dac-ChIP-peaks (S4 Fig), suggesting that Dac might indeed collaborate with CiA to enhancer expression of its targets. In addition, we cannot exclude other mechanisms of Dac action independent of its role in transcriptional regulation. In fact, although it was not reported in Drosophila tissues, the human Dac homologue DACH1 presents both nuclear and cytoplasmic localization in different tissues [72–75]. Moreover, DACH1 localization shifts from the nucleus in normal tissue to the cytoplasm in ovarian cancer [72]. Dac/DACH1 might be involved in the control of Hh signaling pathway in a subcellular localization manner. Further studies are required to elucidate the mechanism by which Dac affects CiFL accumulation and consequently potentiates Hh signaling.
dac¸ an orchestrator of eye development that promotes Hh signaling dynamics
In the developing eye, dac lies downstream of eye absent (eya) and the dpp signaling pathway [50,76–78] where it regulates multiple events that together coordinate cell proliferation and differentiation in time and space. We demonstrate here that dac potentiates Hh signaling. This together with dac’s role in the transition from proliferating progenitor cells to committed precursor cells together with Dpp signal [69] can account for the pleiotropic and essential roles played by dac (Fig 7). Retinogenesis starts with the formation of the MF and the triggering of two major Hh targets: dpp and ato. The weakening of Hh signaling together with the reduced dpp transcription can account for MF delay, as both signals are required for this morphogenetic event [18,19,23,28–31]. Right at the MF and immediately posterior to it, the cell cycle is tightly regulated. We observe that the expression of the p21/p27 Cdk inhibitor dap is lost in dac- cells (S2E Fig) and that this is accompanied by persistent cycling beyond the SMW (S2A and S2C´ Fig). This cell cycle misregulation, together with the aberrant restriction of ato expression, is the likely cause of the abnormal retinogenesis in dac-mutant tissues that includes ommatidia with variable number of cells. Posterior to the MF, high Hh signaling levels activates the expression of Rdx, which, together with Cul3, targets CiFL to full proteasomal degradation. In doing so, Hh signal becomes attenuated, and this attenuation allows cells to exit the furrowed state. In fact, dac-mutant clones posterior to the MF often remain as apically constricted inpouchings of the disc epithelium with high levels of P-MyOII, accumulated CiFL and expression of CiA target genes (Figs 4 and 5).
Fig 7. Model by which Dac orchestrates retinogenesis.
Schematic of the L3 eye disc spanning from the anterior proliferating progenitors (left) to the differentiated photoreceptors (right), showing the expression domain and activity of Dac, Hh, Ci, Dpp, and Yki/Hth (upper panel) and their molecular interactions to coordinate proliferation and differentiation of the retina (lower panel). Hh expressed by the differentiating PRs diffuses short-range, promoting a gradient of Ci activity. High levels of Ci promote the synchronized re-entrance in the cell cycle in the SMW [25,34–36] and turns off its own signaling pathway via rdx expression [45–48]. Anterior to this domain, low Hh signaling initiates the expression of ato, which promotes the acquisition of a proneural fate [18–22]. In addition, in this domain, Hh signaling upregulates dap, which keeps precursor cells in G1-phase by inhibiting CycE [24–27], and activates dpp expression [12]. Together, Hh and Dpp induce MF formation and high dac expression [50]. In turn Dac, whose expression extends from the FMW to the SMW, straddling the MF, ensures the transition from proliferation to G1 arrest state by inhibiting the transcriptional activity of the Yki-Hth complex in the FMW [69]. Moreover, together with Dpp, Dac represses Hth expression [69]. Finally, we show here that Dac potentiates Hh signaling in the MF, upstream of Ci. This positive feedback loop is essential to control all Hh functions, including the timing of MF formation, the accuracy of cell cycle control, the tissue changes associated to the MF, the correct specification of the founder PR8 cell and the attenuation of Hh signaling posterior to the MF, which allows the progression of the differentiation wave.
This report, together with our previous study [69], places dac as an essential regulator of retinal development, controlling the transitions from proliferating progenitor cells to committed precursor cells first [69] and then from precursors to differentiating retinal cells (this work). The dac expression profile spans the regions of the eye disc where it exerts its functions: the increasing Dac expression in precursor cells approaching the MF ensures that cells transit from proliferation to G1 arrest, while peak levels in the MF secure proper retinogenesis by potentiating Hh signaling here. High dac expression in the MF in addition to its downregulation in differentiating photoreceptors could both contribute to turn off Hh signaling in this domain. This could be achieved, at least in part, by inducing the activation of a negative feedback via rdx expression and by limiting CiFL accumulation downstream or in parallel to Fu, respectively. Whether this function is also carried out by the vertebrate dac homologues, the DACH1 and DACH2 genes, awaits further investigation.
Materials and Methods
Fly strains and genetics
Fly stocks used were dac3, smo3, hhP30 (hhZ), rdx03477 (rdxZ), P{dpp-lacZ.Exel.2}3 (dpp-Z),ciZ [79], UAS-HA:dacF [80], UAS-mCD8GFP, UAS-mCherry (TRIP #35787), UAS-fuEE [81–83], UAS-Cipka [82], ptc-Gal4 [84], ap-Gal4 [85]. Mutant clones for dac3 marked by the absence of GFP were generated through mitotic recombination [86]. The MARCM technique [87] was used to induce clones marked by the presence of GFP, mutant for dac3 or smo3 or smo3 and dac3 or expressing UAS-fuEE or UAS-cipka or mutant for dac3 and expressing UAS-fuEE or UAS-cipka. Larvae were heat-shocked for 1 hour at 37°C between 48 and 72h after egg laying. Gain of function experiments using UAS-HA:dacF were performed using the ptc-Gal4 or ap-Gal4 driver that drives expression in the AP boundary compartment or dorsal region of the wing disc, respectively. Crosses carrying UAS-HA:dacF and the corresponding controls were raised at 18°C, while others were raised at 25°C.
Immunohistochemistry
Imaginal discs were dissected and fixed according to standard protocols. Primary antibodies used were mouse anti-Dac (1:100; mAbdac2.3, DSHB); mouse anti-CycB (1:25; F2F4, DSHB); mouse anti-β-Galactosidase (1:200; Z378B, Promega), rabbit anti-β-Galactosidase (1:1000; 55976, Cappel); rat anti-E-Cad (1:50; DCAD2, DSHB); guinea pig anti-CycE (1:1000; gift from T. Orr-Weaver, Whitehead Institute, Cambridge, USA); rabbit anti-Ato (1:5000; [22]); rabbit anti-pMLC (1:10; 36715, Cell Signaling), which reveals pMyOII; rat anti-Elav (1:1000; 7E8A10, DSHB); mouse anti-Elav (1:100; 9F8A9, DSHB); guinea pig anti-Sens (1:1000; [88]); guinea pig anti-Pros (1:25; [89]); rabbit anti-Sal (1:200; [90]); rabbit anti-PH3 (1:200; 9701, Cell Signaling); mouse anti-Ptc (1:100; Drosophila Ptc (Apa1), DSHB); rat anti-Ci (1:10; 2A1, DSHB). Rhodamine-conjugated (Sigma) and C660–conjugated Phalloidin (Biotium) were used at a concentration of 0.3 μM and 5U/ml, respectively. DAPI was used at a concentration of 1ng/ml. Fluorescently labeled secondary antibodies were from Jackson Immunoresearch, (1:200). Imaging was carried out on Leica SP2 or SP5 confocal microscopy set ups. Plot profiles of fluorescent intensity were obtained using an NIH ImageJ program [91]. Fluorescent intensities were normalized to the maximum intensity for each channel.
In situ hybridization
Anti-mouse-HRP (Sigma) was used for immunoperoxidase staining. Digoxigen labelled dap RNA probe (Roche) was produced from the cDNA clone LP07247 (BDGP). For BrdU incorporation assay, eye-antennal discs were dissected and incubated in 10μM BrdU in PBS for 30min. BrdU was detected with an anti-BrdU antibody (1:400; Roche) after treatment with DNase.
Dac-ChIP Seq
Wandering 3rd instar larvae (Dac:GFP, Bloomington stock 42269) were dissected in cold PBS and imaginal discs were fixed with formaldehyde for 25 minutes. Chromatin was fragmented by sonication till it reached an average size of 500 bp. 20 μl of protein A/G magnetic beads (Merck, Millipore) was added to pre-clean the samples. The anti-GFP Ab (ab290, Abcam) was added to a fixed chromatin aliquot and incubated at 4°C overnight. Immunocomplexes were recovered by adding protein A/G magnetic beads to the sample and incubating for 3 hours at 4°C. Beads were resuspended in elution buffer, RNase was added to the immunoprecipitated chromatin and incubated for 30 minutes at 37°C. ChIP libraries were prepared with the Truseq DNA library prep kit (Illumina) and the samples were sequenced on a HiSeq 2000 (Illumina). The reads were cleaned using fastq-mcf and mapped with bowtie2 to the Drosophila melanogaster genome (Flybase version 5). Dac-ChIP peaks (minus input) were called using macs2 (dac-ChIP-vs-ChIP_Input_peaks.bed). From this bed file all regions lying in a 5’UTR or 1kb upstream of a TSS were removed using bedtools intersectBed, to retain only distal Dac-ChIP-peaks (dac-ChIP-vs-ChIP_Input_peaks_not-in-5UTR_not-in-1kb-up.bed). The distal Dac-ChIP-peaks were loaded into i-cistarget [92], a motif enrichment tool, to obtain a final table (S4 Fig) of potential Dac targets. Access to the whole dataset can be found in the GEO database with the accession number GSE82151 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE82151), used as reference for all subsequent manuscripts referring to these data.
Supporting Information
All panels show standard confocal sections of L3 eye imaginal discs, containing (A-A´´,D-D´) dac3 or (B-B´,E-E´) smo3 single mutant clones or (C-C´,F-F´) smo3, dac3 double mutant clones marked by the presence of GFP (green in A, A´´,B,C,D,E,F) and outlined by dashed line (in A´, A´´,B´,C´,D´,E´,F´). Discs are stained with (A-B´) anti-E-Cad (magenta) or (C-C´) phalloidin (magenta) to outline cell shape or with (D-F’) anti-Elav (magenta) to label differentiating PRs. A´´ corresponds to a merge between the GFP signal in A and more basal sections. Note that while loss of smo function has little effect on MF formation (arrow in B´) and PR differentiation, loss of both smo and dac function completely disrupts MF formation (arrows in C´) and PR differentiation. Scale bars represent 50μm.
(TIF)
All panels show (A-D´) standard confocal sections or (E) standard brightfield image of L3 eye discs, containing dac3 mutant clones marked by the absence of GFP (green in A,B,C,D and brown in E) and outlined by yellow dashed lines (in A’,B´,C´,D´,E). Discs are stained with (A-A´) anti-CycB (magenta), (B-B´) anti-PH3 (magenta), (C-C´) anti-BrdU (magenta), (D-D´) anti-CycE (magenta) and (E) dap mRNA (blue). The positions of the FMW and SMW are indicated by white and yellow arrowheads, respectively. White arrows in B’ and C’ point to the delayed re-entrance in cell cycle posterior to the SMW. Solid lines in D’ delimit the high levels of CycE within the MF in wild type cells. Scale bars represent 50μm.
(TIF)
All panels show standard confocal sections of L3 eye imaginal discs, containing dac3 mutant clones marked by the absence of GFP (green in A,B) and outlined by dashed line in A´, A´´,B´. Discs are stained with anti-Ptc (red) and anti-Elav (blue). Plain lines in A´ and B´ indicate the MF position. Scale bars represent 50μm.
(TIF)
(A) Example gene (nej) with a Dac ChIP peak, 3kb upstream of the transcription start site. (B) Sequence logo of a candidate Dac motif found as the most enriched motif in the distal ChIP-peak. (C) Sequence logo of a Gli consensus binding site (annotated in the transfac database: transfac_pro-M01037) found significantly enriched in the set of distal ChIP-peaks when compared to random background sequences (p<0.001). (D) List of the proposed Dac targets, predicted by the presence of a nearby (<5kb from TSS or intronic) ChIP-seq peak, and the presence of the candidate Dac motif, as predicted by i-cisTarget [92].
(TIF)
(A-C´) Standard confocal sections of L3 eye imaginal discs (B-B´) wild type or (A-A´´´,C-C´) containing dac3 clones labeled with GFP (green in A,C) and outlined by dashed lines in A´-A´´´,C´, stained with (A-A´´´) anti-Elav (blue in A,A´, white in A´´´) and anti—β-Galactosidase to reveal dppZ (red in A-A´ and white in A´´) or (B-B´) anti-β-Galactosidase to reveal ciZ (green) and DAPI (magenta in B) or (C-C´) anti-β-Galactosidase to reveal ciZ (red in C and white in C´) and anti-ELAV (blue in C). (D-F,H-J) Standard confocal sections of L3 wing imaginal discs with anterior to the left and dorsal up in which ptc-Gal4 drives the expression of UAS-mCherry (green in D,E,F,H,I,J) and (H,I-J) UAS-HA::dac. Discs are stained with anti-Dac (red in D,H) and anti-β-Galactosidase to reveal ciZ (blue in D,H and magenta in E-F,I-J). E-E´ and I-I´ are magnifications of the dashed white square in D and H, respectively. F and J correspond to the area delimited by white dashed lines in E and I, respectively. The dashed yellow lines in E´ and I´ outline the AP compartment boundaries. (G,K) Profiles of the mCherry (green lines) and ciZ (magenta lines) intensity signals across the AP axis in F and J, respectively. Scale bars represent 50μm.
(TIF)
Acknowledgments
We thank C.S. Lopes, P. Thérond, G. Mardon, T. Orr-Weaver, the Bloomington Drosophila Stock Center, the Developmental Studies Hybridoma Bank (DSHB), the TRiP at Harvard Medical School (NIH/NIGMS R01-GM084947) and the Drosophila Genomics Resource Center for fly stocks and reagents. The manuscript was improved by the critical comments of Takashi Koyama and Élio Sucena.
Data Availability
Access to the whole Dac-ChIP Seq dataset can be found in the GEO database with the accession number GSE82151 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE82151) as mentioned in the Material and Methods section.
Funding Statement
This work was supported by grants from Fundação para a Ciência e Tecnologia (PTDC/BIA-BCM/71674/2006) to FJ; from BFU2012-34324 and BFU2015- 66040 (MINECO, Spain) to FC and The Research Foundation—Flanders (FWO, www.fwo.be) (grants G.0640.13 and G.0791.14) to SA. CBP was the recipient of fellowship from Fundação para a Ciência e Tecnologia (grant SFRH/BPD/46983/2008) and FJ is the recipient of IF/01031/2012 from Fundação para a Ciência e Tecnologia. JJ is the recipient of a FWO PhD Fellowship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ingham PW, McMahon AP (2001) Hedgehog signaling in animal development: paradigms and principles. Genes Dev 15: 3059–3087. [DOI] [PubMed] [Google Scholar]
- 2.Varjosalo M, Taipale J (2008) Hedgehog: functions and mechanisms. Genes Dev 22: 2454–2472. 10.1101/gad.1693608 [DOI] [PubMed] [Google Scholar]
- 3.Jiang J, Hui CC (2008) Hedgehog signaling in development and cancer. Dev Cell 15: 801–812. 10.1016/j.devcel.2008.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Briscoe J, Therond PP (2013) The mechanisms of Hedgehog signalling and its roles in development and disease. Nat Rev Mol Cell Biol 14: 416–429. 10.1038/nrm3598 [DOI] [PubMed] [Google Scholar]
- 5.Wallace VA (2008) Proliferative and cell fate effects of Hedgehog signaling in the vertebrate retina. Brain Res 1192: 61–75. [DOI] [PubMed] [Google Scholar]
- 6.Treisman JE (2013) Retinal differentiation in Drosophila. Wiley Interdiscip Rev Dev Biol 2: 545–557. 10.1002/wdev.100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amato MA, Boy S, Perron M (2004) Hedgehog signaling in vertebrate eye development: a growing puzzle. Cell Mol Life Sci 61: 899–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee JD, Treisman JE (2002) Regulators of the morphogenetic furrow. Results Probl Cell Differ 37: 21–33. [DOI] [PubMed] [Google Scholar]
- 9.Baker NE (2007) Patterning signals and proliferation in Drosophila imaginal discs. Curr Opin Genet Dev 17: 287–293. [DOI] [PubMed] [Google Scholar]
- 10.Wolff T, Ready DF (1991) The beginning of pattern formation in the Drosophila compound eye: the morphogenetic furrow and the second mitotic wave. Development 113: 841–850. [DOI] [PubMed] [Google Scholar]
- 11.Dominguez M, Hafen E (1997) Hedgehog directly controls initiation and propagation of retinal differentiation in the Drosophila eye. Genes Dev 11: 3254–3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blackman RK, Sanicola M, Raftery LA, Gillevet T, Gelbart WM (1991) An extensive 3' cis-regulatory region directs the imaginal disk expression of decapentaplegic, a member of the TGF-beta family in Drosophila. Development 111: 657–666. [DOI] [PubMed] [Google Scholar]
- 13.Penton A, Selleck SB, Hoffmann FM (1997) Regulation of cell cycle synchronization by decapentaplegic during Drosophila eye development. Science 275: 203–206. [DOI] [PubMed] [Google Scholar]
- 14.Horsfield J, Penton A, Secombe J, Hoffman FM, Richardson H (1998) decapentaplegic is required for arrest in G1 phase during Drosophila eye development. Development 125: 5069–5078. [DOI] [PubMed] [Google Scholar]
- 15.Dong X, Zavitz KH, Thomas BJ, Lin M, Campbell S, et al. (1997) Control of G1 in the developing Drosophila eye: rca1 regulates Cyclin A. Genes Dev 11: 94–105. [DOI] [PubMed] [Google Scholar]
- 16.Lopes CS, Casares F (2010) hth maintains the pool of eye progenitors and its downregulation by Dpp and Hh couples retinal fate acquisition with cell cycle exit. Dev Biol 339: 78–88. 10.1016/j.ydbio.2009.12.020 [DOI] [PubMed] [Google Scholar]
- 17.Heberlein U, Moses K (1995) Mechanisms of Drosophila retinal morphogenesis: the virtues of being progressive. Cell 81: 987–990. [DOI] [PubMed] [Google Scholar]
- 18.Heberlein U, Wolff T, Rubin GM (1993) The TGF beta homolog dpp and the segment polarity gene hedgehog are required for propagation of a morphogenetic wave in the Drosophila retina. Cell 75: 913–926. [DOI] [PubMed] [Google Scholar]
- 19.Ma C, Zhou Y, Beachy PA, Moses K (1993) The segment polarity gene hedgehog is required for progression of the morphogenetic furrow in the developing Drosophila eye. Cell 75: 927–938. [DOI] [PubMed] [Google Scholar]
- 20.Heberlein U, Singh CM, Luk AY, Donohoe TJ (1995) Growth and differentiation in the Drosophila eye coordinated by hedgehog. Nature 373: 709–711. [DOI] [PubMed] [Google Scholar]
- 21.Jarman AP, Sun Y, Jan LY, Jan YN (1995) Role of the proneural gene, atonal, in formation of Drosophila chordotonal organs and photoreceptors. Development 121: 2019–2030. [DOI] [PubMed] [Google Scholar]
- 22.Jarman AP, Grell EH, Ackerman L, Jan LY, Jan YN (1994) Atonal is the proneural gene for Drosophila photoreceptors. Nature 369: 398–400. [DOI] [PubMed] [Google Scholar]
- 23.Dominguez M (1999) Dual role for Hedgehog in the regulation of the proneural gene atonal during ommatidia development. Development 126: 2345–2353. [DOI] [PubMed] [Google Scholar]
- 24.Escudero LM, Freeman M (2007) Mechanism of G1 arrest in the Drosophila eye imaginal disc. BMC Dev Biol 7: 13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Firth LC, Baker NE (2005) Extracellular signals responsible for spatially regulated proliferation in the differentiating Drosophila eye. Dev Cell 8: 541–551. [DOI] [PubMed] [Google Scholar]
- 26.de Nooij JC, Graber KH, Hariharan IK (2000) Expression of the cyclin-dependent kinase inhibitor Dacapo is regulated by cyclin E. Mech Dev 97: 73–83. [DOI] [PubMed] [Google Scholar]
- 27.de Nooij JC, Hariharan IK (1995) Uncoupling cell fate determination from patterned cell division in the Drosophila eye. Science 270: 983–985. [DOI] [PubMed] [Google Scholar]
- 28.Strutt DI, Mlodzik M (1997) Hedgehog is an indirect regulator of morphogenetic furrow progression in the Drosophila eye disc. Development 124: 3233–3240. [DOI] [PubMed] [Google Scholar]
- 29.Greenwood S, Struhl G (1999) Progression of the morphogenetic furrow in the Drosophila eye: the roles of Hedgehog, Decapentaplegic and the Raf pathway. Development 126: 5795–5808. [DOI] [PubMed] [Google Scholar]
- 30.Curtiss J, Mlodzik M (2000) Morphogenetic furrow initiation and progression during eye development in Drosophila: the roles of decapentaplegic, hedgehog and eyes absent. Development 127: 1325–1336. [DOI] [PubMed] [Google Scholar]
- 31.Fu W, Baker NE (2003) Deciphering synergistic and redundant roles of Hedgehog, Decapentaplegic and Delta that drive the wave of differentiation in Drosophila eye development. Development 130: 5229–5239. [DOI] [PubMed] [Google Scholar]
- 32.Escudero LM, Bischoff M, Freeman M (2007) Myosin II regulates complex cellular arrangement and epithelial architecture in Drosophila. Dev Cell 13: 717–729. [DOI] [PubMed] [Google Scholar]
- 33.Corrigall D, Walther RF, Rodriguez L, Fichelson P, Pichaud F (2007) Hedgehog signaling is a principal inducer of Myosin-II-driven cell ingression in Drosophila epithelia. Dev Cell 13: 730–742. [DOI] [PubMed] [Google Scholar]
- 34.Duman-Scheel M, Weng L, Xin S, Du W (2002) Hedgehog regulates cell growth and proliferation by inducing Cyclin D and Cyclin E. Nature 417: 299–304. [DOI] [PubMed] [Google Scholar]
- 35.Vrailas AD, Moses K (2006) Smoothened, thickveins and the genetic control of cell cycle and cell fate in the developing Drosophila eye. Mech Dev 123: 151–165. [DOI] [PubMed] [Google Scholar]
- 36.Baonza A, Freeman M (2005) Control of cell proliferation in the Drosophila eye by Notch signaling. Dev Cell 8: 529–539. [DOI] [PubMed] [Google Scholar]
- 37.Jiang J (2002) Degrading Ci: who is Cul-pable? Genes Dev 16: 2315–2321. [DOI] [PubMed] [Google Scholar]
- 38.Chen Y, Cardinaux JR, Goodman RH, Smolik SM (1999) Mutants of cubitus interruptus that are independent of PKA regulation are independent of hedgehog signaling. Development 126: 3607–3616. [DOI] [PubMed] [Google Scholar]
- 39.Price MA, Kalderon D (1999) Proteolysis of cubitus interruptus in Drosophila requires phosphorylation by protein kinase A. Development 126: 4331–4339. [DOI] [PubMed] [Google Scholar]
- 40.Wang G, Wang B, Jiang J (1999) Protein kinase A antagonizes Hedgehog signaling by regulating both the activator and repressor forms of Cubitus interruptus. Genes Dev 13: 2828–2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alexandre C, Jacinto A, Ingham PW (1996) Transcriptional activation of hedgehog target genes in Drosophila is mediated directly by the cubitus interruptus protein, a member of the GLI family of zinc finger DNA-binding proteins. Genes Dev 10: 2003–2013. [DOI] [PubMed] [Google Scholar]
- 42.Dominguez M, Brunner M, Hafen E, Basler K (1996) Sending and receiving the hedgehog signal: control by the Drosophila Gli protein Cubitus interruptus. Science 272: 1621–1625. [DOI] [PubMed] [Google Scholar]
- 43.Aza-Blanc P, Ramirez-Weber FA, Laget MP, Schwartz C, Kornberg TB (1997) Proteolysis that is inhibited by hedgehog targets Cubitus interruptus protein to the nucleus and converts it to a repressor. Cell 89: 1043–1053. [DOI] [PubMed] [Google Scholar]
- 44.Methot N, Basler K (1999) Hedgehog controls limb development by regulating the activities of distinct transcriptional activator and repressor forms of Cubitus interruptus. Cell 96: 819–831. [DOI] [PubMed] [Google Scholar]
- 45.Kent D, Bush EW, Hooper JE (2006) Roadkill attenuates Hedgehog responses through degradation of Cubitus interruptus. Development 133: 2001–2010. [DOI] [PubMed] [Google Scholar]
- 46.Zhang Q, Zhang L, Wang B, Ou CY, Chien CT, et al. (2006) A hedgehog-induced BTB protein modulates hedgehog signaling by degrading Ci/Gli transcription factor. Dev Cell 10: 719–729. [DOI] [PubMed] [Google Scholar]
- 47.Ou CY, Wang CH, Jiang J, Chien CT (2007) Suppression of Hedgehog signaling by Cul3 ligases in proliferation control of retinal precursors. Dev Biol 308: 106–119. [DOI] [PubMed] [Google Scholar]
- 48.Baker NE, Bhattacharya A, Firth LC (2009) Regulation of Hh signal transduction as Drosophila eye differentiation progresses. Dev Biol 335: 356–366. 10.1016/j.ydbio.2009.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mardon G, Solomon NM, Rubin GM (1994) dachshund encodes a nuclear protein required for normal eye and leg development in Drosophila. Development 120: 3473–3486. [DOI] [PubMed] [Google Scholar]
- 50.Firth LC, Baker NE (2009) Retinal determination genes as targets and possible effectors of extracellular signals. Dev Biol 327: 366–375. 10.1016/j.ydbio.2008.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strutt DI, Mlodzik M (1995) Ommatidial polarity in the Drosophila eye is determined by the direction of furrow progression and local interactions. Development 121: 4247–4256. [DOI] [PubMed] [Google Scholar]
- 52.Strutt DI, Wiersdorff V, Mlodzik M (1995) Regulation of furrow progression in the Drosophila eye by cAMP-dependent protein kinase A. Nature 373: 705–709. [DOI] [PubMed] [Google Scholar]
- 53.Pan D, Rubin GM (1995) cAMP-dependent protein kinase and hedgehog act antagonistically in regulating decapentaplegic transcription in Drosophila imaginal discs. Cell 80: 543–552. [DOI] [PubMed] [Google Scholar]
- 54.Chanut F, Heberlein U (1995) Role of the morphogenetic furrow in establishing polarity in the Drosophila eye. Development 121: 4085–4094. [DOI] [PubMed] [Google Scholar]
- 55.Ma C, Moses K (1995) Wingless and patched are negative regulators of the morphogenetic furrow and can affect tissue polarity in the developing Drosophila compound eye. Development 121: 2279–2289. [DOI] [PubMed] [Google Scholar]
- 56.Wehrli M, Tomlinson A (1995) Epithelial planar polarity in the developing Drosophila eye. Development 121: 2451–2459. [DOI] [PubMed] [Google Scholar]
- 57.Ou CY, Lin YF, Chen YJ, Chien CT (2002) Distinct protein degradation mechanisms mediated by Cul1 and Cul3 controlling Ci stability in Drosophila eye development. Genes Dev 16: 2403–2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim SS, Zhang RG, Braunstein SE, Joachimiak A, Cvekl A, et al. (2002) Structure of the retinal determination protein Dachshund reveals a DNA binding motif. Structure 10: 787–795. [DOI] [PubMed] [Google Scholar]
- 59.Chen R, Amoui M, Zhang Z, Mardon G (1997) Dachshund and eyes absent proteins form a complex and function synergistically to induce ectopic eye development in Drosophila. Cell 91: 893–904. [DOI] [PubMed] [Google Scholar]
- 60.Zhou J, Wang C, Wang Z, Dampier W, Wu K, et al. (2010) Attenuation of Forkhead signaling by the retinal determination factor DACH1. Proc Natl Acad Sci U S A 107: 6864–6869. 10.1073/pnas.1002746107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Basler K, Hafen E (1989) Dynamics of Drosophila eye development and temporal requirements of sevenless expression. Development 107: 723–731. [DOI] [PubMed] [Google Scholar]
- 62.Baker NE, Yu S, Han D (1996) Evolution of proneural atonal expression during distinct regulatory phases in the developing Drosophila eye. Curr Biol 6: 1290–1301. [DOI] [PubMed] [Google Scholar]
- 63.Baonza A, Casci T, Freeman M (2001) A primary role for the epidermal growth factor receptor in ommatidial spacing in the Drosophila eye. Curr Biol 11: 396–404. [DOI] [PubMed] [Google Scholar]
- 64.Li Y, Baker NE (2001) Proneural enhancement by Notch overcomes Suppressor-of-Hairless repressor function in the developing Drosophila eye. Curr Biol 11: 330–338. [DOI] [PubMed] [Google Scholar]
- 65.Frankfort BJ, Mardon G (2002) R8 development in the Drosophila eye: a paradigm for neural selection and differentiation. Development 129: 1295–1306. [DOI] [PubMed] [Google Scholar]
- 66.Voas MG, Rebay I (2004) Signal integration during development: insights from the Drosophila eye. Dev Dyn 229: 162–175. [DOI] [PubMed] [Google Scholar]
- 67.Dominguez M, Wasserman JD, Freeman M (1998) Multiple functions of the EGF receptor in Drosophila eye development. Curr Biol 8: 1039–1048. [DOI] [PubMed] [Google Scholar]
- 68.Ohlmeyer JT, Kalderon D (1998) Hedgehog stimulates maturation of Cubitus interruptus into a labile transcriptional activator. Nature 396: 749–753. [DOI] [PubMed] [Google Scholar]
- 69.Bras-Pereira C, Casares F, Janody F (2015) The retinal determination gene Dachshund restricts cell proliferation by limiting the activity of the Homothorax-Yorkie complex. Development 142: 1470–1479. 10.1242/dev.113340 [DOI] [PubMed] [Google Scholar]
- 70.Li X, Perissi V, Liu F, Rose DW, Rosenfeld MG (2002) Tissue-specific regulation of retinal and pituitary precursor cell proliferation. Science 297: 1180–1183. [DOI] [PubMed] [Google Scholar]
- 71.Frith MC, Fu Y, Yu L, Chen JF, Hansen U, et al. (2004) Detection of functional DNA motifs via statistical over-representation. Nucleic Acids Res 32: 1372–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liang F, Lu Q, Sun S, Zhou J, Popov VM, et al. (2012) Increased expression of dachshund homolog 1 in ovarian cancer as a predictor for poor outcome. Int J Gynecol Cancer 22: 386–393. 10.1097/IGC.0b013e31824311e6 [DOI] [PubMed] [Google Scholar]
- 73.Yan W, Wu K, Herman JG, Brock MV, Fuks F, et al. (2013) Epigenetic regulation of DACH1, a novel Wnt signaling component in colorectal cancer. Epigenetics 8: 1373–1383. 10.4161/epi.26781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhu H, Wu K, Yan W, Hu L, Yuan J, et al. (2013) Epigenetic silencing of DACH1 induces loss of transforming growth factor-beta1 antiproliferative response in human hepatocellular carcinoma. Hepatology 58: 2012–2022. 10.1002/hep.26587 [DOI] [PubMed] [Google Scholar]
- 75.Yan W, Wu K, Herman JG, Brock MV, Zhou Y, et al. (2014) Epigenetic silencing of DACH1 induces the invasion and metastasis of gastric cancer by activating TGF-beta signalling. J Cell Mol Med 18: 2499–2511. 10.1111/jcmm.12325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pappu KS, Mardon G (2004) Genetic control of retinal specification and determination in Drosophila. Int J Dev Biol 48: 913–924. [DOI] [PubMed] [Google Scholar]
- 77.Pappu KS, Chen R, Middlebrooks BW, Woo C, Heberlein U, et al. (2003) Mechanism of hedgehog signaling during Drosophila eye development. Development 130: 3053–3062. [DOI] [PubMed] [Google Scholar]
- 78.Pignoni F, Hu B, Zavitz KH, Xiao J, Garrity PA, et al. (1997) The eye-specification proteins So and Eya form a complex and regulate multiple steps in Drosophila eye development. Cell 91: 881–891. [DOI] [PubMed] [Google Scholar]
- 79.Jiang J, Struhl G (1998) Regulation of the Hedgehog and Wingless signalling pathways by the F-box/WD40-repeat protein Slimb. Nature 391: 493–496. [DOI] [PubMed] [Google Scholar]
- 80.Tavsanli BC, Ostrin EJ, Burgess HK, Middlebrooks BW, Pham TA, et al. (2004) Structure-function analysis of the Drosophila retinal determination protein Dachshund. Dev Biol 272: 231–247. [DOI] [PubMed] [Google Scholar]
- 81.Zhou Q, Kalderon D (2011) Hedgehog activates fused through phosphorylation to elicit a full spectrum of pathway responses. Dev Cell 20: 802–814. 10.1016/j.devcel.2011.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Methot N, Basler K (2000) Suppressor of fused opposes hedgehog signal transduction by impeding nuclear accumulation of the activator form of Cubitus interruptus. Development 127: 4001–4010. [DOI] [PubMed] [Google Scholar]
- 83.Shi Q, Li S, Jia J, Jiang J (2011) The Hedgehog-induced Smoothened conformational switch assembles a signaling complex that activates Fused by promoting its dimerization and phosphorylation. Development 138: 4219–4231. 10.1242/dev.067959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tang CY, Sun YH (2002) Use of mini-white as a reporter gene to screen for GAL4 insertions with spatially restricted expression pattern in the developing eye in drosophila. Genesis 34: 39–45. [DOI] [PubMed] [Google Scholar]
- 85.Calleja M, Moreno E, Pelaz S, Morata G (1996) Visualization of gene expression in living adult Drosophila. Science 274: 252–255. [DOI] [PubMed] [Google Scholar]
- 86.Xu T, Rubin GM (1993) Analysis of genetic mosaics in developing and adult Drosophila tissues. Development 117: 1223–1237. [DOI] [PubMed] [Google Scholar]
- 87.Lee T, Luo L (1999) Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22: 451–461. [DOI] [PubMed] [Google Scholar]
- 88.Nolo R, Abbott LA, Bellen HJ (2000) Senseless, a Zn finger transcription factor, is necessary and sufficient for sensory organ development in Drosophila. Cell 102: 349–362. [DOI] [PubMed] [Google Scholar]
- 89.Charlton-Perkins M, Whitaker SL, Fei Y, Xie B, Li-Kroeger D, et al. (2011) Prospero and Pax2 combinatorially control neural cell fate decisions by modulating Ras- and Notch-dependent signaling. Neural Dev 6: 20 10.1186/1749-8104-6-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Barrio R, de Celis JF, Bolshakov S, Kafatos FC (1999) Identification of regulatory regions driving the expression of the Drosophila spalt complex at different developmental stages. Dev Biol 215: 33–47. [DOI] [PubMed] [Google Scholar]
- 91.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, et al. (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9: 676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Imrichova H, Hulselmans G, Atak ZK, Potier D, Aerts S (2015) i-cisTarget 2015 update: generalized cis-regulatory enrichment analysis in human, mouse and fly. Nucleic Acids Res 43: W57–64. 10.1093/nar/gkv395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dokucu ME, Zipursky SL, Cagan RL (1996) Atonal, rough and the resolution of proneural clusters in the developing Drosophila retina. Development 122: 4139–4147. [DOI] [PubMed] [Google Scholar]
- 94.Baker NE, Yu SY (1997) Proneural function of neurogenic genes in the developing Drosophila eye. Curr Biol 7: 122–132. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
All panels show standard confocal sections of L3 eye imaginal discs, containing (A-A´´,D-D´) dac3 or (B-B´,E-E´) smo3 single mutant clones or (C-C´,F-F´) smo3, dac3 double mutant clones marked by the presence of GFP (green in A, A´´,B,C,D,E,F) and outlined by dashed line (in A´, A´´,B´,C´,D´,E´,F´). Discs are stained with (A-B´) anti-E-Cad (magenta) or (C-C´) phalloidin (magenta) to outline cell shape or with (D-F’) anti-Elav (magenta) to label differentiating PRs. A´´ corresponds to a merge between the GFP signal in A and more basal sections. Note that while loss of smo function has little effect on MF formation (arrow in B´) and PR differentiation, loss of both smo and dac function completely disrupts MF formation (arrows in C´) and PR differentiation. Scale bars represent 50μm.
(TIF)
All panels show (A-D´) standard confocal sections or (E) standard brightfield image of L3 eye discs, containing dac3 mutant clones marked by the absence of GFP (green in A,B,C,D and brown in E) and outlined by yellow dashed lines (in A’,B´,C´,D´,E). Discs are stained with (A-A´) anti-CycB (magenta), (B-B´) anti-PH3 (magenta), (C-C´) anti-BrdU (magenta), (D-D´) anti-CycE (magenta) and (E) dap mRNA (blue). The positions of the FMW and SMW are indicated by white and yellow arrowheads, respectively. White arrows in B’ and C’ point to the delayed re-entrance in cell cycle posterior to the SMW. Solid lines in D’ delimit the high levels of CycE within the MF in wild type cells. Scale bars represent 50μm.
(TIF)
All panels show standard confocal sections of L3 eye imaginal discs, containing dac3 mutant clones marked by the absence of GFP (green in A,B) and outlined by dashed line in A´, A´´,B´. Discs are stained with anti-Ptc (red) and anti-Elav (blue). Plain lines in A´ and B´ indicate the MF position. Scale bars represent 50μm.
(TIF)
(A) Example gene (nej) with a Dac ChIP peak, 3kb upstream of the transcription start site. (B) Sequence logo of a candidate Dac motif found as the most enriched motif in the distal ChIP-peak. (C) Sequence logo of a Gli consensus binding site (annotated in the transfac database: transfac_pro-M01037) found significantly enriched in the set of distal ChIP-peaks when compared to random background sequences (p<0.001). (D) List of the proposed Dac targets, predicted by the presence of a nearby (<5kb from TSS or intronic) ChIP-seq peak, and the presence of the candidate Dac motif, as predicted by i-cisTarget [92].
(TIF)
(A-C´) Standard confocal sections of L3 eye imaginal discs (B-B´) wild type or (A-A´´´,C-C´) containing dac3 clones labeled with GFP (green in A,C) and outlined by dashed lines in A´-A´´´,C´, stained with (A-A´´´) anti-Elav (blue in A,A´, white in A´´´) and anti—β-Galactosidase to reveal dppZ (red in A-A´ and white in A´´) or (B-B´) anti-β-Galactosidase to reveal ciZ (green) and DAPI (magenta in B) or (C-C´) anti-β-Galactosidase to reveal ciZ (red in C and white in C´) and anti-ELAV (blue in C). (D-F,H-J) Standard confocal sections of L3 wing imaginal discs with anterior to the left and dorsal up in which ptc-Gal4 drives the expression of UAS-mCherry (green in D,E,F,H,I,J) and (H,I-J) UAS-HA::dac. Discs are stained with anti-Dac (red in D,H) and anti-β-Galactosidase to reveal ciZ (blue in D,H and magenta in E-F,I-J). E-E´ and I-I´ are magnifications of the dashed white square in D and H, respectively. F and J correspond to the area delimited by white dashed lines in E and I, respectively. The dashed yellow lines in E´ and I´ outline the AP compartment boundaries. (G,K) Profiles of the mCherry (green lines) and ciZ (magenta lines) intensity signals across the AP axis in F and J, respectively. Scale bars represent 50μm.
(TIF)
Data Availability Statement
Access to the whole Dac-ChIP Seq dataset can be found in the GEO database with the accession number GSE82151 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE82151) as mentioned in the Material and Methods section.