Abstract
HIV-1 Gag is the master orchestrator of particle assembly. The central role of Gag at multiple stages of the HIV lifecycle has led to efforts to develop drugs that directly target Gag and prevent the formation and release of infectious particles. Until recently, however, only the catalytic site protease inhibitors have been available to inhibit late stages of HIV replication. This review summarizes the current state of development of antivirals that target Gag or disrupt late events in the retrovirus lifecycle such as maturation of the viral capsid. Maturation inhibitors represent an exciting new series of antiviral compounds, including those that specifically target CA-SP1 cleavage and the allosteric integrase inhibitors that inhibit maturation by a completely different mechanism. Numerous small molecules and peptides targeting CA have been studied in attempts to disrupt steps in assembly. Efforts to target CA have recently gained have considerable momentum from the development of small molecules that bind CA and alter capsid stability at the post-entry stage of the lifecycle. Efforts to develop antivirals that inhibit incorporation of genomic RNA or to inhibit late budding events remain in preliminary stages of development. Overall, the development of novel antivirals targeting Gag and the late stages in HIV replication appears much closer to success than ever, with the new maturation inhibitors leading the way.
Keywords: Gag, HIV, antiretroviral, virus assembly, capsid, matrix, nucleocapsid, allosteric integrase inhibitors, bevirimat
1. INTRODUCTION AND SCOPE OF THIS REVIEW
The development of highly-active antiretroviral therapy for treating HIV infection has been a remarkable success story that has turned a once fatal infection into a chronic, manageable condition [1]. Current FDA-approved antiretroviral drugs target viral reverse transcriptase, protease, integrase, and entry processes (coreceptor or fusion blockade). It is desirable to continue to develop new antiretrovirals directed against alternative targets in the virus lifecycle in order to further optimize therapeutic options, overcome resistance to existing medications, and potentially contribute to elimination of viral reservoirs. There are currently no drugs in clinical use that specifically target HIV assembly, budding, or release. HIV-1 Gag has been the target of a number of attempts to produce antivirals but to date only the protease inhibitors act at late stages of the lifecycle. This review focuses on some new developments in targeting late events in the lifecycle, including HIV-1 assembly, budding, and maturation. The basic mechanisms of HIV-1 assembly will first be discussed in order to provide the reader with a basis for understanding the targeted proteins and processes. We will then turn to the most promising new classes of inhibitors that act during assembly and maturation of the viral core. Other targets for which recent progress has not been robust will only be briefly discussed. The reader is also referred to a number of previously published reviews of inhibitors directed against Gag and the HIV-1 assembly pathway for further background information [2–5].
2. GAG AND HIV-1 PARTICLE ASSEMBLY
HIV-1 is a member of the Lentivirus genus of the family Retroviridae. Retroviral genomes encode gag (group-specific antigen) genes as an essential component of their genome. Retroviral gag genes encode Gag proteins that play a number of critical roles in the viral lifecycle. Gag proteins are perhaps best known as the master directors of the process of virus assembly, and for their roles in generating the immature capsid shell and mature core of the virion (for reviews, see [6–9]). Expression of Gag protein in numerous cell types generates virus-like particle formation in the absence of all other viral gene products, a characteristic that illustrates the central role of Gag in particle formation. For the purposes of this review, we will limit our discussion to the HIV-1 Gag protein, a 55-kilodalton protein also known as Pr55Gag. In the remainder of this text we will usually refer to HIV-1 Pr55Gag as Gag, while making distinctions for individual Gag cleavage products where appropriate.
Gag is translated from unspliced viral RNA on free cytosolic ribosomes. An important fatty acid modification occurs during translation of Gag, the addition of the 14-carbon myristic acid moiety to the N-terminus of Gag by cellular N-myristoyl transferase [10–11]. In the absence of myristoylation, viral assembly is defective and no infectious particles are formed. Myristic acid, together with other signals in MA, directs the normal targeting of Gag to the plasma membrane of the cell, and plays an important role in mediating membrane interactions as further discussed below [12–13]. An important concept in HIV assembly is the role of Gag as a polyprotein precursor. Individual domains of Gag are referred to in the context of their subsequent proteolytic cleavage products, designated from N- to C-terminus as matrix (MA), capsid (CA), spacer peptide-1 (SP1), nucleocapsid (NC), spacer peptide-2 (SP2) and p6. In the context of the full-length, uncleaved precursor Gag polyprotein, functional domains exist within these major regions, but additional functional domains may cross the proteolytic cleavage site, such that major rearrangements and changes in functional characteristics are present in the full-length protein as compared to the subunit cleavage products. Two examples of major conformational and functional changes that occur during Gag cleavage are the N-terminal hairpin of CA, which forms only following cleavage at the MA-CA junction, and the alpha helical CA-SP1 segment that plays an important structural role in the immature core but is lost upon cleavage. The general organization of Gag and its cleavage products is illustrated in Fig. 1.
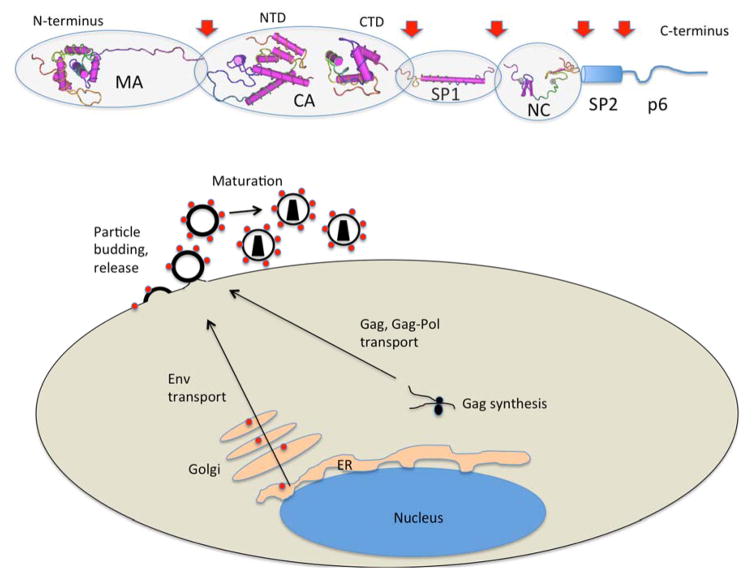
Fig. (1). HIV-1 Gag and Basics of Particle Assembly.
The HIV Gag polyprotein is represented at the top of the figure, using structures of individual regions of the protein. Proteolytic cleavage sites are indicated by red arrows. The structures represented are from [61, 158–161] with PDB IDs from N- to C-terminus: 2HMX (MA), 2GOL (CA NTD), 1A8O (CA CTD), 1U57 (SP1), 1F6U (NC). Structures were prepared using CN3d version 4.3. Below a cell is represented to illustrate Gag protein formation on cytosolic ribosomes and assembly and budding at the plasma membrane. The process of maturation of the core occurs during or immediately following particle budding.
Gag traverses the cytoplasm to reach the plasma membrane by an as-yet poorly understood mechanism. Some studies suggest that Gag is first translated in a pericentriolar location followed by directional outward transport to the plasma membrane [14–15]. It was frequently assumed that Gag must travel in an active, directional manner and that specific transport mechanisms such as motor-driven, cytoskeleton-mediated transport would become apparent. With the exception of reports of the involvement of the kinesin KIF4 in Gag trafficking [16–17], this plausible hypothesis of directed trafficking of Gag has not been well substantiated. Studies with Gag-GFP fusion proteins appear to show diffusion of Gag throughout the cytoplasm, followed by multimerization and assembly at punctate sites on the plasma membrane, rather than a sequential, directional outward movement of Gag [18–20]. In such studies, Gag first appears as a faint but diffuse cytosolic blush that increases with time (and folding of GFP), followed by the development of concentrated puncta representing developing particle buds on the plasma membrane. It is therefore safe to say that the field has not yet clearly identified trafficking pathways for Gag that could serve as targets for antiretroviral therapy.
The way in which Gag interacts with cellular membranes is an area that is better understood than Gag’s mechanism of trafficking. The N-terminal MA domain of Gag is the key determinant of the plasma membrane interactions of the full-length Pr55Gag molecule. An important development in this area was the discovery that MA interacts with phosphatidylinositol (4,5) bisphosphate (PI(4,5)P2) on the cytosolic face of the plasma membrane [21]. The structural basis for this interaction has been well described, and helps to explain the role of myristic acid in the assembly of HIV particles [22–24]. Myristate is normally somewhat sequestered within a pocket on the membrane-binding face of the globular head of MA. Upon interaction with membranes containing PI(4,5)P2, a conformational change allows myristate to interact with the lipid bilayer. PI(4,5)P2 thus acts as a trigger for a “myristoyl switch” that contributes to membrane interactions. Additional electrostatic interactions occur between basic residues on the membrane-binding face of MA and negatively-charged phospholipids of the cytosolic face of the plasma membrane, so that insertion of myristate into the bilayer, together with electrostatic interactions, constitute the primary Gag-membrane interactions [24–25]. In vitro liposome binding studies have confirmed the importance of electrostatic interactions between basic residues on MA and negatively-charged phospholipids and demonstrate enhancement of binding upon addition of PI(4,5)P2 [25–27]. Multimerization of Gag is also required for stable interactions with the membrane, and the avidity of interactions increases as the Gag lattice forms and enlarges during the particle budding process. An interesting recent finding in this area is that RNA interactions with the MA domain can inhibit membrane interactions [28–29]. The dynamics of RNA-mediated inhibition of binding vs. PI(4,5)P2-mediated interactions in vivo are not yet entirely clear, but there is evidence that PI(4,5)P2-containing liposomes can out-compete nucleic acids for MA binding [30]. Another important and related finding resulted from a recent study examining Gag-RNA interactions by crosslinking-immunoprecipitation. This study revealed that while the NC region mediates interactions with viral genomic RNA, the MA region binds to cytosolic tRNAs [31]. Thus the tRNA interaction may play a part in modulating membrane interactions, with displacement of tRNA required for MA-membrane interactions to occur. The interactions of Gag with the plasma membrane have been more extensively summarized in several recent reviews [32–34].
In an infected cell, Gag does not assemble virus particles in isolation, but instead must recruit other viral (and potentially cellular) components to the assembly site. Gag-pol fusion proteins in the form of Pr160Gag-Pol are generated through a ribosomal frameshifting event. Gag-Pol proteins are incorporated into the developing particle bud at a ratio of approximately one Gag-Pol per 20 Gag molecules [35]. During particle budding, concentration of Pol within the developing virion and potentially a cellular membrane component leads to dimerization and activation of the protease domain, which autocatalytically cleaves itself out of the polyprotein and subsequently cleaves Gag and Gag-Pol molecules into their mature constituents in an ordered, sequential manner [36–37]. Overexpression of Gag-Pol causes premature activation of protease, and inhibits particle formation [38]. This tightly regulated process could potentially be targeted by antivirals, and there is evidence outlined below that compounds that bind to Pol proteins can indeed disrupt assembly events (see section on ALLINIs below). The viral genomic RNA is selectively incorporated into the developing particle as a dimer through interactions of the psi region of the RNA with the NC region of Gag [39–40], and this step represents another potential target relevant to Gag’s role in assembly. The site of first interaction of RNA with Gag is unresolved, with some evidence favoring incorporation at the plasma membrane as opposed to locations within the cytosol [41–42]. The HIV envelope glycoprotein complex (Env) is also recruited to the developing particle in a manner that also remains under some study. Env clearly takes a different route than Gag to reach the assembly site, traversing the secretory pathway as a trimer prior to reaching the plasma membrane [43–44]. My group has presented evidence favoring a model in which the Env trimers on the plasma membrane are subsequently internalized and directed to the endosomal recycling compartment, where Rab11-FIP1C and Rab14 subsequently direct their outward movement to the particle budding site [45]. Thus, while Gag directs the overall process of membrane interaction and particle budding, the assembly and budding process is actually a coordinated process in which Gag, Gag-Pol, Env, and viral RNA all come together on a concentrated location on the plasma membrane in order to form infectious particles. Conceivably, Gag itself or any of these Gag interactors could be targets for antiviral drug development.
Gag-Gag multimerization is an essential component of the particle assembly process that deserves additional discussion. Knowledge of the process of Gag-Gag multimerization was greatly facilitated by early studies showing that Gag forms particle-like structures in vitro [46–49]. Under appropriate conditions, purified CA proteins can self-assemble to form organized tubes. These tubes have proven quite useful in defining the structural nature of the mature core of HIV, including recent high-resolution CryoEM studies and X-ray crystallographic studies that together have produced a complete structural model of the mature core, and advanced knowledge of immature core structure [50–54]. Expression of wildtype Pr55Gag or Gag mutants lacking the p6 domain has been utilized extensively to study the formation of the initial immature Gag core. Nucleic acids can trigger Gag-Gag multimerization and spherical particle formation in vitro [46, 55]. Particles formed in vitro that were significantly smaller than the Gag immature cores observed in cells were converted to a more representative size through the addition of an inositol phosphate (IP6), and biophysical studies revealed a change from a more globular to an extended conformation through this addition [56–57]. This observation suggested that Gag is initially folded and more compact, and upon membrane interactions forms an extended conformation necessary for oligomerization. Single-molecule FRET experiments supported this model, again suggesting that there is a conformational switch to an extended conformation that may be triggered by MA interactions with phosphoinositides at the plasma membrane, promoting subsequent oligomerization [58]. These in vitro and single molecule experiments are consistent with cell-based experiments in which three components appear essential to successful particle assembly: membrane interactions (including a role for myristoylation), NC-RNA interactions, and direct Gag-Gag interactions [59–60].
Our current understanding of Gag-Gag multimerization has benefitted from a large and growing number of NMR and crystal structures of Gag protein subunits, as well as structures of assemblies derived from from cryoEM and cryo-electron tomographic studies as already mentioned. A complete discussion of the body of structural data relevant to Gag is beyond the scope of this review. However, discussion of some of the essential information relevant to the current development of inhibitors of assembly is warranted. The CA protein consists of two major domains, an N-terminal domain (NTD) and C-terminal domain (CTD), connected by a flexible linker. Critical interactions occur between helix 2 of the CTD, forming an important dimer interface [61]. A number of additional functional surfaces in CA that are essential to proper core assembly were identified by mutagenesis, infectivity, and EM studies [62]. Interactions are not limited to the CTD but also are critical in the NTD [63]. Recent cryoEM studies of the mature core reveal new molecular interactions that could be targeted by antivirals [53]. Yet another productive development that should be of benefit to the development of anti-Gag compounds is the increasing knowledge of the interactions of Trim5alpha and other innate restriction factors with the viral capsid [64–65]. Structural studies of cellular factors bound to the capsid are increasingly revealing binding pockets that can be similarly targeted by drugs [66], some of which will be described later in this review.
The process of HIV particle assembly has been the subject of a number of reviews in recent years. We refer the interested reader to reviews that more comprehensively describe each of the steps that have been presented briefly here. These include comprehensive reviews of HIV assembly [67–70], reviews of Gag-membrane interactions [32–34], and reviews of the structural basis of HIV assembly [71–72].
3. INHIBITORS TARGETING GAG, ASSEMBLY PROCESSES, AND MATURATION
There are currently 37 licensed antiretroviral drugs or drug combinations available for human use [73]. Of the licensed compounds, only the protease inhibitors can be thought of as acting at the stage of assembly and maturation. The current FDA-approved HIV-1 protease inhibitors are competitive inhibitors of the active site of the enzyme, and function by inhibiting cleavage of Gag and Gag-Pol, resulting in the production of non-infectious virions and the arrest of further replication [36]. Here we will not discuss the well-studied protease inhibitors, but will instead concentrate on newer compounds, focusing on those that directly bind to Gag itself or inhibit other functions of Gag during assembly or post-entry events of the viral lifecycle.
3.1. Maturation Inhibitors targeting CA-SP1 cleavage
The cleavage of Pr55Gag into its subunits and subsequent formation of a conical viral core is known as maturation. Maturation of retroviral particles is an ordered process that occurs simultaneously with or immediately following particle budding [36–37, 74]. During maturation a sequential cleavage of Gag occurs that is regulated by the rate of processing at each individual cleavage site [37]. Initially there is rapid cleavage at the C-terminus of SP1, followed by cleavage between MA and CA. Additional cleavage events then occur, with the final (slowest) reaction separating SP1 from the C-terminus of CA. This final cleavage event is critical in proper virion morphogenesis, as disruption of this cleavage site through mutagenesis leads to aberrant core formation and the generation of non-infectious particles. The CA-SP1 junction lies within a predicted alpha helical segment of Gag within the immature Gag lattice that is thought to stabilize the immature lattice, thus suggesting that cleavage at this site triggers a conformational switch that is critical to subsequent formation of the mature conical core [75–76]. Recent cryoEM data are compatible with the formation of a six-helix bundle by this region of Gag [52]. A derivative of betulinic acid, 3-O-[3′,3′-dimethylsuccinyl]-betulinic acid with the trade name Bevirimat (BVM), previously known as PA-457 or DSB, was found to inhibit HIV replication by inhibiting maturation [77–80]. This compound inhibits cleavage at the CA-SP1 site, resulting in accumulation of a p25 precursor product when studied in vitro [79, 81]. BVM was thought to bind to a pocket near the CA-SP1 junction, based upon resistance mutations that arise in the immediate vicinity of this cleavage site when virus is grown in the presence of the drug [82–83] . BVM and its approximate location of action is indicated in Fig. 2. Incorporation of BVM into immature particles but not mature particles was demonstrated, suggesting that the binding pocket is present in the immature Gag lattice and lost upon proteolytic cleavage [80]. Direct binding to CA-SP1 was confirmed using photoaffinity crosslinking and mass spectrometry, and interestingly also indicated some crosslinking with the major homology region of Gag [84]. Notably, resistance mutations derived in response to PF-46396, a maturation inhibitor of differing chemical structure from BVM, mapped to the CA-SP1 cleavage site, CA residue 201, and also to the major homology region (MHR) [85]. These results are significant because they suggest there is a functional relationship between the MHR and the CA-SP1 region, two domains known to be critical to assembly and maturation, and because they help to define a maturation inhibitor binding pocket that can be the target for further generations of inhibitors.
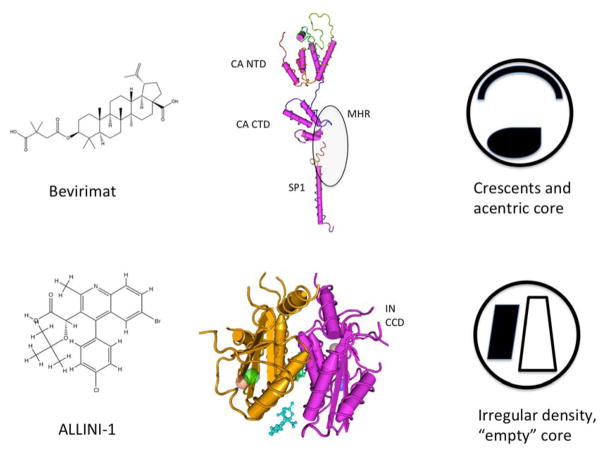
Fig. (2). Distinct mechanism of action of maturation inhibitors.
Bevirimat (top) inhibits CA-SP1 cleavage, binding to a pocket that is not fully elucidated but includes key residues in SP1 and in the CA CTD surrounding the CA-SP1 cleavage site. MHR = major homology region. Disruption of CA-SP1 cleavage results in aberrant particle formation with a membrane crescent of Gag and an acentric core. The ALLINIs act by a very distinct mechanism, binding to the LEDGF binding region at the dimer interface of the IN catalytic core domains (CCDs) and promoting aberrant multimerization. Particles treated with these inhibitors display a displaced density (presumably the viral RNA) and an empty core. Structures shown are from [51, 105, 161] with PDB IDs of 4USN (CA), 1U57 (SP1), and 4GW6 (IN + ALLINI).
One reason that the maturation inhibitors are of great interest is that they represent the first drugs directly targeting Gag that have been taken into clinical trials and proven to inhibit HIV in vivo. BVM was developed and advanced to phase II clinical trials by Panacos Pharmaceuticals (Watertown, MA) [86–90]. The drug was generally safe and well-tolerated, and resulted in significant reductions in viral loads in approximately 50% of patients. The lack of efficacy of BVM in the non-responders was a result of naturally occurring polymorphisms in the patient viral isolates that mapped to sites downstream from the CA-SP1 cleavage site. Follow-up laboratory studies demonstrated that these polymporphisms diminished the ability of BVM to inhibit CA-SP1 cleavage [82, 86–87]. The partial efficacy led to the discontinuation of further product development of BVM itself as a clinical antiretroviral. However, these studies provided proof that maturation inhibitors can work in humans, and have resulted in the development of new derivatives of betulinic acid that are active against BVM-resistant viruses [91–92]. Development of maturation inhibitors for clinical use is therefore progressing. An exciting report from the Conference on Retroviruses and Opportunistic Infections reported that BMS-955176, a second-generation maturation inhibitor, elicited promising declines in viral loads of individuals with wildtype HIV-1 or with HIV-1 encoding the Gag polymorphisms that had led to BVM resistance. This drug is currently in phase 2b testing [93].
3.2. Allosteric integrase inhibitors
The allosteric integrase (IN) inhibitors or ALLINIs (also referred to as LEDGINs) represent a new and exciting class of inhibitors that unexpectedly block replication at the stage of maturation of the viral core. Although these are not strictly targeted against Gag, we will discuss them here because they target steps in core maturation similar to the CA-SP1 binding pocket-based maturation inhibitors described above. However, ALLINIs inhibit maturation through a distinct and fascinating mechanism. Unlike the approved IN inhibitors (raltegravir, dolutegravir, elvitegravir), the ALLINIs were designed not as strand transfer inhibitors but rather as molecules that block the lens epithelium-derived growth factor (LEDGF/p75) binding site on the IN protein. LEDGF/p75 is a host protein that binds to an IN tetramer that itself is bound to viral DNA. LEDGF tethers the IN complex to chromatin sites, predominantly within actively transcribing genes (reviewed in [94–96]). The discovery of LEDGF/p75 itself as a bona fide IN binding protein responsible for attaching the IN complex to chromatin elicited great excitement, and the development of inhibitors of the interaction naturally followed. Initial efforts to develop inhibitors were aided by careful structural studies. Cocrystallization of the integrase binding domain or IBD on the LEDGF molecule bound to an integrase catalytic core domain (CCD) dimer revealed a clear binding pocket at the CCD dimer interface [97]. Peptides based on the LEDGF/p75 IBD were found to enhance IN tetramerization while blocking IN-DNA and IN-LEDGF/p75 binding, suggesting that the IN-LEDGF/p75 interaction could be a unique target for antiretroviral therapy [98–99]. Subsequently two groups identified quinoline compounds that inhibit LEDGF-IN binding and catalytic activity in vitro, arriving at these compounds through very different strategies [100–101]. Christ and colleagues used molecular modeling to identify candidates and then synthesized them and evaluated them for binding to the IN CCD and for antiviral activity, designating their lead quinolines LEDGINs [100]. Fenwick and colleagues discovered similar compounds (now designated as members of Boehringer Ingelheim or BI compounds) using a functional high-throughput screen [101–102]. The lead compounds initially appeared promising by in vitro assays, and further derivatives have improved upon their antiviral activity, with a reported EC50 of <15 nM for some of the BI compounds. Additional studies confirmed that the ALLINIs indeed inhibited integration by promoting IN multimerization in the absence of viral DNA as expected [103–104]. The structural basis of ALLINI interactions within the LEDF-binding pocket between CCDs has been well studied, and is depicted in Fig. 2b [105]. Surprisingly, however, there was more to the antiviral activity of the ALLINIs than could be explained by inhibition of integration. Viruses produced in the presence of ALLINIs were found to be non-infectious, indicating a block during late stages of replication [106–108]. In fact, the principal antiviral effect of the ALLINIs occurs during late stage events [107]. ALLINIs did not alter particle quantity or the incorporation of genomic RNA. Furthermore, ALLINIs did not affect Gag protein processing, in contrast to the betulinic acid-derived maturation inhibitors described previously. Remarkably, particle core morphology was defective, with the majority of viral particles showing an eccentric positioning of electron dense material and often an empty-appearing core. This suggests that the viral ribonucleoprotein complex (RNP) is misplaced to a location outside of the conical core during particle maturation. ALLINIs enhanced intra-virion multimerization of IN, suggesting that the enhanced multimerization is a key factor in disrupting the maturation process [107, 109–110].
The discovery of this new class of antiretrovirals with a truly unique mechanism of action is an exciting development for several reasons. This provides a second very viable means of inhibiting maturation of virions, and one that is unlikely to exhibit cross-resistance with bevirimat or its derivatives. The ALLINIs also act by a unique mechanism that is entirely distinct from the strand transfer inhibitors of IN, so that they may prove useful in combination and should also inhibit HIV-1 mutants that are resistant to the current integrase inhibitors. Perhaps even more valuable is the discovery of a class of inhibitors that appear to act through eliciting abnormal levels or stability of multimerization. The ALLINIs are now entering clinical trials, and it seems likely that they will be an effective addition to the current antiretroviral armamentarium.
3.3. Inhibitors targeting Gag
In addition to the two classes of maturation inhibitors discussed above, there have been efforts to target other steps in the assembly process. Some of these have run their course and are no longer being actively pursued for clinical development, while others continue in preclinical stages of investigation. In the following section we will discuss efforts to target specific target sites on Gag as well as Gag-membrane interactions for the development of antiviral therapy.
3.3.1. Inhibitors of MA-membrane interactions
The interaction of the MA region of Gag with cellular membranes is essential to the assembly process, so efforts to block this interaction have been pursued in hopes of developing effective assembly inhibitors. N-terminal myristoylation of Gag is essential for particle production, and plays important roles in subcellular localization and membrane interactions of Gag. Myristic acid is added to the developing Gag polypeptide co-translationally through the action of cellular N-myristoyl transferase [10–11]. This offered an attractive early target for antiretroviral development, and competitive inhibitors of cellular N-myristoyl transferase were evaluated as early as 1991 [111–112]. Although these inhibitors had reasonable effects on particle production in cell culture and inhibited acylation of Gag with an IC50 of approximately 10 μg/ml [112], they proved too toxic to be practical for drug development. This is not surprising, given the number of important host cellular proteins that are myristoylated.
The interactions of the viral MA protein with the inner leaflet of the plasma membrane offer other potential therapeutic targets. A direct interaction between MA and PI(4,5)P2 plays an important role in myristic acid exposure and membrane interactions [21–22]. Potentially inhibiting this interaction could be targeted, as suggested by cellular studies in which depletion or relocalization of PI(4,5)P2 disrupts normal assembly. One group has pursued efforts to identify small molecules that bind to the PI(4,5)P2 binding site for antiviral development [113–114]. Using in silico screening, these investigators identified a compound that bound MA and inhibited HIV replication with an IC50 value of 7.5–15.6 μM. Mutations surrounding the PI(4,5)P2 binding site reduced binding of one lead compound, providing some evidence that this molecule indeed targets the intended site. It remains to be seen if further optimization will make this strategy therapeutically viable.
3.3.2. Inhibitors targeting CA
CA is potentially an extremely attractive target for the development of inhibitors, as it plays a central role in multimerization of Gag, forms a number of interfaces critical to immature particle formation, forms the crucial conical core of the virus, and is the natural target of potent post-entry restriction factors such as TRIM5α [115] and TRIMCyp [116]. A large body of structural information is now available regarding the CA-CA interactions within the mature conical core [53, 71], providing a basis for rational drug design. Some current evidence favors the complete disassembly of the immature Gag core prior to the reassembly of the mature capsid during maturation [117], which would provide a multitude of interacting sites and transitional stages that might be targeted to inhibit subsequent infectious particle formation. Furthermore, the incoming core interacts with host transportins and nuclear pore proteins such as TNP03 [118], Nup153 and Nup358 [119–120], and CPSF6 [121] prior to and during nuclear entry of the preintegration complex. Altogether these interactions provide a broad number of potentially “druggable” interfaces.
CAP-1 is a small molecule reported by the Summers laboratory in 2003 as the first CA-directed inhibitor of HIV-1 assembly [122]. CAP-1 bound to a surface-exposed region of the NTD of CA in both mature CA and uncleaved Gag, and revealed an NTD binding pocket that has subsequently been identified as the target of other inhibitors. CAP-1 and its derivatives inhibited HIV replication in cell culture and appeared to exert an effect on capsid maturation. Binding affinities and IC50s were relatively low. Detailed structural data for CAP-1 in complex with the CA NTD was reported in 2007 [123], revealing a deep hydrophobic binding pocket into which CAP-1 binds and displaces Phe32 of the NTD while inducing a conformational change in the NTD. Fig. 3A illustrates CAP-1 binding to the CA NTD. Modifications of CAP-1 have been pursued, as another group used CAP-1 as the lead compound to synthesize additional inhibitors using a thiourea scaffold [124]. Remarkably, the CAP-1 binding site on the NTD has been separately identified as the likely binding site for capsid inhibitors in a screen conducted by the Prevelige group [125–126]. These inhibitors had relatively high IC50 values, in the range of 10 μM, and it is unclear if these acyhydrozone-based CAP-1 binding site inhibitors themselves are being further pursued. However, an independent screen by another group yielded benzodiazepine compounds that inhibited CA polymerization in the μM range and targeted the same CAP-1 binding site on the CA NTD [127]. Further work along these lines led to the discovery of a series of benzodiazepines and benzimidazoles that also bind the NTD in a location similar to that of CAP-1 [128] (Fig. 3B). The benzodiazepines and benzimidazoles induce a pocket in the NTD that overlaps the CAP-1 binding site but that also expands the pocket significantly. These inhibitors are much more potent, with EC50s in the 60–70 nM range. Interestingly, they block replication by very different mechanisms. The benzodiazepines derived in this study inhibit assembly and particle release, while the benzimidazoles block formation of the mature capsid. Taking all these studies together, the CAP-1 binding pocket appears to be a viable target on the NTD that has been repeatedly identified in screens for inhibitors of capsid assembly and continues to be targeted for drug development.
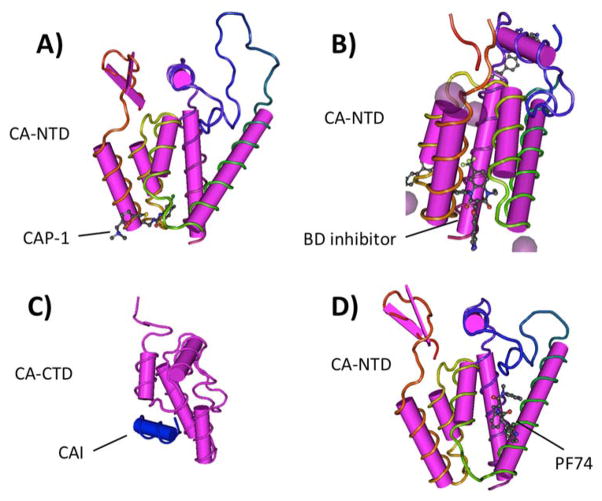
Fig. (3). CA inhibitors and binding pockets.
Binding sites for distinct inhibitors are shown. A) CAP-1 binding pocket on the CA NTD. B) Benzodiazepine (BD) inhibitor is shown also binding in the CAP-1 binding pocket of the NTD; note the NTD orientation is changed from (A) to emphasize the inhibitor binding pocket. C) CAI is a peptide binding to an interface on the CA CTD. D) PF74 binds to a distinct pocket on the CA NTD; orientation of the NTD is similar to that shown in (A). Structures are from [123, 128, 162–163] with PDB IDs of 2JPR (CAP-1), 4E91 (BD), 3DS1 (CAI), and 2XDE (PF74).
A distinct CA binding compound targeting a different site termed capsid assembly inhibitor or CAI was reported in 2005 by the Krausslich group [129]. They identified the CAI peptide through screening a phage display library, and showed that this peptide inhibited in vitro assembly of CA tubes and immature VLPs, suggesting a binding site present in both forms of Gag. There was no inhibition of viral replication in cell culture, as the peptide did not enter cells. However, CAI was suggested as a lead compound for drug development and as a probe of a unique binding site where other compounds might bind. Structural studies of the complex revealed that CAI bound as an amphipathic helix to a conserved hydrophobic groove on the CA CTD, where it may act to disrupt the CTD dimer interface [130] (Fig. 3C). Further development of the basic CAI peptide has been actively pursued. Modifications to the peptide were used to stabilize its alpha helical structure and to convert it to a cell-penetrating peptide. The resulting modified peptide, termed NYAD-1, was able to penetrate cells, bound the CTD with higher affinity, and showed some relatively weak inhibition of particle formation from 293T cells [131–132]. Additional derivatives of this peptide as well as additional peptides designed to inhibit the dimer interface of CA have been developed and tested for in vitro and cell culture effects [133–134]. These studies provide proof-of-principle that rationally-designed inhibitors of key CA-CA interfaces can disrupt particle formation. They are limited thus far in clinical application, however, as the inhibitory activity remains relatively weak and the ability to deliver peptides intracellularly remains challenging.
CA-binding inhibitors of HIV replication can also act at post-entry stage of the virus lifecycle. This is best represented by the Pfizer compound PF-3450074 (PF74)[135] and by the pyrrolopyrazolone HIV-1 inhibitors BI-1 and BI-2 from Boehringer Ingelheim [136]. PF74 and the BI compounds bind to the same binding pocket on the CA NTD, which is distinct from the CAP-1 binding pocket previously discussed. The location of this alternative NTD binding site is indicated in Fig. 3D. PF74 binding was suggested to accelerate uncoating, and its antiviral effect could be partially inhibited by cyclosporine. The binding pocket for PF74 as solved in complex with the CA hexamer lies at the NTD-CTD interface, and overlaps with that of CPSF6 and NUP153 [65]. Thus this drug may be blocking a normal binding site for cellular factors that serve to assist in nuclear import of the capsid. Adding to this very intriguing story, the BI compounds bind the same pocket, but instead of enhancing uncoating, they appear to stabilize the viral capsid and inhibit uncoating. These new inhibitors therefore act at a similar post-entry stage of the viral lifecycle and appear to disrupt interactions with host cell proteins, while exerting opposing effects on capsid stability.
CA is an attractive target because of its relatively high conservation among HIV-1 isolates and its sensitivity to mutagenesis [137–138] A variety of additional inhibitors have been reported that target CA. The interested reader is referred to more comprehensive reviews of this topic [2, 139]. Overall activity in this area appears to be robust and promising. The ability to inhibit HIV replication using small molecules that bind with distinct mechanisms and at distinct sites on CA, along with the identification of key CA binding host factors that target in some cases the identical binding pockets, makes this an area of keen interest. It seems likely that there will be clinically-relevant inhibitors of assembly in the near future that target CA. Furthermore, studies of some of the newer inhibitors will continue to inform our understanding of basic aspects of assembly and uncoating.
3.3.3. Inhibition of NC-RNA interactions
NC plays a number of crucial roles in the virus lifecycle, including genomic RNA packaging, contributing to Gag-Gag multimerization, and acting as a nucleic acid chaperone and promoting reverse transcription. It is not surprising therefore that NC has been the target of attempts to develop antiretroviral drugs. Molecules that inhibit zinc incorporation in the NC zinc fingers will not be discussed here but have been previously reviewed [140–141]. NC has in general proven to be a very difficult drug target to date. An interesting target related to HIV assembly is the genomic RNA binding of NC. The highly structured RNA elements in the 5′ UTR of the RNA can potentially be targeted to prevent interactions with NC. The 5′ UTR includes the transactivation responsive element (TAR), the poly A tract, the primer binding site (PBS), and four stem-loops (SL1-SL4) that make up the packaging domain. TAR has been targeted using anthraquinone compounds in in vitro studies [142]. A variety of peptides that disrupt viral binding to NC have been reported [143–146]. These studies seem to establish that the NC-psi RNA interaction can be specifically inhibited, but these efforts have not progressed past early preclinical proof-of-concept studies. More recently, compounds that disrupt the formation of NC-SL3 complex were identified thorugh a high-throughput screen [147]. Several of these compounds bound to the SL3 loop with μM affinity. Bell and colleagues identified a small molecule inhibitor of Gag-psi interactions that was specific for the intact Pr55GagΔp6 protein [148]. These studies again seem quite early in terms of any clinical application. Perhaps more encouraging are NC inhibitors from Boehringer Ingelheim that have been shown to inhibit HIV replication in cell culture with EC50s in the micromolar range [149]. In summary, efforts to target NC-RNA interactions are being pursued but appear to be at a very early stage of development.
3.3.4. Inhibition of ESCRT-mediated particle release
The C-terminal p6 domain of Gag includes motifs that link Gag to the cellular endosomal sorting complex required for transport (ESCRT) pathway (reviewed in [150–151]). The PTAP motif in p6 binds to ESCRT component TSG101, while the YPXnL motif links to ESCRT component ALIX [152–154]. Depletion of ESCRT components from cells produces a powerful block to HIV particle release [155], as does the use of dominant-negative forms of ESCRT components [156]. The importance of ESCRT pathway interactions to viral replication suggests that targeting the Gag-ESCRT interactions could be an additional productive target for the development of antiretrovirals. To date, however, small molecules or other compounds targeting this pathway as HIV inhibitors have not been reported, with the exception of a cyclic peptide inhibiting Gag-TSG101 binding [157]. This appears to be an area worthy of further work in the future.
4. CONCLUDING REMARKS AND PERSPECTIVES FOR THE FUTURE
The development of antiretrovirals targeting Gag has generally been slow and somewhat disappointing up to this point, especially when compared with the relatively rapid and highly successful development of inhibitors of reverse transcriptase, integrase, and protease. It appears that this is now poised to change. The maturation inhibitors targeting CA-SP1 cleavage appear very promising, and clinical trials with second-generation maturation inhibitors are underway. The ALLINIs are a recent and very exciting category of maturation inhibitors that target maturation through a unique mechanism. CA inhibitors of assembly have been under study for many years without moving much closer to the clinic. Molecules targeting the NTD-CTD junction such as PF74 and the BI compounds appear poised to change this, however, as they have dramatic and unprecedented effects on capsid stability and interfere with a key binding site on the CA hexamer for host proteins. Thus the maturation inhibitors and several new compounds targeting CA provide much reason for optimism that novel drugs targeting late stages of the HIV lifecycle will soon be part of the antiretroviral armamentarium.
Acknowledgments
P. S. acknowledges support from NIH R01 GM111027, R01 AI058858, R01 AI111863 and from the NIH/NICHD-supported IMPAACT and NIH/NIAID VTEU clinical trials networks.
Footnotes
CONFLICT OF INTEREST
The author confirms that he has no conflict of interest with the content of this review.
References
- 1.Palmisano L, Vella S. A brief history of antiretroviral therapy of HIV infection: success and challenges. Ann Ist Super Sanita. 2011;47(1):44–8. doi: 10.4415/ANN_11_01_10. [DOI] [PubMed] [Google Scholar]
- 2.Bocanegra R, Rodriguez-Huete A, Fuertes MA, Del Alamo M, Mateu MG. Molecular recognition in the human immunodeficiency virus capsid and antiviral design. Virus Res. 2012;169(2):388–410. doi: 10.1016/j.virusres.2012.06.016. [DOI] [PubMed] [Google Scholar]
- 3.Jiang Y, Liu X, De Clercq E. New therapeutic approaches targeted at the late stages of the HIV-1 replication cycle. Curr Med Chem. 2011;18(1):16–28. doi: 10.2174/092986711793979751. [DOI] [PubMed] [Google Scholar]
- 4.Prevelige PE., Jr New approaches for antiviral targeting of HIV assembly. J Mol Biol. 2011;410(4):634–40. doi: 10.1016/j.jmb.2011.03.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waheed AA, Freed EO. HIV type 1 Gag as a target for antiviral therapy. AIDS Res Hum Retroviruses. 2012;28(1):54–75. doi: 10.1089/aid.2011.0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wills JW, Craven RC. Form, function, and use of retroviral gag proteins. Aids. 1991;5(6):639–54. doi: 10.1097/00002030-199106000-00002. [DOI] [PubMed] [Google Scholar]
- 7.Swanstrom R, Wills JW. Synthesis, Assembly, and Processing of Viral Proteins. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor (NY): 1997. [PubMed] [Google Scholar]
- 8.Vogt VM. Retroviral Virions and Genomes. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor (NY): 1997. [PubMed] [Google Scholar]
- 9.Freed EO. HIV-1 gag proteins: diverse functions in the virus life cycle. Virology. 1998;251(1):1–15. doi: 10.1006/viro.1998.9398. [DOI] [PubMed] [Google Scholar]
- 10.Gottlinger HG, Sodroski JG, Haseltine WA. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proceedings of the National Academy of Sciences of the United States of America. 1989;86(15):5781–5. doi: 10.1073/pnas.86.15.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bryant M, Ratner L. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proceedings of the National Academy of Sciences of the United States of America. 1990;87(2):523–7. doi: 10.1073/pnas.87.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hermida-Matsumoto L, Resh MD. Localization of human immunodeficiency virus type 1 Gag and Env at the plasma membrane by confocal imaging. Journal of virology. 2000;74(18):8670–9. doi: 10.1128/jvi.74.18.8670-8679.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Provitera P, El-Maghrabi R, Scarlata S. The effect of HIV-1 Gag myristoylation on membrane binding. Biophys Chem. 2006;119(1):23–32. doi: 10.1016/j.bpc.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 14.Poole E, Strappe P, Mok HP, Hicks R, Lever AM. HIV-1 Gag-RNA interaction occurs at a perinuclear/centrosomal site; analysis by confocal microscopy and FRET. Traffic. 2005;6(9):741–55. doi: 10.1111/j.1600-0854.2005.00312.x. [DOI] [PubMed] [Google Scholar]
- 15.Perlman M, Resh MD. Identification of an intracellular trafficking and assembly pathway for HIV-1 gag. Traffic. 2006;7(6):731–45. doi: 10.1111/j.1398-9219.2006.00428.x. [DOI] [PubMed] [Google Scholar]
- 16.Martinez NW, Xue X, Berro RG, Kreitzer G, Resh MD. Kinesin KIF4 regulates intracellular trafficking and stability of the human immunodeficiency virus type 1 Gag polyprotein. Journal of virology. 2008;82(20):9937–50. doi: 10.1128/JVI.00819-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang Y, Winkler U, Freed EO, Torrey TA, Kim W, Li H, Goff SP, Morse HC., 3rd Cellular motor protein KIF-4 associates with retroviral Gag. Journal of virology. 1999;73(12):10508–13. doi: 10.1128/jvi.73.12.10508-10513.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jouvenet N, Bieniasz PD, Simon SM. Imaging the biogenesis of individual HIV-1 virions in live cells. Nature. 2008;454(7201):236–40. doi: 10.1038/nature06998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jouvenet N, Simon SM, Bieniasz PD. Visualizing HIV-1 assembly. J Mol Biol. 2011;410(4):501–11. doi: 10.1016/j.jmb.2011.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hubner W, Chen P, Del Portillo A, Liu Y, Gordon RE, Chen BK. Sequence of human immunodeficiency virus type 1 (HIV-1) Gag localization and oligomerization monitored with live confocal imaging of a replication-competent, fluorescently tagged HIV-1. Journal of virology. 2007;81(22):12596–607. doi: 10.1128/JVI.01088-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ono A, Ablan SD, Lockett SJ, Nagashima K, Freed EO. Phosphatidylinositol (4,5) bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(41):14889–94. doi: 10.1073/pnas.0405596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saad JS, Ablan SD, Ghanam RH, Kim A, Andrews K, Nagashima K, Soheilian F, Freed EO, Summers MF. Structure of the myristylated human immunodeficiency virus type 2 matrix protein and the role of phosphatidylinositol-(4,5)-bisphosphate in membrane targeting. J Mol Biol. 2008;382(2):434–47. doi: 10.1016/j.jmb.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saad JS, Loeliger E, Luncsford P, Liriano M, Tai J, Kim A, Miller J, Joshi A, Freed EO, Summers MF. Point mutations in the HIV-1 matrix protein turn off the myristyl switch. J Mol Biol. 2007;366(2):574–85. doi: 10.1016/j.jmb.2006.11.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saad JS, Miller J, Tai J, Kim A, Ghanam RH, Summers MF. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(30):11364–9. doi: 10.1073/pnas.0602818103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chukkapalli V, Hogue IB, Boyko V, Hu WS, Ono A. Interaction between the human immunodeficiency virus type 1 Gag matrix domain and phosphatidylinositol-(4,5)-bisphosphate is essential for efficient gag membrane binding. Journal of virology. 2008;82(5):2405–17. doi: 10.1128/JVI.01614-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dalton AK, Ako-Adjei D, Murray PS, Murray D, Vogt VM. Electrostatic interactions drive membrane association of the human immunodeficiency virus type 1 Gag MA domain. Journal of virology. 2007;81(12):6434–45. doi: 10.1128/JVI.02757-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dick RA, Goh SL, Feigenson GW, Vogt VM. HIV-1 Gag protein can sense the cholesterol and acyl chain environment in model membranes. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(46):18761–6. doi: 10.1073/pnas.1209408109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chukkapalli V, Inlora J, Todd GC, Ono A. Evidence in support of RNA-mediated inhibition of phosphatidylserine-dependent HIV-1 Gag membrane binding in cells. Journal of virology. 2013;87(12):7155–9. doi: 10.1128/JVI.00075-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Inlora J, Collins DR, Trubin ME, Chung JY, Ono A. Membrane binding and subcellular localization of retroviral Gag proteins are differentially regulated by MA interactions with phosphatidylinositol-(4,5)-bisphosphate and RNA. MBio. 2014;5(6):e02202. doi: 10.1128/mBio.02202-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alfadhli A, Still A, Barklis E. Analysis of human immunodeficiency virus type 1 matrix binding to membranes and nucleic acids. Journal of virology. 2009;83(23):12196–203. doi: 10.1128/JVI.01197-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kutluay SB, Zang T, Blanco-Melo D, Powell C, Jannain D, Errando M, Bieniasz PD. Global changes in the RNA binding specificity of HIV-1 gag regulate virion genesis. Cell. 2014;159(5):1096–109. doi: 10.1016/j.cell.2014.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dick RA, Vogt VM. Membrane interaction of retroviral Gag proteins. Front Microbiol. 2014;5:187. doi: 10.3389/fmicb.2014.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olety B, Ono A. Roles played by acidic lipids in HIV-1 Gag membrane binding. Virus Res. 2014;193:108–15. doi: 10.1016/j.virusres.2014.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vlach J, Saad JS. Structural and molecular determinants of HIV-1 Gag binding to the plasma membrane. Front Microbiol. 2015;6:232. doi: 10.3389/fmicb.2015.00232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jacks T, Power MD, Masiarz FR, Luciw PA, Barr PJ, Varmus HE. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature. 1988;331(6153):280–3. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- 36.Konvalinka J, Krausslich HG, Muller B. Retroviral proteases and their roles in virion maturation. Virology. 2015;479–480C:403–417. doi: 10.1016/j.virol.2015.03.021. [DOI] [PubMed] [Google Scholar]
- 37.Wiegers K, Rutter G, Kottler H, Tessmer U, Hohenberg H, Krausslich HG. Sequential steps in human immunodeficiency virus particle maturation revealed by alterations of individual Gag polyprotein cleavage sites. Journal of virology. 1998;72(4):2846–54. doi: 10.1128/jvi.72.4.2846-2854.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park J, Morrow CD. Overexpression of the gag-pol precursor from human immunodeficiency virus type 1 proviral genomes results in efficient proteolytic processing in the absence of virion production. Journal of virology. 1991;65(9):5111–7. doi: 10.1128/jvi.65.9.5111-5117.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lawrence DC, Stover CC, Noznitsky J, Wu Z, Summers MF. Structure of the intact stem and bulge of HIV-1 Psi-RNA stem-loop SL1. J Mol Biol. 2003;326(2):529–42. doi: 10.1016/s0022-2836(02)01305-0. [DOI] [PubMed] [Google Scholar]
- 40.Webb JA, Jones CP, Parent LJ, Rouzina I, Musier-Forsyth K. Distinct binding interactions of HIV-1 Gag to Psi and non-Psi RNAs: implications for viral genomic RNA packaging. RNA. 2013;19(8):1078–88. doi: 10.1261/rna.038869.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jouvenet N, Simon SM, Bieniasz PD. Imaging the interaction of HIV-1 genomes and Gag during assembly of individual viral particles. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(45):19114–9. doi: 10.1073/pnas.0907364106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuzembayeva M, Dilley K, Sardo L, Hu WS. Life of psi: how full-length HIV-1 RNAs become packaged genomes in the viral particles. Virology. 2014;454–455:362–70. doi: 10.1016/j.virol.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Checkley MA, Luttge BG, Freed EO. HIV-1 envelope glycoprotein biosynthesis, trafficking, and incorporation. J Mol Biol. 2011;410(4):582–608. doi: 10.1016/j.jmb.2011.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Otteken A, Earl PL, Moss B. Folding, assembly, and intracellular trafficking of the human immunodeficiency virus type 1 envelope glycoprotein analyzed with monoclonal antibodies recognizing maturational intermediates. Journal of virology. 1996;70(6):3407–15. doi: 10.1128/jvi.70.6.3407-3415.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qi M, Williams JA, Chu H, Chen X, Wang JJ, Ding L, Akhirome E, Wen X, Lapierre LA, Goldenring JR, Spearman P. Rab11-FIP1C and Rab14 direct plasma membrane sorting and particle incorporation of the HIV-1 envelope glycoprotein complex. PLoS pathogens. 2013;9(4):e1003278. doi: 10.1371/journal.ppat.1003278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Campbell S, Rein A. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. Journal of virology. 1999;73(3):2270–9. doi: 10.1128/jvi.73.3.2270-2279.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gross I, Hohenberg H, Krausslich HG. In vitro assembly properties of purified bacterially expressed capsid proteins of human immunodeficiency virus. Eur J Biochem. 1997;249(2):592–600. doi: 10.1111/j.1432-1033.1997.t01-1-00592.x. [DOI] [PubMed] [Google Scholar]
- 48.Campbell S, Vogt VM. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. Journal of virology. 1995;69(10):6487–97. doi: 10.1128/jvi.69.10.6487-6497.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ehrlich LS, Krausslich HG, Wimmer E, Carter CA. Expression in Escherichia coli and purification of human immunodeficiency virus type 1 capsid protein (p24) AIDS Res Hum Retroviruses. 1990;6(10):1169–75. doi: 10.1089/aid.1990.6.1169. [DOI] [PubMed] [Google Scholar]
- 50.Woodward CL, Cheng SN, Jensen GJ. Electron cryotomography studies of maturing HIV-1 particles reveal the assembly pathway of the viral core. Journal of virology. 2015;89(2):1267–77. doi: 10.1128/JVI.02997-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schur FK, Hagen WJ, Rumlova M, Ruml T, Muller B, Krausslich HG, Briggs JA. Structure of the immature HIV-1 capsid in intact virus particles at 8.8 A resolution. Nature. 2015;517(7535):505–8. doi: 10.1038/nature13838. [DOI] [PubMed] [Google Scholar]
- 52.Bharat TA, Castillo Menendez LR, Hagen WJ, Lux V, Igonet S, Schorb M, Schur FK, Krausslich HG, Briggs JA. Cryo-electron microscopy of tubular arrays of HIV-1 Gag resolves structures essential for immature virus assembly. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(22):8233–8. doi: 10.1073/pnas.1401455111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao G, Perilla JR, Yufenyuy EL, Meng X, Chen B, Ning J, Ahn J, Gronenborn AM, Schulten K, Aiken C, Zhang P. Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics. Nature. 2013;497(7451):643–6. doi: 10.1038/nature12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gres AT, Kirby KA, KewalRamani VN, Tanner JJ, Pornillos O, Sarafianos SG. STRUCTURAL VIROLOGY. X-ray crystal structures of native HIV-1 capsid protein reveal conformational variability. Science. 2015;349(6243):99–103. doi: 10.1126/science.aaa5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Datta SA, Rein A. Preparation of recombinant HIV-1 gag protein and assembly of virus-like particles in vitro. Methods Mol Biol. 2009;485:197–208. doi: 10.1007/978-1-59745-170-3_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Campbell S, Fisher RJ, Towler EM, Fox S, Issaq HJ, Wolfe T, Phillips LR, Rein A. Modulation of HIV-like particle assembly in vitro by inositol phosphates. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(19):10875–9. doi: 10.1073/pnas.191224698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Datta SA, Zhao Z, Clark PK, Tarasov S, Alexandratos JN, Campbell SJ, Kvaratskhelia M, Lebowitz J, Rein A. Interactions between HIV-1 Gag molecules in solution: an inositol phosphate-mediated switch. J Mol Biol. 2007;365(3):799–811. doi: 10.1016/j.jmb.2006.10.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Munro JB, Nath A, Farber M, Datta SA, Rein A, Rhoades E, Mothes W. A conformational transition observed in single HIV-1 Gag molecules during in vitro assembly of virus-like particles. Journal of virology. 2014;88(6):3577–85. doi: 10.1128/JVI.03353-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dou J, Wang JJ, Chen X, Li H, Ding L, Spearman P. Characterization of a myristoylated, monomeric HIV Gag protein. Virology. 2009;387(2):341–52. doi: 10.1016/j.virol.2009.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li H, Dou J, Ding L, Spearman P. Myristoylation is required for human immunodeficiency virus type 1 Gag-Gag multimerization in mammalian cells. Journal of virology. 2007;81(23):12899–910. doi: 10.1128/JVI.01280-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gamble TR, Yoo S, Vajdos FF, von Schwedler UK, Worthylake DK, Wang H, McCutcheon JP, Sundquist WI, Hill CP. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science. 1997;278(5339):849–53. doi: 10.1126/science.278.5339.849. [DOI] [PubMed] [Google Scholar]
- 62.von Schwedler UK, Stray KM, Garrus JE, Sundquist WI. Functional surfaces of the human immunodeficiency virus type 1 capsid protein. Journal of virology. 2003;77(9):5439–50. doi: 10.1128/JVI.77.9.5439-5450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lanman J, Lam TT, Emmett MR, Marshall AG, Sakalian M, Prevelige PE., Jr Key interactions in HIV-1 maturation identified by hydrogen-deuterium exchange. Nat Struct Mol Biol. 2004;11(7):676–7. doi: 10.1038/nsmb790. [DOI] [PubMed] [Google Scholar]
- 64.Ganser-Pornillos BK, Chandrasekaran V, Pornillos O, Sodroski JG, Sundquist WI, Yeager M. Hexagonal assembly of a restricting TRIM5alpha protein. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(2):534–9. doi: 10.1073/pnas.1013426108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bhattacharya A, Alam SL, Fricke T, Zadrozny K, Sedzicki J, Taylor AB, Demeler B, Pornillos O, Ganser-Pornillos BK, Diaz-Griffero F, Ivanov DN, Yeager M. Structural basis of HIV-1 capsid recognition by PF74 and CPSF6. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(52):18625–30. doi: 10.1073/pnas.1419945112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Price AJ, Jacques DA, McEwan WA, Fletcher AJ, Essig S, Chin JW, Halambage UD, Aiken C, James LC. Host cofactors and pharmacologic ligands share an essential interface in HIV-1 capsid that is lost upon disassembly. PLoS pathogens. 2014;10(10):e1004459. doi: 10.1371/journal.ppat.1004459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sundquist WI, Krausslich HG. HIV-1 assembly, budding, and maturation. Cold Spring Harb Perspect Med. 2012;2(7):a006924. doi: 10.1101/cshperspect.a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Balasubramaniam M, Freed EO. New insights into HIV assembly and trafficking. Physiology (Bethesda) 2011;26(4):236–51. doi: 10.1152/physiol.00051.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee SK, Potempa M, Swanstrom R. The choreography of HIV-1 proteolytic processing and virion assembly. J Biol Chem. 2012;287(49):40867–74. doi: 10.1074/jbc.R112.399444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.O’Carroll IP, Soheilian F, Kamata A, Nagashima K, Rein A. Elements in HIV-1 Gag contributing to virus particle assembly. Virus Res. 2013;171(2):341–5. doi: 10.1016/j.virusres.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ganser-Pornillos BK, Yeager M, Pornillos O. Assembly and architecture of HIV. Adv Exp Med Biol. 2012;726:441–65. doi: 10.1007/978-1-4614-0980-9_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ganser-Pornillos BK, Yeager M, Sundquist WI. The structural biology of HIV assembly. Curr Opin Struct Biol. 2008;18(2):203–17. doi: 10.1016/j.sbi.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. [accessed June 15, 2015];FDA-approved HIV medicines. https://aidsinfo.nih.gov/education-materials/fact-sheets/21/58/fda-approved-hiv-medicines.
- 74.Vogt VM. Proteolytic processing and particle maturation. Curr Top Microbiol Immunol. 1996;214:95–131. doi: 10.1007/978-3-642-80145-7_4. [DOI] [PubMed] [Google Scholar]
- 75.Gross I, Hohenberg H, Wilk T, Wiegers K, Grattinger M, Muller B, Fuller S, Krausslich HG. A conformational switch controlling HIV-1 morphogenesis. EMBO J. 2000;19(1):103–13. doi: 10.1093/emboj/19.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liang C, Hu J, Russell RS, Roldan A, Kleiman L, Wainberg MA. Characterization of a putative alpha-helix across the capsid-SP1 boundary that is critical for the multimerization of human immunodeficiency virus type 1 gag. Journal of virology. 2002;76(22):11729–37. doi: 10.1128/JVI.76.22.11729-11737.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fujioka T, Kashiwada Y, Kilkuskie RE, Cosentino LM, Ballas LM, Jiang JB, Janzen WP, Chen IS, Lee KH. Anti-AIDS agents, 11. Betulinic acid and platanic acid as anti-HIV principles from Syzigium claviflorum, and the anti-HIV activity of structurally related triterpenoids. J Nat Prod. 1994;57(2):243–7. doi: 10.1021/np50104a008. [DOI] [PubMed] [Google Scholar]
- 78.Kanamoto T, Kashiwada Y, Kanbara K, Gotoh K, Yoshimori M, Goto T, Sano K, Nakashima H. Anti-human immunodeficiency virus activity of YK-FH312 (a betulinic acid derivative), a novel compound blocking viral maturation. Antimicrobial agents and chemotherapy. 2001;45(4):1225–30. doi: 10.1128/AAC.45.4.1225-1230.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li F, Goila-Gaur R, Salzwedel K, Kilgore NR, Reddick M, Matallana C, Castillo A, Zoumplis D, Martin DE, Orenstein JM, Allaway GP, Freed EO, Wild CT. PA-457: a potent HIV inhibitor that disrupts core condensation by targeting a late step in Gag processing. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(23):13555–60. doi: 10.1073/pnas.2234683100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhou J, Chen CH, Aiken C. The sequence of the CA-SP1 junction accounts for the differential sensitivity of HIV-1 and SIV to the small molecule maturation inhibitor 3-O-{3′ 3′-dimethylsuccinyl}-betulinic acid. Retrovirology. 2004;1:15. doi: 10.1186/1742-4690-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sakalian M, McMurtrey CP, Deeg FJ, Maloy CW, Li F, Wild CT, Salzwedel K. 3-O-(3′,3′-dimethysuccinyl) betulinic acid inhibits maturation of the human immunodeficiency virus type 1 Gag precursor assembled in vitro. Journal of virology. 2006;80(12):5716–22. doi: 10.1128/JVI.02743-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Adamson CS, Ablan SD, Boeras I, Goila-Gaur R, Soheilian F, Nagashima K, Li F, Salzwedel K, Sakalian M, Wild CT, Freed EO. In vitro resistance to the human immunodeficiency virus type 1 maturation inhibitor PA-457 (Bevirimat) Journal of virology. 2006;80(22):10957–71. doi: 10.1128/JVI.01369-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhou J, Chen CH, Aiken C. Human immunodeficiency virus type 1 resistance to the small molecule maturation inhibitor 3-O-(3′,3′-dimethylsuccinyl)-betulinic acid is conferred by a variety of single amino acid substitutions at the CA-SP1 cleavage site in Gag. Journal of virology. 2006;80(24):12095–101. doi: 10.1128/JVI.01626-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nguyen AT, Feasley CL, Jackson KW, Nitz TJ, Salzwedel K, Air GM, Sakalian M. The prototype HIV-1 maturation inhibitor bevirimat binds to the CA-SP1 cleavage site in immature Gag particles. Retrovirology. 2011;8:101. doi: 10.1186/1742-4690-8-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Waki K, Durell SR, Soheilian F, Nagashima K, Butler SL, Freed EO. Structural and functional insights into the HIV-1 maturation inhibitor binding pocket. PLoS pathogens. 2012;8(11):e1002997. doi: 10.1371/journal.ppat.1002997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Knapp DJ, Harrigan PR, Poon AF, Brumme ZL, Brockman M, Cheung PK. In vitro selection of clinically relevant bevirimat resistance mutations revealed by “deep” sequencing of serially passaged, quasispecies-containing recombinant HIV-1. Journal of clinical microbiology. 2011;49(1):201–8. doi: 10.1128/JCM.01868-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lu W, Salzwedel K, Wang D, Chakravarty S, Freed EO, Wild CT, Li F. A single polymorphism in HIV-1 subtype C SP1 is sufficient to confer natural resistance to the maturation inhibitor bevirimat. Antimicrobial agents and chemotherapy. 2011;55(7):3324–9. doi: 10.1128/AAC.01435-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Qian K, Bori ID, Chen CH, Huang L, Lee KH. Anti-AIDS agents 90. novel C-28 modified bevirimat analogues as potent HIV maturation inhibitors. Journal of medicinal chemistry. 2012;55(18):8128–36. doi: 10.1021/jm301040s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Smith PF, Ogundele A, Forrest A, Wilton J, Salzwedel K, Doto J, Allaway GP, Martin DE. Phase I and II study of the safety, virologic effect, and pharmacokinetics/pharmacodynamics of single-dose 3-o-(3′,3′-dimethylsuccinyl)betulinic acid (bevirimat) against human immunodeficiency virus infection. Antimicrobial agents and chemotherapy. 2007;51(10):3574–81. doi: 10.1128/AAC.00152-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Temesgen Z, Feinberg JE. Drug evaluation: bevirimat--HIV Gag protein and viral maturation inhibitor. Current opinion in investigational drugs. 2006;7(8):759–65. [PubMed] [Google Scholar]
- 91.Dang Z, Ho P, Zhu L, Qian K, Lee KH, Huang L, Chen CH. New betulinic acid derivatives for bevirimat-resistant human immunodeficiency virus type-1. Journal of medicinal chemistry. 2013;56(5):2029–37. doi: 10.1021/jm3016969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dang Z, Qian K, Ho P, Zhu L, Lee KH, Huang L, Chen CH. Synthesis of betulinic acid derivatives as entry inhibitors against HIV-1 and bevirimat-resistant HIV-1 variants. Bioorganic & medicinal chemistry letters. 2012;22(16):5190–4. doi: 10.1016/j.bmcl.2012.06.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hwang C, Sevinsky H, Ravindran P, Xiao H, Ray N, Shurmann D, Sobotha C, Krystal M, Dicker I, Lataillade M. Antiviral activity/safety of a second-generation HIV-1 maturation inhibitor. Conference on Retroviruses and Opportunistic Infections; Seattle, WA. 2015. [Google Scholar]
- 94.Christ F, Debyser Z. The LEDGF/p75 integrase interaction, a novel target for anti-HIV therapy. Virology. 2013;435(1):102–9. doi: 10.1016/j.virol.2012.09.033. [DOI] [PubMed] [Google Scholar]
- 95.Engelman A, Cherepanov P. The lentiviral integrase binding protein LEDGF/p75 and HIV-1 replication. PLoS pathogens. 2008;4(3):e1000046. doi: 10.1371/journal.ppat.1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Poeschla EM. Integrase, LEDGF/p75 and HIV replication. Cell Mol Life Sci. 2008;65(9):1403–24. doi: 10.1007/s00018-008-7540-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cherepanov P, Ambrosio AL, Rahman S, Ellenberger T, Engelman A. Structural basis for the recognition between HIV-1 integrase and transcriptional coactivator p75. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(48):17308–13. doi: 10.1073/pnas.0506924102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Al-Mawsawi LQ, Christ F, Dayam R, Debyser Z, Neamati N. Inhibitory profile of a LEDGF/p75 peptide against HIV-1 integrase: insight into integrase-DNA complex formation and catalysis. FEBS Lett. 2008;582(10):1425–30. doi: 10.1016/j.febslet.2008.02.076. [DOI] [PubMed] [Google Scholar]
- 99.Hayouka Z, Rosenbluh J, Levin A, Loya S, Lebendiker M, Veprintsev D, Kotler M, Hizi A, Loyter A, Friedler A. Inhibiting HIV-1 integrase by shifting its oligomerization equilibrium. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(20):8316–21. doi: 10.1073/pnas.0700781104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Christ F, Voet A, Marchand A, Nicolet S, Desimmie BA, Marchand D, Bardiot D, Van der Veken NJ, Van Remoortel B, Strelkov SV, De Maeyer M, Chaltin P, Debyser Z. Rational design of small-molecule inhibitors of the LEDGF/p75-integrase interaction and HIV replication. Nat Chem Biol. 2010;6(6):442–8. doi: 10.1038/nchembio.370. [DOI] [PubMed] [Google Scholar]
- 101.Fenwick C, Amad M, Bailey MD, Bethell R, Bos M, Bonneau P, Cordingley M, Coulombe R, Duan J, Edwards P, Fader LD, Faucher AM, Garneau M, Jakalian A, Kawai S, Lamorte L, LaPlante S, Luo L, Mason S, Poupart MA, Rioux N, Schroeder P, Simoneau B, Tremblay S, Tsantrizos Y, Witvrouw M, Yoakim C. Preclinical profile of BI 224436, a novel HIV-1 non-catalytic-site integrase inhibitor. Antimicrobial agents and chemotherapy. 2014;58(6):3233–44. doi: 10.1128/AAC.02719-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Han YS, Quashie P, Mesplede T, Xu H, Mekhssian K, Fenwick C, Wainberg MA. A high-throughput assay for HIV-1 integrase 3′-processing activity using time-resolved fluorescence. Journal of virological methods. 2012;184(1–2):34–40. doi: 10.1016/j.jviromet.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 103.Christ F, Shaw S, Demeulemeester J, Desimmie BA, Marchand A, Butler S, Smets W, Chaltin P, Westby M, Debyser Z, Pickford C. Small-molecule inhibitors of the LEDGF/p75 binding site of integrase block HIV replication and modulate integrase multimerization. Antimicrobial agents and chemotherapy. 2012;56(8):4365–74. doi: 10.1128/AAC.00717-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tsiang M, Jones GS, Niedziela-Majka A, Kan E, Lansdon EB, Huang W, Hung M, Samuel D, Novikov N, Xu Y, Mitchell M, Guo H, Babaoglu K, Liu X, Geleziunas R, Sakowicz R. New class of HIV-1 integrase (IN) inhibitors with a dual mode of action. J Biol Chem. 2012;287(25):21189–203. doi: 10.1074/jbc.M112.347534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Feng L, Sharma A, Slaughter A, Jena N, Koh Y, Shkriabai N, Larue RC, Patel PA, Mitsuya H, Kessl JJ, Engelman A, Fuchs JR, Kvaratskhelia M. The A128T resistance mutation reveals aberrant protein multimerization as the primary mechanism of action of allosteric HIV-1 integrase inhibitors. J Biol Chem. 2013;288(22):15813–20. doi: 10.1074/jbc.M112.443390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jurado KA, Engelman A. Multimodal mechanism of action of allosteric HIV-1 integrase inhibitors. Expert Rev Mol Med. 2013;15:e14. doi: 10.1017/erm.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jurado KA, Wang H, Slaughter A, Feng L, Kessl JJ, Koh Y, Wang W, Ballandras-Colas A, Patel PA, Fuchs JR, Kvaratskhelia M, Engelman A. Allosteric integrase inhibitor potency is determined through the inhibition of HIV-1 particle maturation. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(21):8690–5. doi: 10.1073/pnas.1300703110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Le Rouzic E, Bonnard D, Chasset S, Bruneau JM, Chevreuil F, Le Strat F, Nguyen J, Beauvoir R, Amadori C, Brias J, Vomscheid S, Eiler S, Levy N, Delelis O, Deprez E, Saib A, Zamborlini A, Emiliani S, Ruff M, Ledoussal B, Moreau F, Benarous R. Dual inhibition of HIV-1 replication by integrase-LEDGF allosteric inhibitors is predominant at the post-integration stage. Retrovirology. 2013;10:144. doi: 10.1186/1742-4690-10-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Balakrishnan M, Yant SR, Tsai L, O’Sullivan C, Bam RA, Tsai A, Niedziela-Majka A, Stray KM, Sakowicz R, Cihlar T. Non-catalytic site HIV-1 integrase inhibitors disrupt core maturation and induce a reverse transcription block in target cells. PLoS One. 2013;8(9):e74163. doi: 10.1371/journal.pone.0074163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Desimmie BA, Schrijvers R, Demeulemeester J, Borrenberghs D, Weydert C, Thys W, Vets S, Van Remoortel B, Hofkens J, De Rijck J, Hendrix J, Bannert N, Gijsbers R, Christ F, Debyser Z. LEDGINs inhibit late stage HIV-1 replication by modulating integrase multimerization in the virions. Retrovirology. 2013;10:57. doi: 10.1186/1742-4690-10-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Devadas B, Lu T, Katoh A, Kishore NS, Wade AC, Mehta PP, Rudnick DA, Bryant ML, Adams SP, Li Q, et al. Substrate specificity of Saccharomyces cerevisiae myristoyl-CoA: protein N-myristoyltransferase. Analysis of fatty acid analogs containing carbonyl groups, nitrogen heteroatoms, and nitrogen heterocycles in an in vitro enzyme assay and subsequent identification of inhibitors of human immunodeficiency virus I replication. J Biol Chem. 1992;267(11):7224–39. [PubMed] [Google Scholar]
- 112.Saermark T, Kleinschmidt A, Wulff AM, Andreassen H, Magee A, Erfle V. Characterization of N-myristoyl transferase inhibitors and their effect on HIV release. Aids. 1991;5(8):951–8. doi: 10.1097/00002030-199108000-00005. [DOI] [PubMed] [Google Scholar]
- 113.Zentner I, Sierra LJ, Fraser AK, Maciunas L, Mankowski MK, Vinnik A, Fedichev P, Ptak RG, Martin-Garcia J, Cocklin S. Identification of a small-molecule inhibitor of HIV-1 assembly that targets the phosphatidylinositol (4,5)-bisphosphate binding site of the HIV-1 matrix protein. ChemMedChem. 2013;8(3):426–32. doi: 10.1002/cmdc.201200577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zentner I, Sierra LJ, Maciunas L, Vinnik A, Fedichev P, Mankowski MK, Ptak RG, Martin-Garcia J, Cocklin S. Discovery of a small-molecule antiviral targeting the HIV-1 matrix protein. Bioorganic & medicinal chemistry letters. 2013;23(4):1132–5. doi: 10.1016/j.bmcl.2012.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427(6977):848–53. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 116.Sayah DM, Sokolskaja E, Berthoux L, Luban J. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature. 2004;430(6999):569–73. doi: 10.1038/nature02777. [DOI] [PubMed] [Google Scholar]
- 117.Keller PW, Huang RK, England MR, Waki K, Cheng N, Heymann JB, Craven RC, Freed EO, Steven AC. A two-pronged structural analysis of retroviral maturation indicates that core formation proceeds by a disassembly-reassembly pathway rather than a displacive transition. Journal of virology. 2013;87(24):13655–64. doi: 10.1128/JVI.01408-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Krishnan L, Matreyek KA, Oztop I, Lee K, Tipper CH, Li X, Dar MJ, Kewalramani VN, Engelman A. The requirement for cellular transportin 3 (TNPO3 or TRN-SR2) during infection maps to human immunodeficiency virus type 1 capsid and not integrase. Journal of virology. 2010;84(1):397–406. doi: 10.1128/JVI.01899-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bichel K, Price AJ, Schaller T, Towers GJ, Freund SM, James LC. HIV-1 capsid undergoes coupled binding and isomerization by the nuclear pore protein NUP358. Retrovirology. 2013;10:81. doi: 10.1186/1742-4690-10-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Matreyek KA, Engelman A. Viral and cellular requirements for the nuclear entry of retroviral preintegration nucleoprotein complexes. Viruses. 2013;5(10):2483–511. doi: 10.3390/v5102483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Price AJ, Fletcher AJ, Schaller T, Elliott T, Lee K, KewalRamani VN, Chin JW, Towers GJ, James LC. CPSF6 defines a conserved capsid interface that modulates HIV-1 replication. PLoS pathogens. 2012;8(8):e1002896. doi: 10.1371/journal.ppat.1002896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tang C, Loeliger E, Kinde I, Kyere S, Mayo K, Barklis E, Sun Y, Huang M, Summers MF. Antiviral inhibition of the HIV-1 capsid protein. J Mol Biol. 2003;327(5):1013–20. doi: 10.1016/s0022-2836(03)00289-4. [DOI] [PubMed] [Google Scholar]
- 123.Kelly BN, Kyere S, Kinde I, Tang C, Howard BR, Robinson H, Sundquist WI, Summers MF, Hill CP. Structure of the antiviral assembly inhibitor CAP-1 complex with the HIV-1 CA protein. J Mol Biol. 2007;373(2):355–66. doi: 10.1016/j.jmb.2007.07.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Li J, Tan Z, Tang S, Hewlett I, Pang R, He M, He S, Tian B, Chen K, Yang M. Discovery of dual inhibitors targeting both HIV-1 capsid and human cyclophilin A to inhibit the assembly and uncoating of the viral capsid. Bioorg Med Chem. 2009;17(8):3177–88. doi: 10.1016/j.bmc.2009.02.051. [DOI] [PubMed] [Google Scholar]
- 125.Jin Y, Tan Z, He M, Tian B, Tang S, Hewlett I, Yang M. SAR and molecular mechanism study of novel acylhydrazone compounds targeting HIV-1 CA. Bioorg Med Chem. 2010;18(6):2135–40. doi: 10.1016/j.bmc.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 126.Prevelige PE., Jr Small molecule inhibitors of HIV-1 capsid assembly. US PATENT. 2006
- 127.Fader LD, Bethell R, Bonneau P, Bos M, Bousquet Y, Cordingley MG, Coulombe R, Deroy P, Faucher AM, Gagnon A, Goudreau N, Grand-Maitre C, Guse I, Hucke O, Kawai SH, Lacoste JE, Landry S, Lemke CT, Malenfant E, Mason S, Morin S, O’Meara J, Simoneau B, Titolo S, Yoakim C. Discovery of a 1,5-dihydrobenzo[b][1,4]diazepine-2,4-dione series of inhibitors of HIV-1 capsid assembly. Bioorganic & medicinal chemistry letters. 2011;21(1):398–404. doi: 10.1016/j.bmcl.2010.10.131. [DOI] [PubMed] [Google Scholar]
- 128.Lemke CT, Titolo S, von Schwedler U, Goudreau N, Mercier JF, Wardrop E, Faucher AM, Coulombe R, Banik SS, Fader L, Gagnon A, Kawai SH, Rancourt J, Tremblay M, Yoakim C, Simoneau B, Archambault J, Sundquist WI, Mason SW. Distinct effects of two HIV-1 capsid assembly inhibitor families that bind the same site within the N-terminal domain of the viral CA protein. Journal of virology. 2012;86(12):6643–55. doi: 10.1128/JVI.00493-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sticht J, Humbert M, Findlow S, Bodem J, Muller B, Dietrich U, Werner J, Krausslich HG. A peptide inhibitor of HIV-1 assembly in vitro. Nat Struct Mol Biol. 2005;12(8):671–7. doi: 10.1038/nsmb964. [DOI] [PubMed] [Google Scholar]
- 130.Ternois F, Sticht J, Duquerroy S, Krausslich HG, Rey FA. The HIV-1 capsid protein C-terminal domain in complex with a virus assembly inhibitor. Nat Struct Mol Biol. 2005;12(8):678–82. doi: 10.1038/nsmb967. [DOI] [PubMed] [Google Scholar]
- 131.Zhang H, Zhao Q, Bhattacharya S, Waheed AA, Tong X, Hong A, Heck S, Curreli F, Goger M, Cowburn D, Freed EO, Debnath AK. A cell-penetrating helical peptide as a potential HIV-1 inhibitor. J Mol Biol. 2008;378(3):565–80. doi: 10.1016/j.jmb.2008.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bhattacharya S, Zhang H, Debnath AK, Cowburn D. Solution structure of a hydrocarbon stapled peptide inhibitor in complex with monomeric C-terminal domain of HIV-1 capsid. J Biol Chem. 2008;283(24):16274–8. doi: 10.1074/jbc.C800048200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhang H, Curreli F, Waheed AA, Mercredi PY, Mehta M, Bhargava P, Scacalossi D, Tong X, Lee S, Cooper A, Summers MF, Freed EO, Debnath AK. Dual-acting stapled peptides target both HIV-1 entry and assembly. Retrovirology. 2013;10:136. doi: 10.1186/1742-4690-10-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhang H, Curreli F, Zhang X, Bhattacharya S, Waheed AA, Cooper A, Cowburn D, Freed EO, Debnath AK. Antiviral activity of alpha-helical stapled peptides designed from the HIV-1 capsid dimerization domain. Retrovirology. 2011;8:28. doi: 10.1186/1742-4690-8-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Shi J, Zhou J, Shah VB, Aiken C, Whitby K. Small-molecule inhibition of human immunodeficiency virus type 1 infection by virus capsid destabilization. Journal of virology. 2011;85(1):542–9. doi: 10.1128/JVI.01406-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lamorte L, Titolo S, Lemke CT, Goudreau N, Mercier JF, Wardrop E, Shah VB, von Schwedler UK, Langelier C, Banik SS, Aiken C, Sundquist WI, Mason SW. Discovery of novel small-molecule HIV-1 replication inhibitors that stabilize capsid complexes. Antimicrobial agents and chemotherapy. 2013;57(10):4622–31. doi: 10.1128/AAC.00985-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Li G, Verheyen J, Rhee SY, Voet A, Vandamme AM, Theys K. Functional conservation of HIV-1 Gag: implications for rational drug design. Retrovirology. 2013;10:126. doi: 10.1186/1742-4690-10-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Rihn SJ, Wilson SJ, Loman NJ, Alim M, Bakker SE, Bhella D, Gifford RJ, Rixon FJ, Bieniasz PD. Extreme genetic fragility of the HIV-1 capsid. PLoS pathogens. 2013;9(6):e1003461. doi: 10.1371/journal.ppat.1003461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Klumpp K, Crepin T. Capsid proteins of enveloped viruses as antiviral drug targets. Current opinion in virology. 2014;5:63–71. doi: 10.1016/j.coviro.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 140.de Rocquigny H, Shvadchak V, Avilov S, Dong CZ, Dietrich U, Darlix JL, Mely Y. Targeting the viral nucleocapsid protein in anti-HIV-1 therapy. Mini Rev Med Chem. 2008;8(1):24–35. doi: 10.2174/138955708783331603. [DOI] [PubMed] [Google Scholar]
- 141.Garg D, Torbett BE. Advances in targeting nucleocapsid-nucleic acid interactions in HIV-1 therapy. Virus Res. 2014;193:135–43. doi: 10.1016/j.virusres.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Sosic A, Frecentese F, Perissutti E, Sinigaglia L, Santagada V, Caliendo G, Magli E, Ciano A, Zagotto G, Parolin C, Gatto B. Design, synthesis and biological evaluation of TAR and cTAR binders as HIV-1 nucleocapsid inhibitors. Medchemcomm. 2013;4(10):1388–1393. [Google Scholar]
- 143.Dietz J, Koch J, Kaur A, Raja C, Stein S, Grez M, Pustowka A, Mensch S, Ferner J, Moller L, Bannert N, Tampe R, Divita G, Mely Y, Schwalbe H, Dietrich U. Inhibition of HIV-1 by a peptide ligand of the genomic RNA packaging signal Psi. ChemMedChem. 2008;3(5):749–55. doi: 10.1002/cmdc.200700194. [DOI] [PubMed] [Google Scholar]
- 144.Druillennec S, Dong CZ, Escaich S, Gresh N, Bousseau A, Roques BP, Fournie-Zaluski MC. A mimic of HIV-1 nucleocapsid protein impairs reverse transcription and displays antiviral activity. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(9):4886–91. doi: 10.1073/pnas.96.9.4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Park MY, Kwon J, Lee S, You J, Myung H. Selection and characterization of peptides specifically binding to HIV-1 psi (psi) RNA. Virus Res. 2004;106(1):77–81. doi: 10.1016/j.virusres.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 146.Pustowka A, Dietz J, Ferner J, Baumann M, Landersz M, Konigs C, Schwalbe H, Dietrich U. Identification of peptide ligands for target RNA structures derived from the HIV-1 packaging signal psi by screening phage-displayed peptide libraries. Chembiochem. 2003;4(10):1093–7. doi: 10.1002/cbic.200300681. [DOI] [PubMed] [Google Scholar]
- 147.Warui DM, Baranger AM. Identification of specific small molecule ligands for stem loop 3 ribonucleic acid of the packaging signal Psi of human immunodeficiency virus-1. Journal of medicinal chemistry. 2009;52(17):5462–73. doi: 10.1021/jm900599v. [DOI] [PubMed] [Google Scholar]
- 148.Bell NM, L’Hernault A, Murat P, Richards JE, Lever AM, Balasubramanian S. Targeting RNA-protein interactions within the human immunodeficiency virus type 1 lifecycle. Biochemistry. 2013;52(51):9269–74. doi: 10.1021/bi401270d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Goudreau N, Hucke O, Faucher AM, Grand-Maitre C, Lepage O, Bonneau PR, Mason SW, Titolo S. Discovery and structural characterization of a new inhibitor series of HIV-1 nucleocapsid function: NMR solution structure determination of a ternary complex involving a 2:1 inhibitor/NC stoichiometry. J Mol Biol. 2013;425(11):1982–98. doi: 10.1016/j.jmb.2013.02.022. [DOI] [PubMed] [Google Scholar]
- 150.Votteler J, Sundquist WI. Virus budding and the ESCRT pathway. Cell Host Microbe. 2013;14(3):232–41. doi: 10.1016/j.chom.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Weiss ER, Gottlinger H. The role of cellular factors in promoting HIV budding. J Mol Biol. 2011;410(4):525–33. doi: 10.1016/j.jmb.2011.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Demirov DG, Orenstein JM, Freed EO. The late domain of human immunodeficiency virus type 1 p6 promotes virus release in a cell type-dependent manner. Journal of virology. 2002;76(1):105–17. doi: 10.1128/JVI.76.1.105-117.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Gottlinger HG, Dorfman T, Sodroski JG, Haseltine WA. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proceedings of the National Academy of Sciences of the United States of America. 1991;88(8):3195–9. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Strack B, Calistri A, Craig S, Popova E, Gottlinger HG. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell. 2003;114(6):689–99. doi: 10.1016/s0092-8674(03)00653-6. [DOI] [PubMed] [Google Scholar]
- 155.Garrus JE, von Schwedler UK, Pornillos OW, Morham SG, Zavitz KH, Wang HE, Wettstein DA, Stray KM, Cote M, Rich RL, Myszka DG, Sundquist WI. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107(1):55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- 156.Demirov DG, Ono A, Orenstein JM, Freed EO. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(2):955–60. doi: 10.1073/pnas.032511899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Tavassoli A, Lu Q, Gam J, Pan H, Benkovic SJ, Cohen SN. Inhibition of HIV budding by a genetically selected cyclic peptide targeting the Gag-TSG101 interaction. ACS Chem Biol. 2008;3(12):757–64. doi: 10.1021/cb800193n. [DOI] [PubMed] [Google Scholar]
- 158.Amarasinghe GK, De Guzman RN, Turner RB, Chancellor KJ, Wu ZR, Summers MF. NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the psi-RNA packaging signal. Implications for genome recognition. J Mol Biol. 2000;301(2):491–511. doi: 10.1006/jmbi.2000.3979. [DOI] [PubMed] [Google Scholar]
- 159.Kelly BN, Howard BR, Wang H, Robinson H, Sundquist WI, Hill CP. Implications for viral capsid assembly from crystal structures of HIV-1 Gag(1–278) and CA(N)(133–278) Biochemistry. 2006;45(38):11257–66. doi: 10.1021/bi060927x. [DOI] [PubMed] [Google Scholar]
- 160.Massiah MA, Starich MR, Paschall C, Summers MF, Christensen AM, Sundquist WI. Three-dimensional structure of the human immunodeficiency virus type 1 matrix protein. J Mol Biol. 1994;244(2):198–223. doi: 10.1006/jmbi.1994.1719. [DOI] [PubMed] [Google Scholar]
- 161.Morellet N, Druillennec S, Lenoir C, Bouaziz S, Roques BP. Helical structure determined by NMR of the HIV-1 (345–392)Gag sequence, surrounding p2: implications for particle assembly and RNA packaging. Protein Sci. 2005;14(2):375–86. doi: 10.1110/ps.041087605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Bartonova V, Igonet S, Sticht J, Glass B, Habermann A, Vaney MC, Sehr P, Lewis J, Rey FA, Krausslich HG. Residues in the HIV-1 capsid assembly inhibitor binding site are essential for maintaining the assembly-competent quaternary structure of the capsid protein. J Biol Chem. 2008;283(46):32024–33. doi: 10.1074/jbc.M804230200. [DOI] [PubMed] [Google Scholar]
- 163.Blair WS, Pickford C, Irving SL, Brown DG, Anderson M, Bazin R, Cao J, Ciaramella G, Isaacson J, Jackson L, Hunt R, Kjerrstrom A, Nieman JA, Patick AK, Perros M, Scott AD, Whitby K, Wu H, Butler SL. HIV capsid is a tractable target for small molecule therapeutic intervention. PLoS pathogens. 2010;6(12):e1001220. doi: 10.1371/journal.ppat.1001220. [DOI] [PMC free article] [PubMed] [Google Scholar]