Abstract
Single stranded oligonucleotides (SSO) represent a novel therapeutic modality that opens new space to address previously undruggable targets. In spite of their proven efficacy, the development of promising SSO drug candidates has been limited by reported cases of SSO-associated hepatotoxicity. The mechanisms of SSO induced liver toxicity are poorly understood, and up to now no preclinical in vitro model has been established that allows prediction of the hepatotoxicity risk of a given SSO. Therefore, preclinical assessment of hepatic liability currently relies on rodent studies that require large cohorts of animals and lengthy protocols. Here, we describe the establishment and validation of an in vitro assay using primary hepatocytes that recapitulates the hepatotoxic profile of SSOs previously observed in rodents. In vitro cytotoxicity upon unassisted delivery was measured as an increase in extracellular lactate dehydrogenase (LDH) levels and concomitant reduction in intracellular glutathione and ATP levels after 3 days of treatment. Furthermore, toxic, but not safe, SSOs led to an increase in miR-122 in cell culture supernatants after 2 days of exposure, revealing the potential use of miR122 as a selective translational biomarker for detection of SSO-induced hepatotoxicity. Overall, we have developed and validated for the first time a robust in vitro screening assay for SSO liver safety profiling which allows rapid prioritization of candidate molecules early on in development.
Introduction
Single stranded oligonucleotides (SSOs) can bind by Watson-Crick hybridization to an RNA target. This binding recruits RNAseH, which induces cleavage and degradation of the target RNA. This mechanism can be applied to specifically alter disease-relevant protein expression. Thus, SSOs are an attractive drug modality that gained increasing interest over the past years. However, despite three decades of SSO drug development, the clinical success has been very limited and led to only two FDA approved drugs [1]. Successes have been hampered initially by insufficient SSO stability and delivery to target tissue. Drug-induced toxicities including hepatotoxicity and pro-inflammatory effects further complicated drug development [1].
Although these toxicities occur in a concentration-dependent manner, there are SSOs which highly accumulate in the liver without causing harm to cells, while other molecules accumulate to a lesser extent, but exert significant hepatotoxicity. The mechanisms of these effects are poorly understood and may be caused by 1) hybridization of the respective SSO to non-intended RNA targets due to similarity (hybridization-dependent off-target toxicity [2, 3]) or 2) binding of the SSO to intracellular proteins, thereby interfering with their function and ultimately leading to toxicity (hybridization independent toxicity [4, 5]).
The major adverse effects caused by SSOs in vivo can be summarized as accumulation and toxicity in liver and kidney as well as inflammatory reactions at the site of injection and flu like-symptoms [5]. Preclinically, whole body autoradiography of SSO-treated mice revealed that the main organs of SSO exposure after i.v. application are liver, kidney and spleen [6, 7], which are also the main organs where SSO-related toxicities are most often observed. SSO-associated liver and renal toxicities as well as immunotoxicity have been assessed early on in development using mainly rodent in vivo tests. Mice and rats treated with toxic SSOs showed significantly increased plasma aminotransferase levels (ALT and AST) and histopathological evidence for necrosis and apoptosis in liver and kidney [8, 9]. Furthermore, toxic SSOs induced lymphoid hyperplasia and organ cell infiltrates in rodents [10]. Early rodent toxicity tests performed to address SSO safety liabilities come with a high attrition rate and require large sets of animal cohorts for screening purposes which pose a significant challenge for SSO development from an ethical, cost, resource and time point of view. Recently it has been shown that certain sequence motifs are associated with hepatotoxicity risk [8, 11], but until now, no in vitro test has been identified that is able to recapitulate and predict the in vivo effect in cellular models [12] and thus is suited to replace in vivo testing.
To address this limitation, we have developed an in vitro screening assay to profile SSOs for liver liability before any animal studies are initiated. We successfully recapitulated the in vivo hepatotoxic profile by 1) testing a panel of SSOs of known in vivo liability, 2) using species-relevant primary hepatocytes, 3) applying SSOs to hepatocytes without the assistance of delivery technology and 4) selecting readouts with in vivo comparators. Remarkably, these experimental conditions successfully recapitulated the in vivo hepatotoxic profile of the tested SSOs. In addition to a decrease in cell viability, we report elevated levels of IL1α, MIP1α and miR-122 in the supernatant of primary hepatocytes exposed to toxic, but not safe SSOs. The assay presented here allows for the first time a rapid, early in vitro assessment of a key liability of SSOs. It can be used to prioritize lead series before initiation of animal studies and has the potential to significantly reduce the hepatotoxicity risk during SSO development.
Materials and Methods
Mouse liver perfusion and hepatocyte and non-parenchymal cell isolation
All procedures were conducted in strict adherence to the Swiss federal ordinance on animal protection and welfare, according to the rules of the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC), and with the explicit approval of the local veterinary authority (Kantonales Veterinäramt Basel-Stadt, Switzerland).
Primary mouse hepatocytes were isolated from 10- to 13-week old male C57Bl6 mice by a retrograde two-step collagenase liver perfusion. Briefly, fed mice were euthanized with sodium pentobarbital (120 mg/kg, i.p.). Perfusion tubing was inserted via the right ventricle into the v. cava caudalis. Following ligation of the v. cava caudalis distal to the v. iliaca communis, the portal vein was cut and the two-step liver perfusion was started. The liver was first perfused for 5 min with a pre-perfusing solution consisting of calcium-free, EGTA (0.5 mM)-supplemented, HEPES (20 mM)-buffered Hank's balanced salt solution, followed by a 12-min perfusion with NaHCO3 (25 mM)-supplemented Hank's solution containing CaCl2 (5 mM) and collagenase (0.2 U/ml; Collagenase Type II, Worthington). Flow rate was maintained at 7 ml/min and all solutions were kept at 37°C. After in situ perfusion, the liver was excised, the liver capsule was mechanically opened, the cells were suspended in William’s Medium E (WME) without phenol red (Sigma W-1878), and filtered through a set of nylon cell straines (40- and 70-mesh). Dead cells were removed by a Percoll (Sigma P-4937) centrifugation step (percoll density: 1.06 g/ml, 50g, 10 min) and an additional centrifugation in WME (50xg, 3 min).
The supernatant from the hepatocytes sedimentation step was kept for the isolation of the non-parenchymal cell (NPC) fraction. Cells were pelleted by a centrifugation step (1200rpm, 10min, Eppendorf 5810R) and resuspended in 12 mL William’s Medium E (WME) without phenol red (Sigma W-1878). NPCs were isolated by a Percoll gradient (23% (upper phase) and 50% (bottom phase); Sigma P-4937) centrifugation step (Percoll density: 1.06 g/ml, 1350g, 10 min, brake off). Cells were washed once in WME, centrifuged (200g, 5 min, Eppendorf 5810R) and resuspended in 13mL William’s Medium E (WME) containing 10% fetal calf serum, penicillin (100 U/ml), streptomycin (0.1 mg/ml).
Cell- culturing and oligonucleotide exposure
Freshly isolated primary mouse or cryopreserved human (BioreclamationIVT, Brussels, Belgium) hepatocytes were suspended in WME supplemented with 10% fetal calf serum, penicillin (100 U/ml), streptomycin (0.1 mg/ml) at a density of approx. 5 x 106 cells/ml and seeded into collagen-coated 96-well plates (Becton Dickinson AG, Allschwil, Switzerland) at a density of 0.25 x 105 cells/well (mouse) or 0.4 x 105 (human). Cells were pre-cultured for 3 to 4h allowing for attachment to cell culture plates before start of treatment with oligonucleotides. Seeding medium was replaced by 90 μl of serum free WME and 10 μl of oligonucleotide stock solutions in PBS were added to the cell culture and left on the cells for 3 days.
Mouse NPCs were plated into collagen-coated 96-well plates (Becton Dickinson AG, Allschwil, Switzerland) at a density of approx. 0.25 x 105 cells/well. For co-cultures primary mouse hepatocytes, suspended in WME supplemented with 10% fetal calf serum, penicillin (100 U/ml), streptomycin (0.1 mg/ml) at a density of 0.25 x 105 cells/well, were seeded after 30 min on top of the NPC layer. Cells were cultured for 3 to 4h allowing for attachment to cell culture plates before start of treatment with oligonucleotides as described above.
HepG2 cells were cultivated at app. 80% confluence in MEM medium with GlutaMax (Gibco#41090), supplemented with 10% fetal calf serum. Cells were detached with Accutase (Sigma #A6964) and seeded into collagen-coated 96-well plates (Becton Dickinson AG, Allschwil, Switzerland) at a density of 0.5 x 105 cells/well. Cells were pre-cultured at 37°C and 5%CO2 overnight, allowing for attachment before start of treatment with oligonucleotides described above.
RNA isolation and qPCR
mRNA purification from mouse hepatocytes was performed using the RNeasy 96 Kit (Qiagen, Hombrechtikon, Switzerland) including an RNAse free DNAse I treatment according to the manufacturer’s instructions. cDNA was synthesized using iScript single strand cDNA Synthesis Kit (Bio-Rad Laboratories AG, Reinach, Switzerland). Quantitative real-time PCR assays (qRT-PCR) were performed using the Roche SYBR Green I PCR Kit and the Light Cycler 480 (Roche Diagnostics, Rotkreuz, Switzerland) with specific DNA primers. Analysis was done by the ΔΔCt threshold method to determine expression relative to RPS12 mRNA. Each analysis reaction was performed in duplicate, with two samples per condition.
LDH, Albumin, GSH and ATP assays
Lactate dehydrogenase (LDH) released into the culture media was determined using a Cytotoxicity Detection Kit (Roche 11644793001, Roche Diagnostics GmbH Roche Applied Science Mannheim, Germany) according to the manufacturer's protocol. Albumin secretion was quantitated using mouse albumin ELISA Kit from Alpco diagnostics (41-ALBMS-E01). Intracellular GSH levels were determined in intact cells by a fluorescent assay using monochlorobimane (Fluka, 69899). In brief after removal of the cell supernatant, 100 μl of Krebs Henseleit Buffer (KHB) was added to the wells and background fluorescence was recorded. Next 50 μl of a 300 μM solution of monochlorobimane dissolved in KHB was added to the cells and incubated for 15 min at 37°C, before the reaction was stopped by removal of the monochlorobimane solution and addition of 100μL of fresh KHB to each well. Fluorescence was measured using a VictorV3 reader. For the determination of cellular ATP levels the CellTiter-Glo® Luminescent Cell Viability Assay (G9242, Promega Corporation, Madison WI, USA) was used according to the manufacturer's protocol.
All Experiments were performed in triplicates.
Apoptosis assay
Caspase-3/7 activity was determined using the Caspase-Glo® 3/7 Assay (Promega Corporation, Madison WI, USA). In brief, Caspase-Glo® 3/7 reagent was added to the cells at indicated time points, incubated for 30 min, before luminescence was determined on an Enspire multi-mode plate reader (Perkin Elmer) according to the manufacturer's instructions.
Cytokine measurement
25 μl of supernatants from hepatocyte/NPC co-cultures were collected after 24, 48 and 64 hours of LNA treatment and stored at -20°C until analysis. For cytokine profiling, supernatants were thawed on ice, diluted 2x in sample dilution buffer (BioRad catalog # M60-009RDPD) and analyzed by multiplex ELISA using a mouse cytokine 23-plex (Bio-Plex Pro Mouse Cytokine 23-plex Assay, catalog # M60009RDPD) and Bio-Plex® 200 Systems (BioRad) according to the manufacturer’s instructions. The 23 analytes were: IL-1α, IL-1β, IL-2, IL-3, IL-4, IL-5, IL-6, IL-9, IL-10, IL-12 (p40), IL-12 (p70), IL-13, IL-17A, Eotaxin, G-CSF, GM-CSF, IFN-γ, KC, MCP-1, MIP-1α, MIP-1β, RANTES and TNF-α. Data of selected analytes are reported as average concentrations and standard deviations (SD) of triplicate wells.
miRNA analysis
To isolate miRNAs, 70 ul of cell culture supernatants (SN) were collected and the RNA was isolated using the miRNeasy Mini Kit (Qiagen, Germany, Catalog number 217004) following the manufacturer’s instructions. A volume of 1 μl of RNA was used for the cDNA synthesis using the cDNA TaqMan® MicroRNA Reverse Transcription kit (Applied Biosystems, Thermo Fischer Scientific, USA, (Catalog number 4366596) and the expression of miR-122 was determined using a TaqMan miRNA assay (hsa-miR-122-5p; Applied Biosystems, Thermo Fischer Scientific, USA). Briefly, each sample was analyzed in duplicates and the reactions were performed using 1.33 μL of cDNA in a final volume of 20 μL with TaqMan® Fast Advanced Master Mix reagents (Applied Biosystems, Thermo Fischer Scientific, USA). The PCR conditions were standardized to 95°C for 20 s followed by 40 cycles of 95°C for 1 s and 60°C for 20 s. The reactions were carried out on a QuantStudio 12K Flex Real-Time PCR System (Applied Biosystems, Lyfe technologies, USA), the EDS files were loaded in to the Quantstudio 12K Flex Software v1.2.2 and the raw Ct values were calculated using automatic baseline and fixed threshold values. The fold changes in microRNA expression were calculated according to the ΔΔCt-method, using spiked-in ath-miR-159 for normalization [13]. Each analysis reaction was performed in duplicate, with two samples per condition.
Results
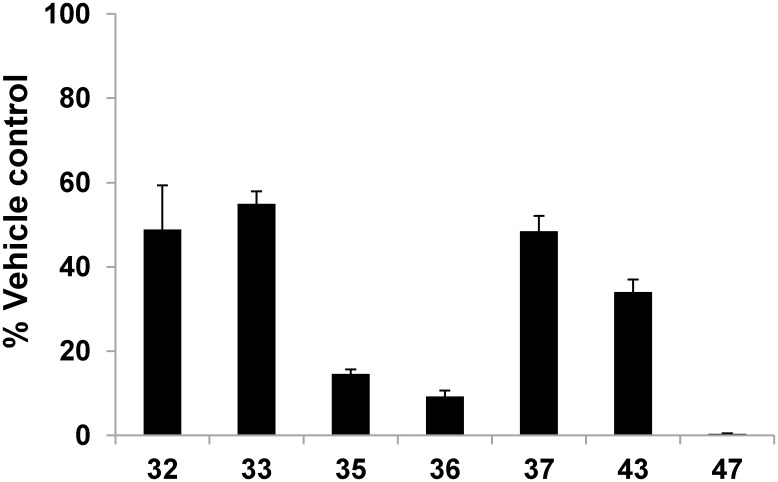
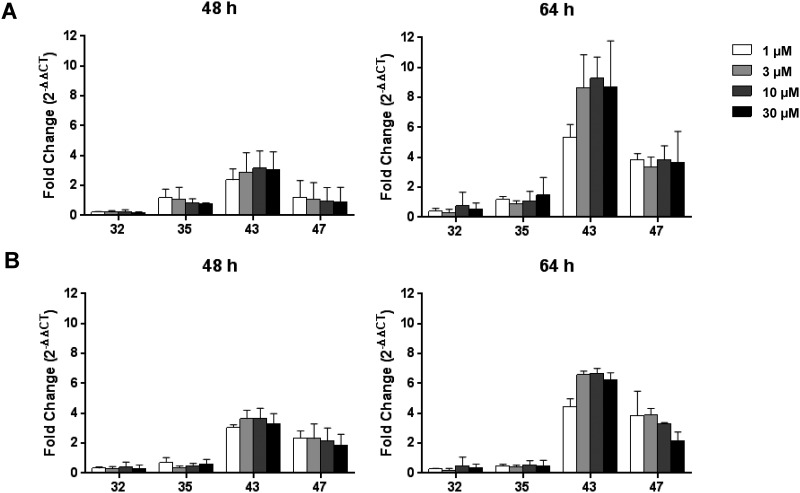
For evaluation and validation of potential predictive in vitro hepatotoxicity assays, a set of SSOs with known mouse in vivo hepatotoxicity was selected. These SSOs had LNA-modified nucleotides in the wings (LNA gapmers) and were directed against mouse myd88 mRNA (Table 1). Oligonucleotides were tested in freshly isolated primary mouse hepatocytes for 2–3 days by gymnotic delivery (without any carrier or transfection reagent; [14, 15]. In order to determine whether the oligonucleotides were taken up by the cells via the productive pathway, target knockdown after 48 hours of treatment was determined. Reduction of myd88 mRNA by 50–90% was observed in primary hepatocytes treated with SSOs 32, 33, 35, 36, 37, 43 and 47 indicating target engagement (Fig 1).
Table 1. Selection of tool SSOs with documented in vivo hepatotoxicity (ALT levels after 2 week treatment in mice with 5 x 15 mg/kg tail vein injection) that were used for the validation of in vitro hepatotoxicity assays.
| SSO | ALT [U/L] | Sequence |
|---|---|---|
| 32 | 64 | 5'- mCS AS AS aS gS gS aS aS aS cS aS cS aSmCS AS T -3' |
| 33 | 59 | 5'- mCS AS AS aS tS gS cS tS gS aS aS aS cS TS AS T -3' |
| 35 | 67 | 5'- mCS TSmCS aS aS cS aS tS cS aS aS gS cS AS GS T -3' |
| 36 | 1889 | 5'- ASmCS TS gS cS tS tS tS cS cS aS cS tSmCS TS G -3' |
| 37 | 2368 | 5'- GSmCSmCS tS cS cS cS aS gS tS tS cS cS TS TS T -3' |
| 43 | 1890 | 5'- GS AS TS gS cS cS tS cS cS cS aS GS TS T -3' |
| 47 | ND | 5'- GS AS cS aS tS tS gS cS cS tSmCS TS A -3' |
ND: no ALT levels were determined, since group had to be sacrificed early due to severe toxicity. Small letters: nucleotides; s: phosphorothioate backbone; Capital letters: beta-oxy LNA, mC: methylated cytosine.
Fig 1. SSO induced target knock down in primary mouse hepatocytes.
Normalized expression of myd88 mRNA after 48 hour treatment of mouse hepatocytes with 30 μM of the respective oligonucleotide. Data are means ± SD.
In order to see, if the in vivo hepatotoxicity phenotype of our selected set of tool compounds can be recapitulated on an in vitro level, we evaluated various parameters in primary mouse hepatocytes treated with SSOs at different time points. Our readouts involved determination of hepatocyte function by measurement of albumin secretion, assessment of cell viability (ATP and LDH), oxidative stress (GSH), apoptosis induction (Caspase 3/7) and finally the secretion of potential biomarkers (miR122) and the influence of co-culturing non parenchymal liver cells as well as their influence on cytokine secretion.
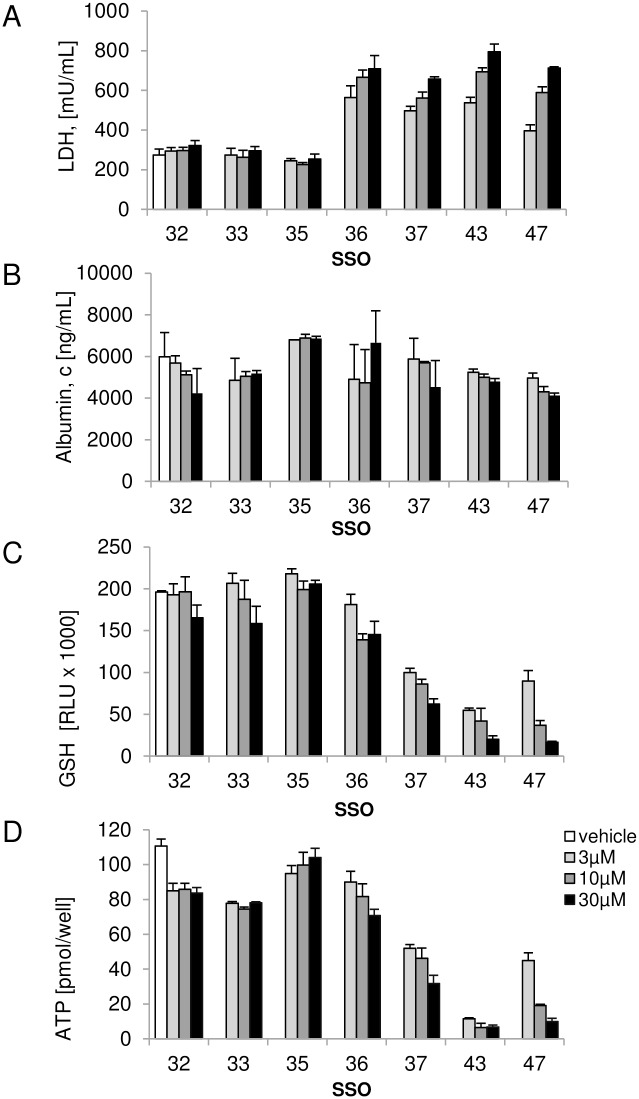
The effects of SSOs on hepatocyte function and cell viability were first investigated in vitro by determination of albumin secretion and LDH levels in the supernatant as well as intracellular GSH levels and cellular ATP content. No significant changes in hepatocyte function or viability was observed 2 days after incubation of cells with all oligonucleotides (data not shown). After 3 days of treatment, LDH levels were significantly increased in supernatants of cells treated with toxic SSOs 36, 37, 43 and 47, while no change in LDH levels was seen with safe oligonucleotides 32, 33 and 35 (Fig 2A). In contrast, albumin secretion was not notably changed compared to vehicle treated hepatocytes for all SSOs tested indicating that albumin is not a sensitive marker for oligonucleotide induced toxicity in this setting (Fig 2B). In order to assess the potential of SSOs to induce oxidative stress, intracellular GSH content was determined after 3 days of exposure. Hepatotoxic SSOs 37, 43 and 47 induced a significant reduction in glutathione levels in mouse hepatocyte cultures while SSO 36 showed a mild but statistically highly significant reduction in GSH at 10 and 30 μM (p<0.001 using two-way ANOVA followed by Dunett’s multiple comparison test). Safe SSOs 32, 33 and 35 did not change GSH levels compared to vehicle (Fig 2C). A similar picture was obtained when assessing intracellular ATP levels which revealed a good differentiation between toxic and safe SSOs. Cellular ATP levels were slightly reduced in hepatocytes treated with safe SSOs 32, 33 and 35 compared to the vehicle group, but this effect was small and not concentration-dependent. Treatment with hepatotoxic SSOs, 37, 43 and 47 led to a marked reduction in ATP while SSO 36 showed a concentration dependent mild reduction in intracellular ATP concentration, which is at 30 μM statistical significant compared to the safe SSO 32 (Fig 2D).
Fig 2. SSO induced toxicity in primary mouse hepatocytes.
Secreted (A) LDH and (B) albumin levels and intracellular (C) GSH and (D) ATP concentrations after 3 day treatment with a tool set of hepatotoxic and non-hepatotoxic SSOs. Data are means ± SD.
When safe and toxic SSOs where tested in a non-primary cell model such as HepG2 using similar readouts, we did not see any of the above described effects (supporting information S1 Fig), underlining the importance of using the right cell model that is capable of gymnotic SSO uptake and trafficking similar to the physiological situation to recapitulate SSO induced toxicity.
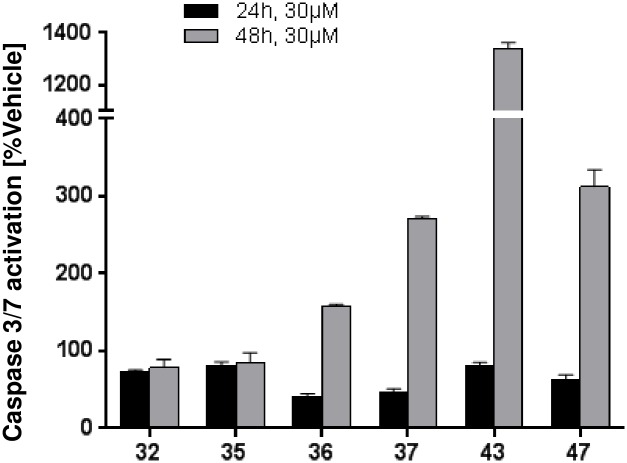
In order to understand earlier events that ultimately result in reduction of cell viability after treatment with hepatotoxic SSOs, SSO-mediated effect on apoptosis by measuring caspase-3/7 activation at 3 different time points was assessed. After 24 hours of incubation, no activation of caspase-3/7 could be detected (Fig 3). At 48 hours, a clear increase in caspase-3/7 activation was observed with toxic SSOs 36, 37, 43 and 47 with the strongest response induced by SSO 43, while no effect was seen with safe SSOs 32 and 35. After 3 days of treatment increases in caspase-3/7 levels were still evident with toxic oligonucleotides (not shown). These signals perfectly correlated with the in vitro cytotoxicity profile of SSOs after 3 day treatment. These results clearly indicate that hepatotoxic SSOs induce apoptosis in mouse hepatocytes, which then finally leads to the changes in cell viability shown in Fig 2.
Fig 3. Apoptosis induced by SSOs in primary mouse hepatocytes.
Changes in caspase-3/7 activation in primary mouse hepatocytes after 24 and 48 h of incubation with the respective SSO at 30 μM. Data are normalized to vehicle treated cells and are means ± SD.
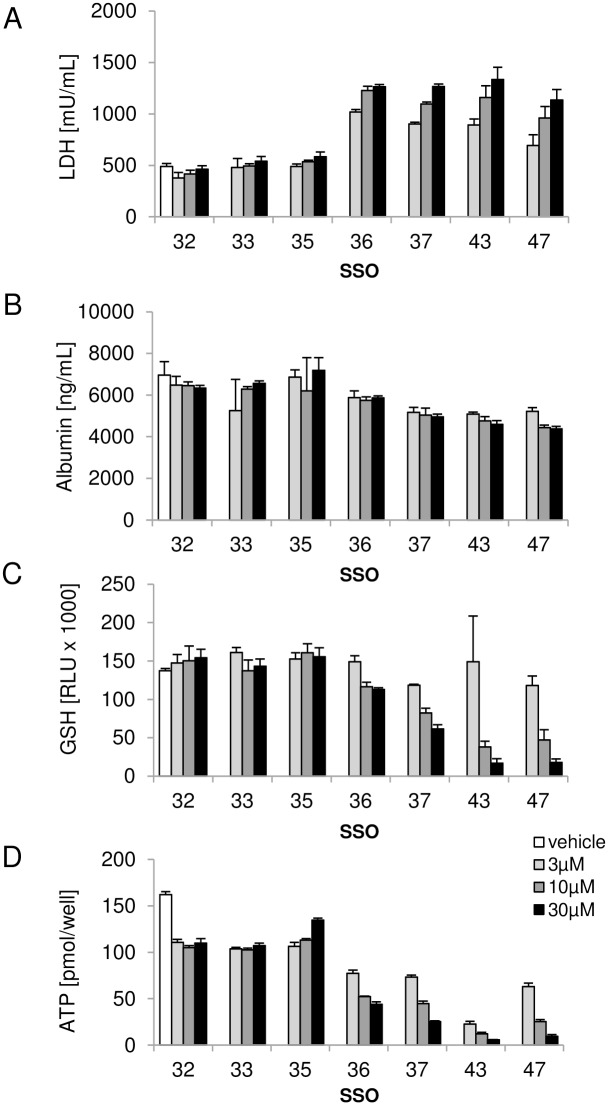
It is well established that SSOs are taken up by non-parenchymal cells (NPC) in the liver [16] and NPCs are described to have a prominent contribution to SSO induced hepatotoxicity in vivo [5]. In order to determine the potential contribution of the non-parenchymal cell fraction to the degree of hepatotoxicity seen with SSOs, another set of in vitro experiments was conducted in mouse hepatocyte-NPC co-cultures. As shown in Fig 4, co-cultures treated with hepatotoxic SSOs 36, 37, 43 and 47 for 3 days showed increased LDH levels (A), slightly decreased albumin secretion (B), and a clear reduction in intracellular GSH levels (C) and ATP content (D) compared to vehicle treated co-cultures. None of these parameters changed after incubation with safe SSOs 32, 33 and 35. Compared to the results obtained with mouse hepatocyte monocultures, the co-cultures displayed similar sensitivity in LDH, GSH and ATP changes towards discrimination of toxic and non-toxic SSOs.
Fig 4. SSO induced toxicity in primary mouse hepatocyte NPC co-cultures.
Secreted (A) LDH and (B) albumin levels and (C) intracellular GSH and (D) ATP concentrations after 3 day treatment of mouse primary hepatocyte NPC co-cultures with a tool set of hepatotoxic and non-hepatotoxic SSOs. Data are means ± SD.
With a subset of 2 safe and 2 toxic SSOs (32, 35, 43 and 47) we evaluated, if the NPC fraction alone is functional and responding to SSO treatment (supporting information S2 Fig). In summary, we observed an increase in LDH levels in the supernatants of NPCs treated with toxic compounds 43 and 47, while no changes were seen with SSOs 32 and 35. These data demonstrate that both mouse primary hepatocytes and non-parenchymal cells are susceptible to SSO induced toxicity.
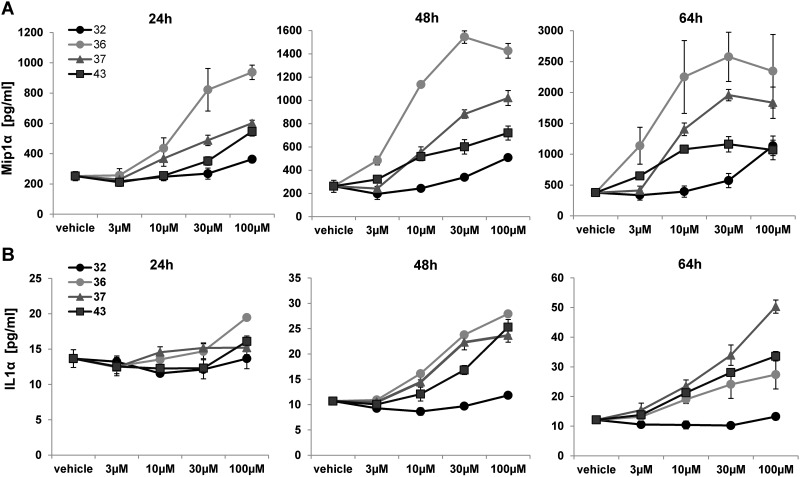
To further elucidate the effect of oligonucleotides on hepatocytes and non-parenchymal co-cultures, changes in secreted cytokines and miR-122, a hepatocyte-specific marker for tissue damage [17, 18] were investigated upon SSO treatment. The cytokine secretion profile of safe (32) and hepatotoxic (36, 37 and 43) SSOs was determined using a multiplex Bio-Plex® assay after 1, 2 and 3 days of SSO treatment. Of the 23 tested analytes (see material and methods), Macrophage Inflammatory Protein 1 alpha (MIP1α) and Interleukin 1 alpha (IL1α) showed a time-dependent response in mouse co-cultures treated with hepatotoxic SSOs. As shown in Fig 5, MIP1α was already induced up to 5 fold after 24 hours of treatment with toxic SSOs 36, 37 and 43. We also observed an increase in IL1α with toxic SSO with a later onset than MIP1α, clearly separating toxic and non-toxic SSO at 48 and 64 hours.
Fig 5. SSO induced cytokine secretion in primary mouse hepatocyte NPC co-cultures.
Secretion of (A) MIP1α and (B) IL1α after 24, 48 and 64 hour treatment of primary mouse hepatocyte NPC co-cultures with safe SSO 32 and hepatotoxic SSOs 36, 43 and 47. Data are means ± SD.
miR-122 release into the supernatants was investigated in mouse hepatocyte monoculture and hepatocyte-NPC co-cultures treated with non-toxic SSO 32 and 35 and toxic SSOs 43 and 47. Increases in miR122 were observed already after 2 days of treatment with toxic SSOs (Fig 6). Although a 4-10-fold increase in miR122 was detected in the supernatants of co-cultures treated with SSOs 43 and 47 compared to vehicle (Fig 6A), the amount was not significantly higher than the miR-122 release observed when the hepatocytes were cultured alone (Fig 6B). Overall, these data further support the idea that liver injury observed in vivo is a direct result of SSO induced damage in each individual exposed liver cell type rather than an event confined to non-parenchymal cells only.
Fig 6. miR-122 release.
(A) Mouse hepatocyte-NPC co-cultures or (B) hepatocyte monocultures were treated with the respective SSOs for 48 or 64 hours and cell-free supernatant was collected. The levels of miR-122 at 48 h and 64 h were assessed using real-time qPCR. miRNA levels were normalized to vehicle treated cells. Data are means ± SD.
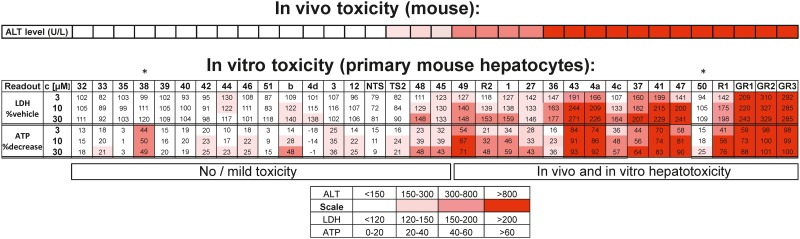
To further validate this novel in vitro assay set up and to determine if the correlation of the in vitro toxicity pattern with mouse in vivo hepatotoxicity seen for our 7 model SSOs translates to a larger set of oligonucleotides, the tool set was extended to 34 SSOs targeting different mRNAs and showing a broad spectrum of mouse in vivo hepatotoxicity. Cellular ATP and secreted LDH were selected as readouts after 3 days of treatment in freshly isolated mouse hepatocyte monocultures, as these two assays turned out to be sufficient to recapitulate the in vivo toxicity pattern with our initial set of tool SSOs. As shown in Fig 7, there is good correlation for 32 out of 34 SSOs between in vivo hepatotoxicity and in vitro readouts. SSOs that induced increases in plasma ALT levels in vivo also clearly showed an increase in LDH and a reduction in ATP levels in vitro in mouse hepatocytes, with only 2 SSOs, where in vivo toxicity was not predicted with the cellular assay. Thus, this assay is suitable to predict the hepatotoxic potential of oligonucleotides prior to moving into first in vivo studies.
Fig 7. In vitro prediction of SSO induced hepatotoxicity in primary mouse hepatocytes.
Increase in secreted LDH levels and reduction in cellular ATP levels after 3 day treatment of primary mouse hepatocytes with 34 SSOs with different in vivo hepatotoxicity as reported by plasma ALT concentration after sub-chronic treatment. *outliers.
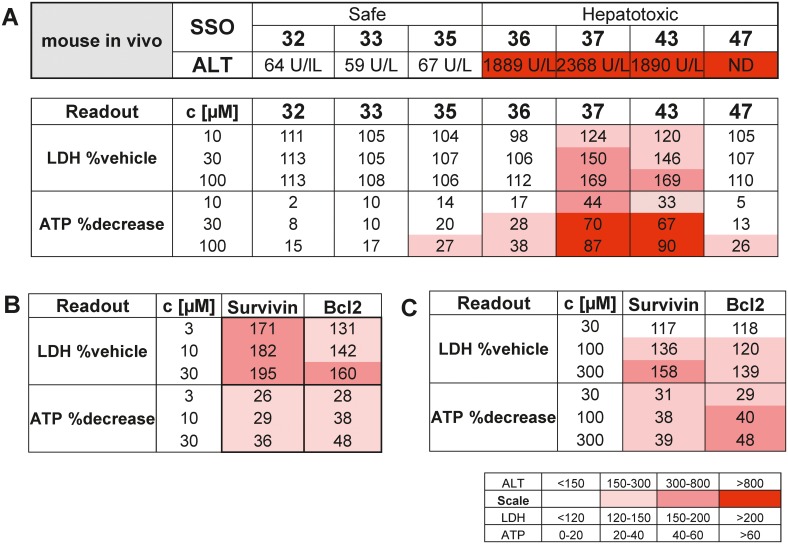
Finally, in an attempt to establish a human relevant in vitro model, a similar protocol was established for human primary hepatocytes. Cytotoxicity readouts were measured 3 days after treatment with the myd88 tool SSOs, initially tested in primary mouse hepatocytes. A clear pattern reflecting in vivo innocuous versus in vivo hepatotoxic SSOs in human hepatocytes was obtained for 5 out of the 7 tested myd88 tool compounds in human hepatocytes (Fig 8A). Toxic SSOs 37 and 43 showed a dose dependent increase in LDH levels and decrease in intracellular ATP levels, whereas SSOs 32, 33 and 35 had no apparent effect. Lastly, in order to see the human relevance of this assay, two historical development SSOs targeting Survivin and Bcl2, with documented pre-clinical as well as clinical liver effects [19–21] were tested in mouse (Fig 8B) and human primary hepatocytes (Fig 8C). With both SSOs, increases in LDH levels and reduction in cellular ATP were observed in mouse and human cellular systems, suggesting reliability of the human hepatocyte assay for estimation of a potential clinical liver toxicity risk.
Fig 8. SSO induced toxicity in cryopreserved human hepatocytes.
(A) Changes in secreted LDH and cellular ATP concentrations after 3 day incubation of human hepatocytes with a tool set of hepatotoxic and non-hepatotoxic myd88 SSOs and changes of ATP and LDH after 3 day treatment of (B) mouse hepatocytes and (C) human hepatocytes with two SSOs targeting Bcl2 and Survivin that have been tested in the clinic. Data are means from 2 experiments in triplicates.
Discussion
We here describe a novel in vitro assay which for the first time is able to capture the hepatotoxic potential of SSO drugs and can be used as a screen to prioritize molecules before in vivo studies are initiated. In order to allow for the detection of hybridization dependent off-target effects as well as aptameric non-hybridization dependent effects, SSOs were delivered to cells by gymnosis, without use of delivery vehicle or molecular conjugation. Using a balanced set of SSO reference molecules with documented in vivo liver toxicity, SSO-induced changes in LDH, GSH, ATP, caspase-3/7 activity, cytokine- and miR122-release were observed and correlated with the in vivo toxicity profile. Assessing this battery of cellular markers in a time- and concentration-dependent manner, a minimal set of required measurements could be defined, which allows to accurately detect hepatotoxic SSOs, namely assessment of LDH and ATP after 3 days of continuous treatment at 1–30 μM (Fig 2) in primary mouse hepatocytes. All seven selected tool SSOs used for assay development showed target reduction, and the extend of mRNA knockdown did not correlate with the toxic effect observed in the same cells or in vivo in mice, ruling out that some SSOs are not toxic in our setting just because of lack of exposure. Data from apoptosis induction experiments suggest a mechanism underlying SSO-induced toxicity involving early activation of this cellular process ultimately leading to cell death. Unlike earlier reports, where cellular caspase-3 activation was described in A549 or HepG2 cells transfected with SSOs, but no correlation between cellular toxicity and toxicities in mice could be observed [12], our in vitro caspase-3/7 activation data perfectly fit with in vivo the hepatotoxicity observed in mice.
As non-parenchymal cells have been shown to preferentially take up SSOs in the liver, it has been speculated that Kupffer cell activation and subsequent inflammatory processes may be initial events triggering hepatotoxicity [5, 16, 22]. In particular, in in vivo studies in mice with SSOs, an elevation of MIP1α and IL1α among other pro-inflammatory cytokines and chemokines in the liver has been reported [23]. We obtained similar results in our in vitro assay, when the effect of SSOs was investigated in hepatocyte NPC co-cultures. A significant increase in secreted MIP1α and IL1α levels was observed in co-cultures treated with toxic SSOs already after one or two days of treatment, respectively. However, when the cellular toxicity profile induced by our tool set of SSOs was compared in hepatocyte mono- and NPC co-cultures, no major difference in sensitivity towards detecting toxic SSOs in vitro was observed (Figs 2 and 4). These data, together with the fact that the established hepatocellular damage marker miR-122 was increased in both mono- and co-cultures (Fig 6) clearly suggest that in addition to an NPC-mediated component to acute hepatic toxicity of SSOs [5] there is a similar direct effect of SSOs on hepatocytes. The model established here not only serves to rapidly screen for bad actors early on in the pipeline, but could also be a model to increase the mechanistic understanding of SSO-induced hepatotoxicity which ultimately should lead to improved design of safe and efficacious drugs. A cornerstone of such a concept, which so far has been lacking in SSO drug development, is translation of pre-clinical findings to human [5]. A series of reports describe changes observed in rodent and non-rodent studies related to SSO-induced toxicity, but as the safest preclinical candidates progress into the clinic, the human relevance of the observed effects remained unclear so far. We therefore aimed to establish a human in vitro system for SSO-induced hepatotoxicity in parallel to the rodent one described, to compare data for our tool SSO set and explore, if a human cell system would be able to correctly predict molecules where the clinical liver profile is known (Fig 8). A good correlation between in vitro mouse and human hepatocyte toxicity was obtained for 5 out of 7 SSOs from our myd88 tool SSO set. Signal strength was generally weaker in human cells and required higher concentrations, but the relative ranking was the same as in the mouse system. If the difference observed between mouse and human hepatocytes for two out of the seven tool SSOs is a real species difference or just a matter of lower sensitivity cannot be answered yet due to the lack of respective clinical data and remains to be investigated. Importantly, the human hepatocyte model correctly reflected the human liver profile of two historical SSOs that showed grade 3 increases in liver enzymes in phase 1 clinical trials [19, 20].
Our results suggest that acute liver toxicity caused by SSOs can be triggered by a direct effect on hepatocytes, which seems reproducible across species albeit some differences were observed. Thus incorporation of rodent and human in vitro hepatotoxicity testing using primary hepatocytes will improve prioritization of SSO that will be moved into in vivo safety testing in rodents and higher species and addresses translation of effects to human.
In summary, we provide for the first time a reliable in vitro assay system which allows selection of SSO candidate drugs which have a lower risk for causing hepatotoxicity early on in development. The cellular system used here is based on primary cells which are able to take up SSO drugs in an unassisted manner thus representing a more in vivo-like model to study uptake, processing and potential hepatotoxicity of novel SSO drugs which is relevant for the in vivo situation.
Supporting Information
Secreted LDH (upper panel) intracellular ATP (lower panel) concentrations after 3 day treatment with a tool set of safe (32, 35) and hepatotoxic (43, 47) SSOs. Data are means ± SD.
(TIF)
LDH levels in the supernatant of primary mouse NPCs after 2 day treatment with safe (32, 35) and toxic (43, 47) SSOs. Data are means ± SD.
(TIF)
Acknowledgments
We thank Nathalie Schaub, Evelyn Durr, Regine Gerard Susanne Fischer for excellent technical support, Stefan Kustermann and Natascha Santacroce for help with the figures and Erich Koller, Marianne Rudolph Hansen and Mark Turner for all fruitful discussions and critical review of the manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
All authors are employees of Roche Innovation Center Basel or Roche Innovation Center Copenhagen A/S, F. Hoffmann-La Roche Ltd. The funder provided support in the form of salaries for authors (SS, FB, AM, CBL, TM, MH, YT, FS, TS, ABR), but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of these authors are articulated in the ‘author contributions’ section.
References
- 1.Andrews L, Ralston S, Blomme E, Barnhart K. A snapshot of biologic drug development: Challenges and opportunities. Hum Exp Toxicol. 2015;34(12):1279–85. Epub 2015/11/29. 10.1177/0960327115603594 . [DOI] [PubMed] [Google Scholar]
- 2.Burel SA, Hart CE, Cauntay P, Hsiao J, Machemer T, Katz M, et al. Hepatotoxicity of high affinity gapmer antisense oligonucleotides is mediated by RNase H1 dependent promiscuous reduction of very long pre-mRNA transcripts. Nucleic Acids Res. 2015. Epub 2015/11/11. 10.1093/nar/gkv1210 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kamola PJ, Kitson JD, Turner G, Maratou K, Eriksson S, Panjwani A, et al. In silico and in vitro evaluation of exonic and intronic off-target effects form a critical element of therapeutic ASO gapmer optimization. Nucleic Acids Res. 2015;43(18):8638–50. Epub 2015/09/05. 10.1093/nar/gkv857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kakiuchi-Kiyota S, Whiteley LO, Ryan AM, Mathialagan N. Development of a Method for Profiling Protein Interactions with LNA-Modified Antisense Oligonucleotides Using Protein Microarrays. Nucleic Acid Ther. 2016;26(2):93–101. Epub 2015/12/09. 10.1089/nat.2015.0576 . [DOI] [PubMed] [Google Scholar]
- 5.Frazier KS. Antisense oligonucleotide therapies: the promise and the challenges from a toxicologic pathologist's perspective. Toxicol Pathol. 2015;43(1):78–89. Epub 2014/11/12. 10.1177/0192623314551840 . [DOI] [PubMed] [Google Scholar]
- 6.Geary RS, Norris D, Yu R, Bennett CF. Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv Drug Deliv Rev. 2015;87:46–51. Epub 2015/02/11. 10.1016/j.addr.2015.01.008 . [DOI] [PubMed] [Google Scholar]
- 7.Straarup EM, Fisker N, Hedtjarn M, Lindholm MW, Rosenbohm C, Aarup V, et al. Short locked nucleic acid antisense oligonucleotides potently reduce apolipoprotein B mRNA and serum cholesterol in mice and non-human primates. Nucleic Acids Res. 2010;38(20):7100–11. Epub 2010/07/10. 10.1093/nar/gkq457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burdick AD, Sciabola S, Mantena SR, Hollingshead BD, Stanton R, Warneke JA, et al. Sequence motifs associated with hepatotoxicity of locked nucleic acid—modified antisense oligonucleotides. Nucleic Acids Res. 2014;42(8):4882–91. Epub 2014/02/20. 10.1093/nar/gku142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stanton R, Sciabola S, Salatto C, Weng Y, Moshinsky D, Little J, et al. Chemical modification study of antisense gapmers. Nucleic Acid Ther. 2012;22(5):344–59. Epub 2012/08/03. 10.1089/nat.2012.0366 . [DOI] [PubMed] [Google Scholar]
- 10.Henry S. P KT-W, Kramer-Stickland K., Zanardi T.A., Fey R.A., Levin A.A. Toxicologic properties of 2’-methoxyethyl chimeric antisense inhibitors in animals and man In: (Ed.) C ST, editor. Antisense drug technology: principles, strategies and applications. 2nd ed ed. Carlsbad: CRC Press; (2008). p. 327–63 [Google Scholar]
- 11.Hagedorn PH, Yakimov V, Ottosen S, Kammler S, Nielsen NF, Hog AM, et al. Hepatotoxic potential of therapeutic oligonucleotides can be predicted from their sequence and modification pattern. Nucleic Acid Ther. 2013;23(5):302–10. Epub 2013/08/21. 10.1089/nat.2013.0436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drygin D, Barone S, Bennett CF. Sequence-dependent cytotoxicity of second-generation oligonucleotides. Nucleic Acids Res. 2004;32(22):6585–94. Epub 2004/12/18. 10.1093/nar/gkh997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. Epub 2002/02/16. 10.1006/meth.2001.1262 . [DOI] [PubMed] [Google Scholar]
- 14.Stein CA, Hansen JB, Lai J, Wu S, Voskresenskiy A, Hog A, et al. Efficient gene silencing by delivery of locked nucleic acid antisense oligonucleotides, unassisted by transfection reagents. Nucleic Acids Res. 2010;38(1):e3 Epub 2009/10/27. 10.1093/nar/gkp841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koller E, Vincent TM, Chappell A, De S, Manoharan M, Bennett CF. Mechanisms of single-stranded phosphorothioate modified antisense oligonucleotide accumulation in hepatocytes. Nucleic Acids Res. 2011;39(11):4795–807. Epub 2011/02/25. 10.1093/nar/gkr089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Butler M, Stecker K, Bennett CF. Cellular distribution of phosphorothioate oligodeoxynucleotides in normal rodent tissues. Lab Invest. 1997;77(4):379–88. Epub 1997/11/14. . [PubMed] [Google Scholar]
- 17.Wang K, Zhang S, Marzolf B, Troisch P, Brightman A, Hu Z, et al. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci U S A. 2009;106(11):4402–7. Epub 2009/02/28. 10.1073/pnas.0813371106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Filipowicz W, Grosshans H. The liver-specific microRNA miR-122: biology and therapeutic potential. Prog Drug Res. 2011;67:221–38. Epub 2010/12/15. . [PubMed] [Google Scholar]
- 19.Tolcher AW P A, Papadopoulos KP, Agnew J, Lokiec FM, Rezai K, Kalambakas S, and Buchbinder. Results of a phase 1, open-label, dose-escalation study evaluating the safety and tolerability of EZN-3042, a survivin mRNA antagonist, administered with docetaxel (D) in adult patients (Pts) with advanced solid tumors. Mol Cancer Ther. 2011;10:A216 [Google Scholar]
- 20.Raetz EA, Morrison D, Romanos-Sirakis E, Gaynon P, Sposto R, Bhojwani D, et al. A phase I study of EZN-3042, a novel survivin messenger ribonucleic acid (mRNA) antagonist, administered in combination with chemotherapy in children with relapsed acute lymphoblastic leukemia (ALL): a report from the therapeutic advances in childhood leukemia and lymphoma (TACL) consortium. J Pediatr Hematol Oncol. 2014;36(6):458–63. Epub 2013/11/28. 10.1097/MPH.0b013e3182a8f58f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frieden M, Orum H. The application of locked nucleic acids in the treatment of cancer. IDrugs. 2006;9(10):706–11. Epub 2006/10/04. . [PubMed] [Google Scholar]
- 22.Juliano RL, Ming X, Carver K, Laing B. Cellular uptake and intracellular trafficking of oligonucleotides: implications for oligonucleotide pharmacology. Nucleic Acid Ther. 2014;24(2):101–13. Epub 2014/01/05. 10.1089/nat.2013.0463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Senn JJ, Burel S, Henry SP. Non-CpG-containing antisense 2'-methoxyethyl oligonucleotides activate a proinflammatory response independent of Toll-like receptor 9 or myeloid differentiation factor 88. J Pharmacol Exp Ther. 2005;314(3):972–9. Epub 2005/05/28. 10.1124/jpet.105.084004 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Secreted LDH (upper panel) intracellular ATP (lower panel) concentrations after 3 day treatment with a tool set of safe (32, 35) and hepatotoxic (43, 47) SSOs. Data are means ± SD.
(TIF)
LDH levels in the supernatant of primary mouse NPCs after 2 day treatment with safe (32, 35) and toxic (43, 47) SSOs. Data are means ± SD.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.