Abstract
Eukaryotic organisms typically express multiple type IV P-type ATPases (P4-ATPases), which establish plasma membrane asymmetry by flipping specific phospholipids from the exofacial to the cytosolic leaflet. Saccharomyces cerevisiae, for example, expresses five P4-ATPases, including Neo1, Drs2, Dnf1, Dnf2, and Dnf3. Neo1 is thought to be a phospholipid flippase, although there is currently no experimental evidence that Neo1 catalyzes this activity or helps establish membrane asymmetry. Here, we use temperature-conditional alleles (neo1ts) to test whether Neo1 deficiency leads to loss of plasma membrane asymmetry. Wild-type (WT) yeast normally restrict most of the phosphatidylserine (PS) and phosphatidylethanolamine (PE) to the inner cytosolic leaflet of the plasma membrane. However, the neo1-1ts and neo1-2ts mutants display a loss of PS and PE asymmetry at permissive growth temperatures as measured by hypersensitivity to pore-forming toxins that target PS (papuamide A) or PE (duramycin) exposed in the extracellular leaflet. When shifted to a semi-permissive growth temperature, the neo1-1ts mutant became extremely hypersensitive to duramycin, although the sensitivity to papuamide A was unchanged, indicating preferential exposure of PE. This loss of asymmetry occurs despite the presence of other flippases that flip PS and/or PE. Even when overexpressed, Drs2 and Dnf1 were unable to correct the loss of asymmetry caused by neo1ts. However, modest overexpression of Neo1 weakly suppressed loss of membrane asymmetry caused by drs2Δ with a more significant correction of PE asymmetry than PS. These results indicate that Neo1 plays an important role in establishing PS and PE plasma membrane asymmetry in budding yeast.
Keywords: ATPase, membrane lipid, phosphatidylethanolamine, phosphatidylserine, transporter, duramycin, P4-ATPase, papuamide A, flippase
Introduction
The plasma membrane is a complex amalgam of proteins, carbohydrates, and lipids with an overall asymmetric distribution of these molecules between the two leaflets (1, 2). For example, the aminophospholipids phosphatidylethanolamine (PE)2 and phosphatidylserine (PS) are mainly concentrated in the cytosolic leaflet, although phosphatidylcholine and sphingolipids are enriched in the extracellular leaflet (3–5). Phospholipid asymmetry is crucial for plasma membrane integrity and several cellular processes such as signal transduction (6), cell division (7, 8), and vesicular transport (9–11). Controlled disruption of phospholipid asymmetry and exposure of PS in the extracellular leaflet stimulates blood coagulation and is required for the recognition and phagocytosis of apoptotic cells (12–14). Plasma membrane asymmetry is established by P4-ATPases, which pump specific phospholipid substrates (e.g. PS and PE) from the extracellular leaflet to the cytosolic leaflet.
P4-ATPases belong to the P-type ATPase superfamily, which has been phylogenetically divided into five subgroups (P1–P5) (15, 16). The well characterized P1-, P2-, and P3-ATPases transport cations (e.g. the Ca2+-ATPase and Na+/K+-ATPase) or heavy metals (e.g. the Menkes disease copper transporter). In contrast, the P4-ATPases evolved the ability to transport bulky phospholipid molecules across biological membranes, and the P5-ATPase substrate is still unknown (17–19). Although there are no crystal structures determined for the P4-ATPases, they share the same domain organization as ion-transporting P-type ATPases, for which multiple crystal structures are available (16). The cytosolic actuator (A), nucleotide-binding (N), and phosphorylation (P) domains catalyze phosphate transfer from ATP to an Asp residue in the P domain and its subsequent dephosphorylation by the A domain. For P4-ATPases, unidirectional phospholipid translocation from the extracellular (or lumenal) leaflet to the cytosolic leaflet is associated with the dephosphorylation step (E2∼P to E1 conformational transition) (20). The membrane domain is usually composed of 10 transmembrane segments, which form transport substrate-binding sites and conduits through the membrane. Most P4-ATPases associate with a β-subunit consisting of a glycosylated ectodomain flanked by two transmembrane segments (21–23).
Eukaryotic organisms usually express multiple P4-ATPase genes. There are 14 mammalian P4-ATPases that are associated with intellectual disability (24), neurodegeneration (25), liver disease (26, 27), hearing loss (28), diabetes (29), immune deficiency (30, 31), anemia (32), and reduced male fertility (33). Budding yeast express the following five members of this subgroup: Dnf1, Dnf2, Dnf3, Drs2, and Neo1. Neo1 is the only P4-ATPase essential for viability, although Drs2 and the Dnf proteins are collectively essential (34). The reason why a single-celled organism requires so many P4-ATPases is unclear. However, gene duplication and subsequent specialization for substrate specificity, subcellular localization, modes of regulation, and binding partners may have contributed to the expansion.
The substrate preferences and transport mechanism are best understood for Drs2 and Dnf1 and least well understood for Neo1 (35). Dnf1 (and the nearly identical Dnf2) interacts with the Lem3 β-subunit, cycles between the plasma membrane, endosomes, and TGN, and has a strong substrate preference for lyso-phosphatidylcholine (lyso-PC) and lyso-PE, phospholipids lacking one fatty acyl chain (21, 23, 34, 36–38). Drs2 is a heterodimer with Cdc50, localizes to the TGN and early endosome, primarily flips di-acylated phosphatidylserine (PS), and has a weaker activity toward PE (17, 23, 35, 37, 39–41). Neo1 has no known β-subunit and localizes broadly throughout the Golgi and endosomes (42–44), and the substrate preference of this essential P4-ATPase has not yet been determined. The substrate transported by mammalian Neo1 orthologs ATP9A and ATP9B is also unknown. However, knockdown of the Caenorhabditis elegans Neo1 ortholog TAT-5 causes exposure of PE in the outer leaflet of the plasma membrane, suggesting that PE may be the preferred substrate (45).
Exposure of aminophospholipids (PS and PE) on the cell surface can be detected using membrane-impermeable cytotoxic peptides that bind to the headgroup and subsequently generate a pore in the membrane. Papuamide A (Pap A) is potent cytotoxic agent that binds specifically to PS, and duramycin binds specifically to PE with affinity in the nanomolar range (46–48). Wild-type (WT) yeast cells are relatively resistant to Pap A and duramycin because most of these aminophospholipids are restricted to the inner cytosolic leaflet of the plasma membrane. In contrast, flippase mutants expose aminophospholipids in the extracellular leaflet and are hypersensitive to these cytotoxic agents. Here, we use thermal inactivation of temperature conditional alleles of NEO1 (neo1ts) to explore the influence of Neo1 on plasma membrane phospholipid asymmetry and the relationship between Neo1, Drs2, and Dnf1 for this function.
Results
Inactivation of Neo1 Causes a Loss of Membrane Asymmetry
To test the influence of Neo1 on membrane asymmetry, we probed neo1 mutants with pore-forming cytotoxic peptides that preferentially bind to PS or PE exposed on the outer leaflet of the plasma membrane. Cells become hypersensitive to Pap A when they lose PS asymmetry and become hypersensitive to duramycin when they lose PE asymmetry. Growth of neo1ts (neo1-1 and neo1-2) mutants over a range of toxin concentrations was compared with WT and drs2Δ, a flippase mutant that is known to display a loss of PS and PE asymmetry. Growth of each strain was plotted relative to WT cells incubated in the absence of toxin (Fig. 1). These neo1-1 and neo1-2 alleles harbor a different set of mutations, and the neo1-1 mutant has a lower restrictive temperature, indicating that the Neo1-1 protein function is more strongly perturbed than Neo1-2 at a given temperature (e.g. 30 °C) (42).
FIGURE 1.
neo1ts cells display a loss of membrane asymmetry. A and B, WT (MTY219RR), neo1-1 (MTY628-15B), neo1-2 (MTY628-34A), and drs2Δ (ZHY615M2D) cells were incubated for 20 h at the permissive growth temperature of 27 °C (A) or the semi-permissive growth temperature of 30 °C (B) with increasing concentrations of papuamide A. C and D, same set of strains was assayed for duramycin sensitivity at 27 °C (C) or 30 °C (D). Growth relative to WT cells in the absence of drug was plotted (n ≥5, error bars ±S.E.M.). Sigmoidal dose-response curve fitting modality was applied when the adjusted R2 values are greater than 0.8.
At the neo1ts permissive growth temperature of 27 °C, neo1-1, neo1-2, and drs2Δ strains grew nearly as well as WT in the absence of Pap A or duramycin, but all three mutants were hypersensitive to both toxins relative to WT cells (Fig. 1, A and C). The neo1-1ts and neo1-2ts mutants showed a similar level of sensitivity to Pap A at 27 °C (IC50 of 2.5–3.5 μg/ml), whereas drs2Δ cells displayed the greatest Pap A sensitivity (IC50 of ∼1.0 μg/ml (Fig. 1A and Table 1)). Thus, drs2Δ had a greater loss of PS asymmetry at this temperature. In contrast, the neo1-1 and drs2Δ mutants were equally sensitive to duramycin at 27 °C, although the weaker neo1-2 mutant was less sensitive (IC50 of ∼10 μm versus ∼20 μm, see Table 1). Even though the temperature-conditional Neo1 mutants retained sufficient activity to support nearly WT growth at 27 °C (Fig. 1, 0 toxin), they clearly displayed a loss of PS and PE asymmetry. Thus, the mutant Neo1 proteins must have reduced activity at 27 °C relative to WT Neo1.
TABLE 1.
Approximate IC50 values for strains treated with cytotoxic peptides at the indicated temperatures
| IC50 |
||
|---|---|---|
| 27 °C | 30 °C | |
| Papuamide Aa | ||
| WT | >6 | >6 |
| neo1-1 | 2.5–3.5 | 2.5–3.5 |
| neo1-2 | 2.5–3.5 | 2.5–3.5 |
| drs2Δ | ∼1.0 | 2.5–3.5 |
| Duramycinb | ||
| WT | 40–50 | 50–60 |
| neo1-1 | ∼10 | ∼2 |
| neo1-2 | ∼20 | 30–40 |
| drs2Δ | ∼20 | 30–40 |
a Values were measured in microgram/ml.
b Values were measured in micromolar.
At 30 °C, the growth defect of the stronger temperature-sensitive mutant, neo1-1, became detectable as the rate of growth was reduced to 75% relative to WT cells in the absence of toxins (Fig. 1, B and D). Sensitivity of neo1-1 to Pap A was modestly enhanced at this semi-permissive temperature, but sensitivity of neo1-2 was unchanged relative to 27 °C (Fig. 1, A and B). The drs2Δ mutant actually became more resistant to Pap A and duramycin at 30 °C relative to 27 °C (Fig. 1, A and B), which likely reflects a greater ability of the remaining P4-ATPases to compensate for the loss of Drs2 at higher temperatures (42). Most striking was the extreme hypersensitivity of neo1-1 to duramycin at 30 °C (IC50 of ∼2 μm) (Fig. 1D and Table 1). At this temperature, neo1-2 and drs2Δ displayed a comparable hypersensitivity to duramycin. Importantly, a sufficient inactivation of neo1-1 to just give a detectable growth defect correlated with a substantial loss of PE asymmetry. These results suggest that the primary role of Neo1 is to establish PE asymmetry, but it also has a significant influence on PS asymmetry.
Exposure of PE and PS on Neo1ts Cells Is Not a Secondary Effect of Drs2/Dnf Mislocalization or Loss of Activity at the Plasma Membrane
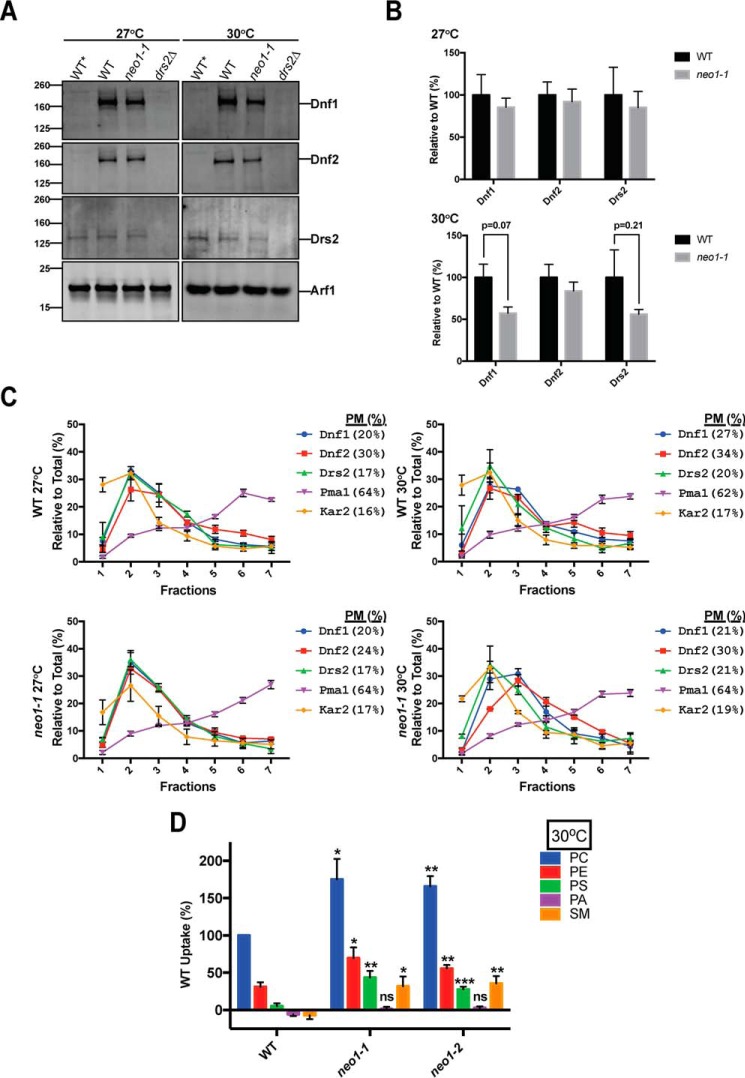
Inactivation of Drs2 or the Dnf1/2 flippases also causes a loss of PS and PE asymmetry of the plasma membrane. Thus, it was possible that the expression or activity of these other P4-ATPases was perturbed in neo1ts mutants, thereby causing the loss of membrane asymmetry. Initially, we tested whether loss of Neo1 activity can influence expression of Drs/Dnf proteins. To test this, we quantified the expression of Dnf1-HA, Dnf2-HA, and Drs2 using Western blotting analysis (Fig. 2A). The HA tags were integrated into the chromosomal loci, and so Dnf1-HA and Dnf2-HA were expressed from their endogenous promoters. WT* cells lack the HA tags, and WT cells express both the tagged Dnf proteins and the WT Neo1 and Drs2. Equal amounts of total protein were loaded in each lane, which was confirmed by probing for Arf1 as a loading control. Although there appeared to be a partial reduction in the amount of Dnf1-HA and Drs2 in the neo1-1 cells relative to WT cells, these differences were not statistically significant (Fig. 2B, n = 3).
FIGURE 2.
Inactivation of Neo1 does not lead to significant changes in the distribution or expression of Drs2/Dnf P4-ATPases but does increase Dnf1 and Dnf2 plasma membrane flippase activity. A, Western blotting analysis of wild-type and neo1ts cells expressing HA-tagged Dnf1 (MTYD1-219RRL and MTYD1-62815BL) and HA-tagged Dnf2 (MTYD2-219RRL and MTYD2-62815BL) at 27 and 30 °C. Equal amounts of total protein (0.75 mg) were loaded from each sample and probed with anti-HA, anti-Drs2, and anti-Arf1. WT* (untagged) and drs2Δ were used as specificity controls. Arf1 levels were used to demonstrate equal loading of the samples. These images are representative of at least three biological replicates. B, quantification of Dnf1-HA, Dnf2-HA, and Drs2 levels in wild-type and neo1ts cells at 27 and 30 °C. Intensity values for neo1ts cells were normalized to those values of wild-type cells at corresponding temperatures (n ≥3, error bars ±S.E.). C, subcellular fractionation was done with wild-type and neo1ts cells expressing Dnf1-HA and Dnf2-HA (same strains as used for A). Kar2 and Pma1 were probed as ER and plasma membrane markers, respectively. Dnf1-HA, Dnf2-HA, Drs2, Kar2, and Pma1 levels for each fraction were plotted as the percentage of total intensity for each protein in all seven fractions. (For example, Dnf1-HA in fraction 7 was divided by the sum of Dnf1-HA in all seven fractions.) The percent in plasma membrane fractions (% PM) is the sum of the percentages in fractions 5–7. D, inactivation of neo1ts alleles leads to increased Dnf1/Dnf2 activities at the plasma membrane. Lipid uptake assays were performed with WT (MTY219RR), neo1-1 (MTY628-15B), and neo1-2 (MTY628-34A) cells grown at 30 °C using fluorescent-labeled (NBD) phospholipids. Lipid uptake activities were plotted as percentage of NBD-PC uptake for wild-type cells. SM, sphingomyelin; ns, not significant. *, p < 0.05; **, p < 0.01; ***, p < 0.001; Student's t test, n ≥9; error bars ±S.E.
We also tested whether the loss of PS and PE plasma membrane asymmetry in neo1ts cells could be explained by loss of Dnf1-HA and/or Dnf2-HA from the plasma membrane using subcellular fractionation. Dnf1 and Dnf2 are localized to Golgi, endosomes, and the plasma membrane, whereas Drs2 is normally localized to the TGN (34, 39). However, loss of Drs2 activity does cause a substantial increase in its transport to the plasma membrane (49). WT and neo1-1 cells were grown at 27 °C, and half of the culture was shifted to 30 °C for 1 h prior to lysis and fractionation in a sucrose gradient (Fig. 2C). This fractionation method does not separate ER, Golgi, and endosomes from each other, but it does separate these organelles from the plasma membrane reasonably well (50).
To assess the success of subcellular fractionation, we probed each fraction for Kar2 as an ER marker and Pma1 as a plasma membrane marker. We defined Pma1-enriched and Kar2-depleted fractions as plasma membrane fractions (fractions 5–7; % PM). In plasma membrane-enriched fractions, Pma1 did not significantly change upon temperature shift in wild-type (64.1 ± 1.5 and 62.5 ± 1.9%, respectively) and neo1ts cells (64.1 ± 1.6 and 64.0 ± 2.0%, respectively) at 27 and 30 °C. In wild-type cells, there is a slight but significant (Student's t test; p = 0.032) increase in the percentage of plasma membrane-enriched Dnf1 upon temperature shift from 20 ± 1.90 to 26.6 ± 0.80%, although the percentage of plasma membrane-enriched Dnf2 (30.3 ± 3.1 and 34.2 ± 2.0%, respectively) did not change significantly (Fig. 2C). In neo1ts cells, the percentage of plasma membrane-enriched Dnf1 remains unchanged upon temperature shift (20.8 ± 5.2 and 20.8 ± 0.6%, respectively), although the percentage of Dnf2 in plasma membrane-enriched fractions increased non-significantly (Student's t test; p = 0.13) from 23.8 ± 1.3 to 30.2 ± 3.1%. Upon temperature shift, we did not observe any significant increase in the percentage of Drs2 in the plasma membrane-enriched fractions in either wild-type (17.2 ± 1.8 and 19.7 ± 3.7%, respectively) or neo1ts cells (17.0 ± 1.3 and 21.3 ± 5.0%), implying Drs2 retained its normal localization in neo1ts cells. In summary, neither the total amount nor the distribution of Drs2/Dnf family flippases between the plasma membrane and internal organelles changed significantly upon inactivation of neo1ts. Based on these results, exposure of PE and PS on neo1ts cells cannot be accounted for by a decrease in the expression or loss of plasma membrane localization for the Drs2/Dnf family flippases.
However, loss of PE and PS asymmetry in neo1ts cells might be due to reduced activities of Dnf1 and Dnf2 at the plasma membrane. To test this, we measured plasma membrane flippase activity, catalyzed primarily by Dnf1 and Dnf2, in WT and neo1ts cells using NBD-labeled PC, PE, PS, phosphatidic acid (PA), and sphingomyelin. A dnf1,2Δ stain was also assayed, and the low level of fluorescent lipid uptake was subtracted as background. Rather than a loss of activity, we found significantly enhanced activity for all substrates except NBD-PA (Fig. 2D). The ability of neo1ts cells to exclude NBD-PA argues against an increase in nonspecific flip-flop. These strains also excluded propidium iodide, which was used to eliminate dead cells from this flow cytometry-based analysis. We conclude that Dnf1 and Dnf2 are present and active at the plasma membrane of neo1ts mutants. Although Drs2 cannot be directly measured using this lipid uptake activity assay, it is unlikely that Drs2 activity is substantially perturbed in neo1ts cells because even partial loss of Drs2 activity in the neo1ts background is lethal (42).
Exposure of PE on Neo1ts Cells Is Not a Secondary Effect of Perturbing COPI Function
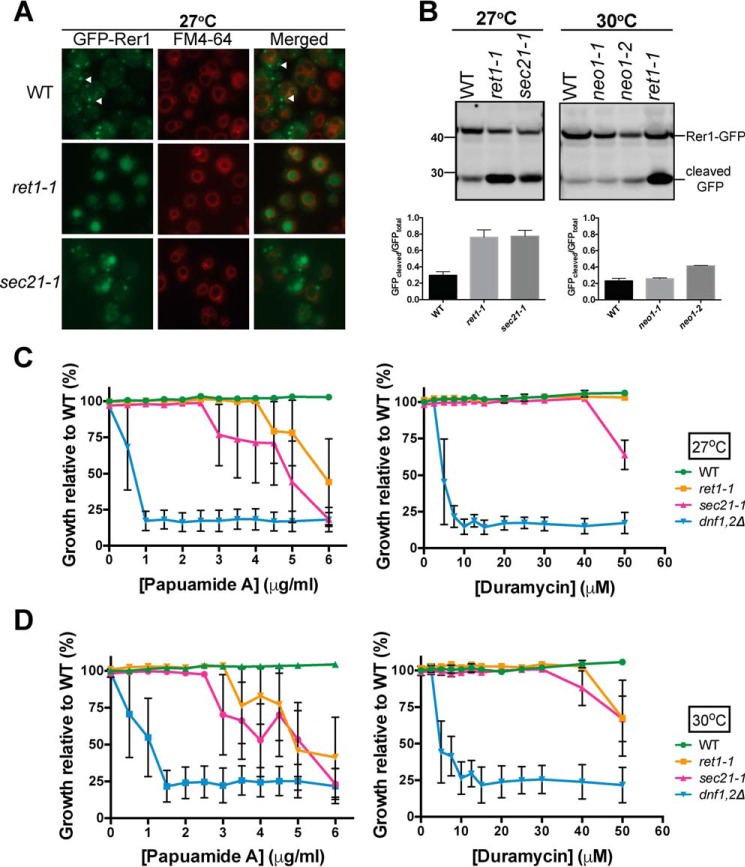
Inactivation of Neo1 causes protein trafficking defects in the early secretory pathway, and the COPI cargo protein GFP-Rer1 is mislocalized from the cis-Golgi to the vacuole lumen (42). It is possible that the loss of plasma membrane asymmetry in neo1ts mutants is a secondary consequence of perturbing COPI-dependent trafficking pathways. To test this, we examined plasma membrane organization in COPI mutants (ret1-1 and sec21-1) at a temperature where the strains grow well but show a strong defect in GFP-Rer1 localization. At 27 °C, most of GFP-Rer1 localized to Golgi punctae in WT cells but was mislocalized to the vacuole, marked by FM4-64, in ret1-1 and sec21-1 (Fig. 3A). Vacuoles are fragmented in neo1ts cells making the mislocalization of GFP-Rer1 difficult to detect by microscopy (42), and so Western blots were used to quantify the amount of GFP-Rer1 mislocalized to the vacuole (51).
FIGURE 3.
neo1ts mutants display a dramatic loss of membrane asymmetry at growth temperatures that do not disrupt protein trafficking. A, Rer1-GFP trafficking in wild-type cells (SEY6210) and copIts alleles (ret1-1 and sec21-1) at 27 °C. FM4-64 staining was done with wild-type and copIts alleles expressing pURA3-GFP-RER1. White arrows indicate the punctate localization for GFP-tagged Rer1. B, quantitative analysis of Rer1-GFP localization by Western blotting analysis for wild-type, copIts, and neo1ts cells grown at 27 °C. Ratios of GFP cleaved versus total GFP were determined for wild-type, copIts, and neo1ts cells; n ≥2. Duramycin and papuamide A sensitivity assays were performed with wild-type and copIts cells at 27 °C (C) and at 30 °C (D). Growth relative to wild-type cells (SEY6210) was plotted; n ≥3. Sigmoidal dose-response curve fitting modality yielded adjusted R2 values smaller than 0.8. Data points were connected to generate the graphs.
Upon arrival in the vacuole, the GFP-Rer1 fusion protein is cleaved by vacuolar proteases to release GFP. In WT cells, ∼30% of GFP-Rer1 is cleaved at steady state, providing a population average of the amount of the fusion protein in the vacuole. Consistent with the imaging result, about 75% of GFP-Rer1 is cleaved in the COPI mutants indicating mislocalization to the vacuole (Fig. 3B). At 30 °C, where neo1-1 displayed substantial sensitivity to duramycin (Fig. 1D) GFP-Rer1 was not mislocalized to the vacuole as indicated by the small percentage (22%) of GFP-Rer1 that was cleaved (Fig. 3B).
The COPI mutants were modestly sensitive to papuamide A and almost completely resistant to duramycin at 27 °C, but became slightly more sensitive to the toxins at 30 °C (Fig. 3, C and D). These results indicate that loss of COPI function does cause a partial loss of PS asymmetry and a relatively minor exposure of PE in the outer leaflet of the plasma membrane. In contrast, neo1ts mutants display a dramatic loss of membrane asymmetry at growth temperatures that do not disrupt protein trafficking. Thus, loss of membrane asymmetry in neo1ts cells cannot be explained by a loss of COPI function.
Relationship between P4-ATPases That Potentially Flip PE
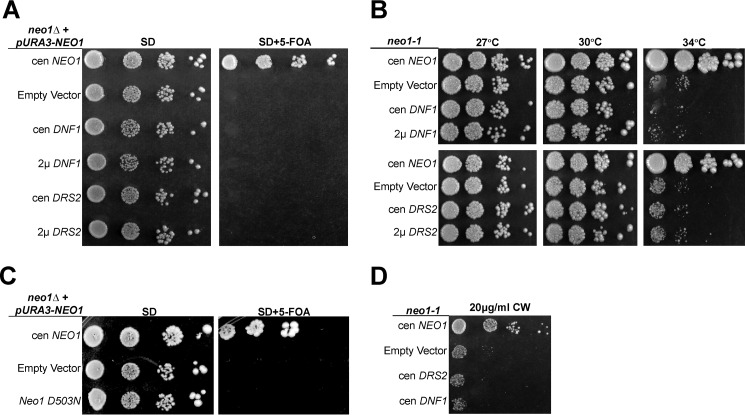
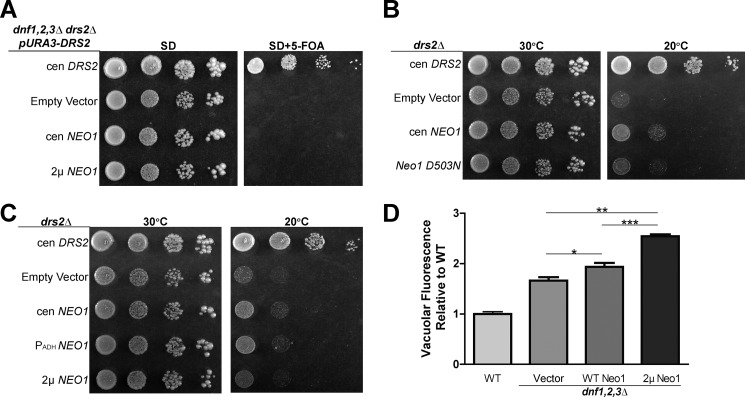
The loss of PE and PS asymmetry in neo1 mutants suggested that Neo1 shares a substrate preference with Drs2 and Dnf1 and that translocation of PE across membranes could be Neo1's essential function. Neo1 is broadly distributed throughout the Golgi and has an overlapping localization with Drs2 and Dnf1 in the late Golgi and endosomal system (34, 43, 44). Disruption of NEO1 in a haploid strain is lethal, indicating that the other P4-ATPases cannot compensate for loss of NEO1 at endogenous levels of expression. However, we hypothesized that overexpression of Drs2 or Dnf1, which are both capable of flipping NBD-phosphatidylethanolamine (NBD-PE), might compensate for loss of Neo1. To test this, we performed a plasmid shuffle assay to replace NEO1 with low copy (cen) or multicopy (2μ) plasmids expressing DNF1 with its β-subunit LEM3 or DRS2 with its β-subunit CDC50. neo1Δ strains covered by NEO1 on a URA3-marked plasmid and harboring DNF1 or DRS2 plasmids were serially diluted on 5-FOA plates to select for colonies having lost the URA3-marked NEO1 plasmid (Fig. 4A). A control strain harboring a second copy of WT NEO1 on a LEU2 plasmid formed colonies on 5-FOA, and all of the strains grew well on minimal medium (SD) lacking 5-FOA, as expected. In contrast, no colonies grew on 5-FOA from the neo1Δ cells expressing additional copies of DNF1 or DRS2. Thus, overexpression of DNF1 or DRS2 with their co-chaperones was unable to suppress neo1Δ lethality.
FIGURE 4.
NEO1 exerts an essential function that cannot be provided by DNF1 or DRS2. A, overexpression of DNF1 or DRS2 with their co-chaperones failed to suppress neo1Δ lethality. neo1Δ + pRS416-NEO1 (YWY10)-expressing single copy (cen) NEO1 (pRS313-NEO1), empty vector (pRS313), single copy or multicopy (2μ) DNF1 or DRS2 (pRS313-DNF1, pRS423-DNF1, pRS315-DRS2, or pRS425-DRS2), and their co-chaperones (pRS425-LEM3 or pRS423-CDC50, respectively) were spotted onto minimal media plates (SD), and the pRS416-NEO1 plasmid was counter-selected on 5-FOA. B, neo1ts mutant (MTY628-15B) was transformed with the same set of plasmids and assayed for growth at the indicated temperatures. C, neo1Δ + pRS416-NEO1 (YWY10)-expressing single copy NEO1 (pRS313-NEO1), empty vector (pRS313), or single copy catalytically dead neo1-D503N (pRS313-neo1-D503N) was spotted onto minimal media plates (SD), and the pRS416-NEO1 plasmid was counter-selected on 5-FOA. D, neo1-1 hypersensitivity to CW at 30 °C was not suppressed by overexpression of DRS2 or DNF1. Images are representative of three independent experiments.
To provide a more sensitive assay for suppression, we next transformed the neo1-1ts mutant with the same set of plasmids harboring DRS2 or DNF1 and their β-subunits. The neo1-1 cells carrying an empty vector grew well at the permissive temperatures of 27 and 30 °C, but they grew very slowly at the semi-permissive temperature of 34 °C relative to the neo1-1 strain complemented with WT NEO1. Overexpression of neither DNF1 nor DRS2 was able to suppress the growth defect of neo1-1 at 34 °C (Fig. 4B). Lack of suppression by DRS2 or DNF1 raised the concern that Neo1 might have an essential function that is independent of its presumed catalytic activity. However, mutation of the aspartic acid predicted to be phosphorylated during the catalytic cycle inactivated Neo1 based on its inability to complement the neo1Δ strain (Fig. 4C, neo1-D503N).
The neo1-1 mutant has previously been shown to also display a defect in Golgi glycosylation events, potentially caused by hyperacidification of this organelle (42, 52). This underglycosylation of cell wall components causes hypersensitivity to the chitin-binding compound calcofluor white (CW) (42). To determine whether overexpression of the other flippases would suppress the glycosylation defects, we tested whether DRS2 or DNF1 overexpression could suppress neo1-1 CW hypersensitivity. We found that DRS2 and DNF1 overexpression both failed to suppress this phenotype (Fig. 4D).
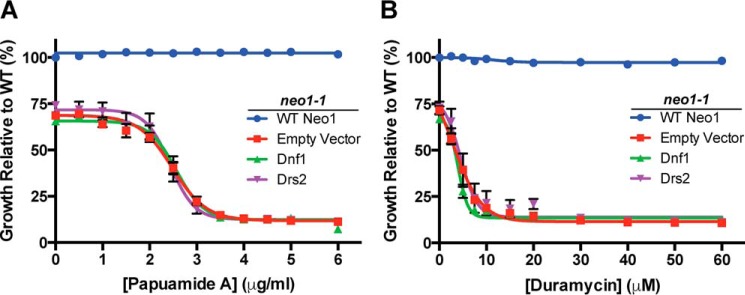
We next tested whether overexpression of Drs2 or Dnf1 could correct the loss of plasma membrane asymmetry caused by inactivating neo1-1. For example, if Neo1 failed to translocate PE to the cytosolic leaflet of early Golgi membranes, Drs2 and/or Dnf1 could potentially flip this substrate as membrane flowed through the TGN or upon arrival at the plasma membrane. However, when assayed at 30 °C, additional copies of Drs2 or Dnf1 failed to suppress the Pap A or duramycin sensitivity of neo1-1. As expected, WT NEO1 fully complemented this phenotype and restored PS and PE asymmetry (Fig. 5, A and B).
FIGURE 5.
Moderate overexpression of DNF1 or DRS2 failed to suppress loss of membrane asymmetry in neo1-1 cells. Pap A (A) and duramycin (B) sensitivity of neo1-1 cells expressing single copy NEO1, DNF1, DRS2, or empty vector at 30 °C; n ≥4.
To further address the relationship between P4-ATPases with potentially overlapping substrates, we tested whether NEO1 could suppress growth and membrane asymmetry defects caused by loss of the Drs2/Dnf group of P4-ATPases. A plasmid shuffle assay was used to show that overexpression of NEO1 from low copy or multicopy vectors failed to suppress the lethality of the drs2Δ dnf1,2,3Δ quadruple mutant (Fig. 6A). Surprisingly, we found that Neo1 overexpression from a low copy plasmid, but not the multicopy plasmid, weakly suppressed the cold-sensitive growth defect of drs2Δ at 20 °C. NEO1 overexpressed from a low copy plasmid, but using the moderate strength ADH promoter (PADH-NEO1) also weakly suppressed drs2Δ (Fig. 6B). To further probe this weak suppression, we compared WT NEO1 with neo1-D503N and found that only WT NEO1 could suppress drs2Δ (Fig. 6C).
FIGURE 6.
NEO1 overexpression weakly suppresses drs2Δ but not dnf1,2,3Δ. A, overexpression of NEO1 failed to suppress dnf1,2,3Δ drs2Δ synthetic lethality. dnf1,2,3Δ drs2Δ pRS416-DNF1 (ZHY704) strains expressing single copy (cen) DRS2 (pRS313-DRS2), empty vector (pRS313), single copy or multicopy (2μ) NEO1 (pRS313-NEO1 or pRS423-NEO1, respectively) were spotted onto minimal media plates (SD), and pRS416-DRS2 was counter-selected on 5-FOA. B, overexpression of NEO1 can partially suppress drs2Δ cold sensitivity. drs2Δ strains (ZHY615M2D) expressing single copy DRS2 (pRS313-DRS2), empty vector (pRS313), or constructs driving increasing NEO1 expression (pRS313-NEO1, pRS413-PADH-NEO1, and pRS423-NEO1, respectively) were spotted onto SD plates and incubated at indicated temperatures. C, catalytically dead neo1-D503N failed to suppress drs2Δ cold sensitivity. D, overexpression of NEO1 exacerbated the hyperacidification of vacuoles in dnf1,2,3Δ cells (PFY3273A). Wild-type cells and dnf1,2,3Δ cells transformed with empty vector (pRS313), single copy NEO1 (pRS313-NEO1), or multicopy NEO1 (pRS423-NEO1) were stained with quinacrine, and vacuolar fluorescence was quantified using flow cytometry relative to WT cells (BY4741) (*, p < 0.01; ***, p < 0.0001, Student's t test, n = 4). Images of colony growth are representative of three independent experiments.
The dnf1,2,3Δ triple mutant does not exhibit a growth defect, but it does display hyperacidic vacuoles, a phenotype also displayed by neo1ts mutants (52). Therefore, we tested whether NEO1 overexpression could suppress this dnf1,2,3Δ phenotype. To assay the relative degree of vacuolar acidification, we stained cells with quinacrine and measured their fluorescence by flow cytometry. Quinacrine accumulation in cells is driven by the pH gradient across the vacuolar membrane, and so the fluorescence intensity is proportional to vacuole acidity. As observed previously, dnf1,2,3Δ cells accumulated more quinacrine in the vacuole than WT cells (Fig. 6D, vector). Surprisingly, instead of suppressing, NEO1 overexpression exacerbated the vacuole hyperacidification phenotype (Fig. 6D, WT and 2μ NEO1). The reason for these negative effects of 2μ NEO1 are unclear, but these data show that moderate overexpression of NEO1 fails to compensate for loss of the Dnf P4-ATPases.
We also examined the influence of NEO1 overexpression on plasma membrane asymmetry defects exhibited by drs2Δ and dnf1,2Δ strains. NEO1 overexpression had no influence on the Pap A or duramycin sensitivity of dnf1,2Δ cells (Fig. 7, A and B). However, we observed a small but statistically significant increase in Pap A and duramycin resistance for drs2Δ cells overexpressing NEO1 from a low copy plasmid relative to the empty vector control (Fig. 7, C and D). The increase in duramycin resistance was more pronounced than the increase is Pap A resistance, again implying that Neo1 has a greater influence on PE than PS asymmetry.
FIGURE 7.
Moderate overexpression of NEO1 can partially suppress loss of PE and PS asymmetry in drs2Δ cells but not in dnf1,2Δ cells. A and B, sensitivity of WT (BY4741) and dnf1,2Δ cells (PFY3275F) expressing empty vector (pRS313) or single copy NEO1 (pRS313-NEO1) to Pap A (A) and duramycin (B) at 30 °C. C and D, sensitivity of drs2Δ cells expressing single copy DRS2 (pRS313-DRS2) and empty vector (pRS313) or single copy NEO1 (pRS313-NEO1) to papuamide A (C) or duramycin (D) at 30 °C. Growth relative to WT cells in the absence of drug was plotted (*, p < 0.01; **, p < 0.001; ***, p < 0.0001 Student's t test; n ≥5). Images are representative of four independent experiments.
Discussion
Membrane asymmetry is a highly conserved feature of the eukaryotic cell plasma membrane, and the P4-ATPases play a crucial role in establishing the differences in phospholipid composition between inner and outer leaflets of the bilayer. Biochemical and genetic data strongly support the proposed flippase activity of several P4-ATPases and demonstrate important differences in substrate preferences for these pumps (11, 17, 18, 35, 40). However, there is a paucity of data for the Neo1/ATP9/TAT-5 branch of the P4-ATPase phylogenetic group with regard to substrate specificity, influence on membrane asymmetry, and functional relationship with other P4-ATPases in the cell.
Here, we show that neo1ts mutants display a loss of plasma membrane asymmetry and aberrantly expose PS and PE in the outer leaflet of the plasma membrane at permissive growth temperatures. Partial thermal inactivation of Neo1ts to produce a mild growth defect led to a substantial increase in PE exposure without significantly increasing exposure of PS relative to cells grown at permissive temperature. This correlation between diminished growth and enhanced exposure of PE suggests that PE translocation by Neo1 could be its essential function. Alternatively, Neo1 could flip PS and PE equally well, but the anionic PS could be preferentially retained in the cytosolic leaflet by interactions with the cytoskeleton or membrane proteins.
Neo1 localizes to cis, medial, and TGN cisternae of the Golgi in addition to late endosomal compartments (42–44). The failure to flip PE and PS from the lumenal leaflet to the cytosolic leaflet of the Golgi could allow exposure of these aminophospholipids in the extracellular leaflet of the plasma membrane as membrane flows by vesicular transport from the Golgi to the plasma membrane. However, the neo1ts mutants express four other P4-ATPases with the ability to flip PE and/or PS that localize downstream of Neo1 to the TGN, early endosomes, and plasma membrane. It is unclear why these P4-ATPases cannot compensate for the Neo1 deficiency and correct the membrane asymmetry defect.
We considered the possibility that loss of asymmetry in neo1ts mutants was a secondary effect of disrupting the function of these other P4-ATPases. However, we could find no evidence that inactivation of Neo1 indirectly perturbed the levels, localization, or activity of Dnf1, Dnf2, or Drs2. We further tested whether there was an inadequate amount of Drs2 and Dnf1 in neo1ts cells by overexpressing these proteins from multicopy plasmids, but there was a striking lack of suppression of either the growth defect or the loss of plasma membrane asymmetry in neo1ts cells. In contrast, we could detect a mild suppression of drs2Δ growth and membrane asymmetry defects by modest overexpression of Neo1. We observed a more substantial correction of PE exposure relative to PS, again suggesting a preference of Neo1 in establishing PE asymmetry. However, the overriding conclusion is that there is very little functional overlap between Neo1 and the Drs2/Dnf group of P4-ATPases.
One possible reason for the lack of functional overlap is that Neo1 flips a different molecular species of PE and PS than does Drs2 or Dnf1. Pap A and duramycin recognize only the headgroups of the lipids and do not appear to distinguish between forms differing in their acyl chain composition. Therefore, we do not know the precise molecular composition of the aminophospholipids exposed in the outer leaflet of neo1ts and drs2Δ mutants. The flippases, in contrast, can distinguish phospholipids by both their headgroup and acyl chain occupancy. For example, we have isolated gain of function mutations in Dnf1 that greatly increase its ability to transport NBD-PS across the plasma membrane, and these mutants fall into two classes (35, 37, 41). The first class of Dnf1 mutants will transport lyso-PS (lacking one acyl chain), but not diacyl-PS, and will not replace the function of Drs2 in vivo. The second mutant class transports lyso-PS and diacyl-PS, and these Dnf1 mutants will replace the function of Drs2 in vivo (37). Similarly, the ability of Neo1 to weakly replace Drs2 function in vivo, but not the function of Dnf1, suggests that Neo1 can flip dually acylated phospholipid. Perhaps there is a significant difference in the preference of Drs2 and Neo1 for phospholipids with different acyl chain lengths or degree of saturation. For example, if Neo1 preferentially flips PE with unsaturated fatty acids, but Drs2 prefers fully saturated PE, we would expect a lack of functional overlap.
What is the function of Neo1 and how does this protein influence membrane asymmetry? The loss of membrane asymmetry in neo1ts mutants supports the idea that Neo1 is a phospholipid flippase, but this is insufficient evidence to prove this point. For example, general defects in membrane biogenesis in neo1 cells could lead to plasma membrane instability and transient ruptures that expose aminophospholipids in the outer leaflet. However, a nonspecific membrane defect would presumably expose PS and PE equivalently, and we observe conditions where PE is preferentially exposed in neo1ts cells relative to PS or preferentially restored to the inner leaflet when Neo1 is overexpressed in drs2Δ cells. Our data also suggest that there is not a general increase in nonspecific lipid flip-flop across the neo1-ts membrane, and the Dnf1 and Dnf2 flippases are active at the plasma membrane.
It was also possible that the loss of membrane asymmetry reflected a secondary consequence of disrupting Neo1 function in protein trafficking or the regulation of organellar pH (42, 52). Prior work has shown neo1ts mutants mislocalize the COPI cargo Rer1 from the Golgi membrane to the vacuole and hyperacidify Golgi and vacuole compartments (42). However, these phenotypes were observed at the nonpermissive temperature of 37 °C, and no significant defect was observed at permissive growth temperatures. In contrast, neo1ts mutants display a loss of asymmetry even at permissive growth temperatures. Moreover, COPI mutants that display a strong defect in Rer1-GFP localization display only a partial loss of PS asymmetry (relative to neo1 or drs2Δ cells) and a negligible loss of PE asymmetry. Together, these data argue that loss of membrane asymmetry is not a secondary consequence of disrupting protein trafficking or organelle acidification in neo1ts cells.
These observations suggest that the primary function of Neo1 is phospholipid translocation with a preference for PE. However, definitive evidence for this biochemical activity requires reconstitution of purified Neo1 into proteoliposomes and detection of a flippase activity. Attempts to reconstitute Neo1 have not yet been successful, but the results reported here will help guide the choice of substrates to test in these assays.
Experimental Procedures
Reagents
5-FOA was purchased from Zymo Research. Papuamide A was purchased from the University of British Columbia Depository. Duramycin, quinacrine dihydrochloride, and calcofluor white were purchased from Sigma. FM4-64 was purchased from Life Technologies, Inc. Monoclonal anti-GFP (clone 1C9A5) and anti-HA antibodies (clone 12CA5) were purchased from Vanderbilt Antibody and Protein Resource. Anti-Arf1 antibody was generated by the Graham laboratory (49). Anti-Pma1 and anti-Kar2 were gifts from Amy Chang (University of Michigan) and Jeff Brodksy (University of Pittsburgh), respectively. IRDye® 800CW goat anti-rabbit IgG (H+L) and 680RD goat anti-mouse IgG (H+L) secondary antibodies were purchased from Li-COR Biosciences.
Strains and Plasmid Construction
Strains and plasmids used in this study are listed in Tables 2 and 3. Yeast strains were grown and transformed using standard media and transformation techniques (53–55). For growth tests on solid media, 0.1 A600 cells were spotted with 10-fold serial dilutions onto synthetic media or synthetic media containing 5-FOA. A NEO1 genomic DNA fragment with 518 bp upstream and 126 bp downstream of the NEO1 open reading frame (ORF) was amplified from yeast genomic DNA using primers containing BamHI and XhoI restriction sites and cloned into pRS416, pRS313, and pRS423 to generate pRS416-NEO1, pRS313-NEO1, and pRS423-NEO1. pRS313-Neo1 D503N and pRS416-Neo1 D503N plasmids were generated by gene splicing overlap extension PCR using Neo1_D503N_For (5′-CGTATTGAATACCTTCTAAGCAATAAAACAGGGACGCTTA-3′) and Neo1_D503N_Rev (5′-TAAGCGTCCCTGTTTTATTGCTTAGAAGGTATTCAATACG-3′) (56).
TABLE 2.
Strains used in this study
| Strain | Genotype | Plasmid | Source |
|---|---|---|---|
| BY4741 | MATa his3Δ1 leu2Δ0 | Invitrogen | |
| ura3Δ0 met15Δ0 | |||
| BY4742 | MATα his3Δ1 leu2Δ0 | Invitrogen | |
| ura3Δ0 lys2Δ0 | |||
| SEY6210 | MATα his3Δ1 leu2Δ0 | 60 | |
| ura3Δ0 lys2Δ0 trp1Δ suc2Δ | |||
| ZHY124-15B1B | MATα his3Δ1 leu2Δ0 | 42 | |
| ura3Δ0 lys2Δ0 trp1Δ suc2Δ | |||
| ade2Δ neo1-1::HIS3-KanMX | |||
| ZHY124-34A2A | MATα his3Δ1 leu2Δ0 | 42 | |
| ura3Δ0 lys2Δ0 trp1Δ suc2Δ | |||
| ade2Δ neo1-2::HIS3-KanMX | |||
| EGY1211-6B | MATα his3Δ1 leu2Δ0 | 61 | |
| ura3Δ0 lys2Δ0 trp1Δ suc2Δ sec21-1 | |||
| EGY101-6D | MATα his3Δ1 leu2Δ0 | 61 | |
| ura3Δ0 lys2Δ0 trp1Δ suc2Δ ret1-1 | |||
| PFY3273A | MATa his3Δ1 leu2Δ0 ura3Δ0 | 34 | |
| met15Δ0 dnf1Δ dnf2Δ dnf3Δ | |||
| PFY3275F | MATa his3Δ1 leu2Δ0 ura3Δ0 | 34 | |
| met15Δ0 dnf1Δ dnf2Δ | |||
| YWY10 | MATa his3Δ1 leu2Δ0 ura3Δ0 | This study | |
| lys2Δ0 neo1Δ::KanMX | pRS416-NEO1 | ||
| ZHY615M2D | MATα his3Δ1 leu2Δ0 ura3Δ0 | 34 | |
| lys2Δ0 drs2Δ | |||
| ZHY704 | MATα his3Δ1 leu2Δ0 ura3Δ0 | ||
| lys2Δ0 dnf1Δ dnf2Δ dnf3ΔΔ | pRS416-DRS2 | 34 | |
| drs2Δ::LEU2 | |||
| MTY219RR | MATa his3Δ1 leu2Δ0 ura3Δ0 | This study | |
| lys2Δ0 neo1Δ::KanMX | pR413-NEO1 | ||
| MTY628-15B | MATa his3Δ1 leu2Δ0 ura3Δ0 | This study | |
| lys2Δ0 neo1Δ::KanMX | pR413-neo1-1 | ||
| MTY628-34A | MATa his3Δ1 leu2Δ0 ura3Δ0 | pR413-neo1-2 | This study |
| lys2Δ0 neo1Δ::KanMX | |||
| MTY219RRL | MATa his3Δ1 leu2Δ0 ura3Δ0 | pRS315-NEO1 | This study |
| lys2Δ0 neo1Δ::KanMX | |||
| MTY628-15BL | MATa his3Δ1 leu2Δ0 ura3Δ0 | pRS315-neo1-1 | This study |
| lys2Δ0 neo1Δ::KanMX | |||
| MTY628-34AL | MATa his3Δ1 leu2Δ0 ura3Δ0 | pRS315-neo1-2 | This study |
| lys2Δ0 neo1Δ::KanMX | |||
| MTYD1-219RRL MATa | his3Δ1 leu2Δ0 ura3Δ0 | pRS315-NEO1 | This study |
| lys2Δ0 neo1Δ::KanMX DNF1::6XHA::His5 | |||
| MTYD1-62815BL MATa | his3Δ1 leu2Δ0 ura3Δ0 | pRS315-neo1-1 | This study |
| lys2Δ0 neo1Δ::KanMX DNF1::6×HA::His5 | |||
| MTYD2-219RRL | DNF2::6XHA::His5 | pRS315-NEO1 | This study |
| MTYD2-62815BL MATa | his3Δ1 leu2Δ0 ura3Δ0 | pRS315-neo1-1 | This study |
| lys2Δ0 neo1Δ::KanMX DNF2::6XHA::His5 | |||
| RBY3901 | ZHY615M2D | pRS313 | 35 |
| RBY3904 | ZHY615M2D | pRS313-DRS2 | 35 |
| MTY40 | ZHY615M2D | pRS313-NEO1 | This study |
| MTY44 | ZHY615M2D | pRS413-ADH-NEO1 | This study |
| MTY48 | ZHY615M2D | pRS423-NEO1 | This study |
| MTY52 | ZHY615M2D | pRS313-Neo1[D503N] | This study |
| MTY300 | ZHY704 | pRS313 | This study |
| MTY303 | ZHY704 | pRS313-DNF1 | This study |
| MTY306 | ZHY704 | pRS313-DRS2 | This study |
| MTY309 | ZHY704 | pRS313-NEO1 | This study |
| MTY312 | ZHY704 | pRS423-NEO1 | This study |
| RBY601 | PFY3275F | pRS313 | 35 |
| RBY604 | PFY3275F | pRS313-DNF1 | 35 |
| MTY104 | PFY3275F | pRS313-NEO1 | This study |
| MTY5001 | MTY219RR | pRS416 | This study |
| MTY5005 | MTY628-15B | pRS416-NEO1 | This study |
| MTY5009 | MTY628-15B | pRS416 | This study |
| MTY5017 | MTY628-15B | pRS416-DNF1 | This study |
| MTY5025 | MTY628-15B | pRS416-DRS2 | This study |
| MTY6000 | MTY628-15B | pRS416; pRS425 | This study |
| MTY6004 | MTY628-15B | pRS416-NEO1; pRS425 | This study |
| MTY6008 | MTY628-15B | pRS416; pRS425 | This study |
| MTY6012 | MTY628-15B | pRS416-DNF1; pRS425-LEM3 | This study |
| MTY6016 | MTY628-15B | pRS426-DNF1; pRS425-LEM3 | This study |
| MTY6020 | MTY628-15B | pRS315-DRS2; pRS426-CDC50 | This study |
| MTY6024 | MTY628-15B | pRS425-DRS2; pRS426-CDC50 | This study |
| MTY400 | BY4741 | pRS313 | This study |
| MTY403 | PFY3273A | pRS313-NEO1 | This study |
| MTY406 | PFY3273A | pRS423-NEO1 | This study |
| MTY700 | YWY10 | pRS313; pRS425 | This study |
| MTY703 | YWY10 | pRS313-NEO1; pRS425 | This study |
| MTY706 | YWY10 | pRS313-DNF1; pRS425-LEM3 | This study |
| MTY709 | YWY10 | pRS423-DNF1; pRS425-LEM3 | This study |
| MTY712 | YWY10 | pRS315-DRS2; pRS423-CDC50 | This study |
| MTY715 | YWY10 | pRS425-DRS2; pRS423-CDC50 | This study |
| MTY718 | YWY10 | pRS313-Neo1[D503N]; pRS425 | This study |
| MTY6210R | SEY6210 | pSKY5/RER1–0 | This study |
| MTY15B1BR | ZHY124–15B1B | pSKY5/RER1–0 | This study |
| MTY34A2AR | ZHY124–34A2A | pSKY5/RER1–0 | This study |
| MTY1211–6BR | EGY1211–6B | pSKY5/RER1–0 | This study |
| MTY101-6DR | EGY1211–6D | pSKY5/RER1–0 | This study |
TABLE 3.
Plasmids used in this study
| Plasmid | Notes | Source |
|---|---|---|
| pR413-NEO1 | cen, PNEO1, HIS3 | 34 |
| pR413-neo1-1 | cen, PNEO1, HIS3 | 42 |
| pR413-neo1-2 | cen, PNEO1, HIS3 | 42 |
| pRS416-NEO1 | cen, PNEO1, URA3 | This study |
| pRS416 | cen, URA3 | 51 |
| pRS425 | cen, LEU2 | 55 |
| pRS313 | cen, HIS3 | 55 |
| pRS423 | 2μ, HIS3 | 34 |
| pRS313-DRS2 | cen, PDRS2, HIS3 | This study |
| pRS313-NEO1 | cen, PNEO1, HIS3 | 34 |
| pRS413-ADH-NEO1 | cen, PADH, HIS3 | 34 |
| pRS423-NEO1 | 2μ, PNEO1, HIS3 | This study |
| pRS313-Neo1[D503N] | cen, PNEO1, HIS3 | This study |
| pRS315-DRS2 | cen, PDRS2, LEU2 | 34 |
| pRS425-DRS2 | 2μ, PDRS2, LEU2 | 34 |
| pRS313-DNF1 | cen, PDNF1, HIS3 | 36 |
| pRS423-DNF1 | 2μ, PDNF1, HIS3 | 34, 37 |
| pRS416-DNF1 | cen, PDNF1, URA3 | 33 |
| pRS426-DNF1 | 2μ, PDNF1, URA3 | 33 |
| pRS423-CDC50 | 2μ, PCDC50, HIS3 | 33 |
| pRS425-LEM3 | 2μ, PLEM3, LEU2 | 35 |
| pSKY5/RER1-0 | cen, PTDH3, URA3 | 62 |
| PYM15 | integrating, HIS5, 6XHA | 63 |
The plasmid pRS416-NEO1 was transformed into BY4743 neo1Δ, and the resulting cells were sporulated. After random sporulation, spore YWY10 (MATa his3 leu2 met15 ura3 lys2 neo1Δ pRS416-NEO1) was selected on SD plates with G418 but lacking uracil. A plasmid shuffling strategy was used to replace the plasmid pRS416-NEO1 in YWY10 with plasmids containing neo1ts alleles. pRS413-neo1-1 and pRS413-neo1-2 were transformed into YWY10, and the resulting cells were selected with 5-FOA to exclude cells containing pRS416-NEO1. The resulting strains transformed with pRS413-neo1-1 and pRS413-neo1-2 were MTY628-15B and MTY628-34A, respectively.
Sensitivity Assays
For Pap A and duramycin treatments, 0.1 A600 of midlog phase cells were seeded in 96-well plates with or without drug. Plates were incubated at 27 or 30 °C for 20 h, and the concentration of cells was measured in A600/ml with a Multimode Plate Reader Synergy HT (Bio-Tek). Relative growth was compared with vehicle-treated WT cells in the assay. For data presentation, the sigmoidal curve fitting modality from GraphPad Prism6 was used when adjusted R2 values were greater than 0.8 for all experimental groups. When curve fitting produced an R2 value of <0.8, data points were simply connected without curve fitting to generate the toxin sensitivity graphs.
Quinacrine Staining
For quinacrine staining, cells were grown to mid-logarithmic phase in enriched media at 30 °C. Cells were resuspended in YPD medium containing 100 mm HEPES, pH 7.6, 200 μm quinacrine with 25 μg/ml propidium iodide and stained for 8 min at 30 °C. Stained cells were washed three times with 100 mm HEPES, pH 7.6, and the average vacuolar fluorescence intensity of 10,000 live cells was measured using the FITC channel of a fluorescence-activated cell sorter (52).
Lipid Uptake Assay
Lipid uptake assays were performed with approximately 1 A600 of wild-type and neo1ts mutant cells grown at 30 °C to mid-logarithmic phase and using 1-palmitoyl-2{6-[(7-nitro-2–1,3-benzoxadiazol-4-yl)amino]hexanoyl)-sn-glycero-3-phospholipids (NBD-PLs) for 30 min as described previously (35). The amount of NBD-PL taken up by dnf1,2Δ cells was measured and subtracted as background from the NBD-PL taken up by WT and neo1ts cells. Lipid uptake activities were plotted as the percentage of NBD-PC lipid uptake activity for wild type cells.
Fluorescence Microscopy
Cells were washed three times prior to imaging with PBS containing 2% glucose. Images were acquired on a Zeiss Axioplan Scope using a 63× Plan-Apochromat oil differential interference contrast objective. An FITC or Cy3 filter was used for visualizing GFP or FM4-64 respectively.
Vacuolar Staining
Cells were grown at the indicated temperatures to mid-logarithmic phase and stained using 32 μm FM4-64 in enriched medium (YPD) for 20 min in the dark. The cells were chased in YPD lacking FM4-64 for 3 h and then washed twice with PBS containing 2% glucose prior to imaging.
Western Blotting Analysis
Cells were grown to mid-logarithmic phase at indicated temperatures. 0.1 A600 eq of cell lysate was subjected to SDS-gel electrophoresis. Western blotting analysis was performed using anti-GFP antibody (1:2000) (57) to distinguish between cleaved GFP (26 kDa) and intact GFP-tagged Rer1 (40 kDa). For quantitation, membranes were scanned at 800 nm using the Odyssey Infrared Imaging System; median GFP intensities were quantified using Odyssey software (LI-COR Biosciences). Ratios of GFPcleaved versus GFPtotal were plotted.
Subcellular Fractionation
Cell lysates were analyzed by subcellular fractionation as described previously (57). Seven fractions in total were collected from the top of the sucrose gradient. Each fraction was immunoblotted against Kar2 (ER marker) (anti-Kar2, 1:2000 (58)) and Pma1 (plasma membrane marker) (anti-Pma1, 1:5000 (59)) to assess the success of the fractionation. In addition, Dnf1, Dnf2, and Drs2 levels were quantified in each fraction by immunoblotting against an HA tag (Dnf1-HA and Dnf2-HA) (anti-HA antibody, 1:1000, Vanderbilt Antibody and Protein Resource) and Drs2 (anti-Drs2, 1:500 (39)). The HA tags were integrated into the chromosomal DNF1 and DNF2 loci so the tagged proteins would be expressed at endogenous levels. Protein levels in each fraction were quantified as the percentage of total amount of that protein in the seven fractions. Subcellular fractionation was performed three independent times.
Protein concentrations of total cell lysates collected for subcellular fractionations were measured using a bicinchoninic acid assay (BCA) according to the manufacturer's instructions. 0.75 mg of total cell lysates from each experimental group was subjected to SDS-gel electrophoresis. Western blotting analysis was performed to measure the levels of endogenously HA-tagged Dnf1, Dnf2, and native Drs2 levels in wild-type and neo1ts cells both at permissive and non-permissive temperatures. Immunoblots were also probed with anti-Arf1 serum (49). Levels of Dnf1, Dnf2, and Drs2 proteins were quantified in neo1ts cells relative to the levels of these proteins in wild-type cells. Three independent transformants from each experimental group was used for Western blotting analysis.
Author Contributions
M. T. conducted most experiments. Y. W. helped with strain and plasmid construction. M. T. and T. R. G. analyzed the data and wrote the paper.
Acknowledgments
We thank Jeff Brodsky for anti-Kar2 and Amy Chang for anti-Pma1 antibodies. Flow cytometry experiments were performed in the Vanderbilt University Medical Center (VUMC) Flow Cytometry Shared Resource. We also thank Hannah M. Hankins for reviewing an earlier version of the manuscript. The VUMC Flow Cytometry Shared Resource is supported by the Vanderbilt Ingram Cancer Center by National Institutes of Health Grant P30 CA68485 and Vanderbilt Digestive Disease Research Center by National Institutes of Health DK058404.
This work was supported by National Institutes of Health Grant GM107978 (to T. R. G.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- PE
- phosphatidylethanolamine
- PS
- phosphatidylserine
- PA
- phosphatidic acid
- PC
- phosphatidylcholine
- NBD-PL
- 1-palmitoyl-2{6-[(7-nitro-2–1,3-benzoxadiazol-4-yl)amino]hexanoyl)-sn-glycero-3-phospholipid
- CW
- calcofluor white
- TGN
- trans-Golgi network
- ER
- endoplasmic reticulum
- 5-FOA
- 5-fluoroorotic acid
- Pap A
- papuamide A.
References
- 1. Sebastian T. T., Baldridge R. D., Xu P., and Graham T. R. (2012) Phospholipid flippases: building asymmetric membranes and transport vesicles. Biochim. Biophys. Acta 1821, 1068–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hankins H. M., Baldridge R. D., Xu P., and Graham T. R. (2015) Role of flippases, scramblases and transfer proteins in phosphatidylserine subcellular distribution. Traffic 16, 35–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bretscher M. S. (1972) Phosphatidyl-ethanolamine: differential labelling in intact cells and cell ghosts of human erythrocytes by a membrane-impermeable reagent. J. Mol. Biol. 71, 523–528 [DOI] [PubMed] [Google Scholar]
- 4. Verkleij A. J., Zwaal R. F., Roelofsen B., Comfurius P., Kastelijn D., and van Deenen L. L. (1973) The asymmetric distribution of phospholipids in the human red cell membrane. A combined study using phospholipases and freeze-etch electron microscopy. Biochim. Biophys. Acta 323, 178–193 [DOI] [PubMed] [Google Scholar]
- 5. Gordesky S. E., Marinetti G. V., and Love R. (1975) The reaction of chemical probes with the erythrocyte membrane. J. Membr. Biol. 20, 111–132 [DOI] [PubMed] [Google Scholar]
- 6. van Meer G., Voelker D. R., and Feigenson G. W. (2008) Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9, 112–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Emoto K., Kobayashi T., Yamaji A., Aizawa H., Yahara I., Inoue K., and Umeda M. (1996) Redistribution of phosphatidylethanolamine at the cleavage furrow of dividing cells during cytokinesis. Proc. Natl. Acad. Sci. U.S.A. 93, 12867–12872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Emoto K., and Umeda M. (2000) An essential role for a membrane lipid in cytokinesis. Regulation of contractile ring disassembly by redistribution of phosphatidylethanolamine. J. Cell Biol. 149, 1215–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Graham T. R. (2004) Flippases and vesicle-mediated protein transport. Trends Cell Biol. 14, 670–677 [DOI] [PubMed] [Google Scholar]
- 10. Xu P., Baldridge R. D., Chi R. J., Burd C. G., and Graham T. R. (2013) Phosphatidylserine flipping enhances membrane curvature and negative charge required for vesicular transport. J. Cell Biol. 202, 875–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Alder-Baerens N., Lisman Q., Luong L., Pomorski T., and Holthuis J. C. (2006) Loss of P4 ATPases Drs2p and Dnf3p disrupts aminophospholipid transport and asymmetry in yeast post-Golgi secretory vesicles. Mol. Biol. Cell 17, 1632–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Segawa K., Kurata S., Yanagihashi Y., Brummelkamp T. R., Matsuda F., and Nagata S. (2014) Caspase-mediated cleavage of phospholipid flippase for apoptotic phosphatidylserine exposure. Science 344, 1164–1168 [DOI] [PubMed] [Google Scholar]
- 13. Yang H., Kim A., David T., Palmer D., Jin T., Tien J., Huang F., Cheng T., Coughlin S. R., Jan Y. N., and Jan L. Y. (2012) TMEM16F forms a Ca2+-activated cation channel required for lipid scrambling in platelets during blood coagulation. Cell 151, 111–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Suzuki J., Umeda M., Sims P. J., and Nagata S. (2010) Calcium-dependent phospholipid scrambling by TMEM16F. Nature 468, 834–838 [DOI] [PubMed] [Google Scholar]
- 15. Kühlbrandt W. (2004) Biology, structure and mechanism of P-type ATPases. Nat. Rev. Mol. Cell Biol. 5, 282–295 [DOI] [PubMed] [Google Scholar]
- 16. Palmgren M. G., and Nissen P. (2011) P-type ATPases. Annu. Rev. Biophys. 40, 243–266 [DOI] [PubMed] [Google Scholar]
- 17. Zhou X., and Graham T. R. (2009) Reconstitution of phospholipid translocase activity with purified Drs2p, a type-IV P-type ATPase from budding yeast. Proc. Natl. Acad. Sci. U.S.A. 106, 16586–16591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Coleman J. A., Kwok M. C., and Molday R. S. (2009) Localization, purification, and functional reconstitution of the P4-ATPase Atp8a2, a phosphatidylserine flippase in photoreceptor disc membranes. J. Biol. Chem. 284, 32670–32679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. van Veen S., Sørensen D. M., Holemans T., Holen H. W., Palmgren M. G., and Vangheluwe P. (2014) Cellular function and pathological role of ATP13A2 and related P-type transport ATPases in Parkinson's disease and other neurological disorders. Front. Mol. Neurosci. 7, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vestergaard A. L., Coleman J. A., Lemmin T., Mikkelsen S. A., Molday L. L., Vilsen B., Molday R. S., Dal Peraro M., and Andersen J. P. (2014) Critical roles of isoleucine-364 and adjacent residues in a hydrophobic gate control of phospholipid transport by the mammalian P4-ATPase ATP8A2. Proc. Natl. Acad. Sci. U.S.A. 111, E1334–E1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Puts C. F., Panatala R., Hennrich H., Tsareva A., Williamson P., and Holthuis J. C. (2012) Mapping functional interactions in a heterodimeric phospholipid pump. J. Biol. Chem. 287, 30529–30540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. van der Velden L. M., Wichers C. G., van Breevoort A. E., Coleman J. A., Molday R. S., Berger R., Klomp L. W., and van de Graaf S. F. (2010) Heteromeric interactions required for abundance and subcellular localization of human CDC50 proteins and class 1 P4-ATPases. J. Biol. Chem. 285, 40088–40096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Saito K., Fujimura-Kamada K., Furuta N., Kato U., Umeda M., and Tanaka K. (2004) Cdc50p, a protein required for polarized growth, associates with the Drs2p P-type ATPase implicated in phospholipid translocation in Saccharomyces cerevisiae. Mol. Biol. Cell 15, 3418–3432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Onat O. E., Gulsuner S., Bilguvar K., Nazli Basak A., Topaloglu H., Tan M., Tan U., Gunel M., and Ozcelik T. (2013) Missense mutation in the ATPase, aminophospholipid transporter protein ATP8A2 is associated with cerebellar atrophy and quadrupedal locomotion. Eur. J. Hum. Genet. 21, 281–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhu X., Libby R. T., de Vries W. N., Smith R. S., Wright D. L., Bronson R. T., Seburn K. L., and John S. W. (2012) Mutations in a P-type ATPase gene cause axonal degeneration. PLoS Genet. 8, e1002853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bull L. N., van Eijk M. J., Pawlikowska L., DeYoung J. A., Juijn J. A., Liao M., Klomp L. W., Lomri N., Berger R., Scharschmidt B. F., Knisely A. S., Houwen R. H., and Freimer N. B. (1998) A gene encoding a P-type ATPase mutated in two forms of hereditary cholestasis. Nat. Genet. 18, 219–224 [DOI] [PubMed] [Google Scholar]
- 27. Klomp L. W., Vargas J. C., van Mil S. W., Pawlikowska L., Strautnieks S. S., van Eijk M. J., Juijn J. A., Pabón-Peña C., Smith L. B., DeYoung J. A., Byrne J. A., Gombert J., van der Brugge G., Berger R., Jankowska I., et al. (2004) Characterization of mutations in ATP8B1 associated with hereditary cholestasis. Hepatology 40, 27–38 [DOI] [PubMed] [Google Scholar]
- 28. Stapelbroek J. M., Peters T. A., van Beurden D. H., Curfs J. H., Joosten A., Beynon A. J., van Leeuwen B. M., van der Velden L. M., Bull L., Oude Elferink R. P., van Zanten B. A., Klomp L. W., and Houwen R. H. (2009) ATP8B1 is essential for maintaining normal hearing. Proc. Natl. Acad. Sci. U.S.A. 106, 9709–9714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dhar M. S., Sommardahl C. S., Kirkland T., Nelson S., Donnell R., Johnson D. K., and Castellani L. W. (2004) Mice heterozygous for Atp10c, a putative amphipath, represent a novel model of obesity and type 2 diabetes. J. Nutr. 134, 799–805 [DOI] [PubMed] [Google Scholar]
- 30. Siggs O. M., Arnold C. N., Huber C., Pirie E., Xia Y., Lin P., Nemazee D., and Beutler B. (2011) The P4-type ATPase ATP11C is essential for B lymphopoiesis in adult bone marrow. Nat. Immunol. 12, 434–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yabas M., Teh C. E., Frankenreiter S., Lal D., Roots C. M., Whittle B., Andrews D. T., Zhang Y., Teoh N. C., Sprent J., Tze L. E., Kucharska E. M., Kofler J., Farell G. C., Bröer S., Goodnow C. C., and Enders A. (2011) ATP11C is critical for the internalization of phosphatidylserine and differentiation of B lymphocytes. Nat. Immunol. 12, 441–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yabas M., Coupland L. A., Cromer D., Winterberg M., Teoh N. C., D'Rozario J., Kirk K., Bröer S., Parish C. R., and Enders A. (2014) Mice deficient in the putative phospholipid flippase ATP11C exhibit altered erythrocyte shape, anemia, and reduced erythrocyte life span. J. Biol. Chem. 289, 19531–19537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xu P., Okkeri J., Hanisch S., Hu R. Y., Xu Q., Pomorski T. G., and Ding X. Y. (2009) Identification of a novel mouse P4-ATPase family member highly expressed during spermatogenesis. J. Cell Sci. 122, 2866–2876 [DOI] [PubMed] [Google Scholar]
- 34. Hua Z., Fatheddin P., and Graham T. R. (2002) An essential subfamily of Drs2p-related P-type ATPases is required for protein trafficking between Golgi complex and endosomal/vacuolar system. Mol. Biol. Cell 13, 3162–3177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Baldridge R. D., and Graham T. R. (2012) Identification of residues defining phospholipid flippase substrate specificity of type IV P-type ATPases. Proc. Natl. Acad. Sci. U.S.A. 109, E290–E298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu K., Hua Z., Nepute J. A., and Graham T. R. (2007) Yeast P4-ATPases Drs2p and Dnf1p are essential cargos of the NPFXD/Sla1p endocytic pathway. Mol. Biol. Cell 18, 487–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Baldridge R. D., Xu P., and Graham T. R. (2013) Type IV P-type ATPases distinguish mono- versus diacyl phosphatidylserine using a cytofacial exit gate in the membrane domain. J. Biol. Chem. 288, 19516–19527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Riekhof W. R., and Voelker D. R. (2006) Uptake and utilization of lyso-phosphatidylethanolamine by Saccharomyces cerevisiae. J. Biol. Chem. 281, 36588–36596 [DOI] [PubMed] [Google Scholar]
- 39. Chen C. Y., Ingram M. F., Rosal P. H., and Graham T. R. (1999) Role for Drs2p, a P-type ATPase and potential aminophospholipid translocase, in yeast late Golgi function. J. Cell Biol. 147, 1223–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Natarajan P., Wang J., Hua Z., and Graham T. R. (2004) Drs2p-coupled aminophospholipid translocase activity in yeast Golgi membranes and relationship to in vivo function. Proc. Natl. Acad. Sci. U.S.A. 101, 10614–10619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Baldridge R. D., and Graham T. R. (2013) Two-gate mechanism for phospholipid selection and transport by type IV P-type ATPases. Proc. Natl. Acad. Sci. U.S.A. 110, E358–E367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hua Z., and Graham T. R. (2003) Requirement for neo1p in retrograde transport from the Golgi complex to the endoplasmic reticulum. Mol. Biol. Cell 14, 4971–4983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wicky S., Schwarz H., and Singer-Krüger B. (2004) Molecular interactions of yeast Neo1p, an essential member of the Drs2 family of aminophospholipid translocases, and its role in membrane trafficking within the endomembrane system. Mol. Cell. Biol. 24, 7402–7418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., and O'Shea E. K. (2003) Global analysis of protein localization in budding yeast. Nature 425, 686–691 [DOI] [PubMed] [Google Scholar]
- 45. Wehman A. M., Poggioli C., Schweinsberg P., Grant B. D., and Nance J. (2011) The P4-ATPase TAT-5 inhibits the budding of extracellular vesicles in C. elegans embryos. Curr. Biol. 21, 1951–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Parsons A. B., Lopez A., Givoni I. E., Williams D. E., Gray C. A., Porter J., Chua G., Sopko R., Brost R. L., Ho C. H., Wang J., Ketela T., Brenner C., Brill J. A., Fernandez G. E., et al. (2006) Exploring the mode-of-action of bioactive compounds by chemical-genetic profiling in yeast. Cell 126, 611–625 [DOI] [PubMed] [Google Scholar]
- 47. Aoki Y., Uenaka T., Aoki J., Umeda M., and Inoue K. (1994) A novel peptide probe for studying the transbilayer movement of phosphatidylethanolamine. J. Biochem. 116, 291–297 [DOI] [PubMed] [Google Scholar]
- 48. Iwamoto K., Hayakawa T., Murate M., Makino A., Ito K., Fujisawa T., and Kobayashi T. (2007) Curvature-dependent recognition of ethanolamine phospholipids by duramycin and cinnamycin. Biophys. J. 93, 1608–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liu K., Surendhran K., Nothwehr S. F., and Graham T. R. (2008) P4-ATPase requirement for AP-1/clathrin function in protein transport from the trans-Golgi network and early endosomes. Mol. Biol. Cell 19, 3526–3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Georgiev A. G., Sullivan D. P., Kersting M. C., Dittman J. S., Beh C. T., and Menon A. K. (2011) Osh proteins regulate membrane sterol organization but are not required for sterol movement between the ER and PM. Traffic 12, 1341–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jarmoszewicz K., L̸ukasiak K., Riezman H., and Kaminska J. (2012) Rsp5 ubiquitin ligase is required for protein trafficking in Saccharomyces cerevisiae COPI mutants. PloS one 7, e39582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Brett C. L., Kallay L., Hua Z., Green R., Chyou A., Zhang Y., Graham T. R., Donowitz M., and Rao R. (2011) Genome-wide analysis reveals the vacuolar pH-stat of Saccharomyces cerevisiae. PLoS One 6, e17619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sherman F. (2002) Getting started with yeast. Methods Enzymol. 350, 3–41 [DOI] [PubMed] [Google Scholar]
- 54. Gietz R. D., and Schiestl R. H. (2007) High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2, 31–34 [DOI] [PubMed] [Google Scholar]
- 55. Sikorski R. S., and Hieter P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Heckman K. L., and Pease L. R. (2007) Gene splicing and mutagenesis by PCR-driven overlap extension. Nat. Protoc. 2, 924–932 [DOI] [PubMed] [Google Scholar]
- 57. Hankins H. M., Sere Y. Y., Diab N. S., Menon A. K., and Graham T. R. (2015) Phosphatidylserine translocation at the yeast trans-Golgi network regulates protein sorting into exocytic vesicles. Mol. Biol. Cell 26, 4674–4685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Brodsky J. L., and Schekman R. (1993) A Sec63p-BiP complex from yeast is required for protein translocation in a reconstituted proteoliposome. J. Cell Biol. 123, 1355–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gong X., and Chang A. (2001) A mutant plasma membrane ATPase, Pma1–10, is defective in stability at the yeast cell surface. Proc. Natl. Acad. Sci. U.S.A. 98, 9104–9109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Robinson J. S., Klionsky D. J., Banta L. M., and Emr S. D. (1988) Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell. Biol. 8, 4936–4948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gaynor E. C., Chen C. Y., Emr S. D., and Graham T. R. (1998) ARF is required for maintenance of yeast Golgi and endosome structure and function. Mol. Biol. Cell 9, 653–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sato K., Sato M., and Nakano A. (2001) Rer1p, a retrieval receptor for endoplasmic reticulum membrane proteins, is dynamically localized to the Golgi apparatus by coatomer. J. Cell Biol. 152, 935–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Janke C., Magiera M. M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., and Knop M. (2004) A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21, 947–962 [DOI] [PubMed] [Google Scholar]