Abstract
We have previously developed technology for multiplexing probes for the detection of transcription of many genes simultaneously within single cells. This has allowed us to determine the spatial localization of multiple genes with respect to each other in the nucleus, and ultimately the expression profile of the cell with respect to surrounding cells in a tissue. Six parameters of transcriptional organization in individual cells from culture and tissue were used to characterize significant differences in intracellular and intercellular expression patterns while preserving cellular morphology and histological context. We found that, unlike yeast, mammalian expression is excluded from the periphery and in addition, a subtle but complex organization underlies the transcriptional activity of these cells, both intra- and intercellularly. The approach has sufficient spatial resolution to be applied to the detection of chromosomal translocations or the identification of cancer cells.
Transcription is an early indicator of cellular response to micro-environmental stimuli, so examining transcription can provide a glimpse into a cell's response and its environment. Furthermore, it has been suggested that the peripheral spatial distribution of transcription underlies gene activity, at least in yeast [Casolari et al., 2004; Cabal et al., 2006; Taddei et al., 2006]. Comparable studies [Roix et al., 2003; Zink et al., 2004; Brown et al., 2006; Fraser and Bickmore, 2007; Heard and Bickmore, 2007; Mahy et al., 2002] have recently been reported for highly expressed genes in mammalian cells. This study extends these analyses using the power of multiplex FISH. Analysis of the location of multiple, active genes have become possible with the development of multiplexed probes in FISH protocols [Levsky et al., 2002]. Here we use multiplexed fluorescence in situ hybridization (FISH) and sophisticated statistical tests applied to large data sets that can rigorously determine the spatial position of 10 expressed genes simultaneously [Levsky et al., 2002]. By assessing many active genes within a single nucleus one can determine rigorously whether a preferred location exists specifically for transcriptional activity and if the microenvironment influences the pattern. We characterized transcriptional organization by measuring the number and relative position of active gene loci, number of active alleles, distance between transcription sites and heterochromatin territories, internuclear radial position of transcription, and similarity of gene expression in neighboring cells. To measure these six parameters we developed automated methods that analyze multiplexed RNA FISH [Levsky and Singer, 2003; Kosman et al., 2004] data acquired using fluorescence microscopy. This technique can be applied to any adherent cell or tissue as the nuclear morphology [Levsky et al., 2002] and tissue context [Capodieci et al., 2005] are preserved. The data show intracellular clustering of transcriptional events for multiple genes revealing several ways in which transcription is organized and previously undiscovered intercellular gene expression relationships.
ANALYSIS OF THE SPATIAL DISTRIBUTION OF TEN GENES
The transcriptional profile of 10 serum-induced genes in DLD-1 human colon adenocarcinoma cells was analyzed using methods previously described [Levsky et al., 2002] (Table I, Fig. 1A–C). A software program recorded the number of active alleles, three-dimensional Cartesian coordinates of each transcription site and position of each cell. These data characterize the expression signature for each cell, accounting for its activity, nuclear organization and correlation of transcriptional activity with neighboring cells.
Table I.
Characterizing Transcriptional Organization
| Gene | Cells expressing gene (%) |
Cells expressing two active alleles (%) |
Mean distance to chromatin boundary (nm) |
Mean radial location (%) |
Standard deviation of radial location distribution |
Clustering factor (inter- cellular) |
|---|---|---|---|---|---|---|
| ACTB | 48 | 21 | 277 | 58 | 19 | 1.02 |
| JUN | 62 | 24 | 140 | 67 | 22 | 1.03 |
| MYC | 17 | 1.7 | — | 62 | 24 | 0.850* |
| CCND1 | 15 | 1.7 | — | 57 | 20 | 0.992 |
| DUSP1 | 28 | 8.8 | 100 | 63 | 24 | 1.09 |
| EGR1 | 67 | 33 | 167 | 62 | 19 | 0.945* |
| ACTG1 | 80 | 41 | 207 | 48 | 18 | 0.998 |
| IL8 | 28 | 6.6 | 162 | 55 | 19 | 1.10* |
| MCL1 | 14 | 2.8 | — | 57 | 15 | 1.09 |
| KLF10 | 78 | 40 | 185 | 58 | 17 | 1.02 |
Summary of five measurements made to characterize transcriptional order. Intranuclear distances between transcription sites are shown in Figure 1H–I. Clustering factor is the mean intercellular distance of cells co-expressing a gene normalized by the mean intercellular distance of every cell (Fig. 3A–B).
Clustering factors have statistically significant intercellular distance frequency distributions (Mann-Whitney P < 0.05).
Figure 1. Internuclear transcriptional organization.
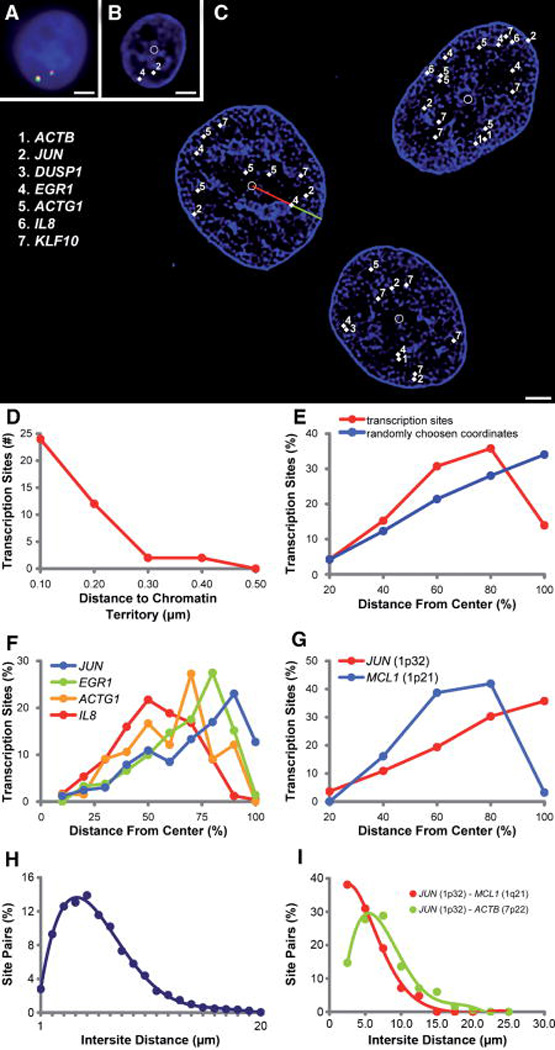
A: Multiplexed FISH detects transcription, the multi-colored foci, in a DAPI-stained nucleus (blue). (Bar = 1 µm). B: The transcription sites, labeled with binary combinations of four dyes, are identified and annotated by software (white diamonds, numbers corresponding to key, inset in C). The deconvolved DAPI data reveal boundaries of heterochromatin domains. Software labels the nuclear geometric centroid (white circle). (Bar = 1 µm). C: Three human adenocarcinoma (DLD-1) G2 nuclei with labeled transcription sites (key inset) and geometric centroids (white circles) and heterochromatin domains (blue). Software measures the relative radial position of each transcription site by normalizing the distance from the nuclear centroid to the transcription site (red line) by the distance of a line from the nuclear centroid to the edge of the nucleus intersecting the transcription site (green and red line combined, see Methods). (Bar = 3 µm). D: Histogram of distances between 40 transcription sites and the closest heterochromatin domain. Ninety percent of sites are within 200 nm of heterochromatin domains. E: The radial distribution of 1,285 transcription sites differs significantly (P = 0.0048) from the distribution of twice the number of points randomly generated within the same nuclear bounds. F: Four significantly distinct radial distributions were associated with the actively transcribing genes ACTG1, IL8, EGR1, and JUN (Mann-Whitney P = 0.0029, 0.0061, 0.0043, respectively). G: Genes from the same chromosome can be resolved and show directionality of the chromatin territories. MCL1, from the q-arm of chromosome 1 is centrally located relative to the locus ofJUN, which resides on the p-arm (P = 0.0044). H: The intranuclear distance between each transcription site was measured and is normally distributed. I: The distances between transcription sites expressed from the same chromosome are shorter than the distances between unlinked sites.
Whether polymerase II transcription is confined to nuclear sub-domains or is clustered randomly remains debatable despite elegant work on the subject [Jackson et al., 1993; Wansink et al., 1993; Kurz et al., 1996; Fay et al., 1997; Abranches et al., 1998; Jackson et al., 1998; Dietzel et al., 1999; Verschure et al., 1999; Wei et al., 1999; Mahy et al., 2002]. Our data indicate RNA expression occurs in selective nuclear domains as 90% of transcription was within 200 nm of condensed chromatin domains (Fig. 1D). Transcription was not observed in densely stained chromatin domains, showing this organization is characteristic for several serum responsive transcripts. This is consistent with observations from studies of single genes [Kurz et al., 1996; Dietzel et al., 1999].
Relative to randomly generated coordinates, the intranuclear radial position of transcription sites clusters preferentially (χ2 P < 0.0001) from half to three-quarters of the distance from the nuclear center which corresponds to ~30% of the nuclear volume (Fig. 1E). The paucity of transcription near the nuclear perimeter may result from chromatin condensed directly inside the envelope. This is consistent with work suggesting gene dense regions are internal [Gilbert et al., 2005]. In contrast, studies in Saccharomyces suggest a preferred peripheral location of expressed genes [Casolari et al., 2004; Cabal et al., 2006; Taddei et al., 2006].
Studies have shown that radial distributions of chromosomes may be unique [Boyle et al., 2001; Cremer et al., 2001]. Our high-resolution analysis provided the diffraction-limited spatial coordinates for each transcription site, which revealed four statistically unique (P < 0.01) radial distributions (Fig. 1F). In one case, genes expressed from a single chromosome had significantly different radial distributions (P = 0.0044) demonstrating that active sub-domains of a chromosome can have discernible and resolvable spatial locations (Fig. 1G).
Multiplexed FISH has the spatial resolution to resolve individual transcription events. We determined that the assignment of gene identities had an error no higher than 7%. Experiments labeling active transcription have suggested that clusters of RNA in euchromatin were in transcriptional “factories” containing numerous species of transcripts. The estimated number of such nuclear clusters varies from hundreds to tens of thousands [Jackson et al., 1993, 1998; Pombo et al., 1998, 2000]. Our analysis did not reveal that the serum-induced genes were localized in “factories”, based on the low probability of genuine overlap (Fig. 1H). Other studies have revealed that co-localization of two linked mouse genes 20 Mb apart on chromosome 7 occurred preferentially during gene activation [Osborne et al., 2004], suggesting clustering of co-regulated linked genes can occasionally occur, perhaps to regulate transcriptional associations or translocations [Branco and Pombo, 2006]. The genes in our study are mostly on different chromosomes, which may explain absence of transcriptional clustering in our data. Alternatively, given the known constraints on chromatin motion [Chubb et al., 2002], there might have been insufficient time after activation by serum, but before fixation for transiently activated loci to diffuse and come together.
DETECTION OF TRANSLOCATIONS BY INTERPHASE MULTIPLEX FISH
The resolution of this multiplex technique was sufficient to determine that expressing genes located on the same chromosome were statistically closer to each other than genes on different chromosomes (Fig. 1I). As an application of this, we could determine that chromosomal translocations in interphase cells could be discerned with high statistical accuracy simply by a rigorous distance analysis, as transcription sites became linked when their chromosome regions formerly in the trans-configuration were brought into a cis-configuration (Fig. 2).
Figure 2. Mapping chromosomal translocations in human samples.
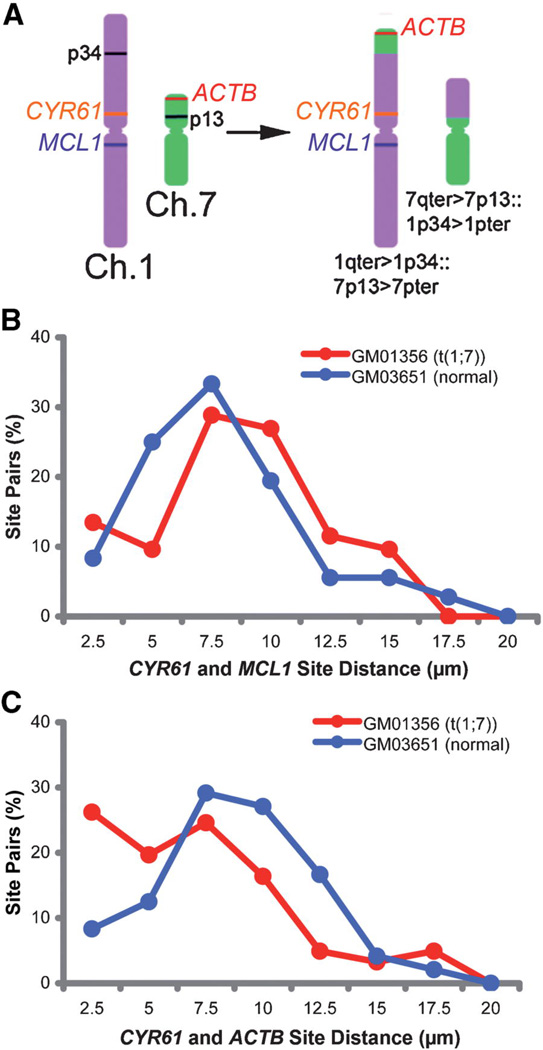
A: Primary-cultured fibroblasts from a patient repository were analyzed by FISH. The GM01356 line had a translocation between Chromosomes 1 and 7 identified by conventional karyotype analyses. B: Transcription site distributions in interphase were assayed for GM01356 and a normal control, GM03651, after serum starvation and stimulation. The CYR61 and MCL1 gene loci, which are not affected by the rearrangement, have the same site distribution in both control and translocation lines. C: In analysis of only 87 interphase nuclei from the translocation, the CYR61 and ACTB genes show a marked alteration in their site-to-site distance profile. These genes have been brought together by the translocation, and now follow a distribution characteristic of same chromosome gene pairs, as shown in Figure 1I.
INTERCELLULAR SPATIAL DISTRIBUTION
The analysis of the transcriptional profiles of individual cells allowed a different kind of spatial organization to be analyzed: that of intercellular organization. We compared the expression profiles of cells relative to their neighbors by calculating an intercellular spatial distribution and clustering factor (Fig. 3). The data demonstrated a significant difference in the intercellular spatial distributions of cells expressing MYC, EGR1, or IL8 (Mann-Whitney P < 0.05, Table II, Fig. 3C). Cells expressing MYC or EGR1 tended to be closer together compared to cells expressing IL8, which were more sparsely distributed. This observation was not correlated to the number of cells expressing the gene (Spearman non-parametric correlation test P = 0.63).
Figure 3. Internuclear transcriptional organization.
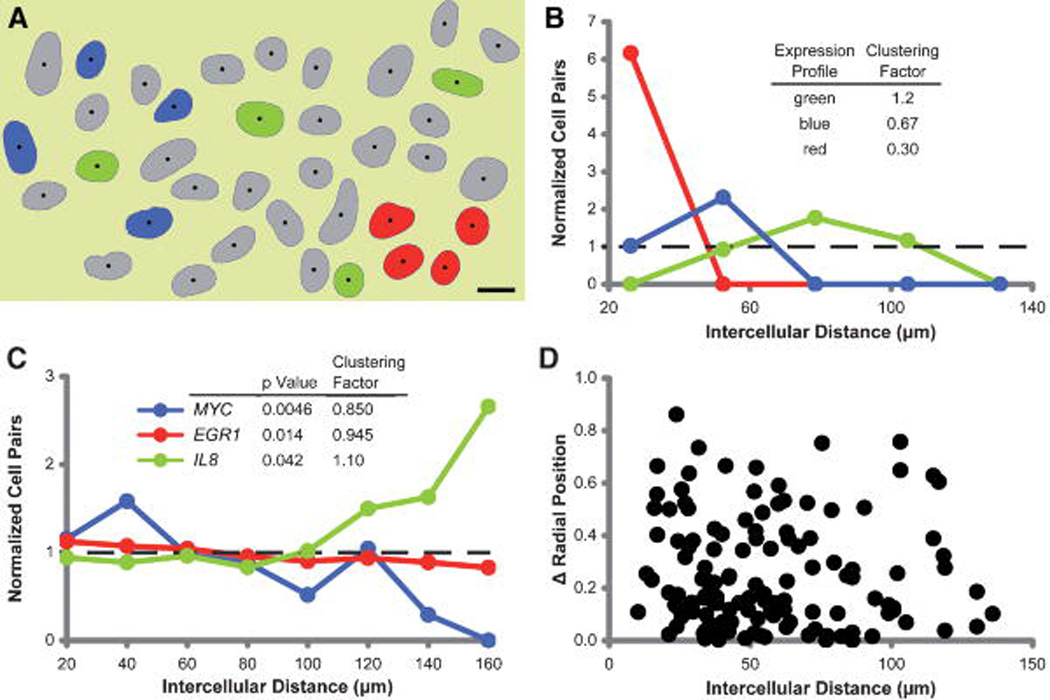
A: Fields of cells with DAPI counter-stained nuclei were automatically segmented (blue outlines) and the geometric centroids were calculated (black dots). Cells with nuclei that were not completely in the imaged field were rejected and are not shown. The nuclei of cells expressing three hypothetical gene expression profiles are marked in green, blue, and red. Empirically, the cells with red or blue nuclei are most clustered and the cells with green nuclei are sparsely distributed. The intercellular distance was measured between the geometric centroids of the nuclei. The number of cell pairs is equal to (n2−n)/2. (Bar = 10 µm). B: The frequency distribution of intercellular distances for each co-expressing cell pair normalized by the distribution of intercellular distances for every cell pair in the population. The expected distribution is indicated by the dashed line. The clustering factor is calculated as the mean intercellular distance for the sub set of cells with shared expression profiles normalized by the mean intercellular distance for all cells. Clustering factors greater than 1 indicate a sparely distributed sub-population of cells and clustering factors less than 1 indicate clustering. To determine if the differences in distributions are significant, the data are subjected to the non-parametric Mann-Whitney test. Analysis of the hypothetical field in (A) shows that cells with red nuclei are most clustered (clustering factor 0.3, peak intercellular distance ~25 µm), cells with blue nuclei are less clustered (clustering factor 0.67, peak intercellular distance ~50 µm), and cells with green nuclei are sparse (clustering factor 1.2, peak intercellular distance at ~80 µm). C: Distributions of intercellular distances for cells co-expressing either MYC (blue, 435 pairs, Mann-Whitney P = 0.0046), EGR1 (red, 7,381 pairs, Mann-Whitney P = 0.014) or IL8 (green, 1,225 pairs, Mann-Whitney P = 0.042) normalized by the intercellular distances of all cells. Cells expressing MYC or EGR1 are clustered (clustering factors, 0.850 and 0.945, respectively). Cells expressing IL8 are sparsely distributed. D: Correlation of the difference in radial position of MYC transcription sites and the intercellular distance for each cell pair co-expressing MYC. There is no correlation between radial distances of expressed c-myc loci and the intercellular distance.
Table II.
Intercellular Spatial Distributions
| Gene | Cells expressing gene (%) |
P-value (Mann- Whitney) |
Clustering factor |
|---|---|---|---|
| MYC | 17 | 0.0046 | 0.850* |
| EGR1 | 67 | 0.014 | 0.945* |
| CCND1 | 15 | 0.055 | 0.992 |
| ACTG1 | 80 | 0.78 | 0.998 |
| ACTB | 48 | 0.81 | 1.02 |
| KLF10 | 78 | 0.96 | 1.02 |
| JUN | 62 | 0.77 | 1.03 |
| DUSP1 | 28 | 0.075 | 1.09 |
| MCL1 | 14 | 0.38 | 1.09 |
| IL8 | 28 | 0.042 | 1.10* |
Expression profiles for 10 genes were interrogated by RNA FISH in DLD-1 human colon adenocarcinoma cells. The clustering factor is the mean intercellular distance of cells co-expressing a gene normalized by the mean intercellular distance of every cell (Fig. 3A–B). The Table is sorted by increasing clustering factor. The clustering factor for each gene is not correlated to expression (Spearman non-parametric correlation test P = 0.63).
Clustering factors for genes with significantly different frequency distributions (P < 0.05, Mann-Whitney).
Similar gene expression profiles in neighboring cells could theoretically result from community effect signaling [Gurdon et al., 1993], synchrony in cell cycle stage or heritability of nuclear architecture through cell division [Gerlich et al., 2003; Walter et al., 2003; Thomson et al., 2004]. We found no evidence of inheritance of intranuclear radial position in clustered cells (Fig. 3D). This indicates that factors such as local signaling and cell cycle stage are more likely to have an epigenetic impact in maintaining tissue transcription patterns than genetically inherited nuclear architecture.
Cells expressing IL8 were sparsely distributed indicating that if one cell expressed the gene, nearby cells were unlikely to be expressers. One explanation for this could be gene imprinting since IL8 has been shown to express one of its two alleles predominantly [Levsky et al., 2002]. Alternatively, anti-correlated transcriptional profiles may result from lateral inhibition, whereby two initially identical cells adopt different differentiation states as one cell inhibits the default fate of its neighbor.
The fact that intercellular organization could be detected in cultured cells raised the possibility that it would be even more evident in tissue which has a superimposed microenvironment. Intercellular spatial distribution analysis was applied to published examples of transcription detected in clinical prostate tissue specimens and augmented [Capodieci et al., 2005]. The intercellular spatial distributions of cells expressing FOLH1 were statistically clustered (P < 0.0001) in human prostate tissue (Fig. 4). The intercellular clustering of cells expressing a gene or group of genes could therefore have significant diagnostic or prognostic value.
Figure 4. Intercellular spatial distributions measured in human prostate tissue.
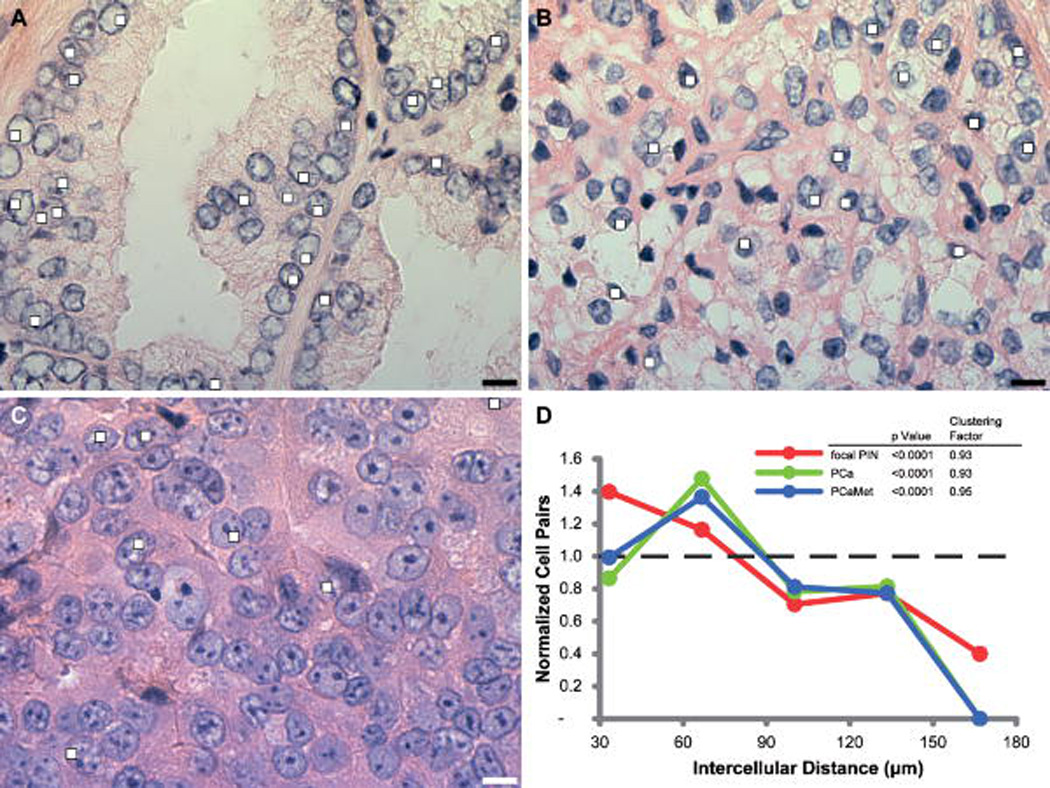
A–C: Hematoxylin and eosin-stained images of epithelial cells from paraffin-embedded human prostate tissue sections with established pathologic morphologies: (A) prostatic intraepithelial neoplasia (PIN), (B) prostate cancer (PCa), and (C) prostate cancer metastasis (PCaMet). The FOLH1 transcription sites are labeled with white boxes. (Bars = 10 µm, images adapted from [Capodieci et al., 2005]). D: Analysis of the intercellular spatial distribution (Fig. 3A–B) of cells expressing FOLH1. The analysis was modified so that the areas void of tissue did not skew the results. Cell pairs that were separated by a region void of tissue were rejected from the analysis so that the analysis was performed on the contiguous portions of tissue only. The data show that cells expressing FOLH1 are significantly clustered in each tissue sample (Mann-Whitney P < 0.0001).
The ensemble of parameters characterizing transcriptional order for the 10 genes studied form the basis for a multi-dimensional, multi-parameter map that was interrogated to make intra- and inter-cellular gene expression comparisons. The preparation of similar maps under different cellular conditions or in different cell lines or tissues will allow for quantification of organizational changes that effect cell states or are in turn affected by the cellular microenvironment. Other cell-type and tissue-specific parameters could be measured to broaden the conclusions drawn. For instance, the cellular milieu surrounding pre-malignant cells could be correlated with their changes in expression pattern. A thorough, quantitative approach to evaluating nuclear organization allows for rigorous investigation of the interaction of gene expression with cellular and tissue structure and environment and ultimately will lead to a more objective means of pathologic diagnosis.
MATERIALS AND METHODS
Cell Culture
Tissue culture for the human colon adenocarcinoma line DLD-1 (American Type Culture Collection CCL-221) was performed according to supplier's guidelines. Cells were subjected to over-night starvation and serum stimulation for 30–45 min, in the presence of 10 µg/ml cycloheximide, according to the established protocols.
FISH
Probe preparation and hybridization were performed as described previously [Levsky et al., 2002] for the following genes: Early Growth Response Protein 1 (EGR1, GenBank accession NM_001964, Chromosome 5q31), β-actin (ACTB, NM_001101, 7p22), γ-actin (ACTG1, NM_001614, 17q25), c-myc (MYC, NM_002467, 8q24), c-jun (JUN, NM_002228, 1p32), Cyclin D1 (CCND1, NM_001758, 11q13), Interleukin-8 (IL8, NM_000584, 4q13), Myeloid Cell Leukemia 1 (MCL1, NM_021960, 1q21), Transforming Growth Factor β Immediate Early Gene 1 (KLF10, NM_005655, 8q22), Dual Specificity Kinase 1/MAP Kinase-Phosphatase (DUSP1, NM_004417, 5q34), and Cysteine-Rich Angiogenic Factor (CYR61, NM_001554).
Microscopy
Three-dimensional stacks of images were acquired using a BX61 microscope with a 60×, 1.4 NA objective (Olympus) and CoolSNAP-HQ CCD camera (Photometrics) using IPLab software (version 3, BD Biosciences Bioimaging). We used filters for DAPI (#31000), FITC (#41001), Cy3 (#SP-102v1), Cy3.5 (#SP-103v1), and Cy5 (#41008) (Chroma Technology).
Statistical Analysis
Statistical tests were performed using Prism (version 4, GraphPad Software).
Measuring Distance Between Transcription Sites and Chromatin Territories
To measure distance between transcription sites and chromatin territories, three-dimensional image data of the DAPI counter-stained nuclei were deconvolved, using a constrained iterative algorithm (Huygens Professional, Scientific Volume Imaging), to reveal condensed chromatin territories and were segmented into binary maps using a threshold. The shortest Cartesian distance between each transcription site and the segmented chromatin territories was calculated. The distance between transcription sites and any other nuclear structure can be similarly measured. Also, the site location relative to a defined cellular axis (e.g., basilar-apical) could be analyzed when biologically relevant.
Measuring the Intranuclear Radial Position of Transcription Sites
The intranuclear radial distribution of each transcription site was measured by normalizing the distance between the nuclear geometric centroid and the transcription site by the length of a line drawn from the geometric centroid of the nucleus to the nuclear boundary intersecting the transcription site (Fig. 1C).
Measuring Intranuclear Transcription Site Clustering
To measure intranuclear transcription site clustering, distances between every pair of transcription sites were compared to distances between the transcription sites of specific genes (Fig. 1H–I).
Measuring Intercellular Spatial Distribution and Clustering Factor
In this analysis, the intercellular spatial distributions of cells expressing a given gene were interrogated for statistically significant deviations that indicated the clustering or sparseness of sub-sets of cells expressing that gene (Fig. 3A–B). The clustering factor is the average intercellular distance of cells expressing a given gene normalized by the average intercellular distance of all cells. In this study, the intercellular spatial distribution and clustering factor was examined independently for each gene. These techniques can be applied to examine more complex expression profiles for instance to identify sub populations in a heterogeneous environment as occurs in tissue.
Software segmented the nuclei of cells in the population and the cellular positions were recorded by finding the geometric centroid of each nucleus. The Cartesian distance between each cell pair was measured in each acquired field. The number of cell pairs is equal to (n2−n)/2. Then the distance between cells expressing each gene was measured. This created 11 distributions of intercellular distances: one distribution for all cell pairs and one distribution for cells expressing each of the 10 genes examined. Using the D'Agostino and Pearson omnibus normality test, these 11 datasets were found not to be normally distributed. The non-parametric Mann-Whitney test was applied to determine significance (Table I). The clustering factor indicated whether cells expressing the gene were either clustered (clustering factor <1) or sparse (clustering factor >1) relative to the total population. The clustering factor is only a ratio of mean intercellular distances used to summarize the amount clustering. Examining the distribution of intercellular distances for genes with statistically different distributions yield more information.
Nearest neighbor analyses of single cell expression profiles provide parameters for quantifying inheritance patterns of alleles and cell-to-cell signaling which direct development.
Acknowledgments
Grant sponsor: NIH to RHS; Grant number: CA-83208.
Footnotes
Authors' Contributions
Jeffrey Levsky acquired, analyzed and organized the data, some of which was originally published in Levsky, et al., Science 297:836–840, 2002 and wrote the first draft. Shailesh Shenoy wrote the algorithms for intercellular analysis and applied them to data in Figs. 3 & 4, performed some statistical analysis, assisted with microscopy and wrote the figure legends and methods describing the software. Jon Chubb provided valuable contextual advice and rewrote some of the text. Charles Hall provided statistical advice. Paola Capodieci provided original data for the analysis in Figure 4 that was originally published in Nature Methods 2:663–665, 2005. Robert H. Singer supervised the work.
References
- Abranches R, Beven AF, Aragon-Alcaide L, Shaw PJ. Transcription sites are not correlated with chromosome territories in wheat nuclei. J Cell Biol. 1998;143:5–12. doi: 10.1083/jcb.143.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle S, Gilchrist S, Bridger JM, Mahy NL, Ellis JA, Bickmore WA. The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Hum Mol Genet. 2001;10:211–219. doi: 10.1093/hmg/10.3.211. [DOI] [PubMed] [Google Scholar]
- Branco MR, Pombo A. Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Bios. 2006;4:e138. doi: 10.1371/journal.pbio.0040138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JM, Leach J, Reittie JE, Atzberger A, Lee-Prudhoe J, Wood WG, Higgs DR, Iborra FJ, Buckle VJ. Coregulated human globin genes are frequently in spatial proximity when active. J Cell Biol. 2006;172:177–187. doi: 10.1083/jcb.200507073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabal GG, Genovesio A, Rodriguez-Navarro S, Zimmer C, Gadal O, Lesne A, Buc H, Feuerbach-Fournier F, Olivo-Marin JC, Hurt EC, Nehrbass U. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature. 2006;441:770–773. doi: 10.1038/nature04752. [DOI] [PubMed] [Google Scholar]
- Capodieci P, Donovan M, Buchinsky H, Jeffers Y, Cordon-Cardo C, Gerald W, Edelson J, Shenoy SM, Singer RH. Gene expression profiling in single cells within tissue. Nat Methods. 2005;2:663–665. doi: 10.1038/NMETH786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 2004;117:427–439. doi: 10.1016/s0092-8674(04)00448-9. [DOI] [PubMed] [Google Scholar]
- Chubb JR, Boyle S, Perry P, Bickmore WA. Chromatin motion is constrained by association with nuclear compartments in human cells. Curr Biol. 2002;12:439–445. doi: 10.1016/s0960-9822(02)00695-4. [DOI] [PubMed] [Google Scholar]
- Cremer M, von Hase J, Volm T, Brero A, Kreth G, Walter J, Fischer C, Solovei I, Cremer C, Cremer T. Non-random radial higher-order chromatin arrangements in nuclei of diploid human cells. Chromosome Res. 2001;9:541–567. doi: 10.1023/a:1012495201697. [DOI] [PubMed] [Google Scholar]
- Dietzel S, Schiebel K, Little G, Edelmann P, Rappold GA, Eils R, Cremer C, Cremer T. The 3D positioning of ANT2 and ANT3 genes within female X chromosome territories correlates with gene activity. Exp Cell Res. 1999;252:363–375. doi: 10.1006/excr.1999.4635. [DOI] [PubMed] [Google Scholar]
- Fay FS, Taneja KL, Shenoy S, Lifshitz L, Singer RH. Quantitative digital analysis of diffuse and concentrated nuclear distributions of nascent transcripts, SC35 and poly(A) Exp Cell Res. 1997;231:27–37. doi: 10.1006/excr.1996.3460. [DOI] [PubMed] [Google Scholar]
- Fraser P, Bickmore W. Nuclear organization of the genome and the potential for gene regulation. Nature. 2007;447:413–417. doi: 10.1038/nature05916. [DOI] [PubMed] [Google Scholar]
- Gerlich D, Beaudouin J, Kalbfuss B, Daigle N, Eils R, Ellenberg J. Global chromosome positions are transmitted through mitosis in mammalian cells. Cell. 2003;112:751–764. doi: 10.1016/s0092-8674(03)00189-2. [DOI] [PubMed] [Google Scholar]
- Gilbert N, Gilchrist S, Bickmore WA. Chromatin organization in the mammalian nucleus. Int Rev Cytol. 2005;242:283–336. doi: 10.1016/S0074-7696(04)42007-5. [DOI] [PubMed] [Google Scholar]
- Gurdon JB, Lemaire P, Kato K. Community effects and related phenomena in development. Cell. 1993;75:831–834. doi: 10.1016/0092-8674(93)90526-v. [DOI] [PubMed] [Google Scholar]
- Heard E, Bickmore W. The ins and outs of gene regulation and chromosome territory organisation. Curr Opin Cell Biol. 2007;19:311–316. doi: 10.1016/j.ceb.2007.04.016. [DOI] [PubMed] [Google Scholar]
- Jackson DA, Hassan AB, Errington RJ, Cook PR. Visualization of focal sites of transcription within human nuclei. EMBO J. 1993;12:1059–1065. doi: 10.1002/j.1460-2075.1993.tb05747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, Iborra FJ, Manders EM, Cook PR. Numbers and organization of RNA polymerases, nascent transcripts, and transcription units in HeLa nuclei. Mol Biol Cell. 1998;9:1523–1536. doi: 10.1091/mbc.9.6.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosman D, Mizutani CM, Lemons D, Cox WG, McGinnis W, Bier E. Multiplex detection of RNA expression in Drosophila embryos. Science. 2004;305:846. doi: 10.1126/science.1099247. [DOI] [PubMed] [Google Scholar]
- Kurz A, Lampel S, Nickolenko JE, Bradl J, Benner A, Zirbel RM, Cremer T, Lichter P. Active and inactive genes localize preferentially in the periphery of chromosome territories. J Cell Biol. 1996;135:1195–1205. doi: 10.1083/jcb.135.5.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levsky JM, Singer RH. Gene expression and the myth of the average cell. Trends Cell Biol. 2003;13:4–6. doi: 10.1016/s0962-8924(02)00002-8. [DOI] [PubMed] [Google Scholar]
- Levsky JM, Shenoy SM, Pezo RC, Singer RH. Single-cell gene expression profiling. Science. 2002;297:836–840. doi: 10.1126/science.1072241. [DOI] [PubMed] [Google Scholar]
- Mahy NL, Perry PE, Gilchrist S, Baldock RA, Bickmore WA. Spatial organization of active and inactive genes and noncoding DNA within chromosome territories. J Cell Biol. 2002;157:579–589. doi: 10.1083/jcb.200111071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, Fraser P. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- Pombo A, Cuello P, Schul W, Yoon JB, Roeder RG, Cook PR, Murphy S. Regional and temporal specialization in the nucleus: A transcriptionally-active nuclear domain rich in PTF, Oct1 and PIKA antigens associates with specific chromosomes early in the cell cycle. EMBO J. 1998;17:1768–1778. doi: 10.1093/emboj/17.6.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pombo A, Jones E, Iborra FJ, Kimura H, Sugaya K, Cook PR, Jackson DA. Specialized transcription factories within mammalian nuclei. Crit Rev Eukaryot Gene Expr. 2000;10:21–29. [PubMed] [Google Scholar]
- Roix JJ, McQueen PG, Munson PJ, Parada LA, Misteli T. Spatial proximity of translocation-prone gene loci in human lymphomas. Nat Genet. 2003;34:287–291. doi: 10.1038/ng1177. [DOI] [PubMed] [Google Scholar]
- Taddei A, Van Houwe G, Hediger F, Kalck V, Cubizolles F, Schober H, Gasser SM. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature. 2006;441:774–778. doi: 10.1038/nature04845. [DOI] [PubMed] [Google Scholar]
- Thomson I, Gilchrist S, Bickmore WA, Chubb JR. The radial positioning of chromatin is not inherited through mitosis but is established de novo in early G1. Curr Biol. 2004;14:166–172. doi: 10.1016/j.cub.2003.12.024. [DOI] [PubMed] [Google Scholar]
- Verschure PJ, van Der Kraan I, Manders EM, van Driel R. Spatial relationship between transcription sites and chromosome territories. J Cell Biol. 1999;147:13–24. doi: 10.1083/jcb.147.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J, Schermelleh L, Cremer M, Tashiro S, Cremer T. Chromosome order in HeLa cells changes during mitosis and early G1, but is stably maintained during subsequent interphase stages. J Cell Biol. 2003;160:685–697. doi: 10.1083/jcb.200211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wansink DG, Schul W, van der Kraan I, van Steensel B, van Driel R, de Jong L. Fluorescent labeling of nascent RNA reveals transcription by RNA polymerase II in domains scattered throughout the nucleus. J Cell Biol. 1993;122:283–293. doi: 10.1083/jcb.122.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Somanathan S, Samarabandu J, Berezney R. Three-dimensional visualization of transcription sites and their association with splicing factor-rich nuclear speckles. J Cell Biol. 1999;146:543–558. doi: 10.1083/jcb.146.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink D, Amaral MD, Englmann A, Lang S, Clarke LA, Rudolph C, Alt F, Luther K, Braz C, Sadoni N, Rosenecker J, Schindelhauer D. Transcription-dependent spatial arrangements of CFTR and adjacent genes in human cell nuclei. J Cell Biol. 2004;166:815–825. doi: 10.1083/jcb.200404107. [DOI] [PMC free article] [PubMed] [Google Scholar]


