Abstract
Improved diagnostic methods are needed for bronchiolitis obliterans syndrome (BOS), a serious complication after allogeneic hematopoietic cell transplantation (HCT) and lung transplantation. For proteins candidate discovery, we compared plasma pools from HCT transplantation recipients with: BOS at onset (n=12), pulmonary infection (n=16), chronic graft-versus-host disease without pulmonary involvement (n=15), and no chronic complications post-HCT (n=15). Pools were labeled with different tags [isobaric Tags for Relative and Absolute Quantification (iTRAQ)], and two software tools identified differentially expressed proteins (≥1.5-fold change). Candidate proteins were further selected using a six-step computational biology approach. The diagnostic value of the lead candidate, matrix metalloproteinase-3 (MMP-3), was evaluated by ELISA in plasma of a verification cohort (n=112) with and without BOS following HCT (n=76) or lung transplantation (n=36). MMP-3 plasma concentrations differed significantly between patients with and without BOS (AUC=0.77). Thus, MMP-3 represents a potential non-invasive blood test for diagnosis of BOS.
1. Introduction
Bronchiolitis obliterans syndrome (BOS) occurs following allogeneic hematopoietic cell transplantation (HCT) when a transplant donor’s immune system attacks lung tissue, ultimately leading to fibrosis of small airways and symptoms of respiratory insufficiency. BOS is a manifestation of pulmonary chronic graft-versus-host disease (cGVHD) and is associated with high morbidity and mortality (1–6). A similar syndrome occurs in lung transplantation recipients. Studies in human lung transplantation recipients and animal models suggest that BOS is initiated by immune recognition of foreign antigens in the lung epithelia, leading to inflammation and fibroproliferation within the small airways (bronchioles), which further results in air trapping and pulmonary insufficiency (1, 7–13). Although little is known about the precise mechanisms underlying the initiation or progression of BOS, perturbations in cellular, cytokine, and protein profiles have been documented in lung tissue and bronchoalveolar lavage (BAL) fluid of lung transplantation recipients, supporting the model of an aberrant immune response (14–22). BOS diagnosis in allogeneic HCT recipients corresponds to a poor prognosis due to a high infection risk and pulmonary failure, with less than 20% survival at 5 years (2). For patients who do survive, progressive lung disease is often accompanied by impaired quality of life, decreased endurance, compromised ability to perform daily activities, and eventual oxygen dependence.
Currently, BOS diagnosis continues to be based on pulmonary function test (PFT) parameters reflecting the obstructive lung defect, particularly a decline in forced expiratory volume in 1 second (FEV1). For HCT recipients, the 2014 National Institutes of Health (NIH) consensus project on diagnostic criteria for BOS were modified from those of the 2005 NIH consensus to increase the specificity for obstructive lung defects and attempt to identify BOS earlier in its course (23). For lung transplantation recipients, similar to HCT recipients, a decline in FEV1 is considered consistent with the onset of BOS and may be confirmed by a lung biopsy showing pulmonary fibrosis (7). However, PFTs have shown limited value for BOS diagnosis by identifying only the most severe cases (7, 23, 24). Therefore, plasma proteins correlated with BOS would be extremely valuable to allow early diagnosis, guide treatment choices, and monitor responses.
Herein, we used a mass spectrometry-based approach as a discovery engine to unambiguously identify candidate plasma proteins associated with BOS in a discovery cohort of 58 HCT recipients. We subsequently used computational biology to characterize a list of high-priority protein candidates. Finally, the diagnostic value of the lead candidate, matrix metalloproteinase-3 (MMP-3, also called stromelysin-1), was evaluated by enzyme-linked immunosorbent assay (ELISA) in the plasma of a verification cohort of 112 patients with and without BOS following HCT or lung transplantation.
2. Methods
2.1. Patients and samples
Three cohorts of patients were included in this study (BOS discovery, BOS verification, and cGVHD post-HCT verification). Patients were treated at the University of Michigan, the Fred Hutchinson Cancer Research Center, and the Indiana University School of Medicine. All patients or their legal guardians provided written informed consent, and the collection of samples for studying post-HCT or post-lung transplantation complications was approved by the institutional review boards of the University of Michigan, the Fred Hutchinson Cancer Research Center, and Indiana University School of Medicine. Heparinized blood samples were collected prospectively, either prior or at the time of the onset of symptoms consistent with BOS, and at matched time points post-HCT or lung transplantation in patients who did not develop BOS. Samples were aliquoted without additives into cryovials and stored at −80°C. BOS diagnosis was established after extensive testing for respiratory infections and PFT criteria for BOS established by the HCT and lung transplantation consensus (7, 23).
2.2. Proteomics analysis
The proteomics methods are detailed in the Supplemental Methods in the Supporting Information.
2.3. Six-step computational biology approach
The six-step computational biology approach is detailed in Supplemental Methods in the Supporting Information.
2.4. ELISAs
MMP-3 antibody pairs were purchased from Bio-techne, and an FN1 ELISA kit from eBioscience. Samples and standards were analyzed in duplicate according to the manufacturer’s protocols. Briefly, capture antibody was coated in 96-well plates (Corning) overnight at room temperature (RT). Standards were prepared and plasma samples diluted (1:25) and added for 2 hours at RT while mixing on an orbital shaker. After washing, biotinylated antibody was added and incubated for 2 hours. Plates were washed and streptavidin-conjugated horseradish peroxidase (SA-HRP) was diluted as recommended and added for 30 minutes. The TMB color substrate was prepared and incubated for 6 minutes. The reaction was terminated by addition of 1.5 M sulfuric acid, and the absorbance immediately determined using a SpectraMax 384plus plate reader (Molecular Devices).
2.5. Statistical analysis
Differences in characteristics between patient groups were assessed with Student t tests for continuous variables and with χ2 tests for categorical variables. Logarithmic (Log2) transformed protein values were used in all analyses because of the skewness of the raw values. Median protein concentrations from individual samples in the verification cohort were compared using the Wilcoxon-Mann-Whitney test. p values were not corrected for multiple comparisons in a priori analyses. AUCs were estimated nonparametrically.
3. Results
3.1. Proteomics discovery
We first performed a discovery proteomics analysis comparing four pools of plasma in the same proteomics experiment. Pool 1 contained plasma from 12 HCT recipients who met BOS criteria and with plasma samples available at the onset. Pool 2 contained plasma from 16 HCT recipients with pulmonary infections who never had BOS during their course. Pool 3 contained plasma from 15 HCT recipients with cGVHD without pulmonary involvement (cGVHD no BOS), and pool 4 contained plasma from 15 HCT recipients with no chronic complications (plasma samples collected at a similar time point post-HCT as BOS samples). Each pool contained 25 µl plasma and was labeled with a unique tag [isobaric Tags for Relative and Absolute Quantification (iTRAQ)] allowing for differential quantification. Plasma pools were compared between patients with BOS versus the other three conditions. The clinical characteristics of patients in this discovery cohort are provided in Table S1.
The acquired liquid chromatography-tandem mass spectrometry (LC-MS/MS) data were analyzed using two approaches and two search engines, Proteome Discoverer™ version 1.4 (Thermo Scientific) and Mascot™ version 2.4 (Matrix Science) (25). In the Proteome Discoverer analysis, a total of 857 proteins were identified with at least one peptide after filtering using a stringent 1% peptide-level false discovery rate (FDR; see methods), and of these, the levels of 855 proteins (Table S2) were quantified. Of these proteins, 21 were upregulated more than 1.5-fold, and 222 proteins were downregulated more than 33% in the BOS pool (pool 1) compared with the pool without chronic complications (pool 4). In the Mascot™ analysis, a total of 682 proteins were identified after filtering with a significance threshold of p<0.01 (see methods). Using the weighted protein abundance ratio in Mascot™, the protein abundance ratios were obtained for 462 (Table S3) of the 682 proteins. Of these proteins, 62 were upregulated more than 1.5-fold, and 65 were downregulated more than 33% in the BOS pool (pool 1) compared with the pool without chronic complications (pool 4). A total of 422 proteins were identified by both approaches, and of these, 5 were upregulated more than 1.5-fold and 52 were downregulated more than 33%.
3.2. Selection of lead candidates from our computational biology approach
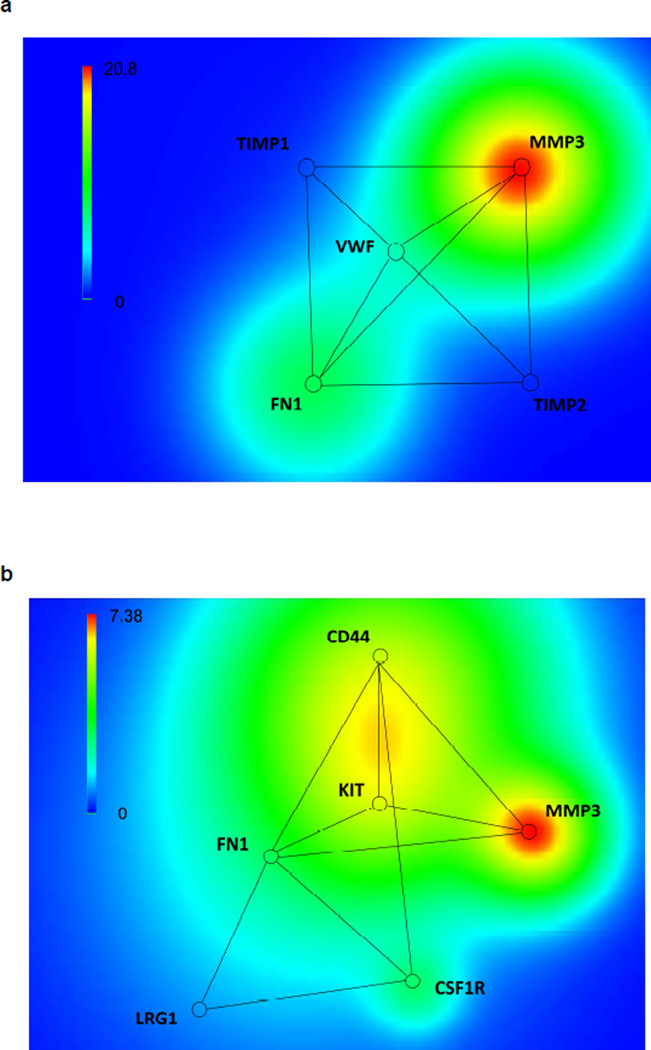
We next applied a computational biology approach to characterize and screen BOS-specific proteins from the two mass-spectrometry identification results. The original number of proteins identified in the Proteome Discoverer™ analysis was 857 (mapped to 822 genes). After six steps of system biology data processing, five BOS-specific proteins were characterized (Table 1a and Tables S4, S6) from formed protein–protein interaction network module (Figure 1a). The original number of proteins identified in the Mascot™ experiment was 462 (mapped to 436 genes). After six steps of computational biology data processing, six BOS-specific proteins were characterized (Table 1b and Tables S5, S7) from formed protein–protein interaction network module (Figure 1b). The discrepancy between the two characterized network modules is due to the complexity of the mass spectrometry techniques and the different protein identification and quantification methods implemented in the two mass spectrometry data analysis software tools (see Discussion). However, two proteins, fibronectin 1 (FN1) and MMP-3, were consistently identified by both approaches, making them potential BOS-specific proteins.
Table 1 .
| a. List of 5 BOS-specific biomarker candidates
selected via six-step computational biology approach from the
Proteome Discoverer™ analysis | ||||||
|---|---|---|---|---|---|---|
| Protein | Description | Gene symbol | Relevant Score(Local) | BOS/cGVHD | Penalty score | Final score |
| MMP3 | Stromelysin-1 | MMP3 | 1.79 | 7.25 | 1.00 | 11.17 |
| FINC | Fibronectin | FN1 | 1.73 | 7.41 | 7.14 | 1.56 |
| VWF | von Willebrand factor | VWF | 1.74 | 2.90 | 7.35 | 0.45 |
| TIMP1 | Metalloproteinase inhibitor 1 | TIMP1 | 2.79 | 0.77 | 32.00 | 0.03 |
| TIMP2 | Metalloproteinase inhibitor 2 | TIMP2 | 0.98 | 0.80 | 28.80 | 0.01 |
| b. List of 6 BOS-specific biomarker candidates selected via six-step computational biology approach from the Mascot™ analysis | ||||||
|---|---|---|---|---|---|---|
| Protein | Description | Gene symbol | Relevant Score(local) | BOS/cGVHD | Penalty score | Final score |
| P08254 | Stromelysin-1 | MMP3 | 0.79 | 7.41 | 1.00 | 5.82 |
| P16070 | CD44 antigen | CD44 | 5.42 | 6.45 | 1.68 | 5.55 |
| P07333 | Macrophage colony-stimulating factor 1 receptor | CSF1R | 2.99 | 0.58 | 2.18 | 1.73 |
| P02751 | Fibronectin 1 | FN1 | 1.79 | 0.67 | 7.14 | 1.61 |
| P10721 | Mast/stem cell growth factor receptor Kit | KIT | 1.89 | 0.79 | 1.87 | 1.51 |
| P02750 | Leucine-rich alpha-2-glycoprotein | LRG1 | 0.36 | 0.82 | 24.00 | 0.02 |
Relevant Score(local): relevance score which reflects the importance of the protein in the network with the 5 candidates using the protein–protein interaction pairs confidence score.
Relevant Score(local): relevance score which reflects the importance of the protein in the network with the 6 candidates using the protein–protein interaction pairs confidence score.
Figure 1. a, Biomarker selection via gene terrain visualization tools shown in a 2D panel obtained with the Proteome Discoverer™ proteomics analysis; b, biomarker selection via gene terrain visualization tools shown in a 2D panel obtained with the Mascot™ proteomics analysis.
MMP-3: matrix metalloproteinase-3; FN1: fibronectin; VWF: von Willebrand factor; TIMP1: Metalloproteinase inhibitor 1; TIMP2: Metalloproteinase inhibitor 2; CD44: CD44 antigen; CSF1R: Macrophage colony-stimulating factor 1 receptor; KIT: Mast/stem cell growth factor receptor Kit; LRG1: Leucine-rich alpha-2-glycoprotein.
3.3. Validation of MMP-3 by ELISA in a verification cohort
We then evaluated the diagnostic potential of the candidate proteins for BOS diagnosis in a verification cohort of 112 transplantation recipients. The clinical characteristics of patients in this verification cohort combining HCT and lung transplantation are described in Table 2. We categorized plasma samples of the verification cohort into two different groups according to the presence or absence of BOS diagnosis: patients after either HCT or lung transplantation presenting with BOS (n=40) and patients without BOS (n=72). Plasma from healthy donors (n=20) was used as the reference. In patients without BOS, samples were obtained at equivalent time points after HCT as from patients with BOS. Recipients of grafts from mobilized peripheral blood (PB) or from donors who were not family members were overrepresented in the BOS group (26). As previously reported (27), patients who developed prior acute GVHD were more likely to develop cGVHD and BOS, but this was not different from patients with cGVHD and no BOS (Table S8).
Table 2.
Characteristics of patients in the verification cohort
| HCT patients | Lung transplanted patients | ||||||
|---|---|---|---|---|---|---|---|
| BOS (n=31) | No BOS (n=45) | p | BOS (n=10) | No BOS (n=26) | p | ||
| Age, years | |||||||
| median | 55 | 54 | ns | 63.5 | 57 | ns | |
| range | (19–73) | (22–72) | (48–70) | (32–66) | |||
| Diagnosis | |||||||
| hematological malignancies | 30 | 45 | - | - | |||
| nonmalignant hematological diseases | 1 | 0 | - | - | |||
| COPD | - | - | 6 | 14 | |||
| IPF | - | - | 3 | 6 | |||
| cystic fibrosis | - | - | 0 | 2 | |||
| primary pulmonary hypertension | - | - | 0 | 1 | |||
| sarcoidosis | - | - | 1 | 0 | |||
| alpha-1 antitrypsin | - | - | 0 | 1 | |||
| scleroderma | - | - | 0 | 2 | |||
| Type of transplantation | |||||||
| HCT with PB | 30 | 35 | 0.02 | - | - | ||
| HCT with BM | 1 | 10 | - | - | |||
| HCT with CB | 0 | 0 | - | - | |||
| single lung transplantation | - | - | 2 | 3 | ns | ||
| bilateral lung transplantation | - | - | 8 | 23 | |||
| HLA match | |||||||
| 6 out of 6 | 20 | 36 | ns | - | - | ||
| < 6 | 11 | 9 | - | - | |||
| HLA-A match ≥ 1 | - - |
- - |
4 | 12 | ns | ||
| HLA-B match ≥ 1 | 7 | 13 | |||||
| HLA-DR match ≥ 1 | - | - | 5 | 6 | |||
| Donor type | |||||||
| related donor | 4 | 30 | <0.0001 | - | - | ns | |
| unrelated donor | 27 | 15 | 10 | 26 | |||
| Conditioning regimen | |||||||
| reduced intensity | 15 | 21 | ns | - | - | ||
| full intensity | 16 | 24 | - | - | |||
| Time post-transplant to BOS diagnosis, days | |||||||
| median | 531 | - | 409 | - | |||
| range | (97–1260) | - | (199–841) | - | |||
| Time post-transplant to sample acquisition, days | |||||||
| median | 550 | 369 | ns | 347 | 373 | ns | |
| range | (89–1233) | (80–3641) | (173–813) | (133–772) | |||
| FEV1 at diagnosis of BOS | |||||||
| 60–75% | 16 | - | 9 | - | |||
| 40–59% | 13 | - | 1 | - | |||
| ≤39% | 2 | - | 0 | - | |||
| Prior acute GVHD 2–4 or concomittant acute rejection | |||||||
| yes | 26 | 27 | 0.03 | 4 | 4 | ns | |
| no | 5 | 18 | 6 | 22 | |||
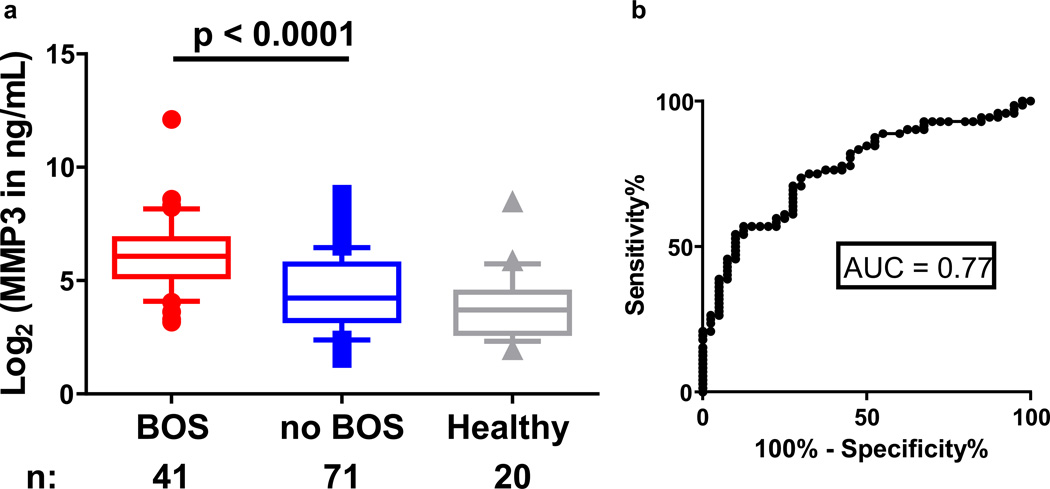
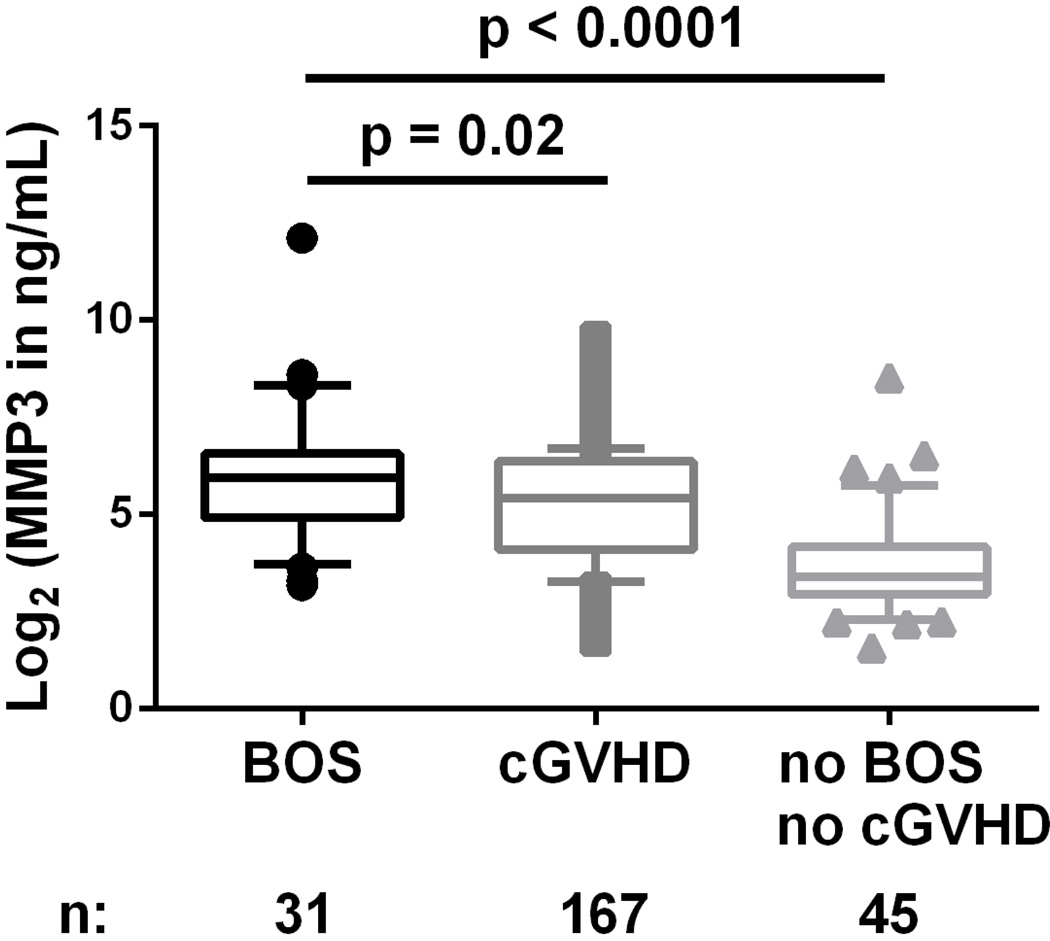
We first analyzed plasma concentrations of both FN1 and MMP3 in a pilot study of 17 BOS cases versus 13 controls without BOS. FN1 was not significantly different between the two groups in the pilot study (Figure S1), and thus, further analysis focused on MMP3. Plasma MMP-3 concentrations were significantly higher in patients with BOS than in patients without BOS and healthy donors (p<0.0001; Figure 2A). We next generated a receiver operating characteristic (ROC) curve, which represents the false and true positive rates for every possible level of MMP3, comparing the patients with and without BOS, and found that the area under the ROC curve (AUC) was 0.77 (95% confidence interval: 0.68–0.86; Figure 2B). These data show that MMP-3 is a sensitive and specific discriminator for the diagnosis of BOS in HCT and lung transplantation recipients who exhibit a decline in FEV1. Because it was also important to show that MMP-3 is a marker of BOS specifically versus cGVHD, we compared MMP-3 plasma concentrations in patients with BOS post-HCT (n=31) versus patients with cGVHD but without lung involvement (n=167). The clinical characteristics of patients in this cohort of only HCT recipients are described in Table S2. MMP-3 concentrations were significantly higher in patients with BOS than in patients with cGVHD without BOS (p=0.02; Figure 3) and in HCT patients without cGVHD (p<0.0001; Figure 3). Of note, our study was not powered to compare concentrations in patients with versus without BOS in the lung transplantation subgroup.
Figure 2. a, MMP-3 plasma concentrations measured by ELISA in the BOS verification cohort; b, ROC curve comparing patients with and without BOS post-transplantation.
a. Individual log2 transformed MMP-3 in ng/ml are plotted for BOS patients (n = 41), no BOS patients (n = 71), and healthy donors (n = 20). Wilcoxon-Mann-Whitney test comparing the median of BOS vs. no BOS, p<0.0001. b. Area under the ROC = 0.77.
Figure 3. MMP-3 plasma concentrations measured by ELISA in an HCT cohort of patients with cGVHD not of the lung and with BOS post-HCT.
Individual log2 transformed MMP-3 in ng/ml are plotted for BOS patients (n = 31), no BOS with cGVHD patients (n = 167), and no BOS no cGVHD patients (n = 45). Wilcoxon-Mann-Whitney test comparing the median: BOS vs. cGVHD, p=0.02; BOS vs. BOS no cGVHD, p<0.0001.
3.4. MMP-3 plasma concentrations prior to the BOS diagnosis
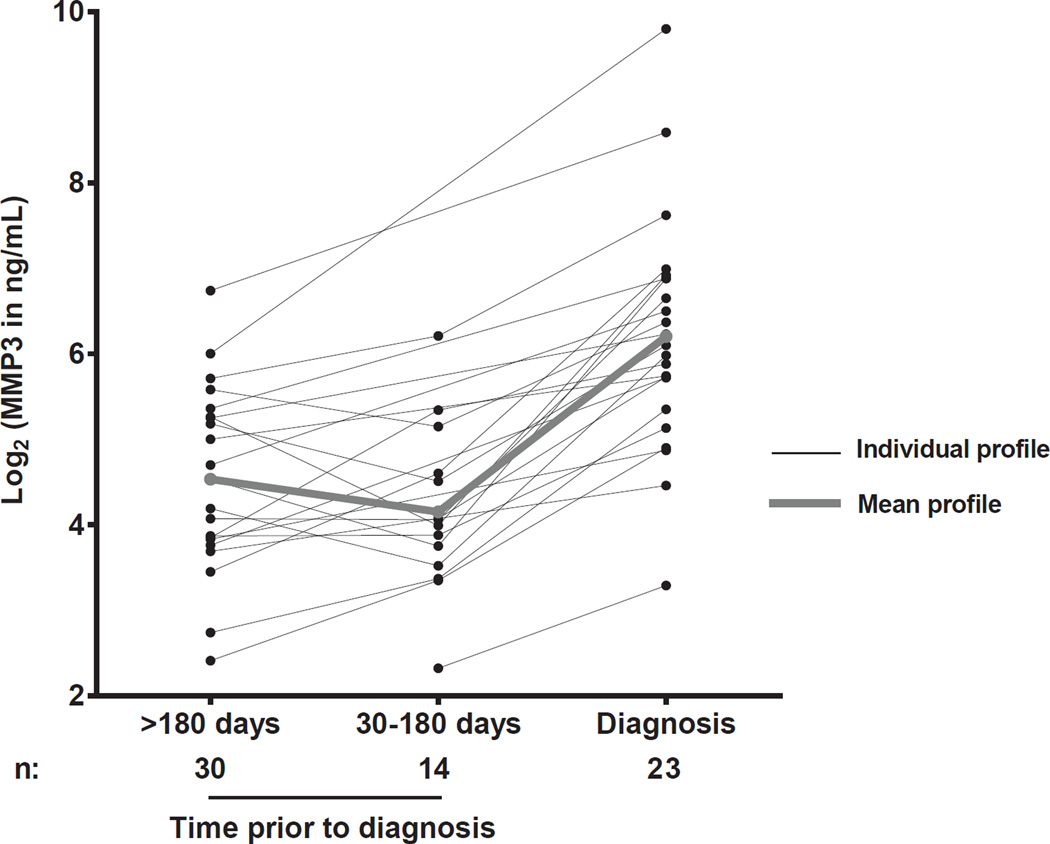
We next measured the plasma concentrations of MMP-3 on timepoints prior BOS diagnosis in 13 HCT and 10 lung transplantation patients with BOS for whom prior onset samples were available, to evaluate MMP-3 plasma concentrations kinetics before the diagnosis of BOS (Figure 4). Measurements in longitudinal samples showed that MMP3 is not elevated in the plasma prior to the onset of BOS as defined by FEV1 criteria, even when the samples were taken closer to the diagnosis (30 to 180 days prior).
Figure 4. MMP-3 plasma concentration kinetics prior to BOS diagnosis in 13 HCT and 10 lung transplantation patients.
Individual log2 transformed MMP-3 in ng/ml are graphed at >180 days, 30 to 180 days prior to and at the BOS diagnosis. The bold line represents the mean of the cohort.
4. Discussion
MMP-3 was identified as a candidate plasma protein specific for BOS after HCT and lung transplantation via an in-depth tandem MS (MS/MS)–based analysis of plasma combined with a multiscale computational biology process: (1) generation of abundance rate from the comparison of plasma samples taken from BOS and cGVHD without lung involvement, (2) generation of protein–protein interactions pairs in the STRING database and selection of the BOS-relevant proteins by the relevance score Rp, (3) computing penalty scores for unspecific proteins found in proteomics experiments for cancer and cardiovascular diseases, (4) pair-wise baseline transformation and comparison of proteins specific for BOS versus pulmonary infection versus cGVHD without BOS, (5) filtering out proteins found in proteomics experiments that are not related to BOS using domain knowledge provided by experts, and (6) visual exploration of the candidate list using a GeneTerrain computational biology visual analytic software tool (28–30). This process enabled us to reduce a list of hundreds of proteins to about a dozen proteins with little human selection bias. Furthermore, we used this approach for two separate proteomics analyses done with different protein search engines and selected proteins reported by both analyses with reliable quantification as potential candidate markers. In the identification of protein–protein interaction modules, it has been challenging to determine which proteins identified from proteomics results represent true positive signals instead of the background as contaminants. One recently developed approach involved generation of the Contaminant Repository for Affinity Purification (the CRAPome) by aggregating negative controls from multiple studies of affinity purification coupled with mass spectrometry (31). By filtering out unspecific proteins using the penalty score, we applied a similar approach that is key to increasing the specific relevance of the proteins identified. With our approach, we found a protein that was not only identified as highly relevant in two separate proteomics analyses but also in a large independent cohort with a high AUC. The technique used for protein detection validation was a sandwich ELISA that is the most relied-on approach for clinical tests because it is highly sensitive and specific due to the extreme affinity of two antibodies recognizing different epitopes from the full length of the protein (32, 33).
In this study, we favored the large scale proteomics approach as compared to the hypothesis-driven approach because 1) an hypothesis-driven approach focusing on MMPs has been performed previously in the context of BOS following lung transplantation (34), and 2) we hypothesize that we could find additional proteins using MS/MS proteomics that will not have been thought of just based on functional hypothesis as we have shown for other models such as acute GVHD markers (i.e. ST2, (35)). The proteomics workflow is used as a discovery tool for potential candidate markers; however, in this ITRAQ experiment, only one protein (MMP3) was validated, which is lower than numbers obtained in our previous IPAS experiments (35–37), in proportion to the number of proteins quantified (approximately 2000 vs. 500, respectively). This suggests that lower sensitivity is achieved with the ITRAQ workflow as compared to the IPAS workflow. The mostly likely explanations for the differences between the ITRAQ workflow vs. the IPAS workflow are: (i) a lower plasma volume was processed (20–50 µl vs. 300 µl), and (ii) single-dimensional fractionation at the tryptic peptide level vs. two-dimensional fractionation only then followed by tryptic digestion. Based on the different biochemical conditions between IPAS and iTRAQ, it is quite possible that some classes/types of proteins could be favored in one method versus the other. Due to the prohibitive cost and labor intensity of multi-dimensional fractionation such as was done in IPAS (480 pools), we do not consider future workflows involving IPAS, and instead we have already taken several steps to improve the sensitivity and quantification of our current MS/MS workflow for future studies that include better depletion of higher abundant proteins, use of Tandem Mass Tag (TMT) labeling, longer LC-MS elution gradient, and optimizing mass spectrometry data collection.
MMPs are prominent contributors to microenvironmental signaling, because these proteolytic enzymes degrade structural components of the extracellular matrix (ECM), permitting tissue remodeling. Additionally, MMPs can release cell-bound inactive precursor forms of growth factors, degrade cell–cell and cell–ECM adhesion molecules, activate precursor zymogen forms of other MMPs, and inactivate inhibitors of MMPs and other proteases (38). MMP-3 in particular has been shown to promote the epithelial–mesenchymal transition (EMT) that can result in tissue fibrosis (39). It is important to note that MMP-3 is not the only protease capable of initiating EMT (39). In our study, MMP-10 (stromelysin-2) was also identified and upregulated in the BOS proteomics experiment but did not pass our six-step computational biology filtering process. In addition, to formally prove that our approach targeted the most biologically relevant proteins, we measured MMP-10 plasma concentration by ELISA and did not observe a difference in MMP-10 levels between patients with and without BOS (data not shown). Elevated plasma levels of MMP-9, and TIMP-1 have been previously associated with BOS after lung transplantation (34), but in our proteomics study, these proteins were identified and found increased in both BOS and cGVHD samples. Thus, they were not selected by our computational biology process. During the acute phase of GVHD, MMPs inhibitors such as TIMP-1 have been found elevated more often than the MMPs per se (40, 41). However, MMPs have been reported to be correlated with inflammation as well, and MMP-9 was shown in a small cohort of HCT patients to be correlated with acute GVHD (42) and that suppression of MMP-9 by a plasmin inhibitor attenuated murine acute GVHD (43). In our knowledge, there is no studies on MMPs correlated with chronic GVHD.
Few other potential biological correlates of BOS following HCT have been proposed using a hypothesis-driven approach such as lung epithelial proteins: Clara cell secretory protein-16 (44) and Serum Krebs Von Den Lungen-6 (45), or BAFF levels and CD19+CD21low B cells (46).
The combination of discovery proteomics analysis and computational biology is an efficient method for reducing a list of thousands of proteins to a short list of candidate proteins that can be validated by ELISA. This method combines various information sources, including protein quantification, protein–protein interaction networks, and domain knowledge from experts, making it a powerful approach for distinguishing proteins related to BOS from those not related. While the method successfully identified candidate proteins in this study, it still has weaknesses. First, the mass spectrometry-based protein identification and quantification analysis is not robust due to the low abundances of candidate proteins and the complexity of data. It is common to identify only one peptide in a candidate protein using bottom-up mass spectrometry because of the low abundance. As a result, “one hit wonders” in protein identification are often reported, which may introduce many false-positive identifications. This was the main reason for the discrepancies between the two protein lists reported by Proteome Discoverer and Mascot. Second, the filtering process heavily relies on domain knowledge of experts, which plays an important role in filtering out proteins that are commonly observed in plasma but are not related to BOS. This reliance on expert domain knowledge makes it difficult to apply this method to diseases that have not been well studied.
Although tested in a relatively small cohort, MMP-3 appears to represent a non-invasive blood test for diagnosis of BOS as defined by FEV1 criteria. MMP-3 plasma concentrations kinetics showed that MMP-3 is not elevated in the plasma prior to the onset of BOS, even when the samples were taken closer to the diagnosis (30 to 180 days prior) suggesting that MMP-3 is augmented only when FEV1 is decreased and thus represent a non-invasive aid for diagnosis rather than a prognostic test. If MMP-3 correlation with BOS is confirmed in future prospective studies and further correlated to risk for poor clinical outcomes, MMP-3 measurement may provide opportunities for future therapeutic intervention to minimize BOS severity or to stabilize the disease. It has recently been shown that Fluticasone, Azithromycin, and Montelukast (FAM) inhalation treatment halted pulmonary decline in new-onset BOS after HCT in the majority of patients and permitted reductions in systemic steroid exposure (47). Indeed, protein levels cutoffs can be used to risk-stratify patients at low- or high-risk for developing BOS. As a next step, a prospective multicenter trial should include intervention and control strategies. High-risk patients are candidates for FAM treatment or additional/different treatment as soon as a diagnosis of BOS is established. In the near future, a targeted therapy should also be available (MMPs inhibitors are currently under development) (48). Low-risk patients could be randomized for a standard intervention versus a novel treatment with lower toxicity or more targeted therapies as proposed above. Plasma proteins measurement following HCT and lung transplantation will enable precision medicine for future clinical trials, just as protein measurement has done for other diseases (49).
Supplementary Material
Acknowledgments
The authors would like to thank the clinicians of all of the institutions who participated in the accrual of samples; the Fred Hutchinson Cancer Research Center data managers, particularly Kate Chilson, for excellent management of the database and biobank; and the members of the Paczesny laboratory. We would like to acknowledge our funding sources including: the National Marrow Donor Program through the Amy Strelzer Manasevit Scholar grant 200513 (to S.P.), NIH R01CA168814 (to S.P.), and NIH R01CA118953 (to S.J.L.), R01HL094260 (to J.A.H), and JSPS KAKENHI 15K19563 (to Y.I.). The Fred Hutch Proteomics Facility is partially funded by Cancer Center Support Grant P30 CA015704 from the National Institutes of Health.
List of abbreviations
- AUC
area under the ROC curve
- BAL
bronchoalveolar lavage
- BOS
bronchiolitis obliterans syndrome
- cGVHD
chronic graft-versus-host disease
- ELISA
enzyme-linked immunosorbent assay
- EMT
epithelial–mesenchymal transition
- FEV1
forced expiratory volume in 1 second
- FN1
fibronectin 1
- HCT
allogeneic hematopoietic cell transplantation
- iTRAQ
isobaric Tags for Relative and Absolute Quantification
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- MMP-3
matrix metalloproteinase-3
- µL
microLiter
- NIH
National Institutes of Health
- n
number of patients
- PFT
pulmonary function test
- PPI
protein–protein interaction
- p
p-value
- ROC
receiver operating characteristic
- Rp
relevance score
Footnotes
Disclosure
The authors of this manuscript have conflicts of interest to disclose as described by the American Journal of Transplantation. S.P. is an inventor on a patent on “Methods of detection of graft-versus-host disease” (US- 13/573,766). The other authors have no conflicts of interest to disclose.
Additional Supporting Information may be found in the online version of this article.
Supplemental Methods
Figure S1: FN1 plasma concentrations measured by ELISA in a pilot set of 17 BOS cases and 13 controls post-HCT. BOS, bronchiolitis obliterans syndrome; cGVHD, chronic graft-versus-host disease; ELISA, enzyme-linked immunosorbent assay; FN1, fibronectin 1; HCT, allogeneic hematopoietic cell transplantation.
Table S1: Characteristics of patients in the discovery cohort
Table S2: A list of 855 proteins with quantification information identified in the Proteome Discoverer™ analysis
Table S3: A list of 462 proteins with quantification information identified in the Mascot™ analysis
Table S4: A list of 133 proteins remaining in the Proteome Discoverer result after pair-wise baseline transformation and comparison
Table S5: A list of 121 proteins remaining in the Mascot result after pair-wise baseline transformation and comparison
Table S6: A list of 84 proteins remaining in the Proteome Discoverer result after constructing the network using protein-protein interaction
Table S7: A list of 73 proteins remaining in the Mascot result after constructing the network using protein-protein interaction
Table S8: Characteristics of patients in the HCT cohort with cGVHD. cGVHD, chronic graft-versus-host disease; HCT, allogeneic hematopoietic cell transplantation.
References
- 1.Barker AF, Bergeron A, Rom WN, Hertz MI. Obliterative bronchiolitis. The New England journal of medicine. 2014;370(19):1820–1828. doi: 10.1056/NEJMra1204664. [DOI] [PubMed] [Google Scholar]
- 2.Williams KM, Chien JW, Gladwin MT, Pavletic SZ. Bronchiolitis obliterans after allogeneic hematopoietic stem cell transplantation. Jama. 2009;302(3):306–314. doi: 10.1001/jama.2009.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chien JW, Duncan S, Williams KM, Pavletic SZ. Bronchiolitis obliterans syndrome after allogeneic hematopoietic stem cell transplantation-an increasingly recognized manifestation of chronic graft-versus-host disease. Biology of blood and marrow transplantation : journal of the American Society for Blood and Marrow Transplantation. 2010;16(1 Suppl):S106–S114. doi: 10.1016/j.bbmt.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dudek AZ, Mahaseth H, DeFor TE, Weisdorf DJ. Bronchiolitis obliterans in chronic graft-versus-host disease: analysis of risk factors and treatment outcomes. Biology of blood and marrow transplantation : journal of the American Society for Blood and Marrow Transplantation. 2003;9(10):657–666. doi: 10.1016/s1083-8791(03)00242-8. [DOI] [PubMed] [Google Scholar]
- 5.Nishio N, Yagasaki H, Takahashi Y, Muramatsu H, Hama A, Tanaka M, et al. Late-onset non-infectious pulmonary complications following allogeneic hematopoietic stem cell transplantation in children. Bone marrow transplantation. 2009;44(5):303–308. doi: 10.1038/bmt.2009.33. [DOI] [PubMed] [Google Scholar]
- 6.Uhlving HH, Buchvald F, Heilmann CJ, Nielsen KG, Gormsen M, Muller KG. Bronchiolitis obliterans after allo-SCT: clinical criteria and treatment options. Bone marrow transplantation. 2012;47(8):1020–1029. doi: 10.1038/bmt.2011.161. [DOI] [PubMed] [Google Scholar]
- 7.Meyer KC, Raghu G, Verleden GM, Corris PA, Aurora P, Wilson KC, et al. An international ISHLT/ATS/ERS clinical practice guideline: diagnosis and management of bronchiolitis obliterans syndrome. The European respiratory journal. 2014;44(6):1479–1503. doi: 10.1183/09031936.00107514. [DOI] [PubMed] [Google Scholar]
- 8.Husain AN, Siddiqui MT, Holmes EW, Chandrasekhar AJ, McCabe M, Radvany R, et al. Analysis of risk factors for the development of bronchiolitis obliterans syndrome. American journal of respiratory and critical care medicine. 1999;159(3):829–833. doi: 10.1164/ajrccm.159.3.9607099. [DOI] [PubMed] [Google Scholar]
- 9.Nicod LP. Mechanisms of airway obliteration after lung transplantation. Proceedings of the American Thoracic Society. 2006;3(5):444–449. doi: 10.1513/pats.200601-007AW. [DOI] [PubMed] [Google Scholar]
- 10.Girnita AL, Duquesnoy R, Yousem SA, Iacono AT, Corcoran TE, Buzoianu M, et al. HLA-specific antibodies are risk factors for lymphocytic bronchiolitis and chronic lung allograft dysfunction. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2005;5(1):131–138. doi: 10.1111/j.1600-6143.2004.00650.x. [DOI] [PubMed] [Google Scholar]
- 11.Palmer SM, Davis RD, Hadjiliadis D, Hertz MI, Howell DN, Ward FE, et al. Development of an antibody specific to major histocompatibility antigens detectable by flow cytometry after lung transplant is associated with bronchiolitis obliterans syndrome. Transplantation. 2002;74(6):799–804. doi: 10.1097/00007890-200209270-00011. [DOI] [PubMed] [Google Scholar]
- 12.Panoskaltsis-Mortari A, Tram KV, Price AP, Wendt CH, Blazar BR. A new murine model for bronchiolitis obliterans post-bone marrow transplant. American journal of respiratory and critical care medicine. 2007;176(7):713–723. doi: 10.1164/rccm.200702-335OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Srinivasan M, Flynn R, Price A, Ranger A, Browning JL, Taylor PA, et al. Donor B-cell alloantibody deposition and germinal center formation are required for the development of murine chronic GVHD and bronchiolitis obliterans. Blood. 2012;119(6):1570–1580. doi: 10.1182/blood-2011-07-364414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Charpin JM, Valcke J, Kettaneh L, Epardeau B, Stern M, Israel-Biet D. Peaks of transforming growth factor-beta mRNA in alveolar cells of lung transplant recipients as an early marker of chronic rejection. Transplantation. 1998;65(5):752–755. doi: 10.1097/00007890-199803150-00027. [DOI] [PubMed] [Google Scholar]
- 15.DiGiovine B, Lynch J, Martinez F, Flint A, Whyte R, Iannettoni M, et al. Bronchoalveolar lavage neutrophilia is associated with obliterative bronchiolitis after lung transplantation: role of IL-8. J Immunol. 1996;157(9):4194–5202. [PubMed] [Google Scholar]
- 16.Iacono A, Dauber J, Keenan R, Spichty K, Cai J, Grgurich W, et al. Interleukin 6 and interferon-gamma gene expression in lung transplant recipients with refractory acute cellular rejection: implications for monitoring and inhibition by treatment with aerosolized cyclosporine. Transplantation. 1997;64(2):263–269. doi: 10.1097/00007890-199707270-00015. [DOI] [PubMed] [Google Scholar]
- 17.Jonosono M, Fang KC, Keith FM, Turck CW, Blanc PD, Hall TS, et al. Measurement of fibroblast proliferative activity in bronchoalveolar lavage fluid in the analysis of obliterative bronchiolitis among lung transplant recipients. The Journal of heart and lung transplantation : the official publication of the International Society for Heart Transplantation. 1999;18(10):972–985. doi: 10.1016/s1053-2498(99)00055-8. [DOI] [PubMed] [Google Scholar]
- 18.Magnan A, Mege JL, Escallier JC, Brisse J, Capo C, Reynaud M, et al. Balance between alveolar macrophage IL-6 and TGF-beta in lung-transplant recipients. Marseille and Montreal Lung Transplantation Group. American journal of respiratory and critical care medicine. 1996;153(4 Pt 1):1431–1436. doi: 10.1164/ajrccm.153.4.8616577. [DOI] [PubMed] [Google Scholar]
- 19.Moudgil A, Bagga A, Toyoda M, Nicolaidou E, Jordan SC, Ross D. Expression of gamma-IFN mRNA in bronchoalveolar lavage fluid correlates with early acute allograft rejection in lung transplant recipients. Clinical transplantation. 1999;13(2):201–207. doi: 10.1034/j.1399-0012.1999.130208.x. [DOI] [PubMed] [Google Scholar]
- 20.Riise GC, Kjellstrom C, Ryd W, Schersten H, Nilsson F, Martensson G, et al. Inflammatory cells and activation markers in BAL during acute rejection and infection in lung transplant recipients: a prospective, longitudinal study. The European respiratory journal. 1997;10(8):1742–1746. doi: 10.1183/09031936.97.10081742. [DOI] [PubMed] [Google Scholar]
- 21.Ross DJ, Cole AM, Yoshioka D, Park AK, Belperio JA, Laks H, et al. Increased bronchoalveolar lavage human beta-defensin type 2 in bronchiolitis obliterans syndrome after lung transplantation. Transplantation. 2004;78(8):1222–1224. doi: 10.1097/01.tp.0000137265.18491.75. [DOI] [PubMed] [Google Scholar]
- 22.Whitehead BF, Stoehr C, Wu CJ, Patterson G, Burchard EG, Theodore J, et al. Cytokine gene expression in human lung transplant recipients. Transplantation. 1993;56(4):956–961. doi: 10.1097/00007890-199310000-00034. [DOI] [PubMed] [Google Scholar]
- 23.Jagasia MH, Greinix HT, Arora M, Williams KM, Wolff D, Cowen EW, et al. National Institutes of Health Consensus Development Project on Criteria for Clinical Trials in Chronic Graft-versus-Host Disease: I. The 2014 Diagnosis and Staging Working Group report. Biology of blood and marrow transplantation : journal of the American Society for Blood and Marrow Transplantation. 2015;21(3):389 e381–401 e381. doi: 10.1016/j.bbmt.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bergeron A, Godet C, Chevret S, Lorillon G, Peffault de Latour R, de Revel T, et al. Bronchiolitis obliterans syndrome after allogeneic hematopoietic SCT: phenotypes and prognosis. Bone marrow transplantation. 2013;48(6):819–824. doi: 10.1038/bmt.2012.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20(18):3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 26.Anasetti C, Logan BR, Lee SJ, Waller EK, Weisdorf DJ, Wingard JR, et al. Peripheral-blood stem cells versus bone marrow from unrelated donors. The New England journal of medicine. 2012;367(16):1487–1496. doi: 10.1056/NEJMoa1203517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee SJ, Vogelsang G, Flowers ME. Chronic graft-versus-host disease. Biology of blood and marrow transplantation : journal of the American Society for Blood and Marrow Transplantation. 2003;9(4):215–233. doi: 10.1053/bbmt.2003.50026. [DOI] [PubMed] [Google Scholar]
- 28.You Q, Fang S, Chen JY. GeneTerrain: visual exploration of differential gene expression profiles organized in native biomolecular interaction networks. Information Visualization. 2008:1–12. [Google Scholar]
- 29.Naylor S, Chen JY. Unraveling human complexity and disease with systems biology and personalized medicine. Personalized medicine. 2010;7(3):275–289. doi: 10.2217/pme.10.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu X, Hasan MA, Chen JY. Pathway and network analysis in proteomics. Journal of theoretical biology. 2014;362:44–52. doi: 10.1016/j.jtbi.2014.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mellacheruvu D, Wright Z, Couzens AL, Lambert JP, St-Denis NA, Li T, et al. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nature methods. 2013;10(8):730–736. doi: 10.1038/nmeth.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paczesny S. Discovery and validation of graft-versus-host disease biomarkers. Blood. 2013;121(4):585–594. doi: 10.1182/blood-2012-08-355990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clarke W. Contemporary Practice in Clinical Chemistry. 2nd. AACC Press; 2011. [Google Scholar]
- 34.Smith GN, Jr, Mickler EA, Payne KK, Lee J, Duncan M, Reynolds J, et al. Lung transplant metalloproteinase levels are elevated prior to bronchiolitis obliterans syndrome. American journal of transplantation : official journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2007;7(7):1856–1861. doi: 10.1111/j.1600-6143.2007.01850.x. [DOI] [PubMed] [Google Scholar]
- 35.Vander Lugt MT, Braun TM, Hanash S, Ritz J, Ho VT, Antin JH, et al. ST2 as a marker for risk of therapy-resistant graft-versus-host disease and death. The New England journal of medicine. 2013;369(6):529–539. doi: 10.1056/NEJMoa1213299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ferrara JL, Harris AC, Greenson JK, Braun TM, Holler E, Teshima T, et al. Regenerating islet-derived 3-alpha is a biomarker of gastrointestinal graft-versus-host disease. Blood. 2011;118(25):6702–6708. doi: 10.1182/blood-2011-08-375006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paczesny S, Braun TM, Levine JE, Hogan J, Crawford J, Coffing B, et al. Elafin is a biomarker of graft-versus-host disease of the skin. Science translational medicine. 2010;2(13):13ra12. doi: 10.1126/scitranslmed.3000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Page-McCaw A, Ewald AJ, Werb Z. Matrix metalloproteinases and the regulation of tissue remodelling. Nature reviews Molecular cell biology. 2007;8(3):221–233. doi: 10.1038/nrm2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Radisky DC, Levy DD, Littlepage LE, Liu H, Nelson CM, Fata JE, et al. Rac1b and reactive oxygen species mediate MMP-3-induced EMT and genomic instability. Nature. 2005;436(7047):123–127. doi: 10.1038/nature03688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paczesny S, Krijanovski OI, Braun TM, Choi SW, Clouthier SG, Kuick R, et al. A biomarker panel for acute graft-versus-host disease. Blood. 2009;113(2):273–278. doi: 10.1182/blood-2008-07-167098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salmela MT, Karjalainen-Lindsberg ML, Jeskanen L, Saarialho-Kere U. Overexpression of tissue inhibitor of metalloproteinases-3 in intestinal and cutaneous lesions of graft-versus-host disease. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2003;16(2):108–114. doi: 10.1097/01.MP.0000051681.43441.82. [DOI] [PubMed] [Google Scholar]
- 42.Tagami K, Yujiri T, Takahashi T, Kizuki N, Tanaka Y, Mitani N, et al. Increased serum levels of matrix metalloproteinase-9 in acute graft-versus-host disease after allogeneic haematopoietic stem cell transplantation. International journal of hematology. 2009;90(2):248–252. doi: 10.1007/s12185-009-0358-6. [DOI] [PubMed] [Google Scholar]
- 43.Sato A, Nishida C, Sato-Kusubata K, Ishihara M, Tashiro Y, Gritli I, et al. Inhibition of plasmin attenuates murine acute graft-versus-host disease mortality by suppressing the matrix metalloproteinase-9-dependent inflammatory cytokine storm and effector cell trafficking. Leukemia. 2015;29(1):145–156. doi: 10.1038/leu.2014.151. [DOI] [PubMed] [Google Scholar]
- 44.Mattsson J, Remberger M, Andersson O, Sundberg B, Nord M. Decreased serum levels of clara cell secretory protein (CC16) are associated with bronchiolitis obliterans and may permit early diagnosis in patients after allogeneic stem-cell transplantation. Transplantation. 2005;79(10):1411–1416. doi: 10.1097/01.tp.0000158354.39635.ab. [DOI] [PubMed] [Google Scholar]
- 45.Gassas A, Schechter T, Krueger J, Craig-Barnes H, Sung L, Ali M, et al. Serum Krebs Von Den Lungen-6 as a Biomarker for Early Detection of Bronchiolitis Obliterans Syndrome in Children Undergoing Allogeneic Stem Cell Transplantation. Biology of blood and marrow transplantation : journal of the American Society for Blood and Marrow Transplantation. 2015;21(8):1524–1528. doi: 10.1016/j.bbmt.2015.04.021. [DOI] [PubMed] [Google Scholar]
- 46.Kuzmina Z, Krenn K, Petkov V, Kormoczi U, Weigl R, Rottal A, et al. CD19(+)CD21(low) B cells and patients at risk for NIH-defined chronic graft-versus-host disease with bronchiolitis obliterans syndrome. Blood. 2013;121(10):1886–1895. doi: 10.1182/blood-2012-06-435008. [DOI] [PubMed] [Google Scholar]
- 47.Williams KM, Cheng GS, Pusic I, Jagasia M, Burns L, Ho VT, et al. Fluticasone, Azithromycin, and Montelukast Treatment for New-Onset Bronchiolitis Obliterans Syndrome after Hematopoietic Cell Transplantation. Biology of blood and marrow transplantation : journal of the American Society for Blood and Marrow Transplantation. 2015 doi: 10.1016/j.bbmt.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vandenbroucke RE, Libert C. Is there new hope for therapeutic matrix metalloproteinase inhibition? Nature reviews Drug discovery. 2014;13(12):904–927. doi: 10.1038/nrd4390. [DOI] [PubMed] [Google Scholar]
- 49.Jameson JL, Longo DL. Precision Medicine - Personalized, Problematic, and Promising. The New England journal of medicine. 2015 doi: 10.1056/NEJMsb1503104. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.