Abstract
Mixed phenotype acute leukemia (MPAL) is a heterogeneous group of poor-prognosis leukemias with immunophenotypic features of at least two cell lineages. The full spectrum of genetic mutations in this rare disease has not been elucidated, limiting our understanding of disease pathogenesis and our ability to devise targeted therapeutic strategies. We sought to define the mutational landscape of MPAL by performing whole exome sequencing on samples from 23 adult and pediatric MPAL patients. We identified frequent mutations of epigenetic modifiers, most notably mutations of DNMT3A in 33% of adult MPAL patients. Mutations of activated signaling pathways, tumor suppressors and transcription factors were also frequent. Importantly, many of the identified mutations are potentially therapeutically targetable with agents currently available or in various stages of clinical development. Therefore, the mutational spectrum we identified provides potential biological insights and is likely to have clinical relevance for patients with this poor-prognosis disease.
Introduction
Mixed phenotype acute leukemia (MPAL) is a relatively rare and difficult to treat neoplasm with immunophenotypic features of at least two cell lineages (typically myeloid with B– or T–lymphoid). This duality led to our interest in examining patient samples using whole exome sequencing (WES) for mutations associated with those arising in stem cells, as well as for myeloid and lymphoid regulators.
MPAL accounts for approximately 2–5% of acute leukemias1. The World Health Organization (WHO) recently redefined biphenotypic and bilineal leukemias under MPAL, requiring specific lineage-defining immunophenotypic criteria to standardize the diagnosis2–4. MPAL can exhibit either two distinct affected cell populations of different lineages, one clonal cell population with characteristics of two lineages, or a combination of both. Markers that are pathognomonic to each of two lineages are required for diagnosis. A previous study of MPAL assessed mutations in 17 genes of 31 patients5. Here, we examined 23 MPAL patients aged 2–74 years with WES, capturing a broader array of mutations.
Methods
To perform WES, genomic DNA was extracted from frozen blood or bone marrow cell pellets (12 T/myeloid, 10 B/myeloid, and 1 B/T-lymphoid). The median age was 35 years, including 5 pediatric patients <18 years old). Under IRB approval, pathology and flow cytometry records were reviewed by a hematopathologist to ensure samples met WHO 2008 MPAL criteria. Nine samples were from relapse. Four of the samples were cytogenetically normal, 5 had monosomy 5 and/or 7, two had an MLL rearrangement, one was positive for the Philadelphia chromosome, and 11 had other cytogenetic abnormalities (Supplemental Table 1).
WES libraries were constructed and annealed in-solution to the HGSC VCRome2.1 design (42Mb, NimbleGen) as described6. Paired-end sequencing (2 × 101 base pairs) using the Illumina HiSeq 2000 or 2500 platforms was performed. Samples achieved >96% of the targeted exome bases covered to a depth of ≥20× or (average 120×). Data processing, mutation calling and annotation was performed as described6. Rigorous stepwise filtering was applied to identify likely somatic and pathogenic variants (Suppl. Fig. 1s). Briefly, only nonsynonymous variants (nonsense, missense, frameshift, at splice site) with ≥4 supporting sequencing reads and a variant allele fraction of ≥0.05 were selected. Potential germline SNPs were eliminated by comparison to common SNPs (with a minor allele frequency ≥0.005) in dbSNP build 139, 1000 Genome phase III dataset, the NHLBI GO Exome Sequencing Project (ESP6500), and the Exome Aggregation Consortium (ExAC) datasets. In the absence of germline DNA samples, we primarily focused on a set of 562 genes included in the cancer gene census (http://cancer.sanger.ac.uk/census/) and previously reported in leukemias. The COSMIC database (http://cancer.sanger.ac.uk/cosmic) was used to highlight known cancer mutations. Six commonly used mutation prediction algorithms (SIFT, Polyphen2, LRT, LR, Mutation Assessor, Mutation Taster) were used to predict each mutation’s functional importance. Only known cancer hotspot mutations and likely pathogenic mutations predicted to be “damaging” by at least 3 algorithms were selected. The selected mutations were further prioritized based on the mutation type, variant allele fraction, COSMIC site frequency, and gene function. Finally, 54 mutations were selected (Supplemental Table 2).
Results/Discussion
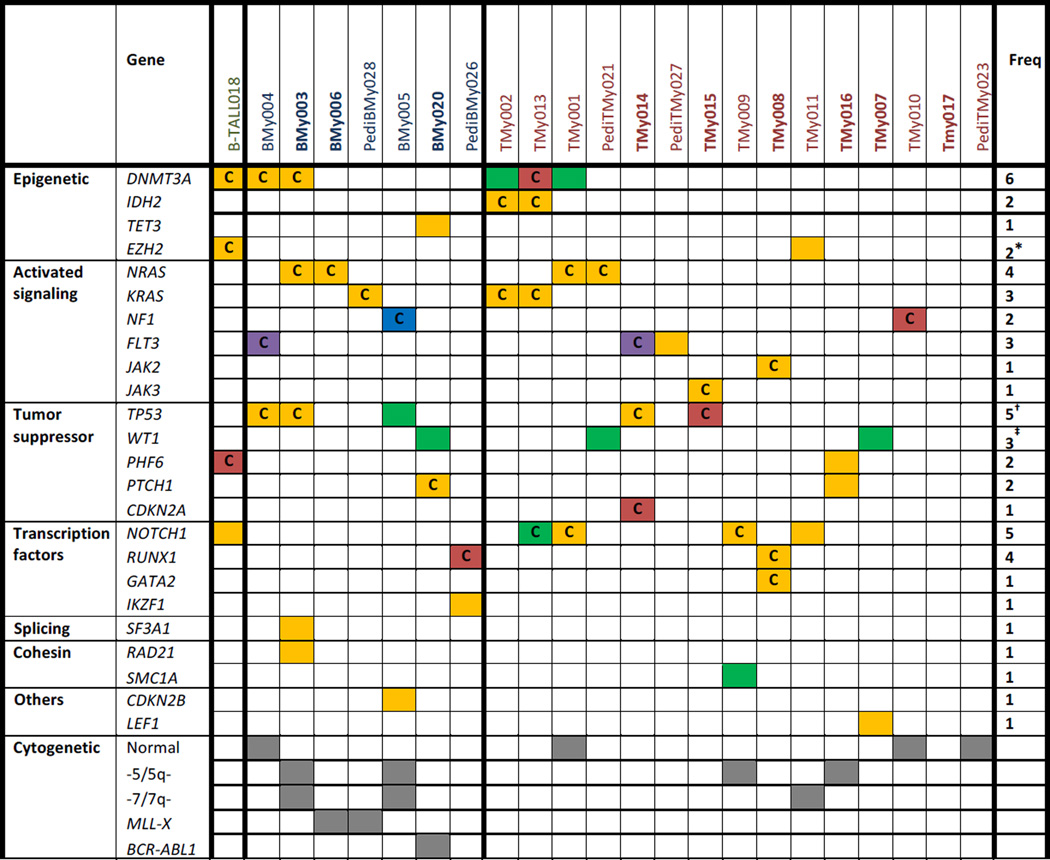
A previous study evaluated 31 patients with MPAL for mutations in 17 genes recurrently mutated in acute leukemias identified genomic lesions in only 12 (39%) of the patients, including alterations of IKZF1, EZH2, TET2, ASXL1, and NOTCH1. They identified no mutations in the other probed genes including DNMT3A, FLT3, NPM1, RUNX1 and WT15. The authors concluded that the mutational spectrum of MPAL may differ from other acute leukemias. To more fully evaluate the mutational landscape of MPAL, we performed whole exome sequencing in 23 patients with this rare disease, identifying mutations in 21 patients (91%) (Table 1).
Table 1.
Summary of mutations from whole exome sequencing of MPAL samples.
Eight of the 23 samples (35%) had mutations in epigenetic regulatory genes (Table 1). The de novo DNA methyltransferase, DNMT3A, was the most frequently mutated gene with 6 patients (26%) harboring mutations. All the DNMT3A mutations involved the methyltransferase domain, 3 of which were missense mutations at Arg882, the hotspot common in acute myeloid leukemia (AML). DNMT3A mutations occurred in all immunophenotypic subtypes examined, and similar to reports in AML, MPAL patients with a mutation in DNMT3A trended towards being older and more likely to have normal cytogenetics when compared to patients without a DNMT3A mutation (Suppl. Table 3)7,8. Other mutations in epigenetic regulatory genes were found in IDH2 (9%), TET3 (4%), and EZH2 (9%).
We also identified mutually exclusive mutations of activated signaling genes in 61% of patients. Mutations of the RAS pathway were particularly common, including mutations of NRAS (17%), KRAS (13%) and NF1 (9%). Three patients had FLT3 mutations (one missense mutation and two internal tandem duplication ITD mutation), and 2 had JAK pathway mutations including a JAK2 V617F mutation in a patient with a history of pre-existing essential thrombocythemia (ET).
Tumor suppressors were also frequently mutated, most notably five patients harbored mutations of TP53 (22%). Three samples (13%) also had four exon 1 or 7 frameshift mutations of WT1, consistent with a small prior study showing WT1 mutations in up to 20% of biphenotypic and undifferentiated acute leukemia9. We also identified recurrent mutations in a number of transcription factors. NOTCH1 mutations were present in 5 of the 16 patients (32%) with T lymphoblastic subtypes (T/myeloid and B/T ALL) and none of the patients with B/myeloid disease. Mutations of RUNX1 and GATA2 were found in 2 and 1 patient, respectively. One of the RUNX1 mutations occurred in a patient with secondary MPAL arising from pre-existing ET, consistent with the reported increased risk of progression to acute leukemia in JAK2 V617F mutant MPN patients with co-occurrence of a RUNX1 mutation10,11. The other patient with a RUNX1 mutation was a pediatric patient who developed therapy-related MDS/MPAL after treatment for neuroblastoma. RUNX1 mutations are common in patients with therapy-related MDS12, and mutations of RUNX1 in MDS are associated with an increased risk of progression to acute leukemia13. Therefore it is likely that in these two patients, the acquisition of a RUNX1 mutation was a critical driver of their disease.
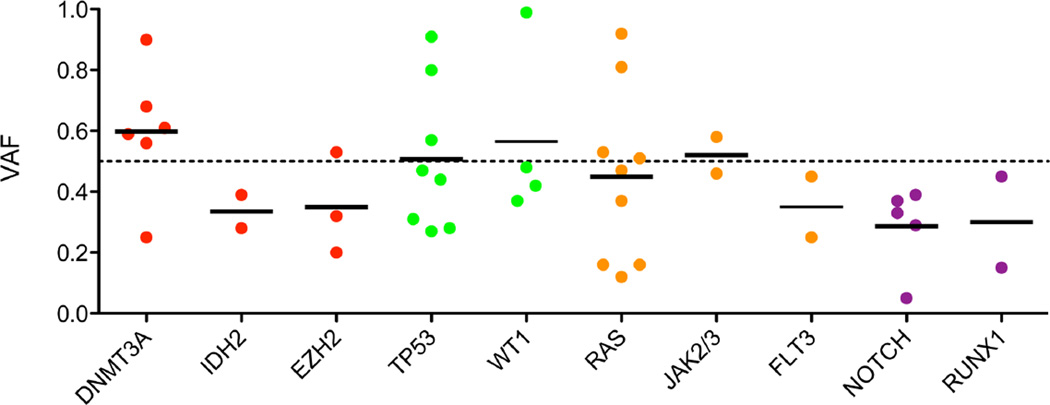
To assess possible clonal origin of mutations, we calculated the variant allele frequency of individual mutations (Fig. 1). Mutations of DNMT3A and tumor suppressors consistently showed high VAFs suggesting these mutations arise early in disease development. Mutations of lineage specific transcription factors tended to occur at lower VAFs indicting they may arise in a smaller sub-clonal population. The VAFs of mutations involving activated signaling pathways were more widely distributed indicating these mutations can arise at varying stages of disease development. Of the DNMT3A-mutant patients, 4/6 had a concomitant NRAS/KRAS mutation and 3/6 had a NOTCH1 mutation, with 2/6 having both a NRAS/KRAS and a NOTCH1 mutation. Evaluation of the VAF of these patients suggests that the DNMT3A mutation was present in the founding pre-leukemic clone, which then acquired a driver mutation of the RAS-pathway, with a T lineage-specific NOTCH1 mutation subsequently arising in a subordinate clone. For example, the VAFs of the DNMT3A, KRAS, and NOTCH1 mutations of patient TMy013 were 0.56. 0.37, and 0.29, respectively (Suppl. Table S2).
Figure 1. Variant allele frequency (VAF) of selected mutations in MPAL.
Quantitative measure of variant containing reads estimates the abundance of these mutations divided into epigenetic modifiers (red), tumor suppressors (green), tyrosine-kinase signaling genes (orange), and transcription factors (purple).
MPAL is defined by the co-expression of lymphoid and myeloid surface markers, suggesting a leukemia arising from developmental arrest of a precursor cell capable of differentiating into both lineages, such as a stem or progenitor cell. Consistent with this view is the finding of frequent mutation in genes encoding epigenetic modifiers, particularly DNMT3A (33% of the adult patients), which are also found in hematologic malignancies of both lymphoid and myeloid origin as well as age-related clonal hematopoiesis8,14–19. Supporting the stem cell association of mutations in these genes is data from animal models, in which loss of these genes leads to increased hematopoietic stem cell self-renewal and increased risk of hematopoietic disorders of either lineage20–22.
We also observed mutations in many genes previously associated primarily with one or the other branch of hematopoietic differentiation. For example, RUNX1 and GATA2 mutations are typically associated with myeloid disease. Similarly, we see lymphoid disease-associated mutations such as NOTCH1. In addition, we note the conspicuous absence of mutations in NPM1, which have not yet been found in any lymphoid malignancy, and the absence of PAX5 and IL7R mutations, which are almost exclusively associated with lymphoid disease, although the sample size is too small for definitive conclusions. In addition, the sample number prevents statistical analysis of differences between T- and B-ALL-type MPAL, but we observe NOTCH1 mutations associated exclusively with the T/myeloid, as might be expected. Thus, this constellation of mutations is interesting in light of the mixed phenotype of this malignancy, and may reflect mutations acquired during specific stages of differentiation, and/or permissive for development of specific lineages.
In summary, we identified a spectrum of genetic abnormalities with potential biological and clinical implications in a cohort of MPAL patients. Strikingly, one-third of adult patients harbored DNMT3A mutations, supporting a likely stem cell origin of this disease. Importantly, a number of the mutations we identified are potentially targetable by agents that are currently available or are being testing in clinical trials including epigenetically targeted agents, tyrosine kinase pathway inhibitors, and NOTCH1 inhibitors. Therefore, our data support the use of genomic sequencing in patients with this poor-prognosis disease toward a goal of precision medicine.
Supplementary Material
Acknowledgments
This work was supported by DK092883, CA183252, CA126752, CA125123, the Edward P Evans Foundation, The Samuel Waxman Cancer Research Foundation, Henry Malvin Helis Foundation, CPRIT, and the Lester and Sue Smith Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
OE conceived the study and drafted the manuscript. OE and LW generated data, JNP reviewed pathology analyses on cases, OE, LW, RER, DW, and MAG analyzed and discussed the data, SMK and MA provided samples, all authors commented commented on the manuscript; RER and MAG supervised the study and edited the manuscript.
Conflicts of Interest
The authors have no conflicts of interest to declare.
References
- 1.Weinberg OK, Arber DA. Mixed-phenotype acute leukemia: historical overview and a new definition. Leukemia. 2010;24:1844–1851. doi: 10.1038/leu.2010.202. [DOI] [PubMed] [Google Scholar]
- 2.Borowitz MJ. Mixed phenotype acute leukemia. Cytometry B Clin Cytom. 2014;86:152–153. doi: 10.1002/cyto.b.21155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Swerdlow S, Campo E, Harris N. Acute Leukemias of Ambiguous Lineage. 2008 [Google Scholar]
- 4.van den Ancker W, Westers TM, de Leeuw DC, et al. A threshold of 10% for myeloperoxidase by flow cytometry is valid to classify acute leukemia of ambiguous and myeloid origin. Cytometry B Clin Cytom. 2013;84:114–118. doi: 10.1002/cyto.b.21072. [DOI] [PubMed] [Google Scholar]
- 5.Yan L, Ping N, Zhu M, et al. Clinical, immunophenotypic, cytogenetic, and molecular genetic features in 117 adult patients with mixed-phenotype acute leukemia defined by WHO-2008 classification. Haematologica. 2012;97:1708–1712. doi: 10.3324/haematol.2012.064485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang L, Yamaguchi S, Burstein MD, et al. Novel somatic and germline mutations in intracranial germ cell tumours. Nature. 2014;511:241–245. doi: 10.1038/nature13296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thol F, Damm F, Ludeking A, et al. Incidence and prognostic influence of DNMT3A mutations in acute myeloid leukemia. J Clin Oncol. 2011;29:2889–2896. doi: 10.1200/JCO.2011.35.4894. [DOI] [PubMed] [Google Scholar]
- 8.Ley TJ, Ding L, Walter MJ, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.King-Underwood L, Pritchard-Jones K. Wilms' tumor (WT1) gene mutations occur mainly in acute myeloid leukemia and may confer drug resistance. Blood. 1998;91:2961–2968. [PubMed] [Google Scholar]
- 10.Beer PA, Delhommeau F, LeCouedic JP, et al. Two routes to leukemic transformation after a JAK2 mutation-positive myeloproliferative neoplasm. Blood. 2010;115:2891–2900. doi: 10.1182/blood-2009-08-236596. [DOI] [PubMed] [Google Scholar]
- 11.Ding Y, Harada Y, Imagawa J, Kimura A, Harada H. AML1/RUNX1 point mutation possibly promotes leukemic transformation in myeloproliferative neoplasms. Blood. 2009;114:5201–5205. doi: 10.1182/blood-2009-06-223982. [DOI] [PubMed] [Google Scholar]
- 12.Harada H, Harada Y, Niimi H, Kyo T, Kimura A, Inaba T. High incidence of somatic mutations in the AML1/RUNX1 gene in myelodysplastic syndrome and low blast percentage myeloid leukemia with myelodysplasia. Blood. 2004;103:2316–2324. doi: 10.1182/blood-2003-09-3074. [DOI] [PubMed] [Google Scholar]
- 13.Tsai SC, Shih LY, Liang ST, et al. Biological Activities of RUNX1 Mutants Predict Secondary Acute Leukemia Transformation from Chronic Myelomonocytic Leukemia and Myelodysplastic Syndromes. Clin Cancer Res. 2015;21:3541–3551. doi: 10.1158/1078-0432.CCR-14-2203. [DOI] [PubMed] [Google Scholar]
- 14.Walter MJ, Ding L, Shen D, et al. Recurrent DNMT3A mutations in patients with myelodysplastic syndromes. Leukemia. 2011;25:1153–1158. doi: 10.1038/leu.2011.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neumann M, Heesch S, Schlee C, et al. Whole-exome sequencing in adult ETP-ALL reveals a high rate of DNMT3A mutations. Blood. 2013;121:4749–4752. doi: 10.1182/blood-2012-11-465138. [DOI] [PubMed] [Google Scholar]
- 16.Couronne L, Bastard C, Bernard OA. TET2 and DNMT3A mutations in human T-cell lymphoma. N Engl J Med. 2012;366:95–96. doi: 10.1056/NEJMc1111708. [DOI] [PubMed] [Google Scholar]
- 17.Jaiswal S, Fontanillas P, Flannick J, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371:2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Genovese G, Kahler AK, Handsaker RE, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371:2477–2487. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie M, Lu C, Wang J, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014;20:1472–1478. doi: 10.1038/nm.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanaka S, Miyagi S, Sashida G, et al. Ezh2 augments leukemogenicity by reinforcing differentiation blockage in acute myeloid leukemia. Blood. 2012;120:1107–1117. doi: 10.1182/blood-2011-11-394932. [DOI] [PubMed] [Google Scholar]
- 21.Challen GA, Sun D, Jeong M, et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat Genet. 2012;44:23–31. doi: 10.1038/ng.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moran-Crusio K, Reavie L, Shih A, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20:11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.