Abstract
This protocol describes the application of the MS2 system to the yeast Saccharomyces cerevisiae. ASH1 mRNA tagged with six MS2 repeats (6MBSs) is used to follow the localization of the ASH1 mRNA particles to the bud tip of a haploid yeast cell. W303 yeast cells transformed with pG14-MS2-GFP and pGAL-lacZ-MS2-ASH1 are grown on select medium lacking tryptophan and leucine. RNA expression is induced by the addition of galactose, and a time-lapse movie is then acquired.
MATERIALS
Reagents
Concanavalin A (Sigma)
Galactose (30%)
SD-Leu-Trp medium containing 2% glucose
SD-Leu-Trp medium containing 2% raffinose
Yeast cells (W303) transformed with pG14-MS2-GFP and pGAL-lacZ-MS2-ASH1
Many factors can lead to cells with high levels of autofluorescence, rendering them unsuitable for the studies described here. Cells should always be grown in minimal medium (e.g., SD medium) because YPD medium produces autofluorescence. Yeast cells grown to high density (OD600 >1.5) will also show high autofluorescence. A culture started from an older plate of cells should be grown for 2 d to allow the cells to lose their vacuoles and other inclusions. Do not use yeast strains harboring mutations in the ADE1 or ADE2 genes, because those cells accumulate a pigment that increases autofluorescence.
Equipment
Centrifuge
Erlenmeyer flask (250 mL)
Fluorescence microscope with appropriate attachments
Although it is simple to generate a targeted knock-in or deletion in yeast strains, it is challenging to image them, because yeast cells are smaller than a mammalian cell nucleus. It is important to use proper optics and cameras, otherwise the size of a pixel on the camera will be close to the size of a yeast nucleus (the diameter of haploid yeast cell nucleus is <2 μm). At least a 100X objective (with a NA ≥ 1.4) is required and the camera should have pixel sizes as small as possible. A high-magnification objective (Olympus 150X 1.45 NA TIRF) allows the use of cameras with a larger pixel size. These usually have faster read-out times, in order to track single molecules with good time resolution.
Glass coverslips or glass-bottom dishes suitable for live-cell imaging (e.g., Delta T Dishes, Bioptechs, Inc.)
The Delta T system provides the best z-stability and is easy to handle. It also allows one to use the same microscope setup as for mammalian cells.
Incubator (preset to 30°C), with rotating wheel/shaking platform
Spectrophotometer
Tube (conical, 50-mL)
Vacuum aspirator (optional; see Step 9)
Vortex mixer
METHOD
Inoculate a Preculture
1. Inoculate 5 mL of SD-Leu-Trp containing 2% glucose with a single yeast colony in a 50-mL conical tube.
2. Incubate the culture overnight on a rotating wheel at 30°C.
Start the Imaging Culture
3. The next morning, remove the culture from the incubator and leave it on the bench. This is the starter culture, which should not be too dense (OD600 <2).
If the OD600 is too high, too many cells were inoculated or the culture was incubated for too long.
4. In midafternoon, collect the cells by centrifugation at 1000g for 5 min.
5. Wash the cells by adding 5 mL of SD-Leu-Trp containing 2% raffinose.
6. Repeat Step 4.
7. Add 5 mL of SD-Leu-Trp containing 2% raffinose, and resuspend the cells by vortexing. Measure the OD600 of the culture and dilute to OD600 = 0.01 in a 250-mL Erlenmeyer flask containing 50 mL of SD-Leu-Trp with 2% raffinose.
8. Incubate the culture on a shaking platform at 200 rpm overnight at 30°C.
In the morning, the cells should be ready to image. They should be in the early exponential phase of growth, with OD600 <1.
Prepare Imaging Dishes
One critical step to image yeast cells is to keep them stably attached to the glass surface of the dish. To avoid having cells float away during a time-lapse movie, use a lectin to noncovalently attach them to the glass.
9. Add 500 μL of concanavalin A at 1 μg/μL to the Delta T dish and incubate for 5 min at room temperature. Remove liquid using a vacuum aspirator or a pipette.
A small film of concanavalin A will stay on the glass.
10. Allow the dish to dry. (Put in a fume hood to accelerate evaporation.) Use dishes immediately or store for several weeks at room temperature.
11. Combine 40 μL of yeast cells from Step 8 with 960 μL of SD-Leu-Trp containing 2% raffinose (preheated to 30°C) and add to the concanavalin A-coated Delta T dish. Incubate for 30 min at 30°C.
Cells will settle to the bottom of the dish and attach to the surface.
12. Place the Delta T dish on the stage. To avoid evaporation of the medium, place a lid over the dish.
Induce ASH1 mRNA Expression and Localize with a Time-Lapse Movie
13. Add 100 μL of 30% galactose very slowly to the cells by adding the solution to the side wall of the dish. This will induce ASH1 mRNA expression within 10 min.
Add the galactose slowly to the medium, to prevent the cells from detaching.
14. Acquire 4D datasets using at least 0.2- to 0.3-μm z-steps. Ten z-steps are sufficient to ensure that the particle remains in the imaging planes during acquisition. ASH1 mRNA particles are bright and move at a speed around 0.2-0.4 μm/sec. To obtain good tracks, it is therefore necessary to acquire at least 3 images/sec.
Most microscopes will not allow 4D imaging at this speed and movies must be acquired in a single imaging plane. As a result of imaging in a single plane, many movies will miss particles in some frames as they move in the axial plane. To track a particle over a longer timescale, acquire many movies to find one where the signal stays in focus. See Troubleshooting.
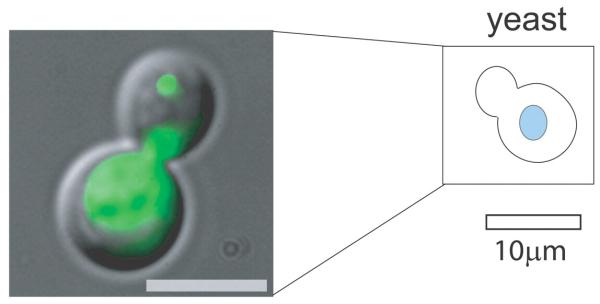
Figure 1 shows localization of the daughter-cell-specific ASH1 mRNA to the bud tip of a yeast daughter cell.
Figure 1.
Localization of the daughter-cell-specific ASH1 mRNA to the bud tip of a daughter cell in S. cerevisiae. Bar, 5 μm. (Reprinted from Bertrand et al. 1998, with permission from Elsevier © 1998.)
TROUBLESHOOTING
Many of the points described in Imaging Real-Time Gene Expression in Mammalian Cells with Single-Transcript Resolution (PMID: 21356977) are also valid in yeast. This includes phototoxicity, bleaching of xFP signals, z-stage stability, and camera settings. Therefore, only yeast-specific troubleshooting points are addressed here.
Problem: No RNA particles are observed.
[Step 14]
Solution: Consider the following:
1. GFP- or RNA-expressing plasmids may not be present. Be sure that cells are always grown under correct selective conditions. Without selection, plasmids will be lost.
2. Cells may have been grown with the wrong sugar source. Galactose induction only works when no glucose is present in the medium.
3. The RNA-expressing plasmid may have lost MBSs.
Problem: There is strong autofluorescence.
[Step 14]
Solution: Consider the following:
1. Cells may have accumulated metabolites responsible for the strong autofluorescence. Dilute the cells and grow for at least 1 d. Ensure that cells never reach OD600 >1. Rapidly dividing cells will lose autofluorescence over time.
2. Do not use strains that have mutations in ADE1 or ADE2 genes, which will cause cells to accumulate a red pigment.
Problem: Cells detach from the glass surface.
[Step 14]
Solution: Consider the following:
1. The concanavalin A-coated Delta T dishes may be too old. Make freshly coated dishes.
2. Incubate cells for longer times to allow them to settle and attach to the dish.
DISCUSSION
The MS2 system was developed and used for the first time in yeast (Bertrand et al. 1998). Imaging MS2-labeled mRNAs in yeast is not very different from what has been described for mammalian cells, but there are yeast-specific factors to be considered. As with mammalian cells, expressing the right concentration of the MCP-xFP fusion protein is important. Vectors expressing MCP-GFP with or without an NLS should be used, depending on what step of the mRNA expression pathway the experiment addresses. Selected MCP-GFP expression vectors for yeast are shown here:
| Vector | MCP- fusion |
Promoter Selection | Reference | |
|---|---|---|---|---|
| pG14- | NLS- | |||
| MS2- | MCP- | ampR, LEU, 2 | Bertrand et al. (1998) | |
| GFP | GFP | GDP | μm | |
| GFP- | Beach et al. (1999) | |||
| pCP-GFP | MCP | MET25 | ampR, HIS,CEN | |
The pG14-MS2-GFP construct constitutively expresses an NLS-MS2-GFP from a strong glyceraldehyde-3-phosphate dehydrogenase (GPD) promoter, leading to very high nuclear MCP-GFP accumulation (Bertrand et al. 1998). The strong nuclear signal does not allow mRNA detection in the nucleus, but this construct is ideal for cytoplasmic RNA localization studies. An MCP-GFP fusion without an NLS is expressed from the pCP-GFP vector, leading to MCP-GFP expression all over the cell. This makes it more difficult to detect single GFP tagged mRNAs and careful titration of GFP levels is necessary. To allow different levels of GFP expression, the MCP-GFP fusion is expressed from an inducibleMET25 promoter. Expression can be induced by changing to a medium lacking methionine. Variation in expression levels is controlled by the time the cells are grown in medium lacking methionine. Therefore, low MCP-GFP concentrations can be achieved with shorter induction times in medium lacking methionine.
ACKNOWLEDGMENTS
This work is supported by the National Institutes of Health grants AR41480 (R.H.S.) and 5P01 CA100324 (J.S.C.).
Footnotes
For basic yeast manipulations, see Methods in Yeast Genetics: A Cold Spring Harbor Laboratory course Manual (Amberg et al. 2005) and http://www.yeastgenome.org/.
References
- Amberg DC, Burke D, Strathern JN. Methods in yeast genetics: A Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2005. 2005. [Google Scholar]
- Beach DL, Salmon ED, Bloom K. Localization and anchoring of mRNA in budding yeast. Curr. Biol. 1999;9:569–578. doi: 10.1016/s0960-9822(99)80260-7. [DOI] [PubMed] [Google Scholar]
- Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. Localization of ASH1 mRNA particles in living yeast. Mol. Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]