Abstract
S100A1 has been suggested as a therapeutic agent to enhance myocyte Ca2+ cycling in heart failure, but its molecular mode of action is poorly understood. Using FRET, we tested the hypothesis that S100A1 directly competes with calmodulin (CaM) for binding to intact, functional ryanodine receptors type I (RyR1) and II (RyR2) from skeletal and cardiac muscle, respectively. Our FRET readout provides an index of acceptor-labeled CaM binding near donor-labeled FKBP (FK506-binding protein 12.6) on the cytoplasmic domain of RyR in isolated sarcoplasmic reticulum vesicles. S100A1 (0.01–400 μm) partially inhibited FRET (i.e. CaM binding), with Ki > 10 μm, for both RyR1 and RyR2. The high [S100A1] required for partial effects on FRET indicates a lack of competition by S100A1 on CaM/RyR binding under normal physiological conditions. High-resolution analysis of time-resolved FRET detects two structural states of RyR-bound CaM, which respond to [Ca2+] and are isoform-specific. The distribution of these structural states was perturbed only by high micromolar [S100A1], which promoted a shift of bound CaM to a lower FRET orientation (without altering the amount of CaM bound to RyR). Thus, high micromolar S100A1 does alter the CaM/RyR interaction, without involving competition. Nevertheless, submicromolar S100A1 can alter RyR function, an effect that is influenced by both [Ca2+] and [CaM]. We conclude that CaM and S100A1 can concurrently bind to and functionally modulate RyR1 and RyR2, but this does not involve direct competition at the RyR CaM binding site.
Keywords: calcium channel, calmodulin (CaM), fluorescence resonance energy transfer (FRET), sarcoplasmic reticulum (SR), structure-function, FKBP12.6, fluorescence lifetime
Introduction
In striated muscle, the ryanodine receptor (RyR)2 is the channel responsible for Ca2+ release from the major intracellular Ca2+ reservoir, the sarcoplasmic reticulum (SR). RyR activity is tightly regulated by endogenous small molecules and proteins that maintain healthy Ca2+ cycling pertaining to relaxation and contraction of cardiac cells and skeletal muscle fibers. Two important modulators of Ca2+ cycling are EF-hand Ca2+-binding proteins, calmodulin (CaM) and S100A1. CaM is well known to directly bind to the RyR isoforms in skeletal (RyR1) and cardiac (RyR2) muscle and modulate channel activity in vitro or in situ (1, 2). S100A1 has also been shown to interact with and functionally modulate RyR1 and RyR2 (3–5). As a modulator of RyR2 activity, CaM is widely considered to be an effective inhibitor of Ca2+ leak through RyR2 (6, 7), and S100A1 is thought to exert a similar effect. Moreover, S100A1-based gene therapy shows promising effectiveness to correct abnormal Ca2+ cycling in heart failure (8, 9). To optimally exploit the therapeutic potential of S100A1, it is important to understand the structural basis of its molecular mode of action on the two principal targets hypothesized to mediate its effect on intracellular Ca2+: the RyR2 channels and the sarco/endoplasmic reticulum Ca2+-ATPase·phospholamban complex (10) that is responsible for active SR Ca2+ uptake. The present study is primarily focused on the S100A1/RyR2 interaction.
Similar to CaM, the tertiary structure of S100A1 and its modulatory action toward RyRs are affected by Ca2+ binding. With [Ca2+]cyto <0.5 μm, cytoplasmic exposure to nanomolar S100A1 has been shown to increase RyR1 activity, whereas ≥7 μm Ca2+ abolished this modulatory action (3, 11). The apo (Ca2+-free)-CaM similarly increases RyR1 activity at nanomolar Ca2+, but the Ca2+-bound CaM decreases activity at high micromolar Ca2+ (1, 2, 12).
The modulatory action of S100A1 on RyR2 appears different from RyR1. In single-channel electrophysiology studies, cytoplasmic exposure to micromolar S100A1 has been shown to decrease the RyR2 channel activity at submicromolar and high micromolar Ca2+ (13). This in vitro action is consistent with reported effects of S100A1 in cardiomyocytes, whereby the addition of 0.1 μm S100A1 to permeabilized myocytes decreases RyR leak in resting conditions, and pressure injection of 0.1 μm S100A1 into cardiomyocytes increases depolarization-induced Ca2+ transient amplitudes (5). Thus, S100A1 and CaM might appear to exert similar effects on the RyR2 channel activity because CaM is well characterized to decrease RyR2 activity at both submicromolar and high micromolar Ca2+ (14, 15). However, measurements of [3H]ryanodine binding to isolated cardiac SR membranes, a convenient index of the open RyR, show activation of RyR2 at both nano- and micromolar calcium (4), which appears inconsistent with some of the channel and myocyte measurements (5, 6, 13). This discrepancy indicates that the in vivo effect of S100A1 on RyR activity depends on interactions with intracellular partners or conditions that are insufficiently understood.
Treves et al. (3) identified three potential S100A1 binding sites on RyR1, but the fact that a single residue mutation (L3625D) in murine RyR1 abolishes the modulatory action of S100A1 on the activity of single RyR1 in lipid bilayers implicates one functional binding site (11). This mutation resides in a well characterized CaM-binding domain of RyR that has also been shown to bind S100A1 (with micromolar affinity) when this RyR region is isolated as a peptide (11, 16). This is consistent with the finding that high micromolar CaM displaces RyR1, in rabbit skeletal muscle SR vesicles, from S100A1-linked Sepharose resin and vice versa (16, 17).
Given the high sequence identity between the CaM binding domains of RyR1 and RyR2, the S100A1 binding sites have been inferred to also be conserved between RyR1 and RyR2 (13). Taken together, these results have led to the hypothesis that S100A1 competes with CaM for the same binding site in RyR1 and RyR2. However, most studies that suggest competition between CaM and S100A1 for RyR use micromolar S100A1 and CaM to demonstrate this behavior. Moreover, S100A1 has micromolar affinity for the RyR peptide (16), which is inconsistent with the nanomolar [S100A1] shown to maximally modulate RyR activity and Ca2+ cycling in myocytes (4).
Here, we used established functional and state-of-the-art structural analysis approaches (18–20) to directly test the hypothesis that S100A1 competes with CaM binding to RyR1 or RyR2 in SR vesicles isolated from porcine skeletal muscle or hearts, respectively (Fig. 1). Our FRET toolkit is uniquely capable of resolving RyR-specific readouts from SR samples, in which RyRs are known to only be a small fraction (∼25%) of the S100A1 targets in resting cardiomyocytes (6). Results largely contradict the tested hypothesis and show that S100A1 and CaM can concurrently bind to and regulate RyR1 and RyR2.
FIGURE 1.
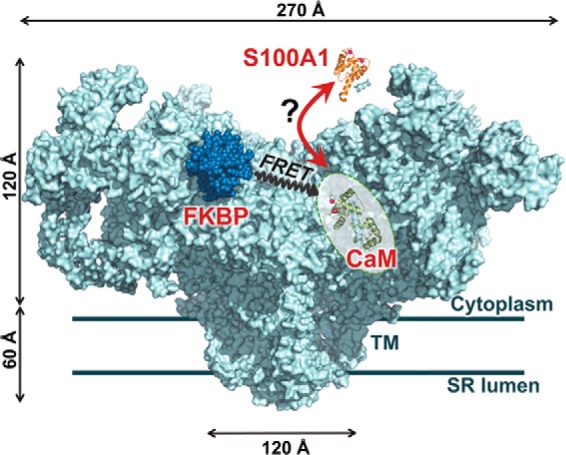
Hypothesis tested and RyR-targeted FRET molecular toolkit. S100A1 (orange) competes with CaM (green) binding to RyR (light blue). The RyR1 model (cyan) is based on a published cryo-EM density map (Protein Data Bank code 3J8H) (42) with docked FKBP12 (dark blue) and suggested location of the CaM binding site (18) indicated (shaded oval). FKBP and CaM N-lobe bind to locations that are within ∼50 Å from each other in the RyR complex, a feature that is exploited by our FRET-based system to investigate the RyR structure and binding events in vitro and in situ (18–20).
Results
Effect of S100A1 and CaM on [3H]Ryanodine Binding to Cardiac and Skeletal SR
Because we are testing the hypothesis of competition between S100A1 and CaM for the same RyR binding site, we carried out all studies using SR preparations treated to remove residual CaM (as described under “Experimental Procedures”), thus tightly controlling any potential interference of CaM with S100A1 binding. In agreement with previous reports (21, 22), purified WT-S100A1 consisted of a mixture of molecules that had either retained or lost the N-terminal methionine. In addition, the S100A1 molecules were largely in the reduced, monomeric form, as determined from using electrospray ionization mass spectrometry. Although residual CaM is highly variable, it has not been controlled in the few previous studies that have assessed the modulatory action of S100A1 on RyR1 (3) and RyR2 (4, 23) activity in isolated SR membranes, where it may have led to confounding results. To investigate whether CaM alters the functional S100A1/RyR interaction, we evaluated the effect of 0.01–100 μm S100A1 on [3H]ryanodine binding to CaM-stripped skeletal or cardiac SR, in the presence and absence of saturating (800 nm) CaM. We carried out these studies at 30 nm and 30 μm free Ca2+, thus reflecting cytosolic conditions in relaxed and contracting muscle cells. Despite the capacity for Ca2+ binding, the addition of 100 μm S100A1 marginally altered nanomolar and micromolar free Ca2+ by <20%. In previous FRET studies, we have shown that 800 nm CaM saturates the CaM binding sites on RyR (18, 20). Therefore, by using saturating CaM in these [3H]ryanodine binding assays, we gauged the functional effect of S100A1 on RyR that is devoid of or fully loaded with CaM. In accord with previous studies (1, 2, 12), CaM significantly increased [3H]ryanodine binding to skeletal SR (RyR1) in nanomolar Ca2+ by 10 ± 1-fold but decreased it in micromolar Ca2+ by 72 ± 1% (Fig. 2A). Saturating CaM lowered [3H]ryanodine binding to cardiac SR (RyR2) in both 30 nm and 30 μm Ca2+ by 15 ± 1 and 29 ± 1% (Fig. 2A), respectively, which is also consistent with previous studies (14, 15).
FIGURE 2.
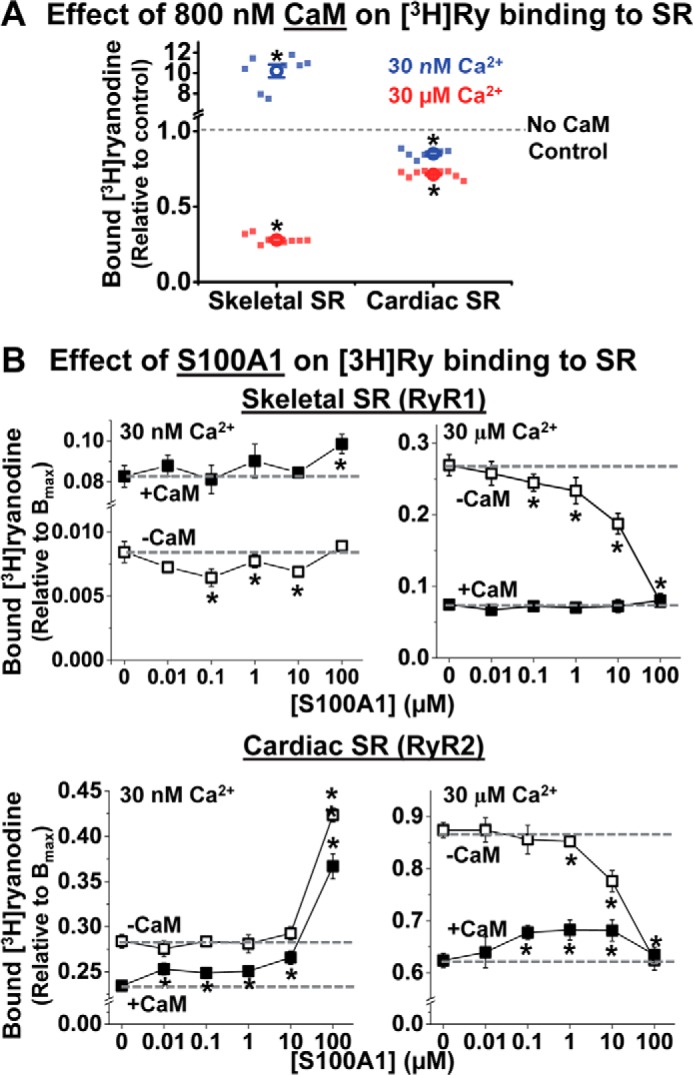
CaM alters the effect of S100A1 on [3H]ryanodine binding to RyR1 and RyR2. SR vesicles from skeletal or cardiac muscle were incubated with S100A1 (0–100 μm) and CaM (0 or 800 nm) at 30 nm or 30 μm Ca2+ in the presence of [3H]ryanodine. A, as a control, the functional effect of 800 nm CaM was tested on [3H]ryanodine binding to skeletal (left) and cardiac (right) SR vesicles in 30 nm (blue) or 30 μm (red) Ca2+. Data are shown as individual data points (square) and mean ± S.E. (open circle). The fraction of [3H]ryanodine binding is relative to “no CaM” control. B, effect of S100A1 on [3H]ryanodine binding to skeletal SR (top panels) and cardiac SR (bottom panels) at 30 nm Ca2+ (left) and 30 μm Ca2+ (right) and in the absence of CaM (hollow squares) and in the presence of 800 nm CaM (solid squares). Control levels (absence of S100A1) are indicated by dashed lines. The fraction of [3H]ryanodine bound to SR vesicles is relative to maximum binding capacity (Bmax). Data are shown as mean ± S.E. (n = 6–8). *, significant differences relative to no-S100A1 controls; p < 0.05, as determined by paired Student's t test.
We tested the functional effect of S100A1 on RyR1 in skeletal SR vesicles. In 30 nm Ca2+ and in the absence of CaM, 0.1–10 μm S100A1 inhibited [3H]ryanodine binding in a biphasic profile, with maximum inhibition at 0.1 μm S100A1. With saturating CaM, however, [3H]ryanodine binding was further increased by S100A1, but this tendency became significant only at 100 μm S100A1 (Fig. 2A, left). Thus, in the absence of CaM, S100A1 has the opposite effect of CaM on RyR1, whereas S100A1 may synergize with saturating CaM to enhance RyR1 activation in nanomolar Ca2+.
In micromolar Ca2+ and in the absence of CaM, ≤1 μm S100A1 had statistically significant, but modest, inhibitory effects on RyR1 [3H]ryanodine binding, whereas 10 and 100 μm S100A1 had robust inhibitory effects (Fig. 2A, right). With saturating CaM, however, S100A1 had no effect on [3H]ryanodine binding (Fig. 2A, right).
The functional effect of S100A1 on RyR2 differs from its effect on RyR1. In nanomolar Ca2+ and in the absence of CaM (Fig. 2B, left), up to 10 μm S100A1 had negligible effects on cardiac SR [3H]ryanodine binding, but a robust increase (59 ± 5%) was seen in 100 μm S100A1. In saturating CaM, however, a statistically significant enhancement of [3H]ryanodine binding was revealed for all tested S100A1 concentrations. In fact, the inhibitory effect of CaM on RyR2 was completely overcome at 1 μm S100A1 and became a net RyR2 activation at 100 μm S100A1 (Fig. 2B, left). This is consistent with previously reported [3H]ryanodine RyR2-binding results (4).
In micromolar Ca2+ and in the absence of CaM, we found inhibition of [3H]ryanodine binding to RyR2, which is statistically significant only at high [S100A1] (Fig. 2B, right; 10 and 100 μm S100A1). With saturating CaM, we found a biphasic effect, activation of [3H]ryanodine binding plateauing in 0.1–10 μm S100A1 (Fig. 2B, right). This, too, is consistent with previously reported [3H]ryanodine RyR2 binding results (4).
Overall, these results establish that the S100A1 used in this study modulates RyR in concentrations ranging from 0.01 to 100 μm, which cover the range reported for striated muscle tissue (3, 24). Whether this modulation is via orthosteric competition with CaM is explored further in this report.
S100A1 Competition of AF568-labeled CaM (A-CaM) Binding to SR
Two studies previously reported that micromolar CaM competes with S100A1-linked Sepharose for binding to skeletal SR vesicles and vice versa (16, 17), and these findings have been the primary basis of the hypothesis that these two proteins compete for overlapping binding sites on RyR1 and RyR2. However, given that S100A1 has other targets found in native SR preparations, there is a strong need to determine whether competition is specifically for RyR1. Furthermore, it is important to determine whether this mechanism also applies to CaM and S100A1 binding to RyR2, particularly given the strong therapeutic potential of S100A1 for heart failure (10, 23, 25–27). There are few data indicative of S100A1 interacting at the CaM binding site on RyR2. It was indirectly indicated in a report (13) that an RyR2 mutation (L3591D) abolishes both CaM- and S100A1-mediated modulation of RyR2 in nanomolar Ca2+. However, this is not definitive evidence of a direct interaction. For comparison with previous binding studies (16, 17) and with experiments shown below that examine competition of S100A1 with RyR·CaM binding, we first quantified the level of S100A1 competing with CaM binding to all of its SR-associated targets (as opposed to only RyR). We utilized 0.1 μm A-CaM, to quantify, via fluorescence intensity of AF568 probe, the level of CaM co-sedimented with skeletal or cardiac SR vesicles following incubation with 0–400 μm S100A1. As a control, we show that a 200-fold excess of unlabeled CaM largely competes with the A-CaM retained by SR vesicles from both skeletal and cardiac muscle (Fig. 3, triangles). This control indicates that A-CaM binds to CaM binding sites endogenous to SR membranes (i.e. to all CaM-binding proteins in SR). A-CaM bound to skeletal SR was also significantly reduced upon incubation with ≥100 μm S100A1, but this effect was more pronounced at 300 μm Ca2+ than at 30 nm Ca2+ with 17 and 45%, respectively, of A-CaM bound (Fig. 3A). Conversely, preincubation of S100A1 equally reduced A-CaM binding to cardiac SR at both nano- and micromolar Ca2+, by as high as 33 and 42%, respectively (Fig. 3B).
FIGURE 3.
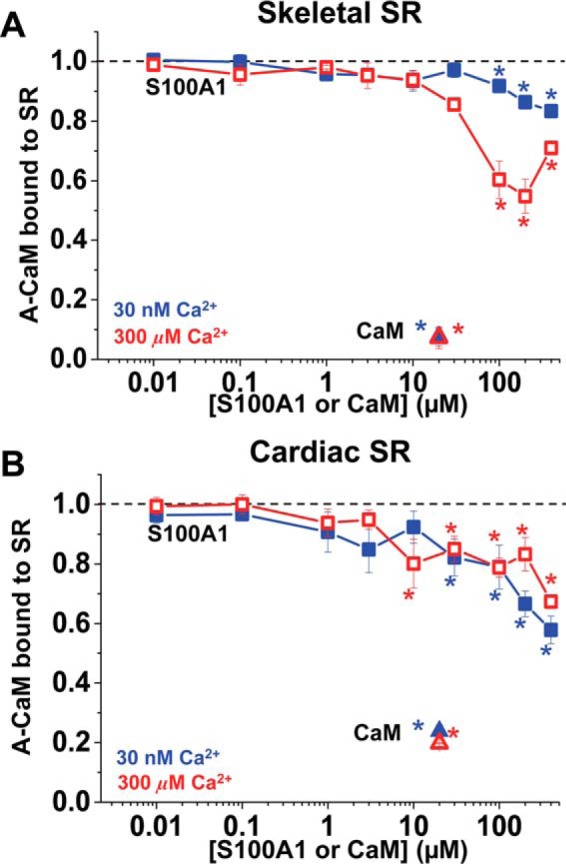
Competitive inhibition of CaM binding to SR vesicles by S100A1. SRs from skeletal (A) or cardiac (B) muscle were preincubated with WT-S100A1 or WT-CaM with 30 nm (blue solid symbol) or 300 μm (red open symbol) Ca2+ and then incubated with 0.1 μm A-CaM before SR membrane sedimentation by centrifugation. Data are normalized to no-S100A1 or no-CaM controls (black dotted lines) and expressed as means ± S.E. (n = 4). *, significantly different from control, p < 0.05 as determined by paired Student's t test.
Overall, results shown in Fig. 3 are consistent with a significant overlap between the S100A1 and CaM binding sites within SR samples, particularly in micromolar Ca2+ (for skeletal muscle), and therefore they are in agreement with results from the competition assays that used S100A1- or CaM-conjugated Sepharose to retain rabbit SR vesicles (16, 17). Next, we tested whether S100A1 and CaM bind at overlapping sites on RyR1 and RyR2.
S100A1 Competition of A-CaM Binding to RyRs
To resolve CaM binding specifically to RyR (as opposed to all CaM binding sites within SR), we measured FRET from donor-labeled FKBP (D-FKBP) to A-CaM, with the donor and acceptor labels attached at residue 49 and 34, respectively. FRET between D-FKBP and A-CaM was measured as a decrease in donor fluorescence intensity, as we have shown previously (18–20, 28). We have previously established that this D-FKBP variant remains >90% bound to RyR over the time course of the experiment (18), which makes it ideal for providing an accurate and precise FRET readout. Furthermore, we have shown that these labeled FKBP and CaM variants have similar functional effects as WT-FKBP12.6 and WT-CaM on [3H]ryanodine binding to skeletal SR vesicles (18, 19). In the representative fluorescence spectra (Fig. 4, A and B) of skeletal SR samples labeled with D-FKBP, the decrease in fluorescence intensity due to FRET in the presence of 0.1 μm A-CaM was prevented by incubation with 20 μm unlabeled WT-CaM. By comparison, a much larger S100A1 concentration (>10 μm) S100A1 was required to elicit a decrease in the apparent A-CaM binding to RyR1. It is important to note that A-CaM is competed by WT-CaM with a Ki (∼0.1 μm) that is equivalent to the [A-CaM] used in this assay (Fig. 4), indicating that A-CaM and WT-CaM bind to the RyR1 with similar affinities. As shown in Fig. 4A, FRET to A-CaM in skeletal SR membranes is slightly reduced by preincubation of >30 μm S100A1 in micromolar, but not in nanomolar, Ca2+. A reduction in FRET could be attributed to reduced binding or to a structural change within the D-FKBP·A-CaM·RyR1 macromolecular complex. However, the reduction in FRET that we observed is marginal and inconsistent with inhibition of CaM binding via the direct competition for the CaM binding site hypothesized based on studies using heavy skeletal SR (16, 17). Because these previous reports used SR from rabbit muscle, as opposed to the porcine SR used here, we tested the hypothesis of a species-dependent S100A1/CaM/RyR1 interaction by undertaking similar FRET experiments using rabbit skeletal SR and found that the effect of S100A1 on FRET is conserved between rabbit and pig skeletal SR (i.e. there is no competition between S100A1 and CaM for RyR1 binding) (data not shown).
FIGURE 4.
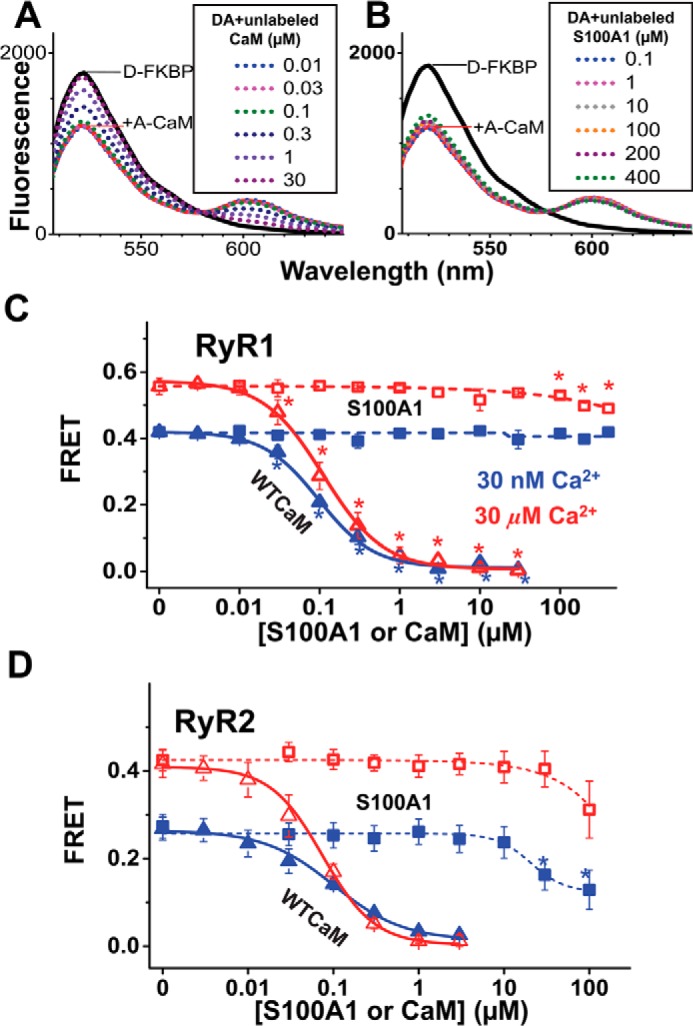
Competitive inhibition of A-CaM binding to RyR1 and RyR2 by S100A1. RyR in native SR membranes isolated from skeletal or cardiac muscle was labeled with D-FKBP, preincubated with S100A1 (0–400 μm), and then incubated with 0.1 μm A-CaM (labeled via T34C mutation). The inhibition of CaM bound to RyR was calculated as the fractional decrease of FRET between D-FKBP and A-CaM, based on the fluorescence intensity readout. Shown are representative fluorescence spectra of samples preincubated with unlabeled CaM (A) or S100A1 (B). Such measurements are plotted as competition curves for samples consisting of skeletal (C) and cardiac (D) SR. Data are shown as mean FRET ± S.E. (n = 4). *, significantly different from no-S100A1 controls, as determined by paired Student's t test.
Using cardiac SR, we observed that ≥30 μm S100A1 fractionally decreases FRET between D-FKBP and A-CaM, and this effect is larger at nanomolar Ca2+ relative to micromolar Ca2+ (Fig. 4D). Given that FRET is subject to both the level of A-CaM bound to the RyR and the distance between FRET probes, the S100A1-mediated reduction in FRET may be attributed to a reduction in CaM association with RyR relative to FKBP and/or to a structural change affecting the donor-acceptor distance. Experiments discussed further under “TR-FRET Analysis of the S100A1 Effects on RyR·CaM Structure and Binding” were designed to discern between these two mechanisms.
CaM Competition of A-S100A1 Binding to RyRs
To quantify S100A1 binding specifically to RyR, we measured FRET from d-FKBP to A-S100A1. To prepare A-S100A1, we used the WT single Cys (Cys-85) and also engineered several single-Cys S100A1 mutants, with the labeling site at T6C, T42C, and at the N and C termini via Ala-Cys-Ala tags at either end of S100A1. In the single-Cys S100A1 mutants, we substituted the single native Cys of WT-S100A1 (Cys-85) with either Ala (C85A) or Ser (C85S), as indicated (Fig. 5). In testing A-S100A1 labeled at its C terminus (AF568-S100A1-C85S-ACA), we were pleased to observe substantial FRET from D-FKBP to saturating [A-S100A1] in both RyR1 and RyR2 samples at 30 μm Ca2+ (Fig. 5, A and B). In 30 nm Ca2+, however, we observed insignificant FRET in the RyR1 sample and modest FRET in the RyR2 sample (Fig. 5, A and B). Saturation curves indicate A-S100A1/RyR binding with micromolar affinity. To our surprise, however, FRET was not perturbed by the addition of a molar excess of WT-S100A1, which indicates that this A-S100A1 construct is unlikely to be representative of WT-S100A1 binding. Furthermore, despite the lack of competition with WT-S100A1, FRET was fully abolished by the addition of a molar excess of WT-CaM. Moreover, we observed this trend (of selective competition of A-S100A1 by WT-CaM but not by WT-S100A1) for all labeled single-Cys S100A1 constructs tested (six different A-S100A1 constructs, as indicated in Fig. 5C). Explicitly, FRET from D-FKBP to nearly saturating A-S100A1 (2 μm), on both RyR1 and RyR2, was abolished by 20 μm WT-CaM, partially reduced by 20 μm unlabeled corresponding single-Cys S100A1 construct, and unaffected by 20 μm WT-S100A1 (Fig. 5, C and D). Given that our FRET readout, in this case, reflects binding of S100A1 to RyR in the proximity of D-FKBP, the lack of FRET inhibition by the addition of WT-S100A1 indicates that our A-S100A1 constructs do not reflect WT-S100A1 binding to both RyR1 and RyR2. Consequently, we did not pursue further studies with A-S100A1 in this project.
FIGURE 5.
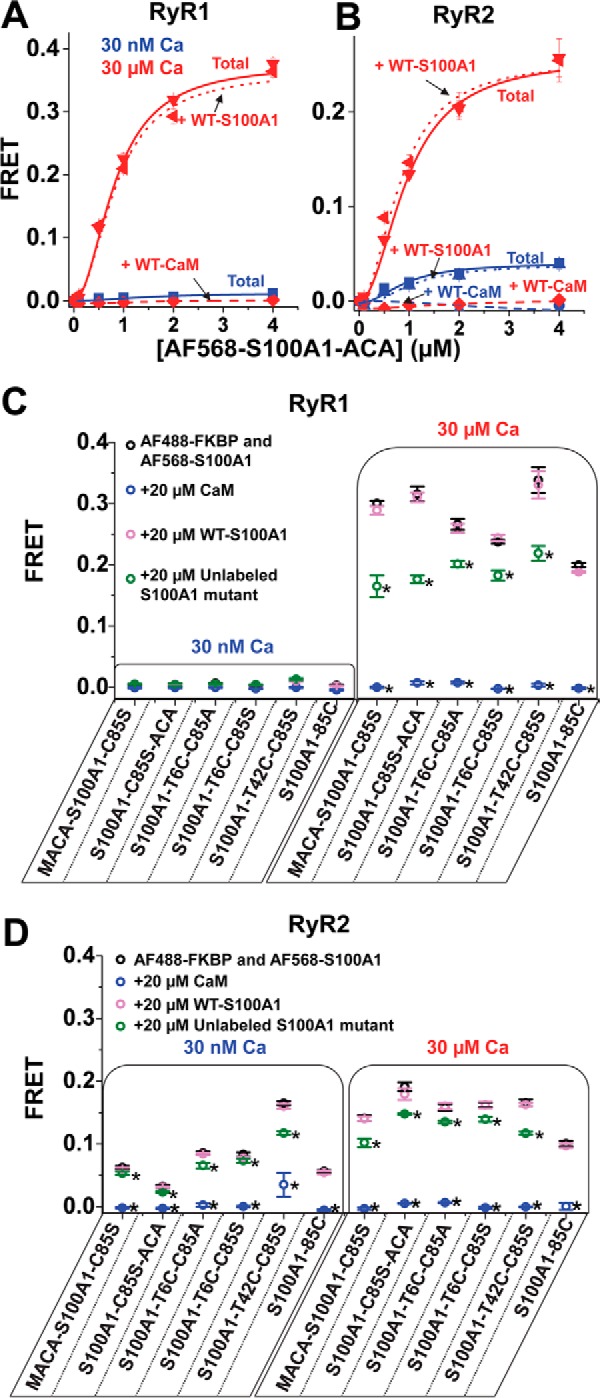
Competition of A-S100A1 by unlabeled S100A1 and CaM for binding to RyR. RyRs in SR membranes isolated from skeletal or cardiac muscle were labeled with D-FKBP and incubated with A-S100A1. Binding specificity of A-S100A1 to RyR was investigated by the addition of a >10-fold molar excess of unlabeled S100A1 or CaM. Saturation binding of AF568-S100A1-C85S-ACA to RyR from skeletal (A) and cardiac (B) SR is indicated as Total (solid lines) at 30 nm Ca2+ (blue) and 30 μm Ca2+ (red). Total FRET is compared with the saturation binding curve in the presence of 40 μm WT-S100A1 (dotted lines) or 40 μm WT-CaM (dashed lines). Binding specificities of several A-S100A1 variants to RyR from skeletal SR (C) or cardiac SR (D) were tested by the addition of 20 μm CaM (blue circles), WT-S100A1 (pink circles), or unlabeled S100A1 (green circles). Data are expressed as means ± S.E. (n = 4). *, significantly different from respective control (black circles), p < 0.05, as determined by analysis of variance with Fisher's post hoc test.
TR-FRET Analysis of the S100A1 Effects on RyR·CaM Structure and Binding
The D-FKBP to A-CaM FRET changes due to the addition of Ca2+ and/or S100A1 (Fig. 4) could be due to changes in either CaM binding or structure of the RyR·CaM complex. To resolve this ambiguity, we used TR-FRET, which allows simultaneous detection of both RyR·CaM binding and structure. The acceptor was attached at either CaM residue 34 (AF568 attached via CaM N-lobe; AN-CaM) or 110 (AF647 attached via CaM C-lobe; AC-CaM) of the full-length CaM, and the concentration of these acceptor-labeled CaMs was adjusted to give maximal FRET (18, 20). We tested the effect of 200 μm S100A1 because this concentration significantly altered FRET between D-FKBP and A-CaM in the competition experiments above (Fig. 4).
TR-FRET data were first analyzed to determine the fraction of donors participating in FRET (i.e. the fraction of D-FKBPs that are near an occupied CaM binding site). In all TR-FRET measurements, the fraction of uncoupled donors (i.e. donors not participating in FRET) was 10 ± 1%, and the addition of 200 μm S100A1 did not perturb this value, thus confirming that S100A1 does not compete with D-FKBP12.6 binding to RyRs. The 10% fraction of uncoupled donor is in accord with the previously reported slow D-FKBP/RyR dissociation rate (18).
Another advantage of TR-FRET is its ability to directly resolve multiple distances (i.e. structural states) within a protein complex as well as the population of probes in each structural state (29, 30). With AN-CaM, the TR-FRET data corresponding to both RyR isoforms (Fig. 6, A and B) fit best to a two-state Gaussian distribution model, characterized by ∼47 Å (R1) and 80–90 Å (R2) distances between the FKBP-attached donor and the CaM-attached acceptor at both nanomolar and micromolar Ca2+ (Fig. 6, C and D).
FIGURE 6.
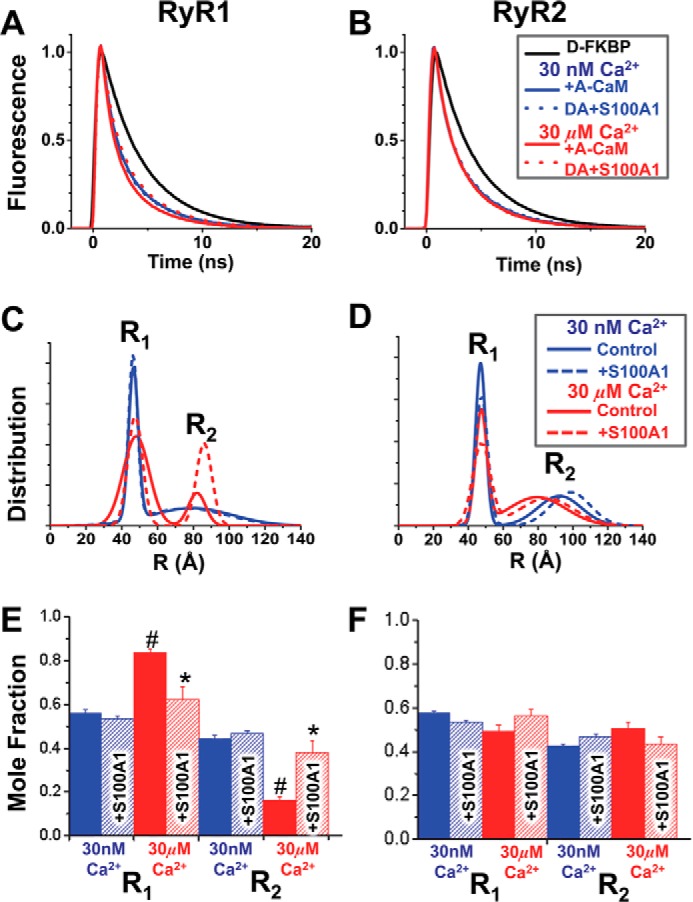
TR-FRET detection of S100A1 structural effect on the CaM·RyR complex. SR membranes from skeletal (A, C, and E) or cardiac (B, D, and F) muscle were labeled with D-FKBP, preincubated with 200 μm S100A1, and then incubated with 800 nm CaM labeled with acceptor probe at the N-lobe residue via T34C (AN-CaM). A and B, fluorescence decays of D-FKBP with AN-CaM in low (blue) and high Ca2+ (red), with (dotted lines) and without (solid lines) S100A1. C and D, multiexponential analysis of the TR-FRET data yielded a two-distance (R1 and R2) Gaussian distribution model of the separation between D-FKBP and AN-CaM within RyR. E and F, molar fractions of the populations associated with each of the two structural states characterized by distances R1 and R2. The S100A1 effects are shown in hatched bars. *, significantly different from the no-S100A1 control. #, significantly different from 30 nm Ca2+, p < 0.05, as determined by analysis of variance with Fisher's post hoc test.
For RyR1 in nanomolar Ca2+, we observe a narrow R1 component and a much broader (more disordered) R2 component (Fig. 6C, blue). In micromolar Ca2+, the R1 component broadens somewhat, suggesting increased disorder, whereas the R2 component sharply narrows, suggesting a more ordered underlying structure (Fig. 6C (red) and Table 1). In nanomolar Ca2+, the conformations corresponding to the R1 and R2 distances are almost equally populated, whereas in micromolar Ca2+, R1 is dominant (>80%; Fig. 6E). Only at micromolar Ca2+ does S100A1 (200 μm) balance the molar fractions of the R1 and R2 conformations (decreased R1, increased R2; Fig. 6E) and narrows the R1 full width at half-maximum (FWHM; Table 1). These results are consistent with previous studies indicating that the RyR1 binding site of the apo-CaM N-lobe is different from that of Ca2+-CaM (31, 32).
TABLE 1.
Fitting parameters from multiexponential fitting of TR-FRET between D-FKBP and full-length A-CaM labeled at its N- or C-lobe; effect of S100A1 and Ca2+
Data are expressed as means ± S.E. from experimental variation of fits (n = 4 experiments).
| RyR | [Ca2+] | [S100A1] | Acceptor at N-lobe site |
Acceptor at C-lobe site |
||||
|---|---|---|---|---|---|---|---|---|
| R1 | FWHM(R1) | R2 | FWHM(R2) | R3 | FWHM(R3) | |||
| μm | μm | Å | Å | Å | ||||
| RyR1 | 0.03 | 0 | 47 ± 0.4 | 8 ± 0.8 | 84 ± 4 | 59 ± 11 | 96 ± 0.6 | 35 ± 2 |
| 200 | 47 ± 0.3 | 8 ± 0.8 | 85 ± 4 | 60 ± 10 | 96 ± 0.6 | 34 ± 2 | ||
| 30 | 0 | 48 ± 0.1 | 18 ± 1.5a | 84 ± 2 | 19 ± 4a | 81 ± 0.1a | 35 ± 1 | |
| 200 | 48 ± 0.2 | 11 ± 1.5b | 85 ± 3 | 25 ± 11a | 81 ± 0.2a | 38 ± 3 | ||
| RyR2 | 0.03 | 0 | 47 ± 0.6 | 8 ± 1.0 | 92 ± 1 | 27 ± 1 | 95 ± 2 | 45 ± 7 |
| 200 | 48 ± 0.2 | 7 ± 0.6 | 95 ± 2 | 28 ± 8 | 94 ± 2 | 35 ± 7 | ||
| 30 | 0 | 47 ± 0.2 | 8 ± 1.3 | 79 ± 1a | 24 ± 7 | 81 ± 0.4a | 35 ± 4 | |
| 200 | 48 ± 0.2 | 17 ± 2.7a,b | 86 ± 1a,b | 30 ± 5 | 84 ± 1a | 49 ± 6 | ||
a Significant effect of Ca2+ (p < 0.05), as determined by analysis of variance with Fisher's post hoc test.
b Significant effect of S100A1 (p < 0.05), as determined by analysis of variance with Fisher's post hoc test.
For RyR2 in nanomolar Ca2+, we observe a behavior similar to that of RyR1 (Fig. 6, D versus C) (i.e. narrow R1 and broader R2 conformations) (blue). In contrast to RyR1, however, this pattern changes only marginally for RyR2 in micromolar Ca2+, with a shortening of R2, from 96 to 79 Å, being the only significant effect of Ca2+. This indicates a Ca2+-driven structural change of either CaM or RyR2 itself. S100A1 causes a lengthening of R2 that is statistically significant in micromolar Ca2+ (from 79 to 86 Å) and only a trend in nanomolar Ca2+ (Fig. 6F and Table 1). For RyR2, the R1 and R2 conformations are roughly equally populated under all tested conditions (Fig. 6F). Next, we used TR-FRET to examine the structural effect of S100A1 on the C-lobe of full-length CaM bound to RyR. We have previously demonstrated that the CaM N-lobe is proximal to FKBP (strong FRET), and the C-lobe is distal (weak FRET) when bound to either RyR1 (18) or RyR2 (20). To accommodate this difference in distances, we used a FRET pair (AF568-AF647) with a longer Förster distance (R0 = 82 Å versus 62 Å for the AF488-AF568 pair) for labeling D-FKBP and AC-CaM. Donor fluorescence decays, detected via direct waveform recording, were used to resolve the distribution of distances separating the D-FKBP and AC-CaM (labeled at the C-lobe via T110C) probes. In this case, the TR-FRET data fit best a one-distance Gaussian distribution model. For both RyR1 and RyR2, this distance (R3) displayed a Ca2+-dependent shortening of ∼15 Å (Table 1), but S100A1 did not alter this distance (Table 1). Thus, S100A1 does not appear to structurally alter the interaction between RyR and the C-lobe of full-length CaM.
Discussion
Using FRET methods that specifically resolve CaM binding to RyRs, we have tested the hypothesis that S100A1 modulates RyR activity via an interaction with the CaM binding site. This hypothesis has emerged based on the apparent competition of micromolar CaM or S100A1 with retention of RyR1 (in isolated skeletal SR vesicles) on Sepharose derivatized with either S100A1 or CaM, respectively (16, 17). Further supporting the tested hypothesis, S100A1 was shown to bind to an isolated 12-mer peptide that corresponds to RyR1 residues 3616–3627, which is a subsection of the 3614–3643 CaM binding domain 2 (16, 17), and mutation of one residue in this site (L3625D in RyR1 and L3591D in RyR2) can abolish binding of S100A1 to the RyR peptide (11) and RyR channel modulation by S100A1 (11, 13). However, none of these studies have demonstrated that S100A1 binds specifically at the CaM binding site of full-length RyRs. Indeed, semiquantitative analysis in a recent report indicated that increased expression of S100A1 in rat cardiomyocytes does not perturb CaM/RyR2 association (6). Our D-FKBPs exclusively bind to RyRs in skeletal and cardiac SR vesicles and, via FRET, they enable quantitative detection of A-CaM binding (within ≤100 Å of D-FKBP) to intact, functional RyR1 or RyR2 in native SR membranes (18–20). Using this FRET system, we determined the structural influence of S100A1 on the CaM interaction with RyR1 and RyR2. To observe the reverse relationship (effect of CaM on the S100A1·RyR complex) we created an extensive array of A-S100A1 for intended FRET studies with D-FKBP. All six different A-S100A1 constructs tested appear to bind to sites on RyR1 and RyR2 within ≤100 Å of D-FKBP, given the FRET readout. However, all A-S100A1 binding sites differ from the endogenous S100A1 binding sites but overlap with the CaM binding site (Fig. 5, C and D). In addition to FRET-based quantification of CaM/RyR binding, we also used [3H]ryanodine binding to native SR membranes to determine whether CaM modulates the functional effect of S100A1 on RyR function (i.e. whether CaM and S100A1 may coexist within the RyR complex).
S100A1 Marginally Affects CaM Binding to RyR1 and RyR2
We found that FRET from D-FKBP to subsaturating A-CaMs is modestly reduced by a large (300–4000-fold) excess of S100A1 relative to A-CaM (Fig. 4). This small effect may be explained by a reduction in A-CaM binding or by a change in the RyR-bound A-CaM conformation caused by S100A1 binding that changes donor-acceptor distance relationships.
Our TR-FRET results indicate that AN-CaM adopts two structural states (conformations characterized by distances R1 and R2) when bound to RyRs, each state exhibiting measurable disorder (Fig. 6, C and D). These two structural states are detected for both RyR1 and RyR2, in nanomolar or micromolar Ca2+. The short distance, R1, is similar (47–48 Å) for both isoforms and calcium conditions. S100A1 does not significantly affect this parameter, but it does change the width (i.e. disorder) of the distribution in micromolar Ca2+ (FWHM(R1) in Table 1) and in opposite directions for RyR1 (narrower, less disorder) and RyR2 (broader, more disorder).
The long distance, R2, is affected neither by Ca2+ nor by S100A1, for RyR1 (84–85 Å), but it becomes sharply narrower (less disordered) in micromolar Ca2+, and S100A1 somewhat broadens this state (FWHM(R2) in Table 1). The R2 of RyR2-bound AN-CaM in nm Ca2+ (92–95 Å) is significantly longer than for RyR1, whereas in micromolar Ca2+, it trends shorter (79 Å), and S100A1 lengthens it to 86 Å.
The single-Gaussian distance distribution, R3, was resolved based on TR-FRET between D-FKBP and the C-lobe of full-length CaM (using AC-CaM). R3 is similar in RyR1 and RyR2 and is significantly shortened by Ca2+, but S100A1 does not affect this state (Table 1).
Taken together, these results indicate that the N-lobe of CaM bound to RyR probes (pivots between) two structural states while anchored by a C-lobe in a single state. Measurable disorder, as indicated by FWHM (Table 1), is detected in all three distance measurements. Between RyR1 and RyR2, these states show significant, albeit small, differences both in their structural response to Ca2+ and to S100A1. However, TR-FRET data analysis detected no effect of S100A1 on the fraction of CaM associated with RyR1 or RyR2, as indicated by the unchanged fraction of uncoupled donor.
These results were surprising because it is thought that S100A1 modulates RyR function via the CaM binding site (16, 17). Several factors could explain this discrepancy between these and previous results. For the most part, high concentrations of Sepharose-linked S100A1 or CaM could have retained RyR1 indirectly due to its interaction with other SR membrane proteins. We show evidence of this in micromolar Ca2+ with >30 μm S100A1 reducing CaM co-sedimentation with skeletal and cardiac SR membrane proteins (Fig. 3). An additional or alternative explanation, perhaps even more likely, for the competition observed by Wright et al. (17) is indicated by the behavior of A-S100A1 illustrated in Fig. 5, where several different A-S100A1 constructs are competed from RyRs by WT-CaM but not by WT-S100A1. This indicates that covalently modifying S100A1 (e.g. by attaching it to Sepharose, as done in Ref. 17) may change its binding specificity (relative to WT, unmodified S100A1), enabling the S100A1-Sepharose to attach at the CaM binding site of RyR1 or a site that is greatly impacted by CaM binding to RyR, rather than the true S100A1 binding site. This surprising behavior was replicated with several labeling sites, including Cys-85, Thr-6, Thr-42, and each terminus, thus suggesting that the specificity of S100A1 for a particular binding site on RyR is prone to change in response to covalent modification.
Evidently, these A-S100A1 constructs cannot be used to locate S100A1 on the RyR map. However, they show strong FRET that is specific for the CaM binding site (Fig. 5). This behavior could prove useful for monitoring the location of the 12-mer peptide (included in the main CaM binding site) that is hypothesized to represent their binding site on RyR (16). For this application, the difference in FRET efficiency with A-CaM bound to RyR1 (E ≈ 0.3) versus RyR2 (E ≈ 0.16) is intriguing and is reminiscent of the isoform differences suggested via cryo-EM analysis in an elegant previous report (33).
In light of the marginal influence of S100A1 on CaM binding to RyR, it is surprising that a single-residue mutation within the CaM binding domain of RyR1 (L3625D) and RyR2 (L3591D) abolished their S100A1-mediated functional modulation (11, 13). Our results suggest that residues within the RyR CaM binding domain affect the S100A1/RyR interaction through an allosteric mechanism rather than through orthosteric, direct physical contact with S100A1.
CaM Governs the Modulatory Effect of S100A1 on RyR Function
We show that nanomolar S100A1 modestly but significantly modulates RyR1 and RyR2 function (Fig. 2B), despite our FRET results demonstrating that nanomolar S100A1 does not significantly alter the structural basis of CaM/RyR binding (Fig. 4). This indicates that nanomolar S100A1 does not functionally modulate RyR activity via the CaM binding site. Moreover, CaM does affect the S100A1-mediated functional regulation of RyR1 and RyR2, albeit in slightly different ways.
Our functional results (Fig. 2) suggest that there are at least two S100A1 binding sites on RyR1 at low Ca2+: a nanomolar affinity inhibitory binding site that might be blocked by CaM/RyR1 binding and a micromolar affinity activatory binding site that is apparent when the inhibitory site is blocked by the presence of CaM. It also appears that S100A1 only mimics the action of CaM at micromolar Ca2+, not nanomolar Ca2+, indicating that S100A1 may bind to RyR1 at a site that overlaps with the CaM binding site in micromolar Ca2+, but not in nanomolar Ca2+. Given that S100A1 altered the conformation of the N-lobe, but not the C-lobe, of CaM binding to RyR (Table 1), it is plausible that micromolar S100A1 interferes with the CaM N-lobe interaction with RyR1 without significantly affecting the overall CaM/RyR1 apparent affinity, as previously speculated (34).
A report using saponin-skinned murine skeletal muscle fibers reported a dose-dependent increase in Ca2+-induced SR Ca2+ release when human recombinant S100A1 protein was added in the range from 0.01–1 μm, whereas 10 μm S100A1 tended to decrease isometric contractile force again (35). The inhibitory action of S100A1 is concurrent with our own observations of S100A1 reducing [3H]ryanodine binding to RyR1, albeit with submicromolar S100A1 (Fig. 2B). This is noticeable in both nanomolar and micromolar Ca2+ with CaM absent, which suggests that S100A1 may compensate for an absence of CaM in a skeletal muscle fiber, although the integration of signals from various accessory proteins probably contributes to the increased Ca2+-induced SR Ca2+ release observed by Most et al. (35).
Before this study, there was a general appreciation for the modulatory effect of S100A1 on RyR, although there were notable discrepancies in the literature, which probably reflect the complex and multifaceted interplay of RyR regulators. In 10 nm Ca2+, the addition of submicromolar S100A1 has been shown to increase [3H]ryanodine binding to skeletal SR membranes (RyR1), although this effect was abolished at 100 μm Ca2+ (3). As mentioned, we demonstrate that submicromolar S100A1 decreases [3H]ryanodine binding to RyR1, but this is restricted to the absence of CaM. Our data indicate (Fig. 2B) that this apparent discrepancy could be partially accounted for by residual endogenous CaM. Additionally, unaccounted impact of other accessory proteins as well as variation in the post-translationally modified state of RyR1 may also be responsible for the apparent discrepancies between studies.
At nanomolar Ca2+, [3H]ryanodine binding to RyR2 has been previously shown to be increased by 0.1 μm S100A1 but decreased by >1 μm S100A1 (4), whereas Yamaguchi et al. (13) have shown an inhibitory effect of submicromolar S100A1 on recombinant RyR2 in lipid bilayers. A possible cause for the discrepancy between reports and with our own data presented in this study is, again, the variable amount of residual endogenous CaM in SR membrane preparations, which we show greatly impacts the functional effect of S100A1 (Fig. 2).
Our results suggest that CaM binding to RyR2 enables an S100A1-mediated activation of RyR2 at both nanomolar and micromolar Ca2+, which can override the inhibitory effect of CaM. This activation could be attributable to S100A1 competing with the inhibitory effect of CaM or to an allosteric interaction of CaM on S100A1-mediated modulation of RyR2. Given that micromolar S100A1 only modestly alters the CaM binding conformation, it appears more likely that the latter is occurring. Either way, these results require both S100A1 and CaM to concurrently bind to RyR2. Numerous studies in electrically stimulated and quiescent cardiomyocytes have indicated that S100A1 indeed has a dual effect on the activity of the macromolecular RyR2 complex by improving both systolic opening and diastolic closure of the channel when intracellular S100A1 protein levels are elevated above normal (4, 6, 25). Increasing the amount of S100A1 bound to the RyR2 significantly reduced the calcium spark rate in normal cardiomyocytes and rescued the arrhythmogenic diastolic RyR2 dysfunction in failing cells. As such, these studies in intact cardiomyocytes partly resemble our experimental findings for [3H]ryanodine binding gleaned from cardiac SR vesicle preparations when applied [S100A1] exceeded [CaM]. Overall, our results support the notion of S100A1 action being integrated in vivo within a complex context of modulatory signals from accessory molecules.
Functional and Structural Effect of S100A1 under Diastolic and Systolic Ca2+
Our results indicate that the RyR1-S100A1 functional and structural interactions are more Ca2+-sensitive than RyR2-S100A1 (Fig. 6 and Table 1). Based on the reported low Ca2+ affinity of S100A1 (KCa approximately low millimolar) (36), S100A1 is likely to remain in the apo-state in the concentrations tested. Thus, it is likely that the Ca2+ dependence of the S100A1 structural effect on CaM·RyR1 is due to changes within RyR rather than S100A1. However, it is also possible that upon binding to RyR, the Ca2+ affinity of S100A1 is drastically enhanced, as shown for CaM when it binds to RyR1 (37).
Post-translational modification of the only Cys of S100A1 (residue 85) has been shown to enhance Ca2+ affinity to micromolar, which would facilitate the transition from apo- to Ca-S100A1 states. However, post-translational modifications are unlikely to be involved in our results, given that our mass spectrometry analyses indicated that S100A1 was unmodified before experiments, and the experimental conditions included the reducing agents GSH and DTT.
Possible S100A1 Binding Site(s) on RyR
Our results indicate a new model of S100A1/RyR interaction that involves S100A1 binding to more than one site on RyR and affecting the CaM·RyR structure but not the fractional occupancy of CaM binding sites on RyR. This is congruent with a report by Treves et al. (3) that proposed three S100A1 binding sites on RyR, including residues in RyR regions 1861–2155, 3774–3874, and 4425–4621. This encourages reassessment of these regions as candidates of S100A1 binding sites on RyR.
Despite some indications of competition between CaM and S100A1 binding to SR membranes and RyR, it should be understood that this is only observed when [S100A1] is in large (∼1000-fold) molar excess. This is not entirely physiologically relevant when one considers a study indicating that the protein level of S100A1 is 9-fold greater than CaM in human hearts (38), with total [CaM] around 6 μm in the myocyte (39).
It may be interpreted from the lack of competition with CaM binding that S100A1 does not bind to RyR in our SR preparations. Despite not demonstrating a clear direct physical interaction between WT-S100A1 and RyR in our FRET-based assays, we clearly demonstrate that [3H]ryanodine binding is influenced by nanomolar [S100A1], which thus indicates that S100A1 can functionally modulate RyR (Fig. 2A). In addition, further evidence of a physical interaction between RyR2 and S100A1 has been recently provided via proximity ligation assays and immunoprecipitation (6).
Conclusion
Although the hypothesis that CaM and S100A1 compete for the same RyR binding site is consistent with previous biochemical studies, our structural analysis of the RyR·CaM complex using TR-FRET reveals that S100A1 does not directly interfere with CaM binding to RyRs (Fig. 4) in the concentration range where CaM-dependent functional effects of S100A1 are observed (Fig. 2). Instead, S100A1 perturbs the structure of the RyR·CaM complex (Fig. 6). Thus, the structural mechanism for the interplay between CaM and S100A1 in regulation of RyR is more complex than previously thought.
Experimental Procedures
Isolation of SR Vesicles
SR membrane vesicles were isolated from porcine longissimus dorsi muscle and porcine cardiac ventricle by differential centrifugation of homogenized tissue, in accordance with Ref. 14. Heavy SR, rich in RyR1, was isolated by fractionation of skeletal crude SR vesicles using a discontinuous sucrose gradient (14). All vesicles were flash-frozen and stored at −80 °C. Immediately before the fluorescence binding studies described below, the SR vesicles were stripped of residual endogenous CaM by incubation with myosin light chain kinase-derived CaM binding peptide, followed by sedimentation, in accordance with Ref. 12.
Expression, Purification, and Fluorescence Labeling of FKBP and CaM
Single-cysteine mutants of FKBP12.6 (C22A/R49C/C76I, termed FKBP) and CaM (T34C or T110C) were separately expressed in Escherichia coli BL21(DE3)pLysS (Agilent Technologies), purified, and labeled with fluorescent probes Alexa Fluor 488 C5 maleimide (AF488), Alexa Fluor 568 C5 maleimide (AF568), or Alexa Fluor 647 C2 maleimide (AF647) (Life Technologies, Inc.), as described previously (19, 37). All fluorescently labeled proteins were dialyzed against a solution containing 30 mm NaCl and 20 mm MOPS (pH 7) and stored at −80 °C. Stoichiometric labeling of fluorescent CaM and FKBP to ≥95% was determined from the absorbance of bound dye relative to protein concentration determined via a bicinchoninic acid assay (Thermo Fisher Scientific) and SDS-PAGE densitometry. Essentially complete labeling was confirmed by electrospray ionization mass spectrometry.
Expression, Purification, and Fluorescence Labeling of S100A
Single-cysteine variants (wild type and mutants: T6C/C85S, T6C/C85A, T42C/C85S, ACA-S100A1-C85S, and S100A1-C85S-ACA) of human S100A1 were expressed in E. coli BL21(DE3) and purified using an adapted protocol (40). A pRSETb T7 expression vector plasmid inserted with human S100A1 cDNA was transformed into E. coli BL21(DE3)pLysS, and the cells were grown at 37 °C until optical density at 600 nm reached ∼1. At that point, protein expression was induced by the addition of 0.4 mm isopropyl β-d-thiogalactoside and a further 3–4 h of incubation. The cells were spun, resuspended in a homogenization solution (2 mm EDTA, 1 mm EGTA, 1 mm TCEP, 1 mm PMSF, 1 μg/ml aprotinin/leupeptin, 1 mm benzamidine, 0.1% Tween 20, 31 μg/ml DNase I, 5 mm MgCl2, and 50 mm Tris-HCl, pH 7.5), and briefly sonicated on ice with a Branson Ultrasonics Sonifier 250 (Danbury, CT). The homogenate was spun at 10,000 × g for 30 min. The supernatant was incubated for 20 min at 4 °C with 50% saturated ammonium sulfate. Following centrifugation, the ammonium sulfate saturation was increased to 80%, and the supernatant was incubated for 20 min at 4 °C and then centrifuged. S100A1 in the supernatant was precipitated by decreasing the pH to 4.1 and then pelleted by centrifugation. The pellet was resuspended in a solution containing 300 mm NaCl and 100 mm MOPS (pH 7) and dialyzed against 30 mm NaCl and 10 mm MOPS (pH 7). The S100A1 was further purified by loading onto a column containing phenyl-Sepharose CL-4B (GE Healthcare Bio-Sciences AB, Uppsala, Sweden) with 10 mm CaCl2 and eluting using elution solution (500 mm NaCl, 5 mm EGTA, and 50 mm Tris, pH 7.5). S100A1 was dialyzed against a solution containing 30 mm NaCl and 20 mm MOPS (pH 7.5) and concentrated using an Amicon stirred cell with a 3-kDa ultrafiltration disc (EMD Millipore, Billerica, MA). Protein purity and concentration were determined from SDS-PAGE and a bicinchoninic acid assay (Thermo Fisher Scientific), respectively. S100A1 was identified by electrospray ionization mass spectrometry using a QSTAR XL (AB Sciex, Framingham MA). In agreement with previous reports (21, 22), the WT-S100A1 sample contained a mixture of molecules that had either retained or lost the N-terminal methionine. In addition, the S100A1 molecules were largely in the reduced, monomeric form. A small fraction of protein contained the initiator N-formylmethionine, which has been observed previously (22).
Single-Cys variants of S100A1 were labeled with AF568 and purified using the same protocol as described previously for labeling of CaM (22). All fluorescently labeled S100A1 variants were dialyzed against a solution containing 30 mm NaCl and 20 mm MOPS (pH 7) and stored at −80 °C. Stoichiometric labeling of fluorescent S100A1 to ≥95% was determined from the absorbance of bound dye relative to protein concentration determined via a bicinchoninic acid assay (Thermo Fisher Scientific) and SDS-PAGE densitometry. Essentially complete labeling was confirmed by electrospray ionization mass spectrometry.
[3H]Ryanodine Binding to SR Vesicles
Skeletal and cardiac SR membranes (1 and 3 mg/ml, respectively) were preincubated with the indicated range of [S100A1], with and without 800 nm CaM, for 30 min at 4 °C in a solution containing 150 mm KCl, 5 mm GSH, 1 μg/ml aprotinin/leupeptin, 1 mm DTT, 1 mm EGTA, 65 μm CaCl2 (30 nm free Ca, as determined by MaxChelator), or 1.02 mm CaCl2 (30 μm free calcium, as determined by MaxChelator), 0.1 mg/ml of BSA, and 20 mm K-PIPES (pH 7.0). Media containing S100A1 (100 μm) had a slightly shifted (<20%) free Ca2+, relative to the no-S100A1 controls, as determined using the Fura-2 calcium-sensitive dye or a calcium-sensitive electrode (Thermo Scientific, Beverly, MA) in solutions calculated for both nanomolar and micromolar free Ca2+, respectively. The assay solution at 30 nm Ca2+ additionally contained 5 mm Na2ATP, and 5 mm caffeine was also added to cardiac SR assays. Binding of [3H]ryanodine (10 or 15 nm) was determined following a 3-h incubation at 37 °C and filtration through grade GF/B Glass Microfiber filters (Brandel Inc., Gaithersburg, MD) using a Brandel Harvester. In 4 ml of Ecolite scintillation mixture (MP Biomedicals, Solon, OH), the 3H retained on the filter was measured using a Beckman LS6000 scintillation counter.
Fluorescently Labeled CaM Binding to SR Vesicles
Skeletal SR (0.5 mg/ml) or cardiac SR (1 mg/ml) membranes were preincubated with the indicated [WT-CaM] or [S100A1] for 120 min, at 22 °C in binding media (150 mm KCl, 5 mm GSH, 1 μg/ml aprotinin/leupeptin, 1 mm EGTA, 0.1 mg/ml BSA, and 20 mm K-PIPES, pH 7.0) supplemented with 65 μm CaCl2 to give 30 nm free Ca or with 1.338 mm CaCl2 to give 300 μm free Ca2+ (as determined by MaxChelator). The binding of A-CaM to the treated SR membranes was measured following a 30-min incubation at 22 °C with 0.2 μm A-CaM, centrifugation at 100,000 × g for 25 min, and resuspension in a solution containing 5% SDS, 50 mm NaCl, 1 mm EGTA, and 20 mm Na-PIPES (pH 7.0). Bound A-CaM was determined from the fluorescence intensity at 600 nm (560-nm excitation, using a 570-nm emission long pass filter) (18).
FRET Measurements
Skeletal or cardiac SR (0.5 mg/ml) membranes were preincubated with 60 nm AF488- or AF568-labeled FKBP (donor, D-FKBP), for 90 min, at 37 °C, in a solution containing 150 mm KCl, 5 mm GSH, 0.1 mg/ml BSA, 1 μg/ml aprotinin/leupeptin, 1 mm DTT, and 20 mm PIPES (pH 7.0). To remove unbound D-FKBP, the SR membranes were spun at 110,000 × g for 20 min and then resuspended to 3 mg/ml. These samples were then incubated with the indicated range of [WT-CaM] or [WT-S100A1] for 120 min at 22 °C in binding media containing 30 nm free Ca2+ or 1.02 mm CaCl2 to give 30 μm free Ca2+ (calculated by MaxChelator). The binding of AF568- or AF647-labeled CaM (acceptor, A-CaM) or AF568-labeled S100A1 (A-S100A1) to RyR in the proximity of D-FKBP was measured following a 30-min incubation at 22 °C with 0.1 μm A-CaM or 2 μm A-S100A1, unless otherwise indicated. Steady-state fluorescence spectra were acquired from samples in a 384-well, optical bottom, black wall plate using a Gemini EM microplate fluorometer (with a 490-nm excitation wavelength and a 495-nm long pass filter in the emission channel). FRET efficiency was measured as the fractional decrease of donor fluorescence intensity due to the presence of acceptor fluorophore, according to Ref. 19.
For TR-FRET measurements, we used time-correlated single-photon counting (41) or direct waveform recording, as described previously (29, 30). Global multiexponential analysis of the TR-FRET data was used to test a series of structural models, as described extensively elsewhere (30, 41)
This mode of analysis resolves both binding and structural information from TR-FRET data. SR membranes labeled with D-FKBP were incubated with 200 μm S100A1 for 2 h at 22 °C before a 30-min incubation at 22 °C with 800 nm A-CaM.
Statistics
Sample means are from four or more independent experiments, and numbers of observations (n) are given in the figure legends. Each experiment was carried out using at least two independent SR preparations, isolated from different animals. Average data are provided as mean ± S.E. Statistical significance was evaluated by use of either paired or unpaired Student's t test or analysis of variance with Fisher's post hoc test, as appropriate.
Author Contributions
R. T. R., D. D. T., D. M. B., and R. L. C. designed the study and wrote the paper. F. R. N. purified fluorescent proteins and undertook steady-state FRET experiments with cardiac SR membranes. D. R. and P. M. provided purified recombinant S100A1 protein and DNA, assisted with critical evaluation of the results, and edited the manuscript. R. T. R. purified unlabeled protein; labeled S100A1; and performed time-resolved and steady-state FRET studies, sedimentation assays, and [3H]ryanodine binding assays using skeletal and cardiac SR membranes. All authors critically evaluated and approved the final version of the manuscript.
Acknowledgments
Spectroscopy studies were performed at the Biophysical Technology Center, University of Minnesota. We thank Dr. Ji Li for assistance with the Ca2+ indicator measurements and Octavian Cornea for preparing the manuscript for submission.
This work was supported in part by National Institutes of Health Grants R01HL092097 (to R. L. C. and D. M. B.), R01GM027906 (to D. D. T.), and T32HL069764 (to F. R. N.) and American Heart Association Grants 14POST20310024 (to R. T. R.) and 15GRNT25610022 (to R. L. C.). P. M. holds patents on the therapeutic use of S100A1 in cardiovascular diseases. P. M. is a shareholder of UniQure N.K. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- RyR
- ryanodine receptor
- CaM
- calmodulin
- FKBP
- FK506-binding protein
- TR-FRET
- time-resolved FRET
- SR
- sarcoplasmic reticulum
- A-CaM
- AF568/acceptor-labeled CaM
- D-FKBP
- donor-labeled FKBP
- A-S100A1
- acceptor-labeled S100A1
- AN-CaM
- AF568 attached via CaM N-lobe
- AC-CaM
- AF647 attached via CaM C-lobe
- R
- distance
- FWHM
- full width at half-maximum.
References
- 1. Buratti R., Prestipino G., Menegazzi P., Treves S., and Zorzato F. (1995) Calcium dependent activation of skeletal muscle Ca2+ release channel (ryanodine receptor) by calmodulin. Biochem. Biophys. Res. Commun. 213, 1082–1090 [DOI] [PubMed] [Google Scholar]
- 2. Tripathy A., Xu L., Mann G., and Meissner G. (1995) Calmodulin activation and inhibition of skeletal muscle Ca2+ release channel (ryanodine receptor). Biophys. J. 69, 106–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Treves S., Scutari E., Robert M., Groh S., Ottolia M., Prestipino G., Ronjat M., and Zorzato F. (1997) Interaction of S100A1 with the Ca2+ release channel (ryanodine receptor) of skeletal muscle. Biochemistry 36, 11496–11503 [DOI] [PubMed] [Google Scholar]
- 4. Kettlewell S., Most P., Currie S., Koch W. J., and Smith G. L. (2005) S100A1 increases the gain of excitation-contraction coupling in isolated rabbit ventricular cardiomyocytes. J. Mol. Cell Cardiol. 39, 900–910 [DOI] [PubMed] [Google Scholar]
- 5. Völkers M., Loughrey C. M., Macquaide N., Remppis A., DeGeorge B. R. Jr., Wegner F. V., Friedrich O., Fink R. H., Koch W. J., Smith G. L., and Most P. (2007) S100A1 decreases calcium spark frequency and alters their spatial characteristics in permeabilized adult ventricular cardiomyocytes. Cell Calcium 41, 135–143 [DOI] [PubMed] [Google Scholar]
- 6. Ritterhoff J., Völkers M., Seitz A., Spaich K., Gao E., Peppel K., Pleger S. T., Zimmermann W. H., Friedrich O., Fink R. H., Koch W. J., Katus H. A., and Most P. (2015) S100A1 DNA-based inotropic therapy protects against proarrhythmogenic ryanodine receptor 2 dysfunction. Mol. Ther. 23, 1320–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Oda T., Yang Y., Uchinoumi H., Thomas D. D., Chen-Izu Y., Kato T., Yamamoto T., Yano M., Cornea R. L., and Bers D. M. (2015) Oxidation of ryanodine receptor (RyR) and calmodulin enhance Ca release and pathologically alter, RyR structure and calmodulin affinity. J. Mol. Cell Cardiol. 85, 240–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Marks A. R. (2013) Calcium cycling proteins and heart failure: mechanisms and therapeutics. J. Clin. Invest. 123, 46–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bers D. M. (2014) Cardiac sarcoplasmic reticulum calcium leak: basis and roles in cardiac dysfunction. Annu. Rev. Physiol. 76, 107–127 [DOI] [PubMed] [Google Scholar]
- 10. Most P., Pleger S. T., Völkers M., Heidt B., Boerries M., Weichenhan D., Löffler E., Janssen P. M., Eckhart A. D., Martini J., Williams M. L., Katus H. A., Remppis A., and Koch W. J. (2004) Cardiac adenoviral S100A1 gene delivery rescues failing myocardium. J. Clin. Invest. 114, 1550–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yamaguchi N., Prosser B. L., Ghassemi F., Xu L., Pasek D. A., Eu J. P., Hernández-Ochoa E. O., Cannon B. R., Wilder P. T., Lovering R. M., Weber D., Melzer W., Schneider M. F., and Meissner G. (2011) Modulation of sarcoplasmic reticulum Ca2+ release in skeletal muscle expressing ryanodine receptor impaired in regulation by calmodulin and S100A1. Am. J. Physiol. Cell Physiol. 300, C998–C1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fruen B. R., Black D. J., Bloomquist R. A., Bardy J. M., Johnson J. D., Louis C. F., and Balog E. M. (2003) Regulation of the RYR1 and RYR2 Ca2+ release channel isoforms by Ca2+-insensitive mutants of calmodulin. Biochemistry 42, 2740–2747 [DOI] [PubMed] [Google Scholar]
- 13. Yamaguchi N., Chakraborty A., Huang T. Q., Xu L., Gomez A. C., Pasek D. A., and Meissner G. (2013) Cardiac hypertrophy associated with impaired regulation of cardiac ryanodine receptor by calmodulin and S100A1. Am. J. Physiol. Heart Circ. Physiol. 305, H86–H94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fruen B. R., Bardy J. M., Byrem T. M., Strasburg G. M., and Louis C. F. (2000) Differential Ca2+ sensitivity of skeletal and cardiac muscle ryanodine receptors in the presence of calmodulin. Am. J. Physiol. Cell Physiol. 279, C724–C733 [DOI] [PubMed] [Google Scholar]
- 15. Xu L., and Meissner G. (2004) Mechanism of calmodulin inhibition of cardiac sarcoplasmic reticulum Ca2+ release channel (ryanodine receptor). Biophys. J. 86, 797–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Prosser B. L., Wright N. T., Hernãndez-Ochoa E. O., Varney K. M., Liu Y., Olojo R. O., Zimmer D. B., Weber D. J., and Schneider M. F. (2008) S100A1 binds to the calmodulin-binding site of ryanodine receptor and modulates skeletal muscle excitation-contraction coupling. J. Biol. Chem. 283, 5046–5057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wright N. T., Prosser B. L., Varney K. M., Zimmer D. B., Schneider M. F., and Weber D. J. (2008) S100A1 and calmodulin compete for the same binding site on ryanodine receptor. J. Biol. Chem. 283, 26676–26683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cornea R. L., Nitu F., Gruber S., Kohler K., Satzer M., Thomas D. D., and Fruen B. R. (2009) FRET-based mapping of calmodulin bound to the RyR1 Ca2+ release channel. Proc. Natl. Acad. Sci. U.S.A. 106, 6128–6133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cornea R. L., Nitu F. R., Samsó M., Thomas D. D., and Fruen B. R. (2010) Mapping the ryanodine receptor FK506-binding protein subunit using fluorescence resonance energy transfer. J. Biol. Chem. 285, 19219–19226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guo T., Fruen B. R., Nitu F. R., Nguyen T. D., Yang Y., Cornea R. L., and Bers D. M. (2011) FRET detection of calmodulin binding to the cardiac RyR2 calcium release channel. Biophys. J. 101, 2170–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bolewska K., Kozłowska H., Goch G., Mikołajek B., and Bierzyński A. (1997) Molecular cloning and expression in Escherichia coli of a gene coding for bovine S100A1 protein and its Glu32 → Gln and Glu73 → Gln mutants. Acta Biochim. Pol. 44, 275–283 [PubMed] [Google Scholar]
- 22. Zhukova L., Zhukov I., Bal W., and Wyslouch-Cieszynska A. (2004) Redox modifications of the C-terminal cysteine residue cause structural changes in S100A1 and S100B proteins. Biochim. Biophys. Acta 1742, 191–201 [DOI] [PubMed] [Google Scholar]
- 23. Pleger S. T., Shan C., Ksienzyk J., Bekeredjian R., Boekstegers P., Hinkel R., Schinkel S., Leuchs B., Ludwig J., Qiu G., Weber C., Raake P., Koch W. J., Katus H. A., Müller O. J., and Most P. (2011) Cardiac AAV9-S100A1 gene therapy rescues post-ischemic heart failure in a preclinical large animal model. Sci. Transl. Med. 3, 92ra64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Remppis A., Most P., Löffler E., Ehlermann P., Bernotat J., Pleger S., Börries M., Reppel M., Fischer J., Koch W. J., Smith G., and Katus H. A. (2002) The small EF-hand Ca2+ binding protein S100A1 increases contractility and Ca2+ cycling in rat cardiac myocytes. Basic Res. Cardiol. 97, I56–I62 [DOI] [PubMed] [Google Scholar]
- 25. Brinks H., Rohde D., Voelkers M., Qiu G., Pleger S. T., Herzog N., Rabinowitz J., Ruhparwar A., Silvestry S., Lerchenmüller C., Mather P. J., Eckhart A. D., Katus H. A., Carrel T., Koch W. J., and Most P. (2011) S100A1 genetically targeted therapy reverses dysfunction of human failing cardiomyocytes. J. Am. Coll. Cardiol. 58, 966–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Most P., Seifert H., Gao E., Funakoshi H., Völkers M., Heierhorst J., Remppis A., Pleger S. T., DeGeorge B. R. Jr., Eckhart A. D., Feldman A. M., and Koch W. J. (2006) Cardiac S100A1 protein levels determine contractile performance and propensity toward heart failure after myocardial infarction. Circulation 114, 1258–1268 [DOI] [PubMed] [Google Scholar]
- 27. Pleger S. T., Most P., Heidt B., Voelkers M., Hata J. A., Katus H. A., Remppis A., and Koch W. J. (2006) S100A1 gene transfer in myocardium. Eur. J. Med. Res. 11, 418–422 [PubMed] [Google Scholar]
- 28. Svensson B., Oda T., Nitu F. R., Yang Y., Cornea I., Chen-Izu Y., Fessenden J. D., Bers D. M., Thomas D. D., and Cornea R. L. (2014) FRET-based trilateration of probes bound within functional ryanodine receptors. Biophys. J. 107, 2037–2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gruber S. J., Cornea R. L., Li J., Peterson K. C., Schaaf T. M., Gillispie G. D., Dahl R., Zsebo K. M., Robia S. L., and Thomas D. D. (2014) Discovery of enzyme modulators via high-throughput time-resolved FRET in living cells. J. Biomol. Screen. 19, 215–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Guhathakurta P., Prochniewicz E., and Thomas D. D. (2015) Amplitude of the actomyosin power stroke depends strongly on the isoform of the myosin essential light chain. Proc. Natl. Acad. Sci. U.S.A. 112, 4660–4665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rodney G. G., Moore C. P., Williams B. Y., Zhang J. Z., Krol J., Pedersen S. E., and Hamilton S. L. (2001) Calcium binding to calmodulin leads to an N-terminal shift in its binding site on the ryanodine receptor. J. Biol. Chem. 276, 2069–2074 [DOI] [PubMed] [Google Scholar]
- 32. Samsó M., and Wagenknecht T. (2002) Apocalmodulin and Ca2+-calmodulin bind to neighboring locations on the ryanodine receptor. J. Biol. Chem. 277, 1349–1353 [DOI] [PubMed] [Google Scholar]
- 33. Huang X., Fruen B., Farrington D. T., Wagenknecht T., and Liu Z. (2012) Calmodulin-binding locations on the skeletal and cardiac ryanodine receptors. J. Biol. Chem. 287, 30328–30335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bers D. M. (2011) Calmodulin and S100A1 fine tune skeletal muscle E-C coupling: focus on “modulation of sarcoplasmic reticulum Ca2+ release in skeletal muscle expressing ryanodine receptor impaired in regulation by calmodulin and S100A1”. Am. J. Physiol. Cell Physiol. 300, C974–C975 [DOI] [PubMed] [Google Scholar]
- 35. Most P., Remppis A., Weber C., Bernotat J., Ehlermann P., Pleger S. T., Kirsch W., Weber M., Uttenweiler D., Smith G. L., Katus H. A., and Fink R. H. (2003) The C terminus (amino acids 75–94) and the linker region (amino acids 42–54) of the Ca2+-binding protein S100A1 differentially enhance sarcoplasmic Ca2+ release in murine skinned skeletal muscle fibers. J. Biol. Chem. 278, 26356–26364 [DOI] [PubMed] [Google Scholar]
- 36. Goch G., Vdovenko S., Kozłowska H., and Bierzyñski A. (2005) Affinity of S100A1 protein for calcium increases dramatically upon glutathionylation. FEBS J. 272, 2557–2565 [DOI] [PubMed] [Google Scholar]
- 37. Fruen B. R., Balog E. M., Schafer J., Nitu F. R., Thomas D. D., and Cornea R. L. (2005) Direct detection of calmodulin tuning by ryanodine receptor channel targets using a Ca2+-sensitive acrylodan-labeled calmodulin. Biochemistry 44, 278–284 [DOI] [PubMed] [Google Scholar]
- 38. Pedrocchi M., Hauer C. R., Schäfer B. W., Erne P., and Heizmann C. W. (1993) Analysis of Ca2+-binding S100 proteins in human heart by HPLC-electrospray mass spectrometry. Biochem. Biophys. Res. Commun. 197, 529–535 [DOI] [PubMed] [Google Scholar]
- 39. Maier L. S., Ziolo M. T., Bossuyt J., Persechini A., Mestril R., and Bers D. M. (2006) Dynamic changes in free Ca-calmodulin levels in adult cardiac myocytes. J. Mol. Cell. Cardiol. 41, 451–458 [DOI] [PubMed] [Google Scholar]
- 40. Ehlerman P., Remppis A., Most P., Bernotat J., Heizmann C. W., and Katus H. A. (2000) Purification of the Ca2+-binding protein S100A1 from myocardium and recombinant Escherichia coli. J. Chromatogr. B Biomed. Sci. Appl. 737, 39–45 [DOI] [PubMed] [Google Scholar]
- 41. Dong X., and Thomas D. D. (2014) Time-resolved FRET reveals the structural mechanism of SERCA-PLB regulation. Biochem. Biophys. Res. Commun. 449, 196–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yan Z., Bai X. C., Yan C., Wu J., Li Z., Xie T., Peng W., Yin C. C., Li X., Scheres S. H., Shi Y., and Yan N. (2015) Structure of the rabbit ryanodine receptor RyR1 at near-atomic resolution. Nature 517, 50–55 [DOI] [PMC free article] [PubMed] [Google Scholar]


