Abstract
The epigenetic determinants driving the responses of CD4 T cells to antigen are currently an area of active research. Much has been done to characterize helper T cell subsets and their associated genome-wide epigenetic patterns. In contrast, little is known about the dynamics of histone modifications during CD4 T cell activation and the differential kinetics of these epigenetic marks between naïve and memory T cells. In this study we have detailed the dynamics of genome-wide promoter H3K4me2 and H3K4me3 over a time course during activation of human naïve and memory CD4 T cells. Our results demonstrate that changes to H3K4 methylation occur relatively late after activation (5 days) and reinforce activation-induced upregulation of gene expression, affecting multiple pathways important to T cell activation, differentiation, and function. The dynamics and mapped pathways of H3K4 methylation are distinctly different in memory cells, which have substantially more promoters marked by H3K4me3 alone, reinforcing their more differentiated state. Our study provides the first data examining genome-wide histone modification dynamics during CD4 T cell activation, providing insight into the cross talk between H3K4 methylation and gene expression, and underscoring the impact of these marks upon key pathways integral to CD4 T cell activation and function.
Keywords: H3K4 methylation, epigenetics, CD4 T cell activation, immune memory, ChIP-Seq
Introduction
CD4 T cells are an essential arm of the adaptive immune system. Produced in the thymus during fetal development, mature cells are antigen-specific and capable of responding to foreign invaders, differentiating into an effector pool after antigenic stimulation. Once an infection clears, these effector cells undergo a contraction phase, where lingering antigen-specific cells with increased receptor affinity for their antigen have developed cellular ‘memory.’ Memory CD4 T cells are capable of rapidly responding to antigen by proliferating and producing cytokines more quickly and in larger amounts than naïve cells.1 However, the molecular processes of differentiation and epigenetic alterations to the genome that occur in a dynamic fashion to determine the nature of CD4 memory during these phases after antigenic stimulation remain poorly understood.
In recent years, epigenetic modifications have received much attention for their contributions to the complex regulation of cellular gene expression.2 DNA methylation, histone modifications, and chromatin regulators all confer different levels of gene regulation and play critical roles in cellular differentiation that are currently ill-defined. Additionally, a variety of regulatory regions, such as promoters, enhancers, super-enhancers, gene bodies, and intergenic regions can be affected in different ways by epigenetic modifications, and often multiple modifications occur for one gene across these different regions of the genomic landscape. In particular, histone modifications have largely been explored with respect to their effect upon transcription, especially when located within promoter and enhancer regions. Two histone lysine modifications, H3K4me3 and H3K4me2, are often scrutinized due to their associations with active, highly transcribed promoters.3 Most of the literature has focused upon embryonic stem (ES) cells and cancer cells with respect to understanding these modifications (reviewed in 4-6 and 7), although several papers have explored the role of epigenetic modifications in neuronal differentiation8 and T cell differentiation.9,10,11
The interplay between H3K4 methylation and gene regulation is complex and involves numerous transcription factors, assembly of complexes to modify histones, and other signal networks to achieve their results. Reports in both yeast and in mammals have demonstrated that promoter H3K4me3 is often deposited in response to an increase in transcriptional activity and expanded due to a feed-forward mechanism.12-15 When located near the transcriptional start site (TSS), this modification also promotes active transcription.14, 16, 17 H3K4me2 is often found alongside H3K4me3 in actively transcribed promoters, but when found alone, particularly in non-CpG containing promoters, this modification is thought to confer a ‘poised’ state to differentiated cells, i.e. not actively transcribed, but prepared for rapid transcriptional activation.3, 18 The role of poising genes in naïve CD4 T cells could be facilitating antigen-driven differentiation of effector subsets (Th1, Th2, Th17), while poising genes in memory CD4 T cells might be a mechanism for enhancing the response upon re-exposure to cognate antigens.
Work in CD4 T cells has been largely confined to quantifying differences in enrichment for these modifications with respect to subset differentiation. Barski and colleagues were the first to profile human CD4 T cells using ChIP-Seq, mapping several histone modifications, variant histones, and RNA polymerase II in resting CD4 T cells.9 Additional work with this data set demonstrated various combinatorial patterns of histone modifications found throughout the genome.19 Wei et al. demonstrated that five different subsets of mouse CD4 T cells expanded in culture exhibited distinct sets of H3K4me3 and H3K27me3 unique to their cellular phenotypes. However, one observation from this work was that a number of genes retained some amount of plasticity, with key transcription factors and cytokines maintaining permissive chromatin marks in non-corresponding lineages.10 Zhang et al. explored several histone modifications alongside transcription during thymic development of CD4 T cells in mice and ultimately found that histone marks are dynamic and reversible throughout T cell development.20 Two papers published in 2009 examined histone modifications after short term activation of CD4 T cells (i.e. 4 h and 18 h), concluding that few significant changes in histone dynamics had occurred at these time points.21, 22 Allan and colleagues demonstrated that SUV39H1, an H3K9 methyltransferase, was key to silencing Th1 genes during Th2 differentiation.23 A recent study in mouse CD8 T cells examined the dynamics of H3K4me3 and H3K27me3 in a mouse model of viral infection.11 They found distinctly different patterns of histone modifications in naïve, effector, and memory cells, which correlated with functions specific to these subsets
Though histone modification profiles of static helper T cell lineages have been mapped, no studies to date have examined their dynamics in CD4 T cell activation over a time course. Here we examine the kinetics of promoter H3K4me2 and H3K4me3 marks and evaluate expression changes, comparing naïve and memory CD4 T cells over a time course 1 day, 5 days, and 2 weeks after activation via T cell receptor crosslinking and CD28-mediated co-stimulation. We also correlate alterations in these modifications to transcriptional changes throughout activation using deep RNA sequencing. Our profiles of these marks delineate epigenetic regulation of gene expression essential for numerous pathways related to T cell activation and immune function. Taken together, these data reveal many new avenues for additional exploration of what establishes a memory CD4 T cell and regulates subset differentiation. As such, we provide a rich data set for other investigators to analyze and advance their own work in this context.
Results
ChIP-Seq Quality Control
To examine the dynamics of histone modifications over time, naïve (CD45RA+CD45RO−) and memory (CD45RA−CD45RO+) CD4 T cells were isolated from the peripheral blood of four healthy human donors and activated with anti-CD3/anti-CD28 beads and cultured in rIL2-supplemented media for 1 day, 5 days, and 2 weeks. Purity of each subset was > 94% for all donors (Supplementary Figure S1). ChIP-Seq for H3K4me2 and H3K4me3 was performed on cells from all conditions and time points using antibodies specific to each histone modification with minimal cross-reaction, alongside RNA-Seq for the same conditions. The raw data for both ChIP-Seq and RNA-Seq from each condition are available at the NIH Gene Expression Omnibus site (accession # GSE73214). Promoter results for each modification demonstrated that peaks were generally found within 1 kb of the TSS (Supplementary Figure S2), so promoter analysis was conducted within this radius.
Importantly, p-value distributions for H3K4me2 and H3K4me3 clearly demonstrated that these modifications vary predominantly by cell type and time point rather than by the individual donors (Figure 1A-B), showing that donor variability did not significantly impact our results. To determine if there was significant variation in signal-to-background ratios across samples, we plotted the distribution of coverage depths across the genome for each sample (Supplementary Figure S3). While different antibodies have different coverage distributions, samples with the same antibody and consistent signal-to-background ratios will follow the same curve. A higher fraction of low-coverage bases in a sample relative to others of the same type is indicative of a lower signal-to-background ratio. The majority of samples have consistent signal-to-background ratios. Principal coordinate analysis24 revealed that the biological effects of interest were visible in the first 4 principal coordinates, demonstrating that technical confounding factors were not an issue (Supplementary Figure S4).
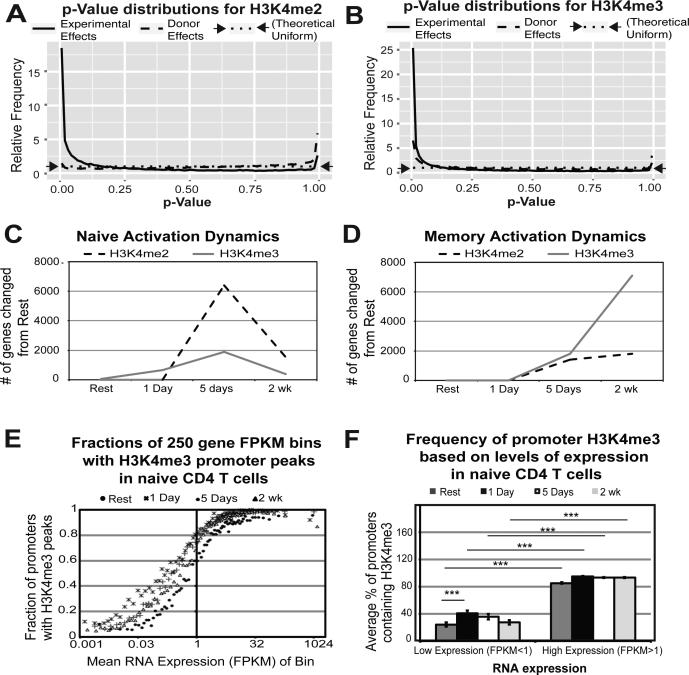
Figure 1. Differential promoter H3K4me2 and H3K4me3 is dependent on condition (i.e. time point and subset) and is dynamic throughout CD4 T cell activation.
For H3K4me2 A) and H3K4me3 B), the distribution of p-values for two different tests is plotted: an ANOVA test for differences between all combinations of experimental factors (cell type and time point) while treating the donor as a batch effect (solid line), and an ANOVA between donors while treating experimental factors as batch effects (dashed line). A uniform distribution (indicated by the dotted line) would indicate no evidence of differential enrichment by donor, while the strength of the spike near p=0 indicates significant differential enrichment by the experimental design. H3K4me enrichment is dynamic throughout activation for both naïve C) and memory D) subsets. E) Promoter H3K4me3 peaks correlate with high RNA expression throughout activation. Bins of 250 genes were plotted for mean RNA expression and % of promoters containing H3K4me3 peaks at all time points examined. F) Highly expressed genes have significantly more promoters with H3K4me3 peaks. Genes were divided into low (FPKM<1) and high (FPKM>1) expression and plotted against their average promoter H3K4me3 peak percentage for all time points examined (resting, 1 day, 5 days, and 2 weeks) in naïve CD4 T cells. Error bars represent variance from one-way ANOVA. Reported p-values are from Tukey's HSD post-hoc analysis (***p<0.0001).
In order to ensure that called peaks were robust in the face of technical and biological variation, we used the IDR framework developed for the ENCODE project. While ENCODE typically uses 2 biological replicates to assess biological reproducibility, we have 4 replicates for each group, which results in 6 pairwise comparisons for each group. For each of the 6 pairs, the IDR method produces a count of peaks that can be confidently called at our chosen threshold of reproducibility, IDR=2%. For each group, we selected the maximum out of these 6 counts, as recommended by the ENCODE documentation for datasets of more than 2 samples. Then we selected that number of peaks from the top of the list of peaks called on the combined dataset of all 4 replicates. The number of peaks selected for each group is shown in Supplementary Figure S5. Since the combined peak calls are based on 4 times the sequencing depth, the peaks are called more confidently than in the individual replicates, so the 2% threshold represents an upper bound on the fraction of irreproducible peaks. The IDR thresholds were generally consistent across all replicates, and variations in number of peaks called by IDR do not associate with low signal-to-background ratios in the coverage depth distributions. Thus, the IDR threshold of 2% is well above the technical noise floor imposed by any issues with ChIP efficiency.
It is also possible that differences in ChIP efficiency could confound estimates of fold changes between sample groups. In order to assess this possible technical limitation, we looked at the MA plots of each sample versus all the other samples of the same type after normalization (Supplementary Figures S6-S7). Each MA plot generally has a low-abundance “noise mode” (log2(CPM) ≈ 2), representing the promoters with only background noise, and a high-abundance “signal mode” (log2(CPM) ≈ 7) representing the promoters containing ChIP signal. In the analyzed samples, the signal mode is centered vertically within 0.5 log2 fold difference of zero, indicating that the edgeR normalization used is effectively controlling for global differences in ChIP efficiency and other technical artifacts. The few samples for which the signal mode is far away from zero are samples with lower sequencing depth, which means that these samples receive less weight in the model fitting stage. Given these observations, variations in ChIP efficiency are not responsible for any perturbation larger than approximately 0.5 in reported log2 fold changes.
Promoter H3K4 methylation is dynamic throughout T cell activation
The premise of the present study is that CD4 T cell activation is a dynamic process and needs to be studied as a function of time after activation (i.e. kinetic analysis). It is well established that changes to RNA transcription during activation are rapid and occur as early as 30 minutes.25 During our time course, 8655 genes in naïve and 7644 genes in memory changed RNA expression by two-fold or more 1 day after activation in both naïve and memory CD4 T cells (FDR < 0.05; Supplementary Tables S1 and S2). In striking contrast, differences in promoter H3K4me2 and H3K4me3 enrichment were primarily documented only later at 5 days after activation in both naïve and memory (Figure 1C-D).
However, in naïve cells a subset of 611 promoters showed a change in promoter H3K4me3 by 1 day after activation (2-fold change in either direction; FDR <0.1). 178 genes (29%) changed RNA expression by this time point (2-fold change in either direction; FDR <0.05; Supplementary Table S3). Only 15 gene promoters out of 611 increased their H3K4me3 enrichment by 1 day, and 10/15 (67%) of these show a significant increase in RNA expression, suggesting the biological significance of these changes. This list includes IL2RA, which is critical for early T cell activation, as well as IRF4, which has been demonstrated as an important regulator of IL-10 production.26 Pathway mapping of the entire set of 611 genes with early promoter H3K4me3 changes reveals genes linked to many important and established T cell pathways including PI3K-AKT and JAK/STAT, WNT signaling,27 interferon signaling,28 and networks for histone demethylation (Table 1). Most of the genes in this list decreased in promoter H3K4me3 by 1 day and were either concurrently unexpressed or decreasing in RNA expression by 1 day, suggesting portions of these pathways were being inhibited. Interestingly, 55 olfactory receptor genes lost the H3K4me3 mark and decrease their RNA expression by this time point. These genes are either unexpressed or lowly expressed in T cells, which is likely reinforced by the loss of H3K4me3, but the relevance of these targets to T cell activation remains unexplained. In contrast to naïve cells, memory cells showed no promoters changing H3K4me3 enrichment 1 day after activation.
Table 1.
Top pathways obtained via Signaling Pathway Impact Analysis (SPIA) for genes changing in promoter H3K4me3 in naïve CD4 T cells 1 day after activation.
| Database | Pathway | # of genes in pathway | # of genes differentially enriched at 24 h | % of pathway | p-value |
|---|---|---|---|---|---|
| Reactome | Signaling by GPCR | 1002 | 227 | 23% | 3.08E-41 |
| KEGG | Olfactory transduction | 397 | 156 | 39% | 3.06E-65 |
| Reactome | Immune System | 1075 | 129 | 12% | 0.0000135 |
| KEGG | PI3K-AKT signaling pathway | 334 | 52 | 16% | 0.000366 |
| Reactome | Cytokine Signaling in Immune system | 303 | 45 | 15% | 0.000193 |
| PANTHER | WNT signaling pathway | 215 | 35 | 16% | 0.00138 |
| Reactome | Interferon Signaling | 189 | 32 | 17% | 0.000151 |
| Reactome | Ion channel transport | 169 | 30 | 18% | 0.00154 |
| KEGG | JAK-STAT signaling pathway | 156 | 29 | 19% | 0.00072 |
| PANTHER | Integrin signalling pathway | 109 | 22 | 20% | 0.00111 |
| Reactome | Cell-Cell communication | 106 | 21 | 20% | 0.00125 |
| Reactome | Stimuli-sensing channels | 94 | 20 | 21% | 0.001247 |
| Reactome | Meiotic recombination | 75 | 18 | 24% | 0.000176 |
| Reactome | PKMTs methylate histone lysines | 59 | 17 | 29% | 0.000009713 |
| Reactome | HDMs demethylate histones | 43 | 13 | 30% | 0.000115 |
| Reactome | RNA Polymerase I Promoter Opening | 52 | 12 | 23% | 1.52E-07 |
| NCI | Internalization of ErbB1 | 32 | 11 | 34% | 0.000316 |
| KEGG | Taste transduction | 44 | 10 | 23% | 0.00125 |
| Reactome | Antigen activates B Cell Receptor (BCR) leading to generation of second messengers | 31 | 10 | 32% | 0.00134 |
| BioCarta | 41bb-dependent immune response | 6 | 2 | 33% | 7.12E-06 |
Despite the dynamic nature of both RNA expression and H3K4me3 as a function of time, promoter H3K4me3 consistently correlates with high RNA expression throughout activation (Figure 1E-F). Thus, even in our time-based dynamic analysis, H3K4me3 correlates with high gene expression as T cell activation progresses from rest to two weeks after activation, and a high percentage of highly expressed genes in activated CD4 T cells have promoter sites enriched for this mark (Figure 1E). These results confirm and extend previously reported evidence for higher RNA expression in genes with H3K4me3 peaks in their promoters.10, 29, 30 We further validated these conclusions by correlating H3K4me3 peak presence to low and high RNA expression and conducting a one-way ANOVA to demonstrate significance (Figure 1F).
Genes with promoter H3K4 methylation peaks in resting naïve and memory CD4 T cells map to pathways important for T cell activation and cytokine signaling
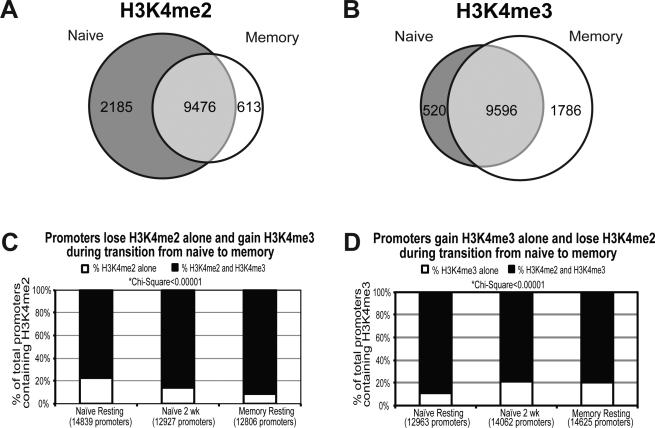
Memory CD4 T cells demonstrate a more rapid and robust response to antigen than naïve cells. We compared the genome-wide H3K4 methylation status of naïve and memory cells at rest in order to elucidate whether these marks were epigenetic contributors to the phenotypic differences. Naïve cells showed more unique H3K4me2 promoter peaks than memory cells at rest (2185 vs. 613), while memory cells contained more H3K4me3 peaks unique to their subset than naïve (1786 vs. 520) (Figure 2A-B). Pathway mapping demonstrated that genes marked with H3K4me2 in naïve cells but not memory CD4 populated canonical pathways related to critical activation events, including T cell receptor signaling, NFAT, PI3K, phospholipase and CD28 co-stimulation. On the other hand, promoters marked with H3K4me3 exclusively in memory cells populated pathways for IL2 signaling, G protein signaling, and CXCR4 signaling. A different set of genes from those marked with H3K4me2 in naïve also mapped to NFAT signaling (Table 2, Supplementary Table S4).
Figure 2. Naïve and memory CD4 T cells have different promoter H3K4me2 and H3K4me3 content.
Venn diagrams of H3K4me2 A) and H3K4me3 B) promoter peaks found in naïve and memory CD4 T cells at rest prior to activation. Promoters in CD4 T cells gain H3K4me3 and lose H3K4me2 during transition from naïve to memory phenotype. C) The percentage of promoters with H3K4me2 peaks that contain H3K4me3 is shown in naïve cells at rest and 2 weeks after activation, and in memory cells at rest. D) The percentage of promoters with H3K4me3 peaks that contain H3K4me2 is also shown for these three conditions.
Table 2.
Canonical pathway mapping of genes marked with H3K4me at rest in naïve and memory CD4 T cells using Ingenuity Pathway Analysis. Specific gene lists for each pathway are reported in Supplementary Table S4.
| Pathway # | H3K4me2 in Naïve Only | H3K4me3 in Memory Only |
|---|---|---|
| 1 | T Cell Receptor signaling | G protein signaling |
| 2 | Phospholipase C signaling | CXCR4 signaling |
| 3 | PI3K signaling | Role of NFAT in the immune response |
| 4 | Role of NFAT in the immune response | IL-2 signaling |
| 5 | CD28 signaling in T helper cells | IL-1 signaling |
H3K4me2 and H3K4me3 peaks are often found in combination when located in promoters, but they can also occur in isolation. Naïve cells at rest display a higher percentage of promoters with H3K4me2 peaks alone than memory (22.3% vs. 8.7%; 3,298 vs. 1,112), while memory cells at rest display a higher percentage with H3K4me3 alone than naive (20% vs. 11%; 2,931 vs. 1,422). By 2 weeks after activation of naïve cells these marks transition to a CD4 memory-like pattern, with fewer isolated H3K4me2 peaks and more H3K4me3 peaks (Figure 2C-D).
H3K4 methylation status at rest delineates key pathways in naïve CD4 T cells
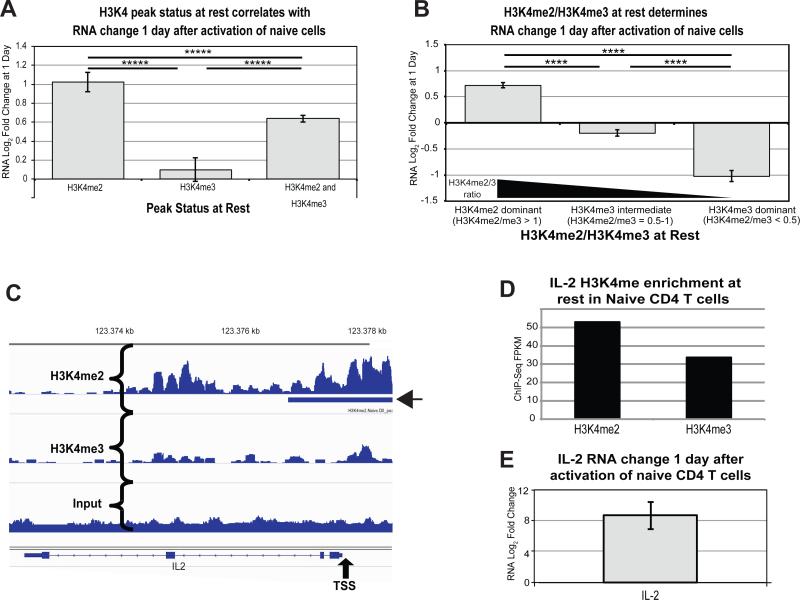
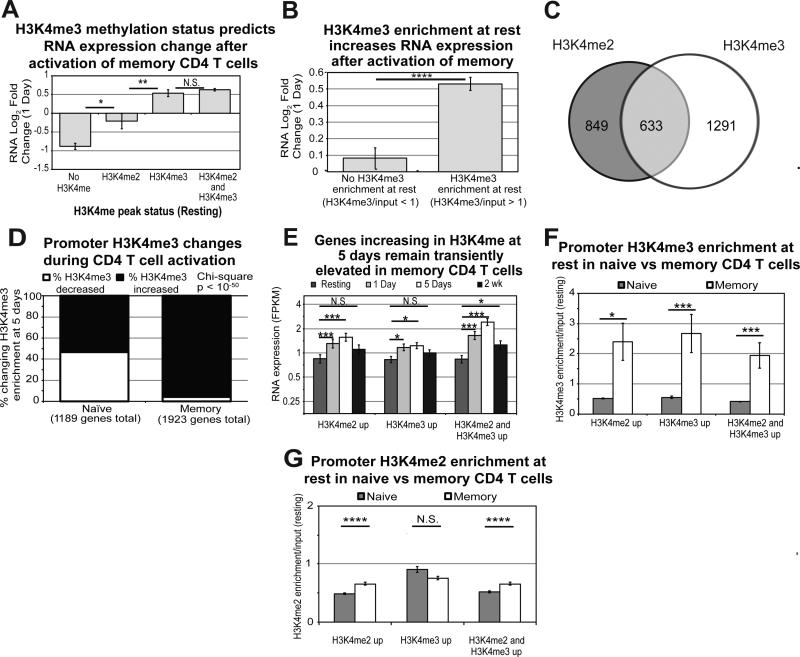
We hypothesized that promoter H3K4me3 at rest would promote increases in RNA expression after activation, allowing us to predict early gene expression. Therefore, we evaluated changes in RNA expression 1 day after activation for genes containing peaks of H3K4me2 alone, H3K4me3 alone, or both in promoters in resting naïve cells. Against our expectations, the results clearly show that H3K4me3 alone is not predictive of increasing gene expression 1 day after activation in naïve cells (Figure 3A). Surprisingly, genes containing only H3K4me2 peaks at rest showed a robust increase in expression. Gene promoters containing both modifications increased RNA expression, but to an intermediate degree compared to H3K4me2 alone.
Figure 3. H3K4me2 dominance at rest promotes transcriptional upregulation 1 day after activation of naïve CD4 T cells.
A) Mean increase in RNA expression (Log2 Fold Change) 1 day after activation of naïve CD4 T cells is plotted for genes with H3K4me2 peaks alone, H3K4me3 peaks alone, or both H3K4me2 and H3K4me3 peaks. Error bars represent standard error of the mean. Reported p-values are from Tukey's HSD post-hoc analysis following one-way ANOVA (*****p<1×10−6) B) Mean increase in RNA expression 1 day after activation is plotted for genes with a continuum of H3K4me2/H3K4me3 at rest in naïve cells. Error bars represent standard error of the mean. Reported p-values are from Tukey's HSD post-hoc analysis following one-way ANOVA (****p<1×10−5). C) Genome viewer landscape of H3K4me2 (upper panel) and H3K4me3 (lower panel) in the promoter of IL2. A called peak for H3K4me2 is denoted by the arrow head. TSS = Transcriptional Start Site. D) ChIP-Seq FPKM for H3K4me2 and H3K4me3 in the IL2 promoter of resting naïve CD4 T cells 1 kb on either side of the TSS. E) The RNA increase in IL2 transcript 1 day after activation of naïve CD4 T cells as measured by RNA-Seq.
To validate these results, we performed an orthogonal analysis by examining overall enrichment for these modifications across the promoters and calculating changes to gene expression for genes with various ratios of H3K4me2/H3K4me3 (Figure 3B, Supplementary Figure S8A). Using these ratios, we found that genes with a higher enrichment of promoter H3K4me3 relative to H3K4me2 (i.e. H3K4me3 dominant) demonstrated decreased expression by 1 day, with a higher frequency of genes decreasing in expression overall. Conversely, genes with higher H3K4me2 enrichment at rest relative to H3K4me3 (i.e. H3K4me2 dominant), increased gene expression at 1 day. As an example, IL-2, which shows a large increase in RNA expression upon CD4 T cell activation, is H3K4me2 dominant at rest in naïve cells (Figure 3C-E).
One hypothesis is that genes already marked at rest with H3K4me3 have high baseline expression, so the changes with activation are minimal. However, H3K4me3 dominant genes displayed significantly lower mean baseline RNA expression than the H3K4me2 dominant genes (Supplementary Figure S8B-C). Overall, resting H3K4me2 marks are more predictive of genes set to increase expression after activation of naïve CD4 T cells, and co-marking with H3K4me3 reduces activation-induced expression.
H3K4me2 and H3K4me3 work in tandem during activation to influence key pathways in naïve CD4 T cells
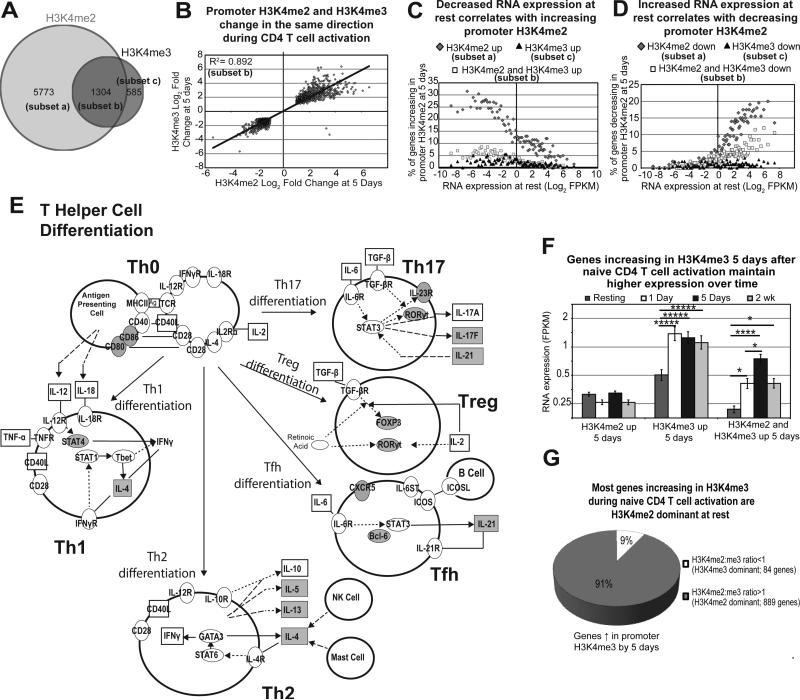
Most of the activation-induced changes to H3K4me2 and H3K4me3 enrichment are seen late in activation (5 days) (Figure 1C-D). Naïve cells have 7077 promoters that change H3K4me2 enrichment by 5 days (Figure 4A). Most of these gene promoters (subset a: Figure 4A) change H3K4me2 enrichment without a change in H3K4me3 (5773 promoters; 81%). However, a total of 1889 promoters change for H3K4me3, the majority of which change for both H3K4me3 and H3K4me2 (subset b = 1304; 69%). Both modifications change in the same direction during this period, and the correlation is highly statistically significant (R2 = 0.89; Figure 4B). Additionally, as shown in Figure 4C-D, changes to promoter H3K4me2 by 5 days in naïve cells correlate strongly to resting RNA expression. These results and immune pathway mapping for genes increasing in H3K4me2 during naïve CD4 T cell activation (Figure 4E) are consistent with the hypothesis that resting RNA expression impacts activation-induced changes to H3K4me2 marks, which in turn regulate the RNA expression of this subset of genes.
Figure 4. Promoter H3K4me2 and H3K4me3 act in tandem to regulate RNA expression during naïve CD4 T cell activation and promote Th differentiation.
A) Venn diagram of promoters changing in H3K4me2 and H3K4me3 enrichment 5 days after activation of naïve CD4 T cells. B) Promoters changing in both H3K4me2 and H3K4me3 enrichment 5 days after activation are changing in the same direction. Pearson's correlation is pictured. C) Bins of 200 genes were plotted for RNA expression at rest (x-axis), and the percentage of genes increasing in H3K4me2 alone (dark grey diamonds), H3K4me3 alone (black triangles), or both (white squares) at 5 days (y-axis). D) Bins of 200 genes were plotted for RNA expression at rest and the percentage of genes decreasing in H3K4me2 alone (dark grey diamonds), H3K4me3 alone (black triangles), or both (white squares) at 5 days. E) Th cell differentiation is one of the top pathways influenced by increased promoter H3K4me2 in naïve CD4 T cells during activation. Shaded molecules represent gene promoters that significantly increase H3K4me2 enrichment 5 days after activation of naïve CD4 T cells. F) RNA expression for all time points examined was plotted for genes increasing in promoter H3K4me2 alone, H3K4me3 alone, and both H3K4me2 and H3K4me3 at 5 days. Error bars represent standard error of the mean. ****p<1×10−5, *p<0.01 from Fisher's LSD post-hoc analysis following one-way ANOVA. G) 91% of genes increasing in H3K4me3 during activation were H3K4me2 dominant at rest in naïve cells.
Given these dynamic changes in H3K4 methylation as a function of time after activation, we next focused on the correlations with the equally dynamic changes in RNA expression (resting, 1 day, 5 days, and 2 weeks; Figure 4F, Supplementary Figure S8D). These results revealed three distinct patterns of H3K4 methylation changes correlating with activation-induced gene expression. First, genes that increased H3K4me2 alone between rest and 5 days demonstrated low levels of RNA expression at all time points, while those with decreasing H3K4me2 exhibited high RNA expression. Second, genes that increased their H3K4me3 enrichment alone by 5 days showed a significant increase in RNA expression from rest to 1 day that is sustained for the entire 2 weeks. Third, if H3K4me2 and H3K4me3 increased together from rest to 5 days, a significant increase in gene expression from low resting levels is demonstrated and maintained out to two weeks. These results suggest that increased H3K4me3 by 5 days associates with reinforcement of earlier activation-induced RNA expression changes. Interestingly, 91% of genes increasing in promoter H3K4me3 5 days after activation of naïve cells are H3K4me2 dominant at rest (Figure 4G), suggesting that the increased expression associated with H3K4me2 dominance early activation of naïve cells is also associated with the addition of H3K4me3 later in activation.
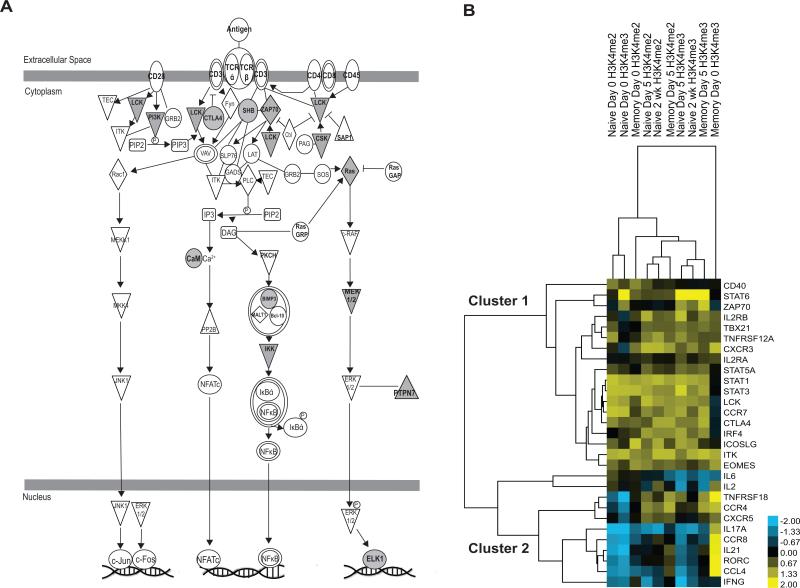
As shown previously, promoter H3K4me2 dominance in naïve CD4 T cells at rest predicts which genes are set to increase after activation. Our data demonstrate that multiple pathways important for CD4 T cell activation are associated with H3K4me2 dominance at rest in naïve cells. Examples include cytokine signaling, such as IL-2, IL-4, IL-6, and IL-17 as well as TNF receptor, 4-1BB, MAPK, NFkB, Jak-Stat pathway and OX40 pathway signaling (Table 3). H3K4me2 dominance at rest also correlates with T cell receptor (Figure 5A) and CD40 signaling. More general cellular pathways are also affected, such as the DNA damage response by BRCA1 (Supplementary Table S5). Interestingly, hierarchical clustering of promoter H3K4me2 and H3K4me3 for a subset of 29 genes chosen as important for CD4 T cell function demonstrates that these genes form two clusters: one with predominantly T cell receptor cascade components and STAT family members (cluster 1), and another containing genes predominantly associated with T cell effector function and differentiation, such as cytokines and master transcriptional regulators (cluster 2) (Figure 5B).
Table 3.
Activation-induced immunologic pathways guided by H3K4me2 dominance at rest in naïve CD4 T cells as reported by ImmuneMap.
| Pathway | Total Genes | H3K4me2 dominant genes | % population |
|---|---|---|---|
| IL21 | 34 | 13 | 38.24% |
| 4-1BB | 29 | 9 | 31.03% |
| TNF superfamily pathway | 71 | 19 | 26.76% |
| GITR | 27 | 7 | 25.93% |
| CD40 | 48 | 12 | 25.00% |
| NFKB pathway | 84 | 20 | 23.81% |
| IL4 | 56 | 13 | 23.21% |
| IL1R1 | 55 | 12 | 21.82% |
| TLR signaling | 102 | 22 | 21.57% |
| IL6 | 44 | 9 | 20.45% |
| MAPK | 174 | 35 | 20.11% |
| IL17 | 50 | 10 | 20.00% |
| IL12 | 83 | 16 | 19.28% |
| Vitamin D | 90 | 17 | 18.89% |
| IL2 | 48 | 9 | 18.75% |
| General JAK / STAT | 155 | 29 | 18.71% |
| OX40 | 55 | 9 | 16.36% |
| Innate immunity signaling | 136 | 22 | 16.18% |
| TCR signaling | 92 | 14 | 15.22% |
| CTLA4 | 66 | 10 | 15.15% |
Figure 5. Promoter H3K4me2 enrichment influences T cell activation and differentiation in naïve CD4 T cells.
A) T cell receptor signaling is a major pathway influenced by H3K4me2 dominance in naïve cells. Shaded symbols represent genes that are H3K4me2 dominant at rest in naïve cells and have increased RNA expression 1 day after activation. B) Hierarchical clustering of H3K4me2 and H3K4me3 enrichment for a subset of genes important to T cell function reveals genes cluster according to their roles in activation and/or differentiation.
Resting memory cells were particularly different for H3K4me3 in loci defining effector function of Th subtypes, including CCR8 (Tregs, Th2)31, IFNG (Th1), IL17A and RORC (Th17). These genes all had significantly higher RNA expression 1 day after activation of memory CD4 T cells vs. naïve (Supplementary Figure S8E). Genes increasing in H3K4me3 in naïve cells populate networks for cellular maintenance and hematologic system development/differentiation. These networks all center around NFκB (Supplementary Figure S8F). In contrast, cellular functions correlating with H3K4me2 at rest and maintained by H3K4me3 after activation map to pathways regulating cell proliferation, cell death, necrosis, and apoptosis, suggesting that these basic cellular functions are regulated by both H3K4me2 and H3K4me3 during naïve CD4 T cell activation.
H3K4me3 at rest predicts increased gene expression during memory CD4 T cell activation
Promoter H3K4 methylation in memory CD4 T cells exhibits different dynamics and different correlations with activation-induced gene expression than naïve cells. In memory CD4, H3K4me2 does not correlate with or predict increased RNA expression after activation, in contrast to what we found in naïve cells. However, H3K4me3 peaks at rest predict increased RNA expression after activation (Figure 6A). Promoter H3K4me3 enrichment analysis supports this conclusion, showing that RNA expression of genes with H3K4me3 increases significantly more one day after activation than for those without H3K4me3 (Figure 6B). Interestingly, memory cells exhibit only a fraction of the H3K4me2 changes observed in naïve by 5 days, while they have a greater number changing for H3K4me3 alone (Figure 6C). Additionally, 96% of those changing promoter H3K4me3 in memory cells are increasing enrichment (Figure 6D).
Figure 6. H3K4me3 enrichment plays a greater role in transcriptional upregulation in memory CD4 T cells during activation.
A) RNA Log2 Fold Changes at 1 day for genes containing no H3K4me, H3K4me2 alone, H3K4me3 alone, or both H3K4me2 and H3K4me3 at rest in memory cells. Error bars represent standard error of the mean. P-values are for two-tailed student t-test (*p<0.01, **p<001). B) RNA Log2 Fold Changes at 1 day for genes with H3K4me3 compared to no enrichment at rest in memory. Error bars represent standard error of the mean. P-values are for two-tailed student t-test (****p<1×10−5). C) Venn diagram for genes changing H3K4me2 and H3K4me3 enrichment 5 days after activation of memory CD4 T cells. D) Frequency plots for genes increasing or decreasing in H3K4me3 enrichment in naïve and memory CD4 T cells at 5 days. E) RNA expression for all time points examined was plotted for genes changing in H3K4me2 alone, H3K4me3 alone, and both at 5 days in memory cells. Error bars represent standard error of the mean. ***p<0.0001, *p<0.01 from Fisher's LSD post-hoc analysis following one-way ANOVA. F) Average enrichment for H3K4me3 across the promoter is plotted for the three classes of genes changing in H3K4 methylation during activation of naïve and memory CD4 T cells. Error bars represent standard error of the mean. P-value represents two-tailed student t-test (*p<0.01, ***p<0.0001). G) Average enrichment for H3K4me2 across the promoter is plotted for the three classes of genes changing in H3K4 methylation during activation of naïve and memory CD4 T cells. While differences in enrichment are statistically significant, the differences are modest and the biological significance is questionable. Error bars represent standard error of the mean. P-value represents two-tailed student t-test (****p<1×10−5).
As in naïve cells, changes to H3K4me2 and H3K4me3 peak at 5 days in memory cells. However, unlike naïve cells, no relationship exists between changes to promoter H3K4me2 and resting RNA expression. On the other hand, increased H3K4me3 correlates with increased RNA expression at 1 and 5 days (Figure 6E). As shown in Figure 6F, memory cells contain significantly higher promoter H3K4me3 than naïve at rest for all 3 subsets of gene promoters increasing in H3K4 methylation. These results are consistent with the premise that memory CD4 obtained the epigenetic marks necessary for this early activation response during differentiation from naïve cells. In contrast, the difference in baseline H3K4me2 enrichment of these two subsets is negligible (Figure 6G). Multiple immunologically critical pathways mapped to H3K4me3 marks overlapped with those epigenetically influenced by H3K4me2 in naïve cells: T cell receptor signaling, TNF receptor signaling, OX40 signaling, and IL-2 receptor signaling (Table 4).
Table 4.
Activation induced pathways guided by H3K4me2 dominance in naïve CD4 T cells and H3K4me3 enrichment in memory CD4 T cells reported by Ingenuity Pathway Analysis. Gene symbols in bold represent genes common to both gene sets in each pathway.
Multiple upstream regulators are also shared in common by pathways under the influence of H3K4 methylation in naïve and memory cells. Not surprisingly, these include the T cell receptor, CD3, and the co-stimulatory molecules, CD40LG and CD28, but they also include p53, E2F4, E2F1, MYC, and other important signaling molecules (Supplementary Table S6). Thus, immunological pathways and cellular functions pertaining to the cell cycle, transcription, and DNA repair are correlated with H3K4 methylation in CD4 T cells.
CpG islands impact H3K4 methylation dynamics after CD4 T cell activation
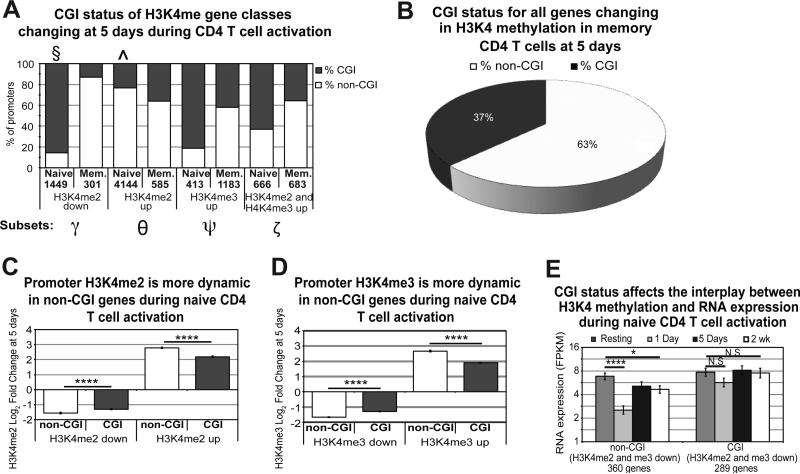
Genes containing CpG islands (CGIs) were demonstrated to have different dynamics for H3K4 methylation during differentiation of hematopoietic stem cells than non-CGI containing genes.18 Therefore, we examined the percentage of CpG islands as a function of the previously described gene subsets marked by H3K4me2 and H3K4me3 (Figure 7A). Surprisingly, the percentage of CGI-containing genes was markedly different between naïve and memory cells. In naïve cells, 88% of promoters decreasing H3K4me2 enrichment contained a CGI (Figure 7A, denoted with §), while nearly 80% of those increasing were without a CGI (denoted with ^). Memory cells displayed the opposite pattern from naïve for several gene sets, including genes increasing in H3K4me3 (Ψ), genes decreasing in H3K4me2 (γ), and genes increasing in both H3K4me2 and H3K4me3 (ζ). Note that 63% of the genes changing for H3K4 methylation in memory cells did not contain a CpG island while 60% of gene promoters genome-wide contain a CGI32 (Figure 7B).
Figure 7. CGI status affects H3K4 methylation and accounts for differences in naïve and memory H3K4 dynamics.
A) Differences in CGI content are seen in the previously defined gene subsets between naïve and memory. Numbers listed in the x-axis represent the number of promoters in the denoted subset. Non-CGI (white) and CGI (grey) promoters show different dynamics between naïve and memory when examining each subset of genes including: H3K4me2 decreased (γ), H3K4me2 increased (θ), H3K4me3 increased (Ψ), and H3K4me2/me3 increased (ζ). Naïve cells in particular show a propensity for genes decreasing in H3K4me2 to contain CGI sites (§) and those increasing in H3K4me2 to be non-CGI site-containing (^). Less than 10 genes showed a pattern of H3K4me3 decrease in memory cells and are therefore not included. B) Genes changing in H3K4 methylation in memory cells are predominantly non-CGI. C) and D) non-CGI promoters show more change (up and down) in enrichment for H3K4me2 (C) and H3K4me3 (D) 5 days after activation. Error bars represent standard error of the mean. P-value represents two-tailed student t-test (****p< p<1×10−5). E) Decreases in H3K4me2 and H3K4me3 correlate with sustained decreases in RNA expression only in non-CGI genes. Error bars represent standard error of the mean. P-value represents two-tailed student t-test (****p<1×10−5, *p<0.01).
Comparing H3K4 methylation dynamics of genes with and without CGI promoters during activation revealed significantly greater changes in non-CpG promoters for both H3K4me2 (Figure 7C) and H3K4me3 (Figure 7D). Additionally, while loss of H3K4me2 and H3K4me3 did not seem to impact RNA expression of genes 2 weeks after activation (Supplementary Figure S8D), evaluation of CGI status showed that non-CGI containing genes had a sustained decrease in RNA expression 2 weeks after activation for this subset, while CGI-containing genes did not (Figure 7E). Therefore, non-CGI containing genes are more prone to changes in promoter H3K4 methylation, and these increased fluctuations correlate with changes in RNA expression after activation.
Discussion
Our results clearly show that promoter H3K4 methylation is an important and dynamic epigenetic contributor to CD4 T cell activation and differentiation. H3K4me3 in promoter sequences is most commonly associated with actively transcribed genes, consistent with the concept of a permissive mark.33 On the other hand, there is also published data demonstrating that H3K4me3 is often found in promoters of genes that are unexpressed in mammalian cells.13, 34, 35 Our new data support the paradigm that the H3K4 methylation landscape at rest can predict transcriptional changes after activation, though we find that this is true for some but not all marked genes. However, we also find that the landscape is shaped by activation-induced gene expression in CD4 T cells. This reveals a more complicated model than H3K4me3 as a simple ‘activating’ mark.
In a technical context, we performed two types of analysis for the ChIP-Seq results. Many ChIP-Seq studies focus upon peak analysis, but our parallel analysis for differential enrichment adds a powerful additional approach supported by other studies.36-38 Differential enrichment analysis can reveal significant histone enrichment in a much larger sequence frame, and at the global level it provides a more complete view of possible histone regulated promoters. For example, our analysis for differential enrichment investigated the landscape over a 2 kb stretch of the regions surrounding the TSS on either side (i.e. 1 kb up- and downstream of the TSS). There are often multiple histone peaks called within these regions, though overall differential enrichment identifies more significant changes to the promoter landscape than peak calling.
Taking the H3K4 methylation data at rest and the RNA expression data at 1 day after activation, we propose a mechanism for naïve CD4 T cells where the presence of H3K4me2 is a dominant early regulator of increased RNA transcription. Enrichment for H3K4me2 alone results in maximal change in activation-induced gene expression (Figure 3A-B). Consistent with this result, previous reports show that over 90% of transcription factor binding regions overlap with H3K4me2 peaks,39 so one possibility is that H3K4me2 specifically plays a role in transcription factor recruitment. Surprisingly, if the promoter at rest also contains H3K4me3, then H3K4me3 acts as a “governor” in naïve cells to reduce the levels but not prevent the gene transcription upregulated by activation. We also demonstrate that differential enrichment for promoter H3K4me2 is a function of the RNA expression found prior to activation in naïve cells and supports the role for H3K4me2 as a ‘poising’ promoter mark to our model.
While H3K4me2 and H3K4me3 are mutually exclusive marks on each individual histone molecule, these marks can be located within the same nucleosome given its octamer structure.30 The presence of two copies of the H3 histone in each octomer can allow for one H3 histone to be marked in one pattern with the second marked by a different pattern even at same site and in the same cell. Unfortunately, while the question of how commonly this occurs might be answered by single cell ChIP analysis, this technology is still in its infancy and requires proprietary instruments not available to us.40 Additionally, the multicellular and/or polyclonal nature of our cell populations, even though already purified into naïve and memory CD4 cell subsets, also likely contributes to the overlap observed between these two marks in our data. Our results show that genes containing both H3K4me2 and H3K4me3 peaks exhibit the highest RNA expression when measured at each time point based on a static read-out of RNA expression at a single point in the evolution of T cell activation (Supplementary Figure S8C). These results are not to be confused with the observation that H3K4me2 is associated with the greatest differential RNA change after activation of naïve cells as measured by log2fold change from one time point to another (i.e. 1 to 5 days).
Additionally, we found that 89% of gene promoters containing H3K4me3 in resting naïve cells also contain H3K4me2 peaks (Figure 2D). Thus, it is plausible that the ‘active’ conformation commonly ascribed to promoters containing H3K4me3 is actually due to the combinatorial effects of H3K4me2 and H3K4me3. This observation was previously reported in the literature for mouse hematopoietic stem cells (HSC's).18 However, one key difference with our results is that there were genes containing H3K4me3 peaks alone in mouse HSC's, while in human resting naive CD4 T cells 1,422 out of 12,963 (11%) H3K4me3-containing promoters did not contain H3K4me2 peaks. The discordance could be explained by the different radii surrounding the TSS used in the analyses (i.e. 1 kb in our data versus 7 kb in Orford et al.), differences by species, or differences between hematopoietic stem cells and CD4 T cells.
Resting memory cells contain far more promoters with H3K4me3 peaks alone (2,931 out of 14,625 H3K4me3-positive promoters; 20%). In contrast, only 8.7% of H3K4me2-positive promoters contain H3K4me2 peaks alone. Thus, differentiation of human CD4 to memory cells results in increased numbers of genes with only promoter H3K4me3 and a loss of the ‘poised’ promoters containing only H3K4me2 peaks reported for HSC's. If genes containing H3K4me2 peaks alone truly represent a poised state as proposed by Orford et al., it is logical that the loss of these poised genes over time is one component of cellular differentiation, where poised genes are replaced with purposeful epigenetic marks to reinforce chosen cellular agendas. The Epigenomics Roadmap Consortium also made a similar observation when they analyzed 111 human epigenomes derived from healthy human subjects and representing multiple cell types, including fetal progenitors.41 Based on the ENCODE data, Das et al. recently observed that a loss of H3K4me2 and gain of H3K4me3 occurred during differentiation from embryonic to non-embryonic stem cells,42 reinforcing the concept that H3K4me2 is converted to H3K4me3 during differentiation.
Since H3K4me2 appears to be a poising mark responsible for providing a transitional framework for further differentiation, one hypothesis is that during thymocyte development this mark is added to genes crucial for T cell differentiation, preparing them for further development upon activation. Previous work examining H3K4me2 during thymic development in mice suggested that the transcription factors PU.1 and GATA3 were likely responsible for H3K4 methyltransferase recruitment during thymic development.20 NFκB is also known to play a crucial role during T cell development43 in addition to activation, and our network analysis suggests that the H3K4 methyltransferases might interact with this transcription factor (Supplementary Figure S8F), thus potentially being another mechanism for targeting H3K4me2 to differentiation-related pathways during development. Further mechanistic studies will be required to explore this possibility.
Our report is the first to demonstrate that changes to the H3K4me2 landscape 5 days after naïve CD4 T cell activation conform very closely to the RNA expression of these same genes measured prior to activation (Figure 4C-D). Enrichment of this promoter mark also clusters according to activation status at each successive time point in both naïve and memory CD4 (Figure 5A), indicating the dynamic changes in H3K4me2 during activation. On the other hand, genes with increasing promoter H3K4me3 marks also increase in RNA expression earlier in activation and display a sustained increase in expression out to 2 weeks (Figure 4F). Additionally, genes that are increasing in both H3K4me2 and H3K4me3 begin at a lower resting expression than those increasing for H3K4me3 alone, suggesting that the H3K4me2 increases during naïve CD4 T cell activation might help to drive further increases in RNA expression by acting as a scaffold for the tri-methylation of H3K4.
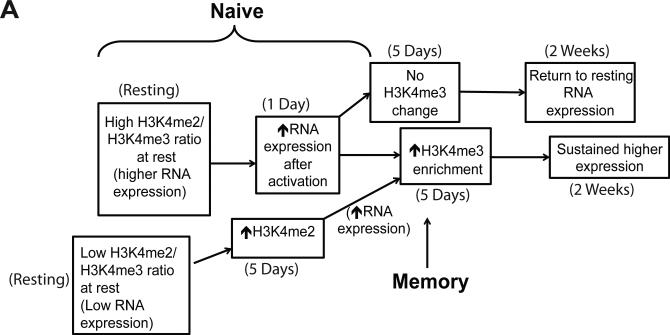
Therefore, we propose a model where promoter H3K4me2 at rest propels RNA expression after activation through recruitment of transcription factors and methyltransferase complexes. The dramatic promoter H3K4me2 changes we observed during naïve CD4 T cell activation prime the cells for further epigenetic modifications critical for differentiation driven by transcription factor recruitment (Figure 8). A key point in the model is that this H3K4me2-based priming step leading to an increase in H3K4me3, and recruitment of transcription factors is not necessary in memory cells, as their previous exposure to antigen has already achieved differentiation through this mechanism. That explains why there is no association between H3K4me2 at rest and an increase in RNA expression after activation for this subset. Our conclusion is further supported by pathway mapping for promoter H3K4me2 increases in naïve cells, which demonstrates that T helper cell differentiation is one of the top canonical pathways influenced by changes to this modification (Figure 4E).
Figure 8. Model for the role of H3K4 methylation during CD4 T cell activation.
A) In naïve cells, H3K4me2 dominance results in upregulation of expression. Low expression genes increase in H3K4me2, allowing H3K4me3 marks to be deposited in order to sustain increased expression. Genes whose H3K4me3 enrichment is not increased return to baseline expression. Memory cells generally bypass these mechanisms, starting with significantly higher promoter H3K4me3 at baseline. Time points at which these changes were recorded are documented.
Another key epigenetic determinant delineating naïve and memory CD4 T cell promoters is CpG methylation.44 Previous work has demonstrated that DNA methylation and H3K4 methylation tend to be mutually exclusive.33, 45 DNA methyltransferase 3a (DNMT3a), which is responsible for de novo CpG methylation, binds to H3K4me0 and cannot bind DNA in regions containing H3K4me2/3. COMPASS-like complexes that methylate H3K4 include CXXC domains responsible for binding unmethylated CpG residues on DNA and that binding acts to couple H3K4 methylation to unmethylated DNA.15, 45 DNA methylation also more commonly impacts non-CGI promoters, and memory cells tend to be hypomethylated in promoters compared to naïve cells.44 Therefore, it is possible that differential CpG methylation between naïve and memory CD4 T cells provides one explanation for their differing H3K4 methylation dynamics.
A novel result revealed by our work supports the hypothesis described above that non-CGI promoters have differential impacts. Thus, gene promoters with and without CGIs exhibited different dynamics after activation with respect to H3K4 methylation enrichment in both naïve and memory cells. Non-CGI promoters demonstrate much greater up- and down-regulation in H3K4me2 and H3K4me3 enrichment after activation (Figure 7C-D). Therefore, CGI may stabilize promoters by reducing fluctuations in H3K4 methylation. On the other hand, the overall H3K4me enrichment of non-CGI promoters remains lower than that of promoters containing CGI as activation evolves. Changes to H3K4 methylation in non-CGI promoters appear to be more prevalent in memory cells (Figure 7A-B). Additionally, the only correlations between decreasing promoter H3K4me3 levels and gene expression during activation of naïve cells are observed for non-CGI promoters (Figure 7E). Therefore, the CpG content of promoters influences the dynamics as well as the overall effects of H3K4 methylation during CD4 T cell activation. Memory cells primarily change H3K4 methylation enrichment in non-CGI promoters when compared to naïve. These results suggest that the evolutionary selection for genes with CGI and non-CGI promoters assigns more roles for non-CGI gene networks in differentiated CD4 memory T cells and more CGI gene networks for naïve cells.
We have offered this first work as a resource and acknowledge some limitations to the present study. First, our experimental designs do not involve selective perturbations in the system such as siRNA or small molecule inhibition of H3K4 methylation mechanisms, which would provide more mechanistic support for our conclusions. However, it is also the necessary first study of the dynamics of activation-induced changes in naïve vs. memory CD4 T cells to inform the hypotheses and properly design the next phase of studies. Second, we limited our analysis to promoter regions. Enhancers, gene bodies, and intergenic regions are also important to the enrichment landscape for these histone modifications and likely impact the regulation of gene expression. Enhancers in particular have been most heavily studied in CD4 T cells with respect to histone modifications and clearly inform differentiation pathways in these cells.46 While examination of these additional regions was outside the scope of this initial report, analyses of these data are currently underway.
Another limitation is that we have evaluated the role of 2 histone modifications out of over 100 that are currently documented.47 Combinations of these different modifications can often be found within nucleosomes and modify the core histones that comprise them. These modifications may act in combination with other epigenetic alterations, including DNA methylation and chromatin modifying complexes.19, 41, 48
In summary, this study demonstrates that there are distinct and dynamic differences in the methylation states of H3K4 histones between naïve and memory CD4 T cells that can be correlated with resting RNA expression and post-activation gene transcription. Moreover, these differences map to multiple important immune networks that are critical for CD4 T cell function and differentiation. We have shown that histone modifications are highly dynamic during immune activation; these post-activation dynamics differ markedly between naïve and memory cells, contributing to the differences in their activation dynamics and phenotypes. The question remains whether these dynamic, activation-induced epigenetic processes are an absolute mechanistic requirement for T cell differentiation and function or represent complex epigenetic changes associated with activation but not actually regulating the process. This question, as well as the different mechanisms regulating these dynamic changes in H3K4 methylation in naïve vs. memory cells, must be addressed in future work.
Materials and Methods
Ethics Statement
All the studies in this manuscript were covered by Human Subjects Research Protocols approved by the Institutional Review Board of The Scripps Research Institute. Informed written consent was obtained from all study subjects in the study.
Isolation and activation of human lymphocytes
Peripheral blood was collected from 4 healthy donors, and peripheral blood mononuclear cells (PBMC) were purified by centrifugation through a histopaque (Sigma, #10071) gradient. CD4 T cells were negatively selected using the EasySep™ Human Naive CD4+ T Cell Enrichment Kit (Stemcell Technologies, #19155) or the EasySep™ Human Memory CD4 T Cell Enrichment Kit (Stemcell Technologies, #19157). Cell purity was assessed by flow cytometry staining with antibodies specific for CD4 (SK3, eBioscience, #12-0047-42), CD45RA (HI100, eBioscience, #11-0458-42), and CD45RO (UCHL1, eBioscience, #25-0457-42). Data acquisition was conducted on an LSR-II (BD Biosciences) and analysis was performed using FlowJo (Treestar). Live cells were gated based on forward by side scatter area. Doublets were excluded based on forward scatter height by forward scatter width and side scatter height by side scatter width. Live cells were then gated on CD4 staining and cell purity following isolation was determined by CD45RA vs. CD45RO staining of the CD4+ population. Cell purity for all donors was > 94%.
CD4 T cells were cultured in RPMI 1640 (Mediatech, #SH30027.07) supplemented with 100 U/ml Penicillin, 100 μg/ml Streptomycin and 10% FBS at 37 °C and 5% CO2. T cells were activated with DynaBead Human T-Activator CD3/CD28 (Invitrogen, #111.32D) for 1 and 5 days. Cells maintained in culture out to 2 weeks received 30 U/mL of human recombinant IL-2 (NIH repository) beginning at day 5 after activation. Samples for ChIP-Seq and RNA-Seq were collected from the 4 donors at rest, 1 day, 5 days, and 2 weeks after activation. For chromatin isolation from each sample, 3 × 107 cells were fixed for 10 min in cell culture medium with 1% formaldehyde at room temperature. Fixation was quenched with 10% glycine for 5 min at room temperature. Cell pellets were flash frozen in liquid nitrogen and stored at −80 °C until chromatin isolation. Cells harvested for RNA (5 × 106 cells) were washed 3 times in 1 mL of PBS without calcium and magnesium (Corning, #21-040-CV), flash frozen in liquid nitrogen, and stored at −80 °C until RNA purification. RNA for RNA-Seq was isolated from purified cells using an All Prep kit (QIAGEN, #80004) following the manufacturer's instructions (www.qiagen.com).
Preparation of sequencing libraries and ChIP-Seq and deep RNA sequencing
For RNA-Seq, purified total RNA was converted to cDNA using the Ovation RNA-Seq system (NuGEN) followed by S1 endonuclease digestion (Promega, M5761) as previously described.49 Digested cDNA libraries were then end-repaired and A-tailed. Indexed adapters were ligated, and ligation product was purified on Agencourt AMPure XP beads (Beckman Coulter Genomics, #A83880) followed by size selection from 2% agarose. Purified product was amplified with 15 cycles of PCR followed by size selection from 2% agarose. Libraries were assessed on an Agilent Bioanalyzer using a DNA chip and quantitated using the Quant-iT ds DNA BR Assay kit (Invitrogen, #Q32853) and a Qubit Fluorimeter (Invitrogen). Cluster generation and sequencing on an Illumina HiSeq system was conducted directly with purified libraries following manufacturer's instructions (www.illumina.com). 100 bp single-end reads were generated for naïve and memory CD4 T cells from 4 donors with 2 samples per lane.
For ChIP-Seq, 10 ng of purified DNA from individual chromatin IPs were end repaired and A-tailed. Indexed adapters were ligated, and ligation product was purified on Agencourt Ampure XP beads and processed as described for RNA-Seq products.
Chromatin Immunoprecipitation
Chromatin was isolated from cell pellets using the ChIP-It Express Enzymatic Shearing Kit (Active Motif, #53035) per the manufacturer's instructions (www.activemotif.com). For immunoprecipitation, 7.5 μg of chromatin was diluted in a total of 1 mL of low salt wash buffer (0.1% SDS, 1.0% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl (pH 8.1), 500 mM NaCl) with protease inhibitor cocktail and pre-cleared with 30 μL of Dynal Protein G magnetic beads (Invitrogen, #10004D) for 2 h at room temperature with rotation. 20 μL of pre-cleared chromatin was saved for input analysis and stored at −80 °C. Remaining chromatin was incubated with 2 μL of anti-H3K4me2 antibody (Millipore, #07-030) or 4 μL of anti-H3K4me3 antibody (Millipore, #07-473) overnight at 4 °C with rotation. The lots for each antibody used were validated as specific for each respective histone modification by the company. 30 μL of Dynal Protein G beads were added to each ChIP and incubated at 4 °C with rotation for 2 h. Beads were washed three times in low salt wash buffer and then 2 times with high salt wash buffer (0.1% SDS, 1.0% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl (pH 8.1), 500 mM NaCl) with 5 min of rotation at 4 °C for each wash. Beads were resuspended in 150 μL of elution buffer (1% SDS, 0.1M NaHCO3) and incubated in a thermomixer at 65 °C for 30 minutes at 1,200 rpm to reverse cross-linking. 2 μL of Proteinase K (Invitrogen, #AM2546) was added to each sample, and 6 μL of 5 M NaCl was added for a total concentration of 200 mM. Samples were then incubated in a thermomixer at 65 °C overnight at 1,200 rpm. Eluted samples were removed from the beads and purified using the Qiaquick PCR purification kit (QIAGEN, #28104) per the manufacturer's instructions (www.qiagen.com).
ChIP-Seq analysis
Because there was no precedent for sample size for ChIP-Seq differential binding analysis at the beginning of this project, statistical methods did not inform the number of donors we used for analysis. However, we found that the 4 donors we used gave us ample statistical significance for our ChIP-Seq analysis as demonstrated in the P-value plots in our results (Figure 1A-B).
Reads were aligned to hg19 using bowtie2.50 Sample normalization factors adjusting for sequencing depth and compositional bias for each histone mark were determined by un-weighted Trimmed Mean of M-values (TMM) on 10 kb bin read counts as described in.51 These normalization factors were used in all differential binding analyses described below. For each condition, peaks were called independently for each of the four donors using the MACS peak caller52 on the aligned reads. Then reproducible consensus peaks were determined using the Irreproducible Discovery Rate framework with a threshold of 0.02.53 The same process was repeated with all aligned reads from all conditions to obtain a single set of condition-independent consensus peaks for cross-condition comparison using an IDR threshold of 0.01 because the larger set of data allowed a more stringent threshold. This whole procedure was repeated for H3K4me2 and H3K4me3 samples to obtain condition-specific and condition-independent peak sets for all three histone marks. For each histone mark, reads whose 5-prime mapping location overlapped each condition-independent consensus peak were counted for each sample. Counts were analyzed for differential binding between conditions using edgeR's quasi-likelihood F-test,54 with a model including the condition as the main effect and donor as a batch effect. P-values were adjusted for multiple testing by computing False Discovery Rate (FDR) using the method of Benjamini & Hochberg.55
In order to determine the effective promoter radius for each histone mark, the distance from each unique TSS annotated in UCSC genes to the distance to the nearest peak was determined for each condition's consensus peak set. The distribution of these distances was plotted. It was assumed that these histone marks would be uniformly distributed throughout most of the genome, but enriched in promoters. Visual inspection revealed, for all histone marks and conditions, a peak at small distances and flat background level at larger distances, consistent with the assumption of enrichment of peaks near promoters. For both H3K4me2 and H3K4me3, the distribution flattens out to the background level near 1kb. These distances were used as the effective promoter radius when defining the promoter region associated with each transcriptional start site (TSS). The promoter region of each annotated transcript was defined by extending a region upstream and downstream from the TSS by the determined radius. Overlapping promoters from the same gene were merged into one. The number of reads overlapping each promoter in each sample was counted. Promoter counts were analyzed using the same model as for the peak analysis. As a negative control, the ChIP-Seq input samples were also analyzed in the same way to verify that the differential binding test did not give false positive results. For each IP type, principal coordinates analysis (PCoA) was performed using the plotMDS function in edgeR, and the samples were plotted in the first four principal coordinates.24
Because dispersions were found to vary with time point, each test for differential binding between conditions was conducted using dispersions estimated from only the samples from time points associated with the conditions being tested, which results in a conservative test according to the edgeR author (personal communication). For example, for the comparison memory resting vs. memory 5 days, all samples from rest and 5 days were used for estimating dispersions. Results for differential binding were filtered with an FDR cut-off of < 0.1 and a fold enrichment cut-off of log2 fold change > 1 or < −1.
RNA-Seq analysis
RNA-Seq reads were aligned to the UCSC hg19 transcriptome and genome using Tophat 2.56 The number of reads aligning unambiguously to each gene in each sample was computed. Genes without at least 5 reads assigned in at least one sample were considered not detected and were discarded. Normalization factors were computed in nonstandard fashion in order to mitigate batch effects since two time points (resting and 5 days) were prepped and sequenced separately from the other two (1 day and 2 wk). First, ordinary normalization factors were computed using TMM, and an intercept-only model was fit to the data using these normalization factors. The genes were split into 100 bins by average CPM, and the 20 genes with the lowest estimated biological coefficients of variation in that bin were selected, yielding a total of 2000 low variability genes distributed across the full range of expression values (approximately 10% of all expressed genes). TMM normalization factors were then computed using only these genes, and the resulting factors had a much smaller correlation with the two batches. These normalization factors were used to normalize the full dataset for all further differential expression analysis and quantification. Gene counts were analyzed for differential expression using the same model as for the peak analysis. Gene expression levels for each sample were quantified as FPKM (fragments per kilobase per million fragments sequenced), using the length of the longest transcript isoform for each gene. Batch-corrected average gene expression levels for each condition were quantified by back-transforming the fitted model coefficients for each condition onto a raw count scale and then normalizing to FPKM as for the sample counts. Cut-offs imposed for differential expression analysis included an FDR of < 0.05 and log2 fold change > 1 or < −1.
ChIP-Seq and RNA-Seq combined analysis
Tests were performed for correlation between presence of a ChIP-Seq peak at a given experimental condition and either RNA-Seq expression level (FPKM values) at a given experimental condition or expression log2 fold change between two conditions, for all genes in the genome. First, genes were partitioned by promoter peak presence or absence, and then a Kolmogorov-Smirnoff test (a non-parametric test for distributional differences) was performed to test for significant differences in the RNA-Seq statistic of interest between the partitions, and 95% confidence intervals for the difference in means were constructed (based on an assumption of a normal distribution). The raw data for both ChIP-Seq and RNA-Seq are available at the NIH Gene Expression Omnibus site (accession # GSE73214).
Code availability
The codes used for ChIP-Seq and RNA-Seq analysis can be accessed at https://github.com/DarwinAwardWinner/cd4-histone-paper-code.
Pathway analysis
Pathway mapping used several different tools: ImmuneMap (developed in our laboratory), as well as a Signaling Pathway Impct Analysis (SPIA) tool-based workflow57 to map multiple databases (Panther, KEGG, Biocarta, Reactome, and the National Cancer Institute Pathway Interaction Database (NCI)). We also used Ingenuity Pathway Analysis (IPA, QIAGEN, www.qiagen.com/ingenuity). The premise for this approach is that each tool has its values and no single tool available today for mapping pathways is sufficiently comprehensive. Functional pathway mapping included p-values and log fold changes for differential enrichment or differential expression.
Supplementary Material
Acknowledgments
This research was supported by funds from the National Institutes of Health: U19 AI063603 (DRS), 1TL1 TR001113-01 (SAL), T32 DK007022-30, Postdoctoral Juvenile Diabetes Research Foundation fellowship (HKK), and the Verna Harrah Research Funds supporting the Salomon laboratory.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.McKinstry KK, Strutt TM, Swain SL. The potential of CD4 T-cell memory. Immunology. 2010;130(1):1–9. doi: 10.1111/j.1365-2567.2010.03259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rothbart SB, Strahl BD. Interpreting the language of histone and DNA modifications. Biochim Biophys Acta. 2014;1839(8):627–43. doi: 10.1016/j.bbagrm.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet. 2011;12(1):7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 4.Lessard JA, Crabtree GR. Chromatin regulatory mechanisms in pluripotency. Annu Rev Cell Dev Biol. 2010;26:503–32. doi: 10.1146/annurev-cellbio-051809-102012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tee WW, Reinberg D. Chromatin features and the epigenetic regulation of pluripotency states in ESCs. Development. 2014;141(12):2376–90. doi: 10.1242/dev.096982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vastenhouw NL, Schier AF. Bivalent histone modifications in early embryogenesis. Curr Opin Cell Biol. 2012;24(3):374–86. doi: 10.1016/j.ceb.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neff T, Armstrong SA. Chromatin maps, histone modifications and leukemia. Leukemia. 2009;23(7):1243–51. doi: 10.1038/leu.2009.40. [DOI] [PubMed] [Google Scholar]
- 8.Burney MJ, Johnston C, Wong KY, Teng SW, Beglopoulos V, Stanton LW, et al. An epigenetic signature of developmental potential in neural stem cells and early neurons. Stem Cells. 2013;31(9):1868–80. doi: 10.1002/stem.1431. [DOI] [PubMed] [Google Scholar]
- 9.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 10.Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30(1):155–67. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Russ BE, Olshanksy M, Smallwood HS, Li J, Denton AE, Prier JE, et al. Distinct epigenetic signatures delineate transcriptional programs during virus-specific CD8(+) T cell differentiation. Immunity. 2014;41(5):853–65. doi: 10.1016/j.immuni.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, et al. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419(6905):407–11. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 13.Gu B, Lee MG. Histone H3 lysine 4 methyltransferases and demethylases in self-renewal and differentiation of stem cells. Cell Biosci. 2013;3(1):39. doi: 10.1186/2045-3701-3-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wozniak GG, Strahl BD. Hitting the 'mark': interpreting lysine methylation in the context of active transcription. Biochim Biophys Acta. 2014;1839(12):1353–61. doi: 10.1016/j.bbagrm.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 15.Clouaire T, Webb S, Skene P, Illingworth R, Kerr A, Andrews R, et al. Cfp1 integrates both CpG content and gene activity for accurate H3K4me3 deposition in embryonic stem cells. Genes Dev. 2012;26(15):1714–28. doi: 10.1101/gad.194209.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, et al. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131(1):58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 17.van Nuland R, Schram AW, van Schaik FM, Jansen PW, Vermeulen M, Marc Timmers HT. Multivalent engagement of TFIID to nucleosomes. PLoS One. 2013;8(9):e73495. doi: 10.1371/journal.pone.0073495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orford K, Kharchenko P, Lai W, Dao MC, Worhunsky DJ, Ferro A, et al. Differential H3K4 methylation identifies developmentally poised hematopoietic genes. Dev Cell. 2008;14(5):798–809. doi: 10.1016/j.devcel.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40(7):897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang JA, Mortazavi A, Williams BA, Wold BJ, Rothenberg EV. Dynamic transformations of genome-wide epigenetic marking and transcriptional control establish T cell identity. Cell. 2012;149(2):467–82. doi: 10.1016/j.cell.2012.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lim PS, Hardy K, Bunting KL, Ma L, Peng K, Chen X, et al. Defining the chromatin signature of inducible genes in T cells. Genome Biol. 2009;10(10):R107. doi: 10.1186/gb-2009-10-10-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barski A, Jothi R, Cuddapah S, Cui K, Roh TY, Schones DE, et al. Chromatin poises miRNA- and protein-coding genes for expression. Genome Res. 2009;19(10):1742–51. doi: 10.1101/gr.090951.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Allan RS, Zueva E, Cammas F, Schreiber HA, Masson V, Belz GT, et al. An epigenetic silencing pathway controlling T helper 2 cell lineage commitment. Nature. 2012;487(7406):249–53. doi: 10.1038/nature11173. [DOI] [PubMed] [Google Scholar]
- 24.Gower JC. Some Distance Properties of Latent Root and Vector Methods Used in Multivariate Analysis. Biometrika. 1966;53(3):325–338. [Google Scholar]
- 25.Li Y, Chen G, Ma L, Ohms SJ, Sun C, Shannon MF, et al. Plasticity of DNA methylation in mouse T cell activation and differentiation. BMC Mol Biol. 2012;13:16. doi: 10.1186/1471-2199-13-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee CG, Hwang W, Maeng KE, Kwon HK, So JS, Sahoo A, et al. IRF4 regulates IL-10 gene expression in CD4(+) T cells through differential nuclear translocation. Cell Immunol. 2011;268(2):97–104. doi: 10.1016/j.cellimm.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 27.Staal FJ, Luis TC, Tiemessen MM. WNT signalling in the immune system: WNT is spreading its wings. Nat Rev Immunol. 2008;8(8):581–93. doi: 10.1038/nri2360. [DOI] [PubMed] [Google Scholar]
- 28.Turner MD, Nedjai B, Hurst T, Pennington DJ. Cytokines and chemokines: At the crossroads of cell signalling and inflammatory disease. Biochim Biophys Acta. 2014;1843(11):2563–2582. doi: 10.1016/j.bbamcr.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 29.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448(7153):553–60. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kimura H. Histone modifications for human epigenome analysis. J Hum Genet. 2013;58(7):439–45. doi: 10.1038/jhg.2013.66. [DOI] [PubMed] [Google Scholar]
- 31.Soler D, Chapman TR, Poisson LR, Wang L, Cote-Sierra J, Ryan M, et al. CCR8 expression identifies CD4 memory T cells enriched for FOXP3+ regulatory and Th2 effector lymphocytes. J Immunol. 2006;177(10):6940–51. doi: 10.4049/jimmunol.177.10.6940. [DOI] [PubMed] [Google Scholar]
- 32.Meng H, Cao Y, Qin J, Song X, Zhang Q, Shi Y, et al. DNA Methylation, Its Mediators and Genome Integrity. Int J Biol Sci. 2015;11(5):604–617. doi: 10.7150/ijbs.11218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo H, Zhu P, Yan L, Li R, Hu B, Lian Y, et al. The DNA methylation landscape of human early embryos. Nature. 2014;511(7511):606–10. doi: 10.1038/nature13544. [DOI] [PubMed] [Google Scholar]
- 34.Pan G, Tian S, Nie J, Yang C, Ruotti V, Wei H, et al. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell. 2007;1(3):299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 35.Zhao XD, Han X, Chew JL, Liu J, Chiu KP, Choo A, et al. Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell. 2007;1(3):286–98. doi: 10.1016/j.stem.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 36.Pekowska A, Benoukraf T, Ferrier P, Spicuglia S. A unique H3K4me2 profile marks tissue-specific gene regulation. Genome Res. 2010;20(11):1493–502. doi: 10.1101/gr.109389.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Young MD, Willson TA, Wakefield MJ, Trounson E, Hilton DJ, Blewitt ME, et al. ChIP-seq analysis reveals distinct H3K27me3 profiles that correlate with transcriptional activity. Nucleic Acids Res. 2011;39(17):7415–27. doi: 10.1093/nar/gkr416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chakraborty AK, Weiss A. Insights into the initiation of TCR signaling. Nat Immunol. 2014;15(9):798–807. doi: 10.1038/ni.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y, Li X, Hu H. H3K4me2 reliably defines transcription factor binding regions in different cells. Genomics. 2014;103(2-3):222–8. doi: 10.1016/j.ygeno.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 40.Rotem A, Ram O, Shoresh N, Sperling RA, Goren A, Weitz DA, et al. Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nat Biotechnol. 2015;33(11):1165–72. doi: 10.1038/nbt.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roadmap Epigenomics C, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518(7539):317–30. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Das S, Das P, Mitra S, Dasgupta M, Chakrabarti J, Larsson E. Epigenetic transfiguration of H3K4me2 to H3K4me3 during differentiation of embryonic stem cell into non-embyronic cells. RNA and Transcription. 2015;1(3):18–33. [Google Scholar]
- 43.Gerondakis S, Banerjee A, Grigoriadis G, Vasanthakumar A, Gugasyan R, Sidwell T, et al. NF-kappaB subunit specificity in hemopoiesis. Immunol Rev. 2012;246(1):272–85. doi: 10.1111/j.1600-065X.2011.01090.x. [DOI] [PubMed] [Google Scholar]
- 44.Komori HK, Hart T, LaMere SA, Chew PV, Salomon DR. Defining CD4 T cell memory by the epigenetic landscape of CpG DNA methylation. J Immunol. 2015;194(4):1565–79. doi: 10.4049/jimmunol.1401162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hashimoto H, Vertino PM, Cheng X. Molecular coupling of DNA methylation and histone methylation. Epigenomics. 2010;2(5):657–69. doi: 10.2217/epi.10.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vahedi G TH, Nakayamada S, Sun H, Sartroelli V, Kanno Y, O'Shea JJ. STATs Shape the Active Enhancer Landscape of T Cell Populations. Cell. 2013;151(5):981–993. doi: 10.1016/j.cell.2012.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zentner GE, Henikoff S. Regulation of nucleosome dynamics by histone modifications. Nat Struct Mol Biol. 2013;20(3):259–66. doi: 10.1038/nsmb.2470. [DOI] [PubMed] [Google Scholar]
- 48.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21(3):381–95. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Head SR, Komori HK, Hart GT, Shimashita J, Schaffer L, Salomon DR, et al. Method for improved Illumina sequencing library preparation using NuGEN Ovation RNA-Seq System. Biotechniques. 2011;50(3):177–80. doi: 10.2144/000113613. [DOI] [PubMed] [Google Scholar]
- 50.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature methods. 2012;9(4):357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lun AT, Smyth GK. De novo detection of differentially bound regions for ChIP-seq data using peaks and windows: controlling error rates correctly. Nucleic Acids Res. 2014;42(11):e95. doi: 10.1093/nar/gku351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Feng J, Liu T, Zhang Y. Baxevanis Andreas D., editor. Using MACS to identify peaks from ChIP-Seq data. Current protocols in bioinformatics. 2011 doi: 10.1002/0471250953.bi0214s34. Chapter 2: Unit 2 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li QB, James B. Huang Haiyan; Bickel, Peter J. Measuring reproducibility of high-throughput experiments. The Annals of Applied Statistics. 2011;5(3):1752–1779. [Google Scholar]
- 54.Lund SP, Nettleton D, McCarthy DJ, Smyth GK. Detecting differential expression in RNA-sequence data using quasi-likelihood with shrunken dispersion estimates. Stat Appl Genet Mol Biol. 2012;11(5) doi: 10.1515/1544-6115.1826. [DOI] [PubMed] [Google Scholar]
- 55.Benjamini YH, Yosef Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995;57(1):289–300. [Google Scholar]
- 56.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tarca AL, Draghici S, Khatri P, Hassan SS, Mittal P, Kim JS, et al. A novel signaling pathway impact analysis. Bioinformatics. 2009;25(1):75–82. doi: 10.1093/bioinformatics/btn577. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.