Abstract
Inflammation and endoplasmic reticulum (ER) stress are associated with many neurological diseases. ER stress is brought on by the accumulation of misfolded proteins in the ER, which leads to activation of the unfolded protein response (UPR), a conserved pathway that transmits signals to restore homeostasis or eliminate the irreparably damaged cell. We provide evidence that inhibition or genetic haploinsufficiency of protein kinase R-like endoplasmic reticulum kinase (PERK) can selectively control inflammation brought on by ER stress without impinging on UPR-dependent survival and adaptive responses or normal immune responses. Using astrocytes lacking one or both alleles of PERK or the PERK inhibitor GSK2606414, we demonstrate that PERK haploinsufficiency or partial inhibition led to reduced ER stress-induced inflammation (IL-6, CCL2, and CCL20 expression) without compromising prosurvival responses. In contrast, complete loss of PERK blocked canonical PERK-dependent UPR genes and promoted apoptosis. Reversal of eIF2α-mediated translational repression using ISRIB potently suppressed PERK-dependent inflammatory gene expression, indicating that the selective modulation of inflammatory gene expression by PERK inhibition may be linked to attenuation of eIF2α phosphorylation and reveals a previously unknown link between translational repression and transcription of inflammatory genes. Additionally, ER-stressed astrocytes can drive an inflammatory M1-like phenotype in microglia, and this can be attenuated with inhibition of PERK. Importantly, targeting PERK neither disrupted normal cytokine signaling in astrocytes or microglia nor impaired macrophage phagocytosis or T cell polarization. Collectively, this work suggests that targeting PERK may provide a means for selective immunoregulation in the context of ER stress without disrupting normal immune function.
Keywords: astrocyte, chemokine, cytokine, microglia, unfolded protein response (UPR)
Introduction
Normal inflammatory responses are transient, protective, and essential to overall health. However, chronic inflammation is associated with a multitude of common maladies, including obesity, diabetes, cancer, and neurodegeneration. Left unchecked, inflammation can cause cellular dysfunction and tissue damage from the prolonged production of reactive oxygen/nitrogen species and proteases, among others, potentially exacerbating the disease state (1). Broadly targeting inflammatory processes with drugs such as glucocorticoids can be difficult in chronic conditions because of serious side effects (2) and may impair normal inflammatory processes that are essential for tissue repair and resolution (3, 4). The delicate balance between beneficial and detrimental inflammation is of profound importance in the CNS, where damage can have dire and long-lasting effects. Within the CNS, glial cells, particularly astrocytes and microglia, have a key role in controlling the inflammatory environment (5, 6). Glial cells are in an inflammatory (gliotic) state in most neurological diseases. As with peripheral inflammatory responses, gliosis, when properly executed and resolved, is protective and promotes homeostasis (7). However, chronic glia-derived inflammation may contribute to neurodegeneration (8). Conversely, a reduction of immunological function and surveillance in the CNS can be equally disruptive and may worsen injuries, impair cognitive function, and contribute to neurological diseases (9, 10). Thus, a major challenge is to identify processes driving chronic damaging inflammation that can be targeted without disruption of normal beneficial immune function.
Cell and tissue damage can elicit an inflammatory response (11). In neurodegenerative diseases, cells are likely damaged through several processes, including the accumulation of misfolded proteins, leading to endoplasmic reticulum (ER)3 stress (12). ER stress activates the unfolded protein response (UPR), a homeostatic response to the accumulation of misfolded proteins in the ER. The UPR is mediated by trans-ER membrane proteins that include PERK, inositol-requiring enzyme 1 (IRE1), and activating transcription factor (ATF) 6. This response promotes reduced protein synthesis through PERK-dependent phosphorylation of eukaryotic initiation factor 2α (eIF2α) and selective up-regulation of transcriptional regulators and molecular chaperones to restore homeostasis (13). Apoptotic pathways are also readied should the adaptive response fail. In addition to intracellular signaling in response to ER stress, there is also extracellular communication via cytokine and chemokine production (14). The ER stress pathway is coupled to inflammatory responses through activation of key signaling networks such as NF-κB and JAK/STAT, leading to the production of cytokines, chemokines, and reactive species (15, 16). In chronic diseases, ER stress and inflammation may collaborate to drive sustained inflammation (14). By targeting ER stress signaling, it may be possible to break this cycle. PERK is a particularly attractive target because it is a key player in ER stress-induced inflammation, and disruption of PERK has been shown to be beneficial in mouse models of Alzheimer disease and prion disease (17, 18).
In this study, we tested the hypothesis that ER stress-induced inflammation could be selectively regulated without disrupting normal inflammatory responses. We modulated ER stress signaling by disrupting PERK function genetically or pharmacologically with the small-molecule PERK inhibitor GSK2606414 (19) to demonstrate that an intermediate level of PERK activity greatly reduces inflammatory responses without compromising ER stress-induced adaptive responses or normal immunological function. Overall, this study provides in vitro proof of concept for PERK as a selective immunoregulatory target.
Results
PERK Activation Promotes Inflammatory Gene Expression
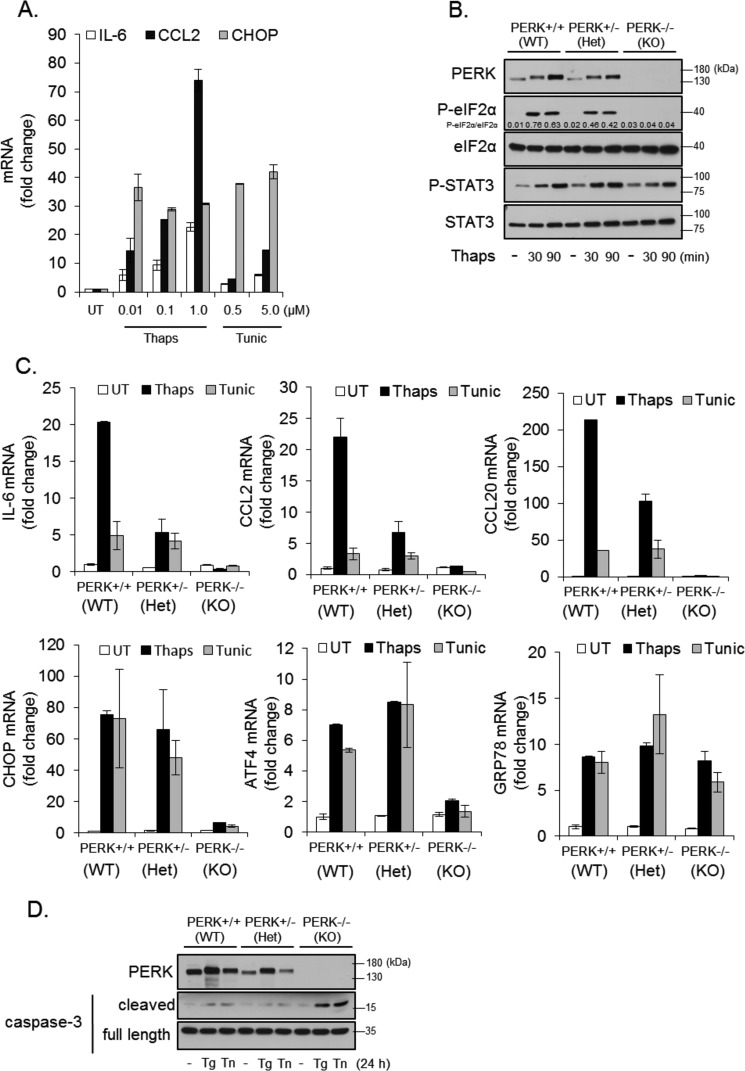
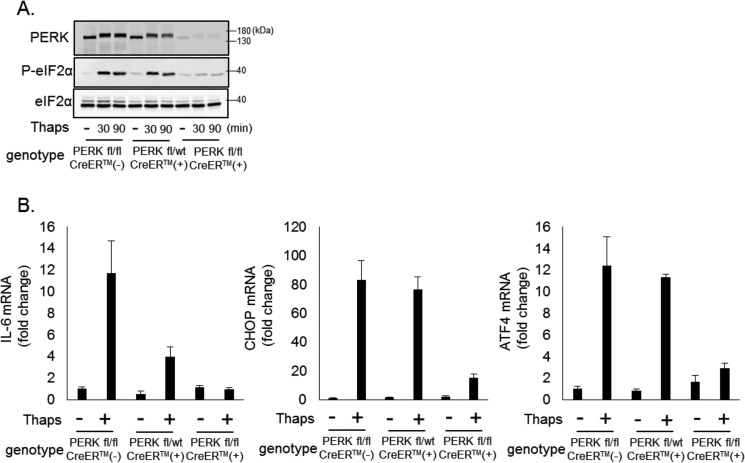
PERK has emerged as a critical signaling node linking ER stress and inflammation (14, 20). Consistent with previous reports, ER stress drives the expression of inflammatory cytokines and chemokines, including IL-6 and CCL2, and the ER stress-selective transcription factor CHOP (Fig. 1A). To verify the importance of PERK in driving inflammatory responses, we isolated primary astrocytes from wild-type, PERK heterozygous (+/−), and PERK knock-out (−/−) embryos and induced ER stress in these cells. As shown in Fig. 1B, wild-type and PERK+/− cells responded to ER stress with increased phosphorylation of eIF2α and STAT3. However, as expected, eIF2α phosphorylation was reduced in PERK+/− cells and absent in PERK−/− cells. STAT3 phosphorylation was not reduced in PERK+/− cells and was attenuated in PERK−/− cells. Wild-type astrocytes increased IL-6, CCL2, and CCL20 in response to ER stress, and this was reduced in PERK+/− astrocytes and abolished in PERK−/− cells (Fig. 1C). In contrast, PERK haploinsufficiency did not reduce ER stress-induced expression of the prototypical UPR genes CHOP, ATF4, and GRP78. In PERK−/− cells, induction of CHOP and ATF4 was blocked, whereas up-regulation of GRP78, which is ATF6-dependent (21), remained unaffected (Fig. 1C). These data indicate that, with partial loss of PERK, cells are still able to mount an adaptive survival response, albeit with reduced inflammatory cytokine and chemokine production. Consistent with this, wild type and PERK+/− cells exposed to ER stress for 24 h showed little apoptosis, as measured by caspase 3 cleavage, whereas PERK−/− had a marked increase in cleaved caspase 3 (Fig. 1D). To validate these findings, we used an additional genetic approach to acutely knock out PERK. PERK floxed (fl/fl) mice (22) were crossed with the global tamoxifen-inducible cre line, CAGG-CreERTM. Astrocytes were isolated from littermates and treated for 24 h with 4-hydroxytamoxifen to delete one or both alleles of PERK. Seventy-two hours after removal of tamoxifen, astrocytes were treated with thapsigargin. As shown in Fig. 2A, PERK expression and eIF2a phosphorylation were modestly reduced in CreERTM-positive PERKfl/wt astrocytes and absent in CreERTM-positive PERKfl/fl cells. Consistent with the previous data, deletion of one PERK allele reduced ER stress-induced IL-6 expression but not CHOP or ATF4. Deletion of both PERK alleles abrogated IL-6, CHOP, and ATF4 expression (Fig. 2B). These data indicate that, although PERK is essential for survival during ER stress, an intermediate level of PERK will suffice. From these data, we hypothesized that it may be possible to target PERK with partial inhibition as a means to control ER stress-induced inflammation without disrupting adaptive responses to ER stress or normal immunological function.
FIGURE 1.
PERK-dependent modulation of inflammatory and survival responses. A, primary astrocytes were untreated (UT or −) or treated with the indicated concentration of thapsigargin (thaps) or tunicamycin (tunic) for 4 h, followed by qPCR analysis. B, primary astrocytes were isolated from wild-type, PERK+/−, or PERK−/− astrocytes and treated with thaps (1 μm) for 30 or 90 min, followed by immunoblotting. C, astrocytes were treated with thaps (1 μm) or tunic (5 μm) for 4 h, followed by qPCR analysis. D, astrocytes were treated with thaps (Tg, 1 μm) or tunic (Tn, 5 μm) for 24 h, followed by immunoblotting. Het, heterozygous.
FIGURE 2.
Inducible deletion of PERK regulates ER stress-induced gene expression. A, astrocytes were isolated from PERKfl/fl, PERKfl/wt × CAGG-CreERTM and PERKfl/fl × CAGG-CreERTM littermates. Astrocytes were treated with 4-hydroxytamoxifen (1 μm) for 24 h to delete the floxed PERK alleles in the Cre-expressing cells. Seventy-two hours after removal of the tamoxifen, cells were treated with thaps (1 μm) for the indicated times, followed by immunoblotting. B, astrocytes were isolated and treated with 4-hydroxytamoxifen as in A and then treated with thaps (1 μm) for 4 h, followed by qPCR analysis.
Small-molecule Inhibition of PERK Selectively Suppresses Inflammatory Gene Expression
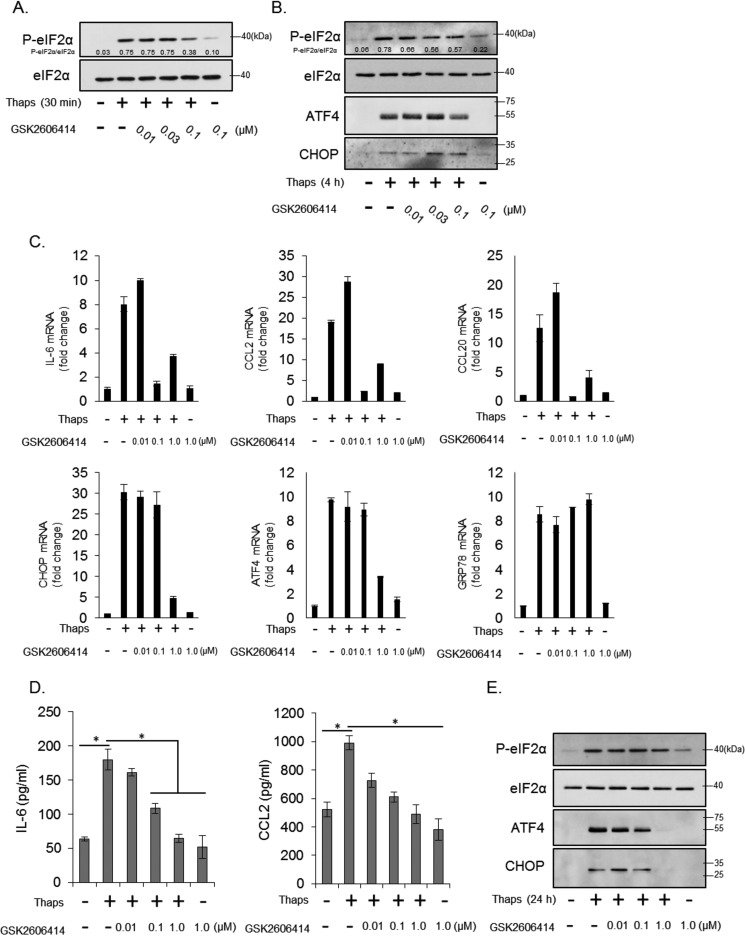
To test this hypothesis, we used the small-molecule inhibitor of PERK GSK2606414. As shown in Fig. 3A, 0.1 μm GSK2606414 partially inhibited, by ∼50%, thapsigargin-induced eIF2α phosphorylation at 30 min, indicating partial inhibition of PERK. By 4 h, the inhibitory effect of GSK2606414 was still apparent, although diminished. This level of inhibition led to a minor (∼20%) decrease in ATF4 protein expression and did not inhibit the low level of CHOP expression (Fig. 3B). However, it was effective at blocking ER stress-induced IL-6, CCL2, and CCL20 but not CHOP, ATF4, and GRP78 mRNA (Fig. 3C), similar to that observed in PERK+/− cells. Only at high concentrations (1 μm) was GSK2606414 able to block CHOP and ATF4 mRNA expression (Fig. 3C), similar to that observed in PERK−/− cells. GSK2606414 also significantly inhibited ER stress-induced IL-6 and CCL2 protein after 24 h, as measured by ELISA (Fig. 3D). At this time point, GSK2606414 was no longer effective at inhibiting eIF2α phosphorylation but, at high concentrations, did prevent ATF4 and CHOP expression (Fig. 3E). To extend these findings, we examined the inhibitory effect of GSK2606414 on ER stress-induced cytokine and chemokine expression by multiplex ELISA. In addition to the significant inhibition of IL-6 and CCL2, we also observed inhibition of ER stress-induced CXCL10, CXCL1, and CXCL2 (Fig. 4). These data demonstrate that modest PERK inhibition can reduce some inflammatory molecules stimulated by ER stress.
FIGURE 3.
Differential modulation of inflammatory and adaptive gene expression by inhibition of PERK. A and B, astrocytes were treated with thaps (1 μm) for 30 min (A) or 4 h (B) in the presence of increasing concentrations of the PERK inhibitor GSK2606414, followed by immunoblotting. C, astrocytes were treated with thaps (1 μm) for 4 h in the presence of increasing concentrations of GSK2606414, followed by qPCR analysis. D, astrocytes were treated with thaps (1 μm) for 24 h in the presence of increasing concentrations of GSK2606414. Cytokine production was measured by ELISA. n = 3, *, p < 0.05. E, astrocytes were treated with thaps (1 μm) for 24 h in the presence of increasing concentrations of GSK2606414, followed by immunoblotting.
FIGURE 4.
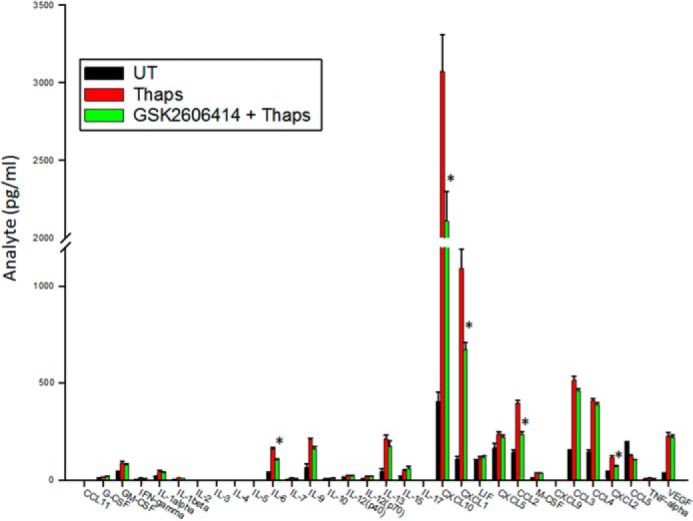
PERK-dependent regulation of cytokines and chemokines. Astrocytes were treated with thaps (1 μm) for 24 h in the absence or presence of GSK2606414 (0.1 μm). Cell culture supernatants were collected and analyzed by multiplex ELISA. n = 3, *, p < 0.05.
ER Stress Augments Cytokine-induced Gene Expression in a PERK-dependent Fashion
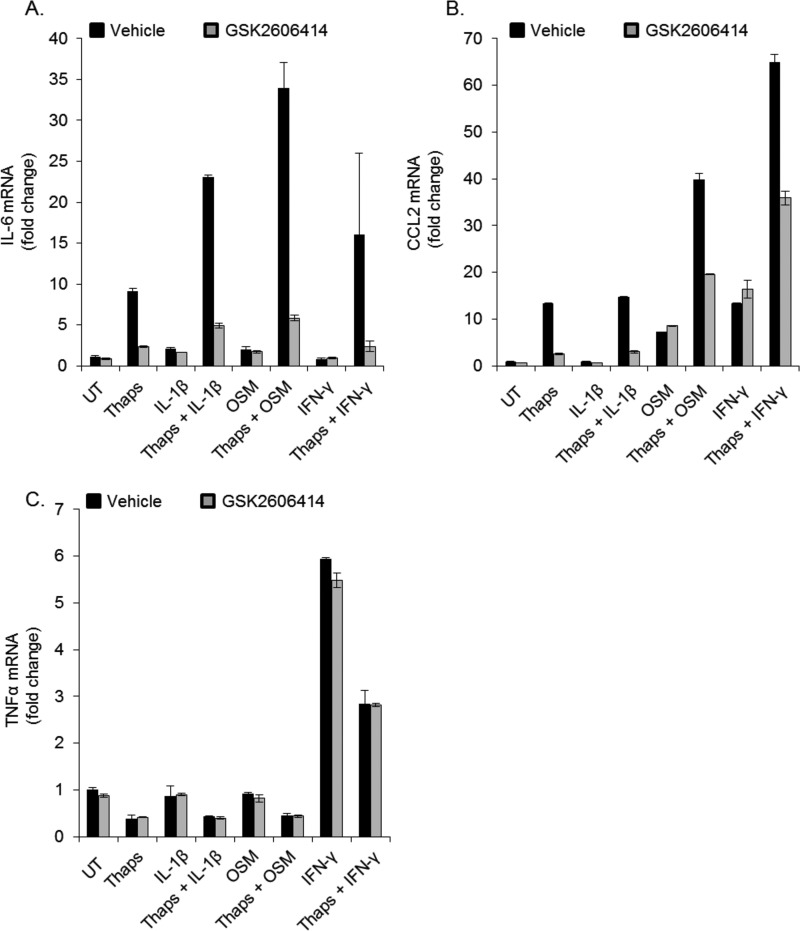
Not only does ER stress stimulate inflammation, it can also alter the response to other inflammatory signals (16, 23, 24). To test whether PERK inhibition could modulate this effect, astrocytes were treated with the cytokines IL-1β, OSM, or IFN-γ individually or in the presence of thapsigargin. ER stress enhanced the expression of IL-6 in response to IL-1β, OSM, and IFN-γ (Fig. 5A) and, similarly, enhanced the expression of CCL2 in response to IFN-γ and OSM (Fig. 5B). Cytokines did not affect thapsigargin-induced CHOP expression (data not shown). To determine whether the enhanced inflammatory gene expression was sensitive to PERK inhibition, astrocytes were treated with cytokines and thapsigargin in the absence or presence of the PERK inhibitor. GSK2606414 did not affect gene expression induced by cytokines alone but did attenuate the enhancement by ER stress (Fig. 5, A and B). Additionally, ER stress did not enhance, but instead suppressed, IFN-γ-induced TNFα expression, and this was unaffected by PERK inhibition (Fig. 5C). These data indicate that PERK is dispensable in canonical cytokine signaling and that PERK inhibition can block ER stress-induced augmentation of cytokine-mediated gene expression.
FIGURE 5.
ER stress and cytokines interact in a PERK-dependent fashion. Astrocytes were pretreated with thaps (1 μm) for 30 min, followed by stimulation with IL-1β (100 pg/ml), OSM (1 ng/ml), or IFN-γ (10 ng/ml) in the absence or presence of GSK2606414 (0.1 μm) for 4 h, followed by qPCR analysis for IL-6 (A), CCL2 (B), and TNFα (C). UT, untreated.
Inflammatory Gene Expression Is Dependent on the Phospho-eIF2α-induced Translation Block but Independent of ATF4
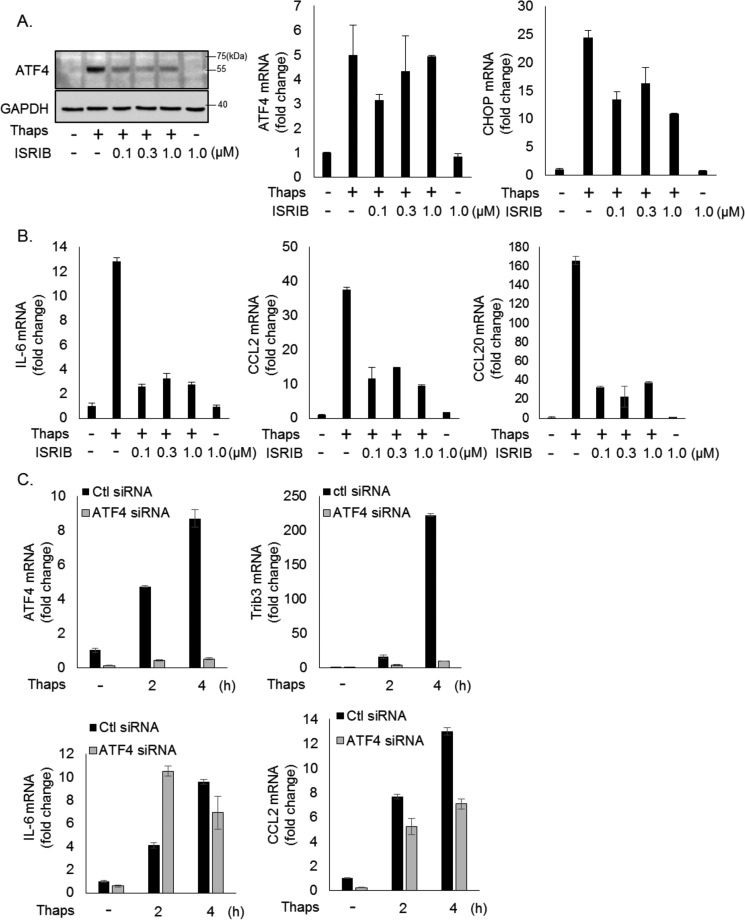
Our previous work demonstrated that ER stress-induced IL-6 and CCL2 in astrocytes is dependent on JAK1 (16). However, our data showing that deletion of one PERK allele attenuates IL-6 and CCL2 but does not reduce STAT3 phosphorylation suggests that additional mechanisms contribute to the sensitivity of these genes to PERK inhibition. Many of the PERK-mediated effects are dependent on eIF2α phosphorylation and subsequent expression of ATF4 (25). Therefore, we tested whether the eIF2α-induced translational inhibition was involved in the regulation of these inflammatory genes using the recently described compound ISRIB (26). This compound reverses the translational block brought on by phosphorylation of eIF2α through an agonistic effect on EIF2B (27). As expected, ISRIB suppressed thapsigargin-induced ATF4 protein expression, had no effect on ATF4 mRNA, and modestly reduced CHOP mRNA (Fig. 5A). Interestingly, this highlights that PERK controls ATF4 at two levels through distinct mechanisms. ATF4 protein is dependent on eIF2α-induced translational inhibition, whereas ATF4 mRNA is PERK-dependent but independent of eIF2α. These data are consistent with data showing that PERK-NRF2 signaling drives ATF4 transcription (28). Concomitant with suppression of ATF4 protein, ISRIB potently suppressed thapsigargin-induced IL-6, CCL2, and CCL20 expression (Fig. 6B). This suggests that ER stress-induced inhibition of translation promotes transcription of inflammatory molecules such as IL-6, CCL2, and CCL20. Importantly ISRIB does not broadly block inflammatory gene expression, as it had no effect on cytokine-induced IL-6 or CCL2 expression (data not shown). To test whether ATF4 connects translational repression to inflammatory gene expression, we used siRNA to knock down ATF4. As shown in Fig. 6C, ATF4 expression is reduced, and thapsigargin-induced expression of the ATF4 target gene tribbles 3 (Trib3) (29) is abrogated by ATF4 siRNA. This confirms functional silencing of ATF4. Knockdown of ATF4 did not inhibit thapsigargin-induced IL-6 and partially reduced CCL2 expression (Fig. 6C). Collectively, these data indicate that the eIF2α-induced translational block is essential for inflammatory gene expression but is largely independent of ATF4.
FIGURE 6.
ER stress-induced inflammatory gene expression is dependent on eIF2α-mediated translational repression. A and B, astrocytes were treated with thaps (1 μm) for 4 h in the presence or absence of increasing concentrations of ISRIB, followed by immunoblotting or qPCR analysis. C, astrocytes were transfected with control (Ctl) or ATF4 siRNA for 48 h. The cells were then treated with thaps (1 μm) for the indicated times, followed by qPCR analysis.
PERK Activation in ER-stressed Astrocytes Influences the Microglial Phenotype
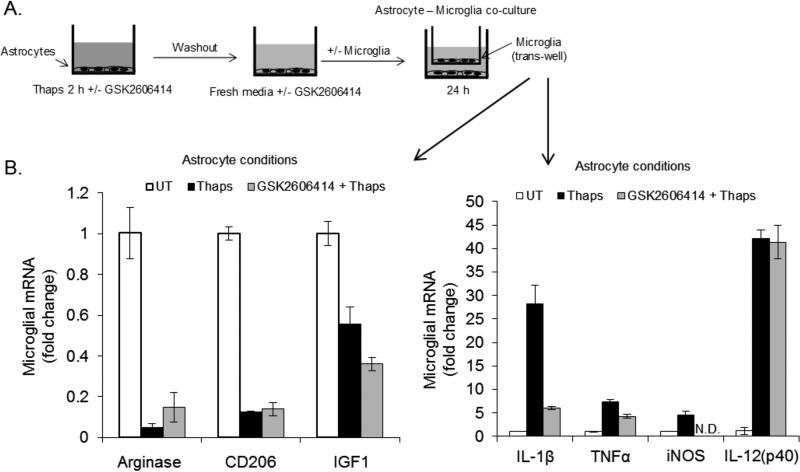
We previously demonstrated that PERK-dependent communication between ER-stressed astrocytes and naïve microglia resulted in the activation of microglia (16). Therefore, we tested whether PERK inhibition could also modulate astrocyte-microglia interactions. Astrocytes were treated transiently (2 h) with thapsigargin in the absence or presence of GSK2606414, washed thoroughly to remove the thapsigargin, and cultured in fresh medium with or without GSK2606414. Following the addition of fresh medium, primary microglia were added in a Transwell (Fig. 7A). The microglia were cultured with astrocytes for 24 h. As shown in Fig. 7B, astrocytes under ER stress promoted a proinflammatory M1-like phenotype in microglia that included down-regulation of the M2 markers arginase, CD206, and IGF1 and up-regulation of inflammatory genes, including IL-1β, TNFα, inducible nitric-oxide synthase (iNOS), and IL-12p40. PERK inhibition had no effect on the down-regulation of M2 markers but did reduce the levels of IL-1β, TNFα, and inducible nitric-oxide synthase. The expression of IL-12p40 was unaffected by PERK inhibition (Fig. 7B). These data indicate that ER-stressed astrocytes promote a proinflammatory phenotype in microglia and that this can be attenuated by inhibiting PERK.
FIGURE 7.
ER-stressed astrocytes promote inflammatory microglia. A, astrocytes were treated with thaps (1 μm) for 2 h in the absence or presence of GSK2606416 (0.1 μm). The astrocytes were then washed three times to remove thaps and continued in fresh medium with or without GSK2606414. Microglia were added in a Transwell after removal of thaps-containing medium. The astrocyte:microglia co-culture was incubated for 24 h, followed by qPCR analysis of the microglia (B). UT, untreated.
PERK Inhibition Does Not Affect Normal Immune Responses in Vitro
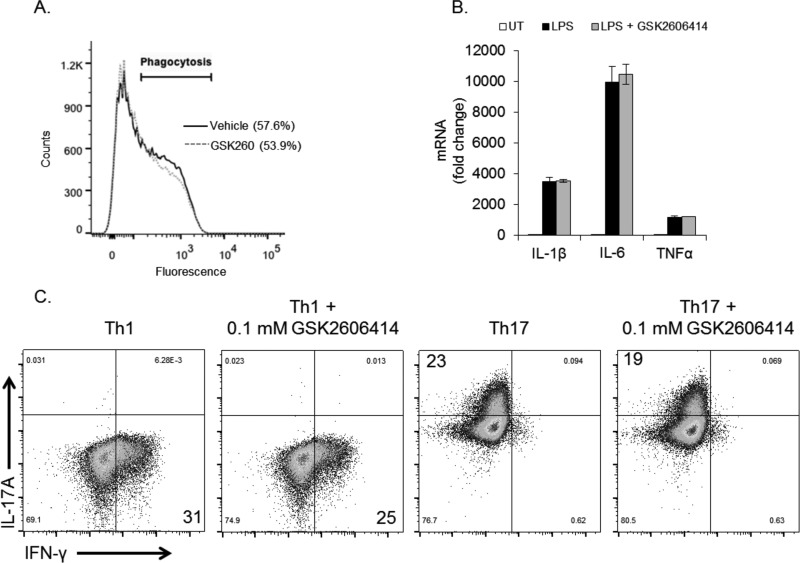
A major challenge in managing inflammation is to suppress chronic, pathological inflammation while leaving normal beneficial immunological functions intact. Our observation that PERK inhibition suppresses ER stress-induced inflammatory gene expression but not cytokine-induced gene expression (Fig. 5) suggests that targeting PERK may not disrupt normal immune function. To test this, we used several primary cell types to assess the immunological impact of PERK inhibition. Macrophage-mediated phagocytosis is a key component in innate immunity. Treatment of bone marrow-derived macrophages for 24 h with GSK2606414 did not cause toxicity (data not shown) or impair phagocytosis (Fig. 8A). Next, we examined the inflammatory response in primary microglia to the bacterial component LPS. Similar to our findings with cytokines, LPS stimulated potent inflammatory gene expression that was not affected by PERK inhibition (Fig. 8B). Differentiation of naïve CD4+ T cells into various effector T cell subsets is paramount in establishing an appropriate adaptive immune response. As such, we tested whether PERK inhibition could alter CD4+ T cell polarization. T cells were cultured under Th1- or Th17-polarizing conditions in the absence or presence of GSK2606414 for 3 days. As shown in Fig. 8C, GSK2606414 had little effect on Th1 or Th17 polarization, as assessed by their hallmark cytokines IFN-γ and IL-17A, respectively. These data indicate that, although PERK inhibition can suppress inflammation under ER stress conditions, targeting PERK does not have a broad impact on overall immunological function. In line with these results, PERK has also been shown to be dispensable for B cell development and immunoglobulin secretion (30).
FIGURE 8.
PERK inhibition does not impair immunological responses in vitro. A, phagocytosis was measured in bone marrow-derived macrophages treated without or with GSK2606414 (0.1 μm) for 24 h prior to the assay. B, primary microglia were treated with LPS (10 ng/ml) for 4 h in the absence or presence of GSK2606414 (0.1 μm), followed by qPCR analysis. UT, untreated. C, naïve CD4+ T cells were isolated and polarized to Th1 or Th17 phenotypes in the absence or presence of GSK2606414 (0.1 μm) for 3 days, followed by analysis by flow cytometry.
Discussion
Neurological diseases are a growing public health problem. Decades of research have greatly expanded our knowledge of neuroscience but have yet to lead to effective therapies, largely because of the complex nature of these diseases. We now appreciate that critical non-neuronal factors, such as neuroinflammation, are involved in neurological diseases. Specifically, inflammation is thought to contribute to the chronic non-resolving pathology common to neurodegenerative diseases (5). Selectively disrupting disease-driven inflammation while leaving normal immune function intact could provide a new therapeutic avenue. Unfortunately, targets to achieve such specificity are currently unknown. In this study, we provide evidence that small molecule inhibition or genetic haploinsufficiency of the UPR kinase PERK can selectively control inflammation brought on by ER stress without impinging on UPR-dependent survival and adaptive responses.
In healthy tissue, an inflammatory response is finely tuned and exquisitely effective at restoring homeostasis. A key step in the restoration of homeostasis is the resolution of inflammation. In chronic diseases, ER stress may contribute to the sustained production of inflammatory mediators, thus obstructing homeostatic mechanisms (31, 32). The cellular response to ER stress is multifaceted, with pathways activated to promote adaptation and survival as well as apoptosis and inflammation (14, 20). Perturbations in the signaling molecules controlling the UPR can lead to functional changes in outcome. For example, we have shown previously (16) and show in Fig. 1 that astrocytes are highly resistant to ER stress. However, knockout of PERK reduces this resistance, as shown by increased caspase 3 cleavage following ER stress. We found that partial loss of PERK function did not impair PERK-dependent survival responses but did modulate other pathways. Specifically, we observed that reduced PERK function attenuated some ER stress-induced inflammatory responses. The PERK-dependent pathways that are attenuated by PERK haploinsufficiency, leading to reduced inflammatory gene expression, appear to involve eIF2α-dependent translational repression. This is consistent with a previous study showing that eIF2α phosphorylation is required for Yersinia infection-induced TNFα expression (33). However, the exact mechanism linking eIF2α to transcription of inflammatory genes remains to be fully elucidated. This study and our previous work indicate that at least IL-6, CCL2, and CCL20 expression are dependent on both JAK1 and translational repression in response to ER stress. These pathways may be working in parallel, or this could indicate a functional relationship between JAK1 and eIF2α. Additionally, this could suggest that stress that causes an intermediate level of eIF2α phosphorylation is perceived by the cell as survivable, and inflammatory responses are kept low, although intense stressors that cause maximal eIF2α phosphorylation may be perceived as a danger signal, and inflammatory responses are initiated to recruit and activate phagocytic, antigen-presenting, and other immune cells to re-establish tissue homeostasis. Additional studies are needed to formally test these hypotheses.
Alterations in microglia number, morphology, and phenotype are observed in most neurodegenerative diseases. The microglial phenotype is highly plastic, encompassing a continuum of functions ranging from restorative to damaging, depending on cell-cell interactions and environmental cues within the CNS (34). Our findings indicate that ER-stressed astrocytes can influence microglia to adopt an inflammatory M1-like phenotype in a PERK-dependent fashion. In co-culture experiments (Fig. 7), only astrocytes were exposed to ER stress, whereas both astrocytes and microglia were exposed to the PERK inhibitor. Accordingly, it is possible that the PERK inhibitor influenced the microglia response to the ER-stressed astrocytes. However, based on our data, this is unlikely, as PERK inhibition potently suppressed astrocyte-induced IL-1β expression in microglia but had no effect on LPS-induced IL-1β expression. This suggests that it may be possible to shift microglial function in response to specific stimuli (i.e. ER stress) without affecting overall function.
The selective blockade of inflammation by PERK inhibition may have therapeutic relevance, particularly in the context of neurodegeneration, where ER stress is prevalent. Indeed, targeting the PERK-eIF2α pathway using GSK6206414, ISRIB, or genetic approaches has been shown to attenuate disease in mouse models of prion and Alzheimer disease (17, 18, 35, 36). One mechanism the authors uncovered for the beneficial actions of PERK inhibition is the restoration of critical synaptic proteins such as SNAP25 and PSD95. The levels of these proteins are restored because the PERK-dependent translational block is removed. Moreover, there was a concomitant reduction in astrogliosis (18, 35). Although inflammation was not examined in these studies, based on our findings, we would speculate that the selective attenuation of ER stress-induced inflammation may be an additional beneficial effect of GSK2606414 in prion and other neurodegenerative diseases. Overall, our in vitro findings suggest that PERK may be a selective immunotherapeutic target in chronic diseases involving ER stress.
Experimental Procedures
Reagents
TaqMan primers and Silencer® select predesigned siRNAs were purchased from Life Technologies. ELISAs were purchased from BioLegend, and mouse cytokine/chemokine multiplex assays were purchased from EMD Millipore. The following antibodies were purchased from Cell Signaling Technology: PERK (3192), phospho-eIF2α (3398), eIF2α (5324), ATF4 (11815), caspase 3 (9665), phospho-STAT3 (9145), and STAT3 (12640). GADD153/CHOP (SC-7351) was from Santa Cruz Biotechnology, and GAPDH (MAB374) was from EMD Millipore. HRP-conjugated anti-mouse (NA931) and anti-rabbit (NA934) secondary antibodies were from GE Life Sciences. Recombinant IFN-γ, IL-1β, and Oncostatin M (OSM) were purchased from R&D Technologies.
Mice and Primary Cell Preparations
C57Bl/6, PERK+/−, PERK floxed, and CAGG-CreERTM mice were purchased from The Jackson Laboratory and bred and housed in the animal facility at the University of Alabama at Birmingham or at West Virginia University under the care of the animal resources program. Primary murine astrocyte or microglial cultures were prepared as described previously (37). This protocol was modified to obtain PERK−/− astrocytes. PERK+/− mice were bred, and cerebrums were isolated from individual embryonic day 18.5 embryos and used for astrocyte cultures. This provided cultures with each genotype (PERK+/+, PERK+/−, and PERK−/−) as littermates. Astrocytes or microglia were cultured in DMEM with 10% FBS, 16 mm HEPES, 1× non-essential amino acids, 2 mm l-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, and 50 μg/ml gentamicin. Astrocytes were separated from microglia by shaking at 200 RPM for 1.5 h, and astrocyte cultures contained >90% glial fibrillary acid protein-positive (GFAP+) cells as determined by immunofluorescent microscopy. Following shaking, microglia containing supernatants were centrifuged at 400 × g for 5 min, and microglia were resuspended in fresh medium and plated. CD4+ T cells were isolated from the spleens of C57Bl/6 mice using anti-CD4 DynaBeads (Life Technologies) according to the protocol of the manufacturer.
Immunoblotting
Cells were washed twice with PBS and lysed with lysis buffer (20 mm Tris (pH 7.5), 150 mm NaCl, 2 mm EDTA, 2 mm EGTA, and 0.5% Nonidet P-40) containing 1 mm phenylmethanesulfonyl fluoride, 25 μg/ml leupeptin, 25 μg/ml aprotinin, and 1× phosphatase inhibitor mixture (Pierce) as described previously (38). Protein concentrations were determined using the BCA assay (Pierce). Equal amounts of protein from each sample were solubilized in Laemmli sample buffer (2% SDS) and heated for 5 min at 95 °C. Proteins were separated by SDS-polyacrylamide gel electrophoresis and transferred to nitrocellulose, and then the membranes were blocked in 5% milk, followed by an overnight incubation at 4 °C with primary antibody diluted in 5% BSA or milk according to the recommendation of the manufacturer. Horseradish peroxidase-conjugated donkey anti-rabbit or donkey anti-mouse (1:4000 dilution) secondary antibodies were incubated for 1 h at room temperature, followed by detection with enhanced chemiluminescence. Films were digitized with an Epson Perfection V300 photo scanner or imaged digitally using a ChemiDoc Touch (Bio-Rad). Densitometry was performed using ImageJ.
Quantitative RT-PCR
RNA was isolated using TRIzol (Sigma-Aldrich) as described previously (39). RNA was quantified using a NanoDrop (NanoDrop Technologies), and 1 μg of RNA was used for cDNA synthesis using Moloney murine leukemia virus reverse transcriptase (Promega). The cDNA was analyzed by quantitative PCR performed using TaqMan gene expression assays according to the instructions of the manufacturer in an ABI Step One Plus (Applied Biosystems). Reactions were carried out in 20 μl and analyzed using the ΔΔCt method.
ELISA
Culture supernatants (100 μl, undiluted) were collected and assayed by ELISA for murine IL-6 and CCL2 (BioLegend) or by multiplex ELISA (Millipore) according to the protocol of the manufacturer.
Transfections
Primary astrocytes were transfected with the indicated siRNA (50 pmol/35-mm well) using Lipofectamine RNAiMAX (Life Technologies) according to the protocol of the manufacturer. Cells were used for experiments 48–72 h after transfection.
Phagocytosis Assay
Bone marrow-derived macrophages were isolated and cultured as described previously (40). Cells were plated in 6-well plates and treated as indicated. Cells were then incubated with pHrodo Escherichia coli particles for 30 min at 37 °C. Cells were then scraped and collected on ice. Samples were run on a LSRII FACSCalibur, and data were analyzed using FlowJo software.
T Cell Polarization
CD4+ T cells were cultured in RPMI 1640 medium with 10% FBS, 100 units/ml penicillin, 100 μg/ml streptomycin, 1× nonessential amino acids, 1 mm sodium pyruvate, 2.5 μm 2-mercaptoethanol, and 2 mm l-glutamine. T cells were activated by the addition of anti-CD3 and anti-CD28 (10 μg/ml each, BD Biosciences) and irradiated splenocytes (antigen-presenting cells) at a ratio of 5:1 antigen-presenting cells:T cells. Cells were polarized toward either Th1 by the addition of IL-12 (10 ng/ml, R&D Systems) and anti-IL-4 (10 μg/ml, BioLegend) or Th17 by the addition of TGF-β (5 ng/ml), IL-6 (20 ng/ml), IL-23 (10 ng/ml) (R&D Systems), anti-IFN-γ (10 μg/ml), and anti-IL-4 (10 μg/ml) (BioLegend), modified from Ref. 41. Cells were cultured under polarizing conditions for 3 days and then analyzed by FACS analysis.
Statistics
qPRC data are the means of two to three technical replicates and are representative of at least three independent experiments. ELISA data are the means of multiple experiments and significance, where p < 0.05, was determined by one-way analysis of variance with post hoc analysis.
Author Contributions
G. P. M. oversaw the project, designed the study, conducted experiments, and wrote the manuscript. L. N. G. conducted experiments and edited the manuscript. K. A., E. S. P., N. T. S., S. A. G., B. C. M., R. R., and A. L. R. conducted experiments. E. N. B. oversaw the project and edited the manuscript.
Acknowledgments
We thank Dr. Chad Steele (University of Alabama at Birmingham) for assistance with multiplex ELISA and Dr. Bandi Talkington (West Virginia University) for editorial assistance. We also thank Savannah Sims and John Nowery for technical assistance.
This work was supported in part by National Multiple Sclerosis Society Career Transition Award TA3050-A-1 (to G. P. M.), National Multiple Sclerosis Society Collaborative Award CA-1059-A-13 (to E. N. B.), and National Institutes of Health Grants NS57563 and CA158534 (to E. N. B.). Support was also provided by the West Virginia Clinical and Translational Science Institute and NIGMS, National Institutes of Health Grant U54GM104942. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- ER
- endoplasmic reticulum
- UPR
- unfolded protein response
- PERK
- protein kinase R-like endoplasmic reticulum kinase
- ATF
- activating transcription factor
- OSM
- Oncostatin M
- qPCR
- quantitative PCR
- thaps
- thapsigargin
- tunic
- tunicamycin
- CHOP
- C/EBP homologous protein
- C/EBP
- CCAAT/enhancer-binding protein.
References
- 1. Medzhitov R. (2008) Origin and physiological roles of inflammation. Nature 454, 428–435 [DOI] [PubMed] [Google Scholar]
- 2. Coutinho A. E., and Chapman K. E. (2011) The anti-inflammatory and immunosuppressive effects of glucocorticoids, recent developments and mechanistic insights. Mol. Cell Endocrinol. 335, 2–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Buckley C. D., Gilroy D. W., Serhan C. N., Stockinger B., and Tak P. P. (2013) The resolution of inflammation. Nat. Rev. Immunol. 13, 59–66 [DOI] [PubMed] [Google Scholar]
- 4. Raposo C., Graubardt N., Cohen M., Eitan C., London A., Berkutzki T., and Schwartz M. (2014) CNS repair requires both effector and regulatory T cells with distinct temporal and spatial profiles. J. Neurosci. 34, 10141–10155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ransohoff R. M., and Brown M. A. (2012) Innate immunity in the central nervous system. J. Clin. Invest. 122, 1164–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Burda J. E., and Sofroniew M. V. (2014) Reactive gliosis and the multicellular response to CNS damage and disease. Neuron 81, 229–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Verkhratsky A., Parpura V., Pekna M., Pekny M., and Sofroniew M. (2014) Glia in the pathogenesis of neurodegenerative diseases. Biochem. Soc. Trans. 42, 1291–1301 [DOI] [PubMed] [Google Scholar]
- 8. Heneka M. T., Kummer M. P., and Latz E. (2014) Innate immune activation in neurodegenerative disease. Nat. Rev. Immunol. 14, 463–477 [DOI] [PubMed] [Google Scholar]
- 9. Bush T. G., Puvanachandra N., Horner C. H., Polito A., Ostenfeld T., Svendsen C. N., Mucke L., Johnson M. H., and Sofroniew M. V. (1999) Leukocyte infiltration, neuronal degeneration, and neurite outgrowth after ablation of scar-forming, reactive astrocytes in adult transgenic mice. Neuron 23, 297–308 [DOI] [PubMed] [Google Scholar]
- 10. Schwartz M., Kipnis J., Rivest S., and Prat A. (2013) How do immune cells support and shape the brain in health, disease, and aging? J. Neurosci. 33, 17587–17596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen G. Y., and Nuñez G. (2010) Sterile inflammation: sensing and reacting to damage. Nat. Rev. Immunol. 10, 826–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hetz C., and Mollereau B. (2014) Disturbance of endoplasmic reticulum proteostasis in neurodegenerative diseases. Nat. Rev. Neurosci. 15, 233–249 [DOI] [PubMed] [Google Scholar]
- 13. Ron D., and Walter P. (2007) Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 8, 519–529 [DOI] [PubMed] [Google Scholar]
- 14. Zhang K., and Kaufman R. J. (2008) From endoplasmic-reticulum stress to the inflammatory response. Nature 454, 455–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kitamura M. (2011) Control of NF-κB and inflammation by the unfolded protein response. Int. Rev. Immunol. 30, 4–15 [DOI] [PubMed] [Google Scholar]
- 16. Meares G. P., Liu Y., Rajbhandari R., Qin H., Nozell S. E., Mobley J. A., Corbett J. A., and Benveniste E. N. (2014) PERK-dependent activation of JAK1 and STAT3 contributes to endoplasmic reticulum stress-induced inflammation. Mol. Cell Biol. 34, 3911–3925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ma T., Trinh M. A., Wexler A. J., Bourbon C., Gatti E., Pierre P., Cavener D. R., and Klann E. (2013) Suppression of eIF2α kinases alleviates Alzheimer's disease-related plasticity and memory deficits. Nat. Neurosci. 16, 1299–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Moreno J. A., Halliday M., Molloy C., Radford H., Verity N., Axten J. M., Ortori C. A., Willis A. E., Fischer P. M., Barrett D. A., and Mallucci G. R. (2013) Oral treatment targeting the unfolded protein response prevents neurodegeneration and clinical disease in prion-infected mice. Sci. Transl. Med. 5, 206ra138. [DOI] [PubMed] [Google Scholar]
- 19. Axten J. M., Medina J. R., Feng Y., Shu A., Romeril S. P., Grant S. W., Li W. H., Heerding D. A., Minthorn E., Mencken T., Atkins C., Liu Q., Rabindran S., Kumar R., Hong X., et al. (2012) Discovery of 7-methyl-5-(1-{[3-(trifluoromethyl)phenyl]acetyl}-2,3-dihydro-1H-indol-5-yl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine (GSK2606414), a potent and selective first-in-class inhibitor of protein kinase R (PKR)-like endoplasmic reticulum kinase (PERK). J. Med. Chem. 55, 7193–7207 [DOI] [PubMed] [Google Scholar]
- 20. Martinon F., and Glimcher L. H. (2011) Regulation of innate immunity by signaling pathways emerging from the endoplasmic reticulum. Curr. Opin. Immunol. 23, 35–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yoshida H., Haze K., Yanagi H., Yura T., and Mori K. (1998) Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins: involvement of basic leucine zipper transcription factors. J. Biol. Chem. 273, 33741–33749 [DOI] [PubMed] [Google Scholar]
- 22. Zhang P., McGrath B., Li S., Frank A., Zambito F., Reinert J., Gannon M., Ma K., McNaughton K., and Cavener D. R. (2002) The PERK eukaryotic initiation factor 2 α kinase is required for the development of the skeletal system, postnatal growth, and the function and viability of the pancreas. Mol. Cell Biol. 22, 3864–3874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Martinon F., Chen X., Lee A.-H., and Glimcher L. H. (2010) TLR activation of the transcription factor XBP1 regulates innate immune responses in macrophages. Nat. Immunol. 11, 411–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu Y.-P., Zeng L., Tian A., Bomkamp A., Rivera D., Gutman D., Barber G. N., Olson J. K., and Smith J. A. (2012) Endoplasmic reticulum stress regulates the innate immunity critical transcription factor IRF3. J. Immunol. 189, 4630–4639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wek R. C., and Cavener D. R. (2007) Translational control and the unfolded protein response. Antioxid. Redox. Signal 9, 2357–2371 [DOI] [PubMed] [Google Scholar]
- 26. Sidrauski C., Acosta-Alvear D., Khoutorsky A., Vedantham P., Hearn B. R., Li H., Gamache K., Gallagher C. M., Ang K. K., Wilson C., Okreglak V., Ashkenazi A., Hann B., Nader K., Arkin M. R., et al. (2013) Pharmacological brake-release of mRNA translation enhances cognitive memory. eLife 2, e00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sidrauski C., Tsai J. C., Kampmann M., Hearn B. R., Vedantham P., Jaishankar P., Sokabe M., Mendez A. S., Newton B. W., Tang E. L., Verschueren E., Johnson J. R., Krogan N. J., Fraser C. S., Weissman J. S., et al. (2015) Pharmacological dimerization and activation of the exchange factor eIF2B antagonizes the integrated stress response. eLife 4, e07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Afonyushkin T., Oskolkova O. V., Philippova M., Resink T. J., Erne P., Binder B. R., and Bochkov V. N. (2010) Oxidized phospholipids regulate expression of ATF4 and VEGF in endothelial cells via NRF2-dependent mechanism: novel point of convergence between electrophilic and unfolded protein stress pathways. Arterioscler. Thromb. Vasc. Biol. 30, 1007–1013 [DOI] [PubMed] [Google Scholar]
- 29. Han J., Back S. H., Hur J., Lin Y.-H., Gildersleeve R., Shan J., Yuan C. L., Krokowski D., Wang S., Hatzoglou M., Kilberg M. S., Sartor M. A., and Kaufman R. J. (2013) ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat. Cell Biol. 15, 481–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gass J. N., Jiang H.-Y., Wek R. C., and Brewer J. W. (2008) The unfolded protein response of B-lymphocytes: PERK-independent development of antibody-secreting cells. Mol. Immunol. 45, 1035–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nathan C., and Ding A. (2010) Nonresolving inflammation. Cell 140, 871–882 [DOI] [PubMed] [Google Scholar]
- 32. Chovatiya R., and Medzhitov R. (2014) Stress, inflammation, and defense of homeostasis. Mol. Cell 54, 281–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shrestha N., Bahnan W., Wiley D. J., Barber G., Fields K. A., and Schesser K. (2012) Eukaryotic initiation factor 2 (eIF2) signaling regulates proinflammatory cytokine expression and bacterial invasion. J. Biol. Chem. 287, 28738–28744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Prinz M., Priller J., Sisodia S. S., and Ransohoff R. M. (2011) Heterogeneity of CNS myeloid cells and their roles in neurodegeneration. Nat. Neurosci. 14, 1227–1235 [DOI] [PubMed] [Google Scholar]
- 35. Moreno J. A., Radford H., Peretti D., Steinert J. R., Verity N., Martin M. G., Halliday M., Morgan J., Dinsdale D., Ortori C. A., Barrett D. A., Tsaytler P., Bertolotti A., Willis A. E., Bushell M., and Mallucci G. R. (2012) Sustained translational repression by eIF2α-P mediates prion neurodegeneration. Nature 485, 507–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Halliday M., Radford H., Sekine Y., Moreno J., Verity N., le Quesne J., Ortori C. A., Barrett D. A., Fromont C., Fischer P. M., Harding H. P., Ron D., and Mallucci G. R. (2015) Partial restoration of protein synthesis rates by the small molecule ISRIB prevents neurodegeneration without pancreatic toxicity. Cell Death Dis. 6, e1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Meares G. P., Qin H., Liu Y., Holdbrooks A. T., and Benveniste E. N. (2013) AMP-activated protein kinase restricts IFN-γ signaling. J. Immunol. 190, 372–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Meares G. P., Zmijewska A. A., and Jope R. S. (2004) Heat shock protein-90 dampens and directs signaling stimulated by insulin-like growth factor-1 and insulin. FEBS Lett. 574, 181–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ma X., Reynolds S. L., Baker B. J., Li X., Benveniste E. N., and Qin H. (2010) IL-17 enhancement of the IL-6 signaling cascade in astrocytes. J. Immunol. 184, 4898–4906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Qin H., Holdbrooks A. T., Liu Y., Reynolds S. L., Yanagisawa L. L., and Benveniste E. N. (2012) SOCS3 deficiency promotes M1 macrophage polarization and inflammation. J. Immunol. 189, 3439–3448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Laurence A., Tato C. M., Davidson T. S., Kanno Y., Chen Z., Yao Z., Blank R. B., Meylan F., Siegel R., Hennighausen L., Shevach E. M., and O'Shea J. J. (2007) Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity 26, 371–381 [DOI] [PubMed] [Google Scholar]