Abstract
Ragweed allergens affect several million people in the United States and Canada. To date, only two ragweed allergens, Amb t 5 and Amb a 11, have their structures determined and deposited to the Protein Data Bank. Here, we present structures of methylated ragweed allergen Amb a 8, Amb a 8 in the presence of poly(l-proline), and Art v 4 (mugwort allergen). Amb a 8 and Art v 4 are panallergens belonging to the profilin family of proteins. They share significant sequence and structural similarities, which results in cross-recognition by IgE antibodies. Molecular and immunological properties of Amb a 8 and Art v 4 are compared with those of Bet v 2 (birch pollen allergen) as well as with other allergenic profilins. We purified recombinant allergens that are recognized by patient IgE and are highly cross-reactive. It was determined that the analyzed allergens are relatively unstable. Structures of Amb a 8 in complex with poly(l-proline)10 or poly(l-proline)14 are the first structures of the plant profilin in complex with proline-rich peptides. Amb a 8 binds the poly(l-proline) in a mode similar to that observed in human, mouse, and P. falciparum profilin·peptide complexes. However, only some of the residues that form the peptide binding site are conserved.
Keywords: allergen, allergy, crystal structure, epitope mapping, protein stability, IgE binding, peptide binding, pollen
Introduction
The designation of “panallergen” usually refers to minor allergens, present in most eukaryotic cells, that are responsible for IgE cross-reactivity to a variety of allergenic sources (1). In addition to being ubiquitous, they share highly conserved sequences and three-dimensional structures, which subsequently satisfies the requirements for cross-recognition by IgE (1). Due to this cross-recognition, panallergens are thought to be indicators of sensitization to several pollens (2, 3). Therefore, it is possible that panallergens could be used to predict cross/poly-sensitization to a variety of pollen allergens (4).
Only a few protein families contain panallergens, including calcium-binding proteins (4), cyclophilins (5), nonspecific lipid transfer proteins, pathogenesis-related protein (PR-10), polcalcins, and profilins (1). Profilins, which are the focus of this study, are small (12–15 kDa), mostly eukaryotic proteins that are involved in regulating the actin cytoskeleton (6). They are capable of binding actin and poly(l-proline) (7–9) and are also able to interact with membrane phospholipids, such as phosphatidylinositol 4-phosphate and phosphatidylinositol 4,5-bisphosphate (10–12). The profilin family shares highly conserved amino acid sequences that, in some instances, have over 75% identity, even between distantly related sources. Based on their source, profilins are described as originating from mammals, plants, other eukaryotes, and viruses (13). Plant profilins can be further divided into pollens, food, and products. Pollens include trees, grasses, and weeds; food includes fruits, legumes, nuts/seeds, and vegetables; and the product subclassification includes latex (1).
The profilins found in weed pollens are of particular interest due to their increasing presence as sources of seasonal allergies (14, 15). Weeds encompass a heterogeneous group of plants that are generally unwanted and invasive; thus, the term “weed” does not represent any particular botanical group or family (16). However, the clinically relevant allergens found in weed pollen, thus far, have been identified in plants belonging to only five different families, one of which is the Asteraceae family. The Asteraceae family includes thousands of plant species, but the important allergenic members are ragweed (Ambrosia), mugwort (Artemisia), feverfew (Parthenium), and sunflower (Helianthus) (15).
In this study, we structurally characterized Amb a 8, a ragweed profilin found in short or common ragweed (Ambrosia artemisiifolia), as well as the mugwort (Artemisia vulgaris) profilin Art v 4. Short ragweed, as well as giant ragweed, acts as a major elicitor of type 1 allergenic reactions in late summer and early fall, affecting over 15 million people in the United States and Canada (15). Amb a 8 has been found to have an 89% amino acid sequence identity with Art v 4 (4). In addition to their sequence identity, both Art v 4 and Amb a 8 have comparable IgE cross-reactivity, suggesting that either allergen could be used as a marker for profilin sensitization in weeds (4). Moreover, we compared molecular and immunological properties of Amb a 8 and Art v 4 with those of Bet v 2, a birch pollen profilin.
Results
Clustering Analysis of the Profilin Family
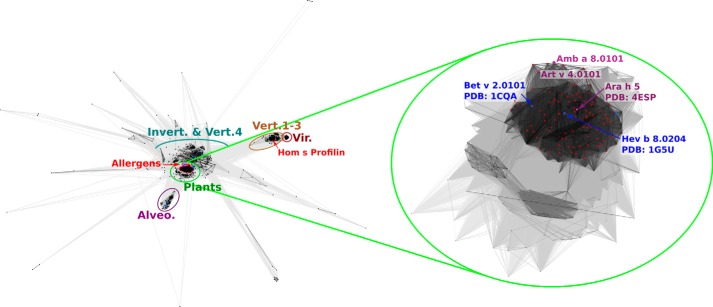
Sequences of proteins from the Pfam profilin family (Pfam00235), including sequences of the allergens classified in the AllFam database (17) of allergen families (AF051), were retrieved from the Pfam database (18). Clustering of the sequences was carried out to identify groups of similar sequences and to determine the distribution of known allergens and proteins with known structures within the data set. Clustering was performed based on their pairwise BLAST similarity scores (19) using a p value threshold of 1e−6. Clustering at more stringent p values gave similar results that are in agreement with the high similarity of profilins from various sources with only a few subgroups clearly separated from the obtained supercluster (Fig. 1).
FIGURE 1.
Two-dimensional projection of the CLANS clustering results. Proteins are indicated by dots. Lines indicate sequence similarity detectable with BLAST and are colored by a spectrum of shades of gray according to the BLAST p value. Sequences corresponding to structures in the PDB are indicated by blue dots, and sequences of known allergens are indicated by red dots.
Plant profilin sequences, including all of the allergen sequences classified in Allfam (with the exception of human profilin that is classified as an autoallergen), clustered together in a dense group in the central part of the diagram (represented by structures 1G5U (Hav b 8), 1A0K (Arabidopsis thaliana), and 1CQA (Bet v 2)). The remaining part of the supercluster (represented by structures 1ACF (Acanthamoeba castellanii), 1F2K (A. castellanii), 1K0K (Saccharomyces cerevisiae), 3D9Y (Schizosaccharomyces pombe), and 1PRQ (A. castellanii)) is composed of sequences from invertebrates and vertebral profilin 4. Furthermore, vertebral profilin 4 is more similar to invertebrate profilins than it is to vertebral profilins 1–3 (13) because it does not contain the poly(l-proline) binding site and does not interact with actin (20). Less similar sequences from different taxonomic groups, mostly Protists (e.g. Amoebozoa), are located peripherally but with clear connections to the central supercluster.
There are only two groups that are clearly separated. First, sequences of profilins from Alveolata, represented by structures of profilins from Apicomplexan parasites Toxoplasma gondii (PDB code 3NEC)2 and Plasmodium falciparum (PDB code 2JKF), formed a subgroup clearly separated but still closely connected to the central supercluster (Alveolata). A second separate cluster is formed by human profilins 1, 2, and 3 and the corresponding profilins from other Vertebrata (Vertebrata 1–3) (represented by structures 2V8C (Mus musculus), 2VK3 (Rattus norvegicus), 1AWI (Homo sapiens), 1PNE (Bos taurus), and 1D1J (H. sapiens)) with a very closely connected but clearly distinguishable subgroup of sequences from viruses.
Structural Analysis of Methylated Amb a 8
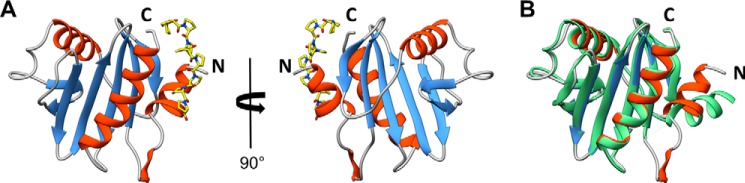
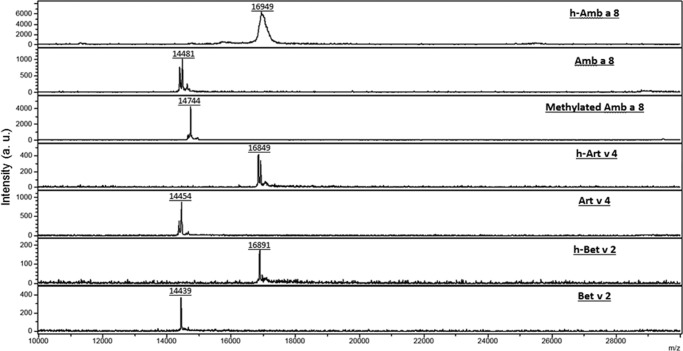
Full-length, native Amb a 8.0101 (UNP Q2KN24) is composed of 133 residues with a molecular mass of 14,245 Da. The construct created for these studies contained a start methionine followed by a His6 tag, tobacco etch virus cleavage site, and spacer region of residues GSG. The start methionine of Amb a 8 was removed in the construct, and the N-terminal region that is visible in the electron density is GSG followed by a serine, which corresponds to residue 2 of Amb a 8. Residue −2, the first glycine of the tag linker, to residue 133, a methionine, are visible in the electron density with average B factors of 11.9 Å2. The overall fold of Amb a 8 (Fig. 2) is similar to that of other profilins in that it is composed of two terminal α-helices, one short α-helix, and a β hairpin that sandwich a central five-stranded antiparallel β sheet. Amb a 8, which includes eight lysine residues, was subjected to reductive methylation, which was confirmed via MALDI TOF/TOF (Fig. 3). Electron density reveals that seven (residues 37, 45, 73, 88, 89, 97, and 98) of eight lysine residues were dimethylated. The eighth lysine (residue 54) has a disordered side chain that cannot be modeled. Amb a 8 also contains three cysteine residues, two of which participate in a disulfide bond (residues 95 and 117), and the third cysteine has clear electron density, indicating that it has been modified by β-mercaptoethanol (residue 13). Additionally, it appears that benzoic acid is bound to Amb a 8 at the surface of the molecule with an average B factor of 14.9 Å2. The source of the benzoic acid is unknown; however, its placement is based entirely on the electron density, which unquestionably fits that molecule.
FIGURE 2.
A, a structure of Amb a 8·poly(l-Pro)14 complex. Poly(l-Pro) is shown in a stick representation. B, superposition of Amb a 8 and Bet v 2 (PDB entry 1CQA).
FIGURE 3.
Mass spectrometry results for Amb a 8, Art v 4, and Bet v 2. The experiment was performed with a Bruker Ultraflex II MALDI TOF/TOF and processed with flex control software and flexAnalysis version 3.3. Intensity (arbitrary units (a.u.)).
Structural Analysis of Amb a 8 Bound with Poly(l-Proline)14
Unlike methylated Amb a 8, native Amb a 8 in the presence of poly(l-proline)14 crystallized in the P21 space group and contains two subunits in the asymmetric unit cell. Clear electron density is visible for residues −1 to 133 in chain A and residues 0–133 in chain B. The poly(l-proline)14 chains that are bound to chains A and B contain 9 ordered proline residues. Both chain A and chain B have a cysteine residue that has been modified by β-mercaptoethanol at position 13, like the one that is seen for methylated Amb a 8; however, chain A has electron density for two positions of the modified cysteine residue. Chains A and B differ from one another in that chain A has a disulfide bridge between residues 95 and 117, like that of the methylated Amb a 8, whereas no disulfide bridge exists between 95 and 117 of chain B. Both chains A and B help stabilize the poly(l-proline) chains through interactions with the side chain residues from Trp-3, Tyr-6, His-10, Trp-35, and Tyr-127.
Structural Analysis of Amb a 8 Bound with Poly(l-Proline)10
Native Amb a 8 in the presence of poly(l-proline)10 also crystallized in the P21 space group; however, it contains one subunit per asymmetric unit cell, unlike that of Amb a 8·poly(l-proline)14. Clear electron density is visible for residues 2–133 in chain A as well as all 10 proline residues of poly(l-proline)10. Similar to that of the Amb a 8·poly(l-proline)14 structure, the Amb a 8·poly(l-proline)10 structure also contains a disulfide link between Cys-95 and Cys-117. A superposition of Amb a 8·poly(l-proline)10 on Amb a 8·poly(l-proline)14 revealed an RMSD value of 0.46 Å. There are no significant differences between the Amb a 8·poly(l-proline)14 and the Amb a 8·poly(l-proline)10 structures. One minor difference between the two structures is the position of Gly-17. In the Amb a 8·poly(l-proline)14 structure, Gly-17 is positioned toward the surface of the subunit, whereas in the Amb a 8·poly(l-proline)10 structure, Gly-17 is 3.7 Å closer to the core of the protein. Due to the overall similarity and an RMSD value of 0.5 Å, for the remainder of the study, we will refer to the Amb a 8·poly(l-proline)14 structure for analysis.
Structural Analysis of Art v 4
Full-length, native Art v 4.0101 (UNP Q8H2C9) is also composed of 133 residues but has a molecular mass of 14,207 Da. Modifications made to the N terminus of Amb a 8, described above, were also applied to the construct of Art v 4. The first visible residue of Art v 4 is serine of the spacer region, followed by a glycine of the spacer region and then serine, which corresponds to residue 2 of native Art v 4. Art v 4 has the same overall fold as other profilins, with a total of three α-helices and a seven-stranded antiparallel β sheet. Residues −1, serine of the tag linker, to 133, a methionine, are visible in the electron density with average B factors of 17.9 Å2. Art v 4 also contains three cysteine residues, two of which form a disulfide bond (Cys-95 and -117), and one cysteine, like that of Amb a 8, that is modified by β-mercaptoethanol. Cys-95 is modeled at two positions with 117; however, it is apparent that Cys-95 may be modeled at a third position that does not allow for the formation of a disulfide bond. Due to software restrictions, this third conformation of Cys-95 has been omitted. Furthermore, the structure of Art v 4 contains one molecule of HEPES from the crystallization condition that is located at the surface of the protein.
Comparison of Amb a 8 and Art v 4
A superposition of Art v 4 on methylated Amb a 8 results in an RMSD of 0.6 Å. Overall, the two profilin structures superpose with very few structural differences. A slight difference between the two structures is located at the N terminus. Modified lysine residues 45, 88, and 89 of methylated Amb a 8 (PDB code 5EM1) have positions different from the lysine residues in the Art v 4 structure. Additionally, residue 89 has visible electron density for modeling at two positions. Another difference between Amb a 8 and Art v 4 is ligand location. Whereas both benzoic acid of methylated Amb a 8 and HEPES of Art v 4 bind at the surface of their respective structures, benzoic acid is located near residues 45 and 46 of methylated Amb a 8, whereas HEPES is located closest to residues 3 and 6 of Art v 4.
Comparison of Methylated Amb a 8 and Amb a 8 Bound with Poly(l-Proline)14
A superposition of chain A from methylated Amb a 8 on chain A from Amb a 8 with bound poly(l-proline)14 resulted in an RMSD value of 0.7 Å. The largest difference is seen at the N terminus; this difference is due to the binding of the poly(l-proline). The N terminus of methylated Amb a 8 is located toward the surface of the molecule, whereas the N terminus of the Amb a 8 with bound poly(l-proline) is swung out to allow the proline chain to bind. The same changes are seen when comparing chain A from methylated Amb a 8 with chain B from Amb a 8 with bound poly(l-proline)14.
Structurally Similar Profilins
A Dali search was performed using the structures of either methylated Amb a 8 or Art v 4 as search models to determine what proteins are structurally similar to both profilins. Both searches, using methylated Amb a 8 or Art v 4 as the query, resulted in practically identical lists of similar structures. Also, since Amb a 8 and Art v 4 are so similar, all superpositions of related structures were performed using Amb a 8 as the reference structure. Profilin I from A. thaliana (PDB code 1A0K) was the most structurally related profilin that superposed on Amb a 8 with an RMSD value of 0.9 Å, over 130 aligned Cα atoms, and a sequence identity of 63%. Aside from A. thaliana profilin I, profilins that are registered allergens from latex (Hev b 8, PDB code 1G5U), peanut (Ara h 5, PDB code 4ESP), and birch (Bet v 2, PDB code 1CQA) were the next most structurally related proteins to both Amb a 8 and Art v 4; Heb v 8 superposed on Amb a 8 with an RMSD value of 1.0 Å, over 129 aligned Cα atoms, and a sequence identity of 69%; Ara h 5 superposed on Amb a 8 with an RMSD value of 1.0 Å, over 124 aligned Cα atoms, and a sequence identity of 63%; Bet v 2 superposed on Amb a 8 with an RMSD value of 1.1 Å, over 117 aligned Cα atoms, and a sequence identity of 74%.
Comparison of Mugwort Profilin Art v 4, Ragweed Profilin Amb a 8, and Birch Pollen Profilin Bet v 2
Birch pollen profilin (Bet v 2) was compared with the plant profilins described in this study. A superposition of 1CQA on Art v 4 revealed an RMSD value of 1.1 Å, over 118 aligned Cα atoms and a sequence identity of 77%, whereas a superposition of 1CQA on Amb a 8 revealed an RMSD value of 1.08 Å, over 117 aligned Cα atoms, and a sequence identity of 74%. The region with the largest structural differences is located at the N-terminal α-helix (residues 1–12); both Art v 4 and Amb a 8 have the α-helix located toward the core of the protein, whereas the α-helix of Bet v 2 is swung out toward the periphery of the protein (Fig. 2). Another major difference between the weed pollen profilins and the tree pollen profilin described here is that Bet v 2 is missing residues 13–21, whereas Art v 4 and Amb a 8 have clear electron density for these residues. The remainder of the Bet v 2 protein is similar to Art v 4 and Amb a 8 with minor shifts located from residue 41 to 62, which includes a loop-α-helix-loop motif, a loop region consisting of residues 68–73, and the C-terminal α-helix consisting of residues 125–133.
Comparison of Plant Profilins and Profilins from H. sapiens
Profilin II from humans (PDB code 1D1J) was compared with the plant profilins described here; a superposition of 1D1J on Amb a 8 (PDB ID 5EM1) revealed an RMSD value of 1.80 Å, over 122 aligned Cα atoms, and a sequence identity of 23%. The areas with the largest differences are located at the loop regions between α1 and β1, β4, and β5, and β5 and β6. Aside from profilin II from humans, we also compared plant profilins with human platelet profilin (PDB codes 1AWI and 1CF0) and human profilin-1 (PDB codes 3CHW and 2PAV) (Fig. 4). All four of the aforementioned structures contain polyproline peptides that are bound to the profilin molecule. A superposition of each individual human profilin on Amb a 8 with bound poly(l-proline)14 (PDB code 5EV0) reveals RMSD values in the range of 1.7–1.8 Å over 122 aligned Cα atoms and sequence identities all around 24%. Superposition revealed that all proline-rich peptides are located at the surface of the profilin in the same location as the poly(l-prolines) that bound to Amb a 8.
FIGURE 4.
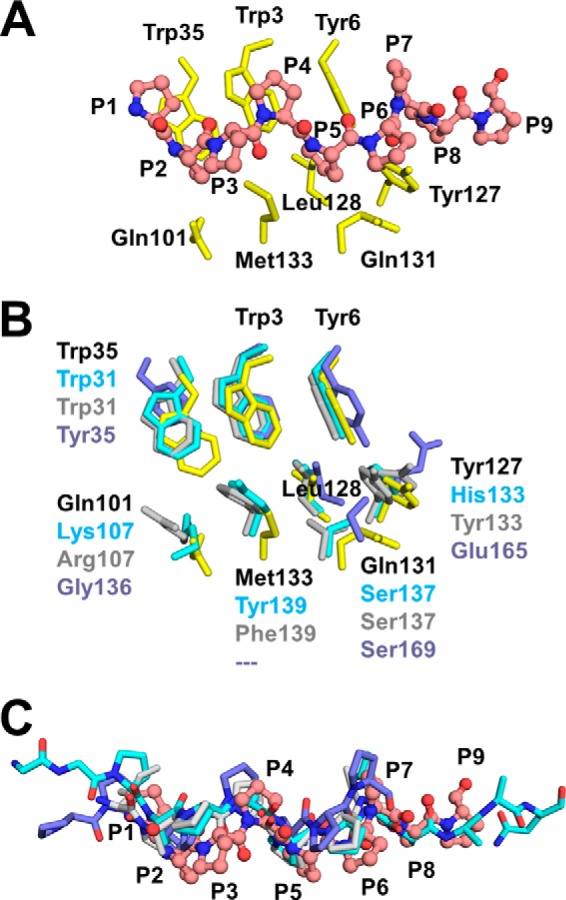
A, interactions between Amb a 8 and poly(l-Pro)14. Amb a 8 residues are shown as yellow sticks, whereas poly(l-Pro) is shown in a ball-and-stick representation and labeled using one-letter codes. B, superposition of profilin complexes with proline-rich peptides. Only side chains of profilin residues involved in binding of proline-rich peptides are shown. The superposition involves Amb a 8 (yellow sticks), human profilin (cyan sticks; PDB entry 3CHW), mouse profilin (gray sticks; PDB entry 2V8F), and profilin from P. falciparum (purple sticks; PDB entry 2JKG). C, superposition of profilin complexes with proline-rich peptides. Only poly(l-Pro) and proline-rich peptide are shown. Poly(l-Pro) from the Amb a 8·poly(l-Pro)14 complex is shown in ball-and-stick representation.
Profilin Stability Determined Using Differential Scanning Fluorimetry
Thermal stability of Amb a 8, Art v 4, and Bet v 2 in the presence of l-proline or poly(l-proline) peptides was checked using differential scanning fluorimetry. For proline screening (Fig. 5), melting temperatures (Tm) were compared with protein stability in gel filtration buffer (10 mm Tris, pH 7.4, 150 mm NaCl) with no proline or proline oligomers. h-Amb a 8 was most stable in the presence of Pro6 and Pro10 (Tm increase of 5 °C) and did not show decreased stability in any of the conditions. Amb a 8 was most stable in the presence of Pro10 (Tm increase of 5 °C) and least stable with Pro14 (Tm decrease of 1 °C). h-Art v 4 was most stable in the presence of Pro10 (Tm increase of 2 °C) and least stable in the presence of l-proline and Pro6 (Tm decrease of 8 °C). Art v 4 was most stable in the presence of Pro10 and Pro14 (Tm increase of 4 °C) and least stable in the presence of Pro6 (Tm decrease of 1 °C). h-Bet v 2 was most stable in the presence of l-proline (Tm increase of 8 °C) and did not show decreased stability in any of the conditions. Bet v 2 did not show an increase in stability in any of the conditions and was most unstable in the presence of Pro6 and Pro10 (Tm decrease of 3 °C). In addition to l-proline or poly(l-proline) peptide screening, profilin stability was also measured in different concentrations of urea: 0, 0.5, 1.0, 1.5, 2.0, 2.5, 3.0, 3.5, 4.0, 5.0, 6.0, or 8.0 m urea (Fig. 6). In the 0 m urea condition, protein was mixed 1:1 with gel filtration buffer (10 mm Tris, pH 7.4, 150 mm NaCl), which was used as the control. Fig. 8 indicates protein stability for 0, 0.5, and 1.0 m urea only; urea concentrations greater than 1.0 m displayed high initial fluorescence, followed by a sharp decrease, which is indicative of denatured protein and prohibits accurate determination of protein Tm. h-Amb a 8 showed increased stability in 0.5 m urea (Tm increase of 3 °C) and was least stable in 1.0 m urea (Tm decrease of 1 °C). Amb a 8 was most stable in the presence of 0.5 m urea (Tm increase of 1 °C) and least stable in 1.0 m urea (Tm decrease of 1 °C). h-Art v 4 did not show increased stability in the presence of urea and was least stable in 1.0 m urea (Tm decrease of 11 °C). Art v 4 showed increased stability in 0.5 m urea (Tm increase of 2 °C) and did not show decreased stability in 1.0 m urea. h-Bet v 2 was most stable in 0.5 m urea (Tm increase of 2 °C) and least stable in 1.0 m urea (Tm decrease of 15 °C). Bet v 2 was most stable in 1.0 m urea (Tm increase of 1 °C) and least stable in 0.5 m urea (Tm decrease of 2 °C). Furthermore, profilin stability was also measured in different urea concentrations at pH 7.5; however, no significant differences were observed between urea concentrations that did not have their pH adjusted and concentrations that were adjusted to pH 7.5 (data not shown).
FIGURE 5.
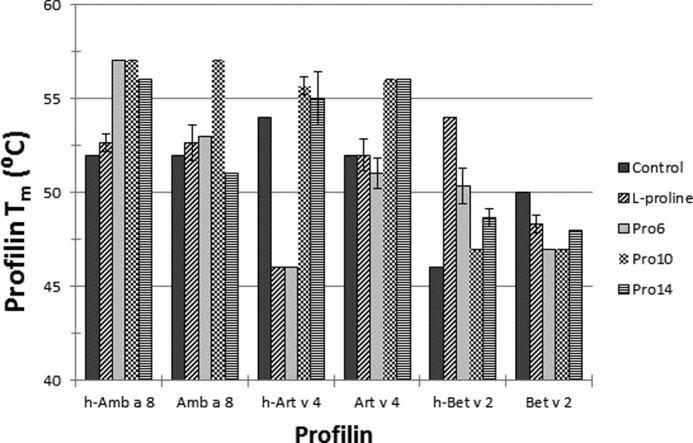
DSF profilin proline screening. Amb a 8, Art v 4, Bet v 2, and their His-tagged counterparts were screened against 10.0 mm l-proline, Pro6, Pro10, or Pro14 to determine change in protein melting temperature (Tm). The control was profilin stability in gel filtration buffer (10 mm Tris, pH 7.4, 150 mm NaCl). Error bars, S.D.
FIGURE 6.
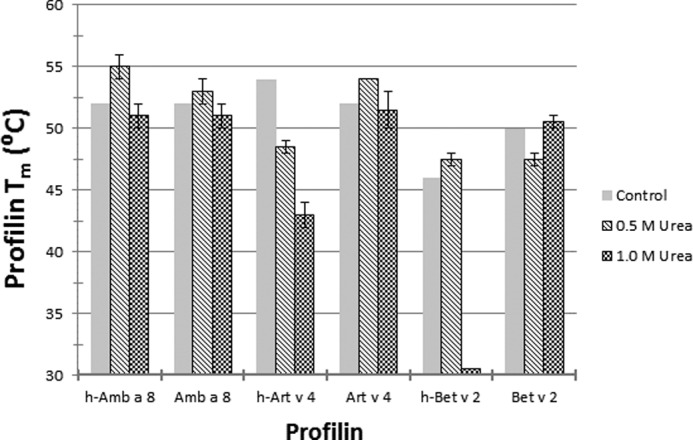
DSF profilin urea screening. Amb a 8, Art v 4, Bet v 2, and their His-tagged counterparts were screened against different concentrations of urea to determine change in protein melting temperature (Tm). The control was profilin stability in gel filtration buffer (10 mm Tris, pH 7.4, 150 mm NaCl). Error bars, S.D.
FIGURE 8.
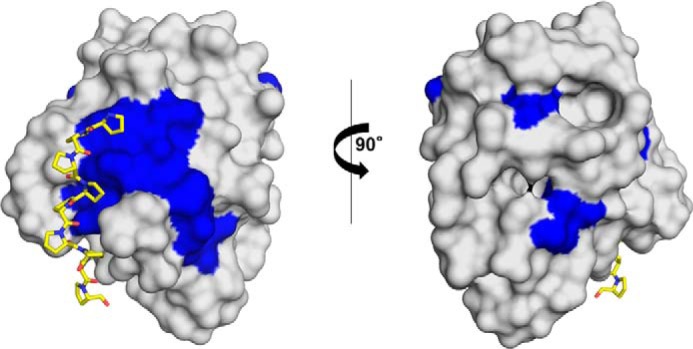
Surface representation of Amb a 8. Residues marked in blue are identical in Amb a 8 and human profilins 1. The poly(l-proline) is shown in a stick representation.
Immunologic Properties of Amb a 8, Art v 4, and Bet v 2
Sera from patients allergic to seasonal allergens, including trees, grasses, and weeds, were screened for the presence of anti-Art v 4-specific IgE antibody using ELISA. Sera from three patients with high IgE immunoreactivity to Art v 4 were selected for the comparative studies. All three sera contained IgE reacting with all three profilins: Art v 4, Amb a 8, and Bet v 2. The intensity of IgE reactivity to Art v 4 was more similar to that to Amb a 8 than Bet v 2. (Fig. 7 and Table 1). This was supported by an ELISA inhibition assay using soluble Art v 4 or Bet v 2 as an inhibitory allergen, and both were used at a concentration allowing for maximum inhibition (Fig. 8 and Table 2). Application of soluble Art v 4 completely abolished IgE binding to all three allergens tested. Although the application of soluble Bet v 2 completely abolished IgE binding to Bet v 2, it was able to inhibit IgE binding to Art v 4 or Amb a 8 in 19–49% only. No significant inhibition using an irrelevant allergen (Der p 1) could be demonstrated.
FIGURE 7.
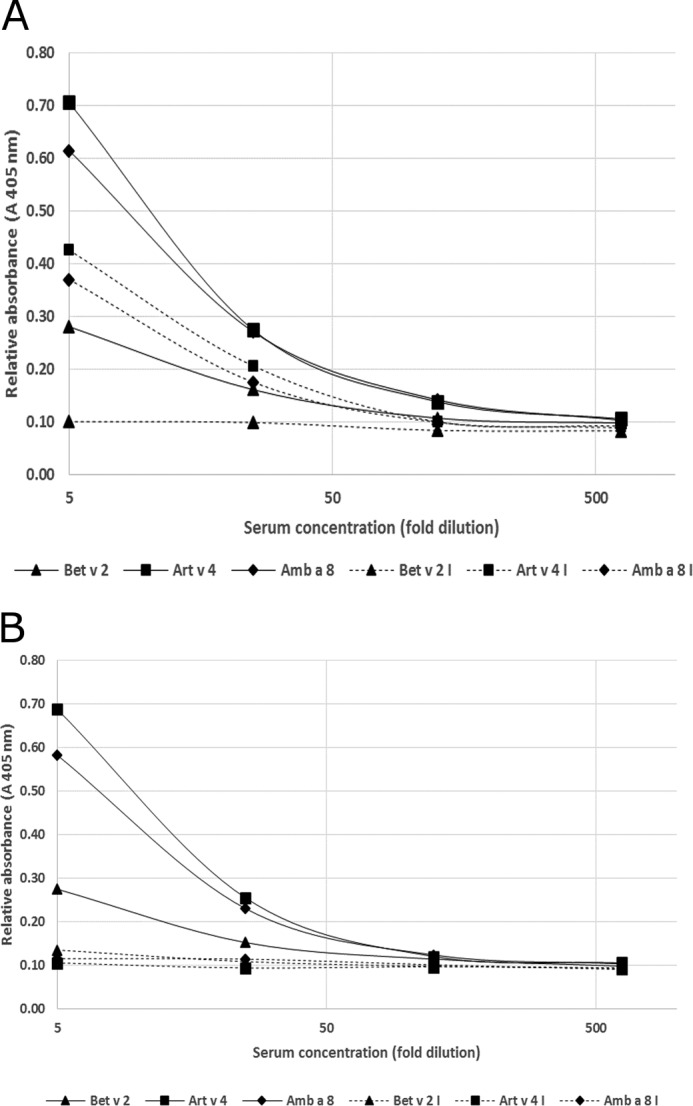
A, representative inhibition ELISA results with Bet v 2 used as an inhibiting allergen. Bet v 2, Bet v 2 coated on a plate; Art v 4, Art v 4 coated on a plate; Amb a 8, Amb a 8 coated on a plate; I, inhibition by Bet v 2 (30 μg/ml). B, representative inhibition ELISA results using Art v 4 as an inhibiting allergen. Bet v 2, Bet v 2 coated on a plate; Art v 4, Art v 4 coated on a plate; Amb a 8, Amb a 8 coated on a plate; I, inhibition by Art v 4 (30 μg/ml).
TABLE 1.
ELISA results of IgE binding to Art v 4, Amb a 8, and Bet v 2
Sera from three patients with seasonal allergic rhinitis were used. The results are presented as OD normalized for background OD (ELISA run on wells not coated with any allergen). The results represent means ± S.D. of three determinations.
| Patient | Art v 4 | Amb a 8 | Bet v 2 |
|---|---|---|---|
| P1 | 2.128 ± 0.128 | 1.937 ± 0.133 | 1.625 ± 0.148 |
| P2 | 2.242 ± 0.173 | 1.132 ± 0.064 | 0.890 ± 0.056 |
| P3 | 0.606 ± 0.076 | 0.515 ± 0.03 | 0.181 ± 0.01 |
TABLE 2.
ELISA inhibition results for individual patients (n = 3) using either Art v 4 or Bet v 2 as inhibiting allergen
% inhibition, (OD for binding without inhibition − OD for binding with inhibiting allergen)/OD for binding without inhibition. All OD values were normalized for the background OD (for no allergen coated on a plate).
| Soluble allergen for inhibition | Plates coated with allergen |
||
|---|---|---|---|
| Art v 4 | Amb a 8 | Bet v 2 | |
| % inhibition | |||
| Art v 4 | 97 | 92 | 81 |
| 96 | 91 | 87 | |
| 99 | 97 | 88 | |
| Bet v 2 | 49 | 45 | 98 |
| 37 | 29 | 90 | |
| 23 | 19 | 99 | |
Discussion
Analysis of profilins' sequences revealed several distinct profilin groups (Fig. 1). The major cluster of sequences may be divided into two subclusters, and one of them contains all plant profilins, including all currently registered profilin allergens. Taking into account the high sequence similarity of plant profilins, it is not surprising that these allergens are highly cross-reactive (14, 21–23). However, it is quite surprising that currently the only source of allergenic profilins is from plants. One could expect that at least some proteins that form the second part of the major cluster (i.e. invertebrates and vertebral profilin 4 due to their relatively high sequence similarity to allergenic plant profilins) should also be identified as allergens. Our clustering results are in agreement with previous studies of evolutionary origins of profilins showing that profilins 4 are most similar to invertebrate profilins and originated before vertebrate evolution (13). The proteins that were investigated here (Amb a 8.0101, Art v 4.0101, and Bet v 2.0101) have relatively low levels of sequence identity and similarity when compared with human profilins, and this fact correlates well with the presence of a separate cluster containing human proteins (Vertebrata 1–3) and other vertebrate profilins 1, 2, and 3. In terms of sequence, the plant profilins are also clearly distinct from profilins originating from Alveolata, represented, for example, by profilins from important human pathogens like T. gondii and P. falciparum. Although the profilins are often considered to be exclusively eukaryotic proteins, there is a group of profilins that originate from viruses. These proteins were shown to be similar to profilins 3 (13), and our results also reveal a distinct cluster of viral proteins next to the cluster Vertebrata 1–3.
Amb a 8.0101 and Art v 4.0101 have almost 90% sequence identity, and their structures are very similar as well (Fig. 2). Although crystallization of Art v 4.0101 did not require any protein modification, crystallization of Amb a 8 required methylation of lysine side chains or the addition of poly(l-proline). Comparison of crystal structures of Amb a 8, Art v 4, and Bet v 2 revealed a significant difference in conformation of the N-terminal fragment of Bet v 2 relative to the same fragments in the weed profilins. Such a difference may be caused by crystal packing; however, it is also possible that it illustrates a conformational change that is important for protein function. The second possibility is quite likely because the N-terminal part of the profilin participates in binding of proline-rich peptides. Besides Bet v 2 and Amb a 8 and Art v 4 (reported here), there are only two other allergenic profilins that have their structures determined: Ara h 5 and Hev b 8. All of these allergens have similar structures and sequences, which further explains high cross-reactivity between proteins belonging to this group of allergens.
The structures reported here of Amb a 8 in complex with poly(l-Pro)10 or poly(l-Pro)14 are the first structures of a plant profilin in complex with a proline-rich peptide. Previously reported profilin·proline-rich peptide complexes were formed by human, mouse, or P. falciparum proteins (Fig. 4). Analysis of the available complexes shows that the peptide binding site may be divided into two regions. In Amb a 8, the first region is formed by N-terminal residues (Trp-3, Tyr-6, and Trp-35), and the second region is formed by the C-terminal part of the protein (Gln-101, Tyr-127, Gln-131, and Met-133). Although the N-terminal region is highly conserved (Fig. 4B), with Trp-3 and Tyr-6 being present in all currently available complexes, the C-terminal regions are more variable. Additional insight into the binding of proline-rich peptides may be obtained by a comparison of structures of profilins in free and complexed forms. For example, the Hev b 8 molecule has the same conformation of the N-terminal helix as in the case of Amb a 8 and Art v 4; however, the conformation of Ara h 5 closely resembles an “open” conformation, as is observed in the Bet v 2 structure. Closer inspection of the two possible conformations observed for allergenic profilins clearly indicated that although a “closed” conformation of Amb a 8, Art v 4, and Hev b 8 is compatible with the formation of interactions necessary for the binding of proline-rich peptides, the “open” conformation most likely cannot mediate formation of the complex with the peptide. This is related to the fact that the conserved residues, Trp-3 and Tyr-6, are displaced by 6–8 Å (as measured by the change of Cα positions) in the “open” conformation and therefore are not able to participate in the formation of the groove that binds the proline-rich peptide. Such a groove has to have a shape compatible with the characteristic shape of the type II helix formed by the polyproline fragments. In fact, despite significant differences in the amino acid composition of the peptide binding site, the central part of the peptide is bound in a very similar mode by the ragweed, human, mouse, and P. falciparum profilins.
Despite the fact that profilins are widely distributed in pollens and food, most often they are minor allergens that cause relatively mild reactions. These mild reactions are mainly correlated to the low stability of profilins that are susceptible to denaturation in conditions used during food processing/preparation. Only profilins that do not undergo high temperature treatment, like those in raw fruits, are considered to be the major allergens. Because the profilins studied in this paper originate from, and are present in, plant pollens, we decided to test their stability. The stability of Amb a 8, Art v 4 a, and Bet v 2 was studied using differential scanning fluorimetry (DSF), and we observed the influence of various compounds on the thermal stability of the proteins. Because our proteins were produced in a recombinant form, we also decided to check the influence of the His tag on protein stability. In all cases, the presence or absence of the purification tag affected protein stability; however, the effects depended on the protein tested (Figs. 5 and 6). Generally, polyproline peptides stabilized or, at least, did not destabilize Amb a 8 and Art v 4. The effect of the peptides on stability of h-Bet v 2 were less pronounced; however, Bet v 2 was clearly destabilized by l-proline and the poly(l-peptides). It is worth noting that the presence of free proline was detected in pollen of Asteraceae species (24). All proteins denatured in 1.5 m urea, which indicates that all proteins are quite unstable in the presence of this compound. Interestingly, h-Amb a 8, Amb a 8, Art v 4, and h-Bet v 2 were mildly stabilized by 0.5 m urea, and Bet v 2 was marginally stabilized by 1.0 m urea. Comparison of the recombinant profilins shows that h-Amb a 8 and h-Bet v 2 seem to be more stable than h-Art v 4 in the presence of l-proline or poly(l-proline), whereas Bet v 2 tends to be less stable than Amb a 8 or Art v 4. The latter observation may be explained by the fact that both Amb a 8 and Art v 4 may be additionally stabilized by a disulfide bridge formed by Cys-95 and Cys-117 (Amb a 8 numbering). There is no such bond in Bet v 2.0101 because the Cys-95 is replaced by Thr. Both Cys residues are conserved in Amb a 8.0101, Amb a 8.0102, Art v 4.0101, and Art v 4.0201. However, during the analysis of the crystal structures reported here, it was noticed that the formation of the disulfide linkage is not observed in all molecules. It is possible that it is caused by the E. coli strain used for protein production and that the formation of the disulfide bridges is not complete or that the disulfide bridges are easily damaged during the diffraction experiment (25).
The relatively low stability, as well as insufficient information on folding of profilins, may pose another problem. Namely, many researchers during purification of natural or recombinant profilins use a method that combines an affinity chromatography (binding to polyproline) that is later followed by elution with a solution of concentrated urea and refolding of the denatured profilin (7, 26). However, only rarely is the refolding followed by use of CD spectroscopy or other experimental techniques that may confirm that the protein is properly folded (27). In most cases, the researchers simply assume that the refolding was successful and use the protein for immunological studies (14, 22, 28–34). The use of unfolded protein with destroyed conformational epitopes may lead to errors, for example, in the antigenic characterization of proteins.
Profilins were identified as novel allergens over 25 years ago, and the first profilin that was shown to be an allergen is Bet v 2 (birch pollen allergen) (35). Currently, there are almost 50 profilins recognized as allergens, and they represent one of the largest groups of panallergens (1). There is still an ongoing discussion of whether profilins are relevant allergens from the clinical point of view (1, 22, 36–38). Currently, it is clear that some profilins may be not only relevant pollen allergens but also important food allergens in some cases. For example, Cit s 2 (profilin from orange) and Cuc m 2 (profilin from melon) are relevant and major food allergens (31, 39). In the case of Cit s 2 and Cuc m 2, the fact that oranges and melons most often are consumed in a raw form may be a very important factor because the allergens do not undergo denaturation before contact with a sensitized individual. The same is most likely also true in the case of profilins found in pollens; despite relatively poor stability in comparison with other food allergens, Amb a 8, Art v 4, and Bet v 2 may be intact upon entry into the human respiratory system.
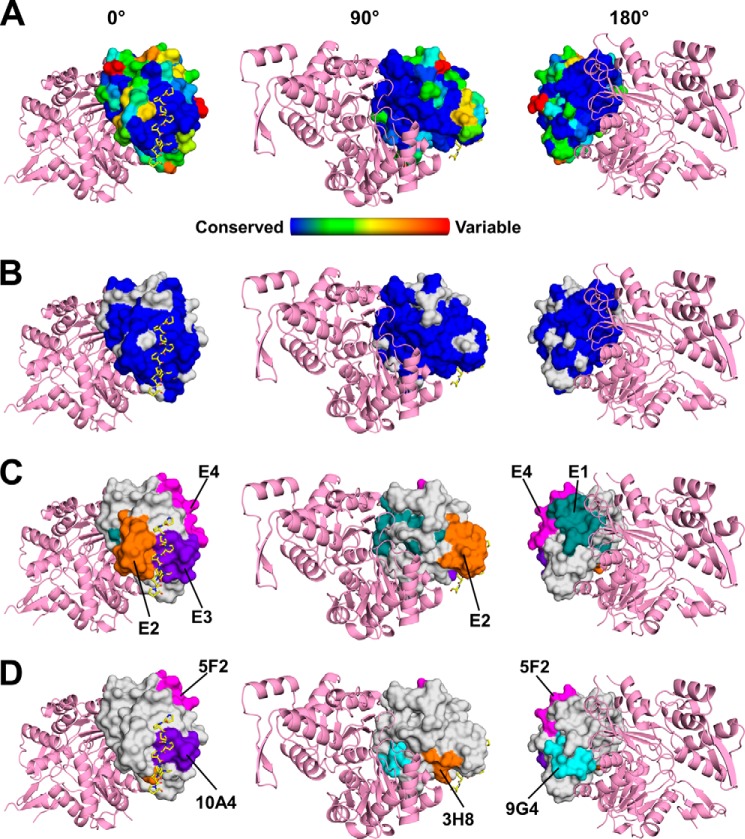
The high level of sequence identity of allergenic profilins and common co-sensitization or co-recognition make the investigation of the role of individual profilins in induction of sensitization and triggering allergic symptoms difficult. In addition, studies of the molecular basis of profilin allergenicity are additionally complicated by the fact that profilins form complexes not only with actin, but also with many other proteins by binding proline-rich peptides (6, 40–42). The residues participating in the formation of protein·protein complexes are highly conserved, as is illustrated in Fig. 9A. Fig. 9B indicates residues that are identical between Amb a 8, Art v 4, and Bet v 2, which are also highly conserved among protein·protein complexes. For Cuc m 2 and Hel a 2 (profilin from sunflower pollen), it was experimentally demonstrated that the IgE biding epitopes overlap significantly with areas of the molecules participating in formation of the protein·protein complexes (Fig. 9, C and D) (43, 44). The residues forming these epitopes in Cuc m 2 and Hel a 2 are also highly conserved in Amb a 8, Art v 4, and Bet v 2. The presence of IgE epitopes E3 (Cuc m 2) and 10A4 (Hel a 2) is somewhat surprising because the residues participating in the binding of proline-rich peptides are also conserved in human profilins (Figs. 8 and 9). However, it is possible that the conserved region only partially overlaps with IgE epitopes.
FIGURE 9.
A, sequence conservation of profilins calculated with ConSurf is mapped on the surface of Amb a 8. Actin (pink) was modeled using the structure of human profilin 1 in complex with actin (PDB entry 3CHW). Poly(l-Pro) is shown in a stick representation with carbon atoms marked in yellow. The figure shows three different orientations of the molecules corresponding to 0, 90, and 180° rotation along the y axis. B, residues that are identical in Amb a 8, Art v 4, and Bet v 2 are shown in blue. Residues that are not 100% conserved in these allergens are marked in gray. C, epitopes identified in Cuc m 2. The epitopes are labeled using a convention used in the original paper reporting these epitopes (44, 74). D, epitopes identified in Hel a 2.0101. The epitopes are labeled using a convention used in the original paper reporting these epitopes (43).
The recombinant profilins were recognized by IgE from some allergic patients, which is consistent with previous reports (21, 27, 45, 46). Several studies have demonstrated that IgE from patients sensitized to one profilin binds to profilins from all other sources (40, 41). In many cases, detection of IgE directed to one plant profilin indicates sensitization to other plant profilins (47, 48). However, other studies indicated only partial IgE cross-reactivity between profilins from different sources, and in some cases, sensitization to one profilin is not associated with sensitization to profilins from other sources (49, 50). This may be related to structural differences between individual profilins and may also depend on the repertoire of IgE directed to species-specific epitopes of individual profilins (27).
Detected in the current study, differences in IgE binding between Art v 4, Amb a 8, and Bet v 2 indicate the existence of unique, species-specific epitopes in those profilins. Moreover, phylogenetically distant species, such as mugwort and birch, seem to contain more species-specific IgE-binding epitopes than closely related species, such as mugwort and ragweed. The inhibition of IgE binding to Art v 4 or Amb a 8 by Bet v 2 is less efficient than inhibition of IgE binding to nPhl p 12 (native Timothy pollen profilin) that was reported previously (27). Interestingly, the tested sera were collected from patients living in the area where mugwort and birch, but not ragweed, pollens are present. Therefore, it can be argued that similar intensity of IgE reactivity to Art v 4 and Amb a 8 does not reflect similar environmental exposure to those allergens but rather true cross-reactivity related to structural similarities between Art v 4 and Amb a 8 proteins. This is consistent with epidemiological studies that indicate that only a single allergenic source, probably that providing the highest profilin exposure, may be responsible for triggering IgE response to profilins (23). Moreover, IgE cross-reactivity to ragweed allergen Amb a 1 of sera derived from mugwort allergic patients living in the area with no ragweed exposure has been reported (46). In summary, our study provides evidence for the existence of a specific IgE response to well characterized profilins Art v 4 and Amb a 8 and demonstrates the differences in IgE binding between weed profilins and tree profilins, which are related to the structural differences of those proteins.
Experimental Procedures
Cloning, Expression, and Purification
Recombinant Amb a 8.0101, Art v 4.0101, and Bet v 2.0101 were ordered from DNA 2.0 (Menlo Park, CA) in expression vector pJexpress411 with kanamycin resistance. The recombinant DNA was designed with a cleavable polyhistidine tag (MHHHHHHSSGVDLGTENLYFQS↓GSG, where the arrow marks a tobacco etch virus cleavage site) for ease of purification and cleavability to aid in crystallization. This plasmid was then transformed into E. coli strain BL21 (DE3). Cells were grown in LB broth at 37 °C to an A600 of 0.6–0.8 and then induced with isopropyl β-d-1-thiogalactopyranoside to a final concentration of 400 μm. Following induction, the cells were incubated overnight with shaking at 16 °C. Cells were harvested by centrifugation for 30 min at 10,500 × g at 4 °C and then resuspended in 50 mm Tris, pH 7.4, 500 mm NaCl, 10 mm imidazole, 2% glycerol, and 20 mm β-mercaptoethanol (lysis buffer) supplemented with protease inhibitor tablets (Pierce) used according to the manufacturer's directions and then disrupted by sonication using a Branson Sonifier 450 sonicator. Homogenate was centrifuged for 30 min at 10,500 × g, and a clear, yellow supernatant was separated from the pelleted cellular debris. The crude extract was then loaded onto a 12 × 1.5-cm, 5-ml bed volume column of nickel-nitrilotriacetic acid-agarose medium (Pierce) previously equilibrated in wash buffer (50 mm Tris, pH 7.4, 500 mm NaCl, 30 mm imidazole, 2% glycerol, and 20 mm β-mercaptoethanol). Following a washing step with wash buffer, protein was eluted from the medium using elution buffer (50 mm Tris, pH 7.4, 500 mm NaCl, 250 mm imidazole, 2% glycerol, and 20 mm β-mercaptoethanol). Fractions were analyzed by SDS-PAGE, and those fractions containing protein were pooled and dialyzed against 10 mm Tris, pH 7.4, 150 mm NaCl, and 5 mm β-mercaptoethanol overnight at 4 °C. Protein was concentrated using an Amicon Ultra concentrator (Millipore) with a 10,000 Da molecular mass cut-off and then purified on a Superdex 200 column attached to an ÄKTA Pure FPLC system (GE Healthcare). A solution composed of 10 mm Tris-HCl and 150 mm NaCl at pH 7.4 was used for gel filtration of both proteins. Fractions containing protein were pooled, and the concentration was determined using the Bradford method (51).
His tag cleavage was achieved by supplementing pure protein (1–2 mg/ml) with tobacco etch virus protease in a 1:100 (w/w) protease/protein ratio and then dialyzed overnight at 4 °C. In this work, recombinant proteins with a His tag attached are referred to as h-profilin (i.e. h-Amb a 8, h-Art v 4, h-Bet v 2), and recombinant proteins with the His tag cleaved off are referred to with the name of the profilin (i.e. Amb a 8, Art v 4, and Bet v 2, respectively). Following dialysis, the protein-tobacco etch virus solution was loaded onto nickel-nitrilotriacetic acid-agarose medium equilibrated in wash buffer and allowed to incubate with the medium for 30 min at 4 °C. Subsequent to incubation, cleaved protein was eluted with wash buffer, concentrated, and then loaded onto a Superdex 200 column attached to an ÄKTA Pure FPLC gel filtration system (GE Healthcare) in a buffer containing 10 mm Tris-HCl and 150 mm NaCl at pH 7.4. Fractions containing cleaved protein were pooled and concentrated to ∼10 mg/ml. For studies of profilin bound to poly(l-proline), each profilin was titrated with l-Pro14 or l-Pro10 (NeoBiolabs, Cambridge, MA) in a 1:15 ratio at 30 °C with stirring for 5 h.
Crystallization, Data Collection, and Processing
Crystallization experiments for Amb a 8, Amb a 8·poly(l-proline)14 after titration, Amb a 8·poly(l-proline)10 after titration, and Art v 4 were performed at 293 K using the sitting-drop vapor diffusion method and MRC 2-drop 96-well crystallization plates (Molecular Dimensions, Altamonte Springs, FL). A solution of recombinant, His tag-free protein was mixed with well solution (0.1 m HEPES, pH 6.5–8.5, and 0.5–1.44 m sodium citrate) in a 1:1 ratio. Crystals of native Art v 4 appeared within 48 h; however, no native Amb a 8 crystals appeared. Only after subjecting native Amb a 8 to reductive methylation, according to previous protocols (52, 53), or titrating Amb a 8 with poly(l-proline)14 or poly(l-proline)10 did crystallization occur. The level of methylation was confirmed using a Bruker Ultraflex II MALDI TOF/TOF mass spectrometer (Billerica, MA) (Fig. 3). Crystals were cryo-protected with well solution followed by immediate cryo-cooling in liquid N2. Data for Amb a 8, Amb a 8·poly(l-proline)14, Amb a 8·poly(l-proline)10, and Art v 4 were collected from single crystals at 100 K at the Southeast Regional Collaborative Access Team (SER-CAT) 22BM, SER-CAT 22ID, Structural Biology Center 19BM (54) and the Life Sciences Collaborative Access Team (LS-CAT) 21-ID-G beamlines, respectively, at the Advanced Photon Source, Argonne National Laboratory (Argonne, IL). Data were processed with the HKL-2000 software package (55). Methylated Amb a 8 crystallized in the primitive orthorhombic space group P212121, Amb a 8 with bound poly(l-proline)14 and poly(l-proline)10 crystallized in the primitive monoclinic space group P21, and Art v 4 crystallized in the primitive orthorhombic space group of P21212. Data collection statistics are reported in Table 3.
TABLE 3.
Crystallographic data and refinement statistics for Amb a 8, Amb a 8·poly(l-Pro)14, Amb a 8·poly(l-Pro)10, and Art v 4
Values in parenthesis refer to the highest resolution shell. AU, asymmetric unit.
| PDB code | 5EM1 | 5EV0 | 5EVE | 5EM0 |
|---|---|---|---|---|
| Structure | Amb a 8 | Amb a 8 + poly (l-Pro)14 | Amb a 8 + poly (l-Pro)10 | Art v 4 |
| Data collection | ||||
| Wavelength (Å) | 0.98 | 0.98 | 1.0 | 0.98 |
| Unit cell parameters | ||||
| a, b, c (Å) | 32.2, 58.5, 60.6 | 55.1, 40.3, 60.63 | 35.5, 40.4, 42.0 | 58.1, 59.2, 32.6 |
| α,β, γ(degrees) | α = β = γ = 90 | α = γ = 90, β = 104.4 | α = γ = 90, β = 91.9 | α = β = γ = 90 |
| Space group | P212121 | P21 | P21 | P21212 |
| Solvent content (%) | 39 | 46 | 42 | 37 |
| Protein chains in AU | 1 | 2 | 1 | 1 |
| Resolution range (Å) | 50.0–1.45 | 50.0–2.10 | 50.0–2.55 | 50.0–1.10 |
| Highest resolution shell (Å) | 1.48–1.45 | 2.14–2.10 | 2.59–2.55 | 1.12–1.10 |
| Unique reflections | 20,925 (1015) | 14,967 (717) | 3959 (179) | 44,768 (2084) |
| Redundancy | 7.2 (4.3) | 3.4 (3.0) | 3.4 (2.8) | 4.3 (4.0) |
| Completeness (%) | 99.9 (98.0) | 98.4 (92.4) | 97.1 (91.3) | 96.4 (90.8) |
| Rmerge | 0.065 (0.350) | 0.079 (0.427) | 0.081 (0.452) | 0.051 (0.724) |
| Rpim | 0.026 (0.185) | 0.050 (0.285) | 0.51 (0.301) | 0.028 (0.407) |
| Rmeas | 0.070 (0.398) | 0.094 (0.515) | 0.096 (0.545) | 0.058 (0.835) |
| Average I/σ(I) | 32.2 (2.5) | 22.7 (2.7) | 23.3 (2.5) | 36.8 (2.1) |
| Refinement | ||||
| Rwork (%) | 14.2 | 19.3 | 20.0 | 16.5 |
| Rfree (%) | 16.5 | 23.4 | 23.3 | 19.0 |
| Mean B value (Å2) | 14.6 | 36.1 | 39.4 | 14.5 |
| B from Wilson plot (Å2) | 15.2 | 36.6 | 59.6 | 11.5 |
| RMSD bond lengths (Å) | 0.02 | 0.02 | 0.01 | 0.02 |
| RMSD bond angles (degrees) | 2.3 | 1.8 | 1.2 | 2.1 |
| No. of amino acid residues | 135 | A = 134, B = 133 | 132 | 134 |
| No. of water molecules | 201 | 80 | 8 | 213 |
| Ramachandran plot | ||||
| Most favored regions (%) | 96.6 | 97.5 | 97.1 | 96.9 |
| Additional allowed regions (%) | 3.4 | 2.5 | 2.9 | 3.1 |
Structure Determination and Refinement
Both Amb a 8 and Art v 4 were solved by molecular replacement using models of each respective protein generated from SWISS-MODEL (56) using birch pollen profilin (PDB code 1CQA) as a template. The structure of Amb a 8·polyproline was solved by molecular replacement using the native structure determined here as a start model. Molecular replacement was performed with HKL-3000 (57) integrated with MOLREP (58) and selected programs from the CCP4 package (59). Models were rebuilt using COOT (60) and refined with REFMAC (61). TLS was used in the final stages of refinement, and TLS groups were determined using the TLSMD server (62). MOLPROBITY (63) and ADIT (64) were used for structure validation.
DSF
Enzyme stability was observed following the ThermoFluor method (65), using a CFX96 TouchTM real-time PCR detection system (Bio-Rad) (66). Sypro Orange (excitation, 280 and 450 nm; emission, 610 nm) in DMSO (Sigma-Aldrich) was added to the profilins (in 10 mm Tris, 150 mm NaCl, pH 7.4) at 1.0 mg/ml in a 1:1000 ratio. Protein (with His tag and without His tag) was screened against 10.0 mm l-proline, poly(l-proline)6, poly(l-proline)10, or poly(l-proline)14 and against a urea gradient from 0 to 8.0 m urea. The screening condition and protein were mixed in a 1:1 ratio in a 96-well hard shell PCR plate (Bio-Rad). The plates were sealed with Microseal® “B” adhesive seals (Bio-Rad) and heated in a CFX96 real-time PCR instrument (Bio-Rad) from 30 to 90 °C in 2.0 °C increments for 1 min. Proline and polyproline screening was run in triplicate, and urea screening was run in duplicate.
Sequence Similarity-based Clustering
The sequences of Profilin family proteins representing Pfam00235 were downloaded from the Pfam database. To visualize pairwise similarities between profilins and identify groups of more closely related sequences, we used CLANS (cluster analysis of sequences), a Java utility that applies a version of the Fruchterman-Reingold graph layout algorithm (67). CLANS uses the p values of highly scoring segment pairs obtained from an N × N BLAST search to compute attractive and repulsive forces between each sequence pair in a user-defined data set. A two-dimensional representation of sequence subgroups was achieved by randomly seeding the sequences in the arbitrary distance space and then moving them within this environment according to the force vectors resulting from all pairwise interactions; the process was repeated until convergence.
Other Computational Calculations
The SSM algorithm (68) in COOT was used to superpose protein models. All figures containing protein structures were prepared with PyMOL (69) or Chimera (70). PDBePISA was used to calculate probable quaternary structure (71). The Dali server (72) was used to identify structurally similar proteins. The ConSurf server was used to map sequence conservation on the structure of Amb a 8 (73).
ELISA
Fifty mugwort, grass, and/or birch allergic patients participated in a study approved by the Local Bioethics Committee (R-I-002/36/2015), and all patients signed the informed consent. Their sera were screened for IgE reactivity to recombinant Art v 4. The patients selected for the experiment (n = 3) were allergic to mugwort allergen Art v 4, and none of the studied patients were allergic to ragweed by skin prick testing.
Ninety-six-well MaxiSorp microtiter plates (Nunc, Roskilde, Denmark) were coated with 100 μl of Art v 4, Amb a 8, or Bet v 2, each at a final concentration of 10 μg/ml diluted in coating buffer (25 mm carbonate/bicarbonate buffer, pH 9.4), and incubated overnight at 4 °C. After washing with TBST buffer, the plates were blocked with TBST + 1% BSA for 1 h at room temperature. Subsequently, the plates were washed in TBST, and serial dilutions of patients' sera in TBST + 1% BSA (1:5, 1:25, 1:125, and 1:625) were loaded on the plates and incubated for 2 h at room temperature. The plates were thoroughly washed in TBST, and a monoclonal anti-human IgE antibody conjugated to alkaline phosphatase diluted 1:1000 in TBST + 1% BSA (BD Pharmingen, Heidelberg, Germany) was applied and incubated for 2 h at room temperature. Following washing, bound antibodies were detected by incubation with p-nitrophenyl phosphate (Sigma Fast p-nitrophenyl phosphate tablet sets, Sigma-Aldrich). The reaction was quantified by measuring color intensity at 405 nm. To evaluate specificity of the test in all experiments, negative controls were used: no allergen immobilized on the plate, no serum applied, irrelevant allergen immobilized on the plate, or no anti-IgE added. All samples were run in triplicate, and the mean value was used for analysis. The cut-off for positive values was determined using OD values of 10 non-allergic patients' sera and calculated from their mean OD plus three S.D. values.
For the ELISA inhibition assay, in addition to regular ELISA performed as described above, serial dilutions of patients' sera were preincubated for 30 min at room temperature with an allergen dissolved in TBST + 1% BSA at concentrations up to 30 μg/ml. The concentration of soluble allergens was well above the minimal concentration necessary for maximum inhibition as determined by preliminary dose-response studies (not shown). The samples were then loaded on a plate, and the procedure was continued as described for the ELISA above.
Author Contributions
L. R. O. and C. R. S. purified recombinant profilins. L. R. O., C. R. S., M. L. P., J. Z. H., and J. G. crystallized the profilin allergens discussed in this study. L. R. O. performed mass spectrometry experiments, determined the crystal structures, deposited structures to the PDB, and also performed the calculations described under “Other Computational Calculations.” K. A. M. performed sequence-based clustering, C. R. S. performed DSF experiments, K. K. performed the ELISA experiments, W. T. B. worked on biochemical characterization of profilins, and M. C. supervised experiments and participated in analysis of results. All authors contributed to writing the manuscript with a final review by M. C.
Acknowledgments
The structural results shown here are derived from work performed at Argonne National Laboratory, at the Southeast Regional Collaborative Access Team (SER-CAT) and the Life Sciences Collaborative Access Team (LS-CAT) of the Advanced Photon Source. Supporting institutions for SER-CAT may be found on the SER-CAT website. Use of the Advanced Photon Source was supported by the United States Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract W-31-109-Eng-38.
This work was supported in part by an ASPIRE III grant from the Office of the Vice President of Research at the University of South Carolina, and by the NIAID of the National Institutes of Health under Award Number R01AI077653. Partial support was also obtained from Medical University of Bialystok, Poland. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The atomic coordinates and structure factors (codes 5EM1, 5EV0, 5EVE, and 5EM0) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- PDB
- Protein Data Bank
- RMSD
- root mean square deviation
- DSF
- differential scanning fluorimetry
- TLS
- translation/libration/screw.
References
- 1. Hauser M., Roulias A., Ferreira F., and Egger M. (2010) Panallergens and their impact on the allergic patient. Allergy Asthma Clin. Immunol. 6, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mari A. (2001) Multiple pollen sensitization: a molecular approach to the diagnosis. Int. Arch. Allergy Immunol. 125, 57–65 [DOI] [PubMed] [Google Scholar]
- 3. Tinghino R., Twardosz A., Barletta B., Puggioni E. M., Iacovacci P., Butteroni C., Afferni C., Mari A., Hayek B., Di Felice G., Focke M., Westritschnig K., Valenta R., and Pini C. (2002) Molecular, structural, and immunologic relationships between different families of recombinant calcium-binding pollen allergens. J. Allergy Clin. Immunol. 109, 314–320 [DOI] [PubMed] [Google Scholar]
- 4. Wopfner N., Gruber P., Wallner M., Briza P., Ebner C., Mari A., Richter K., Vogel L., and Ferreira F. (2008) Molecular and immunological characterization of novel weed pollen pan-allergens. Allergy 63, 872–881 [DOI] [PubMed] [Google Scholar]
- 5. Ghosh D., Mueller G. A., Schramm G., Edwards L. L., Petersen A., London R. E., Haas H., and Gupta Bhattacharya S. (2014) Primary identification, biochemical characterization, and immunologic properties of the allergenic pollen cyclophilin cat R 1. J. Biol. Chem. 289, 21374–21385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carlsson L., Nyström L. E., Sundkvist I., Markey F., and Lindberg U. (1977) Actin polymerizability is influenced by profilin, a low molecular weight protein in non-muscle cells. J. Mol. Biol. 115, 465–483 [DOI] [PubMed] [Google Scholar]
- 7. Lindberg U., Schutt C. E., Hellsten E., Tjäder A. C., and Hult T. (1988) The use of poly(l-proline)-Sepharose in the isolation of profilin and profilactin complexes. Biochim. Biophys. Acta 967, 391–400 [DOI] [PubMed] [Google Scholar]
- 8. Schutt C. E., Myslik J. C., Rozycki M. D., Goonesekere N. C., and Lindberg U. (1993) The structure of crystalline profilin-β-actin. Nature 365, 810–816 [DOI] [PubMed] [Google Scholar]
- 9. Tanaka M., and Shibata H. (1985) Poly(l-proline)-binding proteins from chick embryos are a profilin and a profilactin. Eur. J. Biochem. 151, 291–297 [DOI] [PubMed] [Google Scholar]
- 10. Hansson A., Skoglund G., Lassing I., Lindberg U., and Ingelman-Sundberg M. (1988) Protein kinase C-dependent phosphorylation of profilin is specifically stimulated by phosphatidylinositol bisphosphate (PIP2). Biochem. Biophys. Res. Commun. 150, 526–531 [DOI] [PubMed] [Google Scholar]
- 11. Lassing I., and Lindberg U. (1985) Specific interaction between phosphatidylinositol 4,5-bisphosphate and profilactin. Nature 314, 472–474 [DOI] [PubMed] [Google Scholar]
- 12. Machesky L. M., Goldschmidt-Clermont P. J., and Pollard T. D. (1990) The affinities of human platelet and Acanthamoeba profilin isoforms for polyphosphoinositides account for their relative abilities to inhibit phospholipase C. Cell Regul. 1, 937–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Polet D., Lambrechts A., Vandepoele K., Vandekerckhove J., and Ampe C. (2007) On the origin and evolution of vertebrate and viral profilins. FEBS Lett. 581, 211–217 [DOI] [PubMed] [Google Scholar]
- 14. Valenta R., Duchene M., Ebner C., Valent P., Sillaber C., Deviller P., Ferreira F., Tejkl M., Edelmann H., and Kraft D. (1992) Profilins constitute a novel family of functional plant pan-allergens. J. Exp. Med. 175, 377–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gadermaier G., Hauser M., and Ferreira F. (2014) Allergens of weed pollen: an overview on recombinant and natural molecules. Methods 66, 55–66 [DOI] [PubMed] [Google Scholar]
- 16. Gadermaier G., Dedic A., Obermeyer G., Frank S., Himly M., and Ferreira F. (2004) Biology of weed pollen allergens. Curr. Allergy Asthma Rep. 4, 391–400 [DOI] [PubMed] [Google Scholar]
- 17. Radauer C., Bublin M., Wagner S., Mari A., and Breiteneder H. (2008) Allergens are distributed into few protein families and possess a restricted number of biochemical functions. J. Allergy Clin. Immunol. 121, 847–852.e7 [DOI] [PubMed] [Google Scholar]
- 18. Finn R. D., Bateman A., Clements J., Coggill P., Eberhardt R. Y., Eddy S. R., Heger A., Hetherington K., Holm L., Mistry J., Sonnhammer E. L., Tate J., and Punta M. (2014) Pfam: the protein families database. Nucleic Acids Res. 42, D222–D230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., and Lipman D. J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Behnen M., Murk K., Kursula P., Cappallo-Obermann H., Rothkegel M., Kierszenbaum A. L., and Kirchhoff C. (2009) Testis-expressed profilins 3 and 4 show distinct functional characteristics and localize in the acroplaxome-manchette complex in spermatids. BMC Cell Biol. 10, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ganglberger E., Radauer C., Wagner S., Ríordáin G., Beezhold D. H., Brehler R., Niggemann B., Scheiner O., Jensen-Jarolim E., and Breiteneder H. (2001) Hev b 8, the Hevea brasiliensis latex profilin, is a cross-reactive allergen of latex, plant foods, and pollen. Int. Arch. Allergy Immunol. 125, 216–227 [DOI] [PubMed] [Google Scholar]
- 22. Scheurer S., Wangorsch A., Nerkamp J., Skov P. S., Ballmer-Weber B., Wüthrich B., Haustein D., and Vieths S. (2001) Cross-reactivity within the profilin panallergen family investigated by comparison of recombinant profilins from pear (Pyr c 4), cherry (Pru av 4) and celery (Api g 4) with birch pollen profilin Bet v 2. J. Chromatogr. B Biomed. Sci. Appl. 756, 315–325 [DOI] [PubMed] [Google Scholar]
- 23. Alvarado M. I., Jimeno L., De La Torre F., Boissy P., Rivas B., Lázaro M. J., and Barber D. (2014) Profilin as a severe food allergen in allergic patients overexposed to grass pollen. Allergy 69, 1610–1616 [DOI] [PubMed] [Google Scholar]
- 24. Mondal A. K., Parui S., and Mandal S. (1998) Analysis of the free amino acid content in pollen of nine Asteraceae species of known allergenic activity. Ann. Agric. Environ. Med. 5, 17–20 [PubMed] [Google Scholar]
- 25. Weik M., Ravelli R. B., Kryger G., McSweeney S., Raves M. L., Harel M., Gros P., Silman I., Kroon J., and Sussman J. L. (2000) Specific chemical and structural damage to proteins produced by synchrotron radiation. Proc. Natl. Acad. Sci. U.S.A. 97, 623–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kaiser D. A., Goldschmidt-Clermont P. J., Levine B. A., and Pollard T. D. (1989) Characterization of renatured profilin purified by urea elution from poly-l-proline agarose columns. Cell Motil. Cytoskeleton 14, 251–262 [DOI] [PubMed] [Google Scholar]
- 27. Radauer C., Willerroider M., Fuchs H., Hoffmann-Sommergruber K., Thalhamer J., Ferreira F., Scheiner O., and Breiteneder H. (2006) Cross-reactive and species-specific immunoglobulin E epitopes of plant profilins: an experimental and structure-based analysis. Clin. Exp. Allergy 36, 920–929 [DOI] [PubMed] [Google Scholar]
- 28. Barderas R., Villalba M., Pascual C. Y., Batanero E., and Rodríguez R. (2004) Profilin (Che a 2) and polcalcin (Che a 3) are relevant allergens of Chenopodium album pollen: isolation, amino acid sequences, and immunologic properties. J. Allergy Clin. Immunol. 113, 1192–1198 [DOI] [PubMed] [Google Scholar]
- 29. Bonura A., Trapani A., Gulino L., Longo V., Valenta R., Asero R., and Colombo P. (2014) Cloning, expression in E. coli and immunological characterization of Par j 3.0201, a Parietaria pollen profilin variant. Mol. Immunol. 57, 220–225 [DOI] [PubMed] [Google Scholar]
- 30. Cases B., Pastor-Vargas C., Dones F. G., Perez-Gordo M., Maroto A. S., de las Heras M., Vivanco F., and Cuesta-Herranz J. (2010) Watermelon profilin: characterization of a major allergen as a model for plant-derived food profilins. Int. Arch. Allergy Immunol. 153, 215–222 [DOI] [PubMed] [Google Scholar]
- 31. López-Torrejón G., Crespo J. F., Sánchez-Monge R., Sánchez-Jiménez M., Alvarez J., Rodriguez J., and Salcedo G. (2005) Allergenic reactivity of the melon profilin Cuc m 2 and its identification as major allergen. Clin. Exp. Allergy 35, 1065–1072 [DOI] [PubMed] [Google Scholar]
- 32. López-Torrejón G., Ibáñez M. D., Ahrazem O., Sánchez-Monge R., Sastre J., Lombardero M., Barber D., and Salcedo G. (2005) Isolation, cloning and allergenic reactivity of natural profilin Cit s 2, a major orange allergen. Allergy 60, 1424–1429 [DOI] [PubMed] [Google Scholar]
- 33. Tawde P., Venkatesh Y. P., Wang F., Teuber S. S., Sathe S. K., and Roux K. H. (2006) Cloning and characterization of profilin (Pru du 4), a cross-reactive almond (Prunus dulcis) allergen. J. Allergy Clin. Immunol. 118, 915–922 [DOI] [PubMed] [Google Scholar]
- 34. Westphal S., Kempf W., Foetisch K., Retzek M., Vieths S., and Scheurer S. (2004) Tomato profilin Lyc e 1: IgE cross-reactivity and allergenic potency. Allergy 59, 526–532 [DOI] [PubMed] [Google Scholar]
- 35. Valenta R., Duchêne M., Pettenburger K., Sillaber C., Valent P., Bettelheim P., Breitenbach M., Rumpold H., Kraft D., and Scheiner O. (1991) Identification of profilin as a novel pollen allergen; IgE autoreactivity in sensitized individuals. Science 253, 557–560 [DOI] [PubMed] [Google Scholar]
- 36. Asero R., Monsalve R., and Barber D. (2008) Profilin sensitization detected in the office by skin prick test: a study of prevalence and clinical relevance of profilin as a plant food allergen. Clin. Exp. Allergy 38, 1033–1037 [DOI] [PubMed] [Google Scholar]
- 37. Asero R., Wopfner N., Gruber P., Gadermaier G., and Ferreira F. (2006) Artemisia and Ambrosia hypersensitivity: co-sensitization or co-recognition? Clin. Exp. Allergy 36, 658–665 [DOI] [PubMed] [Google Scholar]
- 38. Vieths S., Scheurer S., and Ballmer-Weber B. (2002) Current understanding of cross-reactivity of food allergens and pollen. Ann. N.Y. Acad. Sci. 964, 47–68 [DOI] [PubMed] [Google Scholar]
- 39. Crespo J. F., Retzek M., Foetisch K., Sierra-Maestro E., Cid-Sanchez A. B., Pascual C. Y., Conti A., Feliu A., Rodriguez J., Vieths S., and Scheurer S. (2006) Germin-like protein Cit s 1 and profilin Cit s 2 are major allergens in orange (Citrus sinensis) fruits. Mol. Nutr. Food Res. 50, 282–290 [DOI] [PubMed] [Google Scholar]
- 40. Baek K., Liu X., Ferron F., Shu S., Korn E. D., and Dominguez R. (2008) Modulation of actin structure and function by phosphorylation of Tyr-53 and profilin binding. Proc. Natl. Acad. Sci. U.S.A. 105, 11748–11753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Birbach A. (2008) Profilin, a multi-modal regulator of neuronal plasticity. BioEssays 30, 994–1002 [DOI] [PubMed] [Google Scholar]
- 42. Boopathy S., Silvas T. V., Tischbein M., Jansen S., Shandilya S. M., Zitzewitz J. A., Landers J. E., Goode B. L., Schiffer C. A., and Bosco D. A. (2015) Structural basis for mutation-induced destabilization of profilin 1 in ALS. Proc. Natl. Acad. Sci. U.S.A. 112, 7984–7989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Asturias J. A., Gómez-Bayón N., Arilla M. C., Sánchez-Pulido L., Valencia A., and Martínez A. (2002) Molecular and structural analysis of the panallergen profilin B cell epitopes defined by monoclonal antibodies. Int. Immunol. 14, 993–1001 [DOI] [PubMed] [Google Scholar]
- 44. López-Torrejón G., Díaz-Perales A., Rodríguez J., Sánchez-Monge R., Crespo J. F., Salcedo G., and Pacios L. F. (2007) An experimental and modeling-based approach to locate IgE epitopes of plant profilin allergens. J. Allergy Clin. Immunol. 119, 1481–1488 [DOI] [PubMed] [Google Scholar]
- 45. Ebner C., Hirschwehr R., Bauer L., Breiteneder H., Valenta R., Ebner H., Kraft D., and Scheiner O. (1995) Identification of allergens in fruits and vegetables: IgE cross-reactivities with the important birch pollen allergens Bet v 1 and Bet v 2 (birch profilin). J. Allergy Clin. Immunol. 95, 962–969 [DOI] [PubMed] [Google Scholar]
- 46. Oberhuber C., Ma Y., Wopfner N., Gadermaier G., Dedic A., Niggemann B., Maderegger B., Gruber P., Ferreira F., Scheiner O., and Hoffmann-Sommergruber K. (2008) Prevalence of IgE-binding to Art v 1, Art v 4 and Amb a 1 in mugwort-allergic patients. Int. Arch. Allergy Immunol. 145, 94–101 [DOI] [PubMed] [Google Scholar]
- 47. Rossi R. E., Monasterolo G., Operti D., and Corsi M. (1996) Evaluation of recombinant allergens Bet v 1 and Bet v 2 (profilin) by Pharmacia CAP system in patients with pollen-related allergy to birch and apple. Allergy 51, 940–945 [PubMed] [Google Scholar]
- 48. Villalta D., and Asero R. (2010) Sensitization to the pollen pan-allergen profilin: is the detection of immunoglobulin E to multiple homologous proteins from different sources clinically useful? J. Investig. Allergol. Clin. Immunol. 20, 591–595 [PubMed] [Google Scholar]
- 49. Asero R., Mistrello G., Roncarolo D., Amato S., Zanoni D., Barocci F., and Caldironi G. (2003) Detection of clinical markers of sensitization to profilin in patients allergic to plant-derived foods. J. Allergy Clin. Immunol. 112, 427–432 [DOI] [PubMed] [Google Scholar]
- 50. Vallverdú A., Asturias J. A., Arilla M. C., Gómez-Bayón N., Martínez A., Martínez J., and Palacios R. (1998) Characterization of recombinant Mercurialis annua major allergen Mer a 1 (profilin). J. Allergy Clin. Immunol. 101, 363–370 [DOI] [PubMed] [Google Scholar]
- 51. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 52. Rayment I. (1997) Reductive alkylation of lysine residues to alter crystallization properties of proteins. Methods Enzymol. 276, 171–179 [PubMed] [Google Scholar]
- 53. Walter T. S., Meier C., Assenberg R., Au K. F., Ren J., Verma A., Nettleship J. E., Owens R. J., Stuart D. I., and Grimes J. M. (2006) Lysine methylation as a routine rescue strategy for protein crystallization. Structure 14, 1617–1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rosenbaum G., Alkire R. W., Evans G., Rotella F. J., Lazarski K., Zhang R. G., Ginell S. L., Duke N., Naday I., Lazarz J., Molitsky M. J., Keefe L., Gonczy J., Rock L., Sanishvili R., et al. (2006) The Structural Biology Center 19ID undulator beamline: facility specifications and protein crystallographic results. J. Synchrotron Radiat. 13, 30–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 56. Bordoli L., Kiefer F., Arnold K., Benkert P., Battey J., and Schwede T. (2009) Protein structure homology modeling using SWISS-MODEL workspace. Nat. Protoc. 4, 1–13 [DOI] [PubMed] [Google Scholar]
- 57. Minor W., Cymborowski M., Otwinowski Z., and Chruszcz M. (2006) HKL-3000: the integration of data reduction and structure solution: from diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr. 62, 859–866 [DOI] [PubMed] [Google Scholar]
- 58. Vagin A., and Teplyakov A. (1997) MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 59. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G. W., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., et al. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 61. Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., and Vagin A. A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Painter J., and Merritt E. A. (2006) TLSMD web server for the generation of multi-group TLS models. J. Appl. Crystallogr. 39, 109–111 [Google Scholar]
- 63. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B. 3rd, Snoeyink J., Richardson J. S., and Richardson D. C. (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yang H., Guranovic V., Dutta S., Feng Z., Berman H. M., and Westbrook J. D. (2004) Automated and accurate deposition of structures solved by x-ray diffraction to the Protein Data Bank. Acta Crystallogr. D Biol. Crystallogr. 60, 1833–1839 [DOI] [PubMed] [Google Scholar]
- 65. Pantoliano M. W., Petrella E. C., Kwasnoski J. D., Lobanov V. S., Myslik J., Graf E., Carver T., Asel E., Springer B. A., Lane P., and Salemme F. R. (2001) High-density miniaturized thermal shift assays as a general strategy for drug discovery. J. Biomol. Screen. 6, 429–440 [DOI] [PubMed] [Google Scholar]
- 66. Lavinder J. J., Hari S. B., Sullivan B. J., and Magliery T. J. (2009) High-throughput thermal scanning: a general, rapid dye-binding thermal shift screen for protein engineering. J. Am. Chem. Soc. 131, 3794–3795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Frickey T., and Lupas A. (2004) CLANS: a Java application for visualizing protein families based on pairwise similarity. Bioinformatics 20, 3702–3704 [DOI] [PubMed] [Google Scholar]
- 68. Krissinel E., and Henrick K. (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 [DOI] [PubMed] [Google Scholar]
- 69. DeLano W. L. (2012) The PyMOL Molecular Graphics System, version 1.5.0.1, Schroedinger, LLC, New York [Google Scholar]
- 70. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 71. Krissinel E., and Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 72. Holm L., and Rosenström P. (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ashkenazy H., Erez E., Martz E., Pupko T., and Ben-Tal N. (2010) ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 38, W529–W533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Tordesillas L., Pacios L. F., Palacín A., Cuesta-Herranz J., Madero M., and Díaz-Perales A. (2010) Characterization of IgE epitopes of Cuc m 2, the major melon allergen, and their role in cross-reactivity with pollen profilins. Clin. Exp. Allergy 40, 174–181 [DOI] [PubMed] [Google Scholar]