Abstract
The bacterium Bradyrhizobium japonicum USDA110 does not synthesize siderophores for iron utilization in aerobic environments, and the mechanism of iron uptake within symbiotic soybean root nodules is unknown. An mbfA bfr double mutant defective in iron export and storage activities cannot grow aerobically in very high iron medium. Here, we found that this phenotype was suppressed by loss of function mutations in the feoAB operon encoding ferrous (Fe2+) iron uptake proteins. Expression of the feoAB operon genes was elevated under iron limitation, but mutants defective in either gene were unable to grow aerobically over a wide external ferric (Fe3+) iron (FeCl3) concentration range. Thus, FeoAB accommodates iron acquisition under iron limited and iron replete conditions. Incorporation of radiolabel from either 55Fe2+ or 59Fe3+ into cells was severely defective in the feoA and feoB strains, suggesting Fe3+ reduction to Fe2+ prior to traversal across the cytoplasmic membrane by FeoAB. The feoA or feoB deletion strains elicited small, ineffective nodules on soybean roots, containing few bacteria and lacking nitrogen fixation activity. A feoA(E40K) mutant contained partial iron uptake activity in culture that supported normal growth and established an effective symbiosis. The feoA(E40K) strain had partial iron uptake activity in situ within nodules and in isolated cells, indicating that FeoAB is the iron transporter in symbiosis. We conclude that FeoAB supports iron acquisition under limited conditions of soil and in the iron-rich environment of a symbiotic nodule.
Keywords: bacteria, iron, nitrogen fixation, symbiosis, transport metal, BRADYRHIZOBIUM
Introduction
Iron is an essential nutrient involved in many biological processes, including the TCA cycle, electron transport, detoxification, signal transduction, nucleic acid synthesis, gene regulation, and nitrogen fixation (1). The bioavailability of iron is low in aerobic environments because it is oxidized in the ferric (Fe3+) form and therefore highly insoluble. High affinity iron acquisition systems are expressed under iron limitation to scavenge the metal.
To acquire ferric iron, many bacteria synthesize and secrete low molecular weight iron chelators called siderophores and then import the Fe3+-bound chelate back into the cell (2). In Gram-negative bacteria, this uptake system includes an outer membrane receptor, a periplasmic protein, and an inner membrane ABC transporter (3). Some bacteria can also use other iron chelates such as siderophores produced by other microbes or heme (4, 5). The genes encoding siderophore or heme uptake proteins are induced under iron limitation, and iron uptake under conditions where the metal is replete is not well described.
Rhizobia bacteria live as free living soil organisms or as an endosymbiont within legume root nodule cells, where they fix atmospheric nitrogen to ammonia to fulfill the nitrogen requirement of the host. Iron is essential for rhizobia-legume symbioses for the nitrogenase enzymes and ancillary processes associated with it (6). However, bacterial iron transporters needed to establish symbiosis and function within nodule plant cells have not been elucidated. Some rhizobial species synthesize siderophores and the cognate transport proteins, but mutations in the corresponding genes do not affect the establishment or maintenance of symbiosis (7–10).
Bradyrhizobium japonicum USDA110 does not synthesize siderophores (11), but free living cells can utilize heme as an iron source (12) and also siderophores (xenosiderophores) produced by other microorganisms (13, 14). In addition, many bacteria, including the Rhizobia, grow well aerobically in culture with Fe3+ salts, independent of siderophores, but the mechanism of uptake is unknown in most cases. A ferrichrome receptor mutant of B. japonicum strain 61A152 does not establish an effective symbiosis on soybean, but ferrichrome is a fungal siderophore not available to B. japonicum in nodules (15). Thus, those authors suggest that the fegA gene plays some other role. In addition a fegA mutant of B. japonicum USDA122 has no symbiotic phenotype (14).
Within infected plant cells of nodules, the symbiotic bacteria, called bacteroids, are surrounded by a plant membrane to form the symbiosome (6). Expression of the soybean divalent metal transporter GmDMT1 is enhanced in the symbiosome membrane (16), and the soybean symbiosome membrane has ferric reductase activity (17). This suggests that iron traverses the symbiosome membrane in the ferrous (Fe2+) form in situ.
Ferrous iron (Fe2+) is soluble and stable under anaerobic and microaerobic conditions. FeoB is a ferrous iron transporter that is widely distributed among bacteria and archaea (18). The feoB gene is usually found in an operon with feoA, which encodes an auxiliary protein necessary for ferrous iron uptake, but its exact role is unknown (18). As expected, bacterial feoB mutants have a growth phenotype in low O2 environments, but not under aerobic conditions (18–23).
In the present study, we found that FeoAB is essential for aerobic growth and Fe3+ utilization in free living cells over a wide external iron concentration range and for symbiosis with soybean. Thus, B. japonicum employs a single iron transporter to adapt to diverse environmental conditions.
Results
Identification of Secondary Site Mutations That Suppress the Growth Phenotype of an mbfA bfr Double Mutant
MbfA is the major iron exporter in B. japonicum (24), and Bfr is an iron storage protein needed to control iron toxicity in that organism (25). In a previous study, we demonstrated that an mbfA bfr double mutant has a severe growth phenotype in medium supplemented with a high iron concentration (25). Cells of the mbfA bfr strain grown to mid-log phase in liquid culture did not grow when spotted on solid medium containing 1.65 mm FeSO4, whereas the wild type, bfr, and mbfA strains were able to grow (supplemental Fig. S1A).
In a search for secondary site mutations that suppressed this growth phenotype, mbfA bfr cells were plated onto solid medium containing 1.65 mm FeSO4 and selected for colonies that arose spontaneously. Ten colonies were scored, and four were pursued further by genotyping. The four colonies were rescored using the spotting assay as described above. The suppressor strains grew similarly to the wild type and single mutants (supplemental Fig. S1A). Interestingly, suppressors 6, 8, and 9 were unable to grow on plates containing 50 μm FeSO4 (supplemental Fig. S1B), suggesting that those mutants not only tolerated high iron but required it. Suppressors 6, 8, 9, and 11 were able to grow in medium containing 100 nm heme as the iron source, and therefore all strains were grown in this manner for maintenance and for primary cultures used in inoculations.
The Suppressor Mutants Harbor Loss of Function Alleles in the Ferrous Iron Transporter Genes feoA or feoB
The wild type strain USDA110, the mbfA bfr double mutant, and the four suppressor strains were genotyped by whole genome DNA sequencing, and mutated regions were verified by resequencing PCR-amplified fragments around the lesions. Suppressor strains 6, 8, and 9 have mutations within the coding region of the putative feoB gene (blr6523) encoding the ferrous (Fe2+) cytoplasmic (inner) membrane transporter FeoB (supplemental Fig. S2 A). The mutations include an in-frame deletion, a transposon insertion, and a nonsense mutation (supplemental Fig. S2 B). Suppressor 11 contains a missense mutation within the feoA gene resulting in a Glu-40 to Lys substitution. The feoA gene is immediately upstream of feoB that likely forms an operon (supplemental Fig. S2A). FeoA is an accessory protein also involved in Fe2+ uptake, but its exact function is not known (18, 26).
The feoB(Y454term) and feoA(E40K) alleles of suppressor 9 and 11, respectively, were reconstructed in an mbfA bfr background, and the suppressor phenotypes were confirmed (supplemental Fig. S3). Both of these gene variants were shown to be loss of function alleles based on the following: Partial diploids were constructed in an mbfA bfr background that harbored both the wild type and the mutant feo alleles (S9+ and S11+). Both strains grew in media with 50 μm FeSO4, but not with 1.65 mm FeSO4, (supplemental Fig. S3), similar to the mbfA bfr parent strain. Thus, the wild type feo genes were dominant over the suppressor alleles. In addition, feoA and feoB deletions mutants were constructed in the mbfA bfr background, and each mutant recapitulated the suppressor phenotype of growth in media containing 1.65 mm FeSO4 (supplemental Fig. S3). It is likely that a defect in iron uptake activity counteracted iron toxicity of the mbfA bfr mutant in high iron medium leading to the suppressor phenotype.
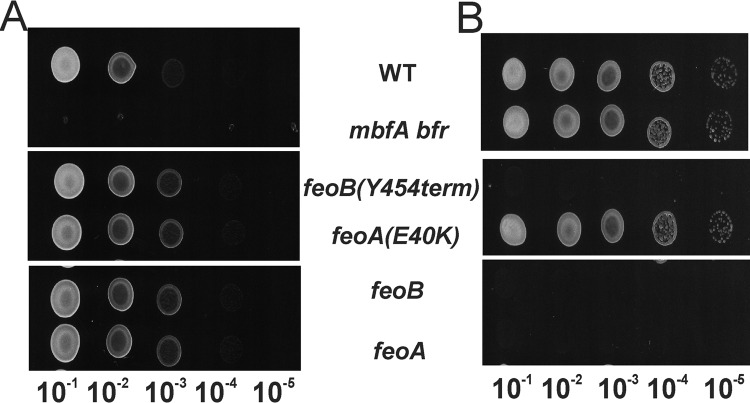
The feoA and feoB deletions, as well as the feoB(Y454term) and feoA(E340K) alleles, were each constructed in wild type (mbfA+ bfr+) backgrounds and characterized by spotting assays (Fig. 1). feoA, feoB, and feoB(Y454term) grew in solid medium containing 1.65 mm FeSO4, but not 50 μm FeSO4, which suggests a defect in iron acquisition. The feoA(E40K) mutant did not have a discernible growth phenotype on plates in the wild type background but did in the mbfA bfr background.
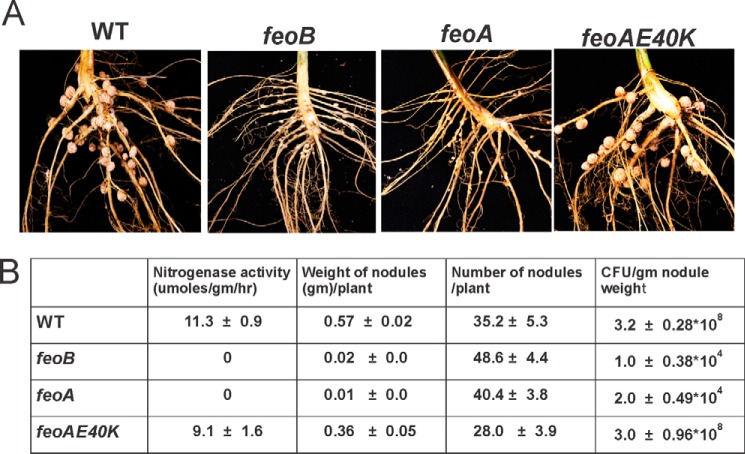
FIGURE 1.
Growth of mutants harboring feoA or feoB suppressor alleles or deletions in the wild type background. Cells of mutant strains harboring feoB(Y454term), feoA(E40K) ΔfeoA, and ΔfeoB grown in liquid cultures supplemented with 100 nm heme were serial diluted 10−1–10−5-fold, and 10 μl was spotted on plates containing either 1.65 mm FeSO4 (A) or 50 μm FeSO4 (B). Each panel represents a single plate, and the image was separated for clarity of presentation.
feoA and feoB Are Required for Aerobic Growth with Ferric Iron in the Growth Medium over a Wide Concentration Range of Iron
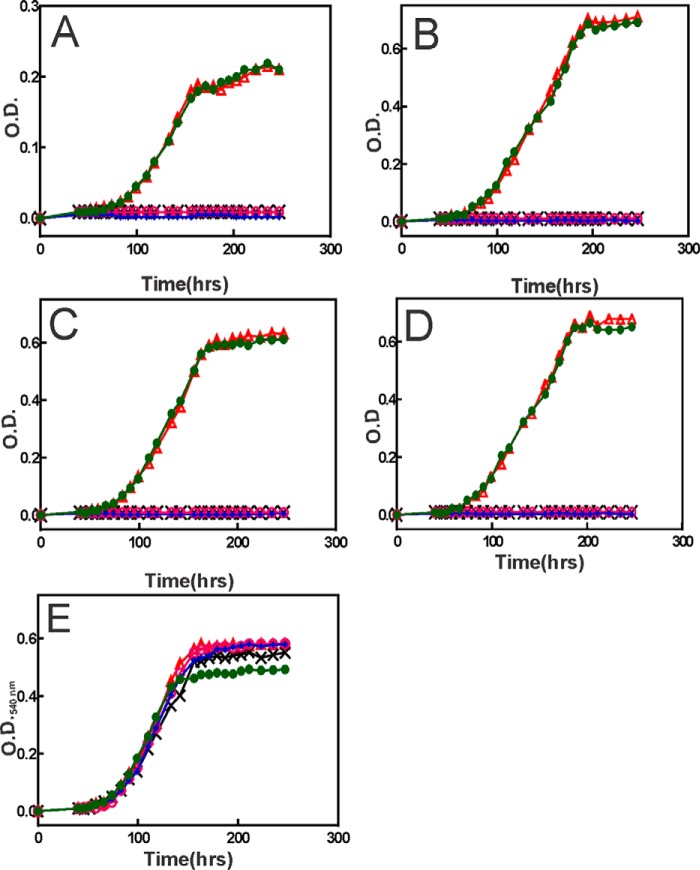
Iron is predominantly in the ferric (Fe3+) form in aerobic environments, and all experiments in this study were carried out aerobically. Therefore, it was not expected that the selection would yield mutations in a ferrous iron transport system. Although reduced iron (FeSO4) was used in the plates (supplemental Figs. S1 and S2 and Fig. 1), we assumed that it would oxidize in air. Thus, we measured aerobic growth of the wild type and mutant strains in minimal medium using ferric iron (FeCl3) as the nutritional iron source (Fig. 2). The feoA and feoB mutants examined were constructed in wild type backgrounds.
FIGURE 2.
Requirement of feoA and feoB for aerobic growth with ferric iron in the growth medium over a wide concentration range of iron. Growth medium was inoculated with 1 × 106 cells/ml of the wild type (green, closed circles), feoB (magenta, open circles), feoA (blue, closed diamonds), feoB(Y454term) (black, crosses), and feoA(E40K) (red, open triangles) strains. Strains were grown in minimal medium supplemented with either no added iron (A), 5 μm FeCl3 (B), 50 μm FeCl3 (C), 500 μm FeCl3 (D), or 0.5 μm heme (E). Aliquots were taken at the indicated time points, and the optical density was measured at 540 nm (A540).
The wild type grew in minimal medium with no supplemented iron, but only to an A540 of ∼0.2. By contrast, the feoA, feoB, and feoB(Y454term) mutants showed no growth in that medium (Fig. 2A). The feoA(E40K) mutant grew similar to the wild type, suggesting a leaky phenotype that will be discussed further below. Similar results were observed when cells were grown in GSY medium, except that the wild type and feoA(E40K) cells grew to a higher A540, presumably because of contaminating iron (0.3 μm) in the medium (supplemental Fig. S4). The addition of 5, 50, or 500 μm FeCl3 to the growth medium did not rescue growth of the feoA, feoB, or feoB(Y454term) mutants, although the wild type grew well with iron supplementation (Fig. 2, B–D). Heme can be used as an iron source in B. japonicum (12), is known to be taken up by the Hmu system (27), and does not utilize FeoAB. Accordingly, the mutants grew well in medium supplemented with 500 nm heme (Fig. 2E).
We conclude that the ferrous iron transport system FeoAB is required for ferric iron utilization. Moreover, FeoAB appears to be the major transporter over a wide concentration of ferric iron in the medium. This latter point is noteworthy because high affinity iron transport systems are usually expressed under iron limitation, but not under iron replete conditions (14, 28). By contrast, feoA and feoB are needed under severe iron limitation (Fig. 2A), as well as under iron-replete conditions (Fig. 2, B–D).
feoA and feoB Are Required for Iron Uptake with Both Ferrous and Ferric Iron as Substrates
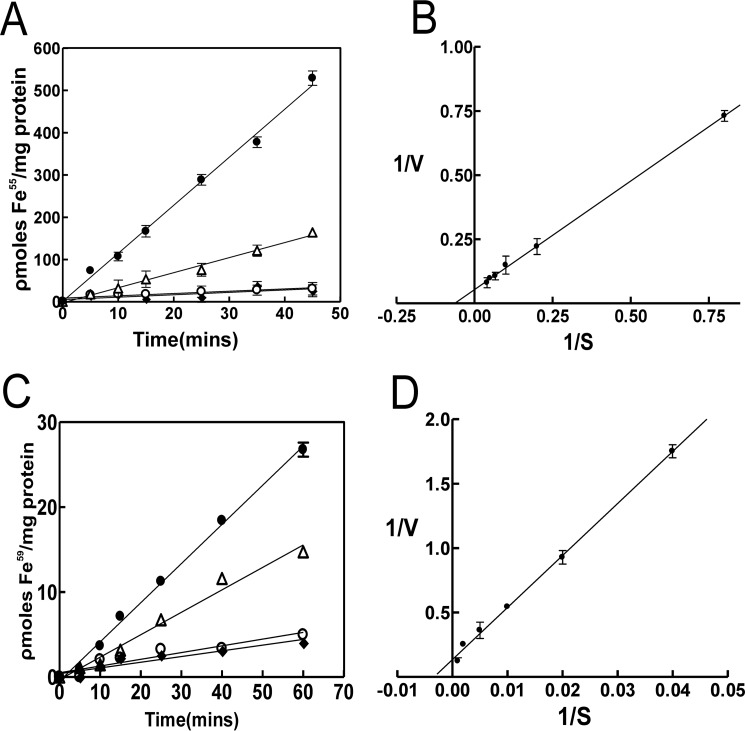
The feoB gene is widespread among prokaryotes, and FeoB is known to be an inner membrane Fe2+ transporter (18). However, it is shown here to be required for growth with ferric iron. Thus, we measured iron uptake activity in the wild type and mutants using Fe2+ or Fe3+ as substrate (Fig. 3).
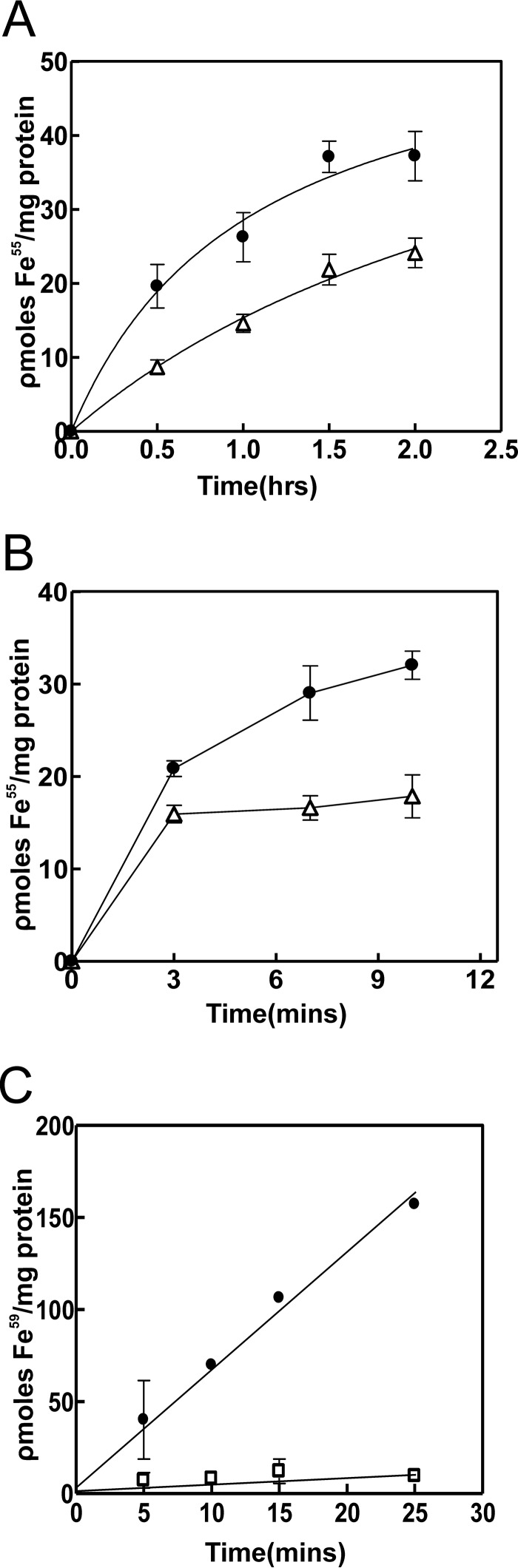
FIGURE 3.
Requirement of feoA and feoB for Fe2+ and Fe3+ uptake by B. japonicum. A, cells of the wild type (closed circles), feoB (open circles), feoA (closed diamonds), and feoA(E40K) (open triangles) strains were grown in modified GSY medium with 100 nm heme. At time 0, 25 nm 55Fe2+ was added to the assay medium, and aliquots were subsequently taken at various time points and counted. Each time point is the average of three biological replicate samples ± S.D. (error bars). B, Lineweaver-Burk plot of initial velocity−1 (10 min) of uptake of 55Fe2+ by wild type cells as a function of the starting iron concentration−1, ranging from 1.25 to 25 nm substrate. C, cells of the wild type (closed circles), feoB (open circles), feoA (closed diamonds), and feoA(E40K) (open triangles) strains were grown in modified GSY medium with 100 nm heme. At time 0, 50 nm 59Fe3+ was added to the assay medium, and aliquots were subsequently taken at various time points and counted. Each time point is the average of triplicate samples ± S.D. (error bars). D, Lineweaver-Burk plot of initial velocity−1 (10 min) of uptake of 59Fe3+ by wild type cells as a function of the starting iron concentration−1, ranging from 25 to 1000 nm substrate.
55Fe2+ uptake observed in the wild was almost completely abolished in the feoB and the feoA deletion strains (Fig. 3A). Thus, as expected, FeoAB is required for uptake of ferrous iron by B. japonicum. The initial uptake of 55Fe2+ uptake by wild type cells was measured as a function of the initial substrate concentration, and the data are presented as a Lineweaver-Burk plot (Fig. 3B). The average apparent Km value of three trials was 16.8 ± 3.2 nm.
Iron uptake by the wild type was readily detectable with 59Fe3+ as substrate but was severely defective in the feoB and the feoA deletion strains (Fig. 3C). These observations are consistent with the dependence on feoA and feoB for aerobic growth with ferric iron (Fig. 2). Fe3+ is likely reduced to Fe2+ in the medium or in the periplasm and subsequently transported across the inner membrane by FeoB. The initial uptake of 59Fe3+ uptake by wild type cells was measured as a function of the initial substrate concentration, and the data are presented as a Lineweaver-Burk plot (Fig. 3D). The average apparent Km value of three trials was 278 ± 18 nm. This higher value compared with Fe2+ may reflect a reduction step, differences in mechanisms of traversing the outer membrane, or some other variable.
Uptake of 55Fe2+ or 59Fe3+ by the feoA(E40K) strain was diminished but substantially greater than the deletion strains (Fig. 3). This diminished activity is sufficient to support aerobic growth in liquid medium, which likely explains the ability of that mutant to grow in liquid medium (Fig. 2 and supplemental Fig. S4).
The feoA and feoB Genes Are Regulated by Iron
The growth curve data (Fig. 2 and supplemental Fig. S4) suggest that FeoAB is the primary iron transporter in B. japonicum over a wide concentration range of iron. Thus, we monitored feoA and feoB mRNA in cells grown at various iron concentrations to determine whether these genes were regulated by iron (Fig. 4). The transcript levels of feoA and feoB were ∼5-fold higher in cells grown in medium with no added iron when compared with cells grown in 10 or 20 μm FeCl3 (Fig. 4) and dropped even lower in medium with 100 μm FeCl3. Thus, like other high affinity iron transport systems, the FeoAB system is maximally expressed under iron limitation. However, the lower levels of expression of this system at the higher iron concentrations must be sufficient to support growth because these genes are essential under those conditions.
FIGURE 4.
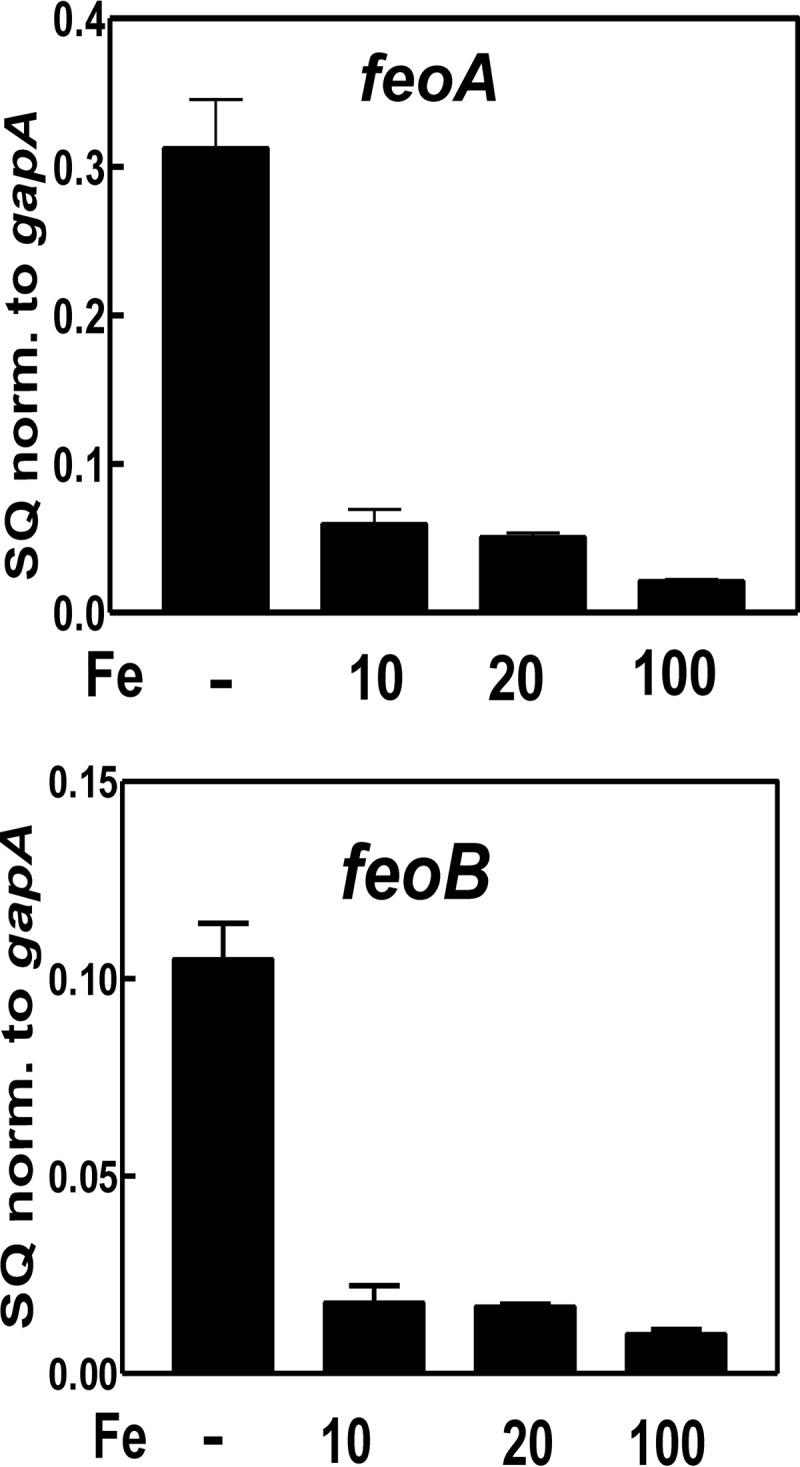
Regulation of feoA and feoB by iron. The steady-state transcript levels of feoA and feoB in the wild type strain grown in modified GSY medium with either no added iron, 10 μm FeCl3, 20 μm FeCl3, or 100 μm FeCl3 were analyzed by qualitative real time PCR. The data are expressed as relative starting quantities (SQ) of respective mRNAs normalized to the housekeeping gene gapA and are presented as averages of three replicates ± S.D. (error bars).
Iron-responsive Gene Expression Is Aberrant in feoA and feoB Mutants
The feoA and feoB deletion mutants grew well in minimal (Fig. 2E) or GSY (supplemental Fig. S4E) medium supplemented with 500 nm heme as the iron source, but not in its absence. We wanted to assess whether the feoA and feoB mutants had other phenotypes associated with iron limitation with heme despite the good growth.
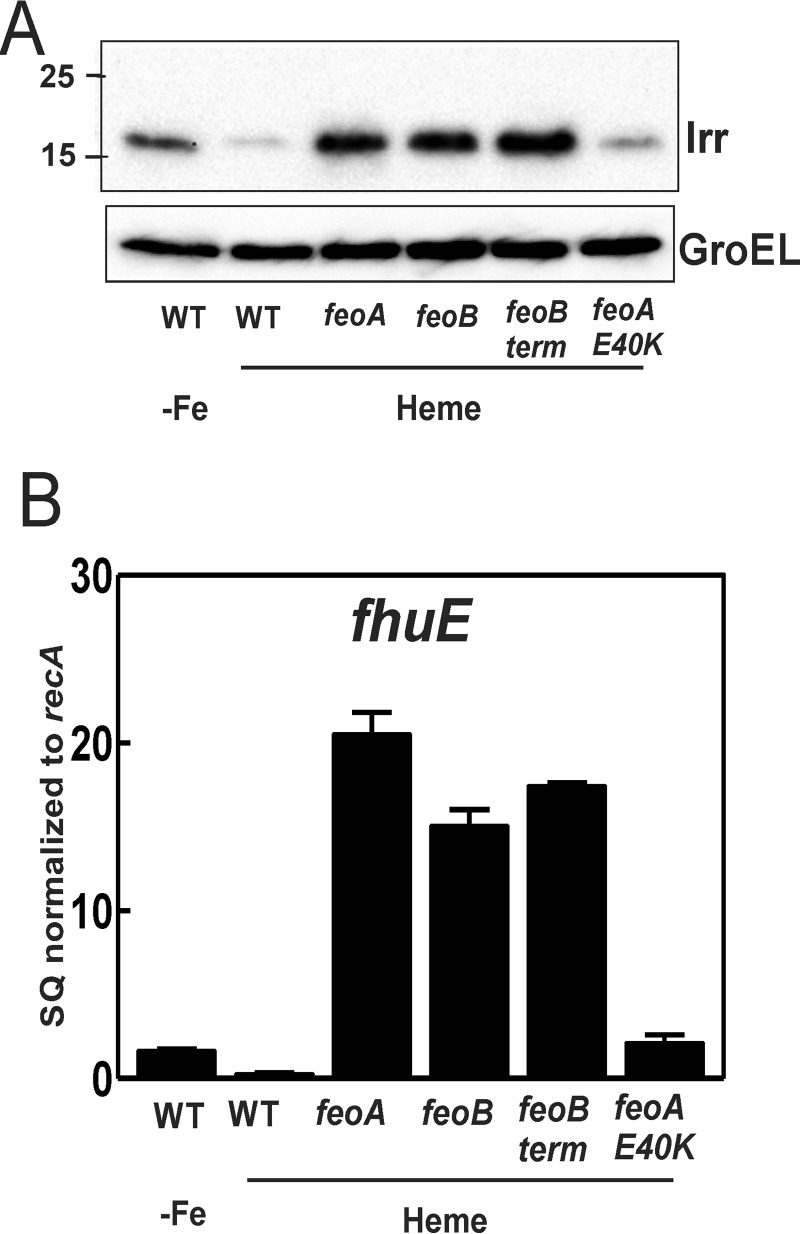
Irr is the global iron-responsive transcriptional regulator in B. japonicum and related bacteria (28). Irr accumulates under iron limitation but degrades in response to the metal in a heme-dependent manner (29). We monitored Irr levels by Western blot analysis in cells of the wild type and mutant strains (Fig. 5A). Irr accumulation of wild type cells grown in modified GSY medium with heme as the only added iron source was diminished compared with cells grown in medium with no iron supplementation, which is expected because iron causes Irr degradation. Cells of the feoA and feoB deletion mutants, as well as feoB(Y454term), grown in heme-supplemented medium accumulated much more Irr than the wild type grown under the same condition, consistent with a lower cellular iron content. Thus, heme enters cells and can rescue growth in the mutants, but the iron level attained is not sufficient to degrade Irr. Clearly, the non-heme contaminating iron in the medium (0.3 μm as determined by atomic absorption) contributes to the cellular iron content in the wild type, but the feo mutants do not have access to it. In previous studies, GSY or similar medium without added iron has been used as the low iron conditions sufficient to induce Irr accumulation in cells (30). The current work suggests that B. japonicum can tolerate substantially lower cellular iron levels. The Irr level in the feoA(E40K) was comparable with the wild type, consistent with other leaky phenotypes of this mutant.
FIGURE 5.
Aberrant regulation of iron-dependent gene expression. A, Western blot analysis of Irr was performed on wild type strain grown in modified GSY medium supplement with no added iron (-Fe) and on wild type, feoB, feoA, feoB(Y454), and feoA(E40K) strains grown in modified GSY medium with 100 nm heme. The protein was detected using anti-Irr antibodies. GroEL was used as a control for a protein not regulated by iron, and it was detected using anti-GroEL antibodies. 15 μg of protein were loaded per lane. B, steady-state transcript levels of fhuE the wild type strain grown in modified GSY medium supplemented with no added iron (-Fe) and the wild type, feoB, feoA, feoB(Y454), and feoA(E40K) strains grown in modified GSY medium with 100 nm heme were analyzed by qualitative real time PCR. The data are expressed as relative starting quantities (SQ) of respective mRNAs normalized to the gene recA and are presented as averages of three biological replicates ± S.D. (error bars).
Irr activates the fhuE gene, and transcript was measured by quantitative real time PCR (Fig. 5B). fhuE levels were over 15-fold greater in the feo mutants that showed elevated Irr levels compared with the wild type. The feoA(E40K) mutant also showed elevated fhuE levels but to a lesser extent. The findings show that FeoAB is needed to maintain iron homeostasis under low iron conditions.
feoA and feoB Are Essential for Symbiosis with Soybean
To determine whether feoA and feoB are essential for symbiosis, soybean seedlings were inoculated with wild type and mutant strains and grown for 28 days in nitrogen-free soil. Strains feoB and feoA incited small, poorly developed nodules on soybean plants (Fig. 6A). The nodules contained few viable bacteria, which could not fix nitrogen as measured by acetylene reduction activity (Fig. 6B). The general appearance of 28-day-old plants inoculated with either feoB or feoA strain were identical to that of the uninoculated plant controls, consistent with the lack of nitrogen fixation activity (data not shown). These observations show that feoA and feoB are essential for establishing symbiosis with soybean.
FIGURE 6.
feoA and feoB are essential for symbiosis with soybean. A, roots of 28-day-old soybean plants, along with nodules incited by either wild type, feoB, feoA, and feoA(E40K) strains. All pictures were taken with lens fixed at the same distance and magnification. B, table listing the symbiotic properties of soybean nodules incited by the wild type, feoB, feoA, and feoA(E40K) strains. The data presented are averages of data obtained from 10 plants from each strain ± S.D.
FeoAB Is Involved in Iron Transport in Symbiosis
The route of iron entry into symbiotic bacteroids is unknown for any Rhizobium-legume symbiosis. The inability of the feoA or feoB strain to establish symbiosis does not directly address this question because those genes may be essential for infection and development of the nodule before symbiosis is established. Accordingly, the lack of bacteria within nodules elicited by the mutants prevents a direct measurement of iron uptake. The feoA(E40K) mutant has diminished but measurable iron uptake activity in free living cells (Fig. 3), grows well in culture (Fig. 2), and shows normal iron-responsive gene expression (Fig. 5). Importantly, it also elicited nitrogen fixing nodules on soybean (Fig. 6), thereby providing a tool to directly address the role of FeoAB on iron transport.
Roots with attached nodules were incubated in medium containing 5 μm Fe2+ and harvested at various time points. The bacteroids were then isolated from the nodules, and radiolabel was measured (Fig. 7A). Radiolabel was taken up by the feoA(E40K) mutant within nodules, but at a lower rate, similar to what was observed in free living cells. Radiolabel found within the plant cytosol fraction was almost identical in nodules elicited by the wild type or feoA(E40K) strain (supplemental Fig. S6), suggesting that the lower rate of iron uptake by feo(E40K) bacteroids within nodules was not limited by entry into nodule plant cells. To address this further, we measured 55Fe2+ uptake by isolated bacteroids and found that the feoA(E40K) strain had lower uptake activity than the wild type (Fig. 7B). We conclude that the FeoAB transport system participates in iron uptake within symbiotic bacteroids.
FIGURE 7.
Bacterial iron transport in nodules in situ and in isolated bacteroids. A, roots with attached nodules incited by either the wild type strain (closed circles) or feoA(E40K) strain (open triangles) were incubated with 5 μm 55Fe2+ at time 0. At each time point, root with attached nodules was harvested, and the radiolabel from isolated bacteroids was counted. B, bacteroids were isolated from nodules incited by either the wild type strain (closed circles) or feoA(E40K) strain (open triangles) and were resuspended in assay medium. At time 0, 25 nm 55Fe2+ was added to the assay medium, and aliquots were subsequently taken at various time points and counted. C, wild type strain grown in modified GSY medium supplemented with 100 nm heme (closed circles) or bacteroids isolated from nodules incited by the wild type strain (open squares) were suspended in assay medium. At time 0, 1 μm 59Fe3+ was added to the assay medium, and aliquots were subsequently taken at various time points and counted. Each time point is the average of three biological replicates ± S.D. (error bars).
We also measured ferric (Fe3+) uptake by isolated bacteroids of the wild type but found almost no activity, even when using 1 μm 59Fe3+ as substrate (Fig. 7C). This differed from free living cells, which utilized Fe3+ (Fig. 3A), suggesting a ferric reductase activity in those cells that is absent in symbiotic bacteroids.
Discussion
B. japonicum resides in the iron-limited conditions of aerobic soils and also within the iron-rich milieu of soybean root nodules. In the present study, we show that the ferrous iron transporter FeoAB supports iron acquisition under these diverse conditions. FeoAB is required for aerobic growth using ferric iron as a nutritional iron source. In addition, B. japonicum does not employ different high affinity and low affinity transporters to support growth under iron-limited and iron-replete conditions, respectively, but instead, FeoAB is the primary iron transporter for over a wide range of external iron concentrations. Moreover, our findings support the conclusion that FeoAB functions as the iron transporter within plant cells of symbiotic root nodules.
B. japonicum does not produce siderophores for ferric iron utilization, and the current study reveals FeoB as the inner membrane component for uptake. The inability of feoA or feoB mutants to grow aerobically in culture or incorporate radiolabel from 59Fe3+ strongly suggests that Fe3+ is reduced to Fe2+ extracellularly or within the periplasm for uptake by FeoB. The inner membrane-bound ferric reductase FrcB was previously identified in B. japonicum, but an frcB mutant does not have a growth phenotype (31). Helicobacter pylori does not synthesize siderophores, and an feoB mutant of that species is deficient in iron uptake using radiolabeled Fe3+ or Fe2+ as substrate, similar to what we observed for B. japonicum (23). However, ferric iron sources support growth of the feoB strain (23). The FtrABCD system of Burkholderia cenocepacia is required for Fe3+ utilization under iron-limiting conditions (32), whereas the homologous systems in Bordetella (33) and Brucella abortus (34) use it for Fe2+ utilization, but it is presumably first oxidized to Fe3+ for uptake across the cytoplasmic membrane.
High affinity iron uptake systems are often induced by iron limitation, and this is also true for B. japonicum. For example, genes needed for the utilization of heme or xenosiderophores (siderophores made by other microbes) are induced in cells grown in low iron medium (28). The feoA and feoB genes were induced under that condition as well (Fig. 4) and were required for growth (Fig. 2 and supplemental Fig. S4). However, the FeoAB transporter is also needed under high iron conditions, and in fact the genes were identified in a selection under high iron conditions (supplemental Fig. S1). feoA and feoB transcripts sharply declined with increasing cellular exposure to iron (Fig. 4), but those levels must be sufficient for iron acquisition. Comparative transcriptome analysis between free living and symbiotic B. japonicum cells suggests that bacteroids have a relatively high iron content (35), which agrees with direct measurements of the peribacteroid space in soybean nodules (36). The feo genes must be able to support viability in the symbiotic milieu. Thus, B. japonicum uses the same iron transporter in the low iron environment expected to occur in soils and the high environment of the nodule.
The importance of iron to nitrogen fixation in Rhizobium-legume symbioses has long been recognized in its role in the nitrogenase enzymes and ancillary functions. Screens for nodulation-deficient B. japonicum mutants have been carried out previously (37, 38), but neither feoA nor feoB were identified. Based on the current work, it is very likely that the population of mutants used to inoculate plants did not include feoA or feoB mutants because they would not be viable in the culture medium used to propagate them. feoA and feoB are widespread among prokaryotes, and homologs are found in numerous bacterial symbionts of legumes, including other Bradyrhizobia, and the β-Proteobacteria Herbaspirillum lusitanum, Cupriavidus taiwanensis, and Burkholderia caribensis. Surprisingly, those genes are not found in the α-Proteobacteria Sinorhizobium meliloti or Rhizobium leguminosarum. The fact that mutant screens for symbiosis failed to identify symbiotic iron transporters in those species raises the possibility the requisite system is also required under routine culture conditions.
The inability of null feoA or feoB mutant to elicit functional nodules shows that those genes are necessary to establish symbiosis, but it does not inform on the status of iron transport into symbiotic bacteroids. Fortuitously, the selection for suppressors identified feoA(E40K), which displayed good growth in culture, displayed partial iron uptake activity in culture, and elicited nodules on soybean. Iron uptake by feoA(E40K) bacteroids within nodules or isolated cell was diminished, as was observed in cultured cells. Those observation strongly implicate FeoAB in iron uptake by symbiotic bacteroids.
Both free living bacteria and isolated bacteroids transported Fe2+ into cells, but only cultured cells incorporated radiolabel from 59Fe3+ (Figs. 3 and 7B). Intact nodules bathed in Fe2+ resulted in radiolabel incorporated into bacteroids in a feo-dependent manner (Fig. 7A). The plant symbiosome membrane surrounding bacteroids has a ferric reductase activity (17) and a divalent metal ion transporter (16). Symbiotic bacteroids may not need a ferric reductase activity because reduction is carried out by the host, whereas free living bacteria would require that activity.
Experimental Procedures
Strains and Media
B. japonicum USDA110 is the parent strain used in this study. B. japonicum strains were routinely grown at 29 °C in glutamate-salts-yeast (GSY) extract medium as described previously (39) (0.3 g/liter KH2PO4, 0.3 g/liter Na2HPO4, 0.1 g/liter MgSO4-7H2O, 0.05 g/liter CaCl2-2H2O, 4 g/liter glycerol, 1 g/liter yeast extract, 0.01 g/liter H3BO3, 0.001 g/liter ZnSO4-2H2O, 0.001 g/liter FeCl3-6H2O, 0.0005 g/liter CuSO4-5H2O, 0.0005 g/liter MnCl2-4H2O, 0.0005 g/liter Na2MoO4-2H2O, 0.0001 g/liter biotin, pH 6.8). Where noted, modified GSY medium containing 0.5 g/liter of yeast extract, instead of 1 g/liter, was used. The actual iron concentration of the unsupplemented medium was 0.3 μm, as determined with a PerkinElmer Life Sciences model 1100B atomic absorption spectrometer.
Identification of Suppressor Mutants
50 ml of culture of B. japonicum strain mbfA bfr double mutant was grown in GSY medium. The cells were washed twice in PBS and were resuspended in GSY medium. A total of 1 × 108 cells were spread across solid medium containing 1.65 mm FeSO4. The plates were incubated for 13–16 days at 29 °C until colonies were observed. All colonies were picked, and their ability to grow in high iron medium was verified by performing spotting assay as described below. Isolation of genomic DNA was performed for four of the suppressor strains using a phenol/chloroform extraction method. Nucleotide mutations in the suppressors were identified using high throughput sequencing using an Illumina HiSeq2500 (New York State Center of Excellence) and comparing suppressor strains to the parent mbfA bfr double mutant strain. Nucleotide mutations and deletions were confirmed using PCR and subsequent DNA sequencing of the PCR product (Roswell Park DNA Sequencing Lab).
Construction of Strains and Plasmids
For creating the feoA and feoB mutant strains, the open reading frame plus 600 bp of flanking DNA was isolated by PCR, using genomic DNA as a template and ligated into pBluescript SK2 to make pSKfeoA and pSKfeoB. A deletion removing only the open reading frame was constructed by inverse PCR as described previously (31), and the product was blunt ligated. The resulting flanks were then restriction digested from pBluescript SK2 and ligated into the vector pLO1 to make pLO1feoA and pLO1feoB, respectively. QuikChange mutagenesis (Stratagene) was performed on pSKfeoA and pSKfeoB to create pSKfeoA(E40K) and pSKfeoB(Y454term), respectively. These modified fragments were then moved into pLO1 to make pLO1feoA(E40K) and pLO1feoB(Y454term). All four pLO1 strains were then introduced into either B. japonicum USDA110 or mbfA bfr double mutant strain, by conjugation. Single recombinants and double recombinants arising from these were selected as described previously (40). All of the mutations in the strains created were confirmed by the size and sequence of the PCR products using primers outside the flanks.
Spotting Assay
The cells were grown in modified GSY medium supplemented with 100 nm heme to mid-log phase. They were then washed, normalized to A540 nm of 0.35, serial diluted, and spotted on plates containing either 50 μm FeSO4 or 1.65 mm FeSO4.
Growth Curve Analysis
The cells were grown in minimal medium (0.3 g/liter KH2PO4, 0.3 g/liter Na2HPO4, 0.25 g/liter NH4Cl, 4 ml/liter glycerol, 1 ml/liter 10% MgSO4, 1 ml/liter 5% CaCl2, 1 ml/liter trace elements, 1 ml/liter vitamin stock) or modified GSY medium. The medium was supplemented with either no added iron, different concentrations of FeCl3, or 500 nm heme as described in the text. Growth rates were analyzed by measuring the optical density of cells at 540 nm every 6 h until they reached stationary phase.
Ferrous Iron Uptake Assay
Mid-log phase cultures were spun down, washed, and resuspended in uptake buffer (0.2 m MOPS, 2% (w/v) glycerol, pH 6.8) to A540 nm of 0.3. 30 ml of cell solution was placed into a 125-ml Erlenmeyer flask and preincubated for 15 min with 100 μm sodium ascorbate at 28 °C, with shaking. At time 0, 55FeCl3 premixed with ascorbate was added to the cell solution. 1-ml aliquots were removed at various time points and added to ice-cold quench buffer (0.1 m Tris, 1 mm ascorbate, and 100 μm FeSO4, pH 6.0). The cells were collected immediately after quenching on 0.45-μm filters, presoaked in quench buffer. The cells were washed with 3 ml of ice-cold quench buffer and radioactive 55Fe content was counted using a scintillation counter. Internalized 55Fe levels were normalized to the protein levels in the cell. For determination of initial velocities, 55Fe2+ uptake was measured within the linear range for uptake.
Ferric Iron Uptake Assay
The cells were grown to mid-log phase, centrifuged, washed twice, and resuspended in uptake buffer (0.2 m MOPS, 20 mm citrate, and 2% (w/v) glycerol, pH 6.8) to an A540 nm of 0.3. 30 ml of cells were placed into a 125-ml flask and preincubated at 28 °C with shaking. At time 0, 59FeCl3 was added to the cell suspension. 1-ml aliquots were removed at various time points and added to 3 ml of quench buffer precooled on ice. The quench buffer contained 0.1 m Tris, 0.1 m succinate, and 10 mm EDTA, pH 6.0. The quenched cells were collected immediately on 0.45-mm filters presoaked in quench buffer containing 40 μm Fe-EDTA and then counted on a Wallac 1480 WizardTM 3′ automatic γ counter. Internalized 59Fe levels were normalized to protein levels in the cell. For determination of initial velocities, 59Fe3+ uptake was measured within the linear range for uptake.
Western Blot Analysis
The cells were harvested by centrifugation at 13,000 × g for 7 min, washed twice in phosphate-buffered saline (10 mm Na2HPO4, 2 mm KH2PO4, 137 mm NaCl, and 2.7 mm KCl, pH 7.4), and resuspended in the same buffer. The protein concentrations were measured by the BCA protein assay (Pierce). 15 μg of protein from each sample were boiled in SDS loading buffer and loaded on 15% polyacrylamide gel, and immunoblotting was carried out. Anti-Irr antibodies were raised in rabbits as described previously (41) and used at a dilution of 1:2500. Anti-GroEL (Enzo Life Sciences, catalog no. ADI-SPS-875-F, lot no. 12051424) was used at a dilution of 1:8000. HRP-conjugated goat anti-rabbit IgG (Southern Biotech, Birmingham, AL, catalog no. 4010-05, lot no. E5606-P020E) was used as secondary antibody, and the blot was detected using the Immobion chemiluminescence system (Millipore).
Quantitative Real Time PCR
The cells were grown to mid-log phase, and RNA was isolated by the hot phenol method as described previously (28). 1 μg of RNA from each strain were used to make cDNA using Bio-Rad cDNA synthesis kit. Quantitative PCRs were performed as described previously (28). The data were normalized to recA or gapA, as mentioned in the figure and are expressed as average of triplicates, with S.D. represented by the error bars.
Growth and Harvest of Soybean
Soybeans (Glycine max cv. Essex) were grown in Conviron growth chamber under a 16-h light/8-h dark regime at 25 °C. Germinated seedlings were either not inoculated with any strain or inoculated with strains USDA 110, feoA, feoB, or feoA(E40K) grown in vermiculite containing N-free supplemental nutrients. Nodules were harvested from 25–28-day-old plants. To determine viable cell counts in nodules, the nodules were weighed, surface sterilized in 95% ethanol, crushed in 40 mm Tris (pH 7.4), and plated in serial dilutions on GSY medium containing 15 μm hemin. Acetylene reduction assays were carried out as described previously (42).
Iron Uptake by Isolated Bacteroids
10 g of nodules were collected and immediately crushed using a motor and pestle in 10 ml of either ferrous or ferric uptake buffer (described above), filtered through cheese cloth, and centrifuged at 5000 × g for 15 min. The pellet was washed twice and normalized to final A540 nm of 0.35, and uptake assay was carried out using radioactive iron as described above for free living cells.
In Situ Iron Uptake by Bacteroids in Nodules
Roots with attached nodules were taken and washed in ferrous uptake buffer to remove the soil. Then they were submerged in 200 ml of uptake buffer and maintained at 28 °C with slight shaking. At time 0, 5 μm final concentration of iron ascorbate was added. At each time point, root containing nodules was removed and quenched immediately in quench buffer (0.1 m Tris, 1 mm ascorbate, and 100 μm FeSO4, pH 6.0), and all the nodules were removed. Nodules were then crushed using motor and pestle in 5 ml of quench buffer, filtered through a cheese cloth, and centrifuged at 5000 × g for 15 min. A portion of the supernatant containing the plant cytosol fraction was collected, and radioactive counts were measured. The pellet fraction containing the bacteroids were resuspended in quench buffer and were collected immediately on 0.45-μm filters presoaked in quench buffer. Bacteroids were then washed with 3 ml of ice-cold quench buffer, and radioactive iron content was counted using scintillation counter. A portion of bacteroids and plant cytosol was collected for protein measurement. Internalized 55Fe levels were normalized to protein levels in the respective fractions.
Author Contributions
S. S. and M. R. O. conceived and designed the experiments and wrote the manuscript. S. S. carried out the experiments.
Supplementary Material
Footnotes
This work was supported by National Institutes of Health Grant R01 GM099667 (to M. R. O.) The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Figs. S1–S5.
References
- 1. Andrews S. C., Robinson A. K., and Rodríguez-Quiñones F. (2003) Bacterial iron homeostasis. FEMS Microbiol. Rev. 27, 215–237 [DOI] [PubMed] [Google Scholar]
- 2. Chu B. C., Garcia-Herrero A., Johanson T. H., Krewulak K. D., Lau C. K., Peacock R. S., Slavinskaya Z., and Vogel H. J. (2010) Siderophore uptake in bacteria and the battle for iron with the host: a bird's eye view. Biometals 23, 601–611 [DOI] [PubMed] [Google Scholar]
- 3. Faraldo-Gómez J. D., and Sansom M. S. (2003) Acquisition of siderophores in Gram-negative bacteria. Nat. Rev. Mol. Cell Biol. 4, 105–116 [DOI] [PubMed] [Google Scholar]
- 4. Wandersman C., and Stojiljkovic I. (2000) Bacterial heme sources: the role of heme, hemoprotein receptors and hemophores. Curr. Opin. Microbiol. 3, 215–220 [DOI] [PubMed] [Google Scholar]
- 5. Genco C. A., and Dixon D. W. (2001) Emerging strategies in microbial haem capture. Mol. Microbiol. 39, 1–11 [DOI] [PubMed] [Google Scholar]
- 6. Brear E. M., Day D. A., and Smith P. M. (2013) Iron: an essential micronutrient for the legume-rhizobium symbiosis. Front. Plant Sci. 4, 359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stevens J. B., Carter R. A., Hussain H., Carson K. C., Dilworth M. J., and Johnston A. W. (1999) The fhu genes of Rhizobium leguminosarum, specifying siderophore uptake proteins: fhuDCB are adjacent to a pseudogene version of fhuA. Microbiology 145, 593–601 [DOI] [PubMed] [Google Scholar]
- 8. Yeoman K. H., Wisniewski-Dye F., Timony C., Stevens J. B., deLuca N. G., Downie J. A., and Johnston A. W. (2000) Analysis of the Rhizobium leguminosarum siderophore-uptake gene fhuA: differential expression in free-living bacteria and nitrogen-fixing bacteroids and distribution of an fhuA pseudogene in different strains. Microbiology 146, 829–837 [DOI] [PubMed] [Google Scholar]
- 9. Persmark M., Pittman P., Buyer J. S., Schwyn B., Gill P. R., and Neilands J. B. (1993) Isolation and structure of rhizobactin 1021, a siderophore from the alfalfa symbiont Rhizobium meliloti 1021. J. Am. Chem. Soc. 115, 3950–3956 [Google Scholar]
- 10. Lynch D., O'Brien J., Welch T., Clarke P., Cuív P. O., Crosa J. H., and O'Connell M. (2001) Genetic organization of the region encoding regulation, biosynthesis, and transport of rhizobactin 1021, a siderophore produced by Sinorhizobium meliloti. J. Bacteriol 183, 2576–2585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Guerinot M. L., Meidl E. J., and Plessner O. (1990) Citrate as a siderophore in Bradyrhizobium japonicum. J. Bacteriol. 172, 3298–3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Noya F., Arias A., and Fabiano E. (1997) Heme compounds as iron sources for nonpathogenic Rhizobium bacteria. J. Bacteriol. 179, 3076–3078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Plessner O., Klapatch T., and Guerinot M. L. (1993) Siderophore utilization by Bradyrhizobium japonicum. Appl. Environ. Microbiol. 59, 1688–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Small S. K., Puri S., Sangwan I., and O'Brian M. R. (2009) Positive control of ferric siderophore receptor gene expression by the Irr protein in Bradyrhizobium japonicum. J. Bacteriol. 191, 1361–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Benson H. P., Boncompagni E., and Guerinot M. L. (2005) An iron uptake operon required for proper nodule development in the Bradyrhizobium japonicum-soybean symbiosis. Mol. Plant Microbe Interact. 18, 950–959 [DOI] [PubMed] [Google Scholar]
- 16. Kaiser B. N., Moreau S., Castelli J., Thomson R., Lambert A., Bogliolo S., Puppo A., and Day D. A. (2003) The soybean NRAMP homologue, GmDMT1, is a symbiotic divalent metal transporter capable of ferrous iron transport. Plant J. 35, 295–304 [DOI] [PubMed] [Google Scholar]
- 17. LeVier K., Day D. A., and Guerinot M. L. (1996) Iron uptake by symbiosomes from soybean root nodules. Plant Physiol. 111, 893–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cartron M. L., Maddocks S., Gillingham P., Craven C. J., and Andrews S. C. (2006) Feo-transport of ferrous iron into bacteria. Biometals 19, 143–157 [DOI] [PubMed] [Google Scholar]
- 19. Kammler M., Schön C., and Hantke K. (1993) Characterization of the ferrous iron uptake system of Escherichia coli. J. Bacteriol. 175, 6212–6219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Naikare H., Palyada K., Panciera R., Marlow D., and Stintzi A. (2006) Major role for FeoB in Campylobacter jejuni ferrous iron acquisition, gut colonization, and intracellular survival. Infect. Immun. 74, 5433–5444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Robey M., and Cianciotto N. P. (2002) Legionella pneumophila feoAB promotes ferrous iron uptake and intracellular infection. Infect. Immun. 70, 5659–5669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Thomas-Charles C. A., Zheng H., Palmer L. E., Mena P., Thanassi D. G., and Furie M. B. (2013) FeoB-mediated uptake of iron by Francisella tularensis. Infect Immun 81, 2828–2837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Velayudhan J., Hughes N. J., McColm A. A., Bagshaw J., Clayton C. L., Andrews S. C., and Kelly D. J. (2000) Iron acquisition and virulence in Helicobacter pylori: a major role for FeoB, a high-affinity ferrous iron transporter. Mol. Microbiol. 37, 274–286 [DOI] [PubMed] [Google Scholar]
- 24. Sankari S., and O'Brian M. R. (2014) A bacterial iron exporter for maintenance of iron homeostasis. J. Biol. Chem. 289, 16498–16507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sankari S., and O'Brian M. R. (2016) Synthetic lethality of the bfr and mbfA genes reveals a functional relationship between iron storage and iron export in managing stress responses in Bradyrhizobium japonicum. PLoS One 11, e0157250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Su Y. C., Chin K. H., Hung H. C., Shen G. H., Wang A. H., and Chou S. H. (2010) Structure of Stenotrophomonas maltophilia FeoA complexed with zinc: a unique prokaryotic SH3-domain protein that possibly acts as a bacterial ferrous iron-transport activating factor. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 66, 636–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nienaber A., Hennecke H., and Fischer H. M. (2001) Discovery of a haem uptake system in the soil bacterium Bradyrhizobium japonicum. Mol. Microbiol. 41, 787–800 [DOI] [PubMed] [Google Scholar]
- 28. Yang J., Sangwan I., Lindemann A., Hauser F., Hennecke H., Fischer H. M., and O'Brian M. R. (2006) Bradyrhizobium japonicum senses iron through the status of haem to regulate iron homeostasis and metabolism. Mol. Microbiol. 60, 427–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Qi Z., Hamza I., and O'Brian M. R. (1999) Heme is an effector molecule for iron-dependent degradation of the bacterial iron response regulator (Irr) protein. Proc. Natl. Acad. Sci. U.S.A. 96, 13056–13061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jaggavarapu S., and O'Brian M. R. (2014) Differential control of Bradyrhizobium japonicum iron stimulon genes through variable affinity of the iron response regulator (Irr) for target gene promoters and selective loss of activator function. Mol. Microbiol. 92, 609–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Small S. K., and O'Brian M. R. (2011) The Bradyrhizobium japonicum frcB gene encodes a diheme ferric reductase. J. Bacteriol. 193, 4088–4094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mathew A., Eberl L., and Carlier A. L. (2014) A novel siderophore-independent strategy of iron uptake in the genus Burkholderia. Mol. Microbiol. 91, 805–820 [DOI] [PubMed] [Google Scholar]
- 33. Brickman T. J., and Armstrong S. K. (2012) Iron and pH-responsive FtrABCD ferrous iron utilization system of Bordetella species. Mol. Microbiol. 86, 580–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Elhassanny A. E., Anderson E. S., Menscher E. A., and Roop R. M. 2nd. (2013) The ferrous iron transporter FtrABCD is required for the virulence of Brucella abortus 2308 in mice. Mol. Microbiol. 88, 1070–1082 [DOI] [PubMed] [Google Scholar]
- 35. Pessi G., Ahrens C. H., Rehrauer H., Lindemann A., Hauser F., Fischer H. M., and Hennecke H. (2007) Genome-wide transcript analysis of Bradyrhizobium japonicum bacteroids in soybean root nodules. Mol. Plant Microbe Interact. 20, 1353–1363 [DOI] [PubMed] [Google Scholar]
- 36. Wittenberg J. B., Wittenberg B. A., Day D. A., Udvardi M. K., and Appleby C. A. (1996) Siderophore-bound iron in the peribacteroid space of soybean root nodules. Plant Soil 178, 161–169 [Google Scholar]
- 37. Regensburger B., Meyer L., Filser M., Weber J., Studer D., Lamb J. W., Fischer H. M., Hahn M., and Hennecke H. (1986) Bradyrhizobium japonicum mutants defective in root nodule bacteroid developent ad nitrogen fixation. Arch. Microbiol. 144, 355–366 [Google Scholar]
- 38. So J. S., Hodgson A. L., Haugland R., Leavitt M., Banfalvi Z., Nieuwkoop A. J., and Stacey G. (1987) Transposon-induced symbiotic mutants of Bradyrhizobium japonicum: isolation of two gene regions essential for nodulation. Mol. Gen. Genet. 207, 15–23 [DOI] [PubMed] [Google Scholar]
- 39. Frustaci J. M., Sangwan I., and O'Brian M. R. (1991) Aerobic growth and respiration of a δ-aminolevulinic acid synthase (hemA) mutant of Bradyrhizobium japonicum. J. Bacteriol. 173, 1145–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Escamilla-Hernandez R., and O'Brian M. R. (2012) HmuP is a co-activator of Irr-dependent expression of heme utilization genes in Bradyrhizobium japonicum. J. Bacteriol. 194, 3137–3143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hamza I., Chauhan S., Hassett R., and O'Brian M. R. (1998) The bacterial Irr protein is required for coordination of heme biosynthesis with iron availability. J. Biol. Chem. 273, 21669–21674 [DOI] [PubMed] [Google Scholar]
- 42. O'Brian M. R., Kirshbom P. M., and Maier R. J. (1987) Tn5-induced cytochrome mutants of Bradyrhizobium japonicum: effects of the mutations on cells grown symbiotically and in culture. J. Bacteriol. 169, 1089–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.