Abstract
The chloroplast-localized proteins play roles in plant salt stress response, but their mechanisms remain largely unknown. In this study, we screened a yellow leaf mutant, yl1-1, whose shoots exhibited hypersensitivity to salt stress. We mapped YL1 to AT3G57180, which encodes a YqeH-type GTPase. YL1, as a chloroplast stroma-localized protein, could be markedly reduced by high salinity. Upon exposure to high salinity, seedling shoots of yl1-1 and yl1-2 accumulated significantly higher levels of Na+ than wild type. Expression analysis of factors involved in plant salt stress response showed that the expression of ABI4 was increased and HKT1 was evidently suppressed in mutant shoots compared with the wild type under normal growth conditions. Moreover, salinity effects on ABI4 and HKT1 were clearly weakened in the mutant shoots, suggesting that the loss of YL1 function impairs ABI4 and HKT1 expression. Notably, the shoots of yl1-2 abi4 double mutant exhibited stronger resistance to salt stress and accumulated less Na+ levels after salt treatment compared with the yl1-2 single mutant, suggesting the salt-sensitive phenotype of yl1-2 seedlings could be rescued via loss of ABI4 function. These results reveal that YL1 is involved in the salt stress response of seedling shoots through ABI4.
High salinity is a serious factor that influences plant productivity. It affects various aspects of plant physiology and metabolism by inducing osmotic stress and ion toxicity1. The early-occurring osmotic stress triggers physiological changes, such as membrane interruption in roots and reduction of water absorption capacity in plants. Ion over-accumulation, which is the second phase of salt stress, can induce severe Na+/K+ imbalance and toxic effects2,3,4.
Plants have evolved different molecular mechanisms to adapt to hyperionic stress4,5. The calcium-responsive salt overly sensitive (SOS) regulatory pathway, which is mainly for ion homeostasis, has been established in Arabidopsis6. SOS3, a myristoylated calcium-binding protein, recognizes salt-elicited cytosolic calcium signals and then interacts with and activates SOS2 for signal transmission7,8. SOS1, a plasma membrane Na+/H+ anti-porter and regulated by SOS3 and SOS2, aids in sodium transport from root cells back into the soil or from epidermal cells into the xylem9. Overexpression of SOS1 confers plant salt stress resistance10. High-affinity K+ transporter (HKT1), an important regulator that is independent of the SOS pathway and exhibits significant functions in root and shoot ion homeostasis, has been extensively studied11,12. HKT1, as a K+/Na+ symporter, is highly expressed in the root stele and leaf vasculature and retrieves sodium from the root-to-shoot xylem sap in Arabidopsis4,13. AtHKT1 knock-out mutant leaves exhibit high sensitivity to salt stress because of excessive sodium accumulation14, and AtHKT1 overexpression in roots enhances the salt tolerance of the entire plant15. In addition, the tonoplast-localized Na+ (K+)/H+ exchanger NHX1 confers Na+ or K+ storage into vacuoles16,17. AtNHX1 overexpression could reduce Na+ stress through enhancing intracellular K+/Na+ ratios in tomato18.
The phytohormone abscisic acid (ABA) exerts a significant function for coping with salt stress3. The ABA-deficient mutants aba1, aba2, and aba3 show a readily wilting phenotype under salt or drought stress. ABSCISIC ACID INSENSITIVE (ABI) 4 was first isolated from a screen for ABA-insensitive mutants during seed germination19. ABI4, as a member of the plant-specific AP2/EREBP family, is involved in many signal transduction pathways, such as sugar signaling and mitochondrial/chloroplast retrograde signaling20,21,22. The abi4 mutant exhibits salt stress resistance because less sodium is accumulated in plant shoots. ABI4-overexpressing (dexamethasone-induced) plants show increased salt sensitivity because ABI4 downregulates HKT1 expression by directly binding to the promoter ABE-element GC(C/G)GCTT(T)23.
It is generally accepted that, high salinity can cause photosynthesis inhibition in plants, and leaf growth is very sensitive to salt stress. This phenomenon may be attributed to the disruption of chloroplast development24,25. CO2 fixation is sensitive to environmental stresses. Therefore, salt stress can inhibit the repair of PS II via the ROS-induced suppression of PS II protein synthesis, which in turn triggers an imbalance between the photo-damage and repair rates of PS II26,27. Moreover, recent studies have suggested that the chloroplast proteins also play roles in plant salt stress response28,29,30. However, the mechanisms are largely unclear. In this study, we screened the yellow leaf 1-1 (yl1-1) mutant. The shoots of yl1-1 showed evident salt stress-sensitive phenotypes. We demonstrated that YL1, as a chloroplast protein, is involved in the high salinity response of seedling shoots through ABI4.
Results
Phenotypes of yl1-1 Mutant
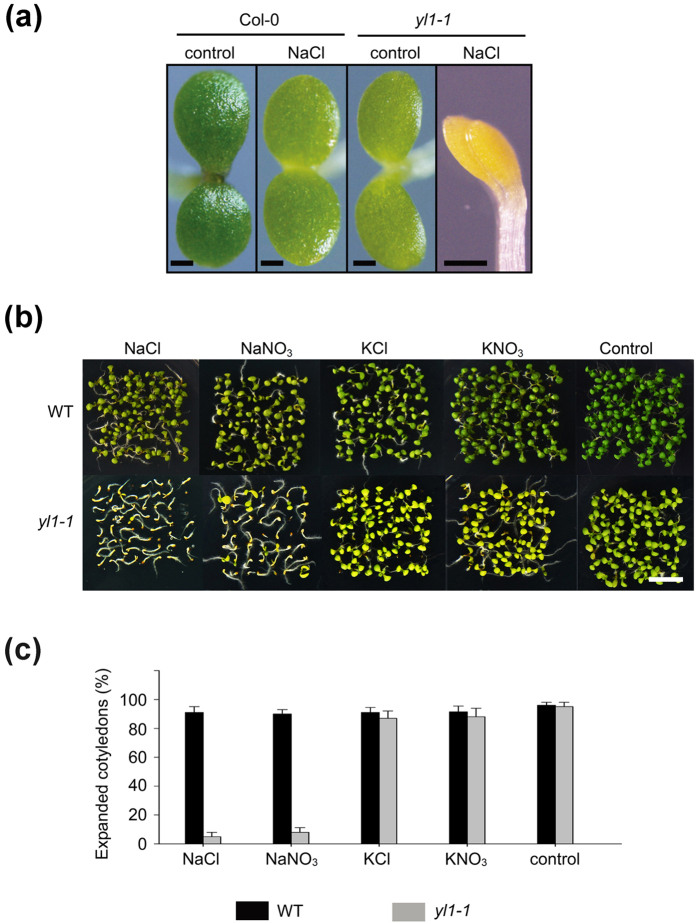
Arabidopsis seedling shoots usually exhibit pale coloration and stunted phenotypes under salt stress conditions (Fig. 1a). We are interested in mutants that the seedling shoots exhibit extremely sensitive phenotypes under salt stress. The mutant yl1-1 was isolated from approximately 30,000 ethane methylsulfonate (EMS)-mutagenized Col-0 M2 seedlings, which conferred a pale-green shoot phenotype under normal growth conditions (Figs 1a and S1). However, under salt stress conditions, shoot of yl1-1 showed evidently stunted phenotype compared with wild type (Figs 1a and S1), while little differences in root development could be observed (Fig. S1). Three additional salts (NaNO3, KCl, or KNO3) were used in seedling growth experiments to understand the phenotypes of yl1-1 hypersensitivity to salt stress better. The results showed that the percentages of the fully expanded cotyledons of yl1-1 seedlings were significantly lower in growth conditions with NaCl or NaNO3 than with KCl or KNO3 (Fig. 1b,c). By contrast, wild type seedlings did not exhibit clear differences under these different salt treatments (Fig. 1b,c). These observations suggest that Na+ toxicity leads to stunted yl1-1 shoot phenotypes.
Figure 1. Salt stress sensitive phenotypes of yl1-1 seedling shoots.
(a) Phenotypes of seedlings under control or salt stress conditions. Each lane represents the corresponding seedling grown for 5 days after germination under the indicated condition (NaCl, 150 mM). Three independent experiments were performed with similar results and one representative is showed. Scale bars = 0.5 mm. (b) Phenotypes of wild type or yl1-1 plants grown in different salinity stresses (NaCl, NaNO3, KCl, KNO3, and control; 150 mM) for 5 days after germination. Three independent experiments were performed with similar results and one representative is showed. Scale bar = 5 mm. (c) The percentages of the fully expanded seedling cotyledons showed in (b). Values are means ± SE of three independent replicates.
Positional Cloning of yl1-1
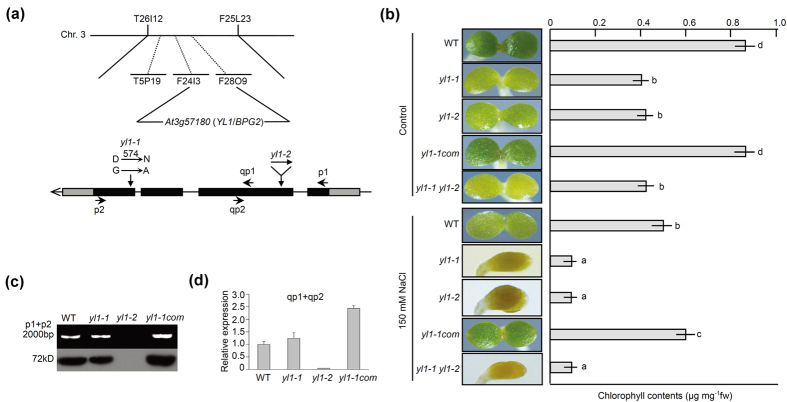
A map-based cloning approach was applied to identify the target gene(s) responsible for the yl1-1 phenotypes (Fig. 2a). F1 plants were generated by crossing yl1-1 with Landsberg (an Arabidopsis wild ecotype) and self-fertilization to generate the F2 population. We mapped YL1 to At3g57180 (Brassinazole Insensitive Pale Green 2, BPG2), positioned it between the BAC markers F24I3 and F28O9, and identified a G to A base substitution, which caused a D to N change in position 574 of the amino acid sequence (Fig. 2a).
Figure 2. Positional cloning of yl1-1.
(a) Map position of yl1-1 on chromosome 3. Some of the used BAC markers are indicated. Gene structure of YL1(At3g57180, BPG2) is shown. The yl1-1 mutated site of G to A (amino acid of D to N) and the inserted position of a YL1 T-DNA insertion mutant, yl1-2, are all indicated. Short arrows represent primers used in (c,d). (b) Phenotypes of wild type, yl1-1, yl1-2, yl1-1com, and yl1-1 yl1-2 (cross F1 line) seedling grown under control or 150 mM NaCl condition for 5 days after germination. Scale bars = 0.5 mm. Endogenous chlorophyll (a,b) contents of the corresponding plants are shown. Data are mean values of three replicates ± SE. Statistical significant differences are indicated by different lowercase letters (P < 0.01). (c) Full-length YL1 gene amplification from cDNA and immunoblot analysis for YL1 in wild type, yl1-1, yl1-2, and yl1-1 complementary plants (yl1-1com). Primers p1 and p2 showed in a were used. (d) qRT-PCR analysis of YL1 expression levels in wild type, yl1-1, yl1-2, and yl1-1 complementary plant yl1-1com. Primers qp1 and qp2 showed in a were used. The internal control gene was Actin2.
Previous studies reported that BPG2, as a YqeH-type GTPase, participates in chloroplast rRNA maturation31,32. To determine whether the lack of YL1 is responsible for the phenotypes of yl1-1, we introduced the full-length open reading frame of YL1 to the yl1-1 mutant under the control of the 35S promoter. The complemented plant (yl1-1com) showed a normal leaf phenotype (Figs 2b and S2a). A mutant of At3g57180, yl1-2 (bpg2-2, salk_068713), was obtained from ABRC stock (Figs 2b and S2a). Western blot, gene full-length amplification PCR from cDNA, and real-time quantitative PCR (qRT-PCR) analyses showed that YL1 expression was extremely low in yl1-2, which suggested a knock-out mutant (Fig. 2c,d). The yl1-1, yl1-2, and cross F1 line (yl1-1 yl1-2) exhibited evident pale-green leaf phenotypes under normal growth conditions (Figs 2b and S2a). Under salt stress treatment, the yl1-1, yl1-2, and cross F1 line showed extremely stunted cotyledon phenotypes, whereas the wild type and yl1-1com plants exhibited normal phenotypes except for the decrease of chlorophyll contents at day 5 after germination (Fig. 2b). In addition, the Fv/Fm and endogenous contents of chlorophyll in the mature plants of yl1-1, yl1-2, and cross F1 line were significantly lower than those in the wild type and yl1-1com (Fig. S2b,c). These results indicate that the yl1-1 phenotypes are caused by YL1 function loss.
Subcellular Localization and Expression Pattern of YL1
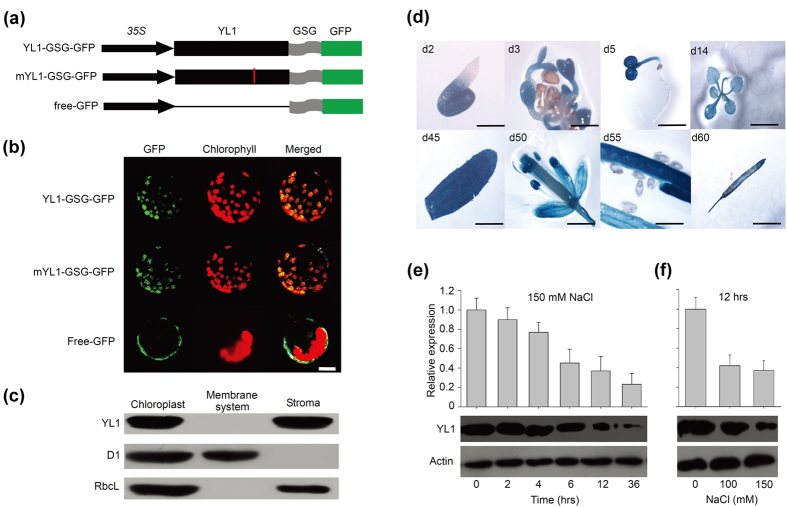
Previous studies stated that YL1 mainly localizes in chloroplast31,32. To further determine details of YL1 subcellular localization, we constructed a YL1-GSG-GFP plasmid containing a flexible peptide (GGGSSSSGGG) between the YL1 and GFP gene sequences (Fig. 3a). GFP fluorescence was clearly detected in the chloroplasts of YL1-GSG-GFP transgenic plant protoplasts (Fig. 3b). Immunoblot analysis of the stroma and membrane fractions from Percoll-purified chloroplasts demonstrated that YL1 protein is localized in the stroma, and not in the membrane (Fig. 3c). Here, the chloroplast stroma protein RbcL and thylakoid membrane protein D1 were used as control (Fig. 3c). In addition, the localization of the mutated YL1 (mYL1) in yl1-1 is not changed (Fig. 3b), further confirming that the mutation elicits protein function defects.
Figure 3. Subcellular Localization and Expression Patterns of YL1.
(a) The structures of plasmids used for YL1 protein subcellular localization experiment. The black arrows indicate the 35S promoter; the black cuboids indicate the CDS of YL1; the gray curving cuboids indicate the flexible peptide (GSG) and the green cuboids indicate the GFP CDS. For plasmid of mYL1-GSG-GFP, the red break line indicates the mutant site (G to A) of YL1 in yl1-1 mutant. (b) Localization of YL1 protein within the chloroplast by GFP assay. The green fluorescence indicates the GFP, the red indicates the chlorophyll fluorescence. Bar = 5 μm. (c) YL1 localizes in the stroma fractions. Intact chloroplasts were isolated from cotyledons of 5-d-old yl1-1 seedlings, then separated into stroma and membrane fractions, which were used for immunonlot analysis. D1 is mainly associated with membrane system while RbcL localizes in stroma. (d) GUS staining analysis of YL1 in a 2-d-old germinating peeled seed (scale bar = 1 mm), expanded cotyledons of 3-d-old seedlings (scale bar = 5 mm), 5-d-old seedling cotyledons (scale bar = 5 mm), 14-d-old plant leaves (scale bar = 10 mm), a green cauline leaf from a 45-d-old plant (scale bar = 5 mm), a flower with full organs (scale bar = 5 mm), immature green seeds from a 55-d-old plant (scale bar = 1 mm), and a silique containing mature seeds from a 60-d-old plant (scale bar = 1 mm). (e) qRT-PCR and immunoblot analysis of YL1 expression levels under salt stress. Total RNA or protein was isolated from 5-d-old seedling cotyledons treated with 150 mM NaCl for different time. (f) qRT-PCR and immunoblot analysis of YL1 expression levels under different NaCl concentrations. Total RNA or protein was isolated from cotyledons of 5-d-old seedlings treated with 0 mM, 100 mM, or 150 mM NaCl for 12 h. For immunoblot analysis, each lane was loaded on 20 μg cotyledon protein. For transcriptional analysis, the internal control gene was Actin2. Three independent experiments were performed with similar results. One representative experiment is showed.
The expression pattern analysis used in β-glucuronidase (GUS) staining showed that the YL1 gene is expressed in most of the green tissues throughout the plant growth cycle, even in germinating seeds (Fig. 3d). However, the GUS activity is extremely low in roots and mature seeds (Fig. 3d), suggesting low expression of YL1 in these tissues. To detect the effect of salt stress on the expression of YL1, total RNA was isolated from 5-d-old wild type seedling shoots harvested after salt treatment. The results of qRT-PCR showed that the levels of YL1 transcripts were evidently reduced with increasing salt concentration or treatment time, which were further confirmed by immunoblot and GUS activity analysis (Figs 3e,f and S3). When plants were treated with 150 mM NaCl for 12 h, YL1 expression decreased to less than half of that of the non-treated plants (Fig. 3e,f). In addition, no matter treated with or without NaCl, YL1 transcripts were hardly detected in roots (Fig. S4). These results indicate that the YL1 could be significantly downregulated by salt stress in seedling shoots.
Loss of YL1 Function Causes Shoot Na+ Accumulation under Salt Stress
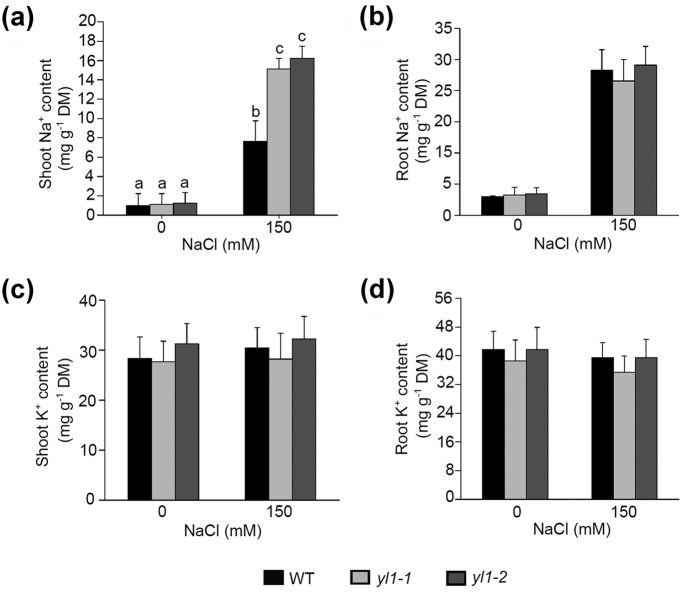
Basing on the results presented in Fig. 1b, we speculate that Na+ homeostasis is disrupted in yl1-1 shoot tissues under salt stress conditions. Thus, Na+ and K+ levels were measured using atomic absorption spectrophotometry. The Na+ contents in the shoots were substantially higher in yl1-1 and yl1-2 than in wild type plants treated with 150 mM NaCl for 2 days (Fig. 4a). By contrast, no differences in Na+ contents were observed in roots among these genotypes (Fig. 4b). K+ levels were similar among the three genotypes regardless of tissue type (Fig. 4c,d). Thus, we conclude that Na+ is over-accumulated in the shoots of yl1 seedlings under salt stress.
Figure 4. Na+ and K+ contents in seedling shoots or roots.
(a) Na+ contents in 5-d-old seedling shoots of wild type, yl1-1, and yl1-2 treated with 0 mM or 150 mM NaCl for 2 d on 1/2 MS medium. Data represent means ± SE of three independent experiments. One-way ANOVA (Duncan’s multiple range test) was performed, and statistical significant differences are indicated by different lowercase letters (P < 0.01). (b) Na+ contents in roots of 5-d-old seedlings described in (a). (c) K+ contents in shoots of 5-d-old seedlings described in (a). (d) K+ contents in roots of 5-d-old seedlings described in (a).
Loss of YL1 Function Affects ABI4 Expression in Seedling Shoots
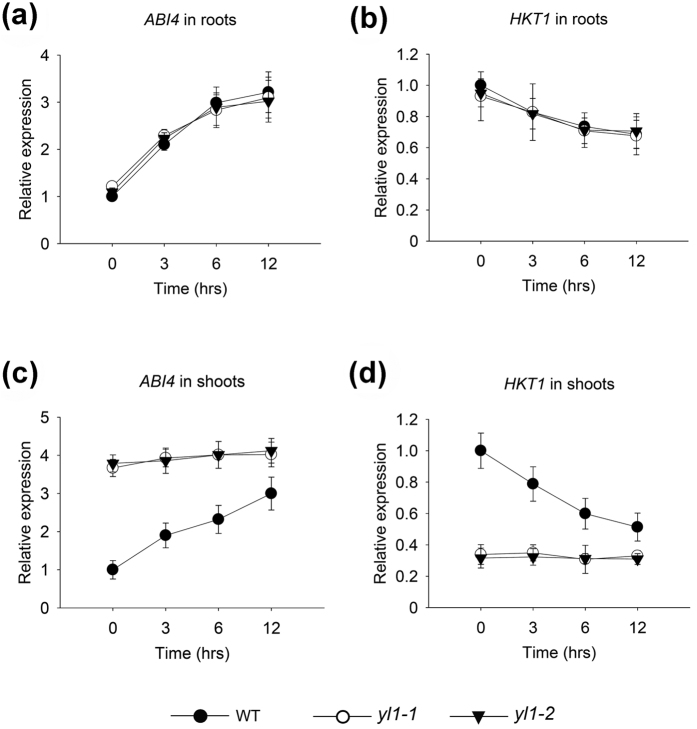
To identify the factor(s) that possibly play roles in Na+ over-accumulation in the shoots of yl1-1 and yl1-2, transcripts of several genes involved in plant salt stress response were detected. The results showed that, regardless of treatment with or without salt, no clear differences were observed in the expression levels of the SOS pathway genes, ABA biosynthesis pathway genes, or NHX1 among the studied genotypes (Figs S5 and S6). Similar results of ABI4 or HKT1 transcription levels were found in the roots of the detected genotypes (Fig. 5a,b). However, in the shoots of the mutant seedlings, ABI4 expression was three- to fourfold higher, whereas HKT1 transcription levels were substantially lower than those in the wild type grown under normal growth conditions (Fig. 5c,d). Moreover, after 12 h of salt stress treatment, evident inductions of ABI4 and reductions in HKT1 transcripts were observed in the shoots of wild type and roots of all detected genotypes (Fig. 5), but no significant changes of ABI4 or HKT1 could be observed in the mutant shoots (Fig. 5c). These results suggest that loss of YL1 function impairs ABI4 and HKT1 expression in seedling shoots. A previous study stated that ABI4-overexpressed plants are sensitive to salt stress because of HKT1 depression23. Based on these evidences, the salt-sensitive phenotype of the yl1 mutant shoot is possibly tightly associated with the high levels of ABI4.
Figure 5. Relative expression analysis of ABI4 and HKT1.
(a) Transcript abundance analysis of ABI4 in roots of wild type, yl1-1, and yl1-2 5-d-old seedlings treated with 150 mM NaCl for 0, 3, 6, or 12 h. (b) Transcript abundance analysis of HKT1 in 5-d-old seedling roots of wild type, yl1-1, and yl1-2 treated with 150 mM NaCl for 0, 3, 6, or 12 h. (c) Transcript abundance analysis of ABI4 in shoots of wild type, yl1-1, and yl1-2 5-d-old seedlings treated with 150 mM NaCl for 0, 3, 6, or 12 h. (d) Transcript abundance analysis of HKT1 in 5-d-old seedling shoots of wild type, yl1-1, and yl1-2 treated with 150 mM NaCl for 0, 3, 6, or 12 h. The internal control gene was Actin2. Data represent means ± SE of three independent experiments.
ABI4 Functions in Salt Stress Response of yl1 Mutant Seedlings
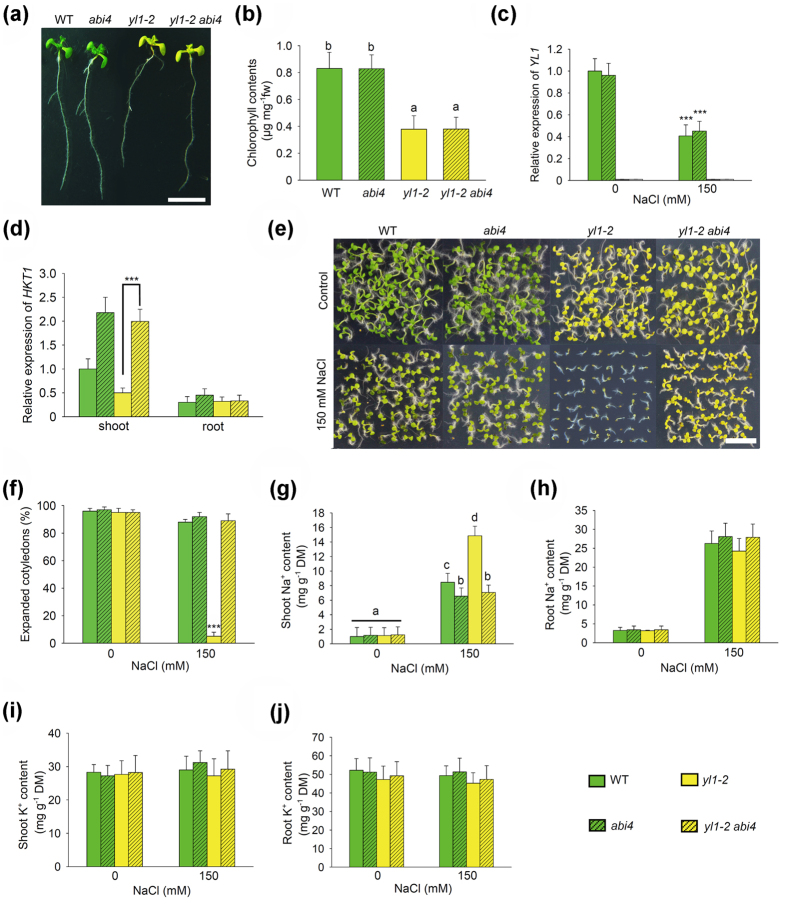
To further confirm the above issue, a double mutant, yl1-2 abi4, was created. The 10-d-old seedling shoot of yl1-2 abi4 showed a pale-green phenotype due to the low chlorophyll contents, revealing that the YL1 roles in leaf coloration are independent of ABI4 (Fig. 6a,b). YL1 transcripts were not clearly influenced by the abi4 mutant, indicating that YL1 expression was also ABI4-independent (Fig. 6c). However, HKT1 mRNA levels were significantly higher in the shoots of yl1-2 abi4 than in the yl1-2 single mutant, which was similar to that in abi4 (Fig. 6d). Furthermore, under high-salinity growth conditions, the percentages of the fully expanded cotyledons of yl1-2 abi4 seedlings were similar to those of the wild type and abi4 (Fig. 6e,f). The seedling shoots of salt-treated yl1-2 abi4 also accumulated lower levels of Na+ than the yl1-2 single mutant, whereas K+ contents did not considerably change in all detected genotypes (Fig. 6g–j). Thus far, the abovementioned results clearly indicate that the salt stress-sensitive phenotype of yl1-2 shoot could be completely rescued by the loss of ABI4 function.
Figure 6. Loss of ABI4 rescues salt stress sensitivity of yl1 seedlings.
(a) Phenotypes of 10-d-old wild type, abi4, yl1-2, and double mutant yl1-2 abi4 grown under normal growth conditions. Scale bar = 10 mm. (b) Chlorophyll contents (chlorophyll a & b) of 10-d-old plants showed in (a). (c) Transcript abundance analysis of YL1 in wild type, abi4, yl1-2, and double mutant yl1-2 abi4 5-d-old seedlings treated with 150 mM NaCl for 12 h or not. (d) Transcript abundance of HKT1 in 5-d-old seedling shoots and roots of genotypes showed in (a). (e) 5-d-old seedling phenotypes of wild type, abi4, yl1-2, and double mutant yl1-2 abi4 grown in 0 mM or 150 mM NaCl. Scale bar = 5 mm. (f) The percentages of the fully expanded cotyledons of plants showed in (e). (g) Na+ contents in 5-d-old seedling shoots of wild type, abi4, yl1-2, and double mutant yl1-2 abi4 treated with 0 mM or 150 mM NaCl for 2 d. (h) Na+ contents in 5-d-old seedling roots described in (g). (i) K+ contents in 5-d-old seedling shoots described in (g). (j) K+ contents in 5-d-old seedling roots described in (g). For (b,g), one-way ANOVA (Duncan’s multiple range test) was performed, and statistical significant differences are indicated by different lowercase letters (P < 0.01). For (c,d,f,) values are means ± SE of three independent replicates (asterisk indicates P < 0.001, Student’s t-test). For (c,d) the internal control gene was Actin2.
Discussion
Hyperionic stress causes plant growth inhibition or plant death by disturbing the physiological functions of the shoots and roots3,5,25. This study focused on shoot phenotypes to elucidate the mechanisms of plant response to salt stress. yl1-1, a mutant with severely stunted shoot phenotype under salt stress conditions, was isolated from an EMS-mutagenized library in Col-0 background (Fig. 1a). YL1, which is mainly localized in the chloroplast stroma (Fig. 3b,c), is a YqeH-type GTPase. GTPase, including Obg, Era, YlqF, YqhC, YsxC, YqeH, and YloQ-type families, exists in almost all organisms and is involved in regulating diverse cellular processes33,34. However, the YqeH is only conserved in bacteria and plant chloroplast35. In Bacillus subtilis, YqeH is involved in 30S ribosome subunit biogenesis and 16S rRNA maturation36,37. In plants, BPG2 (YL1) plays roles in chloroplast ribosome RNA maturation31. Kim et al. proposed that BPG2 (YL1) binds to plastid rRNA for chloroplast translation apparatus assembly32. AtNOA1/RIF1, a YqeH-type GTPase homolog in Arabidopsis, confers an atypical nitric oxide synthase activity, plastid ribosome function, and root growth behavior control38,39,40. OsNOA1 RNAi rice seedlings show chlorotic phenotype41. Nevertheless, no study has investigated the involvement of YqeH-type proteins in environmental stress response.
Na+ accumulation was high in the shoots of yl1-1 and yl1-2 under salt stress conditions (Fig. 4a), suggesting that the mechanisms of Na+ exclusion were interrupted because of the loss of YL1 functions. Na+ exclusion broadly refers to two mechanisms: Na+ efflux from root epidermis and decrease of Na+ delivery from root to shoot4. SOS pathway genes play vital roles in Na+ efflux in roots9,42. Homologs of SOS1 from rice, wheat, and tomato have been characterized in controlling Na+ transport43,44,45. YL1 was difficult to detect in roots, and the expression levels of SOS pathway genes were not altered in the yl1 mutants (Figs 3a and S6). Moreover, loss of YL1 functions did not clearly affect root development or ion homeostasis under controlled or high salinity conditions (Figs 4 and S1). These results suggest that the salt hypersensitivity of mutant shoots is not caused by the reduced Na+ efflux from root epidermis but probably by the increase of Na+ delivery from root to shoot. To our knowledge, Na+ delivery from root to shoot mainly depends on the transpiration stream in the xylem4,46,47. HKT1 could directly retrieve Na+ from the xylem sap back to the phloem of the shoot and unload it in the root, which in turn reduces Na+ accumulation in the shoot13,15. In wheat, TmHKT1;4-A2 is expressed in leaf sheath and reduces Na+ concentration in the shoot48. ABI4 has been recently reported to act as a negative regulator that could directly bind to the promoter region and inhibit HKT1 expression in Arabidopsis23. In the shoots of yl1 mutants, ABI4 was highly expressed, and the HKT1 transcription levels were approximately half of those of the wild type (Fig. 5). By contrast, in the shoots of the yl1-2 abi4 double mutant, the transcripts of HKT1 were evidently higher than those in yl1-2 (Fig. 6d). Furthermore, the loss of ABI4 function could rescue yl1-2 shoot salt-sensitive phenotypes (Fig. 6e,f). These observations suggest that HKT1 is depressed by high levels of ABI4 in shoots of yl1 mutants.
ABI4, which binds to the coupling element 1 (CE1, CACCG) in vitro, is involved in regulating a large number of genes49,50. ABI4 plays key roles in sugar, ABA, and pathogen-response signaling pathways through binding to S-box or G-box sequences51,52. Furthermore, ABI4 expression could be regulated by many factors. During seed maturation, germination, and the early-stage of seedling development, ABI4 could be induced by ABA and cytokinin and be repressed by auxin53,54. ABI4 could also directly bind to its own promoter CE1-like element and activate gene expression55. Previous studies also reported that ABI4 is significantly downregulated in the wrky18 and wrky40 mutant seedlings56. Moreover, the transcription factor SCR was also proven to directly bind to ABI4 promoter and negatively regulate ABI4 expression57.
At the seedling stage, similar to YL1, ABI4 expression is readily detected in shoots55. Our results showed that ABI4 was more highly expressed in the shoots of yl1 mutants compared with in the shoots of the wild type (Fig. 5c), whereas in the roots of yl1-1 or yl1-2, ABI4 expression was almost unchanged (Fig. 5a). Consistent with previous study23, ABI4 transcripts could be clearly increased by salt stress signaling in the wild type (Fig. 5c), which is contrary to YL1 (Fig. 3e,f). However, in shoots of yl1, the salinity effect on ABI4 expression was clearly weakened (Fig. 5c). The abovementioned results suggest that YL1 may be a potential factor that acts upstream of ABI4 in salt stress signaling because the YL1 expression is independent of ABI4 (Fig. 6c). Together with the phenotype of yl1-2 abi4 under salt stress treatment (Fig. 6e), we reveal that ABI4 plays roles in seedling shoot salt-sensitive phenotype of yl1 mutants. Considering that the YL1 protein localizes in the chloroplast and ABI4 is a nucleus-localized transcription factor, several signals may be derived from the chloroplast and effectively transmitted to the nucleus when YL1 dysfunctions. Several studies stated that ABI4 plays key roles in chloroplast retrograde signaling. ABI4, which could bind the Lhcb family gene promoter CCAC-motif, acts downstream of GUN1 and PTM and negatively regulates target gene transcriptions20,22,58. Thus, whether YL1 regulates ABI4 through retrograde signaling need to be identified in future studies.
Recent studies have identified several genes are involved in plant shoot response to salt stress46,47. Nevertheless, the regulatory mechanisms of plant shoot salt tolerance remain unclear. yl1-1 was investigated, and the results suggested that chloroplast proteins such as YL1 could be involved in plant salt stress response through nuclear stress-responsive factors. We speculate that the reduction of YL1 and induction of ABI4 under high salinity conditions may be an adaptive mechanism to achieve Na+ equilibrium in the entire plant, which needs to be investigated further. Although the signaling pathway is unclear, our results may open a new insight into the association of plant salt stress response and the chloroplast.
Methods
Plant Materials and Growth Condition
All wild type plants in the study are Arabidopsis thaliana Col-0. The mutants, yl1-1, yl1-2 (salk_068713), and abi4 (salk_080095) are in the Col-0 genetic background. NaClO (0.1%) and ethanol (70%) are used for seed sterilization. Seedlings are grown in solid 1/2 Murashige and Skoog (MS) medium containing 1% sucrose.
Constructs and plant transformation
For construct used in yl1-1com, CDS sequence of YL1 was subcloned into pBI121 vector with recognition sites for the restriction enzymes BamHI and SalI. For YL1-GSG-GFP (mYL1-GSG-GFP) construct, a flexible peptide GSG (GGGSSSSGGG) was added between YL1 (mYL1) CDS sequence of termination coden free and GFP sequence. The YL1-GSG-GFP (mYL1-GSG-GFP) sequence was also cloned into pBI121 vector. Constructs were introduced into Agrobacterium tumefaciens GV3101 for plant transformation. Transgenic plants were selected on 1/2 MS medium containing 50 mg L−1 kanamycin. Plants were selfed twice and T3 homozygous plants were used. Primers are listed in Table S1.
Stress Treatment and Phenotypes Recording
Surface sterilized seeds were plated in solid 1/2 MS medium described above containing 150 mM NaCl or not. Plated seeds were 4 °C-dark treated for 2 d, then transferred to incubator (22 °C, 16/8 h light/dark) and grown for 5 d. The number of plants with (n) or without (m) fully expanded cotyledons was recorded. Then the percentages of fully expanded seedling cotyledons were calculated with (n/n + m) *100%.
Transcript Analysis
For gene transcription analysis under salt conditions, 4-d-old seedlings grown under normal conditions were transferred to the MS medium containing 0, 100, or 150 mM NaCl and then treated for different times (2–36 h). The shoots and roots were separately harvested and total RNA was extracted with Trizol (invitrogen). cDNA synthesis was performed using PrimeScript reverse transcriptase (RT) with oligo dT primer using the PrimeScript RT master mix kit (Takara). qRT-PCR experiments were carried out by an CFX96 real-time PCR system (Bio-Rad, C1000) using SYBR green real-time PCR master mix (Takara). Detection for each gene transcript was performed for at least three biology replicates and each bio-replicate containing three technical replicates. All used primers are listed in Table S1.
YL1 Promoter Construction and GUS staining
The pYL1-GUS was constructed by subcloning a 1.5 kb fragment upstream of the YL1 translation start site into pBI122 binary vector. GUS staining analysis for different tissues and GUS activity analysis in seedlings for different treatment were according to Yan59. Fluorescence of GUS was measured with a Microplate Spectrofluorometer (IC Measurement Acc for FL Solutions, F-4500, HITACHI). Primers are listed in Table S1.
Immunoblot Analysis
Plant materials were prepared according to the methods described in Transcript analysis. Total proteins of such as seedling shoots were extracted using Plant Protein Extraction Kit (CWBIO) and measured using a nano-drop instrument (Nano-Drop, ND-1000 Spectrofluorometer). The polyclonal antibody of YL1 was obtained from rabbits by Abmart (China). The RbcL, D1 and Actin antibodies were all purchased from Agrisera. Proteins after electrophoresis were blotted to nitrocellulose membranes and then probed with specific antibodies. The nitrocellulose membranes were visualized with an enhanced Lumi-Light Western Blotting Substrate kit (Thermo Scientific).
Ion Content Measurement
For measurement to Na+ and K+. the 3-d-old seedlings grown under normal conditions were transferred to the MS medium containing 0 or 150 mM NaCl and then grown for 2 d. The shoots and roots from these 5-d-old plant were separately harvested, dried for 48 h at 80 °C and then ground to powder. The same mass tissue powder was digested in concentrated (69%, v/v) HNO3 for 24 h at room temperature for elemental extraction. Na+ and K+ concentrations was determined by atomic absorption spectrophotometry (novAA300, analytikjena).
Chloroplast Isolation
5-d-old seedling cotyledons were harvested and grounded in isolation buffer (20 mM HEPES/KOH, pH 8.0, 0.3 M sorbitol, 5 mM MgCl2, 5 mM EGTA, 5 mM EDTA, and 10 mM NaHCO3). The homogenate was filtered and centrifuged at 3000 g for 3 min. The pellet was resuspended in 1 mL isolation buffer. The intact chloroplasts are isolated through Percoll gradient method60. Thylakoid membranes and stroma proteins were prepared from isolated intact chloroplasts.
Chlorophyll content measurement
Cotyledons or leaves of plant were collected, fresh weighed and washed in distilled water. Chlorophyll was extracted in 80% (v/v) acetone at 25 °C in darkness for 24 h, and concentration of chlorophyll a/b was determined according to Komatsu31.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL data libraries under the following accession numbers: YL1, At3g57180; ABI4, At2g40220; HKT1, At4g10310; SOS1, At2g01980; SOS2, At5g35410; SOS3, At5g24270; NHX1, At5g27150; ABA1, At5g67030; ABA2, At1g52340; ABA3, At1g16540; AAO3, At2g27150; NCED3, At3g14440.
Additional Information
How to cite this article: Li, P.-C. et al. Arabidopsis YL1/BPG2 Is Involved in Seedling Shoot Response to Salt Stress through ABI4. Sci. Rep. 6, 30163; doi: 10.1038/srep30163 (2016).
Supplementary Material
Acknowledgments
We sincerely thank the two research groups of Komatsu T. and Kim B.-H for the great previous work on BPG2 (YL1). The present work was supported by the National Basic Research Program (Grant No. 2012CB114200), the National Natural Science Foundation (Grant No. 31370305), the Genetically Modified Organisms Breeding Major Projects (Grant No. 2014ZX08009-003-002), and the Natural Science Foundation of Shandong Province (Grant No. ZR2015CQ012) in China.
Footnotes
Author Contributions P.-C.L., J.-G.H., C.-A.W. and C.-C.Z. designed the study. P.-C.L., S.-W.Y., Y.-Y.L. and P.S. performed the research. P.-C.L. and J.-G.H. analyzed the data. P.-C.L. and C.-C.Z. contributed to writing the article. All authors reviewed the manuscript.
References
- Myouga F. et al. A heterocomplex of iron superoxide dismutases defends chloroplast nucleoids against oxidative stress and is essential for chloroplast development in Arabidopsis. Plant Cell 20, 3148–3162 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail A., Takeda S. & Nick P. Life and death under salt stress: same players, different timing? J. Exp. Bot. 65, 2963–2979 (2014). [DOI] [PubMed] [Google Scholar]
- Deinlein U. et al. Plant salt-tolerance mechanisms. Trends Plant Sci. 19, 371–379 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.-L. & Shi H. Physiological and molecular mechanisms of plant salt tolerance. Photosynth. Res. 115, 1–22 (2013). [DOI] [PubMed] [Google Scholar]
- Gupta B. & Huang B. Mechanism of salinity tolerance in plants: physiological, biochemical, and molecular characterization. Int. J. Genomics 2014, 701596 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J.-K. Genetic analysis of plant salt tolerance using Arabidopsis. Plant Physiol. 124, 941–948 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfter U., Ishitani M. & Zhu J.-K. The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc. Natl. Acad. Sci. USA 97, 3735–3740 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Ishitani M., Halfter U., Kim C.-S. & Zhu J.-K. The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc. Natl. Acad. Sci. USA 97, 3730–3734 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Ishitani M., Kim C. & Zhu J.-K. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. USA 97, 6896–6901 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Lee B. H., Wu S. J. & Zhu J. K. Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana. Nat. Biotechnol. 21, 81–85 (2003). [DOI] [PubMed] [Google Scholar]
- Rus A. et al. AtHKT1 facilitates Na+ homeostasis and K+ nutrition in planta. Plant Physiol. 136, 2500–2511 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthomieu P. et al. Functional analysis of AtHKT1 in Arabidopsis shows that Na+ recirculation by the phloem is crucial for salt tolerance. EMBO J. 22, 2004–2014 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunarpi et al. Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na+ unloading from xylem vessels to xylem parenchyma cells. Plant J. 44, 928–938 (2005). [DOI] [PubMed] [Google Scholar]
- Mäser P. et al. Altered shoot/root Na+ distribution and bifurcating salt sensitivity in Arabidopsis by genetic disruption of the Na+ transporter AtHKT1. FEBS Lett. 531, 157–161 (2002). [DOI] [PubMed] [Google Scholar]
- Møller I. S. et al. Shoot Na+ exclusion and increased salinity tolerance engineered by cell type–specific alteration of Na+ transport in Arabidopsis. Plant Cell 21, 2163–2178 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apse M. P., Aharon G. S., Snedden W. A. & Blumwald E. Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285, 1256–1258 (1999). [DOI] [PubMed] [Google Scholar]
- Barragan V. et al. Ion exchangers NHX1 and NHX2 mediate active potassium uptake into vacuoles to regulate cell turgor and stomatal function in Arabidopsis. Plant Cell 24, 1127–1142 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidi E. O. et al. The AtNHX1 exchanger mediates potassium compartmentation in vacuoles of transgenic tomato. Plant J. 61, 495–506 (2010). [DOI] [PubMed] [Google Scholar]
- Finkelstein R. R., Li Wang M., Lynch T. J., Rao S. & Goodman H. M. The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA2 domain protein. Plant Cell 10, 1043–1054 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koussevitzky S. et al. Signals from chloroplasts converge to regulate nuclear gene expression. Science 316, 715–719 (2007). [PubMed] [Google Scholar]
- Giraud E., Van Aken O., Ho L. H. & Whelan J. The transcription factor ABI4 is a regulator of mitochondrial retrograde expression of ALTERNATIVE OXIDASE1a. Plant Physiol. 150, 1286–1296 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X. et al. A chloroplast envelope-bound PHD transcription factor mediates chloroplast signals to the nucleus. Nat. Commun. 2, 477 (2011). [DOI] [PubMed] [Google Scholar]
- Shkolnik-Inbar D., Adler G. & Bar-Zvi D. ABI4 downregulates expression of the sodium transporter HKT1;1 in Arabidopsis roots and affects salt tolerance. Plant J. 73, 993–1005 (2013). [DOI] [PubMed] [Google Scholar]
- Hasegawa P. M., Bressan R. A., Zhu J.-K. & Bohnert H. J. Plant cellular and molecular responses to high salinity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 463–499 (2000). [DOI] [PubMed] [Google Scholar]
- Munns R. & Tester M. Mechanisms of Salinity Tolerance. Annu. Rev. Plant Biol. 59, 651–681 (2008). [DOI] [PubMed] [Google Scholar]
- Takahashi S. & Murata N. How do environmental stresses accelerate photoinhibition? Trends Plant Sci. 13, 178–182 (2008). [DOI] [PubMed] [Google Scholar]
- Murata N., Takahashi S., Nishiyama Y. & Allakhverdiev S. I. Photoinhibition of photosystem II under environmental stress. BBA-Bioenergetics 1767, 414–421 (2007). [DOI] [PubMed] [Google Scholar]
- Krasensky J., Broyart C., Rabanal F. A. & Jonak C. The redox-sensitive chloroplast trehalose-6-phosphate phosphatase AtTPPD regulates salt stress tolerance. Antioxid. Redox Sign. 21, 1289–1304 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana N., Chauhan H. & Khurana P. Characterization of a chloroplast localized wheat membrane protein (TaRCI) and its role in heat, drought and salinity stress tolerance in Arabidopsis thaliana. Plant Gene 4, 45–54 (2015). [Google Scholar]
- Tan J. et al. A novel chloroplast-localized pentatricopeptide repeat protein involved in splicing affects chloroplast development and abiotic stress response in rice. Mol. plant 7, 1329–1349 (2014). [DOI] [PubMed] [Google Scholar]
- Komatsu T. et al. The chloroplast protein BPG2 functions in brassinosteroid-mediated post-transcriptional accumulation of chloroplast rRNA. Plant J. 61, 409–422 (2010). [DOI] [PubMed] [Google Scholar]
- Kim B.-H., Malec P., Waloszek A. & Arnim A. Arabidopsis BPG2: a phytochrome-regulated gene whose protein product binds to plastid ribosomal RNAs. Planta 236, 677–690 (2012). [DOI] [PubMed] [Google Scholar]
- Anand B., Surana P. & Prakash B. Deciphering the catalytic machinery in 30S ribosome assembly GTPase YqeH. PLoS One 5, e9944 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand B., Verma S. K. & Prakash B. Structural stabilization of GTP-binding domains in circularly permuted GTPases: implications for RNA binding. Nucleic Acids Res. 34, 2196–2205 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynaud E. G. et al. Human Lsg1 defines a family of essential GTPases that correlates with the evolution of compartmentalization. BMC Biol. 3, 21 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uicker W. C., Schaefer L., Koenigsknecht M. & Britton R. A. The essential GTPase YqeH is required for proper ribosome assembly in Bacillus subtilis. J. Bacteriol. 189, 2926–2929 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh P. C., Morimoto T., Matsuo Y., Oshima T. & Ogasawara N. The GTP-binding protein YqeH participates in biogenesis of the 30S ribosome subunit in Bacillus subtilis. Genes Genet. Syst. 82, 281–289 (2007). [DOI] [PubMed] [Google Scholar]
- Zhao X. et al. NITRIC OXIDE-ASSOCIATED PROTEIN1 (AtNOA1) is essential for salicylic acid-induced root waving in Arabidopsis thaliana. New Phytol. 207, 211–224 (2015). [DOI] [PubMed] [Google Scholar]
- Guo F. Q., Okamoto M. & Crawford N. M. Identification of a plant nitric oxide synthase gene involved in hormonal signaling. Science 302, 100–103 (2003). [DOI] [PubMed] [Google Scholar]
- Gas E., Flores-Perez U., Sauret-Gueto S. & Rodriguez-Concepcion M. Hunting for plant nitric oxide synthase provides new evidence of a central role for plastids in nitric oxide metabolism. Plant Cell 21, 18–23 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. et al. OsNOA1/RIF1 is a functional homolog of AtNOA1/RIF1: implication for a highly conserved plant cGTPase essential for chloroplast function. New Phytol. 187, 83–105 (2010). [DOI] [PubMed] [Google Scholar]
- Zhu J.-K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53, 247–273 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Atienza J. et al. Conservation of the salt overly sensitive pathway in rice. Plant Physiol. 143, 1001–1012 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H. et al. Functional characterization of a wheat plasma membrane Na+/H+ antiporter in yeast. Arch. Biochem. Biophys. 473, 8–15 (2008). [DOI] [PubMed] [Google Scholar]
- Olias R. et al. The plasma membrane Na+/H+ antiporter SOS1 is essential for salt tolerance in tomato and affects the partitioning of Na+ between plant organs. Plant Cell Environ. 32, 904–916 (2009). [DOI] [PubMed] [Google Scholar]
- Jiang C. et al. ROS-mediated vascular homeostatic control of root-to-shoot soil Na delivery in Arabidopsis. EMBO J. 31, 4359–4370 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C., Belfield E. J., Cao Y., Smith J. A. C. & Harberd N. P. An Arabidopsis soil-salinity–tolerance mutation confers ethylene-mediated enhancement of sodium/potassium homeostasis. Plant Cell 25, 3535–3552 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Spielmeyer W., Lagudah E. S. & Munns R. Comparative mapping of HKT genes in wheat, barley, and rice, key determinants of Na+ transport, and salt tolerance. J. Exp. Bot. 59, 927–937 (2008). [DOI] [PubMed] [Google Scholar]
- Leon P. & Gregorio J. & Cordoba, E. ABI4 and its role in chloroplast retrograde communication. Front. Plant Sci. 3, 304 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y. F. et al. Binding of ABI4 to a CACCG motif mediates the ABA-induced expression of the ZmSSI gene in maize (Zea mays L.) endosperm. J. Exp. Bot. 63, 5979–5989 (2012). [DOI] [PubMed] [Google Scholar]
- Adie B. A. et al. ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. Plant Cell 19, 1665–1681 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeekens S. & Hellmann H. A. Sugar sensing and signaling in plants. Front. Plant Sci. 5, 113 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price J., Li T. C., Kang S. G., Na J. K. & Jang J. C. Mechanisms of glucose signaling during germination of Arabidopsis. Plant Physiol. 132, 1424–1438 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shkolnik-Inbar D. & Bar-Zvi D. Expression of ABSCISIC ACID INSENSITIVE 4 (ABI4) in developing Arabidopsis seedlings. Plant Signal. Behav. 6, 694–696 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossi F. et al. The Arabidopsis ABA-INSENSITIVE (ABI) 4 factor acts as a central transcription activator of the expression of its own gene, and for the induction of ABI5 and SBE2.2 genes during sugar signaling. Plant J. 59, 359–374 (2009). [DOI] [PubMed] [Google Scholar]
- Shang Y. et al. The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell 22, 1909–1935 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H., Hao Y. & Kong D. SCARECROW has a SHORT-ROOT-independent role in modulating the sugar response. Plant Physiol. 158, 1769–1778 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott A., Jung H.-S., Koussevitzky S. & Chory J. Plastid-to-Nucleus retrograde signaling. Annu. Rev. Plant Biol. 57, 739–759 (2006). [DOI] [PubMed] [Google Scholar]
- Yan K. et al. Stress-induced alternative splicing provides a mechanism for the regulation of MicroRNA processing in Arabidopsis thaliana. Mol. Cell 48, 521–531 (2012). [DOI] [PubMed] [Google Scholar]
- Stöckel J. & Oelmüller R. A novel protein for photosystem I biogenesis. J. Biol. Chem. 279, 10243–10251 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.