Abstract
A defining feature of the mammalian liver is polyploidy, a numerical change in the entire complement of chromosomes. The first step of polyploidization involves cell division with failed cytokinesis. Although polyploidy is common, affecting ~90% of hepatocytes in mice and 50% in humans, the specialized role played by polyploid cells in liver homeostasis and disease remains poorly understood. The goal of this study was to identify novel signals that regulate polyploidization, and we focused on microRNAs (miRNAs). First, to test whether miRNAs could regulate hepatic polyploidy we examined livers from Dicer1 liver-specific knockout mice, which are devoid of mature miRNAs. Loss of miRNAs resulted in a 3-fold reduction in binucleate hepatocytes, indicating that miRNAs regulate polyploidization. Secondly, we surveyed age-dependent expression of miRNAs in wild-type mice and identified a subset of miRNAs, including miR-122, that is differentially expressed at 2–3 weeks, a period when extensive polyploidization occurs. Next, we examined Mir122 knockout mice and observed profound, life-long depletion of polyploid hepatocytes, proving that miR-122 is required for complete hepatic polyploidization. Moreover, the polyploidy defect in Mir122 knockout mice was ameliorated by adenovirus-mediated over-expression of miR-122, underscoring the critical role miR-122 plays in polyploidization. Finally, we identified direct targets of miR-122 (Cux1, Rhoa, Iqgap1, Mapre1, Nedd4l and Slc25a34) that regulate cytokinesis. Inhibition of each target induced cytokinesis failure and promoted hepatic binucleation.
Conclusion
Our data demonstrate that miR-122 is both necessary and sufficient in liver polyploidization. Among the different signals that have been associated with hepatic polyploidy, miR-122 is the first liver-specific signal identified. These studies will serve as the foundation for future work investigating miR-122 in liver maturation, homeostasis and disease.
Keywords: hepatocyte, polyploidy, binucleation, miR-122 microRNA-122, postnatal development
Introduction
Polyploidy, a numerical change in the complement of chromosomes, is a defining feature of the liver. Most easily observed as binucleate hepatocytes in liver sections, polyploidy is determined by the number of nuclei per cell and the DNA content of each nucleus. Diploid hepatocytes are exclusively mononucleate and polyploid hepatocytes (e.g., tetraploid, octaploid) are mono- or binucleate. Polyploidization begins early in life when a subset of proliferating diploid hepatocytes fails to undergo cytokinesis, resulting in binucleate tetraploids with two diploid nuclei (1, 2). A binucleate tetraploid then enters the cell cycle and, following successful cytokinesis, generates a pair of mononucleate tetraploid hepatocytes. A proliferating mononucleate tetraploid then undergoes cytokinesis failure and generates a binucleate octaploid, which, in turn, proliferates to yield a pair of mononucleate octaploids after successful cytokinesis. The process repeats, generating hepatocytes with even greater ploidy values. In this way, the liver develops with a combination of mononucleate (≥ diploid) and binucleate (≥ tetraploid) hepatocytes. Approximately half of hepatocytes in adult human livers are polyploid (3) and as many as 90% of adult mouse hepatocytes are polyploid (4). It is unclear whether polyploid hepatocytes play a specialized role in liver homeostasis (reviewed in (5)). Nearly a dozen factors have been shown to regulate hepatic polyploidy, including p53 (6), E2F1/7/8 (7–9), cMyc (10), insulin (11) and Rb (12). How all of these diverse signals are integrated is unknown, and it remains to be seen if there is a liver-specific regulator that drives polyploidization.
MicroRNAs (miRNA) are ~22 nucleotide-long, single-stranded, non-coding RNAs that typically negatively regulate expression of protein coding genes. When associated with Argonaute in the miRNA-induced silencing complex, mature miRNAs bind to miRNA-specific “seed sequences” in the 3’ untranslated region (3’UTR) of target genes and facilitate mRNA degradation or impair translation (reviewed in (13)). A single miRNA can attenuate expression of numerous target genes/proteins. In the liver, miRNAs contribute to diverse cellular functions, including hepatic differentiation, regeneration from injury and metabolism. miRNAs are involved in progression of liver diseases, including viral hepatitis, steatosis, cirrhosis and liver cancers. miRNA-122 (miR-122) is a liver-abundant miRNA that accounts for over 70% of total miRNAs in hepatocytes (14, 15) and is also expressed at low levels in the heart (16). During liver development, expression of miR-122 initiates around embryonic day 12.5 (E12.5) and increases continuously until adulthood (17, 18); thus, miR-122 expression closely associates with hepatic maturation. In diet-induced obesity animal models, transient depletion of miR-122 in adults by antisense oligonucleotides significantly inhibits hepatic lipogenesis and improves hepatic steatosis (19). In contrast, long-term deletion of Mir122 in knockout mice promotes early onset of steatohepatitis, fibrosis and eventually leads to spontaneous hepatocarcinogenesis (20, 21), which is facilitated after exposure to the hepatocarcinogen diethylnitrosamine (22). Although the duration and timing of miR-122 deletion leads to differences in hepatic phenotype for reasons that are not fully understood, these reports highlight a critical role for miR-122 in liver homeostasis and disease.
We set out to identify signals that control polyploidization in the liver, and we focused on cytokinesis regulators (reviewed in (23)). No individual signal functions as the dominant driver of hepatic polyploidization, which suggests that initiation/promotion of binucleation is coordinated by the combined activity of multiple effectors. Since miRNAs can coordinate the expression of sets of genes/proteins, we hypothesized that one or more miRNAs could alter expression of key cytokinesis effectors, induce cytokinesis failure and initiate/promote hepatic binucleation. Consistent with this hypothesis, miR-15 family members have been shown to regulate binucleation of cardiac myocytes (24). Here, we identified miRNAs differentially expressed in livers of mice at 2 and 3 weeks of life, ages that correspond to rapid hepatic binucleation. We demonstrate that miR-122 is necessary and sufficient for hepatic polyploidization. Mir122 knockout livers are depleted of polyploid hepatocytes, and this defect is rescued by miR-122 over-expression. Furthermore, we identified the mechanism of miR-122-regulated polyploidy: miR-122 antagonizes the expression of the pro-cytokinesis effectors Cux1, Rhoa, Mapre1, Iqgap1, Nedd4l and Slc25a34, which leads to cytokinesis failure and expansion of binucleate hepatocytes.
Materials and Methods
Mouse strains
The Institutional Care and Use Committees of the University of Pittsburgh, The Ohio State University and Children’s Hospital of Philadelphia approved all mouse experiments. The following inbred strains were used: wild-type (WT) C57BL/6 (Jackson Labs, Bar Harbor, ME), Mir122 germline knockout (“Mir122-KO”) (20), Mir122 liver-specific knockout (“Mir122-LKO;” Mir122loxP/loxP Albumin-Cre+) (20) and Dicer1 liver-specific knockout (“Dicer1-LKO;” Dicer1loxP/loxP Alfp-Cre+) (25). Control mice for Mir122 deletion studies were Mir122+/+ littermates or Mir122loxP/loxP Albumin-Cre−. Control mice for Dicer1 deletion studies were Dicer1loxP/+or loxP/loxP Alfp-Cre−. Unless noted, male mice were used for experiments.
Quantification of mono- and binucleate hepatocytes in liver sections
Mono- and binucleate hepatocytes were quantified in frozen liver sections stained with Phalloidin/Alexa Fluor 488 (Life Technologies, Carlsbad, CA) + Hoechst 33342 or in paraffin sections stained with anti-β-catenin antibody (Santa Cruz, Dallas, TX) + hematoxylin or Hoechst 33342. The binucleation curve in Fig. 1A was derived from C57BL/6 livers ages 8, 10, 12, 14, 18, 20, 29, 42 and 56d (mixed gender, 8–12d; males, 14–56d). Livers from mice 8–20d-old were imaged with an Olympus Fluoview 1000 confocal microscope using 0.2 µm Z-stacks, and livers >20d-old were imaged with Nikon TiU by manually focusing from the top to bottom of each cell. A minimum of 500 hepatocytes/sample was scored.
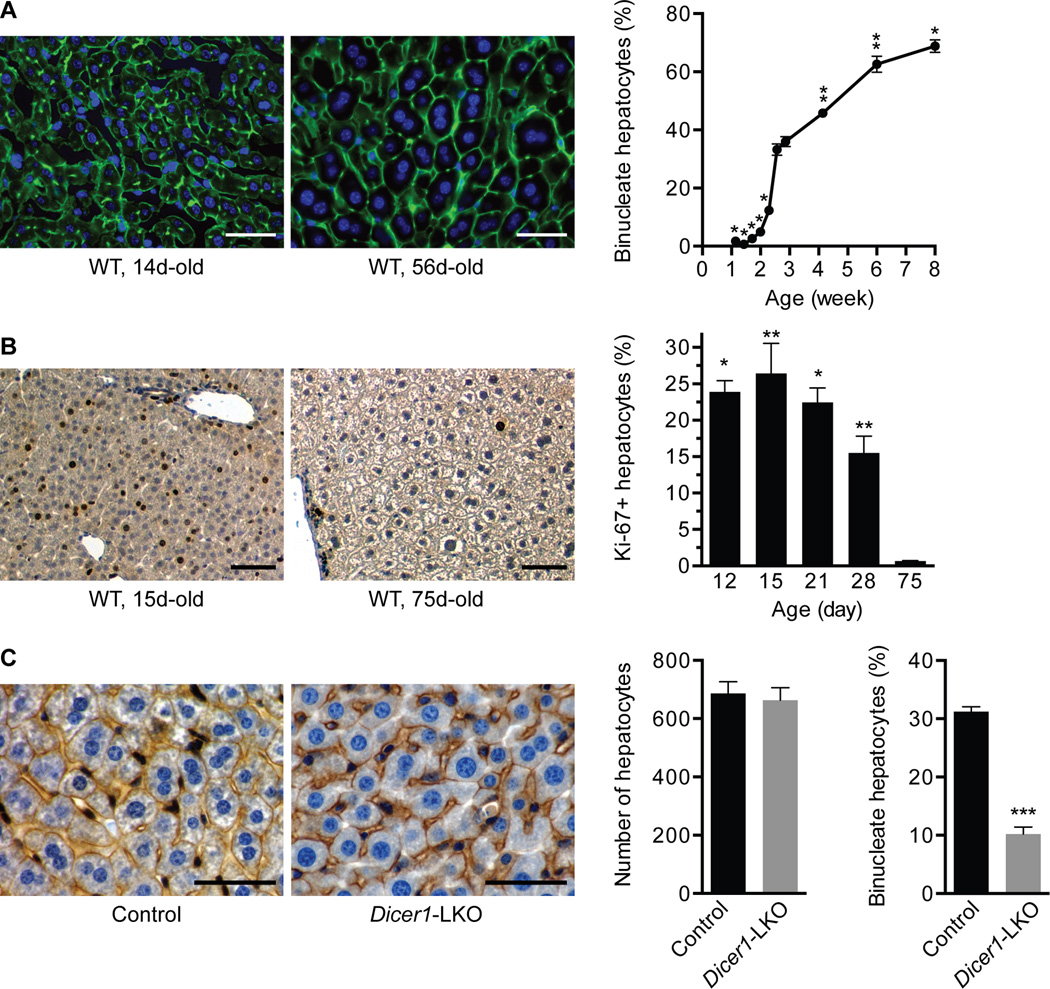
Fig. 1. Hepatic binucleation begins at 14d and is impaired in Dicer1-deleted livers.
(A) Livers from WT C57BL/6 mice ages 8–56d (n = 3) were stained with phalloidin (green) to mark cell boundaries and Hoechst 33342 (blue) to mark nuclei and the number of mono- and binucleate hepatocytes counted. Representative images show a 14d-old liver with small hepatocytes that are predominantly mononucleate and a 56d-old liver with large hepatocytes that are mono- or binucleate (left). The percentage of binucleate hepatocytes at each age is indicated (right). * P ≤ 0.003, ** P ≤ 0.02, compared to 18 and 20d. (B) Livers from WT C57BL/6 mice ages 12–75d (n = 3) were stained with the proliferation marker Ki-67 (brown) and hematoxylin (blue) for nuclei. Representative images show Ki-67+ hepatocytes in 15 and 75d-old livers (left). The percentage of Ki-67+ hepatocytes is indicated (right). P values at 12–28d are compared to 75d. (C) Livers from control or Dicer1-LKO mice ages 22–28d (n = 5) were stained for β-catenin (brown) to mark cell membranes and hematoxylin (blue) for nuclei. Representative images are shown (left). Control and Dicer1-LKO livers had equivalent numbers of cells per 200× field-of-view (middle), and binucleate hepatocytes were reduced 3-fold in Dicer1-LKO livers (right). *** P ≤ 0.0001. Graphs show mean ± SEM. Scale bars are 50 µm.
Microscopy
Unless otherwise noted, fluorescent images were captured with a Nikon TiU fluorescent microscope equipped with a monochrome camera; non-fluorescent images were captured with a Nikon TS100 microscope equipped with a color camera. Images were processed with Nikon NIS Elements Basic Research software and Adobe Photoshop CS6.
Statistical analysis
Statistical significance was calculated using 2-tailed Student’s t test unless otherwise noted. P values < 0.05 were considered significant.
Additional methods
Please refer to Supporting Information.
Results
miRNAs are necessary for hepatic binucleation
Binucleate hepatocytes were previously shown to emerge at the time of weaning in rats (11), but the timing of hepatic binucleation differs in mice. As early as 20–21d (prior to weaning), 30–40% of hepatocytes in WT C57BL/6 mice are binucleate and tetraploid (4), indicating that binucleation initiates before 3 weeks (wk) of life. To determine the kinetics of hepatocyte binculeation in mice, we counted the number of mono- and binucleate hepatocytes in liver sections (Fig. 1A). Between 8 and 14 days (d) the percentage of binucleate hepatocytes was <3% and then from 14–18d binucleate hepatocytes rapidly expanded to ~35%. Binucleate cells continued to increase slowly after 21d to comprise ~70% of hepatocytes by 8wk, consistent with the number of binucleate cells seen in suspensions of hepatocytes isolated by collagenase perfusion (4). The data indicate that a specialized mitotic program between 14–18d promotes rapid polyploidization of the liver. Postnatal liver development is characterized by robust hepatocyte proliferation, as measured by the cell cycle marker Ki-67 (Fig. 1B) and reported by others (26), so the burst in binucleate hepatocytes is likely a consequence of enhanced proliferation by diploid hepatocytes predisposed for cytokinesis failure.
To test whether miRNAs affect liver binucleation, we examined livers in Dicer1 liver-specific knockout mice (Dicer1-LKO) (25). Cells lacking Dicer1, an RNAse III-type enzyme that cleaves miRNA precursors, are devoid of mature miRNAs. Livers from 22–28d-old control and Dicer1-LKO mice contained the same number of cells per section, but loss of Dicer1 led to 60–70% reduction in binucleate hepatocytes (Fig. 1C). Loss of Dicer1 leads to a 10–20-fold increase in the percentage of Ki-67+ hepatocytes at 28d of age, compared to controls, indicating that Dicer1-LKO livers are highly proliferative during postnatal development (25); thus, the loss of binucleate hepatocytes in Dicer1-LKO livers is not simply explained by failure to proliferate. We were unable to track binucleation in adults because Dicer1-LKO livers are spontaneously repopulated by Dicer1+ hepatocytes as early as 6 weeks (27). Together, the data support the idea that functional miRNAs are required for polyploidization in the liver.
A subset of miRNAs is differentially expressed during postnatal development
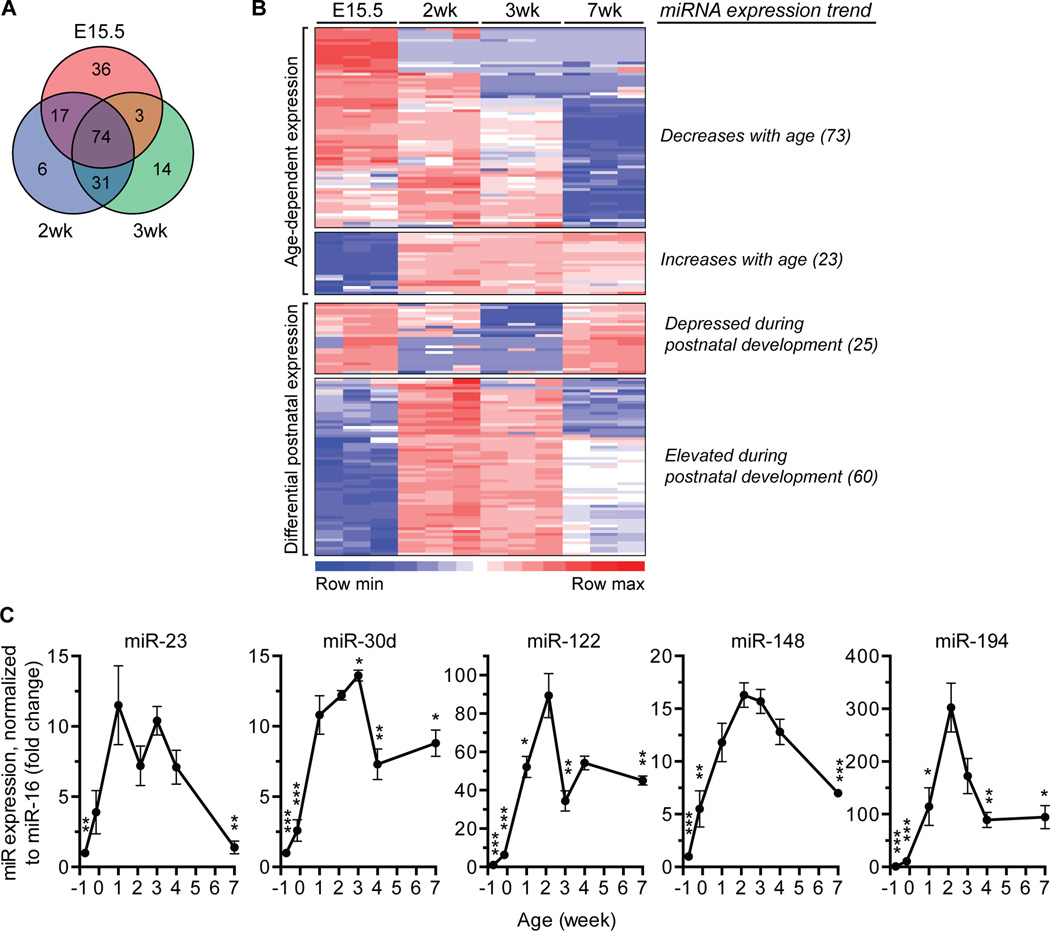
We performed a screen to identify miRNAs differentially expressed in 2–3wk-old livers, ages when rapid binucleation takes place. We compared expression of ~600 miRNAs using the NanoString nCounter Mouse miRNA Expression Assay in embryonic livers (embryonic day 15.5, E15.5), postnatal livers (2 and 3wk) and adult livers (7wk). A total of 181 miRNAs were differentially expressed in embryonic and postnatal livers compared to the adult (Fig. 2A). As expected, miRNA expression was age-dependent (Fig. 2B and Supporting Table 1): 73 miRNAs were highly expressed in embryonic liver and decreased into adulthood; in contrast, 23 miRNAs were lowly expressed in embryos and increased with age. Additionally, nearly half of the miRNAs were differentially expressed during postnatal development: 25 miRNAs were down-regulated at 2 and/or 3wk, whereas 60 miRNAs were up-regulated.
Fig. 2. miRNAs are temporally expressed in the mouse liver.
(A,B) miRNA expression was assessed in livers from WT C57BL/6 mice at ages embryonic day 15.5 (E15.5), 2-, 3- and 7wk (n = 3) using the nCounter Mouse miRNA Expression Assay from NanoString Technologies. Among the ~600 miRNAs examined, 181 were differentially expressed at E15.5, 2- and 3wk compared to 7wk-old livers (P < 0.05), as indicated in the Venn diagram (A). The heat map illustrates 4 major expression patterns (B). The number of miRNAs in each category is indicated in parentheses. Detailed expression profiles are in Supporting Table 1. (C) Expression of select miRNAs (elevated during postnatal development) was tracked in WT C57BL/6 livers at ages E15.5, E19.5, 1-, 2-, 3-, 4- and 7wk (n = 3–4) by quantitative reverse transcriptase PCR (qRT-PCR). Peak expression of miR-23, -30d, -122, -148 and -194 occurred at 1–3wks. Expression values were normalized to miR-16. * P < 0.04, ** P ≤ 0.009, *** P < 0.0008, compared to 2wk. Graphs show mean ± SEM.
To validate the NanoString data, we interrogated expression of a subset of miRNAs that are enriched in the liver: miR-23, -30d, -122, -148 and -194 (28). Consistent with the NanoString observations, these miRNAs were specifically up-regulated during the postnatal time period (Fig. 2C and Supporting Fig. 1). For example, miR-122 was previously shown to be ~50-fold up-regulated in adult livers compared to E15.5 (17, 18). We also observed robust expression of miR-122 in adults, but peak miR-122 expression occurred specifically at 2wk. Although RNU6b is frequently used for normalization, we found that miR-16 was more consistently expressed at all ages (Supporting Fig. 1A). Regardless of the normalization method used, however, similar expression patterns were seen for each miRNA (Fig. 2C and Supporting Fig. 1B).
We speculated that one or more of the miRNAs could antagonize pro-cytokinesis genes, thereby promoting cytokinesis failure and the emergence of binucleate hepatocytes. To determine which miRNAs were predicted to target cytokinesis genes, we screened all 60 of the miRNAs with elevated expression at 2 and/or 3 weeks. The 3’UTRs of cytokinesis-associated genes (defined as the 323 genes classified by Ingenuity Pathway Analysis, IPA, as “affecting” or “activating” cytokinesis) were screened using TargetScan 6.2 for seed matching sequences to each of the miRNAs. As seen in Supporting Table 2, most of the miRNAs are predicted to target multiple genes. Together, these data indicate that mouse liver miRNAs are expressed in an age-dependent manner, a subset is differentially expressed during postnatal development and numerous miRNAs are predicted to antagonize cytokinesis-associated genes.
miR-122 is required for complete hepatic polyploidization
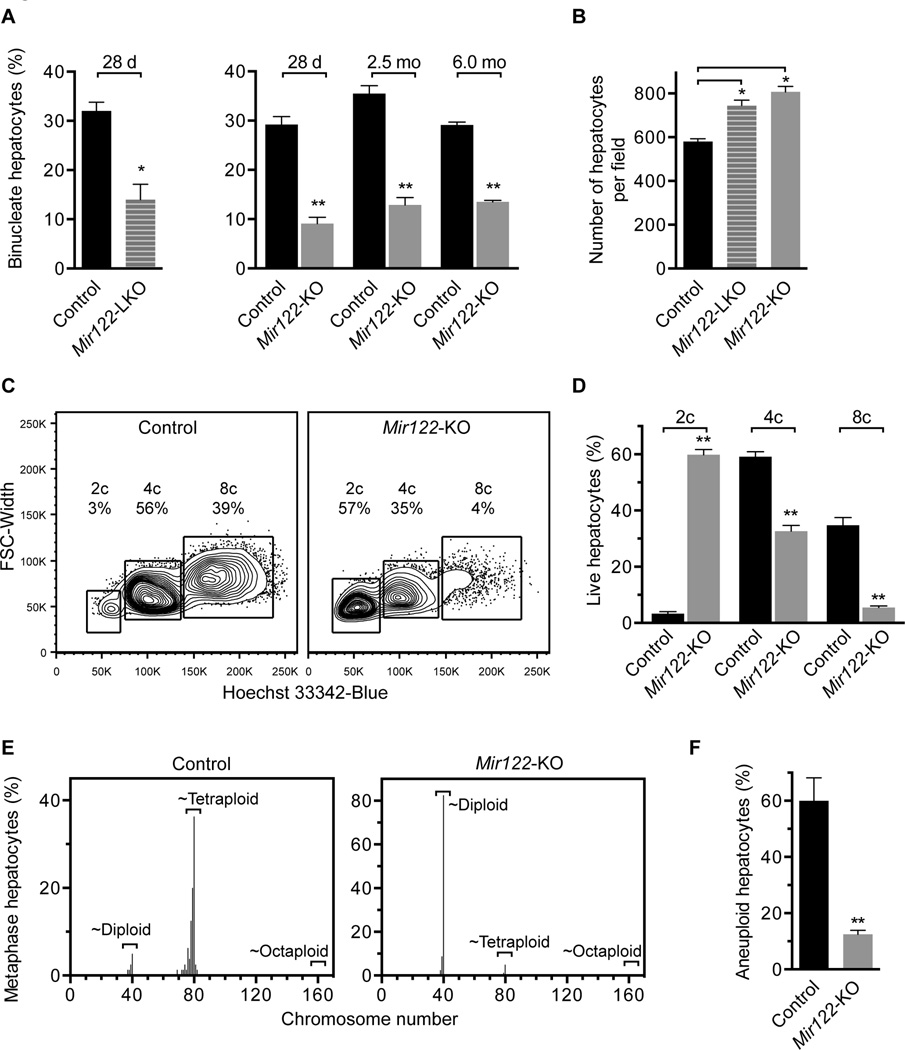
We decided to focus on miR-122, which is predicted to target 38 of 323 cytokinesis-associated genes (Supporting Table 2). miR-122 is a compelling candidate because Mir122-deficient livers were previously noted to have fewer binucleate hepatocytes compared to controls (Hsu and Ghoshal, unpublished observations 2012), which suggested that miR-122 could contribute to polyploidization. Moreover, as a liver-specific miRNA, we hypothesized that miR-122 could serve as a tissue-specific regulator of polyploidy in the liver. To determine how loss of miR-122 affects nuclearity, ploidy and aneuploidy we utilized liver-specific Mir122 knockout mice (Mir122-LKO) and germline Mir122 knockout (Mir122-KO) mice (20). First, we counted the number of binucleate hepatocytes in Mir122-LKO and -KO livers (Supporting Fig. 2A). At 28d, following the period of rapid binucleation, Mir122-LKO and -KO livers displayed 60–70% reduction in the number of binucleate hepatocytes (Fig. 3A). Consistent with enrichment for mononucleate hepatocytes that are smaller than binucleate polyploids, Mir122-LKO and -KO livers contained 20–25% more hepatocytes per liver section than controls (Fig. 3B). Livers from 2.5- and 6.0-month (mo)-old Mir122-KO mice also contained 60–70% fewer binucleate hepatocytes (Fig. 3A), indicating that the binucleation defect caused by miR-122 loss persists throughout life. Aside from the binucleation defect, Mir122-KO livers were nearly identical to controls in terms of proliferation, cell cycle arrest and apoptosis (Supporting Fig. 2B–D).
Fig. 3. Mir122-deficient livers are enriched for diploid, euploid hepatocytes.
(A,B) Livers from control, Mir122-LKO and Mir122-KO livers were stained with phalloidin and Hoechst 33342, as shown in Supporting Fig. 2A (n = 4–5 mice/age/genotype). The percentage of binucleate hepatocytes was reduced 2–3-fold at 28d (LKO and KO), 2.5mo (KO) and 6.0mo (KO) (A). 28d-old Mir122-LKO and Mir122-KO livers had 20–25% more hepatocytes per 150× field-of-view (B). (C,D) Single cell suspensions of hepatocytes were stained with a viability marker and Hoechst 33342, allowing identification of live cells with 2c, 4c and 8c nuclear content, corresponding to diploid, tetraploid and octaploid hepatocytes, respectively. Representative ploidy profiles are shown (C) and the percentage of each population quantified (n = 4; 2.5mo) (D). (E,F) Metaphase cytogenetic analysis revealed skewed chromosome counts (hypo-diploid, hypo-tetraploid and hyper-tetraploid) in control hepatocytes but not Mir122-KO hepatocytes (n = 4) (E). The overall degree of aneuploidy in Mir122-KO hepatocytes was reduced 4–5-fold compared to controls (F). See Supporting Fig. 4 individual karyotypes. * P ≤ 0.03, ** P ≤ 0.0001. Graphs show mean ± SEM.
Secondly, we measured hepatocyte ploidy. In adult mice that are predominantly quiescent (4, 29), hepatic ploidy is accurately determined by flow cytometric quantification of nuclear content. We isolated hepatocytes from 2.5mo-old mice and determined ploidy of live hepatocytes (Supporting Fig. 3). As expected, only a small fraction of control hepatocytes were 2c (3%) and the majority were 4c (59%) or 8c (35%) (Fig. 3C,D). In contrast, the ploidy spectrum of Mir122-KO hepatocytes was significantly altered. Most Mir122-KO hepatocytes were 2c (60%) and many fewer cells were 4c (33%) or 8c (6%).
Finally, we measured aneuploidy by metaphase cytogenetic analysis. Chromosome counts for control hepatocytes clustered at ~40 (diploid) or ~80 (tetraploid), but there were also numerous cells with hypo-diploid, hypo-tetraploid and hyper-tetraploid chromosome counts (Fig. 3E and Supporting Fig. 4). Similar to previous studies (4, 30), 60% of control hepatocytes contained random whole chromosome gains and/or losses, chromosome losses outnumbered chromosome gains and structural rearrangements were uncommon (Fig. 3F and Supporting Fig. 4). Loss of miR-122 dramatically altered hepatic aneuploidy. Mir122-KO hepatocytes were enriched for diploid chromosome counts, and, for each sample analyzed, only 2–3 aneuploid karyotypes were identified per 20 cells (12.5%), a level that, in our experience, is slightly above background. Collectively, the data show that Mir122-deficient livers are predominantly diploid and euploid, which indicates that miR-122 is necessary for complete hepatic polyploidization.
miR-122 promotes hepatic binucleation
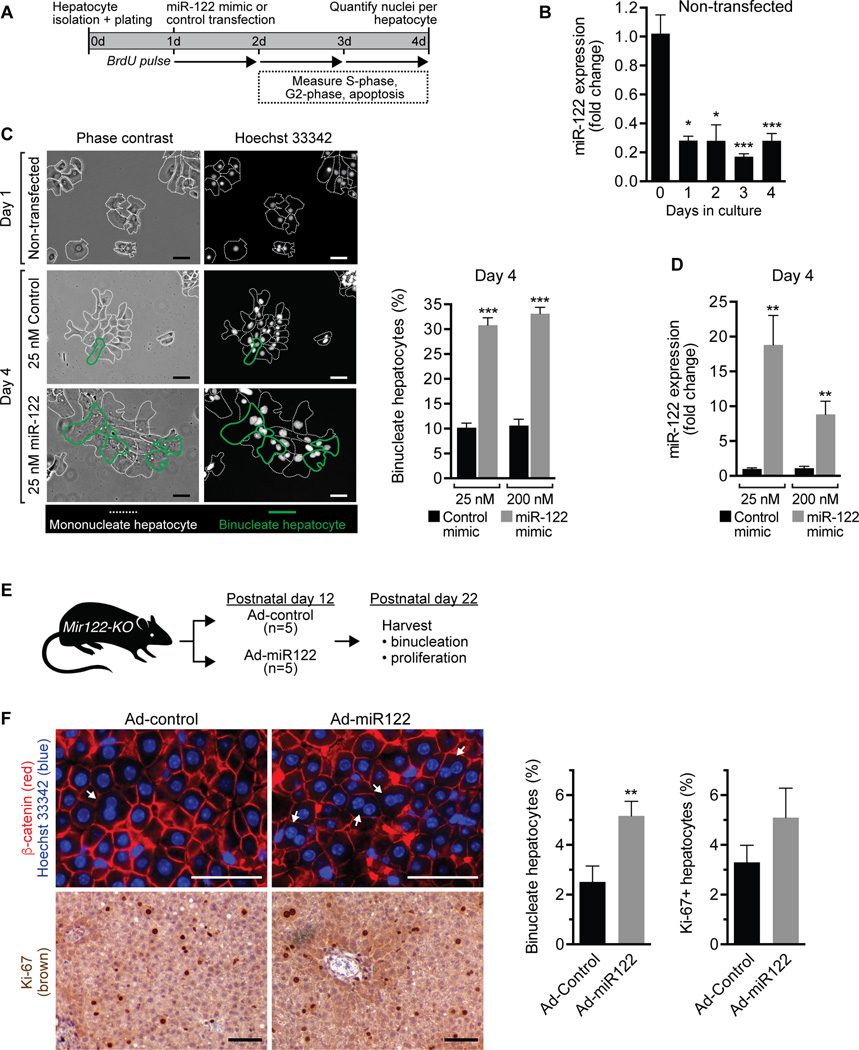
We next hypothesized that miR-122 activity is sufficient to induce hepatic binucleation. To test this idea, we over-expressed miR-122 in WT diploid hepatocytes and tracked binucleation in vitro (Fig. 4A). Although miR-122 is robustly expressed by WT hepatocytes, expression decreases 75–80% after 1d in culture and remains depressed for additional days (Fig. 4B). We isolated hepatocytes from 14d-old WT livers that are ≥97% diploid, transfected with synthetic miR-122 mimic or control mimic (25 nM or 200 nM) and monitored proliferation. On day 1, prior to transfection, hepatocytes appeared spread-out and were predominantly mononucleate (Fig. 4C). Proliferation kinetics were similar between control- and miR-122-transfected cells, but there was a trend toward delayed S-phase at days 2 and 3 in hepatocytes over-expressing miR-122 (Supporting Fig. 5A,B). Cell death was minimal (Supporting Fig. 5C). Hepatocytes migrated together during the culture period, became smaller and formed tightly packed clusters. By day 4, miR-122 was induced 10–20-fold in miR-122-transfected cells (Fig. 4D). Hepatocyte cultures were comprised of a heterogeneous mixture of mono- and binucleate cells, and multinucleate cells were rarely observed (<1%) (Fig. 4C). It was difficult to identify individual cells in the tightly packed clusters; therefore, we harvested samples by trypsinization and prepared cytospins, which allowed identification of single cells and quantification of the number of nuclei per cell (Supporting Fig. 6). Binucleate hepatocytes were seen in 30–35% of miR-122-transfected hepatocytes compared to 10% of control-transfected hepatocytes (Fig. 4C). Thus, the data show that miR-122 promotes hepatic binucleation.
Fig. 4. miR-122 induces binucleation in vitro and in vivo.
(A–D) Diploid hepatocytes isolated from 14d-old WT C57BL/6 mice were plated on day 0, transfected with miR-122 mimic or control mimic (25 nM or 200 nM) on day 1 and proliferation/nucleation tracked until day 4. (A) Experimental design. (B) Primary non-transfected hepatocytes rapidly lost miR-122 expression in culture, as measured by qRT-PCR and normalized to RNU6b (n = 3). (C) Cultured non-transfected hepatocytes were predominantly mononucleate on day 1, and by day 4 hepatocytes transfected with 25 nM or 200 nM miR-122 mimic contained 3-fold more binucleate hepatocytes compared to controls. Representative images are shown (left). Dashed white lines indicate mononucleate hepatocytes, and bold green lines mark binucleate hepatocytes. Graph shows the percentage of binucleate hepatocytes observed at day 4 (n = 3–6) (right). (D) Hepatocytes transfected with miR-122 mimic contained 10–20-fold more miR-122 at day 4. miR-122 expression was normalized to RNU6b (n = 3 from 2 independent experiments). (E,F) Mir122-KO mice were injected with Ad-miR122 or Ad-control (1 × 109 viral particles) on postnatal day 12 (n = 5) and livers harvested on postnatal day 22. (E) Cartoon showing the experimental design is shown. (F) Livers were stained with β-catenin (red) to mark membranes and Hoechst 33342 (blue) to mark nuclei. Arrows indicate binucleate hepatocytes. Additionally, livers were stained with Ki-67 (brown) to identify proliferating cells. Representative images are shown (left). The percentage of binucleate hepatocytes and Ki-67+ hepatocytes is shown in the graphs (right). * P < 0.04, ** P < 0.02, *** P < 0.007. Graphs show mean ± SEM. Scale bars are 50 µm.
To complement the in vitro approach, we next tested whether exogenous miR-122 could promote hepatic binculeation in Mir122-KO mice. We generated an adenovirus encoding four tandem copies of miR-122 (Ad-miR122), which robustly induces miR-122 expression in vitro (Supporting Fig. 7). Mir122-KO mice were injected with Ad-miR122 or control adenovirus (Ad-control) on postnatal day 12 and livers harvested at postnatal day 22 (Fig. 4E). We rationalized that exogenous miR-122 activity would promote cytokinesis failure and the emergence of binucleate hepatocytes. At postnatal day 22, we failed to detect elevated levels of miR-122 in Ad-miR122-injected mice (data not shown). This is not surprising because expression by non-integrating viruses is diluted and frequently lost in proliferating cells (31), and hepatocytes proliferate extensively between postnatal days 12 and 22 (Fig. 1B). However, we observed a significant change in phenotype on postnatal day 22. Mir122-KO mice infected with Ad-miR122 had nearly twice as many binucleate cells compared to Mir122-KO livers infected by Ad-control (Fig. 4F). There was also a trend toward increased proliferation, as measured by Ki-67 staining, but the difference was not significant (Fig. 4F). Together with the in vitro observations, these data show that exogenous miR-122 rescues the binucleation defect in Mir122-KO mice, indicating that miR-122 is sufficient to induce binucleation during postnatal liver development.
miR-122 directly regulates genes involved in cytokinesis
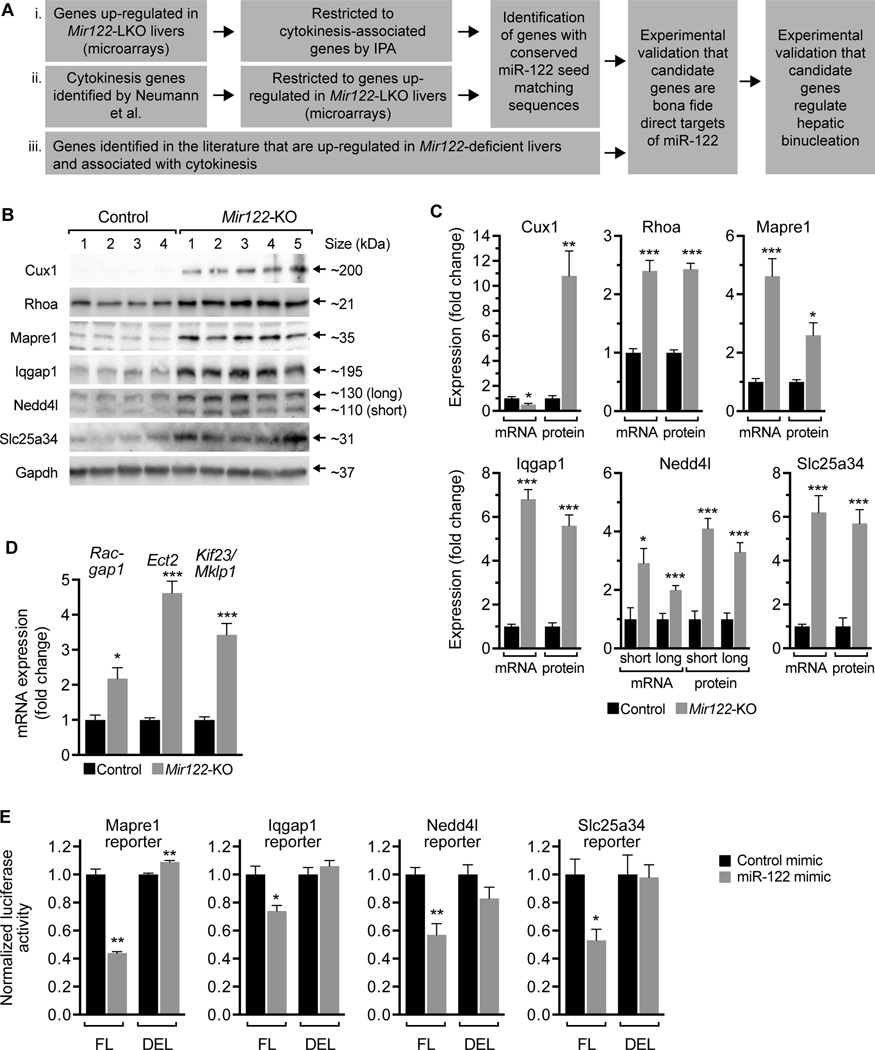
Control and Mir122-KO livers have similar cell cycle kinetics (Supporting Fig. 2B–D), so differences in proliferation, arrest or apoptosis cannot explain the ploidy defect caused by miR-122 loss. Based on these observations, we hypothesized that miR-122 regulates hepatic binucleation by antagonizing genes involved in cytokinesis. To identify candidate cytokinesis target genes with miR-122 seed matching sequences we used bioinformatic and literature-based screening approaches (Fig. 5A). First, using published expression profiles of Mir122-LKO and control livers (20), IPA revealed 63 cytokinesis-associated genes that were differentially expressed in Mir122-LKO livers (Supporting Table 3). We screened 3’UTRs and identified 4 genes with conserved seed matching sequences that were up-regulated ≥1.3-fold: Mapre1, Nedd4l, Rhoa and Vps4a (Table 1 and Supporting Table 4). Secondly, in an unbiased screen for genes involved in cell division, Neumann et al. identified 427 genes involved in binucleation (32). Comparison of these genes with the Mir122-LKO/control microarray data revealed 31 genes that were up-regulated in Mir122-LKO livers (Supporting Table 5). We screened 3’UTRs and narrowed the list to 3 genes with conserved miR-122 seed matching sequences that were most highly up-regulated (≥1.3-fold): Galc, Nedd4l and Slc25a34 (Table 1). Finally, we searched the literature for genes involved in cytokinesis that were also validated miR-122 targets. We identified Cux1 (17, 33), Iqgap1 (20, 34, 35), Mapre1 (36, 37) and Rhoa (2, 20, 38, 39), genes previously implicated in cytokinesis and demonstrated up-regulated in Mir122-deficient livers (Table 1). Three of the targets, Rhoa, Mapre1 and Nedd4l, were identified by multiple methods, which supports our screening approach. Thus, among the cytokinesis-associated genes identified, we decided to focus on the most promising candidates for further analysis: Cux1, Rhoa, Mapre1, Iqgap1, Nedd4l and Slc25a34.
Cux1 is a transcription factor that promotes expression of centralspindlin components Racgap1, Ect2 and Kif23/Mklp1 (33). The centralspindlin complex regulates cleavage furrow ingression and abscission (reviewed in (40)). Inhibition of Racgap1, Ect2 and Kif23/Mklp1 blocks cytokinesis in a variety of cell types, and these proteins have been associated with liver binucleation (2, 9). We found 10-fold up-regulation in Cux1 protein in Mir122-KO livers but not at the mRNA level (Fig. 5B,C), indicating that miR-122 controls Cux1 translation and not mRNA stability (41). Although Racgap1, Ect2 and Kif23/Mklp1 lack miR-122 seed matching sequences in their 3’UTR (data not shown), these genes were up-regulated 2–5-fold in Mir122-KO livers (Fig. 5D). Thus, the data indicate that miR-122 regulates the centralspindlin complex via the transcription factor Cux1.
Rhoa is a small GTPase that promotes formation of the actomyosin ring and facilitates myosin II constriction to induce cleavage furrow ingression (38). Cytokinesis failure in hepatocytes was previously associated with loss and/or mislocalization of active Rhoa (2). In agreement with earlier work (20), we found 2.4-fold up-regulation of Rhoa in Mir122-KO livers (Fig. 5B,C).
Mapre1 is a microtubule-tip associated protein that performs multiple roles, including cell division. During cytokinesis, Mapre1 binds and stabilizes Aurora B kinase (36, 37), which phosphorylates centralspindlin complex member Kif23/Mklp1 and promotes cytokinesis completion (42). Similar to previous results (20, 43), we found that Mapre1 was up-regulated 3–5-fold in Mir122-KO livers (Fig. 5B,C).
Iqgap1 is a Ras GTPase-activating-like protein that localizes to the spindle midbody during cytokinesis where it promotes abscission and completion of cytokinesis (35). Depletion of Iqgap1 leads to a dramatic increase in the number of bi- and multinucleate cells (35). Consistent with earlier work (20, 34), Iqgap1 was up-regulated 6–7-fold in Mir122-KO livers (Fig. 5B,C).
Nedd4l is an E3 ubiquitin ligase that facilitates proteasome-mediated degradation (reviewed (44)). Identified by Neumann et al. (32), Nedd4l inhibition blocked cytokinesis and induced formation of binucleate cells; however, it is unknown how Nedd4l affects cytokinesis. Nedd4l has two isoforms (a “short” 110 kDa form and a “long” 130 kDa form) that are encoded as distinct transcripts from the same gene (45). Both isoforms were significantly elevated in 2–4-fold in Mir122-KO livers (Fig. 5B,C).
Slc25a34 is a member of the Slc25 family and is believed to facilitate transport over the mitochondrial membrane (46). Slc25a34 was also identified in the Neumann et al screen, and it is unclear how this gene affects cytokinesis. We found that Slc25a34 was up-regulated ~6-fold in Mir122-KO livers (Fig. 5B,C).
Fig. 5. miR-122 regulates expression of cytokinesis-related targets.
(A) Strategy for identifying miR-122 target genes that regulate cytokinesis. (B,C) Cux1, Rhoa, Mapre1, Iqgap1, Nedd4l and Slc25a34 were up-regulated in 28d-old Mir122-KO livers compared to the controls at the protein and/or mRNA levels (n = 4–5). Western blots are shown for 4 control and 5 Mir122-KO livers; Gapdh was used as a loading control (B). Graphs show relative mRNA and protein expression levels normalized to Gapdh (C). (D) Centralspindlin complex members Racgap1, Ect2 and Kif23/Mklp1 were also up-regulated in 28d-old Mir122-KO livers (n = 5). The graph shows mRNA expression normalized to Gapdh. (E) NIH 3T3 cells were transfected with Mapre1, Iqgap1, Nedd4l or Slc25a34 reporter plasmids containing predicted miR-122 3’UTR seed sequences (full length, FL) or with the seed sequences deleted (DEL). Reporter constructs are detailed in Supporting Fig. 8. Cells were co-transfected with control or miR-122 mimic (25 nM) and luciferase activity measured after 48h. For each set of reporters, miR-122 expression reduced reporter activity 26–47% in the presence of FL reporters but not DEL reporters. Graphs show normalized luciferase activity (Renilla luciferase activity controlled by FL or DEL normalized to firefly luciferase activity expressed by the same plasmid) (n = 3 replicates from 2–3 independent experiments). * P < 0.03, ** P < 0.008, *** P < 0.0008. Graphs show mean ± SEM.
Table 1.
Cytokinesis-associated genes with conserved miR-122 seedmatching sequences in the 3'UTR
| Accession no. | Symbol | Gene name | Alias | Identification Method |
|---|---|---|---|---|
| NM_009986 | Cux1 | Cut-like homeobox 1 | Cutl1 | c |
| NM_008079 | Galc | Galactosylceramidase | b | |
| NM_016721 | Iqgap1 | IQ domain-containing GTPase-activating protein 1 | c | |
| NM_007896 | Mapre1 | Microtubule-associated protein, RP/EB family, member 1 | Eb1, Bim1p | a, c |
| NM_031881 | Nedd4l | Neural precursor cell expressed, developmentally down-regulated gene 4-like | Nedd4b, Nedd4-2 | a, b |
| NM_016802 | Rhoa | Ras homolog gene family, member A | Arha, Arha1, Arha2 | a, c |
| NM_001013780 | Slc25a34 | Solute carrier 25 family, member a34 | Gm1369 | b |
| NM_126165 | Vps4a | Vacuolar protein sorting 4a (yeast) | a |
IPA analysis (JCI 2012;122:2871)revealed 63 cytokinesis-associated genes(P < 0.05) differentially expressed in Mir122-LKO livers. Among these, 4 genes up-regulated ≥ 1.3-fold are indicated with conserved miR-122 seed matching sequences as determined by TargetScan and/or MiRanda algorithms: Mapre1, Nedd4l, Rhoa and Vps4a. See Supporting Tables 3 and 4 for additional information.
Neumann et al. (Nature 2010;464:721) characterized 427 genes involved in binucleation, and we showed that 31 genes were up-regulated in Mir122-LKO livers (P < 0.05) (JCI 2012;122:2871). Among these, 3 genes up-regulated ≥ 1.3-fold are listed with conserved miR-122 seed matching sequences as determined by TargetScan and/or MiRanda algorithms: Galc, Nedd4l and Slc25a34. See Supporting Table 5 for additional information.
Genes identified in the literature that were implicated in cytokinesis and up-regulated in Mir122-deficient livers: Cux1, Iqgap1, Mapre1 and Rhoa.
Cux1, Rhoa and Mapre1 were previously shown to function as direct miR-122 targets (17, 20, 39). They contain miR-122 seed sequences in their 3’UTR that facilitate miR-122-induced degradation. To confirm whether Iqgap1, Nedd4l and Slc25a34 were also direct targets of miR-122 we performed luciferase reporter assays. The 3’UTR of each gene (either containing the predicted seed sequence(s) or with the seed sequence deleted) was cloned downstream of Renilla luciferase in psiCHECK2 reporter vector (Supporting Fig. 8A). NIH 3T3 cells with undetectable levels of miR-122 were then co-transfected with “full length” (FL) or deleted (DEL) reporter constructs plus synthetic miR-122 mimic or control mimic. Luciferase activity is expected to be high in all conditions except when exogenous miR-122 interacts with a bona fide seed sequence and represses Renilla luciferase expression. As a positive control we started with Mapre1, which has a single predicted miR-122 seed sequence (Supporting Fig. 8B). Indeed, miR-122 repressed luciferase activity ~50% in the FL reporter but had minimal effect on the DEL reporter, confirming that Mapre1 is a direct miR-122 target (Fig. 5E). We identified 1–5 putative miR-122 seed sequences in the 3’UTRs of Igqap1, Nedd4l and Slc25a34 (Supporting Fig. 8B) and generated FL and DEL reporter constructs for each gene. Renilla luciferase activity was repressed 25–50% in a miR-122-dependent manner for each FL reporter but not when the seed sequences were deleted (Fig. 5E). The data show that Cux1, Mapre1, Iqgap1, Nedd4l and Slc25a34 are direct targets of miR-122.
Depletion of miR-122 targets promotes formation of binucleate hepatocytes
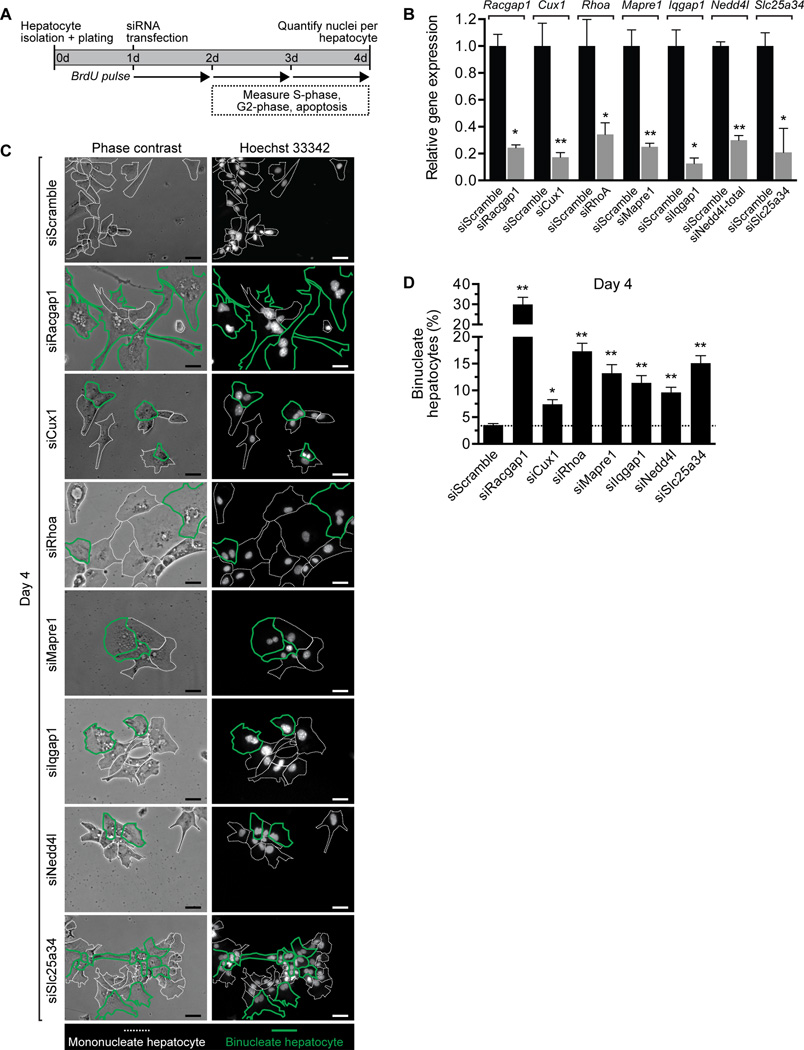
Cux1, Rhoa, Mapre1, Iqgap1, Nedd4l and Slc25a34 are all direct targets of miR-122 that are up-regulated in Mir122-KO livers (Fig. 5). To determine how these genes affect cell division in hepatocytes we tracked proliferation in vitro. Hepatocytes from 14–15d-old WT livers that are ≥97% diploid were transfected with siRNAs to each of the target genes (Fig. 6A). In addition to the target genes, we also depleted diploid hepatocytes of Racgap1, a member of the centralspindlin complex that is essential for successful cytokinesis (47). Approximately 75% of the cells were successfully transfected, and targets were down-regulated 70–90% by day 4 (Fig. 6B and Supporting Fig. 9). We tracked proliferation on days 2–4 using BrdU incorporation and phospho-histone H3 staining (Supporting Fig. 10A,B). Proliferation by target-depleted hepatocytes varied day-by-day, but there was a trend toward decreased proliferation by cells depleted of Cux1, Mapre1 and Slc25a34. Cell death was < 2% in all of the cultures (Supporting Fig. 10C). By day 4, cultures contained mono- and binucleate hepatocytes, and multinucleate cells were rare (<1%) (Fig. 6C and Supporting Fig. 11). Cultures transfected with scrambled siRNA contained 3% binucleate hepatocytes. As expected, Racgap1-depleted hepatocytes were highly enriched (30%) for binucleate cells. Depletion of the other targets also resulted in a significant increase in binucleate hepatocytes: Cux1 (7%), Rhoa (17%), Mapre1 (13%), Iqgap1 (11%), Nedd4l (10%) and Slc25a34 (15%) (Fig. 6D). Taken together, the data indicate that Cux1, Rhoa, Mapre1, Iqgap1, Nedd4l and Slc25a34 are necessary for complete cytokinesis in hepatocytes.
Fig. 6. Inhibition of miR-122 target genes promotes formation of binucleate hepatocytes.
(A) Experimental design for tracking diploid hepatocytes depleted of target genes. Diploid hepatocytes isolated from 14–15d-old WT C57BL/6 mice were plated on day 0, transfected with 20–40 nM siRNAs on day 1 and proliferation/nucleation tracked until day 4. (B) Hepatocytes transfected with scrambled siRNA or siRNAs to Racgap1, Cux1, Rhoa, Mapre1, Iqgap1, Nedd4l or Slc25a34 showed reduced target expression on day 4 (n = 2–4 replicates from 2 independent experiments). The graph shows relative gene expression measured by qRT-PCR normalized with Gapdh. Total Nedd4l was measured using primers detecting both the short and long forms. Protein levels are shown in Supporting Fig. 9. (C,D) The number of nuclei/hepatocyte was determined on day 4. Representative images illustrate a mixture of mono- and binucleate hepatocytes (C), and the percentage of binucleate hepatocytes calculated (n = 3–6) (D). Dashed white lines indicate mononucleate hepatocytes, and bold green lines mark binucleate hepatocytes. The dashed line on the graph indicates the percentage of binucleate cells in cultures of hepatocytes transfected with scrambled siRNA. * P < 0.04, ** P < 0.009. Graphs show mean ± SEM. Scale bars are 50 µm.
Discussion
In this work, we investigated the role of miRNAs in hepatic polyploidy. Compared to livers with a full complement of miRNAs, Dicer1 knockout livers had 60–70% fewer binucleate hepatocytes at 22–28d. Loss of Dicer1 is also accompanied with increased hepatic proliferation (25); thus, our data suggest that miRNAs affect cytokinesis fidelity as well as proliferation. To determine which miRNAs were involved, we identified 85 miRNAs differentially expressed at 2 or 3 weeks, ages associated with extensive binucleation. miR-122 and miR-194, for instance, were expressed at low levels during embryonic development and then increased 50–100-fold in adults. Intriguingly, miR-122 and miR-194 expression spiked around 2wk at a level 2–3 times higher than is typically seen in adults. The transient differential expression of numerous miRNAs is not surprising because livers undergo profound changes during postnatal development, including acquisition of “adult” drug-metabolizing and metabolic processes, expansion of hepatocytes and bile duct epithelial cells and loss fetal hematopoietic cells (26, 48). Hence, the changes in miRNA expression we observed could be related to many different processes. We focused on miR-122, a pleiotropic miRNA among the most abundantly expressed miRNAs in the liver (14, 15). Loss of miR-122 led to 60–70% reduction in binucleate hepatocytes at 28d, and this defect persisted throughout life. The ploidy spectrum was also dramatically altered in Mir122-KO livers – diploid hepatocytes were enriched nearly 20-fold. Proliferating polyploid hepatocytes can undergo chromosome segregation errors and generate aneuploid daughters. Consistent with increased numbers of diploid hepatocytes and corresponding decrease in polyploids, the degree of aneuploidy in Mir122-KO livers was minimal. In addition to being required for polyploidy, miR-122 actively promotes polyploidy. Adenovirus-mediated expression of miR-122 in Mir122-KO livers promoted hepatic binucleation and rescued the binucleation defect; similarly, miR-122 over-expression induced binculeation in cultured WT diploid hepatocytes. Taken together, the data demonstrate that miR-122 is necessary and sufficient for polyploidization of the liver.
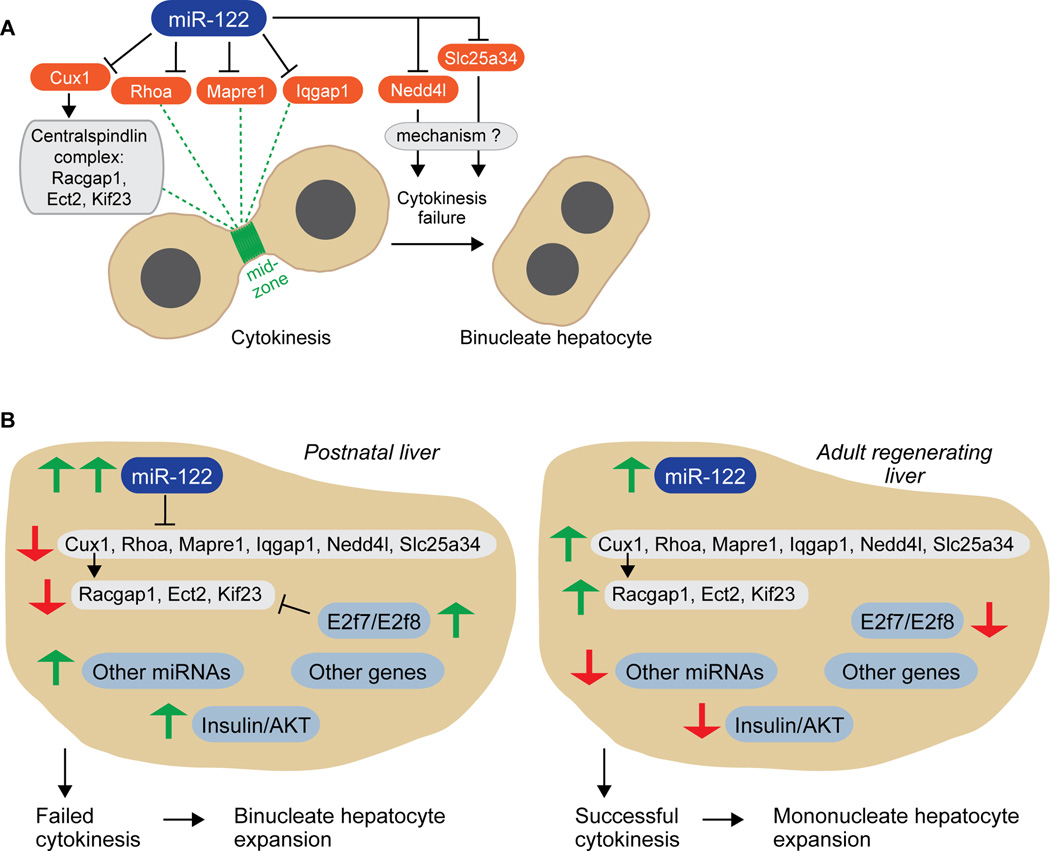
Proliferating diploid hepatocytes can go through mitosis, generate a pair of diploid nuclei and then experience cytokinesis failure, which leads to the formation of a binucleate tetraploid hepatocyte with two diploid nuclei (1, 2). We reasoned that miR-122 could control hepatic binculeation by directly targeting genes involved in cytokinesis. We identified and characterized miR-122 targets that influence cytokinesis progression: Cux1, Rhoa, Mapre1, Iqgap1, Nedd4l and Slc25a34 (Fig. 7A). Among these targets, Cux1 (17), Rhoa (39) and Mapre1 (20) were previously characterized as direct targets of miR-122, and we showed that Iqgap1, Nedd4l and Slc25a34 were also direct miR-122 targets via seed sequences within their 3’UTRs. As expected, all of the targets were notably up-regulated in Mir122-KO livers. We finally tested how deletion of each target affects hepatocyte proliferation. Consistent with the targets promoting successful cytokinesis, cultures of primary diploid hepatocytes depleted of each target generated populations of binucleate hepatocytes.
Fig. 7. A network of signals regulates hepatic binucleation.
(A) The model details a mechanism of miR-122-regulated cytokinesis. miR-122 directly antagonizes pro-cytokinesis targets (shown in orange), induces cytokinesis failure and emergence of binucleate hepatocytes. (B) The cartoon illustrates signaling networks involved in cytokinesis failure in postnatal developing livers and successful cytokinesis in adult regenerating livers. Green arrows indicate elevated expression; red arrows indicate reduced expression.
We propose a model in which miR-122 collaborates with other signals to regulate hepatic binucleation and polyploidy (Fig. 7B). During postnatal liver development, miR-122 is expressed at exceptionally high levels (2–3-fold greater than in adults), allowing miR-122 to directly down-regulate targets involved in cytokinesis, including Cux1, Rhoa, Mapre1, Iqgap1, Nedd4l and Slc25a34. Down-regulation of Cux1, in turn, reduces centralspindlin complex members Racgap1, Ect2 and Kif23/Mklp1. Although miR-122 is an important regulator of hepatic polyploidy, loss of miR-122 does not completely block binculeation, which suggests that other signals/networks are involved. For example, transcription factors E2F7 and E2F8 are robustly expressed during postnatal development, and they also antagonize Racgap1, Ect2 and Kif23/Mklp1 (the same set of centralspindlin targets indirectly regulated by miR-122) (9). Other signals may also contribute, including insulin/AKT signaling (11), unknown genes and even other miRNAs differentially expressed during postnatal development (e.g., miR-148, miR-194). Thus, the combined activity of all the signals in the network leads to inhibition of cytokinesis machinery, failure of cytokinesis and expansion of binucleate hepatocytes. The opposite situation occurs in adults where hepatocyte proliferation following partial hepatectomy involves successful cytokinesis (49). In adult livers, miR-122, E2F7, E2F8 and possibly other genes/miRNAs have reduced expression (compared to postnatal livers). Consequently, pro-cytokinesis signals are expressed appropriately, cytokinesis machinery remains functional and proliferating hepatocytes generate mononucleate daughter cells.
Despite our understanding of the myriad signals that have been shown to affect hepatic polyploidy, a major unanswered question is why polyploidy is so common, affecting 50% and 90% of adult hepatocytes in humans and mice, respectively (3, 4). Is there an evolutionarily conserved, liver-specific driver of polyploidy? We speculate miR-122 is a likely candidate. miR-122 is expressed nearly exclusively in the liver (16), and we hypothesize that it functions as a tissue-specific signal, which provides an explanation for the high percentage of polyploid cells in the liver. Consistent with this idea, it is intriguing to consider that cardiac myocytes become binucleate via a failed cytokinesis mechanism (50) and miR-122 is expressed (albeit at low levels) in the heart (16). Future studies should examine a role for miR-122 in cardiac myocyte polyploidy.
In summary, we identified a novel function for miR-122 and showed for the first time that miR-122 is required for complete hepatic polyploidization. Up-regulation of miR-122 during postnatal liver development, in collaboration with other signals, promotes a cell division program associated with failed cytokinesis. We propose that miR-122 regulates polyploidy in two major ways: first, by tissue-specific expression in the liver and, secondly, by inducing cytokinesis failure in an age-dependent manner.
Supplementary Material
Acknowledgments
We thank Lynda Guzik (McGowan Institute Flow Cytometry Core at University of Pittsburgh) for flow cytometry assistance; Nichole Owen, Muhsen Al-Dhalimy and Susan Olson (Cytogenetics Research Service Laboratory at Oregon Health and Science University) for mouse karyotypes; and the University of Pittsburgh Center for Biologic Imaging for confocal microscopy. Thanks to Frances Alencastro, Matthew Weirich and Patrick Wilkinson (University of Pittsburgh) for technical support and critical suggestions.
Financial Support
This work was supported by grants to AWD from the NIH (R01 DK103645) and the Commonwealth of Pennsylvania (4100068505) and to KG from the NIH (R01 CA193244).
Abbreviations
- miRNA
microRNA
- 3’UTR
3’ untranslated region
- miR-122
microRNA-122
- WT
wild-type
- Dicer1-LKO
Dicer1 liver-specific knockout
- Mir122-LKO
Mir122 liver-specific knockout
- Mir122-KO
Mir122 germline knockout
- mo
month
- wk
week
- d
day
- h
hour
- min
minute
- IPA
Ingenuity Pathway Analysis
- LKO
liver-specific knockout
- KO
knockout
- 2c
diploid cellular content
- 4c
tetraploid cellular content
- 8c
octaploid cellular content
- FL
Full-length 3’UTR reporter
- DEL
deleted 3’UTR reporter
- kDa
kilodalton
- SEM
standard error of mean
- qRT-PCR
quantitative reverse transcriptase polymerase chain reaction
Contributor Information
Shu-hao Hsu, Email: hsush1@ntu.edu.tw.
Evan R. Delgado, Email: evd7@pitt.edu.
P. Anthony Otero, Email: pao19@pitt.edu.
Kun-yu Teng, Email: teng.40@osu.edu.
Huban Kutay, Email: huban.kutay@osumc.edu.
Kolin M. Meehan, Email: kolin.mmeehan@gmail.com.
Justin B. Moroney, Email: jbm63@pitt.edu.
Jappmann K. Monga, Email: jkm59@pitt.edu.
Nicholas J. Hand, Email: njhand@mail.med.upenn.edu.
Joshua R. Friedman, Email: jfried12@its.jnj.com.
Kalpana Ghoshal, Email: kalpana.ghoshal@osumc.edu.
Andrew W. Duncan, Email: duncana@pitt.edu.
References
- 1.Guidotti JE, Bregerie O, Robert A, Debey P, Brechot C, Desdouets C. Liver cell polyploidization: a pivotal role for binuclear hepatocytes. J Biol Chem. 2003;278:19095–19101. doi: 10.1074/jbc.M300982200. [DOI] [PubMed] [Google Scholar]
- 2.Margall-Ducos G, Celton-Morizur S, Couton D, Bregerie O, Desdouets C. Liver tetraploidization is controlled by a new process of incomplete cytokinesis. J Cell Sci. 2007;120:3633–3639. doi: 10.1242/jcs.016907. [DOI] [PubMed] [Google Scholar]
- 3.Duncan AW, Hanlon Newell AE, Smith L, Wilson EM, Olson SB, Thayer MJ, Strom SC, et al. Frequent aneuploidy among normal human hepatocytes. Gastroenterology. 2012;142:25–28. doi: 10.1053/j.gastro.2011.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duncan AW, Taylor MH, Hickey RD, Hanlon Newell AE, Lenzi ML, Olson SB, Finegold MJ, et al. The ploidy conveyor of mature hepatocytes as a source of genetic variation. Nature. 2010;467:707–710. doi: 10.1038/nature09414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duncan AW. Aneuploidy, polyploidy and ploidy reversal in the liver. Semin Cell Dev Biol. 2013;24:347–356. doi: 10.1016/j.semcdb.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 6.Kurinna S, Stratton SA, Coban Z, Schumacher JM, Grompe M, Duncan AW, Barton MC. p53 regulates a mitotic transcription program and determines ploidy in normal mouse liver. Hepatology. 2013;57:2004–2013. doi: 10.1002/hep.26233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen HZ, Ouseph MM, Li J, Pecot T, Chokshi V, Kent L, Bae S, et al. Canonical and atypical E2Fs regulate the mammalian endocycle. Nat Cell Biol. 2012;14:1192–1202. doi: 10.1038/ncb2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conner EA, Lemmer ER, Sanchez A, Factor VM, Thorgeirsson SS. E2F1 blocks and c-Myc accelerates hepatic ploidy in transgenic mouse models. Biochem Biophys Res Commun. 2003;302:114–120. doi: 10.1016/s0006-291x(03)00125-6. [DOI] [PubMed] [Google Scholar]
- 9.Pandit SK, Westendorp B, Nantasanti S, van Liere E, Tooten PC, Cornelissen PW, Toussaint MJ, et al. E2F8 is essential for polyploidization in mammalian cells. Nat Cell Biol. 2012;14:1181–1191. doi: 10.1038/ncb2585. [DOI] [PubMed] [Google Scholar]
- 10.Baena E, Gandarillas A, Vallespinos M, Zanet J, Bachs O, Redondo C, Fabregat I, et al. c-Myc regulates cell size and ploidy but is not essential for postnatal proliferation in liver. Proc Natl Acad Sci U S A. 2005;102:7286–7291. doi: 10.1073/pnas.0409260102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Celton-Morizur S, Merlen G, Couton D, Margall-Ducos G, Desdouets C. The insulin/Akt pathway controls a specific cell division program that leads to generation of binucleated tetraploid liver cells in rodents. J Clin Invest. 2009;119:1880–1887. doi: 10.1172/JCI38677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mayhew CN, Bosco EE, Fox SR, Okaya T, Tarapore P, Schwemberger SJ, Babcock GF, et al. Liver-specific pRB loss results in ectopic cell cycle entry and aberrant ploidy. Cancer Res. 2005;65:4568–4577. doi: 10.1158/0008-5472.CAN-04-4221. [DOI] [PubMed] [Google Scholar]
- 13.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 15.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang X, Gal J, Zhuang X, Wang W, Zhu H, Tang G. A simple array platform for microRNA analysis and its application in mouse tissues. RNA. 2007;13:1803–1822. doi: 10.1261/rna.498607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu H, He JH, Xiao ZD, Zhang QQ, Chen YQ, Zhou H, Qu LH. Liver-enriched transcription factors regulate microRNA-122 that targets CUTL1 during liver development. Hepatology. 2010;52:1431–1442. doi: 10.1002/hep.23818. [DOI] [PubMed] [Google Scholar]
- 18.Hand NJ, Master ZR, Eauclaire SF, Weinblatt DE, Matthews RP, Friedman JR. The microRNA-30 family is required for vertebrate hepatobiliary development. Gastroenterology. 2009;136:1081–1090. doi: 10.1053/j.gastro.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 20.Hsu SH, Wang B, Kota J, Yu J, Costinean S, Kutay H, Yu L, et al. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J Clin Invest. 2012;122:2871–2883. doi: 10.1172/JCI63539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsai WC, Hsu SD, Hsu CS, Lai TC, Chen SJ, Shen R, Huang Y, et al. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J Clin Invest. 2012;122:2884–2897. doi: 10.1172/JCI63455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsu SH, Wang B, Kutay H, Bid H, Shreve J, Zhang X, Costinean S, et al. Hepatic Loss of miR-122 Predisposes Mice to Hepatobiliary Cyst and Hepatocellular Carcinoma upon Diethylnitrosamine Exposure. Am J Pathol. 2013;183:1719–1730. doi: 10.1016/j.ajpath.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Normand G, King RW. Understanding cytokinesis failure. Adv Exp Med Biol. 2010;676:27–55. doi: 10.1007/978-1-4419-6199-0_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Porrello ER, Johnson BA, Aurora AB, Simpson E, Nam YJ, Matkovich SJ, Dorn GW, 2nd, et al. MiR-15 family regulates postnatal mitotic arrest of cardiomyocytes. Circ Res. 2011;109:670–679. doi: 10.1161/CIRCRESAHA.111.248880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hand NJ, Master ZR, Le Lay J, Friedman JR. Hepatic function is preserved in the absence of mature microRNAs. Hepatology. 2009;49:618–626. doi: 10.1002/hep.22656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Septer S, Edwards G, Gunewardena S, Wolfe A, Li H, Daniel J, Apte U. Yes-associated protein is involved in proliferation and differentiation during postnatal liver development. Am J Physiol Gastrointest Liver Physiol. 2012;302:G493–503. doi: 10.1152/ajpgi.00056.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sekine S, Ogawa R, Ito R, Hiraoka N, McManus MT, Kanai Y, Hebrok M. Disruption of Dicer1 induces dysregulated fetal gene expression and promotes hepatocarcinogenesis. Gastroenterology. 2009;136:2304–2315. e2301–e2304. doi: 10.1053/j.gastro.2009.02.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu CG, Calin GA, Meloon B, Gamliel N, Sevignani C, Ferracin M, Dumitru CD, et al. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc Natl Acad Sci U S A. 2004;101:9740–9744. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fausto N, Campbell JS. The role of hepatocytes and oval cells in liver regeneration and repopulation. Mech Dev. 2003;120:117–130. doi: 10.1016/s0925-4773(02)00338-6. [DOI] [PubMed] [Google Scholar]
- 30.Duncan AW, Hanlon Newell AE, Bi W, Finegold MJ, Olson SB, Beaudet AL, Grompe M. Aneuploidy as a mechanism for stress-induced liver adaptation. J Clin Invest. 2012;122:3307–3315. doi: 10.1172/JCI64026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang L, Wang H, Bell P, McMenamin D, Wilson JM. Hepatic gene transfer in neonatal mice by adeno-associated virus serotype 8 vector. Hum Gene Ther. 2012;23:533–539. doi: 10.1089/hum.2011.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neumann B, Walter T, Heriche JK, Bulkescher J, Erfle H, Conrad C, Rogers P, et al. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature. 2010;464:721–727. doi: 10.1038/nature08869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seguin L, Liot C, Mzali R, Harada R, Siret A, Nepveu A, Bertoglio J. CUX1 and E2F1 regulate coordinated expression of the mitotic complex genes Ect2, MgcRacGAP, and MKLP1 in S phase. Mol Cell Biol. 2009;29:570–581. doi: 10.1128/MCB.01275-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with 'antagomirs'. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 35.Lian AT, Hains PG, Sarcevic B, Robinson PJ, Chircop M. IQGAP1 is associated with nuclear envelope reformation and completion of abscission. Cell Cycle. 2015:1–17. doi: 10.1080/15384101.2015.1044168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Banerjee B, Kestner CA, Stukenberg PT. EB1 enables spindle microtubules to regulate centromeric recruitment of Aurora B. J Cell Biol. 2014;204:947–963. doi: 10.1083/jcb.201307119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun L, Gao J, Dong X, Liu M, Li D, Shi X, Dong JT, et al. EB1 promotes Aurora-B kinase activity through blocking its inactivation by protein phosphatase 2A. Proc Natl Acad Sci U S A. 2008;105:7153–7158. doi: 10.1073/pnas.0710018105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoshida S, Bartolini S, Pellman D. Mechanisms for concentrating Rho1 during cytokinesis. Genes Dev. 2009;23:810–823. doi: 10.1101/gad.1785209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang SC, Lin XL, Li J, Zhang TT, Wang HY, Shi JW, Yang S, et al. MicroRNA-122 triggers mesenchymal-epithelial transition and suppresses hepatocellular carcinoma cell motility and invasion by targeting RhoA. PLoS One. 2014;9:e101330. doi: 10.1371/journal.pone.0101330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.White EA, Glotzer M. Centralspindlin: at the heart of cytokinesis. Cytoskeleton (Hoboken) 2012;69:882–892. doi: 10.1002/cm.21065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kojima K, Takata A, Vadnais C, Otsuka M, Yoshikawa T, Akanuma M, Kondo Y, et al. MicroRNA122 is a key regulator of alpha-fetoprotein expression and influences the aggressiveness of hepatocellular carcinoma. Nat Commun. 2011;2:338. doi: 10.1038/ncomms1345. [DOI] [PubMed] [Google Scholar]
- 42.Guse A, Mishima M, Glotzer M. Phosphorylation of ZEN-4/MKLP1 by aurora B regulates completion of cytokinesis. Curr Biol. 2005;15:778–786. doi: 10.1016/j.cub.2005.03.041. [DOI] [PubMed] [Google Scholar]
- 43.Hsu SH, Yu B, Wang X, Lu Y, Schmidt CR, Lee RJ, Lee LJ, et al. Cationic lipid nanoparticles for therapeutic delivery of siRNA and miRNA to murine liver tumor. Nanomedicine. 2013;9:1169–1180. doi: 10.1016/j.nano.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goel P, Manning JA, Kumar S. NEDD4-2 (NEDD4L): the ubiquitin ligase for multiple membrane proteins. Gene. 2015;557:1–10. doi: 10.1016/j.gene.2014.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fu J, Akhmedov D, Berdeaux R. The short isoform of the ubiquitin ligase NEDD4L is a CREB target gene in hepatocytes. PLoS One. 2013;8:e78522. doi: 10.1371/journal.pone.0078522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haitina T, Lindblom J, Renstrom T, Fredriksson R. Fourteen novel human members of mitochondrial solute carrier family 25 (SLC25) widely expressed in the central nervous system. Genomics. 2006;88:779–790. doi: 10.1016/j.ygeno.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 47.Lekomtsev S, Su KC, Pye VE, Blight K, Sundaramoorthy S, Takaki T, Collinson LM, et al. Centralspindlin links the mitotic spindle to the plasma membrane during cytokinesis. Nature. 2012;492:276–279. doi: 10.1038/nature11773. [DOI] [PubMed] [Google Scholar]
- 48.Apte U, Zeng G, Thompson MD, Muller P, Micsenyi A, Cieply B, Kaestner KH, et al. beta-Catenin is critical for early postnatal liver growth. Am J Physiol Gastrointest Liver Physiol. 2007;292:G1578–1585. doi: 10.1152/ajpgi.00359.2006. [DOI] [PubMed] [Google Scholar]
- 49.Miyaoka Y, Ebato K, Kato H, Arakawa S, Shimizu S, Miyajima A. Hypertrophy and unconventional cell division of hepatocytes underlie liver regeneration. Curr Biol. 2012;22:1166–1175. doi: 10.1016/j.cub.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 50.Engel FB, Schebesta M, Keating MT. Anillin localization defect in cardiomyocyte binucleation. J Mol Cell Cardiol. 2006;41:601–612. doi: 10.1016/j.yjmcc.2006.06.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.