Abstract
Sterol regulatory element-binding proteins (SREBPs) are key transcription factors regulating cholesterol and fatty acid biosynthesis. SREBP activity is tightly regulated to maintain lipid homeostasis, and is modulated upon extracellular stimuli such as growth factors. While the homeostatic SREBP regulation is well studied, stimuli-dependent regulatory mechanisms are still elusive. Here we demonstrate that SREBPs are regulated by a previously uncharacterized mechanism through TGF-β activated kinase 1 (TAK1), a signaling molecule of inflammation. We found that TAK1 binds to and inhibits mature forms of SREBPs. In an in vivo setting, hepatocyte-specific Tak1 deletion upregulates liver lipid deposition and lipogenic enzymes in the mouse model. Furthermore, hepatic Tak1 deficiency causes steatosis pathologies including elevated blood triglyceride and cholesterol levels, which are established risk factors for the development of hepatocellular carcinoma (HCC) and are indeed correlated with Tak1-deficiency-induced HCC development. Pharmacological inhibition of SREBPs alleviated the steatosis and reduced the expression level of the HCC marker gene in the Tak1-deficient liver. Thus, TAK1 regulation of SREBP critically contributes to the maintenance of liver homeostasis to prevent steatosis, which is a potentially important mechanism to prevent HCC development.
Keywords: TAK1, SREBP, lipogenesis, liver, steatosis, hepatocellular carcinoma
INTRODUCTION
Sterol regulatory element-binding proteins (SREBPs) are a basic helix-loop-helix-leucine zipper (bHLH-LZ) transcription factor family, which are the central regulators of lipogenesis by transcriptionally activating a number of genes involved in fatty acids, triglycerides and cholesterol synthesis.1 Mammalian SREBP family has three isoforms, SREBP1a, SREBP1c and SREBP2.2 SREBP1a and SREBP1c are splicing variants and are derived from Srebf1 gene, and SREBP2 is encoded by Srebf2. SREBP1a and SREBP1c play predominant roles in fatty acid and triglyceride synthesis by upregulating a series of enzymes including acetyl CoA carboxylase, fatty acid synthase (FASN), and steadily CoA denatures (SCD). While SREBP1a and SREBP1c are functionally overlapped, SREBP1c is known to be the predominant form in the liver. SREBP2 is selectively involved in cholesterol synthesis and uptake by upregulating enzymes such as HMG CoA synthase, HMG CoA reductase and mevalonate kinase (MVK). SREBP2 can play compensatory roles to upregulate fatty acid and triglyceride synthesis when SREBP1 isoforms are genetically deleted.3 Homeostatic SREBP activity is controlled by its unique sterol-dependent feedback loop.1 Inactive precursor SREBPs are localized onto the endoplasmic reticulum (ER) membrane and interact with polytopic SREBP cleavage activated protein (SCAP) and insulin-induced gene protein (Insig).4 When cholesterol levels drop, SREBPs are released from ER to the Golgi apparatus where SREBP undertakes protein processing to release the N-terminal bHLH-LZ domain. The mature bHLH-LZ SREBP enters the nucleus and binds as a dimer to SREBP responsive element (SRE) within promoters of its target genes. As SREBP2 activates cholesterol synthesis, cholesterol gradually builds up in the cell and activates negative feedback loop in which cells restore Insig-SCAP interaction to terminate the SREBP pathway.5 Mature SREBPs are constantly degraded through a ubiquitin-proteasome system,6, 7 which prevents sustained transcriptional activation of lipogenic genes and ensures a quick response to the negative feedback regulation.
In addition to the sterol-dependent feedback loop regulation, SREBP activity is modulated in response to extracellular stimuli. Growth factors enhance the activity of SREBP through the AKT-mTOR pathway, which provides membrane lipids as cell building materials to support cell growth.8–12 Unfolded protein-induced stress conditions, ER stress, are also known to activate SREBP.13, 14 in which upregulated lipogenesis supplies the ER membrane to alleviate ER stress conditions.15, 16 Conversely, protein kinase AMPK, which is activated by nutrient stress (low cellular ATP), is reported to directly phosphorylate and inhibit SREBP, which reduces lipogenesis to restore the ATP level by limiting energy expenditure.17 While the mechanism of sterol feedback loop regulation of SREBP is extensively studied, stimuli-induced regulatory mechanisms of SREBP are still elusive.
TAK1 is a member of the mitogen activated protein kinase kinase kinase (MAP3K) family, and is activated by inflammatory cytokines such as IL-1, TNFα or Toll-like receptor ligands.18 TAK1 signaling promotes inflammatory responses through its downstream targets including but not limited to transcription factors NF-κB and AP-1. Deletion of Tak1 causes TNFα-induced cell death followed by tissue damage and animal mortality in the epithelial and endothelial-specific Tak1-deficiet mouse models.19–21 Hepatocyte-specific deletion of Tak1 causes hepatocyte death resulting in liver injury,22–24 which is also due to TNFα-induced cell death.23, 25 However, unlike epithelial- or endothelial-specific deletion of Tak1, hepatocyte-specific Tak1 deletion does not cause acute animal mortality but induces hepatocellular carcinoma (HCC) around 2 months of age.22–24 Several cancer-associated abnormalities have been implicated in Tak1 deficiency-induced HCC, which include TGF-β-induced fibrosis 26 and hyper-proliferation of hepatocytes.24 One reasonable expiation of why Tak1 deficiency induces HCC is compensatory hyper-proliferation of hepatocytes, which is initiated by TNFα-induced cell death. However, while ablation of TNFα signaling effectively rescues cell death,23, 25 it does not abolish development of HCC as described in this report. Thus, TNFα-induced cell death is not solely the cause of HCC, and the mechanism by which Tak1 deficiency induces HCC is still elusive.
HCC in the Tak1-deficient liver develops spontaneously and very quickly within several weeks, which is different from any abnormalities that are observed in mouse models harboring liver-specific deletion of NF-κB or AP-1 pathways.24, 27, 28 For example, deletion of NF-κB slowly induces HCC at over 12 months of age.24, 29, 30 Deletion of AP-1 activators, JNKs, in hepatocytes enhances chemical-induced HCC but no spontaneous HCC has been reported.27 Thus, we postulate that TAK1 deficiency induces HCC predominantly not through impaired NF-κB or AP-1. Here we report an identification of SREBPs as new TAK1 targets. We show that Tak1 deficiency caused hepatic steatosis through dysregulation of SREBP. Steatosis is recognized as a liver lesion, which can progress to or facilitate HCC. Our results demonstrate that TAK1 regulation of SREBP is critical in liver homeostasis by preventing hepatic steatosis, which could further associate with the prevention of HCC.
RESULTS
Tnfr1 deletion does not effectively block tumorigenesis in Tak1-deficient liver
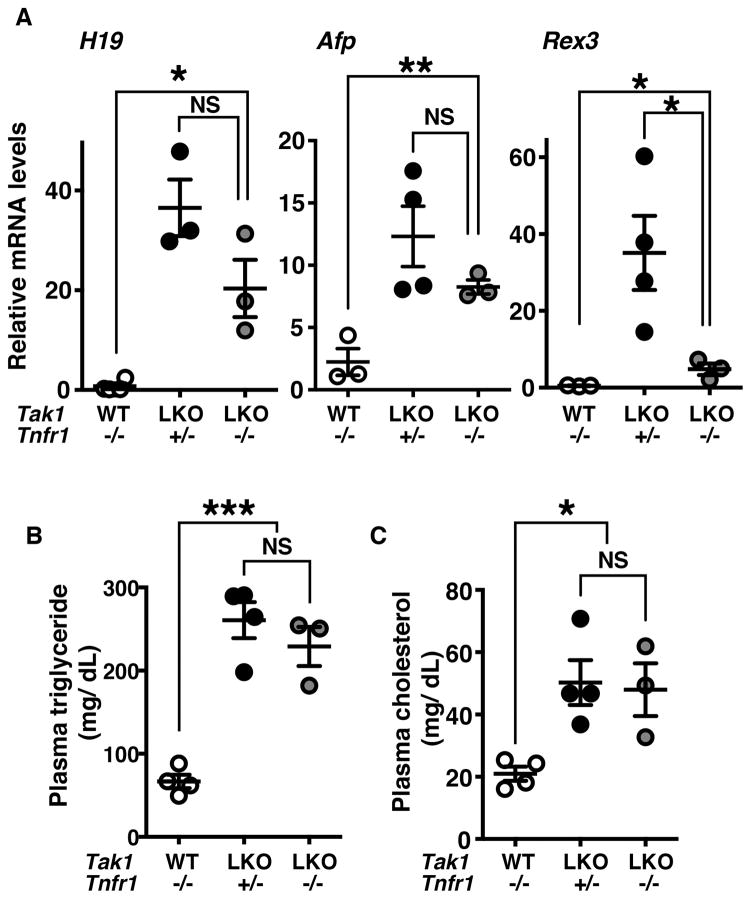
Hepatocyte-specific Tak1-deficient mice have been generated and characterized in three independent studies,22–25 which demonstrated that deletion of Tak1 in the liver induces liver lesions characterized by hepatocyte death and early onset of HCC within 2 months of age. Ablation of TNFα signaling by deletion of Tnfr1 has been demonstrated to abolish hepatocyte death in Tak1-deficient liver,23, 25 indicating that TNFα is the cause of Tak1-deficient hepatocyte death. Cell death is known to induce compensatory proliferation, which is implicated in tumorigenesis.31 Thus, we initially anticipated that TNFα-induced cell death is the cause of HCC. However, we unexpectedly observed liver tumors in mice having liver-specific deletion of Tak1 in the Tnfr1−/− background (Tak1LKO Tnfr1−/−) at 5–8 months old (supplementary Fig. S1A). To validate this observation, we examined the levels of fetal liver gene expression, including a non-cording RNA, H19, alpha-fetoprotein (Afp) and Rex3, which are known to be upregulated at the early stage of HCC.32–34 We previously reported that the Tak1LKO liver at 4–5 weeks of age does not have visually detectable tumors, but these HCC markers are highly increased.22 In the Tak1LKO Tnfr1−/− livers, the levels of the HCC markers were still upregulated compared to the wild type littermate livers, although the levels were slightly or moderately lower compared to littermate mice having functional TNF signaling (Tak1LKO Tnfr1+/−) (Fig. 1A). At 4–5 months of age, the H19 RNA level was highly varied but still upregulated in the Tak1LKO Tnfr1−/− liver compared to the control littermate livers (supplementary Fig. S1B). Thus, TNFα-induced hepatocyte death is not solely the cause of tumorigenesis in the Tak1-deficient liver.
Figure 1. Deletion of Tnfr1 does not prevent tumorigenesis or steatosis in the Tak1-deficient liver.
(A) The liver was isolated from Tak1LKO (LKO) or control no-Cre (WT) littermate in the background of Tnfr1−/− or Tnfr1+/− mice at 6 weeks of age, and RNA was analyzed by the quantitative real time PCR. All data points and means ± SEM of H19, Afp and Rex3 mRNA levels relative to Gapdh are shown. *, p < 0.05; **, p < 0.01; NS, not significant (two-tailed unpaired Student’s t test). (B, C) Blood was collected from the facial vein of the same mice as described (A), and the levels of plasma triglyceride and cholesterol were determined. All data points and means ± SEM are shown. *, p < 0.05; ***, p < 0.001, NS, not significant (one-way ANOVA).
Tak1 deficiency causes liver steatosis through a previously unidentified mechanism
Tak1-deficient liver also exhibited typical features of steatosis including lipid depositions (supplementary Fig. S2), which is consistent with a recent report by Seki’s group.35 The levels of both blood triglycerides and cholesterol were upregulated in Tak1LKO mice (Fig. 1B, 1C supplementary Fig. S3). Importantly, we found that deletion of Tnfr1 did not reduce the levels of blood triglycerides or cholesterol (Fig. 1B and 1C). Given that Tnfr1 deletion completely blocks the hepatocyte death in Tak1LKO mice,25 the hepatic steatosis is independent on TNF-induced cell death in Tak1LKO liver. Many cellular processes not limited to excess lipogenesis or impaired breakdown of lipids have been implicated in hepatic steatosis.36, 37 Accordingly, a diverse set of cell signaling molecules are known to be involved in the development of steatosis including well-known targets of TAK1, i.e. NF-κB and JNK. Liver-specific deletion of NF-κB or JNK causes hepatic steatosis.29, 38 AMP-activated protein kinase (AMPK) is also a target of TAK1,39 and was reported to inhibit lipogenesis.17 An earlier study reports that deletion of Tak1 reduces AMPK activation in cultured hepatocytes.35 Thus, we next examined whether these targets of TAK1 were impaired in the Tak1-deficient liver in vivo. Unexpectedly, we found that activities of NF-κB and JNK were not pronouncedly altered by deletion of Tak1 in the liver (supplementary Fig. S4). Furthermore, the level of AMPK activity was not reduced but rather increased in the Tak1-deficient liver (supplementary Fig. S4). Thus, these TAK1 targets are unlikely to be involved in the steatosis phenotype of the Tak1-deficient liver. These results lead us to explore new targets of TAK1.
TAK1 interacts with and phosphorylates SREBPs
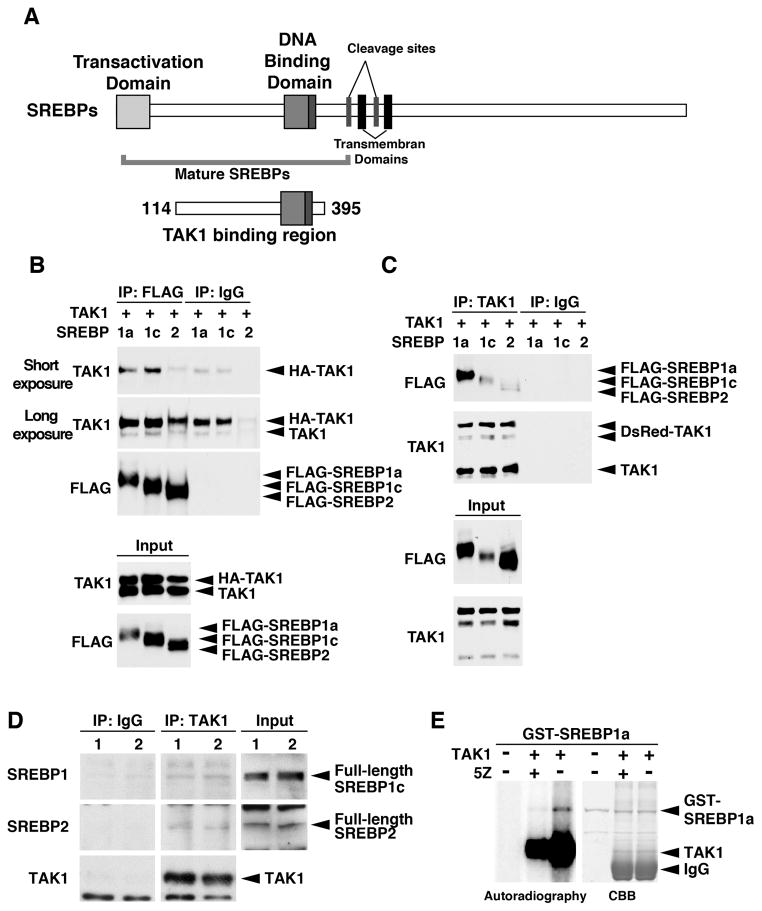
We previously performed a series of yeast-two hybrid screenings using TAK1 protein as a bait.40, 41 Among the identified but not yet characterized binding partners of TAK1, we focused on SREBP2 as a potential target of TAK1, which could be involved in liver steatosis in the Tak1-deficient liver. The N-terminal region of mouse SREBP2 amino acid residues positioned between 114 and 395 was isolated as a binding region of TAK1, which is well conserved among SREPB1a, SREBP1c and SREBP2 (45% identical) (Fig. 2A). The TAK1 binding region of SREBP was mapped between the transactivation domain and the cleavage sites of SREBP, and included the entire region of bHLH-ZF domain, which covers most parts of the mature SREBPs (Fig. 2A). Thus, we examined whether mature forms of SREPB1a, SREBP1c and SREBP2 interact with TAK1 using the overexpression system in human embryonic kidney 293 cells. Exogenously expressed TAK1 was efficiently pulled down by immunoprecipitation against SREBPs (Fig. 2B, top panel), whereas endogenous TAK1 was moderately precipitated (2nd panel). Endogenous TAK1 may be pre-occupied with endogenous TAK1 binding partners including SREBPs, which might cause the less efficient co-precipitation of endogenous TAK1 with exogenously expressed SREBPs. Reciprocally, all matured forms of SREBP isoforms were pulled down with TAK1 (Fig. 2C), although SREBP2 co-precipitated TAK1 with a lesser efficiency compared to SREBP1a and SREBP1c. To examine the interaction between TAK1 and SREBPs under the physiological settings, we also conducted co-immunoprecipitation analysis using proteins from a human HCC cell line, HepG2, and the mouse liver. Endogenous full-length SREBPs were co-precipitated with TAK1 (Fig. 2D and supplementary Fig. S5,). These results suggest that TAK1 interacts with SREBP in cells, and raise the possibility that TAK1 participates in modulation of SREBPs. Because TAK1 is a protein kinase, TAK1 potentially modulates SREBPs through phosphorylation. To test whether TAK1 phosphorylates SREBPs, we conducted an in vitro kinase assay. Activated TAK1 was prepared from HEK293 cells overexpressing the catalytic domain of TAK1 (TAK1ΔC) together with TAK1-binding protein 1 (TAB1), which is an activating subunit of TAK1.42 Bacterially purified recombinant GST-SREBP1a (mature form) was incubated with the TAK1-TAB1. SREBP1a was phosphorylated when incubated with activated TAK1, which was reduced in the presence of a selective inhibitor of TAK1, 5Z-7-Oxozeaenol (5Z) (Fig. 2E). Thus, TAK1 is a potential modulator of SREBPs through binding to and phosphorylating SREBPs.
Figure 2. TAK1 interacts with SREBP.
(A) Structure of SREBPs. The region of TAK1 binding domain isolated from the yeast two-hybrid screening is denoted by encompassing amino acid residues of SREBP2. (B, C) HEK293 cells were transfected with expression vectors for HA- (B) or DsRed- (C) tagged TAK1 and FLAG-tagged mature forms of SREBPs (N-terminal region), and proteins from cell lysates were immuneprecipitated by anti-FALG (B) or anti-TAK1 (C) as well as non-immunized control IgG. Immunoprecipitates were analyzed by immunoblotting. The amounts of input proteins are also shown in bottom two panels. Shorter (B, top panel) and longer (B, second panel) exposures are shown to visualize less efficiently co-precipitated HA-TAK1 (3rd lane) and endogenous TAK1 (D) Cytoplasmic fractions including cytoplasmic organelle fractions from the mouse liver were immunoprecipitated by anti-TAK1 or control IgG. Immunoprecipitates were analyzed by immunoblotting. Each lane (lane 1 and 2) represents a sample from an individual mouse. (E) HEK293 cells were transfected with expression vectors for HA-tagged catalytic domain of TAK1 (TAK1ΔC) together with TAB1. HA-TAK1ΔC was purified from protein extracts by immunoprecipitation, and incubated with bacterially purified GST-tagged mature SREBP1a in the presence (+) or absence (−) of 5Z-7oxozeaenol (5Z) (500 nM). Phosphorylation of the proteins was detected by autoradiography. The protein amounts are shown by coomassie brilliant blue (CBB) staining.
TAK1 inhibits SREBPs
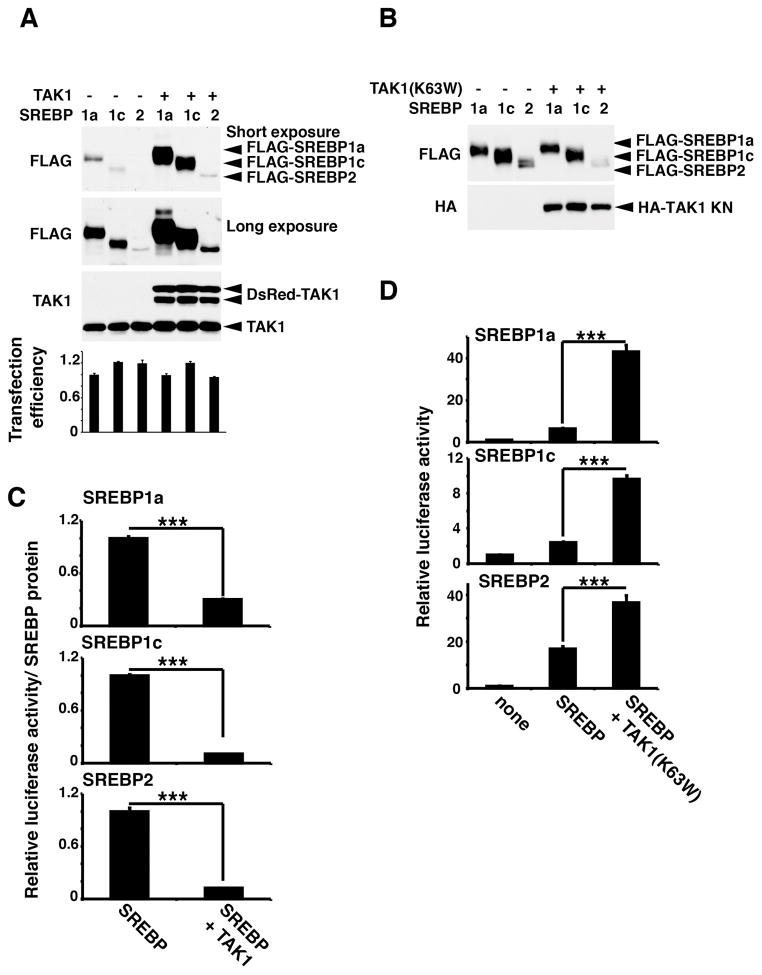
We next examined whether TAK1 modulates activity of SREBPs. We co-expressed TAK1 or the kinase-negative form of TAK1(K63W) together with mature forms of SREBPs. We found that the protein levels of all SREBP isoforms were greatly increased by co-expression of catalytically active TAK1 (Fig. 3A), while kinase-dead TAK1, TAK1(K63W), did not alter the expression levels of SREBPs (Fig. 3B). The transfection efficiencies monitored by co-transfected a constitutively expressed reporter were comparable in all samples, indicating that increased amounts of SREBPs were not due to a higher transfection efficiency (Fig. 3A, bottom graph). An earlier study demonstrates that inhibition of the transcriptional activity of SREBP blocks proteasome-dependent degradation of SREBP,6 which is one of the feedback regulations to maintain the homeostatic level of SREBP activity. Thus, catalytically active TAK1 may inhibit SREBP activity. Indeed, we found that transcriptional activity of SREBPs monitored by a sterol regulatory element (SRE)-driven reporter expression was greatly diminished (Fig. 3C). By contrast, expression of catalytically inactive TAK1 highly increased transcriptional activity of all SREBPs (Fig. 3D). TAK1(K63W) is known to act as a dominant negative inhibitor by preventing autophosphorylation of TAK1.42, 43 Thus, endogenous TAK1 is likely to play an inhibitory role in SREBP activation. These results demonstrate that TAK1 binds to and inhibits mature SREBPs, and this regulation depends on its kinase activity. To obtain mechanistic insights into TAK1 regulation of SREBPs, we tested the possibility that TAK1 inhibits nuclear localization of mature SREBPs. HEK293 cells expressing the mature form of SREBP1a with and without TAK1 were fractionated into the nuclear and non-nuclear cytoplasmic fractions, and the localization of SREBP was analyzed by immunoblotting (supplementary Fig. S6A). We note here that, since TAK1 highly upregulates the amounts of SREBPs as shown in Fig. 3A, the vector amounts used for expression of SREBP with and without TAK1 co-expression were adjusted to achieve the similar expression levels of SREBP between samples. The mature form of SREBP1a was mostly localized to the nucleus, and overexpression of TAK1 did not alter the SREBP localization (supplementary Fig. S6A). We also examined the localization of SREBP2 in wild-type and Tak1LKO liver. SREBP2 was predominantly localized to the nucleus, and we found no observable alteration in the localization of SREPB2 by Tak1 deficiency (supplementary Fig. S6B). These results indicate that TAK1 inhibits transcriptional activity of SREBPs not by blocking their nuclear localization.
Figure 3. TAK1 inhibits transcription activity of SREBP.
HEK293 cells were transfected with the SREBP reporter (SRE-luciferase) and expression vectors for TAK1 (DsRed-TAK1) (A and C) or a kinase-dead form of TAK1 [TAK1(K63W)] (B and D) and mature forms of SREBP together with a constitutive promoter-driven renilla luciferase reporter. (A, B) The levels of transfected SREBPs and TAK1 were analyzed by immunoblotting. (C) SREBP-dependent transcriptional activity was determined by luciferase activity. Relative SREBP activity normalized to the protein levels of transfected SREBPs are shown. Means ± SD. n = 3; ***, p<0.001 (two-tailed unpaired Student’s t test). (D) SREBP-dependent transcriptional activity normalized to renilla luciferase activity is shown. Means ± SD ; n = 3; ***, p < 0.001 (one-way ANOVA).
TAK1 is a regulator of SREBP
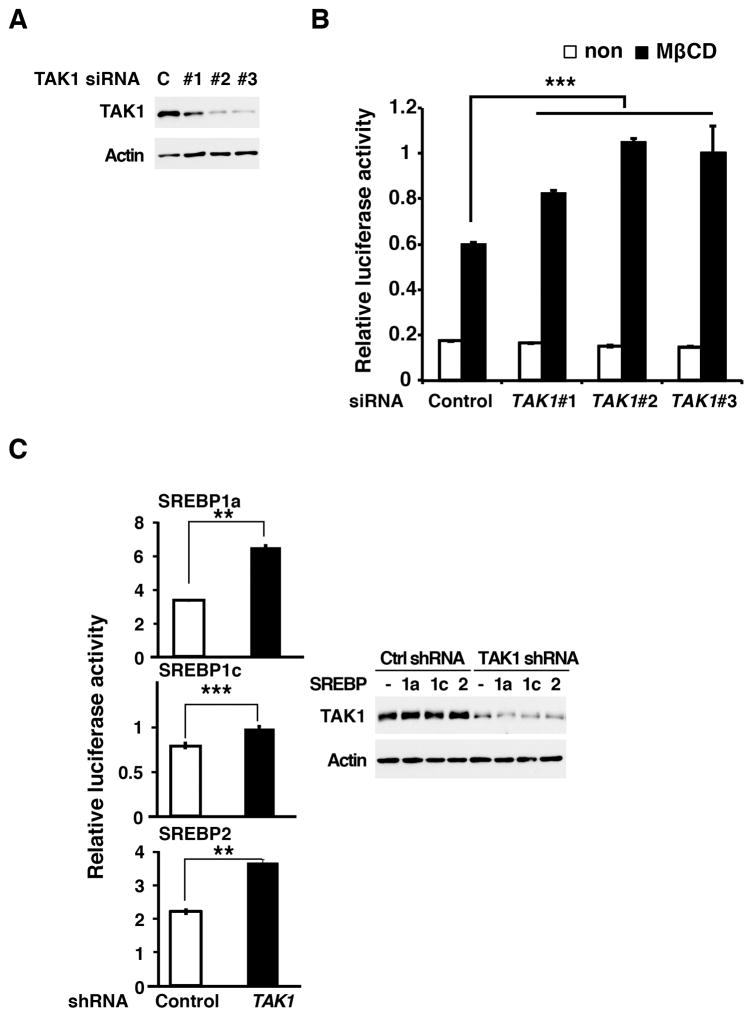
To investigate whether TAK1 modulates SREBP in the physiological settings, we examined whether downregulation of Tak1 alters endogenous SREBP activity. We lowered the TAK1 protein level by utilizing three independent siRNAs targeted against TAK1 mRNA, and transcriptional activity of endogenous SREBP was examined in 293 cells. The knockdown of TAK1 reduced the TAK1 protein ranging from moderately to effectively (Fig. 4A). Under the normal culture condition, SREBP activity was found to be low and was not detectably altered by knockdown of TAK1 (Fig. 4B). However, when endogenous SREBP was activated by depleting cellular cholesterol using methyl-β-cyclodextrin, SREBP activity was augmented at higher levels in TAK1-knockdown cells compared to control cells (Fig. 4B). This suggests that endogenous TAK1 inhibits SREBP. To further examine whether TAK1 modulates SREBP in hepatocytes, the human HCC cell line, HepG2, was utilized. TAK1 was knocked down using an shRNA targeted against TAK1 (Fig. 4C, right panels), and transcriptional activity of exogenously expressed SREBPs was monitored by the reporter assay. Ablation of TAK1 was found to upregulate SREBP activity in HepG2 cells (Fig. 4C). These data demonstrate that endogenous TAK1 suppresses SREBP activity.
Figure 4. Ablation of TAK1 augments SREBP activity.
(A, B) HEK293 cells were treated with three independent siRNAs targeted against TAK1, and SRE-luciferase and control EF1α-renilla luciferase vectors were subsequently transfected at one-day post siRNA introduction. Cells were left untreated or treated with methyl-β-cyclodextrin 6 h before the cell harvesting. Proteins were analyzed by immunoblotting (A). Luciferase activity is normalized to renilla luciferase activity (B). Means ±SD; n = 3; ***, p < 0.001 (one-way ANOVA). (C) A vector expressing small hairpin RNA targeted against TAK1 was transfected into HepG2 cells. Cells were subsequently transfected with the expression vectors for mature SREBPs and the same reporters used in (A, B). Proteins were analyzed by immunoblotting to determine the protein level of TAK1 (right panes). The level of TAK1 was not altered by the 2nd transfections. Luciferase activity is normalized to renilla luciferase activity. Means ±SD; n = 3; **, p <0 .01; ***, p < 0.001 (two-tailed unpaired Student’s t test).
TAK1 suppresses SREBP in the liver
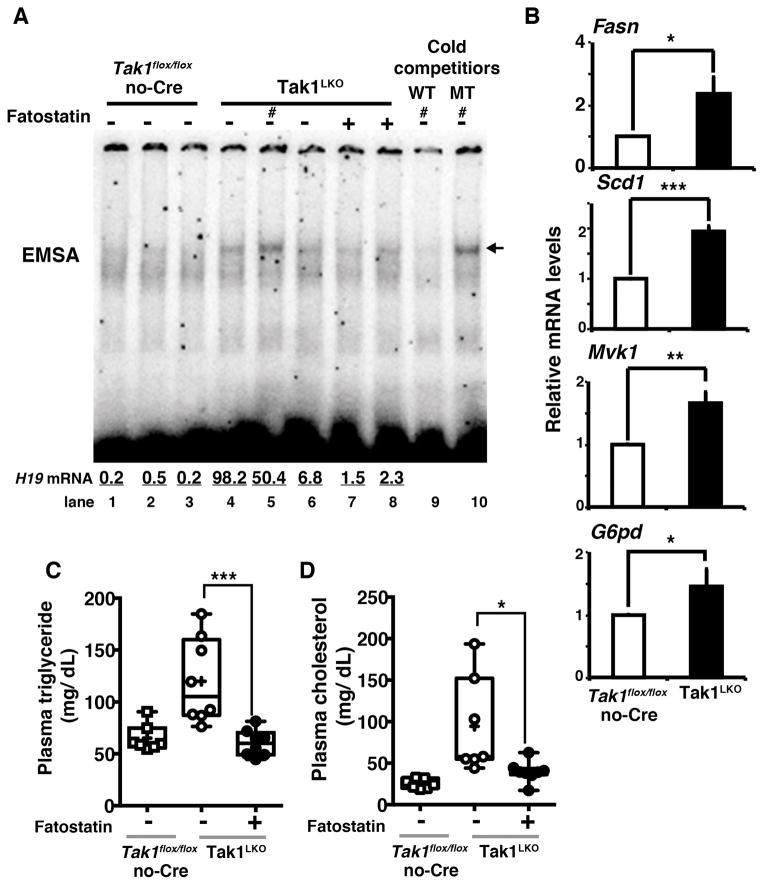
Taking the fact that the liver is the central tissue of lipid synthesis, our results above prompted us to connect between TAK1 regulation of SREBP and Tak1 deficiency-induced hepatic steatosis. We examined whether impaired TAK1-dependent SREBP suppression is associated with steatosis in the Tak1-deficient liver. We first examined endogenous liver SREBP activity by monitoring the specific binding ability to the sterol regulatory element consensus sequence (electron mobility shift assay). The SREBP DNA binding activity was increased in Tak1LKO liver compared to control littermate livers (Fig. 5A lanes 1–6). Consistent with the increased SREBP activity, SREBP target genes including Fasn, Scd1, Mvk1 and glucose-6-phosphate dehydrogenase (G6pd) were upregulated in the Tak1-deficient liver (Fig. 5B). SREBP is known to be self-regulated, and SREBP gene (Srebf) expression showed a trend of increase in Tak1-deficient liver (Supplementary Fig. S7). To investigate whether increased SREBP activity is causally associated with steatosis in the Tak1-deficient liver, we treated Tak1LKO mice with an inhibitor of SREBP, fatostatin, which binds to SCAP and inhibits translocation of SREBP from the endoplasmic reticulum to the Golgi apparatus.44 Treatment of fatostatin in Tak1LKO mice reduced the levels of blood triglyceride and cholesterol (Fig. 5C and 5D). These results suggest that TAK1 regulation of SREBP is critical to prevent hepatic steatosis. Finally, we ask whether the hepatic steatosis by activated SREBP is causally associated with HCC in Tak1LKO mice. We examined the level of HCC marker, H19, expression in the fatostatin treated mouse livers. We found that the reduced SREBP activity by fatostatin was correlated with the lower expression levels of the HCC marker (see Fig. 5A, bottom numbers, lanes 7 and 8), which suggest a potential causal association between activation of SREBP and HCC in Tak1LKO mice.
Figure 5. Deletion of Tak1 upregulates SREBP in the liver.
(A) 4–6 weeks old control (Tak1flox/flox no-Cre) and Tak1LKO mice were treated with or without fatostatin for 30 days. Liver extracts were analyzed by electron mobility shift assay (EMSA). 100 fold excess cold wild type (WT) or mutant (MT) oligonucleotides described in Materials and Methods were added to the reaction mixture to examine the specific binding of the band (lane 9 and 10). Lanes indicated by a symbol # used the protein extract from the same mouse. The relative mRNA levels of H19 relative to the levels of Gapdh are shown in the bottom of the panels. (B) Livers were isolated from 5–6 month old control (Tak1flox/flox no-Cre) and Tak1LKO mice, and the mRNA levels of SREBP target genes were analyzed. Means ± SEM; n = 3; *, p < 0.05; **, p < 0.01; ***, p < 0.001 (two-tailed unpaired Student’s t test). (C, D) Blood was collected from the control (Tak1flox/flox no-Cre) and Tak1LKO mice used in the experiment shown in (A). Blood triglyceride and cholesterol were determined. All data points are shown in the box and whisker plots: median and distribution of 50% of values are shown in the box: whiskers indicate distribution of minimum and maximum values. Asterisks indicate means. Control (Tak1flox/flox no-Cre), n = 7; Tak1LKO, n = 7; Tak1LKO fatostatin treated, n = 8; *, p < 0.05; ***, p < 0.001 (one-way ANOVA).
DISCUSSION
SREBP is the master regulator of lipogenesis, and plays an indispensable role to maintain the proper levels of cellular lipids including triglycerides and sterols though the highly sophisticated proteolytic processing mechanism.1 SREBP is also modulated upon growth stimulation, in which growth factor-PI3K-mTOR pathway upregulates SREBP activity through promoting nuclear localization of SREBP.8–12 Activated SREBP provides triglycerides and sterols to build new cell membranes. In the current study, we show that TAK1 inhibits mature SREBP, and impairment of this regulation induces hepatic steatosis. TAK1 is the inflammatory signaling intermediate, and is activated when cells are exposed to danger signals including inflammatory cytokines and microorganisms.18 Inflammation and lipid homeostasis is closely associated.45 Chronically inflamed conditions induce lipolysis in the adipose tissue and upregulate lipid biogenesis in the liver.46, 47 We show here that ablation of TAK1 disrupts normal lipid homeostasis by increasing SREBP activity. The liver is at the front line to constantly deal with exogenous stressors such as moieties from microorganisms and chemicals, many of which are inflammatory stimuli and activators of TAK1. Thus, hepatic TAK1 is likely to be constantly activated by these stressors, which may be one of the critical mechanisms by which normal liver limits their lipogenesis.
In the current study, we identified SREBPs as novel binding partners of TAK1, and demonstrate that TAK1 inhibits SREBP activity. Although the detailed molecular mechanism is still elusive, our study shows several interesting aspects in TAK1 regulation of SREBP. Conventionally, maturation (processing) and subsequent nuclear localization of SREBP are the keys to SREBP activation.1 However, we found that SREBP nuclear localization is not affected by either activation or ablation of TAK1 (supplementary Fig. S6). In contrast, we observed increased ability of SREBP to bind sterol-responsive element DNA sequence in the Tak1-deficient liver (Fig. 5A), suggesting that TAK1 modulates SREBP DNA binding. We also demonstrate that TAK1 can phosphorylate SREBP using an in vitro kinase assay. Furthermore, we found that endogenous TAK1 binds to full-length SREBP. Collectively, one possible scenario of TAK1 regulation of SREBP is that TAK1 binds to and phosphorylates ER- or Golgi-localized full-length SREBP in the cytoplasm, which does not prevent SREBP maturation or translocation into the nucleus, but the TAK1 modulated (phosphorylated) SREBPs possess diminished transcriptional activity due to their reduced ability of DNA binding. Alternatively or additionally, TAK1 modulation may block transcriptional activity of SREBP by modulating interaction between SREBPs and transcriptional co-factors such as CBP and p300.48, 49 Further investigations to determine the mechanism for TAK1 regulation of SREBP will enhance our understanding in the molecular link between inflammation and lipogenesis.
Hepatic steatosis is a common liver disorder, which is induced by excess alcohol consumption, hepatitis viruses and metabolic syndromes. The mechanisms for development of hepatic steatosis are not yet entirely clear, but are clearly associated with elevated and chronic exposures to stressors. As protein kinase signaling pathways are generally regulated by negative feedback loops mediated by activation or induction of protein phosphatases,50 TAK1 signaling is normally negatively regulated by sustained stimulation.51, 52 Thus, chronic exposures to stressors may not chronically activate but rather impair TAK1 signaling. Given that ablation of TAK1 causes hepatic steatosis, one possible mechanism for hepatic steatosis is impaired TAK1 signaling under chronically stressed conditions, which warrants further investigations of potential causal relationships among chronic stress, TAK1 activity and hepatic steatosis.
Hepatic steatosis is recognized as a liver lesion, which could lead to cirrhosis, fibrosis and ultimately HCC. These pathogenic processes from steatosis to HCC normally take a considerable time in humans, and excess lipogenesis alone by overexpression of mature SREBP1c does not result in HCC in the mouse model.53–55 Hyper-activation of PI3K-mTOR pathway by liver-specific deletion of PTEN effectively induces steatosis through activation of SREBP but develops HCC very slowly, which takes more than 9 months in the mouse model.56 These suggest that not only activation of SREBP but also some additional aberrations are likely to be required for effective development of HCC. Tak1-deficient liver develops steatosis, fibrosis and HCC within 2 months,23, 24 which is exceptionally effective; even more effective compared to the HCC models induced by transgenic expression of oncogenes such as K-Ras and β-catenin.57 Thus, it is unlikely that steatosis alone is the cause of HCC in Tak1-deficient liver. Constantly occurring cell death induces inflammation, which is associated with genetic instability and quick development of HCC.58 Tak1 deficiency induces TNFα-dependent cell death in the liver.23, 25 Combination of steatosis and chronic injury due to constantly occurring cell death may be the cause of HCC in Tak1-deficient liver. However, while ablation of TNFα signaling could rescue hepatocyte death in Tak1-deficient liver, it did not abolish HCC development (see Fig. 1A). Thus, TNFα-induced cell death facilitates but is not causally associated with HCC. It is noteworthy that a non-coding RNA H19 is highly expressed in Tak1-deficient liver even at 4–5 weeks of age when HCC is not yet clearly observed. Importantly, the level of H19 is only moderately reduced by deletion of Tnfr1, suggesting that H19 expression is not the result of cell death. H19 is considered as a marker of cell de-differentiation, which is known to be associated with tumor metastasis.32, 33, 59 Thus, TAK1 may be involved in regulatory processes of hepatocyte differentiation. Nonetheless, TAK1-deficiency induced HCC is unique, which develops quickly but still shares key features of human HCC including steatosis and fibrosis, and warrants future studies.
MATERIALS AND METHODS
Mice and cell culture
Tak1-floxed (Tak1flox/flox) mouse was previously described.60 Alb.Cre transgenic and Tnfr1−/− C57BL/6 mice were obtained from the Jackson Laboratories.61, 62 All strains used were backcrossed at least 7 times to C57BL/6. Both male and female mice were used. Inhibition of SREBP was achieved by intraperitoneal (ip) injection of fatostatin (Millipore) (10 mg/kg mouse weight per day) for 30 consecutive days. All animal experiments were conducted with the approval of the North Carolina State University Institutional Animal Care and Use Committee. All efforts were made to minimize animal suffering. Human embryonic kidney (HEK) 293 and HepG2 cells were cultured in DMEM with 10% bovine growth serum (Hyclone) and penicillin-streptomycin at 37°C in 5% CO2.
Antibodies, reagents, plasmids and siRNAs
Anti-TAK1 antibody was described previously.63 Anti-SREBP1 (2A4) (Santa Cruz Biotechnology), anti-SREBP2 (ab30682) (Abcam), anti-AMPK (Cell Signaling Technology), anti-phospho-AMPK (Thr172) (40H9) (Cell Signaling Technology), anti-JNK1/2 (sc571) (Santa Cruz Biotechnology), anti-phospho-JNK (Thr183/Tyr185) (Cell Signaling Technology), anti-phpspho-p38 (Thr180/Tyr182) (D3F4) (Cell Signaling Technology), anti-p38 (N-20) (Santa Cruz Biotechnology), anti-p65 NF-κB (C20) (Santa Cruz Biotechnology), anti-phospho-p65 (Ser276) (Cell Signaling Technology), anti-β-actin (AC15) (Sigma), anti-lamin B (Ab-1) (Millipore), anti-lactate dehydrogenase (AB1222) (Millipore), anti-HA (HA.11) (Covance) and anti-FLAG (Sigma, M2) were used. The reagents used were methyl-β-cyclodextrin (Sigma), lipopolysaccharide (LPS) (Sigma, from Salmonella minnesota) and Oil-red-O (Sigma). DsRed-tagged TAK1 expression vector and other TAK1 expression plasmids were described previously 25. FALG-tagged SREBP expression vectors were from Addgene.64 To generate GST-tagged SREBP1a expression plasmid, FLAG-SREBP1a was subcloned into a pGEX vector (GE Healthcare). SRE-driven luciferase reporter (SRE-luciferase) and control EF1α promoter-driven renilla luciferase (EF1a-renilla luciferase) constructs were used for the transcription reporter assay.65, 66 A retrovirus vector expressing shRNA targeted against TAK1 (the same targeted sequence as TAK1 siRNA#1 described below) was described previously.67 TAK1 and non-targeting control siRNAs were obtained from Sigma (TAK1 siRNA#1, 5'-GAGAUCGACUACAAGGAGA-3'; TAK1 siRNA#2, 5'-GGCAAAGCAACAGAGUGAAUCUGGA-3'; TAK1 siRNA#3, 5’-GAGUGCUGACAUGUCUGAAAUAGAA-3' and Non-targeting siRNA, 5’-UUCUCCGAACGUGUCACGU-3').
Immunoblotting
Whole cell extracts were prepared using an extraction buffer containing 20 mM HEPES (pH 7.4), 150 mM NaCl, 12.5 mM β-glycerophosphate, 1.5 mM MgCl2, 2 mM EGTA, 10mM NaF, 2 mM DTT, 1 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride, 20 μM aprotinin, and 0.5% Triton X-100. Cell extracts were resolved on SDS-PAGE and transferred to Hypond-P membranes (GE Healthcare). The membranes were immunoblotted with various antibodies, and the bound antibodies were visualized with horseradish peroxidase-conjugated antibodies against rabbit or mouse IgG using the ECL Western blotting system (GE Healthcare).
Immunoprecipitation
HEK293 cells were transfected with plasmids using the standard calcium phosphate method. Cell lysates were prepared in extraction buffer described above. DsRed-tagged TAK1, FLAG-tagged SREBP were immunoprecipitated with anti-TAK1 or anti-FLAG antibodies for overnight. The resulting immune complexes were washed with extraction buffer three times. Samples were resolved on SDS-PAGE and exposed to autoradiographic films.
Subcellular fractionation
Nuclear Extract Kit (Active Motif) was used for cytoplasmic and nuclear fractionation. Briefly, cells were lysed in an ice-cold hypotonic buffer, and the lysates were centrifuged and separated into the supernatant (the cytoplasmic fraction including organelles) and the pellets. Nuclear proteins were extracted form the nuclei-containing pellet.
In vitro kinase assay
HEK293 cells were transfected with expression vectors for HA-tagged catalytic domain of TAK1 (TAK1ΔC) together with TAB1 using the standard calcium phosphate method. Cell lysates were prepared in the extraction buffer described above. HA-TAK1ΔC-TAB1 complex was immunoprecipitated with anti-HA antibody for 2 h at 4°C. TAK1 was pre-activated by incubation in a kinase buffer (10 mM Hepes, pH 7.4, 1 mM DTT, and 5 mM MgCl2) with 1 mM ATP for 30 min at 30°C and washed once with the kinase butter. The TAK1ΔC protein and bacterially purified GST-tagged mature form of SREBP1a were mixed and incubated in the kinase buffer supplemented with 200 μM ATP and 5 μCi γ-[32P]ATP for 30 min at 30°C. Samples were resolved on SDS-PAGE. Proteins were visualized by coomassie brilliant blue staining and the gel was exposed to autoradiographic films.
Quantitative real time PCR Analysis
Total RNA was isolated from the liver using an RNeasy kit (Qiagen) and transcribed into cDNA using MultiScribe™ reverse transcriptase (Life Technologies). Expression levels of Tak1, Srebp and SREBP target genes were determined by quantitative real time PCR (qPCR) and normalized to the level of Gapdh. The following primers were used:
Srebp1a-forward, 5'-GGCCGAGATGTGCGAACT-3'; Srebp1a-reverse, 5’-TTGTTGATGAGCTGGAGCATGT-3’;
Srebp1c-forward, 5'-GGAGCCATGGATTGCACATT-3'; Srebp1c-reverse, 5’-GGCCCGGGAAGTCACTGT-3’;
Srebf2-forward, 5'-GGATCCTCCCAAAGAAGGAG-3'; Srebf2-reverse, 5’-TTCCTCAGAACGCCAGACTT-3’;
Scd1-forward, 5'-CTGACCTGAAAGCCGAGAAG-3'; Scd1-reverse, 5’-GCGTTGAGCACCAGAGTGTA-3’;
Fasn-forward, 5'-AAGGCTGGGCTCTATGGATT-3'; Fasn-reverse, 5’-GGAGTGAGGCTGGGTTGATA-3’;
vk1-forward, 5'-GGGACGATGTCTTCCTTGAA-3'; Mvk1-reverse, 5’-GAACTTGGTCAGCCTGCTTC-3’;
G6pd-forward, 5'-CCTACCATCTGGTGGCTGTT-3'; G6pd-reverse, 5’-TGGCTTTAAAGAAGGGCTCA-3’;
Tak1 exon 2-forward 5’-AGGTTGTCGGAAGAGGAGCT-3’; Tak1 exon 2-reverse 5’-CTCCACAATGAAAGCCTTCC-3’; and
Gapdh-forward, 5'-GAAGGTCGCTGTGAACGGA-3'; Gapdh-reverse, 5'-GTTAGTGGGGTCTCGCTCCT-3'.
Measurement of plasma Triglyceride and Cholesterol level
Peripheral blood was collected from the facial vein into 1.5 ml tube containing 1 μl of 0.5 M EDTA and centrifuged at 1000g for 10 min. Supernatant was kept as plasma. Plasma triglyceride and cholesterol amount was determined by triglyceride measurement kit (Wako) and cholesterol measurement kit (Life Technologies), respectively. The assay was conducted according to the manufactured protocols.
Oil-Red-O staining
Oil-Red-O staining was conducted on frozen section. Briefly, freshly cut frozen sections were dried using hair-dryer for 30 min. Sections were then fixed with 4% paraformaldehyde followed by wash with H2O. After the rinse with 60% isopropanol, sections were incubated in Oil-Red-O solution (5 mg/ml in 60% isopropanol) for 15 min. Samples were washed with 60% isopropanol and were counter stained with Hematoxylin.
Electrophoresis mobility shift assay (EMSA)
Oligonucleotides for NF-κB (5′-AGTTGAGGGGACTTTCCCAGG-3′) was purchased from Promega. Oligonucleotides for sterol regulatory element (SRE), wild type (5’-TTTGAAAATCACCCCACTGCAAACTCC-3’) and mutant (5’-TTTGAAAGTCAAACCGTTGCAAACTCC-3’) sterol regulatory element (SRE) were synthesized.68, 69 The binding reaction contained 32P radiolabeled oligonucleotide probe, 10 μg of cell extracts, 4% glycerol, 1mM MgCl2, 0.5mM EDTA, 0.5 mM DTT, 50 mM NaCl, 10 mM Tris-HCl (pH 7.5), 500 ng of poly (dI-dC) (GE Healthcare), and 10 μg of bovine serum albumin to a final concentration of 15 μl. The reaction mixture were incubated at 25oC for 30 min, separated by 5% (w/v) polyacrylamide gel, and visualized by autoradiography. The NF-κB band was confirmed by super-shift using anti-p65 NF-κB. Since antibodies against SREBPs are not suitable for super-shift of mouse SREBP, the SREBP band was confirmed by a completion with wild type and mutant SRE.
Immunohistochemistry
Immunohistochemistry was performed on formalin-fixed liver sections using anti-SREBP2 (Abcom). The bound antibodies were visualized with biotin-conjugated secondary antibody to rabbit IgG using the ABC/DAB reagents according to the manufacturer's protocol (Vector Laboratories). Sections incubated with non-immunized IgG are served as controls. Images were acquired on an upright microscope (BX41; Olympus). Scale bars, 20 μm.
Statistics
Results are shown as the mean ± standard error or standard deviation as indicated. No statistical methods were used to predetermine sample size. Statistical significance was determined using a two-tailed unpaired Student’s t test or one-way ANOVA (Tukey's post hoc test).
Supplementary Material
Tak1LKO Tnfr1−/− liver still develops tumors.
(A) The number of visually detectable tumors was counted at 5–8 month of age in wild type (WT) (Tak1flox/flox no-Cre), Tak1LKO (LKO) including both Tnfr1+/+ and Tnfr1+/− and Tak1LKO Tnfr1−/− livers. The livers having many tumors approximately more than 30 tumors are indicated at the top of graph. All data points are shown. (B) Livers were isolated from wild type, Tak1LKO, Tnfr1−/− or Tak1LKO Tnfr1−/− mice at 4–5 months of age, and RNA was analyzed by quantitative real time PCR. All points and means ± SEM of H19 mRNA levels relative to Gapdh are shown. No statistically significant differences among the groups are observed (one-way ANOVA).
Lipid deposition in Tak1LKO liver.
Oil-red O staining of 6-month old Tak1LKO and control (Tak1flox/flox no-Cre) littermate livers. Scale bars, 40 μm.
Blood triglyceride and cholesterol levels in 5–6 month old Tak1LKO and control including Tak1flox/flox no-Cre and Tak1flox/flox no-Cre Tnfr1+/− mice. The triglyceride level in 5–6 month old Tak1LKO mice was lower than 5–6 weeks old Tak1LKO mice (see Fig. 1B), but it was significantly higher than control. All data points are shown in the box and whisker plots: median and distribution of 50% of values are shown in the box: whiskers indicate distribution of minimum and maximum values. Asterisks indicate means. *, p < 0.05 (two-tailed unpaired Student’s t test).
AMPK, JNK, p38 and NF-κB are not reduced by Tak1 deficiency in the liver.
Liver protein extracts were prepared from 3–4 months old control (Tak1flox/flox no-Cre Tnfr1−/−) and Tak1LKO Tnfr1−/− mice and analyzed by immunoblotting and electron mobility shift assay (EMSA). For NF-κB EMSA, RAW 264.7 cells treated with 50 ng/ml of TNFα for 30 min was used as a positive control. NF-κB band was determined by a super-shift assay using anti-p65 NF-κB. Anti-p65 binds to DNA-NF-κB complexes, which were migrated slower (outside of the visualized area), and the intensity of DNA-NF-κB complexes was reduced (arrows). Positions of molecular weight markers are shown in the immunoblotting. Asterisk in EMSA indicates a non-specific band.
Interaction between TAK1 and SREBPs in HepG2 cells. Whole cell protein extracts of HepG2 cells were immunoprecipitated by anti-TAK1 or non-immunized control IgG. Immunoprecipitates were analyzed by immunoblotting.
TAK1 does not inhibit nuclear localization of SREBP.
(A) HEK293 cells were transfected with expression vectors for FLAG-SREBP1a (mature form), 0.05 μg for co-expression of FLAG-SREBP1a and HA-TAK1 or 0.5 μg for FLAG-SREBP1a alone. Cells were fractionated into the nuclear and cytoplasmic fractions, and the localization of SREBP was analyzed by immunoblotting. Lactate dehydrogenase (LDH) and lamin B were used as markers for cytoplasmic and nuclear fractions, respectively. C, cytoplasmic fraction; N, nuclear fraction. (B) Frozen sections of livers were prepared from control (Tak1flox/flox no-Cre) and Tak1LKO mice at 4–5 months of age, and immunohistochemistry was performed with anti-SREBP2 or control IgG. The sections were counter-stained with hematoxylin. Scale bars, 20 μm.
Livers were isolated from 5–6 month old control (Tak1flox/flox no-Cre) and Tak1LKO mice, and the mRNA levels of SREBPs were analyzed. Means ± SEM; n = 3; ***, p < 0.001: NS, not significant (two-tailed unpaired Student’s t test).
Acknowledgments
We thank NC State biological animal facility for technical support, Dr. Akira for Tak1-loxed mice, and Simmons, A. for critical reading. This work was supported by National Institutes of Health Grant GM068812 (to J.N-T.). The authors have no conflict of interest with any financial organization regarding the studies described in this manuscript.
Footnotes
AUTHOR CONTRIBUTIONS
SM, KS, EO, YI and JNT performed the experiments and analyzed the data. SM, KM and JNT designed the experiments and wrote the manuscript.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
References
- 1.Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 3.Shimano H, Shimomura I, Hammer RE, Herz J, Goldstein JL, Brown MS, et al. Elevated levels of SREBP-2 and cholesterol synthesis in livers of mice homozygous for a targeted disruption of the SREBP-1 gene. J Clin Invest. 1997;100:2115–2124. doi: 10.1172/JCI119746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang T, Espenshade PJ, Wright ME, Yabe D, Gong Y, Aebersold R, et al. Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell. 2002;110:489–500. doi: 10.1016/s0092-8674(02)00872-3. [DOI] [PubMed] [Google Scholar]
- 5.Goldstein JL, DeBose-Boyd RA, Brown MS. Protein sensors for membrane sterols. Cell. 2006;124:35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 6.Sundqvist A, Ericsson J. Transcription-dependent degradation controls the stability of the SREBP family of transcription factors. Proc Natl Acad Sci U S A. 2003;100:13833–13838. doi: 10.1073/pnas.2335135100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sundqvist A, Bengoechea-Alonso MT, Ye X, Lukiyanchuk V, Jin J, Harper JW, et al. Control of lipid metabolism by phosphorylation-dependent degradation of the SREBP family of transcription factors by SCF(Fbw7) Cell Metab. 2005;1:379–391. doi: 10.1016/j.cmet.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 8.Peterson TR, Sengupta SS, Harris TE, Carmack AE, Kang SA, Balderas E, et al. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell. 2011;146:408–420. doi: 10.1016/j.cell.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laplante M, Sabatini DM. mTORC1 activates SREBP-1c and uncouples lipogenesis from gluconeogenesis. Proc Natl Acad Sci U S A. 2010;107:3281–3282. doi: 10.1073/pnas.1000323107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell. 2010;39:171–183. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Porstmann T, Santos CR, Griffiths B, Cully M, Wu M, Leevers S, et al. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab. 2008;8:224–236. doi: 10.1016/j.cmet.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yecies JL, Zhang HH, Menon S, Liu S, Yecies D, Lipovsky AI, et al. Akt stimulates hepatic SREBP1c and lipogenesis through parallel mTORC1-dependent and independent pathways. Cell Metab. 2011;14:21–32. doi: 10.1016/j.cmet.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ye J, Rawson RB, Komuro R, Chen X, Dave UP, Prywes R, et al. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell. 2000;6:1355–1364. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- 14.Kammoun HL, Chabanon H, Hainault I, Luquet S, Magnan C, Koike T, et al. GRP78 expression inhibits insulin and ER stress-induced SREBP-1c activation and reduces hepatic steatosis in mice. J Clin Invest. 2009;119:1201–1215. doi: 10.1172/JCI37007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schuck S, Prinz WA, Thorn KS, Voss C, Walter P. Membrane expansion alleviates endoplasmic reticulum stress independently of the unfolded protein response. J Cell Biol. 2009;187:525–536. doi: 10.1083/jcb.200907074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ron D, Hampton RY. Membrane biogenesis and the unfolded protein response. J Cell Biol. 2004;167:23–25. doi: 10.1083/jcb.200408117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y, Xu S, Mihaylova MM, Zheng B, Hou X, Jiang B, et al. AMPK phosphorylates and inhibits SREBP activity to attenuate hepatic steatosis and atherosclerosis in diet-induced insulin-resistant mice. Cell Metab. 2011;13:376–388. doi: 10.1016/j.cmet.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mihaly SR, Ninomiya-Tsuji J, Morioka S. TAK1 control of cell death. Cell Death Differ. 2014;21:1667–1676. doi: 10.1038/cdd.2014.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morioka S, Inagaki M, Komatsu Y, Mishina Y, Matsumoto K, Ninomiya-Tsuji J. TAK1 kinase signaling regulates embryonic angiogenesis by modulating endothelial cell survival and migration. Blood. 2012;120:3846–3857. doi: 10.1182/blood-2012-03-416198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Omori E, Matsumoto K, Sanjo H, Sato S, Akira S, Smart RC, et al. TAK1 is a master regulator of epidermal homeostasis involving skin inflammation and apoptosis. J Biol Chem. 2006;281:19610–19617. doi: 10.1074/jbc.M603384200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kajino-Sakamoto R, Inagaki M, Lippert E, Akira S, Robine S, Matsumoto K, et al. Enterocyte-derived TAK1 signaling prevents epithelium apoptosis and the development of ileitis and colitis. J Immunol. 2008;181:1143–1152. doi: 10.4049/jimmunol.181.2.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ikeda Y, Morioka S, Matsumoto K, Ninomiya-Tsuji J. TAK1 binding protein 2 is essential for liver protection from stressors. PLoS ONE. 2014;9:e88037. doi: 10.1371/journal.pone.0088037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inokuchi S, Aoyama T, Miura K, Osterreicher CH, Kodama Y, Miyai K, et al. Disruption of TAK1 in hepatocytes causes hepatic injury, inflammation, fibrosis, and carcinogenesis. Proc Natl Acad Sci U S A. 2010;107:844–849. doi: 10.1073/pnas.0909781107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bettermann K, Vucur M, Haybaeck J, Koppe C, Janssen J, Heymann F, et al. TAK1 suppresses a NEMO-dependent but NF-κB-independent pathway to liver cancer. Cancer Cell. 2010;17:481–496. doi: 10.1016/j.ccr.2010.03.021. [DOI] [PubMed] [Google Scholar]
- 25.Morioka S, Broglie P, Omori E, Ikeda Y, Takaesu G, Matsumoto K, et al. TAK1 kinase switches cell fate from apoptosis to necrosis following TNF stimulation. J Cell Biol. 2014;204:607–623. doi: 10.1083/jcb.201305070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang L, Inokuchi S, Roh YS, Song J, Loomba R, Park EJ, et al. Transforming growth factor-beta signaling in hepatocytes promotes hepatic fibrosis and carcinogenesis in mice with hepatocyte-specific deletion of TAK1. Gastroenterology. 2013;144:1042–1054. e1044. doi: 10.1053/j.gastro.2013.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Das M, Garlick DS, Greiner DL, Davis RJ. The role of JNK in the development of hepatocellular carcinoma. Genes Dev. 2011;25:634–645. doi: 10.1101/gad.1989311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wagner EF, Nebreda AR. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer. 2009;9:537–549. doi: 10.1038/nrc2694. [DOI] [PubMed] [Google Scholar]
- 29.Luedde T, Beraza N, Kotsikoris V, van Loo G, Nenci A, De Vos R, et al. Deletion of NEMO/IKKgamma in liver parenchymal cells causes steatohepatitis and hepatocellular carcinoma. Cancer Cell. 2007;11:119–132. doi: 10.1016/j.ccr.2006.12.016. [DOI] [PubMed] [Google Scholar]
- 30.Maeda S, Kamata H, Luo JL, Leffert H, Karin M. IKKbeta couples hepatocyte death to cytokine-driven compensatory proliferation that promotes chemical hepatocarcinogenesis. Cell. 2005;121:977–990. doi: 10.1016/j.cell.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 31.Karin M. Nuclear factor-kappa B in cancer development and progression. Nature. 2006;441:431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 32.Sohda T, Iwata K, Soejima H, Kamimura S, Shijo H, Yun K. In situ detection of insulin-like growth factor II (IGF2) and H19 gene expression in hepatocellular carcinoma. J Hum Genet. 1998;43:49–53. doi: 10.1007/s100380050036. [DOI] [PubMed] [Google Scholar]
- 33.Matouk IJ, DeGroot N, Mezan S, Ayesh S, Abu-lail R, Hochberg A, et al. The H19 non-coding RNA is essential for human tumor growth. PLoS ONE. 2007;2:e845. doi: 10.1371/journal.pone.0000845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Braeuning A, Jaworski M, Schwarz M, Kohle C. Rex3 (reduced in expression 3) as a new tumor marker in mouse hepatocarcinogenesis. Toxicology. 2006;227:127–135. doi: 10.1016/j.tox.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 35.Inokuchi-Shimizu S, Park EJ, Roh YS, Yang L, Zhang B, Song J, et al. TAK1-mediated autophagy and fatty acid oxidation prevent hepatosteatosis and tumorigenesis. J Clin Invest. 2014;124:3566–3578. doi: 10.1172/JCI74068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cohen JC, Horton JD, Hobbs HH. Human fatty liver disease: old questions and new insights. Science. 2011;332:1519–1523. doi: 10.1126/science.1204265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Browning JD, Horton JD. Molecular mediators of hepatic steatosis and liver injury. J Clin Invest. 2004;114:147–152. doi: 10.1172/JCI22422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sabio G, Cavanagh-Kyros J, Ko HJ, Jung DY, Gray S, Jun JY, et al. Prevention of steatosis by hepatic JNK1. Cell Metab. 2009;10:491–498. doi: 10.1016/j.cmet.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Momcilovic M, Hong SP, Carlson M. Mammalian TAK1 activates Snf1 protein kinase in yeast and phosphorylates AMP-activated protein kinase in vitro. J Biol Chem. 2006;281:25336–25343. doi: 10.1074/jbc.M604399200. [DOI] [PubMed] [Google Scholar]
- 40.Kajino T, Omori E, Ishii S, Matsumoto K, Ninomiya-Tsuji J. TAK1 MAPK kinase kinase mediates transforming growth factor-beta signaling by targeting SnoN oncoprotein for degradation. J Biol Chem. 2007;282:9475–9481. doi: 10.1074/jbc.M700875200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takaesu G, Kishida S, Hiyama A, Yamaguchi K, Shibuya H, Irie K, et al. TAB2, a novel adaptor protein, mediates activation of TAK1 MAPKKK by linking TAK1 to TRAF6 in the IL-1 signal transduction pathway. Mol Cell. 2000;5:649–658. doi: 10.1016/s1097-2765(00)80244-0. [DOI] [PubMed] [Google Scholar]
- 42.Kishimoto K, Matsumoto K, Ninomiya-Tsuji J. TAK1 mitogen-activated protein kinase kinase kinase is activated by autophosphorylation within its activation loop. J Biol Chem. 2000;275:7359–7364. doi: 10.1074/jbc.275.10.7359. [DOI] [PubMed] [Google Scholar]
- 43.Scholz R, Sidler CL, Thali RF, Winssinger N, Cheung PC, Neumann D. Autoactivation of transforming growth factor beta-activated kinase 1 is a sequential bimolecular process. J Biol Chem. 2010;285:25753–25766. doi: 10.1074/jbc.M109.093468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kamisuki S, Mao Q, Abu-Elheiga L, Gu Z, Kugimiya A, Kwon Y, et al. A small molecule that blocks fat synthesis by inhibiting the activation of SREBP. Chem Biol. 2009;16:882–892. doi: 10.1016/j.chembiol.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 45.Glass CK, Olefsky JM. Inflammation and lipid signaling in the etiology of insulin resistance. Cell Metab. 2012;15:635–645. doi: 10.1016/j.cmet.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang HH, Halbleib M, Ahmad F, Manganiello VC, Greenberg AS. Tumor necrosis factor-alpha stimulates lipolysis in differentiated human adipocytes through activation of extracellular signal-related kinase and elevation of intracellular cAMP. Diabetes. 2002;51:2929–2935. doi: 10.2337/diabetes.51.10.2929. [DOI] [PubMed] [Google Scholar]
- 47.Fon Tacer K, Kuzman D, Seliskar M, Pompon D, Rozman D. TNF-α interferes with lipid homeostasis and activates acute and proatherogenic processes. Physiol Genomics. 2007;31:216–227. doi: 10.1152/physiolgenomics.00264.2006. [DOI] [PubMed] [Google Scholar]
- 48.Oliner JD, Andresen JM, Hansen SK, Zhou SL, Tjian R. SREBP transcriptional activity is mediated through an interaction with the CREB-binding protein. Genes Dev. 1996;10:2903–2911. doi: 10.1101/gad.10.22.2903. [DOI] [PubMed] [Google Scholar]
- 49.Osborne TF. Sterol regulatory element-binding proteins (SREBPs): key regulators of nutritional homeostasis and insulin action. J Biol Chem. 2000;275:32379–32382. doi: 10.1074/jbc.R000017200. [DOI] [PubMed] [Google Scholar]
- 50.Junttila MR, Li SP, Westermarck J. Phosphatase-mediated crosstalk between MAPK signaling pathways in the regulation of cell survival. FASEB J. 2008;22:954–965. doi: 10.1096/fj.06-7859rev. [DOI] [PubMed] [Google Scholar]
- 51.Kim SI, Kwak JH, Wang L, Choi ME. Protein phosphatase 2A is a negative regulator of transforming growth factor-β1-induced TAK1 activation in mesangial cells. J Biol Chem. 2008;283:10753–10763. doi: 10.1074/jbc.M801263200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kajino T, Ren H, Iemura S, Natsume T, Stefansson B, Brautigan DL, et al. Protein phosphatase 6 down-regulates TAK1 kinase activation in the IL-1 signaling pathway. J Biol Chem. 2006;281:39891–39896. doi: 10.1074/jbc.M608155200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shimomura I, Hammer RE, Richardson JA, Ikemoto S, Bashmakov Y, Goldstein JL, et al. Insulin resistance and diabetes mellitus in transgenic mice expressing nuclear SREBP-1c in adipose tissue: model for congenital generalized lipodystrophy. Genes Dev. 1998;12:3182–3194. doi: 10.1101/gad.12.20.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nakayama H, Otabe S, Ueno T, Hirota N, Yuan X, Fukutani T, et al. Transgenic mice expressing nuclear sterol regulatory element-binding protein 1c in adipose tissue exhibit liver histology similar to nonalcoholic steatohepatitis. Metabolism. 2007;56:470–475. doi: 10.1016/j.metabol.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 55.Takahashi Y, Soejima Y, Fukusato T. Animal models of nonalcoholic fatty liver disease/nonalcoholic steatohepatitis. World J Gastroenterol. 2012;18:2300–2308. doi: 10.3748/wjg.v18.i19.2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Campbell JS, Hughes SD, Gilbertson DG, Palmer TE, Holdren MS, Haran AC, et al. Platelet-derived growth factor C induces liver fibrosis, steatosis, and hepatocellular carcinoma. Proc Natl Acad Sci U S A. 2005;102:3389–3394. doi: 10.1073/pnas.0409722102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heindryckx F, Colle I, Van Vlierberghe H. Experimental mouse models for hepatocellular carcinoma research. Int J Exp Pathol. 2009;90:367–386. doi: 10.1111/j.1365-2613.2009.00656.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Farazi PA, DePinho RA. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat Rev Cancer. 2006;6:674–687. doi: 10.1038/nrc1934. [DOI] [PubMed] [Google Scholar]
- 59.Zhang L, Yang F, Yuan JH, Yuan SX, Zhou WP, Huo XS, et al. Epigenetic activation of the MiR-200 family contributes to H19-mediated metastasis suppression in hepatocellular carcinoma. Carcinogenesis. 2013;34:577–586. doi: 10.1093/carcin/bgs381. [DOI] [PubMed] [Google Scholar]
- 60.Sato S, Sanjo H, Takeda K, Ninomiya-Tsuji J, Yamamoto M, Kawai T, et al. Essential function for the kinase TAK1 in innate and adaptive immune responses. Nat Immunol. 2005;6:1087–1095. doi: 10.1038/ni1255. [DOI] [PubMed] [Google Scholar]
- 61.Postic C, Shiota M, Niswender KD, Jetton TL, Chen Y, Moates JM, et al. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic β cell-specific gene knock-outs using Cre recombinase. J Biol Chem. 1999;274:305–315. doi: 10.1074/jbc.274.1.305. [DOI] [PubMed] [Google Scholar]
- 62.Pfeffer K, Matsuyama T, Kundig TM, Wakeham A, Kishihara K, Shahinian A, et al. Mice deficient for the 55 kd tumor necrosis factor receptor are resistant to endotoxic shock, yet succumb to L. monocytogenes infection. Cell. 1993;73:457–467. doi: 10.1016/0092-8674(93)90134-c. [DOI] [PubMed] [Google Scholar]
- 63.Ninomiya-Tsuji J, Kishimoto K, Hiyama A, Inoue J, Cao Z, Matsumoto K. The kinase TAK1 can activate the NIK-IκB as well as the MAP kinase cascade in the IL-1 signalling pathway. Nature. 1999;398:252–256. doi: 10.1038/18465. [DOI] [PubMed] [Google Scholar]
- 64.Toth JI, Datta S, Athanikar JN, Freedman LP, Osborne TF. Selective coactivator interactions in gene activation by SREBP-1a and -1c. Mol Cell Biol. 2004;24:8288–8300. doi: 10.1128/MCB.24.18.8288-8300.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hua X, Nohturfft A, Goldstein JL, Brown MS. Sterol resistance in CHO cells traced to point mutation in SREBP cleavage-activating protein. Cell. 1996;87:415–426. doi: 10.1016/s0092-8674(00)81362-8. [DOI] [PubMed] [Google Scholar]
- 66.Uemura N, Kajino T, Sanjo H, Sato S, Akira S, Matsumoto K, et al. TAK1 is a component of the Epstein-Barr virus LMP1 complex and is essential for activation of JNK but not of NF-kappa B. J Biol Chem. 2006;281:7863–7872. doi: 10.1074/jbc.M509834200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Morioka S, Omori E, Kajino T, Kajino-Sakamoto R, Matsumoto K, Ninomiya-Tsuji J. TAK1 kinase determines TRAIL sensitivity by modulating reactive oxygen species and cIAP. Oncogene. 2009;28:2257–2265. doi: 10.1038/onc.2009.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smith JR, Osborne TF, Goldstein JL, Brown MS. Identification of nucleotides responsible for enhancer activity of sterol regulatory element in low density lipoprotein receptor gene. J Biol Chem. 1990;265:2306–2310. [PubMed] [Google Scholar]
- 69.Ringseis R, Rauer C, Rothe S, Gessner DK, Schutz LM, Luci S, et al. Sterol regulatory element-binding proteins are regulators of the NIS gene in thyroid cells. Mol Endocrinol. 2013;27:781–800. doi: 10.1210/me.2012-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tak1LKO Tnfr1−/− liver still develops tumors.
(A) The number of visually detectable tumors was counted at 5–8 month of age in wild type (WT) (Tak1flox/flox no-Cre), Tak1LKO (LKO) including both Tnfr1+/+ and Tnfr1+/− and Tak1LKO Tnfr1−/− livers. The livers having many tumors approximately more than 30 tumors are indicated at the top of graph. All data points are shown. (B) Livers were isolated from wild type, Tak1LKO, Tnfr1−/− or Tak1LKO Tnfr1−/− mice at 4–5 months of age, and RNA was analyzed by quantitative real time PCR. All points and means ± SEM of H19 mRNA levels relative to Gapdh are shown. No statistically significant differences among the groups are observed (one-way ANOVA).
Lipid deposition in Tak1LKO liver.
Oil-red O staining of 6-month old Tak1LKO and control (Tak1flox/flox no-Cre) littermate livers. Scale bars, 40 μm.
Blood triglyceride and cholesterol levels in 5–6 month old Tak1LKO and control including Tak1flox/flox no-Cre and Tak1flox/flox no-Cre Tnfr1+/− mice. The triglyceride level in 5–6 month old Tak1LKO mice was lower than 5–6 weeks old Tak1LKO mice (see Fig. 1B), but it was significantly higher than control. All data points are shown in the box and whisker plots: median and distribution of 50% of values are shown in the box: whiskers indicate distribution of minimum and maximum values. Asterisks indicate means. *, p < 0.05 (two-tailed unpaired Student’s t test).
AMPK, JNK, p38 and NF-κB are not reduced by Tak1 deficiency in the liver.
Liver protein extracts were prepared from 3–4 months old control (Tak1flox/flox no-Cre Tnfr1−/−) and Tak1LKO Tnfr1−/− mice and analyzed by immunoblotting and electron mobility shift assay (EMSA). For NF-κB EMSA, RAW 264.7 cells treated with 50 ng/ml of TNFα for 30 min was used as a positive control. NF-κB band was determined by a super-shift assay using anti-p65 NF-κB. Anti-p65 binds to DNA-NF-κB complexes, which were migrated slower (outside of the visualized area), and the intensity of DNA-NF-κB complexes was reduced (arrows). Positions of molecular weight markers are shown in the immunoblotting. Asterisk in EMSA indicates a non-specific band.
Interaction between TAK1 and SREBPs in HepG2 cells. Whole cell protein extracts of HepG2 cells were immunoprecipitated by anti-TAK1 or non-immunized control IgG. Immunoprecipitates were analyzed by immunoblotting.
TAK1 does not inhibit nuclear localization of SREBP.
(A) HEK293 cells were transfected with expression vectors for FLAG-SREBP1a (mature form), 0.05 μg for co-expression of FLAG-SREBP1a and HA-TAK1 or 0.5 μg for FLAG-SREBP1a alone. Cells were fractionated into the nuclear and cytoplasmic fractions, and the localization of SREBP was analyzed by immunoblotting. Lactate dehydrogenase (LDH) and lamin B were used as markers for cytoplasmic and nuclear fractions, respectively. C, cytoplasmic fraction; N, nuclear fraction. (B) Frozen sections of livers were prepared from control (Tak1flox/flox no-Cre) and Tak1LKO mice at 4–5 months of age, and immunohistochemistry was performed with anti-SREBP2 or control IgG. The sections were counter-stained with hematoxylin. Scale bars, 20 μm.
Livers were isolated from 5–6 month old control (Tak1flox/flox no-Cre) and Tak1LKO mice, and the mRNA levels of SREBPs were analyzed. Means ± SEM; n = 3; ***, p < 0.001: NS, not significant (two-tailed unpaired Student’s t test).