Abstract
The human APOBEC3 family of DNA cytosine deaminases serves as a front-line intrinsic immune response to inhibit the replication of diverse retroviruses. APOBEC3F and APOBEC3G are the most potent factors against HIV-1. As a countermeasure, HIV-1 viral infectivity factor (Vif) targets APOBEC3s for proteasomal degradation. Here, we report the crystal structure of the Vif-binding domain in APOBEC3F and a novel assay to assess Vif-APOBEC3 binding. Our results point to an amphipathic surface that is conserved in APOBEC3s as critical for Vif susceptibility in APOBEC3F. Electrostatic interactions likely mediate Vif binding. Moreover, structure-guided mutagenesis reveals a straight ssDNA-binding groove distinct from the Vif-binding site, and a novel ‘aromatic switch’ is proposed to explain DNA substrate specificities across the APOBEC3 family. This study opens new lines of inquiry that will further our understanding of APOBEC3-mediated retroviral restriction and provides an accurate template for structure-guided development of inhibitors targeting the APOBEC3-Vif axis.
INTRODUCTION
The human intrinsic immune system has developed complex responses to prevent the spread of retroviral pathogens. The apolipoprotein B mRNA-editing catalytic polypeptide 3 (APOBEC3, denoted here as A3)1–3 family of single-stranded (ss) DNA cytosine deaminases provides a critical post-entry obstruction to retroviral replication. In humans, seven family members (A3A, A3B, A3C, A3D, A3F, A3G and A3H) are encoded on chromosome 22 (Fig. 1A)4. A common characteristic of all A3 enzymes is the presence of either one or two zinc-coordinating, DNA cytosine deaminase domains, typically labeled as CD1 and CD24. Each cytosine deaminase domain contains the canonical (C/H)-(A/V)-E-(X23–28)-P-C-X2-C motif and belongs to one of three classes: Z1 (A3A and the CD2 of A3B and A3G), Z2 (A3C, the CD1 of A3B and A3G, and both CD1 and CD2 of A3D and A3F) or Z3 (A3H haplotype II)5. Z2-cytosine deaminase domains can be further divided into three subgroups: A3F-CD1 (CD1 of A3B, A3D and A3F), A3F-CD2 (A3C, and CD2 of A3D and A3F) and A3G-CD1 subgroups (Fig. 1A).
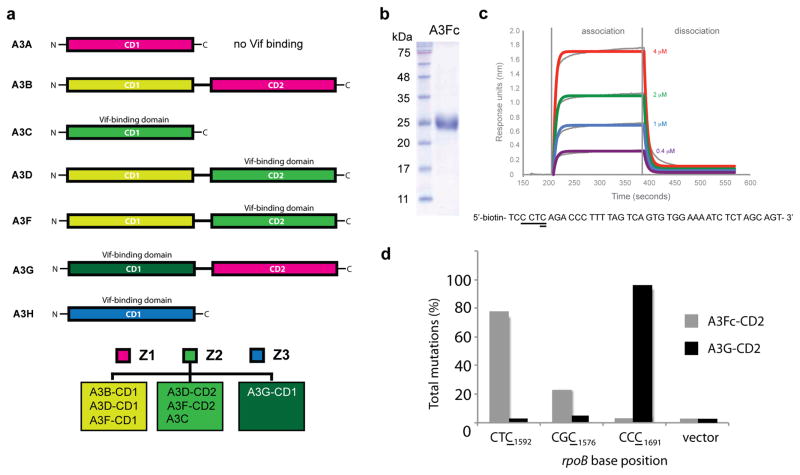
Figure 1. APOBEC3 and characterization of A3Fc-CD2.
(a) Schematic of the seven A3 intrinsic immune restriction factors (A3A, A3B, A3C, A3D, A3F, A3G and A3H). The three classes of DNA cytosine deaminase domains, Z1, Z2 and Z3 are colored pink, green, blue, respectively. The Z2-cytosine deaminase domains are further classified into three subgroups based on sequence similarity. A3F and A3G are the two most potent A3 proteins and exhibit disparate Vif binding sites (CD1 for A3G and CD2 for A3F). (b) SDS-polyacrylamide gel electrophoresis of the purified A3Fc-CD2 after final size exclusion chromatography. (c) Biolayer interferometry kinetic analysis of A3Fc-CD2 binding to ssDNA. Biotin-labeled ssDNA was coupled to streptavidin-coated biosensors and monitored for binding to purified A3Fc-CD2 at 0, 0.4, 1, 2 and 4 μM concentrations. The data was analyzed based on a 1:1 binding model using the BLItz Pro software, with the fitted curves shown as grey lines. The ssDNA sequence used in the assay is shown below the sensorgram, with the A3Fc-CD2 deamination site and target DNA cytosine underlined and double underlined, respectively. (d) RifR mutation profile of E. coli expressed A3Fc-CD2 and A3G-CD2. Histogram showing the percent mutation on specific rpoB nucleotide sequences for A3Fc-CD2 and A3G-CD2. Results are expressed as the percentage of total mutations from six independent experiments with at least 20 RifR colonies sequenced for both A3Fc-CD2 and A3G-CD2.
Experiments with cultured cells demonstrate that only A3D, A3F, A3G and A3H restrict HIV-16, and that A3F and A3G are the most abundant and potent inhibitors7–9. Both A3F and A3G are double-domain DNA cytosine deaminases, and are expressed and packaged into viral particles as oligomers bound to viral RNA and structural proteins10. During reverse transcription, A3 deaminates retroviral DNA cytosines. This conversion of dC to dU results in the incorporation of dA rather than dG in the positive-strand DNA, leading to premature stop codons and detrimental mutations in viral proteins. Of course, HIV-1 has developed countermeasures to antagonize this intrinsic host defense response. HIV-1 viral infectivity factor (Vif) is a late-onset viral protein that recruits an E3 ubiquitin ligase complex (elongin B/C, core binding factor-β, cullin 5, and Rbx) to polyubiquitinate A3, leading to its proteasomal degradation and loss of packaging into nascent virions11,12. In addition, Vif has been shown to impede A3 mRNA translation, virion encapsidation and deamination activity13–15. The degradation of A3F and A3G is dependent on recognition and binding of HIV-1 Vif. Interestingly, while A3F and A3G share a high primary sequence similarity of 50.9%, disparate regions in these proteins are involved in binding HIV-1 Vif16,17. In A3F, the CD2 domain mediates both DNA cytosine deamination and binding to HIV-1 Vif, whereas in A3G, the enzymatically inactive CD1 domain is responsible for binding Vif18.
Current antiretroviral drug treatments have led to significant improvements in quality of life for those infected with HIV-1 or living with AIDS. However, error-prone HIV-1 replication produces high genetic variation that leads to the natural selection of drug-resistant variants of HIV-1, thus necessitating the development of alternate strategies. The A3F- or A3G-Vif interface is an attractive target as a fresh anti-HIV therapeutic approach19. Inhibition of HIV-1 Vif binding to A3F or A3G would allow the reactivation of effective host innate immune responses to HIV-1. More importantly, inhibitors targeting the host side of the A3F- or A3G-Vif interface will be less sensitive to viral mutations and will reduce the probability of HIV-1 variants developing resistance to drug treatments.
Here, we report the crystal structure of the Vif-binding domain in A3F-CD2. Given that A3F and A3G are the most potent A3 restriction factors, this is a key structure needed for the development of new classes of inhibitors to enhance intrinsic immunity against HIV. We perform site-directed mutagenesis to map out the ssDNA-binding site, which reveals a straight groove for DNA binding, as well as a novel aromatic switch that confers nucleotide preferences. Moreover, we develop the first biophysical HIV-1 Vif-APOBEC3 binding assay, and this allows us to identify a negatively charged Vif-binding surface distinct from the ssDNA-binding groove.
RESULTS
A soluble chimeric A3F C-terminal domain
A3 family members are difficult to express and purify in soluble, stable and homogenous quantities. This has hindered structural studies on this important class of macromolecules. Expression of the full-length, CD1 or CD2 A3F domains in E. coli cells resulted in the formation of inclusion bodies. Multiple E. coli cell lines and expression temperatures were tried, but to no avail. Re-engineering of these proteins is necessary to impart solubility and stability. We noticed that residues 195–220 of the A3F-CD2 domain are rich in hydrophobic residues, leading us to hypothesize that these amino acids may lead to protein aggregation. In contrast, residues 197–221 of A3G-CD2 are comparatively less hydrophobic. To generate a soluble construct of A3F-CD2, residues 195–217 of A3F-CD2 were replaced with residues 197–221 from A3G-CD2. This chimeric A3F (A3Fc-CD2) construct (Supplementary Fig. S1) expressed as soluble protein in multi-milligram quantities (Fig. 1B).
The A3Fc-CD2 was shown by in vitro biochemical and biophysical assays to be fully functional. Using biolayer interferometry (BLI), we found that A3Fc-CD2 binds ssDNA with an equilibrium dissociation constant (Kd) of 7 μM (Fig. 1C), and as expected, does not bind double-stranded DNA. Previous chemical shift perturbation and gel shift assays have shown that A3A and A3G-CD2 bind ssDNA with Kd values in the micromolar range20–22, consistent with A3Fc-CD2. In addition, a heterologous E. coli DNA cytosine deaminase mutational assay18 revealed that A3Fc-CD2 was enzymatically active and preferentially targets a 5′-CCTC-3′ sequence in the E. coli RNA polymerase (rpoB) gene (Fig. 1D), consistent with previous studies that show A3F deamination within 5′-TC-3′ dinucleotide signatures in the rpoB gene18.
Crystal structure of A3Fc-CD2
A3Fc-CD2 was crystallized using a unique microseeding, discontinuous vapor diffusion method to reduce protein precipitation and improve crystal growth. Diffraction of the crystals presented additional challenges, as most crystals diffracted between 5 to 10 Å resolution. In the end, only one crystal out of 100 diffracted to ~3.1 Å resolution. The crystal structure of A3Fc-CD2 was determined by molecular replacement and the final refined model consists of A3Fc-CD2 residues 218–373 and a 25-residue N-terminal extension from the corresponding A3G sequence. Data collection and refinement statistics are presented in Table 1.
Table 1.
Data collection and refinement statistics
| A3Fc-CD2 | |
|---|---|
| Data collection | |
| Space Group | I422 |
| Cell dimensions | |
| a,b,c (Å) | 164.0, 164.0, 135.3 |
| α,β,γ (°) | 90 |
| Resolution (Å) | 43.5-3.0 |
| Rmerge (%)a | 10.0 (79.6)b |
| I/σ(I) | 7.2 (1.3)b |
| Completeness (%) | 98.5 (100.0)b |
| Redundancy | 9.6 (10.5)b |
| Refinement | |
| Resolution (Å) | 43.5-3.1 |
| No. unique reflections | 18,499 (1,839)b |
| Rwork (%)c | 23.6 |
| Rfree (%)d | 27.7 |
| No. atoms | 5775 |
| protein | 366 |
| ions | 2 |
| B-factors | |
| overall | 97.2 |
| ions | 75.9 |
| R.m.s. deviations | |
| bonds (Å) | 0.011 |
| angles (°) | 1.5 |
Rmerge = ΣΣj | Ij − <I> |/Σ | <I> |, where Ij and <I> represent the diffraction intensity values of the individual measurements and the corresponding mean values. The summation is over all unique measurements.
Values given in parentheses refer to reflections in the outer resolution shell: 3.11-3.00 Å
, where Fobs and Fcalc are the observed and calculated structure factors, respectively.
For Rfree, the sum is extended over a subset of reflections (5%) excluded from all stages of refinement.
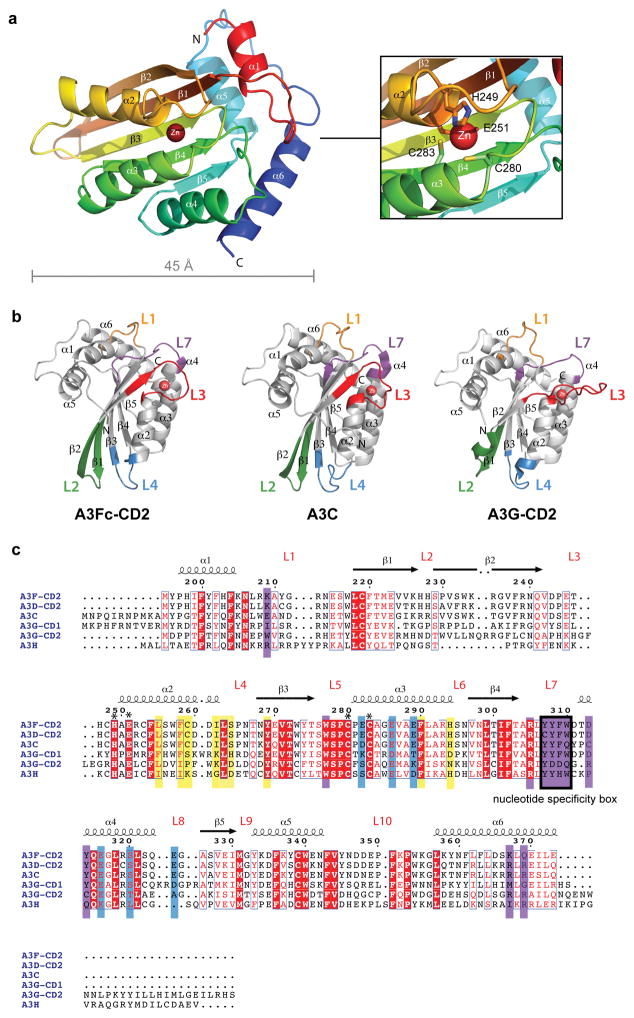
The crystal structure of A3Fc-CD2 consists of a five-stranded, mixed β-sheet core flanked by three α-helices on either side (Fig. 2A). Our overall A3Fc-CD2 structure resembles A3C (PDB: 3VOW) and A3G-CD2 structures (PDB: 3EIU, 2JYW, 2KBO and 3IR2)21–25. Superimposition of A3Fc-CD2 with A3C and A3G-CD2 revealed an overall Cα root mean squared deviation of 2.3 Å and 1.3 Å, respectively. The significant resemblance in the overall fold of A3Fc-CD2 to A3G-CD2 and A3C suggests that this is a conserved structural core for the A3 family.
Figure 2. Crystal structure of A3Fc-CD2.
(a) Ribbon diagram of A3Fc-CD2. The chain is colored in a rainbow gradient from red (N-terminus) to blue (C-terminus). The catalytic zinc atom is labeled and shown as a grey sphere. The inset box shows a zoomed view of the catalytic site. (b) Structural difference between A3Fc-CD2, A3G-CD2 and A3C. All APOBEC3 cytosine deaminase domains have 10 loops. The largest structural deviations reside in loops L1, L2, L3, L4 and L7, colored orange, green, red, blue and purple, respectively. (c) Multiple sequence alignment of A3F-CD2, A3C, A3D-CD2, A3G-CD1, A3G-CD2 and A3H. Numbering of the sequences and depiction of secondary structural elements are based on A3Fc-CD2, and is shown above the sequences. Strictly conserved residues are highlighted in red. Putative A3Fc-CD2 ssDNA-binding and Vif-binding residues are outlined in purple and blue boxes, respectively. Previously identified residues in the hydrophobic V-shaped groove formed by the α2 and α3 helices are outlined in yellow. Catalytic site residues are denoted with an asterisk above its sequence. The sequence alignment was produced using Clustal W51,52 and the alignment graphics were generated using the program ESPript53.
In A3Fc-CD2, the catalytic zinc atom is sequestered by H249, C280 and C283 within the canonical (C/H)-(A/V)-E-(X23–28)-P-C-X2-C cytidine deaminase motif (Fig. 2A). The active site residues superimpose well in all A3 structures. In A3G-CD2 and A3C, a water molecule is coordinated to the catalytic zinc to complete a tetrahedral geometry. This water molecule is activated to become a nucleophile for deamination of the target deoxycytidine nucleotide. In A3Fc-CD2, the catalytic water molecule is not observed due to the moderate resolution of the electron density map.
While the overall structure and active site are similar among all A3 proteins, significant differences are observed in the loops that surround the structural core. Loops have higher conformational flexibility than secondary structural elements, and are often involved in crystal packing interactions. All ten A3Fc-CD2 loops that surround the α/β core superimpose well with those from A3C, suggesting any loops involved in forming the crystal lattice are not significantly affected. In contrast, superimposition of A3Fc-CD2 and A3C to A3G-CD2 revealed structural differences in loops 1, 2, 3, 4 and 7, with the largest displacements in loops 2 and 3 (Fig. 2B). Loop 2 resides between the β1 and β2 strands. The β2 strand in A3Fc-CD2 and A3C forms a 12-residue continuous strand, whereas in A3G-CD2, a bulge disrupts β2 into two four-residue strands (referred to β2 and β2′). Previous NMR and crystallographic experiments showed that the bulge is not an experimental artifact, but rather an intrinsic feature of A3G-CD225. The β2 strand of CD1 was proposed in modeling studies to pair with the β2 strand of CD2 to form an extended β–sheet in full-length A3 structures26–28. If this were the case, differences in β2 would likely result in disparities at the intermolecular interface between the two catalytic domains of A3F and A3G. However, recent studies have contradicted this model24,29 and further studies are required to characterize the interface between CD1 and CD2. Loop 3 is formed between the β2 strand and the α2 helix, and a four-residue insertion in loop 3 of A3G-CD2 produces an extended structure compared to A3C and A3Fc-CD2. In A3G, loop 3 may be involved in Zn-mediated oligomerization25, whereas in A3Fc-CD2 and A3C, loop 3 is shorter and no such protein-protein interactions were detected.
ssDNA-binding site of A3Fc-CD2
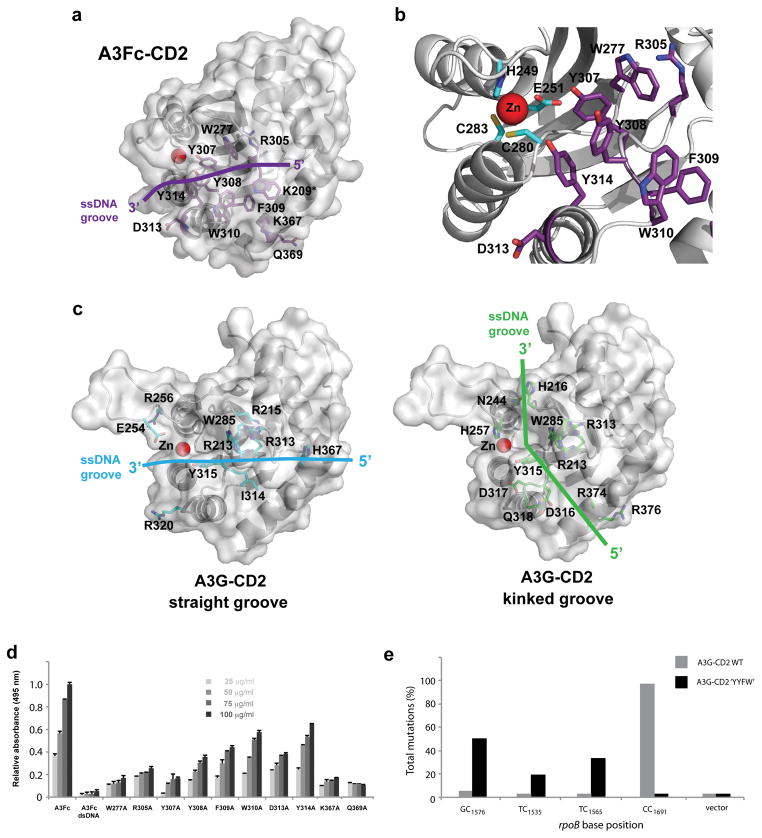
In A3Fc-CD2, the molecular surface reveals a single, partial groove that leads into the catalytic site. This groove is formed between loops 1 and 3 on one side and loop 7 on the opposite side. A number of aromatic (W277, Y307, Y308, F309, W310 and Y314), and charged residues (K209, R305, and D313) line the channel (Fig. 3A and 3B). The positively charged R305 and aromatic W277 and Y307 residues are conserved between A3F-CD2 and A3G-CD2, suggesting a key role in ssDNA binding (Fig. 2C). Based on the overall active site structure, W277 and Y307 are in close proximity to the catalytic residues and zinc atom (Fig. 3B). These two residues are in excellent position to form stabilizing pi-pi interactions to the target nucleotide base. Residue R305 is located directly behind W277 in the active site and forms pi-cation interactions capable of stabilizing W277. Residues Y308, F309, W310 and Y314 line one face of the ssDNA-binding groove and may form base stacking interactions with other bases on the nucleotide chain (for example the -1 and -2 positions of the 5′-TTCA-3′ A3F deamination motif).
Figure 3. Identification of the A3Fc-CD2 ssDNA-binding site.
(a) Molecular surface of the putative A3Fc-CD2 ssDNA-binding site. Key residues proposed for DNA binding are shown in purple. Aromatic and positively charges residues line the wall of a straight groove. This DNA binding groove is consistent with Model #1 in panel (c). Note: Residue W209 belongs to the A3G solubilization linker attached at the N-terminus of A3Fc-CD2. The native residue in A3Fc-CD2 is a lysine, and this residue is in position to accommodate the negative charges fro the phosphate backbone of the ssDNA strand. (b) A3Fc-CD2 active site. Catalytic and ssDNA binding residues are shown in cyan and purple sticks, respectively. (c) Two models of A3G-CD2 ssDNA binding. Model #1 is proposed by Furukawa, et al. and Chen, et al. and is based on a NMR titration analysis of ssDNA21,22. DNA-binding residues were found to line a straight DNA binding groove. Model #2 is proposed by Holden, et al. and displays the DNA binding residues along a kinked groove23. Both A3G-CD2 molecular surfaces are shown in the same orientation as panel (a). (d) Nucleic acid-protein interaction ELISA assay. Alanine scanning mutagenesis of selected A3Fc-CD2 ssDNA-binding site residues. dsDNA was used as a negative control. Results are expressed as the mean relative absorbance (+ standard deviation of the population) of three replicates. (e) RifR mutation profile of E. coli expressed WT A3G-CD2 and A3G-CD2 ‘YYFW’. Histogram showing the percent total mutation on specific rpoB nucleotide sequences. Results are expressed as the percentage of total mutations from six independent experiments with at least 20 colonies sequenced.
To study the roles of each residue in the putative ssDNA-binding groove, alanine-mutants were generated for each position and characterized by BLI and ELISA. These two techniques allow cross-validation of the results and determination of equilibrium binding constants. ELISA studies showed that alanine substitutions at positions 277, 305, 307, 308, 309, 310, 313, 314, 367 and 369 resulted in reduced affinity for ssDNA (Fig. 3D, Table 2 and Supplementary Fig. S2). Kinetic measurements with BLI revealed that alanine substitutions at W277, Y307, D313, K367 and Q369 resulted in the greatest decrease in affinity. Given the importance of W277 and Y307 in coordination of the target deoxycytidine, mutations to these residues, not surprisingly, displayed the largest decreases in ssDNA binding. Mutations distal to the active site (Y308A, F309A, W310A and Y314A) revealed smaller effects on ssDNA binding. The stability of each mutant was also assessed by circular dichroism spectroscopy (results presented in Supplementary Information). In summary, mutations in the ssDNA-binding site did not affect the overall structure of A3Fc-CD2.
Table 2.
A3Fc-CD2 ssDNA-binding site mutations
| A3Fc-CD2 variant | Expression level | Kd (μM) | Tm (°C) |
|---|---|---|---|
| WT | +++++ | 7.2 | 60 |
| W277A | ++ | n.d. | 57 |
| R305A | + | n.d. | - |
| Y307A | +++++ | 410 | 58 |
| Y308A | +++++ | 18 | 58 |
| F309A | +++++ | 31 | 55 |
| W310A | ++++ | 27 | 55 |
| D313A | +++++ | 220 | 58 |
| Y314A | +++++ | 68 | 52 |
| K367A | +++++ | n.d. | 56 |
| Q369A | +++ | 760 | 58 |
n.d.- not determined; too low binding
Implication of an aromatic switch for substrate specificity
In the A3Fc-CD2 structure, a series of aromatic residues in loop 7 (Y307, Y308, F309 and W310) line one side of the ssDNA-binding groove and are in the vicinity of the active site (Fig. 3B). Residues in A3Fc-CD2 loop 7 (Y307, Y308, F309 and W310) are capable of forming favorable stacking interactions with the nucleotide bases at positions -1 and -2 in A3F 5′-TTCA-3′ sequence element. Equivalent residues in A3A were shown by molecular docking studies to be involved in such interactions with the two nucleotides preceding the target cytidine20. These aromatic residues are not conserved across the A3 family (Fig. 2C), thus we predict differences here may determine target DNA specificity. In A3F-CD2, this region is aromatic-rich, with a ‘YYFW’ motif (termed the nucleotide specificity box). However, in A3C and A3G-CD2, the ‘YYFW’ motif is replaced by ‘YYFQ’ and ‘YDDQ’ motifs, respectively. We propose that this ‘aromatic switch’ in the nucleotide specificity box determines substrate specificities in A3s.
To test our hypothesis, we employed a heterologous bacterial DNA cytosine deaminase assay18 to determine whether we are able to switch the substrate preference of A3G to A3F. The rpoB gene contains multiple mutable cytosines in various nucleotide sequence contexts. Thus, identification of rpoB mutational hotspots allows us to analyze substrate preferences at the nucleotide level. In agreement with previous studies, wild-type A3G-CD2 has a strong preference for 5′-CC-3′ dinucleotides (Fig. 3E)18,30,31. Mutating the ‘YDDQ’ nucleotide specificity box in A3G-CD2 to the aromatic rich ‘YYFW’ counterpart in A3F-CD2 resulted in a complete change in substrate preference from 5′-CC-3′ to 5′-TC-3′ or 5′-GC-3′, suggesting four residues within the nucleotide specificity box is all that is necessary to confer substrate specificity (Fig. 3E). Moreover, the rpoB hotspots in A3G-CD2 ‘YYFW’ are in excellent agreement with those preferred by A3F.
A direct HIV-1 Vif-A3F binding assay
The analysis of A3 proteins to HIV-1 Vif have largely been limited to cell-based biochemical assays. To our knowledge, we have now developed the first direct in vitro binding assay, using biolayer interferometry to measure the binding kinetics between full length HIV-1 Vif and A3Fc-CD2. HIV-1 Vif is expressed in E. coli and refolded from inclusion bodies (Supplementary Fig. S3A). Refolded HIV-1 Vif was soluble and stable at low concentrations (<0.1 mg/ml). A circular dichroism wavelength scan of the refolded Vif revealed a β-character protein (Supplementary Fig. S3B), consistent with previous studies32. A3Fc-CD2 bound to biotinylated full-length HIV-1 Vif with a dissociation equilibrium constant (Kd) of 1.1 μM (Supplementary Fig. S4). Our Vif binding assay enabled us to map the HIV-1 Vif binding interface at single-residue resolution, and provides a foundation for the development of new high throughput assays.
HIV-1 Vif binding sites in A3Fc-CD2
Sequence analysis of HIV-1 Vif reveals a unique molecule with an abundance of positively charged (predicted pI >10) and hydrophobic residues. Previous studies have shown that a number of positively charged and hydrophobic motifs on HIV-1 Vif (11Wx2SLVK26,14DRMR17, 55VxIPLx4L64, 69YxxL72, 74TGERxW79, 81LGxGx2IxW89, 96TQx5ADx2I107 and 171EDRW174) are involved in binding to A3F17,33–36. Therefore, a complementary binding surface on A3F-CD2 would be negatively charged and hydrophobic.
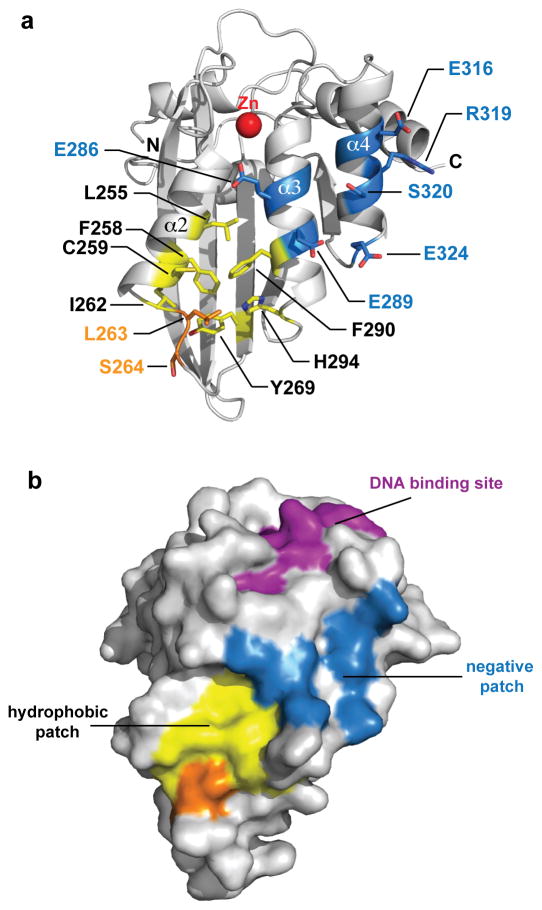
Structural analysis of A3Fc-CD2 revealed a large negatively charged surface formed by the α3 and α4 helices, which is also conserved in A3C and A3D-CD2. Two negatively charged residues, E289 and E324, were previously identified to be involved in Vif binding16,24,37. However, the α3 and α4 helices also have a rich clustering of negatively charged residues (E286, E289, E316, S320 and E324) that we hypothesize to be important to Vif interaction (Fig. 4). Substitution of any of these residues to alanine or glutamine completely abolished binding of A3Fc-CD2 to HIV-1 Vif (Table 3). Mutation to a neighboring arginine residue (R319A) had only minor effects on Vif binding, suggesting that only negatively charged or polar residues are directly involved in Vif binding.
Figure 4. A3Fc-CD2 HIV-1 Vif-binding site.
(a) Biolayer interferometry kinetic analysis of A3Fc-CD2 binding to refolded full length HIV-1 Vif. Biotin-labeled HIV-1 Vif was coupled to streptavidin-coated biosensors and monitored for binding to purified A3Fc-CD2 at 0, 0.4, 1, 2 and 4 μM concentrations. The data were analyzed based on a 1:1 binding model, as only one A3F deamination motif is found on the ssDNA. The calculated fitted curves are shown as grey lines. (b) Putative Vif-binding residues. A number of aromatic and hydrophobic residues, colored in yellow, (L255, F258, L263, Y269 and F290) are buried at the A3Fc-CD2 core. Residues L263 and S264 (shown in orange) were identified in this study to be not important for Vif binding.
Table 3.
A3Fc-CD2 Vif-binding site mutations
| A3Fc-CD2 variant | Expression level | Kd (μM) | Tm (°C) |
|---|---|---|---|
| Vif hydrophobic binding site | |||
| WT | +++++ | 1.1 | 60 |
| L255A | + | n.b. | unstable |
| L255D | ++ | n.b. | 62 |
| F258A | + | n.b. | unstable |
| C259K | ++ | n.b. | 62 |
| C259S | +++++ | n.b. | 54 |
| I262A | + | n.b. | 62 |
| L263A | + | 1.1 | unstable |
| S264D | +++++ | 1.3 | 54 |
| Y269A | + | n.b. | 50 |
| F290A | + | n.b. | 53 |
| F290K | + | n.b. | 62 |
| H294A | +++ | n.b. | 55 |
| H294D | ++ | n.b. | 60 |
| Vif negative binding site | |||
| E286A | +++++ | n.b. | 56 |
| E289Q | +++++ | n.b. | 56 |
| E316Q | +++++ | n.b. | 56 |
| R319A | +++++ | 0.5 | 50 |
| S320A | +++++ | n.b. | 53 |
| E324Q | ++++ | n.b. | 57 |
n.b.- no detectable binding
Recent biochemical studies suggested that a hydrophobic V-shaped groove formed by the α2 and α3 helices of A3F participates in Vif binding17,24,37. This putative hydrophobic Vif-binding site is composed of residues L255, F258, C259, I262, L263, S264, Y269, F290 and H294 (Fig. 4A and 4B). To characterize the role of these hydrophobic residues, alanine- or charge-introducing (aspartate or lysine) mutations were generated. Using our BLI assay, we showed that mutations to L255, F258, C259, I262, Y269, F290 and H294 resulted in loss of Vif binding (Table 3), consistent with previous biochemical studies24. However, the L263A and S264D mutations, which were previously shown to confer 100% resistance to Vif24, resulted in Kd values similar to wild-type A3Fc-CD2, questioning the importance of L263 and S264 in Vif binding (Table 3 and Supplementary Fig. S4).
DISCUSSION
The acquired immunodeficiency syndrome that results from HIV-1 infection remains a global health threat. Interactions between host restriction factors and viral antagonists, such as APOBEC3 and Vif, respectively, represent intriguing targets for the development of drugs to restrict viral replication and dissemination. In this study, we present the crystal structure of a chimeric A3F C-terminal domain containing the Vif-interaction interface. Our combinatorial approach encompassing structural, biochemical and biophysical studies provides insights into the molecular determinants of ssDNA binding, substrate specificity and, importantly, HIV-1 Vif binding.
How the A3 proteins recognize single-stranded nucleic acids has been a controversial question. Structures of A3G-CD2 from NMR and X-ray crystallographic studies yielded at least two distinct models of ssDNA binding (Fig. 3C)21–23. Based on NMR data, Furukawa et al. and Chen, et al. proposed that the DNA-binding site forms a straight groove, with loops 1 and 3 forming one side of the groove and loops 5 and 7 on the other (“Straight model”). The crystal structure of A3G-CD2 and mutational data suggest an alternative mode of ssDNA binding, where the nucleic acid resides in a kinked groove (“Kinked model”)23. In the kinked model, DNA contacts made through residues R213, H216, N244, Y315, D316, D317, Q318, R320, R374, R376 and W285 are proposed to create a bend in the groove (Fig. 3C). In A3Fc-CD2, the direction of the ssDNA-binding groove is most consistent with the “straight model” of DNA binding (Fig. 3C). There are no grooves on the surface of A3Fc-CD2 that correspond to the A3G kinked channel. Detailed comparison of the A3Fc-CD2 and A3G-CD2 crystal structures reveals conformational differences in loops 1 and 7. In A3Fc-CD2, residue W209 and W310 in loop 1 and 7, respectively, appear to block the entrance of the kinked DNA channel. Mutations to residues along the A3F-CD2 straight ssDNA-binding groove (W277, R305, Y307, Y308, F309, W310, Y314) confirm their importance in ssDNA binding. Interestingly, mutations in A3Fc-CD2 that are equivalent to residues proposed to contact ssDNA according to the A3G kinked model (D313, K367, and Q369) also resulted in a significant decrease in ssDNA binding (Fig. 3D and Table 2). While our apo A3Fc-CD2 structure clearly shows a straight ssDNA-binding groove, a substrate-induced conformation change may bring these residues into proximity to bind ssDNA. In addition, it should be emphasized that our current structural understanding of A3 proteins is limited to the analysis of a single domain of A3 proteins. It may also be possible that ssDNA binds to residues identified in both the “straight” and “kinked” models of DNA binding in the context of full-length or higher-ordered oligomeric A3 protein structures.
The A3 cytosine deaminase domains adopt similar structures; however, each has distinct substrate preferences. A3F-CD2 deaminates DNA cytosine within a 5′-TC-3′ sequence, whereas A3G recognizes 5′-CC-3′38. A3A and A3C target both 5′-TC-3′ and 5′-CC-3′ dinucleotides20,39. Our results suggest that the ‘YDDQ’ nucleotide specificity box in A3G-CD2 confers specificity for 5′-CC-3′ dinucleotides, whereas the ‘YYFW’ box in A3Fc-CD2 targets both 5′-TC-3′ and 5′-GC-3′ motifs. There is a clear trend; as the nucleotide specificity box motif becomes more negatively charged and polar, there is an increased preference for cytosine bases. This is consistent with the physiochemical differences between cytosine and thymine/guanine. The cytosine base is three times more hydrophilic than thymine40. The 4-amino group on the cytosine base can form hydrogen bonds to the aspartate or glutamine acceptors found in the A3G-CD2 ‘YDDQ’ nucleotide specificity box. An aromatic rich ‘YYFW’ nucleotide specificity box in A3F-CD2 provides a more complementary environment for the hydrophobic thymidine nucleotides. A3C, which has only one glutamine in its ‘YYFQ’ nucleotide specificity box, recognizes a degenerate sequence– it is able to accommodate the substrates of both A3F (5′-TC-3′) and A3G (5′-CC-3′)39. Our study clearly demonstrates the importance of four residues in the nucleotide specificity box and a switch in hydrophobicities in determining substrate specificities in A3 proteins.
Our current crystal structure and biophysical characterization of A3Fc-CD2 has revealed key determinants of Vif binding. The A3Fc-CD2 Vif-binding interface forms a shallow amphipathic cavity composed of residues from three helices (α2, α3 and α4 helix). Based on our Vif-A3F interaction assay, hydrophobic residues (L255, F258, C259, I262, Y269, F290, and H294) and negatively charged amino acids (E286, E289, E316, and E324) in A3Fc-CD2 were shown to be directly involved in Vif binding. However, solvent accessibility calculations revealed that residues in the hydrophobic Vif-binding groove, L255, F258, I262, Y269 and F290, have a side chain exposed surface area of less than 20% relative to random coil, suggesting that these residues are buried in the protein core and play a role in maintaining structural integrity. Alanine mutations to many of the hydrophobic residues affected the relative contents of secondary structural elements, as determined by CD spectroscopy (Supplementary Discussion and Supplementary Fig. S5), and resulted in a significant decrease in overall protein expression levels (Table 3). In contrast, the mutations to the negatively charged residues did not affect expression levels and CD wavelength scans were superimposable (Supplementary Fig. S6), suggesting no effects on overall structural stability. We suggest that the hydrophobic site is involved in maintaining the structural integrity and stability of A3Fc-CD2. However, it may be possible that the hydrophobic Vif-binding site plays a role in both protein stability and Vif binding. Full characterization of the role of the hydrophobic site in Vif binding awaits the structural determination of an A3-Vif complex.
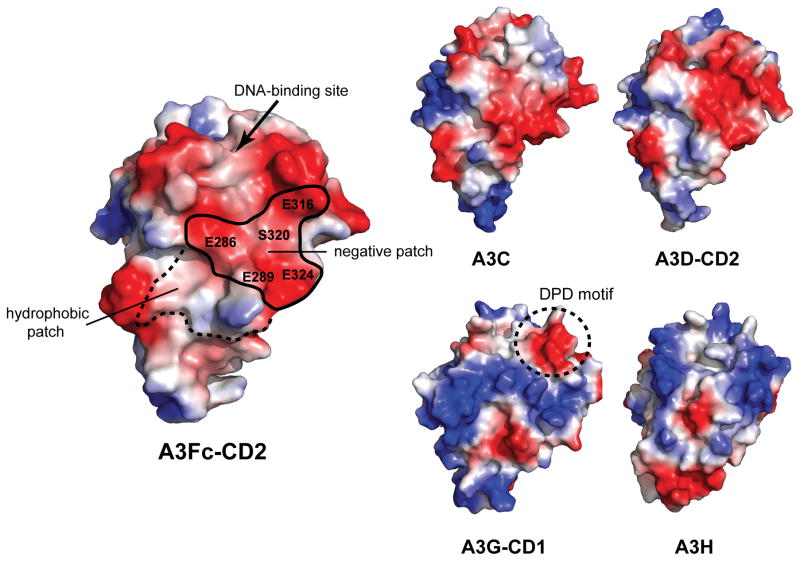
The negatively charged Vif-binding site identified in this study now presents an attractive surface for the development of novel inhibitors capable of disrupting the A3-Vif interface. This negatively charged Vif-binding site is well conserved in A3C and a homology model of A3D-CD2 (Fig. 5), suggesting a common mode of Vif binding for the A3F-CD2 Z2-type DNA cytosine deaminase domains. While the Vif-binding A3G-CD1 domain also belongs to the Z2-class of DNA cytosine deaminase domains, it shares only 43% sequence identity with A3F-CD2 and the residues implicated in Vif-binding are not conserved. The electrostatic potential surface of A3G-CD1 reveals a replacement of the negatively charged patch in A3Fc-CD2 with a positively charged surface (Fig. 5). Moreover, a ‘DPD’ motif in loop 7 of A3G-CD1 previously implicated in Vif binding41 does not overlap with the negatively charged surfaces in A3F-CD2, A3C, and A3D-CD2. This suggests that A3G-CD1 adopts a unique Vif-binding interface that is distinct from other A3F-like Z2-cytosine deaminase domains. The electrostatic surface of A3H, a Z3-DNA cytosine deaminase, is different from all other A3 proteins (Fig. 5). Based on the current A3 structural models, there are likely multiple A3 interfaces that can interact with Vif.
Figure 5. Surface electrostatic potential of APOBEC3 proteins.
A negatively charged surface conserved with other Z2-cytosine deaminase domains is proposed to be important for Vif-binding (shown in dashed lines). Electrostatic potential mapped onto the molecular surface of A3C (PDB: 3VOW), and the homology models of A3D-CD2, A3G-CD1 and A3H. The proposed footprints of the A3 negative and hydrophobic patch involved in Vif binding are shown by the solid and dashed lines, respectively. A previously characterized ‘DPD’ motif involved in A3G Vif binding is displayed for the A3G-CD1 homology model. Red and blue colored regions denote negative and positive charges, respectively. Note: the ssDNA-binding site is at the top of the depicted A3 molecules and has no overlap with the hydrophobic or negatively charged Vif-binding site.
Our crystal structure presented in this manuscript has now identified well-defined sites on A3Fc-CD2 involved in ssDNA and Vif binding. We have also identified novel structural determinants that explain the differences in substrate specificities between A3 family members. Collectively, our studies and findings will be invaluable to the A3 community by providing the relevant structural scaffold for the development of effective HIV-1 inhibitors aimed at selectively disrupting the A3F-Vif interface.
Footnote: While this manuscript was under review, structures of A3A20 and a C-terminal domain of A3F42 (with 11 amino acid substitutions required for solubilization; termed A3F11X-CD2) were reported. The structures of A3A and A3F11X-CD2 align well with A3Fc-CD2. Interestingly, no conformational changes in the chimeric region of A3Fc-CD2 exist.
METHODS
Expression and purification of APOBEC3F
The DNA corresponding to A3F (residues 218–373) with an A3G solubilization linker (residues 197–221) at the N-terminus was codon-optimized and whole-gene synthesized (Biobasic Inc). This A3Fc-CD2 gene was subsequently cloned, according to manufacturer’s protocol, into a pET46 Ek/LIC vector (EMD Millipore) containing a N-terminal His-tag, and thrombin and TEV cleavage sites (Supplementary Fig. S1). A3Fc-CD2 and mutants were expressed in Rosetta-2 (DE3) E. coli cells. Cell cultures were grown to OD600=0.8 and induced with a final concentration of 0.5 mM IPTG for 18 hours at 25°C. Cells were resuspended in Ni-binding buffer (50 mM Tris-HCl pH 8.0, 300 mM NaCl and 20 mM imidazole) with EDTA-free protease inhibitor cocktail and lysed at 30 kpsi using a hydraulic cell disruption system (Constant Systems TS benchtop). The lysate was centrifuged to remove cellular debris prior to loading onto Ni-NTA resin (Thermo Pierce). A3Fc-CD2 was washed with Ni-NTA binding buffer with 50 mM imidazole. A3Fc-CD2 was eluted by a two-step gradient of Ni-binding buffer with 125 mM imidazole and Ni-binding buffer with 500 mM imidazole. Fractions that contained A3Fc-CD2 were concentrated and purified on a Superdex-200 10/300 GL column equilibrated in Buffer A (10 mM Tris-HCl pH 7.5, 0.3 M NaCl, 0.05% (w/v) CHAPS and 2 mM TCEP). For structural studies, A3Fc-CD2 was further purified by anion exchange chromatography. Here, A3Fc-CD2 was pooled and loaded onto a MonoQ HR 5/5 column equilibrated in Buffer B (10 mM Tris-HCl pH 7.5, 0.1 M NaCl, 0.05% (w/v) CHAPS and 2 mM TCEP). A3Fc-CD2 was eluted using a linear gradient of 0–100% Buffer B with 1 M NaCl. Purified A3Fc-CD2 was quantified by A280 and concentrated to 20 mg/ml for crystallization.
Crystallization and data collection
Initial crystals were obtained by mixing a 1:1 ratio of A3Fc-CD2 protein and 18% (w/v) PEG 8000, 0.1 M CHES pH 9.0, and 25% (w/v) glucose on a cover slide and equilibrating over a reservoir of 20% (w/v) PEG 8000 and 0.1 M CHES pH 9.0 for 10 minutes. Subsequently, the cover slide was transferred over to a reservoir containing 18% (w/v) PEG 8000, 0.1 M CHES pH 9.0 and 25% (w/v) glucose, and allowed to grow at 20°C. Needle-like crystals appeared over a five-day period and were crushed using a glass pedestal. Random matrix microseeding43 was performed using a Douglas Instruments Oryx 8 to obtain additional crystallization hits. High quality crystals were obtained by mixing a 3:1 ratio of A3Fc-CD2 protein to seed stock, and equilibrating over a reservoir of 0.17 M magnesium formate, 20% (v/v) glycerol and 10% (w/v) glucose. Wedge-like crystals were harvested and directly flash cooled in liquid nitrogen. A complete dataset was collected at the Advanced Photon Source (Argonne National Laboratories, Argonne, IL), Beamline 24-ID-E. Data were indexed, integrated and scaled using d*trek44.
Structural determination
The A3Fc-CD2 structure was determined by molecular replacement, using the program Phaser45 and the A3C structure (PDB code: 3VOW)24 as the search model. Two clear solutions were identified. Iterative rounds of model rebuilding and simulated annealing torsion angle refinement were performed using the program Coot46 and PHENIX.refine47. Identification of proper sequence registry was confirmed with the location of the catalytic zinc site and the presence of bulky aromatic residues. Molprobity48 was used throughout the refinement to ensure that clashes, rotamers, Asn/Gln/His flips and Ramachandran outliers were minimized. Ramachandran plot analysis revealed that 97.2% and 2.8% of residues are in favored and disallowed regions, respectively. Residues in disallowed regions reside in regions with weaker electron density. The final model contains A3F residues 218–373 and A3G residues 197–219. Weak electron density was observed for residues 225–236. Homology models were generated using the Phyre2 Protein Fold Recognition Server49. All molecular ribbon diagrams and vacuum electrostatic calculations were generated using the program MacPyMOL.
Biolayer interferometry DNA-protein interaction studies
The binding of ssDNA (5′-TCC CTC AGA CCC TTT TAG TCA GTG TGG AAA ATC TCT AGC AGT -3′) to A3Fc-CD2 was measured by biolayer interferometry using a BLItz instrument (Pall/ForteBio). The sequence corresponds to the viral U5 end of the 3′-LTR and was shown to bind to A3F50. This ssDNA has one A3F deamination site (5′-CTCA-3′). Biotinylated ssDNA was diluted to 50 μM in kinetics buffer (PBS, 0.1% (w/v) BSA, and 0.002% (v/v) Tween-20) immobilized on a streptavidin dip and read biosensor for 120 seconds. For the A3Fc-CD2 mutants, 20 μM of biotinylated ssDNA was immobilized on the probe. A3Fc-CD2 was diluted into kinetics buffer (0 μM, 0.4 μM, 1 μM, 2 μM, and 4 μM) and association to the biotinylated ssDNA was measured over 300 seconds. Subsequently, the biosensor was immersed in kinetics buffer for 300 seconds to measure dissociation. The Kd was calculated using the BLItz Pro v. 1.1.0.28 software.
DNA-protein interaction ELISAs
Single stranded 42-mer DNA (Fig. 1C) was synthesized (Integrated DNA Technologies). 1.5 pmol of biotinylated ssDNA was immobilized onto a streptavidin-coated ELISA plate (Quidel) and washed three times with PBS with 0.01% (v/v) Tween-20 (PBS-T). Wells were blocked with 3% (w/v) BSA in PBS-T overnight at 4ºC. 25 μg, 50 μg, 75 μg, and 100 μg of A3Fc-CD2 or A3Fc-CD2 mutants were incubated at 22°C for 1.5 hours and subsequently washed three times with PBS-T. A mouse anti-His primary mAb (Roche) diluted 1:3000 in PBS-T was incubated for 1 hour at 22ºC. The plate was washed three times prior to incubation with a goat anti-mouse HRP-conjugated secondary mAb (Pierce) diluted 1:3000. The plates were developed using TMB-One substrate solution (Kem-En-Tec Diagnostics) for 5 minutes. Color development was stopped with 2 N sulfuric acid and measured at 495 nm. As a negative control, the biotinylated sense and antisense DNA strands (3′-AGG GAG TCT GGG AAA ATC AGT CAC ACC TTT TAG AGA TCG TCA -5′) were heated at 95°C for 15 minutes and allowed to cool to 22ºC in annealing buffer (10 mM Tris-HCl pH 7.5, 1 mM EDTA and 50 mM NaCl). The double stranded DNA probe was then immobilized onto the streptavidin-coated ELISA plate and performed as described above. All experiments were performed in triplicate.
Bacterial cytosine deaminase assay
A bacterial E. coli based rifampicin resistant (RifR) mutation assay was used to monitor the degree of DNA cytosine deaminase activity18. Wild-type A3G-CD2 (residues 197–384), A3G-CD2 ‘YYFW’, A3Fc-CD2 or A3Fc-CD2 ssDNA-binding mutants cloned into pET46-Ek/LIC were transformed into BL21-(DE3) E. coli cells. Single colonies were used to inoculate a 10 ml culture of LB supplemented with 100 μg/ml ampicillin. At OD600=0.6, the cultures were induced with 1 mM final concentration of IPTG and grown overnight at 37°C. Cell cultures were normalized (7 ml of cells at OD600=0.5) and plated onto LB-agar plates containing 100 μg/ml rifampicin to select for RifR clones. Single colonies were picked from plates and colony PCR was performed to amplify the bacterial RNA polymerase gene rpoB. The rpoB PCR products were DNA sequenced with an rpoB sequencing primer (5′-GGC-GAA-ATG-GCG-GAA-AAC-3′). A3F or A3G DNA binding motifs in the rpoB gene were analyzed for C to T mutations.
Circular dichroism and thermal melts
Circular dichroism (CD) wavelength scans and thermal titrations were performed on all A3Fc-CD2 proteins at a concentration of 1.0 mg/ml in PBS, 0.05% (w/v) CHAPS and 1 M guanidine-HCl on a Jasco J-810 spectropolarimeter. CD wavelength scans collected between 190–250 nm using a 1 mm quartz cuvette (Helma) were averaged over five scans. Thermal denaturation of A3Fc-CD2 and mutants were performed by increasing the temperature from 20°C–95°C and monitoring the loss in CD signal at 222 nm.
Biolayer interferometry Vif-APOBEC3F interaction studies
HIV-1 Vif was expressed in BL21-(DE3) E. coli cells, purified and refolded from inclusion bodies, as previously described32. Briefly, Vif inclusion bodies were resolubilized in 6 M guanidine-HCl and 10 mM Tris-HCl pH 7.4 overnight at 22°C, and subsequently clarified by centrifugation prior to Ni-NTA purification. The Ni-NTA column was washed with 8 M urea, 100 mM NaH2PO4 and 10 mM Tris-HCl pH 6.9, and Vif was eluted in the same buffer at pH 4.5. Vif was subsequently purified on a prep grade Superdex-75 10/300 column equilibrated in 8 M urea, 100 mM NaH2PO4, 10 mM β-mercaptoethanol and 10 mM Tris-HCl pH 4.5. The peak corresponding to monomeric Vif was collected and diluted to 0.1 mg/ml and dialyzed 100 mM NaH2PO4, 10 mM Tris-HCl, 150 mM NaCl, 20% (v/v) glycerol pH 6.0 with decreasing concentration of urea (6, 4, 2, 1, and 0.5 M). The final refolded HIV-1 Vif was dialyzed against 10 mM NaH2PO4 pH 6.0, 150 mM NaCl and 20% (v/v) glycerol, and biotinylated using the EZ-Link Sulfo-NHS-LC-Biotinylation kit (Thermo Pierce) according to manufacturer’s protocol. Excess biotin was removed by dialysis. Biotinylated Vif was diluted to 40 μg/ml in 10 mM NaH2PO4, 150 mM NaCl, 20% (v/v) glycerol, 1 mg/ml BSA and 0.002% (v/v) Tween-20 and immobilized onto a BLI streptavidin probe for 120 seconds. A3Fc-CD2 mutants were diluted into 10 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.05% (w/v) CHAPS, 1 mg/ml BSA and 0.002% (v/v) Tween-20 and allowed to associate over 80 seconds. Dissociation was monitored over 110 seconds. Purified A3G-CD2 was used as a negative control.
Supplementary Material
Acknowledgments
This work was supported by an Ontario HIV Treatment Network Operating Grant (ROG-G645), University of Toronto start-up funds, Canada Research Chair in Structural Virology, and a Canadian Institutes of Health Research (CIHR) New Investigator Award (MSH-113554) to J.E.L. A CIHR Postdoctoral Fellowship was awarded to K.K.S., and F.C.A. was supported by a University of Toronto Graduate Scholarship and CIHR Masters Graduate Fellowship. We thank members of the Lee laboratory, in particular Halil Aydin and Dr. May Tsai, for fruitful discussions and general help. The authors are indebted to Douglas Instruments (UK) for the generous use of their Oryx 8 crystallization robot for microseeding and Aiping Dong, Profs. Cheryl Arrowsmith and Aled Edwards from the Structural Genomics Consortium (University of Toronto) for access to their X-ray diffraction facility. This work is based upon research conducted at the NE-CAT 24-ID-E beamline (Advanced Photon Source, APS), supported by award RR-15301 from the National Center for Research Resources (NIH). Use of the APS is supported by the U.S. Department of Energy, Office of Basic Energy Sciences, under contract No. W-31-109-ENG-38.
Footnotes
Author contributions: J.E.L. conceived and supervised the project; K.K.S., A.S., and J.E.L. designed the experiments; K.K.S. and J.E.L. performed the protein expression, crystallization, and structure determination; A.S. and J.E.L. performed all ssDNA-binding ELISAs, biolayer interferometry measurements, bacterial RifR DNA cytosine deaminase assays and circular dichroism spectroscopy; F.C.A. optimized the expression, purification and refolding of HIV-1 Vif for binding studies; K.K.S. and J.E.L. wrote the manuscript.
Competing financial interests: The authors declare no competing financial interests.
Accession codes: Atomic coordinates and structure factors for A3Fc-CD2 have been deposited in the Protein Data Bank (PDB) with the accession code 4J4J.
References
- 1.Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 2.Mangeat B, et al. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- 3.Harris RS, et al. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 4.Jarmuz A, et al. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics. 2002;79:285–296. doi: 10.1006/geno.2002.6718. [DOI] [PubMed] [Google Scholar]
- 5.LaRue RS, et al. Guidelines for naming nonprimate APOBEC3 genes and proteins. J Virol. 2009;83:494–497. doi: 10.1128/JVI.01976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hultquist JF, et al. Human and rhesus APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H demonstrate a conserved capacity to restrict Vif-deficient HIV-1. J Virol. 2011;85:11220–11234. doi: 10.1128/JVI.05238-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dang Y, Wang X, Esselman WJ, Zheng YH. Identification of APOBEC3DE as another antiretroviral factor from the human APOBEC family. J Virol. 2006;80:10522–10533. doi: 10.1128/JVI.01123-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Refsland EW, et al. Quantitative profiling of the full APOBEC3 mRNA repertoire in lymphocytes and tissues: implications for HIV-1 restriction. Nucleic Acids Res. 2010;38:4274–4284. doi: 10.1093/nar/gkq174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng YH, et al. Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J Virol. 2004;78:6073–6076. doi: 10.1128/JVI.78.11.6073-6076.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burnett A, Spearman P. APOBEC3G multimers are recruited to the plasma membrane for packaging into human immunodeficiency virus type 1 virus-like particles in an RNA-dependent process requiring the NC basic linker. J Virol. 2007;81:5000–5013. doi: 10.1128/JVI.02237-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conticello SG, Harris RS, Neuberger MS. The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr Biol. 2003;13:2009–2013. doi: 10.1016/j.cub.2003.10.034. [DOI] [PubMed] [Google Scholar]
- 12.Sheehy AM, Gaddis NC, Malim MH. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat Med. 2003;9:1404–1407. doi: 10.1038/nm945. [DOI] [PubMed] [Google Scholar]
- 13.Feng Y, Love RP, Chelico L. HIV-1 Vif alters processive single-stranded DNA scanning of the retroviral restriction factor APOBEC3G. J Biol Chem. 2013;288:6083–6094. doi: 10.1074/jbc.M112.421875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mercenne G, et al. HIV-1 Vif binds to APOBEC3G mRNA and inhibits its translation. Nucleic Acids Res. 2010;38:633–646. doi: 10.1093/nar/gkp1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stopak K, de Noronha C, Yonemoto W, Greene WC. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol Cell. 2003;12:591–601. doi: 10.1016/s1097-2765(03)00353-8. [DOI] [PubMed] [Google Scholar]
- 16.Smith JL, Pathak VK. Identification of specific determinants of human APOBEC3F, APOBEC3C, and APOBEC3DE and African green monkey APOBEC3F that interact with HIV-1 Vif. J Virol. 2010;84:12599–12608. doi: 10.1128/JVI.01437-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Russell RA, Pathak VK. Identification of two distinct human immunodeficiency virus type 1 Vif determinants critical for interactions with human APOBEC3G and APOBEC3F. J Virol. 2007;81:8201–8210. doi: 10.1128/JVI.00395-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hache G, Liddament MT, Harris RS. The retroviral hypermutation specificity of APOBEC3F and APOBEC3G is governed by the C-terminal DNA cytosine deaminase domain. J Biol Chem. 2005;280:10920–10924. doi: 10.1074/jbc.M500382200. [DOI] [PubMed] [Google Scholar]
- 19.Smith JL, Bu W, Burdick RC, Pathak VK. Multiple ways of targeting APOBEC3-virion infectivity factor interactions for anti-HIV-1 drug development. Trends Pharmacol Sci. 2009;30:638–646. doi: 10.1016/j.tips.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Byeon IJ, et al. NMR structure of human restriction factor APOBEC3A reveals substrate binding and enzyme specificity. Nat Commun. 2013;4:1890. doi: 10.1038/ncomms2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen KM, et al. Structure of the DNA deaminase domain of the HIV-1 restriction factor APOBEC3G. Nature. 2008;452:116–119. doi: 10.1038/nature06638. [DOI] [PubMed] [Google Scholar]
- 22.Furukawa A, et al. Structure, interaction and real-time monitoring of the enzymatic reaction of wild-type APOBEC3G. EMBO J. 2009;28:440–451. doi: 10.1038/emboj.2008.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holden LG, et al. Crystal structure of the anti-viral APOBEC3G catalytic domain and functional implications. Nature. 2008;456:121–124. doi: 10.1038/nature07357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kitamura S, et al. The APOBEC3C crystal structure and the interface for HIV-1 Vif binding. Nat Struct Mol Biol. 2012;19:1005–1010. doi: 10.1038/nsmb.2378. [DOI] [PubMed] [Google Scholar]
- 25.Shandilya SM, et al. Crystal structure of the APOBEC3G catalytic domain reveals potential oligomerization interfaces. Structure. 2010;18:28–38. doi: 10.1016/j.str.2009.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huthoff H, Autore F, Gallois-Montbrun S, Fraternali F, Malim MH. RNA-dependent oligomerization of APOBEC3G is required for restriction of HIV-1. PLoS Pathog. 2009;5:e1000330. doi: 10.1371/journal.ppat.1000330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bulliard Y, et al. Functional analysis and structural modeling of human APOBEC3G reveal the role of evolutionarily conserved elements in the inhibition of human immunodeficiency virus type 1 infection and Alu transposition. J Virol. 2009;83:12611–12621. doi: 10.1128/JVI.01491-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Autore F, Bergeron JR, Malim MH, Fraternali F, Huthoff H. Rationalisation of the differences between APOBEC3G structures from crystallography and NMR studies by molecular dynamics simulations. PLoS One. 2010;5:e11515. doi: 10.1371/journal.pone.0011515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krzysiak TC, Jung J, Thompson J, Baker D, Gronenborn AM. APOBEC2 is a monomer in solution: implications for APOBEC3G models. Biochemistry. 2012;51:2008–2017. doi: 10.1021/bi300021s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liddament MT, Brown WL, Schumacher AJ, Harris RS. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr Biol. 2004;14:1385–1391. doi: 10.1016/j.cub.2004.06.050. [DOI] [PubMed] [Google Scholar]
- 31.Rausch JW, Chelico L, Goodman MF, Le Grice SF. Dissecting APOBEC3G substrate specificity by nucleoside analog interference. J Biol Chem. 2009;284:7047–7058. doi: 10.1074/jbc.M807258200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gallerano D, et al. Biophysical characterization of recombinant HIV-1 subtype C virus infectivity factor. Amino Acids. 2011;40:981–989. doi: 10.1007/s00726-010-0725-x. [DOI] [PubMed] [Google Scholar]
- 33.Dang Y, Wang X, Zhou T, York IA, Zheng YH. Identification of a novel WxSLVK motif in the N terminus of human immunodeficiency virus and simian immunodeficiency virus Vif that is critical for APOBEC3G and APOBEC3F neutralization. J Virol. 2009;83:8544–8552. doi: 10.1128/JVI.00651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He Z, Zhang W, Chen G, Xu R, Yu XF. Characterization of conserved motifs in HIV-1 Vif required for APOBEC3G and APOBEC3F interaction. J Mol Biol. 2008;381:1000–1011. doi: 10.1016/j.jmb.2008.06.061. [DOI] [PubMed] [Google Scholar]
- 35.Dang Y, Davis RW, York IA, Zheng YH. Identification of 81LGxGxxIxW89 and 171EDRW174 domains from human immunodeficiency virus type 1 Vif that regulate APOBEC3G and APOBEC3F neutralizing activity. J Virol. 2010;84:5741–5750. doi: 10.1128/JVI.00079-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pery E, Rajendran KS, Brazier AJ, Gabuzda D. Regulation of APOBEC3 proteins by a novel YXXL motif in human immunodeficiency virus type 1 Vif and simian immunodeficiency virus SIVagm Vif. J Virol. 2009;83:2374–2381. doi: 10.1128/JVI.01898-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Albin JS, et al. A single amino acid in human APOBEC3F alters susceptibility to HIV-1 Vif. J Biol Chem. 2010;285:40785–40792. doi: 10.1074/jbc.M110.173161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu Q, et al. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat Struct Mol Biol. 2004;11:435–442. doi: 10.1038/nsmb758. [DOI] [PubMed] [Google Scholar]
- 39.Langlois MA, Beale RC, Conticello SG, Neuberger MS. Mutational comparison of the single-domained APOBEC3C and double-domained APOBEC3F/G anti-retroviral cytidine deaminases provides insight into their DNA target site specificities. Nucleic Acids Res. 2005;33:1913–1923. doi: 10.1093/nar/gki343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shih P, Pedersen LG, Gibbs PR, Wolfenden R. Hydrophobicities of the nucleic acid bases: distribution coefficients from water to cyclohexane. J Mol Biol. 1998;280:421–430. doi: 10.1006/jmbi.1998.1880. [DOI] [PubMed] [Google Scholar]
- 41.Huthoff H, Malim MH. Identification of amino acid residues in APOBEC3G required for regulation by human immunodeficiency virus type 1 Vif and Virion encapsidation. J Virol. 2007;81:3807–3815. doi: 10.1128/JVI.02795-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bohn MF, et al. Crystal structure of the DNA cytosine deaminase APOBEC3F: the catalytically active and HIV-1 Vif-binding domain. Structure. 2013;21:1042–1050. doi: 10.1016/j.str.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.D’Arcy A, Villard F, Marsh M. An automated microseed matrix-screening method for protein crystallization. Acta Crystallogr D Biol Crystallogr. 2007;63:550–554. doi: 10.1107/S0907444907007652. [DOI] [PubMed] [Google Scholar]
- 44.Pflugrath JW. The finer things in X-ray diffraction data collection. Acta Crystallographica. 1999;D55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- 45.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallographica. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 47.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 50.Mbisa JL, Bu W, Pathak VK. APOBEC3F and APOBEC3G inhibit HIV-1 DNA integration by different mechanisms. J Virol. 2010;84:5250–5259. doi: 10.1128/JVI.02358-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goujon M, et al. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 2010;38:W695–699. doi: 10.1093/nar/gkq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 53.Gouet P, Courcelle E. ENDscript: a workflow to display sequence and structure information. Bioinformatics. 2002;18:767–768. doi: 10.1093/bioinformatics/18.5.767. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.