Abstract
The kinetochore is the primary site of interaction between chromosomes and microtubules of the mitotic spindle during chromosome segregation. The kinetochore is a complex of more than 100 proteins that transiently assemble during mitosis at a single defined region on each chromosome, known as the centromere. Kinetochore assembly and activity must be tightly regulated to ensure proper microtubule interaction and faithful chromosome segregation because perturbation of kinetochores often results in aneuploidy and cell lethality. As such, cell free and reconstituted systems to analyze kinetochore formation and function are invaluable in probing the biochemical activities of kinetochores. In vitro approaches to studying kinetochores have enabled the manipulation of kinetochore protein structure, function, interactions and regulation that are not possible in cells. Here we outline a cell-free approach for the assembly of centromeres and recruitment of functional kinetochores that enables their manipulation and analysis.
Keywords: Kinetochore, in vitro assembly, chromatin, centromere, Xenopus egg extract
1. Introduction
Upon entry into mitosis more than 100 different proteins assemble at the centromeric region of chromosomes to form a kinetochore complex (1). The proper assembly of these proteins gives rise to the activities that are essential for chromosomal attachment to the mitotic spindle, metaphase chromosome alignment and segregation of the genome at anaphase. Once mitosis is complete, the kinetochore is then disassembled until re-assembly at the next entry into mitosis.
Biochemical and genetic studies have identified many centromere and kinetochore proteins yet studying the specific functions of these proteins in cells is challenging for a number of reasons. First and foremost, the kinetochore complex is essential to cell viability, so that perturbing individual protein components often results in cell death without informing on the detailed function of the protein. Second, the kinetochore assembles only at the centromeric region of a chromosome during mitosis. In metazoan cells, the DNA of the centromere is highly repetitive and spans many hundreds of kilobases, thus manipulation of centromeres and analysis of the underlying DNA presents a major challenge (2). Finally, because cells possess a single kinetochore per chromosome and because extracting intact, functional kinetochores from the chromosome has so far proven difficult studies of kinetochore function in vitro have been limited.
Despite these difficulties a number of successful approaches have been employed to study kinetochores. These can generally be classed into two groups, in vitro studies – where components are produced recombinantly, and ex vivo studies – where components are either extracted from cells or studied within cells but typically through tethering components to exogenous locations (3 – 8).
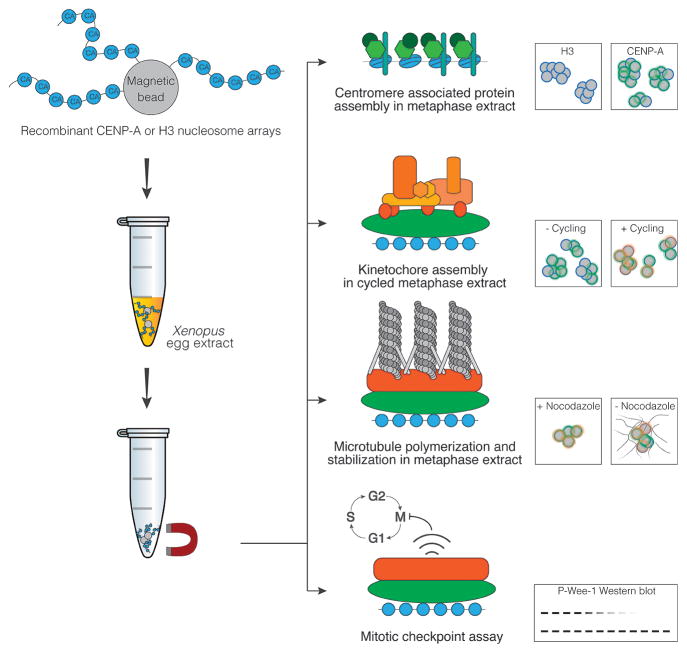
In this chapter we present a detailed method for the cell free assembly of centromeres and kinetochores that circumvents several of the major difficulties associated with studying endogenous kinetochores and enables specific perturbation and analysis of centromere and kinetochore components (9, 10). Our system takes advantage of Xenopus egg extract, a highly concentrated cytoplasmic extract that can execute chromosome segregation in vitro and in which we can manipulate the cell cycle state and protein composition of the extract (Figure 1). We couple these extracts with reconstituted chromatin substrates that resemble endogenous centromeres to build new centromeres and kinetochores on the added chromatin from the proteins present in the extract.
Figure 1. Schematic overview of the in vitro kinetochore assembly process.
Recombinant nucleosome arrays containing CENP-A nucleosomes can recruit functional centromere and kinetochore components when incubated in Xenopus egg extract. Prior attachment of the arrays to magnetic beads allow for easy reisolation of chromatin arrays and any bound complexes for further downstream analysis. Four example experiments utilizing this setup for the analysis of centromere and kinetochore complexes are summarized. These experiments are described in detail in the text.
This system provides a major advantage in that it permits manipulation of the DNA and protein composition of the chromatin template and the protein composition of the extract to study the assembly of functional, microtubule-binding kinetochores. By labeling the input chromatin with biotin we can recover and study near-native kinetochore complexes after their assembly in the extract. A second major advantage of this system is that it is possible to release the extract from metaphase into interphase with the addition of calcium. By cycling the extract through multiple rounds of interphase and metaphase this technique supports the study of cell cycle-associated changes to centromere and kinetochore components.
The method presented in this chapter is an adapted version of a method we have published previously and some steps are identical to the previous version while others have been updated or modified (9, 10). Here, we split the method into a number of broad steps for clarity. The first sections (Sections 3.1 – 3.9) detail the preparation of biotinylated nucleosome arrays and Xenopus egg extract. Four sections (3.10 – 3.13) then describe separate procedures that may be used independently for the analysis of centromere, kinetochore and related complexes in this experimental system. The final methods section (3.14) describes the preparation of samples for analysis by immunofluorescence and subsequent data analysis.
2. Materials
W buffer
50 mM Tris-HCl (pH 7.6), 100 mM NaCl, 1 mM EDTA, 5 mM 2-mercaptoethanol and 0.2 mM PMSF.
Freshly prepare this buffer and add 2-mercaptoethanol and PMSF right before use.
TW buffer
W buffer with 0.1% (vol/vol) Triton X-100.
Freshly prepare this buffer and add 2-mercaptoethanol and PMSF right before use.
Unfolding buffer
7 M guanidine-HCl, 20 mM Tris-HCl (pH 7.5) and 10 mM DTT.
Freshly prepare this buffer for each protein preparation.
Lysis buffer for CENP-A/H4 tetramer purification
(10 ml per cell pellet from 1 liter of bacterial cell culture) 20 mM phosphate buffer (pH 6.8) (for 1 liter of buffer use 9.94 ml of 1 M K2HPO4 and 10.06 ml of 1 M KH2PO4), 1 M NaCl, 5 mM 2-mercaptoethanol, 1 mM PMSF, 1 mM benzamidine, 0.05% NP-40 and 0.2 mg ml−1 lysozyme.
Prepare fresh and add the 2-mercaptoethanol, benzamidine and PMSF right before use; store on ice.
Dialysis buffer for CENP-A/H4 tetramer purification (2 × 4 liters)
10 mM Tris-HCl (pH 7.4), 0.75 M NaCl, 10 mM 2-mercaptoethanol and 0.5 mM EDTA.
Freshly prepare this buffer and add 2-mercaptoethanol right before use.
Urea buffer A
6 M deionized urea, 200 mM NaCl, 10 mM Tris-HCl (pH 8), 1 mM EDTA, 5 mM 2-mercaptoethanol and 0.1 mM PMSF.
Prepare fresh for each protein preparation and add 2-mercaptoethanol and PMSF right before use.
Urea buffer B
6 M deionized urea, 1 M NaCl, 10 mM Tris-HCl (pH 8), 1 mM EDTA, 5 mM 2-mercaptoethanol and 0.1 mM PMSF.
Freshly prepare for each protein preparation and add 2-mercaptoethanol and PMSF right before use.
Refolding buffer
2 M NaCl, 10 mM Tris-HCl (pH 7.6), 1 mM EDTA and 5 mM 2-mercaptoethanol.
Freshly prepare and add 2-mercaptoethanol and PMSF right before use.
HA buffer A
20 mM potassium phosphate (pH 6.8) and 5 mM 2-mercaptoethanol.
This buffer can be prepared in advance and kept indefinitely at 4 °C. Add 2-mercaptoethanol right before use.
HA buffer B
20 mM potassium phosphate (pH 6.8), 5 mM 2-mercaptoethanol and 3.5 M NaCl.
This buffer can be prepared in advance and kept indefinitely at 4 °C. Add 2-mercaptoethanol right before use.
HA column storage buffer
0.5 M potassium phosphate (pH 6.8).
Store at 4 °C indefinitely.
HA column cleaning buffer
0.5 M KOH.
Store at 4 °C indefinitely.
S column (HiTrap) buffer A
10 mM Tris-HCl (pH 7.4), 10 mM 2-mercaptoethanol and 0.5 mM EDTA.
Freshly prepare and add 2-mercaptoethanol immediately before use.
S column (HiTrap) buffer B
10 mM Tris-HCl (pH 7.4), 10 mM 2-mercaptoethanol, 0.5 mM EDTA and 2 M NaCl.
Freshly prepare and add 2-mercaptoethanol immediately before use.
PEG (MW 4,000 Da)
0.5 M NaCl, 20% (wt/vol) Dissolve 20 g of PEG and 2.9 g of NaCl in water to a final volume of 100 ml.
Filter-sterilize and store at room temperature (RT) indefinitely.
TE plus 2.5 mM NaCl
10 mM Tris-HCl (pH 8.0), 0.25 mM EDTA and 2.5 mM NaCl.
Store at RT indefinitely.
TE, 10×
100 mM Tris-HCl (pH 7.5) and 2.5 mM EDTA.
Filter-sterilize or autoclave, and store at RT indefinitely.
High-salt (HS) assembly buffer
10 mM Tris-HCl, 0.25 mM EDTA and 2 M NaCl in cold H2O; adjust the pH to 7.5 (while cold) with HCl.
Make fresh for each assembly.
Low-salt (LS) assembly buffer
10 mM Tris-HCl, 0.25 mM EDTA and 2.5 mM NaCl in cold H2O; adjust the pH to 7.5 (while cold) with HCl.
Make fresh for each assembly.
Tris-acetate, 1M
Dissolve 12.1 g of Tris in 75 ml of water; adjust the pH to 7.9 with acetic acid. Bring the solution to a final volume of 100 ml with water.
Store at RT indefinitely.
Array digestion (AD) buffer, 10×
500 mM potassium, 200 mM Tris-acetate, 10 mM DTT and 5 mM MgCl2 (pH 7.9).
Store in aliquots at − 20 °C for several years.
TBE, 5×
445 mM Tris base, 445 mM boric acid and 10 mM EDTA (pH 8.0).
Filter-sterilize TBE and store at RT for several weeks (until substantial precipitation of the buffer has occurred).
APS, 10% (wt/vol)
Dissolve 1 g of APS in water to a final volume of 10 ml.
Store at 4 °C for several weeks.
Native acrylamide gel, 5% (wt/vol)
Mix 1.5 ml of 40% (wt/vol) acrylamide, 4.5 ml of water, 6 ml of 1× TBE, 5–10 μl of TEMED and 100 μl of 10% (wt/vol) APS and pour it into a preassembled polyacrylamide gel stand. Add the comb immediately (no stack gel is required) and let the gel solidify before use.
Can be stored at 4 °C for a few days if kept moist, although typically used immediately.
Glycerol, 80% (vol/vol)
Mix 80 ml of glycerol with 20 ml of water.
Filter-sterilize and store at RT indefinitely.
DNA, 19 × 601 (at ~2.5 μg μl−1 or higher)
For preparation see Section 3.5.
Store at − 20 °C indefinitely
H3/CENP-A nucleosomes on biotinylated 601 array DNA, ~2 μM (calculated for 1 × 601 DNA)
For preparation see Section 3.6.
Store at 4 °C for up to a few months.
Bead buffer, 4×
200 mM Tris-HCl (pH 8.0), 1 mM EDTA, 300 mM NaCl and 0.2% (vol/vol) Triton X-100.
Store at RT for several months.
Bead buffer, 1×
50 mM Tris-HCl (pH 8.0), 0.25 mM EDTA, 75 mM NaCl and 0.05% (vol/vol) Triton X-100.
Store at RT for several months.
Array buffer
10 mM Tris-HCl (pH 7.5), 2.5 mM NaCl and 0.25 mM EDTA.
Store at RT indefinitely.
Polyvinyl alcohol, 7.5% (wt/vol)
Dissolve 7.5 g of PVA (MW 30,000–70,000 Da) in water to a final volume of 100 ml.
Filter-sterilize and store at RT indefinitely.
CaCl2, 12 mM (to release Xenopus egg extracts from CSF arrest)
Mix 120 μl of 1 M CaCl2 with 9.88 ml of water.
Filter-sterilize and store in small aliquots at − 20 °C indefinitely.
Marc’s modified Ringer’s solution (MMR), 25×
150mM Na-HEPES (pH 7.8), 2.5 mM EDTA, 2.5 M NaCl, 50 mM KCl, 25 mM MgCl2 and 50 mM CaCl2. For 4 liters, dissolve 142.99 g of HEPES, 584.4 g of NaCl, 14.91 g of KCl, 20.33 g of MgCl2 and 29.4 g of CaCl2; add 20 ml of 0.5 M EDTA and adjust the pH to 7.8 with NaOH.
Store at RT for several months.
Gelatin, 5% (wt/vol)
Mix 20 g of gelatin with 80 ml of water in a glass bottle.
Autoclave and store at RT indefinitely. Before use, liquefy the gelatin by microwaving briefly.
Sucrose, 2 M
Dissolve 684.6 g of sucrose in water and adjust the final volume to 1 liter.
Filter-sterilize and keep the solution at RT; in small aliquots it can be stored at − 20 °C for several years.
K-HEPES, 1 M, pH 7.7
Dissolve 119.15 g of HEPES in water; adjust the pH to 7.7 with KOH and the final volume to 500 ml.
Filter-sterilize and store at RT indefinitely.
K-EGTA, 0.5 M, pH 7.7
Dissolve 95.09 g of EGTA in water; adjust the pH to 7.7 with KOH and the final volume to 500 ml.
Filter-sterilize and store at RT indefinitely.
Extract buffer (XB) salt stock, 20×
2 M KCl, 20 mM MgCl2 and 2 mM CaCl2.
Filter-sterilize and keep it at RT for several months.
CSF-XB buffer, 1×
1× XB salt (50 ml of 20× stock), 50 mM sucrose, 10 mM K-HEPES (pH 7.7), 5 mM K-EGTA, 1 mM MgCl2 and 110 μl of 10 M NaOH.
Freshly prepare this buffer before use.
Protease inhibitors, 1,000×
Mix 10 mg ml−1 of leupeptin, pepstatin A and chymostatin in DMSO.
Aliquot and store at − 20 °C indefinitely.
CSF-XB plus protease inhibitors, 1×
Take 50 ml of freshly prepared 1× CSF-XB buffer and add 50 μl of 1,000× stock protease inhibitors.
Prepare right before use.
CSF-XB plus 0.05% (vol/vol) Triton X-100, 1×
Freshly prepare this solution.
Energy mix, 20×
150 mM creatine phosphate, 20 mM ATP and 20 mM MgCl2.
Aliquot into 500 μl aliquots and keep at − 20 °C for several months.
Cytochalasin D, 1,000×
Prepare stock at a concentration of 10 mg ml−1 in DMSO.
Make 50 μl aliquots and keep them at − 20 °C for several months.
Cytochalasin B, 100×
Prepare stock at a concentration of 10 mg ml−1 in DMSO.
Make 50 μl aliquots and keep them at − 20 °C for several months.
Dejellying buffer
Mix 2% (wt/vol) l-cysteine in 1× MMR at pH 7.8 (adjusted with 10 M NaOH).
Freshly prepare and dissolve l-cysteine right before use.
Antibody dilution buffer (AbDil)
150 mM NaCl, 20 mM Tris-HCl (pH 7.4), 0.1% (vol/vol) Triton X-100 and 2% (wt/vol) BSA.
Store it at 4 °C for a few weeks.
Nocodazole, 1 mg ml−1
Dissolve nocodazole at a concentration of 1 mg ml−1 in DMSO.
Store it in small aliquots at − 20 °C indefinitely.
BRB80, 5×
400 mM PIPES (pH 6.8), 5 mM MgCl2 and 5 mM K-EGTA.
Filter-sterilize and store the solution at 4 °C for several months.
Cushion
Mix 1× BRB80 and 40% (wt/vol) glycerol.
Filter-sterilize and store at RT for several months.
Dilution buffer
1× BRB80, 0.05% (vol/vol) Triton X-100 and 30% (wt/vol) glycerol.
Filter-sterilize and store at RT for several months.
CAPS transfer buffer
10 mM CAPS (pH 11.3), 1% (wt/vol) SDS and 20% (vol/vol) methanol (optional).
Freshly prepare; it is also possible to store the used buffer at 4 °C and reuse it a few times within 1 week.
Milk in PBS-Tween, 5% (wt/vol)
Keep at 4 °C and use within 1 week.
Coverslips coated in poly-l-lysine
Wash coverslips in 1 M HCl overnight at 50–60 °C. Thoroughly rinse coverslips with water until the pH returns to neutral. Add the coverslips to a solution of 1 mg ml−1 poly-l-lysine in distilled water and incubate on a rocker at RT for at least 1 h. Remove the poly-l-lysine solution (this can be stored frozen and reused a few times). Rinse the coverslips thoroughly in ten changes of water. Rinse the coverslips in ethanol.
Store coverslips submerged in 70% (vol/vol) ethanol at RT indefinitely.
Mounting medium
20 mM Tris-HCl (pH 8.8), 90% (wt/vol) glycerol and 0.5% (wt/vol) p-phenylenediamine. Prepare the solution of Tris-HCl (pH 8.8) and glycerol and bring it to the final volume with water. Add the p-phenylenediamine solid, and dissolve it by bubbling nitrogen gas through the solution for a few hours. The solution may turn light brown. Draw into a syringe, minimizing the amount of air trapped in the syringe.
Store in the sealed syringe at − 20 °C. Discard the medium once the solution turns dark brown (typically a month or two).
3. Method
3.1 H3/H4 tetramer or H2A/H2B dimer expression
Transform histone expression plasmids (H2A, H2B, H3 and H4) and a negative control (water) into BL21(DE3) Codon Plus RIPL bacteria.
Plate ~20 – 100 μl of the transformation (including the cells-only control) onto LB/chloramphenicol/carbenicillin plates and incubate overnight at 37 °C.
Pick a single colony and inoculate into 5 ml of LB/chloramphenicol/carbenicillin overnight at 37 °C.
Dilute the starter culture to an optical density at 600 nm (OD600) of 0.005 (approximately 1:1,000) in 2 liters of 2×YT medium.
Continue growth at 37 °C, 200 r.p.m., to an OD600 of 0.5–0.6. Remove 1 ml of sample for an analytical gel in Section 3.3, Step 6 as a control for uninduced expression.
Add IPTG to a final concentration of 0.2 mM.
Grow the culture for a further 3 h at 37 °C.
Pellet cells in 1 liter portions by centrifugation at 2,500g at 4 °C for 15 min.
Discard the supernatant and resuspend each pellet in 25 ml of W buffer.
Combine cells for a single portion of each histone and flash freeze in liquid nitrogen.
3.2 H3/H4 tetramer or H2A/H2B dimer purification
Thaw cells in a 37°C water bath with constant shaking.
Sonicate six times using a probe sonicator with 30 s pulses separated by 1 min intervals at 200 W. Keep the cells on ice when possible.
Split the lysate into two tubes and centrifuge at 25,000g for 20 min at 4 °C.
Discard the supernatant (save 50 μl for analysis in Section 3.3, Step 6) and resuspend the pellet in each tube in 25 ml of TW buffer. For this and the following steps, ensure that the pellet is thoroughly resuspended. Using a spatula to cut and crush the insoluble material and a vortex to aid resuspension.
Sonicate for 30 s on maximum power.
Centrifuge at 25,000g for 20 min at 4 °C.
Discard the supernatant and resuspend the pellet in each tube in 25 ml of TW buffer.
Repeat Steps 6 and 7 three more times, resuspending first in TW buffer and in W buffer the second and third times.
Repeat Step 6 and add 175 μl of DMSO to the pellet in each tube, and then combine both portions and use a spatula to finely crush the pellet.
Add 13.2 ml of unfolding buffer and incubate for 1 h at RT with gentle shaking.
Homogenize in a Dounce homogenizer, until no more large pieces remain.
Centrifuge at 25,000g for 20 min at 4 °C.
Remove the supernatant, but save it in a fresh tube.
Resuspend the pellet again in 13.2 ml of unfolding buffer, centrifuge again as described in Step 12, and combine the supernatant with the supernatant from Step 13.
Dialyze this supernatant overnight into 2 liters of urea buffer A, stirring at 4 °C. The following day, exchange the dialysis buffer to 2 liters of fresh buffer A, and dialyze for a further 3 h at 4 °C. (A white precipitate may form during the dialysis.)
Centrifuge the solution at 12,000g for 20 min at 4 °C.
Measure the conductivity of the supernatant relative to that of the urea buffer. If necessary, dilute the supernatant with fresh urea buffer until the conductivity of the supernatant is less than 1.25 times that of the buffer alone.
Connect a 5 ml HiTrap Q column and a 5 ml HiTrap S column in series on an FPLC system.
Wash columns with three column-volumes (3 CV) of water (30 ml total), then 3 CV of urea buffer A.
Load the supernatant from Step 17 over the columns.
Wash with 2 CV of urea buffer A.
Remove the Q column, keeping the S column attached to the FPLC. The Q column can be reused after elution with urea buffer B.
Elute the S column with a linear gradient from 0 to 37.5% urea buffer B (remainder urea buffer A) over 3 CV, and then with 100% urea buffer B for 3 CV, collecting 1 ml fractions.
Run 10 μl of the peak fractions on a 20% (wt/vol) SDS-PAGE gel. Also run the analytical gel samples taken earlier on this gel for analysis.
Pool the histone-containing fractions and dialyze against 4 liters of water containing 0.1 mM PMSF and 5 mM 2-mercaptoethanol. Refresh the dialysis with fresh water 3 times to ensure complete dialysis. A white precipitate may form during the dialysis.
Clarify the protein preparation by centrifugation at 25,000g for 20 min at 4 °C.
Measure the concentration of the supernatant by Bradford assay.
Lyophilize the supernatant in aliquots (typically 5 mg each).
3.3 H3/H4 tetramer or H2A/H2B dimer assembly
Resuspend lyophilized histone directly into unfolding buffer at a concentration of 10 mg ml−1.
Mix ~10 mg of H3 and an equimolar amount of H4, or ~10 mg of H2A and an equimolar amount of H2B.
Dialyze against refolding buffer, with three changes of 4 liters.
If the volume increased to more than 5 ml during dialysis, concentrate in an Amicon conical concentrator, 10,000 MWCO, to obtain a final volume of ≤5 ml.
Run over a Superdex 200 16/60 column (equilibrated to refolding buffer) at 1 ml min−1; collect 1 ml fractions.
Run fractions on a 20% (wt/vol) denaturing polyacrylamide gel, pool the fractions that have approximately equal amounts of H3 and H4 (or H2A and H2B).
Centrifuge the pooled histone solution for 10 min at 10,000g at 4 °C.
Mix 5 μl of the supernatant with 5 μl of 8 M guanidine-HCl to denature the protein, and use a spectrophotometer to obtain the 280 nm absorption.
Use the theoretical extinction coefficient ε and the calculated molecular weight of each histone to precisely determine the protein concentration in milligrams per milliliter. To calculate protein concentration in mg ml−1 divide the protein absorbance at 280 nm by the extinction coefficient and multiply by the protein’s molecular weight.
Aliquot the remaining pooled H3/H4 tetramer or H2A/H2B dimer in ~100 pmol aliquots, flash freeze in liquid nitrogen and store at − 80 °C.
3.4 CENP-A/H4 tetramer expression and purification
Repeat Steps 1 and 2 from Section 3.1 using a CENP-A/H4 pST39 bicistronic expression plasmid.
Pick a single bacterial colony, inoculate into 5 ml of LB/chloramphenicol/carbenicillin and grow at 37 °C for 8–10 h.
Dilute the culture into 100 ml in LB/chloramphenicol/carbenicillin and continue growth overnight.
Dilute cultures 1:50–1:100 in 6 liters of 2× YT medium.
Grow at 37 °C, with shaking, to an OD600 of 0.2 and then grow at RT until an OD600 of 0.5–0.6.
Add IPTG to a final concentration of 0.2 mM.
Continue growth at RT for ~6 h with shaking.
Pellet cells in 1 liter portions by centrifugation at 2,500g at 4 °C for 15 min.
Freeze cell pellets in liquid nitrogen and store at − 80 °C.
Equilibrate a 60 ml HA column by washing with 1 CV of 20 mM potassium phosphate (pH 6.8).
Clean with ~1 CV (60–100 ml) 0.5 M KOH.
Equilibrate with 2 CV (~120 ml) of 20 mM potassium phosphate (pH 6.8).
Prepare HA column buffers A and B without 2-mercaptoethanol; filter and store at 4 °C.
Thaw cell pellets from Step 9 by resuspending them in cold lysis buffer. Use 10 ml of lysis buffer for each liter of bacterial culture.
Sonicate 6 × 30 s at 200 W with 1 min intervals. Keep on ice between the cycles. (Optional: take an ~1 μl sample for an analytical gel in Step 21.)
Centrifuge at 110,000g for 1 h. (Optional: take an ~2 μl sample for an analytical gel in Step 21.)
Load the supernatant onto the pre-equilibrated HA column at a flow rate of 1–2 ml min−1 using a peristaltic pump or FPLC. Save the flow-through. (Optional: take an ~2 μl sample for an analytical gel in Step 21.)
Attach the HA column to a FPLC system if not already attached.
Wash the column with 6 CV (~400 ml) of 28% HA column buffer B (corresponds to 1 M NaCl). Retain the wash. (Optional: take an ~4 μl sample for an analytical gel in Step 21.)
Run a linear elution gradient from 28% HA column buffer B (remainder HA buffer A) to 100% buffer B over 2 CV (120 ml) and collect 6 ml fractions.
Run fractions of this eluate (~20 μl) and other expression and purification fractions on a denaturing 20% polyacrylamide gel). Relevant fractions will contain visible bands at ~18 kDa (hsCENP-A) and 13 kDa (H4).
Pool fractions containing histones CENP-A and H4 and dialyze overnight against 4 liters of dialysis buffer.
Change the dialysis buffer and dialyze for another 2–4 h.
Load the sample onto 1 ml of HiTrap SP FF column at a flow rate of 1 ml min−1. Save the flow-through. (Optional: take a ~20 μl sample for an analytical gel in Step 27.)
Wash with 10 CV of 37% S column buffer B.
Run a linear gradient from 37% S column buffer B (remainder S column buffer A) to 100% S column buffer B over 20 CV to elute soluble CENP-A/H4 tetramer. Collect 0.5 ml fractions.
Run fractions (~20 μl) on a denaturing 20% (wt/vol) polyacrylamide gel to determine which fractions contain CENP-A and H4 at high purity. Pool fractions containing a roughly equal ratio of CENP-A and H4 that are also free of other contaminating proteins.
Centrifuge the pooled histone solution for 10 min at 10,000g at 4 °C.
Mix 5 μl of the supernatant with 5 μl of 8 M guanidine-HCl to denature the protein, and use a spectrophotometer to obtain the 280 nm absorption.
Use the theoretical extinction coefficient ε and the calculated molecular weight of each histone to precisely determine the protein concentration in milligrams per milliliter. To calculate protein concentration in mg ml−1 divide the protein absorbance at 280 nm by the extinction coefficient and multiply by the protein’s molecular weight.
Aliquot the remaining pooled CENP-A/H4 tetramer into ~100 pmol aliquots, flash freeze in liquid nitrogen and store at − 80 °C.
3.5 Preparation of biotinylated DNA
Transform the 19 × 601 plasmid into SURE2 bacteria (also use SURE2 cells by themselves as a control). Plate 20–100 μl of the transformation or the cells-only control, onto a LB/chloramphenicol/carbenicillin plate and incubate at 37 °C overnight. The following day, pick a single colony and use to inoculate a 5 ml starter culture in LB medium with chloramphenicol/carbenicillin. Grow for 8 h at 37 °C. Dilute the starter culture 1:1,000 into 2 liters of LB medium supplemented with chloramphenicol and carbenicillin and allow it to grow overnight at 37 °C.
Purify plasmid DNA with a Qiagen plasma giga kit tip-10000 (or other comparable method). Typically yields are 7–10 mg of purified plasmid DNA.
Digest 2 × 500–750 μg of plasmid DNA with EcoRI, XbaI, DraI and HaeII (200 U of each enzyme) in a final volume of 1 ml (per reaction) at 37 °C overnight. Check digestion by running 1 μl of the digests on a 0.7% (wt/vol) agarose gel. The 19 × 601 fragment runs at 3.8 kb, the digested backbone appears as four bands <1 kb.
Add 5 M NaCl to the digest to a final concentration of 0.5 M NaCl.
Add 20% (wt/vol) PEG and 0.5 M NaCl to a final concentration of 5% (wt/vol) PEG, 0.5 M NaCl.
Centrifuge at 5,000g for 10 min at RT.
Transfer the supernatant into a fresh tube (take 1 μl of sample for analysis on an agarose gel in Step 10) and keep the original tube in a rack at RT.
Add 20% (wt/vol) PEG and 0.5 M NaCl to the transferred supernatant to obtain a final concentration of 5.5% (wt/vol) PEG, 0.5 M NaCl.
Repeat Steps 6–8, adding additional PEG (in 0.5% (wt/vol) final concentration increments) and centrifuging until the final PEG concentration reaches 10%.
Load each 1 μl sample onto a 0.7% (wt/vol) agarose gel to identify fractions containing the 19 × 601 fragment. Typically, the 19 × 601 DNA fragment precipitates between 7 and 9% final PEG concentration.
Resuspend precipitated DNA (from appropriate tubes stored in Step 7) in TE (pH 8.0) and pool fractions. The volume of TE for resuspension must be determined empirically with the aim of resuspending precipitated DNA at 2–3 μg μl−1.
Dialyze resuspended DNA in 50–200 μl dialysis buttons into TE (pH 8.0) overnight.
Refresh the dialysis buffer and continue dialysis for another 3–4 h.
Determine the concentration of the DNA using a spectrophotometer to measure the OD260. Typically, the digestion of 1 mg of 19 × 601 plasmid DNA yields 300–400 μg digested and purified 19 × 601 DNA fragment.
Analyze 1 μl of a 1:10 dilution of the DNA on a 0.7% (wt/vol) agarose gel to check the purity of the sample.
A concentration of ~2.5 μg μl−1 or higher is ideal. If required, sodium acetate/ethanol-precipitate DNA and resuspend in a smaller volume of TE (pH 8.0).
Mix 500 μg of digested and purified 19 × 601 DNA in a final volume of 400 μl with biotin- 14-dATP, α-thio-dGTP, α-thio-dTTP and dCTP at a final concentration of 35 μM for each nucleotide, as well as 24 μl of Klenow fragment. Also add an appropriate amount of the reaction buffer concentrate provided with the Klenow fragment.
Incubate for 3 h at 37 °C.
Purify biotinylated 19 × 601 DNA using Qiagen PCR purification columns, according to the manufacturer’s instructions. Elute with 30 μl of EB buffer (Qiagen) or water per column, pool the eluted fractions and precipitate using sodium acetate/ethanol precipitation. Alternatively, purify large reactions using G-50 gel filtration resin to remove the unincorporated nucleotides.
Resuspend in 100 μl of TE (pH 8.0) and measure the concentration at OD260 using a spectrophotometer. A final concentration of 3–5 μg μl−1 is ideal. Typically, the input of 500 μg of digested 19 × 601 DNA into the biotinylation reaction yields ~300 μg of biotinylated 19 × 601 DNA.
To analyze the biotinylation efficiency, incubate 500 ng of biotinlyated 19 × 601 DNA with and without 0.5 μl of Streptavidin-FITC and non-biotinylated 19 × 601 DNA with 0.5 μl of streptavidin-FITC in TE (pH 8.0) and 2.5 mM NaCl for 20 min at RT.
Run reactions on a 0.7% (wt/vol) agarose gel. Analyze the DNA by imaging with ethidium bromide stain and the biotinylation by FITC fluorescence. Biotinylation causes the 19 × 601 DNA to migrate in a single band more slowly in the presence of streptavidin than in its absence.
Store biotinylated DNA at − 20 °C.
3.6 Chromatin array assembly
Estimate the required ratios of H3/H4 and CENP-A/H4 tetramers to nucleosome positioning sites on the DNA arrays for equal stoichiometry, and use a slight excess of H2A/H2B dimers (2.2 times the molar DNA amount) (Note 1).
Prepare 500 ml of HS assembly buffer and 2 liters of LS assembly buffer; adjust the pH to 7.5 and store at 4 °C.
Set up chromatin arrays assembly reactions in 1.5 ml tubes on ice. Typically, we use 50 μl reactions or 200 μl reactions at a final concentration of nucleosome positioning sites of 2 μM, and make both individual H3 and CENP-A nucleosome arrays. Mix appropriate volumes of H2O, 10× TE and 5 M NaCl to achieve a final concentration of 1× TE and 2 M NaCl. Add, in the following order, biotinylated DNA, H2A/H2B dimers and CENP-A/H4 or H3/H4 tetramers.
Moisten the dialysis membrane of a small, fixed volume dialysis unit in HS assembly buffer. Pipette the assembly mixture into the dialysis unit.
Submerge dialysis units in the HS assembly buffer.
Transfer the dialysis beaker to a magnetic stir plate at 4 °C. Set the stirring speed to very low.
Connect a 1 ml plastic pipette to one end of two pieces of a peristaltic pump tubing, clip both pieces of the tubing with the plastic pipettes at the same end into the peristaltic pump. Set the flow speed to 0.5 ml min−1. Put one pipette into the 2-liter beaker containing LS assembly buffer and attach the loose end of the tubing with a binder clip to the rim of the dialysis beaker. Attach the second pipette to the rim of the dialysis beaker and put the loose end into a liquid waste container or the sink.
When the buffer exchange is complete (~67 h later), exchange dialysis buffer one more time with fresh LS assembly buffer and dialyze for an additional 4 h.
Transfer the assembly reaction into a 1.5 ml tube on ice. Determine the volume of each assembly and adjust the expected concentration accordingly.
Assembled arrays can be stored for up to ~3 months at 4 °C, although the nucleosome saturation should be tested within a few days of use.
3.7 Testing nucleosome array saturation
Combine 500 ng of assembled array DNA with 1 μl of AvaI in AD buffer supplemented with 1 mM DTT and 1× BSA in a final volume of 20 μl at RT overnight. Use 500 ng of biotinylated DNA (no nucleosomes) as a control (Note 2).
Add Glycerol (80%, vol/vol) to obtain a final concentration of 20% (6.7 μl per 20 μl reaction).
Run the whole reaction (~27 μl) of digested arrays and DNA on a 5% (wt/vol) native acrylamide/bis-acrylamide/0.5×TBE gel in 0.5× TBE as a running buffer. Also run a small volume (0.5 – 1 μl) of DNA ladder on this gel.
Mix 200 ml of 0.5× TBE with 15 μl of Sybr Gold and pour it into a gel-staining container.
Using a spatula, carefully detach the gel from the glass plate so that it rolls slowly into the container. Stain the gel while gently rocking or shaking for 20 min at RT.
Wash the gel three times with H2O to remove residual stain.
Transfer the gel to a UV-transmittable glass plate.
Analyze the gel by fluorescence imaging. Assembly of a nucleosome onto a nucleosome positioning sequence within the array causes a band shift from ~200 bp (DNA control) to ~700 bp (mononucleosomes) (Note 2).
3.8 Preparation of chromatin beads
Wash 45 μl of Dynabeads in ~300 μl of 1× bead buffer three times, using a magnetic tube rack to retain beads between washes (Note 3). Split the beads into three tubes (15 μl per tube: one for CENP-A arrays, one for H3 arrays and one for the bead-only control), place them onto magnetic tube rack and aspirate the supernatant.
To each tube, add 7.5 μl of ~2 μM biotinylated nucleosome arrays (CENP-A or H3 saturated chromatin) or 10 μl of array buffer (for the bead only control) in a final volume of 50 μl bead buffer containing 2.5% (wt/vol) PVA. The volume of this reaction should be scaled according to the amount of beads used. Typically, we couple 1.5 μl of chromatin arrays (~2 μM) to 3 μl of Dynabeads for each sample.
Attach chromatin arrays to beads by incubating at RT for at least 30 min with gentle agitation. Ensure that the shaking speed is enough to keep the beads in solution without spitting all over the inside of the tube (Note 4).
Wash the chromatin-bound beads in 200–300 μl of bead buffer and store at 4 °C.
3.9 Xenopus CSF extract preparation
Prime female frogs with 0.5 ml of PMSG using a 1-ml syringe and a 25-gauge needle. Primed frogs can be used for egg laying for 2 weeks. For optimal egg laying, prime frogs again with 0.25 ml of PMSG 2 d after the first priming.
To induce egg laying, inject three frogs with 0.5 ml of hCG (500 U) 16–18 h before the extract will be prepared.
After injection, separate frogs by placing each into an individual plastic container containing at least 2 liters of 1× MMR, made with deionized water. Keep at 16 °C overnight.
Rinse all glassware with deionized water and prepare the required buffers (2 liters of 1× MMR, 200 ml of dejellying buffer with the cysteine added immediately before use, 500 ml of CSF-XB buffer). Store buffers at 16 °C.
Pour ~10–20 ml hot liquid gelatin solution into a water-filled glass dish and 100-ml glass beaker, immediately swirl the containers to coat the sides with gelatin. Keep the water/gelatin solution in the containers and store at RT until further use.
Remove frogs from their containers, rinse frog eggs twice with ~500 ml of 1× MMR and sort eggs using a glass Pasteur pipette to remove strings of eggs, puffy eggs and activated eggs (Note 5).
Prepare 50 ml of CSF-XB. Transfer 2 ml into a separate tube, add cytochalasin B and transfer 1 ml into two SW-55 centrifuge tubes.
Pour out the water/gelatin solution from the Pyrex glass dish and rinse with ~200 ml of 1× MMR. Carefully pour the eggs into the Pyrex dish and wash twice with 200–300 ml of 1× MMR. Add the cysteine to the dejellying buffer and mix until dissolved. Pour out the 1× MMR from the Pyrex dish containing the eggs and add ~50 ml of the dejellying buffer. Swirl gently, remove the dejellying buffer and add in the remaining 150 ml of the dejellying buffer. Gently swirl again and remove any eggs that show contraction of the dark pigments. The dejellying process is complete when the eggs become closely packed when the Pyrex dish is tilted to one side. This typically takes 5–10 min and should be carefully monitored, as eggs left in dejellying solution for too long will lyse.
Pour off the dejellying buffer and wash the eggs three or four times with 200–300 ml of CSF-XB.
Empty the water/gelatin solution from the 100 ml beaker, add a few milliliters of CSF-XB buffer then carefully add the eggs.
Pour off the CSF-XB buffer and add ~10 ml of CSF-XB with protease inhibitors. Swirl gently, then remove the buffer and pour in the remaining 40 ml of CSF-XB with protease inhibitors.
Transfer the eggs into the SW-55 centrifuge tubes from step 7 above using a fire-polished pipette.
Aspirate excess buffer and gently slide the tubes into 13 ml Falcon tubes without caps.
Centrifuge at RT for ~30 s at ~300 g, and then for ~30 s at ~500 g.
Remove the tubes, aspirate any excess liquid and centrifuge for 15 min at 16 °C at 9,480 g in a SW-55 swinging bucket rotor.
Remove the tubes from the centrifuge buckets and store on ice.
Clean the outside of the tubes with ethanol and pierce with a 1-ml syringe and a 16-gauge needle to collect the straw-colored cytoplasmic fraction (middle layer).
Measure the volume of extract collected and store in a fresh tube on ice.
Add the appropriate amount of protease inhibitors (from 1,000× stock), cytochalsin D (from 1,000× stock), cold sucrose (1:40) and energy mix (1:20) to the extract and mix well. Use extract within 1–2 h as quality declines over time.
3.10 Centromere associated protein assembly in metaphase extracts
Wash chromatin beads (prepared above) twice with 200–300 μl of CSF-XB supplemented with 0.05% (vol/vol) Triton X-100.
Use a magnetic tube rack to collect the beads, discard the supernatant and add 100 μl of fresh CSF extract (prepared above) on ice. Mix the extract-bead mixture by pipetting gently and transfer to an 18–20 °C water bath.
Keep the tubes at 18–20 °C for 60 min. Flick them every 15 min to keep chromatin beads in solution.
Use a magnetic tube rack to collect the beads. This may take a few minutes if the extract is particularly viscous.
Remove the extract and wash the chromatin beads three times in 200–300 μl of cold CSF-XB buffer supplemented with 0.05% (vol/vol) Triton X-100.
Resuspend chromatin beads in 100 μl of CSF-XB plus 0.05% (vol/vol) Triton X-100 and add formaldehyde to a final concentration of 2%. Incubate for 5 min at RT.
Wash three times with 200–300 μl of AbDil and adjust the concentration to 1 μl chromatin beads per 10 μl AbDil. Fixed samples can be stored in AbDil for 1–2 d at 4 °C.
3.11 Kinetochore assembly in cycled extracts
Wash chromatin beads (prepared above) twice with approximately 200–300 μl of CSF-XB supplemented with 0.05% (vol/vol) Triton X-100.
Discard the supernatant and mix chromatin beads with 50 μl of fresh CSF extract (prepared above) on ice. Pipette the extract-bead mixture up and down two or three times and transfer to a 18–20 °C water bath.
After 5–10 min, add CaCl2 to a final concentration of 0.6 mM and incubate for 80 min at 18–20 °C. Flick the tubes every 15 min to keep chromatin beads in solution. (Optional) Take a sample for a western blot to monitor the cell cycle stage (Note 6).
Add one volume (50 μl per tube) of fresh CSF extract and incubate for 90 min to drive the extract into mitosis. Flick the tubes every 15 min to keep chromatin beads in solution. If analyzing the assembly of spindle assembly checkpoint proteins, use CSF extract that has been premixed with nocodazole (or DMSO as a control) at a final concentration of 10 μg ml−1.
Resuspend chromatin beads in 100 μl of CSF-XB plus 0.05% (vol/vol) Triton X-100 and add formaldehyde to a final concentration of 2%. Incubate for 5 min at RT.
Wash three times with 200–300 μl of AbDil and adjust the concentration to 1 μl chromatin beads per 10 μl AbDil.
3.12 Microtubule polymerization and stabilization in metaphase extracts
Wash chromatin beads (prepared above) twice with 200–300 μl of CSF-XB supplemented with 0.05% (vol/vol) Triton X-100.
Use a magnetic tube rack to pellet the chromatin beads, discard the supernatant and mix 2 μl of beads with 20–50 μl of freshly prepared CSF extract per spindle assembly reaction. Mix reactions by pipetting up and down two or three times, transfer to a 0.5-ml Eppendorf tube and incubate on ice (Note 7).
Transfer samples to an 18–20 °C water bath and incubate for 75–90 min. Flick the tubes gently every 15 min to keep the chromatin beads in solution. (Optional) Add 100 ng μl−1 of nocodazole to the reaction or transfer the reaction to 4 °C for 10 min to induce microtubule depolymerization.
Pipette the reaction into 1 ml of dilution buffer supplemented with 2.5% (wt/vol) formaldehyde and fix at 18–20 °C for 10 min. Use a wide tip and pipette gently to avoid breaking the assembled spindles. Mix samples by inverting the tubes.
Place custom-made adaptors into Corex tubes to provide a flat surface. Place a poly-l-lysine–coated coverslip on top and add 5 ml of cushion.
Layer fixed samples on top of the cushion. Use a wide tip and pipette gently to avoid shearing the assembled spindles.
Centrifuge for 20 min at 3,290 g.
Remove the coverslips and postfix in ice-cold methanol for 5 min.
Wash the coverslips 3 × 5 min in AbDil.
Block in AbDil for 30 min at RT.
Incubate with tubulin-specific antibodies (DM1α) at 1:2,000 for 1 h at RT or at 4 °C overnight.
Wash coverslips 3 × 5 min in AbDil.
Stain using appropriate fluorophore-conjugated secondary antibodies for 30 min at RT.
Wash coverslips 3 × 5 min in AbDil.
Stain the coverslip with Hoechst stain (10 μg ml−1 in AbDil) for 10 min at RT.
For mounting and subsequent microscopic analysis, proceed to the immunofluorescence and data analysis section below.
3.13 Mitotic checkpoint assay
For each sample, wash 2 μl of chromatin beads (prepared above) twice with 200–300 μl of CSF-XB supplemented with 0.05% (vol/vol) Triton X-100.
Take 5 μl of CSF extract, mix with 45 μl of SDS-PAGE sample buffer and store at −20 °C for subsequent Western blot analysis of the cell cycle stage.
Add 20 μl of fresh CSF extract to each sample from Step 1 in a 0.7 ml Eppendorf tube. Pipette up and down to mix and incubate for 5–10 min at 18–20 °C in a water bath.
Add CaCl2 to a final concentration of 0.6 mM and mix by flicking the tube (Note 6).
Incubate for 80 min at 18–20 °C, flicking the tubes every 15 min.
Take 5 μl of sample and mix it with 45 μl of SDS-PAGE sample buffer for western blotting, store at −20 °C. To prevent taking beads, do not flick the tubes for 15–20 min before taking the sample so that the chromatin beads sink.
To the remaining 15 μl of sample, add an equal volume of fresh CSF extract supplemented with either nocodazole at 10 μg ml−1 or DMSO as a control. Mix by pipetting up and down and avoid the addition of air bubbles.
Incubate for 90 min at 18–20 °C, flicking the tubes every 15 min.
Take 5 μl of sample (t = 0′) and mix it with 45 μl of SDS-PAGE sample buffer for western blotting, store at −20 °C. Prevent taking beads, as described in Step 6.
Add CaCl2 to the remaining 25 μl of sample to a final concentration of 0.6 mM and mix by flicking the tubes.
Incubate for 10 min at 18–20 °C, take 5 μl of sample (t = 10′) and mix with 45 μl of SDS-PAGE sample buffer for western blotting and leave at −20 °C. Again, avoid taking beads, as described in Step 6.
Repeat Step 11 for t = 20′, t = 30′ and t = 40′ samples. Store samples in SDS sample buffer at − 20 °C for a few weeks.
Heat western blotting samples at 95 °C for 10 min.
Load 20 μl of each sample (equivalent to ~2 μl of extract) on a 12.5% (wt/vol) SDS-PAGE system.
Transfer separated proteins onto PVDF membrane in CAPS transfer buffer for 1 h 45 min at 400 mA.
Block the membrane for 30 min in 5% (wt/vol. milk in PBS-T.
Incubate with primary antibodies (e.g. P-Wee-1–specific, 1:1,000; tubulin-specific (DM1α), 1:2,000. at 4 °C overnight.
Wash 3 × 5 min in PBS-T.
Incubate for 30 min at RT with fluorescently labeled secondary antibodies at a dilution of ~1:500 in 5% (wt/vol) milk in PBS-T.
Wash 3 × 5 min in PBS-T.
Wash two times with PBS and analyze the western blot by fluorescence detection (Note 8).
3.14 Immunofluorescence and data analysis
Dry poly-l-lysine–coated coverslips in a humidified chamber.
Pipette 20 μl of chromatin beads in AbDil (equivalent to ~ 2 μl of chromatin beads) onto each coverslip and let the beads attach to the coverslip for 15 min at RT, without letting them dry out.
Block the coverslips in AbDil by pipetting 30–50 μl on top of the attached beads and incubating for 30 min at RT.
Aspirate the AbDil and incubate each coverslip with antibodies to proteins of interest (diluted in AbDil) for 1 h at RT or overnight at 4 °C (Note 9).
Wash coverslips for 3 × 5 min in AbDil.
Stain the coverslips using appropriate fluorophore-conjugated secondary antibodies for 30 min at RT.
Wash coverslips 3 × 5 min in AbDil.
When using two antibodies raised in the same animal, one must be directly conjugated to a fluorophore. Block with whole IgG of that species for 45 min before incubation with a directly labeled antibody for 1 h at RT.
Wash coverslips 3 × 5 min in AbDil.
Incubate with 30–40 μl of propidium iodide (1:1,000 in AbDil) for 10 min at RT to stain DNA.
Wash coverslips 3 × 5 min in AbDil.
Wash one or two times in PBS.
Add ~ 5 μl of mounting medium to a labeled microscope slide, aspirate the coverslips, and place upside down onto the slide. Carefully remove excess mounting medium and seal the sides of the coverslip to the slide with clear nail polish. Store slides at − 20 °C until analysis.
Image at least three fields of beads per slide, with 0.2 μm section intervals through the beads. Image a few hundred beads in total for each sample, imaging each relevant wavelength at each section. Determine exposure times for each wavelength to ensure that the camera does not saturate. Keep exposure times the same for different samples stained with the same marker.
Ensure images are in a format readable by the ‘imread’ function in MATLAB (e.g., TIFF). Do not convert images to a format that uses lossy compression (e.g., JPEG-compressed image formats) or alters the bit depth of the images.
Quantify the fluorescence signal of beads in each image (Note 8). We have provided a MATLAB script that is freely available at http://straightlab.stanford.edu/software. This script reads TIFF images where each color channel is in a separate file, finds beads using the DNA-stained images and quantifies the intensity of each bead in each color channel (Note 10).
Footnotes
Arrays are easily over- or undersaturated if the amount of histones used for nucleosome assembly was not quite correct. We recommend a careful titration of both tetramer:dimer ratio and the histone:DNA ratio to achieve 90–100% array saturation. Typically, we will test DNA:tetramer ratios of 1:0.8, 1:0.9, 1:1.0, 1:1.1 and 1:1.2 to determine the ideal ratio for high nucleosome saturation. The ratio of H2A/B dimers to DNA is usually more forgiving and is usually kept at 2.2:1 in nucleosome assembly reactions.
In the 19 × 601 array DNA, each positioning sequence is separated by an AvaI restriction site; thus, digestion with AvaI generates 19 ~200-bp fragments. Migration of the mono-nucleosome fragment is dependent on the precise positioning of the core histone complex with respect to the DNA.
A volume of 45 μl of Dynabeads is sufficient to analyze the binding of four different centromere/kinetochore proteins to CENP-A arrays versus H3 arrays. It will also ensure enough for the H4 staining and the bead-only control (no chromatin arrays coupled) coverslips, both of which are required for subsequent quantification by fluorescence microscopy.
Coupling of nucleosome arrays to beads often causes the beads to clump slightly. This effect appears to be histone-dependent and is not observed with naked DNA.
Frogs that lay particularly low quality eggs (where strings of eggs, puffy eggs or activated eggs comprise more than 10% of the whole batch) typically suffer from three major complaints. They may have been ovulated too frequently and must be rested for longer time periods between ovulations. They may be aged, in which case they must be replaced. Else they may be stressed, and treatment would involve removing potential stressors (such as exposure to noise or poor water quality).
Extract that is of poor quality may not exit from mitosis upon calcium addition. Equally, insufficient calcium addition may also prevent mitotic exit. In either case, repeating the experiment with fresh extract and slightly more calcium usually circumvents these problems. Making sure the water temperature does not exceed 16–18 °C during incubations has also been found to help. Extract that does not arrest in mitosis as expected may occur for two reasons. Firstly, the water temperature may be too high, in which case make sure the temperature stays between 16–18 °C throughout the incubation. The second possibility is that kinetochore assembly was inefficient, in which case it is necessary to retry using fresh extract and adding more calcium.
As a control for spindle assembly, it is possible to prepare a sample with 250 Xenopus sperm nuclei per 1 μl of extract. Demembranated sperm is prepared exactly as described by Murray (11).
If no kinetochore or spindle checkpoint proteins are observed this may be for two reasons. The extract quality may have been poor, or kinetochore assembly was inefficient due to the quality of the nucleosome array. Repeating with fresh extract and new arrays typically solves such issues.
In our hands, efficient histone H4 staining at 1:100 can only be obtained with incubation at 4 °C overnight.
For a more detailed description of this MATLAB script see Guse et al (2012) (10).
Contributor Information
Matthew D D Miell, Department of Biochemistry, Stanford University School of Medicine, Stanford, California, USA.
Aaron F Straight, Department of Biochemistry, Stanford University School of Medicine, Stanford, California, USA.
References
- 1.Westhorpe FG, Straight AF. Functions of the centromere and kinetochore in chromosome segregation. Current Opinion in Cell Biology. 2013;25:334–340. doi: 10.1016/j.ceb.2013.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fukagawa T, Earnshaw WC. The Centromere: Chromatin Foundation for the Kinetochore Machinery. Dev Cell. 2014;30:496–508. doi: 10.1016/j.devcel.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mitchison TJ, Kirschner MW. Properties of the kinetochore in vitro. I. Microtubule nucleation and tubulin binding. J Cell Biol. 1985;101:755–765. doi: 10.1083/jcb.101.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitchison TJ, Kirschner MW. Properties of the kinetochore in vitro. II. Microtubule capture and ATP-dependent translocation. J Cell Biol. 1985;101:766–777. doi: 10.1083/jcb.101.3.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kingsbury J, Koshland D. Centromere-dependent binding of yeast minichromosomes to microtubules in vitro. Cell. 1991;66:483–495. doi: 10.1016/0092-8674(81)90012-x. [DOI] [PubMed] [Google Scholar]
- 6.Akiyoshi B, Nelson CR, Ranish JA, Biggins S. Quantitative proteomic analysis of purified yeast kinetochores identifies a PP1 regulatory subunit. Genes Dev. 2009;23:2887–2899. doi: 10.1101/gad.1865909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lechner J, Carbon J. A 240 kd multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell. 1991;64:717–725. doi: 10.1016/0092-8674(91)90501-o. [DOI] [PubMed] [Google Scholar]
- 8.Sorger PK, Severin FF, Hyman AA. Factors required for the binding of reassembled yeast kinetochores to microtubules in vitro. J Cell Biol. 1994;127:995–1008. doi: 10.1083/jcb.127.4.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guse A, Carroll CW, Moree Ben, et al. In vitro centromere and kinetochore assembly on defined chromatin templates. Nature. 2011;477:1–7. doi: 10.1038/nature10379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guse A, Fuller CJ, Straight AF. A cell-free system for functional centromere and kinetochore assembly. Nat Protoc. 2012;7:1847–1869. doi: 10.1038/nprot.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murray AW. Cell cycle extracts. In: Kay BK, Peng HB, editors. Xenopus laevis: Practical Uses in Cell and Molecular Biology. Methods in Cell Biology. Vol. 36. Academic Press Inc; New York: 1991. pp. 581–605. [Google Scholar]