Abstract
Cryptococcus neoformans (Cn) is a common facultative intracellular pathogen that can cause life-threatening fungal meningitis in immunocompromised individuals. Shortly after infection, Cn is detectable as both extra- and intracellular yeast particles, with Cn being capable of establishing long-lasting latent infections within host macrophages. Although recent studies have shown that shed capsular polysaccharides and intact extracellular Cn can compromise macrophage function through modulation of NF-κB signaling, it is currently unclear whether intracellular Cn also affects NF-κB signaling. Utilizing live cell imaging and computational modeling, we find that extra- and intracellular Cn support distinct modes of NF-κB signaling in cultured murine macrophages. Specifically, in RAW 264.7 murine macrophages treated with extracellular glucuronoxylomannan (GXM), the major Cn capsular polysaccharide, LPS-induced nuclear translocation of p65 is inhibited, whereas in cells with intracellular Cn, LPS-induced nuclear translocation of p65 is both amplified and sustained. Mathematical simulations and quantification of nascent protein expression indicate that this is a possible consequence of Cn-induced “translational interference,” impeding IκBα resynthesis. We also show that long term Cn infection induces stable nuclear localization of p65 and IκBα proteins in the absence of additional pro-inflammatory stimuli. In this case, nuclear localization of p65 is not accompanied by TNFα or inducible NOS (iNOS) expression. These results demonstrate that capsular polysaccharides and intact intracellular yeast manipulate NF-κB via multiple distinct mechanisms and provide new insights into how Cn might modulate cellular signaling at different stages of an infection.
Keywords: computational biology, host-pathogen interaction, inflammation, macrophage, NF-kappa B (NF-KB), Cryptococcus neoformans, intracellular pathogen
Introduction
Cryptococcus neoformans (Cn)3 is a spore-forming, intracellular pathogenic yeast found in soil and other niches worldwide (1). Inhalation of spores is thus extremely common, allowing Cn to infect ∼70% of children in urban environments (2). In most cases Cn infections do not present immediate clinical symptoms; rather, Cn is able to enter a chronic latent state that may last for many years by residing within host macrophages. However, in immune compromised individuals, an acute Cn infection or re-emergence of an existing infection can result in life-threatening conditions such as pneumonia or fungal meningitis. A recent estimate suggests these and other Cn-related illnesses cause ∼600,000 deaths per year (3).
A key regulator of this host-pathogen interaction is the Cn polysaccharide capsule. The capsule, primarily composed of glucuronoxylomannan (GXM, ∼80% by mass (4)), is produced and shed during growth and is essential for virulence (5, 6). It promotes Cn survival during the initial stage of infection by acting as an anti-phagocytic coating, thereby preventing ingestion of yeast in a naïve host during the short-lived acute stage of infection (∼7 days after exposure (7)). An escalating humoral response then leads to the phagocytosis of Cn. Unfortunately, the macrophage phagolysosome is a surprisingly permissive environment for Cn growth (8), making Cn a facultative intracellular pathogen (reviewed in Ref. 9). Intracellular Cn continue to produce and shed GXM, resulting in the accumulation of polysaccharide-filled vesicles within the host cell cytosol (8). Although the effects of intracellular GXM are unclear, extracellular GXM is known to disrupt macrophage behaviors and limit inflammation (10, 11). These effects can be explained, at least in part, by the ability of GXM to subvert the activity of the NF-κB signaling pathway (reviewed in Ref. 12).
NF-κB is a family of dimeric transcription factors that regulate the inflammatory response (reviewed in Ref. 13) and includes the ubiquitously expressed p65·p50 heterodimer. The activity of p65-containing dimers is controlled by cytoplasmic sequestration through association with ΙκΒ proteins, primarily IκBα (14). NF-κB activity in macrophages can be triggered by a range of different stimuli, including LPS from Gram-negative bacteria. LPS is detected by Toll-like receptor-4 (TLR4) at the macrophage cell surface, stimulating a cascade of intracellular signaling events that culminate in the phosphorylation of NF-κB·IκB complexes by IκB kinase (IKK). This leads to the proteasomal degradation of IκB proteins and subsequent nuclear accumulation of p65-containing transcription factors (15, 16). Within the nucleus, these p65-containing transcription factors regulate the expression of genes associated with the immune response, including pro-inflammatory cytokines like TNF-α (17–19) and inducible nitric oxide synthase (iNOS) (20, 21), the latter of which is associated with M1 or “classical” activation of macrophages. Crucially, p65 also stimulates resynthesis of IκBα. This action constitutes a delayed autoregulatory loop that promotes relocalization of NF-κB dimers to the cytosol and eventually restores the pathway to an inactive state once stimuli are withdrawn (22, 23). The behaviors produced by this and the various other feedback loops that regulate NF-κB activity in macrophages are highly non-linear and show substantial cell-to-cell variation (24). This has led to increased utilization of complementary computational modeling and live cell imaging to study NF-κB signaling dynamics (23–26).
Previous investigations into the effects of GXM on inflammatory NF-κB signaling, which have predominantly utilized extracellular Cn or purified GXM, have produced conflicting results. Through interaction with exogenous TLR4 receptors in CHO cells, GXM was able to promote NF-κB nuclear localization and chromatin binding (27). Meanwhile, others have shown that GXM can block TLR4-induced inflammation in a LPS-induced mouse model of endotoxic shock. When studied further in cultured murine macrophages, GXM was shown to interact with surface FcγRIIb receptors, blocking LPS-induced Akt and IKK activity via SHIP, thereby abrogating NF-κB induction and downstream inflammation (11). No studies to date have addressed the effects of phagosomal Cn or intracellular GXM on NF-κB signaling. This constitutes a significant gap in our understanding of the Cn-macrophage relationship, and Cn-associated disease as Cn is predominantly intracellular during chronic infection.
In this current study, we present detailed single-cell analysis of NF-κB signaling dynamics in cultured macrophages and show that purified extracellular GXM and intracellular GXM-positive Cn have contrasting effects on the behavior of the pathway in the context of LPS-induced activation. Our results show that GXM pretreatment inhibits the nuclear accumulation of p65 proteins in response to LPS. We also show for the first time that GXM-producing phagosomal Cn modulate host NF-κB signaling in a manner that differs from that of extracellular GXM. Specifically, the duration of LPS-induced p65 nuclear accumulation is significantly increased in macrophages containing relatively low numbers of ingested GXM-expressing Cn but not GXM-null Cn. A combination of computational modeling and experimental methods indicated that this extended response was caused by translational interference in cells infected with GXM-expressing Cn, which acts to decrease the strength of negative feedback. We also found that extended intracellular occupancy of Cn in the absence of LPS promotes the accumulation of both p65 and IκBα in the nucleus, leading to the repression of the NF-κB-regulated target genes TNFα and iNOS. These results suggest that Cn may employ distinct mechanisms of NF-κB modulation at different stages of an infection to promote its own survival within the host.
Results
GXM Attenuates LPS-induced Nuclear Translocation of NF-κB
The anti-inflammatory capacity of Cn capsular polysaccharide was first illustrated by its ability to block LPS-induced TNFα production by monocyte-derived macrophages (10). Subsequent studies showed that GXM activates SHIP via surface FcγRIIb receptors. Active SHIP then blocks the recruitment of MyD88 (myeloid differentiation primary response gene 88) and Akt, thereby impeding IκBα phosphorylation and degradation (11, 12). These data imply that GXM would block LPS-induced nuclear accumulation of p65-containing NF-κB dimers, thereby preventing the NF-κB-dependent production of pro-inflammatory cytokines. To investigate this in detail, we employed a previously described murine macrophage NF-κB reporter cell line comprising RAW 264.7 cells stably expressing p65-EGFP from the RelA (p65) promoter and a destabilized mCherry reporter of TNFα gene transactivation (NF-κB reporter cells) (24). These cells were cultured in the presence or absence of disease-relevant concentrations of GXM, and the temporal dynamics of p65 nuclear translocation were determined by live cell fluorescence imaging (Fig. 1, A–D).
FIGURE 1.
Exposure to GXM inhibits LPS-induced nuclear accumulation of p65. A, fluorescence microscopy images of p65-EGFP (green) localization in RAW 264.7 NF-κB reporter cells exposed to 100 ng/ml LPS with and without a 1-h pretreatment with 200 μg/ml GXM. The scale bar represents 20 μm. B–D, single cell trajectories of the nuc:cyto ratio of p65-EGFP fluorescence after LPS stimulation without (B) and with (C) GXM pretreatment for four representative cells and (D) the population average for GXM, LPS, and GXM + LPS cells. E, quantification of the average maximum amplitude, time to achieve maximum amplitude, and response duration. Error is represented as the S.E. Statistical significance is indicated as follows: *, p < 0.05; **, p < 0.01; and ***, p < 0.001. The data from >100 cells were collected per condition across three independent biological repeats. F, a diagram linking model parameters to biological events. G, P19, the parameter describing the magnitude of MyD88-dependent IKK activity, was decreased to 0.75 and 0.50 of the nominal value, and the model was simulated in MATLAB as described under “Numerical Experiments.” The predicted ratio in the concentration of nuclear to cytoplasmic p65 was plotted as a function of time after LPS stimulation.
We observed that GXM significantly attenuated LPS-induced nuclear accumulation of p65-EGFP and caused it to occur slightly later than in control cells without affecting the overall duration of the response (Fig. 1E). However, treatment with GXM alone had no effect on p65 localization (Fig. 1D). To test whether these data were consistent with GXM blocking MyD88-dependent IκBα phosphorylation and degradation via SHIP activation, we utilized a mathematical model of LPS-induced NF-κB signaling developed by Sung et al. (24) from the original Hoffmann NF-κB model (36). We found that modest reductions (25%) in the magnitude of the initial wave of MyD88-dependent IKK activity, parameter 19 (P19) in the model, recapitulated the decreased amplitude and delay in nuclear p65 accumulation observed in our experiments.
Development of a Strategy to Infect Macrophages with C. neoformans without Perturbing NF-κB Signaling
Having verified that GXM pretreatment can attenuate LPS-induced nuclear translocation of p65 in macrophages, we then hypothesized that GXM produced by phagocytosed, intracellular Cn may have a similar effect. To test this, we developed a methodology to infect macrophages with Cn without compromising measurements of NF-κB activity.
Typically, the phagocytosis of encapsulated Cn by macrophages in vitro is inefficient unless (i) the macrophages are “activated” and (ii) Cn have been opsonized. Treatments that induce macrophage activation (typically co-treatment with LPS and IFN-γ) can confound studies involving NF-κB because they will also induce significant NF-κB activity (Fig. 2A). To circumvent this problem, we transiently stimulated the reporter cells with IFN-γ alone (16 h; Fig. 2B), which enhances activation and Cn clearance in vivo (37) without inducing NF-κB in macrophages (38). This was confirmed by live imaging of NF-κB reporter cells exposed to 500 units/ml IFN-γ (Fig. 2A). As expected, complement, which is commonly used to opsonize Cn, was found to induce NF-κB activity in our reporter cell line. We therefore used 18B7 mouse monoclonal antibodies to capsular GXM (39) as our opsonization agent because they do not induce NF-κB signaling (Fig. 2A).
FIGURE 2.
Effect of macrophage-activating stimuli and Cn opsonins on p65-EGFP localization in RAW 264.7 cells. A, RAW 264.7 NF-κB reporter cells were treated as indicated and imaged by live cell microscopy for periods of up to 5 h. Conditions that induced p65-EGFP nuclear localization (nuc:cyto p65-EGFP ≥ 1) in cells were classified as “activating.” Conditions that caused no apparent accumulation of p65-EGFP were classified as “not activating.” B, schematic of the Cn-macrophage infection protocol. Timeline for the preparation of Cn-infected RAW 264.7 NF-κB reporter cells for live cell imaging.
Intracellular C. neoformans Delays and Extends LPS-induced NF-κB Signaling
Because chronic Cn infection primarily involves intracellular Cn, we investigated whether Cn can modulate the NF-κB pathway after phagocytosis by infecting macrophages with a virulent strain of Cn (H99S) that produces significant quantities of GXM (28).
The phagocytosis of individual Cn by our macrophage reporter cell line did not elicit an immediate NF-κB response (data not shown). However, we found that phagosomal Cn altered the temporal dynamics of LPS-induced NF-κB signaling (Fig. 3, A–C). Notably, intracellular Cn appeared to alter NF-κB signaling in a manner that differed from that of purified, extracellular GXM. Although pretreatment with purified GXM resulted in a ∼2-fold decrease in the amplitude of the LPS-induced p65 response (as defined by the maximal nuc:cyto p65 ratio), cells containing intracellular Cn showed a slight but significant increase in amplitude (Fig. 3C). This was accompanied by a pronounced delay in the time taken to achieve maximal nuclear p65-EGFP levels (Fig. 3C), which also increased with Cn burden (the number of intracellular Cn per macrophage). Furthermore, and in contrast with GXM-dependent effects, the duration of LPS-induced p65 nuclear occupancy was substantially extended in macrophages containing intracellular Cn compared with controls (Fig. 3C). Similar results were obtained in macrophages infected with 24067, a serotype D strain of GXM-expressing Cn (Fig. 3D), with infected cells also exhibiting a delayed and extended NF-κB response to LPS stimulation.
FIGURE 3.
Macrophages containing intracellular Cn exhibit a delayed and sustained NF-κB response to LPS stimulation. A, fluorescence microscopy images of p65-EGFP (green) localization in RAW 264.7 NF-κB reporter cells exposed to 100 ng/ml LPS 2 h after infection with Cn. The scale bar represents 20 μm. Intracellular Cn are indicated with arrows. B, single cell trajectories of the nuc:cyto ratio of p65-EGFP fluorescence after LPS stimulation for cells that do not (Ctrl) and do contain >3 intracellular H99S Cn (3+ H99S) for four representative cells. C–E, the population average trajectory of the nuc:cyto ratio of p65-EGFP fluorescence after LPS stimulation for cells that do not (Ctrl) and do contain intracellular Cn with quantification of the average maximum amplitude, time to achieve maximum amplitude, and response duration for H99S (C), 24067 (D), and Cap59 (E)-infected macrophages. Error is represented as the S.E. Statistical significance is indicated as follows: *, p < 0.05; **, p < 0.01; and ***, p < 0.001 (ANOVA, p < 0.05). Data from >35 cells were collected per condition across a minimum of six independent biological repeats.
To test whether these effects were GXM-dependent, we infected macrophages with Cap59, a serotype A mutant Cn strain that has a defect in capsule synthesis and does not produce capsular GXM (40), and then treated with LPS. In this case no differences were seen in the amplitude, timing, or duration of p65-EGFP nuclear localization (Fig. 3E), suggesting that the ability of intracellular Cn to modulate LPS-induced NF-κB signaling is GXM-dependent.
Intracellular C. neoformans-induced Changes in Macrophage NF-κB Dynamics Can Be Simulated in a Mathematical Model of Macrophage LPS-NF-κB Signaling
To investigate the possible causes of intracellular Cn-induced changes in the dynamics of the NF-κB response to LPS, we again utilized our NF-κB mathematical model modified from that of Sung et al. (24). This enabled us to simulate possible known and suspected effects of intracellular Cn proliferation (Fig. 4A), including (i) translational interference (P9); (ii) inhibition of the rate of IκB nuclear import or export of p65·IκB complexes (P14 and P12, respectively); and (iii) an increase in TIR domain-containing adapter-inducing interferon-β (TRIF)-dependent IKK activation at Cn-containing phagosomes (P20). Regarding mechanism (i) above, live (but not heat-killed) intracellular Cn has been shown to alter protein translation rate in both murine peritoneal macrophages and J774.16 macrophage-like cells (33). We hypothesized that this could prolong the length of time that NF-κB spends in the nucleus by reducing the rate of IκBα synthesis. Regarding mechanism (ii) above, we hypothesized that the large Cn-containing phagosome, which tends to be positioned at the nuclear periphery, and Cn-induced organelle crowding around the nucleus (41, 42) could potentially reduce the rate of p65 and IκBα trafficking across the nuclear membrane, as suggested by spatial models of NF-κB signaling (43), thereby altering the temporal dynamics of NF-κB signaling. Regarding mechanism (iii) above, although GXM has been shown to reduce MyD88-induced IκBα phosphorylation at the cell surface (11), there is a potential for TLR4 to signal through TRIF from the Cn-containing phagosomes, possibly enhancing the NF-κB response to LPS (44).
FIGURE 4.

Numerical experiments using the model of Sung et al. (24) to evaluate potential mechanisms through which intracellular Cn modulates NF-κB signaling. A, diagram linking model parameters to biological events. B–E, select model parameters were varied about a nominal value as indicate in the figure legends and the model was simulated in MATLAB as described under “Numerical Experiments.” The predicted ratio in the concentration of nuclear to cytoplasmic p65 was plotted as a function of time after LPS stimulation.
We evaluated these mechanisms by performing numerical experiments in which the associated parameters were varied individually (Fig. 4, B–E). Our results support the hypotheses that a reduction in the rate of IκBα synthesis (P9), possibly caused by translational interference, or an increase in the strength of TRIF-dependent IKK activation could explain the sustained nuclear localization of NF-κB that we have observed in Cn-infected cells after LPS stimulation (Fig. 4, B and C). Specifically, both a 50% reduction in the rate of IκBα synthesis and a 50% increase in the strength of TRIF-dependent IKK activation were able to extend the duration of NF-κB in the nucleus while modestly increasing and delaying the peak response (Fig. 4, B and E). In both cases, the sustained nuclear localization of p65 may be accomplished through the merging of the early MyD88- and late TRIF-dependent responses. On the other hand, 50% reductions in the rate of nuclear import of IκB or that of the export of the IκB·p65 complex did not significantly alter the dynamics of p65 nuclear localization in response to LPS (Fig. 4, C and D).
Partial Inhibition of Macrophage Protein Translation Caused by Intracellular C. neoformans or Cycloheximide Can Increase the Duration of the p65 Nuclear Occupancy after LPS Stimulation
To investigate the possibility that translational interference was responsible for the change in the dynamics of LPS-induced p65-EGFP nuclear translocation, we first tested whether partial inhibition of protein translation using the ribosome inhibitor, cycloheximide (CHX), could recapitulate the extended response observed in Cn-containing cells. We observed that although complete inhibition of protein synthesis in RAW 264.7 cells with high doses of CHX (10 μm) caused stable nuclear localization of p65-EGFP, partial inhibition achieved using lower doses (1 μm) produced an extended response similar to that observed in cells infected with GXM-positive Cn (Fig. 5A).
FIGURE 5.
Live GXM-positive Cn causes translational interference in RAW 264.7 macrophages. A, quantification of the nuclear:cytoplasmic ratio of p65-EGFP fluorescence in LPS-treated live RAW 264.7 NF-κB-reporter cells pretreated with vehicle (Ctrl) or the indicated doses of CHX. Data from >40 cells were collected per condition across a minimum of two independent biological repeats. B, fluorescence microscopy images of RAW 264.7 cells treated with 100 μm CHX or infected with live or heat-killed H99S Cn. Nascent protein synthesis was detected by staining with OPP-647 (red in merge) and genomic DNA was stained using NuclearMask Blue (blue in merge). Arrows indicate Cn-infected macrophages. C, quantification of OPP-647 staining in non-infected RAW 264.7 cells (No Cn) and cells infected with live (Live; also separated in to low (1–2 Cn; LB) and high burden (3+ Cn; HB) infected cells) or heat-killed (HK) H99S Cn. D, quantification of ribopuromycylation (RPM) staining of H99S-infected RAW264.7 cells. E, OPP-647 staining was quantified as described in B and C for Cap59 infected RAW 264.7 cells. Error is represented as the S.E. Statistical significance is indicated as follows: *, p < 0.05; **, p < 0.01; and ***, p < 0.001 (ANOVA, p < 0.05). Data from >40 cells were collected per condition across a minimum of three independent biological repeats. The scale bar represents 20 μm.
Having established that partially blocking translation could produce an extended NF-κB response to LPS, we measured changes in nascent protein synthesis in RAW 264.7 cells infected with GXM-positive H99S and GXM-negative Cap59 Cn yeast. We found that live but not heat-killed H99S Cn infection caused a statistically significant decrease in nascent protein production in two of three experiments, as measured using Click-iT O-propargyl-puromycin (OPP) staining (Fig. 5, B and C), and the magnitude of this effect increased with intracellular Cn burden. Because statistically significant decreases in translation were also obtained using ribopuromycylation assays (Fig. 5D), this suggests that Cn somehow interferes with translation. However, infection with Cap59 had no significant effect on OPP staining, indicating that this GXM-negative strain was not capable of causing translational interference (Fig. 5E), suggesting that translational interference is GXM-dependent and possibly explaining the inability of intracellular Cap59 to alter the dynamics of LPS-induced NF-κB signaling in macrophages (Fig. 3E).
Proliferation of Intracellular C. neoformans Alone Can Promote Nuclear Accumulation of p65-containing Dimers
During our initial experiments where we investigated altered TLR4 signaling in Cn-infected cells, we noticed that a small number of macrophages containing Cn were found to exhibit nuclear p65-EGFP (nuc:cyto > 1) within 2 h of initial exposure to Cn, prior to LPS addition. This effect was not seen in surrounding uninfected cells. Therefore, these cells were classified as showing CIN-p65 (Cn-induced nuclear accumulation of p65). The proportion of cells showing CIN-p65 increased over time and cells with more intracellular Cn were more likely to exhibit nuclear p65.
To further investigate the effects of proliferating phagosomal Cn alone on NF-κB, we infected macrophages with opsonized Cn, allowed 2 h for phagocytosis of Cn, washed off extracellular Cn (and polysaccharide-containing media), and imaged macrophages containing phagosomal Cn for an additional 24 h.
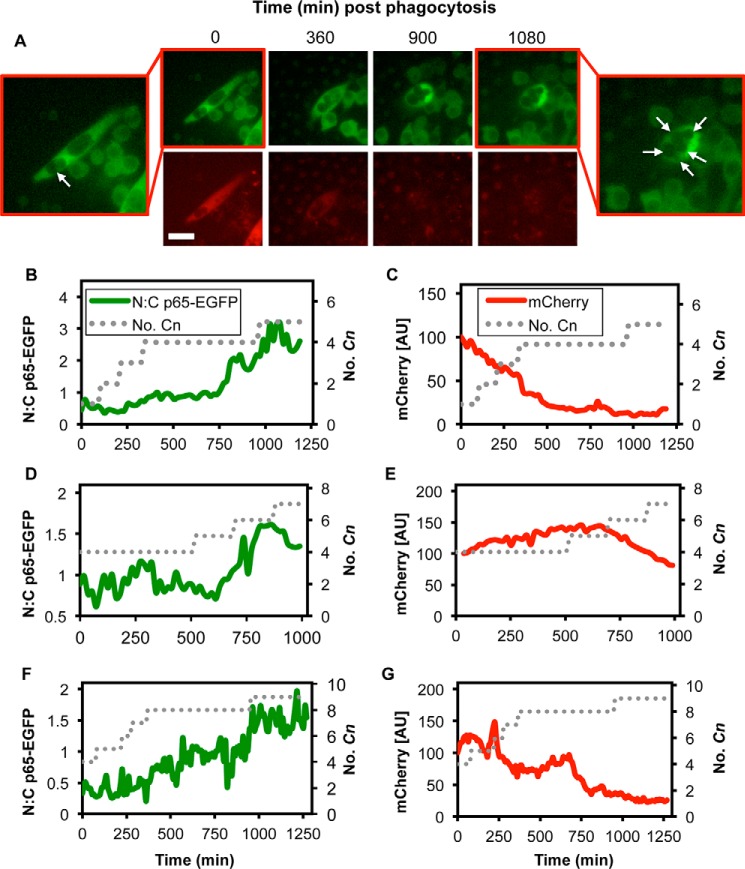
Consistent with our previous results, we found that phagosomal proliferation of Cn alone sometimes promoted nuclear accumulation of p65-EGFP (seen in 2.8% of Cn-containing cells at 2 h and 6.8% of Cn-containing cells at 24 h after Cn exposure, with cells containing an average of 2.7 and 5.6 Cn, respectively). Unlike the transient nuclear localization produced by LPS, which occurs rapidly and is followed by the swift return of p65-containing dimers to the cytosol, Cn-induced nuclear accumulation of p65-EGFP occurred gradually and produced long-lasting nuclear localization (Fig. 6, A and B). This slow accumulation of p65-containing dimers within the nucleus appeared to be influenced by macrophage intracellular Cn “burden,” which gradually increased over time as phagocytosed Cn began to proliferate within individual macrophages (Fig. 6B). Unexpectedly, the nuclear accumulation of p65-EGFP was not accompanied by mCherry expression from the Tnf promoter, despite abundant nuclear p65 (Fig. 6, A and C). This effect was observed in multiple cells (Fig. 6, D–G).
FIGURE 6.
High intracellular Cn burden stimulates nuclear accumulation of p65 without gene expression. A, fluorescence microscopy images of p65-EGFP (green) localization and destabilized mCherry (red) expression in RAW 264.7 NF-κB reporter cells after phagocytosis of opsonized H99S Cn. The scale bar represents 20 μm. Intracellular Cn are indicated with arrows. B and C, quantification of the nuc:cyto ratio of p65-EGFP fluorescence (B) and mCherry fluorescence (C) for the cell depicted in A. The approximate number of intracellular Cn is represented by the dotted line. D–G, representative single cell trajectories of the nuc:cyto ratio of p65-EGFP fluorescence and mCherry fluorescence for three representative cells.
Phagosomal C. neoformans Suppresses TNFα and iNOS Production in Cells Exhibiting Nuclear p65-EGFP
Because Cn-induced nuclear p65 accumulation was not accompanied by NF-κB-regulated gene expression, we speculated that intracellular Cn may also render these cells incapable of responding to additional NF-κB-activating stimuli. To test this, we exposed non-infected and CIN-p65 cells to 100 ng/ml LPS and measured both mCherry levels (reporting TNFα gene transactivation), nuc:cyto p65-EGFP, and total p65-EGFP levels, because the RELA promoter is also positively regulated by NF-κB at high LPS doses (24).
Our data indicate CIN-p65 macrophages were largely refractory to exogenous NF-κB-activating stimuli, because LPS treatment failed to promote either the return of p65-EGFP to the cytosol or expression of TNFα (Fig. 7A). Although neighboring non-infected macrophages exhibited a “normal” NF-κB response to LPS (i.e. a rapid but transient nuclear accumulation of p65-EGFP), LPS seemingly had little to no effect on p65-EGFP distribution in CIN-p65 macrophages (Fig. 7, A and B). Additionally, and in stark contrast to non-infected control cells, the NF-κB-dependent transcriptional output in CIN-p65 macrophages was seemingly negligible with no significant increase observed in either total p65-EGFP or mCherry levels as measured at the population level (Fig. 7, C and D).
FIGURE 7.
High intracellular Cn burden alters the NF-κB response to LPS. A, fluorescence microscopy images of p65-EGFP (green) localization and destabilized mCherry (red) expression in RAW 264.7 NF-κB reporter cells. Cells with Cn (H99S)-induced nuclear p65-EGFP and control cells were imaged for 2 h prior to treatment with 100 ng/ml LPS (T = 0 min). Intracellular Cn are indicated with arrows. B, single cell trajectories of the nuc:cyto ratio of p65-EGFP fluorescence before and after LPS stimulation for representative cells with (Cn +ve) and without (Cn −ve) intracellular Cn as indicated in A. C and D, change in total p65-EGFP (C) and mCherry fluorescence (D) after LPS stimulation for cells exhibiting Cn-induced nuclear p65 (Cn +ve) and control cells that do not contain Cn (Cn −ve). Error is represented as the S.E. Statistical significance is indicated as follows: *, p < 0.05; **, p < 0.01; and ***, p < 0.001. Data from >8 Cn-infected cells (containing an average of 4.5 ± 2.8 Cn) were collected across five independent biological repeats. E, RAW 264.7 NF-κB reporter cells were fixed at the indicated times after stimulation with 100 ng/ml LPS or after infection with H99S and stained with Hoechst 33342 (blue) and immunostained with anti-iNOS antibodies (red). Fluorescence from p65-EGFP is represented in green. The large white circle demarcates a representative Cn-infected macrophage, and the smaller white circle indicates the nucleus of this cell. This result was consistent across all Cn-infected cells (12 cells) from four independent biological repeats. These cells contained an average of 4.3 ± 1.6 Cn. The scale bars represent 20 μm.
Because mCherry expression is regulated by an exogenous copy of the Tnf promoter and may not accurately reflect the regulation of endogenous NF-κB target genes, we also immunostained control and CIN-p65 NF-κB reporter cells for iNOS, a bona fide NF-κB target gene and marker of M1 activation in macrophages. As expected, LPS stimulation of uninfected macrophages induced a small increase in iNOS protein levels at 5 h and high levels of expression by 15 h. However, in Cn-containing macrophages, iNOS was found to be consistently at basal levels 24 h after infection despite stable nuclear localization of p65 (Fig. 7E). Taken together, these data indicated that CIN-p65 was either transcriptionally incompetent or was capable of repressing NF-κB target genes.
Stable Nuclear Localization of p65 in C. neoformans-containing Macrophages Is Caused by Altered Trafficking of IκBα
To investigate how intracellular Cn accumulation may cause stable nuclear localization of p65, we again turned to the mathematical model. We simulated the effect of varying parameters (Fig. 4A) in the absence of LPS (Fig. 8, A–C). As before, we noticed that a sustained decrease in the rate of IκBα synthesis (P9; Fig. 8A), as might be caused by translational interference, could promote stable nuclear localization of p65. However, the model also suggested that a decrease in the rate of nuclear export of NF-κB·IκBα complexes could have a similar effect (Fig. 8B). Changes to other model parameters, such as the rate of IκBα nuclear import (P12; Fig. 8C), did not cause stable nuclear localization of NF-κB.
FIGURE 8.
Numerical experiments using the model of Sung et al. (24) to evaluate potential mechanisms through which intracellular Cn can induce stable nuclear localization of p65. A–C, select model parameters were varied about a nominal value as indicate in the figure legends, and the model was simulated in MATLAB as described under “Numerical Experiments.” The predicted ratio in the concentration of nuclear to cytoplasmic p65 was plotted as a function of time in the absence of LPS stimulation. D, RAW 264.7 NF-κB reporter cells were fixed 24 h after infection with H99S and stained with Hoechst 33342 (blue) and immunostained with anti-IκBα antibodies (red). Fluorescence from p65-EGFP is represented in green. The arrow indicates the nucleus of a representative Cn-infected macrophage. This result was consistent across all Cn-infected cells (11 cells) from six independent biological repeats. These cells contained an average of 6.0 ± 2.2 Cn. The scale bars represent 20 μm.
To discriminate between the two possibilities that stable nuclear p65 was caused by inhibition of either (i) IκBα synthesis or (ii) nuclear export of NF-κB·IκBα complexes, we measured IκBα levels in CIN-p65 cells. We found that all CIN-p65 cells exhibited high levels of nuclear IκBα staining (Fig. 8D), which was consistent with our second hypothesis. Furthermore, because interaction between IκBα and p65 in the nucleus prevents effective chromatin binding (45–48), these data are also consistent with the absence of TNFα and iNOS expression in these cells.
Discussion
The nature of host-pathogen interactions are typically highly dynamic, a metaphorical arms race of move and countermove in which the host and pathogen employ a changing repertoire of strategies to destroy or evade the other. As a key regulator of immune cell function and survival, it is perhaps unsurprising that the NF-κB pathway is a frequent target of human pathogens. Although Yersinia inhibits NF-κB in naive macrophages (49), triggering their apoptosis, the intracellular pathogens Mycobacterium tuberculosis and Shigella both exploit pro-survival activities of the NF-κB pathway during the intracellular phase of infection, buying time to replicate before either actively triggering host cell apoptosis or permitting cell death to escape and disseminate (reviewed in Ref. 50).
Over the past 15 years, a number of studies utilizing both purified capsular components and intact yeast have shown that Cn may also subvert immune cell NF-κB signaling during acute, extracellular infections (10, 11, 27, 51). Perhaps because of differences in experimental design, there is still disagreement within the field, on the precise mechanism(s) and consequences of NF-κB modulation.
Because Gram-negative bacteremia can occur in small numbers of Cn infections (52) and GXM has been shown to block LPS-induced inflammation (11), we utilized a simple cell culture model to study Cn-dependent NF-κB modulation in the context of LPS activation. We assumed that during extracellular infections, macrophages would be more likely to encounter free GXM than intact Cn. This would seem plausible because GXM is shed by proliferating yeast in vivo, accumulates in the serum and tissue of infected individuals to μg/ml levels, and remains for weeks or months before it is cleared (53). Our data suggest that GXM attenuates LPS-induced NF-κB activation by inhibiting the accumulation of p65-containing dimers in the nucleus. This is consistent with reports indicating GXM suppresses phosphorylation and degradation of IκB by promoting SHIP activity (11).
Although the chronic, intracellular phase of Cn infection may last for years or even decades, it has remained unclear whether phagosomal Cn continue to modulate NF-κB signaling. This important question has remained open because of numerous technical challenges. Of these, perhaps the most fundamental is the low rate at which Cn phagocytosis occurs in vitro unless macrophages are first activated by pro-inflammatory stimuli (typically IFN-γ in combination with LPS or TNFα), which could mask the effects of ingested Cn on NF-κB signaling or at least make it extremely difficult to measure using standard biochemical techniques and end point assays.
In this current study, by utilizing a live cell imaging approach, we identified and tracked those cells within an “unactivated” macrophage population that had ingested Cn to measure the dynamics of canonical NF-κB signaling and downstream gene expression. In contrast to the effects of purified GXM, we saw that infection of macrophages with GXM-expressing Cn strains (H99S and 24067) significantly increased the duration of the canonical NF-κB response to LPS. However, this effect was absent when cells were infected with the GXM-deficient Cn strain, Cap59, suggesting that the ability of intracellular Cn to modulate NF-κB signaling was GXM-dependent and that the modulatory effects for GXM differ depending on the mode of presentation (i.e. whether it is extra- or intracellular).
To determine how intracellular Cn might perturb the kinetics of the p65 response to LPS, we employed a mathematical model of NF-κB signaling (24). Simulations indicated that a small reduction in the rate of IκBα synthesis, effectively decreasing the strength of negative feedback, would extend the duration of p65 nuclear occupancy after LPS stimulation. This model-generated hypothesis was plausible because (i) intracellular Cn has been previously shown to affect protein translation in murine peritoneal macrophages and J774.16 macrophage-like cells (33), and (ii) we were able to demonstrate that partial inhibition of translation using CHX extended p65 nuclear occupancy after LPS stimulation in RAW 264.7 macrophages. Measurement of nascent protein production in Cn-infected cells using two different methods showed that only live, GXM-expressing Cn were able to partially inhibit protein synthesis, providing a possible explanation for the differences in LPS-induced NF-κB signaling dynamics observed between GXM-positive and -negative Cn strains.
In this study, we were also able to show that phagosomal proliferation of Cn to high burden was sufficient to induce stable nuclear localization of p65 in the absence of exogenous pro-inflammatory stimuli. Contrary to expectations, this was not accompanied by expression from the NF-κB-regulated Tnf promoter or expression of iNOS, a well studied NF-κB target gene and a marker of M1 macrophage activation. As before, we utilized the mathematical model to identify potential mechanisms to explain the stable nuclear localization of p65 in these cells in the absence of stimuli and again found that this could be caused by decreases in the rate of IκBα synthesis but also by a reduction in the rate of nuclear export of NF-κB·IκBα complexes. These two competing hypotheses were tested by immunostaining cells for IκBα. We found that CIN-p65 cells contained large quantities of nuclear IκBα, which was consistent with the second hypothesis. This finding, although unexpected, is not without precedent and may help to explain the absence of TNFα and iNOS expression in these cells.
We and others have previously shown that extended nuclear occupancy of p65 may not necessarily result in effective interactions with chromatin (22, 47, 48, 54). Trapping of p65 in the nucleus through the use of nuclear export inhibitors (i.e. leptomycin B) may be accompanied by the loss of p65 post-translational modifications, such as phosphorylation of Ser536, that are associated with activity (22). Newly synthesized IκBα in these cells will complex with nuclear p65, decreasing association with chromatin (47, 48). It has also been shown in LPS and leptomycin B co-treated macrophages that IκBα is selectively recruited with p65 to various NF-κB-regulated promoters, including the Tnf, IL-1β, and IL-6 gene promoters, repressing gene transcription (54). Although these data are consistent with our findings, the causes of nuclear p65 and IκBα accumulation and the absence of TNFα and iNOS expression in Cn-infected macrophages remain unclear. One possible explanation is that p65·IκBα complexes are being trapped in the nucleus as a consequence of Cn-induced post-translational modifications or through inhibitory interaction with other nuclear proteins. One such candidate would be protein methyltransferase 2, which has been shown to control inflammatory TLR4·NF-κB signaling in macrophages by directly associating with IκBα in the nucleus, blocking its chromosome region maintenance 1 (CRM1)-dependent nuclear export and decreasing TNFα and IL-6 expression (55, 56). It is also possible that in Cn-containing cells, nuclear p65 may associate with co-repressors such as histone deacetylases 1 and 2 or other negative regulators of p65 activity like SIRT1 (57, 58). Indeed, other pathogens have been shown to utilize SIRT1 activity in macrophages as part of intracellular survival strategies (59). These and other possible mechanisms will be explored in future studies.
In summary, in the context of LPS-induced activation, our data show that Cn is able to alter the way in which macrophages encode information about the inflammatory signaling environment in the dynamics of p65 transcription factors, ultimately suppressing the ability of macrophages to participate in an effective immune response through the production of pro-inflammatory cytokines or the expression of genes required for M1 activation. We also propose that the mechanisms utilized by Cn to modulate NF-κB signaling change as the pathogen transitions from an extracellular to an intracellular lifestyle.
Experimental Procedures
Culture and Opsonization of Cn
Cn serotype A, strains H99S (28), Cap59 and serotype D strain 24067 were grown in yeast peptone dextrose broth for 48 h at 37 °C with shaking (Cap59 was grown at 30 °C for similar amounts of time). After 48 h, the cells were washed three times with PBS and counted. For GXM-expressing strains, 1 × 107 cells were opsonized with 10 μg/ml 18B7 mAb (a kind gift from Dr. Arturo Casadevall, John Hopkins School of Public Health) at 37 °C for either 30 min (H99S) or 1 h (24067) and then washed again (three times with PBS) to remove excess antibody and infected at a 10:1 multiplicity of infection. For experiments requiring heat-killed Cn, yeast were incubated at 60 °C for 30 min prior to opsonization. All cultures were started from frozen stock to ensure that no microevolution occurred.
GXM Purification and Validation
To ensure there was no contaminating galactoxylomannan present, GXM was prepared using the Cn serotype A, galactoxylomannan-negative strain Uge1 (29). Uge1 was cultured in 200 ml of defined minimal medium (15 mm glucose, 10 mm MgSO4-7H2O, 29.4 mm KH2PO4, 0.13 mm glycine, 3 μm thiamine HCl, pH 6.0) for 5 days. Yeast cells were centrifuged using an ultracentrifuge (Beckman Coulter Inc., Pasadena, CA) at 11,000 × g (8000 rpm) for 20 min. GXM was purified from the supernatant by precipitation with 0.2 m NaCl and 0.05% cetyltrimethylammonium bromide as described by Cherniak et al. (30) followed by additional washing steps with 2% acetate in ethanol, 90% ethanol, and absolute ethanol (31). Although not tested specifically in these experiments, this GXM purification protocol has been shown to preserve the O-acetylation of GXM (30). The solution was centrifuged again at 11,000 × g (8000 rpm) for 10 min, the supernatant was removed, and the remaining pellet was dried and weighed. LPS was removed from all samples using a Polymyxin B column in LPS-free distilled water. The samples were then lyophilized and resuspended in LPS-free water. The purified GXM was validated by ELISA using IgG1 mAb 18b7 and a colorimetric sugar assay employing H2SO4 and phenol (32). Purified GXM was negative for endotoxin/LPS contamination, as tested by limulus amebocyte lysate assay (<0.017 EU/ml).
Macrophage Culture and Phagocytosis Conditions
RAW 264.7 (TIB 71) murine macrophages were obtained from ATCC (Manassas, VA). RAW 264.7 stably expressing p65-EGFP fusion proteins from an endogenous reporter and a destabilized mCherry reporter of Tnf promoter activation (“NF-κB reporter cells”) were a kind gift from Dr. Iain Fraser (National Institutes of Health) and have been previously described (24). Both cell lines were cultured in DMEM containing 10% FBS, 200 mm l-glutamine, 100 units/ml penicillin and streptomycin, and 50 μg/ml gentamicin and maintained at 37 °C in a humidified 5% CO2 atmosphere.
For live cell microscopy, 2 × 105 NF-κB reporter cells were seeded into 35-mm glass-bottomed dishes (Cellvis) 24 h prior to imaging in 2 ml of medium. Approximately 16 h prior to Cn infection, the cells were “primed” with IFN-γ (500 units/ml) to increase the proportion of macrophages capable of phagocytosing Cn.
Time Lapse Microscopy and Immunofluorescence
Cells plated in 35-mm glass-bottomed dishes were imaged using a Nikon Ti-Eclipse wide field microscope equipped with a CFI Plan Fluor 40× oil immersion NA 1.30 objective, Intensilight epifluorescence illuminator, computer-controlled stage (Nikon), CoolSNAP MYO camera (Photometrics), and a full environmental enclosure with CO2, humidity, and temperature control (InVivo Scientific). The microscope was controlled using Nikon Elements Software (Nikon).
Images were acquired at 3-min intervals for the indicated durations. Typically, cells were imaged for up to 1 h prior to infection with Cn and then an additional 1 h before cells were exposed to pro-inflammatory stimuli (i.e. LPS). EGFP was excited through a 465–495-nm excitation filter, and emitted light was detected through a 515–555-nm barrier filter reflected from a 505-nm dichroic mirror. mCherry was excited through a 535–550-nm excitation filter, and emitted light was detected through a 610–675-nm barrier filter reflected from a 565-nm dichroic mirror.
In cases where Cn infection induced stable nuclear localization of p65-EGFP, the cells were fixed with ice cold methanol for 5 min and then immunostained with a primary antibody raised against either iNOS (1:100 dilution; catalog no. 13120S, Cell Signaling) or IκBα (1:100 dilution; catalog no. sc-371, Santa Cruz Biotechnology), followed by an anti-rabbit Alexa Fluor 647-labeled secondary antibody (1:200 dilution; catalog no. ab150075, Abcam) and then stained with Hoechst 33342. Stained cells were also imaged using a Nikon Ti-Eclipse wide field microscope. Alexa Fluor 647 was excited through a 590–650-nm excitation filter, and emitted light was detected through a 663–738-nm barrier filter reflected from a 660-nm dichroic mirror. Hoechst 33342 was excited through a 340–380-nm excitation filter, and emitted light was detected through a 435–485-nm barrier filter reflected from a 400-nm dichroic mirror.
Protein Translation Assays
RAW 264.7 cells were seeded at a density of 2 × 105/well into tissue culture-treated 6-well plates (USA Scientific, Ocala, FL) containing no. 1.5 (0.17-mm thick) glass coverslips. The cells were activated with 500 units/ml IFN-γ and 1 μg/ml LPS ∼16 h prior to infection with Cn and again with 1 μg/ml LPS concurrent with Cn infection. The cells were allowed to phagocytose Cn for 2 h (10:1 multiplicity of infection). Extracellular Cn was washed away with RAW264.7 culture medium, and macrophage ribosome activity was determined by two separate methods: ribopuromycylation, adapted from Coelho et al. (33), and staining with O-propargyl-puromycin using the Click-iT Plus OPP protein synthesis assay kit Alexa Fluor 647 (Life Technologies) in accordance with the manufacturer's guidelines. For the ribopuromycylation assay, the cells were incubated for 15 min at 37 °C in a humidified 5% CO2 atmosphere with culture medium supplemented with 91 μm puromycin and 208 μm emetine, with one well not including puromycin as a negative control. The cells were permeabilized with 50 mm Tris-HCl, 10 units/ml RNaseOut, 0.015% digitonin, cOmpleteTM Mini Protease Inhibitor Mixture tablet (Sigma-Aldrich), fixed with 3% paraformaldehyde, and blocked with 1× PBS, 0.05% saponin, 10 mm glycine, and 5% goat serum. The cells were immunostained with a primary antibody raised against puromycin, PMY-2A4 (1:100 dilution, developed by Dr. Jonathan Yewdell (National Institutes of Health) (34), and obtained from the Developmental Studies Hybridoma Bank, created by the NICHD, National Institutes of Health, and maintained at the University of Iowa Department of Biology), detected using Alexa Fluor 555-labeled anti-mouse secondary antibodies (1:200). The cells were co-stained with Hoechst 33342 nuclear stain. With the Click-iT Plus OPP kit, one well was not infected with Cn and treated with 100 μm of the ribosome inhibitor CHX for 30 min. Stained cells were imaged using a Zeiss LSM700 laser scanning confocal microscope. Alexa Fluor 647 fluorescence was excited using a 555-nm laser and detected through a LP640 filter. NuclearMask Blue fluorescence was excited using a 405-nm laser, and fluorescence was detected through a SP 555-nm filter.
Image Analysis
Post-acquisition, 14-bit Nikon nd2 images were analyzed using FIJI (35). Typically, individual field time-courses were thresholded for each recorded fluorescence channel. Mean nuclear and cytoplasmic p65-EGFP fluorescence was quantified for individual cells and expressed as the nuc:cyto ratio. For each cell analyzed, this ratio was used to generate values for (i) amplitude, reported as the maximum nuc:cyto ratio reached during the time course; (ii) time to maximum, reported as minutes after LPS stimulation until the first maximum nuc:cyto ratio was achieved; and (iii) duration, which utilized nuc:cyto normalized to the maximum value and was reported as minutes between nuc:cyto > 0.5 to nuc:cyto < 0.5. Whole cell integrated mCherry fluorescence was determined for each cell and normalized to the pretreatment or initial value. For experiments involving infection of cells with Cn, data were produced for both Cn-containing (Cn +ve) and an equal or greater number of cells that did not contain Cn (Cn −ve) as internal controls. The initial number of Cn present within the cell was recorded and in some cases tracked throughout experiments. For the protein translation assays, 16-bit Zeiss czi images were also analyzed using FIJI. The images were background subtracted, and mean whole cell Alexa Fluor 555 was quantified for each cell for ribopuromycylation straining, and mean nuclear Alexa Fluor 647 fluorescence was quantified for each cell for the Click-iT Plus OPP staining. To avoid bias, all non-infected control cells in every field containing Cn-infected cells were analyzed.
Numerical Experiments
A previous model of NF-κB signaling in macrophages (Sung et al. (24)) was adapted to investigate potential mechanisms through which Cn modulates LPS-induced NF-κB signaling. Specifically, we adapted the original model to account for the ratio of nuclear to cytoplasmic volume, which we took as 0.3 (25). For example, if a chemical species, X, is present in both the nucleus and the cytoplasm, then the mathematical model tracks the concentration in both compartments. Let Xn be the concentration in the nucleus, Xc be the concentration in the cytoplasm, Vn be the volume of the nucleus, and Vc be the volume of the cytoplasm. If α is the rate at which Xc decreases because of the movement of X out of the cytoplasm and into the nucleus, so that dXc/dt = −α, then Xn increases at rate (Vc/Vn)α, so that dXn/dt = … + (Vc/Vn)α. Similarly, when X moves from the nucleus to the cytoplasm, the resulting rate of change of Xc is −Vn/Vc that of Xn. To provide a better qualitative fit to our data, we also lowered the rate of IκB synthesis by a factor of 0.2 and the maximum rate at which IKK is activated through the MyD88- and TRIF-dependent pathways by factors of ½ and , respectively. Aside from these adjustments, the equations are as in the original model (24). A Matlab program was developed to (i) vary the model parameters that control the initial (p19) and delayed (p20) waves of IKK activity, the rate of IκB translation (p9), the rate of nuclear import of IκB (p12), and the rate of nuclear export of IκB-p65 (p14); and (ii) visualize how the timing and magnitude of the p65 response varies with these parameters. Because the mathematical model consists of a system of delay differential equations, we generated a solution history for each numerical experiment; the solver was initialized using the constant history provided in Sung et al. (24). The solution was then simulated over a long time interval (20 h) to eliminate transient behavior. In case our intent was to investigate the effect of changing a select kinetic parameter from the downstream pathway (i.e. p9, p12, or p14), at the end of this time interval the parameter of interest was altered (to reflect a potential effect of Cn infection), and the solution was simulated over an additional time interval (20 h) before LPS stimulation was initiated by changing the associated parameters (p19 and p20) to nonzero values. After LPS stimulation the solution was simulated for an additional 10 h (model driver file in supplemental materials).
Statistical Analysis
For the purified GXM experiments, differences in time to maximum amplitude were analyzed using analysis of variance (ANOVA), whereas differences in the maximum amplitude were analyzed using Wilcoxon Rank sums because the data did not fall into a normal distribution, and Wilcoxon Rank sums is a more conservative test. For the intracellular Cn experiments, the data for time to maximum amplitude, maximum amplitude, and length of time p65 was in the nucleus (duration) were transformed into a normal distribution and then analyzed using multivariate ANOVA with simple effects to test for differences between different amounts of intracellular Cn. Differences in translation were analyzed using ANOVA for assays using the Click-iT OPP staining kit and Wilcoxon Rank sums for the ribopuromycylation assays. To determine whether macrophages with high intracellular burden had decreased TNFα expression after the addition of LPS, the data were analyzed using an unpaired Student's t test. For all tests, p < 0.05 was considered significant.
Author Contributions
L. E. H. performed and analyzed the experiments shown in Fig. 1. J. B. H. performed and analyzed the experiments in Figs. 2, 3, and 6–8. W. D. and R. N. L. performed and interpreted the numerical experiments in Figs. 1, 4, and 8. L. M. S. performed and analyzed the experiments in Fig. 5. E. E. M. and D. E. N. conceived, designed, and coordinated the study. J. B. H., W. D., R. N. L., E. E. M., and D. E. N. wrote the paper. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We appreciate the gift of the RAW 264.7 NF-κB reporter cell line from Iain Fraser (National Institutes of Health). We thank Jonathan Logan Bowling (Middle Tennessee State University) for technical assistance, Kirsten Cunningham and Devyn Hayes for help with data analysis, and Drs. Anthony Farone (Middle Tennessee State University) and Eric Batchelor (National Institutes of Health) for useful discussions.
This work was supported by start-up funds from Middle Tennessee State University (to D. E. N. and E. E. M.), by Middle Tennessee State University Faculty Research and Creative Committee (FRCAC) and the Middle Tennessee State University Foundation (Special Projects) funds (to D. E. N.), and scholarships from Middle Tennessee State University Undergraduate Research Center (URECA) and Sigma Xi Grants-in-Aid of Research Program (GIAR) (to L. E. H.). The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental data.
- Cn
- C. neoformans
- CHX
- cyclohexamide
- EGFP
- enhanced GFP
- GXM
- glucuronoxylomannan
- IKK
- IκB kinase
- iNOS
- inducible nitric-oxide synthase
- SHIP
- SH2-containing inositol phosphatase 1
- TLR
- Toll-like receptor
- TRIF
- TIR domain-containing adapter-inducing interferon-β
- TIR
- Toll-interleukin receptor
- OPP
- O-propargyl-puromycin
- nuc:cyto
- nuclear:cytoplasmic
- ANOVA
- analysis of variance.
References
- 1. Lin X., and Heitman J. (2006) The biology of the Cryptococcus neoformans species complex. Annu. Rev. Microbiol. 60, 69–105 [DOI] [PubMed] [Google Scholar]
- 2. Goldman D. L., Khine H., Abadi J., Lindenberg D. J., Pirofski L., Niang R., and Casadevall A. (2001) Serologic evidence for Cryptococcus neoformans infection in early childhood. Pediatrics 107, E66. [DOI] [PubMed] [Google Scholar]
- 3. Park B. J., Wannemuehler K. A., Marston B. J., Govender N., Pappas P. G., and Chiller T. M. (2009) Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS 23, 525–530 [DOI] [PubMed] [Google Scholar]
- 4. Cherniak R., and Sundstrom J. B. (1994) Polysaccharide antigens of the capsule of Cryptococcus neoformans. Infect. Immun. 62, 1507–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chang Y. C., and Kwon-Chung K. J. (1994) Complementation of a capsule-deficient mutation of Cryptococcus neoformans restores its virulence. Mol. Cell. Biol. 14, 4912–4919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fromtling R. A., Shadomy H. J., and Jacobson E. S. (1982) Decreased virulence in stable, acapsular mutants of Cryptococcus neoformans. Mycopathologia 79, 23–29 [DOI] [PubMed] [Google Scholar]
- 7. Feldmesser M., Kress Y., Novikoff P., and Casadevall A. (2000) Cryptococcus neoformans is a facultative intracellular pathogen in murine pulmonary infection. Infect. Immun. 68, 4225–4237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tucker S. C., and Casadevall A. (2002) Replication of Cryptococcus neoformans in macrophages is accompanied by phagosomal permeabilization and accumulation of vesicles containing polysaccharide in the cytoplasm. Proc. Natl. Acad. Sci. U.S.A. 99, 3165–3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Coelho C., Bocca A. L., and Casadevall A. (2014) The intracellular life of Cryptococcus neoformans. Annu. Rev. Pathol. 9, 219–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Monari C., Bistoni F., Casadevall A., Pericolini E., Pietrella D., Kozel T. R., and Vecchiarelli A. (2005) Glucuronoxylomannan, a microbial compound, regulates expression of costimulatory molecules and production of cytokines in macrophages. J. Infect. Dis. 191, 127–137 [DOI] [PubMed] [Google Scholar]
- 11. Piccioni M., Monari C., Kenno S., Pericolini E., Gabrielli E., Pietrella D., Perito S., Bistoni F., Kozel T. R., and Vecchiarelli A. (2013) A purified capsular polysaccharide markedly inhibits inflammatory response during endotoxic shock. Infect. Immun. 81, 90–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vecchiarelli A., Pericolini E., Gabrielli E., Kenno S., Perito S., Cenci E., and Monari C. (2013) Elucidating the immunological function of the Cryptococcus neoformans capsule. Future Microbiol. 8, 1107–1116 [DOI] [PubMed] [Google Scholar]
- 13. Hayden M. S., and Ghosh S. (2012) NF-κB, the first quarter-century: remarkable progress and outstanding questions. Genes Dev. 26, 203–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Baeuerle P. A., and Baltimore D. (1988) IκB: a specific inhibitor of the NF-κB transcription factor. Science 242, 540–546 [DOI] [PubMed] [Google Scholar]
- 15. Scherer D. C., Brockman J. A., Chen Z., Maniatis T., and Ballard D. W. (1995) Signal-induced degradation of IκBα requires site-specific ubiquitination. Proc. Natl. Acad. Sci. U.S.A. 92, 11259–11263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen Z., Hagler J., Palombella V. J., Melandri F., Scherer D., Ballard D., and Maniatis T. (1995) Signal-induced site-specific phosphorylation targets IκBα to the ubiquitin-proteasome pathway. Genes Dev. 9, 1586–1597 [DOI] [PubMed] [Google Scholar]
- 17. Shakhov A. N., Collart M. A., Vassalli P., Nedospasov S. A., and Jongeneel C. V. (1990) κB-type enhancers are involved in lipopolysaccharide-mediated transcriptional activation of the tumor necrosis factor α gene in primary macrophages. J. Exp. Med. 171, 35–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Collart M. A., Baeuerle P., and Vassalli P. (1990) Regulation of tumor necrosis factor α transcription in macrophages: involvement of four κB-like motifs and of constitutive and inducible forms of NF-κB. Mol. Cell. Biol. 10, 1498–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pahl H. L. (1999) Activators and target genes of Rel/NF-κB transcription factors. Oncogene 18, 6853–6866 [DOI] [PubMed] [Google Scholar]
- 20. Morris K. R., Lutz R. D., Choi H. S., Kamitani T., Chmura K., and Chan E. D. (2003) Role of the NF-κB signaling pathway and κB cis-regulatory elements on the IRF-1 and iNOS promoter regions in mycobacterial lipoarabinomannan induction of nitric oxide. Infect. Immun. 71, 1442–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hughes J. E., Srinivasan S., Lynch K. R., Proia R. L., Ferdek P., and Hedrick C. C. (2008) Sphingosine-1-phosphate induces an antiinflammatory phenotype in macrophages. Circ. Res. 102, 950–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nelson D. E., Ihekwaba A. E., Elliott M., Johnson J. R., Gibney C. A., Foreman B. E., Nelson G., See V., Horton C. A., Spiller D. G., Edwards S. W., McDowell H. P., Unitt J. F., Sullivan E., Grimley R., et al. (2004) Oscillations in NF-κB signaling control the dynamics of gene expression. Science 306, 704–708 [DOI] [PubMed] [Google Scholar]
- 23. Nelson D. E., Sée V., Nelson G., and White M. R. (2004) Oscillations in transcription factor dynamics: a new way to control gene expression. Biochem. Soc. Trans. 32, 1090–1092 [DOI] [PubMed] [Google Scholar]
- 24. Sung M. H., Li N., Lao Q., Gottschalk R. A., Hager G. L., and Fraser I. D. (2014) Switching of the relative dominance between feedback mechanisms in lipopolysaccharide-induced NF-κB signaling. Sci. Signal. 7, ra6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ashall L., Horton C. A., Nelson D. E., Paszek P., Harper C. V., Sillitoe K., Ryan S., Spiller D. G., Unitt J. F., Broomhead D. S., Kell D. B., Rand D. A., Sée V., and White M. R. (2009) Pulsatile stimulation determines timing and specificity of NF-κB-dependent transcription. Science 324, 242–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Williams R., Timmis J., and Qwarnstrom E. (2014) Computational models of the NF-κB signalling pathway. Computation 2, 131–158 [Google Scholar]
- 27. Shoham S., Huang C., Chen J. M., Golenbock D. T., and Levitz S. M. (2001) Toll-like receptor 4 mediates intracellular signaling without TNF-α release in response to Cryptococcus neoformans polysaccharide capsule. J. Immunol. 166, 4620–4626 [DOI] [PubMed] [Google Scholar]
- 28. Janbon G., Ormerod K. L., Paulet D., Byrnes E. J. 3rd, Yadav V., Chatterjee G., Mullapudi N., Hon C. C., Billmyre R. B., Brunel F., Bahn Y. S., Chen W., Chen Y., Chow E. W., Coppée J. Y., et al. (2014) Analysis of the genome and transcriptome of Cryptococcus neoformans var. grubii reveals complex RNA expression and microevolution leading to virulence attenuation. PLoS Genet. 10, e1004261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moyrand F., Lafontaine I., Fontaine T., and Janbon G. (2008) UGE1 and UGE2 regulate the UDP-glucose/UDP-galactose equilibrium in Cryptococcus neoformans. Eukaryot. Cell 7, 2069–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cherniak R., Reiss E., and Turner S. H. (1982) A galactoxylomannan antigen of Cryptococcus neoformans serotype A. Carbohydrate Res. 103, 239–250 [Google Scholar]
- 31. Villena S. N., Pinheiro R. O., Pinheiro C. S., Nunes M. P., Takiya C. M., DosReis G. A., Previato J. O., Mendonça-Previato L., and Freire-de-Lima C. G. (2008) Capsular polysaccharides galactoxylomannan and glucuronoxylomannan from Cryptococcus neoformans induce macrophage apoptosis mediated by Fas ligand. Cell. Microbiol. 10, 1274–1285 [DOI] [PubMed] [Google Scholar]
- 32. Dubois M., Gilles K., Hamilton J. K., Rebers P. A., and Smith F. (1951) A colorimetric method for the determination of sugars. Nature 168, 167. [DOI] [PubMed] [Google Scholar]
- 33. Coelho C., Souza A. C., Derengowski L. D., de Leon-Rodriguez C., Wang B., Leon-Rivera R., Bocca A. L., Gonçalves T., and Casadevall A. (2015) Macrophage mitochondrial and stress response to ingestion of Cryptococcus neoformans. J. Immunol. 194, 2345–2357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. David A., Dolan B. P., Hickman H. D., Knowlton J. J., Clavarino G., Pierre P., Bennink J. R., and Yewdell J. W. (2012) Nuclear translation visualized by ribosome-bound nascent chain puromycylation. J. Cell Biol. 197, 45–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J. Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., and Cardona A. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hoffmann A., Levchenko A., Scott M. L., and Baltimore D. (2002) The IκB-NF-κB signaling module: temporal control and selective gene activation. Science 298, 1241–1245 [DOI] [PubMed] [Google Scholar]
- 37. Hardison S. E., Ravi S., Wozniak K. L., Young M. L., Olszewski M. A., and Wormley F. L. Jr. (2010) Pulmonary infection with an interferon-γ-producing Cryptococcus neoformans strain results in classical macrophage activation and protection. Am. J. Pathol. 176, 774–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cheshire J. L., and Baldwin A. S. Jr. (1997) Synergistic activation of NF-κB by tumor necrosis factor α and γ interferon via enhanced IκBα degradation and de novo IκBβ degradation. Mol. Cell. Biol. 17, 6746–6754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Casadevall A., Cleare W., Feldmesser M., Glatman-Freedman A., Goldman D. L., Kozel T. R., Lendvai N., Mukherjee J., Pirofski L. A., Rivera J., Rosas A. L., Scharff M. D., Valadon P., Westin K., and Zhong Z. (1998) Characterization of a murine monoclonal antibody to Cryptococcus neoformans polysaccharide that is a candidate for human therapeutic studies. Antimicrob. Agents Chemother. 42, 1437–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. García-Rivera J., Chang Y. C., Kwon-Chung K. J., and Casadevall A. (2004) Cryptococcus neoformans CAP59 (or Cap59p) is involved in the extracellular trafficking of capsular glucuronoxylomannan. Eukaryot. Cell 3, 385–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Al-Mehdi A. B., Pastukh V. M., Swiger B. M., Reed D. J., Patel M. R., Bardwell G. C., Pastukh V. V., Alexeyev M. F., and Gillespie M. N. (2012) Perinuclear mitochondrial clustering creates an oxidant-rich nuclear domain required for hypoxia-induced transcription. Sci. Signal. 5, ra47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Murata T., Goshima F., Daikoku T., Inagaki-Ohara K., Takakuwa H., Kato K., and Nishiyama Y. (2000) Mitochondrial distribution and function in herpes simplex virus-infected cells. J. Gen. Virol. 81, 401–406 [DOI] [PubMed] [Google Scholar]
- 43. Ohshima D., Inoue J., and Ichikawa K. (2012) Roles of spatial parameters on the oscillation of nuclear NF-κB: computer simulations of a 3D spherical cell. PLoS One 7, e46911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kagan J. C., Su T., Horng T., Chow A., Akira S., and Medzhitov R. (2008) TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon-β. Nat. Immunol. 9, 361–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Arenzana-Seisdedos F., Thompson J., Rodriguez M. S., Bachelerie F., Thomas D., and Hay R. T. (1995) Inducible nuclear expression of newly synthesized IκBα negatively regulates DNA-binding and transcriptional activities of NF-κB. Mol. Cell. Biol. 15, 2689–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rodriguez M. S., Thompson J., Hay R. T., and Dargemont C. (1999) Nuclear retention of IκBα protects it from signal-induced degradation and inhibits nuclear factor κB transcriptional activation. J. Biol. Chem. 274, 9108–9115 [DOI] [PubMed] [Google Scholar]
- 47. Bergqvist S., Alverdi V., Mengel B., Hoffmann A., Ghosh G., and Komives E. A. (2009) Kinetic enhancement of NF-κBxDNA dissociation by IκBα. Proc. Natl. Acad. Sci. U.S.A. 106, 19328–19333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sung M. H., Salvatore L., De Lorenzi R., Indrawan A., Pasparakis M., Hager G. L., Bianchi M. E., and Agresti A. (2009) Sustained oscillations of NF-κB produce distinct genome scanning and gene expression profiles. PLoS One 4, e7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mukherjee S., Keitany G., Li Y., Wang Y., Ball H. L., Goldsmith E. J., and Orth K. (2006) Yersinia YopJ acetylates and inhibits kinase activation by blocking phosphorylation. Science 312, 1211–1214 [DOI] [PubMed] [Google Scholar]
- 50. Ashida H., Mimuro H., Ogawa M., Kobayashi T., Sanada T., Kim M., and Sasakawa C. (2011) Cell death and infection: a double-edged sword for host and pathogen survival. J. Cell Biol. 195, 931–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ben-Abdallah M., Sturny-Leclère A., Avé P., Louise A., Moyrand F., Weih F., Janbon G., and Mémet S. (2012) Fungal-induced cell cycle impairment, chromosome instability and apoptosis via differential activation of NF-κB. PLoS Pathogens 8, e1002555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rajasingham R., Williams D., Meya D. B., Meintjes G., Boulware D. R., and Scriven J. (2014) Nosocomial drug-resistant bacteremia in 2 cohorts with cryptococcal meningitis, Africa. Emerg. Infect Dis. 20, 722–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Powderly W. G., Cloud G. A., Dismukes W. E., and Saag M. S. (1994) Measurement of cryptococcal antigen in serum and cerebrospinal fluid: value in the management of AIDS-associated cryptococcal meningitis. Clin. Infect. Dis. 18, 789–792 [DOI] [PubMed] [Google Scholar]
- 54. Ghosh C. C., Ramaswami S., Juvekar A., Vu H. Y., Galdieri L., Davidson D., and Vancurova I. (2010) Gene-specific repression of proinflammatory cytokines in stimulated human macrophages by nuclear IκBα. J. Immunol. 185, 3685–3693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dalloneau E., Pereira P. L., Brault V., Nabel E. G., and Hérault Y. (2011) Prmt2 regulates the lipopolysaccharide-induced responses in lungs and macrophages. J. Immunol. 187, 4826–4834 [DOI] [PubMed] [Google Scholar]
- 56. Ganesh L., Yoshimoto T., Moorthy N. C., Akahata W., Boehm M., Nabel E. G., and Nabel G. J. (2006) Protein methyltransferase 2 inhibits NF-κB function and promotes apoptosis. Mol. Cell. Biol. 26, 3864–3874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yeung F., Hoberg J. E., Ramsey C. S., Keller M. D., Jones D. R., Frye R. A., and Mayo M. W. (2004) Modulation of NF-κB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 23, 2369–2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ashburner B. P., Westerheide S. D., and Baldwin A. S. Jr. (2001) The p65 (RelA) subunit of NF-κB interacts with the histone deacetylase (HDAC) corepressors HDAC1 and HDAC2 to negatively regulate gene expression. Mol. Cell. Biol. 21, 7065–7077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Moreira D., Rodrigues V., Abengozar M., Rivas L., Rial E., Laforge M., Li X., Foretz M., Viollet B., Estaquier J., Cordeiro da Silva A., and Silvestre R. (2015) Leishmania infantum modulates host macrophage mitochondrial metabolism by hijacking the SIRT1-AMPK axis. PLoS Pathogens 11, e1004684. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.