Summary
Epigenetic regulation of lineage-specific genes is important for the differentiation and function of T cell. Ten-eleven translocation (Tet) proteins catalyze 5-methylcytosine (5mC) conversion to 5-hydroxymethylcytosine (5hmC) to mediate DNA demethylation. However, the roles of Tet proteins in the immune response are unknown. Here, we characterized the genome-wide distribution of 5hmC in CD4+ T cells and found 5hmC marks putative regulatory elements in signature genes associated with effector cell differentiation. Moreover, Tet2 protein was recruited to 5hmC-containing regions, dependent on lineage-specific transcription factors. Deletion of the Tet2 gene in T cells decreased their cytokine expression, associated with reduced p300 recruitment. In vivo, Tet2 plays a critical role in the control of cytokine gene expression in autoimmune disease. Collectively, our findings suggest that Tet2 promotes DNA demethylation and activation of cytokine gene expression in T cells.
Introduction
Upon activation by antigens, naive CD4+ T cells differentiate into one of several lineages of helper T (Th) cells, including Th1, Th2, Th17 and iTreg cells, defined by their patterns of cytokine production and immune function (Zhu et al., 2010). Th differentiation depends on the complex network of cytokine signaling imposed on lineage-specific transcription factors (Kanno et al., 2012). In addition to transcription factors, growing evidence shows that epigenetic mechanisms are crucial for controlling Th differentiation (Kanno et al., 2012). Wei et al. reported that histone modifications on the lineage-specific genes correlate with genes expression in Th differentiation (Wei et al., 2009). Moreover, we previously reported the recruitment of histone acetyltransferase (p300) and H3K27 demethylase (JMJD3) in the Il17a and Il17f loci in a Th17-specific manner (Wang et al., 2012). Together, these findings suggest that histone modification is an important mechanism for Th differentiation.
DNA methylation at the 5-position of cytosine (5-methylcytosine; 5mC) is one of the key epigenetic mechanisms in development and gene regulation (Bird, 2002), and the alterations in DNA methylation patterns have been implicated in various diseases (Robertson, 2005). The 5-hydroxymethylcytosine (5hmC) was first identified in the T-even bacteriophage and was later found in several tissues (Shen and Zhang, 2013). 5hmC exists in mouse, bovine and rabbit zygotes as well as mouse embryonic stem cells, and accumulates specifically in the paternal pronucleus coinciding with a reduction in 5mC (Shen and Zhang, 2013), implying a potential biological function of 5hmC and a role of DNA demethylation in early development. Recently, several studies identified the Ten-Eleven-Translocation (TET) proteins TET1, TET2 and TET3 as a new family of a-ketoglutarate and Fe2+-dependent enzymes that alter the methylation status of DNA by converting 5mC into 5hmC (Pastor et al., 2013). Functional analyses using Tet-deficient cells have demonstrated their crucial roles in diverse biological processes (Pastor et al., 2013). Although it is becoming increasingly clear that Tet-mediated 5mC oxidation at functional genomic elements is physiologically an important epigenetic process in mammals, the roles of 5hmC and Tet proteins in the immune system remain to be understood.
Here, we for the first time generated genome-wide maps of 5hmC in various Th cells and found 5hmC exists at putative regulatory elements of lineage-specific genes in appropriate Th cells. Tet2 was associated with 5hmC-containing regions; deletion of Tet2 inhibited cytokine expression by Th1 and Th17 cells, resulting in reduction of 5hmC and key transcription factors binding. Finally, we confirmed Tet2 function in regulating the cytokines expression in vivo. Collectively, our findings prove that Tet-mediated active DNA demethylation is an essential epigenetic mechanism for regulation of Th cell function.
Results
Genome-wide distribution of 5hmC in CD4+ T cells
To study the role of DNA demethylation and particularly 5hmC during CD4+ T cell-mediated immune responses, we conducted global mapping of 5hmC via the DNA immunoprecipitation coupled with high-throughput sequencing (DIP-seq) (Ku et al., 2011). As a control, 5mC was analyzed in the same manner. We assessed these modifications in five types of CD4+ T cells: freshly isolated CD4+CD25−CD44lowCD62Lhigh (naive) T cells as well as naïve CD4+ T cells cultured under Th1, Th2, Th17, and TGFβ-induced Treg (iTreg) cell differentiation conditions. The appropriate polarization of each Th subset was confirmed by intracellular staining as well as qPCR. We found enrichment of lineage signature genes in polarized Th cells (Figure S1A). Furthermore, we observed approximately 90% of cells expressing lineage-specific transcription factors in polarized Th cells, although the percentages of signature cytokine-producing cells varied from 7% (Th2) to 50% (Th1 and Th17) under various conditions (Figure S1B). Immunoprecipitated samples were amplified and subjected to Illumina sequencing. A total of 172 million short reads from all samples were aligned onto the mouse genome (mm9 Build 37 assembled by NCBI). In all Th subsets, we identified 372,892 5hmC-occupied peaks and 260,315 5mC-occupied peaks (Table S1). To determine whether our sequencing depth can cover the size of the 5mC and 5hmC libraries, we performed a scaling analysis. Randomly sampled fractions of reads from each sample were subjected to peak identification using SICER program (Zang et al., 2009). We found that about 15 or 6 million reads were sufficient to identify most of peaks for 5mC or 5hmC samples, respectively (Figure S1C). Therefore, our sequencing depth was able to cover the size of all libraries. We then analyzed the distribution of these peaks in the mouse genome in four kinds of regions: promoter (1 kb upstream and downstream of transcription start site), exon, intron, and intergenic regions, according to the annotation of “known genes” from the University of California, Santa Cruz (UCSC) Genome Browse (Kent et al., 2002). As shown in Figure 1A and Table S1, each Th subset displayed similar patterns of 5hmC and 5mC distribution. The majority of 5hmC- and 5mC-associated regions were found in introns and intergenic regions, ranging from 39% to 57%, suggesting that they may potentially function in enhancers. In contrast, only around 4% of 5hmC- and 5mC-occupied peaks were localized to promoters and less than 7% in exons (Figure 1A and Table S1), slightly than the presence of these two regions in the whole mouse genome (Table S1).
Figure 1. Comparison of genome-wide 5hmC and 5mC distribution in each T cell subset.
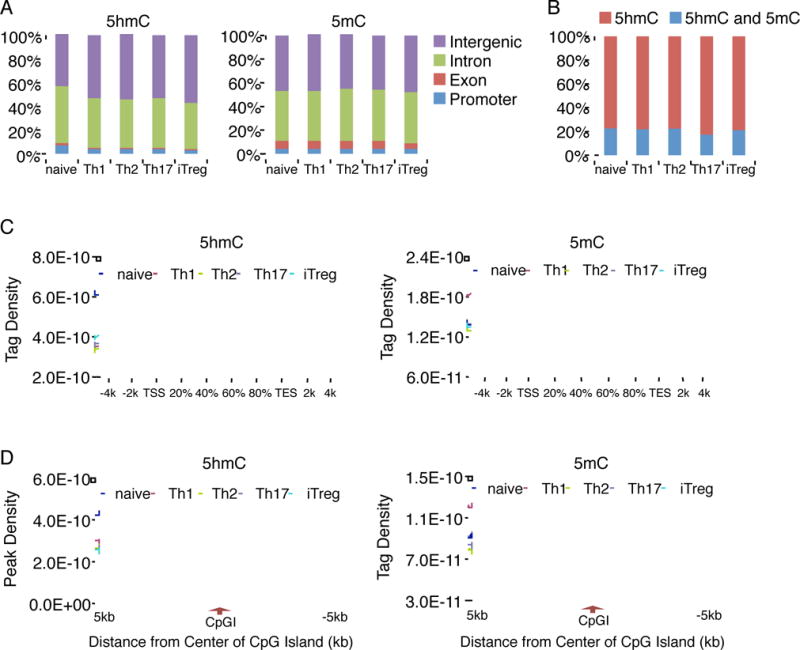
(A) The distribution of 5hmC and 5mC modifications was analyzed on the basis of location: promoter (within 1 kb upstream from the transcription start site), exon, intron and intergenic regions. (B) The bar graph showing the percentage of peaks uniquely associated with 5hmC, or both of 5hmC and 5mC. (C) The normalized tag density profiles for 5hmC (left) and 5mC (right) across gene body ±5 kb flanking regions with 200 bp resolution are shown. (D) The normalized tag density profiles for 5hmC (left) and 5mC (right) around ±5 kb regions flanking CpGI centers with 100 bp resolution are shown. “See also Figure S1 and Table S1 and S2.”
It is unclear whether 5hmC and 5mC colocalize in the same genomic regions during Th differentiation or rather function independently. We then searched for regions with binary marks of both 5hmC and 5mC. 17–23% of 5hmC-occupied regions in all Th subsets share both marks (Figure 1B and Table S2), indicating a potentially active DNA demethylation process occurred in these regions. However, the majority of conversion from 5mC to 5hmC has been maintained in Th subsets.
We next analyzed the distribution of 5hmC and 5mC within gene bodies and 5 kb regions 5′ and 3′ of them. Although similar amounts and chromosomal distribution of 5mC were found in all Th subsets, the highest amounts of 5hmC in extended genic regions were found in naïve T cells (Figure 1C). Moreover, this analysis also revealed the depletion of 5hmC and 5mC islands from regions proximal to the transcription start sites (TSS) (Figure 1C), consistent with the observation that only a minority of the 5hmC and 5mC islands was located in promoter regions (Figure 1A).
In mammal, the majority of CpG dinucleotides are methylated, whereas unmethylated CpGs are found primarily in those regions of DNA with relatively high density of CpG, so call CpG islands (CpGIs) (Bird, 1986). We therefore analyzed the distribution of 5hmC and 5mC among CpGIs. Our results showed that 5hmC was depleted from the centers of CpGIs in all Th subsets, while the 5mC was depleted only at the regions upstream and downstream of the centers of CpGIs (Figure 1D). Similarly, naïve T cells have the highest amounts of 5hmC in CpGIs, suggesting that the change in 5hmC around TSS and CpGIs may play a critical role during Th cell differentiation.
5hmC associates with lineage-specific signature genes
We next examined whether 5hmC and 5mC presented in lineage-specific genes in each Th subset. Firstly, we assessed 5hmC and 5mC patterns in Ifng, Il4, and Il17 cytokine genes, which serve as the defining lineage markers for Th1, Th2, and Th17 cells, respectively. As shown in Figure 2A, 5hmC was strongly associated with Ifng, Il4 and Il17 genes, particularly in some of the evolutionarily conserved non-coding sequences (CNSs) and some promoter regions. Furthermore, we confirmed the distribution of 5hmC and 5mC in naïve, Th1 and Th17 cells by qPCR after immunoprecipitation of 5hmC or 5mC. Consistent with sequencing analysis, the CNS(-6) at Ifng gene, known as an enhancer (Hatton et al., 2006), was highly hydroxymethylated in Th1 cells but hypermethylated in other Th cells (Figure S2A). Similarly, the CNS2, Il17a and Il17f promoters of the Il17 locus were strongly hydroxymethylated in Th17 cells but were hypermethylated in other Th cells (Figure S2B). In addition to lineage-specific cytokines, we also analyzed Il10 gene that is expressed by virtually every Th subsets (Ouyang et al., 2011). As expected, 5hmC was closely marked with some CNSs of Il10 gene in Th1, Th2 and Th17 cells and naïve T cells showed strong 5mC peaks in these regions (Figure 2A and Figure S2C). On the other hand, we could not detect substantial IL-10 production or augmented 5hmC signals in iTreg cells (Figure 2A and data not shown). It was also obvious that many of 5hmC peaks were shared by several lineages, while some lineage-specific peaks were associated with the promoter and CNS regions of lineage-specific genes such as Ifng, Il17a and Il17f (Table S3). As we mentioned above, cells cultured with in vitro polarized conditions are heterogeneous population regarding cytokine production. To assess whether the existence of non-cytokine producing cells affect the results of 5hmC mapping, we used cytokine gene reporter mice (Ifngfyp, Il4gfp, and Il17fCreRosa26yfp) to purify fully polarized Th cells. DIP-PCR analysis revealed that DNA hydroxymethylation on signature genes was very similar before and after cell sorting (Figure S3).
Figure 2. 5hmC and 5mC modifications of signature cytokine genes and transcription factor genes in Th cells.
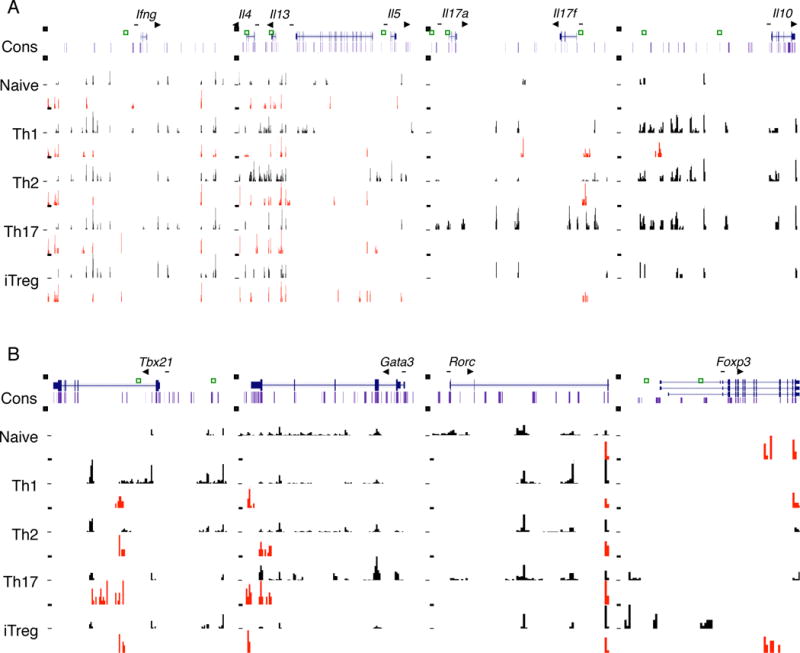
(A, B) Distribution of 5hmC and 5mC in the Ifng (Chr10; 117810000-117940000), Il4, Il13, Il5 (Chr11; 53420500-53553500), Il17a, Il17f (Chr1; 20713500-20787300), Il10 (Chr1; 132884100-132923100), Tbx21 (Chr11; 96958500-96987500), Gata3 (Chr2; 9777000-9802000), Rorc (Chr3; 94175000-94191200) and Foxp3 (ChrX; 7153000-7170500) genomic regions in each T cell subset is shown. All figures with views of 5hmC and 5mC distribution are labeled such that the arrow represents the direction of gene transcription. Gene structure is downloaded from UCSC Genome Browser, and only tags on islands are shown. The islands labeled in black represent 5hmC. The islands labeled in red represent 5mC. Scales are kept constant among cell types. Unique peaks are highlighted by green squares. “See also Figure S2 and S3 and Table S3.”
Lineage-specific transcription factors such as T-bet, GATA3, RORγt, and Foxp3 are well known as master regulators essential for development and function of Th1, Th2, Th17 and Treg cells, respectively. Therefore, we next examined the distribution of 5hmC and 5mC in genes encoding these key transcription factors. The Tbx21 and Foxp3 genes were indeed associated with high 5hmC in Th1 and iTreg cells, respectively, implying regulation of these genes by active DNA demethylation (Figure 2B). Especially, CNS2 in Foxp3 gene was intensely demethylated in iTreg cells. This is consistent with a report on hypomethylation of CNS2 in Foxp3 expression (Ohkura et al., 2012). However, prominent 5hmC peaks were located in Rorc and Gata3 gene in even non-expressing cell lineages (Figure 2B). Thus, these observations suggest that the expression of Gata3 and Rorc may not be regulated by active DNA demethylation.
Collectively, our data prove that 5mC and 5hmC at lineage-specific signature cytokine genes seem to correlate with their expression in Th subsets and suggest that active DNA demethylation may be one important mechanism for gene regulation during Th development.
5hmC marks transcriptional regulatory regions
Epigenetic control of gene expression involves dynamic regulation of DNA methylation and histone modifications. Although several studies assessed the regulation of DNA methylation by histone modification patterns (Cedar and Bergman, 2009), the relationship between active DNA demethylation and histone modification remain unclear. To correlate DNA demethylation and histone modifications, we used published chromatin data from Th cell subsets available in database (Wei et al., 2009). In silico analysis revealed that 5hmC peaks in Th cell-specific signature genes partly colocalized with H3K4me3 modification, a histone marker associated with transcriptionally active state (Figure S4A). These findings support previous reports that 5hmC is enriched in euchromatin in cerebellar cell types (Mellen et al., 2012) and that enhanced H3K4me3 appeared to be correlated with CpG hypomethylation within the Treg cell-associated genes (Ohkura et al., 2012). Yet, the overall picture of H3K4me3 distribution pattern seemed not to match that of 5hmC distribution pattern in each Th subset (Figure S4B). As previously reported, the majority of H3K4me3 is enriched near TSS (Wei et al., 2009), whereas the majority of 5hmC is depleted from TSS (Figure 1 and Figure S4B). In addition, we sought to calculate the total number of peaks that are overlapped between modifications. Expectedly, H3K4me3 preferentially overlapped with 5hmC rather than 5mC. However, only around 10% to 20% of H3K4me3 sites were shared with 5hmC islands in each Th subsets (Figure S4C). Thus, generally speaking, a minority of these two modifications was overlapped in each Th subsets. Collectively, although active DNA demethylation is correlated with the enhancement of H3K4me3, this relationship seems to be limited to certain regions of linage-specific signature gene loci.
To further characterize whether 5hmC contribute to the transcriptional regulation during Th differentiation, we then analyzed whether 5hmC was colocalized with binding sites for active enhancer defining factor p300. The genome-wide p300 binding sites have been previously defined by Vahedi et al in Th1 cells (Vahedi et al., 2012) as well as by Ciofani et al in Th17 cells (Ciofani et al., 2012). As shown in Figure 3A, 26–34% of p300 binding sites colocalized with 9–10% of 5hmC peaks in Th1 and Th17 cells, while only 2–8% of p300 binding sites colocalized with 1–4% of 5mC peaks in these cells. Taken together, our data demonstrate a genome-wide enrichment of 5hmC modification on the active enhancers in effector T cells.
Figure 3. Colocalization of 5hmC with active enhancers bound by p300 and key transcription factors in Th1 and Th17 cells.
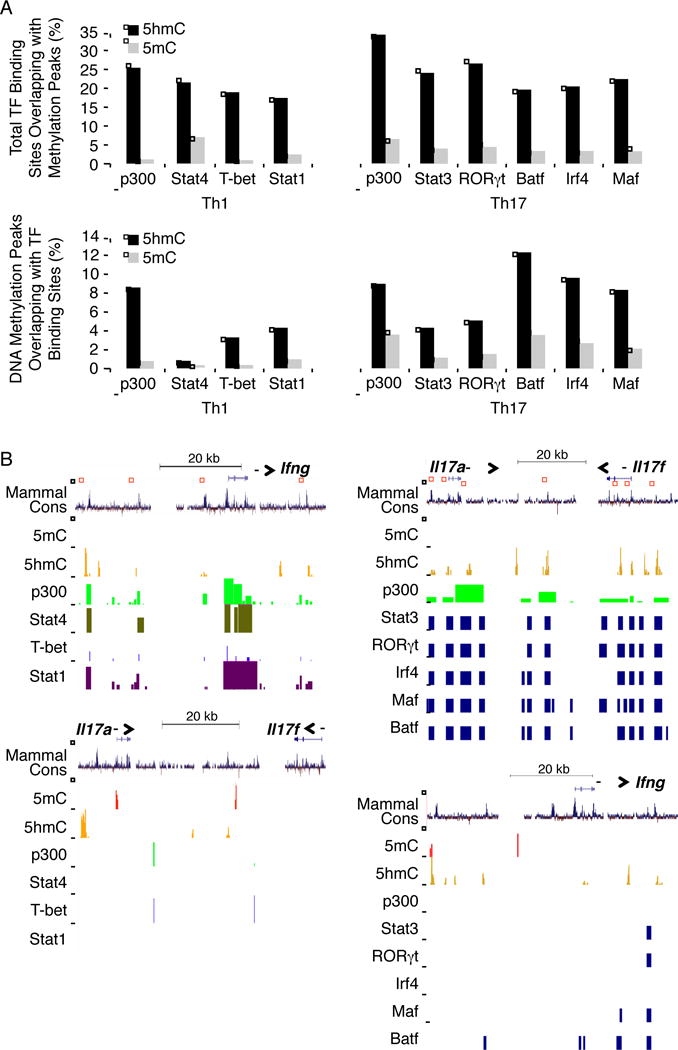
(A) upper; The percentages of total p300 and transcription factor binding sites colocalized with 5mC (grey) and 5hmC (black) peaks was shown in Th1 and Th17 cells. lower; The percentages of 5mC (grey) and 5hmC (black) peaks that were co-localized with p300, Stat4, T-bet, and Stat1 binding sites in Th1 cells (left), as well as with p300, Stat3, Rorc, Batf, Irf4, and Maf binding sites in Th17 cells (right) is shown. (B) Distribution of 5hmC, 5mC, p300 and transcription factors binding sites along Ifng, Il17a and Il17f genomic region in Th1 (Stat4, T-bet, Stat1) (left) and Th17 (Stat3, RORγt, Irf4, Maf, Batf) (right) cells are shown. The regions of overlapping peaks are highlighted by red squares. “See also Figure S4.”
Although our data indicated that 5hmC was depleted from TSS and proximal promoter as well as the centers of CpGIs, we found that the binding sites of key transcription factors as well as evolutionarily conserved enhancers marked by p300 colocalized with 5hmC but not 5mC on lineage-specific signature gene loci in Th1 and Th17 cells (Figure 3B). We therefore explored the colocalization between 5hmC and binding sites of key transcription factors governing Th1 and Th17 development, particularly, Stat4, T-bet, Stat1 in Th1 cells (Vahedi et al., 2012), as well as Stat3, RORγt, Batf, Irf4, and Maf (Ciofani et al., 2012). As expected, 17–26% of these transcription factors binding sites were localized at 5hmC occupied genomic regions, while only 1–7% of them co-localized with 5mC peaks (Figure 3A). Moreover, enrichment of 5hmC modification was found on the binding sites of all tested transcription factors (Figure 3A).
Tet2 recruitment to the cytokine genes is dependent on lineage-specific key transcription factors
Recently, it has been reported that proteins of Tet family, TET1, TET2 and TET3, can regulate gene transcription by converting 5mC to 5hmC. As a first step to verify the role of Tet proteins in Th development, we examined the mRNA expression of Tet genes in each Th subset. Consistent with previous report (Ko et al., 2010), naïve CD4+ T cells expressed all Tet family members in high amounts (Figure 4A). Although the expression of all Tet members was down-regulated following TCR mediated-activation, Tet2 was highly expressed in each Th subset compared to other Tet genes. This result led us to a hypothesis that Tet2 may play a crucial role in Th differentiation.
Figure 4. The recruitment of Tet2 to the signature cytokine gene loci is dependent on lineage-specific transcription factors.
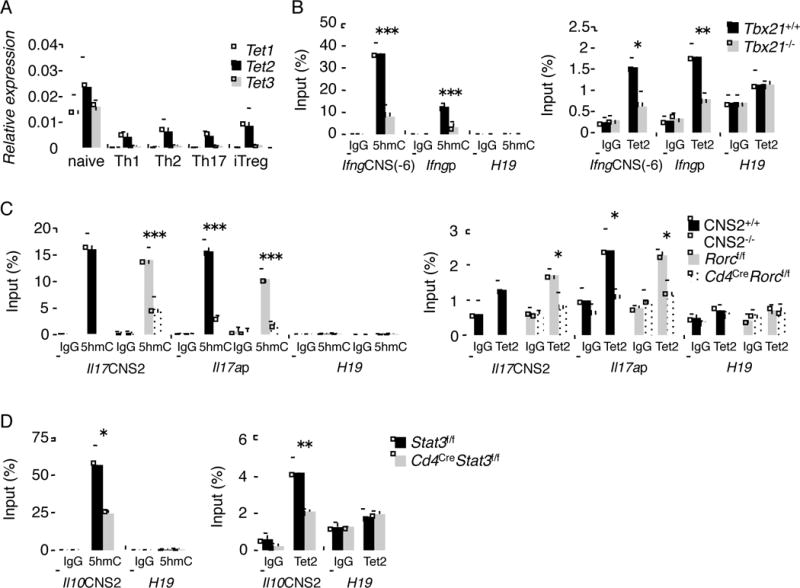
(A) Naïve T cells were cultured under differentiation conditions for each Th subset. Tet family mRNA expression was analyzed by real-time RT-PCR. (B) Naïve T cells from WT and Tbx21−/− were cultured under Th1 conditions for 4 days. 5hmC (left) and Tet2 accessibility (right) in Ifng gene were analyzed by hMeDIP- and ChIP-PCR, respectively. (C) Naïve T cells from littermate control and CNS2−/− or CD4CreRORγf/f mice were cultured under Th17 conditions for 4 days. 5hmC (left) and Tet2 accessibility (right) in Il17 gene were analyzed by hMeDIP- and ChIP-PCR, respectively. (D) Naïve T cells from CD4CreSTAT3f/f and littermate control mice were cultured under Th17 conditions for 4 days. 5hmC (left) and Tet2 accessibility (right) in Il10 gene were analyzed by hMeDIP- and ChIP-PCR, respectively. (A–D) The data represents the average of at least three independent experiments and shown as mean ± SD. “See also Figure S5.”
Since we have identified 5hmC-associated regions, we analyzed whether Tet2 is recruited to these loci in various Th lineages by chromatin-immunoprecipitation (ChIP) assay. As expected, in Th1 cells, Tet2 was found recruited to CNS(-6) and promoter regions of Ifng locus that contain 5hmC, but not to promoter region of H19 imprinted gene (Figure 4B). Tet2 needs co-factor(s) for binding to DNA, since it does not have CXXC DNA binding domain (Pastor et al., 2013). Previously, one group reported that T-bet was bound to CNS(-6) and promoter regions of Ifng locus in Th1 cells (Hatton et al., 2006). Thus, we decided to assess the role of T-bet in Tet2 recruitment. By hMeDIP and ChIP analysis using Tbx21-deficient T cells, 5hmC and Tet2 recruitment in Ifng gene were reduced in Tbx21-deficient Th1 cells (Figure 4B). Thus, these results suggest that Tet2 induces DNA demethylation at Ifng locus in a T-bet-dependent manner in Th1 cells.
Similarly, we examined the recruitment of Tet2 to Il17a and Il17f locus in Th17 cells. We previously reported that CNS2 region of Il17a and Il17f locus is essential for the function of ROR factors in Il17a expression (Wang et al., 2012). Thus, we examined the association of Tet2 in CNS2 and Il17a promoter region. As shown in Figure 4C, Tet2 associated with CNS2 and Il17a promoter regions, which correlated with existence of 5hmC at these sites. Furthermore, 5hmC and Tet2 recruitment were diminished in the absence of CNS2 region or RORγt (Figure 4C), indicating that Tet2 induces DNA demethylation at Il17 locus in a RORγt-dependent manner in Th17 cells. In contrast, although 5hmC peaks of lineage signature genes were observed in Th2 and iTreg cells, we could not detect the strong recruitment of Tet2 to these regions (Figure 2 and Figure S5A and B).
Previous study indicated that Th17 cells produce IL-10 in a STAT3-dependent manner (Stumhofer et al., 2007). Thus, we also analyzed Il10 locus under the Th17 condition by use of STAT3-deficient T cells or their controls. Tet2 was found recruited to CNS2 region of Il10 locus, which contained 5hmC, in a manner dependent on STAT3 (Figure 4D). Collectively, these results suggest that Tet2 induces active DNA demethylation of signature cytokine genes in particular Th subset and its action is dependent on lineage-specific transcription factors.
Tet2 regulates Th1 and Th17 cell differentiation
The above analysis has suggested 5hmC and Tet2 in Th differentiation. To further assess the role of Tet2 in Th cells, we analyzed the differentiation of each Th subset using Tet2-deficient CD4+ T cells (Tet2−/−) derived from Cd2CreTet2f/f mice (Moran-Crusio et al., 2011) in vitro. In accordance with previous reports (Ko et al., 2011), Cd2CreTet2f/f mice had normal development of CD4+ and CD8+ T cells and B cells (data not shown). In support of Figure S5A and B, both intracellular staining and qPCR analysis revealed that the differentiation of Th2 and iTreg cells were not affected in Tet2−/− T cells (Figure S5C–E, G and H). Additionally, 5hmC marks in signature genes were similar between Tet2+/+ and Tet2−/− T cells (Figure S5F and I). In contrast, deletion of Tet2 led to 80–83% reduction in genome-wide 5hmC peaks in Th1 and Th17 cells (Figure S6A). Most of the 20% residual 5hmC peaks in Tet2−/− cells existed in wild-type Th1 and Th17 cells, suggesting in addition to Tet2, other Tet family members may be also responsible for maintaining DNA demethylation process in Th1 and Th17 cells. We also analyzed the general difference in tag density of 5hmC around gene promoters, CpGIs, as well as all 5hmC sites appeared in wild-type Th cells. As shown in Figure S6A, the depletion of 5hmC around TSS and CpGIs was only slightly reduced in Tet2-deficient Th1 cells. However, the depletion of 5hmC in TSS and CpGIs in WT Th17 cells was totally reversed in Tet2-deficient Th17 cells. Moreover, the reduction of 5hmC tag density as a result of Tet2 deficiency was more profound in Th17 cells than in Th1 cells, suggesting that Tet2 deficiency may have more global effect on Th17 cells than on Th1 cells.
Under Th1 polarization condition, Tet2−/− T cells showed a marked reduction in IFN-γ at mRNA and protein expression, but Tbx21 expression and cell proliferation were not affected (Figure 5A and Figure S6B–D). Furthermore, 5hmC marks in Ifng gene were decreased by Tet2 deficiency in Th1 cells (Figure 5B). In contrast, we found that 5hmC status in Tbx21 gene was not changed by Tet2 deficiency (Figure S6E). Moreover, Tet2 did not bind to this region in Th1 cells (Figure S6F), suggesting the involvement of other Tet family proteins. The recruitment of T-bet in Ifng gene was also substantially decreased by Tet2 deficiency in Th1 cells (Figure 5C). Since Tet2 recruitment to Ifng locus is dependent on T-bet (Figure 4B), Tet2 and T-bet may reciprocally regulate the function of each other in Ifng expression. To globally identify the target genes of Tet2 in Th1 cells, we next performed DNA microarray analysis using Tet2+/+ and Tet2−/− Th1 cells. Tet2 target genes were then identified based on microarray and hMeDIP-seq data. A 1.5-fold change (WT vs KO) was used as a cutoff for significance. Although total 229 Th1-specific genes (genes expressed at least 2 fold higher in Th1 than Th17) had 5hmC in Th1 cells, surprisingly, only the expression of Ifng, Il3, and Prdm1 were changed due to Tet2 deficiency (1.3%, 3/229) (Figure 5D). On the other hand, we found 51 genes with at least 1.5-fold difference in gene expression between Tet2+/+ and Tet2−/− Th1 cells, 26 of which have 5hmC peaks on their promoters and/or gene bodies (Table S4), suggesting that Tet2 may regulate the expression of genes without 5hmC modification.
Figure 5. Tet2 is important for Th1 differentiation in vitro.
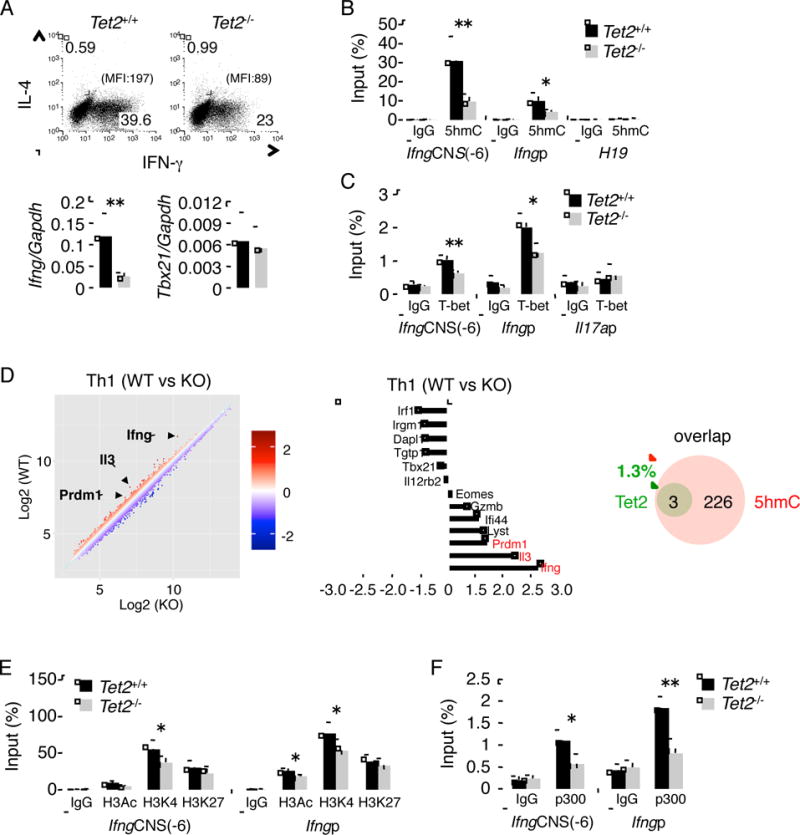
(A) upper; Intracellular cytokine staining following 5 days of Th1 differentiation from Tet2f/f and Cd2CreTet2f/f mice. Data is a representative of at least three individual experiments. lower; the expression of Ifng and Tbx21 mRNA by Tet2f/f and Cd2CreTet2f/f Th1 cells. (B, C) The hMeDIP (B) and ChIP (C) on Ifng gene were performed in Th1 cells from Tet2f/f and Cd2CreTet2f/f mice. (D) Microarray analysis comparing Tet2+/+ versus Tet2−/− Th1 cells. The bar graph showed representative genes. Venn diagram showing the number of 5hmC-containing genes affected or unaffected by Tet2 deficiency. 5hmC-containing genes were identified if at least one peak of α-5hmC binding was present on their promoter plus gene body regions (−3K to TTS). (E, F) The ChIP of Ifng gene was performed in Th1 cells from Tet2f/f and Cd2CreTet2f/f mice. (A–C, E, F) The data represent the average of at least three independent experiments. All the data are shown as mean ± SD. “See also Figure S5 and S6 and Table S4.”
As mentioned above, 5hmC partly correlates with H3K4me3 (Figure S4). Recently, it has been reported that Tet2 facilitates GlcNAcylation and H3K4 methylation (Deplus et al., 2013). Thus, to clarify whether Tet2 regulates the chromatin modification in Th cells, we analyzed H3Ac, H3K4me3 and H3K27me3 modifications at Ifng gene in Tet2−/− Th1 cells by ChIP assay. As shown in Figure 5E, in CNS(-6) and Ifng promoter regions, permissive histone marker H3Ac and H3K4me3 were modestly reduced in Tet2−/− Th1 cells. Additionally, to further understand how Tet2 might regulate chromatin accessibility, we also evaluated the recruitment of p300 to Ifng locus. The deficiency of Tet2 resulted in marked inhibition of the binding of p300 to CNS(-6) and promoter region of Ifng gene (Figure 5F).
Next, we sought to examine the role of Tet2 in Th17 cells in the same manner. Under Th17 differentiation conditions, Tet2−/− T cells showed a marked suppression in IL-17 mRNA and protein expression compared to Tet2+/+ T cells, but the expression of IL-17F, IL-2 and RORγt as well as cell expansion were normal (Figure 6A and Figure S6G–I). In addition, we examined the effect of Tet2 on 5hmC peaks and RORγt recruitment in Il17 locus. As expected, the deficiency of Tet2 led to a reduction of 5hmC and RORγt binding at CNS2 and Il17a promoter regions in Th17 cells (Figure 6B and C), implying a reciprocal regulation of Tet2 and RORγt function in Il17a expression. To assess the target genes of Tet2 in Th17 cells, we also performed microarray analysis using Tet2+/+ and Tet2−/− Th17 cells. Based on the analysis of microarray and MeDIP-seq results, only six genes (Ccl20, Il10, Gpr83, Emp1, Rbl2 and Il17a) were found as Tet2-regulated genes from total 197 Th17 specific genes (genes that are expressed at least 2 fold higher in Th17 than Th1 cells) with 5hmC in Th17 cells (3%, 6/197) (Figure 6D). Similar to the founding in Th1 cells, about 50% of genes with at least 1.5 fold difference in gene expression between Tet2+/+ and Tet2−/− Th17 cells had 5hmC peaks on their promoter and/or gene body regions (Table S4). To evaluate the effect of Tet2 on chromatin remodeling at Il17 locus, we examined histone modifications and p300 recruitment in Tet2−/− Th17 cells. As shown in Figure 6E, H3Ac and H3K4me3 were reduced in Tet2−/− Th17 cells along with increased repressive histone marker H3K27me3 at CNS2 and Il17a promoter. Importantly, the deficiency of Tet2 led to considerable inhibition in binding of p300 to CNS2 and Il17a promoter regions (Figure 6F).
Figure 6. Tet2 controls Th17 cell development in vitro.
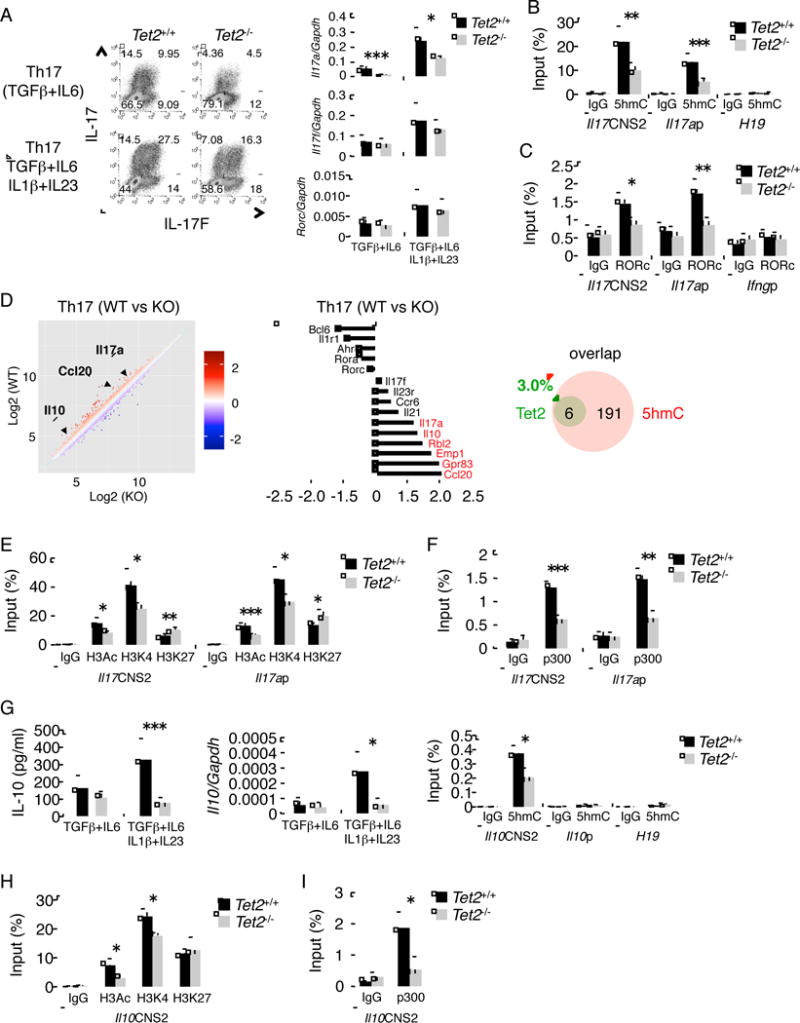
(A) left; Intracellular cytokine staining following 4 days of Th17 cell differentiation from Tet2f/f and Cd2CreTet2f/f mice. Data is a representative of at least three individual experiments. right; the expression of Il17a, Il17f and Rorc mRNA in Th17 cells from Tet2f/f and Cd2CreTet2f/f mice. (B, C) The MeDIP (B) and ChIP (C) on Il17 gene were performed in Th17 cells from Tet2f/f and Cd2CreTet2f/f mice. (D) Microarray analysis comparing Tet2+/+ versus Tet2−/− Th17 cells. The bar graph showed representative genes. Venn diagram showing the number of 5hmC-containing genes affected or unaffected by Tet2 deficiency. 5hmC-containing genes were identified if at least one peak of α-5hmC binding was present on their promoter plus gene body regions (-3K to TTS). (E, F) The ChIP of Il17 gene was performed in Th17 cells from Tet2f/f and Cd2CreTet2f/f mice. (G) left; ELISA analysis of supernatants from Th17 cell differentiation on Day 4. All groups are analyzed in triplicate. middle; the expression of Il10 mRNA in Th17 cells from Tet2f/f and Cd2CreTet2f/f mice. right; a hMeDIP of Il10 and H19 loci was performed in Th17 cells from Tet2f/f and Cd2CreTet2f/f mice. (H, I) The ChIP of Il10 gene was performed in Th17 cells from Tet2f/f and Cd2CreTet2f/f mice. (A–C, E–I) The data represent the average of at least three independent experiments. All the data are shown as mean ± SD. “See also Figure S6 and Table S4.”
Microarray and hMeDIP-seq analysis suggested that Tet2 regulates IL-10 production in Th17 cells. Indeed, under Th17 condition, Tet2−/− T cells displayed a considerable repression of IL-10 production compared with Tet2+/+ T cells (Figure 6G). In addition, we also evaluated 5hmC status of Il10 gene in Tet2−/− T cells. As expected, the ablation of Tet2 resulted in a reduction of 5hmC at CNS2 region but not Il10 promoter (Figure 6G). To elucidate the impact of Tet2 on chromatin structure of Il10 gene locus, we analyzed histone modifications and p300 recruitment in CNS2 region of Il10 gene in Tet2−/− Th17 cells. H3Ac and H3K4me3 marks as well as p300 recruitment were diminished in the absence of Tet2 (Figure 6H and 6I). As previously reported (Ouyang et al., 2011), IL-10-producing IFN-γ+ Th1 are present in a variety of infections and they play an important task of protecting against severe immune-mediated pathology. Thus, we also assessed the role of Tet2 in IL-10 production from Th1 cells. Similar to Th17 cells, the induction of IL-10 and 5hmC mark in CNS2 region of Il10 gene was strongly abrogated by Tet2 deficiency in Th1 cells (data not shown).
Collectively, our results demonstrate that Tet2 promotes the development of Th1 and Th17 cells by regulating specifically the expression of signature cytokines through active DNA demethylation and its deficiency had some effect on chromatin remodeling, especially p300 recruitment to the target genes.
Tet2 controls IL-10, IFN-γ and IL-17 production in autoimmunity
To examine whether Tet2 is critical in Th differentiation in vivo, we first immunized mice with MOG35–55 peptide and analyzed the spleens of Tet2f/f and Cd2CreTet2f/f mice on day 7. Similar to in vitro results, we found that the MOG-specific production of IFN-γ and IL-10 were decreased in Cd2CreTet2f/f mice (Figure S7A). There was no or slight difference in the production of MOG-specific IL-17F and IL-2 between Tet2f/f and Cd2CreTet2f/f mice (Data not shown). However, in conflict with in vitro results, the production of MOG-specific IL-17 was comparable between Tet2f/f and Cd2CreTet2f/f mice (Figure S7A). It has been known that IL-10 plays a critical role in suppressing autoimmunity and inflammatory responses. Previous study demonstrated that IL-10 produced by Th17 cells restrains the pathologic effects of Th17 (McGeachy et al., 2007). Thus, to clarify whether the unexpected high production of IL-17 in Cd2CreTet2f/f mice was caused by the reduced IL-10 production, we treated mice with IL-10 receptor 1-specific blocking antibody (α-IL-10R Ab). After treating with α-IL-10R Ab, Cd2CreTet2f/f mice showed the marked reduction of MOG-specific IL-17 and IFN-α production (Figure 7A), implying that Tet2 plays a similar role in Th1 and Th17 cell differentiation in vitro and in vivo.
Figure 7. Tet2 regulates cytokines production in autoimmunity.
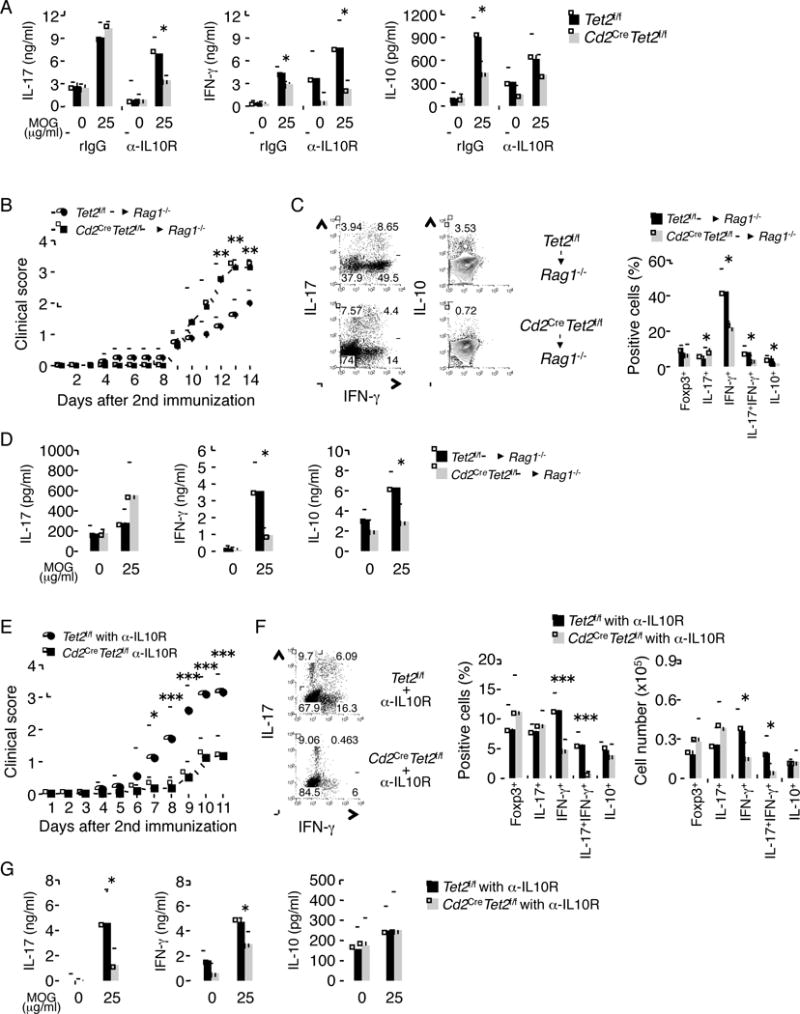
(A) Age-matched Tet2f/f and Cd2CreTet2f/f mice (n = 4) were immunized with MOG35–55 peptides on day 0 and then analyzed on day 7 for IL-17, IFN-γ and IL-10 production. For neutralization of IL-10, α-IL-10R or α-Rat IgG1 was intraperitoneally injected on day 0 and day 4. Data are a representative of at least two individual experiments. (B) Rag1-deficient mice were reconstituted with CD4+ T cells derived from Tet2f/f and Cd2CreTet2f/f mice. Clinical scores were monitored daily following EAE induction with MOG35–55. (C) left; the IL-17, IFN-γ and IL-10 was determined in the infiltrates of CNS of immunized mice. right; statistic of the frequency. (D) IL-17, IFN-γ and IL-10 was measured by ELISA in the splenocytes of immunized mice, after restimulation with indicated concentration of MOG peptide (μg/ml). (E) Age-matched Tet2f/f and Cd2CreTet2f/f mice were immunized twice with MOG35–55. For neutralization of IL-10, 500 μg α-IL-10R or isotype control antibody was administered intraperitoneally at every 4 days after immunization. Shown here is the combinational result of two individual EAE experiments (the total number of mice used: WT = 8, KO = 10). (F) left; the IL-17 and IFN-γ was determined in the infiltrates of CNS of immunized mice with α-IL-10R. right; statistic of the frequency and absolute number. (G) IL-17, IFN-γ and IL-10 was measured by ELISA in the splenocytes of immunized mice with α-IL-10R, after restimulation with indicated concentration of MOG peptide (μg/ml). Data are a representative of at least two individual experiments. All the data are shown as mean ± SD. “See also Figure S7.”
To further address the function of Tet2 in autoimmune responses, both Tet2f/f and Cd2CreTet2f/f mice were subjected to MOG peptide-induced experimental autoimmune encephalomyelitis (EAE)- a mouse model for human multiple sclerosis (Baxter, 2007). The severity of EAE diseases was enhanced in Tet2-deficient mice (Figure S7B). The frequency and absolute number of IL-17+ T cells in central nervous system (CNS) on day 13 after 2nd immunization were strongly elevated in Cd2CreTet2f/f mice, whereas the frequencies and absolute numbers of IFN-γ+, IFN-γ+IL-17+, IL-10+ and IL-10+IL-17+ T cells in CNS were decreased in Cd2CreTet2f/f mice (Figure S7C and D). Coincide with in vitro, the frequency and absolute number of Foxp3+ T cells in CNS were similar between Tet2f/f or Cd2CreTet2f/f mice (Figure S7D). The Cd2-Cre transgenic mice are useful for generating conditional mutations not only in T cells but also in B cells (de Boer et al., 2003). To confirm the CD4+ T cell intrinsic function of Tet2 in vivo, we transferred Tet2f/f or Cd2CreTet2f/f CD4+ T cells into Rag1−/− mice and immunized them with MOG peptide. The mice transferred with Tet2f/f CD4+ T cells developed disease score around 2.0, whereas the mice transferred with Cd2CreTet2f/f CD4+ T cells had more severe symptoms (score around 3.0) (Figure 7B). The frequencies of IFN-γ+, IFN-γ+IL-17+ and IL-10+ T cells in CNS on day 14 after 2nd immunization, but not IL-17+ and Foxp3+ T cells, were much lower in the mice that received Cd2CreTet2f/f CD4+ T cells than Tet2f/f CD4+ T cells (Figure 7C). Furthermore, we found that a marked reduction of IFN-γ and IL-10 secretion, but not IL-17, were also observed by ELISA assays in the splenocytes of mice that received Cd2CreTet2f/f CD4+ T cells (Figure 7D). These results indicate that Tet2 expression in CD4+ T cells plays an important role in the suppression of disease severity of EAE.
To address the mechanism whereby Tet2 restrains the disease severity in EAE, we did EAE experiments in Tet2f/f and Cd2CreTet2f/f mice with α-IL-10R Ab treated at every 4 days during EAE. As a result of the treatment, the severity of EAE diseases was reduced in α-IL-10R Ab treated Cd2CreTet2f/f mice (Figure 7E). The enhancement of IL-17+ T cells in Cd2CreTet2f/f mice was abolished by neutralization of IL-10 (Figure 7F). Furthermore, the frequencies and absolute numbers of IFN-γ+ and IFN-γ+IL-17+ T cells but not Foxp3+ and IL-10+ T cells in CNS on day 11 after 2nd immunization were decreased in α-IL-10R Ab treated Cd2CreTet2f/f mice (Figure 7F). In addition, the strong reduction of IFN-γ and IL-17 secretion was also observed in splenocytes of α-IL-10R Ab-treated Cd2CreTet2f/f mice (Figure 7G).
Taken together, we conclude that Tet2 plays an important role in regulating the expression of IL-10, IL-17 and IFN-γ in vivo and T cell-mediated autoimmune diseases.
Discussion
Epigenetic mechanisms have been proposed to regulate the specification and plasticity of Th cell lineages (Wei et al., 2009). Active DNA demethylation by Tet proteins has been recently identified with critical roles in many physiological processes. Here we provide the first evidence that 5hmC and Tet2 play important roles in T cell function, particularly cytokine expression.
In this study, we first generated genome-wide maps of 5mC and 5hmC modifications in Th cell subsets differentiated in vitro by use of high-throughput DIP-Seq approach. Consistent with other studies, we found that 5hmC is enriched in gene body and enhancer regions. However, a decrease of the overall amount of 5hmC was found when naïve CD4+ T cells differentiated into effector cells. 5hmC cannot be recognized by DNMT1, therefore, cannot be maintained during DNA replication and cell proliferation (Valinluck and Sowers, 2007). A proliferation dependent passive DNA demethylation, suggested by the decrease of 5hmC during differentiation of naïve CD4+ T cells into effectors, may be crucial for the relief of methylation-dependent transcriptional repression in naïve T cells. More interestingly, the lineage-specific cytokine as well as some of the master transcription factors were tightly regulated by DNA methylation in naïve T cells. The repression of these genes expression by 5mC was only removed in the lineage specified by the factors, but was maintained in other lineages, strongly suggesting that the transition from 5mC to 5hmC was an active (replication independent) DNA demethylation process.
It is unclear whether DNA demethylation-dependent gene regulation is controlled by lineage-defining transcription factors. Our data suggest that a proportion of key transcription factor as well as p300 binding sites co-localize with 5hmC but not 5mC in Th1 and Th17 cells, as well as Th2 cells, but not iTreg cells (data not shown). More importantly, the recruitment of Tet2 as well as DNA demethylation is dependent upon key transcription factors such as T-bet, RORγt, and Stat3, indicating a critical role of these transcription factors in the active DNA demethylation and chromatin restructuring process during T cell differentiation. Although the regulation of gene expression by Tet2 seems to be gene specific, the molecular mechanism by which these transcription factors control gene-specific recruitment of Tet2 remains to be determined. The key transcription factors for Treg cells does not colocalize with 5hmC or 5mC (data not shown), suggesting a differential mechanism by which iTreg is epigenetically regulated at the methylation level.
Increasing evidence suggests that DNA methylation is intimately linked to histone modifications. However, precisely how Tet-mediated DNA demethylation and chromatin modifications cooperatively contribute to gene expression has not yet been clearly defined. Here, the integrative analysis of our DIP-seq results and published histone modification data indicated that 5hmC and H3K4me3 modifications were partly colocalized at the regulatory elements of signature genes in Th cells. In support of our findings, recent studies reported that 5hmC and Tet proteins are enriched in euchromatin (Mellen et al., 2012; Deplus et al., 2013). In addition, we found that Tet2 deficiency decreased the recruitment of p300 to promoters and enhancer regions. These results suggest possible regulation of chromatin configuration by Tet proteins.
Our genome-wide analysis revealed a 80–83% reduction of 5hmC modification in Tet2−/− Th1 and Th17 cells as compared to control, suggesting in addition to signature cytokines, Tet2 may also control 5hmC on many other loci across the whole genome in Th1 and Th17 cells. While, about 20% 5hmC were not affected by Tet2 deficiency. Thus, other Tet family members may control demethylation process on these loci in Th1 and Th17 cells. In deed, despite the possession of lineage-specific 5hmC, Tbx21 in Th1 cells and IL-17F in Th17 cells were unaffected by Tet2 depletion. Furthermore, the integrated analysis of Microarray and hMeDIP-seq indicated that the vast majority of expressed genes bearing 5hmC in Th1 and Th17 cells are not affected by Tet2 deficiency in vitro. In addition, we found that the alteration in cytokine expression in Tet2-deficient cells was partial. These results led to a hypothesis that Tet2 function might be compensated by other Tet family members. Moreover, we also found that Tet2 deficiency did not affect Th2 and iTreg differentiation or 5hmC peaks on their signature genes. Thus, further assessment using genetically modified mice of other Tet proteins may yield additional insight into the roles of Tet-mediated active DNA demethylation in Th cell differentiation.
Among all genes with at least 1.5 fold differences in their expression between Tet2+/+ and Tet2−/− Th1 and Th17 cells, 50% of them do not have 5hmC peaks associated with their proximal promoters and gene body. Therefore, these results suggest that Tet2 may regulate these genes indirectly. Alternatively, several reports recently demonstrated that 5hmC could be further oxidized to 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) by Tet proteins (Pastor et al., 2013). Moreover, active DNA demethylation pathways through these modifications have actually been shown in mESCs (Pastor et al., 2013). Hence, we do not exclude the possibility that 5hmC in these genes may already be oxidized to 5fC/5caC. The roles of these oxidants in Th cells differentiation will need to be studied in future studies.
We observed that Tet2-mediated active DNA demethylation controls cytokines production in vivo. Importantly, IL-10 production from CD4+ T cells by Tet2 plays a crucial role in prevention of excessive inflammation in EAE. Type 1 regulatory T (Tr1) cells predominantly produce IL-10 and are differentiated by IL-27 and TGFβ (Awasthi et al., 2007). We found that development of Tr1 cells, which express IFN-γ and IL-10, were inhibited in the absence of Tet2 (data not shown), suggesting that Tet2 plays a crucial role in Tr1 cell development. Although the source of IL-10 production among Th subsets in vivo remains to be elucidated, several papers have proposed that IL-10 from Th1, Tr1 and Th17 cells play a crucial role in limiting the pathologic effects of Th17 during EAE (Ouyang et al., 2011). Thus, we believe that Tet2-mediated IL-10 production from Th1, Tr1 and Th17 cells is a key mechanism for prevention from exacerbation of EAE symptoms.
In summary, our results not only provide a global view of a novel epigenetic modification that occurs during Th differentiation, but also uncover a functional role for Tet2-mediated active DNA demethylation in the function of Th cells both in vitro and in vivo. Further assessment of TET proteins and 5mC oxidants will advance our understanding on the molecular mechanisms underlying dynamic changes of DNA methylation in T cell development and will provide new therapeutic targets for innovative treatments of autoimmune diseases.
Experimental Procedures
Mice
Cd2CreTet2f/f mice have been described previously (Moran-Crusio et al., 2011). Cd2CreTet2f/f mice and wild-type littermates on the mixed background were used in experiments. CD4CreRorcf/f mice and Il17FCre mice were generated in our lab (manuscript in preparation). Il17fCre mice and Rosa26yfp mice were crossed to generate Il17fCreRosayfp mice. Ifngyfp mice were generously provided by R.M. Locksley (University of California, San Francisco). C57BL/6 mice, Tbx21−/− mice (on the B6 background), Il4gfp mice, Rosa26yfp mice and Rag1−/− mice (on the B6 background) were from the Jackson Laboratory. All the mice were housed in the SPF animal facility at the M.D. Anderson Cancer Center and the animal experiments were performed at the age of 6–12 weeks with the use of protocols approved by the Institutional Animal Care and Use Committee.
Naïve T cell Preparation and Differentiation
Naive T cells were isolated by sorting CD4+CD25−CD62LhighCD44low cells from spleens and lymph nodes, differentiated under several Th cell conditions, and analyzed as described (Yang et al., 2008). The naïve T cells (5 × 105 cells/well) were stimulated with the plate-bound α-CD3 (1 μg/ml) and the soluble α-CD28 (1 μg/ml). For Th0 differentiation, the cells were treated with 5 μg/ml α-IFN-γ (XMG1.2; BioXCell), 5 μg/ml α-IL-4 (11B11; BioXCell) and 30 U/ml IL-2. For Th1 differentiation, the cells were treated with 10 ng/ml mIL-12 (Peprotech), 5 μg/ml α-IL-4 and 30 U/ml IL-2. For Th2 differentiation, the cells were treated with 10 ng/ml mIL-4 (Peprotech), 5 μg/ml α-IFN-γ. For iTreg differentiation, the cells were treated with 1 ng/ml TGF-β1 (Peprotech), 5 μg/ml α-IFN-γ, 5 μg/ml α-IL-4 and 30 U/ml IL-2. For Th17 differentiation, the cells were treated with 0.5 ng/ml TGF-β1, 10 ng/ml IL-6 (Peprotech), 5 μg/ml α-IFN-γ, and 5 μg/ml α-IL-4. When indicated, 10 ng/ml IL-23 and 10 ng/ml IL-1β (Peprotech) were used for optimal Th17 cell differentiation.
DIP-sequencing and Data Analysis
The genomic DNA was purified and sonicated. DNA fragments (4 μg) were denatured and incubated with antibody against 5mC (Eurogentec), 5hmC (Active Motif) or control IgG at 4°C overnight. The IPed DNA fragments were prepared with Dynabeads Protein G (Life Technologies) and amplified using the Illumina ChIP-seq DNA preparation kit (IP-202-1012), and sequenced on the Illumina GA II and HiSeq2500 sequencing platforms.
The unique reads for 5mC and 5hmC were mapped into non-overlapping 200 bp windows of mouse genome (mm9). The peaks were called using SICER (Zang et al., 2009), with the Input DNA sample as a control and a FDR of 0.01 as the cutoff. To determine whether the sequencing depth was enough to cover 5mC and 5hmC libraries, we carried out a scaling analysis. The number of peaks identified from fraction of randomly sampled reads in each library using SICER was plotted. An estimation of the total reads beyond which no more peaks would be identified was determined. To calculate the tag density of 5mC and 5hmC, the number of reads was first summed in 5 or 100-bp windows within the regions of 5 kb upstream and downstream of TSS or CpGIs or within the regions of 2 kb upstream and downstream of the centers of all p300/TF binding sites. All tag counts were then normalized by the total number of bases within the windows and the total number of reads in the given library.
Statistical Analysis
The statistical significance was determined by Student’s paired two-tailed t test (*p < 0.05; **p < 0.01; ***p < 0.005). All error bars shown in this article represent standard deviations.
Supplementary Material
Acknowledgments
We thank our colleagues for their help and suggestions. The work was supported in part by grants from NIH to C.D. (AR050772) and grants from National Basic Research Program of China (2013CB967001 and 2015CB964601) to L.W.. K.I. receives JSPS postdoctoral fellowships for research abroad. C.D. is a Bayer Chair Professor at Tsinghua University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers
The DIP-Seq and microarray data are deposited in GEO. Accession number is GSE66268.
Conflict of Interest
The authors declare no competing financial interests.
Author Contributions
C.D. and K.I. designed the research and analyzed the data. P.Z. co-supervised some of the experiments. K.I. performed the most of experiments, and X. W., B.-S.K. and S.T. participated in specific experiments, T.C., Y.D., and Y.Z. carried out Illumina sequencing experiments. D.N.-L. and I.A. provided Cd2CreTet2f/f mice. X.Y., Q.T., T.C. and L.W. analyzed microarray and DIP-seq data. K.I., L.W. and C.D. prepared the manuscript.
References
- Awasthi A, Carrier Y, Peron JP, Bettelli E, Kamanaka M, Flavell RA, Kuchroo VK, Oukka M, Weiner HL. A dominant function for interleukin 27 in generating interleukin 10-producing anti-inflammatory T cells. Nat Immunol. 2007;8:1380–1389. doi: 10.1038/ni1541. [DOI] [PubMed] [Google Scholar]
- Baxter AG. The origin and application of experimental autoimmune encephalomyelitis. Nat Rev Immunol. 2007;7:904–912. doi: 10.1038/nri2190. [DOI] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- Ciofani M, Madar A, Galan C, Sellars M, Mace K, Pauli F, Agarwal A, Huang W, Parkurst CN, Muratet M, et al. A validated regulatory network for Th17 cell specification. Cell. 2012;151:289–303. doi: 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer J, Williams A, Skavdis G, Harker N, Coles M, Tolaini M, Norton T, Williams K, Roderick K, Potocnik AJ, Kioussis D. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur J Immunol. 2003;33:314–325. doi: 10.1002/immu.200310005. [DOI] [PubMed] [Google Scholar]
- Deplus R, Delatte B, Schwinn MK, Defrance M, Mendez J, Murphy N, Dawson MA, Volkmar M, Putmans P, Calonne E, et al. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. Embo J. 2013;32:645–655. doi: 10.1038/emboj.2012.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatton RD, Harrington LE, Luther RJ, Wakefield T, Janowski KM, Oliver JR, Lallone RL, Murphy KM, Weaver CT. A distal conserved sequence element controls Ifng gene expression by T cells and NK cells. Immunity. 2006;25:717–729. doi: 10.1016/j.immuni.2006.09.007. [DOI] [PubMed] [Google Scholar]
- Kanno Y, Vahedi G, Hirahara K, Singleton K, O’Shea JJ. Transcriptional and epigenetic control of T helper cell specification: molecular mechanisms underlying commitment and plasticity. Annu Rev Immunol. 2012;30:707–731. doi: 10.1146/annurev-immunol-020711-075058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M, Bandukwala HS, An J, Lamperti ED, Thompson EC, Hastie R, Tsangaratou A, Rajewsky K, Koralov SB, Rao A. Ten-Eleven-Translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proc Natl Acad Sci U S A. 2011;108:14566–14571. doi: 10.1073/pnas.1112317108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku CS, Naidoo N, Wu M, Soong R. Studying the epigenome using next generation sequencing. J Med Genet. 2011;48:721–730. doi: 10.1136/jmedgenet-2011-100242. [DOI] [PubMed] [Google Scholar]
- McGeachy MJ, Bak-Jensen KS, Chen Y, Tato CM, Blumenschein W, McClanahan T, Cua DJ. TGF-beta and IL-6 drive the production of IL-17 and IL-10 by T cells and restrain T(H)-17 cell-mediated pathology. Nat Immunol. 2007;8:1390–1397. doi: 10.1038/ni1539. [DOI] [PubMed] [Google Scholar]
- Mellen M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151:1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, Figueroa ME, Vasanthakumar A, Patel J, Zhao X, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20:11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkura N, Hamaguchi M, Morikawa H, Sugimura K, Tanaka A, Ito Y, Osaki M, Tanaka Y, Yamashita R, Nakano N, et al. T cell receptor stimulation-induced epigenetic changes and Foxp3 expression are independent and complementary events required for Treg cell development. Immunity. 2012;37:785–799. doi: 10.1016/j.immuni.2012.09.010. [DOI] [PubMed] [Google Scholar]
- Ouyang W, Rutz S, Crellin NK, Valdez PA, Hymowitz SG. Regulation and functions of the IL-10 family of cytokines in inflammation and disease. Annu Rev Immunol. 2011;29:71–109. doi: 10.1146/annurev-immunol-031210-101312. [DOI] [PubMed] [Google Scholar]
- Pastor WA, Aravind L, Rao A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat Rev Mol Cell Biol. 2013;14:341–356. doi: 10.1038/nrm3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- Shen L, Zhang Y. 5-Hydroxymethylcytosine: generation, fate, and genomic distribution. Curr Opin Cell Biol. 2013;25:289–296. doi: 10.1016/j.ceb.2013.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- Stumhofer JS, Silver JS, Laurence A, Porrett PM, Harris TH, Turka LA, Ernst M, Saris CJ, O’Shea JJ, Hunter CA. Interleukins 27 and 6 induce STAT3-mediated T cell production of interleukin 10. Nat Immunol. 2007;8:1363–1371. doi: 10.1038/ni1537. [DOI] [PubMed] [Google Scholar]
- Vahedi G, Takahashi H, Nakayamada S, Sun HW, Sartorelli V, Kanno Y, O’Shea JJ. STATs shape the active enhancer landscape of T cell populations. Cell. 2012;151:981–993. doi: 10.1016/j.cell.2012.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valinluck V, Sowers LC. Endogenous cytosine damage products alter the site selectivity of human DNA maintenance methyltransferase DNMT1. Cancer Res. 2007;67:946–950. doi: 10.1158/0008-5472.CAN-06-3123. [DOI] [PubMed] [Google Scholar]
- Wang X, Zhang Y, Yang XO, Nurieva RI, Chang SH, Ojeda SS, Kang HS, Schluns KS, Gui J, Jetten AM, Dong C. Transcription of Il17 and Il17f is controlled by conserved noncoding sequence 2. Immunity. 2012;36:23–31. doi: 10.1016/j.immuni.2011.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, Cui K, Kanno Y, Roh TY, Watford WT, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–167. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XO, Pappu BP, Nurieva R, Akimzhanov A, Kang HS, Chung Y, Ma L, Shah B, Panopoulos AD, Schluns KS, et al. T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma. Immunity. 2008;28:29–39. doi: 10.1016/j.immuni.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang C, Schones DE, Zeng C, Cui K, Zhao K, Peng W. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25:1952–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations (*) Annu Rev Immunol. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


