Abstract
One of the most efficient mechanisms to keep animal lineages separate is a difference in ploidy level (number of whole genome copies), since hybrid offspring from parents with different ploidy level are functionally sterile. In the freshwater fish family Botiidae, ploidy difference has been held responsible for the separation of its two subfamilies, the evolutionary tetraploid Botiinae and the diploid Leptobotiinae. Diploid and tetraploid species coexist in the upper Yangtze, the Pearl River and the Red River basins in China. Interestingly, the species ‘Botia’ zebra from the Pearl River basin combines a number of morphological characters that otherwise are found in the diploid genus Leptobotia with morphological characters of the tetraploid genus Sinibotia, therefore the aim of the present study is to test weather ‘B.’ zebra is the result of a hybridisation event between species from different subfamilies with different ploidy level. A closer morphological examination indeed demonstrates a high similarity of ‘B.’ zebra to two co-occurring species, the diploid Leptobotia guilinensis and the tetraploid Sinibotia pulchra. These two species thus could have been the potential parental species in case of a hybrid origin of ‘B.’ zebra. The morphologic analysis further reveals that ‘B.’ zebra bears even the diagnostic characters of the genera Leptobotia (Leptobotiinae) and Sinibotia (Botiinae). In contrast, a comparison of six allozyme loci between ‘B.’ zebra, L. guilinensis and S. pulchra showed only similarities between ‘B.’ zebra and S. pulchra, not between ‘B.’ zebra and L. guilinensis. Six specimens of ‘B.’ zebra that were cytogenetically analysed were tetraploid with 4n = 100. The composition of the karyotype (18% metacentric, 18% submetacentric, 36% subtelocentric and 28% acrocentric chromosomes) differs from those of L. guilinensis (12%, 24%, 20% and 44%) and S. pulchra (20%, 26%, 28% and 26%), and cannot be obtained by any combination of genomes from L. guilinensis and S. pulchra. Phylogenetic reconstructions based on sequence data of the mitochondrial cytochrome b gene and the nuclear RAG-1 gene invariably places ‘Botia’ zebra as sister species to S. pulchra, while L. guilinensis is only distantly related. The presented combination of genetic data demonstrates that ‘B.’ zebra is not the result of a hybridisation, but a species of tetraploid genus Sinibotia with a striking morphological evolution towards an enormous similarity with a co-occurring, but not directly related species. The complete lack of knowledge of the ecology of these species, their main predators or their ecological interactions hampers any conclusion regarding the evolutionary advantage of such adaptation.
Introduction
One of the most efficient barriers for horizontal gene flow between vertebrate animals is a difference in ploidy level [1,2]. While it might not prevent an original hybridisation event and in many cases not the viability of the F1-offspring, it generally terminates the reproduction line of such hybrids by sterility of the offspring [3,4,5]. In some exceptional cases, the resulting hybrids can make it through with clonal and/or asexual reproduction [1], but due to the absence of gene flow and recombination, offspring of such lineages resemble F1 hybrid individuals and such lineages often are not long lasting. This general rule has been observed in plants as well as in animals, and exceptional cases are very rare, especially among animals. Therefore, any evolutionary successful case of a hybridisation between parental species that differ in ploidy level would provide an interesting model to study the limits of polyploidy as barrier for horizontal gene flow.
Freshwater fishes of the family Botiidae (Cobitoidea: Cypriniformes) are widespread across East, Southeast and South Asia [6,7]. Many species are valued as ornamental fishes worldwide and as tasty food fishes in the area of occurrence. The monophyly of the family has been demonstrated by morphological as well as genetic data [7–11]; and phylogenetic reconstructions of the family revealed two major, long-time separated lineages, which are referred to as subfamilies Leptobotiinae and Botiinae [11,12]. The most remarkable difference between the two subfamilies comes from cytogenetics: all studied Leptobotiinae are diploid with a chromosome number of 2n = 50, while all Botiinae are tetraploid with 4n = 98–100 [12,13]. It has been hypothesised [12] that the difference in ploidy level has played an important role in the separation of the two lineages, since it represents an efficient barrier for hybridisation between the lineages. Both subfamilies have a similar number of species, which was used to claim that there is no obvious difference in the evolutionary success of diploid or tetraploid animals [14]. Leptobotiinae occur in the northern half of the total distribution area (north of the Mekong basin—China, Japan, eastern Russia, northern Vietnam), while most Botiinae live in the southern half of the total distribution area (Mekong and areas south and west of Mekong—from Pakistan to Laos, Malay Peninsula, Indonesia) [7]. However, Leptobotiinae and Botiinae co-occur in the upper Yangtze, the Pearl and the Red River basins, where the genus Sinibotia (belonging to Botiinae) is distributed with five recognised species in the area that otherwise is inhabited by Leptobotiinae (a sixth species of Sinibotia in the upper Mekong lives outside the range of Leptobotiinae) [7,15].
At least seven species of the genera Leptobotia, Parabotia and Sinibotia occur in the River Li, a northern tributary of the River Xi, Pearl River basin, in southern China [16,17], with two of them being endemic to this river: Leptobotia guilinensis Chen, 1980 and ‘Botia’ zebra Wu, 1939. Since the latter is bearing the diagnostic character of the genus Leptobotia, a simple suborbital spine (versus bifid in all other genera of Botiidae), and generally shows a close similarity to the sympatric Leptobotia guilinensis, ‘Botia’ zebra was placed into Leptobotia [16]. However, in a phylogenetic analysis basing on the mitochondrial cytochrome b gene, ‘B’. zebra was found to be more closely related to the genus Sinibotia, especially to a species that occurs in the River Li, S. pulchra [18]. One of the possible explanations for a strong discrepancy between morphological and mitochondrial characters, respectively, is mitochondrial introgression, a process where an initial hybridisation is followed by repeated back-crossing events with the paternal species; leading to a morphology that is closer to the paternal species, but a mitochondrial genome that is close to the maternal species. In ‘Botia’ zebra the morphology is similar to Leptobotia guilinensis, but the mitochondrial genome close to Sinibotia pulchra and all three species co-occur in the upper River Li (Fig 1). We therefore hypothesise that the evolutionary history of ‘Botia’ zebra included a hybridisation event between L. guilinensis and S. pulchra and test this hypothesis in the present study. Such a hybridisation event between a diploid and a tetraploid species would refute the general assumption that differences in ploidy level represent an efficient barrier against hybridisation and would be of general interest for evolutionary biology.
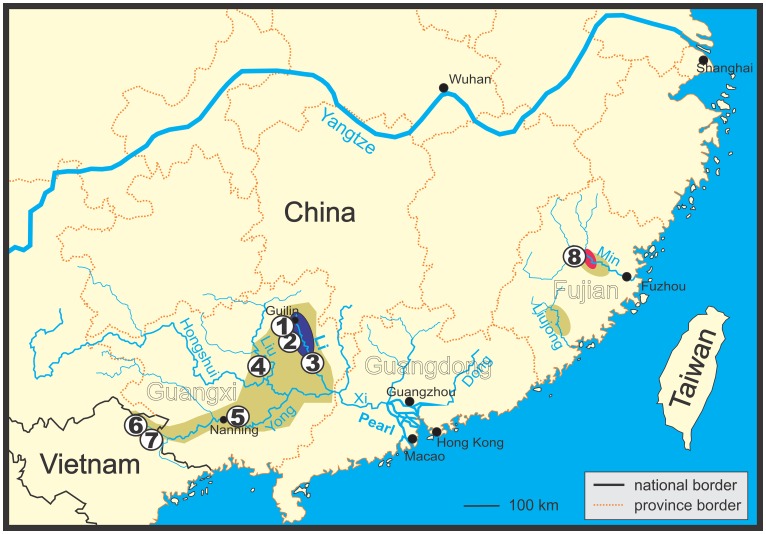
Fig 1. Map of Pearl River basin, Liujong River and Min River in southern China.
Distribution areas of botiid species indicated as follows: Green: S. pulchra, violet: joint distribution area of L. guilinensis and ‘B.’ zebra, pink: new record of ‘B.’ zebra. Circles with numbers indicate geographical origin of analysed samples. Numbers correspond to locality numbers in Table 1.
In the present study, we compare L. guilinensis, S. pulchra and ‘Botia’ zebra using morphologic, cytogenetic, allozyme variability as well as mitochondrial and nuclear DNA sequence characters to test if the later reveals any trace of a past hybridisation between the first two species.
Material & Methods
Specimens
Live individuals of L. guilinensis (eight individuals), S. pulchra (two individuals) and S. zebra (six individuals) were obtained together in one mixed group from an import for the ornamental fish trade (Aquarium Glaser, 63110 Rodgau, Germany). Live fishes were kept in the fish housing facilities in Institute of Animal Physiology and Genetics, 277 21 Liběchov, Czech Republic in 60 l tanks with flow-through water of 22°C at a light;dark cycle of 10:14 h. Fishes were fed ad libitum with life Tubifex worms. Ethanol or formalin fixed specimens were obtained from local food markets in Guilin (25°16’N, 110°17’E), Mengshan (24°12’N, 110°31’E), Fuzhou (26°04’N, 119°18’E) and Nanning (22°48’N, 108°21’E) in the provinces Guangxi and Fuxien in southern China. A total number of 108 individuals of the three species from 9 localities across the whole distribution area of the species has been analysed (Table 1). Additional 33 specimens of 26 other species as comparative material were obtained from the ornamental fish trade (AquaGlobal, 16356 Werneuchen, Germany). Vouchers are deposited in the collection of the Laboratory of Fish Genetics, IAPG AS CR, Liběchov. All experimental procedures involving fishes during this study were approved by the Institutional Animal Care and Use Committee of the Institute of Animal Physiology and Genetics of the Academy of Sciences of the Czech Republic, according with directives from the State Veterinary Administration of the Czech Republic, permit number 155/2012, and by permit number CZ 02386 from the Ministry of Agriculture of the Czech Republic.
Table 1. Number of specimens and geographical origin of Leptobotia guilinensis, ‘B.’ zebra and Sinibotia pulchra analysed in the present study.
| Locality number | Species | n | Sample ID | Locality | River | Drainage | Country | Province |
|---|---|---|---|---|---|---|---|---|
| 1 | L. guilinensis | 23 | A8861-8883 | market in | upper Li | Pearl | China | Guangxi |
| L. guilinensis | 3 | A8901-8903 | Guilin | River | River | |||
| L. guilinensis | 2 | A1798-1799 | (漓江) | (珠江) | ||||
| B. zebra | 2 | A8905-8906 | ||||||
| S. pulchra | 3 | A8889-8891 | ||||||
| S. pulchra | 1 | A8900 | ||||||
| S. pulchra | 6 | A1782-1787 | ||||||
| 2 | L. guilinensis | 10 | A5267-5276 | ornamental | upper Li | Pearl | China | Guangxi |
| B. zebra | 6 | A5277-5282 | fish import | River (漓江) | River | |||
| S. pulchra | 2 | A5286-5287 | (珠江) | |||||
| 3 | B. zebra | 5 | A8604-8608 | market in Mengshan | upper Li River (漓江) | Pearl River (珠江) | China | Guangxi |
| 4 | S. pulchra | 1 | A8993 | Liu River above Yizhou | middle Liu River (柳江) | Pearl River (珠江) | China | Guangxi |
| 5 | S. pulchra | 5 | A9102-9106 | market in Nanning | Yong River (邕江) | Pearl River (珠江) | China | Guangxi |
| 6 | S. pulchra | 1 | A8395 | Bang Giang at Cao Bang city | Bang Giang (Sông Bằng) | Pearl River (珠江) | Vietnam | Cao Bang |
| 7 | S. pulchra | 4 | A8397-8400 | stream in Hoa An district | Bang Giang (Sông Bằng) | Pearl River (珠江) | Vietnam | Cao Bang |
| 8 | B. zebra | 1 | A8614 | unknown | Min River | Min River | China | Fujian |
| S. pulchra | 24 | A8615-8638 | (閩江) | (閩江) | ||||
| 9 | L. guilinensis | 3 | A0205-0208 | ornamental | Details | unknown | ||
| S. pulchra | 2 | A3681-3682 | fish import | |||||
| S. pulchra | 4 | A0015-0018 |
All listed specimens were used in the morphological analyses, but not all for the other methods. Locality number matches the numbers given in Fig 1; n = number of specimens; sample ID is the individual number given to each specimen (collection numbers IAPG).
Morphology
Morphological measurements were taken from 26 specimens of L. guilinensis, S. pulchra and S. zebra with digital callipers point-to-point according [19]. Important morphologic characters and pigmentation were estimated from nearly all specimens either directly in the case of fixed specimens or from photos taken of live specimens. Preparations of suborbital spines of nine specimens were carried out under an Olympus SZX7 stereomicroscope equipped with a u-Eye camera.
Chromosome analysis
Mitotic chromosomes were obtained from regenerated fin tissue as described by [20,21] with slight modifications. Briefly, the posterior margin of the caudal fin was cut off and three weeks later, the regenerated tissue of the fin was collected to be incubated in Ringer solution with 0,025% colchicine for about two hours at room temperature. Cells were fixed in a mixture of methanol and acetic acid (3:1) at 4°C for 25 min. This step was repeated three times. The fixed tissue was minced in 50% acetic acid and drops of the resulting suspension were placed on pre-heated slides (50°C) and sucked back after 20 sec. The slide was dried at room temperature and stained for 10 min in 5% Giemsa solution (pH 6.8) (Merck, Darmstadt, Germany) before examination of metaphase plates with an Olympus AX70 light microscope. From 17 live individuals that were available for the analyses, results with satisfying quality were obtained from nine individuals (five L. guilinensis, two S. pulchra, two ‘B.’ zebra). The number of chromosomes of at least 20 metaphase plates per individual was counted. Chromosome morphology was classified as m—metacentric, sm—submetacentric, st—subtelocentric, a—acrocentric [22].
Allozyme analysis
Fin tissue was homogenised with an equal amount of buffer (0.1 mol/l Tris-HCl pH 8.5) and centrifuged for clarifying. All manipulations with tissue were carried out on ice. Electrophoresis on starch gel was carried out in a refrigerator. Six allozyme loci (glucosephosphate isomerase Gpi-A, aspartate amino transferase s-Aat, malate dehydrogenase s-Mdh A, lactate dehydrogenase Ldh A and Ldh B, phosphoglucomutase Pgm) were stained [23,24]. Altogether 18 individuals were studied. Loci Gpi-A and Pgm were analysed in three and two buffer systems, respectively (F [25], MC2 [26], V [27]).
DNA sequence analyses
Genomic DNA was isolated from fin tissue samples using the DNeasy Tissue kit (Qiagen, Hilden, Germany) according to manufacturer’s instructions. The mitochondrial cytochrome b (cyt b) was amplified and sequenced using the primers Glu-L.Ca14337-14359: 5’- GAA GAA CCA CCG TTG TTA TTC AA– 3’ and Thr-H.Ca15568-15548: 5’- ACC TCC RAT CTY CGG ATT ACA– 3’ [12]. An approximately 970 bp long portion of RAG-1 was amplified using the primers RAG-1F (5’-AGCTGTAGTCAGTAYCACAARATG-3’ [28]) and RAG-RV1 (TCCTGRAAGATYTTGTAGAA-3’ [10]) or RAG-8R (5’-CGC CAC ACA GGY TTC ATC T-3’ [28]. Same primers were used also for sequencing reactions. PCR amplifications were performed in 25 μl reaction volumes of 10 mM Tris-HCl, 50 mM (NH4)2SO4, 0.1% of Triton X-100, 1.5 mM MgCl2, 2 mM TMA oxalate (PCR enhancer), containing 5 nmol of each nucleotide, 1.25 U of Taq polymerase (all chemicals Top-Bio, Prague, Czech Republic) and 12.5 pmol of each primer.
The PCR reaction profile (MJ Research thermocycler) included 5 min of initial denaturation at 95°C, touch-down profile of 1 min at 94°C, 1 min 30 s at 60–55°C (1°C/cycle) and 2 min at 72°C, followed by 30 cycles with annealing temperature held at 54°C. The reaction was completed by final extension at 72°C for 7 min.
PCR products were purified by QIAquick PCR Purification Kit (Qiagen). Forward and reverse sequencing reactions were performed with BigDye™ Terminator Cycle Sequencing Kit v.1.1 (PE Applied Biosystems, Darmstadt, Germany) according to manufacturer’s instructions and products purified with DyeEx Spin Kit (Qiagen). Sequencing was performed on ABI Prism 3130 (Applied Biosystems).
Chromatograms were assembled using SeqMan Pro 10.1.2 of the LaseGene software package (DNASTAR). The sequences were aligned and manually revised in BioEdit 7.0.5.3 [29] and evaluated based on their amino acid translation.
The newly obtained data were deposited in GenBank under the accession numbers KU517025-KU517132.
We have analysed molecular data from altogether 102 individuals of Botiidae representing 29 species currently considered as valid. Based on the former studies on Cobitoidea [10,30], we have selected Gyrinocheilus aymonieri as outgroup.
The molecular datasets were analysed using the Bayesian inference in MrBayes 3.2.2 [31]. The two genes were analysed separately with the aim to see potential discrepancies between mitochondrial and nuclear markers. The datasets were partitioned into codon positions. Prior to the analyses, the MEGA 5.10 software [32] was used to estimate the most suited model for each gene partition under the Bayesian information criterion (BIC). The Bayesian analyses were performed in two independent runs of 5 million generations, each using six Markov Chains, starting with random trees and sampling frequency set to 100 generations. The parameter settings corresponded to the best-fit models. The log-likelihood score distribution was examined in order to assess if stationarity was reached. The first 5000 saved trees were discarded as the burn-in and a 50% majority rule consensus of the remaining trees was computed. Statistical support of clades was assessed by posterior probabilities.
Results
Morphology
Morphometry
- In 13 out of 33 morphometric and meristic characters there was no overlap of measurements between L. guilinensis and S. pulchra (Category A, Table 2), while in further 12 characters, the overlap was small (Category B). In the remaining eight characters the overlap was large (Category C); therefore these characters were unsuited to evaluate a morphological similarity between ‘B.’ zebra and the two potential parental species. However, in one of these ‘uninformative’ characters in Category C (Number of branched dorsal-fin rays), seven out of eight specimens of ‘B.’ zebra showed a character state that was observed in neither L. guilinensis nor S. pulchra, indicating an autapomorphy of ‘B.’ zebra. When comparing ‘B.’ zebra with L. guilinensis and S. pulchra, it shared the range of measurements with S. pulchra in two characters of Category A and in two characters of Category B, with L. guilinensis in six characters of Category A and in eight characters of Category B, while its range was intermediate between the two species in five characters of Category A and in two characters of Category B.
Table 2. Morphometric comparison of Leptobotia guilinensis, Sinibotia pulchra and ‘Botia’ zebra.
| Leptobotia guilinensis | ‘Botia’ zebra | Siibotia pulchra | Comparison | |
|---|---|---|---|---|
| n = 10 | n = 8 | n = 10 | ||
| A. Characters without overlap between Leptobotia guilinensis and Sinibotia pulchra | ||||
| Pre-pelvic length | 51–54 | 56–59 | 55–59 | zebra = pulchra |
| Preanal length | 74–77 | 78–80 | 78–80 | zebra = pulchra |
| Dorsal head length | 16–20 | 20–22 | 21–24 | zebra intermediate |
| Snout length | 6–8 | 10–11 | 12–14 | zebra intermediate |
| Pre-anus length | 63–71 | 70–73 | 73–76 | zebra intermediate |
| Lateral head length | 21–24 | 24–26 | 27–30 | zebra intermediate |
| Head depth at eye | 8–9 | 10–11 | 11–12 | zebra intermediate |
| Head depth at nape | 11–13 | 12–14 | 15–16 | zebra = guilinensis |
| Maximum body depth | 11–16 | 13–17 | 19–23 | zebra = guilinensis |
| Body depth at dorsal origin | 11–17 | 13–17 | 18–23 | zebra = guilinensis |
| Maximum head width | 8–10 | 8–10 | 11–13 | zebra = guilinensis |
| Head width at nares | 4–5 | 4–6 | 6–9 | zebra = guilinensis |
| Body width at anal origin | 4–7 | 5–6 | 8–10 | zebra = guilinensis |
| B. Characters with slight overlap between Leptobotia guilinensis and Sinibotia pulchra | ||||
| Predorsal length | 49–58 | 55–60 | 55–62 | zebra = pulchra |
| Number of pectoral-fin rays | 11–13 | 13–15 | 13–15 | zebra = pulchra |
| Interorbital width | 3–4 | 4–4 | 4–6 | zebra intermediate |
| Length of caudal peduncle | 14–18 | 13–16 | 12–14 | zebra intermediate |
| Length of upper caudal lobe | 16–21 | 18–20 | 20–25 | zebra = guilinensis |
| Length of pectoral fin | 11–14 | 12–14 | 14–19 | zebra = guilinensis |
| Length of lower caudal lobe | 18–21 | 18–21 | 20–26 | zebra = guilinensis |
| Body width at dorsal origin | 6–10 | 6–9 | 10–15 | zebra = guilinensis |
| Depth of caudal peduncle | 10–13 | 10–13 | 13–14 | zebra = guilinensis |
| Length of pelvic fin | 10–12 | 10–11 | 12–15 | zebra = guilinensis |
| Length median caudal rays | 7–10 | 7–9 | 9–15 | zebra = guilinensis |
| Total length | 116–121 | 116–120 | 119–127 | zebra = guilinensis |
| C. Characters with broad overlap between Leptobotia guilinensis and Sinibotia pulchra | ||||
| Eye diameter | 2–3 | 2–3 | 3–3 | |
| Depth of anal fin | 13–15 | 12–15 | 14–17 | |
| Postorbital length | 12–14 | 13–14 | 13–15 | |
| Height of dorsal fin | 11–16 | 11–14 | 12–18 | |
| Branched dorsal-fin rays | 8 ½ | 7(8) ½ | 8 ½ | zebra speciality |
| Branched caudal-fin rays | 9+8 | 9+8 | 9+8 | |
| Branched anal-fin rays | 5 | 5 | 5 | |
| Number of pelvic-fin rays | 8 | 8 | 8 | |
Values give range as % of standard length. Under ‘Comparison’ is indicated if ‘Botia’ zebra has values like one of the potential parental species or if it is intermediate.
Pigmentation pattern
- The pigmentation pattern of S. pulchra is much like that of all other members of the genus Sinibotia: Broad dark brown bars run from one body side across the back to the other side, reaching nearly always below lateral midline and regularly to level of pelvic fin origin (Fig 2). In small and medium sized individuals 6–10 bars are present, much broader than interspaces, but in larger individuals each bar might split into two. On the dorsal side of the head run two dark stripes from the snout to the neck and one on each side of the head from the snout through the eye. Between the dark stripes are two prominent light stripes, a long one from the snout to the end of the operculum and a short along the dorsal midline of the head. In Leptobotia guilinensis¸ body and head are homogenously light to dark brow with a lighter belly. Prominent light blotches are present along the dorsal midline, but often only visible behind the base of the dorsal fin. Dark saddles are sometimes visible between the light blotches, but usually too faint to figure out the precise number and outline. A thin black stripe runs from the snout to the eye, but no light stripes are present. In ‘B.’ zebra the body sides are uniformly brown like in L. guilinensis, but usually in lighter brown. On the back, faint saddles are sometimes visible, often hard to see, never reaching to lateral midline. In some specimens the saddles are present only in the anterior part of the body, but if present along whole back their number is higher than ten. Between the saddles are light blotches, very similar to the light blotches in L. guilinensis, often merging into a line in the anteriormost part of the dorsum. On the head are dark stripes from the snout to the neck and from the snout through the eye two and prominent light stripes between them as described for S. pulchra. In general, ‘B.’ zebra combines the head pigmentation of S. pulchra with the body pigmentation of L. guilinensis.
Fig 2. Habitus of Leptobotia guilinensis, Sinibotia pulchra and ’B.’ zebra.
Specimens of (a) Leptobotia guilinensis, (b) ‘Botia’ zebra and (c) Sinibotia pulchra in lateral and dorsal view. ‘Botia’ zebra shares the general body shape and pigmentation pattern, the smaller head size and the shorter, round fins with L. guilinensis, but the pigmentation of the head with Sinibotia pulchra.
Suborbital spine
–The suborbital spine is an erectable spine formed by the lateral ethmoid bone and located in a skin pocket below each eye. It is present in all members of the family Botiidae as well as in the distantly related families Cobitidae and Serpenticobitidae and its shape is of taxonomical value. In all species of Leptobotia, including L. guilinensis, this spine is simple, meaning it has a single branch and tip [7]. In all other Botiidae, including S. pulchra, the spine is double, meaning it has a main and a side branch and two tips [7]. In ‘B.’ zebra, the spine turned out also to be simple, like in Leptobotia (Fig 3).
Fig 3. Suborbital spines of Leptobotia guilinensis, Sinibotia pulchra and ’B.’ zebra.
Right suborbital spine of (a) Sinibotia pulchra (A1783, 70.5 mm SL), (b) ‘Botia’ zebra (A8607, 61.9 mm SL) and (c) Leptobotia guilinensis (A8573, 70.6 mm SL) in dorsal view. The spine bears a dorsal branch in S. pulchra, but this branch is missing in L. guilinensis and ‘B.’ zebra. A simple (= unbranched) suborbital spine is the diagnostic character for the genus Leptobotia. Scale bar is 1 mm.
Mental lobes
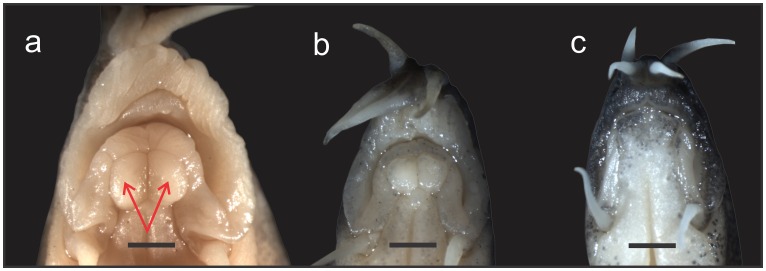
–In many species of Botiidae the lower lip develops two median extensions, called mental lobes, and presence and shape of these extensions are important taxonomic characters [7]. In all species of Sinibotia the extensions are present, large and of oval or kidney-like shape [7,15]. This shape of the mental lobes is considered a diagnostic character for the genus Sinibotia [7]. In all analysed specimens of S. pulchra the mental lobes were present, large and had the shape characteristic for Sinibotia, while in all analysed specimens of L. guilinensis no mental lobes were present. In all analysed specimens of ‘B.’ zebra mental lobes were present, large and had the shape characteristic for Sinibotia (Fig 4).
Fig 4. Mouthes of Leptobotia guilinensis, Sinibotia pulchra and ’B.’ zebra.
Mouth of a) Sinibotia pulchra, A 9102, 73.0 mm SL; b) ‘B.’ zebra, A 8904, 63.1 mm SL and c) Leptobotia guilinensis, A8868, 71.6 mm SL in ventral view. The presence of two prominent buttons on the lower lip (arrows in S. pulchra) are a diagnostic character of the genus Sinibotia. Scale bar is 1 mm.
The literature names additional characters to distinguish between Leptobotia and Sinibotia, namely the presence of scales on the cheeks and of a pario-frontal fontanelle in Leptobotia (vs. both characters absent in Sinibotia) [7,33,34]. Since both turned out to be absent in five dissected specimens of L. guilinensis, these characters are not truly diagnostic and were unsuited for the comparison in the given case.
Chromosome analysis
Metaphases of suited quality for further analyses were obtained from five individuals of L. guilinensis, two ‘B’ zebra and two S. pulchra. The diploid chromosome number of all analysed L. guilinensis was 2n = 50, proving the diploid status of these individuals, while all individuals of ‘B’ zebra and S. pulchra were tetraploid with a chromosome number of 4n = 100 (Table 3, Fig 5). Karyotypes of all analysed species were composed of comparatively small chromosomes, slightly decreasing in size. Especially in the tetraploid species chromosomes were generally very small, with their centromere positions gradually ranging from median to nearly terminal making the borderlines between formal chromosomal categories questionable in a small subset of chromosomal pairs.
Table 3. Chromosme numbers and karyotype composition of Leptobotia guilinensis, Sinibotia pulchra and ’B.’ zebra.
| n | m | sm | st | a | |
|---|---|---|---|---|---|
| Leptobotia guilinensis | 50 | 6 | 12 | 10 | 22 |
| Sinibotia pulchra | 100 | 20 | 26 | 28 | 26 |
| ‘Botia’ zebra | 100 | 18 | 18 | 36 | 28 |
Chromosomal characteristics of Sinibotia pulchra, ‘Botia’ zebra and Leptobotia guilinensis from the upper Li River (Pearl River basin) including diploid chromosome number (2n) and karyotype description.
Fig 5. Karyotypes of Leptobotia guilinensis, Sinibotia pulchra and ’B.’ zebra.
Karyotypes of the diploid species Leptobotia guilinensis (a) and the tetraploid species ‘Botia’ zebra (b) and Sinibotia pulchra (c). Bar = 10 μm.
Allozyme analysis
In three (s-Aat, Ldh A, Ldh B) of the six analysed loci alleles were shared between the three analysed taxa and therefore were not informative for the given study. In all informative loci (Gpi-A, s-Mdh A, Pgm), S. pulchra shared alleles with ‘B’. zebra, but both did not share alleles with L. guilinensis (Table 4). Therefore the allozyme data suggest a high similarity between S. pulchra and ‘B.” zebra, while L. guilinensis appears to be more distantly related.
Table 4. Allozymes of Leptobotia guilinensis, Sinibotia pulchra and ’B.’ zebra.
| Species | n | Gpi-A | Gpi A | Gpi A | s-Aat | s-Mdh A | Ldh-A | Ldh-B | Pgm | Pgm |
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | V | MC 2 | F | MC 2 | MC 2 | V | V | V | F | |
| Sinibotia pulchra | 4 | 055 | 037 | 039 | 096 | A, C | 030, 136 | 053, 067 | 083 | 080 |
| ‘Botia’ zebra | 6 | 055 | 037 | 039 | 096 | A | 030 | 053, 067 | 083 | 080 |
| Leptobotia guilinensis | 8 | 065, 070,083, 099 | 050, 064,078, 095 | 060, 078, 096 | 084, 096 | B | 136 | 053, 067 | 107 | 100 |
Presence of six allozymes in Sinibotia pulchra, ‘Botia’ zebra and Leptobotia guilinensis.
DNA sequence analysis
Table 5 summarises the species and individuals analysed in this study including the novel sequences generated as well as those that were obtained from GenBank.
Table 5. Specimens of Botiidae used in the DNA analyses.
Species, number code, and Genbank accession numbers of Botiidae used in the DNA sequence analyses, cytogenetic analysis and allozyme analysis. Individuals with GenBank assession number have been included in the DNA sequence analyses, those marked a were used in allozyme analyses and those marked with k were karyotyped.
Altogether we have analysed 102 specimens of Botiidae including 49 and 59 novel sequences of cyt b (1121 bp) and RAG1 (971 bp), respectively. Into the cytochrome b dataset, 14 sequences of L. guilinensis, 19 sequences of S. pulchra and 12 sequences of ‘B.’ zebra were included; in the RAG dataset it were 14, 10 and six sequences, respectively.
The models selected for each partition (codon position) based on BIC score were following: TN93+G+I, HKY+G and GTR+G for the 1st, 2nd and 3rd codon positions of cyt b, respectively, and HKY+G, JC+I and JC for the 1st, 2nd and 3rd codon positions of RAG 1, respectively. Those were taken into account for the subsequent Bayesian analyses.
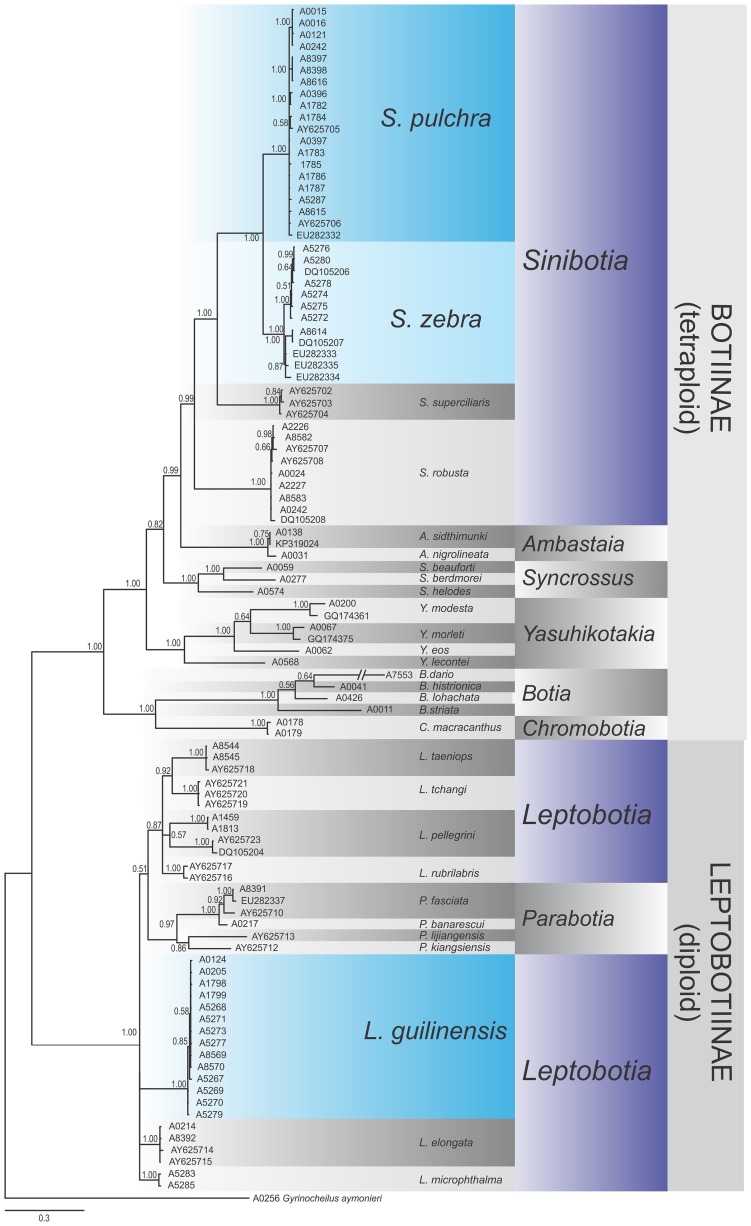
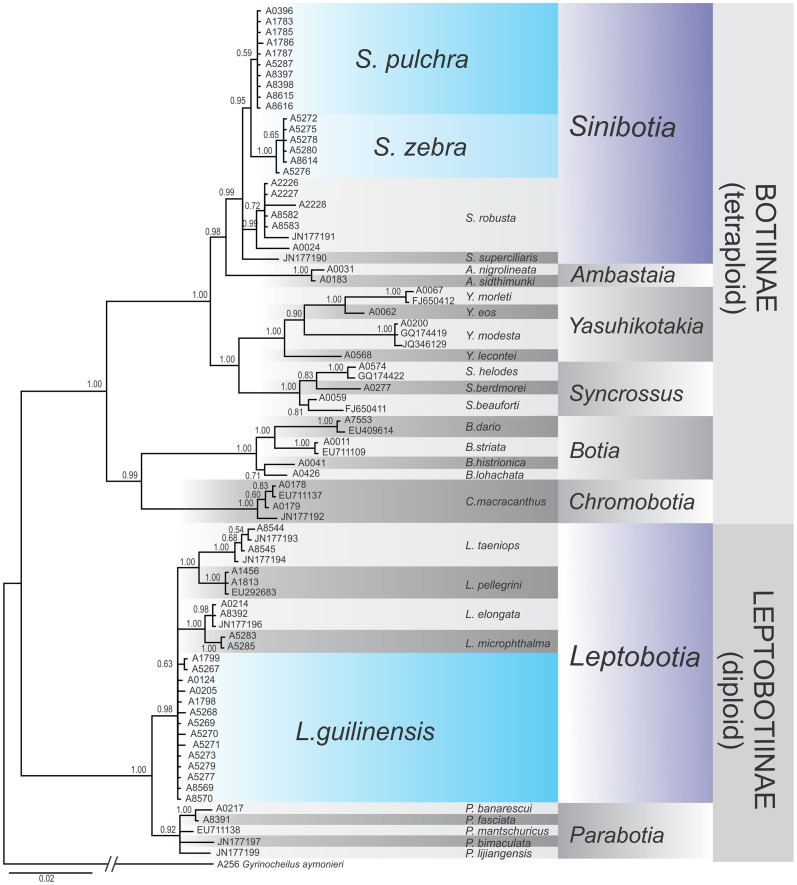
Phylogenetic reconstructions of both analysed genes provide generally congruent genealogies: the major split within Botiidae separated the diploid subfamily Leptobotiinae from the tetraploid subfamily Botiinae (Figs 6 and 7). Both datasets identified all described genera as monophyletic lineages with high statistic support except Leptobotia in the RAG dataset. These observations are well in agreement with former observations [11,12]. In general, the slower evolving RAG gene brought a better resolution at the older genealogic events, while the faster evolving cytochrome b gene had a better resolution around the tips of the trees, which is a well-know characteristics of these two genes. No discrepancies that would indicate a potential hybridisation event were detected.
Fig 6. Phylogenetic relationships of Botiidae as revealed by the Cytochrome b dataset.
Phylogenetic relationships among freshwater fishes of the family Botiidae as revealed by a Bayesian analyses of the mitochondrial cytochrome b gene. The values at the nodes represent the Bayesian posterior probabilities. Sinibotia zebra and Sinibotia pulchra are sister species, while Leptobotia guilinensis is only distantly related.
Fig 7. Phylogenetic relationships of Botiidae as revealed by the RAG-1 dataset.
Phylogenetic relationships among freshwater fishes of the family Botiidae as revealed by a Bayesian analyses of the nuclear RAG-1 gene. The values at the nodes represent the Bayesian posterior probabilities. Sinibotia zebra and Sinibotia pulchra are sister species, while Leptobotia guilinensis is only distantly related.
All specimens of ‘B.´zebra, L. guilinensis and S. pulchra form own monophyletic groups, confirming that the three species are unambiguously identifiable by these markers. The lineages of ‘B.´zebra and S. pulchra show a sister relationship and together are embedded into the comparative material of Sinibotia, while the lineage of L. guilinensis is closely related to all comparative samples of Leptobotia, but only distantly related to the lineages formed by B.´zebra and S. pulchra.
Discussion
Our results demonstrate that ‘B.´zebra shows a high morphological similarity to L. guilinensis, but also shares characters with S. pulchra and in some characters presents an intermediate morphotype. Moreover, except having 7½ -8½ branched dorsal-fin rays (versus 8½ in to L. guilinensis and S. pulchra) it reveals only characters that are also found in either of the two species. The prevalence of synapomorpies with either one of the two other species or an intermediate character state strongly supports the hypothesis of a hybrid origin of ‘B.´zebra. Most important in this respect are the diagnostic characters: the diagnostic character for the genus Leptobotia is the simple suborbital spine. In ‘B.’ zebra, the spine also is simple; therefore it bears the diagnostic character of the genus Leptobotia (Fig 3). Consequently, ‘B.’ zebra was placed into Leptobotia [7,16,35,36]. The diagnostic character of the genus Sinibotia is the presence of a pair of mental lobes in a button-like; and ‘B.’ zebra bears these buttons, meaning it also carries the diagnostic character of the genus Sinibotia (Fig 4). This result offers two potential explanations: either ‘B.´zebra is of hybrid origin or the described characters are not diagnostic. As mentioned above, the pigmentation pattern of ‘B.´zebra includes the head pigmentation of Sinibotia and the body pigmentation of L. guilinensis, further strengthening the assumption of a hybrid origin (Fig 2). Therefore all morphological data suggest that ‘B.’ zebra is a mixture of these two species; that means the product of a hybridisation. As stated above, the different ploidy level of the diploid L. guilinensis and the tetraploid S. pulchra should represent an efficient barrier against any horizontal gene flow between these two lineages.
However, a first hybridisation step would be possible, but potentially formed F1 hybrids would be excluded from further reproduction. In order to test the F1 hybrid status of 'B.' zebra, their ploidy level was investigated. All six analysed individuals of 'B.' zebra were tetraploid with a chromosome number of 4n = 100. Consequently, these individuals were no F1 hybrids, inducing strong doubts against the postulate of the efficiency of ploidy level differences as barrier against gene flow. Moreover, the composition of the karyotype of ‘B.’ zebra turned out to be very similar to that of S. pulchra, but did not reveal any trace of introduction of one or two chromosome sets of L. guilinensis into its karyotype. Due to the high number of uni-armed chromosomes in L. guilinensis the number of uni-armed chromosomes in ‘B.’ zebra would have elevated considerably in comparison to S. pulchra. Nevertheless, the number of uniarmed chromosomes is slightly increased in 'B.' zebra when compared to S. pulchra. Theoretically, this could be the result of a number of back-crossings of the original hybrid with S. pulchra that brought the karyotype composition of ‘B.’ zebra closer to that of S. pulchra, while some chromosomes of L. guilinensis are still present, but not distinguishable from Sinibotia chromosomes with the given Giemsa staining technique. In such case, comparisons of proteins and molecular genetic markers could still reveal a genetic introgression by L. guilinensis.
However, the allozyme comparison did not reveal any sign of L. guilinensis genome, but all analysed specimens of ‘B.’ zebra were in all of the informative proteins undistinguishable from S. pulchra. These results provide evidence that no genetic introgression by L. guilinensis has occurred.
Finally, both phylogenetic reconstructions, one on base of a mitochondrial gene and the other on base of a nuclear gene, suggested with high statistical support that ‘B.’ zebra is the sister lineage to S. pulchra, while all specimens of Leptobotia were only distantly related.
At the end, we report a strong discrepancy between morphological and genetic data with the former suggesting gene flow between the diploid Leptobotia and the tetraploid Sinibotia in the upper River Li basin, while the later did not reveal any sign of genetic introgression of Leptobotia into the evolutionary history of ‘B.’ zebra. Since the amount of information taken from the genetic analyses was high and the different methods that have been applied in the present study have analysed a wide range of genetic data (chromosomes, allozymes, mitochondrial and nuclear DNA sequences), it is very unlikely that a hybridisation event would have stayed undetected.
We finally conclude that ‘B.’ zebra is not the result of a hybridisation event, but a species of Sinibotia that underwent an outstanding example of evolution that has changed its morphology in the way that it strikingly matches the morphology of the co-occurring species L guilinensis. This seems at least surprising, since evolutionary theory pronounces that a strong selection exists against the co-occurrence of highly similar species (competitive exclusion, Gaus’s law) [37,38]. We therefore have to assume that there exists an evolutionary advantage for S. zebra in looking so similar to L. guilinensis. The most common mechanism to achieve such an advantage is mimicry; which helps to reduce the predation pressure on one (Batesian mimicry) or both (Mullerian mimicry) similar species [39,40]. Since all species of Botiidae have a suborbital spine as anti-predator weapon and six out of eight species of Botiidae in the River Li share a pattern of broad bands on the body, the possibility exists that they represent a case of Mullerian mimicry. The only species with a different pattern are L. guilinensis and B. zebra; which does not fit to the assumption of Mullerian mimicry. Nothing is known about ecology and microhabitat of S. zebra and L. guilinensis, but the their frequent occurrence in the same lot on local markets and ornamental fish imports indicate that they live very close to each other, making their case of ‘mimicry’ an interesting topic for further research.
Another result of the present study is the first record of S. zebra from outside the upper River Li basin and even outside the Pearl River basin. Specimen A8614 was found among specimens of S. pulchra from the Min River in Fujian province (Fig 1). It bears the characteristic pigmentation of S. zebra (head like S. pulchra, body like L. guilinensis) as well as the diagnostic combination of mental lobes and a simple suborbital spine and has the S. zebra specific character state of 7½ branched dorsal-fin rays. Interestingly, no species of Leptobotia has been recorded from this basin up to now; therefore no partner for any co-evolution as discussed above would be available. However, our phylogenetic reconstructions based on the mitochondrial and nuclear genes show that specimen A8614 from the Min basin is very closely related to their conspecifics from the Li basin. The same is true for S. pulchra; the specimens from the Min basin bear very similar haplotypes as specimens from the Li River. We consequently assume their presence in the Min basin to be the result of a very recent range extension. Range extensions along the southeaster Chinese coast were possible during the Pleistocene glacial maxima, when the lowered global sea level led to prolongation and joining of coastal rivers. However, no botiid species is known to occur on Taiwan, which also was connected to the Chinese coast during the glacial maxima in Pleistocene and therefore shares several freshwater species with the coastal rivers of China [41,42]. It is possible that the presence of botiid fishes in the Min River basin is even younger than the last glacial maximum and might be the result of human activity.
Acknowledgments
We are deeply thankful for the help of Z. Hang, H. Hengwei, H.F. Hong, L. Kalous, P. Laoobuth, F. Li, H. Mian, E. Ming, M. Petrtyl and K. Udomritthiruj with sample collection. Valuable help with the laboratory work came from T. Dvořák, J. Kopecká and Š. Pelikanová. We thank Larry Page and an anonymous reviewer for their comments on a former draft of the manuscript. The study was supported by grant 13–37277 S of the Czech Science Foundation (GAČR).
Data Availability
All relevant data are within the paper.
Funding Statement
The study was supported by the Czech Science Foundation (GAČR, grant 13-37277 S). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Comai L. The advantages and disadvantages of being polyploidy. Nat Rev Genet. 2005;6: 836–846. [DOI] [PubMed] [Google Scholar]
- 2.Otto SP, Whitton J. Polyploid incidence and evolution. Ann Rev Genet. 2000;34, 401–437. [DOI] [PubMed] [Google Scholar]
- 3.Arnold ML. Natural hybridization and evolution New York: Oxford University Press; 1997. [Google Scholar]
- 4.Benfey TJ. The physiology and behaviour of triploid fishes. Rev Fish Sci 1999;7: 39–67. [Google Scholar]
- 5.Piferrer F, Beaumont A, Falguiere JC, Flajšhans M, Haffray P, Kolombo L. Polyploid fish and shellfish: production, biology and applications to aquaculture for performance improvement and genetic containment. Aquaculture. 2009;239: 125–156. [Google Scholar]
- 6.Bănărescu P. Zoogeography of fresh waters Volume 2 Distribution and dispersal of freshwater animals in North America and Eurasia. Wiesbaden: Aula-Verlag; 1992. [Google Scholar]
- 7.Nalbant TT. Sixty million years of evolution: Part one: Family Botiidae (Pisces: Ostariophysi: Cobitoidea). Trav Mus Natl Hist Nat Grigore Antipa. 2002;44: 309–333. [Google Scholar]
- 8.Saitoh K, Sado T, Mayden RL, Hanzawa N, Nakamura K, Nishida M, et al. Mitogenomic Evolution and Interrelationships of the Cypriniformes (Actinopterygii: Ostariophysi): The first evidence toward resolution of higher-level relationships of the world’s largest freshwater fish clade based on 59 whole mitogenome sequences. J Mol Evol. 2006;63: 826–841. [DOI] [PubMed] [Google Scholar]
- 9.Sawada Y. Phylogeny and zoogeography of the superfamily Cobitoidea (Cyprinoidei, Cypriniformes). Mem Fac Fish Hokkaido Univ. 1982;28: 65–223. [Google Scholar]
- 10.Šlechtová V, Bohlen J, Tan HH. Families of Cobitoidea (Teleostei; Cypriniformes) as revealed from nuclear genetic data and the position of the mysterious genera Barbucca, Psilorhynchus, Serpenticobitis and Vaillantella. Mol Phylogenet Evol. 2007;44: 1358–1365. [DOI] [PubMed] [Google Scholar]
- 11.Tang QY, Xiong B, Yang X, Liu H. Phylogeny of the East Asian botiine loaches (Cypriniformes, Botiidae) inferred from mitochondrial cytochrome b gene sequences. Hydrobiologia. 2005;544: 249–258. [Google Scholar]
- 12.Šlechtová V, Bohlen J, Freyhof J, Ráb P. Molecular phylogeny of the Southeast Asian freshwater fish family Botiidae (Teleostei: Cobitoidea) and the origin of polyploidy in their evolution. Mol Phylogenet Evol. 2006;39: 529–541. [DOI] [PubMed] [Google Scholar]
- 13.Suzuki A, Taki Y. Tetraplodization in the cobitid subfamily Botinae (Pisces, Cypriniformes). Cytobios. 1996;85: 229–245. [Google Scholar]
- 14.Zhan SH, Glick L, Tsigenopoulos CS, Otto SP, Mayrose I. Comparative analysis reveals that polyploidy does not decelerate diversification in fish. J Evol Biol. 2014;27: 391–403. 10.1111/jeb.12308 [DOI] [PubMed] [Google Scholar]
- 15.Yang JX, Chen YR. Revision of the subgenus Botia (Sinibotia) with description of a new species (Cypriniformes: Cobitidae). Ichthyol Explor Freshw. 1992;2: 341–349. [Google Scholar]
- 16.Chen JX. A study on the classification of the botiid fishes of China. Zool Res. 1980;1: 3–26. [Google Scholar]
- 17.Wu HW. On the fishes of Li-Kiang. Sinensia. 1939;10: 92–142. [Google Scholar]
- 18.Tang Q-Y, Yu D, Liu H-Z. Leptobotia zebra should be revised as Sinibotia zebra (Cypriniformes: Botiidae). Zool Res. 2008;29: 1–9. [Google Scholar]
- 19.Kottelat M. Indochinese nemacheilines. A revision of nemacheiline loaches (Pisces: Cypriniformes) of Thailand, Burma, Laos, Cambodia and southern Viet Nam. München: Pfeil; 1990. [Google Scholar]
- 20.Völker M, Sonnenberg R, Ráb P, Kullmann H. Karyotype differentiation in Chromaphyosemion killifishes (Cyprinodontiformes, Nothobranchiidae). II: Cytogenetic and mitochondrial DNA analysesdemonstrate karyotype differentiation and its evolutionary direction in C. riggenbachi. Cytogenet Genome Res. 2006;115: 70–83. [DOI] [PubMed] [Google Scholar]
- 21.Völker M, Ráb P. Direct chromosome preparation from regenerating fin tissue p.37–41 In: Ozouf-Costaz C, Pisano E, Foresti F, de Almeida Toledo LF, editors. Fish Cytogenetic Techniques: Ray-Fin Fishes and Chondrichthyans. Enfield: CRC Press, 2015. [Google Scholar]
- 22.Levan A, Fredga K, Sandberg AA. Nomenclature for centromeric position on chromosomes. Hereditas 1964; 52: 201–220. [Google Scholar]
- 23.Šlechtová V, Šlechta V, Lusková V, Lusk S, Berrebi P. Genetic variability of common barbel, Barbus barbus populations in the Czech Republic. Folia Zool. 1998;47 (Suppl. 1): 21–33. [Google Scholar]
- 24.Šlechtová V, Lusková V, Šlechta V, Lusk S, Halačka K, Bohlen J. Genetic differentiation of two diploid-polyploid complexes of spined loach, genus Cobitis (Cobitidae), in Czech Republic, involving C. taenia, C. elongatoides and C. spp. Allozyme interpopulation and interspecific differences. Folia Zool. 2000;49 (Suppl. 1): 67–78. [Google Scholar]
- 25.Ferguson KA, Wallace ALC. Starch-gel Electrophoresis of Anterior Pituitary Hormones. Nature. 1968;190: 629–630. [DOI] [PubMed] [Google Scholar]
- 26.Clayton JW, Tretiak DN. Amine-citrate buffers for pH control in starch gel electrophoresis. J Fish Res Board Can. 1972;29: 1169–1172. [Google Scholar]
- 27.Valenta M, Hyldgaard-Jensen J, Jensen SE. Interaction of veronal, pyrophosphate, citrate and protein with lactate dehydrogenase isoenzyme determination and kinetics. Acta Vet Scand. 1971;12: 15–35. [PubMed] [Google Scholar]
- 28.Perdices A, Doadrio I, Bermingham E. Evolutionary history of the synbranchid eels (Teleostei: Synbranchidae) in Central America and the Caribbean islands inferred from their molecular phylogeny. Mol Phylogenet Evol. 2005;37: 460–473. [DOI] [PubMed] [Google Scholar]
- 29.Hall T. BioEdit. Biological sequence alignment editor for Win95/98/NT/2K/XP/7. Version 7.2.5; 2003. Available: http://www.mbio.ncsu.edu/BioEdit/bioedit.html
- 30.Bohlen J, Šlechtová V. Phylogenetic position of the fish genus Ellopostoma (Teleostei: Cypriniformes) using molecular genetic data. Ichthyol Explor Freshw. 2009;20: 157–162. [Google Scholar]
- 31.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- 32.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol. 2011;28: 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taki Y. Botia eos, a new spiny loach from Thailand and Laos, with notes on some related forms in Asia. Japanese Journal of Ichthyology. 1972;19: 63–81. [Google Scholar]
- 34.Fang PW. Study on the botoid fishes of China. Sinensia. 1936;7: 1–49. [Google Scholar]
- 35.Li J, Li X-H, Chen X-L. A new species of the genus Leptobotia from Guangxi, China (Cypriniformes Cobitidae). Acta Zootaxon. Sinica 2008;33: 630–633. [Google Scholar]
- 36.Kottelat M. Botia kubotai, a new species of loach (Teleostei: Cobitidae) from the Ataran River basin (Myanmar), with comments on botiine nomenclature and diagnosis of a new genus. Zootaxa 2004;40:1–18. [Google Scholar]
- 37.Gause GF. The struggle for existence. Baltimore: Williams & Wilkins; 1934. [Google Scholar]
- 38.Hardin G. The Competitive Exclusion Principle. Science. 1960;131: 1292–1297. [DOI] [PubMed] [Google Scholar]
- 39.Edmunds M. Defence in animals: a survey of anti-predator defences. Essex: Longman; 1974. [Google Scholar]
- 40.Wickler W. Mimicry in plants and animals. New York: Mc Graw-Hill; 1968. [Google Scholar]
- 41.Huang JP, Lin CP. Lineage-specific late pleistocene expansion of an endemic subtropical gossamer-wing damselfly, Euphaea formosa, in Taiwan. BMC Evol Biol. 2011;11: 94 10.1186/1471-2148-11-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang JQ, Tang WQ, Liao TY, Sun Y, Zhou ZC, Han CC, et al. Phylogeographical Analysis on Squalidus argentatus Recapitulates Historical Landscapes and Drainage Evolution on the Island of Taiwan and Mainland China. Int J Mol Sci. 2012;13: 1405–1425. 10.3390/ijms13021405 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.