Abstract
Non-protein amino acids, particularly isomers of the proteinogenic amino acids, present a threat to proteome integrity if they are mistakenly inserted into proteins. Quality control during aminoacyl-tRNA synthesis reduces non-protein amino acid incorporation by both substrate discrimination and proofreading. For example phenylalanyl-tRNA synthetase (PheRS) proofreads the non-protein hydroxylated phenylalanine derivative m-Tyr after its attachment to tRNAPhe. We now show in Saccharomyces cerevisiae that PheRS misacylation of tRNAPhe with the more abundant Phe oxidation product o-Tyr is limited by kinetic discrimination against o-Tyr-AMP in the transfer step followed by o-Tyr-AMP release from the synthetic active site. This selective rejection of a non-protein aminoacyl-adenylate is in addition to known kinetic discrimination against certain non-cognates in the activation step as well as catalytic hydrolysis of mispaired aminoacyl-tRNAPhe species. We also report an unexpected resistance to cytotoxicity by a S. cerevisiae mutant with ablated post-transfer editing activity when supplemented with o-Tyr, cognate Phe, or Ala, the latter of which is not a substrate for activation by this enzyme. Our phenotypic, metabolomic, and kinetic analyses indicate at least three modes of discrimination against non-protein amino acids by S. cerevisiae PheRS and support a non-canonical role for SccytoPheRS post-transfer editing in response to amino acid stress.
Keywords: amino acid, aminoacyl tRNA synthetase, protein synthesis, transfer RNA (tRNA), translation, Quality control
Introduction
The units of genetic information contained in mRNA codons need to be precisely translated at the ribosome into corresponding amino acid sequences in proteins. Because protein function is dictated by structure and, thus, amino acid sequence, mistakes in translation of the genetic code can yield drastic alterations in the properties of products of mistranslation. Quality control mechanisms exist that help maintain translation fidelity, limiting mistranslation to one mistake in 103-104 codons under normal growth conditions (1). The aminoacyl-tRNA synthetase (aaRS)2 enzymes, which pair amino acids and tRNAs for participation as substrates in protein synthesis, are among the primary determinants of translational fidelity. In the event that a non-cognate amino acid or tRNA is utilized as an aaRS substrate, the resulting mispaired aminoacyl-tRNA (aa-tRNA) species may lead to an error in decoding at the ribosome, resulting in mistranslation of the genetic code. To limit these errors, aaRS enzymes have evolved strict quality control mechanisms, which typically confer high substrate specificity and contribute to an estimated rate of mispaired aa-tRNA production limited to roughly 1 per 3000 aa-tRNAs produced (2), although deviations exist between species and specific aaRSs (1, 3, 4). Some aaRS enzymes hydrolyze or selectively release (5) aminoacyl-adenylate (aa-AMP) species resulting from non-cognate amino acid activation in a quality control step before transfer of the aminoacyl moiety to tRNA. This “pretransfer” editing can be tRNA-dependent (6) or tRNA-independent (7). Approximately half of aaRS enzymes bear a “post-transfer” editing domain distinct from the synthetic domain (8). The active sites of these editing domains serve to recognize misacylated tRNA species and hydrolyze mispaired aa-tRNA ester linkages. aaRSs also rely heavily on substrate discrimination in the synthetic active site. Amino acids or tRNAs with incompatible biophysical geometry in an aaRS active site may bind weakly if at all. As a result, poor substrates for aaRS catalysis are limited in their participation in aa-tRNA synthesis and, thus, ribosomal translation.
Translational fidelity mediated by aaRS enzymes can be a context-dependent phenomenon. In particular, a shift in the abundance of an aaRS cognate or non-cognate substrate may result in unusually high misacylation of tRNAs. Illustrative of this effect is the observation that Chinese hamster ovary cells starved for tyrosine exhibit a high degree of mistranslation of Tyr codons as Phe (9). Rather than relying on editing activity, tyrosyl-tRNA synthetase exhibits high kinetic discrimination between Tyr and Phe at the amino acid activation step. In an environment depleted for its cognate Tyr, an unusually high degree of near-cognate Phe utilization by TyrRS and mistranslation of Tyr codons as Phe are observed. Oxidative stress also plays an important role in non-cognate aa-tRNA synthesis either by generating non-protein amino acids, which must then be proofread (10), or by direct inactivation of such proofreading (10–12). Oxidative stress effects an increase in reactive oxygen species, which may directly damage amino acids. For example, Phe may be oxidatively converted to one of several derivatives. Among these are the proteinogenic amino acid p-Tyr and the non-protein amino acids 3,4-dihydroxy-l-phenylalanine (l-DOPA), ortho-tyrosine (o-Tyr), and meta-tyrosine (m-Tyr) (Fig. 1). Of these, only m-Tyr is a threat to translational fidelity in an Escherichia coli (E. coli) phenylalanyl-tRNA synthetase (EcPheRS) mutant strain defective in post-transfer editing activity (10). The post-transfer editing activity of the wild-type enzyme can hydrolyze m-Tyr-tRNAPhe species that are synthesized as m-Tyr accumulates under conditions of oxidative stress. p-Tyr, which is present at levels necessary for typical protein synthesis under normal conditions, is the most obvious candidate as a non-cognate threat to PheRS product specificity; surprisingly, p-Tyr presents no such challenge to EcPheRS. Without wild-type EcPheRS post-transfer editing activity, E. coli grows as well as wild type, even when challenged with an excess of p-Tyr. Kinetic analysis revealed a high degree of EcPheRS discrimination in the amino acid activation step against p-Tyr but not m-Tyr. Taken together, these observations show that normal EcPheRS editing activity is critical, not under typical growth conditions, but to protect E. coli from cytotoxic mistranslation of Phe codons in conditions that favor intracellular accumulation of m-Tyr (10).
FIGURE 1.
Potential non-cognate amino acid substrates of PheRS. Shown are hydroxylated phenylalanine derivatives that may accumulate as the result of oxidative damage, metabolic conversion, or import.
In contrast to the E. coli enzyme, Saccharomyces cerevisiae cytoplasmic PheRS (SccytoPheRS) exhibits poor discrimination in the amino acid activation step against p-Tyr (∼20-fold lower than EcPheRS), but p-Tyr-tRNAPhe production is limited by post-transfer editing (10). As with EcPheRS, SccytoPheRS post-transfer editing of m-Tyr-tRNAPhe limits cytotoxicity of m-Tyr. Here, we show unexpected phenotypic effects of direct media supplementation of S. cerevisiae with cognate Phe, proteinogenic non-cognate Ala, and non-protein oxidized Phe derivatives in a SccytoPheRS post-transfer quality control mutant. We highlight striking differences in the promiscuity of the yeast and bacterial enzymes for non-protein amino acids and discuss multiple strategies employed by SccytoPheRS to maintain product specificity in the face of conditional threats to translational quality control in a biological context.
Experimental Procedures
Strains and Growth Conditions
We previously constructed a βD243A SccytoPheRS strain with defective post-transfer editing activity (frs1−1), and a wild-type strain constructed from this background, which we refer to as wild type throughout this manuscript (13). Wild type and frs1−1 S. cerevisiae stain were grown in the presence of varying levels of supplemented amino acids as indicated at 30 °C in a 96-well microplate format (xMarkTM Microplate Absorbance Spectrophotometer, Bio-Rad), and optical density at 600 nm was measured at 30-min intervals after brief agitation. To characterize growth under oxidative stress conditions, wild-type and frs1−1 S. cerevisiae strain were grown in minimal media. 50-ml cultures were prepared in triplicate for each condition, and shaken at 240 rpm, 30 °C. Optical density at 600 nm was measured at various time points. Oxidative stress cultures contained 2.0 mm paraquat (methyl viologen dichloride, Sigma). 10-ml samples of exponentially growing culture were passed over Whatman® Protran® BA85 nitrocellulose filter membranes, washed with 10 ml of water, and stored at −80 °C for processing and analysis of intracellular amino acid abundance.
Quantification of Intracellular Amino Acid Abundance
Frozen cell pellets were processed for analysis as previously described (10). Briefly, a mixture of 13C internal standards for Phe, p-Tyr, and l-DOPA were added to each cell pellet before extraction in −20 °C extraction buffer (40% methanol, 40% acetonitrile, 20% water). Solutions were clarified by centrifugation at 16,000 × g for 5 min at 4 °C. Supernatants were transferred to clean tubes and vacuum-dried. Dried samples were re-dissolved in solvent A (water/formic acid, 100/0.1, v/v; 50 μl) and centrifuged (16,000 × g, 5 min), and the supernatant was transferred to LC injector vials. Aliquots of the supernatant (typically 4 μl) were injected onto a reverse phase HPLC column (Phenomenex Kinetex C18, 2.1 × 150 mm, 1.7-μm particle size, 100 Å pore size) equilibrated in solvent A and isocratically eluted (100 μl/m). The effluent from the column was directly connected to an electrospray ion source (Agilent Jet Stream) attached to a triple quadrupole mass spectrometer (Agilent 6460) scanning in the positive multiple reaction monitoring mode with standard resolution settings (FWHM 0.7) using previously optimized conditions for the following transitions: l-DOPA, 198 → 181, 198 → 152; l-[U-13C6]DOPA, 204 → 187, 204 → 152; p-Tyr, 182 → 136; p[U-13C9]Tyr, 191 → 144; Phe, 166 → 120; [U-13C9]Phe, 175 → 128. With each batch of samples a series of standards was simultaneously prepared with the same amount of internal standards and increasing amounts of l-DOPA, m-Tyr and o-Tyr (0, 10, 25, 50, and 100 pmol), p-Tyr (0, 0.5, 1.25, 2.5, and 5 nmol), and Phe (0, 1.5, 3.75, 7.5, and 15 nmol) in duplicate. Typical retention times for l-DOPA, p-Tyr, m-Tyr, o-Tyr, and Phe were 7.3, 10.8, 13.5, 18.1, and 20.6 min, respectively. Peak areas were measured using instrument manufacturer supplied software (Agilent MassHunter). The amount of each analyte in each sample was determined by interpolation from the curves constructed from the standard samples (peak area l-DOPA, p-Tyr, or Phe/peak area l-[U-13C6]DOPA, p-[U-13C9]Tyr, or p-[U-13C9]Phe against the amount of l-DOPA, p-Tyr, or Phe in each standard sample).
Liquid Chromatographic Purification of Amino Acids
The accuracy of kinetic analyses of non-cognate amino acid substrates for enzyme specificity determination depends on the degree of cognate amino acid contamination (14). Therefore, we purified p-Tyr, dl-o-Tyr, and l-DOPA before their use for kinetic analyses. p-Tyr (Regis Chemical Co.) was purified by recrystallization from a minimal volume of boiling water as previously described (15). dl-o-Tyrosine (o-Tyr, Fluka, >96.0% nonaqueous titration) was dissolved in 0.1% trifluoroacetic acid and injected (250 μl aliquots) onto a thoroughly washed semi-preparative reverse phase HPLC column (Keystone Scientific Aquasil C18, 250 × 10 mm) equilibrated and eluted (3 ml/min) with water. 30-s fractions were collected, and aliquots (8 μl) of individual fractions across the peak of 280-nm absorbance eluting between 12.5 and 15 min were screened by LC/MS with 8-μl injections onto an Imtakt Scherzo SS C18 column (100 × 2 mm) equilibrated in eluent A (water/acetonitrile/formic acid, 97/3/0.1) and eluted with an increasing concentration of eluent B (45 mm ammonium formate/acetonitrile 65/35, min/% B; 0/0, 10/0, 40/20, 42/0, 55/0) with the mass spectrometer (Agilent 6460) scanning in the positive ion mode from m/z 100–300. Fractions 27, 28, and 29 each had a signal for o-Tyr (retention time (rt) 24.5 min; 3, 4, and 1 × 108 area counts in the m/z 182 extracted ion chromatograms (EIC), respectively) with no detectable signals for either p-Tyr (rt 16.7 min; m/z EIC 182), m-Tyr (rt 18.4 min; m/z EIC 182), or Phe (rt 24.2 min; m/z EIC 166). With limits of detection for p-Tyr, m-Tyr, and Phe estimated to be below 3.6, 4.5, and 1.4 × 104 area counts, respectively, fractions 27–29 appeared to contain <1 part in 104 of either p-Tyr, m-Tyr, or Phe. Fractions 27–29 were pooled, lyophilized to dryness, weighed (4 mg) and used for the experiments described below. l-DOPA (Sigma) was purified in an identical manner using the same semi-preparative column after it had been thoroughly washed to remove traces of residual analytes. Fractions containing the 280-nm peak of absorption eluting between 11 and 14 min were screened by LC/MS as described above. Fractions 23–28 contained a large peak for l-DOPA with no detectable amount of p-Tyr, m-Tyr, o-Tyr or Phe. These fractions were pooled, lyophilized to dryness (7.2 mg), and used for the experiments described below.
Wild-type and Post-transfer Editing-defective SccytoPheRS Preparation
His6-tagged wild type and post-transfer editing-defective SccytoPheRS were purified from previously constructed strains (16) and dialyzed against PPi to clear the active site of pre-bound Phe as before (10). Active enzyme stock concentrations were determined as previously described (17).
Steady-state Kinetic Analysis of SccytoPheRS
SccytoPheRS kinetic parameters for amino acid activation were determined by measurement of time-course ATP/PPi exchange using standard protocols (8–10, 16). Wild-type SccytoPheRS (5 nm for Phe and dl-o-Tyr, 500 nm for l-DOPA) catalyzed the activation of a given amino acid substrate (25–1000 μm Phe, 100–3000 μm dl-o-Tyr, or 200–1000 μm l-DOPA) in a reaction mix containing 100 mm Na-Hepes (pH 7.2), 30 mm KCl, 10 mm MgCl2, 2 mm NaF, 10 mm β-mercaptoethanol, 2 mm ATP, 2 mm [32P]PPi (∼1–2 cpm/pmol). The reaction was quenched in a solution containing 1% activated charcoal in 5.6% HClO4 and 75 mm PPi, which was then filtered through Whatman No. 3MM filter discs and washed with water. Negative control values (no enzyme present) were subtracted to account for retained PPi. The velocity of [32P]ATP formation as a function of substrate concentration was fitted to the Michaelis-Menten equation by non-linear regression. As a practical consideration, l-DOPA Vmax was not observable. kcat and Km are thus not reported individually for l-DOPA. Δ(v/E)/Δ[l-DOPA] in a linear range was used as a measure of kcat/Km with l-DOPA.
Aminoacylation of tRNA
Transcription of tRNAPheΔA76 in vitro, tRNAPhe ΔA76 radiolabeling with tRNA nucleotidyltransferase, and analysis of the relative efficiency of aminoacylation of tRNAPhe by PheRS were carried out as reported previously (10). The ratio of aa-[32P]AMP and [32P]AMP accumulation over time was used as a measure for the relative efficiency in SccyoPheRS aminoacylation of tRNAPhe with a given amino acid.
Pretransfer Editing
Analysis of pretransfer editing was carried out as previously described (7). Wild-type SccytoPheRS-catalyzed rates of [α-32P]AMP and aa-[α-32P]AMP synthesis were compared. Reactions contained 1 μm SccytoPheRS, 2 mm dl-o-Tyr, 100 mm Na-Hepes (pH 7.2), 30 mm KCl, 10 mm MgCl2, 2 mm NaF, 10 mm β-mercaptoethanol, 50 μm ATP, 10 μm natively purified tRNAPhe (Sigma), and 0.5 μm [α-32P]ATP. Matched no-enzyme control values were subtracted to account for spontaneous hydrolysis of [α-32P]ATP. 1-μl samples of the reaction mix were quenched at various time points with 1.5 μl of 0.5 mm NaOAc (pH 4.2). 1.5 μl of quenched reactions at various time points were spotted onto polyethyleneimine cellulose (Sigma) pre-equilibrated with water. Products were resolved in a mobile phase consisting of 100 mm ammonium acetate and 5% acetic acid. Spots corresponding to o-Tyr-[α-32P]AMP and free [α-32P]AMP were visualized as described previously (10, 18).
Presteady-state Characterization of o-Tyr Transfer
The rate of o-Tyr transfer from o-Tyr-AMP to tRNAPhe was determined with a quench flow RQF-3 KinTek instrument as before (19), with modifications based on unavailability of o-[14C]Tyr. Because we expected o-Tyr-AMP·PheRS complex instability, o-Tyr activation was carried out in situ. Syringe A contained 4 mm ATP, 5 μm post-transfer editing ablated SccytoPhers, 10 mm dithiothreitol, 2 units/ml inorganic pyrophosphatase, 100 mm Na-Hepes (pH 7.2), 30 mm KCl, 10 mm MgCl2, and either 2 mm dl-o-Tyr, 1 mm p-Tyr, or 1 mm Phe. Syringe B contained 1.5 μm [32P]tRNAPhe, 10 mm dithiothreitol, 100 mm Na-Hepes (pH 7.2), 30 mm KCl, and 10 mm MgCl2. Reactions were carried out at 30 °C and quenched with 0.5 m NaOAc. An equal volume of quenched reaction mix and 1:10 diluted S1 were mixed, and the S1 nuclease reaction was carried out at 37 °C for 30 min. 1.5 μl of each time point was spotted on polyethyleneimine cellulose and developed as detailed above. aa-tRNA formation over time was fitted to the single-exponential equation [aa-tRNA] = C + A (1 − ektrans×t).
Measurement of o-Tyr-AMP Stability
o-Tyr-AMP was synthesized under aminoacylation reaction conditions described above in the absence of tRNAPhe using [α-32P]ATP. o-Tyr-AMP was then purified after 15 min via standard acidic phenol-chloroform extraction techniques. The stability of ∼14 μm o-Tyr-AMP under reaction conditions in the absence of enzyme was observed by TLC separation, as above.
Results
SccytoPheRS Post-transfer Editing Limits the Cytotoxicity of Oxidative Stress
After our observations that PheRS post-transfer editing activity is required to protect E. coli from m-Tyr, we examined the role that post-transfer editing activity plays in a eukaryotic species, S. cerevisiae. Having previously demonstrated that p-Tyr and m-Tyr are more toxic to a cytosolic SccytoPheRS post-transfer editing mutant strain (frs1−1) (10), we examined a possible protective effect of editing in the presence of oxidative stress. Methyl viologen (paraquat) is used as a surrogate for oxidative stress, as it exacerbates electron transport leakage from the mitochondrion and single-hydrogen reduction of molecular oxygen. We found that SccytoPheRS post-transfer editing activity limited the toxicity of paraquat (Fig. 2), suggesting a similar protective role of PheRS post-transfer editing in both bacteria and eukarya under oxidative stress.
FIGURE 2.
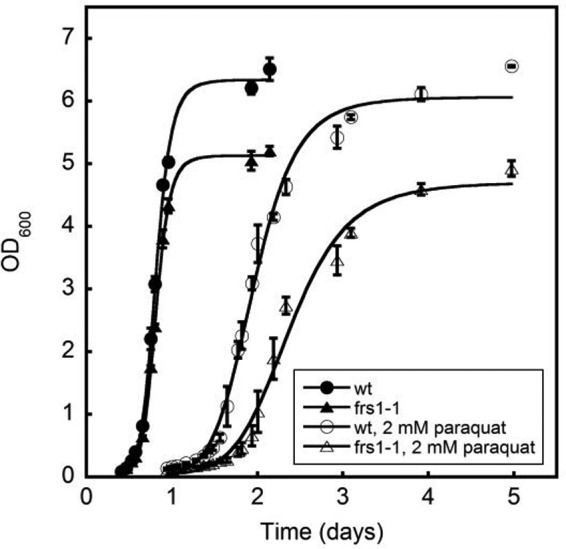
SccytoPheRS post-transfer editing limited the cytotoxicity of oxidative stress. Growth of wild-type (wt) or post-transfer editing ablated SccytoPheRS (frs1−1) grown in minimal media with or without 2.0 mm paraquat. Data represent the average of measurements from three cultures. Error bars represent ± S.E.
Oxidized Phe Derivatives Accumulate Under Oxidative Stress
To test if the protective role that SccytoPheRS post-transfer editing plays in S. cerevisiae is due to a decrease in the availability of intracellular Phe, accumulation of non-cognate PheRS substrates, or some indirect effect, we extracted soluble intracellular metabolites from exponentially growing S. cerevisiae grown in minimal media and examined by LC-MS/MS the relative molar abundance of putative SccytoPheRS substrates (Fig. 3). There are 20 possible hydroxylation states of the phenyl ring of Phe; here, we show the detectable intracellular accumulation of five of them: Phe, p-Tyr, o-Tyr, m-Tyr, and l-DOPA. 2.0 mm paraquat induced the accumulation not only of some oxidized derivatives of Phe but also a doubling of Phe itself. This may represent a sensing mechanism whereby S. cerevisiae compensates for oxidation of its Phe pool by overproducing Phe. Proteinogenic p-Tyr accumulates 5–6-fold under oxidative stress and may do so as a result of a combination of Phe oxidation and programmed biosynthesis. Notably, there is no known hydroxylase in S. cerevisiae that can convert Phe to p-Tyr. o-Tyr accumulates 6–7-fold under oxidative stress, and m-Tyr increases by 2-fold. l-DOPA, which is undetectable in unstressed S. cerevisiae, accumulates to 1–2 orders of magnitude greater than o-Tyr or m-Tyr, which may represent oxidation of intracellular p-Tyr as well as Phe. We also observed that unstressed S. cerevisiae grown in rich media contain levels of l-DOPA similar to those in stressed cultures (not shown). This suggests a degree of l-DOPA contamination in rich media components and subsequent cellular uptake. Because commercial yeast extracts and peptones are undefined and may become oxidized over time, the degree of non-protein amino acid contamination is a challenge to interpretation of results with such undefined media components.
FIGURE 3.
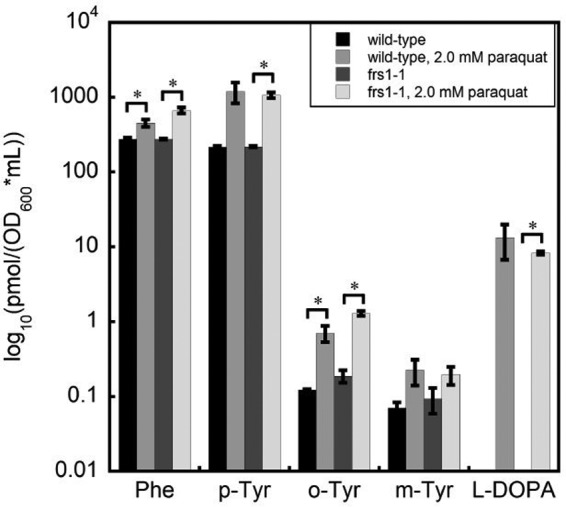
Oxidative stress increased the abundance of non-protein amino acids. Abundance of extracted intracellular amino acids from 10-ml samples of exponentially growing wild-type or frs1−1 grown in minimal media with or without 2.0 mm paraquat. Values are normalized to the optical density at 600 nm of parent cultures at the time of harvest. Samples were taken from triplicate cultures. Error bars represent ± S.E. *, p < 0.05, unpaired t test. One of the three biological samples taken had a total metabolite mass ∼2.5–2.8 times that of the other two replicates, resulting in sufficient variability to not meet the standard conventions of statistical significance for the comparison between unstressed and stressed wild-type cultures for p-Tyr (p = 0.0581), m-Tyr (p = 0.1455), and l-DOPA (p = 0.1127). There was also no significant difference between the amount of m-Tyr in frs1−1 with or without paraquat (p = 0.1867).
SccytoPheRS Post-transfer Editing Has Amino Acid Supplement-dependent Effects on Growth
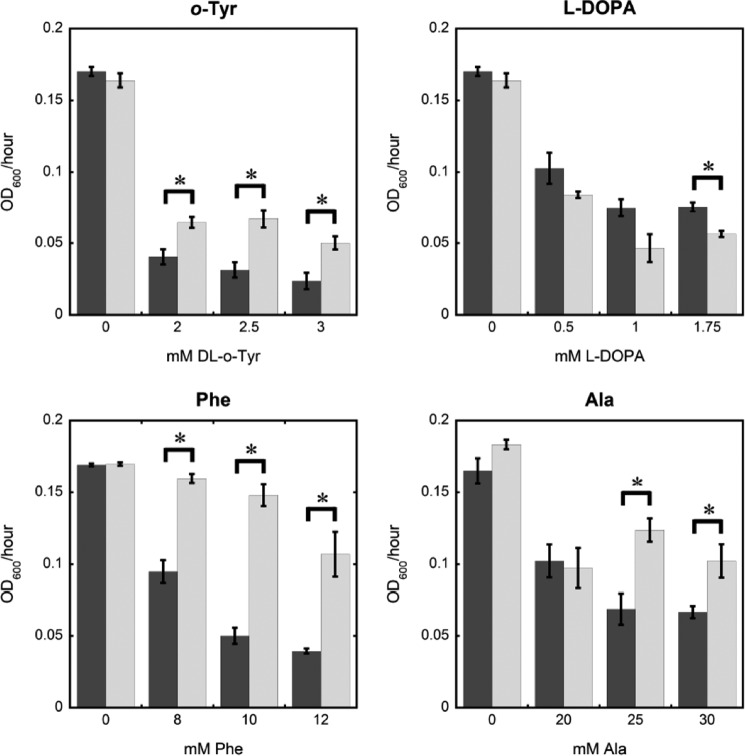
To further investigate the possible effects of non-protein aromatic amino acid accumulation, editing defective SccytoPheRS and wild-type S. cerevisiae were grown with o-Tyr and l-DOPA (Fig. 4, A and B). Given that SccytoPheRS poorly discriminates against p-Tyr relative to the E. coli enzyme, it was not surprising that l-DOPA is more toxic to S. cerevisiae when post-transfer editing activity is ablated. E. coli does not show increased sensitivity to l-DOPA in the absence of EcPheRS post-transfer editing, underscoring an evolutionary divergence in PheRS substrate specificity. Less expected was the complete phenotypic reversal with o-Tyr supplementation. The post-transfer editing-ablated SccytoPheRS strain tolerates o-Tyr better than wild-type S. cerevisiae. When supplemented with either cognate Phe or non-cognate Ala (Fig. 4, C and D), the post-transfer editing mutant also grew better than wild type. The degree of metabolic conversion of o-Tyr or Ala to other substrates is unclear, but the cognate Phe supplementation experiment suggests that the phenotypic effect of post-transfer editing in this condition is independent of its canonical aa-tRNAPhe proofreading activity.
FIGURE 4.
SccytoPheRS post-transfer editing activity had variable effects on growth rate dependent on supplemented amino acids. Exponential phase growth rate of wild-type (dark gray) and frs1−1 (light gray) yeast in minimal media supplemented with indicated amino acids. Data represent the average of measurements from three cultures. Error bars represent ± S.E. *, p < 0.05, unpaired t test.
SccytoPheRS Displays Poor Discrimination against Oxidized Phe Derivatives in Vitro
To examine the substrate specificity of SccytoPheRS, we compared steady-state activation of Phe, o-Tyr, m-Tyr, p-Tyr, l-DOPA, and Ala. No detectable activity was found for 1 mm Ala at 500 nm SccytoPheRS (data not shown), but all oxidized Phe derivatives tested are poorly discriminated against by SccytoPheRS in vitro (Table 1). Values for p-Tyr and m-Tyr are adapted from our previous work (10). o-Tyr, in particular, is only an order of magnitude less efficiently activated than Phe, suggesting a substantial threat to SccytoPheRS product specificity and probable Phe codon mistranslation without an o-Tyr-specific quality control mechanism.
TABLE 1.
Steady-state kinetic parameters for amino acid activation by SccytoPheRS
Data are the average of three replicates ± S.E.
| Amino acid | kcat | Km | kcat/Km | Specificity (Phe/x) |
|---|---|---|---|---|
| s−1 | μm | s−1 × μm−1 | ||
| Phe | 58.2 ± 5.8 | 42.3 ± 7.0 | 1.4 | |
| o-Tyr | 39.4 ± 0.9 | 310.7 ± 48.1 | 0.13 | 11 |
| m-Tyr | 26 ± 4a | 1150 ± 230a | 0.023a | 71a |
| p-Tyr | 0.014a | 120a | ||
| l-DOPA | 7.5 × 10−4 ± 4.4 × 10−5 | 1800 |
a m-Tyr and p-Tyr values adapted from Bullwinkle et al. (10).
An Expanded Form of aaRS Biological Substrate Selectivity
Enzyme specificity for a given substrate is calculated by a comparison of reaction rates.
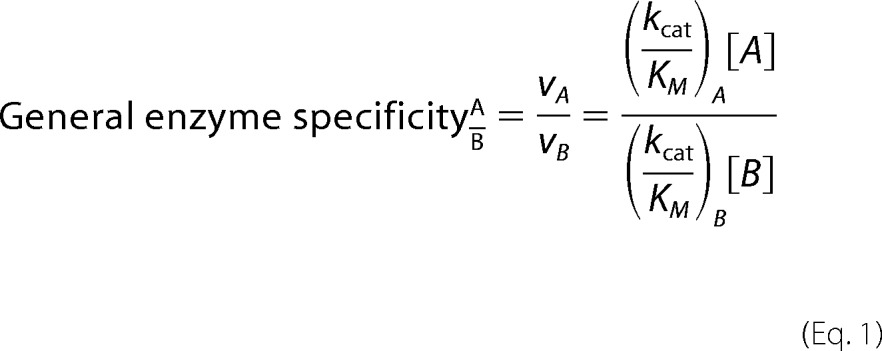 |
For aaRS amino acid specificity calculations, a comparison between two amino acid substrates is typically sufficient, where A is the cognate amino acid, and B is a given near-cognate or non-cognate. In this simple two-substrate comparison, the preference for one substrate versus the other is typically expressed as a direct ratio of catalytic efficiency constants, where [A] = [B],
 |
However, relative reaction rates can change in vivo, as substrate concentrations are often in flux. Proper estimation of cognate specificity relies on measurement of the intracellular concentrations of the relevant substrates, which in turn allows expression of aaRS substrate preference as selectivity in a biological context as follows.
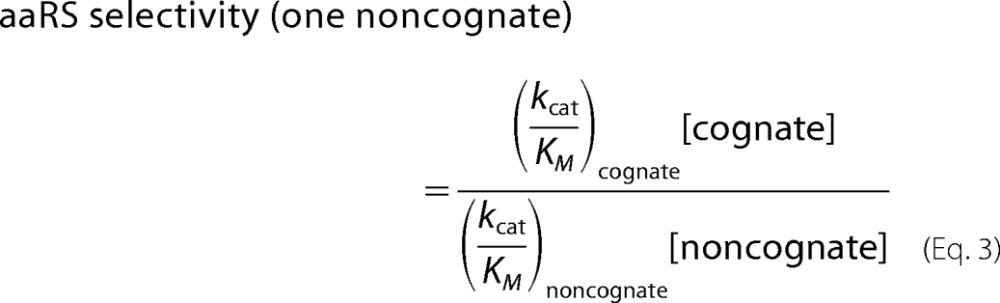 |
Multiple non-cognate amino acids may simultaneously compete for access to the active site of a given aaRS (10), rendering one-to-one reaction rate comparisons insufficient in a biological context. Given the poor individual discrimination that SccytoPheRS exhibits against o-Tyr, m-Tyr, p-Tyr, and l-DOPA, one-to-one comparisons do not adequately describe aaRS cognate selectivity in vivo. Instead, an expanded form is appropriate,
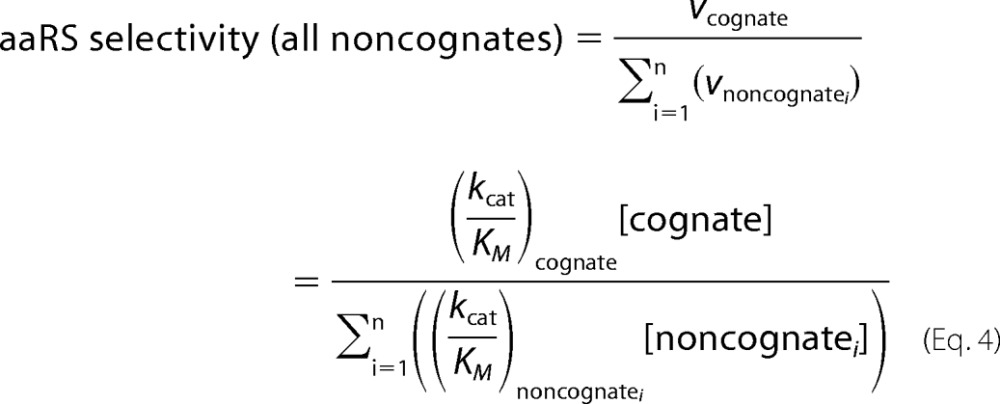 |
Because it is effectively impossible to identify every potential substrate for aaRS activity that may exist, it is practical to simply include terms for known non-cognates and express biological selectivity as less than or equal to the calculated value. For SccytoPheRS, we now include terms for o-Tyr, m-Tyr, p-Tyr, and l-DOPA (Table 2). Of the 19 possible ring-hydroxylated non-cognate Phe derivatives, only 4 were tested here. Moreover, Phe modification need not be limited to hydroxylation of the phenyl ring alone. Phe hydroperoxides, nitrosylated derivatives, or any number of alternatively modified Phe derivatives may be substrates for this enzyme. The intracellular abundance of multiple oxidized Phe derivatives increased under the oxidative stress conditions chosen (Fig. 3). In this case, selectivity is driven primarily by Phe and p-Tyr abundance. When expanded to include all untested SccytoPheRS substrates and variable environmental stresses that cause non-protein amino acids to accumulate, the combined effect of many non-cognates represents a substantial conditional burden to the fidelity of Phe codon translation. The known promiscuity of SccytoPheRS implies that multiple quality control mechanisms address the threat of various non-cognate amino acids that may be misactivated to a greater degree in various environmental contexts.
TABLE 2.
SccytoPheRS selectivity is decreased under oxidative stress
Wild-type and post-transfer editing-defective SccytoPheRS biological selectivity with and without 2.0 mm paraquat is shown. Data are the average of three replicates ± S.E. Wild-type SccytoPheRS selectivity was decreased 3-fold under oxidative stress (p = 0.0003). Post-transfer editing ablated SccytoPheRS selectivity was decreased 2-fold under oxidative stress (p < 0.0001). No statistically significant difference in selectivity exists between strains either without paraquat (p = 0.2668) or with paraquat (p = 0.0584).
| SccytoPheRS biological Phe selectivity | |||
|---|---|---|---|
| Wild type | frs1-1 | Wild type, 2.0 mm paraquat | frs1-1, 2.0 mm paraquat |
| ≤127 ± 1 | ≤124 ± 2 | ≤42 ± 7 | ≤61 ± 2 |
Aminoacylation of tRNAPhe with o-Tyr Is Inefficient
The observed poor discrimination against o-Tyr during amino acid activation suggested that a later quality control mechanism may prevent substantial o-Tyr-tRNAPhe production. The efficiency of o-Tyr-tRNAPhe production was tested in vitro using purified post-transfer editing ablated SccytoPheRS. o-Tyr-tRNAPhe was synthesized to a level of ∼10% of chargeable tRNAPhe, which is substantially lower than the ∼60% charging observed with p-Tyr (Fig. 5). Because the post-transfer editing-defective SccytoPheRS enzyme was used for this aminoacylation, o-Tyr-tRNAPhe limitation is not due to post-transfer editing but must come at a step after activation given the ∼10-fold superior efficiency in o-Tyr activation when compared with p-Tyr (Table 1). No difference in aminoacylation efficiency is observed between wild-type and βD243A SccytoPheRS for either Phe or o-Tyr (supplemental Fig. 1).
FIGURE 5.
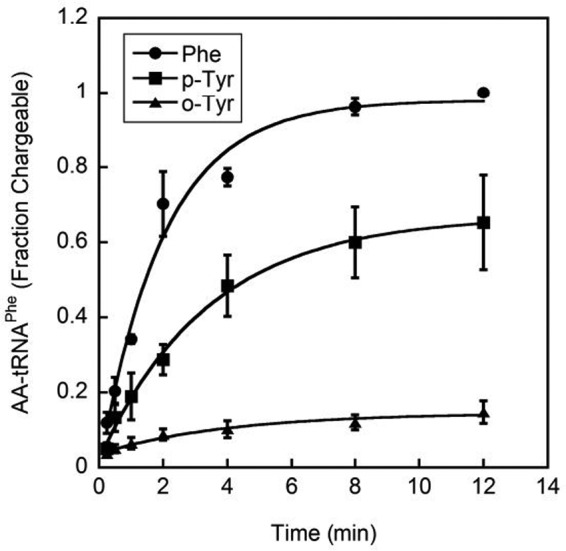
o-Tyr was inefficiently acylated to tRNAPhe. Aminoacylation of tRNAPhe by post-transfer editing ablated SccytoPheRS with 75 μm Phe, 75 μm p-Tyr, and 150 μm dl-o-Tyr, expressed as fraction chargeable tRNAPhe ± S.E. (n = 3). tRNAPhe was transcribed in vitro without A76 before addition of A76 with tRNA nucleotidyltransferase using α-[32P]ATP.
o-Tyr-tRNAPhe Limitation Is Not Due to Hydrolytic PreTransfer Editing
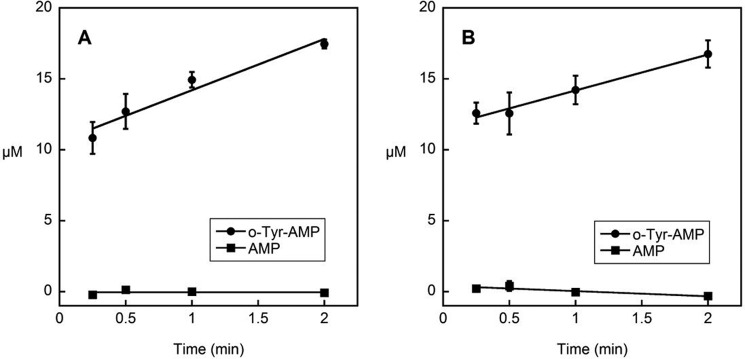
Selective hydrolysis of the products of amino acid misactivation is an established mechanism of aminoacyl-tRNA synthetase quality control. To test if o-Tyr-AMP is selectively hydrolyzed after activation, we observed o-Tyr-AMP and AMP accumulation in vitro. If o-Tyr-AMP were efficiently hydrolyzed after activation, we would see an accumulation of one of the products of this hydrolysis, AMP, faster than the accumulation of o-Tyr-AMP. However, we see that steady-state o-Tyr-AMP accumulation is 3–4 μm/min, whereas AMP does not accumulate appreciably at all by 2 min (Fig. 6), indicating that o-Tyr activation is faster than o-Tyr-AMP hydrolysis. Therefore, rapid pretransfer hydrolysis of o-Tyr-AMP does not provide a likely mechanism to limit o-Tyr-tRNAPhe synthesis by SccytoPheRS.
FIGURE 6.
o-Tyr-AMP was not efficiently hydrolyzed by SccytoPheRS. o-Tyr-AMP accumulates to 17-fold greater than wild-type (panel A) or βD243A (panel B) SccytoPheRS by 2 min. AMP is not appreciably formed. Data are presented as the average of three replicates ± S.E. o-Tyr-AMP is stable in the absence of SccytoPheRS under reaction conditions (supplemental Fig. 3).
o-Tyr-AMP Is Released from the SccytoPheRS Active Site
To test if o-Tyr-tRNAPhe limitation is due to selective release of o-Tyr-AMP from the SccytoPheRS active site, we observed the accumulation of o-Tyr-AMP relative to the concentration of SccytoPheRS in vitro. o-Tyr-AMP accumulates to a concentration 17-fold greater than the concentration of post-transfer editing defective SccytoPheRS by 2 min (Fig. 6), suggesting multiple turnover events in o-Tyr activation as well as release of o-Tyr-AMP from the enzyme active site. Activation of o-Tyr is less efficient in the absence of tRNAPhe (supplemental Fig. 2), which may reflect a more stable activation complex or direct interaction of tRNAPhe in activation chemistry.
o-Tyr-AMP Release Is Due to Slow Transfer
Whereas o-Tyr-AMP has now been shown to be released from the SccytoPheRS active site before transfer, it was unclear if this release is competitive with the rate of o-Tyr transfer from o-Tyr-AMP to tRNAPhe or if there is kinetic discrimination arising from slow o-Tyr transfer. We addressed this ambiguity by comparing the rate of presteady-state transfer for cognate Phe and non-cognate p-Tyr to that of o-Tyr. The rate of Phe transfer from Phe-AMP to tRNAPhe was 2.3 s−1 ± 0.3 s−1, and the rate of p-Tyr transfer was 2.1 s−1 ± 0.2 s−1. o-Tyr transfer from o-Tyr-AMP to tRNAPhe was undetectable over the same time scale, indicating strong kinetic discrimination against o-Tyr in the transfer step. Notably, Phe and p-Tyr transfer are effectively identical, indicating kinetic discrimination at the transfer step is a mechanism specific to o-Tyr and not non-cognate p-Tyr.
Discussion
Limitation of o-Tyr-tRNAPhe by SccytoPheRS
Whereas kinetic discrimination against o-Tyr is poor at the activation step (Table 1), o-Tyr-tRNAPhe is not produced efficiently, independent of post-transfer editing activity (Fig. 5). Despite a 10-fold higher activity in activation when compared with p-Tyr, o-Tyr is much less efficiently acylated to tRNAPhe. These observations suggest a quality control mechanism distinct from established post-transfer editing activity, which acts to limit o-Tyr-tRNAPhe production. Analysis of the products of o-Tyr-stimulated ATPase activity revealed rapid o-Tyr-AMP synthesis (Fig. 6), consistent with activation data (Table 1). AMP accumulation is minimal when compared with o-Tyr-AMP synthesis, suggesting that if hydrolysis of o-Tyr-AMP does contribute to quality control, it is outweighed by the specific rejection of o-Tyr-AMP as the primary effector of pretransfer quality control.
o-Tyr-AMP release may be due to either poor substrate selection in the transfer step resulting in entropic dissociation or to selective adenylate release in competition with an otherwise rapid transfer step. To determine if selective o-Tyr-AMP release is due to inefficiency in the transfer step, we measured the rate of o-Tyr transfer under single-turnover conditions and found that o-Tyr transfers much slower than cognate Phe transfer. Taken together, our data demonstrate that o-Tyr is efficiently activated, but the resulting o-Tyr-AMP is a poor substrate for transfer to tRNAPhe. Much of the o-Tyr-AMP produced is released from the SccytoPheRS synthetic active site, although low level transfer does contribute to a minimal degree of aminoacylation of tRNAPhe with o-Tyr (Fig. 5). This mechanism for preventing a non-protein amino acid from threatening the product specificity of SccytoPheRS is in addition to the high substrate discrimination against most amino acids in the activation step as well as the established post-transfer editing activity that limits p-Tyr-tRNAPhe and m-Tyr-tRNAPhe release. Simultaneous pre- and post-transfer editing activity is not unique to SccytoPheRS (20, 21). Human cytoplasmic LeuRS, like SccytoPheRS, is threatened with multiple non-protein amino acids, requiring distinct mechanisms of quality control by the same enzyme; whereas norvaline is a target for post-transfer editing activity, α-aminobutyrate is primarily a substrate for pretransfer editing by this enzyme (21).
Our findings show that SccytoPheRS bears not only post-transfer editing activity against proteinogenic and non-protein amino acids but both kinetic discrimination against noncognate amino acids at the activation step and kinetic discrimination against a noncognate aminoacyl-adenylate at the transfer step, specifically one derived from a non-protein amino acid. Moreover, the poor discrimination against o-Tyr observed at the activation step yields misleading specificity calculations (Table 1) because the substrate specificity employed by this enzyme is primarily at a step after activation.
Non-protein amino acids are an under-examined threat to translational fidelity due primarily to the only recent advances in chromatographic separation and quantitation of them in the face of large amounts of proteinogenic amino acids with nearly identical biophysical properties in biological samples.
Environmental conditions may drastically alter the intracellular concentration of a given amino acid, skewing the selectivity of the relevant aaRS in vivo. Theoretically, the catalytic efficiency of a given aaRS for any of its substrates may also be context-dependent. However, direct measurement of kcat/Km for every potential substrate in vivo is problematic, particularly when the added dimension of environmentally dependent catalytic efficiency is included. Accounting for conditional substrate abundance, discrimination against m-Tyr is ∼240,000 under typical growth conditions and ∼120,000 in the presence of 2.0 mm paraquat, which would normally indicate no further requirement for quality control. However, SccytoPheRS does bear moderate post-transfer editing activity against m-Tyr-tRNAPhe, suggesting that the requirements for SccytoPheRS product specificity may be higher than the aggregate expected rate of one mispaired aminoacyl-tRNA species in 3000 aa-tRNAs produced (2). Given that the discrimination against o-Tyr is only ∼24,000 under typical growth conditions and ∼6,900 in the presence of 2.0 mm paraquat (much lower discrimination than against m-Tyr), kinetic discrimination against o-Tyr-AMP by SccytoPheRS may play a role in Phe codon quality control at biologically relevant levels, particularly in conditions that favor oxidation of intracellular Phe. Alternatively, the import of exogenous o-Tyr is a plausible scenario, as structurally similar m-Tyr has been shown to be produced and exported by certain plant species in order to shape the ecology of their environments and inhibit the growth of competitor species (22). Production and export of non-protein amino acids is an effective strategy for chemically shaping a competitive environmental niche, provided the producer has the means to prevent the cytotoxic incorporation of these species (for review, see reviewed see Ref. 23).
Extended exposure of S. cerevisiae to low levels of hydrogen peroxide increases the level of protein o-Tyr and m-Tyr in mutants defective in glutathione metabolism (24), although it is unclear whether this reflects oxidation of protein post-translationally or oxidation of intracellular Phe and subsequent mistranslation. Recently, monoclonal antibody production in Chinese hamster ovary cells limited in intracellular Phe has been shown to yield protein products with m-Tyr and o-Tyr at Phe codons (25). Interestingly, p-Tyr is not detected at these codons, suggesting that non-protein amino acid derivatives are a unique threat to the mammalian system. This finding is a surprise because the post-transfer editing activity that limits p-Tyr-tRNAPhe production is apparently insufficient to prevent m-Tyr misincorporation despite m-Tyr-tRNAPhe editing activity in SccytoPheRS. Further testing with mammalian cytoplasmic PheRS is needed to address this apparent discrepancy.
o-Tyr presents only a minor threat at Phe codons throughout the lifespan of a typical yeast cell, but in long-lived cell types, such as terminally differentiated neuronal cells, the cumulative effect of protracted low level o-Tyr incorporation may become a problem. o-Tyr and m-Tyr have long been used as biomarkers of age and oxidative damage in proteins, and oxidative damage of neuronal protein correlates with neurodegeneration, but it is not yet clear if a causal relationship exists or what role persistent low level mistranslation with oxidized amino acids may play in disease states in aging and neurodegenerative disease. S. cerevisiae has evolved at least three distinct mechanisms in part to prevent oxidized Phe derivatives from incorporation at Phe codons despite an apparent minimal need for such stringent quality control under normal conditions. The cumulative effects of long term low level mistranslation and oxidative damage merit further study.
SccytoPheRS Post-transfer Editing Plays a Non-canonical Role in Tolerance to Amino Acid Stress
In E. coli, Phe starvation is detected by sensing elevated uncharged tRNAPhe, a surrogate for low Phe-tRNAPhe levels. In this manner, uncharged tRNAPhe can directly up-regulate Phe biosynthesis. Without PheRS post-transfer editing, mischarged tRNAPhe species, including m-Tyr-tRNAPhe accumulate, mask Phe-tRNAPhe limitation and prevent uncharged tRNAPhe sensing (26). This in turn fails to up-regulate Phe biosynthesis, further compounding Phe limitation relative to EcPheRS non-cognates such as m-Tyr. The yeast amino acid starvation-sensing mechanisms are not as directly tied to individual amino acids, instead relying on general uncharged tRNA sensing via GCN2. GCN2, upon binding uncharged tRNA, phosphorylates eIFα, which causes downstream up-regulation of amino acid biosynthesis genes and simultaneous restriction of general protein synthesis. It is plausible that, as with EcPheRS, a loss of SccytoPheRS editing prevents sensing of Phe starvation due to accumulation of mischarged tRNAPhe species. S. cerevisiae rRNAs and tRNAs are cleaved in a conserved response to oxidative stress. The role of tRNA cleavage products is not well understood, but preliminary work suggests tRNA cleavage as an additional mechanism to limit translation (27, 28). Further limitation of uncharged tRNA pools under oxidative stress highlights a challenge to S. cerevisiae at Phe codons. Not only are Phe pools oxidized, forming additional non-cognates that compete for SccytoPheRS, but a decrease in global tRNA limits the tRNAPhe available for aminoacylation. This may suggest a need for the decreased levels of available tRNAPhe to be charged accurately in this condition of tightly regulated translation, which underscores the importance for SccytoPheRS product specificity mechanisms under oxidative stress.
A loss of post-transfer editing activity is toxic when yeast are exposed to oxidative stress (Fig. 2), but supplementation with specific PheRS non-cognates yields variable toxic effects (Fig. 4). Post-transfer editing-defective S. cerevisiae does not tolerate m-Tyr, p-Tyr, or l-DOPA as well as wild type, but a reversal of these phenotypes is observed upon supplementation with o-Tyr, cognate Phe, or Ala. If post-transfer editing were only responsible for preventing the supplemented amino acids from misacylation of tRNAPhe and cytotoxic insertion at Phe codons, supplementation with cognate Phe should have no effect, particularly because there is no known phenylalanine hydroxylase in S. cerevisiae to convert Phe to non-cognate p-Tyr. Ala, which is not a substrate for PheRS activation, should play no direct role in tRNAPhe charging at all, although the level of metabolic conversion to alternative SccytoPheRS substrates is unclear. These data suggest an additional role in yeast for post-transfer editing in nutrient sensing, analogous to that observed in E. coli. As a result o-Tyr, Phe, and Ala addition must have some indirect metabolic effects that alter typical growth.
Given the limited insight that can be gained by phenotypic analysis alone, it is not yet clear what role post-transfer editing plays in regulation of cellular growth, and the simple answer of conditional mistranslation may not tell the complete story. Particularly, the negative effect cognate Phe and poor activation substrate Ala have on wild-type growth rate suggest that mistranslation plays only a minor role here. Future work will focus on the metabolic profile of post-transfer editing-defective SccytoPheRS yeast to address the role of post-transfer editing in growth under oxidative stress and growth limitation when intracellular amino acid ratios are atypical. In particular, Phe and p-Tyr levels, which seem to be the primary drivers of substrate specificity (Fig. 3), likely shift in abundance when S. cerevisiae is supplemented with various amino acids. Oxidative stress causes accumulation of noncognate species, but the degree to which those species are metabolically converted to alternative SccytoPheRS substrates or to which they interact with alternative metabolic processes is unclear as well. Direct measurement of the levels of mischarged and uncharged aminoacyl-tRNA species will also be a critical step in understanding the role of context-dependent tRNA misacylation in the cell. No matter what the mechanism of growth rate feedback is that describes these phenotypic effects, it is clear that the quality control mechanisms in aminoacyl-tRNA synthesis do not have a role in limiting mistranslation alone.
Author Contributions
A. M., K. F., and M. I. performed the experimental design. A. M., L. H., and K. F. performed the experiments. A. M., L. H., K. F., and M. I. performed the data analysis. A. M. and M. I. wrote the manuscript. A. M., L. H., K. F., and M. I. edited the manuscript. All authors assumed the responsibility of data analysis integrity.
Supplementary Material
Acknowledgment
We are grateful to Dr. Tammy Bullwinkle for valuable input and critical reading of this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant Training Grants T32 GM008512 and GM086252 (to A. M.). This work was also supported by National Science Foundation Grants MCB-1412611 and MCB 1412773. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Fig. 1–3.
- aaRS
- aminoacyl-tRNA synthetase
- aa-tRNA
- aminoacyl-tRNA
- l-DOPA
- 3,4-dihydroxy-l-phenylalanine
- EcPheRS
- E. coli phenylalanyl-tRNA synthetase
- SccytoPheRS
- S. cerevisiae cytoplasmic PheRS.
References
- 1. Loftfield R. B., and Vanderjagt D. (1972) The frequency of errors in protein biosynthesis. Biochem. J. 128, 1353–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Loftfield R. B. (1963) The frequency of errors in protein biosynthesis. Biochem. J. 89, 82–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li L., Boniecki M. T., Jaffe J. D., Imai B. S., Yau P. M., Luthey-Schulten Z. A., and Martinis S. A. (2011) Naturally occurring aminoacyl-tRNA synthetases editing-domain mutations that cause mistranslation in Mycoplasma parasites. Proc. Natl. Acad. Sci. U.S.A. 108, 9378–9383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gomes A. C., Miranda I., Silva R. M., Moura G. R., Thomas B., Akoulitchev A., and Santos M. A. (2007) A genetic code alteration generates a proteome of high diversity in the human pathogen Candida albicans. Genome Biol. 8, R206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hati S., Ziervogel B., Sternjohn J., Wong F. C., Nagan M. C., Rosen A. E., Siliciano P. G., Chihade J. W., and Musier-Forsyth K. (2006) Pre-transfer editing by class II prolyl-tRNA synthetase: role of aminoacylation active site in “selective release” of noncognate amino acids. J. Biol. Chem. 281, 27862–27872 [DOI] [PubMed] [Google Scholar]
- 6. Tan M., Zhu B., Zhou X. L., He R., Chen X., Eriani G., and Wang E. D. (2010) tRNA-dependent pre-transfer editing by prokaryotic leucyl-tRNA synthetase. J. Biol. Chem. 285, 3235–3244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ling J., Peterson K. M., Simonovic I., Söll D., and Simonovic M. (2012) The mechanism of pre-transfer editing in yeast mitochondrial threonyl-tRNA synthetase. J. Biol. Chem. 287, 28518–28525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Roy H., Ling J., Irnov M., and Ibba M. (2004) Post-transfer editing in vitro and in vivo by the β subunit of phenylalanyl-tRNA synthetase. EMBO J. 23, 4639–4648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Raina M., Moghal A., Kano A., Jerums M., Schnier P. D., Luo S., Deshpande R., Bondarenko P. V., Lin H., and Ibba M. (2014) Reduced amino acid specificity of mammalian tyrosyl-tRNA synthetase Is associated with elevated mistranslation of Tyr codons. J. Biol. Chem. 289, 17780–17790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bullwinkle T., Reynolds N. M., Raina M., Moghal A. B., Matsa E., Rajkovic A., Kayadibi H., Fazlollahi F., Ryan C., Howitz N., Faull K. F., Lazazzera B., and Ibba M. (2014) Oxidation of cellular amino acid pools leads to cytotoxic mistranslation of the genetic code. Elife 10.7554/eLife.02501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Netzer N., Goodenbour J. M., David A., Dittmar K. A., Jones R. B., Schneider J. R., Boone D., Eves E. M., Rosner M. R., Gibbs J. S., Embry A., Dolan B., Das S., Hickman H. D., Berglund P., Bennink J. R., Yewdell J. W., and Pan T. (2009) Innate immune and chemically triggered oxidative stress modifies translational fidelity. Nature 462, 522–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ling J., and Söll D. (2010) Severe oxidative stress induces protein mistranslation through impairment of an aminoacyl-tRNA synthetase editing site. Proc. Natl. Acad. Sci. U.S.A. 107, 4028–4033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Reynolds N. M., Ling J., Roy H., Banerjee R., Repasky S. E., Hamel P., and Ibba M. (2010) Cell-specific differences in the requirements for translation quality control. Proc. Natl. Acad. Sci. U.S.A. 107, 4063–4068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cvetesic N., Palencia A., Halasz I., Cusack S., and Gruic-Sovulj I. (2014) The physiological target for LeuRS translational quality control is norvaline. EMBO J. 33, 1639–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Faull K. F., Gan I., Halpern B., Hammond J., Im S., Cotton R. G., Danks D. M., and Freeman R. (1977) Metabolic studies on two patients with nonhepatic tyrosinemia using deuterated tyrosine loads. Pediatr. Res. 11, 631–637 [DOI] [PubMed] [Google Scholar]
- 16. Roy H., Ling J., Alfonzo J., and Ibba M. (2005) Loss of editing activity during the evolution of mitochondrial phenylalanyl-tRNA synthetase. J. Biol. Chem. 280, 38186–38192 [DOI] [PubMed] [Google Scholar]
- 17. Ibba M., Kast P., and Hennecke H. (1994) Substrate specificity is determined by amino acid binding pocket size in Escherichia coli phenylalanyl-tRNA synthetase. Biochemistry 33, 7107–7112 [DOI] [PubMed] [Google Scholar]
- 18. Wolfson A. D., and Uhlenbeck O. C. (2002) Modulation of tRNAAla identity by inorganic pyrophosphatase. Proc. Natl. Acad. Sci. U.S.A. 99, 5965–5970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yadavalli S. S., and Ibba M. (2013) Selection of tRNA charging quality control mechanisms that increase mistranslation of the genetic code. Nucleic Acids Res. 41, 1104–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hendrickson T. L., Nomanbhoy T. K., de Crécy-Lagard V., Fukai S., Nureki O., Yokoyama S., and Schimmel P. (2002) Mutational separation of two pathways for editing by a class I tRNA synthetase. Mol. Cell 9, 353–362 [DOI] [PubMed] [Google Scholar]
- 21. Chen X., Ma J. J., Tan M., Yao P., Hu Q. H., Eriani G., and Wang E. D. (2011) Modular pathways for editing non-cognate amino acids by human cytoplasmic leucyl-tRNA synthetase. Nucleic Acids Res. 39, 235–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bertin C., Weston L. A., Huang T., Jander G., Owens T., Meinwald J., and Schroeder F. C. (2007) Grass roots chemistry: meta-tyrosine, an herbicidal nonprotein amino acid. Proc. Natl. Acad. Sci. U.S.A. 104, 16964–16969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bullwinkle T., Lazazzera B., and Ibba M. (2014) Quality control and infiltration of translation by amino acids outside of the genetic code. Annu. Rev. Genet. 48, 149–166 [DOI] [PubMed] [Google Scholar]
- 24. Poljak A., Dawes I. W., Ingelse B. A., Duncan M. W., Smythe G. A., and Grant C. M. (2003) Oxidative damage to proteins in yeast cells exposed to adaptive levels of H2O2. Redox Rep. 8, 371–377 [DOI] [PubMed] [Google Scholar]
- 25. Popp O., Larraillet V., Kettenberger H., Gorr I. H., Hilger M., Lipsmeier F., Zeck A., and Beaucamp N. (2015) Molecular polygamy: the promiscuity of l-phenylalanyl-tRNA-synthetase triggers misincorporation of meta- and ortho-tyrosine in monoclonal antibodies expressed by Chinese hamster ovary cells. Biotechnol. Bioeng. 112, 1187–1199 [DOI] [PubMed] [Google Scholar]
- 26. Bullwinkle T. J., and Ibba M. (2016) Translation quality control is critical for bacterial responses to amino acid stress. Proc. Natl. Acad. Sci. U.S.A. 113, 2252–2257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Goodarzi H., Liu X., Nguyen H. C., Zhang S., Fish L., and Tavazoie S. F. (2015) Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell 161, 790–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Thompson D. M., Lu C., Green P. J., and Parker R. (2008) tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA 14, 2095–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.