Abstract
Transcriptome remodeling in heart disease occurs through the coordinated actions of transcription factors, histone modifications, and other chromatin features at pathology-associated genes. The extent to which genome-wide chromatin reorganization also contributes to the resultant changes in gene expression remains unknown. We examined the roles of two chromatin structural proteins, Ctcf (CCCTC-binding factor) and Hmgb2 (high mobility group protein B2), in regulating pathologic transcription and chromatin remodeling. Our data demonstrate a reciprocal relationship between Hmgb2 and Ctcf in controlling aspects of chromatin structure and gene expression. Both proteins regulate each others' expression as well as transcription in cardiac myocytes; however, only Hmgb2 does so in a manner that involves global reprogramming of chromatin accessibility. We demonstrate that the actions of Hmgb2 on local chromatin accessibility are conserved across genomic loci, whereas the effects on transcription are loci-dependent and emerge in concert with histone modification and other chromatin features. Finally, although both proteins share gene targets, Hmgb2 and Ctcf, neither binds these genes simultaneously nor do they physically colocalize in myocyte nuclei. Our study uncovers a previously unknown relationship between these two ubiquitous chromatin proteins and provides a mechanistic explanation for how Hmgb2 regulates gene expression and cellular phenotype. Furthermore, we provide direct evidence for structural remodeling of chromatin on a genome-wide scale in the setting of cardiac disease.
Keywords: cardiac hypertrophy, chromatin regulation, epigenetics, gene regulation, heart failure, Ctcf, Hmgb2
Introduction
The genome is organized hierarchically with the functional unit being the nucleosome, a heteromultimeric complex of two copies each of four core histones around which ∼147 base pairs of DNA wrap (1). The histone tails can be modified according to many combinations, resulting in recruitment of chromatin remodeling complexes that in turn determine the spacing of nucleosomes along the genome (2). Additional structural regulation of transcription is conferred by DNA methylation and nucleosome packaging (3) at a larger scale by the formation of chromatin domains (4) and chromosomal territories (5), which controls the interaction of genes with distal regulatory DNA sequences and chromatin-modifying proteins. Chromatin structural proteins in turn regulate these multiple tiers of genomic structure to influence cell type-specific transcription, although the mechanisms for this phenomenon are poorly understood.
Transcriptome remodeling during pathologic stress to the heart has been well documented, as has the role of histone-modifying proteins in this process (6–8). However, chromatin structure remodeling in disease requires coordination with other chromatin features, including DNA methylation (9) and transcription factors (10). How chromatin structure is reorganized in a genome-wide manner to carry out disease-associated gene expression remains poorly understood. In cardiac development, the patterning of histone modifications changes as the cell commits to a lineage, molding the transcriptome for the appropriate phenotype (11, 12). Similar observations have been made in disease; in a transverse aortic constriction model of heart failure in mice, alterations to cis-acting histone post-translational modifications coordinate expression changes of 325 genes (13).
However, chromatin regulation in the setting of cardiac disease involves features more complex than the effects of a local histone modification on gene expression, notably, higher order chromatin structure, which remains unexplored in the heart. Different cell types ought to have different chromatin structure underpinning their different transcriptomes, although most of the knowledge on endogenous chromatin structure (4, 14, 15) and the non-nucleosome proteins that regulate it (16, 17) comes from non-cardiac cells, with some exceptions (18). At the level of the whole nucleus, we have reported a decrease in trimethylation of lysine 9 on histone H3 (H3K9me3),8 a marker of constitutively silenced DNA (19), and an increase in H3K4me3 abundance, a marker of active expression (20), in failing hearts after transverse aortic constriction (21). Similarly, decreased H3K9me2 and increased H3K4me2 in the heart were observed in a mouse model of diabetes with glomerulosclerosis, a condition that can lead to heart disease in humans and induces hypertrophy of the cardiomyocytes in mice (22). Nuclear organization in a larger context is critical for cardiomyocyte function, as evinced by cardiomyopathy resulting from mutations in the nuclear envelope protein lamin (23) and, more recently, by studies of high mobility group nucleosome-binding domain-containing protein 5 (Hmgn5), indicating that chromatin decompaction drives disease by upsetting the normal role of heterochromatin to withstand the forces of myocyte contraction (24). These observations together suggest a more plastic chromatin environment, on a genome-wide scale, underlies gene expression remodeling during heart failure.
CCCTC-binding factor (Ctcf) is an 11 zinc finger protein that organizes higher order chromatin structure by one or more of the following actions: insulating genes from their enhancers (25); orchestrating DNA looping to bring together genes and their regulatory elements (26); and/or localizing to the boundaries between heterochromatin (compact and silenced DNA) and euchromatin (loosely packed and accessible DNA) to prevent heterochromatin spreading (27). Despite the well established role for Ctcf in genome organization, virtually nothing is known about its role in the normal or diseased cardiomyocyte.
High mobility group protein B2 (Hmgb2) is a non-nucleosomal chromatin structural protein, which, by binding to and bending DNA, can alter gene expression (28). We previously found that Hmgb2 abundance is altered in heart disease (21), and in this study we sought to test the molecular mechanisms for its actions at the level of both the chromatin fiber and the entire genome. Herein, we uncover a previously unknown relationship between Hmgb2 and Ctcf, and we use this relationship to explore the role of chromatin architectural proteins in regulating cardiac gene expression in disease.
Results
Hmgb2 and Ctcf Are Inversely Regulated in the Heart
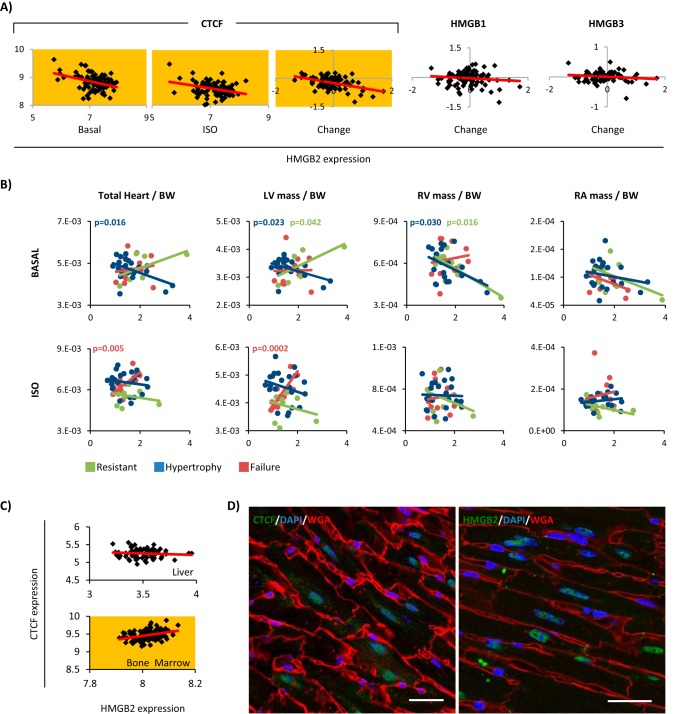
To uncover the contribution of Ctcf and Hmgb2 to the cardiac phenotype, we analyzed microarray data (29) from the hearts of 84 classical inbred and recombinant strains of mice in the basal state and mice that were treated for 3 weeks with isoproterenol, a β-adrenergic agonist that increases the inotropy of the heart and is used to model hypertrophy and failure in animal models (30). In the basal condition, hmgb2 and ctcf mRNA levels are inversely correlated with each other, and this relationship is weakened, but still significant, after isoproterenol treatment (Fig. 1A). This correlation is striking given that 66 of the 84 strains down-regulated Ctcf, whereas the response of Hmgb2 is genotype-dependent (a finding validated in NRVMs, in which Ctcf was down-regulated at the protein level in response to isoproterenol, phenylephrine, or endothelin-1 (data not shown), and the response of Hmgb2 is agonist-dependent (21)). hmgb2 mRNA abundance correlated with that of neither hmgb1 nor hmgb3 in the basal setting or after isoproterenol (Fig. 1A). Strains that respond to isoproterenol by going into heart failure exhibit a significantly direct correlation between the ratio of Ctcf to Hmgb2 and heart size, whereas the mice that are resistant or develop hypertrophy show the opposite trend (Fig. 1B). In support of these observations having functional significance in the heart, microarray data from the same panel of mice taken from other tissues (73 strains analyzed for liver (31) and 98 strains analyzed for macrophages (32)) showed that the relationship between hmgb2 and ctcf levels is organ-dependent (Fig. 1C). Immunohistochemistry to label for Hmgb2 and Ctcf in mouse cardiac tissue sections confirmed that both proteins are expressed in the nuclei of adult myocytes (Fig. 1D).
FIGURE 1.
Ctcf and Hmgb2 are coregulated in the mouse heart. A, hmgb2 and ctcf abundances exhibit an inverse relationship in the basal state that is maintained after isoproterenol treatment. Plotted are microarray data for hmgb2 (x axis) and ctcf (y axis) across 84 mouse strains in the basal state or after treatment with isoproterenol. As controls, we found that hmgb2 abundance had relationship to abundance of neither hmgb1 nor hmgb3 (yellow indicates p value <0.01 (converted from R2 value), red line represents linear regression). B, strains were grouped by their response to isoproterenol: hypertrophy (n = 22), failure (n = 13), minimal change or resistant (n = 9), unassigned (showed traits of different disease states, n = 40, not shown), and their cardiac phenotype compared with hmgb2 and ctcf abundance (x axis ratio of ctcf/hmgb2, y axis heart/heart chamber mass normalized to body weight). Total heart mass and left ventricular mass normalized to body weight showed significant positive correlation with the ratio of ctcf to hmgb2 expression in the isoproterenol-treated hearts for failing mice, but not for hypertrophic or resistant mice (p value <0.05 indicated above each plot and color-coded by strain subset; line represents linear regression). C, unlike the heart, the liver (73 mouse strains) showed no correlation between hmgb2 and ctcf abundance, and the bone marrow (98 mouse strains) had a direct correlation. D, immunohistochemistry demonstrates abundant nuclear expression of Ctcf (left) and Hmgb2 (right) in myocytes in tissue sections from adult mouse heart. Bar, 25 μm.
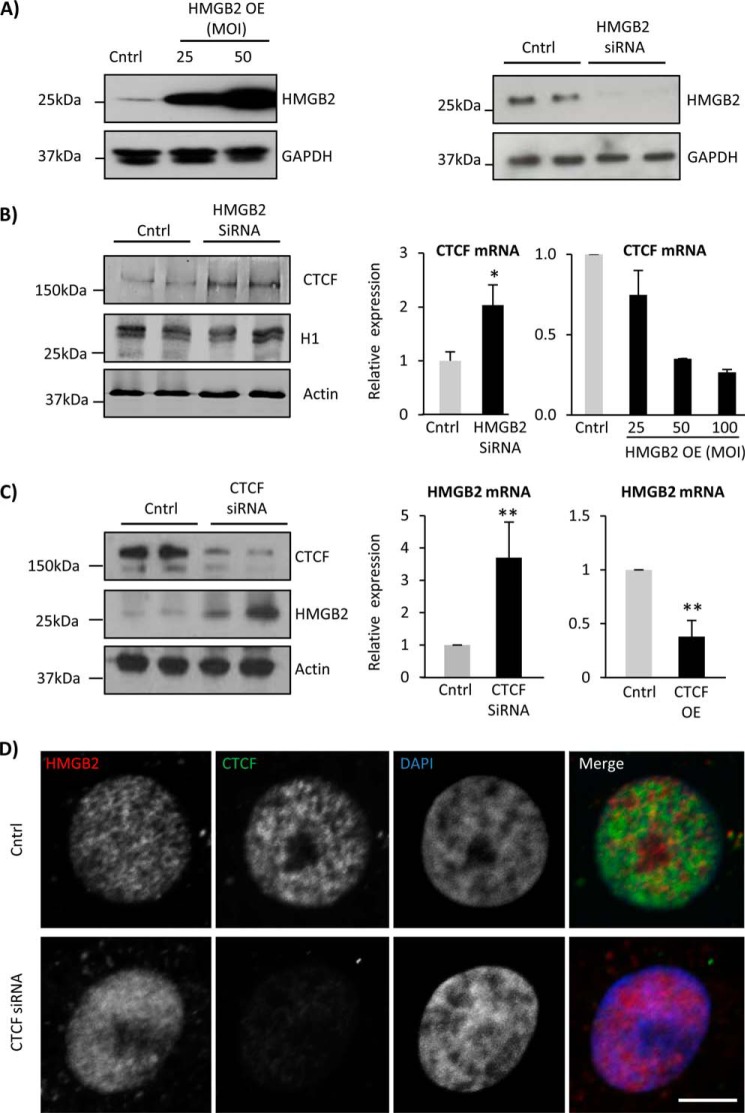
We next sought to functionally validate the inverse regulation between Hmgb2 and Ctcf. We used adenoviruses to overexpress or siRNAs to knock down hmgb2 and ctcf in neonatal rat ventricular myocytes (NRVMs) (Fig. 2). Increased Hmgb2 expression resulted in a dose-dependent reduction in Ctcf at the protein and mRNA level, whereas hmgb2 knockdown caused up-regulation of ctcf at the mRNA level with no change in protein by 72 h (Fig. 2, A and B). hmgb2 knockdown did not affect levels of histone H1, another chromatin structural protein (Fig. 2B). Likewise, ctcf knockdown up-regulated and ctcf overexpression down-regulated Hmgb2 (Fig. 2C). By microscopy, we observed an increase in the overall abundance of Hmgb2 in nuclei depleted of Ctcf (Fig. 2D). Finally, in cardiac-specific ctcf knock-out mice, we observe a doubling (204.9%) in hmgb2 abundance by RNA-seq from isolated adult cardiomyocytes. Together, these findings extend our observation of an endogenous inverse relationship between Ctcf and Hmgb2 levels in the mouse heart by demonstrating that this relationship is dynamic and responsive to experimental perturbation.
FIGURE 2.
Hmgb2 and Ctcf coregulate each other. A, hmgb2 overexpression (hmgb2 virus for 24 h) or knockdown (hmgb2 siRNA for 72 h) was carried out in NRVMs and confirmed by Western blotting. B, qPCR revealed down-regulation of ctcf with hmgb2 overexpression, and up-regulation of ctcf with hmgb2 knockdown (n = 3, * indicates p value <0.05 (t test), error = standard deviation). hmgb2 knockdown caused no change in histone H1. C, similarly, ctcf knockdown caused an up-regulation of Hmgb2 at the mRNA and protein level, whereas ctcf overexpression down-regulated hmgb2 (n = 5 overexpression, n = 3 knockdown, ** indicates p value <0.01, error = standard deviation). hmgb2 overexpression caused up-regulation of ctcf. D, immunolabeling for Hmgb2 and Ctcf in NRVMs confirmed an increase in Hmgb2 abundance after Ctcf knockdown. Bar, 5 μm. All Western blotting assays and qPCR experiments are one representative experiment of at least three. Images are one representative of approximately n = 100.
Hmgb2 and Ctcf Target the Same Loci at Distinct Times
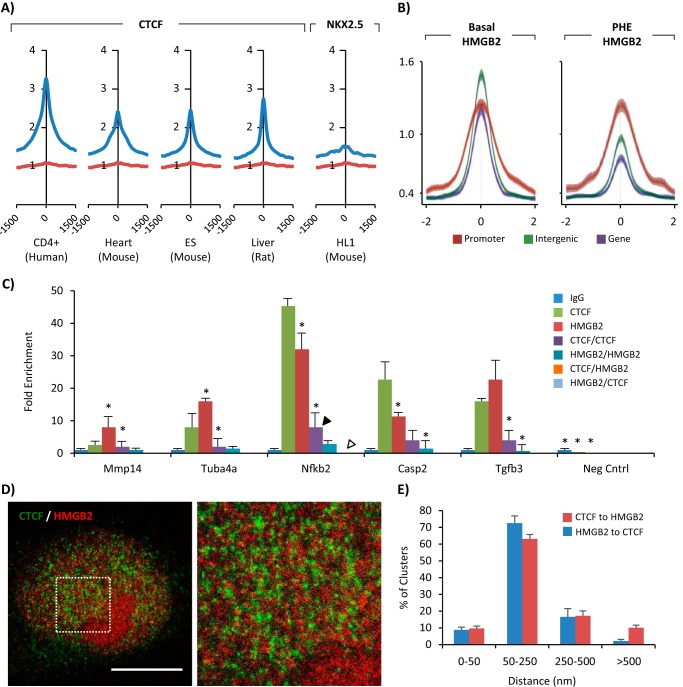
We next examined available Ctcf chromatin immunoprecipitation and DNA sequencing (ChIP-seq) data from human CD4+ cells (gene expression omnibus accession GSM325895), adult mouse heart (UCSC accession wgEncodeEM001684), mouse embryonic stem cells (gene expression omnibus accession GSM69916), and rat liver (33). We performed our own ChIP-seq for Hmgb2 in NRVMs and used liftOver to compare Hmgb2 binding to Ctcf peaks from the other four samples. In all four comparisons, Hmgb2 reads were strongly enriched around Ctcf binding peaks as compared with a randomized set of reads of similar length and number (Fig. 3A). We also compared Ctcf ChIP-seq data in the heart to Hmgb2 binding peaks separated by whether they fell in genes, promoters, or intergenic regions, and we found the greatest enrichment in intergenic regions using basal Hmgb2 ChIP-seq and promoter regions using Hmgb1 ChIP-seq from hypertrophic (phenylephrine-treated) NRVMs (Fig. 3B). For comparison, we mapped Hmgb2 enrichment around peaks for Nkx2.5, a cardiac transcription factor, in HL1 cells ((34) an atrial myocyte cell line) and did not see enrichment (Fig. 3A). Together, these suggest that Hmgb2 and Ctcf bind the same regions of the genome and that a portion of Hmgb2 peaks may be cell type-independent.
FIGURE 3.
Ctcf and Hmgb2 can occupy the same loci but not coincidently. A, ChIP-seq data for Hmgb2 in NRVMs was compared with published ChIP-seq data for Ctcf in multiple tissues. Hmgb2 reads (blue) are enriched around Ctcf binding peaks (point 0 on x axis) but not peaks for the cardiac transcription factor Nkx2.5. Randomly generated reads of the same number and size as the Hmgb2 dataset show no enrichment (red). B, Hmgb2 ChIP-seq peaks were subset by whether they localized to gene bodies, promoters (−2 kb, +500 bp of TSS), or intergenic regions. Ctcf ChIP-seq reads from the adult mouse heart were aligned across Hmgb2 ChIP-seq peaks from basal or phenylephrine-treated NRVMs. C, ChIP-reChIP experiments in NRVMs for Ctcf and Hmgb2 revealed both proteins could bind to the promoter of the five indicated genes; however, there was a loss of DNA recovery at these five loci when immunoprecipitation for Ctcf was followed by immunoprecipitation for Hmgb2 (orange, white arrowhead) but not when followed by a second Ctcf immunoprecipitation (purple, black arrowhead) as compared with Ctcf (green) or Hmgb2 (red) immunoprecipitation alone. This suggests Ctcf does not bind these regions at the same time as Hmgb2. No binding was found in a negative control region, chosen based on absence of reads in the Hmgb2 ChIP-seq data. Data correspond to one representative ChIP assay from a total of two independent assays, each of them with different NRVM isolates. Data are shown as average ± S.D. Statistical significance was assessed by two-tailed Student's t test: *, p < 0.05. D, immunolabeling for Hmgb2 and Ctcf in NRVMs was detected by STED microscopy (zoom in right panel) and confirmed a lack of colocalization of these two proteins (colocalization would appear as yellow). Bar, 5 μm. E, for each Hmgb2 cluster we calculated the distance to the closest Ctcf cluster. Histogram represents distribution of all clusters across six nuclei. The same calculation was repeated for each Ctcf cluster. Only ∼9% of clusters showed colocalization (less than 50 nm, the resolution of the STED microscope).
We validated the Hmgb2 ChIP-seq by selecting 20 peaks that were within 2 kb upstream of the TSS and performing ChIP-PCR in NRVMs. In our analysis, 19 of the 20 peaks showed enriched pulldown over an IgG control. ChIP-PCR for Ctcf showed Ctcf binding five of the 19 promoter regions (Fig. 3C). These five promoters also showed Ctcf binding in the adult mouse heart (UCSC accession wgEncodeEM001684). We then examined whether Hmgb2 and Ctcf co-occupied these promoters in cardiac cells using ChIP-reChIP (Fig. 3C). We immunoprecipitated Ctcf with one antibody, eluted the protein and DNA complex from the beads, and then reimmunoprecipitated for Ctcf using a different antibody. There was some loss of DNA in this process, but in all cases the ChIP-reChIP successfully pulled down the five promoter sequences, serving as a positive control for the assay. We then immunoprecipitated Hmgb2 with one antibody, followed by immunoprecipitation with a second Hmgb2 antibody. In this case, the loss in sample was greater, due to the poor utility of the second Hmgb2 antibody for immunoprecipitation. Despite these limitations, we still achieved enrichment of one of the five Hmgb2 targets. Finally, we immunoprecipitated for Hmgb2 followed by immunoprecipitation for Ctcf (and vice versa, with the first immunoprecipitation for Ctcf followed by immunoprecipitation for Hmgb2). In these experiments, the better performing Hmgb2 antibody was used. However, unlike the control experiments using different Ctcf antibodies against the same protein, here we saw loss of enrichment when immunoprecipitating for both Ctcf and Hmgb2 on the same sample. Together, these experiments indicate that both Ctcf and Hmgb2 bind these five regions, but not at the same time, hence the ability of Ctcf and Hmgb2 to pull down non-overlapping pools of these DNA fragments. Super-resolution imaging of immunolabeled Hmgb2 and Ctcf in NRVM nuclei confirmed the lack of colocalization of Hmgb2 and Ctcf in cardiomyocytes (Fig. 3D); only ∼9% of the Hmgb2 or Ctcf puncta were within 50 nm of each other (we define colocalization as within 50 nm based on the resolution of the microscope, Fig. 3E). HMGB2 and CTCF also did not colocalize in 293T cells (data not shown). By contrast, the ChIP-seq data predicts 20–24% overlap of these two proteins (∼41,000 total Hmgb2 peaks, ∼33,000 total cardiac Ctcf peaks, and ∼8000 overlapping Hmgb2 and Ctcf peaks indicates ∼20% of Hmgb2 peaks and ∼24% of Ctcf peaks should overlap).
Hmgb2 Regulates Ribosomal RNA Transcription
To characterize the phenotypic implications of disrupting the balance of Hmgb2 or Ctcf in myocytes, we analyzed the global effect of Hmgb2 and Ctcf on cardiac gene expression. Previous studies in mouse embryonic fibroblasts have shown that loss of Ctcf increases the nucleolar area; however, in that cell type, the nucleoli number decreased (35). Furthermore, Ctcf depletion in ES cells causes a modest decrease in ribosomal RNA transcripts (35).
In mammals, the ribosome is made up of ∼85 proteins (30–50 for 40S subunit and 40–50 for 60S subunit) and four ribosomal RNAs (rRNAs: 18S, 5S, 5.8S, and 25S) (36). The sequences of DNA encoding these rRNAs colocalize within the nucleus, forming so-called “nucleolar organizing regions” surrounding the nucleolus (37). Mammalian genomes contain >100 repeats of ribosomal DNA units (38). In the rat, the 45S rDNA gene (the precursor for 18S, 5.8S, and 28S) is clustered in repeats on chromosomes 3, 11, and 12, with ∼35 copies per chromosome (NCBI and RefSeq 2012). Like mRNA, rRNA expression is regulated by histone post-translational modifications and DNA methylation (39). The majority of gene copies are silenced in mammalian cells (40).
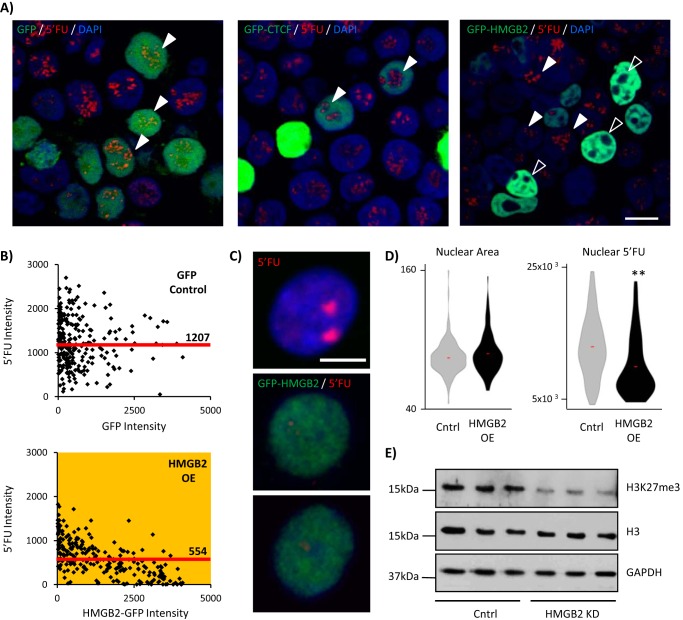
5′-Fluorouridine is a uracil analogue that incorporates into newly transcribed RNA when added to the media of living cells. We overexpressed Gfp alone or Hmgb2 or Ctcf tagged with GFP in 293T cells, incubated them in 5′-fluorouridine for 30 min and then stopped the reaction, fixed the cells, and used immunocytochemistry to detect the localization and intensity of 5′-fluorouridine labeling. We found no overt change to 5′-fluorouridine signal in either the ctcf or Gfp-only overexpressing cells. Similarly, ctcf knockdown in NRVMs had only a modest effect (11% decrease in 5′-fluorouridine intensity, p < 0.001, n = 264 control, n = 287 ctcf knockdown). By contrast, we found a stark absence of transcription in the nucleoli of hmgb2-overexpressing cells (Fig. 4A, open arrowheads). Cells in the same plate that did not actively express the hmgb2 plasmid, and which therefore were not green, also did not show transcriptional inhibition (Fig. 4A, solid arrowheads). We quantified the top 250 cells with the most Gfp expression. There was >50% reduction in the mean 5′-fluorouridine intensity of hmgb2-overexpressing cells (Fig. 4B) and an inverse relationship between 5′-fluorouridine intensity and Gfp intensity in the overexpressing cells (Fig. 4B). Both of these observations were also true when analyzing all (n = 329) hmgb2-overexpressing cells, rather than focusing only on the cells with the greatest overexpression (reduction in median level of 5′FU by 49% and a significant correlation between GFP and 5′FU in overexpressing cells, p value <0.001). Analyses of hmgb2 overexpression in NRVMs showed the same effect (Fig. 4, C and D).
FIGURE 4.
hmgb2 overexpression represses transcription. A, control, hmgb2-gfp-overexpressing or ctcf-gfp-overexpressing 293T cells were treated with 5′FU to label newly transcribed RNA. hmgb2-overexpressing cells (green cells) exhibited a loss of nucleolar transcription (5′FU in red) that was seen with neither the ctcf overexpression nor gfp overexpression alone. Bar, 10 μm. Image representative of n >100; experiment was repeated four times. B, there was a significant (yellow indicates p value <0.001 (Mann-Whitney)) inverse relationship between the intensity of Hmgb2-GFP in the nuclei, and the intensity of the 5′FU signal, which was not seen in the gfp-only treatment (n = top 250 cells with most Gfp intensity; significance was also true when compared with all n = 329 cells measured; median 5′FU intensity is indicated by labeled red line; one representative experiment of three). C, we repeated these analyses in NRVMs after 30 min of 5′FU labeling (red) in control (top panel) and hmgb2-gfp-overexpressing (middle and bottom panels) cells (bar, 5 μm), confirming that hmgb2 overexpression decreases total 5′FU nuclear signal without affecting nuclear size (** indicates p value <0.0001; one representative experiment of three; D). E, Western blotting confirmed a decrease in tri-methylation of lysine 27, a mark of facultative heterochromatin, supporting a role for large scale changes in Hmgb2 concentration to modulate global transcriptional levels (n = 3, blots quantified in ImageJ; H3K27me3 normalized to H3 gave mean signal of 1.03 for Lipofectamine control (S.D. 0.16) versus 0.12 for hmgb2 knockdown (S.D. 0.04), p value = 0.0006 (two-tailed t test; one representative experiment of three).
This global reduction in transcription agreed with our previous finding (21) that hmgb2 knockdown in NRVMs increased the abundance of H3K4 methylation, a modification associated with active promoters and enhancers (20, 41, 42), whereas loss of Hmgb2 decreased the abundance of H3K9me3, a marker of constitutively silenced DNA (19). In this study, we now show that hmgb2 knockdown also decreases the abundance of H3K27me3 (Fig. 4E), a marker of facultative heterochromatin (43), i.e. heterochromatin more likely to be dynamically regulated over the lifetime of the cell.
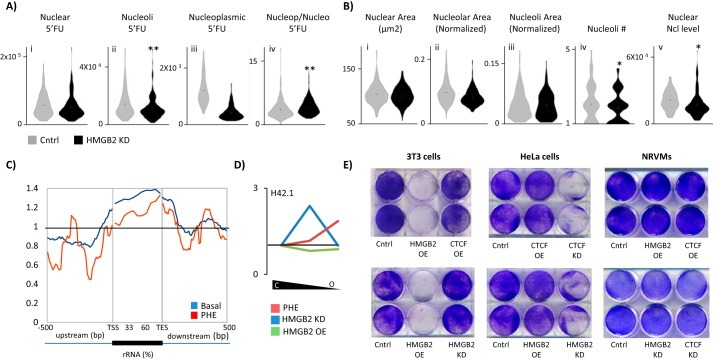
We repeated the global transcription analyses in NRVMs after hmgb2 knockdown. hmgb2 knockdown decreased nucleolar transcription with no change in total transcription (Fig. 5A, panels i–ii), increasing the ratio of nucleoplasmic transcription to nucleolar transcription (Fig. 5A, panel iv). Localization of transcription was determined by costaining for nucleolin to label nucleoli. We tested whether the loss of nucleolar transcription could be due to alterations in the nucleolar structure, and we found no significant difference in nuclear, or nucleolar, size (Fig. 5B, panels i–iii). However, we did find fewer nucleoli on average per nuclei in the knockdown cells (Fig. 5B, panel iv). Additionally, we observed a reduction in the total levels of nucleolin (Fig. 5B, panel v). The ratio of nucleolin localized within the nucleoli versus within the nucleoplasm did not change with knockdown. This suggests that hmgb2 knockdown is disrupting rRNA transcription without disrupting the gross morphology of the nucleoli. Whether the decrease in nucleolin levels with hmgb2 knockdown is a cause or a consequence of alterations in rRNA transcription remains unknown.
FIGURE 5.
Hmgb2 and Ctcf influence nucleolar rRNA transcription. A, control or hmgb2 knockdown NRVMs were treated with 5′FU to label newly transcribed RNA. hmgb2knockdown decreased nucleolar transcription (nucleoli determined by costaining for nucleolin) while increasing the ratio of nucleoplasmic to nucleolar transcription (n = 186 control, 181 knockdown; ** indicates p < 0.01 (Mann-Whitney); one representative experiment of three). B, effect of Hmgb2 on nucleolar transcription could not be explained by changes to nucleolar morphology. (Nucleolar area was determined by nucleolin costaining. Nucleoli area represents area of individual nucleoli, and nucleolar area represents total area of all nucleoli in a nucleus.) However, we found a decrease in the abundance of nucleolin levels (* indicates p < 0.05 (Mann-Whitney); one representative experiment of three). C, Hmgb2 ChIP-seq reads were aligned across ribosomal RNA genes showing Hmgb2 is enriched at these loci in the basal state and 48 h after phenylephrine treatment. D, partial chromatin digestion by micrococcal nuclease was used to isolate euchromatic and heterochromatic DNA in NRVMs followed by qPCR to determine the relative distribution of H42.1, a region of rDNA (see Fig. 7A for schematic). The ratio of intermediately packed chromatin to heterochromatin (center of plot) or euchromatin to heterochromatin (right end of plot) was plotted as a change in the ratio after hmgb2 knockdown or overexpression as compared with basal cells. In the control setting, the majority of H42.1 sequences was in the most heterochromatic fraction. hmgb2 overexpression had little effect on the ratios of heterochromatic and euchromatic rDNA, whereas hmgb2 knockdown increased the ratio of intermediately packed to tightly packed DNA. 48 h after phenylephrine, rDNA was shifted to a more euchromatic environment (average of three separate experiments). E, cell viability was assayed by labeling with crystal violet, which stains living cells. Although the effect of overexpression of hmgb2 was minimal in NRVMs, in dividing 3T3 cells there was a dramatic cell killing effect. In contrast, ctcf or hmgb2 knockdown induced cell death in HeLa cells (representative example of three separate experiments).
During cardiac hypertrophy, both translational efficiency (translational rate of ribosomes) and translational capacity (number of ribosomes) increase to support the elevated protein synthesis necessary for cellular growth (44). rRNA transcription and subsequent synthesis of new ribosomes are necessary for cardiomyocyte hypertrophy induced by phenylephrine (45), and treatment with endothelin-1 and angiotensin II also increases rRNA synthesis (44). Thus, the regulation of rRNA synthesis by Hmgb2 could be one mechanism by which Hmgb2 regulates hypertrophy.
We next sought to understand the observation that both hmgb2 knockdown and overexpression decreased ribosomal transcription. We examined Hmgb2 binding to ribosomal genes and found enrichment across the rDNA gene bodies in NRVMs as compared with the entire genome (Fig. 5C). We hypothesized that these differences could be explained by a concentration-dependent functionality of Hmgb2, such that baseline levels of Hmgb2 are necessary to promote rRNA transcription (in concert with other associated factors); however, an exorbitant amount of Hmgb2 overloads rDNA genes and promotes nonspecific chromatin condensation.
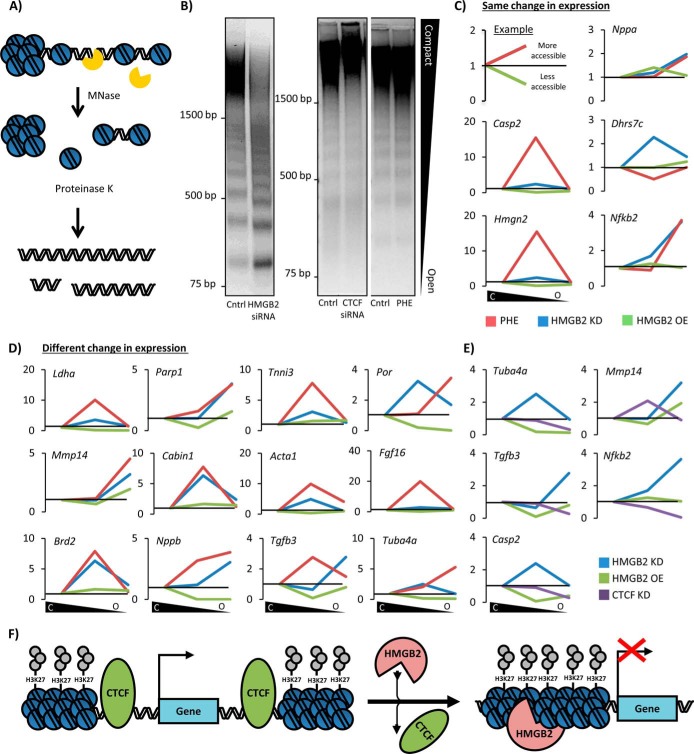
To test this, we partially digested chromatin from NRVMs with micrococcal nuclease, isolated heterochromatic, euchromatic, and intermediately compacted DNA based on the level of digestion, and used qPCR to amplify a region of the rDNA gene (designated H42.1 (35)) (Fig. 5D; see also Fig. 7A for a schematic representation of this assay). We normalized the distribution to the level of the DNA sequence in the most heterochromatic fraction, which accounted for the majority of the cardiac genome. We then compared how this ratio changed with hmgb2 knockdown or overexpression and found that hmgb2 overexpression had minimal effect, whereas phenylephrine treatment increased the ratio of euchromatic to heterochromatic DNA (Fig. 5D). hmgb2 knockdown caused an increase in the ratio of intermediately compacted DNA as compared with heterochromatin (Fig. 5D), illustrating that hmgb2 knockdown and overexpression have opposing effects on rDNA genes.
FIGURE 7.
Hmgb2 and Ctcf control gene accessibility in an antithetic manner at the level of the chromatin fiber. A, chromatinized genomic DNA was treated with MNase (0.001 units), resulting in partial digestion of the genome based on accessibility. B, hmgb2 knockdown increased the abundance of highly digested lower molecular weight fragments of DNA (representing open chromatin), while decreasing the abundance of less digested higher molecular weight fragments (compact chromatin), suggesting a global increase in DNA accessibility. Such dramatic change in global accessibility occurred with neither ctcf knockdown nor phenylephrine treatment (one representative experiment of 6). C, after MNase digestion, DNA fragments were cut from the compact, intermediate, and open regions of the agarose gel and analyzed by qPCR to determine the relative distribution of individual promoter sequences for mRNA-coding genes of interest. Plotted is the change in the ratio of open chromatin to heterochromatin, and intermediate chromatin to heterochromatin, between control and treated cells; as shown in the 1st panel, these ratios are represented in the ensuing graphs as upward inflections for a shift to more accessible chromatin by a given gene and downward deflections for a gene that shifts to more compact DNA. Gene expressions for hmgb2 knockdown and phenylephrine from microarray data in NRVMs (21) were used to distinguish between genes with similar expression changes induced by hmgb2 and phenylephrine (C) or genes regulated differently by hmgb2 and phenylephrine (D). Interestingly, hmgb2 knockdown and phenylephrine showed similar trends for shifting promoter sequences between these categories even when these stimuli (hmgb2 knockdown or phenylephrine) had different effects on the transcription of the gene. E, promoter sequences for the genes bound by Ctcf and Hmgb2 by ChIP were also examined. In four of the five cases, ctcf knockdown shifted these sequences to more compact regions of chromatin. (All MNase data are the average of at least three experiments.) F, model for relationship between Hmgb2 and Ctcf. Ctcf serves as a boundary preventing the spread of heterochromatin, whereas Hmgb2 promotes heterochromatin formation.
It was not previously known that Hmgb2 regulates nucleolar transcription. We thus sought to further explore the specific phenotypes regulated by Hmgb2 acting on rRNA transcription. We hypothesized that the dramatic decrease in rRNA synthesis upon hmgb2 overexpression would disrupt cell growth. We labeled cells with crystal violet and then gently washed the plate to remove dead cells. In 3T3 cells, hmgb2 overexpression was lethal; as predicted, however, this was not the case in HeLa cells, where hmgb2 knockdown but not overexpression resulted in a loss of cells (Fig. 5E). ctcf knockdown was also lethal in HeLa cells (Fig. 5E). In NRVMs, hmgb2 knockdown, hmgb2 overexpression, ctcf knockdown, or ctcf overexpression all had no effect on cell death, possibly because the cells are not dividing, and can therefore better withstand disrupted rRNA synthesis (Fig. 5E). This observation is in agreement with data from COS-1 cells, in which Hmgb2 knockdown suppresses cell division (46). Furthermore, the cell type-specific lethality of Ctcf levels is also in agreement with data showing that Ctcf knock-out is embryonic lethal in mice (47, 48). Ctcf depletion in isolated cells often affects cell division or cell death processes, but in a cell type-dependent manner, potentially due to the cell type-specific localization of Ctcf and arrangement of a higher order structure coordinated by the protein (49).
Interaction of Ctcf and Hmgb2 with Local Chromatin Features to Influence Gene Expression
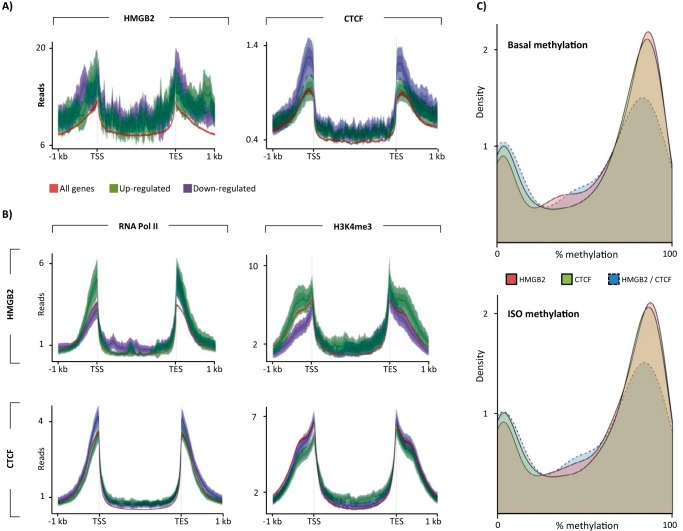
We previously found that hmgb2-regulated genes are enriched in pathways important to cardiac hypertrophy (21). We next asked how Hmgb2 and Ctcf regulate mRNA expression to explain why some hmgb2-regulated genes are up-regulated and others are down-regulated in response to hmgb2 knockdown in cardiomyocytes (21). We used ENCODE date from the adult mouse heart and our Hmgb2 ChIP-seq data. Hmgb2 occupancy was grossly similar between genes up-regulated or down-regulated by hmgb2 knockdown, as was Ctcf enrichment at ctcf-regulated genes (genes up-regulated or down-regulated by ctcf knock-out in the mouse heart; Fig. 6A). Compared with all genes and down-regulated genes, genes that were up-regulated by hmgb2 knockdown had greater levels of the activating marks H3K4me3 and RNA pol II in their promoters in the basal setting (1 kb upstream of TSS; Fig. 6B), suggesting that removing hmgb2 potentiates the transcriptional effect of the local epigenomic code specified through histone post-translational modifications. Remarkably, ctcf-regulated genes showed the opposite pattern, with up-regulated genes depleted in activating marks (Fig. 6B). This suggested that the interaction of these two proteins with local chromatin environment could have opposing effects on gene expression.
FIGURE 6.
Opposing effects of local chromatin environment on Hmgb2 versus Ctcf-regulated genes. A, Hmgb2 reads or Ctcf reads were plotted across genes up-regulated or down-regulated by hmgb2 knockdown or ctcf knock-out (left and right panels, respectively) or across all genes in the genome, revealing that the distribution of Hmgb2/Ctcf is not the major determinant of whether a gene's expression will change with depletion of these proteins (Hmgb2- and Ctcf-regulated genes use expression data averaged from three experiments). B, distribution of ChIP-seq (ENCODE, basal adult mouse heart) reads across hmgb2-regulated genes suggests genes that are up-regulated by knockdown are enriched in the activating marks H3K4me3 and RNA pol II in the basal state as compared with genes that are down-regulated by knockdown (top panel). Strikingly, the opposite is true for genes that are regulated by ctcf (bottom panel; ENCODE data represents one biological replicate). C, we next examined DNA methylation from the mouse heart. Plotted is the bimodal distribution of average DNA methylation across Hmgb2-only and Ctcf-only peaks in intergenic regions. Shared peaks are less likely to have high methylation but rather show low or intermediate level methylation. This is true using DNA methylation data from basal adult mice and mice treated with isoproterenol for 3 weeks (DNA methylation is average of six mice (54)).
We next asked if the local chromatin environment at the shared HMGB2 and CTCF loci correlated with which of the two proteins bound. Ctcf binding has been shown to be sensitive to DNA methylation (50–52) specifically in the context of cell type-dependent binding events (53). We used our bisulfite sequencing data from adult mouse hearts (54) to assess the average DNA methylation status of all CpGs across Hmgb2- and Ctcf-binding sites. Bisulfite sequencing data exhibit a bimodal distribution, with the majority of CpGs either lowly methylated or highly methylated, reflective, we posit, of a relative entrainment of CpG methylation status across the population of sampled cells (i.e. methylated or not). As compared with Hmgb2-only or Ctcf-only intergenic peaks, the shared intergenic please were less likely to be highly methylated and were more likely to have intermediate levels of methylation (Fig. 6C). This is also true when examining methylation data from isoproterenol-treated mouse hearts. We thus speculate that DNA methylation at these peaks might be more dynamic and thus capable of modulating Hmgb2 and Ctcf differential binding.
Others have shown that depleting DNA methylation promotes new Ctcf binding at less than 1.5% of Ctcf's consensus motifs (55), suggesting DNA methylation may be insufficient to regulate differential binding of Ctcf to Hmgb2 and the Ctcf-shared peaks. This previous study also found that the consensus motifs, which did recruit Ctcf as a result of depleted methylation, were sites that would otherwise have bound Ctcf in other cell types and were at newly formed DNase HS sites (55). We thus asked whether the Hmgb2- and Ctcf-shared peaks also had similar features, specifically the ability to bind Ctcf in other cells and an open chromatin phenotype. We combined ENCODE ChIP-seq data for Ctcf from 10 adult mouse organs and We next asked whether the local chromatin environment at the shared Hmgb2 and Ctcf binding loci correlated with either of the two proteins. s to define a set of conserved Ctcf-binding sites. Only 15% of intergenic Ctcf-only peaks in the heart are also conserved Ctcf-binding sites in other organs. By contrast, 64% of Ctcf peaks shared with Hmgb2 overlapped with conserved Ctcf-binding sites (data not shown). On average, Ctcf-only and Hmgb2- and Ctcf-shared peaks both overlap with one DNase HS site in the mouse heart (ENCODE). These findings suggest that other chromatin features may be responsible for the differential binding of Hmgb2 and Ctcf and that DNA methylation could be one such feature. A complementary interpretation is that Hmgb2's ability to promote heterochromatin and decrease accessibility (as evidenced by our MNase assay, Figs. 5 and 7) could prevent Ctcf binding. However, future analyses are necessary to determine the temporal regulation of other chromatin features that influence the differential binding of Hmgb2 or Ctcf to their shared sites in the heart.
Hmgb2 and Ctcf Exert Opposing Effects on Promoter Accessibility
We next sought to investigate direct effects of Hmgb2 and Ctcf to structurally modify local chromatin environment. We isolated nuclei from NRVMs and treated them with 0.001 units of MNase, an enzyme that preferentially digests DNA that is accessible, i.e. not bound by nucleosomes. The digested genome of control NRVMs, when run on an agarose gel, gives bands of multiple sizes as follows: the smallest bands migrate around 150–200 bp (the size of a mono-nucleosome); bands migrating at increasing molecular masses correspond to sections of chromatin which, endogenously, reside in states of greater compaction (Fig. 7, A, concept, and B, data). Electrophoretically less mobile regions at the top of the gel represent DNA that is more compact and heterochromatic; those at the bottom are more open and euchromatic. hmgb2 knockdown shifted the distribution of the genome toward more euchromatic DNA (Fig. 7B), whereas ctcf knockdown or phenylephrine treatment had minimal effect on the global pattern of DNA compaction (Fig. 7B).
To investigate the behavior of individual genes, we repeated this experiment, and we cut the agarose gel containing the digested genome into three sections representing DNA that had been in heterochromatic environments (compact), euchromatic environments (open), or that came from an intermediate region of the genome, and we analyzed by qPCR the distribution of specific promoter sequences for genes of interest. Each plot represents the change in the ratio of intermediate regions or open regions divided by compact regions between basal and treated cells. For all promoters tested (with the exception of that for the gene dhrs7c), the alteration of local accessibility conferred by phenylephrine was very similar to the effect of hmgb2 knockdown (Fig. 7, C and D), i.e. phenylephrine and hmgb2 knockdown made the chromatin more open (although in some cases the effect of hmgb2 knockdown was less pronounced), whereas hmgb2 overexpression had the antithetic effect. This was true even for genes that had distinct transcriptional responses to phenylephrine treatment (56) and hmgb2 knockdown (Fig. 7D) (21). This observation suggests a partial transcription-independent effect on chromatin structure that is shared in both hmgb2 knockdown-induced and phenylephrine-induced hypertrophy, and which we hypothesize can account for the fact that hmgb2 knockdown can prime the genome for hypertrophy in NRVMs by facilitating the actions of other local chromatin features (Fig. 6). Similar to hmgb2 overexpression, ctcf knockdown generally resulted in more heterochromatic packing of the promoter (Fig. 7E), providing chromatin fiber level support for the opposing relationship between these two proteins described thus far in this study. We hypothesize that Ctcf forms chromatin boundaries, with Hmgb2 promoting compaction within these boundaries(Fig. 7F).
Hmgb2 Binding Overlaps Ctcf at Three-dimensional Domain Boundaries and Cardiac Enhancers to Oppositely Regulate mRNA Expression
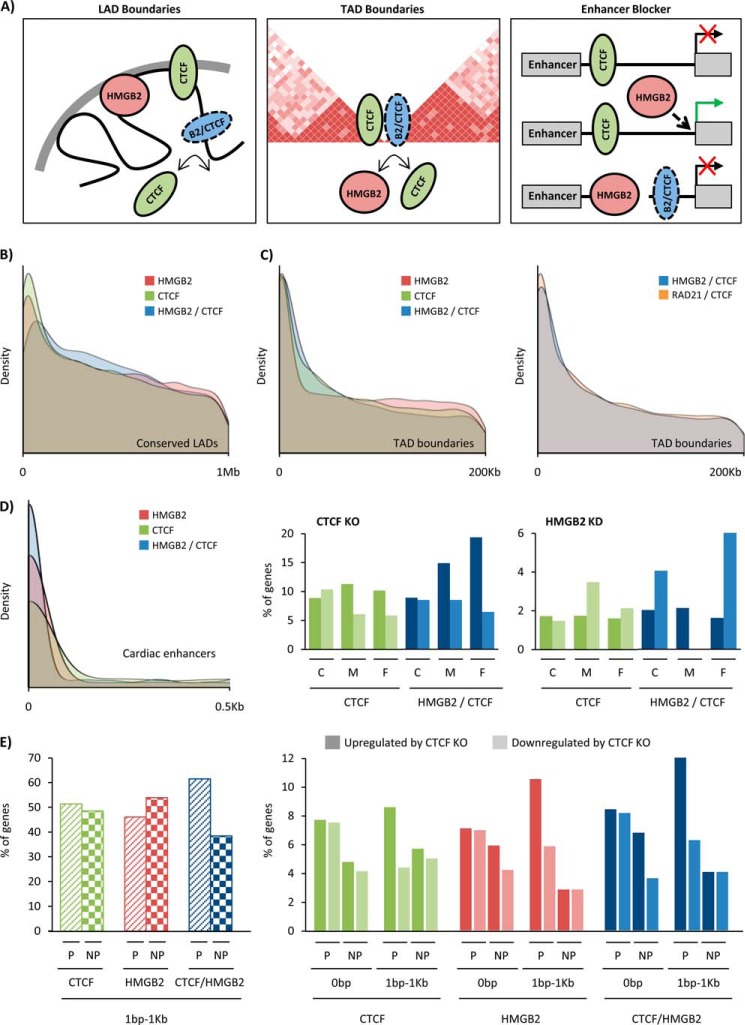
We next investigated the role of Ctcf at the sites that both Ctcf and Hmgb2 can bind. Previous work has shown Ctcf is enriched within 100 kb of lamina-associated domains (LADs, 0.1–10 Mb domains of DNA interacting with the nuclear membrane and associated with repressed transcription (57)). Furthermore, analysis of LADs in four different mouse cell types revealed 33% are shared between cell types (58, 59). We determined the overlap for these four datasets and compared their location to the proximity of Hmgb2 peaks that do not overlap with Ctcf (Hmgb2-only), Ctcf peaks that do not overlap with Hmgb2 (Ctcf-only), and sites than can be bound by either Hmgb2 or Ctcf (Hmgb2- and Ctcf-shared peaks). This analysis confirmed enrichment of Ctcf-only peaks at LAD boundaries and further revealed Hmgb2-only peaks are enriched at boundaries, suggesting possible new functionality for Hmgb2. However, Hmgb2- and Ctcf-shared peaks were not enriched to the same extent, suggesting that the overlapping peaks correspond to a different functional role of Ctcf (Fig. 8, A and B).
FIGURE 8.
Hmgb2 and Ctcf target shared loci near three-dimensional domain boundaries and cardiac enhancers. A, Ctcf is known to be enriched at LAD and TAD boundaries and to serve as a blocker of enhancer promoter interactions. We show Hmgb2- and Ctcf-shared binding sites are also enriched at TAD boundaries and near cardiac enhancers, with opposing effects on the transcription of genes near these enhancers. B, Hmgb2 peaks that do not overlap Ctcf (Hmgb2-only, red), Ctcf peaks that do not overlap Hmgb2 (Ctcf-only, green), and Hmgb2- and Ctcf-shared peaks (blue) were examined to determine their distance to the nearest conserved mouse lamina-associated domain (LAD, conserved across four mouse cell types). Density plots display the distribution of distances, revealing enrichment of both Ctcf and Hmgb2 at LAD boundaries, with overlapping peaks less enriched (highest peak is further from site of LAD, and peak is shorter and more broad). Signal at 0 bp represents peaks that fall within LADs. C, density plots display the distribution of distances between Hmgb2-only, Ctcf-only, and shared peaks to topologically associating domains (TAD, domain boundaries from mouse cortex but we see similar results with boundaries from mESC). All three sets of peaks are enriched at TAD boundaries (left panel). Cohesin (a complex including Rad21) is known to physically interact with Ctcf at TAD boundaries. Hmgb2- and Ctcf-shared peaks show similar levels of enrichment at TAD boundaries as the levels exhibited by Rad21- and Ctcf-shared peaks (right panel). D, Hmgb2-only and Ctcf-only peaks are enriched near cardiac enhancers (defined by H3K27ac from mouse adult cardiomyocytes), with Hmgb2- and Ctcf-shared peaks enriched to a greater degree (left panel). The nearest genes to cardiac enhancers were found and grouped by the distance of the enhancer to a Ctcf peak (green) or shared peak (blue). Close (C) indicates peaks within 30 bp of enhancer, medium (M) indicates 31–381 bp distance, and far (F) indicates 381 bp to 1 kb. Plotted is the percentage of these nearest genes up- or down-regulated by ctcf KO (middle panel) or hmgb2 KD (left panel) at each distance. When Hmgb2- and Ctcf-shared peaks are within 380 bp to 1 kb of an enhancer, loss of ctcf is three times more likely to cause the nearest gene to be up-regulated than down-regulated, whereas the loss of hmgb2 is four times more likely to cause down-regulation. E, for Ctcf-only, Hmgb2-only, and shared peaks that occur within 1 kb of a gene, but not in a gene, we plotted the percentages that were near active genes (P, presence of RNA pol II peak in promoter) or inactive genes (NP, no pol II). Only the shared peaks showed bias toward being near active genes (left panel). When Hmgb2-only, Ctcf-only, or shared peaks are within 1 kb of an active gene, loss of ctcf is two times more likely to cause up-regulation (dark color) than down-regulation (light color), but no bias is seen for inactive genes or peaks within genes. Rad21 and RNA pol II ChIP-seq represent one biological replication from ENCODE. Location of cardiac enhancers (13), LAD boundaries (59), and TAD boundaries (15) come from published work which cite one, two, and multiple (number not provided) biological replicates, respectively.
Topologically associating domains (TADs, megabase-scale domains (median in mESC = 880 kb) of DNA enriched for self-interaction) have Ctcf enriched at their boundaries (15). Furthermore, TAD boundaries are conserved between cell types (∼69% shared between cell types of the same species and ∼65% shared between the same cell type in mouse and human) (15). We compared our peaks to TAD boundaries measured in the mouse cortex and surprisingly found both Hmgb2-only and Hmgb2- and Ctcf-shared peaks were enriched at the boundary (Fig. 8C). We further compared Rad21 (a known component of the cohesin complex) mouse ChIP-seq data with our Ctcf peaks to assess the level of enrichment of Ctcf- and cohesin-shared peaks. As expected, these sites were enriched at TAD boundaries, and the Hmgb2- and Ctcf-shared peaks were enriched to almost the same extent (Fig. 8C). These findings highlight novel functionalities for Hmgb2 (Fig. 8A) to regulate different types of heterochromatin as well as endogenous genomic architecture.
Ctcf has also been shown to act as an enhancer blocker, although recent DNA conformation capture data suggest that whether Ctcf promotes or blocks enhancer interaction with its target gene can depend on whether the interaction occurs within or between TADs (60). We explored whether Ctcf-binding sites regulating enhancer function overlapped with Hmgb2 binding, using cardiac enhancers defined by H3K27ac in adult mouse cardiomyocytes (13). Both Ctcf-only and Hmgb2-only peaks are enriched at these enhancers, with Hmgb2- and Ctcf-shared peaks enriched the most (Fig. 8D). This supports a role for these ubiquitous proteins to regulate cell type-specific gene expression. We then asked for Hmgb2 or Ctcf peaks close to an enhancer (within 30 bp), at a medium distance (31–380 bp) or at a far distance (381 bp to 1 kb), what the effect was on the expression of the nearest gene to the enhancer using RNA-seq data from ctcf knock-out mouse hearts and microarray data from hmgb2 knockdown NRVMs. Loss of ctcf from regions a medium or far distance from an enhancer is biased toward up-regulation of the target gene (although the majority of genes do not change in expression, Fig. 8D), mimicking the expected behavior of an enhancer blocker. The same is seen when ctcf is lost from sites that can also bind Hmgb2. At Hmgb2- and Ctcf-shared peaks, loss of ctcf is three times more likely to result in up-regulation than down-regulation of the nearest gene to the enhancer, whereas loss of hmgb2 is four times more likely to cause down-regulation than up-regulation (Fig. 8D). One possible explanation is that loss of hmgb2 allows Ctcf binding, which promotes enhancer blocking.
We then performed a similar analysis, this time asking how Hmgb2 or Ctcf peaks near genes that regulate the expression of the nearest gene. First, we found that Hmgb2 and Ctcf are just as likely to bind near (designated as within 1 kb of a gene but not within a gene) an active gene as an inactive one, when using the presence of an RNA pol II promoter peak to define active genes (ENCODE ChIP-seq dataset). However, Hmgb2- and Ctcf-shared peaks are biased to binding near the active genes (Fig. 8E, left panel). Second, we found that for Hmgb2-only, Ctcf-only, and Hmgb2- and Ctcf-shared peaks that occur near a gene, loss of ctcf was biased toward up-regulation of the gene if the gene was also active in the basal state (Fig. 8E, right panel). However, there was no bias for genes that did not have RNA pol II nor for genes where Hmgb2 or Ctcf were binding within the gene as opposed to near the gene (Fig. 8A). This finding is similar to our observation that hmgb2 target genes are likely to be up-regulated if bound by RNA pol II in their promoter in the basal state and also is in line with our observations from enhancers, wherein Ctcf binding near, but not in, enhancers or genes is biased to being repressive, a function that is conserved at sites that can also bind Hmgb2.
It remains unknown whether and how Hmgb2 binding can modulate the repressive function of Ctcf on nearby genes. We hypothesize this action to involve mutually exclusive binding of these two proteins, wherein Hmgb2's binding prevents that of Ctcf by changing the chromatin landscape and/or specifically promoting heterochromatin (Fig. 7F).
Additionally, gene ontology analysis of genes with a nearby (within 1 kb) shared Hmgb2 and Ctcf peak are enriched for annotation relating to cytoskeleton, ribosome, and nucleus (enrichment score 4.14, 3.51, and 3.25 respectively, DAVID), whereas gene ontology analysis for the nearest genes to enhancers with Hmgb2- and Ctcf-shared peaks were enriched for transcription (enrichment score 3.62, DAVID), with no enrichment for any disease processes after Bonferroni correction (hypertrophic cardiomyopathy and insulin signaling were the two lowest p values for KEGG pathways, although not significant after correction). These data suggest that although Hmgb2 and Ctcf target cardiac-specific genes and can regulate pathologic pathways (21), their shared binding sites may be more important for regulating gross, rather than stress-responsive, cardiac genome organization.
Discussion
We propose a model (Figs. 7F and 8A) whereby Ctcf, acting in an insulator capacity, serves as a boundary for heterochromatin spreading. In the absence of Ctcf, heterochromatin can spread, silencing nearby regions, a phenomenon that is accompanied by the increased presence of Hmgb2, which maintains the compact environment. Inversely, increased abundance of Hmgb2 can promote heterochromatin spreading and thereby evict Ctcf from specific loci. By disrupting the boundaries of heterochromatin, Hmgb2 and Ctcf can affect multiple genes in a given region, depending on the type and extent of other modifications.
We favor this model, as opposed to one in which Hmgb2 preferentially targets (and differentially regulates) individual genes, because Hmgb2 lacks DNA sequence specificity and is not a cardiac-specific protein. These observations, coupled with our new understanding of the finely regulated balance between Hmgb2 and Ctcf, indicate that although the overall chromatin structure of the myocyte may not be regulated with single gene resolution, this structure is critical for regulating myocyte physiology in health and disease. It is also important to note that the conclusions in this study were made from a combination of in vivo adult mouse models and isolated neonatal ventricular myocytes. Although this approach provides some experimental advantages, there are likely important differences in how adult and neonatal cardiomyocytes package chromatin (related to differences in regenerative, proliferative, and stress response capacities in these cell populations), which will have to be resolved by further experimentation.
An open question when we began these studies was the molecular basis for how Hmgb2 ostensibly promotes the transcription of some genes while inhibiting the expression of others. Previous studies have implicated Hmgb2 in transcriptional activation or repression (61–63), attributing these actions to cooperation with distinct proteins. The predominant hypothesis for the role of Hmgbs in gene expression is that they bend DNA to promote binding by other proteins. Hmgbs may only transiently interact with these client proteins (if at all), thereby acting as promiscuous chaperones at different loci (28). Our data support this model, indicating that Hmgb2 has locus-specific effects on gene expression notwithstanding a conserved effect to compact chromatin in a locus-independent manner. Future studies using locus-specific proteomic analyses will be required to determine which Hmgb2 binding partners encode, in a combinatorial manner, different transcriptional logic. Although informative, this approach may obfuscate the issue of trans effects, particularly through non-coding regions of the genome, by arbitrarily restricting examination of Hmgb2's functions within the physical unit of a gene.
The increased DNA flexibility conferred by Hmgb2 binding (64) that facilitates formation of locus-specific complexes can also more generally facilitate tighter packaging of DNA. Previous reports have shown Hmgb1 is enriched in euchromatic regions of photoreceptor nuclei (65). However, Hmgb1 and Hmgb2 are also bound to highly heterochromatic DNA formed during mitosis (66). Compared with linker histone H1, Hmgb1 also compacts DNA, although to a lesser degree (67), and can directly compete with histone H1 for binding to linker DNA (68). We show that hmgb2 knockdown disrupts global measures of heterochromatin (H3K27me3 abundance) and the chromatin environment at specific genes without altering global levels of histone H1. One potential explanation is that hmgb2 knockdown decreases nucleosome abundance. Others have shown that Hmgb1-deficient mouse embryonic fibroblast cells have reduced nucleosome number, and yeast deficient in the hmgb2 homologue display a shift to euchromatin when assayed by MNase (69), similar to what we observed following hmgb2 knockdown. Indeed, in vitro, Hmgb1 can facilitate nucleosome deposition (70). However, we see no difference in the abundance of histone H3 with Hmgb2 knockdown, although we did not directly measure nucleosome assembly. Hmgb2 can also alter nucleosome distribution by facilitating nucleosome sliding via interaction with SWI/SNF ATP-dependent chromatin remodeling complexes, as shown in vitro (71).
Here, we propose a model where Hmgb2 targeting is partially regulated by the distribution of heterochromatin, such that Ctcf mediates the boundaries between hetero- and euchromatin, and Hmgb2 maintains the integrity of facultative heterochromatin, i.e. genes that are silenced in a given cell type. We reason that the overlap between Hmgb2 ChIP-seq reads from rat cardiomyocytes with Ctcf ChIP-seq peaks in other species and tissues indicates cell type independent functions of the proteins. Indeed, topological domains are largely conserved between cell types and species (15), and Ctcf is both enriched at the topological domains (15) and is critical for maintaining them (72). Unlike transcription factors, Ctcf does not preferentially localize to genes that belong to a similar class (73). However, we see preferential binding of Hmgb2- and Ctcf-shared peaks to genes actively expressed in the heart, suggesting that a subset of the Hmgb2 and Ctcf binding is cell type-specific. Furthermore, the changes to chromatin accessibility at individual promoters induced by hmgb2 knockdown largely mimic the effects of phenylephrine treatment. This is especially interesting given that phenylephrine treatment, unlike hmgb2 knockdown, does not cause global changes in genome accessibility, suggesting that alterations in chromatin packaging, although important for phenotype, may be decoupled from transcriptional changes.
We also find both Ctcf and Hmgb2 regulate nucleolar transcription in multiple cell types, having particular implications for cardiac cells. Nucleolar disruption occurs with cardiac stress (34), and rRNA synthesis is up-regulated in hypertrophy (74). Previous findings identified Hmgb3 as a component of the T-cell nucleolar proteome (75) and found evidence for interaction between Hmgb2 and nucleolin outside of the nucleolus (76).
The relationship between Hmgb2 and Ctcf suggests a mechanism by which both chromatin structural proteins are regulated (in abundance and localization) in part by the chromatin environment. Ctcf, unlike Hmgb2, has DNA binding consensus motifs, and perhaps their coregulation involves sequestering of Ctcf-binding sites into heterochromatin by Hmgb2 and/or is mediated through changes in DNA methylation, although this will take additional experiments to fully elucidate. Our data also indicate that these two proteins confer opposite regulation of chromatin accessibility when they target the same promoters and opposite regulation of gene expression when they target the same cardiac enhancers.
In this model, Ctcf organizes the framework of the genome within which the cell type-specific chromatin factors operate. Hmgb2 also acts within the boundaries of this model to maintain heterochromatic regions (with a high density of Hmgb2 to allow for tight packaging) and facilitate complex formation (with a low density of Hmgb2 priming DNA for binding by other proteins) whose specific functions are dependent on the cell type-specific proteome. In support of this model, we observe a promoter-specific effect of hmgb2 knockdown on transcription but a uniform effect of Hmgb2 to regulate chromatin accessibility at the genomic scale. Thus, in a cell type-dependent way, the nucleus regulates the regions established by Ctcf. However, alterations to balance between Hmgb2 and Ctcf disrupt the boundaries of heterochromatin, undoing the cell type-specific silencing. We hypothesize that the changes in the ratio of Hmgb2 to Ctcf that we observe with cardiac pathology and across genetic backgrounds allow for varied genomic plasticity, supporting a general theory in which global chromatin accessibility is an important component to transcriptome remodeling in disease.
Experimental Procedures
Analysis of the Hybrid Mouse Diversity Panel
Microarray (RNA isolated from the left ventricle) and phenotypic data from 84 classical inbred and recombinant strains of adult (aged 8–10 weeks) female mice in the basal state or after treatment with isoproterenol (Alzet microosmotic pump releasing 30 mg/kg/day) for 3 weeks (29) were analyzed. These data (29) were acquired from female mice for three reasons. First, choosing a single gender removed this as a variable and the scope of this study prohibited repeating all the analyses in both genders. Second, male mice are prone to establishing societal hierarchies when housed in the same cage, which could affect how the mice responded to stress signaling, such as isoproterenol. Third, pilot studies on both genders revealed a more reproducible and pronounced phenotype in female animals. Transcript abundances were correlated to determine an R2 value, which was converted to a p value. Correlation of transcript abundance in liver (31) and bone marrow (32) was also assessed. Separately, the ratio of ctcf to hmgb2 (calculated by subtracting the log-scaled hmgb2 value from the ctcf value) was plotted against cardiac phenotype, with different plots for different subsets of strains. Strains were grouped by whether they went into failure upon isoproterenol treatment (n = 13 strains), developed cardiac hypertrophy (n = 22), showed minimal change (n = 9), or showed phenotypic traits inconsistent with a single disease state (n = 40).
Cell Culture
HEK 293T and HeLa cell lines were grown in DMEM (Gibco, 11965-092) with 10% FBS. Primary NRVMs from 1-day-old rat pups were isolated via enzymatic digestion and plated (1× penicillin, streptomycin, glutamine, 10% horse serum, 5% newborn calf serum, 1.68% M199 salts in DMEM) for 24 h and transferred to DMEM containing 0.1% insulin/transferrin/sodium selenite supplement.
Knockdown was performed with 50 nm total of two siRNAs per mRNA target (Qiagen, hmgb2 mouse, SI01067773 and SI01067759; hmgb2 rat, SI02877252 and SI02877266; ctcf, SI01503187 and SI01503208) suspended in Lipofectamine (Invitrogen, NRVMs, 13778-075; cell lines, 11668-027) at time 0. ctcf siRNA treatment was repeated at 24 h. Cells were assayed at 72 h. Control cells were treated with Lipofectamine alone to control for toxicity.
Overexpression in NRVMs was performed using adenovirus (Vector BioLabs: hmgb2, Adv-290952, or ctcf, Adv-206223, 50 multiplicity of infection) and assayed at 24 h. In cell lines, plasmid constructs with GFP-tagged protein or Gfp alone (Hmgb2, ProSpec, PRO-888; Ctcf and Gfp, pEGFP-Ctcf and pEGFP-C2) were administered via Lipofectamine 2000, and cells were assayed at 24 h.
To model hypertrophy, NRVMs were treated with 10 μm phenylephrine (Sigma, P-6126) at time 0, and cells were assayed at 48 h (21, 77). To visualize cell density, cells were submerged in crystal violet (EMD-Millipore, 192-12) that was diluted (1% in methanol) for 2 min and then gently rinsed.
Western Blotting
Isolated cells were lysed (50 mm Tris, pH 7.4, 10 mm EDTA, 1% SDS, 0.1 mm phenylmethanesulfonyl fluoride/protease inhibitor mixture pellet (Roche Applied Science), 0.2 mm sodium orthovanadate, 0.1 mm sodium fluoride, 10 mm sodium butyrate), sonicated, and separated via SDS-PAGE using Laemmli buffer. Detection was performed on the LI-COR Odyssey. Antibodies were as follows: Ctcf 1:1000 (Active Motif, 61311, rabbit); Hmgb2 1:1000 (Abcam, ab67282, rabbit); histone H1 1:1000 (Abcam, ab4269, mouse); H3K27me3 1:1000 (Abcam, ab6002, mouse); histone H3 1:10,000 (Abcam, ab1791, rabbit); gapdh 1:1000 (Santa Cruz Biotechnology, sc20357, goat); actin 1:1000 (Santa Cruz Biotechnology, sc1616, goat); secondaries 1:10,000 (LI-COR, IRDye conjugated).
Quantitative PCR
Cells were lysed in TRIzol (Ambion, 15596018). cDNA was synthesized using iScript cDNA synthesis kit (Bio-Rad, 170-8891). qPCR was performed using SsoFast EvaGreen Supermix (Bio-Rad, 1725201) on a Bio-Rad C1000 thermocycler. Primers are listed at the end of the “Experimental Procedures.”
Hmgb2 ChIP-seq and Bioinformatics Analysis
Hmgb2 was analyzed by chromatin immunoprecipitation followed by massively parallel DNA sequencing (ChIP-seq). NRVMs were fixed (1% formaldehyde, 10 min), lysed in lysis buffer (50 mm HEPES, pH 7.5, 150 mm NaCl, 1 mm EDTA, pH 8, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, protease inhibitor mixture tablet (Roche Applied Science)), sonicated to fragments of 500 bp, and diluted in RIPA buffer. DNA-bound protein was immunoprecipitated using anti-Hmgb2 (Abcam, ab67282) and precipitated with protein A-conjugated magnetic beads (Millipore, LSKMAGA10). Beads were washed (twice in wash buffer, 0.1% SDS, 1% Triton X-100, 2 mm EDTA, pH 8, 150 mm NaCl, 20 mm Tris-HCl, pH 8; once in 500 mm NaCl in wash buffer). DNA was eluted (1% SDS, 100 mm NaHCO3, 15 min, 30 °C) and phenol/chloroform-purified. Samples were ligated to sequencing adapters with Illumina Paired-End sample prep kit and sequenced on Illumina Genome Analyzer IIx using paired-end sequencing. Reads were aligned to the rat reference genome (rn4) using Bowtie (0.12.7) (78), with a maximum of two allowable mismatches in the seed region (first 28 nucleotides). Randomized reads of the same length and number as the Hmgb2 data set served as control. MACS 1.4.1 (79) was used for peak calling, with a p value cutoff of 10−5. Promoters were defined as 2 kb upstream to 500 bp downstream of the transcription start site (TSS). LiftOver, from UCSC genome browser, was used to convert to mouse and human genomes. ChIP-seq data were deposited in Gene Expression Omnibus (GSE80453).
Hmgb2 data were compared with the following data sets: Ctcf ChIP-seq in human CD4+ cells, gene expression omnibus accession GSM325895; Ctcf ChIP-seq in adult mouse heart, UCSC accession wgEncodeEM001684; Ctcf ChIP-seq in mouse ES cells, gene expression omnibus accession GSM699165; Ctcf ChIP-seq in rat liver (33); cardiac transcription factor ChIP-seq in HL-1 (34); hmgb2 knockdown microarray in NRVMs (21); RNA-seq data for ctcf knock-out mice9; bisulfite sequencing from mouse heart (54); DNase I hypersensitivity sites (DNase HS) from mouse heart, ENCODE, ENCFF001PMI.
We used Genomic Ranges in R to determine the portion of Hmgb2 peaks that were overlapped by Ctcf peaks and designated them Hmgb2- and Ctcf-shared peaks. We then removed these from the other datasets to get Hmgb2-only and Ctcf-only peaks. We compared the proximity of these peaks to other genomic features (lamina-associated domains (57), topologically associating domains (15), and cardiac enhancers (13)) using BED Tools to determine the distance between two features and density plots to visualize the distribution of distances of the Hmgb2 or Ctcf peaks to a genomic feature (set to bp 0). We defined peaks as being close distance (in or within 30 bp), medium distance (31–380 bp), or far distance (381 bp to 1 kb) of a cardiac enhancer and then found the nearest gene (nearest function in Genomic Ranges) and asked how the nearest gene's expression changed in response to ctcf knock-out or hmgb2 knockdown. Bar graphs indicate the percent of genes up- or down-regulated at each distance (with the remaining genes unchanged; not plotted). We also found Hmgb2 or Ctcf peaks within 1 kb of, but not in, a gene and plotted the percent of nearest genes that were active or inactive (determined by the presence of a RNA pol II promoter peak, ENCODE, ENCFF001YAH). For peaks within a gene, or within 1 kb of a gene, we asked how the expression of the gene changed with ctcf knock-out depending on whether the gene was active or inactive in the basal state. Percentage indicates percent of genes up- or down-regulated out of the pool of all genes within that distance of a peak and with the same pol II binding status (promoter peak or not). Ctcf cell type-independent peaks were determined by finding overlapping peaks in 10 adult mouse tissues, including the heart, and mESC cells (ENCODE: ENCFF001YAF, ENCFF001YAC, ENCFF001YAW, ENCFF001YAY, ENCFF001YBA, ENCFF001YAM, ENCFF001YAI, ENCFF001XZU, ENCFF001YBC, ENCFF001YAO, and ENCFF001XZY).
Hmgb2 ChIP-seq and microarray data were compared with Hi-C chromatin conformation capture data (15) to identify genes regulated by Hmgb2 that fell in the same topological domain. Genes regulated by Hmgb2 in rat were found in the genomes of the Hi-C data sets (mm9, hg18). For domains with more than one gene, each gene was compared with every other gene to determine whether they had matching transcriptional responses to hmgb2 knockdown (either both up-regulated or both down-regulated). As control, all RefSeq genes were randomly sampled to create a mock list of genes of the same size as our Hmgb2-regulated list, which were then randomly designated as up or down-regulated, and used to determine the percentage of inter-domain comparisons that matched. This was repeated for a total of 70 times. In all cases, the average percentage that matched plateaued (∼50%) after 5–30 random samples.
Genes regulated by Hmgb2 (microarray) or Ctcf (RNA-seq) were found in the mouse genome and compared with ChIP-seq data in the adult mouse heart from ENCODE datasets as follows: RNA pol II, ENCFF001LKL; H3K4me3, ENCFF001KHV. Alignment of ChIP-seq data across Hmgb2-regulated genes was performed using SeqPlots with the following parameters: anchored features, 10-bp bins, extend targets 1 kb up and downstream. As control, alignment across all genes was determined using RefSeq genes and gene predictions for mm9 downloaded from the UCSC Genome Browser.
ChIP-PCR and ChIP-reChIP
ChIP was performed (80) on 30 million NRVMs fixed (1% formaldehyde, 10 min), lysed (50 mm Tris-HCl, pH 8, 10 mm EDTA, 1% SDS, protease inhibitor mixture Set I Calbiochem I) and sonicated using an EpiShearTM Multi-Sample Sonicator (Active Motif), leading to fragments between 300 and 1000 bp. ChIP was performed using ChIP IT kit (Active Motif, 53040). DNA-bound protein was immunoprecipitated using anti-Hmgb2 (Abcam, ab67282), anti-Ctcf (Active Motif, 61311; Abcam, ab70303), or IgG (Santa Cruz Biotechnology, sc2027). Results from both anti-Ctcf immunoprecipitations were averaged.
For ChIP-reChIP experiments (35) the manufacturer's guidelines for the ChIP kit were followed until elution. Elution was performed in elution buffer shaken at 65 °C for 30 min. The second immunoprecipitation was performed on eluant diluted in IP dilution buffer. ChIP-reChIP experiments used anti-Ctcf (Abcam, ab70303) and anti-Hmgb2 (Abcam, ab67282) antibody except when the second immunoprecipitation was for the same protein as the first, in which case the second immunoprecipitation used anti-Ctcf (Active Motif, 61311) or anti-Hmgb2 (Abcam, ab55169). Primers target the promoter of the indicated genes; negative control was determined from Hmgb2 ChIP-seq. See under “Primers” for the primer sequences.
Immunohistochemistry
Hearts from BALB/c mice (8–10 weeks) were fixed with formalin and paraffin-embedded. Coronal sections (4 μm thickness) were deparaffinized with serial washes as follows: xylene (2 times for 5 min), absolute ethanol (3 times for 3 min), 95% ethanol (2 times for 3 min), 70% ethanol (1 time for 3 min), and distilled water (several rinses and 5 min). For work in isolated cells, samples were fixed with formalin for 10 min. For immunostaining, samples were washed with PBS three times for 5 min and blocked and permeabilized with 5% BSA and 0.1% Triton X-100 in PBS for 30 min. Samples were incubated with primary antibodies overnight at 4 °C (1:100 in 2.5% BSA/PBS; Hmgb2, Abcam ab67282, rabbit or Abcam ab55169, mouse; Ctcf, BD Biosciences 612148, mouse), washed with PBS, and incubated with respective secondary antibodies (confocal, 1:100 in PBS; Alexa Fluor conjugated, Life Technologies, Inc.; STED, 1:100 in PBS; Atto 647N, Sigma for Ctcf and Oregon Green, Life Technologies, Inc., for Hmgb2). DAPI (1:100) was used to demarcate the nucleus, and phalloidin (1:100) was used for cell size analysis, and wheat germ agglutinin (1:100) was used to label the cell membranes. Samples were mounted with Prolong Gold.
5′-Fluorouridine Transcriptional Run-on Assay
Cells were treated with 4 mm 5′FU (Sigma, F5130) for 30 min at 37 °C (81), rinsed with 1× HEPEM wash buffer (65 mm PIPES, 30 mm HEPES, 2 mm MgCl2·6H2O, 10 mm EGTA), and fixed (3.7% formaldehyde, 1× HEPEM, 0.05% Triton X-100, 10 min (NRVM) or 15 min (HEK 293T)). Cells were washed for 5 min with 1× HEPEM (twice), PBS, and 0.05% Tween/PBS and then incubated with primary antibody (BrdU 1:50, Sigma, B8434, mouse) for 2 h at 37 °C. Coverslips were washed with 0.05% Tween/PBS twice and PBS, incubated with secondary antibody (1:100 Alexa Fluor-conjugated, Life Technologies, Inc.) for 1 h at room temperature, washed with PBS (three times for 5 min), and mounted with Prolong Gold. DAPI (1:100) was used to demarcate the nucleus, and nucleolin (1:100, Abcam, ab22758, rabbit) was used to mark the nucleolus. p values are based on Mann-Whitney test.
Microscopy
Images were acquired on a Nikon A1R confocal microscope and analyzed in ImageJ. For colocalization analysis, super-resolution was achieved using dual-color STED microscopy on a custom STED instrument developed at UCLA. Colocalization was measured using in-house software to measure distances between clusters.
MNase Digestion
NRVMs were lysed (10 mm Tris-HCl, pH 7.4, 10 mm NaCl, 2 mm MgCl2, 0.5% Nonidet P-40) and centrifuged at maximum speed for 5 min. Nuclei were washed and resuspended in MNase digestion buffer (10 mm Tris-HCl, pH 7.4, 15 mm NaCl, 60 mm KCl, 1 mm CaCl2) and treated with 0.001 units of micrococcal nuclease (MNase, Worthington, LS004798) for 5 min at 37 °C. Digestion was stopped with 240 μl of MNase digestion buffer, 60 μl of MNase stop buffer (100 mm EDTA, 10 mm EGTA, pH 7.5), 30 μl of 20% SDS, and 9 μl of proteinase K (25 mg/ml). The sample was vortexed, left overnight at 37 °C, and phenol/chloroform-purified.
5 μg of digested DNA was loaded per lane on a 1.5% agarose gel and separated for ∼6 h at ∼50 V at 4 °C. DNA was excised from the gel as follows: 2–20 kb (compact), 700 bp to 1.5 kb (intermediate), and 500–650 bp + 300–500 bp + 100–200 bp (open) (boundaries captured all prominent bands). DNA was purified using QIAquick gel extraction Kit (Qiagen, 28706). Equal volumes of DNA were analyzed by quantitative PCR. See under “Primers.” Expression data for genes whose promoters were analyzed came from microarray for hmgb2 knockdown in NRVMs (21) or phenylephrine treatment in NRVMS (56).
Primers
Quantitative PCR for mRNA is as follows: ctcf F, CCCAGAGTGGTACCATGAAG, and R, ACAGCATCACAGTAGCGACA; hmgb2 F, AAGCCGCGGGGCAAGATGTC, and R, TGCCCTTGGCACGGTATGCA; gapdh F, CCCACTAACATCAAATGGGG, and R, CCTTCCACAATGCCAAAGTT. ChIP-PCR and MNase (primers target promoters of following genes) are as follows: acta1 F, CGCTTGCTCTGGGCCTCGTC, and R, CTGCGGACGCCCACCAACTAC; brd2 F, GCGCGTCCCTGAGCTCCCTT, and R, CCGAGGCAGAGCCTCCAGCA; cabin1 F, CCTGAGCGCGACGGACCAAC, and R, TGCGCGCCAGACACACACAG; casp2 F, AAGGGGCTGATGGCGGCTGA, and R, CGCGGGACCAGGCCAAGAAG; dhrs7c F, TAAGACAGGCAGGACCCAAC, and R, ATCAGTGGTTTCCGATGGTC; fgf16 F, CCCCTTAAGCGCTCCCACCCT, and R, TCCCCTAGTCCCACTCCCCACC; H42.1 F, GACGGAATGAGTGTGTGTGG, and R, CTTGCCTGTCACCCTCTCA; hmgn2 F, TGCGCGACACTGGGCACATC, and R, GCCAGGCCTCGCAAACCCCT; ldha F, CTGGGGTGGAGGTGCAGGGT, and R, CAGGCCCGCCATCCCCCTAA; mmp14 F, AAGGAGGGCATTGGGGCGGG, and R, CGGCGAACTGAGTTGGAAGCCC; nfkb2 F, CTGAACCGGGCCGAAGCCAA, and R, ACCCACTCCCCCACACACCC; nppa F, CAGCTGAGATGCAAGCAGAG, and R, CCTCAGCTGCAAGAGTCACA; nppb F, ACCAGAGTGCCCGGAAGTGGTG, and R, AGGCCCTGCCCGGCTACCAA; parp1 F, CTGCGGCACGAGAGGGAGGA, and R, TGCGGAGCGAGTCCTTGGGG; por F, CCCGCGGTCCTGTAGGTCTCTG, and R, CCGCAGCCTTCTGGTCGGTG; tgfb3 F, CGCGATCCTGGCAGCGGTT, R, CAGAGGGCACCCTCGGCCTT; tnni3 F-AACCCGTGGCCCAGAGAGGG, and R, AGCGACGTCGGACAGGAGCA; tuba4a F, TGGCTCAGGAGGGGGTGCTG, R, GCGCGGGTTGGTGTTAGGGG; negative control F-TGACAATGATGGCCCTAACA, and R, AACCGGGAACACATCATCTC.
Author Contributions
E. M., M. R. G., E. K., H. C., and R. L. carried out the experiments; E. M., M. R. G., E. K., H. C., C. D. R., J. W., S. F., and T. M. V. analyzed the data; S. F. N., E. S., A. J. L., Y. Wu, Y. Wang. S. K. K., and T. M. V. provided reagents, infrastructure, and technical/conceptual support; E. M., M. R. G., and T. M. V. designed the study; E. M., M. R. G., E. K., and T. M. V. wrote the paper. All authors approved of the final manuscript.
Acknowledgments
The ctcf and hmgb2 plasmids used in this work were kindly provided by Dr. Maria Dolores Delgado Villar (Instituto de Biomedicina y Biotecnología de Cantabria (IBBTEC)) and Dr. Kwan Yong Choi (IBB Postech), respectively.
This work was supported in part by National Institutes of Health Grants HL105699 (to T. M. V.), HL115238 (to T. M. V.), HL114437 (to A. J. L. and Y. Wang), and HL129639 (to T. M. V. and Y. Wang), American Heart Association Grant IRG18870056 (to T. M. V.), Thermo Fisher Scientific, and the Department of Anesthesiology at UCLA. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
R. Garrido and T. M. Vondriska, unpublished data.
- H3K9me3
- histone H3 lysine 9 trimethylation
- ChIP seq
- chromatin immunoprecipitation followed by DNA sequencing
- Ctcf
- CCCTC-binding factor
- DNase HS
- DNase I hypersensitivity site
- H3K4me3
- histone H3 lysine 4 trimethylation
- H3K27ac
- histone H3 lysine 27 acetylation
- H3K27me3
- histone H3 lysine 27 trimethylation
- Hmgb2
- high mobility group protein B2
- LAD
- lamina-associated domain
- MNase
- micrococcal nuclease
- NRVM
- neonatal rat ventricular myocyte
- RNA pol II
- RNA polymerase II
- rRNA
- ribosomal RNA
- TAD
- topologically associating domain
- TSS
- transcription start site
- 5FU
- 5′-fluorouridine
- STED
- stimulated emission depletion
- qPCR
- quantitative PCR
- F
- forward
- R
- reverse
- mESC
- mouse embryonic stem cell.
References
- 1. Luger K., Mäder A. W., Richmond R. K., Sargent D. F., and Richmond T. J. (1997) Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260 [DOI] [PubMed] [Google Scholar]
- 2. Strahl B. D., and Allis C. D. (2000) The language of covalent histone modifications. Nature 403, 41–45 [DOI] [PubMed] [Google Scholar]
- 3. Filion G. J., van Bemmel J. G., Braunschweig U., Talhout W., Kind J., Ward L. D., Brugman W., de Castro I. J., Kerkhoven R. M., Bussemaker H. J., and van Steensel B. (2010) Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell 143, 212–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lieberman-Aiden E., van Berkum N. L., Williams L., Imakaev M., Ragoczy T., Telling A., Amit I., Lajoie B. R., Sabo P. J., Dorschner M. O., Sandstrom R., Bernstein B., Bender M. A., Groudine M., Gnirke A., et al. (2009) Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cremer T., Cremer M., Dietzel S., Müller S., Solovei I., and Fakan S. (2006) Chromosome territories–a functional nuclear landscape. Curr. Opin. Cell Biol. 18, 307–316 [DOI] [PubMed] [Google Scholar]
- 6. Rosa-Garrido M., Karbassi E., Monte E., and Vondriska T. M. (2013) Regulation of chromatin structure in the cardiovascular system. Circ. J. 77, 1389–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Backs J., and Olson E. N. (2006) Control of cardiac growth by histone acetylation/deacetylation. Circ. Res. 98, 15–24 [DOI] [PubMed] [Google Scholar]
- 8. Dorn G. W., 2nd. (2011) MicroRNAs in cardiac disease. Transl. Res. 157, 226–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Movassagh M., Choy M. K., Goddard M., Bennett M. R., Down T. A., and Foo R. S. (2010) Differential DNA methylation correlates with differential expression of angiogenic factors in human heart failure. PLoS ONE 5, e8564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gang H., Shaw J., Dhingra R., Davie J. R., and Kirshenbaum L. A. (2013) Epigenetic regulation of canonical TNFα pathway by HDAC1 determines survival of cardiac myocytes. Am. J. Physiol. Heart Circ. Physiol. 304, H1662–H1669 [DOI] [PubMed] [Google Scholar]
- 11. Wamstad J. A., Alexander J. M., Truty R. M., Shrikumar A., Li F., Eilertson K. E., Ding H., Wylie J. N., Pico A. R., Capra J. A., Erwin G., Kattman S. J., Keller G. M., Srivastava D., Levine S. S., et al. (2012) Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell 151, 206–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Paige S. L., Thomas S., Stoick-Cooper C. L., Wang H., Maves L., Sandstrom R., Pabon L., Reinecke H., Pratt G., Keller G., Moon R. T., Stamatoyannopoulos J., and Murry C. E. (2012) A temporal chromatin signature in human embryonic stem cells identifies regulators of cardiac development. Cell 151, 221–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Papait R., Cattaneo P., Kunderfranco P., Greco C., Carullo P., Guffanti A., Viganò V., Stirparo G. G., Latronico M. V., Hasenfuss G., Chen J., and Condorelli G. (2013) Genome-wide analysis of histone marks identifying an epigenetic signature of promoters and enhancers underlying cardiac hypertrophy. Proc. Natl. Acad. Sci. U.S.A. 110, 20164–20169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Simonis M., Klous P., Splinter E., Moshkin Y., Willemsen R., de Wit E., van Steensel B., and de Laat W. (2006) Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat. Genet. 38, 1348–1354 [DOI] [PubMed] [Google Scholar]
- 15. Dixon J. R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J. S., and Ren B. (2012) Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cuddapah S., Schones D. E., Cui K., Roh T. Y., Barski A., Wei G., Rochman M., Bustin M., and Zhao K. (2011) Genomic profiling of HMGN1 reveals an association with chromatin at regulatory regions. Mol. Cell. Biol. 31, 700–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim T. H., Abdullaev Z. K., Smith A. D., Ching K. A., Loukinov D. I., Green R. D., Zhang M. Q., Lobanenkov V. V., and Ren B. (2007) Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 128, 1231–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fedele M., Fidanza V., Battista S., Pentimalli F., Klein-Szanto A. J., Visone R., De Martino I., Curcio A., Morisco C., Del Vecchio L., Baldassarre G., Arra C., Viglietto G., Indolfi C., Croce C. M., and Fusco A. (2006) Haploinsufficiency of the Hmga1 gene causes cardiac hypertrophy and myelo-lymphoproliferative disorders in mice. Cancer Res. 66, 2536–2543 [DOI] [PubMed] [Google Scholar]
- 19. Nakayama J., Rice J. C., Strahl B. D., Allis C. D., and Grewal S. I. (2001) Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292, 110–113 [DOI] [PubMed] [Google Scholar]
- 20. Guenther M. G., Levine S. S., Boyer L. A., Jaenisch R., and Young R. A. (2007) A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130, 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Franklin S., Chen H., Mitchell-Jordan S., Ren S., Wang Y., and Vondriska T. M. (2012) Quantitative analysis of the chromatin proteome in disease reveals remodeling principles and identifies high mobility group protein B2 as a regulator of hypertrophic growth. Mol. Cell. Proteomics 11, M111.014258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gaikwad A. B., Sayyed S. G., Lichtnekert J., Tikoo K., and Anders H. J. (2010) Renal failure increases cardiac histone H3 acetylation, dimethylation, and phosphorylation and the induction of cardiomyopathy-related genes in type 2 diabetes. Am. J. Pathol. 176, 1079–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lu J. T., Muchir A., Nagy P. L., and Worman H. J. (2011) LMNA cardiomyopathy: cell biology and genetics meet clinical medicine. Dis. Model. Mech. 4, 562–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Furusawa T., Rochman M., Taher L., Dimitriadis E. K., Nagashima K., Anderson S., and Bustin M. (2015) Chromatin decompaction by the nucleosomal binding protein HMGN5 impairs nuclear sturdiness. Nat. Commun. 6, 6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bell A. C., West A. G., and Felsenfeld G. (1999) The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98, 387–396 [DOI] [PubMed] [Google Scholar]
- 26. Handoko L., Xu H., Li G., Ngan C. Y., Chew E., Schnapp M., Lee C. W., Ye C., Ping J. L., Mulawadi F., Wong E., Sheng J., Zhang Y., Poh T., Chan C. S., et al. (2011) CTCF-mediated functional chromatin interactome in pluripotent cells. Nat. Genet. 43, 630–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cuddapah S., Jothi R., Schones D. E., Roh T. Y., Cui K., and Zhao K. (2009) Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 19, 24–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Agresti A., and Bianchi M. E. (2003) HMGB proteins and gene expression. Curr. Opin. Genet. Dev. 13, 170–178 [DOI] [PubMed] [Google Scholar]
- 29. Rau C. D., Wang J., Avetisyan R., Romay M. C., Martin L., Ren S., Wang Y., and Lusis A. J. (2015) Mapping genetic contributions to cardiac pathology induced by β-adrenergic stimulation in mice. Circ. Cardiovasc. Genet. 8, 40–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yeager J. C., and Iams S. G. (1981) Isoproterenol-induced cardiac failure in the spontaneously hypertensive rat. Proc. Soc. Exp. Biol. Med. 168, 137–142 [DOI] [PubMed] [Google Scholar]
- 31. Ghazalpour A., Bennett B. J., Shih D., Che N., Orozco L., Pan C., Hagopian R., He A., Kayne P., Yang W. P., Kirchgessner T., and Lusis A. J. (2014) Genetic regulation of mouse liver metabolite levels. Mol. Syst. Biol. 10, 730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Farber C. R., Bennett B. J., Orozco L., Zou W., Lira A., Kostem E., Kang H. M., Furlotte N., Berberyan A., Ghazalpour A., Suwanwela J., Drake T. A., Eskin E., Wang Q. T., Teitelbaum S. L., and Lusis A. J. (2011) Mouse genome-wide association and systems genetics identify Asxl2 as a regulator of bone mineral density and osteoclastogenesis. PLoS Genet. 7, e1002038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schmidt D., Schwalie P. C., Wilson M. D., Ballester B., Gonçalves A., Kutter C., Brown G. D., Marshall A., Flicek P., and Odom D. T. (2012) Waves of retrotransposon expansion remodel genome organization and CTCF binding in multiple mammalian lineages. Cell 148, 335–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. He A., Kong S. W., Ma Q., and Pu W. T. (2011) Co-occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart. Proc. Natl. Acad. Sci. U.S.A. 108, 5632–5637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. van de Nobelen S., Rosa-Garrido M., Leers J., Heath H., Soochit W., Joosen L., Jonkers I., Demmers J., van der Reijden M., Torrano V., Grosveld F., Delgado M. D., Renkawitz R., Galjart N., and Sleutels F. (2010) CTCF regulates the local epigenetic state of ribosomal DNA repeats. Epigenetics Chromatin 3, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nazar R. N. (2004) Ribosomal RNA processing and ribosome biogenesis in eukaryotes. IUBMB Life 56, 457–465 [DOI] [PubMed] [Google Scholar]
- 37. Farley K. I., Surovtseva Y., Merkel J., and Baserga S. J. (2015) Determinants of mammalian nucleolar architecture. Chromosoma 124, 323–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Grummt I. (2003) Life on a planet of its own: regulation of RNA polymerase I transcription in the nucleolus. Genes Dev. 17, 1691–1702 [DOI] [PubMed] [Google Scholar]
- 39. Bártová E., Horáková A. H., Uhlírová R., Raska I., Galiová G., Orlova D., and Kozubek S. (2010) Structure and epigenetics of nucleoli in comparison with non-nucleolar compartments. J. Histochem. Cytochem. 58, 391–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Raska I., Koberna K., Malínský J., Fidlerová H., and Masata M. (2004) The nucleolus and transcription of ribosomal genes. Biol. Cell 96, 579–594 [DOI] [PubMed] [Google Scholar]
- 41. Strahl B. D., Ohba R., Cook R. G., and Allis C. D. (1999) Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proc. Natl. Acad. Sci. U.S.A. 96, 14967–14972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Koch C. M., Andrews R. M., Flicek P., Dillon S. C., Karaöz U., Clelland G. K., Wilcox S., Beare D. M., Fowler J. C., Couttet P., James K. D., Lefebvre G. C., Bruce A. W., Dovey O. M., Ellis P. D., et al. (2007) The landscape of histone modifications across 1% of the human genome in five human cell lines. Genome Res. 17, 691–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R. S., and Zhang Y. (2002) Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298, 1039–1043 [DOI] [PubMed] [Google Scholar]
- 44. Hannan R. D., Jenkins A., Jenkins A. K., and Brandenburger Y. (2003) Cardiac hypertrophy: a matter of translation. Clin. Exp. Pharmacol. Physiol. 30, 517–527 [DOI] [PubMed] [Google Scholar]
- 45. Brandenburger Y., Jenkins A., Autelitano D. J., and Hannan R. D. (2001) Increased expression of UBF is a critical determinant for rRNA synthesis and hypertrophic growth of cardiac myocytes. FASEB J. 15, 2051–2053 [DOI] [PubMed] [Google Scholar]
- 46. Yamazaki F., Nagatsuka Y., Shirakawa H., and Yoshida M. (1995) Repression of cell cycle progression by antisense HMG2 RNA. Biochem. Biophys. Res. Commun. 210, 1045–1051 [DOI] [PubMed] [Google Scholar]
- 47. Fedoriw A. M., Stein P., Svoboda P., Schultz R. M., and Bartolomei M. S. (2004) Transgenic RNAi reveals essential function for CTCF in H19 gene imprinting. Science 303, 238–240 [DOI] [PubMed] [Google Scholar]
- 48. Moore J. M., Rabaia N. A., Smith L. E., Fagerlie S., Gurley K., Loukinov D., Disteche C. M., Collins S. J., Kemp C. J., Lobanenkov V. V., and Filippova G. N. (2012) Loss of maternal CTCF is associated with peri-implantation lethality of Ctcf null embryos. PLoS ONE 7, e34915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Fiorentino F. P., and Giordano A. (2012) The tumor suppressor role of CTCF. J. Cell. Physiol. 227, 479–492 [DOI] [PubMed] [Google Scholar]
- 50. Pant V., Mariano P., Kanduri C., Mattsson A., Lobanenkov V., Heuchel R., and Ohlsson R. (2003) The nucleotides responsible for the direct physical contact between the chromatin insulator protein CTCF and the H19 imprinting control region manifest parent of origin-specific long-distance insulation and methylation-free domains. Genes Dev. 17, 586–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bell A. C., and Felsenfeld G. (2000) Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405, 482–485 [DOI] [PubMed] [Google Scholar]
- 52. Renda M., Baglivo I., Burgess-Beusse B., Esposito S., Fattorusso R., Felsenfeld G., and Pedone P. V. (2007) Critical DNA binding interactions of the insulator protein CTCF: a small number of zinc fingers mediate strong binding, and a single finger-DNA interaction controls binding at imprinted loci. J. Biol. Chem. 282, 33336–33345 [DOI] [PubMed] [Google Scholar]
- 53. Wang H., Maurano M. T., Qu H., Varley K. E., Gertz J., Pauli F., Lee K., Canfield T., Weaver M., Sandstrom R., Thurman R. E., Kaul R., Myers R. M., and Stamatoyannopoulos J. A. (2012) Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome Res. 22, 1680–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chen H., Orozco L. D., Wang J., Rau C. D., Rubbi L., Ren S., Wang Y., Pellegrini M., Lusis A. J., and Vondriska T. M. (2016) DNA methylation indicates susceptibility to isoproterenol-induced cardiac pathology and is associated with chromatin states. Circ. Res. 118, 786–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Maurano M. T., Wang H., John S., Shafer A., Canfield T., Lee K., and Stamatoyannopoulos J. A. (2015) Role of DNA methylation in modulating transcription factor occupancy. Cell Rep. 12, 1184–1195 [DOI] [PubMed] [Google Scholar]
- 56. Bush E., Fielitz J., Melvin L., Martinez-Arnold M., McKinsey T. A., Plichta R., and Olson E. N. (2004) A small molecular activator of cardiac hypertrophy uncovered in a chemical screen for modifiers of the calcineurin signaling pathway. Proc. Natl. Acad. Sci. U.S.A. 101, 2870–2875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Guelen L., Pagie L., Brasset E., Meuleman W., Faza M. B., Talhout W., Eussen B. H., de Klein A., Wessels L., de Laat W., and van Steensel B. (2008) Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453, 948–951 [DOI] [PubMed] [Google Scholar]
- 58. Meuleman W., Peric-Hupkes D., Kind J., Beaudry J. B., Pagie L., Kellis M., Reinders M., Wessels L., and van Steensel B. (2013) Constitutive nuclear lamina-genome interactions are highly conserved and associated with A/T-rich sequence. Genome Res. 23, 270–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Peric-Hupkes D., Meuleman W., Pagie L., Bruggeman S. W., Solovei I., Brugman W., Gräf S., Flicek P., Kerkhoven R. M., van Lohuizen M., Reinders M., Wessels L., and van Steensel B. (2010) Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol. Cell 38, 603–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ong C. T., and Corces V. G. (2014) CTCF: an architectural protein bridging genome topology and function. Nat. Rev. Genet. 15, 234–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Stros M., Ozaki T., Bacikova A., Kageyama H., and Nakagawara A. (2002) HMGB1 and HMGB2 cell-specifically down-regulate the p53- and p73-dependent sequence-specific transactivation from the human Bax gene promoter. J. Biol. Chem. 277, 7157–7164 [DOI] [PubMed] [Google Scholar]
- 62. Zwilling S., König H., and Wirth T. (1995) High mobility group protein 2 functionally interacts with the POU domains of octamer transcription factors. EMBO J. 14, 1198–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lehming N., Le Saux A., Schüller J., and Ptashne M. (1998) Chromatin components as part of a putative transcriptional repressing complex. Proc. Natl. Acad. Sci. U.S.A. 95, 7322–7326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhang J., McCauley M. J., Maher L. J. 3rd, Williams M. C., and Israeloff N. E. (2009) Mechanism of DNA flexibility enhancement by HMGB proteins. Nucleic Acids Res. 37, 1107–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hoppe G., Rayborn M. E., and Sears J. E. (2007) Diurnal rhythm of the chromatin protein Hmgb1 in rat photoreceptors is under circadian regulation. J. Comp. Neurol. 501, 219–230 [DOI] [PubMed] [Google Scholar]
- 66. Pallier C., Scaffidi P., Chopineau-Proust S., Agresti A., Nordmann P., Bianchi M. E., and Marechal V. (2003) Association of chromatin proteins high mobility group box (HMGB) 1 and HMGB2 with mitotic chromosomes. Mol. Biol. Cell 14, 3414–3426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ner S. S., and Travers A. A. (1994) HMG-D, the Drosophila melanogaster homologue of HMG 1 protein, is associated with early embryonic chromatin in the absence of histone H1. EMBO J. 13, 1817–1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Cato L., Stott K., Watson M., and Thomas J. O. (2008) The interaction of HMGB1 and linker histones occurs through their acidic and basic tails. J. Mol. Biol. 384, 1262–1272 [DOI] [PubMed] [Google Scholar]
- 69. Celona B., Weiner A., Di Felice F., Mancuso F. M., Cesarini E., Rossi R. L., Gregory L., Baban D., Rossetti G., Grianti P., Pagani M., Bonaldi T., Ragoussis J., Friedman N., Camilloni G., et al. (2011) Substantial histone reduction modulates genome-wide nucleosomal occupancy and global transcriptional output. PLoS Biol. 9, e1001086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bonne-Andrea C., Harper F., Sobczak J., and De Recondo A. M. (1984) Rat liver HMG1: a physiological nucleosome assembly factor. EMBO J. 3, 1193–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ugrinova I., Pashev I. G., and Pasheva E. A. (2009) Nucleosome binding properties and Co-remodeling activities of native and in vivo acetylated HMGB-1 and HMGB-2 proteins. Biochemistry 48, 6502–6507 [DOI] [PubMed] [Google Scholar]
- 72. Zuin J., Dixon J. R., van der Reijden M. I., Ye Z., Kolovos P., Brouwer R. W., van de Corput M. P., van de Werken H. J., Knoch T. A., van IJcken W. F., Grosveld F. G., Ren B., and Wendt K. S. (2014) Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc. Natl. Acad. Sci. U.S.A. 111, 996–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Phillips J. E., and Corces V. G. (2009) CTCF: master weaver of the genome. Cell 137, 1194–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Brandenburger Y., Arthur J. F., Woodcock E. A., Du X. J., Gao X. M., Autelitano D. J., Rothblum L. I., and Hannan R. D. (2003) Cardiac hypertrophy in vivo is associated with increased expression of the ribosomal gene transcription factor UBF. FEBS Lett. 548, 79–84 [DOI] [PubMed] [Google Scholar]
- 75. Jarboui M. A., Wynne K., Elia G., Hall W. W., and Gautier V. W. (2011) Proteomic profiling of the human T-cell nucleolus. Mol. Immunol. 49, 441–452 [DOI] [PubMed] [Google Scholar]
- 76. Gabellini D., Green M. R., and Tupler R. (2002) Inappropriate gene activation in FSHD: a repressor complex binds a chromosomal repeat deleted in dystrophic muscle. Cell 110, 339–348 [DOI] [PubMed] [Google Scholar]
- 77. Mitchell-Jordan S., Chen H., Franklin S., Stefani E., Bentolila L. A., and Vondriska T. M. (2012) Features of endogenous cardiomyocyte chromatin revealed by super-resolution STED microscopy. J. Mol. Cell. Cardiol. 53, 552–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Langmead B., Trapnell C., Pop M., and Salzberg S. L. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nusbaum C., Myers R. M., Brown M., Li W., and Liu X. S. (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Ubil E., Duan J., Pillai I. C., Rosa-Garrido M., Wu Y., Bargiacchi F., Lu Y., Stanbouly S., Huang J., Rojas M., Vondriska T. M., Stefani E., and Deb A. (2014) Mesenchymal-endothelial transition contributes to cardiac neovascularization. Nature 514, 585–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Rosa-Garrido M., Ceballos L., Alonso-Lecue P., Abraira C., Delgado M. D., and Gandarillas A. (2012) A cell cycle role for the epigenetic factor CTCF-L/BORIS. PLoS ONE 7, e39371. [DOI] [PMC free article] [PubMed] [Google Scholar]